Abstract
Microbes often live in dense, dynamic, multi-species communities whose architecture and function are intimately intertwined. Imaging these complex, three-dimensional ensembles presents considerable technical challenges, however. In this review, I describe light sheet fluorescence microscopy, a technique that enables rapid acquisition of three-dimensional images over large fields of view and over long durations, and I highlight recent applications of this method to microbial systems that include artificial closed ecosystems, bacterial biofilms, and gut microbiota. I comment also on the history of light sheet imaging and the many variants of the method. Light sheet techniques have tremendous potential for illuminating the workings of microbial communities, a potential that is just beginning to be realized.
Graphical Abstract
Main Text
Motivations
Natural microbial communities are spatially complex and temporally dynamic. Spatial organization can be guided by the external environment, for example the nutrient landscape of the gastrointestinal tract or the soil types surrounding plant roots, or by processes driven by the microbes themselves, for example contact-dependent killing [1] or diffusion of secreted metabolites [2]. Temporal variations in species abundance can similarly be driven by changes in external conditions or by microbial processes such as reproduction, migration, and metabolite exchange, giving rise to patterns as simple as monotonic growth and as remarkable as oscillations [3,4]. Understanding microbial communities requires understanding their spatial architecture and its development over time. Conversely, imaging these spatiotemporal dynamics can yield insights into how communities work.
For simple systems, such as bacterial monolayers on surfaces, imaging is straightforward. Many beautiful studies have revealed aspects of inter-bacterial killing [5], stochastic gene expression [6], chemotaxis [7], pilus dynamics and horizontal gene transfer [8,9], antibiotic resistance [10], and more with single-cell resolution. The use of fluorescent probes, especially genetically encoded fluorescent proteins, enables specific labelling of species or molecular structures, which is especially useful in the dense multicellular environments described below.
Most microbial communities of interest, however, are more challenging to image. The most obvious reason is dimensionality. Dense biofilms, gastrointestinal flora, soil microbes, and many other microbial systems are intrinsically three-dimensional in their organization, and so require methods beyond simple widefield imaging. In addition, one often needs large fields of view, fast acquisition speeds, and long imaging durations. Visualizing a microbial community, for example, may require imaging volumes hundreds of microns in extent, with single bacterium (~ 1 micron) resolution. Examining the effects of potentially sudden environmental changes or perturbations can require the rapid capture of 3D snapshots. Observing longer-term population dynamics, or characterizing growth kinetics, can require imaging for durations spanning multiple bacterial generations without appreciable photodamage [11]. These constraints are very difficult to satisfy with standard techniques.
Light Sheet Fluorescence Microscopy
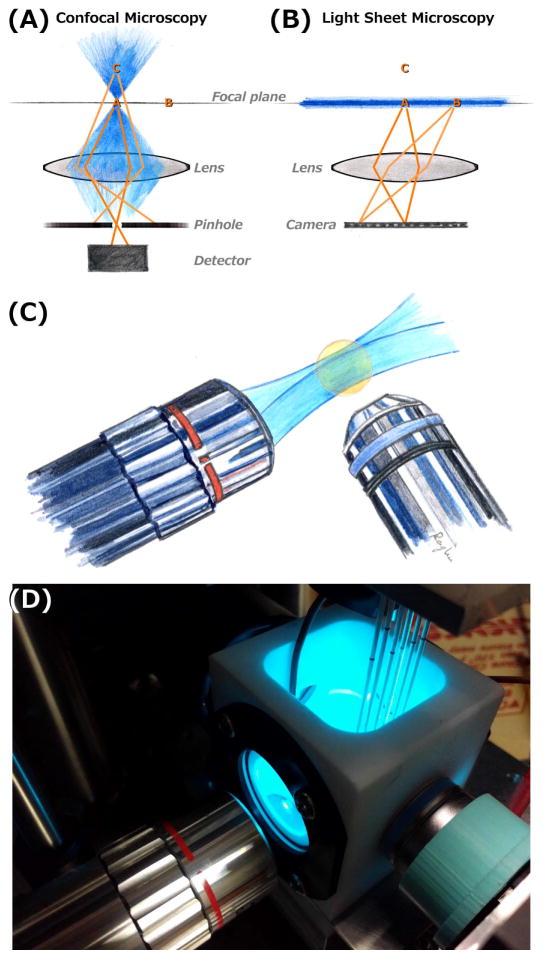
The standard approach to three-dimensional fluorescence microscopy, confocal microscopy, makes use of a pinhole conjugate to the focal point along the axis of an objective lens to discard light from the bulk of a fluorescent specimen (Fig. 1A). While powerful and versatile, it suffers from intrinsic limitations with respect to speed and photodamage. As illustrated, at any instant information is collected from a single point. To construct a three-dimensional image, therefore, this point must be scanned through all three spatial dimensions. This can be parallelized with multiple pinholes, but nonetheless creates a limitation on overall speed. More importantly for many biological applications, a large volume of the specimen is illuminated at any instant, and therefore subject to photobleaching and photodamage, to obtain information about a very small volume (the focal point). As this point is scanned to build up a three-dimensional image, the sample as a whole is redundantly, repeatedly illuminated. In other words, the technique is inefficient in its use of light [12,13].
