Abstract
Compounds based on the 2,3-distyrylindole scaffold were found to exhibit bactericidal properties upon irradiation with white light. At the concentration of 1 μM, the lead compound 1 completely (ca. 109 CFU/mL) eradicated such Gram-positive organisms as S. aureus (MRSA, MSSA), E. faecalis (VRE), S. pyogenes and S. mutans when irradiated with white light for 2 minutes. At the concentration of 5 μM and in the presence of polymyxin E at non-bactericidal 1.25 μg/mL concentration, 1 also showed a 7-log to 9-log reductions in bacterial counts of such Gram-negative organisms as multi-drug resistant (MDR) A. baumannii, MDR P. aeruginosa, E. coli and Klebsiella pneumoniae (CRE: KPC and NDM-1), also when irradiated with white light for 2 minutes. The structure-activity relationship studies revealed that unsubstituted at benzene rings 2,3-distyrylindole 2 was most potent and gave a 5-order of magnitude eradication of a MRSA strain at the concentration of 30 nM upon irradiation with white light. Initial mechanistic experiments revealed the disruption of bacterial cell membrane, but indicated that singlet oxygen production, which is commonly associated with photodynamic therapy, may not play a role in the bactericidal effects of the 2,3-distyrylindoles.
Keywords: Multi-drug resistance, MRSA, VRE, CRE, light-activated, S. mutans
Graphical Abstract

Since the discovery of penicillin, antibiotics have been used globally as life-saving drugs and revolutionized medicine in the 20th century. However, their effectiveness and easy access have led to their overuse in healthcare1 and agriculture2 leading to the build-up of antibiotics in the environment3,4 and the emergence of resistance5 through mutations6 and horizontal gene transfer.7 Despite the implementation of numerous social awareness campaigns8 with the goal of decreasing the inappropriate use of antibiotics in human healthcare and animal husbandry, there is an urgent need for novel or underexplored therapeutic approaches to fight bacterial infectious diseases.
As part of the efforts to address the growing global resistance to antimicrobial medicines, on February 27, 2017 the World Health Organization (WHO) published a list of bacteria that pose the greatest threat to human health and thus require new effective antibiotics.9 The list of bacteria was prioritized based on the current level of resistance, number of deaths they cause, the frequency of infection and the burden they place on health care systems. The list includes Gram-positive organisms, such as vancomycin-resistant enterococci (VRE) and methicillin-resistant Staphylococcus aureus (MRSA), as well as Gram-negative strains, such as Acinetobacter baumannii, Pseudomonas aeruginosa and Carbapenem-resistant Enterobacteriaceae (CRE). Indeed, for example, CRE can kill up to half of the patients who develop bloodstream infections and has been referred to as “nightmare bacteria.”10
One emerging approach for the treatment of drug-resistant bacterial infections is photodynamic inactivation (PDI).11,12 It involves the use of a harmless visible light combined with a light-sensitive dye, referred to as a photosensitizer. When irradiated, the dye is promoted to an excited state and passes the excitation energy to oxygen resulting in the formation of reactive oxygen species. The fact that the ensuing cell death is spatially limited to the areas of where light is applied makes PDI a highly selective treatment approach for localized disease. PDI was originally described in 1900 as an observation that a microorganism Paramecia was killed by a combination of a photosensitizing dye acridine and sunlight.13 Currently, however, the two main applications of PDI are for the treatment of various types of cancer and age-related macular degeneration.14
Starting in the 1990s, significant effort has been applied to develop PDI as an antimicrobial therapy.11,12 An important advantage of PDI over the existing antibiotics is its ability to work equally well against both resistant and sensitive strains.15 Moreover, PDI has been shown to be incapable of inducing resistance in bacteria even after 20 cycles of partial killing with the following regrowth.16,17 Other advantages of PDI over systemically administered antibiotics include: rapid killing of microbial cells, while the systemic antibiotics’ action may take hours to days; rapid achievement of sufficient concentrations at the site of burns or damaged tissue, whose compromised blood supply may prevent the systemic antibiotic from reaching the infected sites; broad spectrum of action resulting in PDI’s quick administration before the infectious agent is identified.18 Furthermore, it is now possible to deliver the photosensitizing agent and light to almost any site in the body through the application of endoscopes and narrow-diameter interstitially inserted needles and fiber optics.19
Although PDI, involving the generation of singlet oxygen (also termed photodynamic therapy (PDT)), is the most well-studied approach for antimicrobial applications, recently, treatment strategies utilizing light activation of antibacterial agents not relying on oxygen or reactive oxygen species have been described. For example, the incorporation of an azobenzene photo-switchable element into the structure of quinolone antibiotics resulted in photo-controlled antibacterial agents active against E. coli.20 In another example, a photo-isomerizable diarylethene moiety was introduced into the backbone of an antimicrobial peptide gramicidin S and the antibacterial activity of the resulting peptidomimetic could be controlled by light.21 Although such approaches indeed allow for spatial control of antibacterial treatment, they do not offer better efficacy at eradicating resistant strains than the original antibiotics from which they are derived.
