Abstract
Phytocannabinoids have well-known cardiovascular implications. For instance, Δ9-tetrahydrocannabinol (Δ9-THC), the principal component of cannabis, induces tachycardia in humans. In order to understand the impact of phytocannabinoids on human cardiovascular health, there is a need to study the metabolism of phytocannabinoids by cardiac cytochromes p450 (CYPs). CYP2J2, the primary CYP of cardiomyocytes, is responsible for the metabolism of the endocannabinoid, anandamide (AEA), into cardioprotective epoxides (EET-EAs). Herein, we have investigated the kinetics of the direct metabolism of six phytocannabinoids (Δ9-THC, Δ8-tetrahydrocannabinol, cannabinol, cannabidiol, cannabigerol, and cannabichromene) by CYP2J2. CYP2J2 mainly produces 1'/1"-OH metabolites of these phytocannabinoids. These phytocannabinoids are metabolized with greater catalytic efficiency compared to the metabolism of AEA by CYP2J2. We have also determined that the phytocannabinoids are potent inhibitors of CYP2J2-mediated AEA metabolism, with Δ9-THC being the strongest inhibitor. Most of the inhibition of CYP2J2 by the phytocannabinoids follow a noncompetitive inhibition model, and therefore dramatically reduce the formation of EET-EAs by CYP2J2. Taken together, these data demonstrate that phytocannabinoids are directly metabolized by CYP2J2 and inhibit human cardiac CYP2J2, leading to a reduction in the formation of cardioprotective EET-EAs.
Keywords: Endocannabinoids, cannabinoids, tetrahydrocannabinol, cannabidiol, cytochrome P450 2J2, anandamide
1. INTRODUCTION
Cannabis sativa has been used for centuries throughout human history for both its psychoactive effects and medicinal properties. Increasingly, legalization of cannabis for medical and recreational use is gaining worldwide support, in conjunction with trends of increased cannabinoid potency. Therefore, studying the effects of cannabinoids derived from cannabis on human health is of medical and scientific interest.
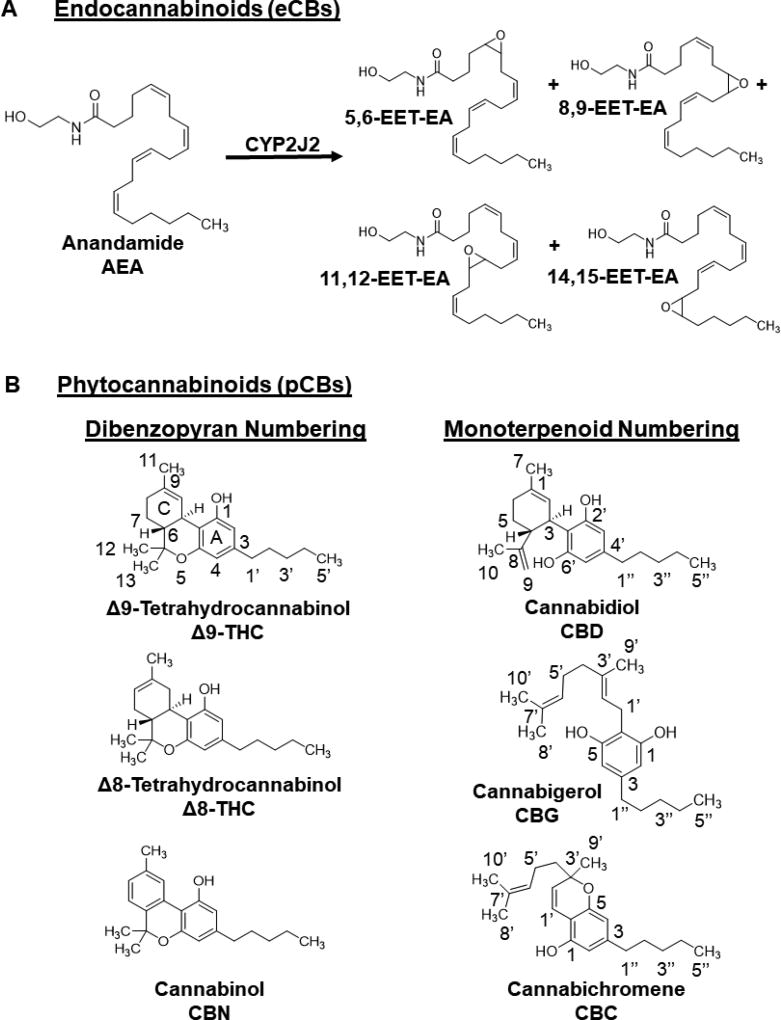
Cannabinoids are broadly classified into three categories depending on their source: (1) endocannabinoids (eCB) that are endogenously produced derivatives of polyunsaturated fatty acids (PUFAs) in animals; (2) phytocannabinoids (pCBs) that are derived from plants; and (3) synthetic cannabinoids. Psychoactive pCBs include Δ9-tetrahydrocannabinol (Δ9-THC), the primary psychoactive component of the plant, Δ8-tetrahydrocannabinol (Δ8-THC), and cannabinol (CBN). Some of the most abundant non-psychoactive pCBs in cannabis include cannabichromene (CBC), cannabidiol (CBD), and cannabigerol (CBG) (Figure 1).
