Abstract
Direct visualization and light control of several cellular processes is a challenge due to spectral overlap of available genetically-encoded probes. Here we report the most red-shifted monomeric near-infrared (NIR) fluorescent protein miRFP720 and the fully NIR Förster Resonance Energy Transfer (FRET) pair miRFP670-miRFP720 that enables design of biosensors compatible with CFP-YFP imaging and blue-green optogenetic tools. We developed a NIR biosensor for Rac1 GTPase and demonstrated its use for multiplexed imaging and light control of RhoGTPase signaling pathways. Specifically, we combined the Rac1 biosensor with CFP-YFP FRET biosensors for RhoA and for Rac1-GDI binding, and then concurrently used LOV-TRAP tool for upstream Rac1 activation. We directly observed and quantified the antagonism between RhoA and Rac1 dependent on the RhoA-downstream effector ROCK, showed that Rac1 activity and GDI binding depend exquisitely on the spatiotemporal coordination between these two molecules, and simultaneously observed Rac1 activity during optogenetic manipulation of Rac1.
Introduction
Multiplexing of fluorescent biosensors and optogenetic systems in live-cell imaging requires spectrally compatible imaging probes. Previously, FRET biosensors based on cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) of the green fluorescent protein (GFP)-like family have been multiplexed with cellular reporters based on near-infrared (NIR) organic dyes 1,2, and with FRET biosensors using orange and red fluorescent proteins (FPs), requiring specialized imaging methods due to substantial spectral overlap with CFP-YFP pair 3–6. CFP-YFP biosensors also cannot be used simultaneously with many optogenetic tools, including light-oxygen-voltage (LOV), cryptochrome-2 (CRY2) and channelrhodopsins 7,8, all of which are activated with blue-green light. Several NIR FPs have been developed 9. However, these spectrally resolvable iRFPs are dimers that limit their use in FRET biosensors. Recently, three monomeric miRFPs were reported 10 but the most blue-shifted and red-shifted monomeric NIR FPs do not represent an efficient FRET pair due to substantial spectral crosstalk. To achieve better FRET compatibility, a more red-shifted monomeric NIR FP is required.
FRET biosensors for RhoGTPases have been instrumental in studying this important class of signaling proteins. RhoGTPases are molecular switches 11 and are activated by release of guanine diphosphate (GDP) and loading of guanine triphosphate (GTP), catalyzed by upstream regulator guanine nucleotide exchange factors (GEF) 12. When on, RhoGTPases bind to their effectors to propagate signals regulating cellular functions. Binding to GTPase activating protein (GAP) accelerates the hydrolysis of GTP to a GDP and turning the GTPases off. The guanine nucleotide dissociation inhibitors (GDI) are negative regulators that bind and chaperone RhoGTPases away into the cytosol 13. Understanding the molecular regulation of RhoGTPases is clearly important because their roles are critical both in normal cellular functions and in many diseases 14. Importantly, RhoGTPases exist in coordinated signaling networks with rapid spatiotemporal dynamics1. To better understand such coordination, new NIR-FRET pairs of FPs are required that enable simultaneous probing of multiple processes and an ability to perturb and observe different RhoGTPase pathways at the same time in a cell.
Here we report the most red-shifted monomeric NIR FP, miRFP720, which is the brightest in mammalian cells among available monomeric NIR FPs developed from bacterial phytochromes. We demonstrated the use of miRFP720 in various expression tags and characterized it as an effective FRET acceptor for the blue-shifted NIR FP, miRFP670 10, making the first fully NIR-FRET pair. Utilizing this pair, we engineered a genetically encoded, single-chain, monomeric NIR-FRET biosensor for Rac1 GTPase with excellent brightness and signal dynamic range. We multiplexed the NIR-Rac1 biosensor with several CFP-YFP FRET biosensors and with LOV-based optogenetic tool for simultaneous detection and perturbation of RhoGTPase activities in single living cells. First, with the NIR-Rac1 biosensor and a CFP-YFP RhoA biosensor 15, we directly visualized a Rac1-RhoA antagonism in motile cells, which depended on the downstream RhoA-ROCK pathway. Second, with the NIR-Rac1 biosensor and a CFP-YFP biosensor for Rac1-GDI binding 2 we found a tight spatiotemporal regulation of Rac1, showed for the first time that activated Rac1 and GDI-bound Rac1 co-existed in specific cellular regions within edge protrusions. We then modified the LOV-TRAP optogenetic tool 16 to achieve higher Rac1 specificity and showed the ability to continuously measure Rac1 activity during photoactivation followed by dark relaxation. Finally, we extended the NIR technology to kinase biosensors targeting protein kinase A (PKA) and c-Jun N-terminal kinase (JNK). We demonstrated that the NIR-FRET pair of FPs are generally applicable to biosensor development, and are easily compatible with existing CFP-YFP biosensors and blue/green optogenetic tools.
Results
Engineering and characterization of miRFP720
To develop a monomeric NIR FP-based FRET acceptor we turned to the most NIR-shifted dimeric FP, iRFP720, engineered from the PAS-GAF domains of the RpBphP2 bacterial phytochrome 9. Because the crystal structure of iRFP720 is not yet determined, we analyzed the structure of the PAS-GAF domains of RpBphP2 17. These domains were crystallized as a dimer with the dimerizing-interface formed by the C-terminal helices of both GAF domains. Specifically, amino acid residues L309, Q312, V313 and W316 (Supplementary Results, Supplementary Fig. 1; here and below the numbering follows that in Supplementary Fig. 1) formed hydrophobic interaction with their counterpart at the other protomer. Importantly, the C-terminal helices of the GAF domains were the same in RpBphP2 and in iRFP720 and likely formed the dimerizing interface in iRFP720. To introduce amino acid substitutions into the interface, we considered amino acids at C-termini of recently reported monomeric miRFPs developed from different bacterial phytochrome RpBphP1. We hypothesized that substituting the five amino acid residues in putative dimerizing interface of iRFP720 with the charged ones 10 should result in a monomeric iRFP720 (Supplementary Fig. 1). The resultant iRFP720/E308K/L309R/Q312E/V313R/W316T mutant was brightly fluorescent and had an absorbance maximum at 702nm and an emission maximum at 720nm (Supplementary Table 1; Supplementary Fig. 2a&b). These maxima are the most NIR-shifted among currently available monomeric NIR FPs (Fig. 1a&b). Further comparison of this mutant with parental iRFP720 and miRFP670 10 using analytical ultracentrifugation confirmed its monomeric state (Supplementary Fig. 3). We termed this mutant miRFP720 and determined that its extinction coefficient is 98,000M−1cm−1, quantum yield is 6.1% and pKa value is 4.5 (Supplementary Table 1; Supplementary Fig. 2c). As expected, monomerization did not affect the properties of miRFP720 compared to parental iRFP720 (Supplementary Table 1) since the introduced mutations are at the C-terminus of the protein, distant from the chromophore pocket (Supplementary Fig. 1).
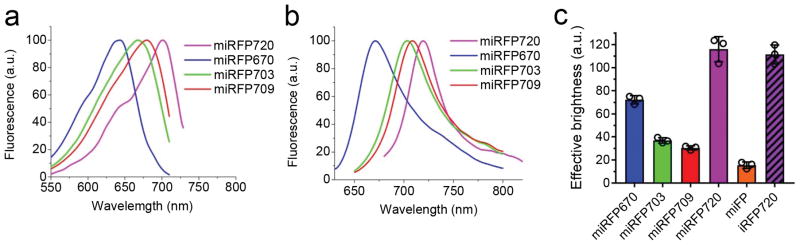
Figure 1. Characterization of the engineered monomeric miRFP720.
(a) Fluorescence excitation and (b) fluorescence emission spectra of miRFP720 (magenta) overlaid with spectra of other monomeric NIR FPs, such as miRFP670 (blue), miRFP703 (green) and miRFP709 (red). (c) NIR fluorescence brightness of live HeLa cells transiently transfected with miRFP720 (magenta, open bar) compared to that of parental dimeric iRFP720 (magenta, shaded bar) and other monomeric NIR FPs, such as miRFP670 (blue), miRFP703 (green), miRFP709 (red) and mIFP (orange), analyzed using flow cytometry. NIR fluorescence intensity was normalized to transfection efficiency (fluorescence intensity of co-transfected EGFP), to excitation efficiency of each NIR FP with 635 nm laser, and to fluorescence signal of each NIR FP in the emission filter. Error bars, S.D. (mean of n=3; independent experiments).
We expressed miRFP720 in HeLa cells and compared its cellular brightness to that of other miRFPs and parental iRFP720 (Fig. 1c). The brightness of cytoplasmic miRFP720 was 1.6-fold higher than that of cells expressing miRFP670, and 3.9-fold higher than cells expressing miRFP709 10. This indicated that miRFP720 retains a high efficiency of binding of biliverdin (BV) chromophore in mammalian cells, which is characteristic of the parental iRFP720 9. miRFP720 was as bright in cells as iRFP720 (Fig. 1c). Photostability of miRFP720 in living cells was high and similar to that of previously reported miRFPs (Supplementary Table 1; Supplementary Fig. 2d). To test performance of miRFP720 as a protein tag, we expressed several fusions in mammalian cells. Both N- and C-terminal fusions localized properly, including with histone 2B which localized appropriately through mitosis and did not affect cell division (Supplementary Fig. 4).
The high extinction coefficient and NIR-shifted excitation and emission spectra make miRFP720 an advantageous FRET acceptor over other miRFPs for a blue-shifted miRFP670 FRET donor (Supplementary Fig. 5a&b). Importantly, red-shifted excitation spectrum of miRFP720 results in minimal direct cross-excitation of this FP in a FRET pair (Supplementary Fig. 5a&b). To test the FRET efficiency of this FP pair, we made a miRFP670-miRFP720 fusion connected by a linker containing the caspase-3 cleavage site -DEVD- and compared its FRET changes upon induction of apoptosis to that of the miRFP670 fusion with miRFP709 (Supplementary Fig. 5c). In transiently transfected HeLa cells, the miRFP670-miRFP720 FRET pair demonstrated 34% change in the donor/FRET ratio after the caspase-3 cleavage whereas the miRFP670-miRFP709 FRET pair resulted in an almost 2-fold less donor/FRET ratio change of 18% (Supplementary Fig. 5d&e). This indicates the superiority of miRFP720 as a FRET acceptor over miRFP709. To compare the miRFP670-miRFP720 FRET pair with standard FRET pairs consisting of the GFP-like FPs we calculated its Förster radius (R0). The calculated R0 of 8.3nm for the miRFP670-miRFP720 FRET pair was 1.5–1.7-fold larger than that of the standard GFP-like FRET pairs, such as CFP-YFP (R0=4.9 nm) or mCerulean/mVenus (R0=5.4nm) 18, suggesting that substantially larger linkers between FPs and sensing parts may be required to design optimal NIR-FRET biosensors. Larger R0 for the miRFP670-miRFP720 FRET pair should also lead to higher FRET efficiency in constructs with similar linkers. The main reason for similar FRET efficiencies experimentally observed in the NIR-FRET pair (Supplementary Fig. 5d) and GFP-like FRET pair 19 is a direct cross-excitation of acceptor. Comparing to GFP-like FPs, NIR FPs have wider spectra that lead to this effect.
