ABSTRACT
Imprinted genes are expressed from one parental allele and regulated by differential DNA methylation at imprinting control regions (ICRs). ICRs are reprogrammed in the germline through erasure and re-establishment of DNA methylation. Although much is known about DNA methylation establishment, DNA demethylation is less well understood. Recently, the Ten-Eleven Translocation proteins (TET1-3) have been shown to initiate DNA demethylation, with Tet1−/− mice exhibiting aberrant levels of imprinted gene expression and ICR methylation. Nevertheless, the role of TET1 in demethylating ICRs in the female germline and in controlling allele-specific expression remains unknown. Here, we examined ICR-specific DNA methylation in Tet1−/− germ cells and ascertained whether abnormal ICR methylation impacted imprinted gene expression in F1 hybrid somatic tissues derived from Tet1−/− eggs or sperm. We show that Tet1 deficiency is associated with hypermethylation of a subset of ICRs in germ cells. Moreover, ICRs with defective germline reprogramming exhibit aberrant DNA methylation and biallelic expression of linked imprinted genes in somatic tissues. Thus, we define a discrete set of genomic regions that require TET1 for germline reprogramming and discuss mechanisms for stochastic imprinting defects.
KEY WORDS: Tet1, Methylcytosine dioxygenase, Genomic imprinting, DNA methylation, Epigenetic reprogramming, Allele-specific expression, Imprinting control region (ICR), Mouse
Summary: Methylcytosine dioxygenase TET1 is a key regulator of imprinting in both the maternal and paternal lineages, with loss resulting in hypermethylation of KvDMR, Peg1 and Peg3 ICRs in sperm and of IG-DMR and H19/Igf2 ICRs in oocytes.
INTRODUCTION
A major factor in determining developmental fate and cellular identity lies in the transcriptional program, which is regulated by chemical modifications of DNA and histone tails (Goldberg et al., 2007). Methyl groups are covalently added to cytosine residues by the DNA methyltransferase (DNMT) enzymes, most often in the context of cytosine guanine dinucleotides (mCpG) (Klose and Bird, 2006). This epigenetic mark is essential for development, as DNMT homozygous null animals exhibit embryonic lethality (Dnmt1, Dnmt3b) or die within the first four weeks of life (Dnmt3a) (Li et al., 1992; Okano et al., 1999). Additionally, dynamic DNA methylation allows a cell to change its transcriptional program and thus differentiate into the many unique cell types that ultimately make up an entire organism (Smith and Meissner, 2013). One such example of dynamic DNA methylation occurs during early mammalian development. After fertilization, DNA methylation patterns that govern sperm and oocyte cell identity must be globally erased to generate a totipotent zygote. This epigenetic reprogramming leads to an almost completely hypomethylated state in the blastocyst and paves the way for subsequent remethylation in cell-specific and tissue-specific patterns as development progresses (Smith et al., 2012).
In the mouse, a second wave of epigenetic reprogramming initiates around embryonic day (E) 7.5 specifically in the primordial germ cells (PGCs) while they begin to proliferate and migrate toward the genital ridge (Leitch et al., 2013). PGCs are recruited from the epiblast and therefore contain a somatic pattern of DNA methylation, which must be erased and reset in germ cells. This PGC-specific demethylation occurs in two phases: first, DNA methylation across the majority of regions in the mouse genome is erased (Seisenberger et al., 2012a). This near global loss of DNA methylation is due to the downregulation of the de novo DNMT genes Dnmt3a/b as well as downregulation and exclusion from the nucleus of UHRF1, which targets the maintenance methyltransferase DNMT1 to the replication fork (Hajkova et al., 2002; Kurimoto et al., 2008). These actions passively dilute DNA methylation with each round of cell division. A second phase of demethylation occurs slightly later, between E11.5 and E13.5, at a subset of repetitive elements as well as at cis-regulatory regions called imprinting control regions (ICRs) (Seisenberger et al., 2012a).
Epigenetic reprogramming of ICRs in the germline is crucial to the appropriate regulation of imprinted genes in offspring. Imprinted genes are uniquely expressed from only one parental allele and play important roles in growth and development (Plasschaert and Bartolomei, 2014). ICRs have DNA methylation on one parental chromosome that controls the monoallelic, parent-of-origin-specific expression of imprinted genes. By E13.5, ICRs are completely demethylated in the germline, which is necessary to allow the acquisition of the parent-of-origin-specific DNA methylation patterns, either at maternally methylated ICRs in the developing oocytes, or at paternally methylated ICRs in the developing sperm (Stewart et al., 2016). This asymmetric DNA methylation of ICRs ensures proper monoallelic expression of imprinted genes in the resultant embryo.
The observation that DNA methylation at ICRs is resistant to the initial phase of demethylation in PGCs is consistent with ICR erasure being an active process. The discovery of the Ten-Eleven Translocation (TET) family of enzymes, which comprises TET1, TET2 and TET3 (also known as tet methylcytosine dioxygenase 1-3) (Tahiliani et al., 2009), implicated their potential relevance to ICR DNA methylation erasure (Hill et al., 2014). All three TETs have the ability to progressively oxidize the methyl group on cytosines (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (Ito et al., 2011; Tahiliani et al., 2009). This iterative oxidation is thought to play an active part in removing DNA methylation in conjunction with replication or base excision repair (Hajkova et al., 2010). In PGCs, Tet1 is the most highly expressed family member, while Tet2 is expressed at a lower level and Tet3 is undetectable (Hackett et al., 2013; Hajkova et al., 2010; Kagiwada et al., 2013; Yamaguchi et al., 2012). Previous studies showed abnormal levels of DNA methylation and abnormal levels of total expression of imprinted genes in Tet1−/− single-knockout and Tet1−/−; Tet2−/− double-knockout mice (Dawlaty et al., 2013; Yamaguchi et al., 2013). However, most experiments in the Yamaguchi et al. (2013) study used a Tet1 allele in which a fusion protein was generated. The catalytic domain of the endogenous TET1 protein was replaced with a β-galactosidase cassette, but the DNA-binding CxxC domain was retained (Yamaguchi et al., 2012). This study also focused primarily on the effects of Tet1 on ICR methylation in the male germline. Additionally, the Tet1−/−; Tet2−/− mouse model did not allow elucidation of the precise contribution of Tet1 to ICR DNA methylation regulation (Dawlaty et al., 2013). Thus, our understanding of the role of Tet1 in the regulation of genomic imprinting is incomplete.
The goal of our study was to define the function of Tet1 in male and female germ cells using a null allele of Tet1 in mice. Furthermore, we aimed to elucidate how loss of Tet1 affects the monoallelic expression of imprinted genes by studying offspring of male and female Tet1 knockout animals. In addition to its requirement in the male germline, our work demonstrates for the first time that Tet1 is required for normal ICR methylation in female germ cells. Moreover, lack of Tet1 in the germline leads to stochastic biallelic expression of imprinted genes across different stages of development in offspring. Importantly, we show that the reliance on TET1-mediated demethylation varies according to the imprinted locus, suggesting a complex relationship between TET1 demethylation and the genomic context.
