ABSTRACT
WASH, a Wiskott–Aldrich syndrome (WAS) family protein, has many cell and developmental roles related to its function as a branched actin nucleation factor. Similar to mammalian WASHC1, which is embryonic lethal, Drosophila Wash was found to be essential for oogenesis and larval development. Recently, however, Drosophila wash was reported to be homozygous viable. Here, we verify that the original wash null allele harbors an unrelated lethal background mutation; however, this unrelated lethal mutation does not contribute to any Wash oogenesis phenotypes. Significantly, we find that: (1) the homozygous wash null allele retains partial lethality, leading to non-Mendelian inheritance; (2) the allele's functions are subject to its specific genetic background; and (3) the homozygous stock rapidly accumulates modifications that allow it to become robust. Together, these results suggest that Wash plays an important role in oogenesis via the WASH regulatory complex. Finally, we show that another WAS family protein, SCAR/WAVE, plays a similar role in oogenesis and that it is upregulated as one of the modifications that allows the wash allele to survive in the homozygous state.
KEY WORDS: Wiskott–Aldrich syndrome, WASH, SCAR/WAVE, WASH regulatory complex, Oogenesis, Genetic assimilation
Summary: Drosophila Wash loss-of-function mutant is not homozygous lethal, but plays essential roles in oogenesis and its loss in the ovary leads to compensation by the related family member Scar.
INTRODUCTION
The Wiskott–Aldrich syndrome (WAS) family of proteins have well characterized roles as branched actin nucleation promoting factors (NPFs) in a wide spectrum of normal cellular and developmental processes, as well as in pathogen infection, disease, and cancer metastasis (Alekhina et al., 2017; Burianek and Soderling, 2013; Massaad et al., 2013; Rottner et al., 2010; Stradal et al., 2004; Takenawa and Suetsugu, 2007). These Class I NPFs share a C-terminal verprolin-homology (WH2), cofilin-homology, and acidic (VCA) domain, which activates the Arp2/3 complex and forms actin meshworks by promoting the nucleation of new F-actin branches upon existing F-actin filaments (Campellone and Welch, 2010; Millard et al., 2004; Rotty et al., 2013). While the C-terminus of this family of proteins is highly conserved, their N-terminal regions show much more diversity among family members and are thought to confer the specific cellular behaviors of the individual proteins within the subfamily (Burianek and Soderling, 2013; Takenawa and Suetsugu, 2007; Veltman and Insall, 2010).
WASH (WASp and SCAR homolog) was initially discovered in a sub-telomeric region of the human genome and subsequently found to be present throughout eukaroytes from entomoeba to humans (Linardopoulou et al., 2007). In Drosophila, we first characterized Wash as an effector of Rho1 GTPase where it functions to prevent premature ooplasmic streaming in the oocyte (Liu et al., 2009). Subsequent work in other organisms and cells has demonstrated that WASH functions as a member of a multiprotein regulatory complex (also containing the proteins FAM21, Strumpellin, SWIP and CCDC53) named the SHRC (WASH regulatory complex), which is structurally analogous to the Wave regulatory complex (Derivery et al., 2009; Duleh and Welch, 2010; Gomez and Billadeau, 2009; Jia et al., 2010; Park et al., 2013). We have previously shown that Drosophila Wash is regulated in a context-dependent manner: it can bind directly to Rho1 GTPase or it can function along with the SHRC (Verboon et al., 2015a). Studies on the SHRC have largely focused on the role of this multiprotein complex in regulating several steps in the recycling endosome pathway, particularly in conjunction with retromer in mammalian cell lines and macrophages (Alekhina et al., 2017; Bartuzi et al., 2016; Buckley et al., 2016; Carnell et al., 2011; Derivery et al., 2009; Duleh and Welch, 2010; Gomez and Billadeau, 2009; Harbour et al., 2012; Monteiro et al., 2013; Park et al., 2013; Piotrowski et al., 2013; Rottner et al., 2010; Ryder et al., 2013; Zech et al., 2011). However, as the knockout of WASH (also known as WASHC1 in mammals) in mice leads to lethality at E7.5 (Gomez et al., 2012; Piotrowski et al., 2013; Xia et al., 2013), it is likely that mammalian WASH has other essential roles in oogenesis and/or very early embryogenesis.
We previously reported that the Drosophila washΔ185 deletion mutants die as late third instar larvae and also exhibit significant defects in oogenesis when reduced maternally (Linardopoulou et al., 2007; Liu et al., 2009). It was recently reported by Bogdan and colleagues that the same washΔ185 deletion allele can be made homozygous, such that it is viable and fertile (Nagel et al., 2017). These mutants (referred to as washΔ185*) lacked Wash expression. Furthermore, the authors did not observe any of the previously described morphological defects in developing washΔ185* egg chambers. Despite the lack of phenotypes during oogenesis, they confirmed our previous studies showing that Wash is expressed in ovaries on immunoblots (Liu et al., 2009; Rodriguez-Mesa et al., 2012). However, while we showed that Wash specifically accumulates at the oocyte cortex in stage 7-9 oocytes, in addition to uniform distribution throughout the cytoplasm of the oocyte, nurse cells and follicle cells, they did not observe cortical accumulation of Wash or differences in staining between the wild type and wash mutants using an affinity-purified version of our Wash P3H3 monoclonal antibody. Taken together, these results led these authors to conclude that the lethality associated with the original washΔ185 allele was due to an unrelated second-site mutation and that Wash is dispensable for oogenesis (Nagel et al., 2017).
Here, we verify that the original washΔ185 allele harbors an unrelated background lethal mutation contributing to the complete lethality of this allele. We separated this unrelated background lethal from the original washΔ185 allele and show that this unrelated lethal does not contribute to any previously described Wash oogenesis phenotypes. Additionally, we find that Wash's survival and phenotypes are subject to background effects: Wash can be made homozygous, but not with the expected Mendelian ratios. If kept in the homozygous state, wash lines will rapidly accumulate modifiers that allow their survival, and in particular, we show that this leads to upregulated expression of another WAS family member, SCAR/WAVE, during oogenesis. We show that wash mutants, as well as RNAi knockdown alleles, exhibit strong and specific oogenesis defects. In addition, we show that Wash protein has a specific pattern of accumulation in the stage 7-9 oocyte cortex, which is lost in wash mutant and RNAi knockdown egg chambers. Furthermore, we found that all four members of the Wash regulatory complex are expressed in oocytes, show specific accumulation in the stage 7-9 oocyte cortex, affect each other's stability and exhibit similar oogenesis phenotypes.
RESULTS
washΔ185 phenotypes are subject to genetic background
To confirm that the washΔ185 allele could be made homozygous, we outcrossed the originally generated washΔ185 line, which has been maintained in the heterozygous state since its generation (Fig. 1A). We kept all outcrossed flies generated containing the Wash deletion or that were homozygous lethal. Overall, we recovered 16 lines containing the washΔ185 deletion allele, of which 9 could be made homozygous and 7 remained inviable, as well as 2 homozygous lethal lines that did not contain the washΔ185 deletion allele (Fig. 1A, Table S1A,B). All washΔ185 deletion lines that were able to be made homozygous were maintained as both homozygous and heterozygous (through continual outcrossing) stocks.
Fig. 1.
Continuously outcrossed washΔ185 flies can be made homozygous, but display sub-Mendelian inheritance. (A) Genetic recombination scheme used to separate and recover the non-related nw† lethal mutation from the washΔ185 allele. (B) Schematic depicting the genetic cross of continuously outcrossed washΔ185hz(outX) heterozygous flies and the expected ratio of heterozygous (het) to homozygous (hz) progeny. (C) Bar plot of the het:hz ratios obtained from the nine washΔ185hz/CyO lines subjected to the genetic cross shown in B (Binomial test; mean±s.d.). Note the sub-Mendelian het:hz ratios in the F1 generation (red line denotes het:hz ratio of 2 expected by Mendelian inheritance). (D,E) Graphs depicting the loss of wash phenotypes at each generation that the washΔ185hz allele is kept homozygous as measured by the average number (mean±s.e.m.) of progeny (D) and percentage of stage 7-9 egg chambers exhibiting actin defects (E). (F) Graph depicting the sterility associated with wild-type, washΔ185hz(outX) and washΔ185hz female flies as assessed by mating single females with wild-type males for 7 days, then scoring the number of offspring produced (two-tailed Student's t-test; mean±s.d.). Sample size (N) is indicated in bar plots in C,E; ns, not significant.
We next performed complementation crosses between all lines generated and found that: (1) the two lethal lines not containing washΔ185 are lethal with each other (Table S1B); (2) these two lines are also lethal with the homozygous inviable washΔ185 deletion lines (Table S1C); and (3) the homozygous inviable washΔ185 lines are not lethal with the homozygous viable washΔ185 lines (Table S1C). Taken together, the results of these crosses show that the original washΔ185 allele contains an unrelated background mutation that causes lethality, and that without this background mutation, the washΔ185 allele can be homozygous viable. Going forward we will refer to the washΔ185 viable alleles as washΔ185hz, the washΔ185 inviable alleles as washΔ185(+nw†), and the non-washΔ185-containing lethal lines as nw† (not Wash lethal). Additionally, using the continually outcrossed heterozygous washΔ185 flies, we created washΔ185hz/washΔ185hz homozygous F1 flies [referred to as washΔ185hz(outX)].
