Abstract
Monophosphoryl lipid A (MPLA) is a clinically utilized toll-like receptor (TLR) 4 agonist that has been found to drive non-specific resistance to infection for up to two weeks. However, the molecular mechanisms conferring protection are not well understood. Here, we found that MPLA prompts resistance to infection, in part, by inducing a sustained and dynamic metabolic program in macrophages that supports improved pathogen clearance. Mice treated with MPLA had enhanced resistance to infection with Staphylococcus aureus and Candida albicans that was associated with augmented microbial clearance and organ protection. Tissue macrophages, which exhibited augmented phagocytosis and respiratory burst after MPLA treatment, were required for the beneficial effects of MPLA. Further analysis of the macrophage phenotype revealed that early TLR4-driven aerobic glycolysis was later coupled with mitochondrial biogenesis, enhanced malate shuttling, and increased mitochondrial ATP production. This metabolic program was initiated by overlapping and redundant contributions of MyD88- and TRIF-dependent signaling pathways as well as downstream mTOR activation. Blockade of mTOR signaling inhibited the development of the metabolic and functional macrophage phenotype and ablated MPLA-induced resistance to infection in vivo. Our findings reveal that MPLA drives macrophage metabolic reprogramming that evolves over a period of days to support a macrophage phenotype highly effective at mediating microbe clearance, and that this results in non-specific resistance to infection.
INTRODUCTION
Nosocomial infections, particularly those caused by antibiotic-resistant organisms, are one of the most pressing threats facing modern healthcare facilities (1). Critically ill and high-risk surgical patients are most vulnerable, although anyone receiving hospital care is at risk. The threat of infection is further compounded by the diversity of nosocomial pathogens, which includes Gram-positive, Gram-negative, and fungal organisms. The best existing defense against nosocomial infection is antibiotic prophylaxis, but antibiotic resistance is spreading so rapidly that strategies beyond antibiotics are needed. Immunotherapy offers a promising approach to address the problem (2). Treatment with TLR4 agonists, such as lipopolysaccharide (LPS), has been recognized for nearly 100 years to improve resistance to infections, but, due to a lack of mechanistic understanding and concerns about toxicity, it has been unclear whether they could be employed safely (3–5). Recent innovations have resulted in the development of TLR4 agonists that possess low toxicity but retain potent immunomodulatory properties. Monophosphoryl lipid A (MPLA), a hydrolyzed derivative of lipid A, is a TLR4 agonist that is employed clinically as a vaccine adjuvant and has the potential for further immunotherapeutic applications (6, 7). We previously demonstrated that (MPLA) improves resistance to Pseudomonas aeruginosa infection as well as polymicrobial sepsis caused by cecal ligation and puncture (8). Further, we found that resistance to infection lasts for at least 15 days after MPLA treatment (9). Thus, MPLA has great potential for use as a safe and effective prophylactic to decrease the incidence and severity of infections in hospitalized patients. However, given that natural TLR4 agonists are derived from Gram-negative bacteria, it was previously unclear whether MPLA confers resistance against common Gram-positive or fungal causes of nosocomial infection such as Staphylococcus aureus and Candida albicans. Furthermore, the molecular mechanisms by which MPLA drives resistance to infection remain unknown.
It has been long recognized that the yeast-derived dectin-1 ligand β-glucan induces resistance to infection in a manner qualitatively similar to that induced by TLR4 agonists (10, 11). Recent research has uncovered that β-glucan-induced resistance to infection is mediated, in part, through sustained metabolic reprogramming of monocytes (12, 13). β-glucan-induced metabolic reprogramming sensitizes monocytes to LPS stimulation such that they have augmented pro-inflammatory cytokine secretion in response to LPS, a phenomenon that has been termed “trained immunity”. This LPS sensitization is hypothesized to underlie the protective phenotype (14). In congruence with this finding, we recently demonstrated that TLR4 agonists similarly induce persistent augmentation of glycolysis in macrophages (9). Yet, in stark contrast with β-glucan, TLR4 agonists induce LPS tolerance, which is defined by suppressed cytokine production in response to LPS challenge (15, 16). Thus, while β-glucan and TLR4 agonists both induce myeloid cell metabolic reprogramming, TLR4-mediated reprogramming likely has a unique and unexplored impact on infection resistance.
Here, we postulated that the mechanism by which TLR4 agonists induce resistance to infection is by driving sustained metabolic reprogramming of macrophages that fuels improved cell-intrinsic antimicrobial responses, rather than by altering LPS sensitivity. We demonstrate that MPLA treatment yields resistance to systemic infection with S. aureus and C. albicans, by inducing a surprisingly dynamic macrophage metabolic program characterized by early aerobic glycolysis followed by sustained glycolysis, mitochondrial biogenesis, increased malate shuttling, and elevated oxygen consumption. Further, we found that the metabolic signaling protein mTOR initiates this phenotype, facilitates persistent macrophage antimicrobial activity and chemokine secretion, and is required for MPLA-induced resistance to infection.
MATERIALS AND METHODS
Mouse Model of Infection
All animal procedures complied with the National Institutes of Health guidelines for the Care and Use of Experimental Animals, and were approved by the Institutional Animal Care and Use Committee at Vanderbilt University. Male 10 to 12-week-old BALB/c mice were purchased from Envigo Laboratories (Indianapolis, IN). Male wild type, RAG2−/− C57/Bl6 and B6.129S4-Ccr2tm1lfc/J mice were purchased from Jackson Laboratories (Bar Harbor, ME).
Staphylococcus aureus was purchased from American Type Culture and Collection (Manassas, VA; ATCC 25923). The culture was grown in tryptic soy broth and resuspended in sterile saline solution prior to inoculation. For intravenous (i.v.) infection, mice were inoculated with 1 × 108 CFU S. aureus diluted in lactated Ringers (LR) solution through the dorsal penile vein. For intraperitoneal (i.p.) infection, mice were inoculated by i.p. injection with 1 × 108 CFU S. aureus diluted in LR solution. Bacterial burden in the spleen, lungs, kidney, and peritoneal lavage fluid was quantified by performing serial dilutions of tissue homogenates or peritoneal lavage fluid, followed by culture on tryptic soy agar overnight (37°C). For survival studies, mice were monitored daily for 15 days following i.v. inoculation with S. aureus.
C. albicans was purchased from American Type Culture and Collection (Manassas, VA; ATCC 10231 Strain 3147 Robin) The culture was grown in yeast extract-peptone-dextrose broth and resuspended in sterile saline solution prior to inoculation. For intravenous (i.v.) infection, mice were inoculated with 1 × 108 CFU S. aureus diluted in LR solution through the dorsal penile vein.
MPLA Treatment
MPLA derived from Salmonella enterica serotype Minnesota Re 595, was purchased from Sigma-Aldrich Corp. (St. Louis, MO) or Invivogen. MPLA was solubilized in 0.2% triethylamine solution (1 mg/ml) or DMSO and sonicated for 1 hour at 40°C. For treatment, MPLA was diluted in LR solution(100 μg/ml) and then administered by i.v. or i.p. injection at 20μg in 0.2 ml. Vehicle treated mice received i.v. or i.p. injection of 0.2 ml LR solution.
Cell Depletion
In neutrophil depletion experiments, mice received i.v. injection with 100 μg of monoclonal anti-mouse 1A8-Ly6G purified blocking antibody or purified rat monoclonal IgG2a, kappa isotype control (eBioscience, San Diego, CA) 24 hours prior to the first MPLA injection. To deplete monocytes/macrophages, mice received i.v. injection with 0.2 ml clodronate-liposomes or PBS control-liposomes (ClodronateLiposomes.com, Amsterdam, Netherlands) 24 hours prior to the first MPLA injection.
Organ injury markers and evaluation of leukocytes in blood
Whole blood was harvested by carotid artery laceration under isoflurane anesthesia and collected in heparinized syringes. Blood was collected in K3EDTA tubes (Greineer Biosciences, San Diego, CA) and stored on ice for complete blood count (CBC), analysis of organ injury markers, and cytokine measurements. CBC measurements were performed using a Forcyte veterinary hematology analyzer (Oxford Science, Oxford, CT). Remaining blood was centrifuged (4750 rpm for 15 minutes at 4°C) to collect plasma for organ injury marker analyses. Blood urea nitrogen (BUN) was measured using the Vet Axcel Chemistry Analyzer (Alfa Wassermann Diagnostic Technologies, LLC, West Caldwell, NJ). The CBC and BUN measurements were analyzed by the Translational Pathology Shared Resource at Vanderbilt University Medical Center.
Cytokine Measurements
Concentrations of IL-6 and TNF-α were measured by Ready-Set-Go ELISA (eBioscience, Inc., San Diego, CA). Concentrations of CCL5, CCL2, CCL3, and CXCL2 were detected by DuoSet ELISA kits from R&D Systems (Minneapolis, MN). All ELISAs were analyzed using a BioTek ELx800 (Biotek, Winooski, VT).
Flow Cytometry
Leukocytes were suspended in cold PBS (1 × 107 cells/ml), incubated with anti-mouse CD 16/32 (eBioscience, San Diego, CA, 1 μl/ml), then flurochrome-conjugated antibodies were added (0.5 μg/106 cells/0.1 ml) at 4°C for 30 minutes. Flow antibodies used for these studies included anti-F4/80-FITC (clone BM8, eBioscience), anti-Ly6G-PE (clone 1A8, BD Biosciences), anti-Ly6C-PE Cy5.5 and appropriate isotype controls (eBioscience and BD Biosciences). Neutrophils are identified as Ly6G+F4/80− cells, macrophages are identified as Ly6C−F4/80+ cells, and monocytes are identified as Ly6C+F4/80+ cells. Samples were run on an Accuri C6 flow cytometer (BD Biosciences, San Diego, CA). Data were analyzed using Accuri C6 software.