Previously, as part of our screening efforts, we described the synthesis of compound 1 (Figure 1) possessing a novel 2,3-distyrylindole scaffold.22 We reported that this compound was devoid of antibacterial, antifungal and anticancer activities in the assays we utilized in our screening program. More recent testing of compound 1, however, led to a serendipitous discovery that its lack of antimicrobial activity is dramatically reversed when it is irradiated with white light. Herein, we describe these recent findings by showing that 1 and its structural analogues represent significant promise as light-activated antimicrobial agents active against resistant Gram-positive and Gram-negative strains, including those that pose the greatest risk to human health as identified by the WHO, as discussed above.
Figure 1.
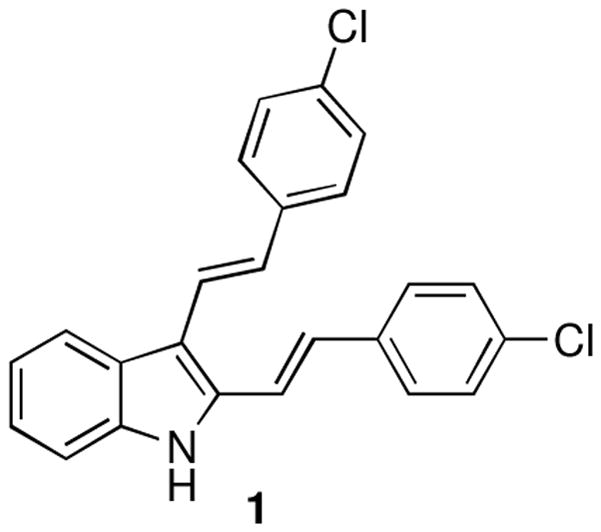
Previously synthesized 2,3-distyrylindole 1.
Our initial experimentation involved the utilization of a multidrug resistant MRSA strain (ATCC BAA-44, HA-MRSA), a clinical isolate from a hospital in Lisbon, Portugal. In addition to beta-lactams, this strain is resistant to antibiotics of many other structural and mechanistic types, such as erythromycin, tetracycline, ciprofloxacin, tobramycin.23,24 Thus, the bacterial isolates (ca. 108 CFU/mL) were incubated with 1 at different concentrations and then irradiated (or left in the dark in the control experiment) with white light from a LumaCare lamp for 2 min. The samples were diluted 10-fold, drip-streaked on TSA plates and incubated overnight at 37 °C. The colony counts were then performed using a dilution that produces a drip-streak with 30–300 colonies, as shown in Figure 2B. The bar graph derived from the colony counts is shown in Figure 2A. It illustrates a strong bactericidal effect of 1, activated with light. A 5-log reduction in bacterial count is observed at the concentration as low as 0.1 μM and a complete killing is achieved at a concentration between 0.5–1 μM.
Figure 2.
Compound 1 eradicates MRSA (ATCC BAA 44) in the presence of light. A: Graph, showing a dose-dependent effect of 1 with and without a 2-minute irradiation with white light. B. Drip-streak plates utilized in the experiment illustrating the CFU counts. The data are derived from a single experiment performed in 3 replicates and serve as an example of 3 independent experiments producing similar results. The stars represent no observable colonies.
As mentioned above, PDI is attractive because it allows for high spatial and temporal resolution. To illustrate this, a bacterial patterning experiment was performed. An agar plate, containing MRSA cells (ca. 108 CFU/mL) and pre-treated with 1 (1 μM), was prepared. A mask was then placed on top of the agar plate and the inoculum was irradiated with white light for 2 min (Figure 3, right). The plate was then incubated at 37 °C overnight and photographed. As can be seen on the photograph shown in Figure 3 (left), bacterial colonies were observed only where the area of the plate was covered to prevent the irradiation. Biological patterning, including the utilization of microorganisms, is an emerging area of research with potential applications in biosensors, drug discovery and diagnostics;25 thus, our results illustrate another area of promise for the light-controlled antimicrobial properties of 2,3-distyrylindoles.