RESULTS
Loss of Tet1 leads to stochastic and variable DNA hypermethylation at ICRs in oocytes
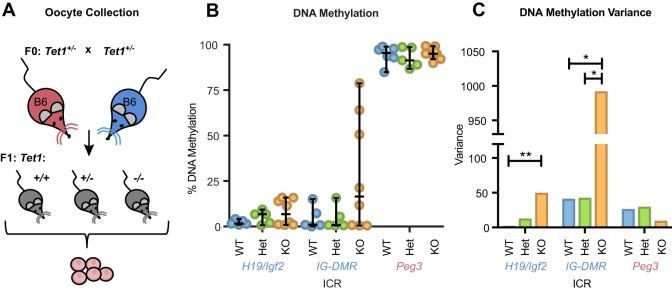
To examine how loss of Tet1 affects DNA methylation (mCpG) in the female germline, we collected germinal vesicle stage oocytes from 3.5-week-old mice generated by mating heterozygous Tet1 mutant mice. One pool of oocytes per mouse was bisulfite treated and ICR mCpG levels were analyzed by pyrosequencing (Fig. 1A). We investigated two of the three known paternally methylated ICRs in the mouse genome: H19/Igf2 and IG-DMR. As expected, Tet1 wild-type (WT) and heterozygous (Het) pools of oocytes had very low levels of mCpG at the H19/Igf2 ICR and IG-DMR, while the maternally methylated Peg3 ICR was nearly completely methylated and served as an internal control for somatic contamination. By contrast, Tet1−/− (knockout, KO) oocyte pools showed stochastic hypermethylation at both H19/Igf2 and IG-DMR, meaning that some KO oocyte pools had mCpG levels comparable to WT and Het pools, whereas other KO oocyte pools had increased levels of mCpG (Fig. 1B). The stochastic nature of the DNA methylation abnormalities was reflected in bimodal distributions in KO oocyte pools at each of the two paternally methylated ICRs (Fig. S1A,B). Affected KO oocyte pools were also variable in the severity of the hypermethylation phenotype, with each oocyte pool exhibiting different levels of abnormal mCpG, particularly at IG-DMR (Fig. 1B, Table S1). Additionally, the variance in mCpG was significantly higher in the KO oocytes at the H19/Igf2 ICR and IG-DMR compared with controls (for WT versus KO, P=0.008 and P=0.026, respectively), whereas there was no difference in mCpG levels and variance at the Peg3 ICR (Fig. 1C). Moreover, there was no correlation between abnormal methylation at IG-DMR and H19/Igf2 in KO oocyte pools (Fig. S1C). Thus, consistent with the hypothesis that Tet1 is required for proper DNA demethylation of ICRs, lack of Tet1 in the female germline leads to stochastic hypermethylation and significant increases in DNA methylation variance at paternally methylated ICRs.
Fig. 1.
Hypermethylation occurs stochastically in Tet1−/− oocytes at the H19/Igf2 ICR and IG-DMR. (A) The breeding scheme used to collect oocytes from 3.5-week-old mice. (B) Average DNA methylation at various ICRs. Individual dots represent the percentage DNA methylation from one pool of oocytes collected from one mouse. ICR labels are color coded according to which parental allele is normally methylated: blue, the paternal allele; pink, the maternal allele. Line, median; bars, 95% confidence interval. WT, wild type; Het, Tet1+/−; KO, Tet1−/−. H19/Igf2 and IG-DMR, WT n=5, Het n=5, KO n=8; Peg3, WT n=6, Het n=5, KO n=9. (C) Calculated variance for each ICR per genotype. *P<0.05, **P<0.01 (Fligner-Killeen test).
Loss of Tet1 leads to biallelic expression and changes in DNA methylation of imprinted genes in the offspring of female KO mice
We speculated that the abnormalities in oocyte methylation might cause developmental defects in the offspring. To address this question, we crossed either Tet1 Het or KO females to WT Tet1 males on the C57BL/6J(CAST7) (C7) background, which have a Mus musculus castaneus chromosome 7 on a C57BL/6J (B6) background (Fig. S2A,B). These F1 hybrid animals harbor single nucleotide polymorphisms (SNPs) on chromosome 7 that allow the parental origin of the RNA to be determined. Het Tet1 females crossed to C7 males are hereafter referred to as mCON, and their offspring are either Het or WT for Tet1. mKO refers to Tet1 KO females crossed to C7 males. mKO offspring are all Het for Tet1. We analyzed the number of live and resorbed embryos and placentas at E10.5 from mCON and mKO matings. By this developmental stage, there was a significant decrease in live embryos (P=0.005) and a significant increase in resorbed embryos (P=0.011) in mKO litters compared with mCON litters (Fig. 2A,B). This indicates that offspring derived from Tet1 KO females are more susceptible to fetal demise than those derived from mCON females at E10.5.
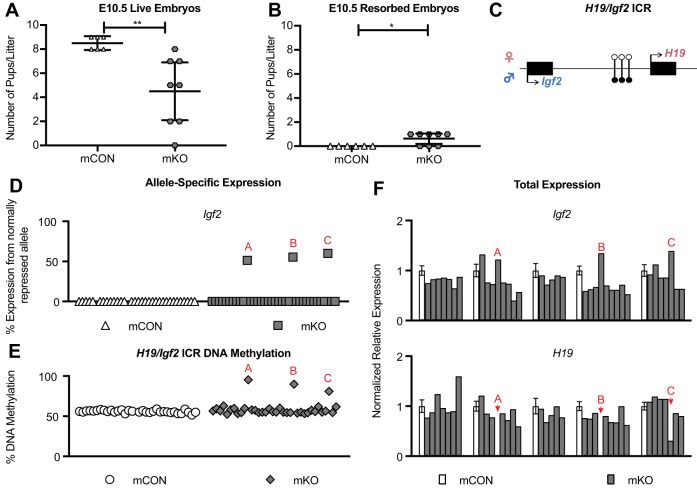
Fig. 2.
Abnormal allele-specific expression and methylation of Igf2 in E10.5 mKO offspring. (A) The number of live embryos per litter at E10.5. **P<0.01, unpaired two-tailed t-test (unequal variance). mCON n=5, mKO n=8 litters. (B) The number of resorbed embryos per litter at E10.5. *P<0.05, unpaired two-tailed t-test (unequal variance). mCON n=5, mKO n=8 litters. (A,B) Line, mean; bars, 95% confidence interval. (C) The mouse H19/Igf2 locus. The maternal allele with maternal-specific H19 expression is represented at the top (pink) and the paternal allele with paternal-specific Igf2 expression is represented at the bottom (blue). Genes, black boxes. Black lollipops, methylated DNA at the ICR; white lollipops, unmethylated DNA at the ICR. (D) Allele-specific expression of Igf2. Abnormal mKO embryos are denoted with red letters, which are consistent between Figs 2 and 3 (embryos are in the same order from left to right in D,E and Fig. 3B). mCON n=32 embryos from 4 litters, mKO n=36 embryos from 7 litters. (E) Average percentage DNA methylation at the H19/Igf2 ICR. mCON n=32 embryos from 4 litters, mKO n=36 embryos from 7 litters. (F) Total expression from qPCR analysis of Igf2 and H19. White columns indicate the average of one of each of the five mCON litters (n=8, 8, 8, 9, 9, respectively, from left to right), which were normalized to the expression of Rplp0, Nono and Rpl13a and set to 1. Error bars indicate s.e.m. The gray columns represent individual mKO embryos that were run on the same plate as the adjacent mCON column. All mCON litters and embryos are in the same order in F as in Fig. 3C. In the H19 plot, H19 expression in embryos A and B is undetectable.
Next, we asked whether abnormal ICR methylation in the germline could affect the allele-specific expression of imprinted genes in the offspring of female Tet1 KO mice. Given that each of the two inherited Tet1−/− parental alleles contributing to an ICR in the offspring has a 50% chance of being unmethylated (previously maternal) or methylated (previously paternal and failed to erase in the germline), a priori we expect that the number of affected offspring would be 50%. Using restriction fragment length polymorphism (RFLP) analysis, we quantified allele-specific expression at the H19/Igf2 locus (Fig. 2C, Table S2). Three mKO-derived embryos from independent litters exhibited biallelic expression of Igf2 (3/36) whereas none of the mCON-derived embryos was affected (0/34) (Fig. 2D). This shows that the imprinting phenotype was neither present in every mKO embryo nor at the expected proportion of 50%. Additionally, in an affected conceptus, Igf2 expression was biallelic in both the embryo and its placenta (Table S2).
To address how mCpG levels at the ICR may correspond to abnormal imprinted gene expression, we analyzed mCpG by pyrosequencing. We found that the mKO embryos with biallelic Igf2 expression also showed DNA hypermethylation at the ICR (Fig. 2E). Additionally, we measured total expression of the imprinted genes Igf2 and H19. Given hypermethylation of the ICR, the three mKO embryos showing biallelic Igf2 expression had low or undetectable levels of H19 expression, as anticipated. Increased total Igf2 expression was also observed in the embryos that expressed Igf2 biallelically and were hypermethylated at the H19/Igf2 ICR, albeit not at 2-fold levels, as might be predicted (Fig. 2F). As expected, H19 expression in both mCON and mKO embryos and placentas was monoallelic or undetectable (Fig. 2F, Table S2). Of note, two of the three affected embryos were female (Tables S2, S3). Together, these results indicate that H19/Igf2 ICR hypermethylated oocytes can contribute to live embryos that have abnormal allele-specific expression of Igf2, which is associated with hypermethylation at the ICR.