NIR-Rac1 FRET biosensor using miRFP670-miRFP720
Using the new miRFP670-miRFP720 FRET pair we developed a monomeric, single-chain biosensor for Rac1 GTPase. We chose Rac1 because of its well-recognized roles within RhoGTPase signaling important in regulating many cellular behaviors. The wide availability of biosensors and optogenetic tools for RhoGTPases utilizing the blue-green-yellow fluorescence will enable simultaneous use of the new NIR-Rac1 biosensor in imaging experiments. The design of this biosensor consists of a NIR-FRET donor (miRFP670) and a NIR-FRET acceptor (miRFP720) with the p21 binding domain (PBD) of p21 activated kinase-1 (PAK1) and a full length Rac1 (Fig. 2a) 20. In addition to miRFP720 we tested previously reported miRFP703/709 10 as FRET acceptors in several orientations within the biosensor. The optimal FRET response was observed with miRFP720 occupying the “FP1” and miRFP670 occupying the “FP2” positions (Supplementary Fig. 6a). To account for larger Förster radius of 8.3nm of this NIR-FRET pair compared to GFP-like FPs, we included an additional flexible linker 15 (denoted “L1”; Fig. 2a) and optimized for a length of 58 amino acids. At and above this length, FRET ratio from both constitutively active and dominant negative versions of the biosensor reached their minimum but the differences between the two mutants were maximized compared to shorter linkers (Supplementary Fig. 7a). A second linker of 10 amino acids (denoted “L2”; Fig. 2a) was included (Supplementary Fig. 7b) to minimize FRET in the dominant negative version of the biosensor. The final design of the NIR-Rac1 biosensor maintains the native Rac1 C-terminus and able to interact with the upstream regulator guanine nucleotide dissociation inhibitor (GDI) (Fig. 2b). The PBD affinity was fine-tuned using an autoinhibitory motif that optimized sensitivity 20, and the use of monomeric components minimized spurious intra/inter-molecular interactions that impact localization and reversibility.
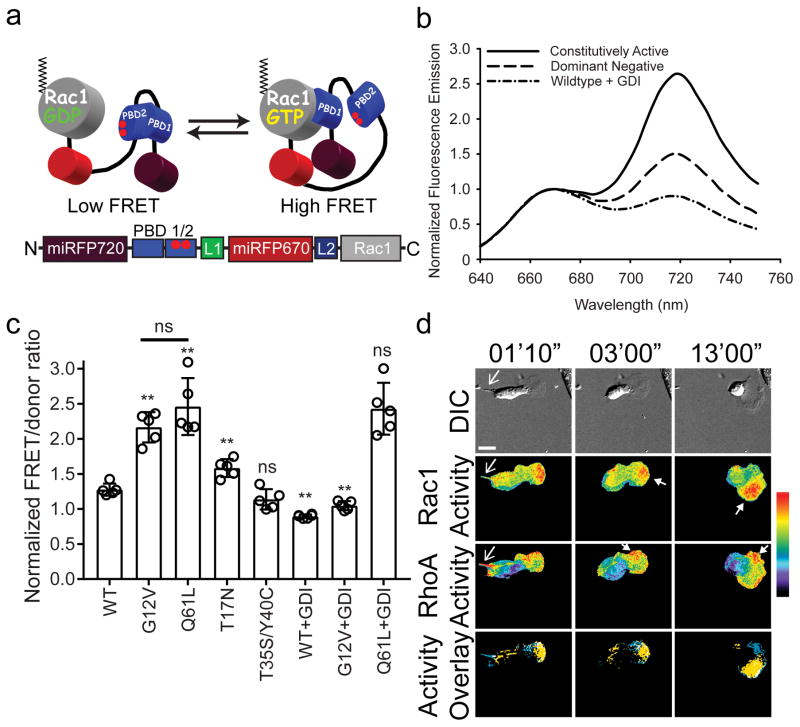
Figure 2. NIR-Rac1 biosensor for live cell imaging.
(a) Schematic of the single-chain NIR-Rac1 biosensor design. From N- to C-terminus: Dark red: miRFP720; PBD1: p21 binding domain 1; PBD2: p21 binding domain containing H83D-H86D GTPase binding deficient mutations; Red: miRFP670; full length Rac1. This orientation enables posttranslational isoprenylation of the C-terminus, thus maintaining the native interaction with appropriate membrane domains and the guanine nucleotide dissociation inhibitor (GDI). GTP versus GDP-loading of Rac1 changes the intramolecular conformation and affects FRET. (b) Representative normalized fluorescence emission spectra of constitutively activated, dominant negative, and wildtype NIR-Rac1 biosensors expressed with excess GDI. Spectra are measured by exciting the NIR-Rac1 sensor mutants expressed in cell suspensions at 600 nm wavelength, and are from the data analyzed and presented in (c), from 5 independent experiments. (c) Wildtype (WT) or mutant versions of NIR-Rac1 biosensor with or without 4× excess GDI. N=5 independent experiments shown with mean ±SEM. Student t-tests are two-tailed. ** WT vs G12V, p=2.626562×10−5; WT vs Q61L, p=2.158669×10−4; WT vs T17N, p=2.122228×10−3; WT vs T35S/Y40C, “ns”=not significant, p=0.1003134; G12V vs Q61L, p=0.1877456; WT vs WT+GDI, p=2.627320×10−6; G12V vs G12V+GDI, p=3.684825×10−6; and Q61L vs Q61L+GDI, p=0.9049651. (d) Representative timelapse panels of imaging RhoA and Rac1 in a single living MEF (additional time points are shown in Supplementary Fig. 11) from 16 cells from 6 independent experiments. 628/32 and 436/20 excitation filters were used for NIR and mCerulean-mVenus FRET excitation, respectively. 480/40 and 535/30 filter pair was used for mCerulean-mVenus FRET signal, whereas 684/24 and 794/160 filter pair was used for miRFP670-miRFP720 FRET signal. Top panels: Differential interference contrast. Middle two panels: Rac1 activity (upper) and RhoA activity (lower). Bottom panels: Localizations of high Rac1 (yellow) and high RhoA (blue) activities are overlaid, where regions of colocalization are shown in white. Regions of high Rac1 and RhoA activities were defined by intensity thresholding the top 2.5 % of pixel ratio values within the image intensity histogram. Regions and features of interest are indicated using matching colored arrowheads. White bar=20μm. Pseudocolor bar corresponds to ratio limits of 1.0 to 1.55 for Rac1 and 1.0 to 1.32 for RhoA activities (black to red).
Fluorometric characterization of NIR-Rac1 biosensor revealed a 2.7-fold difference between the on versus the off state of the biosensor (Fig. 2b). G12V or Q61L constitutively active mutants showed increased FRET compared to the wildtype Rac1 biosensor, but co-expressing GDI attenuated FRET only for the WT and G12V which bind GDI, but not Q61L which cannot bind GDI (Fig. 2c). The T17N dominant negative and the effector-binding mutants (T35S/Y40C) show attenuated FRET (Fig. 2c). Co-expression of the Rac- (Trio) but not the Cdc42-targeting (Intersectin) GEFs result in increased FRET (Supplementary Fig. 6b). Co-expressing p50RhoGAP attenuates FRET, while Rap1GAP which does not target Rac1 has no effect (Supplementary Fig. 6b).
To determine if effectors of Rac1 compete for binding to activated NIR-Rac1 biosensor 15, we performed a PBD-pulldown assay from lysates of cells expressing activated NIR-Rac1 biosensor. The biosensor binds to exogenous PBD only when both PBDs within the biosensor are mutated 20 so that they cannot bind to activated Rac1 (Supplementary Fig. 8a). When expressed in fibroblasts (MEF/3T3), constitutively active (Q61L) versus dominant negative (T17N) NIR-Rac1 biosensor mutants showed approximately 40% difference in FRET (Supplementary Fig. 8b). A slight elevation of FRET in the dominant negative (T17N) mutant is likely attributed to the formation of a stable complex of GTPase and cellular GEF 21, similar to other dominant negative mutants of Rac1 that form stable Rac1-GEF complexes (G15A and D118A) 12, showing same FRET levels (Supplementary Fig. 8c). We applied the synonymous codon modification to the linkers and PBD domains in order to prevent homologous recombination during transfection and transduction 22. We then overexpressed the new NIR-Rac1 biosensor in MEFs together with the original Rac1 biosensor based on CFP-YFP 20,23 to compare the activation patterns of Rac1 in the same cell, using both FRET/donor and subtractive FRET-based ratiometric analyses (Supplementary Fig. 9). Taken together, we have now extended the Rac1 biosensor design 20,23 to enable dynamic measurements of activity using only NIR wavelengths above 600 nm in living cells.
Rac1 and RhoA activities measured simultaneously
Rac1 and RhoA GTPases exist in an antagonism, demonstrated through molecular and biochemical methods 24,25. We decided to use the NIR-Rac1 biosensor with the CFP-YFP RhoA biosensor 15 in single living cells to directly probe this using live-cell imaging. Cells are sensitive to overexpression of biosensors 15; we therefore established stable, tetracycline-inducible cells harboring both RhoA and Rac1 biosensors, with expression levels below those producing effects on cellular motility (Supplementary Fig. 10). A representative, migrating MEF from a time-lapse experiment for RhoA-Rac1 activities is shown in Figure 2d (Supplementary Fig. 11, Video 1). Rac1 activity was predominantly localized in the lamellipodia at the front of the cell as previously shown using another class of Rac1 biosensor 1, and RhoA activity was localized at the retracting tail, in a thin band at the leading edge, or at side edges and at the back of the leading edge protrusion in regions opposite to the direction of cell turning.
NIR-spectra of miRFP670-miRFP720 pair result in negligible overlap with CFP-YFP (Supplementary Fig. 12a), enabling clean separation of two biosensor signals. The rate of photobleach-associated change in FRET/donor from NIR-Rac1 biosensor was minimal over the course of a live-cell experiment compared to the CFP-YFP RhoA biosensor (Supplementary Fig. 12b). This indicated that both donor and acceptor photobleached at similar rates during imaging. We tested if excitation of NIR-Rac1 biosensor at 436nm (excitation wavelength for CFP-YFP biosensor) in addition to 628nm used for excitation of NIR-FRET donor, would affect the photobleaching rate. We measured only a modest effect in FRET/donor ratio when cells were irradiated at 436nm in addition to 628nm excitation (Supplementary Fig. 12b). This effect is likely from differential excitation of miRFPs at the shorter absorption peak near 400nm (Soret-band) that is common to all phytochrome-derived NIR FPs 26 (Supplementary Fig. 12c). Thus, the new NIR-FRET biosensor is compatible with multiplex imaging, only requiring minimal correction for photobleaching using previous methods 27.
