Abstract
Over the past decades, outcomes for children with cancer have improved dramatically through serial clinical trials based in large measure on dose intensification of cytotoxic chemotherapy for children with high-risk malignancies. Progress made through such dose intensification, in general, is no longer yielding further improvements in outcome. With the revolution in sequencing technologies and rapid development of drugs that block specific proteins and pathways, there is now an opportunity to improve outcomes for pediatric cancer patients through mutation-based targeted therapeutic strategies. The Children’s Oncology Group (COG), in partnership with the National Cancer Institute (NCI), is planning a trial entitled the COG-NCI Pediatric Molecular Analysis for Therapeutic Choice (Pediatric MATCH) protocol utilizing an umbrella design. This protocol will have centralized infrastructure and will consist of a biomarker profiling protocol and multiple single-arm phase II trials of targeted therapies. Pediatric patients with recurrent or refractory solid tumors, lymphomas, or histiocytoses with measurable disease will be eligible. The Pediatric MATCH Target and Agent Prioritization (TAP) committee includes membership representing COG disease committees, the Food and Drug Administration, and the NCI. The TAP Committee systematically reviewed target and agent pairs for inclusion in the Pediatric MATCH trial. Fifteen drug-target pairs were reviewed by the TAP Committee, with seven recommended for further development as initial arms of the Pediatric MATCH trial. The current evidence for availability, efficacy, and safety of targeted agents in children for each class of mutation considered for inclusion in the Pediatric MATCH trial is discussed in this review.
Childhood malignancies contain genomic alterations that may predict response to molecularly targeted therapies (1–5). Recurrent genomic alterations occurring in specific cancer histologies typically occur at a frequency of less than 20%, and most occur at a frequency of less than 10% (6). The rare occurrence of pediatric cancers and the low frequency of recurrent genomic alterations make it difficult to design and conduct phase II trials of targeted therapy in a patient population with both a specific diagnosis and a specific genomic alteration. Genomic alterations linked to response to targeted therapy often occur across multiple (and diverse) tumor histologies.
A number of novel clinical trial designs have been suggested to facilitate integration of genomics (7,8) into clinical trials, including umbrella and basket designs, in which patients characterized by the presence of a predictive biomarker are treated on trial arms utilizing the therapy indicated by the identified biomarker. For example, the Molecular Analysis for Therapy Choice (NCI-MATCH) study utilizes a basic strategy of testing patient tumors for molecular targets under an umbrella protocol, then directs patients to one of many separate phase II studies that have molecular eligibility criteria (9). The NCI-MATCH study began enrolling subjects in August 2015; after two months of enrollment, 9% of patients sequenced were found to have an actionable mutation for assignment to one of the 10 treatment arms, a rate likely to increase as additional study arms are opened (10).
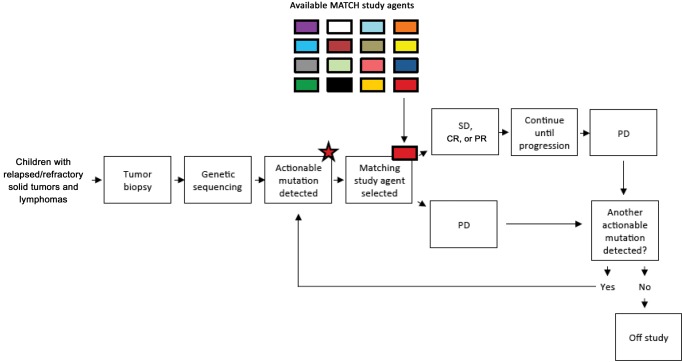
The Children’s Oncology Group (COG) in partnership with the National Cancer Institute (NCI) is planning a trial entitled the COG-NCI Pediatric Molecular Analysis for Therapeutic Choice (Pediatric MATCH) protocol utilizing an umbrella design. This protocol will have centralized infrastructure and consist of a single biomarker profiling (screening) protocol and multiple single-arm phase II trials (subprotocols) of targeted therapies. Pediatric patients with recurrent or refractory solid tumors, histiocytoses, or lymphomas with measurable disease will be eligible (Figure 1).
Figure 1.
Pediatric Molecular Analysis for Therapeutic Choice (MATCH) Trial schema. Subjects with relapsed or refractory solid tumors, lymphomas, and histiocytic disorders are eligible for Pediatric MATCH. Tumor biopsy undergoes sequencing, and if an actionable mutation is detected the subject may be enrolled on a study subarm and receive a “matched” targeted agent. Subjects with stable disease, partial response, or complete response remain on study drug until disease progression. If a subject experiences progressive disease and additional actionable mutations are detected, they may enroll in a second subarm and receive a second targeted agent. If no additional subarm targets are available at the time of progressive disease, the subject goes off-study. CR = complete response; PD = progressive disease; PR = partial response; SD = stable disease.
Given the limited number of children with recurrent malignancies, it is unlikely that every agent of interest will be amenable for study in this patient population and hence there is a need to select or prioritize agent classes for this clinical trial. The Pediatric MATCH Target and Agent Prioritization (TAP) Committee was formed to serve this purpose.
Methods
TAP Committee
The TAP Committee included pediatric oncologists with expertise in cancer genomics and representation from the diversity of COG disease committees, as well as seven members who served as liaisons to the adult NCI MATCH study and organizations and agencies involved in Pediatric MATCH protocol development. The Food and Drug Administration (FDA) and NCI’s Cancer Therapy Evaluation Program (CTEP) and Center for Cancer Research (CCR) were also represented.
Compiling a List of Target-Agent Pairs
The TAP Committee Co-Chairs compiled a comprehensive list of targeted agent classes to be considered for inclusion based on their knowledge of pediatric cancer genomics and a literature review. This list was reviewed by committee members who also recommended additional agents for consideration. A final list of agent classes to be formally reviewed and prioritized was agreed upon by the committee.
Review Process
Each target/agent pair had primary and secondary reviewers, who were assigned to target-agent pairs based on expertise, who expressed interest in a particular target/agent pair, and because of logistical issues (such as availability). The reviewers were asked to define the potential target or biomarker, determine whether the target can be detected with the proposed testing platform, evaluate the frequency of alterations in the target in pediatric malignancies, assess evidence linking target to activity of the agent, consider potential toxicities, and review agents in the class and report on ongoing or planned trials with potential overlap. After conducting this thorough review, the reviewer assigned a priority score (Table 1) and prepared a written report (in standardized format) for the target-agent pair. The committee voted on target-agent pairs following systematic review and discussions. Sources of evidence utilized in conducting reviews included published peer-reviewed literature, abstracts, and unpublished data. Initial reviews were conducted between February 2015 and May 2015.
Table 1.
Summary of TAP committee review of target-agent pairs
| Rank by average TAP score | Agent class | Average TAP score (range) | Example response biomarkers | Example resistance biomarkers | Final priority for pediatric MATCH Trial |
|---|---|---|---|---|---|
| 1 | MTOR inhibitor* | 1.5 (1–2) | TSC1/2 LOF mutations, MTOR mutations, PIK3CA p.H1047R and p.E545K, PTEN deletion | KRAS mutation | Included |
| 2 | MEK inhibitor† | 1.5 (1–2) | NF1 LOF mutation and H/K/NRAS/BRAF-activating mutations | MAPK1, MAPK2, and MEK2 mutations reported to cause resistance | Included |
| 3 | PI3K inhibitor* | 2 (1–3) | Same as mTOR inhibitors | KRAS mutations | Included |
| 4 | PDGFRA inhibitor | 2 (1–3) | PDGFRA amplification, PDGFRA-activating mutation | Unknown | Included |
| 5 | BRAF inhibitor† | 2 (1–3) | BRAF p.V600E mutation and other documented activating mutations, BRAF fusions, amplification WT BRAF | Reported resistance mutations: NRAS Q61, amplification mutant BRAF, MAP2K1 mutations | Included |
| 6 | Extended ALK inhibitor | 2 (1–3) | ROS1 translocations; ALK-activating mutations, ALK translocations | For crizotinib: ALK C1156Y, L1196M, G1123S, L1152R, G1202R. | Included |
| For 2nd/3rd generation: ALK I1171T, V1180L, F1174c, F1245C, R1275Q | |||||
| 7 | TRK inhibitor | 2.5 (1–4) | Translocations involving NTRK1/2/3 | Unknown | Included |
| 8 | BET bromodomain inhibitor | 2.5 (1–4) | MYC or MYCN amplification, MYC translocation, BRD4 translocation | TP53 mutation (early preclinical data suggests possible association w/ resistance) | Not included |
| 9 | CDK4/6 inhibitor | 2.5 (1–4) | CDK 4/6 amplification, CCND2 amplification, SNF5 del | Loss of RB1 expression (no standard assay) | Not included |
| 10 | FGFR inhibitor | 2.5 (1–4) | FGFR-activating mutations, FGFR amplification, FGFR fusions | Depends on agent selected (and range of FGFR selectivity) | Included |
| 11 | 2nd-generation ALK inhibitor | 2.66 (1–5) | ALK-activating mutations, ALK translocations | For crizotinib: ALK C1156Y, L1196M, G1123S, L1152R, G1202R | Not included |
| For 2nd/3rd generation: ALK I1171T, V1180L | |||||
| 12 | AKT inhibitor* | 3 (1–5) | Same as mTOR/PI3K inhibitors | Unknown | Not included |
| 13 | EGFR inhibitor | 3 (1–5) | EGFR-activating mutations, EGFR amplification | Unknown | Not included |
| 14 | IDH 1/2 inhibitors | 3 (2–4) | IDH 1/2 mutations | Unknown | Not included |
| 15 | SMO inhibitor | 3 (1–5) | PTCH1 mutations | GLI2 amplification, SUFU mutations, NMYC amplification | Not included |
| 16 | PARP inhibitor | 3 (2–4) | BRCA1/2 mutation, ATM mutation, EWSR1-FLI1 translocation | Unknown | Not included |
| 17 | ERK inhibitor† | 3.5 (3–4) | Activating MAPK pathway mutations | Unknown | Not included |
Agent classes in the same signaling pathway are identified with PI3K/AKT/mTOR. ALK = anaplastic lymphoma receptor kinase; AKT = activate protein kinase B; EGFR = epidermal growth factor receptor; FGFR = fibroblast growth factor receptor; TAP = Target and Agent Prioritization committee.