Despite being viable, we noticed that washΔ185(+nw†) crossed to washΔ185hz led to fewer than expected homozygous washΔ185-containing flies. In order to explore this observation further, we crossed washΔ185hz heterozygous flies to themselves and found the progeny did not follow Mendelian inheritance. This cross was expected to yield a 2:1 ratio of heterozygous (het):homozygous (hz) flies (Fig. 1B); however, in all 9 washΔ185hz crosses, the ratio of het:hz was >2, showing fewer than expected homozygous mutants, whereas two control crosses had a ratio of ∼2 (Fig. 1C; Table S1A). Importantly, in 7 out of the 9 washΔ185hz lines obtained, this ratio was significantly different to the expected value calculated from the controls. These 9 lines, the products of different recombination events during the original outcross, show significant variability of this ratio, suggesting that the specific genetic background of each line is important, rather than a single modifying locus. While F1 washΔ185hz homozygous progeny from heterozygous parents were initially under-represented and sickly, if subsequently kept as homozygous stocks, they improved quickly (within 5 generations) to become robust stocks, as assayed by number of progeny produced and percentage of egg chambers exhibiting actin defects (Fig. 1D,E; see Materials and Methods). Importantly, after 5 generations, the number of offspring of washΔ185hz lines was not significantly different from the wild type (Fig. 1F). Taken together, our genetic analyses suggest that the washΔ185 allele viability is subject to the genetic background, and can be rapidly and increasingly compensated for by additional modifications.
Wash expression in oocytes is lost in wash mutant and RNAi knockdown backgrounds, but not in the nw† lethal mutation background
We previously demonstrated a role for Wash in Drosophila oogenesis: Wash is enriched at the stage 7-9 oocyte cortex and its loss led to premature ooplasmic streaming (Liu et al., 2009; Rodriguez-Mesa et al., 2012). Bogdan and colleagues were unable to detect this cortical Wash enrichment using the same monoclonal Wash P3H3 antibody after protein A affinity purification and asserted that this staining was an artifact, and combined with the ability of Wash to be homozygous, concluded that Wash was not necessary in oogenesis (Nagel et al., 2017). We were confused by this result because affinity purification of a monoclonal antibody in this manner should return the same IgG while removing cellular proteins and cell culture products only. To understand this inconsistency, we re-characterized the non-affinity purified Wash P3H3 antibody by blotting multiple Drosophila lysates (wild-type embryo whole cell extract, Kc167 whole cell extract, wild-type ovary lysate and ovary lysate from washΔ185hz) and found that the Wash P3H3 monoclonal antisera only recognizes one band at the expected size of 75 kDa (Fig. 2A). Additionally, no bands were detected in washΔ185hz homozygous null ovary lysate, confirming this antibody is specific to Wash (Fig. 2A). We also affinity purified Wash P3H3 antibody (Wash P3H3AP) using protein G, which has a higher affinity for the IgG1 Wash P3H3 antibody than protein A, and the resultant specificity was equivalent to the non-AP antibody (Fig. 2B). Finally, we tested an independent Wash monoclonal antibody (Wash P4D2) which confirms these results (Fig. 2C). Thus, the Wash P3H3 antibody is specific to Wash proteins by western blotting with no detectable recognition of off-target proteins.
Fig. 2.
Wash antibodies are Wash-specific and Wash accumulates at the oocyte cortex. (A-C) Wash antibodies (P3H3, affinity-purified P3H3AP, and P4D2) are specific to Wash in embryo, KC167 cell and wild-type ovary lysate, but absent in washΔ185hz ovary lysate. WCE: whole cell extract. (D-O) Wash is present in the germline and accumulates at the stage 7/8 oocyte cortex in wild type (D,D′,H,H′,L,L′), but not washRNAi (E,E′,I,I′,L,L′) or maternally reduced washΔ185 (F,F′,J,J′,N,N′) and by line plot analysis (G,K,O) when stained with P3H3 (D-F′) P3H3AP (H-J′) and P4D2 (L-N′), respectively. Bars in D-N indicate region measured for the line plots. Red arrows indicate oocyte cortex accumulation of Wash. Red arrowheads indicate Wash cortical accumulation in follicle cells. N=1 representative example. (P,P′) GFP staining at the oocyte cortex in oocytes expressing a GFP-Wash fusion protein. Red arrows indicate oocyte cortex accumulation of GFP-Wash. (Q-S′) Cortical Wash accumulation in oocytes stained with P3H3 in wild type (Q,Q′) and nw† (R,R′), but not in washΔ185hz (S,S′). Red arrows indicate oocyte cortex accumulation of Wash. (T) Line plot showing cortical Wash accumulation in wild type and nw†, but not in washΔ185hz oocytes (corresponding to bars in Q-S). N=1 representative example. (U-W) Micrographs (U-V′) and line plot (W) show that overall staining is decreased by affinity purification. Red arrows indicate oocyte cortex accumulation of Wash. N=1 representative example. (X) Protein gel of 1 µl (1×), 0.5 µl (0.5×), and 0.1 µl (0.1×) of P3H3 and P3H3AP antisera showing that IgG was not lost during affinity purification. Relative quantification of each band is provided below the lane. (Y) Western blots of 1 µl (1×), 0.75 µl (0.75×) and 0.5 µl (0.5×) of in vitro translated Wash probed with P3H3 and P3H3AP showing loss of antibody affinity following affinity purification. Relative quantification of each band is provided below the lane. Scale bars: 50 µm.
As the Wash P3H3 antibody is specific to Wash on blots, we verified our previous immunostaining results in the Drosophila ovary. We stained both wild-type and wash knockdown stage 7-9 egg chambers with Wash P3H3, Wash P3H3AP and Wash P4D2 monoclonal antibodies. We used the wimp mutation to generate reduced wash expression in both the germline and soma (wimp reduces maternal gene expression such that, when in trans to the wash allele, it effectively generates a strong wash hypomorph (Liu et al., 2009; Parkhurst and Ish-Horowicz, 1991). We also knocked down Wash using RNAi constructs expressed in the germline by the UAS-GAL4 system. In control ovaries, Wash is present in the oocyte, nurse cells and somatic follicle cells, and exhibits enrichment at the oocyte cortex when stained with any of the three monoclonal antibodies (Fig. 2D,D′,G,H,H′,K,L,L′,O). In contrast, Wash staining is strongly reduced in the germline of both Wash RNAi and reduced wash egg chambers, as well in the somatic follicle cells of the reduced wash background (Fig. 2E-G,I-K,L-O). As an alternative means to examine Wash localization, we expressed a GFP-Wash fusion allele under the Wash endogenous promoter and observed similar enrichment of the GFP-Wash fusion protein at the oocyte cortex (Fig. 2P,P′). To confirm that this Wash behavior was independent of the non-specific nw† background lethal mutation and absent in the wash homozygous mutants, we stained control, nw† and washΔ185hz ovaries with Wash P3H3 antibodies. Cortical Wash staining was normal in control and the nw† background lethal oocytes, and not detectable in washΔ185hz mutant ovaries (Fig. 2Q-T). Thus, Wash is expressed in the germline, exhibits specific enrichment at the oocyte cortex, and importantly, this localization is not an artifact of the Wash P3H3 antibody. Additionally, the nw† background lethal mutation is completely separate from wash and has no effect on Wash localization.
While these results confirm our initial studies, they do not explain the dichotomy between our results and those of the aforementioned study. One phenomenon we noticed was that staining with Wash P3H3AP antibodies consistently led to reduced signal compared with Wash P3H3 antibody (Fig. 2U-W). One possibility is that incomplete elution of the Wash antibody from the Protein A/G beads leads to a reduction of Wash antibody concentration. To test this, we quantified the IgG heavy chain in Wash P3H3 and Wash P3H3AP with Coomassie stained protein gels loaded with varying amounts of antibody. We observe a similar amount of IgG before and after protein G purification, indicating that antibody is not being lost in the purification process (Fig. 2X). Another possibility is that the antibody recovered from affinity purification has reduced capacity to bind to Wash protein. We probed western blots with varying concentrations of in vitro translated Wash protein with the same concentration of Wash P3H3 and Wash P3H3AP antibody. We observe a 43% reduction in binding of the Wash P3H3AP antibody as compared to that of the Wash P3H3 antibody, suggesting that the antibody binds less effectively post affinity purification (Fig. 2Y). Finally, it is likely that the choice of Protein A or G affects the quality of the antibody recovered since P3H3 as an IgG1 is expected to bind with higher affinity to Protein G. Thus, Wash P3H3 antibody is specific; however, its efficacy may be subject to a loss of affinity by acid elution during affinity purification, may have been inefficiently recovered by using protein A rather than protein G (Wash P3H3 is an IgG1 subclass) during affinity purification, and/or may depend on the process of tissue fixing/preparation for its function in immunostaining experiments.
