This study provided the serologic, genomic, and geospatial investigation of an iatrogenic outbreak that was recognized in rural Cambodia in 2014–2015, showing a massive and spatially localized outbreak of HIV, associated with HCV infections, but a lack of HBV diffusion.
Keywords: Cambodia, HBV, HCV, HIV, iatrogenic outbreak
Abstract
Background
In 2014–2015, 242 individuals aged 2–89 years were newly diagnosed with human immunodeficiency virus type 1 (HIV-1) in Roka, a rural commune in Cambodia. A case-control study attributed the outbreak to unsafe injections. We aimed to reconstruct the likely transmission history of the outbreak.
Methods
We assessed in 209 (86.4%) HIV-infected cases the presence of hepatitis C virus (HCV) and hepatitis B virus (HBV). We identified recent infections using antibody (Ab) avidity testing for HIV and HCV. We performed amplification, sequencing, and evolutionary phylogenetic analyses of viral strains. Geographical coordinates and parenteral exposure through medical services provided by an unlicensed healthcare practitioner were obtained from 193 cases and 1499 controls during interviews.
Results
Cases were coinfected with HCV (78.5%) and HBV (12.9%). We identified 79 (37.8%) recent (<130 days) HIV infections. Phylogeny of 202 HIV env C2V3 sequences showed a 198-sample CRF01_AE strains cluster, with time to most recent common ancestor (tMRCA) in September 2013 (95% highest posterior density, August 2012–July 2014), and a peak of 15 infections/day in September 2014. Three geospatial HIV hotspots were discernible in Roka and correlated with high exposure to the practitioner (P = .04). Fifty-nine of 153 (38.6%) tested cases showed recent (<180 days) HCV infections. Ninety HCV NS5B sequences formed 3 main clades, 1 containing 34 subtypes 1b with tMRCA in 2012, and 2 with 51 subtypes 6e and tMRCAs in 2002–2003.
Conclusions
Unsafe injections in Cambodia most likely led to an explosive iatrogenic spreading of HIV, associated with a long-standing and more genetically diverse HCV propagation.
The World Health Organization (WHO) Global Health Sector Strategy 2016–2021 is paving the road to elimination of human immunodeficiency virus (HIV) and viral hepatitis as major public health threats by 2030, but the burden of new HIV, hepatitis C virus (HCV), and hepatitis B virus (HBV) infections, notably transmitted through unsafe and unnecessary medical injections, is still high [1].
In 2014–2015, one of the largest injection-related outbreaks of HIV occurred in rural Cambodia among residents from the Roka commune (7985 inhabitants) (Figure 1). The details of the outbreak have been reported previously [2]. The Ministry of Health of Cambodia and the National Center for HIV/AIDS, Dermatology and Sexually Transmitted Diseases (NCHADS) conducted a case-control study determining that cases were 5 times more likely to have received therapeutic injections and ruled out associations with commercial sex work, injection drug use, or blood transfusion [3].
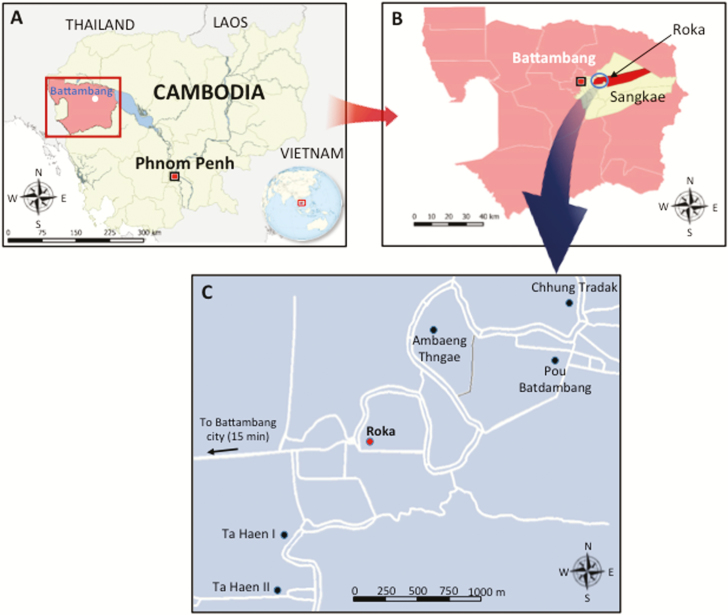
Figure 1.
Geographical location of the iatrogenic human immunodeficiency virus outbreak in the Roka commune, Cambodia, 2014–2015. A, Map showing Cambodia (in yellow) and the Battambang province (in pink). B, Map showing the Roka commune (in red) which is located in the Sangkae district (in yellow). C, Map showing the location of the 6 villages in the Roka commune (7985 inhabitants). Roka village (2338 inhabitants) is indicated by a red circle while the other 5 villages (Ambaeng Thngae [1050 inhabitants], Ta Haen I [1468 inhabitants], Ta Haen II [1050 inhabitants], Pou Batdambang [731 inhabitants], and Chhung Tradak [1348 inhabitants]) are indicated by black circles. Roads are indicated by white lines.
