Abstract
The soil microbiome can produce, resist, or degrade antibiotics and even catabolize them. While resistance genes are widely distributed in the soil, there is a dearth of knowledge concerning antibiotic catabolism. Here we describe a pathway for penicillin catabolism in four isolates. Genomic and transcriptomic sequencing revealed β-lactamase, amidase, and phenylacetic acid catabolon up-regulation. Knocking out part of the phenylacetic acid catabolon or an apparent penicillin utilization operon (put) resulted in loss of penicillin catabolism in one isolate. A hydrolase from the put operon was found to degrade in vitro benzylpenicilloic acid, the β-lactamase penicillin product. To test the generality of this strategy, an E. coli strain was engineered to co-express a β-lactamase and a penicillin amidase or the put operon, enabling it to grow using penicillin or benzylpenicilloic acid, respectively. Elucidation of additional pathways may allow for bioremediation of antibiotic-contaminated soils and discovery of antibiotic-remodeling enzymes with industrial utility.
Introduction
The discovery of antibiotics and their development into an armamentarium against bacterial infections has been one of the great public health success stories of the last century. However, increasing antibiotic resistance in pathogenic bacteria with concomitant decreasing development of new antibiotics threatens a return to the dark ages of the pre-antibiotic era1. Bacterial resistance to antibiotics is ancient2 and ubiquitous in the environment3,4. Moreover, anthropogenic antibiotic use has led to a measurable increase in carriage of antibiotic resistance genes in the environment with the potential to spread to the clinic5.
The ultimate fate of antibiotics in the environment, and what role resistance plays in their mineralization, is unknown. Most antibiotics are natural products, or derivatives thereof, originally isolated from soil bacteria6. Given their soil origin, and the lack of environmental accumulation of these organic compounds, it is natural that some antibiotics are consumed by soil bacteria as carbon or nitrogen sources. This was recognized soon after the mass anthropogenic introduction of antibiotics by studies demonstrating the ability of soil bacteria to mineralize various natural antibiotics including streptomycin7, penicillin G (also known as benzylpenicillin, referred to hereafter simply as penicillin)8, and chloramphenicol9. In the literature, utilization of penicillin has most often focused on Pseudomonas strains, with conflicting evidence for what part of the molecule is used as a carbon source10–12, although catabolism of penicillins in other organisms, such as Klebsiella pneumoniae, has been reported as well13. Little is known about the pathways and enzymes utilized during catabolism, including whether β-lactamase activity is required8,11,14. More recently, the list of antibiotics capable of sustaining bacterial growth has expanded substantially, as has the catalog of bacterial species capable of subsisting on antibiotics14–19. However, controversy still remains over the characterization of resistant, but not metabolizing, versus subsistent growth phenotypes. To date, no specific genes or pathways have been identified that enable bacteria to use antibiotics as a sole carbon source16,20.
Here we provide evidence for a pathway for β-lactam antibiotic catabolism in which amidases, found to be distinct from known penicillin amidase enzymes, link resistance enzymes to central metabolism. We demonstrate that this strategy can be transferred to E. coli, enabling it to grow using penicillin as its sole carbon source. These findings have important implications for antibiotic ecology, bioremediation of antibiotic contaminated sites, and the synthesis of semi-synthetic antibiotics.
Results
Proteobacteria using β-lactams as a sole carbon source
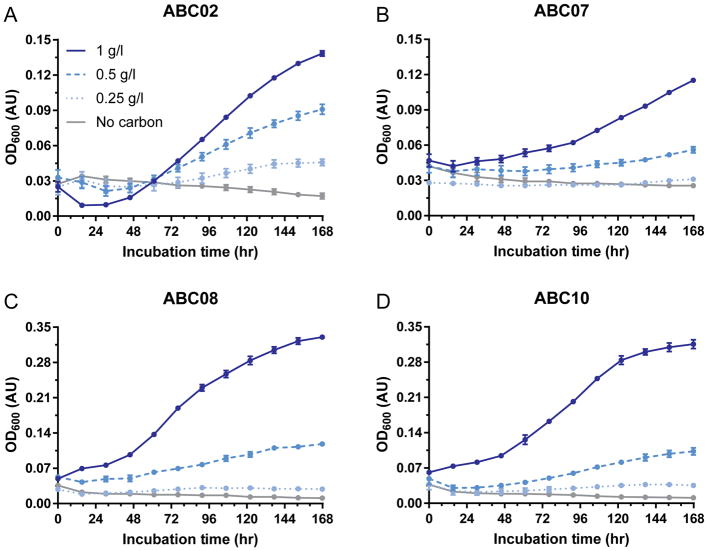
Four Proteobacteria soil isolates of the Burkholderia, Pseudomonas, and Pandoreae genera, termed ABC02, ABC07, ABC08, and ABC10 (Supplementary Table 1; ABC stands for antibiotic catabolizer) were previously isolated from soil by growth in minimal media with penicillin or carbenicillin as carbon sources and were found to be extensively antibiotic-resistant15. To confirm the stability of the antibiotic catabolism phenotypes after extended storage at −80°C, and to control for the unlikely possibility that growth in previous studies was affected by trace amounts of EDTA, we cultured each strain with aeration in M9 media containing 0 g/l, 0.25 g/l, 0.5 g/l, or 1 g/l penicillin as sole carbon source at 25°C and measured culture density over the course of one week (Fig. 1). Each strain grew in a dose-dependent manner, with 1 g/l penicillin supporting the most robust growth.
Figure 1. ABC strains catabolize penicillin as their sole carbon source.
a–d, Growth of strains ABC02 (a), ABC07 (b), ABC08 (c), and ABC10 (d) in minimal media with penicillin as sole carbon source. Figure legend for penicillin carbon source concentration in a applies to all panels. Data points are average of three experiments with SEM error bars.
ABC strains share penicillin-responsive gene regulation
In order to elucidate the enzymes and pathways utilized during penicillin catabolism, we annotated draft genomes21 from all four β-lactam catabolizing strains. In all four genomes we annotated multiple chromosomal β-lactamase genes, including genes annotated as belonging to class A, class C, and class D β-lactamase families (Supplementary Table 2), as well as complete or near-complete phenylacetic acid catabolic pathways22,23. The four ABC strains are able to grow in rich media in the presence of high concentrations (1 g/l) of a variety of β-lactam antibiotics including penicillins (all four ABC strains), cephalosporins (ABC07, ABC08, and ABC10), monobactams (ABC02, ABC08, and ABC10), and, to a limited extent, carbapenems (ABC07, ABC08, and ABC10) (Supplementary Fig. 1).
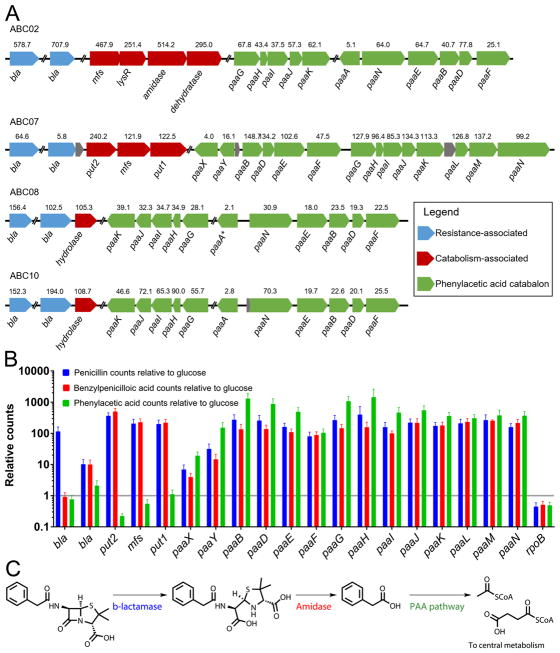
These findings led us to hypothesize that catabolism of penicillin proceeds through canonical hydrolysis of the β-lactam ring to produce benzylpenicilloic acid24,25 which is then processed downstream to central metabolism. We tested this hypothesis using a comparative transcriptomic approach, wherein the four ABC strains were independently cultured in minimal media with a variety of single carbon sources, and their whole-cell gene expression levels were measured and compared under these conditions via RNA-seq. In order to choose substrates with known paths to central metabolism, we initially tested the ability of each of the four strains to utilize 190 distinct carbon sources in a high-throughput phenotypic microarray. Each strain grew on a unique subset of carbon sources (Supplementary Fig. 2), and each strain was cultured with penicillin or a carbon source that feeds directly into central metabolism (glucose for ABC07 and histidine for ABC02, ABC08, and ABC10). Following RNA-seq, we analyzed the transcriptome for differentially expressed genes under these two conditions (see Supplementary Dataset 1 for counts and heat map representation of data). Remarkably, the RNA-seq data suggest that all four strains may utilize a conserved strategy for antibiotic catabolism, consisting of up-regulation of Ambler class A, C, and D β-lactamases23 (Supplementary Table 3), amidases (syntenic with a β-lactamase in strains ABC07, ABC08, and ABC10), and genes involved in phenylacetic acid utilization (Fig. 2a). Interestingly, in the phenotypic microarray assay, all four strains showed robust growth on aromatic substrates, especially phenylalanine and derivatives of phenylacetic acid (Supplementary Fig. 2).
Figure 2. Evidence for shared strategy for penicillin catabolism among ABC strains.
a, Up-regulated open reading frames during growth on penicillin compared to histidine (all but ABC07) or glucose (ABC07) calculated from triplicate cultures. Displayed above each ORF is the relative fold up-regulation while below is the gene name (e.g. bla, β-lactamase; mfs, major facilitator superfamily pump). All genes displayed showed significant up-regulation at adjusted p-value<0.00001 (DESeq) except for paaA of ABC08. b, ABC07 transcriptional response to penicillin, benzylpenicilloic acid, and phenylacetic acid relative to glucose. Relative transcript counts of ORFs identified to be responsive to penicillin are displayed as averages of triplicate RNASeq experiments with SEM error bars. The housekeeping RNA polymerase gene rpoB is present for comparison. c, Hypothesized pathway for penicillin degradation. Penicillin is neutralized by a β-lactamase to produce benzylpenicilloic acid, which acts as substrate for amidases or other hydrolases, releasing phenylacetic acid which is routed to central metabolism as a carbon source by the phenylacetic acid catabolon.