Figure 3.
Bacterial patterning with light. Left: MRSA (ATCC BAA-44) cells pre-treated with 1 (1 μM), spread over an agar plate and irradiated with light through a mask for 2 minutes, and then incubated at 37 °C. NMT: New Mexico Tech. Right: Schematic of the bacterial patterning experiment.
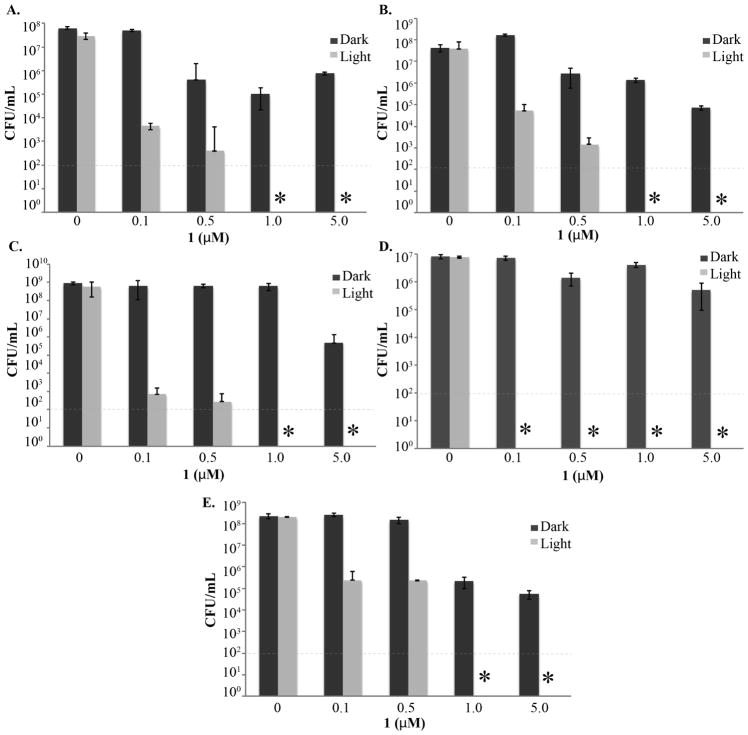
To investigate whether the bactericidal effect of 1 was not limited to HA-MRSA (ATCC BAA-44), we performed experiments with other Gram-positive bacterial strains. The panel included: another multidrug-resistant MRSA strain (ATCC BAA1717, CA-MRSA); drug-sensitive Staphylococcus aureus; vancomycin-resistant Enterococcus faecalis, known as VRE and representing another bacterial genus on the WHO list as discussed above;26 Streptococcus pyogenes, a causative agent of important human diseases, ranging from superficial skin infections to life-threatening systemic infections;27 Streptococcus mutans, a species commonly found in human oral cavity and the main contributor to tooth decay.28 The latter organism is of particular interest due to its resistance to various classes of antibiotics29 and the convenience of light delivery into the oral cavity.
As can be seen in Figure 4, all these Gram-positive organisms are sensitive to the bactericidal effects of 1. While MRSA (ATCC BAA1717), S. aureus, E. faecalis, and S. mutans are completely eradicated at the concentration of 1 μM, S. pyogenes is particularly sensitive and its complete killing is achieved at the concentration of 0.1 μM. Importantly, compound 1 has a significantly lower toxicity in the dark (Figure 4). Indeed, a light-activated agent should possess low toxicity to the target tissue until light is applied. To confirm that the lack of toxicity in the dark is also observed toward mammalian cells, the effects of 1 and several clinically used antibiotics on the viability of MCF-7 breast adenocarcinoma cells were evaluated using the MTT colorimetric assay.30 It was found that 1, with the IC50 of 47.0±2.5 μM, is not any more toxic toward mammalian cells than the clinically used antibiotics, such as tetracycline (IC50 of 41.6±0.3 μM) or norfloxacin (IC50 of 13.8±2.1 μM).
Figure 4.