By 28 February 2015, the identification of new cases had winded down [2]. Among 2045 Roka residents who underwent a free and voluntary HIV screening, 242 HIV-infected cases were newly diagnosed [2], leading to a prevalence of 11.8%, which is approximately 20 times higher than the national average (0.6%) [4]. The sources/origins of the Roka outbreak were unknown. However, it was suspected that the outbreak was iatrogenic, resulting from unsafe injection practices performed by a local unlicensed healthcare practitioner. That lay individual, who did not hold a medical degree, provided medical services to the nearby residents in his home and also made home call visits to clients. By the end of 2014, legal authorities arrested that individual. In December 2015, he underwent trial and was sentenced to 25 years in prison [5].
In this study, we report the likely transmission history of this iatrogenic outbreak by providing unprecedented spatiotemporal insights, a strong association between cases and past injections, as well as a history of HCV and HBV coinfections and the evolutionary phylogenetic analyses of the viral strains.
MATERIALS AND METHODS
Sampling
We conducted our study on 209 HIV-confirmed cases who had sufficient plasma specimens for further investigation. At the time of sampling, all subjects were antiretroviral therapy (ART) naive. For each case, we retrieved from the anonymously generated NCHADS database the following variables: sex, age, place of residence, history of migration to neighboring countries (such as Thailand, Laos, Malaysia, Myanmar, Vietnam), and HIV-1 RNA viral load levels (Abbott RealTime HIV-1 assay, Abbott Molecular, Des Plaines, Illinois).
Diagnostic and Recency Assays
To estimate the recency of HIV infections, all samples were tested with the Sedia Limiting Antigen-Avidity enzyme-linked immunosorbent assay (ELISA) (Sedia Biosciences, Portland, Oregon), using a threshold of 1.5 for normalized optical density values, as recommended to differentiate recent (<130 days) vs nonrecent HIV infections [6]. All samples that were classified as recent by antibody (Ab) avidity testing were further assessed with a HIV-1 Western blot assay (HIV Blot 2.2, MP Diagnostics, Singapore) to define Fiebig stages of HIV-1 infection [7].
All specimens were tested for the presence of HCV Ab and RNA with a rapid diagnostic test (SD Bioline HCV, Standard Diagnostics, Kyonggi-do, Korea) and an in-house polymerase chain reaction (PCR) assay targeting the HCV 5ʹ untranslated region, respectively [8]. Samples that were positive for both HCV Ab and HCV RNA were assessed with an in-house HCV avidity ELISA [9]. We used an avidity index threshold of 43%, as recommended to distinguish recent (<6 months) from nonrecent (>6 months) HCV infections in HCV-viremic patients.
All samples were tested for hepatitis B surface antigen (HBsAg) with the Alere Determine HBsAg rapid diagnostic test (Alere Medical Co, Chiba, Japan). In case of positive HBsAg results, specimens were further tested with ELISA for immunoglobulin M (IgM) antibody to hepatitis B core antigen (HBcIgM Ab) (Bio-Rad, Marnes-la-Coquette, France). A specimen/cutoff ratio >5 was used to identify recent (<6 months) HBV infections, as recently reported [10]. In case of HBsAg negativity, subjects were assessed for hepatitis B surface antibody (HBsAb) titers with ELISA (Bio-Rad, Marnes-la-Coquette, France) to identify those who were protected against HBV. Subjects were considered as negative for HBsAb when titer was <10 IU/L.
Viral Amplification and Sequencing
HIV RNA amplifications were done according to nested reverse transcription-PCR (RT-PCR) procedures established by the French National Agency for Research on AIDS and Viral Hepatitis (ANRS; http://www.hivfrenchresistance.org/) (version January 2015; see Supplementary Table 1). A total of 202 specimens were amplified in the C2V3 region of the HIV env gene (PCR fragment length of 374 base pairs [bp]). Seven missing sequences failed PCR amplification or had low-quality chromatograms. Additionally, 192 samples were amplified in the reverse transcriptase (RT) region (798 bp) and the protease (PR) region (507 bp) of the HIV pol gene. The HCV nonstructural 5B (NS5B) genomic region (371 bp) was amplified from 90 HCV RNA–positive specimens, with a seminested RT-PCR, as described previously [11]. Ten HCV sequences were missing due to failed PCR amplification. A total of 16 samples (11 missing samples due to failed PCR amplification or insufficient volume of plasma) were subjected to HBV DNA amplification in the S-gene (738 bp), as reported elsewhere [12]. All purified PCR products were sequenced using the Big Dye Terminator v3•1 Cycle Sequencing kit (Applied Biosystems).