Notably, the side chain of penicillin (the non-β-lactam region) consists of phenylacetamide, suggesting that growth on penicillin in the ABC strains might proceed through metabolism of this part of the molecule. In order to test this proposal, we performed additional RNA-seq experiments with the Pseudomonas strain ABC07 grown on benzylpenicilloic acid (the product of β-lactamase cleavage of penicillin) or phenylacetic acid as sole carbon sources, in addition to penicillin and glucose as before (Fig. 2b). These data reveal an apparent transcriptional architecture, beginning with a β-lactamase preferentially up-regulated in response to penicillin (compared to glucose: ~65-fold with adjusted p-value 1.6×10−91). This is followed by expression of a putative penicillin utilization operon (put) consisting of four ORFs that appear to be responsive to both penicillin and benzylpenicilloic acid, but not phenylacetic acid: a β-lactamase (~6-fold with adjusted p value 2.2×10−8 compared to glucose), a major facilitator family importer (~122-fold with adjusted p value 2.5×10−128 compared to glucose), and two amidases (termed put1 and put2 here; respectively ~122-fold with adjusted p value 1.3×10−131, and ~240-fold with adjusted p value 1.0×10−126 compared to glucose). Finally, the pathway is apparently completed by up-regulation of the phenylacetic acid catabolon (paa) in response to penicillin, benzylpenicilloic acid, and phenylacetic acid (Fig. 2b). This architecture suggests a conserved catabolic pathway consisting of the following steps: (i) detoxification of penicillin by hydrolysis of the β-lactam ring by a β-lactamase, a canonical β-lactam antibiotic resistance enzyme, (ii) import of the benzylpenicilloic acid product and/or hydrolysis of the amide bond to free the carbon-rich phenylacetic acid side chain, and (iii) processing of phenylacetic acid into acetyl- and succinyl-CoA via the phenylacetic acid catabolon (Fig. 2c).
Growth on penicillin requires paa and put operons
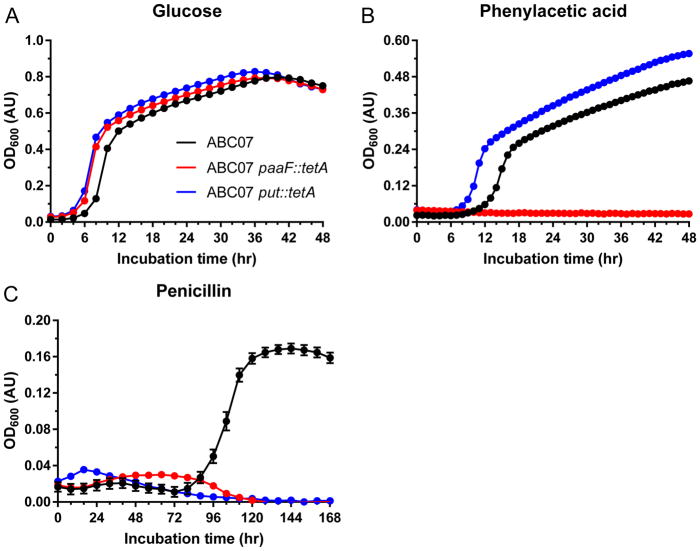
To determine the necessity of the phenylacetic acid catabolon and the put operon in strain ABC07 (Fig. 2), we constructed knock-out strains of ABC07 in which the paaF gene (phenylacetyl-CoA ligase, responsible for the first step of phenylacetic acid catabolism22) and the entire put operon (consisting of a β-lactamase, put2, mfs, and put1) were each replaced chromosomally26 with a tetA open reading frame. These strains, alongside wild-type ABC07, were assayed for their ability to grow in M9 media with 0.4% glucose, phenylacetic acid, or penicillin as their sole carbon sources. In support of our hypothesis, loss of paaF resulted in a strain that, while still able to grow well with glucose (Fig. 3a), was no longer able to sustain growth on phenylacetic acid (Figure 3b). More interestingly, the paaF strain also showed lack of growth using penicillin as sole carbon source, indicating that penicillin catabolism flows through phenylacetic acid (Fig. 3c). Contrasting this is the phenotype of the put knockout, which can grow using either glucose or phenylacetic acid as its carbon source (Fig. 3a,b), indicating little if any role for these four genes in central metabolism or growth on aromatic substrates. Notably though, loss of the put operon results in complete loss of penicillin catabolism (Fig. 3c), indicating the necessity of these genes, along with the paa catabolon, for the antibiotic catabolism phenotype.
Figure 3. paaF and the put operon are necessary for penicillin catabolism in ABC07.
a–c, Growth curves of wild-type ABC07 (black), paaF::tetA (red) and put::tetA (blue) strains in minimal media containing glucose (a), phenylacetic acid (b), or penicillin (c) as the sole carbon source. Measurements are the average of triplicate cultures with standard error of the mean displayed.
Catabolism of the phenylacetamide side chain
Because we found ABC07 to be resistant to a variety of additional β-lactam antibiotics at 1 g/l (Supplementary Fig. 1), we next tested the ability of these compounds, as well as penicillin degradation products, to support growth of ABC07 strains in M9 media as carbon sources at 1 g/l (Supplementary Fig. 3). Of the new carbon sources tested, only one, benzylpenicilloic acid, supported growth of ABC07, but neither the paaF nor the put mutants were capable of growth using it as sole carbon source (Supplementary Fig. 3b), mirroring our observations with penicillin (Fig. 3c and Supplementary Fig. 3a). Interestingly, while phenylacetic acid supports growth of ABC07 and the put strain (Fig. 3b and Supplementary Fig. 3c) the other half of the penicillin structure, the 6-aminopenicillanic acid β-lactam core, does not (Supplementary Fig. 3d), supporting the hypothesis that in ABC07 penicillin catabolism runs almost solely through the phenylacetamide side chain. Further reinforcing this is our observation that none of the ABC07 strains are capable of growth using other β-lactam antibiotics including other penicillin-class drugs (carbenicillin or ampicillin, Supplementary Fig. 3e,f) or the cephalosporin cefuroxime (Supplementary Fig. 3g).
The put operon includes a benzylpenicilloic acid amidase
The presence of amidases and hydrolases that are upregulated by penicillin and benzylpenicilloic acid may suggest the involvement of penicillin amidase (also known as penicillin acylase) enzymes from the N-terminal nucleophile hydrolase enzyme family that are used industrially to hydrolyze the phenylacetamide side chain of penicillin from the β-lactam ring to produce 6-aminopenicillanic acid in the manufacture of semi-synthetic β-lactam antibiotics27. We constructed a phylogenetic tree using the ABC strain amidase/hydrolase sequences, their nearest neighbors in NCBI’s NR database28, and canonical penicillin amidases (EC 3.5.11) downloaded from UniProt29 (Supplementary Fig. 4). The ABC proteins do not appear to cluster with the canonical penicillin amidases and instead group with amidases, amidohydrolases, or hydrolases of unknown specificity (Supplementary Fig. 4).
Because of their limited annotation, catabolic necessity in ABC07 (Fig. 3 and Supplementary Fig. 3), and transcriptional response to penicillin and benzylpenicilloic acid (Fig. 2b), we chose to study the gene products of put1 and put2 in greater detail following heterologous expression in E. coli and purification (Supplementary Fig. 5a). We assayed the resulting purified enzymes for amidase activity by incubation with two chromogenic amide substrates: 6-nitro-3-(phenylacetamido)-benzoic acid (NIPAB) and p-nitroacetanilide. Both substrates have previously been validated as model chromogenic substrates for penicillin30 (NIPAB) or generic amides31 (p-nitroacetanilide). For comparison, we also carried out reactions using commercially available E. coli penicillin amidase enzyme. We found Put1 and Put2 to be functionally distinct from penicillin amidase in their inability to hydrolyze the amide bond in NIPAB while showing activity with p-nitroacetanilide, with penicillin amidase demonstrating the opposite activities (Supplementary Fig. 5b,c). Notably, hydrolysis of the amide bonds in these molecules is functionally equivalent to the hydrolysis of the amide bonds found in penicillin or benzylpenicilloic acid that would result in release of phenylacetic acid.
We next compared the Michaelis-Menten dynamics of Put1, due to its apparent greater activity, and penicillin amidase across four additional substrates including an additional penicillin analog32 (Supplementary Fig. 5d), and three potential peptidase substrates (Supplementary Fig. 5e–g). These assays reveal limited functional overlap between E. coli penicillin amidase and Put1. We therefor undertook a more detailed bioinformatic examination of Put1 in comparison to penicillin amidases. Analysis of the amino acid sequence of Put1 suggests that it lacks a signal peptide found in many penicillin amidases, and Put1 similarly lacks two critical active site residues found in penicillin amidase and N-terminal nucleophile hydrolase family enzymes: a catalytic serine/threonine/cysteine and an asparagine residue in the oxyanion hole33–35 (Supplementary Fig. 6a). Because structure can sometimes more accurately predict enzyme classification than sequence alone (notably in the case of β-lactamases23) we compared the predicted homology-based three-dimensional structure of Put1 to the published crystal structure of E. coli penicillin amidase. While the Put1 alignment to E. coli penicillin amidase reveals no substantial overlap (Supplementary Fig. 6b), a comparison alignment of three authentic penicillin amidase structures shows notable conservation (Supplementary Fig. 6c). Together these findings indicate that Put1 is not a penicillin amidase, and is instead likely an amidase with non-specific activity.