For spatiotemporal quantification, we turned to morphodynamic analysis 1 to analyze the activities of RhoA and Rac1 in the same cell during edge protrusions. Morphodynamic analysis quantifies periodic cycling of cellular protrusions under steady-state conditions 1. The leading edge motion is extracted and activities of RhoA and Rac1 within diffusion-limited “measurement window segments” at the edge and at defined distances away from the edge are quantified (Methods) 1. We used these measurements to determine the temporal cross-correlation between RhoA versus Rac1 activities. We found that the activities are strongly and negatively correlated at the leading edge (within a distance of 0–0.9μm at the edge). This is consistent with Rac1-RhoA-antagonism and it is the first direct characterization of this in the same cell (Fig. 3a). The time difference between the onsets of RhoA and Rac1 activities was approximately 20s, agreeing with previously determined delay calculated from separate imaging of two CFP-YFP biosensors of Rac1 and RhoA 1. Representative morphodynamic maps from the leading edge window segments are shown in Figure 3b (Supplementary Fig. 13a, Video 2&3). Mutually excluded Rac1 and RhoA activities are evident during non-cyclical, large protrusion-retraction of the leading edge (Fig. 3b), and in smaller oscillatory protrusions (Supplementary Fig. 13a). The negative cross-correlation between RhoA and Rac1 activities at the leading edge diminished to below significance in regions away from the edge (>0.9μm) (Fig. 3a), indicating a strong spatial dependence. These results show for the first time, direct measurements of both Rac1 and RhoA activities at the leading edge and quantitative description of the Rac1-RhoA-antagonism, made possible by combining the new NIR-Rac1 biosensor with a CFP-YFP RhoA biosensor 15.
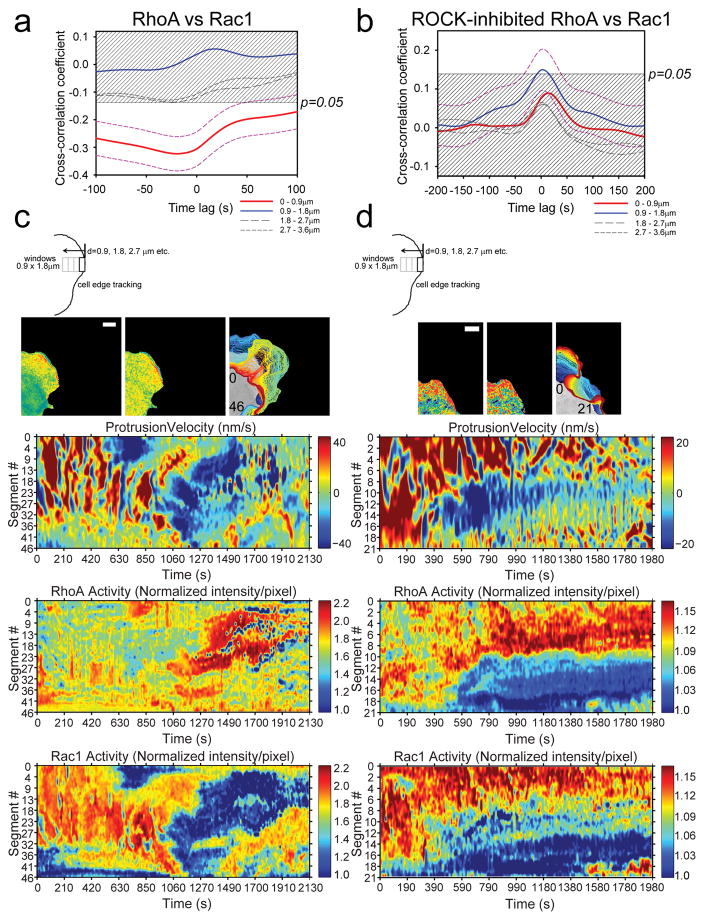
Figure 3. Morphodynamic analysis of RhoA-Rac1 antagonism.
(a) Cross-correlation coefficient as a function of timelags between RhoA and Rac1 activities in protruding edge of MEFs. Dashed magenta lines indicate 95% confidence interval (CI) for the cross-correlation function from the edge region (Red line: 0–0.9μm). The time difference between RhoA versus Rac1 biosensor response was −19.6s ±31.6s (±95% CI); i.e., RhoA activity onset precedes Rac1 activity onset by approximately 19.6s. 95% CI are omitted from the rest of plots for clarity. The shaded region indicates p>0.05. n=1250 individual sampling window segments were measured from 16 cells, from 6 independent experiments. (b) Example edge tracking evolution (top panels) from a single cell and associated morphodynamic maps of the velocity, RhoA and Rac1 activities (lower three panels). The edge velocity, RhoA activity, and Rac1 activity measured within window segments at the leading edge of a single representative cell are compiled on the Y-axes and followed in time on the X-axis. White bar=10μm. Example set taken from total of 16 cells, from 6 independent experiments. (c) Cross-correlation coefficient as a function of timelags between RhoA and Rac1 activities in protruding edge of MEFs with ROCK inhibitor Y-27632 treatment. Dashed magenta lines indicate 95% CI for the cross-correlation function from the region adjacent to the cell edge (Blue line: 0.9–1.8μm). 95% CI are omitted from the rest of plots for clarity. The shaded region indicates p>0.05. n=990 individual sampling window segments were measured from 18 cells, from 3 independent experiments. (d) Example edge tracking evolution (top panels) from a single cell and associated morphodynamic maps of the velocity, RhoA and Rac1 activities (lower three panels). White bar=10μm. Example set taken from 18 cells, from 3 independent experiments. The number inserts in the edge evolution panels in (b) and (d) indicate the orientation of the measurement window segments that correspond to the Y-axes of the morphodynamic maps as shown.
Next, we explored a potential mechanism by which Rac1-RhoA-antagonism was established at the edge. Previously, RhoA-ROCK-myosin was shown to balance the edge protrusion, where this contractile pathway caused stalling of the protrusion during edge cycling 28. The role of this pathway on modulating the RhoA-Rac1 activities was demonstrated using a set of CFP-YFP biosensors for RhoA and Rac1, imaged separately in different cells 28. Thus, we sought to directly visualize this balance of RhoA-Rac1-driven edge protrusions by similarly decoupling the mechanism using the ROCK inhibitor (Y-27632), with direct visualization of Rac1 and RhoA activities in the same cell. Consistent with the previous model 28, the negative cross-correlation between RhoA and Rac1 activities at the edge (0–0.9μm) changed to a positive cross-correlation when ROCK was inhibited, but the strength of correlation was not significant (Fig. 3c). Within 0.9–1.8μm from the edge, RhoA and Rac1 activities are positively correlated and significant (Fig. 3c). In regions further away from the edge, the trend of positive cross-correlation continues but is not significant. The change in cross-correlation between RhoA and Rac1 resulted from the concomitant activation of Rac1 and RhoA which likely prevented the edge from retracting and Rac1 from being deactivated in absence of ROCK-myosin-contractility pathway. This effect is visible in a representative set of morphodynamic maps from a cell that showed non-cyclical, large protrusions-retractions: Rac1-RhoA-antagonism is ablated and two activities are overlapping (Fig. 3b&d) (Supplementary Video 4). In small oscillatory protrusions (Supplementary Fig. 13, Video 3&5), cyclical activation of RhoA (0.9–1.8μm region) continues when ROCK is inhibited, but Rac1 activity remains elevated. These observations suggest that the coordination of RhoA and Rac1 via ROCK is critical in achieving the antagonism, likely pointing to molecular pathways regulated by ROCK being critical for orchestrating this coordination. The simultaneous detection of Rac1 and RhoA activities allowed a direct observation of this phenomenon for the first time.
Multiplex imaging of Rac1 activity and Rac1-GDI binding
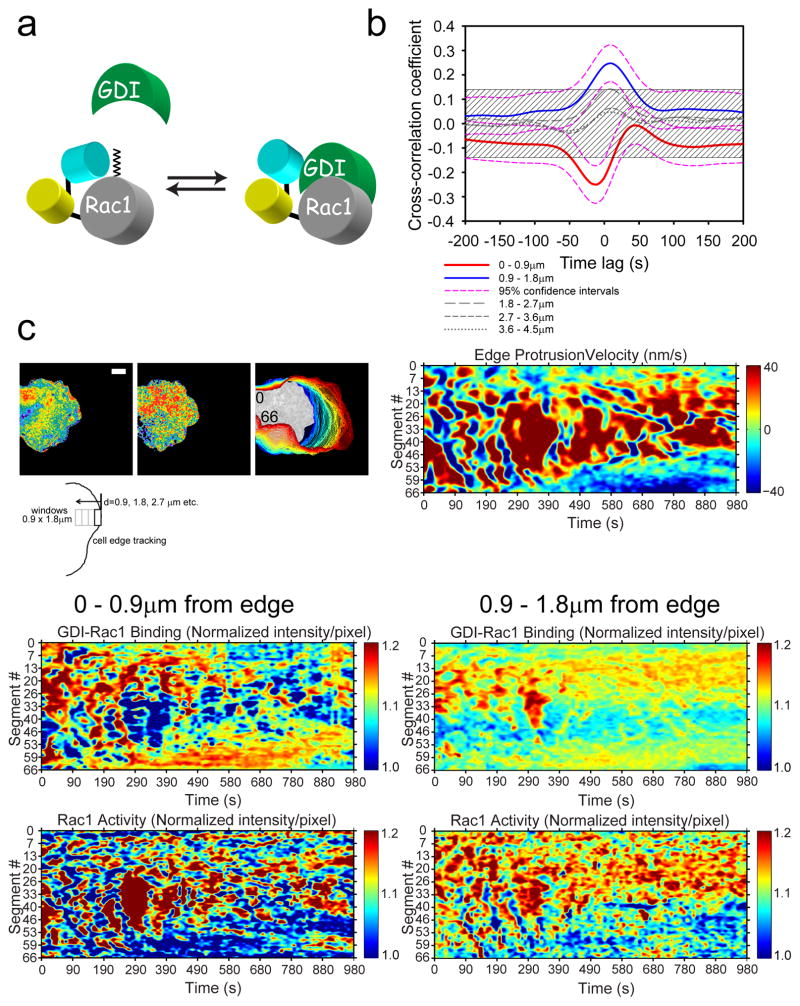
Regulation of RhoGTPases involves three classes of upstream regulators, GEFs, GAPs and GDI 11. The GDI-GTPase interaction is the least characterized of the regulator interactions, but recent studies have revealed spatiotemporal dynamics, regulated in part by cellular kinases 13. We took advantage of a recently reported CFP-YFP biosensor for Rac1-GDI binding (Fig. 4a) 2, and imaged together with NIR-Rac1 biosensor in single living cells. We produced tetracycline-inducible MEFs in which both the NIR-Rac1 and Rac1-GDI biosensors were stably incorporated and live-cell imaging was performed followed by morphodynamic analysis 1. The cross-correlation of Rac1 activity versus Rac1-GDI binding showed a remarkable coordination at the leading edge (0–0.9μm) and at an adjacent region away from the edge (0.9–1.8μm), while distal regions were not correlated (Fig. 4b). At the edge, we observed a strongly negative and significant cross-correlation between Rac1-GDI binding versus Rac1 activation, with approximately 12s difference between Rac1-GDI biosensor response occurring first, followed by NIR-Rac1 biosensor. This indicates that during activation of Rac1, the release from GDI (loss of CFP-YFP-FRET) is followed quickly by activation (gain of NIR-FRET) in the plasma membrane, presumably through encountering a GEF (Supplementary Fig. 14). This timing is similar to Cdc42 activation after the release from GDI 2, pointing to GEF-mediated activation of GTPases occurring rapidly after the release; thus, the release from GDI may be the rate determining step. This negative cross-correlation is visible in representative morphodynamic maps (Fig. 4c) (Supplementary Video 6). Interestingly, at an adjacent region away from the edge (0.9–1.8μm), a strongly positive and significant cross-correlation between Rac1-GDI binding and Rac1 activation was observed (Fig. 4b). This indicates that Rac1 activates in this region at the plasma membrane, but also is permissive to binding by GDI (Supplementary Fig. 14). These results are consistent with a previous observation that activated Rac1 may be removed from signaling pathways by forming a complex with cytoplasmic GDI 29. These observations show a tight coordination, both in space and time, of Rac1 activation and Rac1-GDI interaction for the first time in living cells, made possible by using the CFP-YFP and miRFP670-miRFP720 FRET biosensor combinations, revealing the complexity of spatiotemporal regulation of Rac1 by GDI.