Agent classes in the same signaling pathway are identified with MAPK. Biomarkers of response and resistance are provided as examples; these are selected and do not include all potential variants demonstrated to be associated with response or resistance.
Co-Chair Assessment and Determination of Top Priority Pairs
The TAP Committee Co-Chairs then determined the top priority target-agent pairs to be recommended to the Pediatric MATCH Steering Committee for further development as the initial arms of the trial by assessing the level of evidence linking the biomarker to response to the agent, the ability of the MATCH assay to detect the key biomarker, and the suitability of each target-agent pair for the specific structure and goals of this trial.
Pediatric MATCH investigators provided data for the levels of evidence to be utilized for classifying each arm of the trial as outlined in Table 2. The TAP Committee Co-Chairs then determined the highest level of evidence possible for each agent class evaluated. In applying these levels of evidence, the "clinical end point" and "evidence of clinical activity" are specific to the biomarker-defined population.
Table 2.
Parameters for evaluation of target-agent pairs
| Priority score | Target in pediatrics | Level of evidence (linking target and agent activity) | Specific agent issues (availability, viability, central nervous system penetration) |
|---|---|---|---|
| 1-Must include | Frequent | Clinical trials | No issues |
| 2-Strongly encourage inclusion | Present | Case series or case reports | No issues with at least some agents in class |
| 3-Encourage inclusion | Present | Strong preclinical | Issues present |
| 4-Consider inclusion | Rare | Weak preclinical | Issues with most agents in class |
| 5-Do not include | Not present | No evidence | Issues with all agents in class |
Information regarding the assay to be utilized for the Pediatric MATCH (MATCH assay) was provided to the Co-Chairs of the TAP Committee. For each target-agent pair evaluated by the TAP Committee, the Committee Co-Chairs evaluated whether the MATCH assay would detect the variants anticipated to be present in pediatric malignancies that could predict response to the agent class. The list of seven agent-target pairs to be recommended for inclusion in the initial Pediatric MATCH trial were ultimately determined by the Co-Chairs and approved by the committee.
The Pediatric MATCH study will utilize a version of the ThermoFisher Oncomine Cancer Panel, which has previously been analytically and clinically validated for the adult NCI MATCH clinical trial and has been reviewed and revised to include relevant pediatric cancer gene content. The Oncomine study panel targets a defined set of more than 4000 annotated genomic variants including single-nucleotide variants, insertion/deletions, copy number variants (amplifications), and gene fusions. Of note, the panel is not currently utilized to detect gene deletions (necessitating the use of immunohistochemistry for specific proteins if needed for subprotocol eligibility). In addition, genetic alterations such as complex genetic rearrangements are not routinely detectable by mutation panels, which are also not designed to identify microsatellite instability. The panel will be periodically updated to include additional variants based on emerging genomic and preclinical/clinical data, including novel high-priority variants for pediatric solid tumors, lymphomas, and histiocytoses.
Results
Review and Prioritization of Agent Classes
The final list to be formally reviewed and prioritized contained 15 classes of targeted agents. A number of agent classes were discussed by the committee but ultimately not included in the list of agent classes to be formally reviewed (Table 3). The primary reasons for exclusion varied. In some cases, the frequency of the target or biomarker was uncommon in pediatric malignancies. In other cases, the agent class was deemed to be insufficiently targeted to enable identification of a biomarker predicting agent activity. Other targeted agent classes were excluded because the biomarker predicting agent activity was not yet known or was not detectable by the testing platform (Table 4). Of note, Pediatric MATCH leadership guidance to the committee was that the initial comprehensive review should focus on small molecule inhibitors rather than other classes of novel agents such as engineered cytotoxic T cells.
Table 3.
Levels of evidence for pediatric MATCH trial arms*
| Level | Criteria for levels of evidence |
|---|---|
| Level 1 | The drug is Food and Drug Administration approved for a malignant indication, and there is a molecular abnormality that can serve as a valid predictive marker. The subprotocol will not enroll patients with conditions for which the drug is approved or patients with conditions for which the drug has been shown not to have benefit. |
| Level 2 | The drug is investigational but met a clinical end point (progression-free survival, response) in any malignancy, has evidence of target inhibition, and has evidence of a predictive molecular marker. |
| Level 3 | The drug is investigational but has demonstrated clinical activity in any malignancy and evidence of target inhibition and has demonstrated evidence of a predictive molecular marker. |
MATCH = Molecular Analysis for Therapeutic Choice Trial.
Table 4.
Notable agent classes not reviewed and prioritized by the TAP committee*
| Agent class | Primary reason for exclusion |
|---|---|
| MDM2 inhibitors | Target (MDM2 amplification) uncommon |
| ERBB inhibitors | Target uncommon |
| Met inhibitor | Target (met amplification) uncommon |
| Src/Syk inhibitor | Target uncommon |
| c-Kit inhibitor | Target uncommon |
| Anti-angiogenic (VEGF and Ang/Tie) | Not sufficiently targeted to define biomarker |
| Pan-tyrosine kinase inhibitors | Not sufficiently targeted to define biomarker |
| Aurora kinase inhibitors | Target/biomarker not known |
| Base excision repair inhibitor (TRC102) | Target/biomarker not known |
| ATR kinase inhibitor (VX-970) | Target/biomarker not known |
| FAK inhibitor | Target/biomarker not known |
| CK2 inhibitors | Target/biomarker not defined by genomic alteration |
| IGF1R inhibitors | Target/biomarker not defined by genomic alteration |
TAP = Target and Agent Prioritization committee.
Results of the agent prioritization review are summarized in Box 1. The average priority score for reviewed agent classes ranged from 1.5 to 3.5. The number of committee members submitting a priority score vote ranged from 11 to 17 with an average of 14 members.
Box 1.
Agent classes formally reviewed and prioritized by the TAP committee*
| Agent class |
|---|
|
ALK = anaplastic lymphoma receptor kinase; EGFR = epidermal growth factor receptor; FGFR = fibroblast growth factor receptor; TAP = Target and Agent Prioritization committee.
Several drug classes reviewed by the TAP Committee target different components of the same signaling pathway: the BRAF, MEK, and ERK inhibitors and the PI3K, mTOR, and AKT inhibitors. This fact was acknowledged in the reviews, but each drug class was voted on separately. In addition, for ranking and committee voting, the review of ALK inhibitors was divided into second-generation ALK inhibitors and ALK inhibitors that inhibit additional tyrosine kinases (extended ALK inhibitors), and the review of PI3K/mTOR inhibitors was divided into PI3K and mTOR inhibitors so that the total number of target-agent pairs ranked was 17.
Reviewers raised aspects of study design that impact agent prioritization, including the primary end point to be used to measure agent activity and whether the selected drugs could be studied in combination (with chemotherapy) or only as a single agent. Specifically, if objective response rate is utilized as the primary end point for each phase II trial, then drug classes demonstrated in preclinical studies to have a cytostatic effect would receive a lower priority score. Drug classes demonstrated to have limited single-agent activity but to act synergistically with chemotherapy would receive higher priority if phase II trials combining targeted agents and chemotherapy would be considered for future inclusion in Pediatric MATCH.
Individual Target-Agent Pair Reviews
Evidence supporting a link between genomic alterations and response to therapy for each of the target-agent pairs is discussed below in order of study priority. More complete discussion, including target-agent summaries, level of evidence, biomarker detection, frequency of biomarker in pediatric malignancies, consideration of specific agents, clinical trials planned with biomarker-defined populations, and full summary of TAP Committee comments are provided in the Supplementary Materials and Supplementary Tables 1–5 (available online).