Loss and reduction of Wash leads to defects during oogenesis
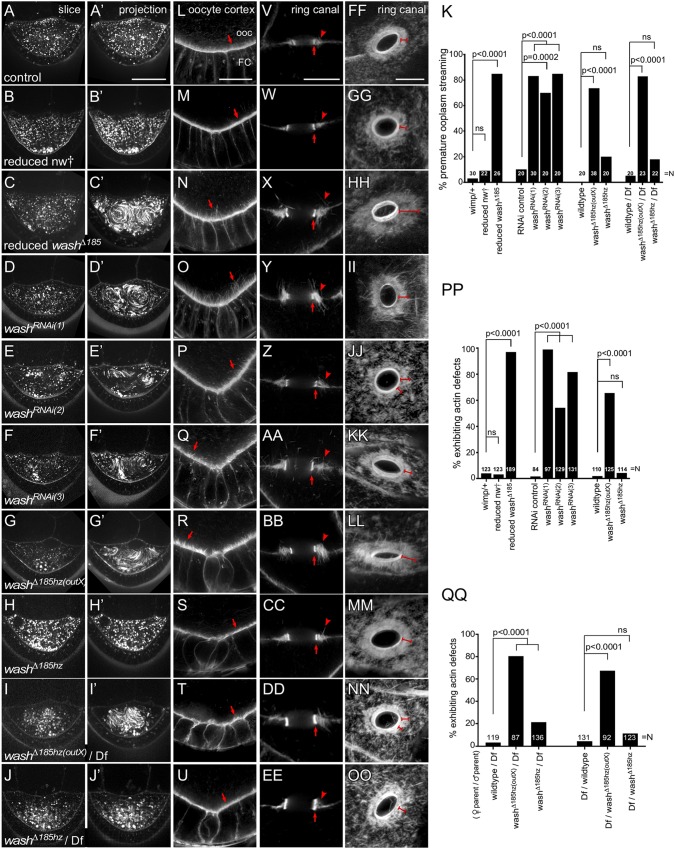
As Wash is expressed and exhibits specific enrichment at the oocyte cortex, we wanted to confirm that the wash-dependent oogenesis defects we had previously characterized were present and independent of the nw† background mutation. We looked at two major phenotypes previously characterized as a consequence of loss of Wash during oogenesis: premature ooplasmic streaming and actin defects at the oocyte cortex and with ring canals (Liu et al., 2009). We examined these phenotypes in stage 7-9 egg chambers from control, reduced nw† (nw† in trans to wimp), reduced wash, three independent Wash RNAi lines expressed in the germline by the UAS-GAL4 system (one RNAi line using short-hairpin and two lines using small interfering RNAs), washΔ185hz(outX) and in washΔ185hz (maintained as a homozygous stock for >5 generations). Control oocytes do not exhibit ooplasmic streaming: yolk granule movement in stage 7/8 oocytes is minimal and uncoordinated (Fig. 3A,A′,K; Movie 1). Reduced nw† oocytes also showed no ooplasmic streaming at stages 7-9, similar to results in the wild type (Fig. 3B,B′,K; Movie 1). In contrast, reduced washΔ185, all three Wash RNAi alleles and washΔ185hz(outX) oocytes all exhibit premature ooplasmic streaming, whereas washΔ185hz displayed no defects (Fig. 3C-H′,K; Movie 1). Finally, we crossed washΔ185hz(outX) and washΔ185hz to a deficiency containing the Wash locus [Df(2R)BSC699; Table S2] and assayed ooplasmic streaming in the resultant progeny (Fig. 3I-J′,K; Movie 1). As expected, washΔ185hz(outX)/Df flies exhibited premature ooplasmic streaming, but washΔ185hz/Df flies did not. Thus, both acute (RNAi knockdown) and chronic (mutant) loss of Wash leads to premature ooplasmic streaming in the stage 7-9 oocyte.
Fig. 3.
Disruption of Wash function results in premature ooplasmic streaming and aberrant actin-based structures. (A-H′) Single time-point and 30 time-point projections of live time-lapse movies of stage 7 oocytes in wild type (A), reduced nw† (line 8) (B), reduced washΔ185 (C), washRNAi(1) (D), washRNAi(2) (E), washRNAi(3) (F), washΔ185hz(outX) (G), washΔ185hz (H), washΔ185hz(outX)/Df (I) and washΔ185hz/Df (J). (K) Quantification of the percentage of stage 7 oocytes exhibiting premature ooplasmic streaming. (L-U) Posterior of stage 7 egg chambers, stained for F-actin (Phalloidin), in wild type (L), reduced nw† (line 8) (M), reduced washΔ185 (N), washRNAi(1) (O), washRNAi(2) (P), washRNAi(3) (Q), washΔ185hz(outX) (R), washΔ185hz (S), washΔ185hz(outX)/Df (T) and washΔ185hz/Df (U). Arrows denote cortical actin in oocyte. Note aberrantly long actin projections at oocyte cortex in L-P (ooc, oocyte; FC, follicle cells). (V-EE) Cross sections of ring canals bridging the oocyte and a nurse cell in stage 7 egg chambers, stained for F-actin (Phalloidin), in wild type (V), reduced nw† (line 8) (W), reduced washΔ185 (X), washRNAi(1) (Y), washRNAi(2) (Z), washRNAi(3) (AA), washΔ185hz(outX) (BB), washΔ185hz (CC), washΔ185hz(outX)/Df (DD) and washΔ185hz/Df (EE). Arrows denote inner actin ring canal and arrowheads denote outer actin ring. Note disrupted outer actin ring structures surrounding intact inner ring canals in X-DD. (FF-OO) Oblique views of ring canals bridging two nurse cells in stage 9 egg chambers, stained for F-actin (Phalloidin), in wild type (FF), reduced nw† (line 8) (GG), reduced washΔ185 (HH), washRNAi(1) (II), washRNAi(2) (JJ), washRNAi(3) (KK), washΔ185hz(outX) (LL), washΔ185hz (MM), washΔ185hz(outX)/Df (NN) and washΔ185hz/Df (OO). Note disorganized outer actin rings in (HH-NN; brackets). (PP) Quantification of the percentage of egg chambers exhibiting actin defects. (QQ) Quantification of the percentage of egg chambers exhibiting actin defects for deficiency crosses generated through both sex/genotype combinations. N for each genotype indicated on bar plots in K, PP and QQ. Two-tailed Fisher's exact test (K,QQ-PP); ns, not significant. Scale bars: 50 µm in A-H′; 25 µm in J-Q; 5 µm in R-GG.
In stage 7-9 wild-type oocytes, F-actin is highly enriched in a tight accumulation at the oocyte cortex and also forms a well-organized outer ring canal which supports the actin-rich inner ring canal function. To assess the integrity of these actin-based structures in control, as well as the varying wash reduction conditions, we fixed stage 7-9 egg chambers then stained them with fluorescently tagged Phalloidin. In wild-type oocytes, cortical actin forms a uniform distribution of similarly sized filaments projecting into the oocyte (Fig. 3L,PP). Reduced nw† oocytes exhibit uniform actin filament organization that is indistinguishable from that in the wild type (Fig. 3M,PP). Reduced washΔ185, all three Wash RNAi alleles and washΔ185hz(outX) oocytes all exhibit disrupted cortical actin organization, leading to abnormally long actin projections and/or tufting of actin filaments at the oocyte cortex, while washΔ185hz oocytes behave similar to control oocytes (Fig. 3N-S,PP). As with premature ooplasmic streaming, washΔ185hz(outX)/Df flies have cortical actin defects in the oocyte, whereas washΔ185hz/Df do not (Fig. 3T,U,QQ). Next, we examined ring canals that connect the oocyte to the nurse cells in a cross-section view and ring canals linking two adjacent nurse cells with a top down view. In oocytes of all genotypes examined, the condition of the inner ring canal is unaffected, consistent with our previous finding that the integrity of this structure is wash-independent (Fig. 3V-OO) (Liu et al., 2009). In wild type and reduced nw† egg chambers, actin forms a well-organized array surrounding the inner ring canal in both the cross-section (Fig. 3V,W) and the planar view of these ring canals (Fig. 3FF,GG). Reduced wash and the three Wash RNAi lines led to a more diffuse and/or disorganized accumulation of actin in the outer array and to large spike-like actin filaments which project away from the area of actin accumulation (Fig. 3X-AA,HH-KK). Similar to our observations with premature ooplasmic streaming, washΔ185hz(outX) and washΔ185hz(outX)/Df oocytes exhibit disrupted cortical actin and actin organization at outer ring canals, while cortical actin and outer ring canal actin organization in washΔ185hz and washΔ185hz/Df oocytes are like those in the wild type (Fig. 3BB-EE,LL-QQ). Interestingly, we note that the washΔ185hz/Df flies that originated with the female of the washΔ185hz genotype has slightly less robust rescue, consistent with a dilution and/or suppression of modifier function or the presence of a small cell biological component/intracellular cue in addition to the genotype (Fig. 3QQ). Thus, the background nw† lethal mutation originally present in the Wash deletion background has no effect on actin cytoskeleton organization oogenesis and we have confirmed our previous findings that Wash reduction and/or loss leads to significant premature ooplasmic streaming, as well as significant actin structural and/or organizational defects using multiple genetic backgrounds and means to perturb Wash function.
SHRC is present in the oocyte and its loss leads to oogenesis defects similar to those seen with knockdown of Wash
Drosophila Wash exhibits complex regulation: it can function as part of the SHRC multiprotein complex (similar to the regulation of SCAR/WAVE proteins), and it can function independently of such a complex (similar to the regulation of WASP/N-WASP) in a context-dependent manner (Verboon et al., 2015a). We have shown that Drosophila Wash functions downstream of Rho1 GTPase signaling in the oocyte, as well as in hemocyte migrations in the embryo (Liu et al., 2009; Verboon et al., 2015a). However, outside of Drosophila, Wash regulation by Rho family GTPases has not yet been described (Jia et al., 2010), and instead, regulation has been characterized solely in the context of its SHRC. To determine if the SHRC is required for Wash functions during oogenesis, we examined the expression and knockdown effects of the SHRC subunits during oogenesis. Previously, we made polyclonal antibodies that recognize each of the SHRC members: CCDC53, Strumpellin, SWIP and FAM21 (Verboon et al., 2015a). We stained stage 7-9 egg chambers and found that all four SHRC proteins are present in the germline and, similar to Wash, accumulate at the oocyte cortex (Fig. 4A-D′).
Fig. 4.

Wash regulatory complex (SHRC) members are expressed in the oocyte. (A-D′) Wild-type stage 7 egg chambers stained for SHRC members CCDC53 (A), Strumpellin (B), SWIP (C) and FAM21 (D). Note enrichment of staining at oocyte cortex (arrows; A′-D′). (E) Western blot showing Wash and SHRC member expression in wild type, washRNAi(1), CCDC53RNAi(1), StrumpRNAi(1), SWIPRNAI(1) and FAM21RNAi(1) ovary lysates. Scale bars: 50 µm.
Mammalian WASH and the SHRC members have been shown to be dependent on interaction with each other for the stability of their multiprotein complex: knocking down Strumpellin, SWIP or FAM21 downregulates expression of WASH and all core SHRC members, whereas knocking down WASH or CCDC53 affects only the expression of each other (Derivery et al., 2009; Jia et al., 2010; Rottner et al., 2010). To determine if Wash and the SHRC are dependent upon each other during Drosophila oogenesis, we examined the expression of Wash and three of the SHRC members (CCDC53, FAM21 and Strumpellin) in ovary lysates generated from wild-type and Wash- or SHRC-knockdown females. In this context, knockdown of Wash or any of the SHRC members downregulates expression of Wash and all other SHRC members (Fig. 4E).
