Abstract
Purpose
Genome-wide association studies have identified multiple genetic variants associated with vertical cup-disc ratio (VCDR). Genetic risk scores (GRS) examine the aggregate genetic effect of individual variants on a trait by combining these separate genetic variants into a single measure. The purpose of this study is to construct GRS for VCDR and determine whether the GRS are associated with VCDR and whether the GRS increase the discriminatory ability for primary open angle glaucoma (POAG) in a Latino population.
Design
Population-based genetic association study.
Participants
A total of 4,018 Latino participants recruited from Los Angeles.
Methods
Weighted and unweighted GRS were constructed using 68 previously reported VCDR single nucleotide polymorphisms (SNPs), as well as SNPs from our own genome-wide association data. Linear and logistic regression analyses examined the associations of GRS with VCDR and POAG, respectively. To evaluate the discriminatory ability of the GRS for POAG, we conducted receiver operating characteristic (ROC) analyses.
Main Outcome Measures
The relationship between GRS and VCDR in Latinos.
Results
The GRS were significantly associated with VCDR (P < 0.0001), after adjusting for age, gender, central corneal thickness, intraocular pressure, and education. The weighted GRS explained an additional 2.74% of the variation in VCDR. Adding the weighted GRS derived from previously reported SNPs resulted in a moderate improvement in the discriminatory ability for POAG during ROC analyses, yielding an AUC of 0.735 (95% CI: [0.701, 0.768]). When our own SNPs were used, the AUC significantly increased to 0.809 (95% CI: [0.781, 0.837], P < 0.0001). We obtained similar results for the unweighted GRS.
Conclusion
To our knowledge, we are the first to report the association between GRS and VCDR, and its improvement in the discriminatory ability of POAG, in a Latino population.
The morphology of the optic disc is commonly assessed during routine ophthalmic examinations to monitor and diagnose multiple ocular diseases, including glaucoma. In particular, the vertical cup-disc ratio (VCDR) is an important clinical measurement to identify glaucomatous damage to the optic nerve. Accordingly, identifying factors that affect VCDR will not only aid in uncovering the biological mechanisms regulating this ocular trait, but may also assist in predicting ocular disease.
Population based epidemiological studies have identified multiple factors associated with VCDR, including higher intraocular pressure (IOP) and lower body mass index (BMI).1–3 Other identified factors, such as being male,1, 2or older age1–4 showed an association with VCDR in several studies and no association in other studies.5, 6 Diastolic blood pressure also has been positively1 and negatively2 associated with VCDR. Despite the identification of conventional risk factors, these systematic and ocular traits account for less than 4% of the variation in VCDR, suggesting other factors may contribute to this ocular trait.2
VCDR also has a demonstrated genetic component, with heritability estimates of 48% to 66% for this trait.7, 8 Genome-wide association studies (GWAS) have identified multiple loci associated with VCDR, including ATOH7, SIX1, CHEK2, and SCYL1.9–12 Despite the identification of genetic variants associated with VCDR, each variant confers only a modest effect and individually has limited predictive power. Genetic risk scores (GRS) examine the aggregate genetic effect by combining these separate genetic variants into a single measure. A previous study identified a polygenetic model for VCDR, and subsequently primary open angle glaucoma (POAG), using various variant significance thresholds.13 This study, however, was conducted in individuals of European descent and may not be generalizable to other ethnic groups. Latinos, a traditionally underrepresented racial group in ocular genetic research, exhibit a high prevalence of POAG.14 As such, examining the association between an aggregate measure of genetic risk and VCDR will further our understanding of the determinants of this trait. Additionally, the generation of GRS for an endophenotype for POAG will enable an opportunity to evaluate whether the addition of this genetic information improves the discriminatory ability for POAG compared to traditional risk factors. To our knowledge, we are the first to report on the association between genetic risk scores and VCDR, and the discriminatory ability for POAG, in a Latino population.
Methods
Ethics Statement
The institutional review board at the University of Illinois at Chicago approved the following research. All clinical investigation was performed according to the principles stated in the Declaration of Helsinki.
Study Sample, VCDR measurement, and Glaucoma Criteria
This research was conducted using previously published data on VCDR11 and POAG.14, 15 The data were collected from the Los Angeles Latino Eye Study (LALES), the largest population-based study of visual impairment and ophthalmic diseases in Latinos. All study participants received detailed ophthalmic examinations described elsewhere.16 Briefly, stereoscopic optic disc photographs were obtained and evaluated using a stereoscopic viewer (Asahi viewer; Pentax, Englewood, CO) to examine the optic nerve. The Humphrey Automated Field Analyzer II (Carl Zeiss Meditech, Dublin, CA) was used to test peripheral vision and a Swedish Interactive Threshold Algorithm standard C24 was used to evaluate the visual field. Using the stereoscopic photographs, a certified ophthalmologist measured the VCDR for the left and right eyes. The average VCDR between the eyes was used for downstream analysis. If one of the measurements was missing, the value from the other eye was used as the final measurement. POAG was determined by agreement of 3 glaucoma specialists using all clinical data with the following criteria: (1) the presence of an open angle and (2) congruent, characteristic, or compatible glaucomatous visual field abnormality and/or (3) evidence of characteristic or compatible glaucomatous optic disc damage in at least one eye. All subjects included in this study were 40 years or older.