Phylogenetic Analysis
Maximum likelihood phylogenies for env C2V3 and PR-RT (following concatenation) HIV, NS5B HCV, and S-gene HBV were inferred with the use of Molecular Evolutionary Genetics Analysis version 6 (MEGA6) software [13]. For reference sequences, we used sequences that were downloaded from the GenBank or Los Alamos database, as well as sequences that were amplified from subjects living in different geographical provinces in Cambodia. Phylogenetic grouping was assessed by bootstrap supports and genetic distances [14]. We also inferred Bayesian temporally resolved phylogenetic trees for HIV and HCV with the use of Bayesian Evolutionary Analysis Sampling Trees (BEAST) software, version 1.8.3 [15]. Reference sequences were amplified from Cambodian subjects infected with HIV or HCV and having a known sampling date, ranging from 1997 to 2014 for HIV, and between 2002 and 2015 for HCV. For each outbreak-associated cluster, we estimated the time to the most recent common ancestor (tMRCA). With HIV env C2V3 sequences, we also investigated the growth rate and lineage-through-time (LTT) of the HIV outbreak (Supplementary Materials).
Geographic HIV Mapping
Through a field study conducted in Roka commune, we measured with a handheld global positioning system (GPS) device the geographic coordinates of 193 HIV-infected cases and 1499 subjects who were found to be HIV-negative during the free and voluntary testing performed between December 2014 and February 2015. For all individuals, we also collected, through face-to-face interviews, information about whether subjects did or did not receive injections from the imprisoned practitioner. All interviewed subjects were superimposed onto a 0.004° latitude × 0.004° longitude grid. The total number of subjects was summed in each cell over the entire study area. For each cell, the prevalence of HIV infection was calculated by dividing the total number of HIV-infected subjects by the total number of tested subjects, and expressed in percentage values. The resulting prevalence in each cell was mapped to the centroid position of each cell. Using the same grids, we generated 1 additional map depicting the interpolated frequency of history of past injections with the informal practitioner. The maps were created using inverse distance weighting, with Quantum Geographic Information Science (QGIS) software version 2.12.3 [16]. The correlation between the 2 maps was assessed using Pearson correlation coefficient (R2) and the t test.
Ethical Committee Approvals
The virological investigation was approved by the Cambodia National Ethical Committee for Health Research (NECHR) (number 353/NECHR). The protocol for GPS mapping was approved by the NECHR (number 244/NECHR), and no sampling of biological material was performed during this field study. All HIV-infected cases and uninfected subjects provided written consent.
RESULTS
Patients’ Characteristics
As summarized in Table 1, the median age of cases was 38.0 years (interquartile range [IQR], 19–54 years; range, 2–89 years), and 129 (61.7%) were female. One hundred fifty-nine (76.1%) were residents in the Roka village. One hundred ninety-nine (95.2%) had never migrated to neighboring countries. The median HIV-1 RNA level was 5.16 (IQR, 4.51–5.61) log10 copies/mL. Forty-one (19.6%) cases were monoinfected with HIV, whereas most (164 [78.5%]) were coinfected with HCV. Cases showing HBV coinfection were uncommon (27 [12.9%]).
Table 1.
Main Characteristics of Human Immunodeficiency Virus (HIV)–Infected Patients During the Iatrogenic HIV Outbreak in Roka, Cambodia, 2014–2015 (N = 209)
| Variable | No. (%) |
|---|---|
| Demographic characteristics | |
| Age, y | |
| Median | 38.0 |
| Interquartile range | 19–54 |
| Range | 2–89 |
| Age distribution | |
| <5 y | 5 (2.4) |
| 5–9 y | 22 (10.5) |
| 10–14 y | 13 (6.2) |
| 15–49 y | 89 (42.6) |
| 50–69 y | 61 (29.2) |
| ≥70 y | 19 (9.1) |
| Female sex | 129 (61.7) |
| Place of residence | |
| Roka | 159 (76.1) |
| Ambaeng Thngae | 42 (20.1) |
| Othersa | 8 (3.8) |
| Migration to neighboring countries | 10 (4.8) |
| Virological characteristics | |
| Plasma HIV-1 RNA viral load (log10 copies/mL) | |
| Median | 5.16 |
| Interquartile range | 4.51–5.61 |
| Full range | 3.29–6.68 |
| Coinfection with HCV and/or HBV | |
| HCV coinfection | 141 (67.5) |
| HBV coinfection | 4 (1.9) |
| HCV and HBV coinfection | 23 (11.0) |
| Recency of HIV, HCV, and HBV infections | |
| Recent HIV infection (<130 d) | 79 (37.8) |
| Recent HCV infection (<6 mo); n = 153b | 59 (38.6) |
| Recent HBV infection (<6 mo); n = 27 | 3 (11.1) |
| Protection against HBV (positive HBsAb); n = 133c | 69 (51.9) |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: HBsAb, hepatitis B surface antibody; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV, human immunodeficiency virus.
aEight cases were living in Ta Haen I (n = 4), Ta Haen II (n = 3), and Pou Batdambang (n = 1).
bEleven samples were not assessed for HCV recency due to insufficient volume.
cHBsAb testing was done in 133 hepatitis B surface antigen–negative subjects who could be further tested (sufficient volume of plasma).