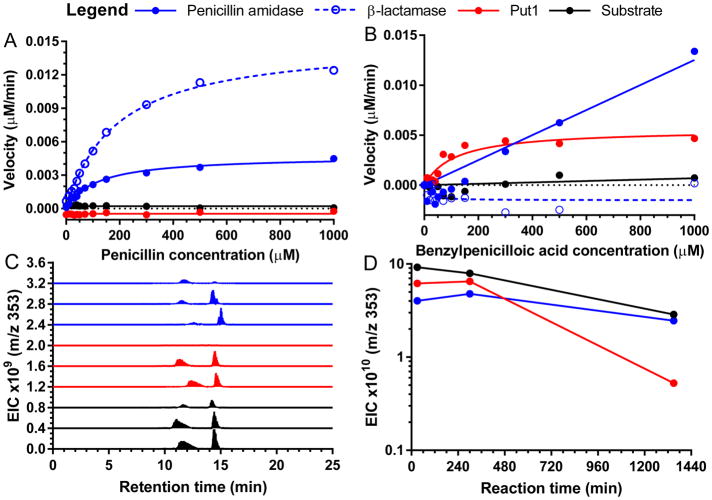
In order to directly evaluate the ability of Put1 to hydrolyze penicillin or benzylpenicilloic acid, as opposed to using chromogenic analogs (Supplementary Fig. 5), we developed a pH-shift kinetic assay36 to compensate for the fact that these two substrates show limited spectroscopic changes during hydrolysis and are, in effect, invisible substrates. While other methods to measure β-lactam hydrolysis rely on specialized instruments such as pH-stat titrators37,38 or indirect measurement and complex modeling using chromogenic competing substrates39, the pH-shift assay can be monitored on a standard UV-vis spectrometer and is directly linked to substrate conversion. Using this assay, we verified that both E. coli penicillin amidase and commercial B. cereus β-lactamase enzymes exhibit Michaelis-Menten kinetics with penicillin as a substrate, as expected. However, we did not observe any substantial activity during incubation of Put1 with penicillin (Fig. 4a), nor did a follow-up liquid chromatography tandem mass spectrometry (LCMS) assay detect any substantial loss of penicillin in a separate reaction (Supplementary Fig. 7a). Notably, the hydrolysis of either the phenylacetamide or β-lactam bonds in penicillin would result in formation of a carboxylic acid and a change in signal in the pH-shift assay. In contrast, when both Put1 and E. coli penicillin amidase were incubated with benzylpenicilloic acid as a substrate, we observed Michaelis-Menten kinetics (Fig. 4b). While the kinetics observed for penicillin amidase and β-lactamase with penicillin could represent the hydrolysis of either the phenylacetamido or β-lactam amide bonds, the only amide bond present in benzylpenicilloic acid is that of the phenylacetamido group, as the β-lactam amide bond is already hydrolyzed. An orthologous assay using the fluorescent derivitization reagent NBD-Cl similarly detected that a new amino group40 is revealed during incubation of Put1 with benzylpenicilloic acid but not penicillin (Supplementary Fig. 7b). These results are consistent with the hydrolysis of benzylpenicilloic acid to form phenylacetic acid. The lack of activity of Put1 with penicillin further supports our hypothesis that Put1 acts downstream of a β-lactamase.
Figure 4. Put1 is a benzylpenicilloic acid-hydrolyzing amidase.
a,b, Hydrolysis activity of E. coli penicillin amidase, B. cereus β-lactamase, and Put1 with penicillin (a) or benzylpenicilloic acid (b) as a substrate was assayed by monitoring absorbance at 404 nm with the colorimetric pH indicators p-nitrophenol and m-nitrophenol, respectively. c, LCMS analysis of benzylpenicilloic acid degradation by E. coli penicillin amidase or Put1 by extracted ion count monitoring. 353 m/z corresponds to singly protonated benzylpenicilloic acid. Within each condition, traces represent (bottom to top) incubation for 30 min, 300 min, and 22.5 hr. Note, benzylpenicilloic acid spontaneously equilibrates to form two diastereomers in aqueous solutions. d, Time course of reactions in c measured by 353 m/z total ion count.
Finally, we directly confirmed benzylpenicilloic acid hydrolysis by Put1 using LCMS. In vitro reactions containing benzylpenicilloic acid and no enzyme, Put1, or penicillin amidase were quenched at three time-points and analyzed for loss of benzylpenicilloic acid (Fig. 4c) (the reaction product, phenylacetic acid, is not detectable by this LCMS method). The Put1 reactions showed substantial benzylpenicilloic acid elimination (Fig. 4c,d), confirming the hydrolysis indicated in the pH-shift (Fig. 4b) and NBD-Cl assays (Supplementary Fig. 7b). These results are in concordance with our hypothesized penicillin catabolic pathway in ABC07, with Put1 acting on benzylpenicilloic acid rather than penicillin, and may also explain the phenotypic and transcriptional data from ABC02, ABC08, and ABC10 (Fig. 2a). In this pathway, the canonical β-lactamase antibiotic resistance enzyme inactivates penicillin to produce benzylpenicilloic acid, which in turn acts as substrate for promiscuous amidase enzymes such as Put1. This results in the release of phenylacetic acid, which is processed by the paa catabolon to produce acetyl- and succinyl-CoA, which feed into central metabolism.
E. coli expressing put operon catabolizes penicillinoids
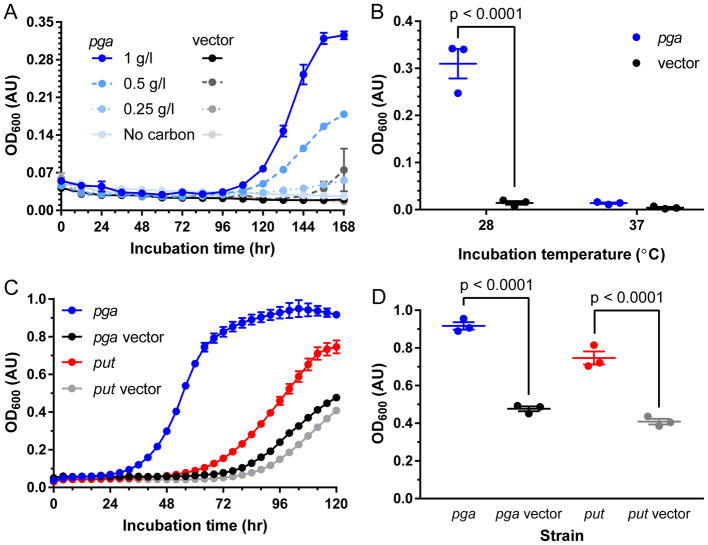
Based on our characterization of the β-lactam catabolic pathways of four soil bacteria, it appears that subsistence on penicillin as a carbon source requires penicillin resistance through β-lactamase activity, the paa catabolon, and an amidase to link these two functions metabolically (Fig. 2c). We set out to test this hypothesis first by engineering an E. coli strain to use penicillin as its sole carbon source. We selected the strain E. coli W (ATCC 9637) due to the presence of a complete and functional paa catabolon in this lineage41 and its use industrially as a source of penicillin amidase27. We confirmed the ability of this strain to catabolize phenylacetic acid in M9 media and cloned an E. coli penicillin amidase gene (pga)42 with truncated signal peptide (necessary due to toxicity issues under constitutive expression) into a β-lactamase-expressing vector. We found that carriage of this plasmid was sufficient to confer the ability to subsist on penicillin as a sole carbon source in a dose-dependent manner (Fig. 5a). Although the pga gene expressed on the vector originates from the E. coli W chromosome, chromosomal expression is insufficient to allow for growth during the time frame investigated as demonstrated by the lack of growth by the vector control strain (Fig. 5a). The importance of amidase activity for penicillin catabolism is further underscored by loss of this phenotype in the pga over-expression strain during growth at 37°C rather than 28°C as a result of inhibition of penicillin amidase post-translational modifications at 37°C27 (Fig. 5b). This experiment demonstrates the potential utility of engineering bacteria for bioremediation of antibiotics.
Figure 5. E. coli expression of penicillin amidase or the put operon gives significantly increased growth on penicillinoids.
a, Growth curve of E. coli W in minimal media with β-lactamase and penicillin amidase (pga, blue traces) expression or vector β-lactamase expression only (black traces) with penicillin as the sole carbon source. b, Final culture densities of E. coli W expressing β-lactamase and penicillin amidase (pga, blue) or β-lactamase only (black) grown at 28°C vs 37°C in minimal media with penicillin as sole carbon source. c, Growth curves of E. coli W expressing penicillin amidase (pga) or vector control (blue and black lines, respectively), or put operon or vector controls (red and grey lines, respectively) in minimal media with benzylpenicilloic acid as sole carbon source. d, Final culture OD600 values of E. coli W expressing penicillin amidase (pga) or vector control (blue and black, respectively) or put operon or vector control (red and grey, respectively). Significance determined by pair-wise ANOVA with Bonferroni correction. All data points are average of triplicate cultures with SEM error shown.
With this proof of principle in hand we set out to test the ability of the put operon to confer increased penicillinoid (i.e. penicillin and its degradation products) catabolism. When expressed in E. coli W, the put operon was not sufficient to give substantially better growth on penicillin compared to vector controls. This is perhaps unsurprising, as penicillin catabolism in ABC07 requires greater than 100-fold over-expression of the put operon (Fig. 2a,b), presumably to augment the low non-specific amidase activity, and it is unclear how levels reached in E. coli might compare. Furthermore, during purification of Put1 from heterologous expression in E. coli we found that this enzyme in particular shows limited stability, with the bulk of the enzyme localizing to the insoluble fraction of cell extracts (Supplementary Fig. 8), indicating poor solubility/compatibility in E. coli. However, we did observe that benzylpenicilloic acid as a carbon source can support growth in the pga over-expressing E. coli strain, and that this growth occurs after a shorter lag-phase compared to growth on penicillin (Fig. 5a,c). When we assayed growth of both engineered E. coli strains using benzylpenicilloic acid as a carbon source we found that expression of the put operon was sufficient to confer a significant improvement in growth compared to its vector control measured both by lag time (Fig. 5c) and culture density reached (Fig. 5d). These results confirm the validity of our hypothesized pathway as well as a role for the put operon in this pathway.