Wash and SHRC inter-dependence for complex stability suggests that the SHRC knockdowns are likely to exhibit phenotypes similar to those in Wash-knockdown flies during oogenesis. To test this, we knocked down each SHRC member in the germline using two independent RNAi lines and assessed their effects on premature ooplasmic streaming and the integrity of actin-based structures in the germline. As anticipated, all four SHRC members show premature ooplasmic streaming phenotypes (Fig. 5A-E; Fig. S1D-G′; Movie 2). Knockdown of the SHRC proteins also leads to disorganized actin at the oocyte cortex (Fig. 5F-I,R; Fig. S1L-O). Similarly, actin structures surrounding the ring canals are diffuse, with increased outer array width and large actin filaments protruding away from the ring canals (Fig. 5J-R; Fig. S1T-W,BB-EE). Thus, during oogenesis, the SHRC may regulate not only Rho signaling, but also Wash.
Fig. 5.
SHRC knockdowns exhibit premature ooplasmic streaming and actin defects. (A-D′) Single time-point and 30 time-point projections of live time-lapse movies of stage 7 oocytes in CCDC53RNAi(1) (A,A′), StrumpRNAi(1) (B,B′), SWIPRNAI(1) (C,C′), FAM21RNAi(1) (D,D′). (E) Quantification of the percentage of stage 7 SHRC RNAi oocytes exhibiting premature ooplasmic streaming. (F-I) Posterior cortex of stage 7 egg chambers stained for F-actin (Phalloidin) in CCDC53RNAi(1) (F), StrumpRNAi(1) (G), SWIPRNAI(1) (H), FAM21RNAi(1) (I). Note aberrantly long actin projections from cortex into oocyte (red arrows). (J-M) Cross sections of ring canals bridging the oocyte and a nurse cell in stage 7 egg chambers, stained for F-actin (Phalloidin) in CCDC53RNAi(1) (J), StrumpRNAi(1) (K), SWIPRNAI(1) (L), FAM21RNAi(1) (M). Red arrows denote inner actin ring canal and red arrowheads denote outer actin ring. Note disrupted actin structures surrounding intact ring canals (yellow arrow in K). (N-Q) Oblique view of ring canals bridging two nurse cells in stage 9 egg chambers, stained for F-actin (Phalloidin), in CCDC53RNAi(1) (N), StrumpRNAi(1) (O), SWIPRNAI(1) (P), FAM21RNAi(1) (Q). Note disorganized outer actin rings (brackets). (R) Quantification of the percentage of egg chambers exhibiting actin defects. Two-tailed Fisher's exact test (E,R). Sample size (N) indicated in bar plots in E and R. Scale bars: 50 µm in A-D′; 25 µm in F-I; 5 µm in J-Q.
SCAR localizes to the oocyte cortex and is upregulated in wash mutants
We were next interested in why in the aforementioned study they did not observe the ovary phenotypes that are highly penetrant with Wash RNAi, maternal reduction of Wash and washΔ185hz(outX) (first generation homozygous washΔ185hz flies). We noticed that wash mutants rapidly lost the strength and penetrance of their oogenesis phenotypes when maintained as a homozygous stock (>5 generations) (Fig. 3H,H′,K,S,CC,MM,PP). One possible explanation for this phenomenon is that other WAS family proteins were either being upregulated or re-localized to perform a subset of essential Wash functions. Consistent with this possibility, in a previous study, we found that both of the other major WAS family proteins in Drosophila, WASp and SCAR, were expressed during oogenesis (Rodriguez-Mesa et al., 2012).
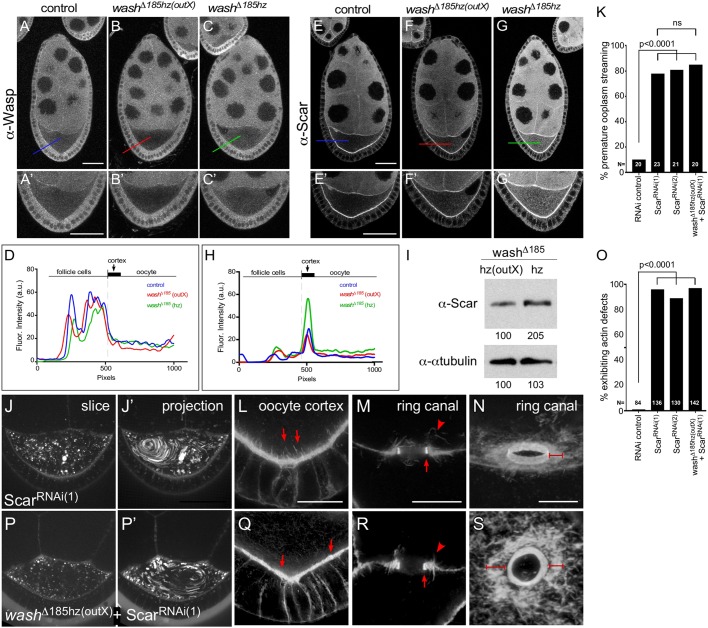
To test whether expression of WASp and/or SCAR proteins changed in response to loss of Wash, we compared WASp and SCAR localization in stage 7-9 oocytes in control, washΔ185hz(outX) and washΔ185hz egg chambers. WASp protein is expressed at a uniform level throughout the nurse cells and follicle cells and is present at lower levels in the oocyte, but with no apparent cortical localization in any of the three conditions (Fig. 6A-D). In contrast to WASp, SCAR exhibits a pattern much more similar to that of Wash: expression throughout nurse cells, decreased expression in follicle cells and expression in the oocyte with enrichment at the oocyte cortex (Fig. 6E,E′,H) (Rodriguez-Mesa et al., 2012; Vartiainen and Machesky, 2004; Zallen et al., 2002). Significantly, we found that cortical SCAR accumulation in wash mutants (washΔ185hz(outX)) recently made homozygous is indistinguishable from that of control egg chambers (Fig. 6F,F′,H). However, in washΔ185hz mutant egg chambers that had been homozygous for multiple generations, we observed an increase in SCAR protein levels with a striking upregulation of SCAR protein accumulation at the oocyte cortex (Fig. 6G-I). This result suggests that SCAR may be able to compensate for the loss of Wash, particularly if selection over several generations is allowed.
Fig. 6.
SCAR accumulates at the oocyte cortex, its loss leads to premature ooplasmic swirling and egg-chamber defects, and it is upregulated in the washΔ185 line when maintained as a homozygous stock. (A-C′) Anti-Wasp (P5E1) staining in stage 7/8 oocytes from wild-type control (A-A′), washΔ185hz(outX) (B-B′) and washΔ185hz (C-C′). (D) Line plot analysis of anti-Wasp (P5E1) staining across the cortex of control, washΔ185hz(outX) and washΔ185hz oocytes (corresponding to bars in A-C). (E-G′) Anti-SCAR (P1C1) staining in stage 7/8 oocytes from control (E,E′), washΔ185hz(outX) (F,F′) and washΔ185hz (G,G′). (E) Line plot analysis of anti-SCAR (P1C1) staining across the cortex of control, washΔ185hz(outX) and washΔ185hz oocytes (corresponding to bars in E-G). (I) Western blot analysis of ovary lysates from washΔ185hz(outX) and washΔ185hz showing increase of Scar protein levels in washΔ185hz egg chambers. Normalized quantification of each band is provided below the lane. (J,J′) Single time-point and 30 time-point projections of live time-lapse movies of stage 7 ScarRNAi(1) oocyte. (K) Quantification of the percentage of stage 7 Scar RNAi and washΔ185hz(outX)+ScarRNAi(1) oocytes exhibiting premature ooplasmic streaming (N for each genotype indicated on graph). (L) Posterior cortex of stage 7 ScarRNAi(1) egg chamber stained for F-actin (Phalloidin) in ScarRNAi(1) stage 7-9 oocyte. Note aberrantly long actin projections from cortex into oocyte (arrows). (M) Cross section of ring canal bridging the oocyte and a nurse cell in stage 7/8 ScarRNAi(1) egg chamber stained for F-actin (Phalloidin). Arrow denotes inner actin ring canal and arrowhead denotes outer actin ring. (N) Oblique view of ring canal bridging two nurse cells in stage 9 ScarRNAi(1) egg chambers stained for F-actin. Note disorganized outer actin ring (bracket). (P,P′) Single time-point and 30 time-point projections of live time-lapse movies of stage 7 washΔ185hz(outX)+ScarRNAi(1) oocyte. (Q) Posterior cortex of stage 7 washΔ185hz(outX)+ScarRNAi(1) egg chamber stained for F-actin (Phalloidin). Note uneven actin projections from cortex into oocyte (arrows). (R) Cross section of ring canal bridging the oocyte and a nurse cell in stage 7/8 washΔ185hz(outX)+ScarRNAi(1) egg chamber stained for F-actin (Phalloidin). Arrow denotes inner actin ring canal and arrowhead denotes outer actin ring. (S) Oblique view of ring canal bridging two nurse cells in stage 9 washΔ185hz(outX)+ScarRNAi(1) egg chambers stained for F-actin. Note uneven outer actin ring (brackets). Two-tailed Fisher's exact test (K,O). Sample size (N) indicated in each figure panel; ns, not significant. Scale bars: 50 µm in A-G′,J,J′,O,O′; 25 µm in L,Q; 5 µm in M-N, R-S.