Genotyping and Quality Control
A total of 4,996 Latinos were genotyped through LALES and the Mexican American Glaucoma Genetic Study (MAGGS) using either the Illumina OmniExpress BeadChip Kit (730,522 markers; Illumina, Inc., San Diego, CA) or the Illumina Hispanic/SOL BeadChip (~2.5 million markers; Illumina, Inc., San Diego, CA). The software Illumina GenomeStudio (v2011.1; Illumina, Inc.) was used to call single nucleotide polymorphisms (SNPs). Genotype quality control and imputation procedures have been described elsewhere.11, 17 Briefly, PLINK (v1.90) was used to perform quality control on the genotype data.18 Additionally, study participants with a genotyping call rate < 97%, inconsistencies between reported and genetically inferred sex, missing VCDR measurements, and duplicates were excluded. Haplotype phasing was conducted using SHAPEIT2,19 and imputation was performed using Minimac320 and the 1000 Genomes Project reference panels.17 Imputed SNPs of low-quality (i.e., Rsq < 0.80) and those with a minor allele frequency < 1% were excluded. After applying these quality control parameters, 4,018 unrelated subjects and over 6.8 million SNPs remained for downstream analysis.
SNP Selection and Construction of Genetic Risk Scores
Unweighted and weighted GRS were constructed based on SNPs previously associated with VCDR.9–12 Risk alleles were defined as alleles associated with an increase in VCDR. If a SNP was reported in multiple studies, the weight from the largest study sample was used. To ensure the weights for SNPs are on the same scale, SNPs from studies using untransformed VCDR values were retained. Additionally, all SNPs, except for rs2159128 (imputation Rsq = 0.67), were well-imputed. This resulted in 68 SNPs to be used for the construction of the GRS. Using a previous candidate gene approach,21 we also constructed unweighted and weighted GRS based on the lead SNP (most significant SNP) from our GWAS results within ± 100kb of the 68 previously reported SNPs. Moreover, unweighted and weighted GRS were generated from our previous genome-wide association data using all independent SNPs (PLINK pruned at r2 = 0.2) with P < 1 × 10−3.11 The unweighted GRS was calculated as the summation of the number of risk alleles across the genetic variants under the assumption that all risk alleles have the same effect. The weighted GRS were constructed by multiplying the VCDR-increasing allele by the effect size as reported in the corresponding study and summing these values together.
Statistical Analysis
Univariate analyses were performed to describe the characteristics of the study sample. Clinical variables (i.e., age, gender, BMI, systolic blood pressure (SBP), central corneal thickness (CCT), IOP, and T2D), potential environmental and socioeconomic confounders (i.e., smoking status, education, and income), and GRS were included in this analysis. Simple and multiple linear regression analyses were conducted to evaluate the association between VCDR and these variables. Since the raw VCDR values were not normally distributed in our dataset, inverse normal transformed VCDR was used for all analyses. Stepwise selection was applied to retain significant covariates at a significance cutoff of P ≤ 0.05. Additional variance of VCDR explained by the GRS were also examined.
To investigate the relationship between GRS and POAG, logistic regression analyses were performed. We created quintiles of the unweighted and weighted GRS to compare individuals with low GRS to individuals with higher GRS on the odds of POAG. Stepwise selection was conducted to retain significant covariates at a significance cutoff of P ≤ 0.05. Receiver operating characteristic (ROC) curve analyses were conducted and the area under the curve (AUC) were calculated to examine the improvement in the discriminatory ability of POAG when the GRS are added into a model with traditional risk factors. All statistical analyses were performed using SAS v9.4 (SAS Inc, Cary, NC) and R v3.3.22
Results
Table 1 presents a summary of the study sample characteristics and simple linear regression results between VCDR and the variables included in this study. The mean (standard deviation, SD) for the untransformed VCDR of the study sample is 0.3 (0.2). Among the study participants, 42.5% are males, 25.0% are diabetics, and 5.7% have POAG. The mean (SD) of age, CCT, IOP, and weighted GRS is 56.7 (10.4) years, 550.1 (33.7) µm, 14.7 (3.0) mmHg, and 0.8 (0.1), respectively. During univariate linear regression, numerous variables are significantly associated with VCDR, including age (P < 0.0001), gender (P = 0.0022), SBP (P < 0.0001), IOP (P < 0.0001), T2D (P = 0.0052), income (P = 0.0479), POAG status (P < 0.0001), and weighted GRS (P < 0.0001). Additionally, BMI, CCT, smoking status, and education are not associated with VCDR.