We next determined if the mCpG and total expression patterns observed at the H19/Igf2 locus were similar at the paternally methylated IG-DMR (Fig. 3A). There was a significant increase in the proportion of E10.5 embryos showing hypermethylation at IG-DMR among mKO as compared with mCON embryos (10/36 versus 1/34, respectively; P=0.007; Fig. 3B). ICR hypermethylation was stochastic, as not every KO embryo was affected and this proportion deviated from the expected 50%. Only one mKO embryo had hypermethylation at both H19/Igf2 and IG-DMR (1/29, embryo ‘A’; Fig. 2C, Fig. 3B), whereas the rest of the affected embryos were hypermethylated at a single locus. This result reflects a lack of correlation between hypermethylation of the two loci in any given mKO animal.
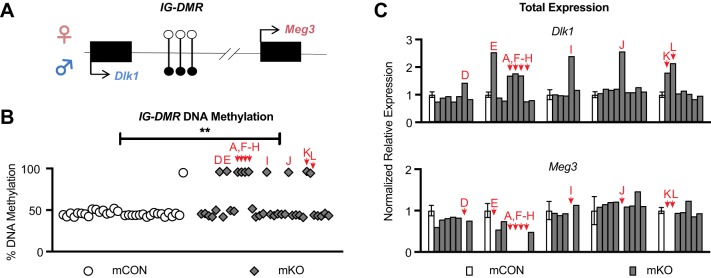
Fig. 3.
Abnormal methylation and total expression at IG-DMR in E10.5 mKO offspring. (A) The mouse IG-DMR locus. The maternal allele and maternal-specific Meg3 expression are represented at the top (pink) and the paternal allele and paternal-specific Dlk1 expression are represented at the bottom (blue). See Fig. 2 legend for details. (B) Percentage DNA methylation at IG-DMR. Embryos are in the same order from left to right as in Fig. 2D,E. mCON n=32 embryos from 4 litters, mKO n=36 embryos from 7 litters. **P<0.01, Fisher's exact test. (C) Total expression from qPCR analysis of Dlk1 and Meg3. All mCON litters and embryos are in the same order as in Fig. 2F. See Fig. 2 legend for details. In the Meg3 plot, expression in embryos A,D-L is undetectable.
Although we could not investigate allele-specific expression in our F1 hybrid model because IG-DMR is on chromosome 12, we were able to measure total expression. Embryos that had abnormal hypermethylation at IG-DMR also had silenced Meg3 expression, as expected given that IG-DMR methylation silences Meg3 expression (Lin et al., 2003). Similar to Igf2 total expression, however, total levels of Dlk1 were variable, ranging from WT levels of expression to 2.5-fold increases in expression compared with controls (Fig. 3C). Of 36 mKO offspring, 12 showed imprinting defects at the H19/Igf2 locus, the IG-DMR locus, or both. Of these 12 affected offspring, nine were females and three were males, indicating a significant sex-biased effect early in development (P=0.033, Table S3) despite the expected Mendelian ratio of male and female embryos. Thus, Tet1 in the maternal germline is also required for proper IG-DMR mCpG levels in offspring and may preferentially affect female offspring.
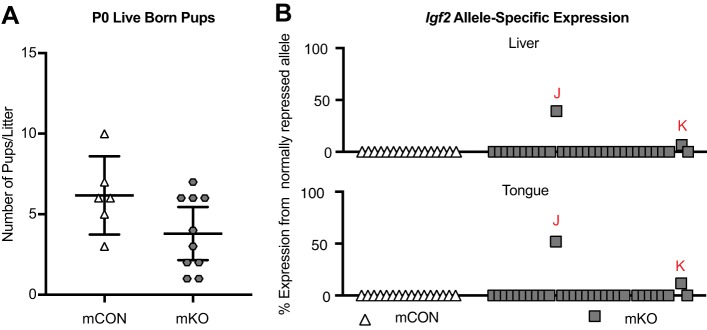
To determine whether the abnormalities observed at the H19/Igf2 ICR during embryogenesis were also present at birth, we isolated tissues from mKO and mCON pups at postnatal day (P) 0 and measured allele-specific expression and mCpG levels. Although the average number of mKO live-born pups was lower than that for mCON pups, this difference was not significant (P=0.067; Fig. 4A). mCON tissues showed no biallelic expression (0/16). However, we observed biallelic expression of Igf2 in some mKO pups (2/31) and this change was consistent between tissues (tongue and liver, Fig. 4B). These affected pups were female (Tables S3, S4). There was no significant difference between the proportion of P0 mKO pups with biallelic Igf2 compared with the proportion of E10.5 mKO embryos with biallelic Igf2 (P=1.00), indicating that these changes are present at the same frequency from mid-gestation through birth.
Fig. 4.
Allelic expression of Igf2 and methylation at the H19/Igf2 ICR is consistent across different tissues in newborn mKO offspring. (A) The number of live pups per litter at P0. mCON n=6 litters, mKO n=10 litters. Line, mean; bars, 95% confidence interval. (B) Biallelic expression of Igf2 in two mKO pups is consistent between tongue and liver. mCON n=16 pups from 3 litters, mKO n=31 pups from 7 litters. Abnormal pups are denoted with red letters and indicate the same pups between graphs. All pups are in the same order from left to right in both the liver and tongue graphs.
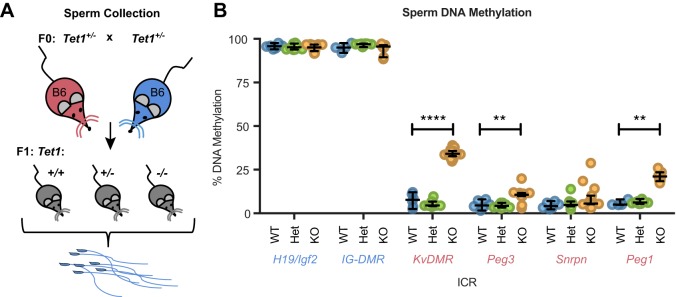
Loss of Tet1 leads to stochastic DNA hypermethylation at ICRs in sperm
Next, we addressed the effects of Tet1 deletion on the male germline. We collected motile sperm from Tet1 WT, Het and KO adult males for mCpG analysis by pyrosequencing (Fig. 5A). WT and Het sperm showed the expected low levels of mCpG at the maternally methylated KvDMR, Peg3, Snrpn and Peg1 (Mest) ICRs. The paternally methylated ICRs H19/Igf2 and IG-DMR had the expected hypermethylation in sperm regardless of the paternal genotype. By contrast, mCpG levels were significantly increased at the KvDMR, Peg1 and Peg3 ICRs in KO samples (for WT versus KO, P<0.0001, P=0.002 and P=0.003, respectively). The Snrpn ICR had neither significant hypermethylation nor significant differences in variance (for WT versus KO, P=0.112 and P=0.91, respectively) (Fig. 5B). The pyrosequencing results were confirmed by bisulfite mutagenesis, followed by cloning and sequencing of the H19/Igf2 and Peg3 ICRs (Fig. S3A,B).
Fig. 5.
Hypermethylation occurs at a subset of ICRs in Tet1−/− sperm. (A) The breeding scheme used to collect mature sperm. (B) Individual dots represent the percentage DNA methylation for sperm collected from one adult mouse. ICR labels are color coded according to which parental allele is normally methylated: blue, the paternal allele; pink, the maternal allele. Line, median; bars, 95% confidence interval. **P<0.01, ****P<0.0001, WT versus KO, two-tailed Mann–Whitney rank-sum test. H19/Igf2, WT n=5, Het n=9, KO n=14; IG-DMR and Peg1, WT n=4, Het n=8, KO n=11; KvDMR and Snrpn, WT n=6, Het n=10, KO n=15; Peg3, WT n=6, Het n=10, KO n=14.
Because the Snrpn ICR was demethylated in the Tet1 KO sperm, we hypothesized that another protein, such as TET2, or a different demethylation pathway could be compensating in the absence of TET1. To test this hypothesis, we examined sperm from Tet1−/−; Tet2−/− double-knockout (DKO) mice and saw no significant difference in DNA methylation level between KO and DKO sperm (Table S5), indicating that TET1 is the primary enzyme responsible for DNA methylation imprint erasure at the KvDMR, Peg1 and Peg3 ICRs but that both TET1 and TET2 are dispensable for the erasure of the Snrpn ICR.