Figure 4. Morphodynamic analysis of Rac1 activity and Rac1-GDI binding.
(a) Schematic drawing of the Rac1-GDI binding biosensor reported previously 2. The wildtype Rac1 is attached to the C-terminus of the FRET “antenna” that selectively detects the binding of Rac1 to GDI. (b) Cross-correlation coefficient as a function of timelags between Rac1-GDI binding and Rac1 activity in protruding edge of MEFs. Dashed magenta lines indicate 95% CI for the cross-correlation function of the edge and the next adjacent region away from the edge (Red line: 0–0.9μm; blue line: 0.9–1.8μm). The time differences between Rac1-GDI versus Rac1 biosensor response was −11.9s ±23s (±95% CI; Rac1-GDI biosensor response preceded Rac1 biosensor response) at the leading edge (Red line) and +9.6s ±25s (±95% CI; Rac1 activity preceded Rac1-GDI biosensor response) at the region adjacent to the edge (Blue line). 95% CI are omitted from the rest of plots for clarity. The shaded region indicates p>0.05. n=827 individual sampling window segments were measured from 10 cells from 3 independent experiments. (c) Example morphodynamic data set of a MEF imaged for Rac1 activity and Rac1-GDI binding in the same cell. The edge tracking evolution (top left panels) from a single cell and associated morphodynamic maps of the edge velocity (top right panel), Rac1-GDI binding at the edge (middle left panel), Rac1-GDI binding at the next adjacent region away from the edge (middle right panel), Rac1 activity at the edge (bottom left panel), and Rac1 activity at the next adjacent region away from the edge (bottom right panel). White bar=10μm. Example set taken from 10 cells from 3 independent experiments. The number inserts in the edge evolution panel in (c) indicate the orientation of the measurement window segments that correspond to the Y-axes of the morphodynamic maps as shown.
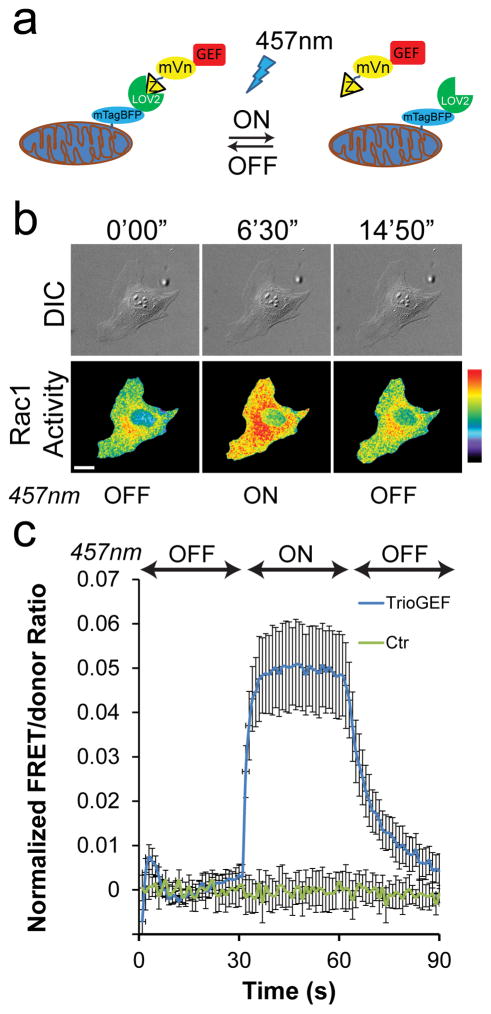
Compatibility of NIR-Rac1 biosensor with LOV-TRAP
To demonstrate the spectral compatibility of the NIR-Rac1 biosensor with optogenetics systems requiring blue-green light, including the Avena sativa phototropin LOV2, we decided to use the recently reported LOV-TRAP system (Fig. 5a) 16. This system consists of two components; mitochondrially-targeted LOV2-Jα and the catalytic domain of a GEF fused to an affinity monobody, which is evolved to bind reversibly to the dark-state of LOV2-Jα 16. Upon irradiation, the Jα-domain becomes labile and the monobody dissociates, releasing the catalytic domain of a GEF from the mitochondrial sequestration. The released GEF-domain activates RhoGTPases with a half-life of approximately 18.5s 16.
Figure 5. Concurrent measurement of Rac1 activity during LOV-TRAP optogenetics.
(a) Schematic drawing of the LOV-TRAP system. Mitochondrially targeted mtagBFP-LOV2wt sequesters the GEF-domain of interest attached to Zdk1 molecule. Upon photoactivation with 457nm light, Zdk1-GEF is released and act on the target GTPase, followed by dark relaxation upon cessation of illumination. (b) An example panel of a MEF undergoing photoactivation of LOV-TRAP and concurrent measurements of Rac1 activity using the NIR-Rac1 biosensor. NIR-Rac1 biosensor image sets were acquired every 10s. 457nm illumination (cycles of 4s-on, 6s-off) was started at 300 s and ended at 600 s time points (additional time points are shown in Supplementary Fig. 15a). White bar=20 μm. Pseudocolor limits are 1.0 to 1.74 (black to red). Example taken from N=17 independent photoactivation experiments. (c) Quantification of Rac1 activity measured concurrently with the LOV-TRAP-TrioGEF photoactivation. During the “on” phase of the 457nm illumination, Rac1 activity levels are significantly elevated compared to the control which received no 457nm illumination. The Student t-test (two-tailed) was used to compare Rac1 activity during photoactivation (300s–600s) and dark relaxation (600s–900s) versus the activity prior to photoactivation (0s–300s), and shown in Supplementary Table 2. N=17 independent photoactivation experiments for LOV-TRAP-TrioGEF, n=10 independent mock-photoactivation experiments for the control condition, all shown with mean ±SEM.
We modified the original LOV-TRAP to specifically target Rac1 activation, by exchanging the catalytic GEF-domain of Vav2, which activates RhoA, Rac1, and Cdc42, to the N-terminal DHPH-SH3 of TrioGEF, which targets Rac1 and another Rac-family member RhoG 30. We produced tetracycline-inducible MEFs expressing NIR-Rac1 biosensor, and transiently transfected N-TOM20-mTagBFP2-LOV2wt and mVenus-Zdk1-TrioDHPH-SH3, at a cDNA ratio of 10:1 16. We imaged Rac1 activity at 10s intervals and photoactivated the LOV-TRAP at the 300s time point using 457nm light (cycling 4s-on/6s-off) for 300s, followed by another 300s of dark relaxation (Fig. 5b) (Supplementary Fig. 15a, Video 7). We observed rapid activation of Rac1 when LOV-TRAP was uncaged, and deactivation following cessation of photoactivation (Fig. 5c). The original LOV-TRAP containing the GEF-domain of Vav2 16 produced similar Rac1 activation compared to the TrioGEF when uncaged, but the decay of Rac1 activity during the dark relaxation appeared slower (Supplementary Fig. 15b). This is consistent with promiscuous targeting of Vav2 towards other RhoGTPases; the cellular GAP may be limiting or other pathways differentially controlling GTPases may have been activated. Interestingly, the LOV-TRAP did not induce changes in leading edge movement as a function of photouncaging (or during dark relaxation), despite Rac1 being activated (Supplementary Fig. 15c). This observation suggests that proper targeting of GEF, via molecular interaction domains that either localizes or facilitates assembly of the downstream effector signaling complex within appropriate cellular compartments, is also required in addition to activated RhoGTPase in order to drive specific cellular phenotype. These results demonstrate that NIR-Rac1 biosensor is complementary to and can be used together with common blue-green light-controlled optogenetics tools, enabling simultaneous detection and modulation of the RhoGTPase activities for the first time.
Development of NIR-kinase biosensors
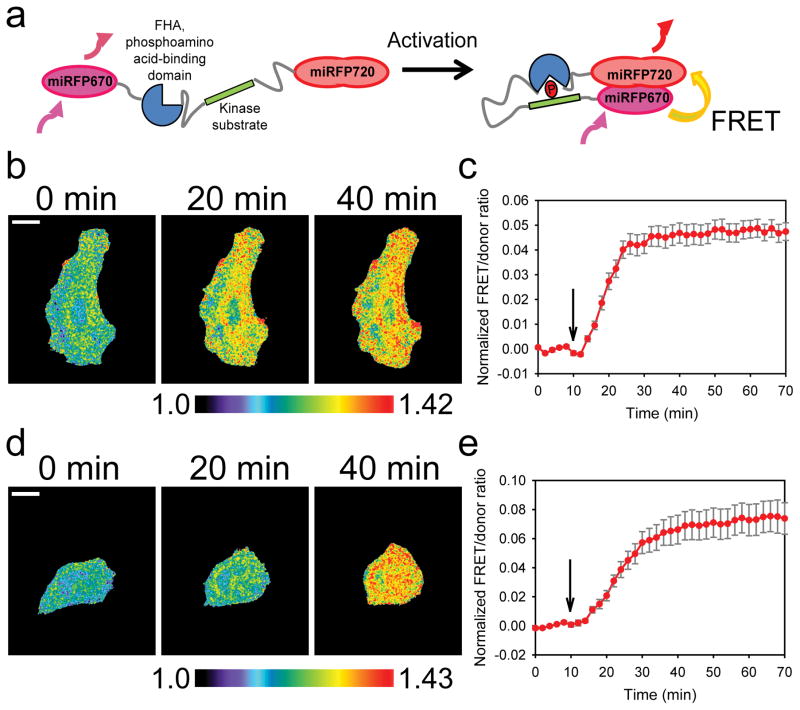
To demonstrate the generalizability of our new NIR-FRET pair of FPs, we extended this approach to biosensors detecting the phosphorylation activity of two kinases, PKA and JNK, AKAR 31 and JNKAR 32, respectively (Fig. 6a). We replaced the original cyan-yellow FPs in the sensors with an optimized backbone 33 with miRFP670 and miRFP720 at N- and C-termini of the sensors, and measured the cellular response to previously used stimulations that induced phosphorylation and increase in FRET upon PKA (Fig. 6b&c) or JNK activation (Fig. 6d&e) 31–33. Upon stimulation, FRET/donor ratio were elevated rapidly and have reached a plateau as previously reported (Fig. 6c&e) (Supplementary Videos 8&9), indicating that the new NIR-FRET pair of FPs we report here, is useful as a general tool to engineer genetically-encoded NIR-FRET biosensors for live-cell imaging.