Table 1.
Summary statistics and simple linear regression results
| Characteristic | Participants (n = 4,018) | P |
|---|---|---|
| VCDR | 0.3 (0.2) | - |
| Age, year | 56.7 (10.4) | <0.0001 |
| Gender, male | 42.5% | 0.0022 |
| BMI, kg/m2 | 30.9 (5.5) | 0.0798 |
| SBP, mmHg | 123.8 (19.1) | <0.0001 |
| CCT, µm | 550.1 (33.7) | 0.8259 |
| IOP, mmHg | 14.7 (3.0) | <0.0001 |
| T2D, yes | 25.0% | 0.0052 |
| Smoking status | 0.9565 | |
| Never | 60.7% | |
| Former | 25.4% | |
| Current | 13.9% | |
| Education, yr | 0.0785 | |
| ≤ 6 | 44.6% | |
| 7–11 | 21.9% | |
| ≥ 12 | 33.4% | |
| IncomeŦ | 0.0479 | |
| < $20,000 | 50.0% | |
| $20,000–$40,000 | 35.9% | |
| > $40,000 | 14.1% | |
| POAG, % | <0.0001 | |
| Cases | 5.7% | |
| Controls | 94.3% | |
| Weighted GRS | 0.8 (0.1) | <0.0001 |
Abbreviations: VCDR, vertical cup-disc ratio; BMI, body mass index; SBP, systolic blood pressure; CCT, central corneal thickness; IOP, intraocular pressure; T2D, type 2 diabetes; POAG, primary open angle glaucoma; GRS, genetic risk score.
Mean and standard deviation are shown for all statistics except for gender, type 2 diabetes, smoking status, education, income, and primary open angle, which are shown as percentages.
Data missing for 513 study participants.
Table 2 displays the SNPs and weights used to construct the GRS from previously reported SNPs. Table 3 presents the multiple linear regression results from model building. With all of the possible risk factors entered into a full linear regression model, only age (P < 0.0001), gender (P = 0.0016), CCT (P = 0.0154), IOP (P < 0.0001), education (P = 0.0106), and the weighted GRS (P < 0.0001) remained in the model at a significance cutoff of P ≤ 0.05 during stepwise selection. The base multiple linear regression model including age, gender, CCT, IOP, and education accounts for 4.30% of the total variance for VCDR. An additional 2.74% of the variance of VCDR is explained by the weighted GRS, yielding a total of 7.04% explained by the model. The unweighted GRS yielded similar results (β = 0.02, P < 0.0001, 2.60% additional variance explained).
Table 2.
Previously reported single nucleotide polymorphisms included in genetic risk scores for vertical cup-disc ratio
| Chr | Nearest Gene | SNP | Alleles | VCDR- increasing allele |
Frequency of VCDR-increasing allele |
Weight | Reference |
|---|---|---|---|---|---|---|---|
| 1 | RERE | rs301801 | T/C | C | 0.21 | 0.008 | 10 |
| 1 | RERE | rs12025126 | T/C | T | 0.59 | 0.011 | 9 |
| 1 | RPE65 | rs1925953 | A/T | T | 0.68 | 0.006 | 12 |
| 1 | CDC7/TGFBR3 | rs1192414 | A/G | A | 0.28 | 0.014 | 12 |
| 1 | CDC7/TGFBR3 | rs4658101 | A/G | A | 0.29 | 0.013 | 12 |
| 1 | F5 | rs10753787 | T/C | C | 0.75 | 0.007 | 12 |
| 3 | FLNB | rs6764184 | G/T | T | 0.29 | 0.007 | 12 |
| 3 | COL8A1 | rs2623325 | C/A | A | 0.23 | 0.016 | 10 |
| 3 | COL8A1 | rs6804624 | T/C | C | 0.37 | 0.008 | 12 |
| 3 | COL8A1 | rs1997404 | T/G | G | 0.26 | 0.008 | 12 |
| 5 | PDZD2 | rs72759609 | T/C | T | 0.91 | 0.012 | 12 |
| 5 | VCAN | rs7717697 | T/C | T | 0.64 | 0.007 | 12 |
| 5 | DUSP1 | rs17658229 | T/C | T | 0.99 | 0.02 | 10 |