PI3K/mTOR Inhibitors
Introduction
Phosphoinositide 3-kinases (PI3Ks) function downstream of receptor tyrosine kinases (RTKs) and G-protein coupled receptors (GPCR) to activate protein kinase B (AKT), which in turn stimulates a number of progrowth and anti-apoptotic pathways within the cell, including regulating mechanistic target of rapamycin (mTOR) activity. Phosphatase and tensin homolog (PTEN) negatively regulates this pathway. mTOR functions in two complexes: TORC1 and TORC2. Allosteric mTOR inhibitors have been developed that selectively inhibit TORC1 activity, while ATP competitive mTOR inhibitors inhibit both TORC1 and TORC2. Activating mutations of this pathway have been identified in osteosarcoma, embryonal rhabdomyosarcoma, and diffuse intrinsic pontine gliomas (3,11,12).
Biomarker and Evidence
There has been extensive preclinical and some clinical evaluation of biomarkers of response to PI3K/mTOR pathway inhibitors, primarily focusing on adult malignancies (13). In preclinical studies in breast cancer, activating mutations of PIK3CA have been shown to confer sensitivity to PI3K inhibitors, AKT inhibitors, allosteric mTOR inhibitors, TORC1/2 inhibitors, and PI3K/mTOR inhibitors (14–21). These findings have been extended to other PIK3CA-mutant tumor models including PI3K inhibition in melanoma, lung, ovarian, prostate, and endometrial cancer (22,23); AKT inhibition in pancreatic, prostate, ovarian, non–small cell lung cancer, and ovarian cancer (24); and PI3K/mTOR inhibition in lung adenocarcinoma (25).
In preclinical studies, the relationship between PTEN deficiency and sensitivity to PI3K pathway inhibitors has been less clear. Some studies have found that some PTEN-deficient cell lines are sensitive to PI3K inhibitors (16,18,20), but others have found that PTEN-deficient cells are preferentially resistant to PI3K inhibitors (22), allosteric mTOR inhibitors (21), and PI3K/mTOR inhibitors (14). Recent studies have suggested that these discrepancies are because PTEN-deficient tumors are specifically dependent on the beta rather than the alpha isoform of PI3K (26–29).
Clinical studies have primarily focused on biomarkers of sensitivity to the allosteric mTOR inhibitors. In general, PIK3CA mutation or PTEN loss of function mutations predicted clinical response to these agents (15,30–33), with one study finding that the H1074R mutation in PIK3CA resulted in a higher response rate than other PIK3CA mutations (32). As an example, of 258 adult patients with advanced cancer treated at a single institution on phase I studies that included various inhibitors of the PI3K/MTOR pathway, 35% (six of 17) of patients with PIK3CA mutations achieved a partial response vs 6% of patients who did not have a PIK3CA mutation (34). Similarly, of 23 patients with PIK3CA-mutant breast, cervical, endometrial, and ovarian cancer treated on various phase I studies of PI3K/MTOR pathway inhibitors at a single institution, 30% had a partial response compared with 10% of patients with the same disease types lacking PIK3CA mutations (35). However, in a randomized study of molecularly targeted therapy for adult patients with advanced cancer, no progression-free survival benefit was seen for everolimus compared with conventional chemotherapy in patients with PI3K/mTOR pathway–activating mutations (36). In several studies, concurrent KRAS or BRAF mutations have been associated with resistance (15,30–32).
Evidence also suggests that downstream pathway mutations confer sensitivity to allosteric mTOR inhibitors. Everolimus was studied in a randomized phase III trial in patients with subependymal giant cell astrocytomas (SEGAs) and a clinical diagnosis of tuberous sclerosis, most of whom were predicted to have loss of function mutations in TSC1 or TSC2; 35% of everolimus-treated patients had at least 50% reduction in SEGA volume, and 53% of everolimus-treated patients had at least 50% reduction in the volume of their concurrent angiomyolipomas vs none in placebo-treated patients (37,38). Additionally, an extraordinary responder with a 14-month complete remission to everolimus and pazopanib has been described as having biallelic-activating mutations in MTOR (39).
Allosteric mTOR inhibitors have demonstrated clinical benefit in pediatric cancers, most recently in a randomized phase II trial when compared with bevacizumab in recurrent rhabdomyosarcoma (RMS) in combination with vinorelbine/cyclophosphamide (40). However, this population was not biomarker selected. Studies in adult malignancies have also shown that patients lacking PIK3CA and PTEN mutations can respond (31). In some studies, but not all, response has been correlated with phosphorylation of the mTORC1 target S6RP (41–46).
Recommendation
The TAP Committee strongly encouraged inclusion of at least one agent from this pathway in the Pediatric MATCH study. The strongest consideration should be given to mTOR or PI3K inhibitors. A combined PI3K/mTOR inhibitor would permit enrollment of all patients with confirmed biomarkers onto one arm, and in preclinical studies ATP-competitive MTOR inhibitors are associated with greater inhibition of downstream targets than rapalogs (47–49). AKT inhibitors were deprioritized because of their earlier stage of clinical development and lack of a defining biomarker that would predict response to AKT inhibition but not PI3K and/or mTOR inhibition. Based on clinical studies in adult patients, patients whose tumors harbor concurrent BRAF or KRAS mutations should be excluded from receiving PI3K, AKT, and mTOR inhibitors (15,30–32).
MEK Inhibitors
Introduction
The RAS–RAF–MEK1/2–ERK1/2 pathway, also known as the classical MAPK pathway, is responsible for controlling multiple key physiological processes (50). The MAPK pathway is one of the most frequently dysregulated signaling cascades in human cancer, and the aberrant activation of this pathway commonly occurs through gain-of-function mutations in genes encoding RAS and RAF family members, as well as by loss of NF1. Despite the low frequency of mutations in the MEK1/2 genes (MAP2K1 and MAP2K2) (51,52), MEK1 and MEK2 have emerged as ideal targets for therapeutic development because of their narrow substrate specificities, their distinctive structure, and their place at the bottleneck in the MAPK signaling pathway. The malignances seen in the pediatric and young adult populations with known MAPK pathway aberrations include hematological and lymphoid malignancies (activating NRAS/KRAS mutations, 20%) (53), rhabdomyosarcoma (activating BRAF, NRAS and PTPN11 mutations, 20%) (54), low-grade glioma (activating mutation or fusion in BRAF, 70%–100%), as well as in glioblastoma multiforme (mutation or deletion of NF1, BRAF mutation, 15%), neuroblastoma (activating mutations in NRAS, PTPN11, 2.9%–3.6%) (55), malignant peripheral nerve sheath tumors (NF1 loss 40%–88%) (56), and melanoma (activating mutation in BRAF, 86%) (57).
Biomarker and Evidence
MEK inhibitors have shown clinical responses in patients with BRAF-mutated melanoma refractory to BRAF inhibitors, leading to FDA approval of trametinib for refractory melanoma both as a single agent (58) as well as in combination with the BRAF inhibitor dabrafenib (59). Similarly, they have also shown clinical responses (20% with PR) in melanoma with NRAS mutation (60). In patients with KRAS-mutant lung cancers, MEK inhibition combined with gemcitabine (61) improves response rate and event-free survival. There is preclinical evidence for activity of MEK inhibitors in NF1-deficient neurofibromas and melanomas, and early results of a phase I trial of selumetinib (AZD6244) have shown clinical responses in more than 50% of pediatric patients with neurofibromatosis-1 (NF-1) with large plexiform neurofibroma (62–64). In uveal melanoma, which is characterized by mutations in GNAQ and GNA11 (G-binding protein alpha subunits that signal via the MAPK pathway), selumetinib results in a higher response rate and prolonged progression-free survival when compared with chemotherapy (65). In summary, there is clinical evidence supporting the following as biomarkers for response: activating N/K/HRAS mutations, activating BRAF mutations (V600E and others), GNAQ and GNA11 activating mutations, inactivating mutations in PTPN11, and inactivation of NF1 through inactivating mutations or insertion/deletion (63).
There are several preclinical studies demonstrating efficacy of MEK inhibitors in pediatric tumors with known RAS-ERK pathway aberrations. The MEK/ERK inhibitor UO126 has been shown to inhibit growth of rhabdomyosarcoma both as a single agent in vivo and in vitro (66) and in combination with the dual PI3K/mTOR inhibitor PI103 (67). In addition, in vitro and in vivo synergy has also been seen between inhibitors of TORC1/2 (AZD8055) and MEK (AZD6244) in embryonal rhabdomyosarcoma (68). Preclinical data also support potential activity for MEK inhibitors against neuroblastoma with MAPK pathway gene mutations (69). Lastly, NF1 deficiency has shown to be predictive of sensitivity to MEK inhibitors in vitro in glioblastoma multiforme (70). In preclinical studies, some MAP2K1 (MEK1) mutations are sensitive to MEK inhibition (71,72).