Figure 1. Chemical structures.
(A) Endocannabinoids (eCBs): anandamide (AEA) and epoxyeicosatrienoyl ethanolamides (EET-EEAs). (B) Phytocannabinoids (pCBs). The numbering for each pCB is given, and Δ8-tetrahydrocannbionol (Δ8-THC) and cannabinol (CBN) follow analogous numbering as Δ9-tetrahydrocannabinol (Δ9-THC). These psychoactive pCBs follow the dibenzopyran numbering system. The non-psychoactive pCBs in cannabis include cannabidiol (CBD), cannabigerol (CBG), and cannabichromene (CBC). They follow a monoterpenoid numbering scheme. The resorcinol ring for each pCB is designated as the A-ring.
Phytocannabinoids have well-known cardiovascular implications that have been difficult to interpret due to variations regarding their effects in different species. For instance, the cardiovascular effects of THC in animals versus humans are contradictory [1, 2]. Δ9-THC induces tachycardia in humans, and only reproduces similar results in conscious monkeys; and prolonged exposure resulted in a reduction in elevated heart rate, as is seen in humans with developed tolerance [3]. In other animal models, Δ9-THC induces bradycardia [4–6]. Interpreting animal model data is further complicated using anesthesia. Experiments using anaesthetized [5] versus non-anaesthetized [4] rats did and did not exhibit tolerance to bradycardia symptoms, respectively, despite increased Δ9-THC administration. This lack of consensus in cross-species studies, changing variables in experimental design, and the psychoactivity of pCBs have obfuscated focus on discerning the exact cardiovascular implications of cannabis. Therefore, in order to understand the impact of pCBs on human cardiovascular health, there is a need to study the metabolism of pCBs by human cardiac enzymes. Of interest are the cytochromes P450 (CYPs), the primary enzymes that are involved in drug metabolism in the human body.
CYPs are known for their ability to metabolize diverse xenobiotics, synthesize steroids, and be involved in fatty acid metabolism [7]. CYPs generally require electrons donated by cytochrome P450 reductase (CPR) in order to oxidize their substrates. Previously, it was demonstrated that pCBs inhibit the metabolism of drugs by microsomal CYPs (1A1 [8, 9], 1A2 [8], 1B1 [10], 2A6 [11], 2B6 [10–12], 2C8 [12], 2C9 [13–15], 2C11 [16], 2C19 [17], 2D6 [18], 3A4 [12, 19], 3A5 [12, 19], and 3A11 [20]). Currently, there is absence of any mechanistic study on the metabolism of pCBs by CYP2J2, the most abundant CYP expressed in the cardiomyocytes of the heart [8, 9].
CYP2J2 is involved in the metabolism of both ω-3 and ω-6 eCBs leading to the formation of eCB epoxides that are vasodilatory, anti-platelet aggregatory, anti-inflammatory, and overall cardioprotective [21]. Anandamide (AEA) was the first eCB discovered. It is derived from the ω-6 fatty acid, arachidonic acid (AA) (Figure 1A) [22]. AEA was shown to be metabolized by several CYPs, including CYP2J2, forming different regioisomers of epoxyeicosatrienoyl ethanolamides (EET-EAs) (Figure 1A) [23, 24]. CYP2J2 has also been shown to metabolize several drugs, and many of which are known to be cardiotoxic [25–28].