| 5 | DUSP1 | rs114503346 | C/T | C | 0.99 | 0.021 | 12 |
| 5 | DUSP1 | rs35084382 | T/C | T | 0.99 | 0.018 | 12 |
| 6 | EXOC2 | rs17756712 | A/G | G | 0.18 | 0.01 | 10 |
| 6 | RREB1 | rs4960295 | G/A | A | 0.51 | 0.007 | 12 |
| 6 | HSF2 | rs868153 | T/G | T | 0.74 | 0.007 | 10 |
| 7 | DGKB | rs10274998 | C/T | T | 0.41 | 0.008 | 12 |
| 8 | CRISPLD1 | rs117598310 | G/T | T | 0.05 | 0.009 | 12 |
| 8 | PSCA | rs2920293 | C/G | C | 0.47 | 0.006 | 12 |
| 9 | CDKN2B | rs1063192 | G/A | A | 0.81 | 0.014 | 9 |
| 9 | CDKN2BAS | rs7865618 | G/A | A | 0.81 | 0.013 | 10 |
| 9 | CDKN2B/AS1 | rs2157719 | C/T | T | 0.81 | 0.013 | 12 |
| 9 | CDKN2B/AS1 | rs1360589 | C/T | T | 0.82 | 0.013 | 12 |
| 10 | ATOH7 | rs7916697 | A/G | G | 0.62 | 0.018 | 12 |
| 10 | ATOH7 | rs7916410 | T/C | C | 0.64 | 0.018 | 12 |
| 10 | ATOH7 | rs1900005 | A/C | C | 0.64 | 0.018 | 10 |
| 10 | ATOH7/PBLD | rs1900004 | C/T | C | 0.64 | 0.013 | 9 |
| 10 | PLCE1 | rs3891783 | C/G | G | 0.37 | 0.007 | 12 |
| 10 | PLCE1 | rs1830890 | A/G | G | 0.28 | 0.006 | 12 |
| 10 | PLCE1 | rs7072574 | G/A | A | 0.28 | 0.009 | 10 |
| 10 | ENO4 | rs1681739 | C/T | T | 0.31 | 0.006 | 12 |
| 11 | SCYL1 | rs17146964 | A/G | A | 0.86 | 0.014 | 9 |
| 11 | SSSCA1 | rs1346 | A/T | A | 0.86 | 0.013 | 12 |
| 11 | ADAMTS8 | rs4936099 | C/A | A | 0.71 | 0.007 | 12 |
| 12 | RPAP3 | rs11168187 | A/G | A | 0.87 | 0.009 | 10 |
| 12 | TMTC2 | rs10862688 | A/G | G | 0.25 | 0.008 | 10 |
| 12 | TMTC2 | rs442376 | T/C | C | 0.60 | 0.011 | 12 |
| 12 | TMTC2 | rs482507 | C/T | T | 0.59 | 0.011 | 12 |
| 12 | TMTC2 | rs324780 | G/A | A | 0.61 | 0.011 | 12 |
| 12 | FAM101A | rs7311936 | G/C | G | 0.67 | 0.006 | 12 |
| 13 | DCLK1 | rs7323428 | G/T | T | 0.31 | 0.007 | 12 |
| 13 | DCLK1 | rs1926320 | T/C | C | 0.32 | 0.012 | 9 |
| 14 | SIX1/6 | rs4901977 | C/T | T | 0.25 | 0.011 | 10 |
| 14 | SIX6 | rs4436712 | G/T | T | 0.34 | 0.009 | 12 |
| 14 | SIX6 | rs8015152 | C/T | T | 0.27 | 0.01 | 12 |
| 14 | SIX1 | rs10483727 | T/C | T | 0.35 | 0.012 | 9 |
| 14 | SIX6 | rs34935520 | G/A | G | 0.35 | 0.009 | 12 |
| 15 | FAM169B | rs6598351 | C/T | T | 0.12 | 0.006 | 12 |
| 15 | ASB7 | rs60779155 | G/A | A | 0.34 | 0.01 | 12 |
| 15 | ASB7 | rs34222435 | C/T | T | 0.33 | 0.01 | 12 |
| 15 | ASB7 | rs4299136 | G/C | C | 0.33 | 0.01 | 12 |
| 16 | SALL1 | rs11646917 | G/T | G | 0.71 | 0.009 | 12 |
| 16 | SALL1 | rs4784295 | C/G | C | 0.18 | 0.009 | 12 |
| 16 | SALL1 | rs1345467 | G/A | G | 0.17 | 0.009 | 12 |
| 17 | BCAS3 | rs8068952 | G/C | C | 0.81 | 0.012 | 9 |
| 19 | ARID3A | rs2159128 | G/T | T | 0.14 | 0.019 | 9 |
| 20 | BMP2 | rs6054374 | C/T | C | 0.46 | 0.007 | 10 |
| 20 | BMP2 | rs6054375 | G/T | G | 0.47 | 0.01 | 12 |
| 20 | BMP2 | rs6107845 | G/A | G | 0.48 | 0.009 | 12 |
| 22 | CHEK2 | rs1547014 | T/C | C | 0.67 | 0.013 | 10 |
| 22 | CHEK2 | rs5762752 | C/G | G | 0.65 | 0.011 | 12 |
| 22 | CHEK2 | rs5752773 | G/C | C | 0.67 | 0.012 | 12 |
| 22 | CHEK2 | rs738722 | T/C | C | 0.69 | 0.012 | 12 |
| 22 | CARD10 | rs2092172 | G/A | A | 0.18 | 0.009 | 12 |
| 22 | CARD10 | rs56385951 | G/A | A | 0.09 | 0.011 | 12 |
| 22 | CARD10 | rs5756813 | G/T | G | 0.41 | 0.008 | 10 |
Table 3.