Immunohistochemistry
Kidney, lung, and spleen were harvested from mice 3 days after i.v. infection with S. aureus. Harvested organs were fixed in 10% neutral buffered formalin overnight. The tissue was processed routinely, embedded in paraffin and sectioned at 5 microns. Slides were placed on the Leica Bond Max IHC stainer and deparaffinized. For F4/80: Enzymatic induced antigen retrieval was performed on the Bond Max using Proteinase K (Dako, North America, Inc) for 5 minutes. Slides were incubated with anti-F4/80 (NB600-404, Novus Biologicals) for one hour at a 1:900 dilution and then incubated in a rabbit anti-rat secondary antibody (BA-4001, Vector Laboratories, Inc.) for 15 minutes at a 1:2000 dilution. The Bond Polymer Refine detection system was used for visualization. For neutrophil: Heat induced antigen retrieval was performed on the Bond Max using their Epitope Retrieval 2 solution for 20 minutes. Slides were incubated with neutrophil marker 6A608 (SC-71674, Santa Cruz, Dallas, TX) for one hour at a 1:250 dilution and then incubated in a rabbit anti-rat secondary antibody (BA-4001, Vector Laboratories, Inc.) for 15 minutes at a 1:2000 dilution. For all slides, the Bond Polymer Refine detection system was used for visualization. All slides were then dehydrated, cleared, and cover-slipped. For optical density quantification, two 10X magnification images from random non-overlapping locations of each organ were analyzed with Immunohistochemistry Image Analysis Toolbox in ImageJ (https://imagej.nih.gov/ij/plugins/ihc-toolbox/index.html)(17). The stock H-DAB model was used to determine the IHC stain followed by a mean gray value measurement for quantification.
Phagocytosis Assay
For in vivo studies, mice were injected i.p. with 500 μg of pHrodo-tagged S. aureus bio-particles (Life Technologies, Carlsbad, CA). After 6 hours, peritoneal leukocytes were harvested by peritoneal lavage with 2mL cold PBS. Following harvest, leukocytes were incubated with surface marker antibodies (anti-Ly6G-FITC, anti-F4/80-FITC, and anti-Ly6C-PE-Cy5.5) and pHrodo mean fluorescence intensity (MFI) was determined by flow cytometry. For in vitro studies, pHrodo-tagged S. aureus was incubated with bone marrow derived macrophages for 2 hours. pHrodo MFI was determined every 15 minutes by a BioTek Synergy MX plate reader (Biotek, Winooski, VT).
Respiratory Burst Assay
Peritoneal leukocytes were assessed via the Respiratory Burst Assay (Cayman Chemical Company, Ann Arbor, MI). In brief, peritoneal leukocytes were obtained by peritoneal lavage with 2 ml cold PBS and incubated with dihydrorodamine-123. ROS were then elicited by 45 minutes of 200nM phorbol 12-myristate 13-acetate (PMA) stimulation. After stimulation, rhodamine 123 MFI was determined via flow cytometry. Neutrophils, macrophages, and monocytes were determined by FSC-A versus SSC-A.
Bone Marrow Derived Macrophages
WT bone marrow derived macrophages were derived from 8–16 week old C57/Bl6 male mice purchased from the Jackson Laboratories. Male MyD88−/− and TRIF−/− mice were purchased from the Jackson Laboratories. Femurs from male TLR4−/− mice were received as a gift from Dr. Brad Grueter and Daniel Kashima of Vanderbilt University. Femurs from male MyD88−/−TRIF−/− double knockout mice were received as a gift from Dr. Doug Golenbock from the University of Massachusetts Medical School. For macrophage generation, femurs were flushed with 5mL cold PBS and bone marrow was incubated on Falcon petri dishes in RPMI 1640 with glutamine and 25mM HEPES (Gibco, Carlsbad, CA) containing 10% certified FBS (Gibco, Carlsbad, CA), 1% antibiotic-antimycotic (Gibco, Carlsbad, CA), and 10ng/mL mouse recombinant macrophage colony stimulating factor (M-CSF, R&D Systems, Minneapolis, MN) for 7 days. At the initiation of the growth period, bone marrow cell concentration was 4×104/ml, as this density has been found to yield mature macrophages (18). M-CSF remained in the culture throughout experimentation. After 7 days of incubation, macrophages were washed with PBS and incubated with media containing various TLR ligands for 24 hours. At 24 hours, macrophages were washed thoroughly with PBS and rested in media for 3 days before assessment. 24hr macrophages were stimulated with MPLA 24hrs prior to assessment.
Measurements of gross metabolism
For ATP assays, BMDMs were plated at 5×105/mL and were lysed with Luciferase Cell Culture Lysis Reagent (Promega, Madison, WI). ATP concentration in the lysate was the incubated with firefly luciferase along with the luminescent ATP Determination Kit (Invitrogen, Carlsbad, CA). Luminescence was then determined by the BioTek Synergy MX plate reader (Biotek, Winooski, VT).
For glucose and lactate measurements, glucose and lactate in the media were measured by the YSI 2300 Stat Plus Glucose & Lactate Analyzer (YSI, Yellow Springs, OH). For nitric oxide secretion assays, nitrites in the media were detected by Griess Reagent Kit (Thermo Fisher Scientific, Waltham, MA). Media was incubated with a 1:1 mixture of N-(1-naphthyl)ethylenediamine and sulfanilic acid for 30 minutes at room temperature and were assessed by spectrophotometric analysis on the BioTek Synergy MX plate reader (Biotek, Winooski, VT). For Mito Tracker staining, MitoTracker Green was purchased from Thermo Fisher (Waltham, MA). BMDMs were incubated in MitoTracker green for 45 minutes and then assessed via flow cytometry.
Seahorse Assay
Cells were plated in a 96-well Seahorse assay plate at 4×104 cells/well in Seahorse Assay Media and assessed on the Seahorse XFe96 Extracellular Flux Analyzer (Agilent Technologies, Santa Clara, CA). For the glycolysis stress test, cells were sequentially treated with 10mM glucose (RPI, Mount Prospect, IL), 1μM oligomycin (Agilent Technologies), and 50mM 2-deoxyglucose (Sigma-Aldrich). For the mitochondrial stress test, Assay media was supplemented with 10mM glucose and cells were sequentially treated with 1μM oligomycin (Agilent Technologies), 1μM FCCP (Agilent Technologies), and 0.5 μM of antimycin A and rotenone (Agilent Technologies).
Mitochondrial DNA Measurement
Mitochondrial DNA (mtDNA) and nuclear DNA (nucDNA) were simultaneously isolated using a DNeasy Blood & Tissue Kit (Qiagen). DNA concentration and quality were verified with a Thermo Scientific NanoDrop 2000 spectrophotometer. Presence of the mitochondrial gene MT-CYB and nuclear gene ACTB were analyzed by Real-Time PCR with the SsoFast EvaGreen Supermix Kit (Bio-Rad, Hercules, CA). Primer sequences for MT-CYB: Forward: GCCACCTTGACCCGATTCT; Reverse: TTCCTAGGGCCGCGATAAT. For ACTB Forward: AGCCATGTACGTAGCCATCCA; Reverse: TCTCCGGAGTCCATCACAATG. Real-Time PCR reactions were run in quadruplicate on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA) and quantification of gene expression was determined by the comparative ΔΔCt method.
Western Blot
Cells were washed and lysed with radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich) containing PhosStop and complete Protease Inhibitor (Roche Diagnostics, Indianapolis, IN, USA). Samples were separated by gel electrophoresis on Mini-Protean precast 4–20% Tris-glycine gels (Bio-Rad). Proteins were then transferred to nitrocellulose membranes (Perkin-Elmer, Boston, MA, USA). Membranes were blocked with 5% fraction V BSA (RPI Corp., Mount Prospect, IL, USA) and incubated with primary antibodies at 4°C overnight. Protein bands were detected with HRP-conjugated secondary antibodies, incubation with ECL reagent (Bio-Rad), and film exposure. Films were processed and analyzed using ImageJ software from the U.S. National Institutes of Health (Bethesda, MD, USA).
Metabolite extraction and gas chromatography - mass spectrometry (GC-MS) analysis of 13C-glucose labeling
BMDMs were primed and placed in glucose-free complete media supplemented with 11mM U-13C6-glucose (Cambridge Isotopes, Tewksbury, MA) and incubated for 24 hours. 1μg/mL MPLA was added to 24hr macrophages for the entire incubation period. BMDMs were then harvested as described in Noguchi et al.(19). Cell metabolism was immediately quenched with pre-cooled −80°C methanol. A biphasic extraction (Folch method) was then used to separate polar metabolites into an aqueous phase and non-polar lipid metabolites into an organic phase (20). The extraction results in mixing free metabolites from all subcellular compartments. Polar metabolites were converted to their tertbutylsilyl derivatives using MBTSTFA + 1% TBDMCS (Thermo Fisher Scientific) (21). Nonpolar extracts were converted to fatty acid methyl esters as described in Young et al.(21). Each derivatized sample was injected into a GC-MS (Agilent 7890A/5975C) equipped with a 30 m HP-5 MS capillary column for analysis of isotopic enrichment. Raw mass isotopologue distributions were corrected for natural isotope abundance as described by Fernandez et al. (22).
13C Metabolic Flux Analysis
GC-MS isotopologue data was analyzed using the MATLAB-based software package INCA (publicly available at http://mfa.vueinnovations.com/mfa)(23). Isotopologue balances were simulated for BMDM metabolism using an elementary metabolite unit (EMU) decomposition of the metabolic reaction network to simulate changes in isotope labeling resulting from variations in intracellular metabolic fluxes(24, 25). The metabolic model provides a detailed description of the TCA cycle and anaplerotic pathways. The complete reaction network and a list of underlying assumptions are provided in Figure S6. The model was constrained by incorporating glucose and oxygen consumption rates (pmole/cell/day) to calculate absolute metabolic fluxes. Oxygen consumption rates were determined by basal oxygen consumption rate measurements in the Seahorse Xfe96. All model fits were over-determined by 32 measurements. Flux estimation was repeated a minimum of 50 times from random initial values to ensure a global minimum was obtained. All results were subjected to a χ2 statistical test to assess goodness-of-fit.
RNA-sequencing
Total RNA was isolated under an RNase-free environment, using the RNeasy Mini Kit (Qiagen). RNA concentration and quality were verified with a Thermo Scientific NanoDrop 2000 spectrophotometer. Total RNA quality was then re-assessed using the 2100 Bioanalyzer (Agilent Technologies). At least 200ng of DNase-treated total RNA with a RNA integrity number greater than 6 was used to generate polyA (mRNA) enriched libraries using TruSeq Stranded mRNA sample kits with indexed adaptors (Illumina). Library quality was assessed using the 2100 Bioanalyzer (Agilent Technologies) and libraries were quantitated using KAPA Library Quantification Kits (KAPA Biosystems). Pooled libraries were subjected to 75 bp paired-end sequencing according to the manufacturer’s protocol (Illumina HiSeq3000). Bcl2fastq2 Conversion Software (Illumina) was used to generate de-multiplexed Fastq files.created by a polyA-selected build. The RNA-seq data was quality controlled following the multi-perspective guideline (26). Quality control metrics were computed from software QC3 (27). No quality issues were observed. Alignment was performed by TopHat 2 (28) on MM10 reference genome. Read count per gene was generated using HTSeq (29). Differential expression analyses were performed using MultiRankSeq (30) which combines results from DEGseq (31), edgeR (32), and baySeq (33). Log2 fold change values shown are derived from DEGseq with an adjusted p<.05. The data from RNA-sequencing have been deposited in NCBI’s Gene Expression Omnibus (34) and are accessible through GEO Series accession number GSE111730 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111730).