Discussion
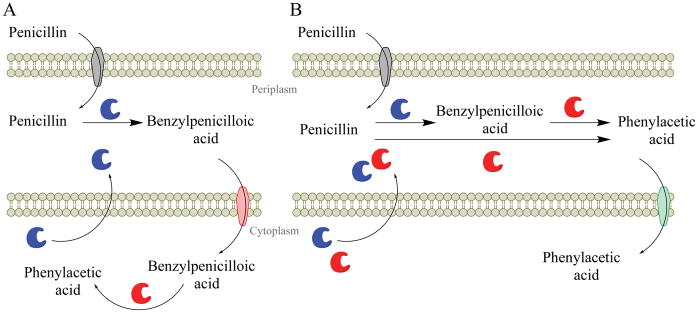
Here we describe the first characterization of an antibiotic catabolism pathway and find it to rely on a novel amidase activity to link β-lactamase and phenylacetic acid catabolism activities (Fig. 2). In strain ABC07, a penicillin utilization operon, put, is regulated by penicillin and its β-lactamase product benzylpenicilloic acid (Fig. 2b) and is necessary for growth on penicillin alongside the phenylacetic acid catabolon (Fig. 3 and Supplementary Fig. 3). The put operon encodes an enzyme with promiscuous amidase activity (Supplementary Fig. 5) that degrades benzylpenicilloic acid via hydrolysis of the amide bond (Fig. 4b–d and Supplementary Fig. 7b). We applied this knowledge to design two strains of E. coli that can consume penicillin as a sole carbon source (Fig. 5). These results led us to develop two related models for penicillin catabolism in bacteria (Fig. 6). The first model (Fig. 6a) describes our hypothesis for penicillin catabolism in strain ABC07. In this model, penicillin enters the periplasm through outer membrane porins where the β-lactam ring is hydrolyzed by β-lactamases to produce benzylpenicilloic acid. Both Put1 and Put2 lack predicted secretion signals43 and are likely localized to the cytoplasm, where they can hydrolyze the amide bond of benzylpenicilloic acid to produce phenylacetic acid and penicic acid. Because there are no known benzylpenicilloic acid inner membrane transporters, we propose that in ABC07 the MFS import pump located in the put operon (Fig. 2a) may fulfill this role. Finally, phenylacetic acid feeds into central metabolism through the phenylacetic acid catabolon. In our second model (Fig. 6b) we propose a modified route for penicillin catabolism in our engineered E. coli strain. In this model, both β-lactamase and penicillin amidase are secreted into the periplasm where phenylacetic acid is produced by penicillin amidase, either from penicillin or benzylpenicilloic acid. In this model, transport of phenylacetic acid into the cytoplasm is mediated by the PaaL permease, part of the phenylacetic acid catabolon.
Figure 6. Schematics illustrating penicillin catabolic strategies.
a, Hypothesized mechanism of penicillin catabolism in ABC07 and studied ABC strains. Outer membrane porins (grey membrane protein) allow uptake of penicillin to the periplasm. In the periplasm, β-lactamases (blue enzyme) rapidly detoxify penicillin to benzylpenicilloic acid. MFS pumps (red membrane protein) transport benzylpenicilloic acid to the cytoplasm where amidases (e.g. Put1, red enzyme) hydrolyze the amide bond to release the phenylacetic acid carbon source. b, Hypothesized mechanism of penicillin catabolism in engineered E. coli. Similar to a, with amide hydrolysis of penicillin or benzylpenicilloic acid occurring in the periplasm via secreted penicillin amidase (red enzyme), and transport of phenylacetic acid to the cytoplasm occurring via PaaL phenylacetic acid permease (green membrane protein).
These two models contrast in their requirements for a β-lactamase enzyme. In the pga over-expressing E. coli model, the β-lactamase acts as a resistance gene only, as E. coli penicillin amidase can use either penicillin or benzylpenicilloic acid as substrates (Fig. 4 and Supplementary Fig. 7a, see also refs. 8,38). In this case, resistance and catabolism could theoretically be decoupled through alternative means of resistance. In contrast, Put1 appears to only act on benzylpenicilloic acid (Fig. 4 and Supplementary Fig. 7) such that in ABC07, and potentially other antibiotic consuming bacteria, β-lactamases may act bifunctionally in resistance and catabolism pathways. This is supported by the presence of penicillin-responsive β-lactamases syntenic to amidases and hydrolases on the chromosomes of ABC07, ABC08, and ABC10 (Fig. 2a). Based on its slow kinetics, it does not appear that Put1 uses penicillin or benzylpenicilloic acid as its natural substrate (Fig. 4b), but the proximity of the put1 and put2 genes to a β-lactamase gene may allow them to be sufficiently up-regulated in the presence of penicillin (Fig. 2b) to overcome catalytic inefficiencies. Notably, because they do not act on penicillin, the low catalytic efficiency of Put1 and Put2 does not endanger the host cell as they are not called upon to act as resistance genes. Synteny of β-lactamases with amidases may therefore present an advantageous pre-condition for penicillin catabolism in the context of a genome containing a phenylacetic acid catabolic pathway. We therefore searched the local genomic context of a library of functionally validated β-lactamase genes for the presence of amidases or genes with related functions. Our analysis found that 2.5% of β-lactamase genes are syntenic (within ca. 1.5 kb) to potential amidases. These pairings fulfill two of three conditions hypothesized in our model for ABC07 (Fig. 6a). A recent genome survey has found that the phenylacetic acid catabolon is present in ~16% of genomes22, implying the coincidence of all three conditions likely occurs in the soil and that the penicillin catabolizing phenotype is far from limited to the four strains analyzed. We propose that antibiotic inactivation followed by carbon source release through a promiscuous enzyme is likely employed by soil bacteria catabolizing other antibiotics as well. We predict that ongoing and future studies will shed further light on these activities.
Antibiotic resistance enzymes are known to be plentiful in soil habitats3, and it is only because of their medical exploitation that antibiotics are treated as privileged molecules not bound by the carbon cycle. Here we have provided ample evidence for the normality of antibiotics in this regard. We characterize a complete antibiotic catabolic pathway that uniquely provides a mechanistic connection between antibiotic producers, antibiotic resistance, and antibiotic catabolism. We have leveraged this mechanistic understanding to engineer E. coli strains that can catabolize penicillin and its degradation product as a sole carbon source. With limited further engineering these strains could be developed as tools for in situ bioremediation of antibiotic-contaminated soils or environments, such as those located near pharmaceutical manufacturers44. These environments are important drivers of antibiotic resistance development45, and their remediation could help prevent the spread of resistance. Of course, the benefits of any such bioremediation program would need to be weighed against the risk of releasing a genetically modified bacterium into the environment and potential spread of antibiotic resistance/degradation genes to other organisms. Finally, antibiotic-catabolizing enzymes have the potential to play an important industrial role in the production of next-generation antibiotics in the same way that the discovery of penicillin amidase spurred the development of semi-synthetic β-lactams through remodeling of natural penicillins. Characterization of hydrolytic enzymes responsible for the catabolism of other antibiotics, such as hypothetical glycosidases acting on aminoglycosides16, could catalyze an explosion of diverse semi-synthetic derivatives in other antibiotic classes. Antibiotic degradation may therefore paradoxically contribute to the development of the next generation of novel antibiotics.
Online Methods
Chemicals
For growth studies and enzymatic assays high purity penicillin G sodium salt (Sigma, P3032), (+)-6-aminopenicillanic acid (Sigma, A70909), phenylacetic acid (Sigma, W287806), and other antibiotics, were purchased. Dextro-(−)-benzylpenicilloic acid hydrate (Sigma, S341967) was purchased from the Sigma-Aldrich collection of rare and unique chemicals. p-Nitroacetanilide (Sigma, 130648), p-nitroaniline (Sigma, 185310), 6-nitro-3-(phenylacetamido)benzoic acid (aka NIPAB, Pfaltz and Bauer, N10625), N-phenylacetyl-p-aminobenzoate (Sigma, P8529), L-glutamate 1-(p-nitroaniline) (Sigma, 49622), L-glutamate γ-(p-nitroaniline) (Sigma, G1135) and Nα-benzoyl-DL-arginine p-nitroaniline (Sigma, B4875) were purchased as enzyme substrates and standards. For the kinetics assays p-nitrophenol (MP Biomedicals, 102461) and m-nitrophenol (Acros organics, 172300100) were purchased as indicators for monitoring of enzyme reaction kinetics in pH-shift assays. All other compounds and buffers used were of standard molecular biology grade.
Bacterial strains and growth conditions
Soil isolates and minimal media
Soil isolates ABC02, ABC07, ABC08, and ABC10 were previously isolated by culturing with the antibiotics penicillin or carbenicillin as carbon sources and were maintained at −80°C as 15% glycerol stocks in single carbon source (SCS) minimal media (SCS media, see below) 15. Isolates were cultured with aeration (taken to be shaking at 220 rpm throughout unless otherwise specified) at 22°C in LB or M9 minimal media (M9 media, see below). Where appropriate, carbon sources were added to various minimal media at 1 g/l. SCS media was prepared by combining 100 ml of 10× YDM-base and 10 ml of 100× YDM-trace metals in 1 liter of water and adjusting the pH to 5.5 with HCl and filter sterilizing. 10× YDM-base consists of (per liter) 50 g (NH4)2SO4, 30 g KH2PO4, 5 g MgSO4 heptahydrate, adjusted to pH 5.5 with NaOH and filter sterilized. 100X-trace metals consists of (per liter) 1.5 g ethylenediaminetetraacetic acid (EDTA), 450 mg ZnSO4 heptahydrate, 100 mg MnCl2 tetrahydrate, 30 mg CoCl2 hexahydrate, 30 mg CuSO4 pentahydrate, 40 mg Na2MoO4 dihydrate, 450 mg CaCl2 dihydrate, 300 mg FeSO4 heptahydrate, and 10 mg KI, filter sterilized 15. While SCS media contains EDTA, a potential alternative source of carbon, the final concentration of this compound, 15 mg/l, is theoretically below the level required for growth. Nevertheless, all growth assays with SCS media were also performed with SCS media without added penicillin or carbenicillin as a negative control to confirm this medium did not support growth. To completely rule out the potential contribution of EDTA to growth, experiments were also repeated in M9 media, which does not include any carbon-containing ingredients, including EDTA. M9 media was prepared by combining (per liter) 200 ml 5× M9 salts (Sigma, M6030), 2 ml 1M MgSO4, and 100 μl 1M CaCl2 in 1 liter of water and adjusting the pH to 7 or 5.5 and filter sterilizing. 5× M9 salts consists of (per liter) 33.9 g Na2HPO4 heptahydrate, 15 g KH2PO4, 5 g NH4Cl, 2.5 g NaCl in a liter of water sterilized by autoclaving. Antibiotic resistance testing was performed in LB in the presence of 1 g/l of the indicated antibiotic. Starter cultures grown for one (ABC07) to two (ABC02, ABC08, and ABC10) days with shaking at room temperature were inoculated (2 μl) into LB media containing antibiotics and cultured aerobically for two days at room temperature prior to reading OD600 reading on a Powerwave HT microplate spectrophotometer (Biotek, Inc.). All growth experiments included triplicate independent cultures and OD600 values were evaluated as averages with standard error unless otherwise noted.