We next looked to see if SCAR was performing similar functions to Wash during oogenesis, as might be expected in order for this compensation to occur so quickly (≤5 generations). Remarkably, RNAi-mediated knockdown of SCAR phenocopies Wash and SHRC knockdowns by exhibiting premature ooplasmic streaming and actin defects at the oocyte cortex and with ring canals (Fig. 6J-O; Fig. S1H,H′,P,X,FF; Movie 2). As Wash and Scar single knockdowns exhibit defective oogenesis phenotypes, these genes are non-redundant. To test whether or not the oogenesis phenotypes worsen upon knockdown of both Wash and Scar, we tested Scar RNAi in a washΔ185hz(outX) background (Fig. 6K,O-S; Movie 2). Interestingly, in this background, oogenesis phenotypes (premature ooplasmic streaming and disrupted actin-rich structures) and their penetrance are similar to those observed with either single reduction: there is no worsening of the phenotypes or penetrance observed. This supports a model wherein proper actin organization/oogenesis can be achieved by either protein, but that a certain threshold of activity of either/both proteins is needed to maintain normal function and that a loss of either or both can cross this threshold. Taken together, these data suggest that SCAR is ideally situated physically and functionally to be upregulated at the oocyte cortex such that it can quickly step in to compensate for the loss of Wash in the oocyte.
DISCUSSION
Drosophila Wash is an important player in many developmental contexts, including oogenesis, immune cell migration, endocytic sorting, epithelial tube length elongation and nuclear architecture (Dong et al., 2013; Linardopoulou et al., 2007; Liu et al., 2009; Nagel et al., 2017; Verboon et al., 2015a,b). Mouse and human WASH orthologs are essential genes that are needed many times throughout development. For example, conditional knockdown of WASH in mouse hematopoietic stem cells results in defective blood production associated with severe cytopenia and anemia (Xia et al., 2014), whereas human WASH genes are at heightened risk for deletions/rearrangements, are upregulated in invasive breast cancers and their overexpression correlates with poor prognosis of esophageal cancers (Huang et al., 2017; Leirdal et al., 2004; Linardopoulou et al., 2007; Nordgard et al., 2008). In addition, the SHRC components of WASH are linked to neurodegenerative disorders, including hereditary spastic paraplegias, Parkinson disease and Hermansky–Pudlak syndrome (Elliott et al., 2013; Freeman et al., 2013; McGough et al., 2014; Ropers et al., 2011; Ryder et al., 2013; Turk et al., 2017; Valdmanis et al., 2007; Vardarajan et al., 2012; Zavodszky et al., 2014a,b). Consistent with the involvement of mammalian WASH in many developmental processes, mouse knockdowns are reported to die at E7.5 (Gomez et al., 2012; Piotrowski et al., 2013; Xia et al., 2013). Similarly, Drosophila Wash was originally reported to be an essential gene required for proper oogenesis and larval development (Linardopoulou et al., 2007; Liu et al., 2009).
This study follows the report by Nagel et al. (2017) that the washΔ185 null allele is viable and that the lethality and oogenesis phenotypes associated with the original washΔ185 allele are due to the presence of an unrelated background mutation. We confirm the presence of an unrelated background lethal in the original washΔ185 line; however, we show that this background mutation does not impact our original conclusions or the oogenesis phenotypes associated with the washΔ185 allele. Accordingly, knockdown of Wash or components of the Wash regulatory complex (SHRC) by RNAi leads to the same oogenesis phenotypes – premature ooplasmic streaming and actin architecture defects in egg chambers – as originally reported for the washΔ185 null allele. The requirement for the SHRC components in oogenesis is exciting, as it provides the first process in which Wash may be regulated by both Rho1 and the SHRC. It will be interesting in future studies to examine the molecular basis of this regulation.
A likely reason for the discrepancy in conclusions between the two studies arises from the striking observation that the washΔ185 allele changes from having distinct, penetrant oogenesis phenotypes when initially homozygous (F1 generation) to being almost indistinguishable from wild type within only a few generations. It is often the case that paralogous genes are able to compensate for each other's loss, and compensation has also been seen in protein families where constituent members have functionally similar roles (Dickinson et al., 2016; White et al., 2013). Indeed, this type of compensation has been shown among WAS family proteins and also between WAS family proteins and formin-based actin nucleation factors (Burke et al., 2014; Davidson et al., 2018; Rotty and Bear, 2014; Zhu et al., 2016). As such, it is not surprising that SCAR could be upregulated such that it can compensate for loss of Wash in oogenesis over time, especially given that it has biochemically similar properties, its reduction leads to similar phenotypes and it already accumulates at similar locations within the egg chamber.
Interestingly, SCAR is not able to compensate for Wash in all contexts. Both Wash and SCAR play important roles in immune cell (hemocyte) developmental migrations, although their reduction leads to very different phenotypes in this tissue and they are involved in different aspects of these stereotypical developmental movements (Evans et al., 2013; Nagel et al., 2017; Verboon et al., 2015a). Not surprisingly then, even after washΔ185 becomes a robust stock in its homozygous viable state, it still exhibits defects in immune cell developmental migrations (Nagel et al., 2017; Verboon et al., 2015a). It will be interesting in the future to uncover the additional ways in which compensation can occur for the loss of Wash in the variety of tissues and developmental contexts in which it functions.
Another interesting facet is how rapidly the phenotypes associated with the washΔ185 allele are lost following its selection as a homozygous stock. Recently, Waddington's work exploring evolution through canalization and assimilation – concluding that stress can lead to the selection and propagation of somatic variation (Waddington, 1942, 1953, 1959) – was re-evaluated (Fanti et al., 2017; Kasinathan et al., 2017). The authors found that the heat shock stress used in the Waddington protocol leads to genetic variation via both de novo mutations and phenocopies that they attributed to epigenetic changes in as few as 4 generations. For instance, a loss of function de novo mutation in trans to the same locus silenced by epigenetic change (phenocopy) allows for selection of a recessive mutation that can become completely genetic in nature within very few generations if selection is applied to the recessive phenotype. Previously, we showed that Drosophila Wash is present in the nucleus and affects both silencing and depression of chromatin depending on the position effect variegation (PEV) system utilized, suggesting that Wash is changing the epigenetic landscape in a complex manner (Verboon et al., 2015b,c). Additionally, loss of Wash leads to fragile chromosomes (Verboon et al., 2015b), and in the salivary gland, leads to the increased presence of DNA damage markers (J.M.V. and S.M.P., unpublished observation), which may suggest increased de novo mutations. Even in the absence of decreased de novo mutations and a presumably isogenized system, the amount of recessive variation that could be complemented by epigenetic changes that we have shown by PEV to occur in wash could potentially lead to many loci for selection to act upon. As previously mentioned, the immune cell migration phenotypes of Wash remain when it is kept in its homozygous viable state. One possible explanation for this observation is a lack of selection in this context: the visible macrophage phenotype identified is decreased lifespan (Nagel et al., 2017), but this phenotype may not confer sensitivity to selective pressure for the selection of modifiers or it may be bred out due to reduced fitness in the context of the population. It will be interesting to challenge the immune cell migration system in Drosophila wash mutants through environmental and/or immune stresses to see if this phenotype can be lost through selection of beneficial modifications, and to uncover the basis and identity of these modifications.
Finally, this study raises some interesting questions of the best way to evaluate and describe gene function. It has been recognized for some time that genetic background can influence phenotypes (Brewer et al., 2015; Dickinson et al., 2016; Doetschman, 2009; Housden et al., 2017; Sibilia and Wagner, 1995; Threadgill et al., 1995). For example, a null allele of the mouse epidermal growth factor receptor (Egfr) locus was shown to die anywhere from the peri-implantation stage to postnatal day 20 depending on genetic background (Sibilia and Wagner, 1995; Threadgill et al., 1995). Even when the genetic background is defined, a recent study has shown that incomplete penetrance and variable expressivity are common (Dickinson et al., 2016). More recently, it has been observed that the means by which gene activities are knocked down or out (e.g. genetic mutation, RNAi, morpholino, CRISPR-Cas9, chemical inhibitor) also influence the resulting phenotypes (cf. Housden et al., 2017): i.e. genetic mutations in the zebrafish egfl7 endothelial extracellular matrix gene were shown to have no obvious phenotypes, whereas knockdown of the egfl7 gene with morpholinos exhibited severe vascular defects (Rossi et al., 2015). These authors went on to show that a set of proteins that could compensate for the deleterious mutation were upregulated in mutants but not in the morphants (Rossi et al., 2015). A recent comprehensive review by Housden et al. (2017) discusses loss-of-function genetic tools for studying biological processes and the parameters for choosing and interpreting the results obtained with the different methods/systems. In the case of the Drosophila washΔ185 allele, unless this loss-of-function mutant under study is the F1 progeny of continuously outcrossed heterozygous flies, many functions of Wash will likely be missed due to compensation by other modifiers. Ironically, the presence of the unrelated lethal nw† mutation in the background of the original washΔ185 background was fortuitous as it kept wash from becoming homozygous, while not impacting its phenotypes. This is of special interest because mammalian WASH is embryonic lethal and because the clinically important phenotypes are most likely represented by the earliest and perhaps least selected upon Wash phenotypes.
MATERIALS AND METHODS
Fly stocks and genetics
Flies were cultured and crossed at 25°C on yeast-cornmeal-molasses-malt extract medium. Flies used in this study are detailed in Table S2. All fly stocks were treated with tetracycline, then tested by PCR to ensure that they did not harbor Wolbachia. RNAi knockdowns were driven maternally by the GAL4–UAS system using the P{matα4-GAL-VP16}V37 driver (Bloomington Drosophila Stock Center, stock #7063). Reduced washΔ185 and nw† were achieved by crossing females of these lines to ru1 h1 Diap11 st1 cu1 RpII140wimp sr1 es ca1/TM3, Sb1 (BDSC, stock #5874) males with the resulting trans-heterozygous females used for experiments (Liu et al., 2009; Parkhurst and Ish-Horowicz, 1991). Outcrossing, recombination and complementation crosses were carried out using standard genetics methods. The presence of the washΔ185 deletion allele was followed by PCR.
washΔ185hz survival over generations, offspring analysis and complementation crosses with Df(2R)BSC699
To determine the number of generations required to generate a robust homozygous washΔ185hz stock, heterozygous washΔ185hz flies were crossed. Ten homozygous females and five homozygous males were subsequently crossed and allowed to lay eggs for 7 days at 25°C. All progeny recovered from the cross were counted, then ten homozygous females and five homozygous males from among the progeny were crossed to obtain the numbers for the subsequent generation. Four biological replicates were performed.