In vitro and in vivo rapamycin treatment
For in vitro studies, rapamycin (Cayman Chemical, Ann Arbor, MI) was reconstituted in DMSO and added to BMDM culture media at 100nM 1 hour prior to the addition of MPLA. Rapamycin was then removed along with MPLA. For in vivo studies, Rapamycin was reconstituted in 100% ethanol and diluted in a vehicle containing90% distilled H2O, 5% Tween 80 (Sigma-Aldrich, St. Louis, MO), and 5% polyethylene glycol 400 (Sigma-Aldrich, St. Louis, MO). Rapamycin or vehicle was then administered at 3mg/kg to mice via i.p. injection 3 hours prior to i.v. injection of MPLA.
Statistics
All data were analyzed with GraphPad Prism 7 (La Jolla, CA) unless otherwise stated. Data from multiple group experiments were analyzed using one-way ANOVA followed by Tukey’s multiple-comparison post-hoc test. All data values are presented as mean ± SEM, except for bacterial counts and body temperature, for which median values alone are designated. A p value of less than or equal to 0.05 was considered statistically significant. For RNA-sequencing studies, Fastq files were analyzed by DESeq. An adjusted p value of 0.05 was used as a cutoff for statistical significance.
RESULTS
MPLA-primed mice are broadly resistant to infection with common nosocomial pathogens
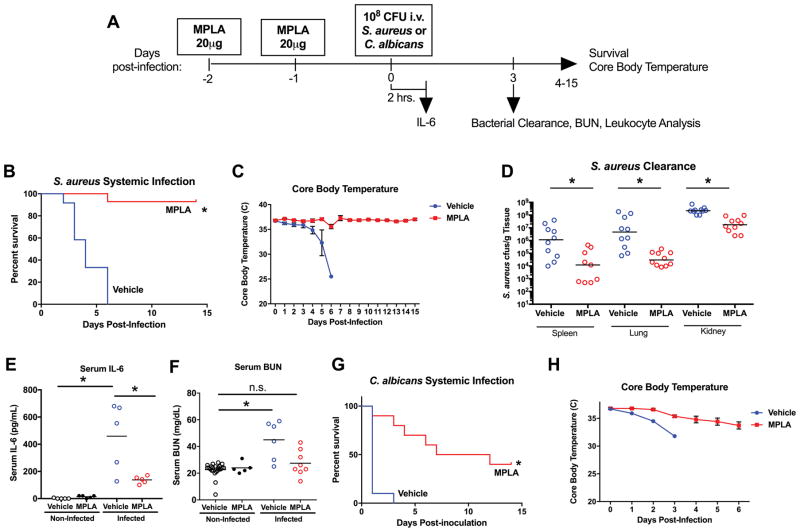
Our previous work showed that MPLA prophylaxis protects against infection with Gram-negative pathogens such as P. aeruginosa and polymicrobial sepsis caused by cecal ligation and puncture(8). Here, we performed further studies to determine whether MPLA-primed mice are resistant to systemic infection with Gram-positive and fungal pathogens that are common causes of nosocomial infection. Mice were primed with intravenous MPLA on 2 consecutive days and challenged intravenously with S. aureus or C. albicans 24 hours later (Figure 1A). Compared to vehicle-primed control mice during S. aureus infection, MPLA-primed mice showed improved survival (Figure 1B), preservation of core body temperature (Figure 1C), lower S. aureus colony forming units in the spleen, lung, and kidney (Figure 1D), lower plasma IL-6 concentrations (Figure 1E) and less kidney injury as indicated by lower plasma BUN concentrations (Figure 1F). Uninfected MPLA-primed mice did not demonstrate differences in IL-6 or BUN compared to uninfected vehicle-primed mice (Figure 1E–F). In a parallel study with intravenous C. albicans infection, MPLA-primed mice showed improved survival and preserved core body temperature (Figure 1G–H).
Figure 1. MPLA induces resistance to S. aureus and C. albicans infection.
A) WT BALB/c mice were injected with intravenous MPLA (20μg/mouse) or vehicle one and two days prior to intravenous inoculation with intravenous 108 cfu S. aureus or 108 cfu C. albicans. B) Kaplan Meier survival curve of vehicle- and MPLA- primed mice. (n=12 mice/group) after S. aureus inoculation. C) Core (rectal) body temperature assessed daily after S. aureus inoculation. D) S. aureus cfu per gram of tissue recoverable from whole spleen, lung, and kidney in vehicle and MPLA-primed mice at 3 days after S. aureus inoculation. E) Concentration of serum IL-6 2 hours after S. aureus inoculation or vehicle. F) Concentration of serum BUN 3 days after S. aureus inoculation or vehicle. G) Kaplan Meier survival curve of vehicle- and MPLA- primed mice. (n=12 mice/group) after intravenous 108 cfu C. albicans inoculation. H) Core (rectal) body temperature after C. albicans inoculation. Data shown as mean +/- SEM. For Kaplan-Meier plots *, p<.05 via log-rank Mantel-Cox test. Otherwise, *, p< .05 as determined by ANOVA with Tukey’s post-hoc multiple comparison test.
Tissue macrophages largely mediate MPLA-induced resistance to S. aureus infection
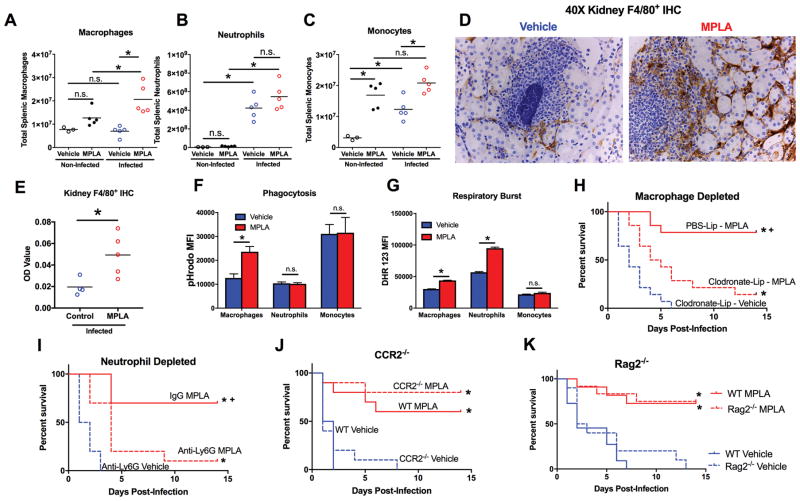
The cellular mechanisms behind MPLA-induced resistance to S. aureus were then explored. While total tissue leukocyte populations were increased in both vehicle and MPLA-primed mice 3 days after S. aureus infection, MPLA-primed mice had significantly increased splenic macrophages and monocytes, but not neutrophils, compared to vehicle-primed mice (Figure 2A–C). Uninfected MPLA-primed mice also demonstrated increased splenic monocytes, a trend towards increasing macrophages, but no difference in neutrophils, compared to uninfected vehicle-primed mice. Immunohistochemical analysis of tissues from mice infected with S. aureus showed a stark increase in F4/80+ macrophages within the kidney of MPLA-primed mice (Figure 2D–E). Macrophages were distributed within the interstitium between renal tubules and examination of S. aureus abscesses within the kidney showed increased infiltration by macrophages in MPLA-treated mice (Figure 2D). Upon assessment of macrophage antimicrobial function, peritoneal macrophages from MPLA-primed mice showed increased phagocytic capacity and respiratory burst activity (Figure 2F–G). Neutrophils and monocytes from MPLA-primed mice did not show increased phagocytic capacity but respiratory burst activity was increased in neutrophils from MPLA-primed mice (Figure 2F–G).
Figure 2. MPLA induces resistance to infection via modulation of tissue macrophages and neutrophils.
A–C) Total splenic A) macrophages, B) neutrophils, and C) monocytes 3 days after S. aureus inoculation or vehicle in MPLA- and vehicle-primed mice. D) Representative image of F4/80+ immunohistochemical staining (brown) in kidney 3 days after S. aureus inoculation in MPLA- and vehicle-primed mice E) Optical density quantification of F4/80+ staining in kidneys. F) Mice were primed with MPLA or vehicle via intraperitoneal injection and 24 hours later pHrodo tagged S. aureus bioparticles were injected into the peritoneal cavity. Peritoneal leukocytes were harvested 6 hours later and assessed for pHrodo MFI via flow cytometry. G) Mice were primed with MPLA or vehicle via intraperitoneal injection and 24 hours later leukocytes were harvested from the peritoneal cavity. Respiratory burst was elicited by 45 minutes of PMA stimulation and DHR 123 MFI of macrophages, neutrophils, and monocytes was assessed by flow cytometry. H) Mice were administered intravenous clodronate-liposomes 24 hours prior to the first MPLA administration. Mice were then inoculated with intravenous 108 cfu S. aureus. Kaplan-Meier survival plot after S. aureus infection (n=15 mice/group). I) Mice were administered intravenous anti-Ly6G antibody 24 hours prior to the first MPLA administration. Mice were then inoculated with intravenous 108 cfu S. aureus. Kaplan-Meier survival plot after S. aureus infection (n=10 mice/group). J) C57BL6 WT and CCR2−/− mice were primed with MPLA or vehicle and inoculated with intravenous 108 cfu S. aureus. Kaplan-Meier survival plot after S. aureus infection (n=10 mice/group). K) WT C57BL6 and RAG2−/− mice were injected with intravenous MPLA or vehicle prior to intravenous inoculation with 108 cfu S. aureus. Kaplan-Meier survival plot (n=10 mice/group). Data shown as mean +/− SEM. For Kaplan-Meier plots *, p<.05 via log-rank Mantel-Cox test. Otherwise, *, p< .05 as determined by ANOVA with Tukey’s post-hoc multiple comparison test
To determine the contribution of leukocyte subsets to MPLA-induced resistance to S. aureus infection, different leukocyte populations were selectively ablated prior to vehicle or MPLA treatment. Tissue macrophage depletion with clodronate-laden liposomes ablated the resistance to S. aureus infection conferred by MPLA treatment (Figure 2H). Likewise, depletion of neutrophils by treatment with anti-Ly6G decreased the survival benefit in MPLA-primed mice challenged with S. aureus (Figure 2I). We then assessed whether inflammatory monocytes contributed to MPLA-induced protection from infection by employing CCR2−/− mice. CCR2 is required for inflammatory monocyte emigration from the bone marrow, and CCR2−/− mice have a diminished circulating Ly6C+ inflammatory monocyte pool (35). Wild type and CCR2−/− mice had similar survival rates during S. aureus infection and MPLA successfully protected both CCR2−/− and wild type mice (Figure 2J). Furthermore, tissue bacterial burden was not different when comparing wild type and CCR2−/− mice treated with MPLA (Supplementary Figure 1A–C). MPLA-primed CCR2−/− mice demonstrated accumulation of F4/80+ macrophages in kidney and lung that was comparable to wild type (Supplementary Figure 1D). Rag2−/− mice were then employed to assess the importance of T and B lymphocytes to MPLA-mediated resistance against S. aureus infection. MPLA treatment successfully protected both Rag2−/− and wild type mice, while vehicle-treated mice in both groups succumbed to infection (Figure 2K). Thus, MPLA protects against S. aureus through a mechanism involving the augmented antimicrobial capacity and recruitment of tissue macrophages, with help from neutrophils, but not inflammatory monocytes or T- and B- lymphocytes.