Despite structural differences between eCBs and pCBs, both of these classes of cannabinoids interact with the endocannabinoid system (ECS) in the body. The ECS system consists of an ensemble of eCBs and eCB-like mediators, their corresponding receptors, and metabolic enzymes involved in ligand formation and degradation [29]. The ECS is involved in homeostatic functions dynamically regulating the functionality of the immune, reproductive, gastrointestinal, and central nervous systems, in addition to that of the brain, liver, and heart [30]. Physiological modulation by the ECS is largely dependent upon the nature and location of the diseased state [31].
The activity of the ECS is primarily elicited via molecular recognition of both pCBs and eCBs by cannabinoid receptors 1 and 2 (CBR1 and CBR2). ECS regulatory roles in the cardiovascular system are complex as the two receptors elicit varied responses [32, 33]. Activation of CBR1 translates to cardiovascular complications by decreasing blood flow and inducing vasoconstriction [34–38]; while CBR2 activity suppresses inflammatory responses of endothelial cells and monocytes [36, 39–41] and is also suggested to provide a protective role against cardiovascular diseases through induced vasodilation [40]. Thus, conflicting natures of CBR1 and CBR2 activation make the ECS a contributor to the generation, as well as amelioration, of cardiovascular disease.
Previously, we showed that CYP2J2 converts AEA into EET-EAs and similar epoxides from ω-3 fatty acid ethanolamides that are derived from DHA and EPA [42]. During inflammation, the levels of these epoxides are comparable to AEA [42]. Inasmuch as CYP2J2 generates cardioprotective epoxides from eCBs and also metabolizes several drugs, CYP2J2 is a prime enzyme for studying the potential effects of pCBs on eCB metabolism. It has been hypothesized that AEA metabolism by unknown CYPs is inhibited by pCBs [43]. Herein, we determine the direct metabolism of selected pCBs by CYP2J2 and evaluate their effects on AEA metabolism. We have determined that the 6 pCBs tested (Δ9-THC, Δ8-THC, CBC, CBD, CBG, and CBN) are all metabolized by CYP2J2 to form new hydroxyl products of the pCBs, with various di-oxygenated products as well. The catalytic efficiencies of the pCB turnovers are similar to or greater than that of AEA. Using CYP2J2-nanodiscs, we measured the kinetics of AEA metabolism by CYP2J2. Further, we have determined that the pCBs are potent inhibitors of AEA metabolism. We determined that CBG is a competitive inhibitor of AEA, whereas Δ9-THC, Δ8-THC, CBD, CBN, and CBC are noncompetitive inhibitors. Of these, Δ9-THC is the strongest inhibitor of AEA metabolism and reduces CYP2J2-mediated AEA metabolism to 20% of the uninhibited activity. Our study demonstrates that the pCBs, especially Δ9-THC, inhibits CYP2J2 and prevent the metabolism of endogenous substrates such as AEA.
2. EXPERIMENTAL PROCEDURES
Materials, CYP2J2 and CPR expression, nanodiscs and Soret titrations can be found in the Supplementary Materials.
2.1 Metabolism of pCBs by CYP2J2
Initial metabolism identification of pCB products were determined using a lipid-reconstituted system as previously described [44]. Briefly, CYP2J2 was reconstituted in 20% POPS using 0.6 µM CYP and 0.6 µM CPR. 60 µM of each pCB was pre-incubated with CYP2J2 and µM CPR for 10 min at 37°C in 0.5 mL of 0.1 M potassium phosphate buffer (pH 7.4). Reactions were initiated upon the addition of 1 mM NADPH and allowed to react for 60 min. Reactions were terminated with 0.1 mL glacial acetic acid and extracted thrice with 0.5 mL ethyl acetate. The samples were centrifuged at 3K RPM and 4°C for 5 min to separate the layers, and the organic layers were combined and dried down under a stream of N2 gas. Metabolites were resuspended in 0.1 mL acetonitrile and were identified using LC-MS/MS analysis as described below.
2.2 Kinetics of pCB metabolism
Kinetics of pCB metabolism were determined using a lipid-reconstituted system as previously described [44]. Metabolites were quantified using an HPLC method (see below). 0.2 µM of CYP2J2 and 0.6 µM of CPR were incubated with varying concentrations of pCBs individually (5–60 µM) for 10 min at 37°C in 0.5 mL of 0.1 M potassium phosphate buffer (pH 7.4). Reactions were initiated with 0.5 mM NADPH and allowed to react for 30 min. Reactions were terminated and extracted using ethyl acetate as stated above. Metabolism of the pCBs was determined to be linear for up to 45 min for each pCB.