Loss of Tet1 leads to biallelic expression and changes in DNA methylation of imprinted genes in the offspring of male KO mice
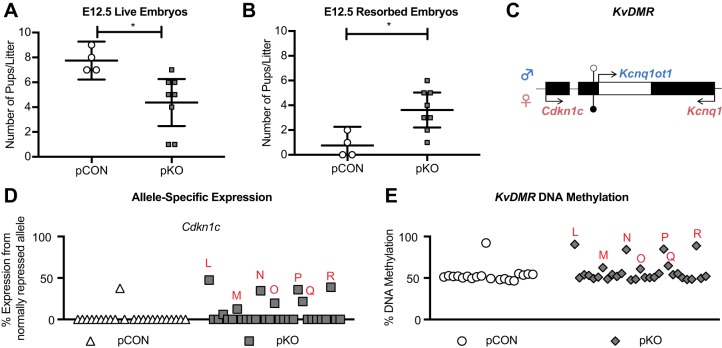
We next asked how hypermethylated sperm affected allele-specific expression of imprinted genes and mCpG levels at ICRs in the offspring of Tet1 KO male mice. Because abnormal methylation at maternally methylated ICRs is thought to be less detrimental to normal development (Kawahara et al., 2007), we hypothesized that changes comparable to maternal Tet1 deficiency would be observed at a slightly later time in gestation in the paternal KO. Therefore, to collect F1 hybrid tissues for analysis, we mated Tet1 Het or KO males with Tet1 WT females on the C7 background to generate paternal control offspring (pCON) or paternal KO offspring (pKO) (Fig. S2C,D). At E12.5, there were significantly fewer live pups (P=0.019) and significantly more resorbed and delayed pups (P=0.011) among the pKO as compared with the pCON offspring (Fig. 6A,B).
Fig. 6.
Biallelic expression of Cdkn1c is prevalent in E12.5 pKO offspring. (A) The number of live embryos per litter at E12.5. (B) The number of resorbed embryos per litter at E12.5. (A,B) pCON n=4 litters, pKO n=5 litters. Line, mean; bars, 95% confidence interval. *P<0.05, unpaired two-tailed t-test. (C) The mouse KvDMR locus. The maternal allele and maternal-specific Cdkn1c and Kcnq1 expression are represented at the top (pink) and the paternal allele and paternal-specific Kcnq1ot1 expression are represented at the bottom (blue). Genes are represented by black boxes, with the exception of Kcnq1ot1, which is represented as a white box within the gene body of Kcnq1. Black lollipops, methylated DNA at the ICR; white lollipops, unmethylated DNA at the ICR. (D) Allele-specific expression of Cdkn1c. pCON n=23 embryos from 3 litters, pKO n=25 embryos from 7 litters. Abnormal embryos are indicated with red letters, which is consistent in D and E. All embryos are in the same order from left to right in D and E. (E) Percentage DNA methylation at KvDMR. pCON n=19 embryos from 3 litters, pKO n=29 embryos from 7 litters.
We then examined changes in allele-specific expression at the KvDMR in E12.5 pCON and pKO embryos (Fig. 6C). pKO embryos were more likely to express Cdkn1c biallelically (7/29) than pCON embryos (1/24) (Fig. 6D). Embryos with biallelic Cdkn1c expression exhibited DNA hypermethylation at the KvDMR (Fig. 6E). Notably, stochastic DNA hypermethylation of the Peg3 ICR was also observed among pKO offspring (Table S6), but no single embryo had both Peg3 and KvDMR hypermethylation (Fig. S4A). Because these loci are at opposite ends of chromosome 7, this result suggests that meiotic recombination might dictate which pups within a litter are affected. The changes in allele-specific expression and DNA methylation were also observed in the corresponding placentas (Fig. S5, Table S6). Unlike what was observed in the maternal offspring, male and female embryos from pKO crosses were equally affected (P=0.665, Tables S3, S6).
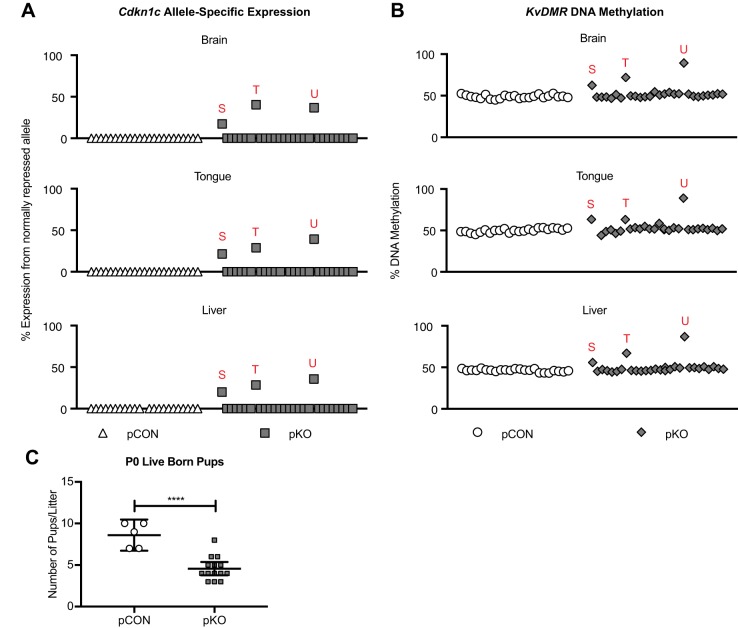
Lastly, to determine if changes in allele-specific expression and mCpG levels at maternally methylated ICRs are detected at birth, we measured imprinted gene expression by RFLP and mCpG levels by pyrosequencing analysis in tissues from pCON and pKO P0 pups. pCON pups exhibited normal monoallelic expression of Cdkn1c, whereas some pKO pups had biallelic Cdkn1c expression (3/29) (Fig. 7A, Table S7). The pups with biallelic expression also had hypermethylation at the KvDMR, which was consistent across three tissue types (Fig. 7B, Table S7). Similar to what was observed at E12.5, litter sizes were significantly smaller for pKO than pCON litters at birth (P<0.0001; Fig. 7C). This suggests that embryonic lethality might contribute to a smaller proportion of affected animals at P0 compared with E12.5. As observed at E12.5, we also noted mutually exclusive hypermethylation of the Peg3 ICR and KvDMR (Fig. S4B). Together, these data show that the sperm hypermethylation in male KO animals was transmitted to a subset of offspring, resulting in defects in imprinted gene expression and ICR methylation.
Fig. 7.
P0 pKO pups show biallelic Cdkn1c expression and methylation defects across tissues. (A) Allele-specific expression of Cdkn1c in P0 brain, tongue and liver. pCON n=23 pups from 4 litters; pKO n=29 pups from 5 litters. Abnormal pups are denoted with unique red letters, which are consistent in A and B. All pups are in the same order from left to right in all graphs in both A and B. (B) Percentage DNA methylation at KvDMR. pCON n=19 pups from 4 litters; pKO n=29 pups from 5 litters. (C) The number of live pups per litter at P0. ****P<0.0001, unpaired two-tailed t-test. pCON n=5 litters; pKO n=13 litters. Line, mean; bars, 95% confidence interval.
DISCUSSION
In the developing germline, DNA methylation at ICRs must be erased so that parent-of-origin-specific imprints can be established according to the sex of the embryo. These sex-specific DNA methylation patterns are necessary for proper imprinted gene expression in the offspring. The importance of this process is highlighted in human imprinting disorders, where loss of imprinted gene expression or changes in DNA methylation at ICRs can lead to disorders such as Beckwith-Wiedemann, Angelman, and Prader-Willi syndromes (Kalish et al., 2014). Although DNA methylation at the global level has been well studied (Guibert et al., 2012; Hajkova et al., 2002; Kagiwada et al., 2013; Seisenberger et al., 2012a; Seki et al., 2005), how imprints are effectively removed to allow imprinted gene expression in subsequent generations remains elusive. Among the recently discovered TET family of proteins (Tahiliani et al., 2009), TET1, the predominantly expressed family member in PGCs, has been implicated in the ICR DNA demethylation process (Dawlaty et al., 2013; Yamaguchi et al., 2013). Until now, the role of TET1 in genomic imprinting was poorly understood, given the paucity of data involving the female germline and lack of allele-specific expression analysis in the offspring. Here, we used a Tet1 null allele to systematically investigate the function of TET1 in DNA demethylation of ICRs in both the maternal and paternal germlines. Furthermore, we determined the effect of Tet1 loss on imprinted gene expression in the offspring of maternal and paternal Tet1 KO mice.