Recency, Phylogenetic Analysis, and Spatial Spread of HIV
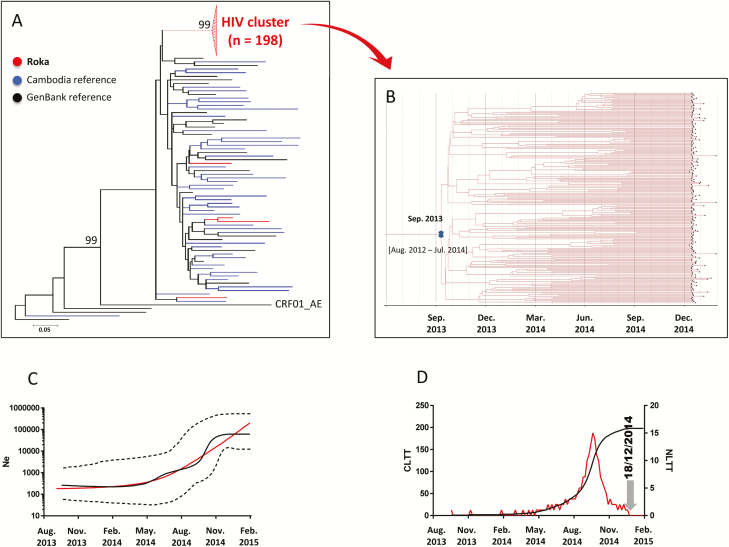
We identified recent HIV infections in 79 (37.8%) cases (Table 1). Western blot analysis classified subjects with recent infections at a Fiebig IV (10 [12.6%]) or V (57 [72.2%]) stage, indicating HIV transmission within the previous 30 or 100 days, respectively. Among 202 HIV C2V3 env sequences that could be analyzed, 198 (98.0%) clustered in a single well-supported monophyletic clade (bootstrap support of 99 and average intracluster nucleotide identity ± standard deviation [SD] of 99.3% ± 0.7) (Figure 2A and Supplementary Table 2). These sequences derived from an ancestral CRF01_AE lineage, and were not closely related to the control sequences. The remaining 4 C2V3 env strains, found outside the outbreak-associated cluster, were more closely related to the control sequences. Additional analysis of 192 concatenated HIV PR-RT sequences supported an identical outbreak-associated clustering pattern (see Supplementary Figure 1). We found that tMRCA of the HIV env outbreak-associated cluster was September 2013 (95% highest posterior density [HDP], August 2012–July 2014) (Figure 2B). The median estimates of the effective population size inferred under the demographic expansion model indicated a pattern of constant population size followed by exponential growth (Figure 2C). The LTT plot depicted from May 2014 a rapid rise of HIV infections with a maximum of 15 infections per day in September 2014, followed by a decline thereafter (Figure 2D). These data were consistent with the date of arrest of the practitioner by the police (end of December 2014).
Figure 2.
Human immunodeficiency virus type 1 (HIV-1) env C2V3 sequences. A, Maximum-likelihood phylogenetic tree for HIV-1 env C2V3 sequences from 202 case sequences (indicated in red), 45 Cambodia sequences (blue), and 24 GenBank reference sequences (black). The Roka outbreak–associated CRF01_AE HIV cluster is represented by a red triangle. Four sequences (amplified from National Center for HIV/AIDS, Dermatology and Sexually Transmitted Diseases [NCHADS] patients 171, 184, 185, and 116) from Roka did not group within the outbreak-associated cluster, and were isolated from individuals presenting nonrecent HIV-1 infections and who were negative for HCV and HBV. The unit for the scale bar of 0.05 is the number of nucleotide substitutions per site. GenBank accession numbers for the Roka HIV env C2V3 sequences were KY570019–KY570220. Accession numbers for other Cambodia sequences were KY570221–KY570265. B, Bayesian time-scaled phylogenetic tree of 198 HIV-1 env C2V3 sequences from the Roka HIV cluster. The branch lengths represent the number of substitutions per site per year. The cross marks the mean estimate of the time to the most recent common ancestor with 95% highest posterior densities (HPDs). C, Effective viral population size over time. The Bayesian skyline plot shows the median viral population size over time (black solid line) and 95% HPDs around the estimate (black dashed lines). Expansion model is indicated by a red line. D, Lineage-through-time plot. CLTT indicates the cumulative lineage through time (black solid line); NLTT indicates the noncumulative lineage through time (red solid line). Abbreviations: HBV, hepatitis B virus; HCV, hepatitis C virus; Ne, effective population size.