2.3 Kinetics of AEA metabolism
AEA metabolism by CYP2J2 was determined in Nanodiscs using 0.2 µM of CYP2J2-NDs and 0.6 µM CPR. AEA was pre-incubated with CYP2J2-ND/CPR for 10 min at 37°C in 0.5 mL of 0.1 M potassium phosphate buffer (pH 7.4). For inhibition experiments, AEA was pre-incubated with the CYP2J2-ND/CPR system along with 30 µM of the tested pCB. Reactions were initiated with 0.5 mM NADPH and allowed to react for 30 min. EET-EAs were extracted using ethyl acetate as stated above, but were resuspended in 180-proof ethanol for LC-MS/MS quantification.
2.4 HPLC quantification of pCB metabolism
Metabolism products were analyzed via high-performance liquid chromatography (HPLC) consisting of an Alliance 2695 analytical separation module (Waters, Milford, MA) and a Waters 996 photodiode diode array detector (Waters). Metabolites were separated in reverse phase using a Phenomenex Prodigy® 5µm ODS-2, 150 × 4.60 mm column (Phenomenex, PN 00F-3300-E0, Torrance, CA). Mobile Phase A consisted of H2O + 0.1% formic acid and Mobile Phase B consisted of acetonitrile + 0.1% formic acid. Metabolites were separated using a linear gradient of 25% Mobile Phase B to 100% Mobile Phase B over 20 min, and then held at 100% Mobile Phase B for an additional 10 min. Δ9-THC, Δ8-THC, and CBD hydroxyl metabolites were monitored at λ = 281 nm; CBG hydroxyl products were monitored at λ = 278 nm; and CBN and CBC at λ = 283 nm. Standard curves for quantification were made using a Δ9-THC-11-OH standard for Δ9-THC-OH, Δ8-THC-OH, and CBD-OH; CBG standard for CBG-OH; CBN standard for CBN-OH; and CBC standard for CBC-OH. There was no significant variation in the absorbance intensities among Δ9-THC, Δ8-THC, and CBD. Elution times are reported in the Section 3.1.1 (Figure 2).
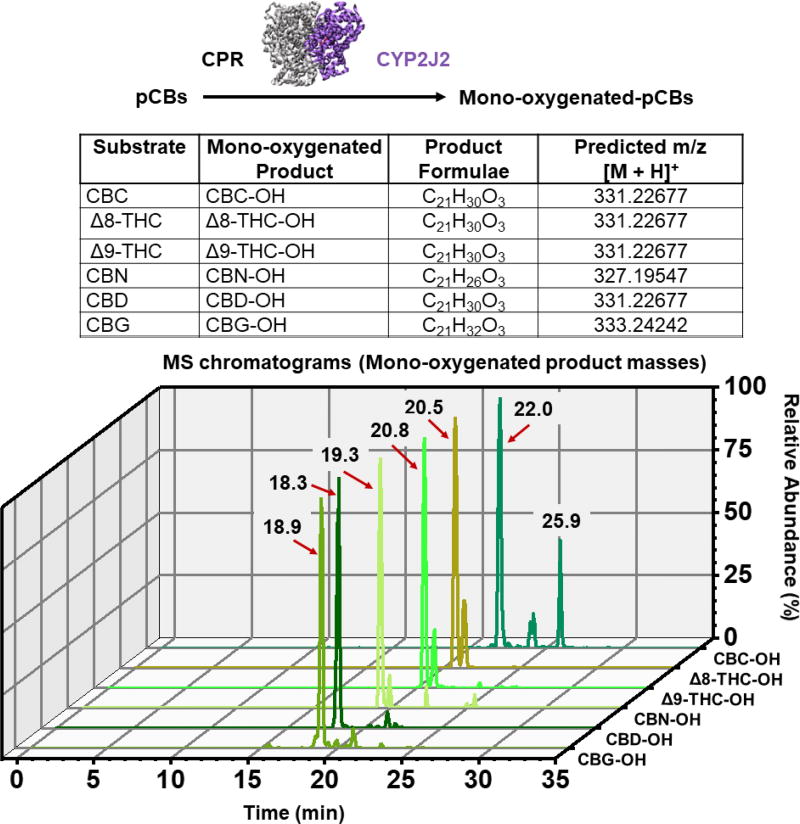
Figure 2. Targeted LC-MS analysis of CYP2J2-mediated pCB metabolism.