Compound 1 is effective against a variety of drug resistant Gram-positive bacterial strains in the presence of light. A. MRSA (ATCC BAA1717). B. S. aureus (ATCC 29213). C. E. faecalis (ATCC 51299). D. S. pyogenes (ATCC 8133). E. S. mutans (Ward’s 85W 2357). The data are derived from a single experiment performed in 3 replicates and serve as an example of 3 independent experiments producing similar results. The stars represent no observable colonies.
The application of the above procedure to Gram-negative bacteria did not lead to the desired killing. Although initially disappointing, this observation is consistent with the literature precedent and explained by the fact that Gram-negative bacteria are typically characterized by an impermeable outer cell membrane that protects the cell from antibiotics, dyes, and detergents. Thus, the outer membrane of Gram-negative bacteria plays an important role in resistance to many antibiotics that are highly effective against Gram-positive bacteria, e.g. macrolides, novobiocin, rifamycin, lincomycin, clindamycin and fusidic acid. Accordingly, for a light-activated therapeutic agent to work, it needs to permeate the outer membrane to get in the cytoplasm. This is believed to be the reason for the higher occurrence of Gram-negative infections in hospital environments.31
A number of approaches to overcome the above obstacle and make a PDI agent active against Gram-negative bacteria have been reported in the literature.32 This can be achieved, for example, by taking advantage of the outer membrane’s intrinsic negative charge due to teichoic acid residues and adding a positive charge to the PDI agent, or coupling it with a positively charged moiety, such as poly-L-lysine, polyethyleneimine and polymyxin nonapetptides.32 These agents are believed to work by disorganizing the outer cell membrane in Gram-negative bacteria and making it penetrable to hydrophobic agents.32 For example, a combination treatment with polymyxin B (PMB), deuteroporphyrin and light resulted in killing of E. coli and P. aeruginosa.33
Thus, a combination including both 1 and polymyxin E (PME) was evaluated against a panel of Gram-negative organisms. This panel included: multidrug resistant Acinetobacter baumannii, a strain that is becoming increasingly important causing hospital-acquired infections and responsive to only a few antibiotics;34 Pseudomonas aeruginosa, also a causative agent of hospital acquired infections, such as ventilator-associated pneumonia and various sepsis syndromes;35 Klebsiella pneumoniae, the most significant member of the Enterobacteriaceae, mostly affecting people with weakened immune system and, as mentioned in the Introduction, is associated with mortality rate of 50%, even with antibiotic therapy;36 Escherichia coli, whose serotypes can cause serious food poisoning.37
As can be seen in Figure 5, treatments involving 1 (5 μM) or PME (1.25 – 2 μg/mL) by themselves had little or no effect either in the dark or with light. However, when 1 and PME were combined at the same concentrations as in the single agent treatments, pronounced effects were observed with irradiation. Indeed, P. aeruginosa and Klebsiella were completely eradicated (Figure 5B and D), while ca. 7-log reductions in bacterial counts were observed in the case of A. baumannii and E. coli (Figure 5A and C). At present, the reason for the dark toxicity of this combination treatment toward E. coli and Klebsiella (Figure 5C and D) is not completely understood and may reside in potentiation of the PME’s action by hydrophobic 1 through the disruption of cells membranes.
Figure 5.
Compound 1 is effective against a variety of drug resistant Gram-negative bacterial strains in the presence of PME and light. A. A. baumannii (ATCC BAA 1797). B. P. aeruginosa (ATCC BAA-2114). C. E. coli (ATCC 29522) D. Klebsiella pneumoniae, CRE (ATCC BAA-2146). The data are derived from a single experiment performed in 3 replicates and serve as an example of 3 independent experiments producing similar results. The stars represent no observable colonies.
To ascertain whether the bactericidal properties are associated with the novel 2,3-distyrylindole scaffold, rather than specifically with compound 1, a group of analogues with differing substitution patterns were synthesized (Figure 6). The synthesis is based on our previously discovered process to access compound 1 by the oxidative Heck reaction22 of p-chlorostyrene with indole. Thus, analogues with alternative positioning of chlorine substituent on the benzene ring 5 and 6 were prepared in acceptable yields (Figure 6). Further, unsubstituted compound 2 and analogues with a different para-substituent 3 and 4 were obtained in acceptable yields as well. Overall, the discovered reaction appears to be useful for the preparation of a large number diverse analogues for the optimization of their antibacterial properties. Importantly, these antibacterial agents are prepared in one step from indole and various substituted styrenes. The spectra of the compounds are included in the supporting information. In a few instances the compounds retained small amounts of solvents even after a prolonged drying. To confirm the structures of the synthesized distyrylindoles, compound 2 was treated with Pd/C under reflux in xylenes to generate known carbazole 7. The 1H and 13C NMR data were fully consistent with those in the published report.38
Figure 6.
a) Synthesis of compounds of 2,3-distyrylindoles 1–6 and b) structure confirmation through the synthesis of known carbazole 7.