Figure 6. NIR-FRET pair of fluorescent proteins in kinase-substrate biosensors.
(a) The basic design principle for the PKA31 and JNK32 sensors, modified from the original cyan-yellow fluorescent protein versions 33 to using the NIR miRFP670 10 and miRFP720 fluorescent proteins. (b) Representative timelapse images of a HeLa cell expressing the NIR-AKAR PKA sensor and undergoing stimulation (additional time points are shown in Supplementary Fig. 16a), from n=3 independent stimulation experiments. 1mM dbcAMP was added at the 10min time point and the FRET/donor ratio was monitored for up to 70min; FRET/donor ratio from NIR-AKR is shown with mean ±SEM (c). White bar, 20μm. Black arrow indicates the time point at which the stimulation was performed. n=3 stimulation experiments. Student t-test was used (two tailed) to determine p-values between FRET/donor ratio before (0–8min) and after stimulation (10–70min), shown in Supplementary Table 3. (d) Representative time-lapse images of a HeLa cell expressing the NIR-JNKAR JNK sensor and undergoing stimulation(additional time points are shown in Supplementary Fig. 16b), from n=3 independent stimulation experiments. 1μg/mL anisomycin was added at the 10 min time point and the FRET/donor ratio was monitored for up to 70 min; FRET/donor ratio from NIR-JNKR is shown with mean ±SEM (e). White bar, 20 μm. Black arrow indicates the time point at which the stimulation was performed. N=3 independent stimulation experiments. Student t-test was used (two tailed) to determine p-values between FRET/donor ratio before (0–8min) and after stimulation (10–70min), shown in Supplementary Table 3.
Discussion
From parental dimeric iRFP720 we engineered the most red-shifted monomeric miRFP720. Moreover, our rationally designed site-specific amino acid substitutions in the dimerizing interface of iRFP720 can be used to monomerize other dimeric NIR FPs 9. The photochemical characteristics of miRFP720 are equal to or exceed those reported for other monomeric NIR FPs 10, making it an advantageous protein tag for multicolor imaging. The red-shift of miRFP720 makes it an ideal FRET acceptor when combined with miRFP670, due to minimal cross-excitation of the acceptor and a good spectral overlap between donor fluorescence and acceptor excitation. Until now, there were no genetically-encoded NIR biosensors based on FRET available. The miRFP670-miRFP720 is the efficient and fully NIR-FRET pair, which can be used as easily as the common CFP-YFP pair in a wide range of FRET applications. Importantly, the miRFP670-miRFP720 pair allows multiplexing with blue-green-yellow channels of fluorescence with practically zero spectral overlap, enabling cross-talk free, simultaneous imaging with CFP-YFP biosensors34 and modulation with blue-green optogenetic tools, such as LOV, CRY2 and channelrhodopsins7,8.
Using the miRFP670-miRFP720 pair, we engineered a NIR biosensor for Rac1 GTPase. We simultaneously imaged Rac1 activity with that of its antagonist RhoA, or its regulation with GDI in the same cell, by combining the NIR biosensor with well-characterized and widely used CFP-YFP biosensors. Rac1-RhoA imaging revealed the antagonistic dynamic of these two canonical GTPases during cell edge movement, dependent on ROCK signaling. Consistent with a previous model 28, ROCK inhibition induced robust protrusions and resulted in activations of both GTPases in the same place at the same time. This agrees with the idea that RhoA-ROCK-myosin pathway effectively acts as a braking mechanism that induces stalling of the leading edge during protrusion-retraction cycles at appropriate timing 28. To achieve this, the antagonism must be established via recruiting GEF-GAP modules that, through feedback mechanism from the downstream molecular pathways, must be activated at appropriate timing and location. Importantly, the multiplexed imaging showed dynamics of Rac1-RhoA activities that appeared to directly correlate Rac1 with protrusions and RhoA with retractions. This is also consistent with measurements made by others using CFP-YFP biosensors in a different fibroblast cell line 28. Previous study has shown that the onset of Rac1 was closer in time to the start of edge retraction whereas that of RhoA was associated with edge protrusion 1. The apparent difference could be attributed to different types of edge protrusions observed in our current iteration of analysis compared to the previous work, evidenced by much longer oscillatory periodicity (90s 1 versus 120–140s in this work; Supplementary Fig. 10). This could stem from the use of different fibronectin and serum that may trigger differential signaling pathways 23, which may change the balance of Rac1-RhoA activities and dynamics of protrusions. Our ability to directly observe both Rac1 and RhoA activities in the same cell as shown here, should facilitate detailed analyses of molecular machinery that coordinate such balance of Rac1-RhoA activities driving cell edge protrusions.
The coordination of GTPase regulation at the leading edge was further demonstrated when we studied the Rac1-GDI interaction and activation, using the NIR-Rac1 biosensor in combination with the CFP-YFP Rac1-GDI biosensor 2. Our experiments showed spatiotemporal coordination of Rac1 activation following its release from GDI at the leading edge. A positive cross-correlation observed at an adjacent region behind the leading edge revealed that activation of Rac1 was followed closely in time by binding to GDI. This is consistent with previous studies in which GDI binds active Rac1 as a mechanism to control Rac1-effector interaction29. More investigation in this area is required to reveal the mechanism of GDI-mediated recycling and control of active Rac1 GTPase but this is the first direct observation of such localized spatial regulation of Rac1 GTPase.
Similar to multiplexing biosensors, it has not yet been possible to use blue/green optogenetic tools together with CFP-YFP biosensors. For LOV2 16, the CFP excitation at 440nm is too close to the photouncaging wavelengths. In the original LOV-TRAP characterization16, either the light or the dark mutant of the LOV-TRAP optogenetic tool was tested against CFP-YFP biosensors for Rho family GTPases 16. Here, we demonstrated that the NIR-Rac1 biosensor can be used to measure Rac1 activity at the same time as using the LOV-TRAP approach 16. The ability to monitor activity and simultaneously perturb a signaling node using optogenetics will be valuable to dissecting signal-pathway interdependency. Now, spectral compatibility with NIR biosensors enables a use of optogenetic tools in direct real-time experiments.
Finally, we demonstrated the generalizability of miRFP720 as an excellent FRET acceptor coupled to miRFP670 to produce genetically-encoded, NIR-FRET biosensors, providing researchers with the NIR-FRET band for direct imaging, quantification and control of several signaling pathways in living cells. We expect these NIR-FRET probes to be suitable for use in vivo in animal imaging modalities due to optimal compatibility with light absorbance characteristics in tissues, enabling deep tissue penetration. Combining functional imaging with non-invasive optogenetic modulation enables relating cell dynamics to metabolic functions at cellular and tissue levels and to behavior at the organ and organismal levels.
Online Methods
Cloning and protein expression
Site-specific mutagenesis of iRFP720 cloned in pBAD/His-B vector was performed with QuikChange mutagenesis kit (Agilent Technologies). LMG194 host cells (Invitrogen) were used for protein expression. A plasmid pWA23h plasmid encoding HO from Bradyrhizobium ORS278 (hmuO) under the rhamnose promoter was co-transformed with a pBAD/His-B plasmid encoding a fluorescent protein 9,35. Bacterial cells were incubated overnight at 37°C in RM minimal medium with ampicillin and kanamycin. To start protein expression, 0.002% arabinose and 0.02% rhamnose were added. After growing for 6–8 h at 37°C, the cells were incubated at 18°C for 24 h. Proteins were purified with Ni-NTA agarose (Qiagen). Protein was eluted with phosphate buffered saline (PBS) containing 100 mM EDTA, instead of imidazole. Then the samples were desalted using PD-10 desalting columns (GE Healthcare).
Protein characterization in vitro
FluoroMax-3 spectrofluorometer (Jobin Yvon) was used for recording of fluorescence spectra, Hitachi U-2000 spectrophotometer was used for absorbance measurements. To determine extinction coefficient, we calculated a ratio between the maximum absorbance of the main peak at Q band and the side peak at Soret band and assumed that extinction coefficient at Soret band corresponds to 39,900 M−1 cm−1 36,37. To determine quantum yield, we measured fluorescence signal of a purified protein in parallel with an equally absorbing Nile blue dye (quantum yield is 0.27 38) and compared the signal at several dilutions. pH titrations were done using a series of buffers (100 mM sodium acetate, 300 mM NaCl for pH 2.5–5.0 and 100 mM NaH2PO4, 300 mM NaCl for pH 4.5–9.0).
To test NIR FPs for oligomeric state, an analytical ultracentrifugation was conducted at 20°C and 58,000 rpm with an Optima XL-I centrifuge (Beckman Coulter) using the AN-60Ti rotor and the absorption optics set to 645 nm. Sednterp v.20120828beta software was used to calculate the partial specific volume of the proteins from their sequence and the density and viscosity of the buffers. The sedimentation parameters were corrected to standard conditions (20,w) using these values. For sedimentation velocity experiments, 350 μl of protein sample and an equal volume of PBS were loaded into two-sector cell assemblies with the protein concentration corresponding to A645 ≈0.9. Fifty scans were collected over the course of a centrifuge run. A subset of scans, beginning with those where a clear plateau was evident between the meniscus and the boundary, was selected for time-derivative analysis using DCDT+ v.2.4.2 software 39,40.
Mammalian plasmids
To test brightness of NIR FPs in HeLa cells they were transiently co-transfected with plasmids encoding EGFP and NIR FP. To construct mammalian expression plasmids, the respective genes of NIR FPs were PCR-amplified as AgeI-NotI fragments and swapped with a gene encoding EGFP in a pEGFP-N1 plasmid (Clontech). For example, the miRFP720 gene was swapped with the EGFP gene, resulting in a pmiRFP720-N1 plasmid. The pmIFP-N1 plasmid was obtained from Addgene (#54620).
For protein tagging and labeling of intracellular structures, a miRFP720 gene was swapped with a mTagBFP2 gene either in C-terminal (for α-tubulin and myosin) or in an N-terminal (for LifeAct, vimentin, LAMP1, H2B and mitochondrial signal) fusions41. For C-terminal fusions, the (SGGGG)n linker was increased to 30 amino acids. For N-terminal fusions the linker length was unchanged.