Figure 1.
(A,B) Schematic illustrations of confocal (A) and light sheet fluorescence (B) microscopies. Excitation light is shown in blue, focused to a point in confocal imaging and shaped into a sheet in light sheet imaging. In each diagram, points A and B lie in the objective lens focal plane, and C is outside the plane. In confocal microscopy, fluorophores outside the focal point are excited, but their emission light is blocked by a confocal pinhole. At any instant, information is collected from only a single point (point A), and three-dimensional images are formed by scanning across all three dimensions. In light sheet microscopy, only fluorophores in the focal plane are excited, and their emission is mapped onto a camera. This provides an image of the entire plane at any instant, and enables three-dimensional imaging by scanning in the one perpendicular direction. (C) Schematic illustration of a typical lens geometry for light sheet fluorescence microscopy. The excitation sheet (blue) is provided by one lens, and emission from its intersection with a specimen (orange) is detected by a perpendicular lens. (D) A photograph of one of the home-built light sheet fluorescence microscopes in the author’s laboratory. The excitation and detection lenses are oriented as in (C). Specimens, in this case larval zebrafish, are mounted in agar gels extruded from glass capillaries, held from above by a computer-controlled stage. The specimens, and the end of the detection lens, are immersed in a temperature-controlled bath.
Light sheet fluorescence microscopy provides an alternative approach to three-dimensional fluorescence imaging that surmounts these limitations. Here, fluorescence excitation is shaped into a thin sheet, emission from which is imaged onto a camera by a perpendicular lens (Fig. 1B). In essence, this is simple widefield imaging, but with the thin sheet providing optical sectioning to illuminate a single plane. Constructing a three-dimensional image only requires scanning in the one perpendicular dimension. Notably, the illuminated volume is roughly equal to the detected volume, minimizing photodamage [12,13]. (The correspondence between the illuminated and imaged volume is not exact due to diffraction of the illumination beam into a bowtie shape, discussed below, but nonetheless the photon efficiency is far greater than is the case for confocal microscopy.) The lateral resolution of the technique (i.e. parallel to the sheet) is simply diffraction limited, and is roughly half the wavelength of light, or a few hundred nanometers in practice. The axial resolution depends on the sheet thickness (and the numerical aperture of the detecting lens), which points to one of the trade-offs inherent in light sheet imaging, the relationship between sheet thickness and field of view. The light sheet is not actually a sheet, but rather a bowtie in cross-section (Figure 1C) due to fundamental aspects of diffraction. The lateral extent of the sheet (the “Rayleigh length”) is proportional to the square of the minimum thickness; a sheet of green light (500 nm wavelength) that is 300 nm thick at its finest point would expand to be 600 nm thick 250 nm away, while a 3 micron thick sheet would have a Rayleigh length 100 times as long. There are clever ways of surmounting this constraint and improving the axial resolution of light sheet microscopy that I will briefly comment on below; in most applications, however, the axial resolution is a few times worse than that of confocal microscopy. For imaging microbial communities, this tradeoff of axial resolution in exchange for imaging speed, field of view, and photodamage is generally well warranted.
Light sheet microscopy is both an old and a new technique. Its first manifestation was as the “ultramicroscope” developed over a century ago by Heinrich Siedentopf and Richard Zsigmondy, in which scattering of a thin sheet of light by colloidal particles was detected with a perpendicular lens [14,15]. This work was impactful: Zsigmondy won the Nobel Prize in Chemistry in 1925, and a 1909 book review in Science noted that “To most chemists and physicists the work of Siedentopf and Zsigmondy is perhaps already familiar” [16]. Its uses for biology were also evident; a fascinating 1915 article in Scientific American, for example, notes its utility for examining organisms “at the limits of microscopic perception,” including several pernicious pathogens [17]. Nonetheless, this microscope geometry seems largely to have been forgotten in subsequent decades. Brief revivals, and the realization that sheets could be used for fluorescence excitation, occurred at various times over the following century, until roughly a decade ago seminal work from Ernst Stelzer and colleagues at the European Molecular Biology Laboratory demonstrated the ability of light sheet fluorescence microscopy to perform whole embryo live imaging with single-cell resolution over many-hour timescales [13], launching the modern renaissance of light sheet microscopy. Refs. [18] and [19] have good discussions of the history of light sheet imaging.