ABC strains carbon source growth studies
For growth on diverse carbon sources, Biolog Phenotype Microarray Plates PM1 and PM2A (Hayward, CA) were used. ABC02, ABC07, ABC08, and ABC10 starter cultures were inoculated directly from frozen stocks and incubated with aeration at room temperature in 5 ml SCS media with 1 g/l carbenicillin (ABC02) or penicillin (ABC07, ABC08, and ABC10) until cultures turned visibly turbid (visible turbidity corresponds roughly to OD600 values ~1.5 to 3 AU, approximately 3 days of growth). Each culture was washed a total of 5 times in SCS media lacking a carbon source by pelleting the cells (3,000 rcf, 5 min) and aspirating supernatants to remove residual glycerol or other potential carbon sources. After the final wash the cultures were resuspended in 20 ml SCS media to an OD600 of 0.1 AU, 100 μl per well was added to the dry Biolog plates, and mixed by repeated pipetting. Plates were incubated at room temperature for 96-hours and growth was determined by subtraction of each well’s individual initial OD600 reading from its final reading as measured on a Powerwave HT microplate spectrophotometer (Biotek, Inc.).
Cultures for growth curves of soil isolates grown in M9 media with β-lactam antibiotics were prepared as described above with media pH adjusted to 5.5 with NaOH. A 96-well plate (COSTAR, 3595) containing 200 μl/well of M9 media with 0.25 g/l, 0.5 g/l, or 1 g/l penicillin were inoculated in triplicate with 2 μl of thrice washed cells and sealed with a Breathe-Easy membrane (Sigma-Aldrich, Z380059). Growth was monitored every 20 minutes at 600 nm in a Powerwave HT microplate spectrophotometer (Biotek, Inc.) at 25°C with constant shaking (medium setting, rpm not available) for 1 week. Growth data were plotted using GraphPad Prism version 7.01 for Windows (GraphPad Software, La Jolla California, USA).
Generation and testing of ABC07 knock-out strains
Generation of the ABC07 paaF and penicillin put operon (bla/put2/mfs/put1) knockout strains was performed following the protocol of Hmelo et al. 26 using the pEXG2 plasmid 46 containing ca. 1,000 base pair flanking regions from around paaF and the put operon separated by a tetA open reading frame (see table S3 for primer sequences). DNA fragments were amplified by polymerase chain reaction with Q5 hotstart master mix polymerase (NEB, M0494L) using ABC07 genomic DNA as template and ligated via Gibson assembly master mix (NEB, E2611S) according to manufacturer’s guidelines. Constructs were confirmed by Sanger sequencing (Genewiz) and introduced into strain ABC07 by biparental mating using E. coli S17 λpir followed by selection on LB agar plates containing gentamicin at 60 μg/ml. Merodiploids were selected for using VBMM agar consisting of 1.5% agar in (per liter) 200 mg MgSO4 heptahydrate, 2 g of citric acid, 10 g of K2HPO4, and 3.5 g of NaNH4HPO4 tetrahydrate and counter-selection was performed on LB agar containing 15% (w/v) sucrose. Loss of the gentamicin resistance cassette marker of pEXG2, loss of the target gene, and gain of tetA were confirmed by routine PCR using 2× ReddyMix master mix (ThermoFisher Scientific AB0575DCLDA) according to the manufacturer’s guidelines. Briefly, inserts were amplified with 25 cycles of denaturing at 94°C for 45 seconds, annealing at 60°C for 45 seconds, and extending at 72°C for 2 minutes.
For the assay, overnight cultures of wild-type, paaF, and put ABC07 strains were grown in LB at 28°C and washed three times in M9 media with no carbon as described above. Glucose, phenylacetic acid, and penicillin in M9 media were initially prepared at a final concentration of 0.4% (w/v) following which pH was adjusted to 5.5 with HCl for penicillin and to pH 7 for glucose and phenylacetic acid with NaOH. Media were aliquoted 200 μl/well into a 96-well plate (COSTAR, 3595) followed by 2 μl of washed cultures and sealed with a Breathe-Easy membrane (Sigma-Aldrich, Z380059). Each strain/condition was set-up in triplicate. Growth was monitored at 600 nm for 1 week with temperature maintained at 28°C with continuous shaking on medium speed for 1 week on a Powerwave HT microplate spectrophotometer (Biotek, Inc.). OD600 measured for each condition were plotted in GraphPad Prism version 7.01 (GraphPad Software, La Jolla California, USA). Further growth studies were performed as above with 1 g/l of the following carbon sources: (at pH 7) phenylacetic acid, glucose, (at pH 5.5) penicillin, benzylpenicilloic acid, 6-aminopenicillanic acid, carbenicillin, ampicillin, and cefuroxime. Optimal culture pH was determined empirically.
E. coli growth conditions
E. coli BL21(DE3) and E. coli DH10β were cultured in LB or Terrific broth (TB, MOBIO, 12105-05 or Fisher Scientific, BP9729-600) with aeration at 37°C with 50 μg/ml kanamycin and/or 100 μg/ml carbenicillin, and 100 to 500 μM isopropyl β-D-1-thiogalactopyranoside (IPTG) when appropriate. E. coli W (ATCC 9637) was purchased from the American Type Culture Collection (ATCC) and propagated according to ATCC instructions or in M9 media prepared as above (adjusted to pH 7.2) with 50 μg/ml kanamycin and 100 μg/ml carbenicillin when appropriate at 28°C. All E. coli strains were maintained as 15% glycerol stocks in LB at −80°C.
Isolate whole transcriptome RNA sequencing
Genome sequencing and open reading frame calling and gene annotation
We previously published whole genome sequences of trains ABC07, ABC08, and ABC10 21. Whole genome sequencing of strain ABC02 was performed exactly as for ABC07, ABC08, and ABC10. Whole genome sequences for ABC02, ABC07, ABC08, and ABC10 (GenBank accessions numbers NGUT00000000, NGUS00000000, NGUR00000000, and NGUQ00000000 respectively) were called for open reading frames and annotated as before 4.
Preparation of cDNA and whole transcriptome RNA sequencing
Strains ABC02, ABC07, ABC08, and ABC10 were prepared for whole transcriptome RNA sequencing (RNAseq) in triplicate for each carbon source as follows. Frozen glycerol stocks with 15% glycerol previously grown in SCS media with 1 g/l penicillin were used to inoculate 5 ml cultures of SCS media containing 1 g/l appropriate carbon source and incubated at room temperature until cultures reached turbidity visually. During this incubation period any residual glycerol is catabolized. Triplicate independent culture flasks containing 100 ml fresh SCS media with 1 g/l carbon source were then inoculated with 100 μl of turbid culture. Strains were grown with aeration at room temperature until early exponential phase at which point cells were harvested by centrifugation (3,000 rcf, 10 min). For storage prior to RNA extraction, cell pellets were resuspended in RNAprotect Bacteria Reagent (QIAGEN, 76506) according to manufacturer’s instructions and stored at −80°C until extraction. Total RNA was extracted via standard bead beating and phenol:chloroform protocol. Ribosomal RNA (rRNA) was depleted from the RNA extract using the Ribo-Zero rRNA Removal kit (Epicentre, MRZMB126) according to the manufacturer’s instructions. Depleted RNA was converted to cDNA for sequencing via SuperScript II (Invitrogen, 18064022) as previously described 47 and sheared in a Covaris S2 (Covaris, MA, USA) to produce 150 bp fragments. Samples were sonicated in 120 μl volumes for 10 min at 10% duty cycle at intensity 5 and 100 cycles per burst.
Sheared, size-selected fragments were end-repaired and ligated with Illumina sequencing adaptors essentially as previously described 47. Single-end 1 × 50bp sequencing was performed at the Genome Technology Access Center (GTAC, Washington University in Saint Louis) using the Illumina HiSeq 2000 platform to a target transcriptome coverage of ca. 10X. Raw reads have been deposited with the Sequence Read Archive at NCBI under BioProject number PRJNA385617. Sequence reads were mapped to sequenced ABC strain genomes using Bowtie 48 with default parameters and raw expression counts were obtained using mrfCounter from the RSeqTools library 49. Normalization and differential expression of open reading frames under each carbon source condition using the binomial test implementation were performed in the R package DESeq 50 following methods described by Anders and Huber for estimating variance. Genes with significantly different expression between each carbon source condition (adjusted p<0.00001, DESeq) were analyzed in more detail. Highly upregulated genes from the ABC07 significant set appearing to correspond to penicillin catabolism (figure 2B) were plotted as normalized reads in triplicate with SEM as the ratio of gene counts during growth on the given substrate to gene counts during growth on glucose using GraphPad Prism version 7.01 for Windows (GraphPad Software, La Jolla California, USA).
Phylogenetic analysis of upregulated amidases, hydrolases, and amidohydrolases
Predicted amidases, hydrolases, and amidohydrolases from ABC02, ABC07, ABC08, and ABC10 that were significantly upregulated during growth on penicillin compared to glucose or histidine were targeted for phylogenetic analysis. Predicted amino acid sequences for the upregulated genes were input into blastp NR database 28 on May 8, 2017 and the top 100 hits were combined and clustered at 70% identity using the cdhit program 51. The ABC strain sequences and their top hits were combined with representative penicillin amidase sequences (E.C. 3.5.11) downloaded from UniProt 29. The combined sequences were aligned by ClustalW 52 in the MEGA7 53 program and used to construct a Maximum Likelihood tree with bootstrap phylogeny test (100 rounds) and default parameters. The final tree was visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/, February 2017).
The amino acid sequence of Put1 was further analyzed for the presence of a signal peptide indicative of secretion, with prediction validation using secreted β-lactamase and penicillin amidase sequences 43. An alignment of Put1 with E. coli pga and pga homologs from other bacteria was generated as above. The three dimensional structure of Put1 was predicted based on homology using the online Phyre tool 54 with intensive settings. The resulting PDB file, as well as other PDB files, were visualized and manipulated using the PyMOL program (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.). PDB files for Put1 and Providencia rettgeri (1CP9) 35 and Kluyvera citrophilia (4PEM) penicillin amidase structures were aligned to the E. coli (1GK9) 34 penicillin amidase structure via the alignto command in PyMOL.