Multiple linear regression results
| Model 1Ŧ | Model 2 | |||
|---|---|---|---|---|
|
|
|
|||
| Characteristic | Beta | P | Beta | P |
| Age | 0.009 | <0.0001 | 0.01 | <0.0001 |
| Gender | 0.103 | 0.0033 | 0.096 | 0.0016 |
| BMI | −0.0045 | NS | - | - |
| SBP | −0.0002 | NS | - | - |
| CCT | −0.0013 | 0.0111 | −0.0011 | 0.0154 |
| IOP | 0.0533 | <0.0001 | 0.0519 | <0.0001 |
| T2D | 0.043 | NS | - | - |
| Smoking Status | −0.0133 | NS | - | - |
| Income | −0.0309 | NS | - | - |
| Education | 0.0417 | 0.0305 | 0.044 | 0.0106 |
| Weighted GRS | 1.767 | <0.0001 | 1.831 | <0.0001 |
Abbreviations: BMI, body mass index; SBP, systolic blood pressure; CCT, central corneal thickness; IOP, intraocular pressure; T2D, type 2 diabetes; GRS, genetic risk score.
Data missing for 513 study participants.
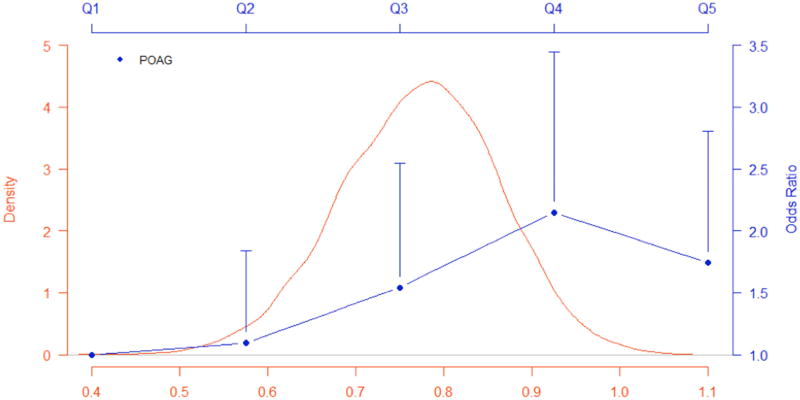
Multiple logistic regression analyses using quintiles of weighted GRS evaluated the association of the weighted GRS on POAG. After performing stepwise regression, age (P < 0.0001), gender (P = 0.0318), CCT (P = 0.0015), IOP (P < 0.0001), SBP (P = 0.0408), and the weighted GRS (P = 0.0011) remained significantly associated with POAG. Figure 1 shows the distribution of the weighted GRS in the study sample and odds ratios of POAG comparing each of the upper GRS quintiles with the lowest, adjusting for age, gender, CCT, IOP, and SBP. Compared to the lowest quintile, both the highest and second highest quintiles had significantly higher odds of POAG, OR = 1.75 (95% CI: [1.09, 2.81]; P = 0.0212) and OR = 2.15 (95% CI: [1.34, 3.45]; P = 0.0015), respectively. Analysis of the unweighted GRS yielded similar estimates and significance levels. The highest and second highest quintiles of the unweighted GRS had significantly higher odds of POAG compared to the lowest quintile, OR = 2.00 (95% CI: [1.24, 3.22]; P = 0.0042) and OR = 1.91 (95% CI: [1.18, 3.10]; P = 0.0087), respectively.
Figure 1. Distribution of weighted genetic risk score from previously reported SNPs and association with primary open angle glaucoma.
Distribution of the weighted GRS and odds ratios of POAG comparing each of the four upper GRS quintiles to the lowest quintile, adjusting for age, gender, CCT, IOP, and SBP. Vertical lines of each point (OR) represents the upper 95% confidence interval.