Evaluation of the synthesized compounds against our original MRSA (ATCC BAA44) strain revealed the initial structure-activity relationship (SAR) in this series of compounds. Specifically, the replacement of chlorine with fluorine at the para-position in compounds 3 and 4 is detrimental for activity and complete killing was not achieved even at a concentration as high as 5 μM (Figure 7B and C, compare with 1 in Figure 2). Moving chlorine to the meta-position, as in compound 6, similarly reduces potency considerably (Figure 7E). Chlorine at the ortho-position, as in 5, appears to retain activity better, but it is still worse than that of compound 1 (Figure 7D). In contrast, analogue 2, with unsubstituted benzene rings, is the most potent so far. The eradication of bacteria by 5 orders of magnitude is observed at concentrations as low as 30 nM (Figure 7A).
Figure 7.
Eradication of MRSA (ATCC BAA44) by analogues of 1. The data are derived from a single experiment performed in 3 replicates and serve as an example of 3 independent experiments producing similar results. The stars represent no observable colonies.
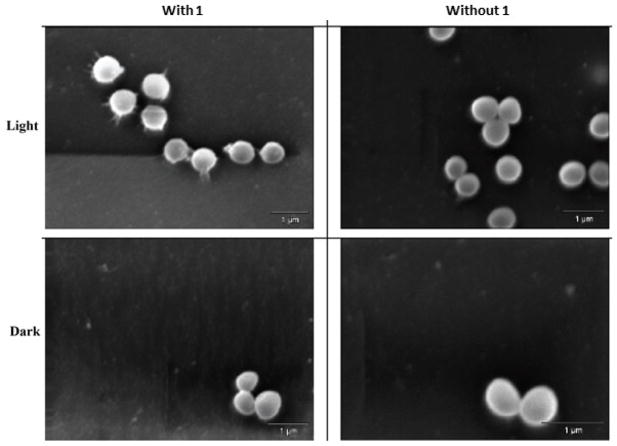
The initial mode of action studies, aimed at understanding how light activation of 2,3-distyrylindoles results in the bactericidal effect, were initiated. Electron microscopy of MRSA cells treated with both 1 and light revealed a morphology involving an apparent cell membrane disruptionand leakage of the intracellular contents. This morphology was not observed in untreated cells or cells treated only with 1 or only with light (Figure 8). To confirm this rationalization of the SEM imaging, a propidium iodide (PI) staining assay was performed. PI is a positively charged fluorescent intercalating agent that can cross the damaged cell membrane and bind to nucleic acids. As can be seen in Figure 9, cells labeled with red fluorescence due to PI are observed only when they were treated with 1 and light together. This red fluorescence is similar to the positive control phenylarsine oxide (PAO), a general cytotoxic agent. In contrast, cells treated with 1 only exhibit green fluorescence due to 1.
Figure 8.
SEM images of MRSA (ATCC BAA44) cells treated with 1.
Figure 9.
Permeability of MRSA (ATCC BAA44) cell membrane probed by PI. Green fluorescence is due to 1. Red fluorescence is indicative of the disruption of cell membrane.
Cell membrane disruption is a characteristic outcome of photodynamic therapy,39 which is based on the generation of singlet oxygen by the irradiated sensitizer, as discussed in the Introduction. Thus, the ability of 1 to generate singlet oxygen was investigated (Figure 10). The reference samples Rubpy and Rose bengal showed singlet oxygen emission at 1270 and 1285 nm, respectively. In contrast, 1 showed a concentration dependent emission at 1210 nm. It is thus unlikely that the bactericidal effect of 1 is due to the production of singlet oxygen. Although matrix effects can shift the singlet oxygen emission by a few nanometers, this significant spectral shift cannot be explained by matrix effects.
Figure 10.
Lack of singlet oxygen generation by 1. Positive controls [Ru(bpy)3](PF6)2 and Rose bengal show emission at 1270–1285 nm likely due to singlet oxygen. 1 shows emission at 1210 nm. Although matrix effects can shift the singlet oxygen emission by a few nanometers, the significant spectral shift by 1 cannot be due matrix effects.