A typical optical arrangement for a light sheet microscope, from the author’s lab, is illustrated schematically and photographically in Figure 1C,D. There is an enormous variety in light sheet designs, in both home-built and commercial instruments. This is accompanied by a proliferation of terminology, which appears to slowly be converging on “light sheet fluorescence microscopy” (LSFM) as a general term, though “selective plane illumination microscopy” (SPIM) is also widely used. Nearly all implementations make use of two lenses oriented perpendicularly to each other, one to transmit the thin excitation sheet and one to detect fluorescence emission. The lenses constrain the orientation of the specimen. In the setup shown in Figure 1D, specimens are introduced vertically; in other setups, horizontal arrangements, including specimens on planar coverslips, can be examined. Before elaborating on design issues, we’ll examine recent imaging applications relevant to microbial communities.
Light Sheet Imaging of Microbial Communities
The aforementioned attributes of sensitive, minimally toxic, three-dimensional imaging with large fields of view make light sheet fluorescence microscopy a valuable tool for exploring microbes in various contexts. Perhaps surprisingly, however, examples are still rare. To the best of my knowledge the first, predating the contemporary re-emergence of light sheet methods, was from Fuchs et al. in 2002 [20], who developed a “Thin Light Sheet Microscope” to look at bacteria in seawater, stained with a fluorescent dye. Beyond this initial proof of concept that demonstrated high-contrast visualization inside aqueous samples, there do not appear to be any published studies building on this work to investigate marine microbes.
More recently, Hekstra, Leibler and colleagues constructed closed ecosystems, in which three species coexist in a sealed chamber, and used light sheet fluorescence microscopy to distinguish and quantify the abundances of three microbial species, a green alga (Chlamydomonas reinhardtii), the bacterium Escherichia coli, and the ciliate Tetrahymena thermophila with multiple replicates each observed for roughly 100 days [21,22]. The high-precision measurements of this unique study enabled analysis of the statistical properties of the population dynamics of this artificial community, especially the existence of correlated fluctuation modes among the species.
Many microbial communities form biofilms, whose structure, composition, and physical properties are important in natural ecosystems as well as artificial environments, such as medical implants. Because of their density, non-superficial characteristics of biofilms are difficult to measure. Very recently, Karampatzakis et al. integrated transient state monitoring, an optical method for measuring oxygen concentration, with light sheet microscopy to measure oxygen abundance inside Pseudomonas aeruginosa biofilms [23]. This allowed determination of the size of anoxic regions, and measurement of the extent of oxygen depletion near micro-colonies. The combination of optical sectioning and optical reporters of chemical activity promises to be a powerful one for interrogating microbial systems.
Höhn et al. used light sheet microscopy to study the dramatic morphological changes of the alga Volvox, which exists as colonies of flagellated individual cells arranged in a spherical shell. As part of its asexual reproduction process, the daughter colony undergoes an “inversion” that turns the structure inside-out, bringing the flagella that were on the interior to the outside of the final structure. Using a home-built imaging system based on the OpenSPIM design [24], Höhn et al. were able to capture three-dimensional images with intervals of 20–300 seconds over few hour durations and, with this data, devise a quantitative model in which curvature, contraction, and elastic stretching drive the overall shape transition. In addition to providing interesting insights into Volvox biology, this work illustrates the potential of direct, three-dimensional observation for understanding the morphology and dynamics of multicellular microbial communities.
My own lab has been using light sheet fluorescence imaging to study the population dynamics of gut microbial communities, using larval zebrafish and their intestinal microbiota as a model system [25,26]. The gastrointestinal microbiota is a topic of enormous contemporary interest, as we increasingly realize that these microbes play major roles in a wide range of physiological processes, and that discordances in microbial community composition are correlated with many diseases. Metagenomic studies, typically based on sequencing DNA from fecal samples, have yielded remarkable insights into the microbial species and genes present in intestinal environments, but due to the inherent lack of spatial and temporal resolution in these approaches, we know almost nothing about the structure and dynamics of these gut communities. This lack of insight is especially important given the large number of microbial species that live in the vertebrate gut, whose coexistence is likely shaped by spatial niches, varied lifestyles, and temporal fluctuations. Zebrafish provide a powerful system in which to uncover both general and specific mechanisms of host-microbe interactions, due to their relative transparency at young ages, their small size, and their amenability to gnotobiotic techniques, meaning that zebrafish can be derived germ-free and exposed to particular, controlled sets of microbial species as potential colonizers. Imaging the larval zebrafish gut environment poses severe challenges for conventional imaging approaches. Its size requires imaging a volume of nearly 1 mm x 0.2 mm x 0.2 mm with single bacterium resolution, with three-dimensional images captured within the roughly 30 second interval between peristaltic contractions, over durations of several hours to investigate bacterial growth and competition dynamics. As described above, this is achievable with light sheet imaging.