Offspring analysis was performed by crossing a single 1-day-old female [wild type, washΔ185hz(outX) or washΔ185hz] with two 1-day-old wild-type males and determining the number of progeny resulting from allowing the female to lay eggs for 7 days. Four biological replicates were performed.
For complementation crosses, wild-type, washΔ185hz(outX) or washΔ185hz flies were crossed with Df(2R)BSC699/SM6a flies (deficiency covering the wash locus) using parents in both genotype orientations (Table S2). Ten females (1-3 days old) were crossed with 5 males (1-3 days old) and the progeny resulting from allowing the female to lay eggs for 7 days were scored for number and genotype. Two biological replicates were performed.
Lysate preparation
Drosophila 0-2 h embryo whole cell extract (WCE) was a gift from Toshi Tsukiyama (Fred Hutchinson Cancer Research Center, Seattle, WA). To make ovary lysate, wild-type females were fattened on yeast for 3 days and then ovaries were dissected into cold PBS. Ovaries were homogenized in HEPES lysis buffer (20 mM HEPES, pH 7, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1% Triton X-100, 5% glycerol, 1 mM DTT) with added protease and phosphatase inhibitors (Roche). Lysate was sonicated 5 times using a Sonic Dismembrator (Model 60; Fisher Scientific) at setting 5 with 10 s per pulse. The extract was centrifuged at 13,500 g for 10 min at 4°C and the supernatant recovered. Supernatant was centrifuged again at max speed for 5 min at 4°C. To make wild-type, washΔ185hz, washRNAi(1), CCDC53RNAi(1), StrumpRNAi(1), SWIPRNAI(1) and FAM21RNAi(1) ovary lysate, females were fattened on yeast for 3 days and then ovaries were dissected into cold PBS. Ovaries were homogenized on ice in Native sample buffer with protease inhibitors [1× Native PAGE Sample Buffer (Invitrogen), 1% DDM]. Lysate was sonicated using a Sonic Dismembrator (Model 60; Fisher Scientific) at setting 3.5 with 10 s per pulse. Lysate was centrifuged at max speed for 30 min at 4°C and supernatant removed. MgCl2 and Benzonase were added to the supernatant to 4 mM and 4%, respectively. Lysate was incubated at room temperature (22°C) for 30 min and then centrifuged at max speed for 30 min at 4°C. Supernatant was removed and stored at –80°C. To make washΔ185hz(outX) and washΔ185hz ovary lysates, females were fattened on yeast for 3 days and then ovaries were dissected into cold PBS. Stage 1-10B ovaries were homogenized in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% Deoxycholate sodium salt, 5 nM EDTA, 50 mM HEPES, pH 7.5) with fresh protease inhibitors added (1× Complete Protease inhibitor; Roche). Lysate was sonicated 5 times using a Sonic Dismembrator (Model 60; Fisher Scientific) at setting 1 with 1 second per pulse. The extract was centrifuged at 13,500 g for 10 min at 4°C and the supernatant recovered.
Drosophila whole-cell Kc167 extracts were made in modified RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% Deoxycholate sodium salt, 5 nM EDTA, 50 mM HEPES, pH 7.5) with fresh protease inhibitors added [1× Complete Protease inhibitor (EDTA free), 1× PhosStop, 2 mM PMSF, 1 mM Na3VO4]. Briefly, cells were grown to confluence in 500 ml spin-flasks, pelleted for 5 min at 1500 rpm, resuspended in 100 ml cold 1× PBS and repelleted for 5 min at 1500 rpm. Cell pellets were flash frozen in liquid nitrogen. Cells were resuspended in RIPA buffer+protease inhibiltors at 100 µl per 1 million cells and incubated on ice for 30 min. Lysate was sonicated using a Sonic Dismembrator (Model 60; Fisher Scientific) at setting 3.5 with 10 s per pulse. Lysate was clarified with a 15 min centrifugation at 15,000 rpm, aliquoted and flash frozen.
Wash antibodies
We previously generated an anti-Wash monoclonal antibody (P3H3) and showed its specificity (Liu et al., 2009; Rodriguez-Mesa et al., 2012; Verboon et al., 2015a). We generated another anti-Wash monoclonal antibody (P4D2) from the same monoclonal fusion that is used in this study. P3H3 was affinity purified by coupling to Protein G beads in 100 mM Tris-HCl, pH 8 buffer for 3 h, nutating at 4°C. Beads were washed once in 100 mM Tris-HCl, pH 8 and then once in 10 mM Tris-HCl, pH 8. Antibody was eluted from the beads using 50 mM glycine, pH 3 and the eluent was immediately neutralized with 1 M Tris-HCl, pH 8.
Western blotting
Lysate samples were normalized to a loading control (β-tubulin E7, 1:2000; Developmental Studies Hybridoma Bank, Iowa City, IA) or (α-tubulin 12G10; 1:5000; DSHB) and then blotted according to standard procedures. The following antibodies were used: anti-Wash mouse monoclonal [P3H3; 1:25 (Liu et al., 2009; Rodriguez-Mesa et al., 2012)]; affinity purified anti-Wash mouse monoclonal (P3H3AP; 1:25; this study); anti-Wash mouse monoclonal (P4D2; 1:1; this study); and anti-Scar mouse monoclonal [P1C1; 1:10 (Liu et al., 2009; Rodriguez-Mesa et al., 2012)]; anti-Strumpellin mouse polyclonal [1:300 (Verboon et al., 2015a)]; anti-CCDC53 mouse polyclonal [1:1000 (Verboon et al., 2015a)]; anti-FAM21 mouse polyclonal [1:300 (Verboon et al., 2015a)]. Quantification of Scar on western blots was performed using ImageJ (National Institutes of Health, Bethesda, MD). A minimum of two biological replicates was performed for each western.
Actin visualization in ovaries
Female flies were fattened on yeast for 2 days and then ovaries were dissected into cold PBS. Ovaries were fixed using 1:6 fix/heptane for 10 min. Fix is: 16.7 mM KPO4 pH 6.8, 75 mM KCl, 25 mM NaCl, 3.3 mM MgCl2, 6% formaldehyde. Ovaries were washed 3 times with PBS plus 0.1% Triton X-100, and then incubated in PBS plus 0.5% Triton X-100 and Alexa Fluor 568-conjugated Phalloidin at 0.005 units/µl (Molecular Probes/Invitrogen, Rockford, IL) at room temperature for 1 h. Ovaries were washed with PTW (1× PBS, 0.1% Tween-20) 10 times for 10 min each, then dissected into individual ovarioles and mounted on slides in Slowfade Gold with DAPI medium (Invitrogen, Rockford, IL). A minimum of two biological replicates were performed for each condition.
Immunostaining of ovaries
Female flies were fattened on yeast for 2 days and then ovaries were dissected and fixed as described above. After 3 washes with PBS plus 0.1% Triton X-100, ovaries were permeabilized in PBS plus 1% Triton X-100 at room temperature for 2 h. Ovaries were washed 3 times with PAT [1× PBS, 0.1% Tween-20, 1% bovine serum albumin (BSA), 0.05% azide] then blocked in PAT at 4°C for 2 h. Antibodies were used at the following concentrations: anti-Wash mouse monoclonal (P3H3; 1:20; Liu et al., 2009; Rodriguez-Mesa et al., 2012); affinity purified anti-Wash mouse monoclonal (P3H3AP; 1:15; this study); anti-Wash mouse monoclonal (P4D2; 1:15; this study); anti-Wasp mouse monoclonal (P5E1; 1:10; Rodriguez-Mesa et al., 2012); anti-SCAR mouse monoclonal (P1C1; 1:50; Rodriguez-Mesa et al., 2012); anti-Strumpellin mouse polyclonal (1:200; Verboon et al., 2015a); anti-CCDC53 mouse polyclonal (1:200; Verboon et al., 2015a); anti-FAM21 mouse polyclonal (1:200; Verboon et al., 2015a); anti-SWIP mouse polyclonal (1:200; Verboon et al., 2015a); anti-GFP mouse monoclonal (1:1000; Sigma-Aldrich, St Louis, MO), and the ovaries incubated for 48 h at 4°C. Ovaries were washed with XNS (1× PBS, 0.1% Tween-20, 0.1% BSA, 4% normal goat serum), then incubated with Alexa Fluor 488-conjugated secondary antibodies (1:1000; Invitrogen) overnight at 4°C. Ovaries were washed with PTW (1× PBS, 0.1% Tween-20), dissected into individual ovarioles, then mounted on slides in Slowfade Gold with DAPI medium (Invitrogen). A minimum of two biological replicates were performed for each condition.
To quantify the Wash or SCAR expression, we measured the averaged fluorescence intensity from 10-pixel-wide sections spanning the indicated regions of the follicle cells and oocyte using ImageJ (NIH). Line profiles were plotted using Prism 7.0a (GraphPad Software Inc.).
Confocal microscopy
Images of fixed tissues were acquired using a Zeiss LSM 780 spectral confocal microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) fitted with a Zeiss 63×/1.4 oil Plan-Apochromat objective. FITC (Alexa Fluor 488) fluorescence was excited with the 488 nm line of an Argon laser, and detection was between 498 and 560 nm. Red (Alexa Fluor 568) fluorescence was excited with the 561 nm line of a DPSS laser and detection was between 570 and 670 nm. Pinhole was set to 1.0 Airy Units. Confocal sections were acquired at 0.25-1.0 µm spacing. Super-resolution images were acquired using an Airyscan detector in Super Resolution mode and captured confocal images were then processed using the Airyscan Processing feature on the Zen software provided by the manufacturer (Carl Zeiss Microscopy GmbH, Jena, Germany).