As the post-antibiotic era is in sight, the possibility of infectious diseases becoming a major cause of lethality can no longer be ignored. Fatal infections are not confined to tropical regions, or even contained in hospitals, they are appearing more and more frequently in the community. Multi-drug resistant Gram-positive organisms, such as vancomycin-resistant enterococci (VRE) and methicillin-resistant Staphylococcus aureus (MRSA), as well as Gram-negative strains, such as Acinetobacter baumannii, Pseudomonas aeruginosa and Carbapenem-resistant Enterobacteriaceae (CRE) are becoming increasingly less and less susceptible to the currently available antibiotics. From this perspective, research described herein is significant. The combination of 2,3-distyrylindoles and light appears to overcome bacterial resistance regardless of the type of bacteria or the mechanism of resistance. Importantly, these compounds are readily prepared via a one-step synthesis, wherein three fragments, indole and two styrene units, are assembled in a convergent manner. Due to its simplicity and functional group tolerance, the synthesis should be amenable to the preparation of a large number of analogues with the desired substitution patterns in either indole or the styrene moieties, enabling the optimization of these compounds during preclinical and possible clinical development. The next step in this research is the elucidation of the MOA. Our preliminary data appear to rule out the singlet oxygen generation as the mechanism responsible for the bactericidal properties of these compounds and this leads us to the exciting process of hypothesis-driven MOA investigations. These studies will be reported in due course.
Supplementary Material
2,3-Distyrylindoles exhibit bactericidal properties upon irradiation with light.
The compounds eradicate Gram-positive organisms when irradiated for 2 minutes.
In the presence of polymyxin E, the compounds eradicate Gram-negative organisms.
Acknowledgments
This project was supported by grants from the National Institute of General Medical Sciences (P20GM103451), SR, LF and ALP acknowledge their NMT Presidential Research Support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goossens H, Ferech M, Vander Stichele R, Elseviers M. The Lancet. 2005;365:579–587. doi: 10.1016/S0140-6736(05)17907-0. [DOI] [PubMed] [Google Scholar]
- 2.Kemper N. Ecol Indic. 2008;8:1. [Google Scholar]
- 3.Martínez JL. Science. 2008;321:365. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 4.Tello A, Austin B, Telfer TC. Environ Health Perspect. 2012;120:1100. doi: 10.1289/ehp.1104650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlet J, Collignon P, Goldmann D, Goossens H, Gyssens IC, Harbarth S, Jarlier V, Levy SB, N'Doye B, Pittet D, Richtmann R. The Lancet. 2011;378:369. doi: 10.1016/S0140-6736(11)60401-7. [DOI] [PubMed] [Google Scholar]
- 6.Martinez JL, Baquero F. Antimicrob Agents Chemother. 2000;44:1771. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies J. Science-AAAS-Weekly. 1994;264:375. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 8.Stockley JM. European antibiotic awareness day 2012: getting smart about antibiotics, a public–professional partnership. 2012 doi: 10.1016/j.jinf.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 9.http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/
- 10.Kuehn BM. Jama. 2013;309:1573. doi: 10.1001/jama.2013.2922. [DOI] [PubMed] [Google Scholar]
- 11.Hamblin MR. Antimicrobial photodynamic inactivation: a bright new technique to kill resistant microbes. Curr Opin Microbiol. 2016;33:67. doi: 10.1016/j.mib.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denis TGS, Hamblin MR. Méndez-Vilas. 2011. pp. 675–683. [Google Scholar]
- 13.Von Tappeiner H. 1900:47. [Google Scholar]
- 14.Moan J, Peng Q. Anticancer Res. 2003;23: 3591. [PubMed] [Google Scholar]