We found variable levels of DNA methylation among Tet1 KO oocyte pools collected from different females at the paternally methylated ICRs H19/Igf2 and IG-DMR. This variability might be attributed to any one of several factors. First, because germ cells are derived from the diploid epiblast, each parental allele has an equal chance of being represented in the gamete. Thus, we expect two distinct populations within a pool of oocytes from a given Tet1 KO female mouse. An allele originally derived from the unmethylated maternal allele will remain unmethylated and thus contribute with no apparent defect to offspring. However, an allele that was originally paternal and methylated may fail to erase in the absence of TET1, in which case these oocytes will be hypermethylated. Therefore, the theoretical maximum DNA methylation level in a bulk pool of oocytes from one mouse will be 50%. However, meiotic defects have been reported in a gene-trap allele of Tet1; these defects were linked to increased levels of oocyte apoptosis during prophase I of meiosis (Yamaguchi et al., 2012). Consistent with this observation, we observe variable levels of hypermethylated oocytes at the paternally methylated ICRs. This hypermethylation includes deviations from the expected 50% levels of DNA methylation, including levels higher than 50%, which can be explained by stochastic oocyte apoptosis. Consistent with variable oocyte apoptosis, we see bimodal distributions of DNA methylation across analyzed oocyte pools (Fig. S1A,B). Second, it is possible that loss of TET1 could affect PGC migration and/or proliferation timing. A number of genes have been reported to affect PGC proliferation and/or migration in a cell-autonomous manner (Saitou and Yamaji, 2012). Lack of Tet1 might directly or indirectly affect the expression of these genes and thus affect the timing or migration patterns of PGCs. If a particular PGC can divide more frequently than another PGC or is delayed in reaching the gonad where proliferation ceases, then these PGCs would lose more DNA methylation, owing to replication in the absence of the DNA methylation machinery, than a PGC that divides more slowly or migrates to the gonad more rapidly. Notably, TET1 binds to the Dazl promoter in ESCs (Williams et al., 2011), a crucial factor in PGC development, survival and differentiation (Haston et al., 2009; Lin and Page, 2005; Ruggiu et al., 1997; Schrans-Stassen et al., 2001). Dazl expression is regulated by promoter methylation, and the promoter is hypermethylated in a PGC-like cell model in which both Tet1 and Tet2 are simultaneously knocked down (Hackett et al., 2013). Lastly, we also see differences in the levels of hypermethylation between H19/Igf2 and IG-DMR (Fig. 1). This further emphasizes that a number of the aforementioned factors may be contributing to the variability at these loci.
We additionally identified ICR hypermethylation alterations in the offspring of female Tet1 KO mice. Notably, the proportion of affected E10.5 mKO embryos mirrors the relative percentage hypermethylation of each ICR in the oocytes. The presence of these abnormalities in both oocytes and E10.5 embryos suggests that abnormalities established in the germline are maintained throughout fertilization and the early epigenetic reprogramming of the preimplantation embryo. Furthermore, offspring with hypermethylation of the H19/Igf2 ICR and/or IG-DMR show close to 100% methylation, indicating that passive loss of methylation or TET2 and TET3 activity is unlikely and/or unable to compensate in the window between gametogenesis and E10.5 at these loci. One mCON animal also exhibited hypermethylation at IG-DMR and this might be because the dam was Het for the Tet1 allele.
In addition to expanding on the role of Tet1 in the regulation of DNA methylation in the female germline, we report for the first time that loss of TET1 leads to biallelic expression of paternally expressed genes when the deletion is inherited from the maternal lineage. At E10.5, we observe completely biallelic expression of Igf2. This level of expression from the normally silent maternal allele corresponds to the level of dysregulation of ICR methylation, which is close to 100% methylated in all of the affected offspring. Biallelic expression of Igf2 is also observed at the newborn (P0) stage, and is consistent across tissues from different germ layers. The proportion of animals with biallelic expression is consistent between the embryonic stage and the newborn stage. These results suggest that alterations at the H19/Igf2 locus are compatible with late gestation development and live birth, consistent with the fact that biallelic Igf2 is detected in a subset of Beckwith-Wiedemann patients (Kalish et al., 2014). It will be interesting to determine if animals with biallelic Igf2 expression develop human imprinting disorder-like phenotypes postnatally. Such a result would indicate that misregulation of Tet1 may be a cause of idiopathic cases of human imprinting disorders.
To date, previous studies regarding Tet1 and genomic imprinting have relied on measuring total expression of imprinted genes. Our data demonstrate that hypermethylation of the H19/Igf2 ICR and IG-DMR is associated with silencing of the maternally expressed genes H19 and Meg3, respectively. However, total levels of expression of the corresponding paternally expressed genes, Igf2 and Dlk1, do not perfectly predict biallelic expression. This is consistent with other reports describing inconsistencies between total and allele-specific expression (Eckersley-Maslin and Spector, 2014). Moreover, our results contrast with Yamaguchi et al. (2013), where decreases in total expression of both maternally and paternally expressed genes at IG-DMR were observed in E19.5 placentas of resorbed embryos. These discrepancies could potentially be due to differences in the quality of the input material from resorption sites.
In this study, we demonstrate that despite equal sex ratios of Tet1 mKO offspring at each developmental stage examined, imprinting phenotypes are biased towards females at the H19/Igf2 ICR and IG-DMR. This result is unexpected, as sex-biased imprinting phenotypes have yet to be described by other groups working with mouse models of Tet1 (Dawlaty et al., 2013; Yamaguchi et al., 2013; Zhang et al., 2016). The nature of this sex-biased effect is unclear. Although there have been previous reports of sex-specific differences in total expression of certain imprinted genes such as H19 and Igf2 in the E14.5 brain, these changes were not observed before sex determination at E10.5 (Faisal et al., 2014). As our bias is observed before sex differentiation, it is likely that differences in sex steroids are not playing a major role in our animals. One possibility is that both male and female mKO offspring have hypermethylation at the H19/Igf2 ICR and IG-DMR, but this abnormal methylation is not maintained in males. It is also possible that imprinting abnormalities at the H19/Igf2 ICR and IG-DMR, as well as defects at another locus (or multiple loci), are both embryonic lethal but affect each sex independently. In this circumstance, we envision that dysregulated imprinting at IG-DMR and the H19/Igf2 ICR is more detrimental in males, but methylation or expression defects elsewhere caused by loss of Tet1 may be more detrimental in females. This scenario is compatible with the early lethality that we observed at E10.5 and is also consistent with the equal sex ratios we observed at this developmental stage. Further studies are warranted to understand this mechanism.
In addition to our female germline and offspring analysis, we also investigated the function of TET1 in the male germline. We demonstrate that lack of TET1 leads to hypermethylation of the maternally methylated ICRs, KvDMR, Peg1, and Peg3. The hypermethylation of KvDMR is consistent with reduced-representation bisulfite sequencing data from Yamaguchi et al. (2013). Additionally, Dawlaty et al. (2013) reported hypermethylation of Peg1, Peg3, and the KvDMR by methylated DNA immunoprecipitation and sequencing analysis. We additionally detected no significant difference in hypermethylation between Tet1 KO and combined Tet1/Tet2 DKO sperm (Table S5) providing support to the role of TET1 as the primary isoform responsible for imprint erasure.
Whereas three of the ICRs investigated in our study demonstrated dependency on TET1 for DNA methylation erasure in sperm, Snrpn was unique in that it was demethylated regardless of the presence of TET1. Both our study and that of Dawlaty et al. (2013) find that the Snrpn ICR is not hypermethylated in the absence of TET1 or with the combined deletion of Tet1 and Tet2. This finding is also consistent with experiments using an aorta-gonad-mesonephros organ culture that recapitulates endogenous PGC ICR demethylation in vitro. Using this system, the investigators inhibited PGC proliferation through the use of a PI3K inhibitor and found that Snrpn was unable to undergo demethylation, whereas the H19/Igf2 ICR did lose DNA methylation (Calvopina et al., 2015). Together, these results indicate that the Snrpn ICR relies on passive demethylation via replication. Although the mechanism underlying TET1-independent demethylation is unknown, the high density of Alu elements belonging to the SINE family of retroelements surrounding the Snrpn ICR might be relevant (Huq et al., 1997). SINEs, unlike other repetitive elements, are not resistant to DNA demethylation in the germline (Seisenberger et al., 2012b) and thus might influence how Snrpn is demethylated. Overall, these results show that DNA demethylation of some maternally methylated ICRs requires TET1 whereas others do not.