Live image acquisition
To obtain live images of oocytes, female flies were first fattened on yeast for 2 days. Females were then injected in the abdomen with 0.4% Trypan Blue (ThermoFisher Scientific, Waltham, MA) diluted 1:5 in PBS, and allowed to sit for 1-2 h. Then ovaries were dissected into individual egg chambers in halocarbon 700 oil (Halocarbon Products, River Edge, NJ) on a cover slip. Images were acquired on a Revolution WD systems (Andor Technology Ltd., Concord, MA) mounted on a Leica DMi8 (Leica Microsystems Inc., Buffalo Grove, IL) with a 63×/1.4 NA objective lens with a 2× coupler and controlled by MetaMorph software. Images were acquired with 561 nm excitation using an Andor iXon Ultra 888 EMCCD camera (Andor Technology Ltd., Concord, MA). Time-lapse images were obtained by taking one single frame acquisition every 10 s for either 5 or 30 min.
Statistical analysis
Stage 7-9 egg chambers stained with Phalloidin were assessed for disrupted actin structures, then binned according to their phenotypes into one of two categories: none/mild and moderate/severe. Gene knockdowns were compared to the appropriate control genotype and dependency was calculated using a two-tailed Fisher's exact test with P<0.01 considered independent and significant.
Stage 7-9 egg chambers were also assessed for whether or not they exhibited premature ooplasmic streaming. These were compared to their appropriate control genotypes (as indicated) and dependency was calculated using a two-tailed Fisher's exact test with P<0.01 considered independent and significant. For the wash offspring analysis and survival over generations, a two-tailed Student's test was used with P<0.01 considered independent and significant.
To evaluate deviation for the expected value of homozygous to heterozygous progeny, two independent +/CyO lines were crossed twice each (10 females to 5 males) and the resultant progeny counted and averaged between the two lines to measure the control expected value (Binomial test). These lines were generated by the outcrosses described previously and harbored neither the washΔ185 or nw allele, and represent the most similar genetic background to wash crosses and progeny were counted. Significance was calculated for each Wash line to the measured expected value and a P<0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank Phil Soriano, Jim Priess, Valera Vasioukhin, Hugo Bellen, Norbert Perrimon, the Bloomington Stock Center, the Kyoto Stock Center, the Harvard Transgenic RNAi Project, the Vienna Drosophila RNAi Center, the Drosophila Genomics Resource Center, and the Developmental Studies Hybridoma Bank for advice, antibodies, DNAs, flies and other reagents used in this study.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.M.V., J.R.D., M.N., S.M.P.; Methodology: J.M.V., J.R.D., M.N., S.M.P.; Validation: J.M.V., J.R.D., M.N., S.M.P.; Formal analysis: J.M.V., J.R.D., M.N., S.M.P.; Investigation: J.M.V., J.R.D., M.N., S.M.P.; Writing - original draft: J.M.V., S.M.P.; Writing - review & editing: J.M.V., J.R.D., M.N., S.M.P.; Visualization: J.M.V., J.R.D., M.N., S.M.P.; Supervision: S.M.P.; Project administration: S.M.P.; Funding acquisition: S.M.P.
Funding
This research was funded in part through the National Institutes of Health National Cancer Institute (NIH/NCI) Cancer Center Support Grant P30 CA015704. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.211573.supplemental
References
- Alekhina O., Burstein E. and Billadeau D. D. (2017). Cellular functions of WASP family proteins at a glance. 130, 2235-2241. 10.1242/jcs.199570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartuzi P., Billadeau D. D., Favier R., Rong S., Dekker D., Fedoseienko A., Fieten H., Wijers M., Levels J. H., Huijkman N. et al. (2016). CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. 7, 10961 10.1038/ncomms10961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer J. R., Molotkov A., Mazot P., Hoch R. V. and Soriano P. (2015). Fgfr1 regulates development through the combinatorial use of signaling proteins. 29, 1863-1874. 10.1101/gad.264994.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley C. M., Gopaldass N., Bosmani C., Johnston S. A., Soldati T., Insall R. H. and King J. S. (2016). WASH drives early recycling from macropinosomes and phagosomes to maintain surface phagocytic receptors. 113, E5906-E5915. 10.1073/pnas.1524532113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burianek L. E. and Soderling S. H. (2013). Under lock and key: spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. 24, 258-266. 10.1016/j.semcdb.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T. A., Christensen J. R., Barone E., Suarez C., Sirotkin V. and Kovar D. R. (2014). Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. 24, 579-585. 10.1016/j.cub.2014.01.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone K. G. and Welch M. D. (2010). A nucleator arms race: cellular control of actin assembly. 11, 237-251. 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell M., Zech T., Calaminus S. D., Ura S., Hagedorn M., Johnston S. A., May R. C., Soldati T., Machesky L. M. and Insall R. H. (2011). Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. 193, 831-839. 10.1083/jcb.201009119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A. J., Amato C., Thomason P. A. and Insall R. H. (2018). WASP family proteins and formins compete in pseudopod- and bleb-based migration. 217, 701-714. 10.1083/jcb.201705160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E., Sousa C., Gautier J. J., Lombard B., Loew D. and Gautreau A. (2009). The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. 17, 712-723. 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Dickinson M. E., Flenniken A. M., Ji X., Teboul L., Wong M. D., White J. K., Meehan T. F., Weninger W. J., Westerberg H., Adissu H. et al. (2016). High-throughput discovery of novel developmental phenotypes. 537, 508-514. 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetschman T. (2009). Influence of genetic background on genetically engineered mouse phenotypes. 530, 423-433. 10.1007/978-1-59745-471-1_23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Kakihara K., Otani T., Wada H. and Hayashi S. (2013). Rab9 and retromer regulate retrograde trafficking of luminal protein required for epithelial tube length control. 4, 1358 10.1038/ncomms2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duleh S. N. and Welch M. D. (2010). WASH and the Arp2/3 complex regulate endosome shape and trafficking. 67, 193-206. 10.1002/cm.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott A. M., Simard L. R., Coghlan G., Chudley A. E., Chodirker B. N., Greenberg C. R., Burch T., Ly V., Hatch G. M. and Zelinski T. (2013). A novel mutation in KIAA0196: identification of a gene involved in Ritscher-Schinzel/3C syndrome in a First Nations cohort. 50, 819-822. 10.1136/jmedgenet-2013-101715 [DOI] [PubMed] [Google Scholar]
- Evans I. R., Ghai P. A., Urbančič V., Tan K.-L. and Wood W. (2013). SCAR/WAVE-mediated processing of engulfed apoptotic corpses is essential for effective macrophage migration in Drosophila. 20, 709-720. 10.1038/cdd.2012.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti L., Piacentini L., Cappucci U., Casale A. M. and Pimpinelli S. (2017). Canalization by selection of de Novo induced mutations. 206, 1995-2006. 10.1534/genetics.117.201079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman C., Seaman M. N. J. and Reid E. (2013). The hereditary spastic paraplegia protein strumpellin: characterisation in neurons and of the effect of disease mutations on WASH complex assembly and function. 1832, 160-173. 10.1016/j.bbadis.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez T. S. and Billadeau D. D. (2009). A FAM21-containing WASH complex regulates retromer-dependent sorting. 17, 699-711. 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez T. S., Gorman J. A., de Narvajas A. A., Koenig A. O. and Billadeau D. D. (2012). Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. 23, 3215-3228. 10.1091/mbc.E12-02-0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour M. E., Breusegem S. Y. and Seaman M. N. J. (2012). Recruitment of the endosomal WASH complex is mediated by the extended ‘tail’ of Fam21 binding to the retromer protein Vps35. 442, 209-220. 10.1042/BJ20111761 [DOI] [PubMed] [Google Scholar]
- Housden B. E., Muhar M., Gemberling M., Gersbach C. A., Stainier D. Y. R., Seydoux G., Mohr S. E., Zuber J. and Perrimon N. (2017). Loss-of-function genetic tools for animal models: cross-species and cross-platform differences. 18, 24-40. 10.1038/nrg.2016.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Lian J., Chen X., Qin G., Zheng Y. and Zhang Y. (2017). WASH overexpression enhances cancer stem cell properties and correlates with poor prognosis of esophageal carcinoma. 108, 2358-2365. 10.1111/cas.13400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Gomez T. S., Metlagel Z., Umetani J., Otwinowski Z., Rosen M. K. and Billadeau D. D. (2010). WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. 107, 10442-10447. 10.1073/pnas.0913293107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinathan B., Ahmad K. and Malik H. S. (2017). Waddington redux: De Novo Mutations underlie the genetic assimilation of stress-induced phenocopies in Drosophila melanogaster. 207, 49-51. 10.1534/genetics.117.205039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leirdal M., Shadidy M., Rosok O. and Sioud M. (2004). Identification of genes differentially expressed in breast cancer cell line SKBR3: potential identification of new prognostic biomarkers. 14, 217-222. 10.3892/ijmm.14.2.217 [DOI] [PubMed] [Google Scholar]