Using a home-built light sheet microscope, my research group has so far explored the dynamics of labelled, commensal bacterial species in the larval zebrafish gut (Figure 2). Even single-species dynamics appear richer than might have been expected; a commensal Aeromonas species, for example, exists both as discrete individuals and as dense clusters whose growth rate is far higher than that of the planktonic cells (Fig. 2B) [25]. More notably, we have examined two-species dynamics between a commensal Vibrio and an Aeromonas species [26], as part of a broad goal of understanding mechanisms of inter-microbial interactions in the gut. Surprisingly, we found that competition between these two took the form of sudden and massive collapses of the Aeromonas population [26]. These collapses are driven by the peristaltic contractions of the gut, to which the densely clustered Aeromonas is susceptible but the planktonic Vibrio is not. These dynamics would be completely unknown if not for imaging, and the high spatial and temporal resolution of light sheet microscopy enabled construction of a quantitative, and experimentally testable, model of stochastic dynamics in this system [26]. More generally, this study revealed that the tempestuous physical environment of the gut can be a major determinant of the population dynamics of gut microbes, a conclusion mirrored in recent studies of human gut microbes in artificial environments [27]. Ongoing work on still more zebrafish commensal microbial species, as well as a variety of chemical and physical perturbations, is continuing to highlight the importance of spatial structure and temporal dynamics.
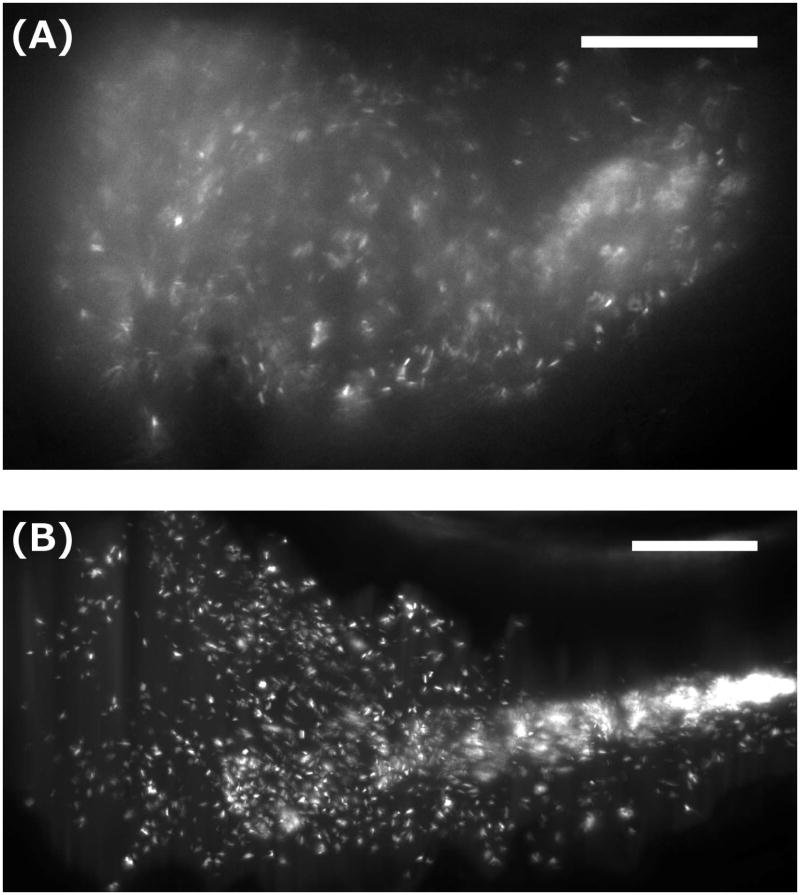
Figure 2.
Light sheet fluorescence microscopy images of bacteria in the larval zebrafish gut. (A) A single optical plane showing a GFP-expressing, commensal Plesiomonas species in the anterior intestinal bulb. Motile individuals (see Supplemental Movie 1) and sparse, likely mucus-rich aggregates are evident. (B) A maximum intensity projection of a three-dimensional image of a commensal Aeromonas species, as in Ref. [Wiles, Jemielita, et al. 2016]. Discrete individuals and a dense, midgut-localized aggregate are evident. (See Supplemental Movie 2 for a rotating 3D representation of the dataset.) Bar = 50 microns, in both panels.