For caspase-3 FRET biosensors, fusions of miRFPP670 with miRFP709 and miRFP670 with miRFP720, containing 11 amino acid linker with the caspase-3 recognition site (GGDEVDGPVAT), were designed. For this, a miRFP670 gene was PCR amplified using primers containing the linker sequence and inserted using the BglII and AgeI restriction sites into pmiRFP709-N1 and pmiRFP720-N1 plasmids, respectively.
For LOV-TRAP optogenetic experiments, pTriEX-N-TOM20-LOV2 and pTriEx-mVenus-Zdk1-VAV2 DHPHC1 16 were a gift from Dr. Klaus Hahn (Addgene plasmid # 81009 and #81133). mTagBFP 42 was cloned into pTriEX-N-TOM20-LOV2 using overlapping PCR with the following primer pairs: 5′-gcagaattatacttggtataccatggtgggccgcaacagcgcgat-3′, 5′-tctagatttaaagttcggatcgctg-3′ to amplify the TOM20 fragment; 5′-cagcgatccgaactttaaatctagaatggtgtctaagggcgaagagctga-3′ and 5′-cctatatgcataattaggatccacttccagaaccattcagcttgtgccccagtttgcta-3′ to amplify mTagBFP2; and 5′-gcaattagcaatgtcaggatccttggctactacacttgaacgtattg-3′ and 5′-gcaatgctaatgcatatattctcgagttaaagttcttttgccgcctcatca-3′ to amplify the LOV2 domain. The full length N-TOM2-mtagBFP-LOV2 was ligated into pTriEX-4 backbone at NcoI/XhoI sites. The full length wildtype TrioGEF was a gift from Dr. Jaap van Buul. Trio DHPH1SH3 (amino acids 1291–1609) 43 was amplified and cloned into pTriEX-mVenus-Zdk1 at the C-terminus using overlapping PCR with the following primer pairs: 5′-gcaagattatatgcatattaccatggtgagcaagggcgaggagctgtt-3′ and 5′-agatccacttccgaattccttgtacagctcgtccatgccgaga-3′ to amplify mVenus; 5′-gtacaaggaattcggaagtggatctatggtggataacaaattcaataaag-3′ and 5′-tttaccagatccaccagatccggatccttttggggcctgggcatcgttca-3′ to amplify Zdk1; and 5′-atccggatctggtggatctggtaaaaggaaagagttcataatggctgagct-3′ and 5′-gcattatagaacaattaatctcgagttaggcgtcattgctggagacggagagc-3′ to amplify the TrioDHPH1SH3 fragment. The product was ligated into pTriEX-4 at NcoI/XhoI sites.
Mammalian cells and transfection
HeLa cells were purchased from the ATCC and were not additionally authenticated or tested for mycoplasma contamination. HeLa cells were grown in a DMEM medium supplemented with 10% FBS, 0.5% penicillin-streptomycin and 2 mM glutamine (Life Technologies/Invitrogen). For microscopy, cells were cultured in 35 mm glass-bottom Petri dishes with no. 1 coverglass (MatTek). Transfections of cells with plasmids were performed using either Effectene (Qiagen) or Mirus LT1 (MirusBio) transfection reagents. LinXE cells (HEK293 derivative; mycoplasma tested) 44 were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 1% Glutamax (Invitrogen) and Penicillin (100 IU/ml)/Streptomycin (100 IU/ml). Mouse embryonic fibroblasts (MEF/3T3 tet-OFF; mycoplasma tested; Clontech) were cultured in DMEM supplemented with 10% FBS, 1% Glutamax (Invitrogen), Penicillin (100 IU/ml)/Streptomycin (100 IU/ml) and neomycin-G418 (1mg/ml), as previously described15.
Fluorescence of cell suspension
Fluorescence of uncleaved and cleaved caspase-3 FRET biosensors was measured in transiently transfected HeLa cells 48 h after the transfection. To activate biosensor cleavage, 2 μM staurosporin was added to cells grown in 6-well plate 6 h before the analysis. Cells were pretreated with 25 μM BV for 24 h before the measurements. Fluorescence spectra of the untreated (uncleaved) and staurosposin-treated (cleaved) cell suspensions were recorded using 610 nm excitation in 150 μl microcuvette and normalized by acceptor fluorescence at 670 nm for comparison.
Fluorescence microscopy
Live HeLa cells were imaged on Olympus IX81 inverted epifluorescence microscope 48 h after transfection with miRFP720 fusions. The microscope was equipped with a 200 W metal halide arc lamp (Lumen220 Pro; Prior), 100× 1.4 NA oil immersion objective lens (UPlanSApo; Olympus) and Cy5.5. filter set (665/45 nm exciter and 725/50 nm emitter). The microscope was operated with SlideBook v.4.1 software (Intelligent Imaging Innovations).
To determine protein photostability, unfused NIR FPs were cytoplasmically expressed in HeLa cells and imaged at determined time periods. Obtained raw data were normalized to corresponding absorbance spectra and extinction coefficients of the proteins, the spectrum of 200 W Me-Ha arc lamp, and the transmission of 665/45 nm photobleaching filter. For determining the FRET efficiency in caspase-3 cleavage experiments, 610 nm wavelength light was used for excitation.
NIR-Rac1 biosensor
The previously published single-chain FRET-biosensor for Rac1, based on mCerulean1 – circularly permutated monomeric Venus 20,23, was used as the backbone for the new NIR-Rac1 biosensor. Briefly, miRFP720 and miRFP670 were synthesized with codon optimization, conservative point mutations to remove internal restriction sites which made them compatible with the original Rac1 biosensor backbone containing a set of unique restriction sites NcoI, BamHI, HindIII, EagI, NotI, EcoRI, and XhoI, and with appropriate terminal restriction site sequences added. The miRFP720 was placed in the FP1 position and miRFP670 in the FP2 position based on the optimization results (Supplementary Fig. 6a). The optimized biosensor backbone contained a miRFP720 as the FRET acceptor, two tandem codon optimized 23 p21 binding domains (PBD) of p21 activated kinase 1 (PAK1, amino acid residues 70–149) to achieve autoinhibitory regulation, separated by a structurally optimized linker (GSGGPPGSGGSG), a unstructured linker (Linker 1: 58 amino acid residues) of optimized length based on a previously described design 45, miRFP670 as FRET donor, an optimized 10 amino acid linker (GSGSGSGGEL; Linker 2), and a full-length wild-type (WT) Rac1 (Fig. 2a). The second PBD contained H83D, H86D point mutations to render it unable to bind to active GTPase. The synonymous modification 22 was applied to the two PBD domains and repeating units within the Linker 1 to prevent homologous recombination and stabilize expression. The resulting biosensor construct was cloned into a pTriEX-4 backbone (Novagen) at NcoI/XhoI sites. For the stable/inducible incorporation of biosensor into cellular genome, pRetro-X system was used. Biosensor cDNA cassette was cloned into the pRetro-X-Hygro and -Puro backbones using the Gateway cloning technique (Invitrogen), following the manufacturer’s protocols. For double positive biosensor transductants, MEFs stably and inducibly harboring the RhoA biosensor 15 or Rac1-GDI biosensor 2 were transduced with retrovirus containing the expression cassette for NIR-Rac1 biosensor under the tetracycline-inducible promoter, but with hygromycin resistance. Methods for production of virus, transduction, followed by selection and induction of biosensor, were described previously 46.
Fluorometric characterization of NIR-Rac1 biosensor
Characterization of biosensor response was performed in LinXE cells (HEK293T derivative) by transient overexpressions of WT or mutant versions of the biosensor, with or without the upstream regulators, as described previously 15. Briefly, LinXE cells were plated on poly-L-lysine (Sigma) coated 6-well plates overnight at 1.25×106 cells/well, and transfected the next day using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocols. Biosensors were co-transfected at ratios of 1:2 with GDI or 1:4 GAP, or 1:1 – 4 for co-transfection with GEFs. The transfection solution additionally contained 25 μM BV. 48 hrs following transfection, cells were washed in PBS, briefly trypsinized and resuspended in cold PBS, and the live cell suspension was placed into a cuvet, and fluorescence emission spectra was measured by spectrofluorometer (Horiba-Jobin-Yvon Fluorolog-3MF2). The fluorescence emission spectra were obtained by exciting the specimen at 600 nm wavelength light, and emission fluorescence was scanned between 640–750 nm. The background fluorescence reading of cells containing an empty vector was used to measure light scatter and autofluorescence, and subtracted from the data. The resulting spectra were normalized to the peak miRFP670 emission intensity at 670 nm to generate the final ratiometric spectra.
NIR-Rac1 biosensor pulldown experiments
LinXE cells were plated at a density of 1.25×106 cells on poly-L-lysine coated 6 well plates. The next day, cells were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocols. 24 h later, cells were lysed in a buffer containing 1% NP-40, 20 mM Tris, 300 mM NaCl, pH 8.0, 1 mM PMSF and protease inhibitor cocktail (Sigma). Lysates were clarified by centrifugation at 14,000 rpm for 10 min at 4°C. After removing an “input” fraction, lysates were incubated with PAK1-PBD beads 47 for 1 h at 4°C with rotation. Samples were washed 3× in lysis buffer, resuspended in final sample buffer and analyzed by Western blotting 20.
Western Blotting
Cell lysates were resolved on 10% SDS-polyacrylamide gels. PVDF membranes were used for blotting. Primary antibody incubations were performed overnight at 4°C (1:1,000 dilution). Secondary fluorescently conjugated antibodies (LiCor) were diluted 1:20,000 and incubated for 1 h at room temperature. Immunoblots were recorded on the Odyssey Imager (LiCor). Primary antibodies used were: β-actin (Santa Cruz Biotechnology; sc-69879 clone AC-15) and Rac1 (MilliporeSigma 05-389; clone 23A).
Live-cell multiplexed FRET imaging
MEF cells expressing the biosensors were plated on 25 mm round #1.5 coverslips (Warner Instruments), coated with 10 μg/ml fibronectin in DPBS for 1hr at room temperature, at 4.5×104 cells per well in standard growth medium containing 25 μM BV on the day of the experiment. Cells were time-lapse imaged beginning 2–3 h post plating. Cells were imaged in Ham’s F-12K medium without phenol red (Crystalgen), sparged with Argon gas to remove dissolved oxygen, and supplemented with 3% fetal bovine serum, 10 mM HEPES, Oxyfluor reagent at 1:100 dilution (Oxyrase Inc.) and 10 mM dl-lactate (Sigma). Imaging medium did not contain exogenous BV during experiments. Cells were imaged at 37°C in a closed imaging chamber 48. We utilized standard widefield microscopy in our live cell FRET experiments, as described previously 49.