The indicated pCBs were incubated with CYP2J2/CPR and the products were extracted with ethyl acetate and analyzed via LC-MS/MS as described in the Section 2.5. Mass peaks corresponding to the predicted mono-oxygenated m/z values ± 5 ppm are shown. Peaks are labelled according to their elution time.
2.5 High-resolution LC-MS and LC-MS/MS analysis of metabolites
Masses of products were determined using the Q-Exactive MS system (Thermo. Bremen, Germany) in the Metabolomics Laboratory of Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign as previously described [44]. Metabolites were separated using the above HPLC method and the Phenomenex Prodigy® 5µm ODS-2, 150 × 4.60 mm column. Negative and positive electrospray ionization was employed. Masses were considered significant if ± 5 ppm of the calculated exact mass.
2.6 LC-MS/MS quantitation of EET-EAs
Samples were analyzed with the 5500 QTRAP LC/MS/MS system (Sciex, Framingham, MA) in Metabolomics Lab of Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign. Software Analyst 1.6.2 was used for data acquisition and analysis. The 1200 series HPLC system (Agilent Technologies, Santa Clara, CA) includes a degasser, an autosampler, and a binary pump. The LC separation was performed on an Agilent Eclipse XDB-C18 (4.6 × 150mm, 5µm) with mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetontrile). The flow rate was 0.4 mL/min. The linear gradient was as follows: 0–2min, 90%A; 8min, 55%A; 13–25min, 40%A; 30min, 30%A; 35min, 25%A; 36–43min, 90%A. The autosampler was set at 10°C. The injection volume was 10 µL. Mass spectra were acquired under positive electrospray ionization (ESI) with the ion spray voltage of +5500 V. The source temperature was 450 °C. The curtain gas, ion source gas 1, and ion source gas 2 were 32, 65, and 50, respectively. Multiple reaction monitoring (MRM) was used for quantitation: m/z 364.2 → m/z 62.0. Internal standard 14,15-EET-EA-d8 was monitored at m/z 372.1 → m/z 63.1.
2.8 Competitive and noncompetitive inhibition equations used in studies
All inhibition studies are best described by a competitive inhibition model (Equation 1) or a noncompetitive inhibition model (Equation 2)
| (Equation 1) |
| (Equation 2) |
where Bmax and K are the Michaelis-Menten kinetic parameters of the substrate [S] without inhibitor, [I] is the concentration of inhibitor, and Ki is the inhibition constant. For kinetic experiments, B and K represent the velocity and Km, respectively; for Soret binding experiments, B and K represent ΔA and KS, respectively.
3. RESULTS
3.1.1 CYP2J2 metabolizes pCBs primarily to 1’/1”-OH metabolites
We first tested the direct endpoint metabolism of the pCBs by CYP2J2-CPR. We determined that the pCBs—Δ9-THC, Δ8-THC, CBD, CBG, CBN, and CBC—are all substrates of CYP2J2 (Figures 2–4). LC-MS/MS analysis indicates that the following classes of oxidation products were produced for each pCB: mono-oxygenation, carboxylation, and di-oxygenation (masses of the products are within 5 ppm of the predicted masses). Chromatograms for the mono-oxygenated products are shown in Figure 2. MS/MS spectra of the major mono-oxygenated products are shown in Figures 3 and 4. Excel files containing all the raw chromatogram and spectra data for the standards and metabolism experiments can be found in the Supplementary Material. A detailed analysis of the MS/MS data is provided in the Supplementary Material.