In vitro enzyme assays
put1 and put2 amplification and cloning
Inserts were amplified from ABC07 genomic DNA using the polymerase chain reaction with Iproof polymerase (BioRad, 172–5330) according to manufacturer instructions. Briefly, inserts were amplified with 30 cycles of denaturing at 98°C for 30 seconds, annealing at 65°C or 60°C for 30 seconds, and extending at 72°C for 1.5 minutes using primers 5093BW and 5094BW for Put1 and 5095BW and 5096BW for Put2 respectively (Table S3). NdeI and SacI restriction sites were introduced during PCR. Inserts and vector pET-28b(+) were digested by SacI and NdeI (New England Biolabs, R0156S and R0111S respectively) overnight at 37°C, heat inactivated at 65°C for 20 minutes and purified by extraction from agarose gels followed by column clean-up (Qiagen QIAquick gel extraction kit, 28704). Inserts were ligated into vector via complementary overhang using the Fast-Link DNA Ligation kit (Epicentre, LK6201H) with a 3:1 insert to vector ratio at room temperature overnight. Ligation reactions were heat inactivated at 70°C for 10 minutes, dialyzed against water, and transformed by electroporation into competent E. coli BL21(DE3) cells using a BioRad Gene Pulser XCell in 1-mm gap cuvettes at 2.0 kV, 200 Ω, and 25 μF. Cells were allowed to recover in 1 ml of rich media (SOC medium, Invitrogen 46-0821) for 1 hr at 37°C then plated on LB agar with kanamycin. Colonies were screened for inserts by colony PCR using Thermo Scientific 2xReady PCR Mix (Thermo Scientific, AB0575DCLDA) according to the manufacturer’s instructions using vector-specific primers TSC54 and TSC55 (Table S3). Successful colonies were cultured in LB with kanamycin and plasmids were extracted by miniprep (Qiagen QIAprep Spin Miniprep kit, 27104) and sequences were verified by Sanger sequencing (Genewiz).
Expression and extraction of Put1 and Put2 enzymes
For heterologous expression and purification of Put1 and Put2 from E. coli, 1.5 ml starter cultures of E. coli BL21(DE3) containing pET-28b(+) with put1 or put2 inserts were grown overnight at 37°C with aeration in TB with kanamycin. Starter cultures were used to inoculate a pair of 2.8 L Fernbach flasks containing 750 ml of TB with kanamycin for each enzyme. Cultures were incubated at 325 rpm at 37°C in a MaxQ 5000 (Thermo Scientific) temperature-controlled incubator until optical densities at 600 nm (OD600) of 0.2 to 0.4 AU were reached (approximately 4 hours). Flasks were held on ice and IPTG was added to a final concentration of 100 (put1) or 500 (Put2) μM before flasks were returned to the shaking incubator now cooled to 25°C. The cultures were shaken overnight at 25°C for approximately 16 hours to induce protein expression after which cells were harvested by centrifugation at 8,000 rcf for 15 minutes at 4°C in a Sorvall Legends XTR centrifuge (Thermo Scientific) in a Fiberlite F14-6×250 LE rotor (Thermo Scientific). Results of attempts to optimize Put1 expression conditions for greater yield of soluble protein can be found in figure S6. Wet cell pellets were weighed and tared on empty centrifuge tubes to find the wet cell mass. All following steps were performed at 4°C or on ice. For cell lysis, pellets were resuspended to 50% w/v in lysis buffer consisting of (per liter) 8.709 g K2HPO4 (50 mM), 29.22 g NaCl (500 mM), 351 μl β-mercaptoethanol (5 mM), 1.36 g imidazole (20 mM), and 100 ml glycerol (10% v/v) brought to a pH of 8 with, for Put1 purifications, addition of protease inhibitor cocktail (Thermo Scientific, A32955) according to manufacturer’s instructions. We did not observe any inhibition of amidase activity due to this addition (assayed by hydrolysis of p-nitroacetanilide). Suspensions were lysed using three cycles of freeze-thawing in ethanol-dry ice baths and sonication using a Branson Sonifier 250 (Branson Ultrasonics) with microtip using the following settings: 50% duty with output adjusted to ~40% (approximately 80 W) on ice twice for 2 minutes until suspension viscosity was reduced. The lysed cell suspensions were clarified by centrifugation at 24,446 rcf for 30 minutes at 4°C in a Sorvall Legends XTR centrifuge (Thermo Scientific) in a Fiberlite F15-8×50 cy rotor (Thermo Scientific). Supernatants were decanted and stored at 4°C until purification.
Purification and storage of Put1 and Put2 enzymes
Protein purifications were carried out by immobilized metal affinity chromatography using nickel resin (Ni-NTA, BioRad, 7800800) and a BioRad Econo system for low-pressure chromatography with a model 2110 fraction collector (BioRad, 7318122). A column was packed with 8 ml of 50% Ni-NTA resin in suspension and equilibrated in lysis buffer at 4.5 ml/min in a 4°C climate controlled room for 9 minutes. Clarified supernatants brought to 50 ml in lysis buffer were loaded onto the column at 4.5 ml/min 25 ml at a time with a lysis buffer wash in between each 25 ml load to decrease clogging. Lysis buffer was run through the column until UV absorbance returned to baseline levels. Proteins were eluted from the column in a gradient consisting of 0% to 100% elution buffer over 30 minutes. Elution buffer consists of lysis buffer with a final concentration of imidazole at 300 mM (20.42 g per liter) and NaCl at 1 M (58.44 g. per liter). Fractions were assayed for the presence of Put1 and Put2 by SDS-PAGE and, for Put1, by p-nitroacetanilide hydrolysis (1 mM substrate concentration) prior to pooling. Briefly, 7.5 μl of eluted solution was combined with 2.5 μl 4× Laemmli sample buffer (BioRad, 161-0747), boiled for 10 minutes, loaded onto a 12% Mini-PROTEAN TGX precast gel (BioRad, 456–1043), and run at 200 V for 40 minutes in 1× Tris/Glycine/SDS buffer (BioRad, 161–0732). Gels were stained using Bio-Safe Coomassie stain (BioRad, 1610796) according to manufacturer’s suggestion and visualized using a Gel-Doc XR+ (BioRad). Fractions containing purified proteins were pooled and concentrated using a 10 kDa molecular weight cut-off filter (Amicon Ultra-15, UFC901024) to ca. 1 ml total volume according to manufacturer’s instructions. Elution buffer was exchanged three times using molecular weight cut-off filters as above for storage buffer consisting of (per liter) 8.709 g K2HPO4 (50 mM), 8.766 g NaCl (250 mM), 1 ml of 1 M dithiothreitol (1 mM) and 50 ml of glycerol (5% v/v) brought to a pH of 7.5. Final protein concentration was determined by Qubit Protein Assay kit (Thermo Scientific, Q33211) and molar enzyme concentration was found using molecular weights predicted by the EXPASY program 55. Enzymes were aliquoted in 55 μl volumes in PCR tubes on dry ice and stored at −80°C until use.
Amidase activity assays
Measurements of amidase activity using the chromogenic penicillin analog 6-Nitro-3-(phenylacetamido)benzoic acid (NIPAB) and chromogenic amide p-nitroacetanilide were conducted as follows. NIPAB and p-nitroacetanilide, both of which have previously been validated as substrates for the detection of penicillin and other amidase activity respectively 30,31, were prepared in DMSO at a concentration of 200 mM and were diluted to 2 mM in phosphate buffered saline pH 7.5 (PBS, 150 mM phosphate and 50 mM NaCl). Each substrate was further diluted in a 96-well plate (COSTAR, 3595) to concentrations from 20 μM to 2000 μM with a volume of 100 μl per well. Commercial penicillin amidase (Sigma, 76427) was diluted in PBS buffer to a concentration of 0.1 units/ml and purified Put1 and Put2 enzymes were diluted in PBS buffer to 4 μM. To initiate reactions, 100 μl of enzyme was added to wells containing 100 μl of substrate and mixed before monitoring at 400 nm every 20 seconds using a Powerwave HT microplate spectrophotometer (Biotek, Inc.) with temperature maintained at 25°C. Change in absorbance over time was converted to μM min−1 using the extinction coefficient for p-nitroacetanilide and NIPAB at pH 7.5 at 400 nm of 11110 M−1 cm−1 (determined empirically) and 9500 M−1 cm−1 30 respectively. The Michaelis-Menten kinetics curve fit equation was solved using GraphPad Prism version 7.01 for Windows (GraphPad Software, La Jolla California, USA). Other chromogenic substrates were similarly assayed at 1 mM concentrations with penicillin amidase (at 0.01 U/ml) and Put1 (at 2 μM) in PBS buffer at 37°C by monitoring at 400 nm (L-glutamate 1-(p-nitroaniline), L-glutamate γ-(p-nitroaniline), and Nα-benzoyl-DL-arginine p-nitroaniline) or at 295 nm (N-phenylacetyl-p-aminobenzoate).
Indirect measurement of penicillin and benzylpenicilloic acid hydrolysis was performed by derivatization of the resulting primary amine by the reagent 4-nitro-7-chloro-benzo-2-oxa-1,3-diazole (NBD-Cl) as previously published 40. Penicillin or benzylpenicilloic acid were diluted to 2 mM in PBS buffer with 5 μM Put1 or 100 U/ml penicillin amidase and allowed to incubate overnight at 37°C. Reactions were quenched by addition of two volumes of cold acetonitrile followed by the addition of 4 mM NBD-Cl in acetonitrile and incubation at 37°C for 2.5 hr. Fluorescence of reactions, 250 μl in a 96-well plate (COSTAR, 3595), was read using a Powerwave HT microplate spectrophotometer (Biotek, Inc.) with excitation at 485 nm and emission at 538 nm with default gain.