Biosensor readouts were measured in cells using a custom Olympus IX81-ZDC microscope, optimized for FRET48. Metamorph v.7.8.13 (Molecular Devices) was used to perform image acquisition. For live cell imaging, images were acquired through a 40× magnification objective lens (Olympus UIS DIC 40× 1.4NA) with 2×2 camera binning, using a custom microscope capable of simultaneous acquisition of FRET (mCerulean-mVenus FRET) and mCerulean emissions through two Coolsnap ES2 cameras (Photometrics) mounted onto the side camera port via an optical beam splitter, containing T505LPXR mirror, ET480/40M for mCerulean emission, and ET535/30M for mVenus-FRET emission (Chroma Technology). The microscope main body prism was replaced with T555LPXR mirror (Chroma Technology), allowing for splitting of longer wavelengths to the bottom mounted camera port. A third camera (Coolsnap HQ2; Photometrics) was mounted on the bottom port of the microscope via a filterwheel and acquired differential interference contrast (DIC) and miRFP670/720 FRET channels which were taken sequentially. The associated error due to motion artifacts during the leading edge (6.36 nm/s +/− 2.6 nm/s (Ave +/− S.D.)) for the control condition in these MEF cells (calculated during Morphodynamic analysis) 1. This translated to, on average, 63.6 nm +/− 26 nm positional uncertainty within the 10 s time interval of an acquisition cycle. This is within the resolution limit of the acquisition condition, with calibrated pixel dimensions at the image plane of 309 nm × 309 nm. In the side-port dual camera system, relative intensities between the FRET and mCerulean channels were balanced by inclusion of a neutral density filter in the FRET channel (ND 0.2 for RhoA biosensor, ND 0.1 for Rac1-GDI biosensor) so that the range of brightness in both channels were similar to each other. Illuminations were all from 100 W Hg arc lamp source. For mCerulean-mVenus FRET, cells were illuminated via ET436/20X bandpass filter (Chroma Technology). For miRFP670-miRFP720 FRET, cells were illuminated via ET628/32X bandpass filter (Chroma Technology). For the emissions, ET480/40M and ET535/30M filter pair was used for mCerulean-mVenus FRET (Chroma Technology), and FF684/24 (Semrock) and FF794/160 (Semrock) filter pair was used for miRFP670 and miRFP720 FRET imaging.
All image channels were aligned prior to ratiometric calculations by a pixel-by-pixel matching using a priori calibration and non-linear coordinate transformation to achieve registration, as previously described 49. Metamorph v.7.8.13 (Molecular Devices) and Matlab (Mathworks) software were used to perform image processing and data analysis. Image processing included flat-field correction, background subtraction, camera-noise subtraction, non-linear coordinate transformation, threshold masking, ratio calculations, and correction for photobleaching, as described previously 49. The detailed optical specifications for the microscope used here, are described elsewhere48. For imaging of biosensors, we adjusted the camera acquisition time duration by targeting to fill approximately 80% of the total digitization range of the CCD circuitry to maximize the dynamic range, using excitation intensities of 0.4–1.0 mW at the specimen plane.
Morphodynamic-mapping and cross-correlation analysis
Morphodynamic-mapping and cross-correlation methods were described previously 1. The primary assumption at the core of this analysis is that the relationship between GTPase activation and the movements of the cell edge during constitutive protrusion-retraction cycles is preserved anywhere within the edge of a single cell, and among all cells within a given population 1. This allows us to evaluate the behavior of a small segment of an edge as an independent sampling entity and then average such measurements from many segments to perform cross-correlational analyses between multiple measurement parameters. Briefly, the analysis algorithm first tracks the leading edge motion by using a previously described method 1. Once the leading edge positions are defined as a function of time from a timelapse series, measurement window “segments” are defined along the cell edge in order to measure the parameters of interest within single window segment entities. In this work, we measured two biosensor activities and the cell edge velocity in sampling window segments of 0.9 × 1.8 μm constructed along the leading edge of cells and tracked the cell edge motion during complete protrusion-retraction cycles. We have previously determined that this particular measurement window dimension was diffusion-limited (minimal diffusional mixing of membrane-bound components between adjacent windows, assuming two-dimensional, membrane-bound diffusion constant: at the cell edge, there would be very little cytoplasm present) within the time interval of imaging (10 s per image), the objective magnification, and the pixel dimension of the imaging system used 1. Thus, by maintaining such a diffusion-limited sampling dimension, we ensure that the readout from individual measurement window segments can be treated as independent sampling entities 1. By constructing many individual, independent measurement window segments along edges in a number of cells from a given population, we achieved sufficient statistical power to determine significance in localization of activities in space and time, as in our previous studies 1,2,23. Next, measurement window segments were moved away from the leading edge in 0.9 μm increments to measure two biosensor readouts at successively distal regions away from the edge. The extent of coupling between changes in the two biosensor activity readouts was determined using cross-correlation function xcov in Matlab and Pearson’s correlation coefficient was used to determine the strength of coupling of two biosensor readouts. In addition to determining the spatial coupling, this approach also measures the temporal relationship between the two measured biosensor readouts. In this work, n=1250 window segments from 16 cells for the control RhoA-Rac1 multiplex measurements, n=990 window segments from 18 cells for ROCK-inhibited RhoA-Rac1 measurements, and n=827 window segments from 10 cells for Rac1-GDI binding and Rac1 activity measurements. The numbers of individual window segments constructed and total number of cells analyzed per cell populations/conditions were similar to previous studies 1,2,23. The resulting cross-correlation functions were compiled and analyzed using the 2000× bootstrapping of the smooth-spline fit functions from the individual window segments to determine the 95% confidence intervals. The autocorrelation function of the leading edge protrusion velocities was used as a measure to indicate the periodicity of the intrinsic protrusion-retraction cycles 1. This analysis indicated that in all cases analyzed, expression of two biosensors did not alter the basic protrusion periodicity from that of the previously published RhoA biosensor MEF cell line 15, imaged under current conditions (Supplementary Fig. 10d). The morphodynamic analysis as used in this work, does not separately parse out the behavior of biosensor activities as a function of edge protrusion or retraction. Instead, it takes the protrusion-retraction as single cycles and produces global cross-correlation functions taking into account many cycles of these processes. Thus, we imaged steady-state cells, taking advantage of the fact that at steady-state cellular edge behaviors are comprised of continuous cycles of protrusion-retraction that occur at random locations along the cell edge. By measuring this, we are able to sample many iterations of protrusion-retraction behaviors and build statistically robust cross-correlation function between the two signaling nodes. While beyond the scope of the current work, future direction could include parsing out the GTPase behaviors during protrusion versus retraction, as well as to analyze the correlative coupling of signaling nodes during acute exogenous stimulations.
In relation to the effect of increasing cell volume in regions away from the leading edge and how accumulating cytoplasmic biosensor could influence the cross-correlation analysis, we expect that the positive cross-correlation observed between Rac1 activity and Rac1-GDI binding at the region adjacent to the leading edge (Fig. 4) to result from their functional importance, as opposed to random response caused by changes in cytoplasmic volume. Because widefield imaging was used, an increase in cytoplasmic volume could cause an increase in FRET signal from the cytoplasmic accumulation of Rac1-GDI biosensor in complex with the cytoplasmic GDI. Conversely, NIR-Rac1 biosensor could show reduced activity from the volume dilution effect (low FRET signal from the inactive and cytoplasmic-partitioned NIR-Rac1 biosensor dilutes the high FRET signal from active biosensor in the plasma membrane: this dilution effect was previously modeled in Spiering et al. 2013) 49. In such a situation where the signal modulation opposes one another, either a negative cross-correlation is expected or no correlation is observed if only one of these volume-associated issues was predominant. Thus, our observation of positive cross-correlation at this location points to both binding of more Rac1 by GDI (and accumulation of Rac1-GDI complex in increased cytoplasmic volume) and activation of Rac1 at this location.
Optogenetic experiments with LOV-TRAP
In experiments involving the LOV-TRAP 16 system to target TrioGEF catalytic domain for Rac1 activation, MEF cells stably and inducibly harboring NIR-Rac1 biosensor was transfected with the required components. Briefly, MEFs were induced to undergo biosensor expression 48 h prior to the experiment in presence of 25 μM biliverdin, following the biosensor induction method described previously 46. 24 h after the induction, cells were briefly trypsinized, pooled and seeded onto 6-well plates at 1×105 cells/well, and transfected using Fugene 6 reagent (Roche), following the manufacturer’s protocols. The DNA ratio of 10:1 was used (pTriEX-N-TOM20-mtagBFP-LOV2 : pTriEX-mVenus-Zdk1-TrioDHPH1SH3), where plasmids were premixed at this ratio prior to making the transfection mixture, following the protocols previously published 16. Cells were imaged 24 h following the transfection. The imaging system was set up to enable whole-field photoactivation using 457 nm light that was pulsed every 10 s (4s-on, 6s-off). Cells positive for the LOV-TRAP expression were visualized and identified using mTagBFP2/mVenus. Cells were then imaged without photoactivation for 31 frames at 10 s interval per frame. The photoactivation sequence was initiated between the frames 31 and 32, and continued at 10s intervals for 30 additional frames concurrent with the biosensor measurements at the same interval. Data were acquired for additional 30 frames after the end of the photoactivation sequence at the same acquisition intervals for a total of 90 frames the beginning to the end.
NIR kinase biosensors
To clone NIR JNKAR and NIR AKAR biosensors, we used a pJNKAR1EV-NES and pAKAR3EV-NES plasmids 33, kindly provided by Kazuhiro Aoki (Kyoto University, Kyoto, Japan). YPet and ECFP in the original AKAR and JNKAR constructs were exchanged for miRFP670 and miRFP720 by using EcoRI/XhoI and NotI/XbaI restriction sites, correspondingly. Fragments encoding NIR sensors were cut out with EcoRI/SalI restriction endonucleases and inserted into pcDNA3.1 plasmid.
For imaging of NIR-AKAR and JNKAR, HeLa cells were seeded on 25 mm round coverslips treated with 10 μg/mL fibronectin (Sigma) in PBS for 1 h at room temperature, at 1×105 cells/coverslip in wells of 6-well plates. Cells were transfected using Fugene 6 reagent (Roche), following the manufacturer’s protocols. 6 h after addition of the transfection mixture to cells, media was supplemented with 25 μM biliverdin. 24 h following the transfection, cells were serum-starved in medium containing 0.1 % BSA without exogenous biliverdin for 8 h. At the end of starvation, cells were mounted in a live-cell imaging chamber atop a microscope stage and imaged at 37 °C under mineral oil. Cells were timelapse imaged at 2 min intervals, acquiring DIC and NIR-FRET/donor image pair at each time point. After establishing the baseline signal response for 10 min in absence of stimuli, stimulations were performed by addition of 1 mM dbcAMP or 1 μg/mL Anisomycin, for AKAR and JNKAR, respectively. Following the stimulation, timelapse imaging was performed for additional 1 h at 2 min intervals. Image processing and analysis of FRET ratio data were performed similar to the NIR-Rac1 biosensor, described above. Although beyond the scope of the current work, further sensor optimization by modulating the dipole coupling angles and changing the lengths and structures of the intramolecular linkers may improve the dynamic range of the sensor response, as we demonstrated here for NIR-Rac1 biosensor.