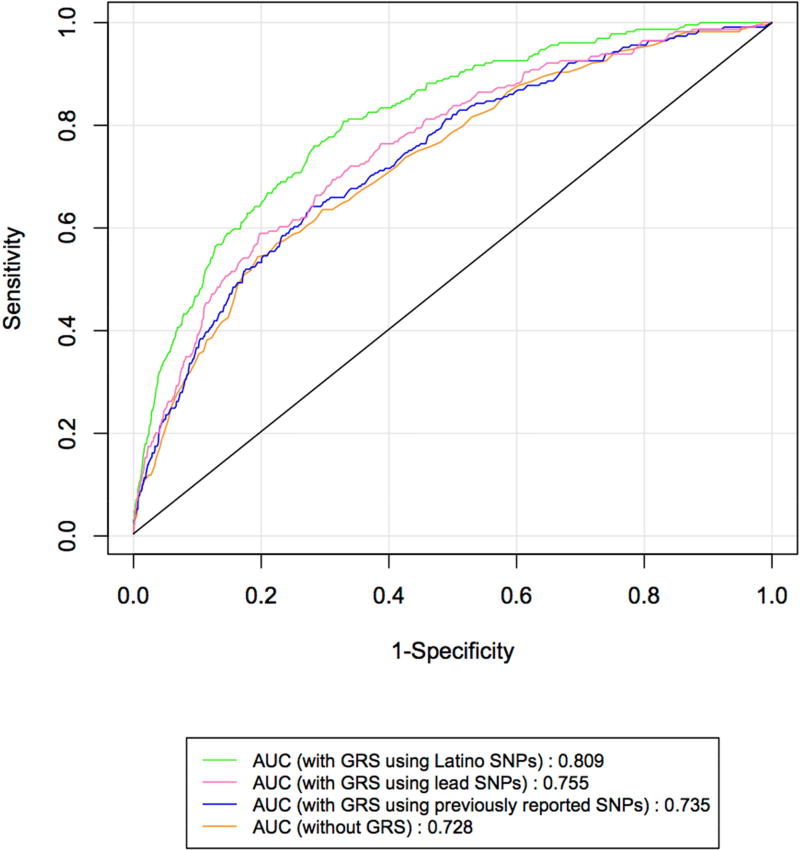
We conducted ROC analyses to examine the discriminatory power of the unweighted and weighted GRS on POAG status. Figure 2 presents the ROC curves for models without and with the weighted GRS constructed from previously reported VCDR SNPs, the weighted GRS generated from lead SNPs, and the weighted GRS derived from our genome-wide association data. Supplementary Table 1 presents the SNPs and corresponding summary statistics used for the construction of the Latino specific GRS. The AUC is 0.728 (95% CI: [0.694, 0.761]) for the model with only age and gender. When the weighted GRS from previously reported SNPs was added into the model, there was a non-significant increase in the AUC to 0.735 (95% CI: [0.701, 0.768]; P = 0.150). When the weighted GRS using the lead SNPs was added to the model, there was a significant increase in the AUC to 0.755 (95% CI: [0.722, 0.787]; P = 0.002). In contrast, the addition of the GRS derived from our own genome-wide association data resulted in a significant increase in the AUC to 0.809 (95% CI: [0.781, 0.837]; P < 0.0001). Similar associations and significance levels were obtained for the unweighted GRS for these analyses (Supplementary Figure 1).
Figure 2. Receiver operating characteristic curves predicting primary open angle glaucoma for weighted GRS.
The curves are based on logistic regression models adjusting for age and gender without and with the weighted GRS constructed from previously reported VCDR SNPs, the weighted GRS generated from lead SNPs, and the weighted GRS derived from our genome-wide association data. AUC represents the area under the curve, with a larger AUC representing better classification of POAG status. The addition of weighted GRS derived from our own genome-wide association data significantly improved the discriminatory ability for POAG (P < 0.0001).
Discussion
In this study, we constructed genetic risk scores based on SNPs previously associated with VCDR and evaluated whether these GRS were associated with VCDR and POAG and increased the discriminatory ability for POAG. We observed significant associations between the GRS and VCDR, indicating a higher GRS was associated with a larger vertical cup-disc ratio. These associations remained significant after the inclusion of traditional risk factors, explaining an additional 2.74% of the variation in VCDR. Moreover, compared to the lowest quintile of the GRS, study participants in the highest two quintiles experienced significantly higher odds of POAG. We show the inclusion of ethnic specific GRS significantly increased the discriminatory power for POAG. Additionally, we obtained similar results for the unweighted GRS. To our knowledge, we are the first to report these associations in a Latino population.
The success of genome-wide association studies in identifying genetic variants indicates that multiple genetic loci, rather than a single gene, contribute to the susceptibility of a given phenotype. Despite the modest effect of individual variants, creating an aggregated score allows for the evaluation of the combined genetic effect of these variants on a trait. The utility of GRS in the fields of public health and medicine has the potential to significantly reduce the incidence of disease by being used as a screening tool to identify individuals at a greater risk of a disorder. GRS can be used to identify subgroups in a population that are at a higher risk for a disorder, so targeted public health interventions can be directed towards them. In a similar manner, GRS aids in the movement towards personalized medicine. By assessing an individual’s GRS before the development of disease, early interventions (e.g., dietary, behavioral, etc.) can be implemented to counterbalance the genetic risk.23 En masse, GRS provide an opportunity to be a useful tool in summarizing an individual’s genetic susceptibility to a trait and may be potentially used for reducing the occurrence of disease.