Recommendation
In view of the high frequency of MAPK pathway aberrations within the pediatric oncology population and promising clinical activity in melanoma as well as in plexiform neurofibroma, TAP Committee members were enthusiastic to include MEK inhibitors as a part of the Pediatric MATCH trial.
PDGFR Inhibitors
Introduction
The platelet-derived growth factor receptors alpha (PRGFRA) and beta (PDGFRB) are expressed in oligodendrocytes and in a variety of cells derived from mesenchymal stem cells including fibroblasts and vascular smooth muscle cells. PDGFRA mutations are found in pediatric high-grade gliomas (HGGs) and diffuse intrinsic pontine gliomas (DIPGs) (73,74), and approximately 25% to 35% of DIPGs have PDGFRA amplification (75). Sarcomas occurring rarely in children, inflammatory myofibroblastic tumors and dermatofibrosarcoma protuberans, have fusions of PDGFRB or PDGFB.
Biomarker and Evidence
Several of the point mutations identified in DIPG and pediatric HGG are transforming in a p53-deficient astrocyte model, and in this model small molecule inhibitors of PDGFRA crenolanib and dasatinib (73) block ligand-independent receptor activation. Phase II studies of imatinib, an inhibitor of PDGFRA, in recurrent gliomas have not shown activity in patients with glioblastoma (76,77). However, PDGFRA amplification and mutation status have not been assessed in these trials, so it is possible that these studies failed because the target population that would benefit from treatment was not adequately identified. In patients with dermatofibrosarcoma protuberans, characterized by a COL1A1-PDGFRB fusion, objective responses are seen in a majority of patients who receive imatinib (78,79). In refractory leukemias with PDGFRB fusions, imatinib therapy has been reported to result in long-term responses (80). In gastrointestinal stromal tumors (GISTs), a subset of the activating PDGFRA mutations predict response to imatinib (81).
Recommendation
If other phase II studies of PDGFR inhibitors in children with PDGFR-mutant HGG and DIPG were not planned, then this would be a reasonable class of agents to include in the pediatric NCI MATCH trial. To date, the only evidence supporting an association between the PDGFR variants most likely to be found in the study patient population is preclinical. This may change as results of the ongoing phase I trial of crenolanib become available. The Pediatric MATCH leadership could consider waiting for the results of that trial to make a decision about including this class of agents.
BRAF Inhibitors
Introduction
Mutations in BRAF that induce constitutive activation of the MAPK pathway arise in approximately 7% of all cancers, including a variety of pediatric malignancies. Activating mutations in BRAF (or genes encoding other MAPK pathway proteins) are observed at a very high frequency in pediatric brain tumors, melanoma, and LCH, with BRAF-V600E being the most common mutation (82).
Biomarker and Evidence
First-generation BRAF inhibitors specifically target BRAF-V600E, BRAF-V600K, or other more rare mutations that induce activation of the BRAF monomer (83). Next-generation agents that target dimeric RAF may impact increased ERK activation induced by BRAF fusion or copy gain alterations (84). Because pathologically activated BRAF acts through phosphorylation of downstream ERK, drugs that inhibit MEK or ERK activity may also be considered for patients with somatic BRAF alterations (see MEK inhibitor and ERK inhibitor sections), either as monotherapy or in combination with RAF inhibitors.
Abundant preclinical evidence strongly supports the function of BRAF-V600E as a driver of pathogenesis across many pediatric diseases (85–87). Phase II and phase III clinical trials demonstrate clinical responses and improved overall survival in adults with advanced metastatic melanoma with the BRAF-V600E mutation treated with vemurafenib (85,88). A randomized study suggested improved efficacy with combination dabrafenib (BRAF inhibition)/trametinib (MEK inhibition) strategy compared with dabrafenib monotherapy (89). Vemurafenib has also been reported to have clinical efficacy in adults with Erdheim-Chester disease and LCH, characterized with sustained responses in most patients and no reports to date of disease progression on therapy (90–92).
Recommendation
Activating mutations in BRAF arise with considerable frequency, and preclinical and clinical evidence strongly support targeting the MAPK pathway as a strategy with potential efficacy for these patients. The early experiences with targeted inhibition of mutant BRAF in melanoma serve as a paradigm for the potential for mutation-directed therapy. However, first-generation BRAF inhibitors have limitations: efficacy only against activated BRAF monomers, considerable side effects, and quick development of resistance, at least in the hypermutated setting of melanoma. Many new agents with more precise targets and combinations of agents at several nodes of the MAPK pathway, or multiple pathways, are in development. The TAP Committee therefore favors inclusion of tumors with BRAF point mutations, fusions, and amplifications in this trial, which would require inclusion of a second-generation BRAF inhibitor.
ALK Inhibitors and Extended ALK Inhibitors
Introduction
ALK encodes the protein anaplastic lymphoma receptor kinase (ALK), which belongs to the insulin receptor superfamily. Germline-activating mutations in ALK result in an increased risk for developing neuroblastoma (NBL) (93,94). ALK is rearranged, mutated, or amplified in several cancers including anaplastic large cell lymphomas (ALCLs), NBL, inflammatory myofibroblastic tumors (IMTs), non–small cell lung cancer (NSCLC), and RMS.
Biomarker and Evidence
ALK rearrangements predict response to crizotinib in NSCLC, IMT, and ALCL (94,95). ALK point mutations are variably sensitive to crizotinib in preclinical models and in clinical trials (96,97,98). ALK amplifications are reported in NBL and RMS, but whether ALK amplification is linked to response to ALK inhibition is not yet known.
ALK rearrangements predict response to crizotinib in NSCLC, IMT, and ALCL (99–101). In a phase III trial in lung cancer with ALK rearrangements, crizotinib therapy produced improved outcomes compared with chemotherapy. In a phase I/II trial in children with recurrent ALCL, there is a very high response rate with crizotinib (100). ALK point mutations are variably sensitive to crizotinib in preclinical models and clinical trials, and in a phase I/II trial of crizotinib in patients with recurrent NBL, occasional radiographic responses are observed (96,97).
Recommendation
Although several trials with first- and second-generation ALK inhibitors are planned in newly diagnosed and recurrent ALCL and NBL, strong consideration should be given to including a second-generation ALK inhibitor or an extended ALK inhibitor in the Pediatric MATCH trial. Despite competing studies there would be an anticipated patient population for this trial. The TAP Committee also recommended that this arm should allow patients with prior crizotinib to enroll. Therefore an additional patient population would be those patients with malignancies with activating ALK mutations who develop resistance to crizotinib.
TRK Inhibitors
Introduction
The TRK family proteins are receptor tyrosine kinases involved in nervous system development. Gene fusions involving each of the NTRK genes have been identified in a wide range of malignancies including several seen in pediatric patients: gliomas, mesoblastic nephroma, and infantile fibrosarcoma (102).
Biomarker and Evidence
The reported TRK fusions occurring in cancer have the 3’ region of TRK including the kinase domain fused to the 5’ sequence from a number of partner genes. For example, the ETV6-NTRK3 fusion identified in mesoblastic nephroma, infantile fibrosarcoma, and other malignancies has varying breakpoints but always contains the kinase domain of NTRK3 and the sterile alpha receptor (SAM) dimerization domain of ETV6. Although NTRK rearrangements were first identified several decades ago, development of TRK inhibitors has been slow, and so only recently has evidence emerged linking the presence of these fusions to response to TRK inhibitors. A patient with lung cancer harboring a MPRIP-NTRK1 translocation had a minor response after crizotinib, a weak TRK inhibitor (103). More recently, a partial response to LOXO-101 in a patient with undifferentiated sarcoma with an LMNA-NTRK1 fusion has been reported (104).
Recommendation
The committee recommended consideration of TRK inhibitors for a second phase of the MATCH trial when more information would be available regarding frequency of TRK fusions in pediatric malignancies and the activity and toxicity profile of the agents being studied.
BET Bromodomain Inhibitors
Introduction
The BET family of bromodomain proteins (BRD2, BRD3, BRD4, BRDT) is a family of actyl-lysine “epigenetic reader” proteins that bind histone tails and modify chromatin accessibility to binding complexes involved in transcription. BET inhibitors directly affect BRD-containing proteins (including BRD3/4 fusions) and the ability of BRD proteins to activate the transcription of oncogenes, such as MYC family proteins (105).
Biomarker and Evidence
The most common biomarkers relevant to pediatric tumors are amplification of MYCN and translocations and amplifications of MYC (56,106,107) in subsets of neuroblastoma, medulloblastoma, and lymphomas. Neuroblastoma cell lines in vitro and in vivo (xenografts and MYCN-transgenic model) with high MYCN levels were selectively sensitive to the BET inhibitor JQ-1 and IBET726, resulting in cell cycle arrest, apoptosis (56,106), and downregulation of MYCN levels and downstream MYC targets. Similar JQ-1 effects were observed in medulloblastoma cell lines, xenografts, and GEMMs with high levels of MYC (107,108). Many studies have also demonstrated similar effects of BET inhibitors JQ1 and OTX015 in preclinical myc-driven leukemias and lymphoma models including MYC translocation–positive Burkitt lymphoma (109,110). Interestingly, in GBM models MYC expression may not correlate with BET inhibitor responsiveness (111).