Direct measurements of penicillin and benzylpenicilloic acid enzymatic hydrolysis were performed by a modification of a pH-dependent, colorimetric, hydrolase assay responsive to cleavage of either amide bonds 36. Briefly, hydrolysis of amide bonds was monitored by color change of the pH indicator p-nitrophenol or m-nitrophenol in response protons released due to the appearance of new carboxylic acid residues following amide hydrolysis (phenylacetic acid in the case of penicillin and benzylpenicilloic acid). Reaction buffer for penicillin assays was prepared by diluting 200 mM p-nitrophenol in DMSO to a final concentration of 550 μM in 5 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer and adjusting to a pH of 7.2 with dilute HCl and NaOH. Penicillin substrate was prepared by diluting 200 mM penicillin in DMSO to 2.4 mM in reaction buffer and adjusting the pH to 7.2 followed by dilution in triplicate in a 96-well plate (COSTAR, 3595) at concentrations ranging from 24 μM to 1.2 mM in 100 μl of p-nitrophenol reaction buffer. Enzymes were diluted into p-nitrophenol reaction buffer and brought to a pH of 7.2. E. coli penicillin G amidase (Sigma, 76427) and B. cereus β-lactamase (Sigma, P0389) were diluted to 0.1 U/ml and 0.2 U/ml respectively and Put1 enzyme was prepared at 4 μM. Reactions were initiated by addition of 100 μl of enzyme to 100 μl of substrates in wells. Kinetics were monitored as before with the exception that data from absorbance at 404 nm was collected. Change in absorbance over time was converted to μM min−1 as described elsewhere 36. Data were fit to Michaelis-Menten curves as before. Benzylpenicilloic acid hydrolysis was assayed as with penicillin with the following alterations. Instead of MOPS media containing p-nitrophenol, a buffer consisting of 10 mM m-nitrophenol at pH 8.4 was used with conversion of absorbance to μM min−1 calculated using an extinction coefficient of 1241.5 M−1 cm−1 at 415 nm (determined empirically). E. coli penicillin G amidase (Sigma, 76427) was used at a final concentration of 5 U/ml and Put1 was used at a final concentration of 2 μM. A higher pH was required due to competition for proton signal from the alkaline amino group of penicic acid 37. Change in signal represents the hydrolysis of the phenylacetamide bond only as the penicillin β-lactam ring is already hydrolyzed in benzylpenicilloic acid. Because benzylpenicilloic acid lacks a β-lactam ring the activity of β-lactamase was not assayed.
Direct degradation of benzylpenicilloic acid and penicillin were followed by liquid chromatography tandem mass spectrometry (LCMS) analysis of quenched enzymatic reactions. Reactions were conducted in 135 μl of PBS buffer at pH 7.5 with 40 μM Put1 or 0.05 U/ml E. coli penicillin G amidase (Sigma, 76427). To initiate reactions, 1 μl of 200 mM penicillin or benzylpenicilloic acid in DMSO was added to each reaction on ice, for a final concentration of ca 1.5 mM substrate. Aliquots of the reaction were quenched after 30 min, 5 hr, and 22.5 hr by removing 25 μl aliquots into 50 μl ice-cold acetonitrile to precipitate enzymes. Quenched samples were analyzed by LCMS at the Donald Danforth Plant Sciences Institute using a Scherzo C18 column with buffers consisting of 0.1% formic acid in water (buffer A) or acetonitrile (buffer B). Analytes were eluted using a gradient method consisting of 4 minutes isocratic with 0% buffer B and a ramp up to 100% buffer B over 12 minutes. Mass spectra were recorded on a Q Exactive mass spectrometer (Thermo Scientific) in polarity switching mode at a resolution of 140,000 (at m/z 200). Phenylacetic acid is not detectable using this method. Benzylpenicilloic acid spontaneously forms a pair of diastereomers that appear as two peaks under these conditions25.
E. coli gain of function assays
E. coli penicillin G amidase and ABC07 put operon cloning
For gain of function experiments, the E. coli W penicillin amidase (pga) open reading frame was cloned into the β-lactamase (bla) containing pZA11 expression vector 56 with its 5′ secretion signal truncated by 6 residues at the N-terminus 42. Inserts were amplified by PCR using Q5 hotstart master mix polymerase (NEB, M0494L) according to manufacturer’s guidelines using primer pair 5777TSC/5779TSC (Table S3). Linear pZA11 was prepared by inverse PCR as above using the primer pair 5780TSC/5781TSC (Table S3). Inserts and vector were gel purified (Qiagen QIAquick gel extraction kit, 28704) and prepared for ligation by overnight double digestion with KpnI-HF and PstI-HF restriction enzymes (NEB, R3142S and R3140S respectively), treatment with Antarctic phosphatase (vector only, NEB, M0289S), and final purification by column clean-up (Qiagen QIAquick PCR purification kit, 28104). Ligation was performed with a 2:1 insert to vector ratio using the Fast-Link DNA Ligation kit (Epicentre, LK6201H) at room temperature for 30 minutes followed by heat inactivated at 70°C for 15 minutes. Transformation was carried out by heat shock at 42°C of 5 μl of ligation product into 50 μl calcium competent E. coli W followed by recovery for 30 minutes at 37°C and plating on to LB agar with 100 μg/ml carbenicillin. Colonies were screened for inserts using colony PCR with Thermo Scientific 2xReady PCR Mix (Thermo Scientific, AB0575DCLDA) according to the manufacturer’s instructions. Briefly, inserts were amplified for 30 cycles of denaturing at 94°C for 45 seconds, annealing at 60°C for 45 seconds, and extending at 72°C for 2 minutes using vector-specific primers 22TSC and 5723TSC (Table S3). Successful colonies were cultured in LB with carbenicillin and plasmids were extracted by miniprep (Qiagen QIAprep Spin Miniprep kit, 27104) and inserts were verified by Sanger sequencing (Genewiz). Following sequencing, a single base deletion was discovered in the first ~8 bp of the pga gene resulting in a frame shift mutation. Expression from a downstream alternate in-frame start codon resulted in a product lacking the first 6 residues of the signal peptide. Attempts to re-clone the wild-type gene failed due to toxicity issues arising from over-expression in pZE21. Cloning of the ABC07 put operon into pZE21 56 was performed similar to pga but via blunt-end ligation using the Fast-Link DNA Ligation kit (Epicentre, LK6201H) according to manufacturer’s instructions. Stocks were maintained in LB with 15% glycerol at −80°C.
E. coli W penicillinoid catabolism assay
E. coli W strains harboring the empty pZA11 vector or pZA11-E. coli pga and empty pZA11/pZE21 or pZA11/pZE21-put operon were cultured in LB supplemented with carbenicillin or carbenicillin and kanamycin overnight at 37°C. Cells were washed three times prior to inoculation in M9 media as before. M9 media containing penicillin or benzylpenicilloic acid as a sole carbon source at 4 g/l was added to a 96-well plate in 200 μl aliquots followed by 2 μl of washed cells in triplicate. Plates were sealed with a Breathe-Easy membrane (Sigma-Aldrich, Z380059) and growth kinetics were monitored at 600 nm every hour using a Powerwave HT microplate spectrophotometer (BioTek, Inc.) at 28°C with constant shaking on medium for 120 hr (5 days). Growth data were plotted and evaluated for significance using GraphPad Prism version 7.01 for Windows (GraphPad Software, La Jolla California, USA) using pair-wise ANOVA tests with Bonferroni correction for multiple comparisons.
Data availability Statement
ABC02 (this manuscript) and ABC07, ABC08, and ABC10 (ref. 21) WGS short-read raw data, assembled genomes, and RNASeq short-read data have been deposited to NCBI under BioProject number PRJNA385617 with BioSample numbers SAMN06915397, SAMN06915398, SAMN06915399, and SAMN06915400 respectively. Whole genome sequences may be found for ABC02, ABC07, ABC08, and ABC10 at DDBJ/ENA/GenBank under the accessions numbers NGUT00000000, NGUS00000000, NGUR00000000, and NGUQ00000000 respectively.
Supplementary Material
Acknowledgments
This work is supported in part by awards to G.D. through the Edward Mallinckrodt, Jr. Foundation (Scholar Award), and from the NIH Director’s New Innovator Award (http://commonfund.nih.gov/newinnovator/), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK: http://www.niddk.nih.gov/), the National Institute of General Medical Sciences (NIGMS: http://www.nigms.nih.gov/), and the National Institute of Allergy and Infectious Diseases (NIAID: https://www.niaid.nih.gov/) of the National Institutes of Health (NIH) under award numbers DP2DK098089, R01GM099538, and R01AI123394, respectively. T.S.C. received support from a National Institute of Diabetes and Digestive and Kidney Diseases Training Grant through award number T32 DK077653 (Phillip I. Tarr, Principal Investigator) and a National Institute of Child Health and Development Training Grant through award number T32 HD049305 (Kelle H. Moley, Principal Investigator). K.J.F. received support from the NHGRI Genome Analysis Training Program (T32 HG000045), the NIGMS Cellular and Molecular Biology Training Program (T32 GM007067), and the NSF as a graduate research fellow (award number DGE-1143954). M.K.G. received support as a Mr. and Mrs. Spencer T. Olin Fellow at Washington University and from the NSF as a graduate research fellow (DGE-1143954). Sequencing through the US Army Edgewood Chemical Biological Center was supported in part through funding provided by the Transformational Medical Technologies Initiative of the Defense Threat Reduction Agency, US Department of Defense. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. We are thankful to Jessica Hoisington-Lopez in the Center for Genome Sciences and Systems Biology at Washington University in St Louis School of Medicine for Illumina sequencing support, Dr. Tim Wencewicz and Dr. Brad Evans for their useful discussions regarding biochemistry and LCMS, and members of the Dantas lab for general helpful discussions regarding the manuscript.
Footnotes
Author contributions
T.S.C., A.S., T.A.G., M.O.A.S., and G.D. conceived of experiments and design of work. T.S.C., B.W., A.S., and T.A.G. performed in vitro, microbial, and transcriptomic experiments. L.A.J., S.M.B., C.N.R., E.W.S., and H.S.G. sequenced strain genomes. T.S.C., A.S., T.A.G., K.J.F, and M.K.G. provided analyses. Article drafting was performed by T.S.C. with critical revision performed by T.S.C., B.W., A.S., K.J.F, M.K.G., M.O.A.S., and G.D.