POAG is a heterogeneous disease, both genetically and phenotypically. As such, investigating quantitative traits and the corresponding genetic variants will aid in understanding the biological mechanisms underlying this disease. We observed significant associations between the GRS and POAG, with higher GRS associated with greater odds of POAG. To further examine the utility of GRS, we performed several ROC analyses to evaluate whether the inclusion of the GRS improved POAG discriminatory ability. We observed a moderate increase in the AUC after including the GRS with traditional risk factors, although the increase was minor, potentially limiting the utility of such genetic risk scores in a clinical setting. A study conducted in a multiethnic Asian population observed a borderline significant improvement in the discriminatory ability for glaucoma when IOP and VCDR GRS were included into a model with traditional risk factors.21 Specifically, the AUC estimate for POAG exhibited a modest improvement when the IOP and VCDR GRS were included with traditional risk factors, increasing from 0.72 to 0.74 (AUC difference = 0.02; P = 0.06).21 In our study of Latinos, we observed similar AUC estimates for both the unweighted and weighted GRS, demonstrating the consistent modest improvement in AUC from previously reported SNPs. Furthermore, using GRS constructed from our own genome-wide association data, we observed significant increases in the AUC with the addition of more SNPs.11 This suggests additional genetic variants, besides those previously reported, with low effect estimates further aid in the discriminating ability for POAG by incorporating additional genetic information into the model. Moreover, ethnic specific weights for the construction of GRS may further aid in improving disease prediction. Together, despite the marginal increases in AUC in both the current and previous studies using published genetic variants, these findings suggest GRS constructed from quantitative traits of POAG can aid in increasing the discriminatory ability for this disease, including variants with lower effect estimates. Moreover, due to the polygenetic nature of POAG, further identification of genetic variants associated with the pathogenesis of POAG may aid in improving the predictive power, and clinical utility, of GRS.
The strengths of this study include the generation of GRS consisting of VCDR SNPs identified to date. Also, we observed significant associations with GRS constructed from SNPs identified primarily in European populations in our study sample consisting of Latinos and as such, these results may be generalizable to other ethnic populations. There are several limitations however. First, we used previously reported genetic variants identified in GWAS thus far, which explain only a small amount of variation in VCDR, which resulted in a moderate improvement in POAG prediction. The GRS used were constructed from a limited number of genetic variants from European and Asian populations, and may not be transferable to other racial groups. Unweighted GRS, however, are preferred to weighted GRS when the existing studies are comprised of different ethnicities compared to the population under study.24 We obtained similar results for both the weighted and unweighted GRS, demonstrating the robustness of our results and the potential transferability of a GRS for VCDR. Additionally, the GRS were constructed based on SNPs identified using traditional GWAS significance thresholds, which may not have captured variants with weaker effect sizes. When we constructed GRS using a larger number of variants weighted by Latino specific estimates, we observed a better classification of POAG, suggesting that increasing the number of SNPs and applying population specific weights can lead to better predictions for POAG.
In summary, we observed GRS composed of 68 previously reported VCDR SNPs were significantly associated with VCDR in a Latino population. Moreover, the GRS were significantly associated with POAG, with individuals with higher GRS experiencing greater odds of POAG. Inclusion of ethnic specific GRS constructed using a larger number of SNPs significantly improved the discriminatory ability for POAG. The application of GRS as a population based evaluation tool can potentially yield significant reductions in disease incidence. By quantifying an individual’s genetic risk before disease development, early interventions can be adopted to counterbalance this genetic risk.
Supplementary Material
Acknowledgments
The authors thank the study participants from LALES and the staff who aided in data collection and processing.
Funding
This research was supported in part by National Institutes of Health (NIH; Bethesda, MD) grants R01EY022651 (to X.G.), P30EY001792 (departmental core grant), and an unrestricted departmental grant from Research to Prevent Blindness. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations and Acronyms
- AUC
area under the curve
- BMI
body mass index
- CCT
central corneal thickness
- GRS
genetic risk score
- GWAS
genome-wide association study
- IOP
intraocular pressure
- LALES
Los Angeles Latino Eye Study
- MAGGS
Mexican American Glaucoma Genetic Study
- POAG
primary open angle glaucoma
- ROC
receiver operating characteristic
- SBP
systolic blood pressure
- SNP
single nucleotide polymorphism
- T2D
type 2 diabetes
- VCDR
vertical cup-disc ratio
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
No conflict of interest to declare.
Xiaoyi Gao is the study chair for the Mexican American Glaucoma Genetic Study.
This article contains additional online-only material. The following should appear online-only: Supplementary Figure 1 and Supplementary Table 1.
We are the first to report the association between GRS and VCDR, and its improvement in the discriminatory ability of POAG, in a Latino population.