BET inhibitors also induce differentiation and growth arrest of NUT midline carcinoma (NMC) cells, which have fusions involving NUT, most commonly partnered with BRD3 or 4 methyltransferase (105,112,113). In medulloblastoma, hedgehog (HH)-driven tumors respond to BET inhibitors via effects on BRD4 binding to the promoters of GLI1 and GLI2 (114,115). Increased levels of BRD3 and 4 (often due to translocation) may also predict activity. There is conflicting data as to whether TP53 missense mutations may promote BET inhibitor resistance (56,107,111). Further preclinical studies are required to determine if all, or subsets of TP53 missense mutations, confer BET inhibitor resistance prior to determining whether these alterations should be used to determine eligibility in clinical trials.
Recommendation
There is preclinical data supporting the use of BET inhibitors for a number of pediatric solid tumors and lymphomas. Biomarkers of activity, such as MYC or MYCN amplification, are often enriched in the poorest prognosis subgroups of neuroblastoma and medulloblastoma patients, and so potential biomarkers of response are expected to be relatively common in the patient population eligible for MATCH. Because of the mechanism of action, this agent class potentially has broad effects on fundamental cellular processes. This likely contributes to the diverse array of biomarkers of response thus far identified in preclinical studies. Consequently, it is still not clear whether there are biomarkers that will predict response across pediatric histologies. In other words, at this early stage of drug development it is not clear whether, outside of rare BRD fusion–positive cancers, BET inhibitors are truly targeted therapies. Phase I trials of this agent class in pediatric patients are ongoing. In summary, while BET inhibitors may be ideally suited to study in the Pediatric MATCH, data from ongoing clinical trials may be needed in order for the TAP Committee to appropriately prioritize this agent class (see section on Co-Chair assessment below).
CDK4/6 Inhibitors
Introduction
CDK4/6 inhibitors are small molecule inhibitors of the cyclin-dependent serine threonine kinases CDK4 and 6, which normally form a complex with cyclin D that phosphorylates the tumor suppressor pRb, preventing its binding to E2F transcription factors, leading to cell cycle progression (116). CyclinD-cdk4/6-INK4a-Rb is one of the most commonly altered pathways in cancers, including amplification and mutations of CCND1, CDK4, or CDK6 in pediatric CNS tumors and NBL, and mutation or deletion of SMARCB1 in rhabdoid tumors and CDKN2A in a wide range of tumors (117–123).
Biomarker and Evidence
Preclinical studies suggest that activating alterations in cyclinD-cdk4/6-INK4a signaling, as well as functionally intact pRb, are required for cdk4/6 inhibitor sensitivity. However, many preclinical studies and early clinical trial results suggest that while necessary, alterations in this pathway are not sufficient to predict response, possibly in part because of redundancies in cyclin/cdk signaling pathways. Nevertheless, in most studies low p16 and intact pRb are required for sensitivity in vitro, but other biomarkers are emerging in specific tumors. For example, NBL sensitivity to the cdk4/6 inhibitor LEE011 correlated with MYCN amplification (124).
There are many relevant preclinical studies in adult tumors, including breast, NSCLC, melanoma, and liposarcomas, that generally demonstrate selective responses in cells in vitro and in vivo, associated in part with activating alterations in cyclinD-cdk4/6-INK4a pathways (reviewed in [125–127]). The preclinical data linking these activating alterations to sensitivity include both shRNA knockdown of relevant targets (eg, cdk4/6) and pharmacologic inhibition. In neuroblastoma shRNA-targeting cdk4 and treatment with LEE011-inhibited growth, inducing cell cycle arrest, senescence, and dose-dependent decreased phosphorylation of pRb and FOXM1 in 12 of 17 cell lines in nanomolar concentrations in vitro and in vivo (124). Although the majority of these lines had hyperactivation of CDK 4/6 signaling, several were not sensitive, but interestingly the presence of MYCN amplification correlated with lower IC50. Palbociclib sensitivity of GBM in vitro and in vivo is associated with deletion of CDKN2A and C as well as low levels of p16 (128–130). Resistance was associated with pRb deletion and/or pRb shRNA knockdown. In one report, nonamplified CDK4 status or high levels of CDK6 conferred sensitivity (129). Treatment with palbociclib (or p16 knockdown) induced growth inhibition and G1 arrest in rhabdoid cell lines and was inversely correlated with p16 expression (131). CDk4 inhibition in rhabdomyosarcoma cell lines with palbociclib also induced G1 arrest and growth inhibition in vitro and in vivo (132); however, a recent study demonstrated that while the growth of most rhabdomyosarcoma cells was inhibited by LEE011 (and CDK4 shRNA), a subset of fusion-positive CDK4-overexpressing cells were resistant (133). These studies highlight the importance of activating alterations that can in part determine cdk4/6 inhibitor sensitivity, but pRb, CDK4, CDK6, and p16 status alone cannot be used to accurately predict the response to cdk4/6 inhibition.
Recommendation
The TAP Committee felt that inclusion of this class of drugs in the Pediatric MATCH trial was dependent on two factors. First, most preclinical studies suggest that these inhibitors are cytostatic and often induce differentiation and/or senescence. Thus, if stable disease or time to progression will not be considered a successful response, then the agent is less likely to be prioritized, especially in the NBL population where Response Evaluation Criteria In Solid Tumors (RECIST) responses may be more difficult to identify in some subsets of patients (eg, patients with only marrow or metaiodobenzylguanidine (MIBG) positivity). Secondly, this agent may be better suited for combination studies with other inhibitors.
FGFR Inhibitors
Introduction
Fibroblast growth factor receptors (FGFRs) bind to fibroblast growth factors that initiate kinase-mediated activation of oncogenic downstream signaling. The FGFR family consists of five receptors named FGFR1 to FGFR5. Amplifications of FGFR1 are seen in 3% of rhabdomyosarma, 10% of breast cancer, and 21% of lung adenocarcinoma, and mutations in FGFR4 have been reported in rhabdomyosarcoma.
Biomarker and Evidence
Deregulated FGFR signaling secondary to amplification, translocations, and point mutations in FGFR1, FGFR2, and FGFR3 is a biomarker that may predict response to FGFR inhibitors (134,135). Supplementary Table 11"4"]?> (available online) lists the common genetic alterations impacting FGFR1-4 in adult and pediatric cancers. Breast cancers that show FGFR1, 2, or 3 amplifications detectable by fluorescence in situ hybridization show sensitivity to FGFR inhibitors, as indicated by a higher response rate to the pan-tyrosine kinase inhibitor dovitinib than seen in those without amplification (136). Multiple myeloma with FGFR3 translocation treated with dovitinib demonstrated stable disease (137). Two patients with GBM with FGFR translocations treated with JNJ-42756493 showed stable disease and minor response (138). The sensitivity of other FGFR genetic alterations to FGFR inhibitors is largely unknown.
Recommendation
The TAP Committee recommended that FGFR inhibitors be included in Pediatric MATCH for tumors characterized by mutations, amplifications, or translocations in FGFR1-4, where the inhibitor has demonstrated activity against the specific FGFR alterations.
Additional Target-Agent Pairs
Discussion of inhibitors against EGFR, IDH, SMO, PARP, and ERK, which were not selected for inclusion in the Pediatric MATCH at this time, is included in the Supplementary Materials (available online).
Co-Chair Assessment
The Co-Chairs deprioritized the CDK4/6 inhibitors and the BET bromodomain inhibitors because the highest possible levels of evidence linking the biomarkers and response were preclinical. MEK, BRAF, PI3K, and mTOR inhibitors were given the highest priority based on inclusion of study arms with these agents in the adult NCI-MATCH trial, suggesting that it would be feasible to plan trial arms for these agents in Pediatric MATCH.
Discussion
The systematic approach undertaken by the Pediatric MATCH TAP Committee produced a prioritized list of targeted agent classes to be considered for inclusion in a basket trial. Prioritization took into account the opinion of Pediatric MATCH stakeholders as well as available evidence. Rapid evolution in novel therapeutics and cancer genomics raises the important question of the optimal manner in which to maintain knowledge during the course of a study such as Pediatric MATCH. The TAP Committee will therefore continue to meet on a quarterly basis during trial development and during the conduct of the trial to evaluate whether additional target-agent pairs should be reviewed by the committee for potential inclusion in Pediatric MATCH.