TLR4 agonists drive a persistent and dynamic metabolic program in macrophages
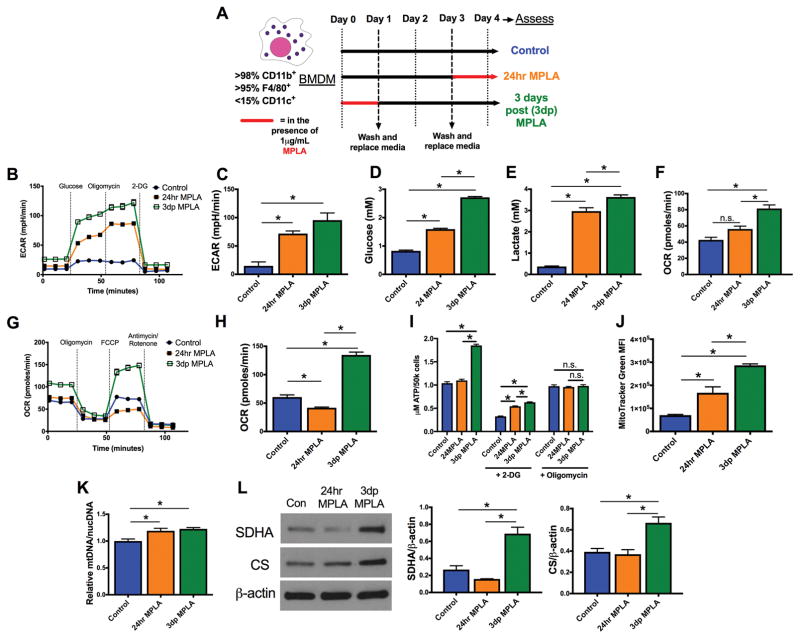
We next sought to determine the molecular mechanisms underlying the persistently augmented macrophage antimicrobial function. Macrophage metabolism drives macrophage antimicrobial function, and we recently reported an increase in macrophage glycolysis at three days following TLR4 agonist exposure (9). Acute exposure of macrophages to TLR agonists (<24hrs) is known to increase glycolysis without changing oxygen consumption (36), a phenomenon termed aerobic glycolysis or the Warburg effect, so we wondered whether the glycolytic phenotype we observed at 3 days after MPLA treatment reflected a persistence of early aerobic glycolysis or an adapted metabolic program. To investigate this, bone marrow derived macrophages (BMDMs) were treated with MPLA for 24 hours, washed, and cultured for an additional 3 days in the absence of MPLA. These macrophages are henceforth referred to as 3 days post-MPLA (3dp) macrophages and are representative of primed macrophages that mediate resistance to infection after MPLA treatment in vivo. We compared 3dp macrophages to classical pro-inflammatory macrophages, defined as macrophages exposed to MPLA for 24 hours (24hr macrophages), and control macrophages that did not receive MPLA treatment. The experimental protocol is defined in Figure 3A. Firstly, the extracellular acidification rate (ECAR) of both 24hr and 3dp macrophages was assessed as an index of glycolytic capacity. The ECAR was significantly elevated in both 24hr and 3dp groups compared to control (Figure 3B–C). The 3dp MPLA and 24hr MPLA macrophages also had increased glucose consumption and lactate production (Figure 3D–E). Interestingly, 3dp macrophages exhibited a significantly elevated basal oxygen consumption rate (OCR) compared to controls, whereas the basal OCR of 24hr macrophages was unchanged (Figure 3F). A mitochondrial stress test showed that 3dp macrophages had significantly elevated maximal OCR compared to controls, whereas maximal OCR of 24hr macrophages was suppressed (Figure 3G–H). The 3dp macrophages produced significantly more ATP than control or 24hr macrophages, and the increase in ATP production could be blocked by the addition of the ATP synthase inhibitor oligomycin and 2-deoxyglucose (2-DG), an inhibitor of glucose metabolism (Figure 3I). LPS induced a similar metabolic phenotype (data not shown).
Figure 3. MPLA drives persistent and dynamic metabolic reprogramming in macrophages.
A) Bone marrow derived macrophages (BMDMs) were primed with 1μg/mL MPLA for 24 hours, washed, and rested for 3 days (3dp macrophages). These macrophages were compared to BMDMs stimulated with 1μg/mL MPLA for 24 hours prior to assessment (24hr macrophages) and untreated BMDMs (control). B) Glycolysis stress test of macrophages as determined by the Seahorse Xfe96. C) Maximal extracellular acidification rate (ECAR) derived after the addition of oligomycin in the glycolysis stress test D) Glucose consumed from BMDMs over 24 hours, as determined by glucose concentration in cell-free medium subtracted by cell-containing medium. E) Lactate produced by BMDMs over 24 hours, as determined by lactate in cell-containing medium subtracted by cell free medium. F) Basal oxygen consumption rate (OCR) determined after the addition of glucose in the glycolysis stress test. G) Mitochondrial stress test of BMDMs as determined by the Seahorse Xfe96. H) Maximal oxidative rate derived after the addition of FCCP in the mitochondrial stress test. I) Intracellular ATP from BMDMs as determined by luminescence assay. Some BMDMs were exposed to 10mM 2-DG or 1μM oligomycin for 3 hours prior to the assay. J) MitoTracker Green staining of BMDMs as determined by flow cytometry. K) Mitochondrial DNA (mtDNA) / nuclear DNA (nucDNA) as determined by qPCR. L) Succinate dehydrogenase (SDHA), citrate synthase (CS), and β-actin as determined by western blot. All bands shown derived from the same samples. Blot cropped to demonstrate relevant bands. Densitometry of SDHA and CS compared to β-actin derived from ImageJ shown. All experiments replicated at least twice. Data shown as mean +/- SEM. *, p< .05 as determined by ANOVA with Tukey’s post-hoc multiple comparison analysis.
Increased basal and maximal oxygen consumption suggests that 3dp macrophages have an altered mitochondrial pool. To assess this, we evaluated mitochondrial content in control and MPLA-primed macrophages. MitoTracker Green MFI was elevated in 24hr and, to an even greater extent, in 3dp macrophages (Figure 3J). Relative mitochondrial DNA (mtDNA) to nuclear DNA (nucDNA) levels were also elevated in 24hr and 3dp macrophages (Figure 3K). Finally, cellular content of the mitochondria-centered enzymes succinate dehydrogenase and citrate synthase was increased in 3dp macrophages compared to control and 24 hour macrophages (Figure 3L). Thus, early glycolysis is sustained following MPLA priming, but early disruptions to oxygen consumption recover in association with mitochondrial biogenesis and increased ATP production.
Malate shuttling sustains elevated mitochondrial activity in MPLA-primed macrophages
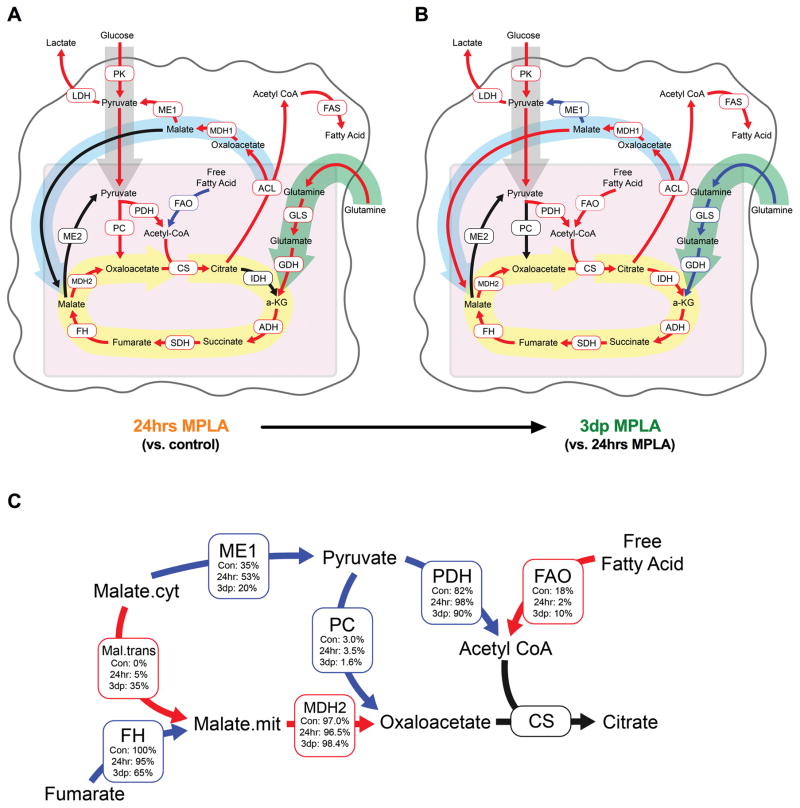
In order to define how MPLA-primed macrophages adapt their metabolic programs over time to fuel late mitochondrial ATP production, we performed U-13C-glucose metabolic flux analysis and modeled the flux rates of metabolic enzymes (Figure 4). Compared to control macrophages, 24hr macrophages expectedly had increased glycolytic flux, along with increased citrate export from the mitochondria to the cytosol, increased fatty acid synthesis (FAS), and increased glutamine anaplerosis compensating for the mitochondrial citrate loss (Figure 4A). Once in the cytosol, citrate is converted to oxaloacetate and acetyl-CoA by ATP citrate lyase (ACL) (37). Cytosolic acetyl-CoA is then used for FAS, whereas cytosolic oxaloacetate is recycled in some manner. Cytosolic oxaloacetate can either be converted to pyruvate resulting in further glycolysis or converted to malate and transported back into the mitochondria resulting in NADH reducing equivalents entering the mitochondria for use in the electron transport chain. Notably, 24hr macrophages recycled all of this cytosolic oxaloacetate back to pyruvate via malic enzyme 1 (Figure 4A). 3dp macrophages, compared to 24hr macrophages, had elevated glycolytic flux and citrate export, but rather than convert cytosolic oxaloacetate to pyruvate, 3dp macrophages significantly increased malate transport to the mitochondria (Figure 4B). The increase in malate transport was further examined by determining the relative flux rates of each reaction that feeds into citrate synthase (Figure 4C). Cytosolic-derived malate, compared to fumarate hydratase-derived malate, constituted a significantly greater proportion of the malate used by malate dehydrogenase 2 in 3dp macrophages. Malate dehydrogenase 2-derived oxaloacetate, compared to pyruvate carboxylase-derived oxaloacetate, constituted a significantly greater proportion of the oxaloacetate used by citrate synthase (Figure 4B). Complete metabolic flux analysis data, along with metabolite enrichments are shown in Supplementary Figure 2 and Supplementary Table 1. Overall, these data suggest that 3dp macrophages adapt to early MPLA-induced disruptions by increasing a malate shuttle to drive cytosolic NADH reducing equivalents into the mitochondria to generate ATP via oxidative phosphorylation.