The deep optical sectioning provided by light sheet microscopy also enables biophysical characterization of the environment experienced by microbial cells. Imaging the dynamics of both passive and magnetically driven probe particles in the zebrafish gut, for example, enables quantification of the viscosity of the lumenal space [28]. One could imagine similar approaches to studying the mechanics of, for example, bacterial biofilms.
As noted earlier, there are few studies making use of light sheet methods to investigate microbial communities, despite a high potential for scientific discovery. In part this is driven by the novelty of the method and the rather bewildering variety of approaches to the technique. I will describe various implementations of light sheet imaging, and then comment further on opportunities for future research.
Light Sheet Designs
Light sheet fluorescence microscopes exist in a dizzying number of forms, and are represented by both home-built setups and commercial instruments (e.g. from Carl Zeiss, Applied Scientific Instruments, Luxendo, LaVision BioTec, and others). In most microscopes, the excitation sheet is formed by rapidly scanning a laser beam [13]. In some, the sheet is created by focusing a wide beam through a cylindrical lens; this approach is sometimes, but not always, referred to as selective plane illumination microscopy (SPIM). The “OpenSPIM” set of instructions and tutorials for building a simple light sheet microscope makes use of the cylindrical lens design [24]. Many variants of light sheet microscopy have been devised to improve resolution or other aspects of optical quality. In the typical two-lens setup (Figure 1), image quality is worse on the “downstream” side of the excitation sheet, and also deteriorates at greater depths relative to the detection lens due to absorption and scattering. This has motivated the widespread use of sample rotation followed by registration and fusion of the different views [29]; the commercial light sheet microscope sold by Carl Zeiss AG makes use of a two-lens with specimen rotation setup. Three- and four-lens setups also exist, in which different lens pairs can be used for excitation and detection, for simultaneous multi-view imaging [30–32]. These methods are effective, but have the drawbacks of increased complexity as well as decreased space for specimens. Even with two lenses, different geometries are possible; in the “iSPIM” [33] and “diSPIM” instruments, available commercially from Applied Scientific Instruments, the two lenses are oriented at 45 degrees from the horizontal, allowing light sheet imaging of specimens on coverslips on standard inverted microscopy setups. Several groups have developed single lens light sheet methods (e.g. [34]), in which off-axis illumination generates an oblique sheet, with emission detected through the rest of the objective lens. Typically, this comes at the cost of reduced fields of view, smaller working distances, or instrumental complexity.
Several instruments have integrated structured illumination with light sheet fluorescence microscopy [35], most notably the lattice light sheet of Betzig and co-workers [36], providing enhanced optical sectioning and super-resolution, though again at the cost of technical complexity. Multi-photon excitation [37], stimulated emission depletion [38], hyperspectral detection [39] and many other embellishments have also been combined with light sheet imaging.
Especially for experiments exploring microbial communities, this author firmly believes that simple, two-lens, unembellished light sheet fluorescence microscopy is sufficient to reveal structure and dynamics at length- and time-scales relevant to these systems. All the examples of microbial community imaging presented above, for example, make use of optically simple methods. In practice, moreover, the main challenges in applying light sheet techniques are not optical, but rather are associated with specimen handling and data analysis. Because of its use of (at least) two orthogonal lenses, standard specimen mounting methods are often not applicable, especially for multiple specimens. With the feasibility of 3D printing and other rapid prototyping and fabrication techniques, however, custom solutions are becoming ever easier to create. Because of its high resolution over large fields of view, the datasets generated by light sheet microscopy can be very large, often requiring considerable effort to analyze to extract quantitative information. There is no generic solution to this challenge, but image processing libraries associated with languages like MATLAB and Python, as well as platforms such as FIJI [40], make custom analysis routines increasingly manageable. In addition, software and analysis pipelines specifically designed for light sheet microscopy data are beginning to emerge, such as multi-view registration modules [41] and a recent suite of protocols from Amat et al. that includes compression and visualization tools [42]. I note, however, that much of the analysis efforts of the light sheet microscopy community are focused on eukaryotic cell tracking, e.g. identifying and monitoring nuclei in developing organisms, and so aren’t necessarily applicable to imaging microbial communities.