Competing financial interests statement
The authors declare that they have no competing financial interests.
References
- 1.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–61. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 3.Crofts TS, Gasparrini AJ, Dantas G. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol. 2017;15:422–434. doi: 10.1038/nrmicro.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forsberg KJ, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–11. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aust MO, et al. Distribution of sulfamethazine, chlortetracycline and tylosin in manure and soil of Canadian feedlots after subtherapeutic use in cattle. Environ Pollut. 2008;156:1243–51. doi: 10.1016/j.envpol.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Wright PM, Seiple IB, Myers AG. The evolving role of chemical synthesis in antibacterial drug discovery. Angew Chem Int Ed Engl. 2014;53:8840–69. doi: 10.1002/anie.201310843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pramer D, Starkey RL. Decomposition of Streptomycin. Science. 1951;113:127. [PubMed] [Google Scholar]
- 8.Kameda Y, Kimura Y, Toyoura E, Omori T. A method for isolating bacteria capable of producing 6-aminopenicillanic acid from benzylpenillin. Nature. 1961;191:1122–3. doi: 10.1038/1911122a0. [DOI] [PubMed] [Google Scholar]
- 9.Abd-El-Malek Y, Monib M, Hazem A. Chloramphenicol, a simultaneous carbon and nitrogen source for a Streptomyces sp from Egyptain soil. Nature. 1961;189:775–6. doi: 10.1038/189775a0. [DOI] [PubMed] [Google Scholar]
- 10.Johnsen J. Utilization of benzylpenicillin as carbon, nitrogen and energy source by a Pseudomonas fluorescens strain. Arch Microbiol. 1977;115:271–5. doi: 10.1007/BF00446452. [DOI] [PubMed] [Google Scholar]
- 11.Beckman W, Lessie TG. Response of Pseudomonas cepacia to beta-Lactam antibiotics: utilization of penicillin G as the carbon source. J Bacteriol. 1979;140:1126–8. doi: 10.1128/jb.140.3.1126-1128.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnsen J. Presence of beta-lactamase and penicillin acylase in a Pseudomonas sp utilizing benzylpenicillin as a carbon source. J Gen Appl Microbiol. 1981;27:499–503. [Google Scholar]
- 13.Wang P, et al. Characterization and mechanism analysis of penicillin G biodegradation with Klebsiella pneumoniae Z1 isolated from waste penicillin bacterial residue. J Ind Eng Chem. 2015;27:50–58. [Google Scholar]
- 14.Barnhill AE, Weeks KE, Xiong N, Day TA, Carlson SA. Identification of multiresistant Salmonella isolates capable of subsisting on antibiotics. Appl Environ Microbiol. 2010;76:2678–80. doi: 10.1128/AEM.02516-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dantas G, Sommer MOA, Oluwasegun RD, Church GM. Bacteria subsisting on antibiotics. Science. 2008;320:100–3. doi: 10.1126/science.1155157. [DOI] [PubMed] [Google Scholar]
- 16.de Bello González TJ, Zuidema T, Bor G, Smidt H, van Passel MWJ. Study of the Aminoglycoside Subsistence Phenotype of Bacteria Residing in the Gut of Humans and Zoo Animals. Front Microbiol. 2015;6:1550. doi: 10.3389/fmicb.2015.01550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xin Z, et al. Isolation, identification and characterization of human intestinal bacteria with the ability to utilize chloramphenicol as the sole source of carbon and energy. FEMS Microbiol Ecol. 2012;82:703–712. doi: 10.1111/j.1574-6941.2012.01440.x. [DOI] [PubMed] [Google Scholar]
- 18.Topp E, et al. Accelerated Biodegradation of Veterinary Antibiotics in Agricultural Soil following Long-Term Exposure, and Isolation of a Sulfamethazine-degrading sp. J Environ Qual. 2013;42:173–8. doi: 10.2134/jeq2012.0162. [DOI] [PubMed] [Google Scholar]
- 19.Tappe W, et al. Degradation of sulfadiazine by Microbacterium lacus strain SDZm4, isolated from lysimeters previously manured with slurry from sulfadiazine-medicated pigs. Appl Environ Microbiol. 2013;79:2572–7. doi: 10.1128/AEM.03636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh F, Amyes SGB, Duffy B. Challenging the concept of bacteria subsisting on antibiotics. Int J Antimicrob Agents. 2013;41:558–563. doi: 10.1016/j.ijantimicag.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 21.Crofts TS, et al. Draft Genome Sequences of Three β-Lactam-Catabolizing Soil Proteobacteria. Genome Announc. 2017;5:8–10. doi: 10.1128/genomeA.00653-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teufel R, et al. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci U S A. 2010;107:14390–5. doi: 10.1073/pnas.1005399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 25.Ghebre-Sellassie I, Hem SL, Knevel AM. Epimerization of benzylpenicilloic acid in alkaline media. J Pharm Sci. 1984;73:125–8. doi: 10.1002/jps.2600730135. [DOI] [PubMed] [Google Scholar]
- 26.Hmelo LR, et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc. 2015;10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valle F, Balbás P, Merino E, Bolivar F. The role of penicillin amidases in nature and in industry. Trends Biochem Sci. 1991;16:36–40. doi: 10.1016/0968-0004(91)90014-m. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–12. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svedas V, Guranda D, van Langen L, van Rantwijk F, Sheldon R. Kinetic study of penicillin acylase from Alcaligenes faecalis. FEBS Lett. 1997;417:414–8. doi: 10.1016/s0014-5793(97)01289-1. [DOI] [PubMed] [Google Scholar]
- 31.Hammond PM, Price CP, Scawen MD. Purification and properties of aryl acylamidase from Pseudomonas fluorescens ATCC 39004. Eur J Biochem. 1983;132:651–5. doi: 10.1111/j.1432-1033.1983.tb07413.x. [DOI] [PubMed] [Google Scholar]
- 32.Szewczuk A, Siewiński M, Słowińska R. Colorimetric assay of penicillin amidase activity using phenylacetyl-aminobenzoic acid as substrate. Anal Biochem. 1980;103:166–169. doi: 10.1016/0003-2697(80)90251-1. [DOI] [PubMed] [Google Scholar]
- 33.Oinonen C, Rouvinen J. Structural comparison of Ntn-hydrolases. Protein Sci. 2000;9:2329–2337. doi: 10.1110/ps.9.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McVey CE, Walsh MA, Dodson GG, Wilson KS, Brannigan JA. Crystal structures of penicillin acylase enzyme-substrate complexes: structural insights into the catalytic mechanism. J Mol Biol. 2001;313:139–150. doi: 10.1006/jmbi.2001.5043. [DOI] [PubMed] [Google Scholar]
- 35.McDonough MA, Klei HE, Kelly JA. Crystal structure of penicillin G acylase from the Bro1 mutant strain of Providencia rettgeri. Protein Sci. 1999;8:1971–81. doi: 10.1110/ps.8.10.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janes LE, Löwendahl AC, Kazlauskas RJ. Quantitative Screening of Hydrolase Libraries Using pH Indicators: Identifying Active and Enantioselective Hydrolases. Chem - A Eur J. 1998;4:2324–2331. [Google Scholar]
- 37.Batchelor FR, Chain EB, Hardy TL, Mansford KR, Rolinson GN. 6-Aminopenicillanic acid. III Isolation and purification. Proc R Soc London Ser B, Biol Sci. 1961;154:498–508. doi: 10.1098/rspb.1961.0047. [DOI] [PubMed] [Google Scholar]
- 38.Margolin AL, Svedas VK, Berezin IV. Substrate specificity of penicillin amidase from E. coli. Biochim Biophys Acta. 1980;616:283–9. doi: 10.1016/0005-2744(80)90145-x. [DOI] [PubMed] [Google Scholar]
- 39.Alkema WB, Floris R, Janssen DB. The use of chromogenic reference substrates for the kinetic analysis of penicillin acylases. Anal Biochem. 1999;275:47–53. doi: 10.1006/abio.1999.4300. [DOI] [PubMed] [Google Scholar]
- 40.Henke E, Bornscheuer UT. Fluorophoric assay for the high-throughput determination of amidase activity. Anal Chem. 2003;75:255–60. doi: 10.1021/ac0258610. [DOI] [PubMed] [Google Scholar]
- 41.Ferrández A, et al. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J Biol Chem. 1998;273:25974–86. doi: 10.1074/jbc.273.40.25974. [DOI] [PubMed] [Google Scholar]
- 42.Schumacher G, Sizmann D, Haug H, Buckel P, Böck A. Penicillin acylase from E. coli: unique gene-protein relation. Nucleic Acids Res. 1986;14:5713–27. doi: 10.1093/nar/14.14.5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 44.Larsson DGJ, de Pedro C, Paxeus N. Effluent from drug manufactures contains extremely high levels of pharmaceuticals. J Hazard Mater. 2007;148:751–755. doi: 10.1016/j.jhazmat.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 45.Pehrsson EC, et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533:212–6. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2005;102:8006–11. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoneda A, Wittmann BJ, King JD, Blankenship RE, Dantas G. Transcriptomic analysis illuminates genes involved in chlorophyll synthesis after nitrogen starvation in Acaryochloris sp CCMEE 5410. Photosynth Res. 2016;129:171–82. doi: 10.1007/s11120-016-0279-1. [DOI] [PubMed] [Google Scholar]
- 48.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Habegger L, et al. RSEQtools: a modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics. 2011;27:281–3. doi: 10.1093/bioinformatics/btq643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S, et al. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:msw054. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelly LA, Mezulis S, Yates C, Wass M, Sternberg M. The Phyre2 web portal for protein modelling, prediction, and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gasteiger E, et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ABC02 (this manuscript) and ABC07, ABC08, and ABC10 (ref. 21) WGS short-read raw data, assembled genomes, and RNASeq short-read data have been deposited to NCBI under BioProject number PRJNA385617 with BioSample numbers SAMN06915397, SAMN06915398, SAMN06915399, and SAMN06915400 respectively. Whole genome sequences may be found for ABC02, ABC07, ABC08, and ABC10 at DDBJ/ENA/GenBank under the accessions numbers NGUT00000000, NGUS00000000, NGUR00000000, and NGUQ00000000 respectively.