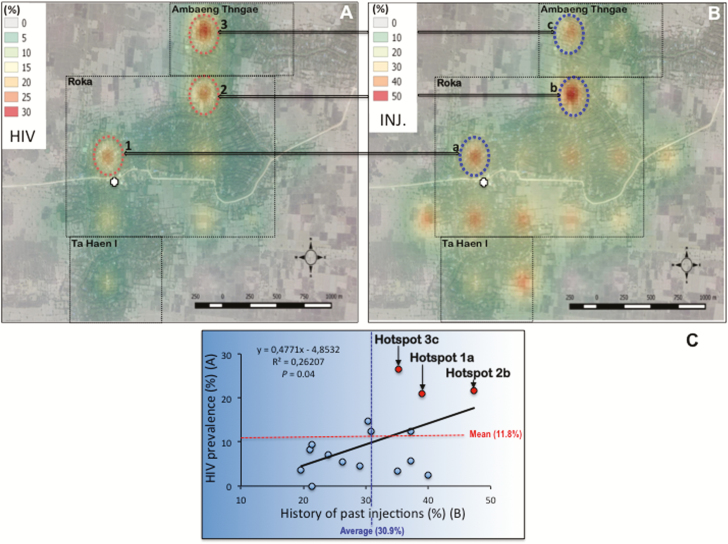
A marked heterogeneity in HIV prevalence was observed in the Roka commune. HIV cases were heavily confined in 3 main geographical microclusters showing HIV prevalence >20% (20.9%, 21.6%, and 26.5% for hotspots 1, 2, and 3, respectively) (Figure 3A). By contrast, some other sectors showed low (ranging between 0 and 5%) HIV prevalence. Regarding the 3 HIV hotspots, 1 was identified in the direct vicinity of the practitioner’s house, while the other 2 were located further, reflecting likely home call visits made by the informal practitioner throughout the investigated area. HIV microclusters matched hotspots of a high frequency of past injections given by the practitioner (39.0%, 47.0%, and 35.3% for hotspots a, b, and c, respectively) (Figure 3B). Overall, we found a significant correlation between HIV prevalence and past parenteral exposure to the practitioner (P = .04) (Figure 3C).
Figure 3.
Geographic variations in interpolated human immunodeficiency virus (HIV) prevalence (A), frequency of history of injections (INJ.) with the unlicensed practitioner (B) across the surveillance area, and correlation (C). Roka, Ambang Thngae, and Ta Haen I villages are delineated by a dotted rectangle. The practitioner’s house is depicted by a white cross. Each hotspot is indicated by a circle, associated with a number (1, 2, or 3 for HIV) or a letter (a, b, or c regarding history of past injections).
Recency and Phylogenetic Analysis of HCV
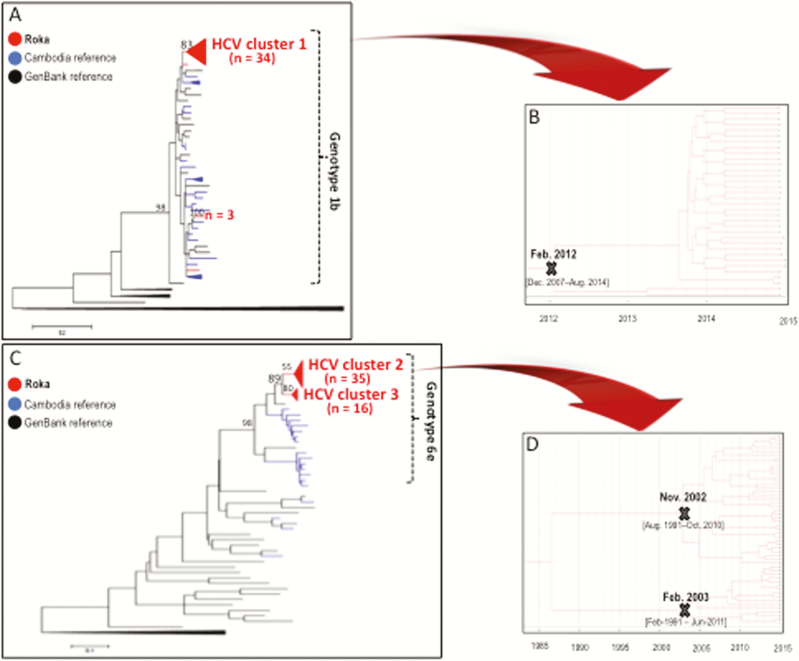
Of 153 subjects coinfected with HCV (11 missing specimens), recent HCV infections were detected in 59 (38.6%) individuals (Table 1), including 10 showing negative HCV Ab/positive HCV RNA patterns, and 49 exhibiting low-avid HCV Ab and positive HCV RNA. Nonrecent HCV infections were diagnosed in 41 (26.8%) subjects with high-avid HCV Ab and positive HCV RNA. Fifty-three (34.6%) persons had cleared HCV (positive HCV Ab but negative HCV RNA). Among the 100 (65.4%) individuals who were HCV viremic, we were able to amplify 90 HCV NS5B sequences. Among these, we found evidence of 3 main outbreak-associated clusters, including 34 HCV subtype 1b isolates in 1 cluster (Figure 4A and Supplementary Table 3), and 51 HCV subtype 6e strains distributed in 2 other clusters (Figure 4C and Supplementary Table 4). The subtype 1b cluster showed an intracluster nucleotide identity ± SD of 99.8% ± 0.4, and emerged in February 2012 (95% HDP, December 2007–August 2014) (Figure 4B). The 35 strains in the subtype 6e cluster 2 showed an intracluster nucleotide identity ± SD of 96.5% ± 1.3%. The 16 isolates in the cluster 3 exhibited a nucleotide identity ± SD of 97.3% ± 1.6%. HCV subtype 6e clusters had older tMRCAs (November 2002, 95% HDP, August 1991–October 2010; and February 2003, 95% HDP, February 1991–June 2011) (Figure 4D).