Future Directions
Light sheet fluorescence microscopy is a powerful and tractable tool for examining microbial communities. Even within the systems described here, opportunities abound for further work, examining more and different microbial species, engineered strains that allow delineation of how specific genetic factors affect community architecture, physical and chemical perturbations to these communities, and more. In addition, a great many environments and organisms have yet to be fully explored via imaging. For example, non-light-sheet imaging, as well as standard colony counting methods, revealed stochasticity inherent in microbial colonization of the gut of the model nematode C. elegans [43]; the dynamics would be fascinating to characterize and correlate with bacterial behaviors. The growth of plant roots has been beautifully imaged by light sheet microscopy [44]; viewing the architecture of the associated microbial communities would be informative. Viral populations, bioreactor communities, food-associated consortia, and more are all microbial systems whose study could benefit from light sheet microscopy, and the future will hopefully yield a steady stream of imaging-based insights into their structure and dynamics.
Supplementary Material
A single optical plane, captured with light sheet fluorescence microscopy, of the anterior bulb region of the gut of a larval zebrafish, colonized with a GFP-expressing Plesiomonas species. The total duration of the movie is 7 seconds, and it should play in real (1x) time.
Rendering of a three-dimensional light sheet fluorescence microscopy image of a commensal Aeromonas species in the gut of a larval zebrafish, as in Ref. [Wiles, Jemielita, et al. 2016].
Highlights.
Light sheet fluorescence microscopy (LSFM) is a powerful 3D imaging technique.
The geometry of LSFM enables imaging large fields of view with low phototoxicity.
LSFM has been applied to several microbial communities in recent years.
Simple LSFM methods have great potential for further microbial studies.
Acknowledgments
The author thanks Edouard Hay, Savannah Logan, and Travis Wiles for providing the images and the bacterial strains represented in Figure 2, and Brandon Schlomann, Travis Wiles, and Karen Guillemin for useful discussions and comments on the manuscript. All zebrafish experiments were performed using protocols approved by the University of Oregon Institutional Animal Care and Use Committee (protocol 15–96).
Research reported in this publication was supported by the NIH as follows: NIGMS award P50GM098911 and NICHD award P01HD22486; the latter provided support for the University of Oregon Zebrafish Facility. The content is solely the responsibility of the authors and does not represent the official views of the NIH. Research reported in this publication was also supported by the National Science Foundation under award numbers 0922951 and 1427957; the M.J. Murdock Charitable Trust; a Scialog Program grant sponsored jointly by Research Corporation for Science Advancement and the Gordon and Betty Moore Foundation through grant to the University of Oregon by the Gordon and Betty Moore and Simons Foundations; and the Kavli Microbiome Ideas Challenge, a project led by the American Society for Microbiology in partnership with the American Chemical Society and the American Physical Society and supported by The Kavli Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McNally L, Bernardy E, Thomas J, Kalziqi A, Pentz J, Brown SP, Hammer BK, Yunker PJ, Ratcliff WC. Killing by Type VI secretion drives genetic phase separation and correlates with increased cooperation. Nature Communications. 2017;8 doi: 10.1038/ncomms14371. ncomms14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harcombe WR, Riehl WJ, Dukovski I, Granger BR, Betts A, Lang AH, Bonilla G, Kar A, Leiby N, Mehta P, et al. Metabolic Resource Allocation in Individual Microbes Determines Ecosystem Interactions and Spatial Dynamics. Cell Reports. 2014;7:1104–1115. doi: 10.1016/j.celrep.2014.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerr B, Riley MA, Feldman MW, Bohannan BJM. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Prindle A, Humphries J, Gabalda-Sagarra M, Asally M, Lee DD, Ly S, Garcia-Ojalvo J, Süel GM. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature. 2015;523:550–554. doi: 10.1038/nature14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basler M, Ho BT, Mekalanos JJ. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell. 2013;152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine JH, Fontes ME, Dworkin J, Elowitz MB. Pulsed Feedback Defers Cellular Differentiation. PLOS Biology. 2012;10:e1001252. doi: 10.1371/journal.pbio.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed T, Shimizu TS, Stocker R. Microfluidics for bacterial chemotaxis. Integr Biol. 2010;2:604–629. doi: 10.1039/c0ib00049c. [DOI] [PubMed] [Google Scholar]