Consistent with DNA methylation results from Tet1 KO sperm, Tet1 pKO offspring also showed hypermethylation of the Peg3 ICR and KvDMR, whereas no changes in DNA methylation were observed at the Snrpn ICR at E12.5 and P0. Hypermethylation at KvDMR was coincident with biallelic expression of Cdkn1c. Unlike the consistent levels of biallelic Igf2 expression observed in offspring of mKO animals, the abnormal maternal expression of Cdkn1c in pKO offspring was variable among affected individuals at both time points investigated. For all developmental stages tested, the variable level of biallelic expression closely mimicked the levels of DNA hypermethylation, and loss of imprinting and DNA hypermethylation were consistent across tissues. Given these findings, the variability in loss of imprinted expression suggests that, at the KvDMR locus, activity of the other TET family members and/or passive dilution may be able to partially compensate for the absence of TET1 between fertilization and the blastocyst stage when the extraembryonic tissues are specified. Moreover, this biallelic expression was more prevalent in the embryonic than newborn stage. Given the smaller litter size of Tet1 pKO offspring compared with pCON offspring at E12.5 and at birth, it is possible that embryonic loss could reduce the number of affected offspring at the later developmental stage.
Similar to our maternal Tet1 KO data, the molecular phenotype of pKO offspring was highly variable. In both male and female offspring, variability in the levels of imprinted genes at the embryonic stages examined could not be explained by changes in mRNA levels of Dnmt1, Dnmt3a, Dnmt3b, Dnmt3l, Uhrf1, Tet2 or Tet3 (data not shown). One potential source of stochasticity could relate to the unique properties of individual imprinted loci. ICRs can differ in size, genomic and chromatin contexts, sequence composition, number and identity of trans-factor binding sites, as well as differences in mechanisms of genomic imprinting. For example, the KvDMR is large, highly enriched in LINE elements, and very CpG rich (Engemann et al., 2000). These inherent differences might underlie some of the stochastic phenotypes observed in our study. Additionally, stochasticity of the imprinting phenotypes might be due to meiotic recombination. Even though some ICRs are on the same parental chromosome, hypermethylation of both loci is not correlated in a given pup (Fig. S4). This observation is incompatible with a model of random segregation of parental alleles. For example, we observe either normal methylation at KvDMR and the Peg3 ICR, or hypermethylation of one locus only, but never hypermethylation at both loci in the same animal. These phenotypes represent three of the four possible recombination states of meiosis on chromosome 7, as mice tend to have one or two recombination events per chromosome (Koehler et al., 2002). Given the proportions of animals we observe with hypermethylation of one of the two loci, the probability of having both hypermethylated loci in one animal is very low. However, we cannot exclude the possibility that hypermethylation of KvDMR and the Peg3 ICR produces a synthetically lethal effect. Lastly, TET1 is a large protein that is known to interact with epigenetic regulators such as SIN3A, HDAC1, HDAC2 and OGT (Vella et al., 2013; Williams et al., 2011), pluripotency factors like NANOG (Costa et al., 2013), and has been suggested to play roles in recruiting the EZH2 subunit of Polycomb Repressive Complex 2 to distinct sites in the genome (Wu et al., 2011). Loss of these interactions may lead to inadequate or inappropriate targeting of these complexes, contributing to further dysregulation of the epigenome in Tet1 KO animals.
In summary, we present a detailed analysis of the previously unexplored maternal germline and describe, for the first time, allele-specific expression abnormalities in Tet1 KO offspring. Together with sperm and paternal KO studies, we determine that Tet1 is an important regulator of genomic imprinting in both the maternal and paternal lineages.
MATERIALS AND METHODS
Animals
Tet1 mice (Dawlaty et al., 2011) were purchased from The Jackson Laboratory (017358; B6;129S4-Tet1tm1.1Jae/J). Mice were backcrossed for at least four generations to C57BL/6J (The Jackson Laboratory, 000664) before analysis, with the exception of sperm samples. Tet1/Tet2 double knockouts have the same allele as the Tet1 single knockouts and the Tet2 allele was originally generated by Li et al. (2011). Tet1 knockout mice were generated by heterozygous mating or by mating Tet1 heterozygotes on an Oct4-GFP heterozygous background (Lengner et al., 2007) (B6;129S4-Pou5f1tm2Jae/J; The Jackson Laboratory, 008214). Tet1 animals were genotyped by lysing ear punches in fast lysis buffer A (25 mM NaOH, 0.2 mM EDTA, pH 8.0) at 95°C for 1 h and then adding an equal volume of fast lysis buffer B (40 mM Tris HCl, pH 8). 2 µl genomic DNA was used for genotyping PCR reactions. CAST7 mice are maintained in our institutional mouse colony. Timed mating was determined by checking for a vaginal sperm plug; E0.5 was taken to be 12.00 h (noon) on the day the plug was observed. Embryos were also visually staged upon dissection. All studies were performed in accordance with procedures approved by the University of Pennsylvania's Institutional Animal Care and Use Committee.
Germ cell collection
Oocytes
Pools of 20-100 germinal vesicle stage oocytes were collected from the ovaries of one 26- to 28-day-old mouse. Cumulus cells were removed from oocytes by mouth pipetting and transferring into clean drops of M2 medium (Sigma-Aldrich, M7167) supplemented with a final concentration of 2.5 μM milrinone. When necessary, oocytes were briefly incubated in a drop of M2 medium containing a final concentration of 0.3 mg/ml hyaluronidase to remove cumulus cells. Oocytes were snap frozen in liquid nitrogen.
Sperm
Adult male mice (>8 weeks of age) were mated with a female for at least two days and then isolated for at least two days. After sacrifice, the epididymis was dissected. Epididymal sperm was collected on a needle and then incubated in room temperature PBS. Motile sperm were collected by removing the supernatant. Sperm were counted on a hemocytometer and then pelleted [10 min at 12,000 rpm (13,523 g)]. The PBS was removed and the sperm pellet was snap frozen in liquid nitrogen.
Tissue homogenization
Embryonic tissue samples were homogenized in tail lysis buffer (50 mM Tris-HCl, 0.5% SDS, 100 mM EDTA, pH 8) with a needle and syringe. P0 brain and tongue were homogenized in tail lysis buffer with a polytron (Kinematica, PT 10-35 GT). P0 livers were divided upon dissection and one half was homogenized with a needle and syringe directly in TRIzol reagent (Thermo Fisher Scientific, 15596026), and the other half was homogenized in tail lysis buffer using the polytron and subsequently processed for DNA.
RNA and cDNA conversion
Tissue lysate was added to TRIzol reagent and mixed thoroughly. TRIzol extraction was performed as specified in the manufacturer's protocol. RNA quantity was determined by NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific) and RNA quality was assessed by running 500 ng on an agarose gel and confirming intact rRNA bands at the expected intensity ratio. Only samples with intact RNA were used for further analysis. 500-1000 ng total RNA was treated with 1.5 µl RQ1 RNase-free DNase (Promega, M6106) for 30 min at 37°C followed by addition of 1.5 µl Stop Solution (Promega) and incubated for 10 min at 65°C. Two-thirds of the treated RNA was placed in a 20 µl cDNA conversion reaction using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific, 18080093) and random primers (Sigma-Aldrich, 11034731001). The remaining third of the treated RNA was used in a negative (no reverse transcriptase) control reaction.
qPCR analysis
A cDNA dilution series was used to make a standard curve from which qPCR primer amplification efficiencies were determined using Power SYBR Green Master Mix (ThermoFisher Scientific, 4368577). For primers and cycling conditions, see Tables S8, S9. cDNA was diluted to 5 ng/well for total expression analysis. Samples were run in triplicate on a 7900 Fast Real-Time PCR System (Applied Biosystems) with no-template controls for each gene. Melting curve analysis was performed to ensure specific amplification. For quality control, any individual Ct value within a triplicate that was >0.5 Ct from the other two was removed. Data were normalized to the geometric mean of three housekeeping genes: Rplp0, Nono and Rpl13a. The control animals were averaged and graphed with the individual Tet1 mutant animals per qPCR plate.