Figure 4.
Hepatitis C virus (HCV) NS5B sequences. A, Maximum-likelihood phylogenetic tree for HCV NS5B sequences from 39 patients with HCV subtype 1b sequences (red), 37 Cambodia sequences (blue), and 29 GenBank reference sequences (black). GenBank accession numbers for the Roka HCV 1b NS5B sequences were KY569650–KY569688. Accession numbers for other Cambodia sequences were KY569689–KY569725. B, Bayesian time-scaled phylogenetic tree of 34 HCV NS5B sequences from the Roka subtype 1b HCV cluster. The branch lengths represent the number of substitutions per site per year. The cross marks the mean estimate of the time to the most recent common ancestor (tMRCA) with 95% highest posterior densities (HPDs). C, Maximum-likelihood phylogenetic tree for HCV NS5B sequences from 51 patients with HCV subtype 6e sequences (red), 18 Cambodia sequences (blue), and 29 GenBank reference sequences (black). The 51 HCV subtype 6e strains were distributed in 2 distinct monophyletic clusters. GenBank accession numbers for the Roka HCV 6e sequences were KY569734–KY569784. Accession numbers for other Cambodia sequences were KY569785–KY569802. D, Bayesian time-scaled phylogenetic tree of 51 HCV NS5B sequences from the Roka genotype 6e clusters (n = 51). The branch lengths represent the number of substitutions per site per year. The mean estimate of the tMRCAs as well as 95% HPDs are indicated with at key internal nodes marked with “X.”
Recency and Phylogenetic Analysis of HBV
Of 27 subjects found positive for HBsAg, only 3 (11.1%) were reactive to HBcIgM (Table 1). From 16 HBV S-gene sequences, 10 (62.5%) were phylogenetically isolated strains, whereas 6 sequences grouped together into 2 distinct clusters of 4 and 2 sequences within HBV-C1 genotype strains. Among 133 HBsAg-negative subjects who could be further tested, 69 (51.9%) showed HBsAb titers ≥10 IU/L (Table 1).
DISCUSSION
This study revealed a massive and spatially localized outbreak of HIV in Cambodia, associated with concomitant HCV infections, but a lack of HBV diffusion. The recency (<4–6 months) of either HIV or HCV was found in approximately one-third of infections, emphasizing the burst of the outbreak.
The monophyletic cluster of CRF01_AE HIV strains with high homology supported the iatrogenic transmission scenario, which was related to a single and recent parenteral source/introduction of HIV into the Roka population. The 3 main HCV clusters showing older origins than the HIV cluster strengthened the iatrogenic scenario, and suggested that HCV likely originated from a long-standing spread of and diverse infectious HCV strains. The geospatial correlation between HIV hotspots and high frequency of injections administered by an unlicensed practitioner highlighted his probable role behind an iatrogenic transmission [17].
This might be the first report of a massive outbreak potentially caused by an unlicensed health practitioner who performed healthcare practices among low-risk subjects in a rural setting. In contrast, similar outcomes have been reported in other massive (>100 cases) injection-related outbreaks among other populations at higher risk of HIV, such as paid blood donors, as reported in Mexico (1986) [18], India (1988) [19], and China (1990–1995) [20]; hospitalized children, as described in Romania (1987–1992) [21, 22], Russia (1988–1989) [23], and Libya (1997–1999) [24, 25]; and persons who inject drugs, as established in Kazakhstan, Kyrgyzstan, and Uzbekistan (2007–2008) [26], Pakistan (2008–2009) [27], and Indiana, United States (2014–2015) [28, 29]. We hypothesize that the Roka outbreak might not be an isolated incident. Other unrecognized iatrogenic outbreaks caused by similar practices could exist, especially in rural areas.
The difference we observed between HIV and HCV dynamics through time is not easy to explain. One might postulate that HIV emerged through 1 unidentified highly viremic HIV-infected Roka resident, who was not treated with ART (or who had interrupted his treatment) and who, unfortunately, received injections from the practitioner in 2013. This source was obviously rare due to the low HIV prevalence and high ART coverage in Cambodia [4]. The HIV outbreak became apparent in late 2014 due to a tuberculosis-associated comorbidity and highlights the importance of HIV testing among TB patients. In contrast, HCV emerged earlier than HIV from at least 3 HCV-independent introductions. However, those infections were unknown given the lack of routine diagnosis and no apparent HCV symptoms [30, 31]. Another possibility to explain this temporal difference may be related to a reduced transmissibility of HIV, with a lower ex vivo viability in needles/syringes (few hours for HIV vs at least 7 days for HCV), as documented by some researchers [32].
To explain the absence of iatrogenic HBV diffusion, 2 reasons could be put forward. First, even if a vast majority of Roka HBV-uninfected cases were unvaccinated (the universal vaccination in children started in 2005 in Cambodia) [33], we showed that more than half of individuals were naturally protected against HBV due to HBsAb positivity, likely resulting from past infections. Second, the proportion of inactive HBsAg carriers, with very low or undetectable serum HBV DNA levels, was likely important among HBV-infected subjects, as previously documented in Cambodia [34]. These 2 specific HBV patterns probably explain the low prevalence of HBV in this population.