- 8.Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M. Direct visualization of horizontal gene transfer. Science. 2008;319:1533–1536. doi: 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- 9.Clarke M, Maddera L, Harris RL, Silverman PM. F-pili dynamics by live-cell imaging. PNAS. 2008;105:17978–17981. doi: 10.1073/pnas.0806786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bos J, Zhang Q, Vyawahare S, Rogers E, Rosenberg SM, Austin RH. Emergence of antibiotic resistance from multinucleated bacterial filaments. PNAS. 2015;112:178–183. doi: 10.1073/pnas.1420702111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laissue PP, Alghamdi RA, Tomancak P, Reynaud EG, Shroff H. Assessing phototoxicity in live fluorescence imaging. Nat Meth. 2017;14:657–661. doi: 10.1038/nmeth.4344. [DOI] [PubMed] [Google Scholar]
- 12.Jemielita M, Taormina MJ, Delaurier A, Kimmel CB, Parthasarathy R. Comparing phototoxicity during the development of a zebrafish craniofacial bone using confocal and light sheet fluorescence microscopy techniques. J Biophotonics. 2013;6:920–928. doi: 10.1002/jbio.201200144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *13.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK. Reconstruction of Zebrafish Early Embryonic Development by Scanned Light Sheet Microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. In this influential paper, Keller et al. demonstrated light sheet fluorescence microscopy of entire zebrafish embryos over the first 24 hours of development. The high spatial and temporal resolution of the imaging enabled tracking of every cell in the embryo over this period. [DOI] [PubMed] [Google Scholar]
- 14.Cahan D. The Zeiss Werke and the Ultramicroscope: The Creation of a Scientific Instrument in Context. Archimedes. 1996 [Google Scholar]
- 15.Siedentopf H, Zsigmondy R. Über Sichtbarmachung und Groessenbestimmung ultramikroskopischer Teilchen, mit beson- derer Anwendung auf Goldrubinglaesern. Annalen der Physik. 1903;10:1–39. [Google Scholar]
- 16.Kahlenberg Louis. Colloids and the Ultramicroscope. Science. 1909;30:184. [Google Scholar]
- 17.The Ultramicroscope and its Application to Modern Biology. Scientific American. 1915;80(Supp):2074–211. author unknown. [Google Scholar]
- 18.Santi PA. Light Sheet Fluorescence Microscopy. Journal of Histochemistry & Cytochemistry. 2011;59:129–138. doi: 10.1369/0022155410394857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keller PJ, Ahrens MB, Freeman J. Light-sheet imaging for systems neuroscience. Nat Meth. 2015;12:27–29. doi: 10.1038/nmeth.3214. [DOI] [PubMed] [Google Scholar]
- 20.Fuchs E, Jaffe JS, Long RA, Azam F. Thin Laser Light Sheet Microscope for Microbial Oceanography. Optics Express. 2002;10:145–154. doi: 10.1364/oe.10.000145. [DOI] [PubMed] [Google Scholar]
- 21.Hekstra DR, Cocco S, Monasson R, Leibler S. Trend and fluctuations: Analysis and design of population dynamics measurements in replicate ecosystems. Phys Rev E. 2013;88:062714. doi: 10.1103/PhysRevE.88.062714. [DOI] [PubMed] [Google Scholar]
- *22.Hekstra DR, Leibler S. Contingency and Statistical Laws in Replicate Microbial Closed Ecosystems. Cell. 2012;149:1164–1173. doi: 10.1016/j.cell.2012.03.040. A fascinating study, monitoring the population fluctuations of closed, 3-species ecosystems and uncovering structure in their statistical dynamics. Arguably overly artificial in its construction, and perhaps not indicative of steady-state dynamics, as all the species decline in abundance over time, the work is nonetheless intriguing, and points to a still unrealized potential for quantitative studies of this sort. [DOI] [PubMed] [Google Scholar]
- 23.Karampatzakis A, Sankaran J, Kandaswamy K, Rice SA, Cohen Y, Wohland T. Measurement of oxygen concentrations in bacterial biofilms using transient state monitoring by single plane illumination microscopy. Biomed Phys Eng Express. 2017;3:035020. [Google Scholar]
- *24.Pitrone PG, Schindelin J, Stuyvenberg L, Preibisch S, Weber M, Eliceiri KW, Huisken J, Tomancak P. OpenSPIM: an open-access light-sheet microscopy platform. Nat Meth. 2013;10:598–599. doi: 10.1038/nmeth.2507. The paper and its associated tutorials and resources, including an email listserv, provide valuable guidance on constructing a light sheet microscope. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jemielita M, Taormina MJ, Burns AR, Hampton JS, Rolig AS, Guillemin K, Parthasarathy R. Spatial and Temporal Features of the Growth of a Bacterial Species Colonizing the Zebrafish Gut. mBio. 2014;5:e01751–14. doi: 10.1128/mBio.01751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *26.Wiles TJ, Jemielita M, Baker RP, Schlomann BH, Logan SL, Ganz J, Melancon E, Eisen JS, Guillemin K, Parthasarathy R. Host Gut Motility Promotes Competitive Exclusion within a Model Intestinal Microbiota. PLOS Biol. 2016;14:e1002517. doi: 10.1371/journal.pbio.1002517. This paper describes apparent competition between two gut microbial species, which imaging reveals is a consequence of their spatial organization and their response to intestinal mechanics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cremer J, Segota I, Yang C, Arnoldini M, Sauls JT, Zhang Z, Gutierrez E, Groisman A, Hwa T. Effect of flow and peristaltic mixing on bacterial growth in a gut-like channel. PNAS. 2016;113:11414–11419. doi: 10.1073/pnas.1601306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taormina MJ, Hay EA, Parthasarathy R. Passive and Active Microrheology of the Intestinal Fluid of the Larval Zebrafish. Biophysical Journal. 2017;113:957–965. doi: 10.1016/j.bpj.2017.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swoger J, Verveer P, Greger K, Huisken J, Stelzer EHK. Multi-view image fusion improves resolution in three-dimensional microscopy. Opt Express. 2007;15:8029–8042. doi: 10.1364/oe.15.008029. [DOI] [PubMed] [Google Scholar]
- 30.Krzic U, Gunther S, Saunders TE, Streichan SJ, Hufnagel L. Multiview light-sheet microscope for rapid in toto imaging. Nature Methods. 2012;9:730–733. doi: 10.1038/nmeth.2064. [DOI] [PubMed] [Google Scholar]
- 31.Schmid B, Shah G, Scherf N, Weber M, Thierbach K, Campos CP, Roeder I, Aanstad P, Huisken J. High-speed panoramic light-sheet microscopy reveals global endodermal cell dynamics. Nat Commun. 2013:4. doi: 10.1038/ncomms3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomer R, Khairy K, Amat F, Keller PJ. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nature Methods. 2012;9:755–763. doi: 10.1038/nmeth.2062. [DOI] [PubMed] [Google Scholar]
- 33.Wu Y, Ghitani A, Christensen R, Santella A, Du Z, Rondeau G, Bao Z, Colón-Ramos D, Shroff H. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2011;108:17708–17713. doi: 10.1073/pnas.1108494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouchard MB, Voleti V, Mendes CS, Lacefield C, Grueber WB, Mann RS, Bruno RM, Hillman EMC. Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms. Nat Photon. 2015;9:113–119. doi: 10.1038/nphoton.2014.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller PJ, Schmidt AD, Santella A, Khairy K, Bao Z, Wittbrodt J, Stelzer EHK. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat Methods. 2010;7:637–642. doi: 10.1038/nmeth.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen B-C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, Liu Z, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat Meth. 2011;8:757–760. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- 38.Friedrich M, Gan Q, Ermolayev V, Harms GS. STED-SPIM: Stimulated emission depletion improves sheet illumination microscopy resolution. Biophys J. 2011;100:L43–45. doi: 10.1016/j.bpj.2010.12.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jahr W, Schmid B, Schmied C, Fahrbach FO, Huisken J. Hyperspectral light sheet microscopy. Nat Commun. 2015;6:7990. doi: 10.1038/ncomms8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preibisch S, Amat F, Stamataki E, Sarov M, Singer RH, Myers E, Tomancak P. Efficient Bayesian-based multiview deconvolution. Nat Meth. 2014;11:645–648. doi: 10.1038/nmeth.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amat F, Höckendorf B, Wan Y, Lemon WC, McDole K, Keller PJ. Efficient processing and analysis of large-scale light-sheet microscopy data. Nat Protocols. 2015;10:1679–1696. doi: 10.1038/nprot.2015.111. [DOI] [PubMed] [Google Scholar]
- 43.Vega NM, Gore J. Stochastic assembly produces heterogeneous communities in the Caenorhabditis elegans intestine. PLOS Biology. 2017;15:e2000633. doi: 10.1371/journal.pbio.2000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *44.Maizel A, von Wangenheim D, Federici F, Haseloff J, Stelzer EHK. High-resolution live imaging of plant growth in near physiological bright conditions using light sheet fluorescence microscopy. The Plant Journal. 2011;68:377–385. doi: 10.1111/j.1365-313X.2011.04692.x. This paper demonstrates the feasibility and the appeal of light sheet imaging of plant root growth. It does not include imaging of microbes, but its methods could be readily applied to this end. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A single optical plane, captured with light sheet fluorescence microscopy, of the anterior bulb region of the gut of a larval zebrafish, colonized with a GFP-expressing Plesiomonas species. The total duration of the movie is 7 seconds, and it should play in real (1x) time.
Rendering of a three-dimensional light sheet fluorescence microscopy image of a commensal Aeromonas species in the gut of a larval zebrafish, as in Ref. [Wiles, Jemielita, et al. 2016].