- 15.Vera DMA, Haynes MH, Ball AR, Dai T, Astrakas C, Kelso MJ, Hamblin MR, Tegos GP. Photochem Photobiol. 2012;88:499. doi: 10.1111/j.1751-1097.2012.01087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maisch T. Photochem Photobiol. 2015;14:1518. doi: 10.1039/c5pp00037h. [DOI] [PubMed] [Google Scholar]
- 17.Giuliani F, Martinelli M, Cocchi A, Arbia D, Fantetti L, Roncucci G. Antimicrob Agents Chemother. 2010;54:637. doi: 10.1128/AAC.00603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreyra DD, Reynoso E, Cordero P, Spesia MB, Alvarez MG, Milanesio ME, Durantini EN. Photochem Photobiol B. 2016;158: 243. doi: 10.1016/j.jphotobiol.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 19.Gad F, Zahra T, Francis KP, Hasan T, Hamblin MR. Photochem Photobiol B. 2004;3:451. doi: 10.1039/b311901g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babii O, Afonin S, Berditsch M, Reiβer S, Mykhailiuk PK, Kubyshkin VS, Steinbrecher T, Ulrich AS, Komarov IV. Angew Chem Int Ed. 2014;53: 3392. doi: 10.1002/anie.201310019. [DOI] [PubMed] [Google Scholar]
- 21.Velema WA, Van Der Berg JP, Hansen MJ, Szymanski W, Driessen AJ, Feringa BL. Nat Chem. 2013;5:924. doi: 10.1038/nchem.1750. [DOI] [PubMed] [Google Scholar]
- 22.Daly S, Hayden K, Malik I, Porch N, Tang H, Rogelj S, Frolova LV, Lepthien K, Kornienko A, Magedov IV. Bioorg Med Chem Lett. 2011;21:4720. doi: 10.1016/j.bmcl.2011.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podoll JD, Liu Y, Chang L, Walls S, Wang W, Wang X. Proc Natl Acad Sci USA. 2013;110:15573. doi: 10.1073/pnas.1310459110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loewen K, Schreiber Y, Kirlew M, Bocking N, Kelly L. Can Fam Physician. 2017;63:512. [PMC free article] [PubMed] [Google Scholar]
- 25.Kim K, Lee BU, Hwang GB, Lee JH, Kim S. Anal Chem. 2010;82:2109. doi: 10.1021/ac9027966. [DOI] [PubMed] [Google Scholar]
- 26.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. Clin Microbiol Infect. 2010;16:541. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 27.Silva-Costa C, Friães A, Ramirez M, Melo-Cristino J. Expert Rev Anti Infect Ther. 2015;13:615. doi: 10.1586/14787210.2015.1023292. [DOI] [PubMed] [Google Scholar]
- 28.Liao Y, Brandt BW, Li J, Crielaard W, Van Loveren C, Deng DM. J Oral Microbiol. 2017;9:1344509. doi: 10.1080/20002297.2017.1344509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leistevuo J, Järvinen H, Österblad M, Leistevuo T, Huovinen P, Tenovuo J. Antimicrob Agents Chemother. 2000;44: 456. doi: 10.1128/aac.44.2.456-457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosmann T. J Immunol Methods. 1983;65:55. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 31.Nikaido H, Vaara M. Microbiol Rev. 1985;49:1. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sperandio FF, Huang YY, Hamblin RM. Recent Pat Antiinfect Drug Discov. 2013;8:108. doi: 10.2174/1574891x113089990012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nitzan Y, Gutterman M, Malik Z, Ehrenberg B. Photochem Photobiol. 1992;55:89. doi: 10.1111/j.1751-1097.1992.tb04213.x. [DOI] [PubMed] [Google Scholar]
- 34.Lynch JP, III, Zhanel GG, Clark NM, Doi Y, Murray GL, Peleg AY, Jones CL, Clancy M, Honnold C, Davis JS, McMillan M. 2017;38:311. [Google Scholar]
- 35.Lynch JP, III, Zhanel GG, Clark NM, Ramírez-Estrada S, Borgatta B, Rello J, Kollef MH, Chastre J, Fagon JY, Tumbarello M, De Pascale G. Seminars in Respiratory and Critical Care Medicine. 2017;38:326–345. doi: 10.1055/s-0037-1602583. [DOI] [PubMed] [Google Scholar]
- 36.Navon-Venezia S, Kondratyeva K, Carattoli A. FEMS Microbiol Rev. 2017;41:252. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 37.Yang SC, Lin CH, Aljuffali IA, Fang JY. Arch Microbiol. 2017:1. doi: 10.1007/s00203-017-1393-y. [DOI] [PubMed] [Google Scholar]
- 38.Banerjee A, Sahu S, Maji MS. Adv Synth Catal. 2017;359:1860. [Google Scholar]
- 39.Huang X, Chen G, Pan J, Chen X, Huang N, Wang X, Liu J. J Mater Chem B. 2016;4:6258. doi: 10.1039/c6tb01122e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.