References
- 1.Kim YJ, Kim JM, Shim SH, et al. Associations between Optic Cup-to-disc Ratio and Systemic Factors in the Healthy Korean Population. Korean J Ophthalmol. 2015;29(5):336–43. doi: 10.3341/kjo.2015.29.5.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amerasinghe N, Wong TY, Wong WL, et al. Determinants of the optic cup to disc ratio in an Asian population: the Singapore Malay Eye Study (SiMES) Arch Ophthalmol. 2008;126(8):1101–8. doi: 10.1001/archopht.126.8.1101. [DOI] [PubMed] [Google Scholar]
- 3.Quigley HA, West SK, Rodriguez J, et al. The prevalence of glaucoma in a population-based study of Hispanic subjects: Proyecto VER. Arch Ophthalmol. 2001;119(12):1819–26. doi: 10.1001/archopht.119.12.1819. [DOI] [PubMed] [Google Scholar]
- 4.Healey PR, Mitchell P, Smith W, Wang JJ. The influence of age and intraocular pressure on the optic cup in a normal population. J Glaucoma. 1997;6(5):274–8. [PubMed] [Google Scholar]
- 5.Ramrattan RS, Wolfs RC, Jonas JB, et al. Determinants of optic disc characteristics in a general population: The Rotterdam Study. Ophthalmology. 1999;106(8):1588–96. doi: 10.1016/S0161-6420(99)90457-8. [DOI] [PubMed] [Google Scholar]
- 6.Varma R, Tielsch JM, Quigley HA, et al. Race-, age-, gender-, and refractive error-related differences in the normal optic disc. Arch Ophthalmol. 1994;112(8):1068–76. doi: 10.1001/archopht.1994.01090200074026. [DOI] [PubMed] [Google Scholar]
- 7.Klein BE, Klein R, Lee KE. Heritability of risk factors for primary open-angle glaucoma: the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 2004;45(1):59–62. doi: 10.1167/iovs.03-0516. [DOI] [PubMed] [Google Scholar]
- 8.Charlesworth J, Kramer PL, Dyer T, et al. The path to open-angle glaucoma gene discovery: endophenotypic status of intraocular pressure, cup-to-disc ratio, and central corneal thickness. Invest Ophthalmol Vis Sci. 2010;51(7):3509–14. doi: 10.1167/iovs.09-4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramdas WD, van Koolwijk LM, Ikram MK, et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010;6(6):e1000978. doi: 10.1371/journal.pgen.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Springelkamp H, Hohn R, Mishra A, et al. Meta-analysis of genome-wide association studies identifies novel loci that influence cupping and the glaucomatous process. Nat Commun. 2014;5:4883. doi: 10.1038/ncomms5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nannini DR, Torres M, Chen YI, et al. A Genome-Wide Association Study of Vertical Cup-Disc Ratio in a Latino Population. Invest Ophthalmol Vis Sci. 2017;58(1):87–95. doi: 10.1167/iovs.16-19891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Springelkamp H, Iglesias AI, Mishra A, et al. New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum Mol Genet. 2017;26(2):438–53. doi: 10.1093/hmg/ddw399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramdas WD, Amin N, van Koolwijk LM, et al. Genetic architecture of open angle glaucoma and related determinants. J Med Genet. 2011;48(3):190–6. doi: 10.1136/jmg.2010.083337. [DOI] [PubMed] [Google Scholar]
- 14.Varma R, Ying-Lai M, Francis BA, et al. Prevalence of open-angle glaucoma and ocular hypertension in Latinos: the Los Angeles Latino Eye Study. Ophthalmology. 2004;111(8):1439–48. doi: 10.1016/j.ophtha.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 15.Gao X, Nannini D, Torres M, Varma R. The Association for Research in Vision and Ophthalmology Annual Meeting. Baltimore, MD: 2017. European ancestry is associated with lower risk of primary open angle glaucoma in Latinos. [Google Scholar]
- 16.Varma R, Paz SH, Azen SP, et al. The Los Angeles Latino Eye Study: design, methods, and baseline data. Ophthalmology. 2004;111(6):1121–31. doi: 10.1016/j.ophtha.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Gao X, Nannini DR, Corrao K, et al. Genome-wide association study identifies WNT7B as a novel locus for central corneal thickness in Latinos. Hum Mol Genet. 2016;25(22):5035–45. doi: 10.1093/hmg/ddw319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CC, Chow CC, Tellier LC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delaneau O, Marchini J Genomes Project C, Genomes Project C. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das S, Forer L, Schonherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–7. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tham YC, Liao J, Vithana EN, et al. Aggregate Effects of Intraocular Pressure and Cup-to-Disc Ratio Genetic Variants on Glaucoma in a Multiethnic Asian Population. Ophthalmology. 2015;122(6):1149–57. doi: 10.1016/j.ophtha.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 22.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 23.Warren HR, Evangelou E, Cabrera CP, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49(3):403–15. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith JA, Ware EB, Middha P, et al. Current Applications of Genetic Risk Scores to Cardiovascular Outcomes and Subclinical Phenotypes. Curr Epidemiol Rep. 2015;2(3):180–90. doi: 10.1007/s40471-015-0046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.