Figure 4. Malate shuttling sustains elevated mitochondrial activity in MPLA primed macrophages.
Control or MPLA-primed BMDMs were exposed to media where the only glucose source was U-13C-glucose for 24 hours. BMDMs were then harvested and metabolites were assessed for incorporation of the 13C isotope by GC-MS. A) Absolute metabolic flux rate map of 24hr MPLA macrophages compared to control macrophages as generated through metabolic flux analysis by INCA (23). Red and blue arrows indicate statistically significant increases or decreases, respectively. B) Absolute metabolic flux rate map of 3dp MPLA macrophages compared to 24hr macrophages. Red and blue arrows indicate statistically significant increases or decreases, respectively. C) Relative enzyme flux rates of control, 24 hr MPLA, and 3dp MPLA macrophages. All enzyme reaction rates were compared to a within-group designation of citrate synthase at 100%. Red and blue arrows indicate statistically significant increases or decreases, respectively, for 3dp MPLA macrophages compared to 24hr MPLA macrophages. Significance for all maps identified by p< .05 as determined by ANOVA with Tukey’s post-hoc multiple comparison analysis. Enrichment labeling can be found in Supplementary Figure 2 and values for each reaction can be found in Supplementary Table 1.
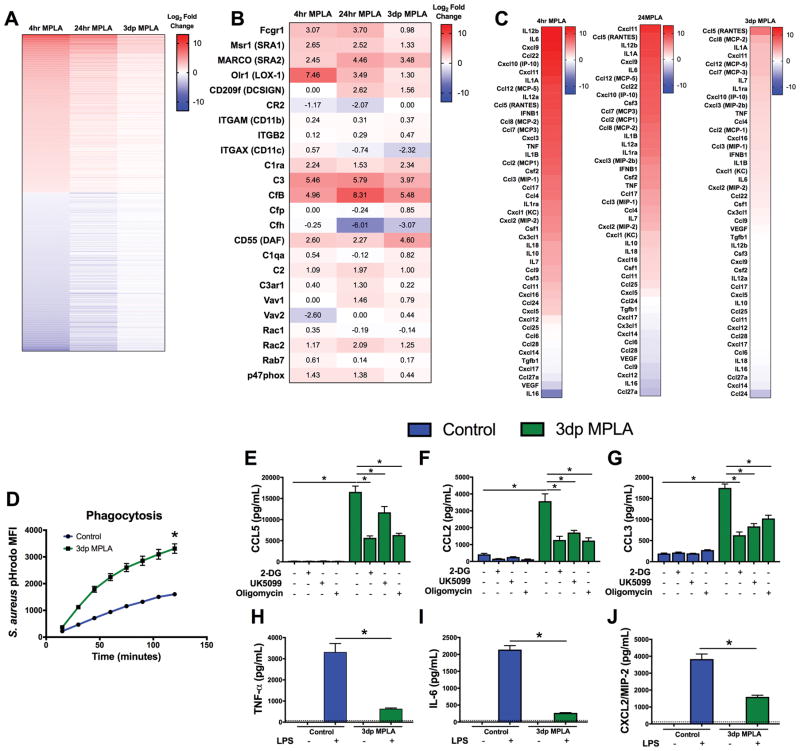
MPLA-primed macrophages retain a phenotype characterized by increased phagocytosis and a persistent chemokine secretion but suppressed production of pro-inflammatory cytokines
We next explored how macrophage gene expression and function changes over time after MPLA exposure. Upon RNA-sequencing, we found significant differential expression of mRNA during the first 4 hours after MPLA priming, as compared to vehicle-treated macrophages, that largely persisted at 24 hours after MPLA priming, but returned near baseline in 3dp macrophages (Figure 5A). However, expression of some gene products persisted in 3dp macrophages. Many of the sustained gene products, such as FcRγ, complement receptors, and scavenger receptors, play an active role in microbial recognition and phagocytosis (Figure 5B). Macrophages also play a crucial role in recruiting leukocytes to sites of infection through the secretion of chemokines, and we previously showed that improved leukocyte recruitment to sites of infection is critical for MPLA prophylaxis (8, 38). We closely examined mRNA expression of chemokine and cytokine genes during MPLA priming. While mRNA of most cytokine genes returned to control levels by 3 days after MPLA treatment, there were some exceptions, such as CCL5 (RANTES), CCL2 (MCP-1) and CCL3 (MIP-1α), which remained elevated in 3dp macrophages (Figure 5C). We hypothesized that the mRNA findings reflected functional alterations of macrophages. Indeed, the persistently augmented phagocytosis-associated genes corresponded not only with a greater S. aureus phagocytic capacity, but also a more rapid induction of phagocytosis in 3dp macrophages (Figure 5D). We previously demonstrated that persistent glycolysis following MPLA exposure supports elevated phagocytosis in macrophages (9). We then assessed basal cytokine secretion over a 6 hour time period while concurrently inhibiting glycolysis with 2-DG, pyruvate transport with UK5099, or ATP synthase with oligomycin (Figure 5E–G). Corresponding with the mRNA, 3dp macrophages were found to exhibit tonic secretion of CCL5, CCL2, and CCL3. Notably, the addition of 2-DG, UK5099, or oligomycin significantly reduced this tonic CCL5, CCL2, and CCL3 secretion. In contrast, 3dp macrophages did not sustain tonic secretion of pro-inflammatory TNFα, IL-6, or CXCL2 (MIP-2), and exhibited reduced TNFα, IL-6, and CXCL2 secretion in response to LPS, reflecting that they are LPS tolerant (Figure 5H–J). Thus, persistent glucose metabolism and mitochondrial activity supports an elevated phagocytic capacity as well as constitutive production of monocyte/macrophage-targeted chemokines, even though pro-inflammatory cytokine production is suppressed.
Figure 5. Reprogrammed mitochondrial metabolism supports a persistent chemokine secretion profile.
A) BMDMs were harvested for mRNA and analyzed via RNA-sequencing. Heat map of the top 1,000 most up-regulated and down-regulated genes after 4hrs of MPLA priming, 24hrs of MPLA priming, and 3 days following the end of priming. Each cell indicates log2 fold change relative to control BMDMs. Genes without significant changes are designated at 0. B) Heat map of phagocytosis-associated gene mRNA after 4hrs of MPLA priming, 24hrs of MPLA priming, and 3 days following the end of priming. Cell values indicate Log2 fold change relative to control BMDMs. C) Heat map list of cytokine mRNA expressed by BMDMs independently ordered by log2 fold change at 4hrs of MPLA priming, 24hrs of MPLA priming, and 3 days following the end of priming. Each cell indicates log2 fold change relative to control BMDMs. D) Phagocytosis of pHrodo-tagged S. aureus bioparticles over 2 hours in control and 3dp MPLA macrophages. E–G) Control or 3dp BMDMs were incubated in the presence of 10mM 2-DG, 80μM UK5099,or 1μM oligomycin. Media was harvested 6 hours after the incubation began. Concentration of E) CCL5 F) CCL2 and G) CCL3 in the media. H–J) Control or 3dp BMDMs were incubated with or without 100ng/mL LPS for 6 hours. H) TNFα, and I) IL-6, J) MIP-2 were determined by ELISA. Data shown as mean +/− SEM. *, p< .05 as determined by ANOVA with Tukey’s post-hoc multiple comparison analysis.
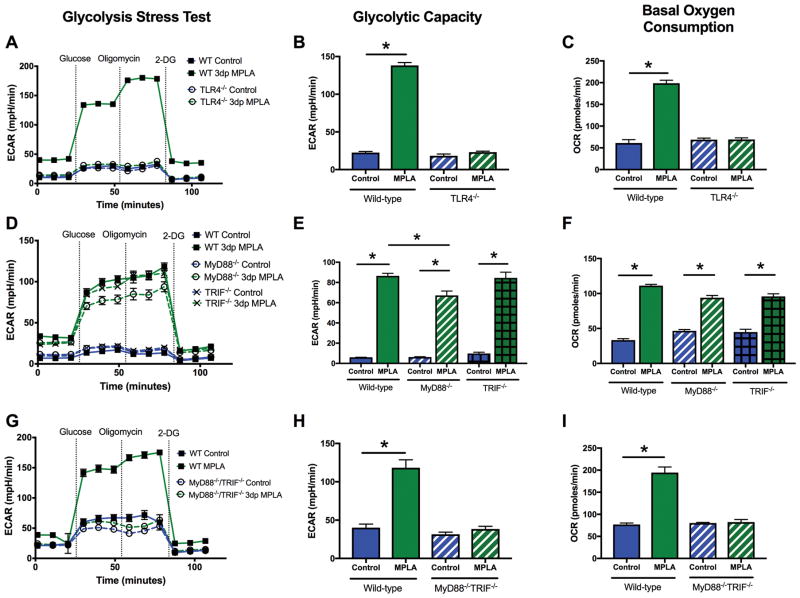
Augmented energy metabolism is orchestrated by MyD88- or TRIF-dependent signaling
The molecular mechanisms that orchestrate MPLA-induced adaptive metabolic reprogramming were then explored. MPLA is a selective TLR4 agonist (39). TLR4 signaling is propagated by the TIR containing adaptor proteins MyD88 and TRIF (40), and, while each adaptor activates separate signaling cascades, there is significant cross-talk between these two pathways (41). As expected, MPLA-induced alterations in OCR and ECAR were ablated in TLR4−/− macrophages (Figure 6A–C). Surprisingly, both MyD88−/− and TRIF−/− 3dp macrophages showed elevations in ECAR and OCR that were comparable to wild type (WT) macrophages (Figure 6D–F). We investigated redundancy of MyD88- and TRIF-dependent signaling for induction of the MPLA-induced metabolic phenotype using MyD88/TRIF double knockout macrophages. MPLA-treated MyD88−/−TRIF−/− 3dp macrophages exhibited an ECAR and OCR that was markedly decreased compared to wild type, and similar to vehicle-treated MyD88−/−TRIF−/− 3dp macrophages and MPLA-treated TLR4−/− macrophages (Figure 6G–I). Thus, either MyD88 or TRIF can induce the metabolic phenotype observed in 3dp macrophages, but at least one adaptor is required.