- Linardopoulou E. V., Parghi S. S., Friedman C., Osborn G. E., Parkhurst S. M. and Trask B. J. (2007). Human subtelomeric WASH genes encode a new subclass of the WASP family. 3, e237 10.1371/journal.pgen.0030237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Abreu-Blanco M. T., Barry K. C., Linardopoulou E. V., Osborn G. E. and Parkhurst S. M. (2009). Wash functions downstream of Rho and links linear and branched actin nucleation factors. 136, 2849-2860. 10.1242/dev.035246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaad M. J., Ramesh N. and Geha R. S. (2013). Wiskott-Aldrich syndrome: a comprehensive review. 1285, 26-43. 10.1111/nyas.12049 [DOI] [PubMed] [Google Scholar]
- McGough I. J., Steinberg F., Jia D., Barbuti P. A., McMillan K. J., Heesom K. J., Whone A. L., Caldwell M. A., Billadeau D. D., Rosen M. K. et al. (2014). Retromer binding to FAM21 and the WASH complex is perturbed by the Parkinson disease-linked VPS35(D620N) mutation. 24, 1670-1676. 10.1016/j.cub.2014.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard T. H., Sharp S. J. and Machesky L. M. (2004). Signalling to actin assembly via the WASP (Wiskott-Aldrich syndrome protein)-family proteins and the Arp2/3 complex. 380, 1-17. 10.1042/bj20040176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P., Rossé C., Castro-Castro A., Irondelle M., Lagoutte E., Paul-Gilloteaux P., Desnos C., Formstecher E., Darchen F., Perrais D. et al. (2013). Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. 203, 1063-1079. 10.1083/jcb.201306162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel B. M., Bechtold M., Rodriguez L. G. and Bogdan S. (2017). Drosophila WASH is required for integrin-mediated cell adhesion, cell motility and lysosomal neutralization. 130, 344-359. 10.1242/jcs.193086 [DOI] [PubMed] [Google Scholar]
- Nordgard S. H., Johansen F. E., Alnaes G. I. G., Bucher E., Syvänen A.-C., Naume B., Borresen-Dale A.-L. and Kristensen V. N. (2008). Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. 47, 680-696. 10.1002/gcc.20569 [DOI] [PubMed] [Google Scholar]
- Park L., Thomason P. A., Zech T., King J. S., Veltman D. M., Carnell M., Ura S., Machesky L. M. and Insall R. H. (2013). Cyclical action of the WASH complex: FAM21 and capping protein drive WASH recycling, not initial recruitment. 24, 169-181. 10.1016/j.devcel.2012.12.014 [DOI] [PubMed] [Google Scholar]
- Parkhurst S. M. and Ish-Horowicz D. (1991). wimp, a dominant maternal-effect mutation, reduces transcription of a specific subset of segmentation genes in Drosophila. 5, 341-357. 10.1101/gad.5.3.341 [DOI] [PubMed] [Google Scholar]
- Piotrowski J. T., Gomez T. S., Schoon R. A., Mangalam A. K. and Billadeau D. D. (2013). WASH knockout T cells demonstrate defective receptor trafficking, proliferation, and effector function. 33, 958-973. 10.1128/MCB.01288-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Mesa E., Abreu-Blanco M. T., Rosales-Nieves A. E. and Parkhurst S. M. (2012). Developmental expression of Drosophila Wiskott-Aldrich Syndrome family proteins. 241, 608-626. 10.1002/dvdy.23742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers F., Derivery E., Hu H., Garshasbi M., Karbasiyan M., Herold M., Nurnberg G., Ullmann R., Gautreau A., Sperling K. et al. (2011). Identification of a novel candidate gene for non-syndromic autosomal recessive intellectual disability: the WASH complex member SWIP. 20, 2585-2590. 10.1093/hmg/ddr158 [DOI] [PubMed] [Google Scholar]
- Rossi A., Kontarakis Z., Gerri C., Nolte H., Holper S., Kruger M. and Stainier D. Y. (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. 524, 230-233. 10.1038/nature14580 [DOI] [PubMed] [Google Scholar]
- Rottner K., Hanisch J. and Campellone K. G. (2010). WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. 20, 650-661. 10.1016/j.tcb.2010.08.014 [DOI] [PubMed] [Google Scholar]
- Rotty J. D. and Bear J. E. (2014). Competition and collaboration between different actin assembly pathways allows for homeostatic control of the actin cytoskeleton. 5, 27-34. 10.1080/19490992.2015.1090670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty J. D., Wu C. and Bear J. E. (2013). New insights into the regulation and cellular functions of the ARP2/3 complex. 14, 7-12. 10.1038/nrm3492 [DOI] [PubMed] [Google Scholar]
- Ryder P. V., Vistein R., Gokhale A., Seaman M. N., Puthenveedu M. A. and Faundez V. (2013). The WASH complex, an endosomal Arp2/3 activator, interacts with the Hermansky-Pudlak syndrome complex BLOC-1 and its cargo phosphatidylinositol-4-kinase type IIalpha. 24, 2269-2284. 10.1091/mbc.E13-02-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibilia M. and Wagner E. F. (1995). Strain-dependent epithelial defects in mice lacking the EGF receptor. 269, 234-238. 10.1126/science.7618085 [DOI] [PubMed] [Google Scholar]
- Stradal T. E. B., Rottner K., Disanza A., Confalonieri S., Innocenti M. and Scita G. (2004). Regulation of actin dynamics by WASP and WAVE family proteins. 14, 303-311. 10.1016/j.tcb.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Takenawa T. and Suetsugu S. (2007). The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. 8, 37-48. 10.1038/nrm2069 [DOI] [PubMed] [Google Scholar]
- Threadgill D. W., Dlugosz A. A., Hansen L. A., Tennenbaum T., Lichti U., Yee D., LaMantia C., Mourton T., Herrup K., Harris R. C. et al. (1995). Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. 269, 230-234. 10.1126/science.7618084 [DOI] [PubMed] [Google Scholar]
- Turk M., Schroder R., Khuller K., Hofmann A., Berwanger C., Ludolph A. C., Dekomien G., Muller K., Weishaupt J. H., Thiel C. T. et al. (2017). Genetic analysis of VCP and WASH complex genes in a German cohort of sporadic ALS-FTD patients. 56, 213.e1-213.e5. 10.1016/j.neurobiolaging.2017.04.023 [DOI] [PubMed] [Google Scholar]
- Valdmanis P. N., Meijer I. A., Reynolds A., Lei A., MacLeod P., Schlesinger D., Zatz M., Reid E., Dion P. A., Drapeau P. et al. (2007). Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. 80, 152-161. 10.1086/510782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardarajan B. N., Bruesegem S. Y., Harbour M. E., Inzelberg R., Friedland R., St George-Hyslop P., Seaman M. N. and Farrer L. A. (2012). Identification of Alzheimer disease-associated variants in genes that regulate retromer function. 33, 2231 e15-2231 e30. 10.1016/j.neurobiolaging.2012.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen M. K. and Machesky L. M. (2004). The WASP-Arp2/3 pathway: genetic insights. 16, 174-181. 10.1016/j.ceb.2004.02.004 [DOI] [PubMed] [Google Scholar]
- Veltman D. M. and Insall R. H. (2010). WASP family proteins: their evolution and its physiological implications. 21, 2880-2893. 10.1091/mbc.E10-04-0372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon J. M., Rahe T. K., Rodriguez-Mesa E. and Parkhurst S. M. (2015a). Wash functions downstream of Rho1 GTPase in a subset of Drosophila immune cell developmental migrations. 26, 1665-1674. 10.1091/mbc.E14-08-1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon J. M., Rincon-Arano H., Werwie T. R., Delrow J. J., Scalzo D., Nandakumar V., Groudine M. and Parkhurst S. M. (2015b). Wash interacts with lamin and affects global nuclear organization. 25, 804-810. 10.1016/j.cub.2015.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon J. M., Sugumar B. and Parkhurst S. M. (2015c). Wiskott-Aldrich Syndrome proteins in the nucleus: aWASH with possibilities. 6, 349-359. 10.1080/19491034.2015.1086051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C. H. (1942). Canalization of development and the inheritance of acquired characters. 150, 563-565. 10.1038/150563a0 [DOI] [Google Scholar]
- Waddington C. H. (1953). Genetic assimilation of an acquired character. 7, 118-126. 10.1111/j.1558-5646.1953.tb00070.x [DOI] [Google Scholar]
- Waddington C. H. (1959). Canalization of development and genetic assimilation of acquired characters. 183, 1654-1655. 10.1038/1831654a0 [DOI] [PubMed] [Google Scholar]
- White J. K., Gerdin A. K., Karp N. A., Ryder E., Buljan M., Bussell J. N., Salisbury J., Clare S., Ingham N. J., Podrini C. et al. (2013). Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. 154, 452-464. 10.1016/j.cell.2013.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Wang S., Du Y., Zhao Z., Shi L., Sun L., Huang G., Ye B., Li C., Dai Z. et al. (2013). WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. 32, 2685-2696. 10.1038/emboj.2013.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Wang S., Huang G., Zhu P., Li M., Ye B., Du Y. and Fan Z. (2014). WASH is required for the differentiation commitment of hematopoietic stem cells in a c-Myc-dependent manner. 211, 2119-2134. 10.1084/jem.20140169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J. A., Cohen Y., Hudson A. M., Cooley L., Wieschaus E. and Schejter E. D. (2002). SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. 156, 689-701. 10.1083/jcb.200109057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E., Seaman M. N. J. and Rubinsztein D. C. (2014a). VPS35 Parkinson mutation impairs autophagy via WASH. 13, 2155-2156. 10.4161/cc.29734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E., Seaman M. N. J., Moreau K., Jimenez-Sanchez M., Breusegem S. Y., Harbour M. E. and Rubinsztein D. C. (2014b). Mutation in VPS35 associated with Parkinson's disease impairs WASH complex association and inhibits autophagy. 5, 3828 10.1038/ncomms4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T., Calaminus S. D. J., Caswell P., Spence H. J., Carnell M., Insall R. H., Norman J. and Machesky L. M. (2011). The Arp2/3 activator WASH regulates alpha5beta1-integrin-mediated invasive migration. 124, 3753-3759. 10.1242/jcs.080986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chai Y., Jiang Y., Li W., Hu H., Li W., Wu J.-W., Wang Z.-X., Huang S. and Ou G. (2016). Functional coordination of WAVE and WASP in C. elegans neuroblast migration. 39, 224-238. 10.1016/j.devcel.2016.09.029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.