The TAP Committee used peer-reviewed publications and published abstracts as their primary sources of evidence for conducting systematic reviews of target-agent pairs. There was a discussion of more extensive use of publically available primary sequencing databanks such as The Cancer Genome Atlas (TCGA), the International Cancer Genome Consortium (ICGC), or the St. Jude PeCan Data Portal. Of note, many available genomic data sets (eg, TCGA) do not include pediatric cancer data. However, there are several limitations to the currently available primary sequencing databanks for pediatric malignancies. Most importantly, almost all of the sequenced samples are newly diagnosed rather than recurrent (post-treatment) samples and therefore may lack relevance to the patient population to be included in the Pediatric MATCH study. The sequencing platforms utilized and the manner in which sequencing data is stored in these databanks will also limit the extent to which these databanks will be informative about the frequency of translocations.
The pediatric malignancy with the greatest number of sequenced samples in ICGC is NBL with 605 cases, but only about 400 of these cases have adequate sequencing data available to assess the frequency of actionable mutations of interest. Given the small number of sequenced cases (1239 pediatric solid tumors in ICGC, 785 pediatric solid tumors in PeCan), there is limited power to detect recurrent mutations occurring at a frequency of less than 10%. Further, for many rare pediatric malignancies no primary sequencing data are available in these databanks. Thus the committee felt that literature review and expert input was an optimal manner in which to assess the frequency of potentially actionable mutations of interest for the purpose of prioritizing target-agent pairs. Additional resources for pediatric-specific cancer sequencing data, such as the recently released Foundation Medicine pediatric database, will be utilized by the committee as they become available (139).
Conclusions
The Pediatric MATCH represents a paradigm shift in approaching refractory and relapsed pediatric cancers with targeted therapeutic approaches based on molecular lesions rather than tumor histology. The “umbrella” approach allows inclusion of children with rare malignancies for whom phase II research opportunities are often limited. The review and prioritization approach described here represents a strategic step toward precision medicine for children with cancer. It is hoped that Pediatric MATCH will establish a dynamic platform from which to gain a better understanding of the genomic landscape of relapsed and refractory cancers and seek efficacy signals of matched therapeutic agents that may improve the outcome for a spectrum of childhood cancers.
Funding
The Children’s Oncology Group has been awarded a grant by the National Cancer Institute (NCI; 1U10CA180886) as a member of the NCI National Clinical Trials Network that will support the Pediatric NCI Molecular Analysis for Therapeutic Choice Trial.
Notes
The funders had no role in the writing of the review or the decision to submit it for publication.
Pediatric MATCH Target and Agent Prioritization Committee: James Amatruda, University of Texas Southwestern Medical Center; Alice Chen, National Cancer Institute; Amar Gajjar, St. Jude Children’s Hospital; Jim Geller, Cincinnati Children's Hospital Medical Center; Richard Gorlick, Montefiore Medical Center; Terzah Horton, Baylor College of Medicine; Javed Khan, National Cancer Institute; Stephen Lessnick, Nationwide Children’s Hospital; Mei Polley, National Cancer Institute; Greg Reaman, Food and Drug Administration; Giles Robinson, St. Jude Children’s Hospital; Malcolm Smith, Cancer Therapy Evaluation Program; Naoko Takebe, Cancer Therapy Evaluation Program.
Supplementary Material
References
- 1. Badalian-Very G, Vergilio JA, Degar BA, et al. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116(11):1919–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mody RJ, Wu YM, Lonigro RJ, et al. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA. 2015;314(9):913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Perry JA, Kiezun A, Tonzi P, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci U S A. 2014;111(51):E5564–E5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pugh TJ, Weeraratne SD, Archer TC, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488(7409):106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor JGt, Cheuk AT, Tsang PS, et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119(11):3395–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Simon R, Roychowdhury S.. Implementing personalized cancer genomics in clinical trials. Nat Rev Drug Discov. 2013;12(5):358–369. [DOI] [PubMed] [Google Scholar]
- 8. Sleijfer S, Bogaerts J, Siu LL.. Designing transformative clinical trials in the cancer genome era. J Clin Oncol. 2013;31(15):1834–1841. [DOI] [PubMed] [Google Scholar]
- 9. Abrams J, Conley B, Mooney M, et al. National Cancer Institute's Precision Medicine Initiatives for the new National Clinical Trials Network. Am Soc Clin Oncol Educ Book. 2014;71–6. [DOI] [PubMed] [Google Scholar]
- 10. Conley BG, Chen A, O'Dweyer P, et al. NCI-molecular analysis for therapy choice (NCI-MATCH) clinical trial: Interim analysis. Cancer Res. 2016;76(14 suppl CT101). [Google Scholar]
- 11. Agaram NP, Chen CL, Zhang L, et al. Recurrent MYOD1 mutations in pediatric and adult sclerosing and spindle cell rhabdomyosarcomas: Evidence for a common pathogenesis. Genes Chromosomes Cancer. 2014;53(9):779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grill J, Puget S, Andreiuolo F, et al. Critical oncogenic mutations in newly diagnosed pediatric diffuse intrinsic pontine glioma. Pediatr Blood Cancer. 2012;58(4):489–491. [DOI] [PubMed] [Google Scholar]
- 13. Weigelt B, Downward J.. Genomic determinants of PI3K pathway inhibitor response in cancer. Front Oncol. 2012;2:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brachmann SM, Hofmann I, Schnell C, et al. Specific apoptosis induction by the dual PI3K/mTor inhibitor NVP-BEZ235 in HER2 amplified and PIK3CA mutant breast cancer cells. Proc Natl Acad Sci U S A. 2009;106(52):22299–22304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Di NF, Arena S, Tabernero J, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest. 2010;120(8):2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O'Brien C, Wallin JJ, Sampath D, et al. Predictive biomarkers of sensitivity to the phosphatidylinositol 3' kinase inhibitor GDC-0941 in breast cancer preclinical models. Clin Cancer Res. 2010;16(14):3670–3683. [DOI] [PubMed] [Google Scholar]
- 18. Sanchez CG, Ma CX, Crowder RJ, et al. Preclinical modeling of combined phosphatidylinositol-3-kinase inhibition with endocrine therapy for estrogen receptor-positive breast cancer. Breast Cancer Res. 2011;13(2):R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Serra V, Markman B, Scaltriti M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68(19):8022–8030. [DOI] [PubMed] [Google Scholar]
- 20. She QB, Chandarlapaty S, Ye Q, et al. Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS One. 2008;3(8):e3065. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Weigelt B, Warne PH, Downward J.. PIK3CA mutation, but not PTEN loss of function, determines the sensitivity of breast cancer cells to mTOR inhibitory drugs. Oncogene. 2011;30(29):3222–3233. [DOI] [PubMed] [Google Scholar]
- 22. Tanaka H, Yoshida M, Tanimura H, et al. The selective class I PI3K inhibitor CH5132799 targets human cancers harboring oncogenic PIK3CA mutations. Clin Cancer Res. 2011;17(10):3272–3281. [DOI] [PubMed] [Google Scholar]
- 23. Yu K, Toral-Barza L, Shi C, et al. Response and determinants of cancer cell susceptibility to PI3K inhibitors: Combined targeting of PI3K and Mek1 as an effective anticancer strategy. Cancer Biol Ther. 2008;7(2):307–315. [DOI] [PubMed] [Google Scholar]
- 24. Meuillet EJ, Zuohe S, Lemos R, et al. Molecular pharmacology and antitumor activity of PHT-427, a novel Akt/phosphatidylinositide-dependent protein kinase 1 pleckstrin homology domain inhibitor. Mol Cancer Ther. 2010;9(3):706–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Engelman JA, Luo J, Cantley LC.. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7(8):606–619. [DOI] [PubMed] [Google Scholar]
- 26. Jia S, Liu Z, Zhang S, et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454(7205):776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wee S, Wiederschain D, Maira SM, et al. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A. 2008;105(35):13057–13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ni J, Liu Q, Xie S, et al. Functional characterization of an isoform-selective inhibitor of PI3K-p110beta as a potential anticancer agent. Cancer Discov. 2012;2(5):425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edgar KA, Wallin JJ, Berry M, et al. Isoform-specific phosphoinositide 3-kinase inhibitors exert distinct effects in solid tumors. Cancer Res. 2010;70(3):1164–1172. [DOI] [PubMed] [Google Scholar]
- 30. Janku F, Tsimberidou AM, Garrido-Laguna I, et al. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther. 2011;10(3):558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garrido-Laguna I, Hong DS, Janku F, et al. KRASness and PIK3CAness in patients with advanced colorectal cancer: outcome after treatment with early-phase trials with targeted pathway inhibitors. PLoS One. 2012;7(5):e38033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Janku F, Wheler JJ, Naing A, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73(1):276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moroney JW, Schlumbrecht MP, Helgason T, et al. A phase I trial of liposomal doxorubicin, bevacizumab, and temsirolimus in patients with advanced gynecologic and breast malignancies. Clin Cancer Res. 2011;17(21):6840–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Janku F, Tsimberidou AM, Garrido-Laguna I, et al. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther. 2011;10(3):558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Janku F, Wheler JJ, Westin SN, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012;30(8):777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Le Tourneau C, Delord JP, Goncalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16(13):1324–1334. [DOI] [PubMed] [Google Scholar]