Figure 6. Augmented energy metabolism is orchestrated by MyD88- or TRIF- dependent signaling.
BMDMs were derived from WT or knockout mice and assessed via the Seahorse Xfe96. Glycolytic capacity was determined after the addition of oligomycin in the glycolysis stress test. Basal OCR was determined after the addition of glucose in the glycolysis stress test. A–C) Glycolysis stress test, glycolytic capacity, and basal OCR of WT or TLR4−/− BMDMs. D–F) Glycolysis stress test, glycolytic capacity, and basal OCR of WT, MyD88−/− or TRIF−/− BMDMs. G–I) Glycolysis stress test, glycolytic capacity, and basal OCR of WT or MyD88 /TRIF double knockout BMDMs. All experiments replicated at least twice. Data shown as mean +/− SEM. *, p< .05 as determined by ANOVA with Tukeys post-hoc multiple comparison analysis.
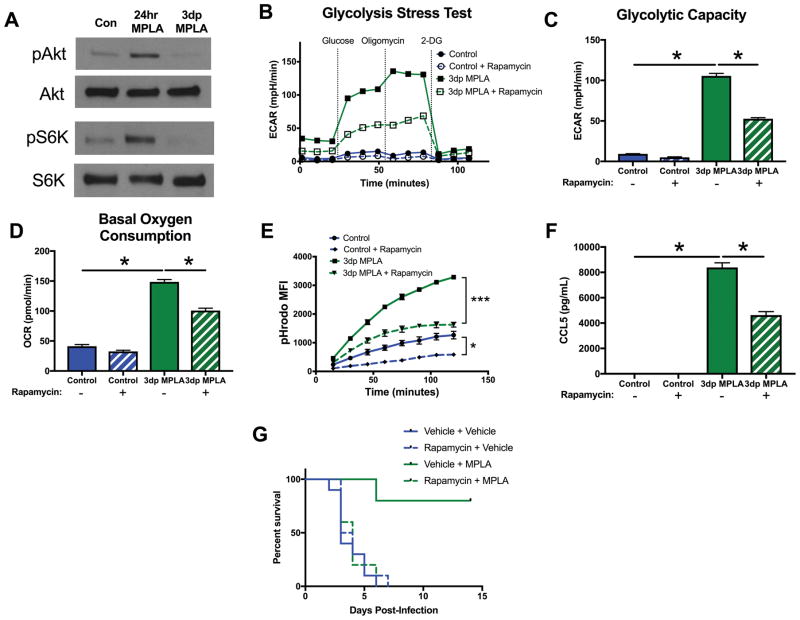
mTOR activation is required for metabolic reprogramming and resistance to infection after MPLA priming
The mammalian target of rapamycin (mTOR) (42), a critical regulator of cellular metabolism, can be activated through both MyD88- or TRIF-dependent mechanisms (43). We explored whether mTOR signaling is required for the development of the 3dp macrophage phenotype. We found that, while mTOR is potently activated by MPLA during priming, Akt phosphorylation and S6K phosphorylation, both indicators of mTOR activation, return to baseline in 3dp macrophages (Figure 7A). However, blockade of mTOR activation with rapamycin during priming significantly attenuated the increased glycolysis (Figure 7B–C) and oxygen consumption (Figure 7D) in 3dp macrophages. Additionally, rapamycin exposure during MPLA priming significantly reduced 3dp macrophage phagocytosis (Figure 7E) and basal CCL5 secretion (Figure 7F). Finally, rapamycin administered to mice 3 hours before MPLA or vehicle treatment completely abolished the survival benefit conferred by MPLA treatment during systemic S. aureus infection (Figure 7G). Thus, mTOR activation during MPLA priming, while not sustained after priming, is required for MPLA to induce macrophage metabolic reprogramming, the macrophage antimicrobial phenotype, and resistance to severe infection in mice.
Figure 7. mTOR-initiated metabolic reprogramming is required for MPLA-induced resistance to infection.
A) Western blot of phosphorylated Akt (pAkt), total Akt (Akt), phosphorylated S6K (pS6K) and total S6K (S6K). Blots representative of 3 repeated experiments. BMDMs were exposed to 100nM rapamycin 1 hour prior to MPLA priming and throughout the 24 hour priming period. Rapamycin was washed out after priming along with MPLA. B) Glycolysis stress test of 3dp macrophages with and without rapamycin during priming. C) Glycolytic capacity of 3dp macrophages with and without rapamycin during priming. D) Basal OCR of 3dp macrophages with and without rapamycin during priming. E) Phagocytosis over time of pHrodo-tagged S. aureus bioparticles by 3dp macrophages with and without rapamycin during priming. F) CCL5 (RANTES) secretion of 3dp macrophages with and without rapamycin during priming after 6 hours of incubation in media. G) Mice were administered 3mg/kg intraperitoneal rapamycin 3 hours prior to the first administration of intravenous MPLA. Kaplan Meier survival plot over 14 days following S. aureus infection (n=10 mice/group). All experiments replicated at least twice. Data shown as mean +/− SEM. *, p< .05 as determined by ANOVA with Tukey’s post-hoc multiple comparison analysis.
DISCUSSION
The key findings of our study are that priming with the TLR4 agonist MPLA induces resistance to systemic infection with S. aureus or C. albicans. We found that this resistance is mediated by increased phagocyte recruitment to infected tissues and persistent improvements to macrophage antimicrobial function. Further, we found that the augmented macrophage antimicrobial function is fueled by dynamic TLR4/mTOR-driven elevations in glycolysis and oxidative phosphorylation, which are facilitated, in part, by delayed alterations in mitochondrial biogenesis and rewiring of malate/NADH shuttling. Finally, we found that this metabolic reprogramming is required for MPLA-induced resistance to infection. To our knowledge, this is the first report to characterize the ability of a TLR4 agonist to persistently alter macrophage metabolism and document the contributions of the metabolic phenotype to macrophage antimicrobial functions.
It has been asserted that TLR4 agonists cause immunoparalysis and susceptibility to infection due to the observation that they induce LPS tolerance (44, 45). While TLR4 agonists indeed cause LPS tolerance, our results consistently demonstrate that LPS tolerance, in and of itself, does not have a causative impact on infection susceptibility (9). Moreover, treatment with TLR4 agonists causes resistance to infection, not susceptibility (46, 47). How, then, do TLR4 agonists drive resistance to infection despite inducing LPS tolerance? Our findings illustrate that TLR4 agonists promote resistance to infection by reprogramming macrophage metabolism in a way that buoys the ability of macrophages to kill diverse pathogenic organisms and recruit additional monocytes and macrophages. It should then be reiterated that our in vivo work suggests TLR4 agonists do not protect animals by inducing LPS tolerance, but rather by persistently augmenting innate antimicrobial functions such as phagocytosis, respiratory burst, and phagocyte recruitment.
Our exploration of the metabolic underpinnings of this macrophage antimicrobial phenotype led us to uncover unique TLR4-driven metabolic phenomena that characterize this phenotype. First, we found that MPLA induces delayed mitochondrial biogenesis that helps support recovery of oxidative metabolism after initial aerobic glycolysis. TLR4 agonists are known to induce mitochondrial biogenesis in parenchymal cells such as hepatocytes, cardiomyocytes, and neurons (48–50). Nevertheless, TLR4 agonists have not previously been demonstrated to induce this phenomenon in macrophages. Mitochondrial biogenesis is an adaptive cellular response to mitochondrial disruption, and it has been noted that patients who survive sepsis undergo mitochondrial biogenesis and ATP recovery, whereas patients who succumb to sepsis do not (51). Thus, we speculate that macrophage mitochondrial biogenesis following inflammatory stimuli is an adaptive process that resolves early disruptions to mitochondria and prepares the cell for upcoming insults.
In addition to mitochondrial biogenesis, we observed dynamic changes to macrophage carbon flux that support the metabolic activity of the expanded mitochondrial pool. Early TLR4-induced aerobic glycolysis is known to induce at least two breaks in the TCA cycle (52). These breaks help fuel the generation of antimicrobial molecules such as nitric oxide and itaconate, but require macrophages to generate lactate, rely on glutamine anaplerosis, and reduce the ability of mitochondria to utilize NADH (52, 53). Mitochondria normally acquire NADH reducing equivalents via malate shuttling (54, 55), yet, it would be futile for macrophages undergoing aerobic glycolysis to shuttle malate into a broken TCA cycle. Rather, macrophages undergoing aerobic glycolysis convert malate to pyruvate via cytosolic malic enzyme 1, a finding confirmed by our flux analysis of 24-hour MPLA macrophages (56, 57). Interestingly, we found that 3 days after MPLA exposure ends, the TCA cycle breaks revert to baseline and macrophages restore malate shuttling, effectively driving NADH reducing equivalents into an expanded mitochondrial pool. Thus, MPLA-primed macrophages generate significant mitochondrial ATP while maintaining the flexibility to utilize glycolytic byproducts for antimicrobial purposes, if needed (58–60). These findings are in concert with evidence from the literature that TLR4 agonist-primed macrophages retain high antimicrobial capacity (16, 61). It still remains unclear how long MPLA-induced metabolic programming lasts, however studies with LPS have shown that innate immunological memory can last for over three weeks (62). We speculate that MPLA-induced metabolic changes in macrophages persist up to 2 weeks after exposure, as this is the time point at which MPLA-induced protection wanes, however the duration of MPLA-induced metabolic changes remains to be determined.