Allele-specific expression analysis
cDNA was diluted to 10 ng/µl and 1 µl was used per PCR reaction. For primers and PCR conditions, see Tables S8, S9. Linear range of amplification was determined for each tissue and developmental stage. Each assay used a different restriction enzyme: H19, Cac8I (NEB, R0579S), 37°C for 3 h, heat inactivated at 65°C for 20 min; Igf2, MluCI (NEB, R0538S), 37°C for 3 h, no heat inactivation; Peg3, MnlI (NEB, R0163S), 37°C for 3 h, heat inactivated at 65°C for 20 min; Cdkn1c, TaqαI (NEB, R0149S), 65°C for 1 h, heat inactivated for 20 min at 80°C; Kcnq1ot1, StuI (NEB, R0187S), 37°C for 3 h, no heat inactivation. Digests were performed using the supplied buffer and 3-8 µl of the PCR reaction in a 20 µl reaction volume. Digests were run on 7% or 12% polyacrylamide gels. Band densitometry was analyzed using ImageJ software. Complete digestion was assessed by running pure parental strain cDNA PCR product digests as controls. Snrpn allele-specific analysis was performed using the LightCycler real-time PCR system (Roche) as described by Mann et al. (2004) with the following modifications: illustra PuReTaq Ready-To-Go PCR Beads were used (GE Healthcare, 27-9559-01) and hybridization probes were purchased from IDT.
DNA extraction
Tissue lysate was incubated overnight with proteinase K (Sigma-Aldrich, P2308) at a final specific activity of 180 U/ml at 37°C. Sperm pellets were resuspended in sperm lysis buffer (20 mM Tris-HCl, 200 mM NaCl, 20 mM EDTA, 4% SDS, pH 8) with the addition of 5 µl β-mercaptoethanol and proteinase K at a final specific activity of 180 U/ml at 55°C overnight. DNA was then phenol-chloroform extracted, followed by ethanol precipitation and resuspended in dH2O or TE buffer (10 mM Tris-HCl, 0.5 mM EDTA, pH 8).
Bisulfite treatment
1 µg sperm or mouse tissue DNA was bisulfite treated using the EpiTect Bisulfite Kit (Qiagen, 59104) and eluted in 20 µl of the supplied EB buffer. Oocyte pools were directly lysed using the LyseAll Lysis Kit (part of the EpiTect Plus Bisulfite Kit) and bisulfite converted using the EpiTect Plus Bisulfite Kit (Qiagen, 59124). Oocyte bisulfite-treated DNA was resuspended in 13 µl of the supplied EB buffer.
Pyrosequencing
Pyrosequencing PCRs and sequencing reactions were carried out as described by de Waal et al. (2014). For primers and PCR conditions, see Tables S8, S9. Briefly, 1-2 µl bisulfite-treated DNA was used to set up pyrosequencing PCRs using the PyroMark PCR Kit (Qiagen, 978703) using locus-specific primers. 2-4 µl of the PCR was used in each pyrosequencing reaction and sequenced on a PyroMark Q96 MD machine (Qiagen). Pyrosequencing peaks were manually inspected for sequencing errors and for matching to the reference expected peaks. CpGs that did not pass these quality control criteria were excluded from the final analysis. Oocyte methylation data were further subjected to the following criteria to ensure there was no somatic cell contamination: for any given sample, two maternally methylated ICRs had to have an average percentage methylation ≥90% and must have amplified at least H19/Igf2 or IG-DMR. Samples that did not meet these criteria were excluded from the final analysis (Table S1).
Bisulfite, clone, and sequencing analysis
Nested PCR reactions were performed using bisulfite-treated DNA (Tables S8, S9) as described (Market-Velker et al., 2010) with the following adaptions: illustra PuReTaq Ready-To-Go PCR Beads were used. 1 µl bisulfite-treated DNA was used for the first round of PCR, then 4 µl of the first-round PCR seeded the second-round PCR reaction. Two independent PCR reactions were set up for both the first and second round. Second-round PCR products were cloned using the StrataClone PCR Cloning Kit (Agilent). At least 20 colonies per plate were picked and analyzed for the insert using EcoRI digestion. Clones containing an insert of the correct size were submitted for Sanger sequencing analysis at the University of Pennsylvania DNA Sequencing Facility. Sequences were analyzed using the QUMA website (Kumaki et al., 2008; quma.cdb.riken.jp/).
Statistics
Analyses were completed using GraphPad Prism software with the following exceptions: variance {σ2=[Σ(xi−x¯)2]/[n−1]} was calculated in Microsoft Excel using the VAR.S function, and the Fligner-Killeen test of homogeneity of variances was calculated in R (www.r-project.org/). Chi-square and Fisher's exact test were calculated on the GraphPad website (https://graphpad.com/quickcalcs/chisquared1.cfm, https://graphpad.com/quickcalcs/contingency1.cfm). Sample size was determined by conducting smaller pilot studies. The investigators were not blinded to the identity of the samples during analysis.
Supplementary Material
Acknowledgements
We thank Dr Guo-Liang Xu and Dr Mingjiang Xu for the Tet1/Tet2 DKO sperm samples; the members of the M.S.B. lab for thoughtful discussions and review of the manuscript, especially Frances Xin, Blake Caldwell and Aimee Juan; Blake Caldwell for experimental assistance; and Anthony Angueira for statistical advice. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.M.S., L.K.A., M.S.B.; Formal analysis: J.M.S., L.K.A.; Investigation: J.M.S., L.K.A.; Resources: M.S.B.; Writing - original draft: J.M.S., M.S.B.; Writing - review & editing: J.M.S., L.K.A., M.S.B.; Visualization: J.M.S.; Supervision: M.S.B.; Funding acquisition: M.S.B.
Funding
This work was supported by the National Institutes of Health [R37GM051279 to M.S.B., T32GM008216 to L.K.A and J.M.S., F31GM119271 to J.M.S.]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.160622.supplemental
References
- Calvopina J. H., Cook H., Vincent J. J., Nee K. and Clark A. T. (2015). The aorta-gonad-mesonephros organ culture recapitulates 5hmC reorganization and replication-dependent and independent loss of DNA methylation in the germline. 24, 1536-1545. 10.1089/scd.2014.0410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y., Ding J., Theunissen T. W., Faiola F., Hore T. A., Shliaha P. V., Fidalgo M., Saunders A., Lawrence M., Dietmann S. et al. (2013). NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. 495, 370-374. 10.1038/nature11925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty M. M., Ganz K., Powell B. E., Hu Y.-C., Markoulaki S., Cheng A. W., Gao Q., Kim J., Choi S.-W., Page D. C. et al. (2011). Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. 9, 166-175. 10.1016/j.stem.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty M. M., Breiling A., Le T., Raddatz G., Barrasa M. I., Cheng A. W., Gao Q., Powell B. E., Li Z., Xu M. et al. (2013). Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. 24, 310-323. 10.1016/j.devcel.2012.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waal E., Mak W., Calhoun S., Stein P., Ord T., Krapp C., Coutifaris C., Schultz R. M. and Bartolomei M. S. (2014). In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies1. 90, 22-22. 10.1095/biolreprod.113.114785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin M. A. and Spector D. L. (2014). Random monoallelic expression: regulating gene expression one allele at a time. 30, 237-244. 10.1016/j.tig.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engemann S., Strödicke M., Paulsen M., Franck O., Reinhardt R., Lane N., Reik W. and Walter J. (2000). Sequence and functional comparison in the Beckwith-Wiedemann region: implications for a novel imprinting centre and extended imprinting. 9, 2691-2706. 10.1093/hmg/9.18.2691 [DOI] [PubMed] [Google Scholar]
- Faisal M., Kim H. and Kim J. (2014). Sexual differences of imprinted genes’ expression levels. 533, 434-438. 10.1016/j.gene.2013.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. D., Allis C. D. and Bernstein E. (2007). Epigenetics: a landscape takes shape. 128, 635-638. 10.1016/j.cell.2007.02.006 [DOI] [PubMed] [Google Scholar]
- Guibert S., Forné T. and Weber M. (2012). Global profiling of DNA methylation erasure in mouse primordial germ cells. 22, 633-641. 10.1101/gr.130997.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J. A., Sengupta R., Zylicz J. J., Murakami K., Lee C., Down T. A. and Surani M. A. (2013). Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. 339, 448-452. 10.1126/science.1229277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W., Walter J. and Surani M. A. (2002). Epigenetic reprogramming in mouse primordial germ cells. 117, 15-23. 10.1016/S0925-4773(02)00181-8 [DOI] [PubMed] [Google Scholar]