- 37. Franz DN, Belousova E, Sparagana S, et al. Efficacy and safety of everolimus for subependymal giant cell astrocytomas associated with tuberous sclerosis complex (EXIST-1): A multicentre, randomised, placebo-controlled phase 3 trial. Lancet. 2013;381(9861):125–132. [DOI] [PubMed] [Google Scholar]
- 38. Kingswood JC, Jozwiak S, Belousova ED, et al. The effect of everolimus on renal angiomyolipoma in patients with tuberous sclerosis complex being treated for subependymal giant cell astrocytoma: Subgroup results from the randomized, placebo-controlled, Phase 3 trial EXIST-1. Nephrol Dial Transplant. 2014;29(6):1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wagle N, Grabiner BC, Van Allen EM, et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov. 2014;4(5):546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mascarenhas L, Meyer WH, Lyden E, et al. Randomized phase II trial of bevacizumab and temsirolimus in combination with vinorelbine (V) and cylcophosphamide (C) for first relapse/disease progression of rhabdomyosarcoma (RMS): A report from the Children's Oncology Group. American Society of Oncology Annual Meeting. J Clin Oncol. 2014;32:5s(suppl; abstr 10003). [Google Scholar]
- 41. Iwenofu OH, Lackman RD, Staddon AP, et al. Phospho-S6 ribosomal protein: A potential new predictive sarcoma marker for targeted mTOR therapy. Mod Pathol. 2008;21(3):231–237. [DOI] [PubMed] [Google Scholar]
- 42. Behbakht K, Sill MW, Darcy KM, et al. Phase II trial of the mTOR inhibitor, temsirolimus and evaluation of circulating tumor cells and tumor biomarkers in persistent and recurrent epithelial ovarian and primary peritoneal malignancies: A Gynecologic Oncology Group study. Gynecol Oncol. 2011;123(1):19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Galanis E, Buckner JC, Maurer MJ, et al. Phase II trial of temsirolimus (CCI-779) in recurrent glioblastoma multiforme: A North Central Cancer Treatment Group Study. J Clin Oncol. 2005;23(23):5294–5304. [DOI] [PubMed] [Google Scholar]
- 44. Tarhini A, Kotsakis A, Gooding W, et al. Phase II study of everolimus (RAD001) in previously treated small cell lung cancer. Clin Cancer Res. 2010;16(23):5900–5907. [DOI] [PubMed] [Google Scholar]
- 45. Cho D, Signoretti S, Dabora S, et al. Potential histologic and molecular predictors of response to temsirolimus in patients with advanced renal cell carcinoma. Clin Genitourin Cancer. 2007;5(6):379–385. [DOI] [PubMed] [Google Scholar]
- 46. Chawla SP, Staddon AP, Baker LH, et al. Phase II study of the mammalian target of rapamycin inhibitor ridaforolimus in patients with advanced bone and soft tissue sarcomas. J Clin Oncol. 2012;30(1):78–84. [DOI] [PubMed] [Google Scholar]
- 47. Chresta CM, Davies BR, Hickson I, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70(1):288–298. [DOI] [PubMed] [Google Scholar]
- 48. Yu K, Toral-Barza L, Shi C, et al. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res. 2009;69(15):6232–6240. [DOI] [PubMed] [Google Scholar]
- 49. Thoreen CC, Kang SA, Chang JW, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284(12):8023–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao Y, Adjei AA.. The clinical development of MEK inhibitors. Nat Rev Clin Oncol. 2014;11(7):385–400. [DOI] [PubMed] [Google Scholar]
- 51. Marks JL, Gong Y, Chitale D, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008;68(14):5524–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Murugan AK, Dong J, Xie J, et al. MEK1 mutations, but not ERK2 mutations, occur in melanomas and colon carcinomas, but none in thyroid carcinomas. Cell Cycle. 2009;8(13):2122–2124. [DOI] [PubMed] [Google Scholar]
- 53. Pui CH, Carroll WL, Meshinchi S, et al. Biology, risk stratification, and therapy of pediatric acute leukemias: An update. J Clin Oncol. 2011;29(5):551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shukla N, Ameur N, Yilmaz I, et al. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18(3):748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bentires-Alj M, Paez JG, David FS, et al. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64(24):8816–8820. [DOI] [PubMed] [Google Scholar]
- 56. Puissant A, Frumm SM, Alexe G, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3(3):308–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lu C, Zhang J, Nagahawatte P, et al. The genomic landscape of childhood and adolescent melanoma. J Invest Dermatol. 2015;135(3):816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Flaherty KT, Robert C, Hersey P, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367(2):107–114. [DOI] [PubMed] [Google Scholar]
- 59. Long GV, Stroyakovskiy D, Gogas H, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371(20):1877–1888. [DOI] [PubMed] [Google Scholar]
- 60. Ascierto PA, Schadendorf D, Berking C, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: A non-randomised, open-label phase 2 study. Lancet Oncol. 2013;14(3):249–256. [DOI] [PubMed] [Google Scholar]
- 61. Janne PA, Shaw AT, Pereira JR, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: A randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14(1):38–47. [DOI] [PubMed] [Google Scholar]
- 62. Jessen WJ, Miller SJ, Jousma E, et al. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J Clin Invest. 2013;123(1):340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nissan MH, Pratilas CA, Jones AM, et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res. 2014;74(8):2340–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Spunt SL, Million L, Anderson JR, et al. Risk-based treatment for nonrhabdomyosarcoma soft tissue sarcomas (NRSTS) in patients under 30 years of age: Children's Oncology Group study ARST0332. J Clin Oncol. 2014;32(5s):10008. [Google Scholar]
- 65. Carvajal RD, Sosman JA, Quevedo JF, et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: A randomized clinical trial. JAMA. 2014;311(23):2397–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Marampon F, Bossi G, Ciccarelli C, et al. MEK/ERK inhibitor U0126 affects in vitro and in vivo growth of embryonal rhabdomyosarcoma. Mol Cancer Ther. 2009;8(3):543–551. [DOI] [PubMed] [Google Scholar]
- 67. Guenther MK, Graab U, Fulda S.. Synthetic lethal interaction between PI3K/Akt/mTOR and Ras/MEK/ERK pathway inhibition in rhabdomyosarcoma. Cancer Lett. 2013;337(2):200–209. [DOI] [PubMed] [Google Scholar]
- 68. Renshaw J, Taylor KR, Bishop R, et al. Dual blockade of the PI3K/AKT/mTOR (AZD8055) and RAS/MEK/ERK (AZD6244) pathways synergistically inhibits rhabdomyosarcoma cell growth in vitro and in vivo. Clin Cancer Res. 2013;19(21):5940–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Eleveld TF, Oldridge DA, Bernard V, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet. 2015;47(8):864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. See WL, Tan IL, Mukherjee J, et al. Sensitivity of glioblastomas to clinically available MEK inhibitors is defined by neurofibromin 1 deficiency. Cancer Res. 2012;72(13):3350–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chakraborty R, Hampton OA, Shen X, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood. 2014;124(19):3007–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Goetz EM, Ghandi M, Treacy DJ, et al. ERK mutations confer resistance to mitogen-activated protein kinase pathway inhibitors. Cancer Res. 2014;74(23):7079–7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Paugh BS, Zhu X, Qu C, et al. Novel oncogenic PDGFRA mutations in pediatric high-grade gliomas. Cancer Res. 2013;73(20):6219–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Puget S, Philippe C, Bax DA, et al. Mesenchymal transition and PDGFRA amplification/mutation are key distinct oncogenic events in pediatric diffuse intrinsic pontine gliomas. PLoS One. 2012;7(2):e30313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zarghooni M, Bartels U, Lee E, et al. Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor alpha and poly (ADP-ribose) polymerase as potential therapeutic targets. J Clin Oncol. 2010;28(8):1337–1344. [DOI] [PubMed] [Google Scholar]
- 76. Raymond E, Brandes AA, Dittrich C, et al. Phase II study of imatinib in patients with recurrent gliomas of various histologies: A European Organisation for Research and Treatment of Cancer Brain Tumor Group Study. J Clin Oncol. 2008;26(28):4659–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Paulsson J, Lindh MB, Jarvius M, et al. Prognostic but not predictive role of platelet-derived growth factor receptors in patients with recurrent glioblastoma. Int J Cancer. 2011;128(8):1981–1988. [DOI] [PubMed] [Google Scholar]
- 78. Kerob D, Porcher R, Verola O, et al. Imatinib mesylate as a preoperative therapy in dermatofibrosarcoma: Results of a multicenter phase II study on 25 patients. Clin Cancer Res. 2010;16(12):3288–3295. [DOI] [PubMed] [Google Scholar]
- 79. Ugurel S, Mentzel T, Utikal J, et al. Neoadjuvant imatinib in advanced primary or locally recurrent dermatofibrosarcoma protuberans: A multicenter phase II DeCOG trial with long-term follow-up. Clin Cancer Res. 2014;20(2):499–510. [DOI] [PubMed] [Google Scholar]
- 80. Weston BW, Hayden MA, Roberts KG, et al. Tyrosine kinase inhibitor therapy induces remission in a patient with refractory EBF1-PDGFRB-positive acute lymphoblastic leukemia. J Clin Oncol. 2013;31(25):e413–e416. [DOI] [PubMed] [Google Scholar]