Upon probing the key signaling proteins responsible for this metabolic programming, we found that the observed metabolic phenotype is induced through overlapping and redundant contributions of MyD88- and TRIF- dependent signaling, but that at least one TLR4 adaptor is required. TLR4 ligation is known to activate both MyD88 and TRIF and, despite reported differences in function of these TLR adaptor proteins, both MyD88 and TRIF can activate the metabolic signaling protein mTOR (42, 63, 64). Further, our results demonstrate that mTOR is an essential node in the initiation of this metabolic programming, and inhibition of mTOR during MPLA priming significantly ablates the protective benefits of MPLA both in vitro and in vivo. This finding demonstrates that the observed metabolic reprogramming is an integral part of MPLA-induced infection resistance. Interestingly, we observed that early mTOR activity does not persist along with the metabolic phenotype, raising the question of how the metabolic phenotype lasts so long. Recent studies have demonstrated that myeloid cells can acquire persistent epigenetic modifications that impact their metabolism (12). Given the prolonged nature of the MPLA-induced phenotype, we speculate that the metabolic phenomena we observed have an epigenetic foundation. However, the role of epigenetic modifications in MPLA-induced resistance to infection remains to be explored.
It is noteworthy that treatment with MPLA induces resistance against Gram negative, Gram positive, and fungal pathogens (65, 66). Thus, although MPLA is derived from Gram negative bacteria, it induces resistance to diverse organisms. This is consistent with studies demonstrating that β-glucan also induces resistance to bacterial and fungal pathogens (11, 67). Likewise, our previous studies show that treatment with products from Gram positive bacteria, such as lipoteichoic acid, induces resistance against both Gram positive and Gram negative bacterial pathogens (68, 69). These findings raise the specter that many diverse microbial products have the ability to induce broad resistance against bacterial and fungal pathogens. As a common feature of this phenomenon is activation of the innate immune system, one could speculate that reprogramming of the innate immune response by microbial products is mediated through a common molecular pathway. However, this contention requires further investigation.
MPLA, and other TLR4 agonists, are strong candidates for translation as agents to reduce the incidence and severity of infection in hospitalized patients at acute risk of infection. MPLA has a proven record of safety in humans, and is currently used as a vaccine adjuvant in multiple FDA-approved vaccines (6, 70, 71). As antimicrobial resistance continues to decrease the efficacy of antibiotics, hospitalized patients will require novel prophylactics to prevent and decrease the severity of nosocomial infection. A temporary reorientation of the metabolic, transcriptional, and antimicrobial landscape of macrophages, such as that provided by MPLA, may significantly improve the immune response of hospitalized patients to diverse nosocomial pathogens.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) Grants RO1 GM121711 (Bohannon, PI), RO1 GM119197 (Sherwood, PI), and RO1 GM104306 (Sherwood, PI), the Vanderbilt University Medical Center Faculty Scholars Award (Bohannon, PI), and the Vanderbilt MSTP T32 GM007347 (Fensterheim). The Translational Pathology Shared Resource is supported by NCI/NIH Cancer Center Support Grant 5P30 CA68485-19 and the Vanderbilt Mouse Metabolic Phenotyping Center Grant 2 U24 DK059637-16. The Agilent Seahorse Extracellular Flux Analyzer is housed and managed within the Vanderbilt High-Throughput Screening Core Facility (an institutionally supported core) and was funded by National Institutes of Health Shared Instrumentation Grant 1S10OD018015.
We would like to thank Dr. Aqeela Afzal for help with the BioTek Synergy MX plate reader. We would also like to thank Dr. Brad Grueter and Daniel Kashima (Vanderbilt University) for providing femurs from TLR4−/− mice and Dr. Doug Golenbock (University of Massachusetts Medical School) for providing femurs from MyD88/TRIF double knockout mice. RNA sequencing was done in collaboration with Dr. Olivia Koues, Dr. Yan Guo, and Dr. Lana Olsen at Vanderbilt University VANGARD and VANTAGE.
Footnotes
Author Contributions: B.A.F., J.K.B., and E.R.S. wrote the manuscript and designed figures. B.A.F., J.K.B. and E.R.S. designed experiments. B.A.F., J.D.Y., L.L., R.R.K., C.L.S., N.K.P., Y.G., I.T., A.H., J.B.F., and J.K.B. performed experiments and acquired data. B.A.F., J.D.Y., L.L., A.G.M., D.L.W., J.K.B., and E.R.S. analyzed data. B.A.F. J.D.Y., A.G.M., J.K.B., and E.R.S. interpreted data. All authors approved the final manuscript.
The authors declare no conflict of interest.
References
- 1.World Health Organization. Antimicrobial resistance: global report on surveillance. World Health Organization; 2014. [Google Scholar]
- 2.Bohannon JK, Hernandez A, Enkhbaatar P, Adams WL, Sherwood ER. The immunobiology of toll-like receptor 4 agonists: from endotoxin tolerance to immunoadjuvants. Shock. 2013;40:451–462. doi: 10.1097/SHK.0000000000000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landy M, Pillemer L. Increased resistance to infection and accompanying alteration in properdin levels following administration of bacterial lipopolysaccharides. Journal of Experimental Medicine. 1956;104:383–409. doi: 10.1084/jem.104.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shilo M. Nonspecific resistance to infections. Annual Reviews in Microbiology. 1959;13:255–278. [Google Scholar]
- 5.Bennett IL, Cluff LE. Bacterial pyrogens. Pharmacological reviews. 1957;9:427–475. [PubMed] [Google Scholar]
- 6.Mata-Haro V, Cekic C, Martin M, Chilton PM, Casella CR, Mitchell TC. The vaccine adjuvant monophosphoryl lipid A as a TRIF-biased agonist of TLR4. Science. 2007;316:1628–1632. doi: 10.1126/science.1138963. [DOI] [PubMed] [Google Scholar]
- 7.Di Paolo D, Lenci I, Cerocchi C, Tariciotti L, Monaco A, Brega A, Lotti L, Tisone G, Angelico M. One-year vaccination against hepatitis B virus with a MPL-vaccine in liver transplant patients for HBV-related cirrhosis. Transpl Int. 2010;23:1105–1112. doi: 10.1111/j.1432-2277.2010.01104.x. [DOI] [PubMed] [Google Scholar]
- 8.Romero CD, Varma TK, Hobbs JB, Reyes A, Driver B, Sherwood ER. The Toll-like receptor 4 agonist monophosphoryl lipid a augments innate host resistance to systemic bacterial infection. Infect Immun. 2011;79:3576–3587. doi: 10.1128/IAI.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fensterheim BA, Guo Y, Sherwood ER, Bohannon JK. The Cytokine Response to Lipopolysaccharide Does Not Predict the Host Response to Infection. J Immunol. 2017;198:3264–3273. doi: 10.4049/jimmunol.1602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Luzio NR, Williams DL, Sherwood ER, Browder IW. Modification of diverse experimental immunosuppressive states by glucan. Surv Immunol Res. 1985;4:160–167. doi: 10.1007/BF02918811. [DOI] [PubMed] [Google Scholar]
- 11.Williams DL, Sherwood ER, Browder IW, McNamee RB, Jones EL, Rakinic J, Di Luzio NR. Effect of glucan on neutrophil dynamics and immune function in Escherichia coli peritonitis. J Surg Res. 1988;44:54–61. doi: 10.1016/0022-4804(88)90122-9. [DOI] [PubMed] [Google Scholar]
- 12.Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JH, Rao NA, Aghajanirefah A, Manjeri GR, Li Y, Ifrim DC, Arts RJ, van der Meer BM, Deen PM, Logie C, O’Neill LA, Willems P, van de Veerdonk FL, van der Meer JW, Ng A, Joosten LA, Wijmenga C, Stunnenberg HG, Xavier RJ, Netea MG. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arts RJ, Novakovic B, ter Horst R, Carvalho A, Bekkering S, Lachmandas E, Rodrigues F, Silvestre R, Cheng S-C, Wang S-Y. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell metabolism. 2016;24:807–819. doi: 10.1016/j.cmet.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Netea MG. Training innate immunity: the changing concept of immunological memory in innate host defence. Eur J Clin Invest. 2013;43:881–884. doi: 10.1111/eci.12132. [DOI] [PubMed] [Google Scholar]
- 15.Madonna G, Peterson J, Ribi E, Vogel S. Early-phase endotoxin tolerance: induction by a detoxified lipid A derivative, monophosphoryl lipid A. Infection and immunity. 1986;52:6–11. doi: 10.1128/iai.52.1.6-11.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong Y, Medvedev AE. Induction of endotoxin tolerance in vivo inhibits activation of IRAK4 and increases negative regulators IRAK-M, SHIP-1, and A20. J Leukoc Biol. 2011;90:1141–1148. doi: 10.1189/jlb.0611273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shu J, Qiu G, Mohammad I. A semi-automatic image analysis tool for biomarker detection in immunohistochemistry analysis. Image and Graphics (ICIG), 2013 Seventh International Conference on; IEEE; 2013. pp. 937–942. [Google Scholar]
- 18.Lee CM, Hu J. Cell density during differentiation can alter the phenotype of bone marrow-derived macrophages. Cell Biosci. 2013;3:30. doi: 10.1186/2045-3701-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noguchi Y, Young JD, Aleman JO, Hansen ME, Kelleher JK, Stephanopoulos G. Effect of anaplerotic fluxes and amino acid availability on hepatic lipoapoptosis. Journal of Biological Chemistry. 2009;284:33425–33436. doi: 10.1074/jbc.M109.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folch J, Lees M, Sloane Stanley G. A simple method for the isolation and purification of total lipids from animal tissues. J biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 21.Young JD, Allen DK, Morgan JA. Isotopomer measurement techniques in metabolic flux analysis II: mass spectrometry. Plant Metabolism: Methods and Protocols. 2014:85–108. doi: 10.1007/978-1-62703-661-0_7. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. Journal of Mass Spectrometry. 1996;31:255–262. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Young JD. INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics. 2014;30:1333–1335. doi: 10.1093/bioinformatics/btu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metabolic engineering. 2007;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young JD, Walther JL, Antoniewicz MR, Yoo H, Stephanopoulos G. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnology and bioengineering. 2008;99:686–699. doi: 10.1002/bit.21632. [DOI] [PubMed] [Google Scholar]