Flow cytometry
Flow cytometry analysis of NIR FPs was performed using a BD LSRII flow cytometer equipped with the 488 nm and 640 nm lasers and a set of emission filters. 20,000–50,000 events for each cell type were analyzed. To quantify cell fluorescence, a mean fluorescent intensity of the double-positive population in the NIR channel was divided by a mean fluorescence intensity of the same population in the green channel, thus normalizing NIR signal to transfection efficiency.
Statistical analysis
All statistical significance based on p-values were calculated using a Student t-test. No vertebrate animals were involved. No statistical methods were used to pre-determine the sample size. No randomizations were used. The investigators were not blinded to allocation during experiments and outcome assessment. Statistical tests used are stated on every figure legend with p-values as appropriate. Data distribution should meet the normal distribution requirements. No estimate of variation. No pre-established criteria were used to determine data inclusion or exclusion.
Data availability
The data that support the findings of this study are available from the corresponding authors on request.
Code availability
All Matlab codes and Metamorph scripts used were previously published elsewhere 1,2,49, but are also available from the corresponding authors on request.
Supplementary Material
Acknowledgments
We thank Michael Brenowitz for the help with analytical ultracentrifugation and Mikhail Baloban for the help with engineering miRFP720. We thank Sara Donnelly for critical reading of the manuscript. This work was supported by grants GM122567, NS099573, NS103573 and CA205262 from the US National Institutes of Health and ERC-2013-ADG-340233 from the EU FP7 program.
Footnotes
Author contributions
D.M.S. and V.V.V. developed the miRFP720 and characterized it in vitro, in cells and as FRET acceptor. L.H., N.C.C. and T.M.H. engineered the NIR-Rac1 biosensor, characterized it in cells and performed live cell imaging. L.H. with the participation of V.V.V. designed the project and planned the experiments and discussed the data with D.M.S. and V.V.V. V.V.V., D.M.S. and L.H. wrote the manuscript. All authors reviewed the manuscript.
Supplementary Information accompanies this paper at http://www.nature.com/
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodgson L, et al. FRET binding antenna reports spatiotemporal dynamics of GDI-Cdc42 GTPase interactions. Nat Chem Biol. 2016;12:802–9. doi: 10.1038/nchembio.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ouyang M, et al. Simultaneous visualization of protumorigenic Src and MT1-MMP activities with fluorescence resonance energy transfer. Cancer Research. 2010;70:2204–12. doi: 10.1158/0008-5472.CAN-09-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant DM, et al. Multiplexed FRET to image multiple signaling events in live cells. Biophys J. 2008;95:L69–71. doi: 10.1529/biophysj.108.139204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demeautis C, et al. Multiplexing PKA and ERK1&2 kinases FRET biosensors in living cells using single excitation wavelength dual colour FLIM. Sci Rep. 2017;7:41026. doi: 10.1038/srep41026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shcherbakova DM, Hink MA, Joosen L, Gadella TW, Verkhusha VV. An orange fluorescent protein with a large Stokes shift for single-excitation multicolor FCCS and FRET imaging. J Am Chem Soc. 2012;134:7913–23. doi: 10.1021/ja3018972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. Annu Rev Biochem. 2015;84:519–50. doi: 10.1146/annurev-biochem-060614-034411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chernov KG, Redchuk TA, Omelina ES, Verkhusha VV. Near-Infrared Fluorescent Proteins, Biosensors, and Optogenetic Tools Engineered from Phytochromes. Chem Rev. 2017;117:6423–6446. doi: 10.1021/acs.chemrev.6b00700. [DOI] [PubMed] [Google Scholar]
- 9.Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods. 2013;10:751–4. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shcherbakova DM, et al. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat Commun. 2016;7:12405. doi: 10.1038/ncomms12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nobes CD, Hall A. Regulation and function of the Rho subfamily of small GTPases. Cur Opin Genetics Dev. 1994;4:77–81. doi: 10.1016/0959-437x(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Mata R, Burridge K. Catching a GEF by its tail. Trends Cell Biol. 2007;17:36–43. doi: 10.1016/j.tcb.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12:493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pai SY, Kim C, Williams DA. Rac GTPases in human diseases. Dis Markers. 2010;29:177–87. doi: 10.3233/DMA-2010-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–72. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, et al. LOVTRAP: an optogenetic system for photoinduced protein dissociation. Nat Methods. 2016;13:755–8. doi: 10.1038/nmeth.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellini D, Papiz MZ. Dimerization properties of the RpBphP2 chromophore-binding domain crystallized by homologue-directed mutagenesis. Acta Crystallogr D Biol Crystallogr. 2012;68:1058–66. doi: 10.1107/S0907444912020537. [DOI] [PubMed] [Google Scholar]
- 18.Muller SM, Galliardt H, Schneider J, Barisas BG, Seidel T. Quantification of Forster resonance energy transfer by monitoring sensitized emission in living plant cells. Front Plant Sci. 2013;4:413. doi: 10.3389/fpls.2013.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson GH, Piston DW. Photobleaching in two-photon excitation microscopy. Biophys J. 2000;78:2159–62. doi: 10.1016/S0006-3495(00)76762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moshfegh Y, Bravo-Cordero JJ, Miskolci V, Condeelis J, Hodgson L. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat Cell Biol. 2014;16:574–86. doi: 10.1038/ncb2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quilliam LA, et al. Identification of residues critical for Ras(17N) growth-inhibitory phenotype and for Ras interaction with guanine nucleotide exchange factors. Molecular & Cellular Biology. 1994;14:1113–21. doi: 10.1128/mcb.14.2.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu B, et al. Synonymous modification results in high-fidelity gene expression of repetitive protein and nucleotide sequences. Genes Dev. 2015;29:876–86. doi: 10.1101/gad.259358.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miskolci V, Wu B, Moshfegh Y, Cox D, Hodgson L. Optical Tools To Study the Isoform-Specific Roles of Small GTPases in Immune Cells. J Immunol. 2016;196:3479–93. doi: 10.4049/jimmunol.1501655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin E, Ouellette MH, Jenna S. Rac1/RhoA antagonism defines cell-to-cell heterogeneity during epidermal morphogenesis in nematodes. J Cell Biol. 2016;215:483–498. doi: 10.1083/jcb.201604015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura F. FilGAP and its close relatives: a mediator of Rho-Rac antagonism that regulates cell morphology and migration. Biochem J. 2013;453:17–25. doi: 10.1042/BJ20130290. [DOI] [PubMed] [Google Scholar]
- 26.Shcherbakova DM, Baloban M, Verkhusha VV. Near-infrared fluorescent proteins engineered from bacterial phytochromes. Curr Opin Chem Biol. 2015;27:52–63. doi: 10.1016/j.cbpa.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodgson L, Nalbant P, Shen F, Hahn K. Imaging and photobleach correction of Mero-CBD, sensor of endogenous Cdc42 activation. Methods Enzymol. 2006;406:140–56. doi: 10.1016/S0076-6879(06)06012-5. [DOI] [PubMed] [Google Scholar]
- 28.Martin K, et al. Spatio-temporal co-ordination of RhoA, Rac1 and Cdc42 activation during prototypical edge protrusion and retraction dynamics. Sci Rep. 2016;6:21901. doi: 10.1038/srep21901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.del Pozo MA, Price LS, Alderson NB, Ren XD, Schwartz MA. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. Embo J. 2000;19:2008–14. doi: 10.1093/emboj/19.9.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellanger JM, et al. The two guanine nucleotide exchange factor domains of Trio link the Rac1 and the RhoA pathways in vivo. Oncogene. 1998;16:147–52. doi: 10.1038/sj.onc.1201532. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, Saulnier JL, Yellen G, Sabatini BL. A PKA activity sensor for quantitative analysis of endogenous GPCR signaling via 2-photon FRET-FLIM imaging. Front Pharmacol. 2014;5:56. doi: 10.3389/fphar.2014.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fosbrink M, Aye-Han NN, Cheong R, Levchenko A, Zhang J. Visualization of JNK activity dynamics with a genetically encoded fluorescent biosensor. Proc Natl Acad Sci U S A. 2010;107:5459–64. doi: 10.1073/pnas.0909671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komatsu N, et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell. 2011;22:4647–56. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piston DW, Kremers GJ. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 2007;32:407–14. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Piatkevich KD, Subach FV, Verkhusha VV. Far-red light photoactivatable near-infrared fluorescent proteins engineered from a bacterial phytochrome. Nat Commun. 2013;4:2153. doi: 10.1038/ncomms3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filonov GS, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–61. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shu X, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–7. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sens R, Drexhage KH. Fluorescence quantum yield of oxazine and carbazine laser dyes. J Luminesc. 1981;24:709–712. [Google Scholar]
- 39.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–46. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 40.Stafford WF., 3rd Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 41.Subach OM, Cranfill PJ, Davidson MW, Verkhusha VV. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One. 2011;6:e28674. doi: 10.1371/journal.pone.0028674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subach OM, et al. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15:1116–24. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Rijssel J, Hoogenboezem M, Wester L, Hordijk PL, Van Buul JD. The N-terminal DH-PH domain of Trio induces cell spreading and migration by regulating lamellipodia dynamics in a Rac1-dependent fashion. PLoS ONE. 2012;7:e29912. doi: 10.1371/journal.pone.0029912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brunet JP, et al. Rotavirus infection induces cytoskeleton disorganization in human intestinal epithelial cells: implication of an increase in intracellular calcium concentration. Journal of Virology. 2000;74:10801–6. doi: 10.1128/jvi.74.22.10801-10806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitlow M, et al. An improved linker for single-chain Fv with reduced aggregation and enhanced proteolytic stability. Protein Eng. 1993;6:989–95. doi: 10.1093/protein/6.8.989. [DOI] [PubMed] [Google Scholar]
- 46.Hodgson L, Pertz O, Hahn KM. Design and optimization of genetically encoded fluorescent biosensors: GTPase biosensors. Methods Cell Biol. 2008;85:63–81. doi: 10.1016/S0091-679X(08)85004-2. [DOI] [PubMed] [Google Scholar]
- 47.Benard V, Bokoch GM. Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods Enzymol. 2002;345:349–59. doi: 10.1016/s0076-6879(02)45028-8. [DOI] [PubMed] [Google Scholar]
- 48.Spiering D, Hodgson L. Multiplex Imaging of Rho Family GTPase Activities in Living Cells. Methods in Molecular Biology. 2012;827:215–34. doi: 10.1007/978-1-61779-442-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spiering D, Bravo-Cordero JJ, Moshfegh Y, Miskolci V, Hodgson L. Quantitative Ratiometric Imaging of FRET-Biosensors in Living Cells. Methods Cell Biol. 2013;114:593–609. doi: 10.1016/B978-0-12-407761-4.00025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors on request.