Our study has some limitations. First, the accuracy of Ab avidity-based results for the identification of recent infections is still debatable, as we could not determine the false recent rate of HCV infections. It is also possible that different HCV genotypes and the occurrence of HIV coinfections could have skewed our estimation of recent HCV infections in either direction [9, 35]. Moreover, the gold standard approach for measuring incidence (ie, negative followed by positive test results through a longitudinal follow-up) cannot be used during outbreak situations. Second, only samples from HIV cases were available for hepatitis testing, as individuals diagnosed with rapid HIV tests were referred for further testing. Therefore, our survey likely missed HCV monoinfections that were part of a larger outbreak of HCV in Roka. This incomplete sampling could have generated a bias in our analysis of HCV clusters and on the timing of introduction of the most recent common ancestor. Last, the legal investigation by police and prosecutors, as well as interviews of cases, could not clearly define the exact modus operandi of the unlicensed practitioner that led to this massive outbreak.
In conclusion, our study provided the detailed serologic, genomic, and geospatial investigation of an iatrogenic outbreak that was recognized in rural Cambodia in 2014–2015. Our unfortunate experience might serve as a global warning, notably for the Southeast Asia region, which is an area of major concern for the iatrogenic risk of blood-borne viruses. Avidity testing and genomic surveillance, in conjunction with spatial analysis, are key milestones to more forcefully advocate for reducing unsafe injection practices and further promoting oral drugs, in countries with concentrated epidemics and also in those with generalized epidemics where outbreaks are probably undiagnosed and/or neglected due to higher background noise.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. F. R., J. N., S. P., D. F., A. R., and V. S. conceived the study. F. R., J. N., D. Z., S. P., C. M., S. M., C. G.-G., G. L., S. K., K. P., A. K., and C. Y. generated and managed the data. F. R., J. N., B. R., A. B., S. P., M. L., L. F., W. K., F. B., M. F., C. M., J.-C. P., and T. B. contributed to the data analysis. F. R., J. N., and A. R. contributed to writing the first draft of the paper.
Acknowledgments. We are indebted to all patients living in the Roka commune and to Sophon Sern for collecting information from exposed residents in Roka; Robert Newman, MD, PhD, who carefully reviewed this manuscript; Ahmed Saadani Hassani, MD, for technical assistance; Patrice Piola, MD, PhD, for statistical support; and Daiana Mir da Silva, MSc, and Gonzalo Bello Bentancor, PhD, for their assistance in phylogenetic analysis.
Disclaimer. The content is solely the responsibility of the authors, and the findings and conclusions in this study do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was supported by grants from the 5% Initiative Expertise France, the Institut Pasteur International Network, and the French National Agency for Research on AIDS and Viral Hepatitis. This research has been also partially been supported by the President’s Emergency Plan for AIDS Relief through the CDC (cooperative agreement number 3U2GGH000989).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. Global hepatitis report, 2017. Available at: http://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/. Accessed 10 May 2017.
- 2. Vun MC, Galang RR, Fujita M, et al. Cluster of HIV infections attributed to unsafe injection practices—Cambodia, December 1, 2014–February 28, 2015. MMWR Morb Mortal Wkly Rep 2016; 65:142–5. [DOI] [PubMed] [Google Scholar]
- 3. Saphonn V, Fujita M, Samreth S, et al. Cluster of HIV infections associated with unsafe injection practices in a rural village in Cambodia. J Acquir Immune Defic Syndr 2017; 75:e82–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vun MC, Fujita M, Rathavy T, et al. Achieving universal access and moving towards elimination of new HIV infections in Cambodia. J Int AIDS Soc 2014; 17:18905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Phnom Penh Post. Doctor gets 25 years for HIV outbreak Available at: http://www.phnompenhpost.com/national/doctor-gets-25-years-hiv-outbreak. Accessed 10 May 2017.
- 6. Duong YT, Kassanjee R, Welte A, et al. Recalibration of the limiting antigen avidity EIA to determine mean duration of recent infection in divergent HIV-1 subtypes. PLoS One 2015; 10:e0114947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 2003; 17:1871–9. [DOI] [PubMed] [Google Scholar]
- 8. Bukh J, Purcell RH, Miller RH. Importance of primer selection for the detection of hepatitis C virus RNA with the polymerase chain reaction assay. Proc Natl Acad Sci U S A 1992; 89:187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaudy-Graffin C, Lesage G, Kousignian I, et al. Use of an anti-hepatitis C virus (HCV) IgG avidity assay to identify recent HCV infection. J Clin Microbiol 2010; 48:3281–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park JW, Kwak KM, Kim SE, et al. Differentiation of acute and chronic hepatitis B in IgM anti-HBc positive patients. World J Gastroenterol 2015; 21:3953–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Budkowska A, Kakkanas A, Nerrienet E, et al. Synonymous mutations in the core gene are linked to unusual serological profile in hepatitis C virus infection. PLoS One 2011; 6:e15871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sitnik R, Pinho JR, Bertolini DA, Bernardini AP, Da Silva LC, Carrilho FJ. Hepatitis B virus genotypes and precore and core mutants in Brazilian patients. J Clin Microbiol 2004; 42:2455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ragonnet-Cronin M, Hodcroft E, Hué S, et al. UK HIV Drug Resistance Database Automated analysis of phylogenetic clusters. BMC Bioinformatics 2013; 14:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 2012; 29:1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. QGIS version 2.14 Available at: http://www.qgis.org/api/2.14/. Accessed 15 January 2017.