- 26.Sheng Q, Vickers K, Zhao S, Wang J, Samuels DC, Koues O, Shyr Y, Guo Y. Multi-perspective quality control of Illumina RNA sequencing data analysis. Brief Funct Genomics. 2016;16:194–204. doi: 10.1093/bfgp/elw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Y, Zhao S, Sheng Q, Ye F, Li J, Lehmann B, Pietenpol J, Samuels DC, Shyr Y. Multi-perspective quality control of Illumina exome sequencing data using QC3. Genomics. 2014;103:323–328. doi: 10.1016/j.ygeno.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S. HTSeq: Analysing high-throughput sequencing data with Python. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, Zhao S, Ye F, Sheng Q, Shyr Y. MultiRankSeq: multiperspective approach for RNAseq differential expression analysis and quality control. BioMed research international. 2014;2014:248090. doi: 10.1155/2014/248090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Feng Z, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardcastle TJ, Kelly KA. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics. 2010;11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol. 2006;7:311–317. doi: 10.1038/ni1309. [DOI] [PubMed] [Google Scholar]
- 36.Galvan-Pena S, O’Neill LA. Metabolic reprograming in macrophage polarization. Front Immunol. 2014;5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 38.Bohannon JK, Luan L, Hernandez A, Afzal A, Guo Y, Patil NK, Fensterheim B, Sherwood ER. Role of G-CSF in monophosphoryl lipid A-mediated augmentation of neutrophil functions after burn injury. J Leukoc Biol. 2016;99:629–640. doi: 10.1189/jlb.4A0815-362R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernandez A, Bohannon JK, Luan L, Fensterheim BA, Guo Y, Patil NK, McAdams C, Wang J, Sherwood ER. The role of MyD88- and TRIF-dependent signaling in monophosphoryl lipid A-induced expansion and recruitment of innate immunocytes. J Leukoc Biol. 2016;100:1311–1322. doi: 10.1189/jlb.1A0216-072R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nature Reviews Immunology. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 41.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitz F, Heit A, Dreher S, Eisenächer K, Mages J, Haas T, Krug A, Janssen KP, Kirschning CJ, Wagner H. Mammalian target of rapamycin (mTOR) orchestrates the defense program of innate immune cells. European journal of immunology. 2008;38:2981–2992. doi: 10.1002/eji.200838761. [DOI] [PubMed] [Google Scholar]
- 43.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Molecular cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, Cheng SC, Ratter J, Berentsen K, van der Ent MA, Sharifi N, Janssen-Megens EM, Ter Huurne M, Mandoli A, van Schaik T, Ng A, Burden F, Downes K, Frontini M, Kumar V, Giamarellos-Bourboulis EJ, Ouwehand WH, van der Meer JW, Joosten LA, Wijmenga C, Martens JH, Xavier RJ, Logie C, Netea MG, Stunnenberg HG. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novakovic B, Habibi E, Wang SY, Arts RJ, Davar R, Megchelenbrink W, Kim B, Kuznetsova T, Kox M, Zwaag J, Matarese F, van Heeringen SJ, Janssen-Megens EM, Sharifi N, Wang C, Keramati F, Schoonenberg V, Flicek P, Clarke L, Pickkers P, Heath S, Gut I, Netea MG, Martens JH, Logie C, Stunnenberg HG. beta-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance. Cell. 2016;167:1354–1368 e1314. doi: 10.1016/j.cell.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wheeler DS, Lahni PM, Denenberg AG, Poynter SE, Wong HR, Cook JA, Zingarelli B. Induction of endotoxin tolerance enhances bacterial clearance and survival in murine polymicrobial sepsis. Shock. 2008;30:267–273. doi: 10.1097/shk.0b013e318162c190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehner MD, Ittner J, Bundschuh DS, van Rooijen N, Wendel A, Hartung T. Improved innate immunity of endotoxin-tolerant mice increases resistance to Salmonella enterica serovar typhimurium infection despite attenuated cytokine response. Infect Immun. 2001;69:463–471. doi: 10.1128/IAI.69.1.463-471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suliman HB, Carraway MS, Welty-Wolf KE, Whorton AR, Piantadosi CA. Lipopolysaccharide stimulates mitochondrial biogenesis via activation of nuclear respiratory factor-1. Journal of Biological Chemistry. 2003;278:41510–41518. doi: 10.1074/jbc.M304719200. [DOI] [PubMed] [Google Scholar]
- 49.Stetler RA, Leak RK, Yin W, Zhang L, Wang S, Gao Y, Chen J. Mitochondrial biogenesis contributes to ischemic neuroprotection afforded by LPS pre-conditioning. J Neurochem. 2012;123(Suppl 2):125–137. doi: 10.1111/j.1471-4159.2012.07951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hickson-Bick DL, Jones C, Buja LM. Stimulation of mitochondrial biogenesis and autophagy by lipopolysaccharide in the neonatal rat cardiomyocyte protects against programmed cell death. Journal of molecular and cellular cardiology. 2008;44:411–418. doi: 10.1016/j.yjmcc.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 51.Carré JE, Orban JC, Re L, Felsmann K, Iffert W, Bauer M, Suliman HB, Piantadosi CA, Mayhew TM, Breen P. Survival in critical illness is associated with early activation of mitochondrial biogenesis. American journal of respiratory and critical care medicine. 2010;182:745–751. doi: 10.1164/rccm.201003-0326OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nature Reviews Immunology. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Neill LA. A broken krebs cycle in macrophages. Immunity. 2015;42:393–394. doi: 10.1016/j.immuni.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 54.Mizuarai S, Miki S, Araki H, Takahashi K, Kotani H. Identification of dicarboxylate carrier Slc25a10 as malate transporter in de novo fatty acid synthesis. Journal of Biological Chemistry. 2005;280:32434–32441. doi: 10.1074/jbc.M503152200. [DOI] [PubMed] [Google Scholar]
- 55.Brunengraber H, Lowenstein JM. Effect of (−)-hydroxycitrate on ethanol metabolism. FEBS letters. 1973;36:130–132. doi: 10.1016/0014-5793(73)80353-9. [DOI] [PubMed] [Google Scholar]
- 56.Kelly B, O’Neill LA. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015;25:771–784. doi: 10.1038/cr.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Infantino V, Iacobazzi V, Palmieri F, Menga A. ATP-citrate lyase is essential for macrophage inflammatory response. Biochemical and biophysical research communications. 2013;440:105–111. doi: 10.1016/j.bbrc.2013.09.037. [DOI] [PubMed] [Google Scholar]
- 58.Jha AK, Huang SCC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, Pearce EJ, Driggers EM, Artyomov MN. Network Integration of Parallel Metabolic and Transcriptional Data Reveals Metabolic Modules that Regulate Macrophage Polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 59.Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell A. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proceedings of the National Academy of Sciences. 2013;110:7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Infantino V, Iacobazzi V, Menga A, Avantaggiati ML, Palmieri F. A key role of the mitochondrial citrate carrier (SLC25A1) in TNFα-and IFNγ-triggered inflammation. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2014;1839:1217–1225. doi: 10.1016/j.bbagrm.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernandes ML, Mendes ME, Brunialti MK, Salomao R. Human monocytes tolerant to LPS retain the ability to phagocytose bacteria and generate reactive oxygen species. Braz J Med Biol Res. 2010;43:860–868. doi: 10.1590/s0100-879x2010007500081. [DOI] [PubMed] [Google Scholar]
- 62.Yoshida K, Maekawa T, Zhu Y, Renard-Guillet C, Chatton B, Inoue K, Uchiyama T, Ishibashi KI, Yamada T, Ohno N, Shirahige K, Okada-Hatakeyama M, Ishii S. The transcription factor ATF7 mediates lipopolysaccharide-induced epigenetic changes in macrophages involved in innate immunological memory. Nat Immunol. 2015;16:1034–1043. doi: 10.1038/ni.3257. [DOI] [PubMed] [Google Scholar]
- 63.Troutman TD, Hu W, Fulenchek S, Yamazaki T, Kurosaki T, Bazan JF, Pasare C. Role for B-cell adapter for PI3K (BCAP) as a signaling adapter linking Toll-like receptors (TLRs) to serine/threonine kinases PI3K/Akt. Proc Natl Acad Sci U S A. 2012;109:273–278. doi: 10.1073/pnas.1118579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Everts B, Amiel E, Huang SC, Smith AM, Chang CH, Lam WY, Redmann V, Freitas TC, Blagih J, van der Windt GJ, Artyomov MN, Jones RG, Pearce EL, Pearce EJ. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKvarepsilon supports the anabolic demands of dendritic cell activation. Nat Immunol. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Romero CD, Varma TK, Hobbs JB, Reyes A, Driver B, Sherwood ER. The Toll-like receptor 4 agonist monophosphoryl lipid a augments innate host resistance to systemic bacterial infection. Infect Immun. 2011;79:3576–3587. doi: 10.1128/IAI.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bohannon JK, Luan L, Hernandez A, Afzal A, Guo Y, Patil NK, Fensterheim B, Sherwood ER. Role of G-CSF in monophosphoryl lipid A-mediated augmentation of neutrophil functions after burn injury. J Leukoc Biol. 2015;99:629–640. doi: 10.1189/jlb.4A0815-362R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tzianabos AO, Cisneros RL. Prophylaxis with the immunomodulator PGG glucan enhances antibiotic efficacy in rats infected with antibiotic-resistant bacteria. Ann N Y Acad Sci. 1996;797:285–287. doi: 10.1111/j.1749-6632.1996.tb52980.x. [DOI] [PubMed] [Google Scholar]
- 68.Murphey ED, Fang G, Varma TK, Sherwood ER. Improved bacterial clearance and decreased mortality can be induced by LPS tolerance and is not dependent upon IFN-gamma. Shock. 2007;27:289–295. doi: 10.1097/01.shk.0000245024.93740.28. [DOI] [PubMed] [Google Scholar]
- 69.Murphey ED, Sherwood ER. Pretreatment with the Gram-positive bacterial cell wall molecule peptidoglycan improves bacterial clearance and decreases inflammation and mortality in mice challenged with Pseudomonas aeruginosa. Microbes Infect. 2008;10:1244–1250. doi: 10.1016/j.micinf.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Didierlaurent AM, Morel S, Lockman L, Giannini SL, Bisteau M, Carlsen H, Kielland A, Vosters O, Vanderheyde N, Schiavetti F, Larocque D, Van Mechelen M, Garcon N. AS04, an aluminum salt- and TLR4 agonist-based adjuvant system, induces a transient localized innate immune response leading to enhanced adaptive immunity. J Immunol. 2009;183:6186–6197. doi: 10.4049/jimmunol.0901474. [DOI] [PubMed] [Google Scholar]
- 71.McKeage K, Romanowski B. Spotlight on AS04-adjuvanted human papillomavirus (HPV) types 16 and 18 vaccine (Cervarix(R)) BioDrugs. 2011;25:265–269. doi: 10.2165/11206830-000000000-00000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.