- 81. Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21(23):4342–4349. [DOI] [PubMed] [Google Scholar]
- 82. Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. [DOI] [PubMed] [Google Scholar]
- 83. Bollag G, Hirth P, Tsai J, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467(7315):596–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Samatar AA, Poulikakos PI.. Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat Rev Drug Discov. 2014;13(12):928–942. [DOI] [PubMed] [Google Scholar]
- 85. Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Berres ML, Lim KP, Peters T, et al. BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups. J Exp Med. 2014;211(4):669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pratilas CA, Xing F, Solit DB.. Targeting oncogenic BRAF in human cancer. Curr Top Microbiol Immunol. 2012;355:83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366(8):707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367(18):1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Haroche J, Cohen-Aubart F, Emile JF, et al. Reproducible and sustained efficacy of targeted therapy with vemurafenib in patients with BRAF(V600E)-mutated Erdheim-Chester disease. J Clin Oncol. 2015;33(5):411–418. [DOI] [PubMed] [Google Scholar]
- 91. Heritier S, Jehanne M, Leverger G, et al. Vemurafenib use in an infant for high-risk langerhans cell histiocytosis. JAMA Oncol. 2015;1(6):836–838. [DOI] [PubMed] [Google Scholar]
- 92. Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;373(8):726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Janoueix-Lerosey I, Lequin D, Brugieres L, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455(7215):967–970. [DOI] [PubMed] [Google Scholar]
- 94. Mosse YP, Laudenslager M, Longo L, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455(7215):930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bresler SC, Weiser DA, Huwe PJ, et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell. 2014;26(5):682–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med. 2012;4(120):120ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Infarinato NR, Park JH, Krytska K, et al. The ALK/ROS1 inhibitor PF-06463922 overcomes primary resistance to crizotinib in ALK-driven neuroblastoma. Cancer Discov. 2016;6(1):96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lovly CM, Gupta A, Lipson D, et al. Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov. 2014;4(8):889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mosse YP, Lim MS, Voss SD, et al. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: A Children's Oncology Group phase 1 consortium study. Lancet Oncol. 2013;14(6):472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368(25):2385–2394. [DOI] [PubMed] [Google Scholar]
- 102. Vaishnavi A, Le AT, Doebele RC.. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015;5(1):25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19(11):1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Doebele RC, Davis LE, Vaishnavi A, et al. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov. 2015;5(10):1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wyce A, Ganji G, Smitheman KN, et al. BET inhibition silences expression of MYCN and BCL2 and induces cytotoxicity in neuroblastoma tumor models. PLoS One. 2013;8(8):e72967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Henssen A, Thor T, Odersky A, et al. BET bromodomain protein inhibition is a therapeutic option for medulloblastoma. Oncotarget. 2013;4(11):2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bandopadhayay P, Bergthold G, Nguyen B, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res. 2014;20(4):912–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mertz JA, Conery AR, Bryant BM, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Boi M, Gaudio E, Bonetti P, et al. The BET bromodomain inhibitor OTX015 affects pathogenetic pathways in preclinical B-cell tumor models and synergizes with targeted drugs. Clin Cancer Res. 2015;21(7):1628–1638. [DOI] [PubMed] [Google Scholar]
- 111. Cheng Z, Gong Y, Ma Y, et al. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19(7):1748–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. French CA. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol. 2012;7:247–265. [DOI] [PubMed] [Google Scholar]
- 113. French CA, Rahman S, Walsh EM, et al. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: Implications for a novel oncogenic mechanism. Cancer Discov. 2014;4(8):928–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Tang Y, Gholamin S, Schubert S, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med. 2014;20(7):732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Long J, Li B, Rodriguez-Blanco J, et al. The BET bromodomain inhibitor I-BET151 acts downstream of smoothened protein to abrogate the growth of hedgehog protein-driven cancers. J Biol Chem. 2014;289(51):35494–35502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sherr CJ, Beach D, Shapiro GI.. Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov. 2015;6(4):353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Bale AE, Yu KP.. The hedgehog pathway and basal cell carcinomas. Hum Mol Genet. 2001;10(7):757–762. [DOI] [PubMed] [Google Scholar]
- 118. Tang JY, Mackay-Wiggan JM, Aszterbaum M, et al. Inhibiting the hedgehog pathway in patients with the basal-cell nevus syndrome. N Engl J Med. 2012;366(23):2180–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Gajjar A, Stewart CF, Ellison DW, et al. Phase I study of vismodegib in children with recurrent or refractory medulloblastoma: A pediatric brain tumor consortium study. Clin Cancer Res. 2013;19(22):6305–6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wang F, Fang Q, Ge Z, et al. Common BRCA1 and BRCA2 mutations in breast cancer families: A meta-analysis from systematic review. Mol Biol Rep. 2012;39(3):2109–2118. [DOI] [PubMed] [Google Scholar]
- 121. Jhuraney A, Velkova A, Johnson RC, et al. BRCA1 Circos: A visualisation resource for functional analysis of missense variants. J Med Genet. 2015;52(4):224–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Williamson CT, Muzik H, Turhan AG, et al. ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors. Mol Cancer Ther. 2010;9(2):347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Smith MA, Hampton OA, Reynolds CP, et al. Initial testing (stage 1) of the PARP inhibitor BMN 673 by the pediatric preclinical testing program: PALB2 mutation predicts exceptional in vivo response to BMN 673. Pediatr Blood Cancer. 2015;62(1):91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Rader J, Russell MR, Hart LS, et al. Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin Cancer Res. 2013;19(22):6173–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. VanArsdale T, Boshoff C, Arndt KT, et al. Molecular pathways: Targeting the cyclin D-CDK4/6 axis for cancer treatment. Clin Cancer Res. 2015;21(13):2905–2910. [DOI] [PubMed] [Google Scholar]
- 126. Dean JL, Thangavel C, McClendon AK, et al. Therapeutic CDK4/6 inhibition in breast cancer: Key mechanisms of response and failure. Oncogene. 2010;29(28):4018–4032. [DOI] [PubMed] [Google Scholar]
- 127. Johnson N, Shapiro GI.. Cyclin-dependent kinase 4/6 inhibition in cancer therapy. Cell Cycle. 2012;11(21):3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wiedemeyer WR, Dunn IF, Quayle SN, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci U S A. 2010;107(25):11501–11506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cen L, Carlson BL, Schroeder MA, et al. p16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro Oncol. 2012;14(7):870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Michaud K, Solomon DA, Oermann E, et al. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70(8):3228–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Katsumi Y, Iehara T, Miyachi M, et al. Sensitivity of malignant rhabdoid tumor cell lines to PD 0332991 is inversely correlated with p16 expression. Biochem Biophys Res Commun. 2011;413(1):62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Saab R, Bills JL, Miceli AP, et al. Pharmacologic inhibition of cyclin-dependent kinase 4/6 activity arrests proliferation in myoblasts and rhabdomyosarcoma-derived cells. Mol Cancer Ther. 2006;5(5):1299–1308. [DOI] [PubMed] [Google Scholar]
- 133. Olanich ME, Sun W, Hewitt SM, et al. CDK4 amplification reduces sensitivity to CDK4/6 inhibition in fusion-positive rhabdomyosarcoma. Clin Cancer Res. 2015;21(21):4947–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Tanizaki J, Ercan D, Capelletti M, et al. Identification of oncogenic and drug-sensitizing mutations in the extracellular domain of FGFR2. Cancer Res. 2015;75(15):3139–3146. [DOI] [PubMed] [Google Scholar]
- 135. Mathur A, Ware C, Davis L, et al. FGFR2 is amplified in the NCI-H716 colorectal cancer cell line and is required for growth and survival. PLoS One. 2014;9(6):e98515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Andre F, Bachelot T, Campone M, et al. Targeting FGFR with dovitinib (TKI258): Preclinical and clinical data in breast cancer. Clin Cancer Res. 2013;19(13):3693–3702. [DOI] [PubMed] [Google Scholar]
- 137. Scheid C, Reece D, Beksac M, et al. Phase 2 study of dovitinib in patients with relapsed or refractory multiple myeloma with or without t(4;14) translocation. Eur J Haematol. 2015;95(4):316–324. [DOI] [PubMed] [Google Scholar]
- 138. Di Stefano AL, Fucci A, Frattini V, et al. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin Cancer Res. 2015;21(14):3307–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Chmielecki JB, He J, Elvin J, et al. Genomic profiling of 1239 diverse pediatric cancers identifies novel discoveries across tumors. Cancer Res. 2016;76(14 suppl):#LB–178. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.