- 17. Janjua NZ, Butt ZA, Mahmood B, Altaf A. Towards safe injection practices for prevention of hepatitis C transmission in South Asia: challenges and progress. World J Gastroenterol 2016; 22:5837–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Avila C, Stetler HC, Sepúlveda J, et al. The epidemiology of HIV transmission among paid plasma donors, Mexico City, Mexico. AIDS 1989; 3:631–3. [DOI] [PubMed] [Google Scholar]
- 19. Bhimani GV, Gilada IS. HIV prevalence in people with no fixed abode: a study of blood donorship patterns and risk determinants [abstract MoC00937]. In: Eighth International AIDS Conference, Amsterdam, the Netherlands, 1992. [Google Scholar]
- 20. Wu Z, Liu Z, Detels R. HIV-1 infection in commercial plasma donors in China. Lancet 1995; 346:61–2. [DOI] [PubMed] [Google Scholar]
- 21. Apetrei C, Loussert-Ajaka I, Collin G, et al. HIV type 1 subtype F sequences in Romanian children and adults. AIDS Res Hum Retroviruses 1997; 13:363–5. [DOI] [PubMed] [Google Scholar]
- 22. Patrascu IV, Dumitrescu O. The epidemic of human immunodeficiency virus infection in Romanian children. AIDS Res Hum Retroviruses 1993; 9:99–104. [DOI] [PubMed] [Google Scholar]
- 23. Bobkov A, Garaev MM, Rzhaninova A, et al. Molecular epidemiology of HIV-1 in the former Soviet Union: analysis of env V3 sequences and their correlation with epidemiologic data. AIDS 1994; 8:619–24. [PubMed] [Google Scholar]
- 24. de Oliveira T, Pybus OG, Rambaut A, et al. Benghazi Study Group Molecular epidemiology: HIV-1 and HCV sequences from Libyan outbreak. Nature 2006; 444:836–7. [DOI] [PubMed] [Google Scholar]
- 25. Yerly S, Quadri R, Negro F, et al. Nosocomial outbreak of multiple bloodborne viral infections. J Infect Dis 2001; 184:369–72. [DOI] [PubMed] [Google Scholar]
- 26. Thorne C, Ferencic N, Malyuta R, Mimica J, Niemiec T. Central Asia: hotspot in the worldwide HIV epidemic. Lancet Infect Dis 2010; 10:479–88. [DOI] [PubMed] [Google Scholar]
- 27. Ansari JA, Salman M, Safdar RM, et al. HIV/AIDS outbreak investigation in Jalalpur Jattan (JPJ), Gujrat, Pakistan. J Epidemiol Glob Health 2013; 3:261–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peters PJ, Pontones P, Hoover KW, et al. HIV infection linked to injection use of oxymorphone in Indiana, 2014–2015. N Engl J Med 2016; 375:229–39. [DOI] [PubMed] [Google Scholar]
- 29. Strathdee SA, Beyrer C. Threading the needle—how to stop the HIV outbreak in rural Indiana. N Engl J Med 2015; 373:397–9. [DOI] [PubMed] [Google Scholar]
- 30. De Weggheleire A, An S, De Baetselier I, et al. A cross-sectional study of hepatitis C among people living with HIV in Cambodia: prevalence, risk factors, and potential for targeted screening. PLoS One 2017; 12:e0183530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Westbrook RH, Dusheiko G. Natural history of hepatitis C. J Hepatol 2014; 61:S58–68. [DOI] [PubMed] [Google Scholar]
- 32. Thompson SC, Boughton CR, Dore GJ. Blood-borne viruses and their survival in the environment: is public concern about community needlestick exposures justified?Aust N Z J Public Health 2003; 27:602–7. [DOI] [PubMed] [Google Scholar]
- 33. Mao B, Patel MK, Hennessey K, Duncan RJ, Wannemuehler K, Soeung SC. Prevalence of chronic hepatitis B virus infection after implementation of a hepatitis B vaccination program among children in three provinces in Cambodia. Vaccine 2013; 31:4459–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamada H, Fujimoto M, Svay S, et al. Seroprevalence, genotypic distribution and potential risk factors of hepatitis B and C virus infections among adults in Siem Reap, Cambodia. Hepatol Res 2015; 45:480–7. [DOI] [PubMed] [Google Scholar]
- 35. Shepherd SJ, McDonald SA, Palmateer NE, et al. HCV avidity as a tool for detection of recent HCV infection: sensitivity depends on HCV genotype. J Med Virol 2018; 90:120–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.