- Hajkova P., Jeffries S. J., Lee C., Miller N., Jackson S. P. and Surani M. A. (2010). Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. 329, 78-82. 10.1126/science.1187945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haston K. M., Tung J. Y. and Reijo Pera R. A. (2009). Dazl functions in maintenance of pluripotency and genetic and epigenetic programs of differentiation in mouse primordial germ cells in vivo and in vitro. 4, e5654 10.1371/journal.pone.0005654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill P. W. S., Amouroux R. and Hajkova P. (2014). DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: An emerging complex story. 104, 324-333. 10.1016/j.ygeno.2014.08.012 [DOI] [PubMed] [Google Scholar]
- Huq A. H. M. M., Sutcliffe J. S., Nakao M., Shen Y., Gibbs R. A. and Beaudet A. L. (1997). Sequencing and functional analysis of the SNRPN promoter: in vitro methylation abolishes promoter activity. 7, 642-648. 10.1101/gr.7.6.642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Shen L., Dai Q., Wu S. C., Collins L. B., Swenberg J. A., He C. and Zhang Y. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. 333, 1300-1303. 10.1126/science.1210597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiwada S., Kurimoto K., Hirota T., Yamaji M. and Saitou M. (2013). Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. 32, 340-353. 10.1038/emboj.2012.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish J. M., Jiang C. and Bartolomei M. S. (2014). Epigenetics and imprinting in human disease. 58, 291-298. 10.1387/ijdb.140077mb [DOI] [PubMed] [Google Scholar]
- Kawahara M., Wu Q., Takahashi N., Morita S., Yamada K., Ito M., Ferguson-Smith A. C. and Kono T. (2007). High-frequency generation of viable mice from engineered bi-maternal embryos. 25, 1045-1050. 10.1038/nbt1331 [DOI] [PubMed] [Google Scholar]
- Klose R. J. and Bird A. P. (2006). Genomic DNA methylation: the mark and its mediators. 31, 89-97. 10.1016/j.tibs.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Koehler K. E., Cherry J. P., Lynn A., Hunt P. A. and Hassold T. J. (2002). Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. 162, 297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaki Y., Oda M. and Okano M. (2008). QUMA: quantification tool for methylation analysis. 36, W170-W175. 10.1093/nar/gkn294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K., Yabuta Y., Ohinata Y., Shigeta M., Yamanaka K. and Saitou M. (2008). Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. 22, 1617-1635. 10.1101/gad.1649908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch H. G., Tang W. W. C. and Surani M. A. (2013). Primordial germ-cell development and epigenetic reprogramming in mammals. 104, 149-187. 10.1016/B978-0-12-416027-9.00005-X [DOI] [PubMed] [Google Scholar]
- Lengner C. J., Camargo F. D., Hochedlinger K., Welstead G. G., Zaidi S., Gokhale S., Scholer H. R., Tomilin A. and Jaenisch R. (2007). Oct4 expression is not required for mouse somatic stem cell self-renewal. 1, 403-415. 10.1016/j.stem.2007.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Bestor T. H. and Jaenisch R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. 69, 915-926. 10.1016/0092-8674(92)90611-F [DOI] [PubMed] [Google Scholar]
- Li Z., Cai X., Cai C.-L., Wang J., Zhang W., Petersen B. E., Yang F.-C. and Xu M. (2011). Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. 118, 4509-4518. 10.1182/blood-2010-12-325241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. and Page D. C. (2005). Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. 288, 309-316. 10.1016/j.ydbio.2005.06.032 [DOI] [PubMed] [Google Scholar]
- Lin S.-P., Youngson N., Takada S., Seitz H., Reik W., Paulsen M., Cavaille J. and Ferguson-Smith A. C. (2003). Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. 35, 97-102. 10.1038/ng1233 [DOI] [PubMed] [Google Scholar]
- Mann M. R. W., Lee S. S., Doherty A. S., Verona R. I., Nolen L. D., Schultz R. M. and Bartolomei M. S. (2004). Selective loss of imprinting in the placenta following preimplantation development in culture. 131, 3727-3735. 10.1242/dev.01241 [DOI] [PubMed] [Google Scholar]
- Market-Velker B. A., Fernandes A. D. and Mann M. R. W. (2010). Side-by-side comparison of five commercial media systems in a mouse model: suboptimal in vitro culture interferes with imprint maintenance. 83, 938-950. 10.1095/biolreprod.110.085480 [DOI] [PubMed] [Google Scholar]
- Okano M., Bell D. W., Haber D. A. and Li E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. 99, 247-257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- Plasschaert R. N. and Bartolomei M. S. (2014). Genomic imprinting in development, growth, behavior and stem cells. 141, 1805-1813. 10.1242/dev.101428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiu M., Speed R., Taggart M., McKay S. J., Kilanowski F., Saunders P., Dorin J. and Cooke H. J. (1997). The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. 389, 73-77. 10.1038/37987 [DOI] [PubMed] [Google Scholar]
- Saitou M. and Yamaji M. (2012). Primordial germ cells in mice. 4, a008375 10.1101/cshperspect.a008375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrans-Stassen B. H. G. J., Saunders P. T. K., Cooke H. J. and de Rooij D. G. (2001). Nature of the spermatogenic arrest in Dazl−/− mice. 65, 771-776. 10.1095/biolreprod65.3.771 [DOI] [PubMed] [Google Scholar]
- Seisenberger S., Andrews S., Krueger F., Arand J., Walter J., Santos F., Popp C., Thienpont B., Dean W. and Reik W. (2012a). The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. 48, 849-862. 10.1016/j.molcel.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S., Peat J. R., Hore T. A., Santos F., Dean W. and Reik W. (2012b). Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. 368, 20110330-20110330. 10.1098/rstb.2011.0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y., Hayashi K., Itoh K., Mizugaki M., Saitou M. and Matsui Y. (2005). Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. 278, 440-458. 10.1016/j.ydbio.2004.11.025 [DOI] [PubMed] [Google Scholar]
- Smith Z. D. and Meissner A. (2013). DNA methylation: roles in mammalian development. 14, 204-220. 10.1038/nrg3354 [DOI] [PubMed] [Google Scholar]
- Smith Z. D., Chan M. M., Mikkelsen T. S., Gu H., Gnirke A., Regev A. and Meissner A. (2012). A unique regulatory phase of DNA methylation in the early mammalian embryo. 484, 339-344. 10.1038/nature10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart K. R., Veselovska L. and Kelsey G. (2016). Establishment and functions of DNA methylation in the germline. 8, 1399-1413. 10.2217/epi-2016-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L. et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. 324, 930-935. 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella P., Scelfo A., Jammula S. G., Chiacchiera F., Williams K., Cuomo A., Roberto A., Christensen J., Bonaldi T., Helin K. et al. (2013). Tet proteins connect the o-linked N-acetylglucosamine transferase ogt to chromatin in embryonic stem cells. 49, 645-656. 10.1016/j.molcel.2012.12.019 [DOI] [PubMed] [Google Scholar]
- Williams K., Christensen J., Pedersen M. T., Johansen J. V., Cloos P. A. C., Rappsilber J. and Helin K. (2011). TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. 473, 343-348. 10.1038/nature10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., D'Alessio A. C., Ito S., Xia K., Wang Z., Cui K., Zhao K., Eve Sun Y. and Zhang Y. (2011). Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. 473, 389-393. 10.1038/nature09934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Hong K., Liu R., Shen L., Inoue A., Diep D., Zhang K. and Zhang Y. (2012). Tet1 controls meiosis by regulating meiotic gene expression. 492, 443-447. 10.1038/nature11709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Shen L., Liu Y., Sendler D. and Zhang Y. (2013). Role of Tet1 in erasure of genomic imprinting. 504, 460-464. 10.1038/nature12805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Xia W., Wang Q., Towers A. J., Chen J., Gao R., Zhang Y., Yen C.-A., Lee A. Y., Li Y. et al. (2016). Isoform switch of TET1 regulates DNA demethylation and mouse development. 64, 1062-1073. 10.1016/j.molcel.2016.10.030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.