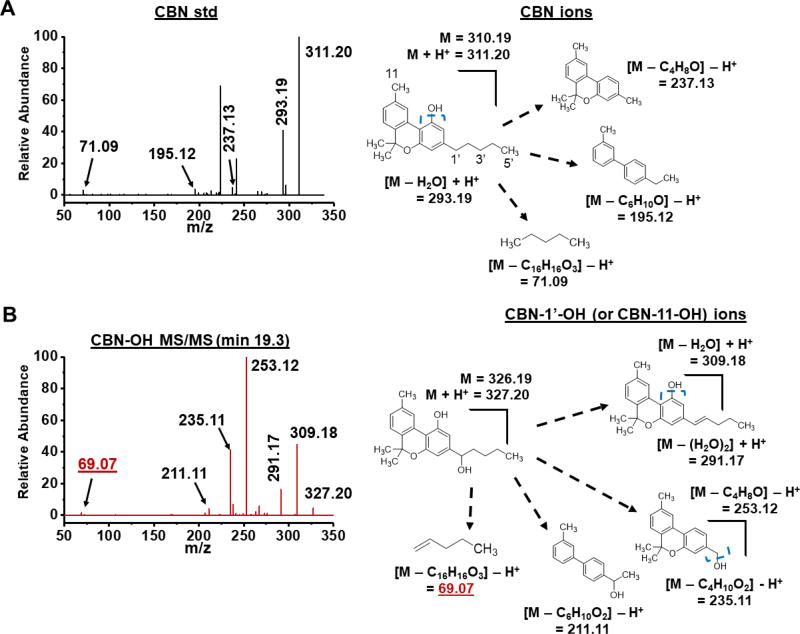
Figure 4. MS/MS spectra of the CBN standard (std) and the major mono-oxygenated CYP2J2 product (CBN-OH).
(A) Spectra of the CBC standard and scheme of major fragmentations. (B) The major CYP2J2 product of CBC and major fragmentations. Elution times of products are given. Blue, dashed brackets indicate sites of fragmentation. Masses that indicate 1’-OH are labelled in red and underlined. Masses were considered significant if within 5 ppm of the predicted masses.
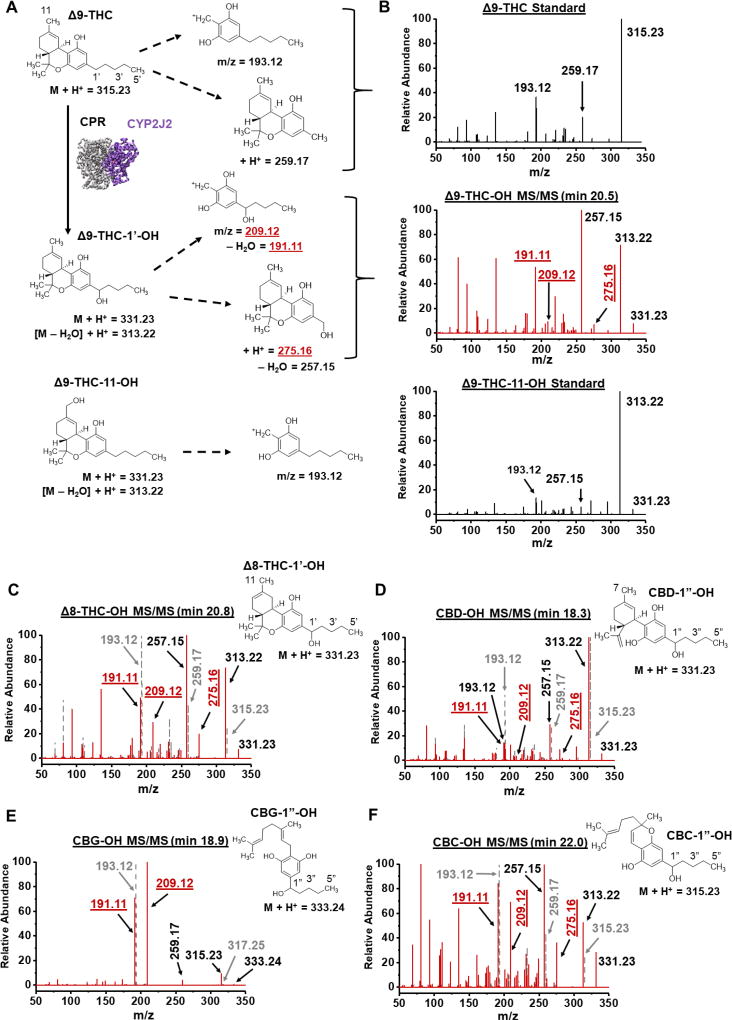
Figure 3. MS/MS spectra of the major mono-oxygenated products of Δ9-THC, Δ8-THC, CBD, CBG and CBC.
(A) Schematic of Δ9-THC fragmentation pathways for the Δ9-THC and Δ9-THC-11-OH standards and the CYP2J2-mediated Δ9-THC-OH product. Analogous ions are identified for the other pCBs. (B) Spectra of the Δ9-THC standard, Δ9-THC-11-OH standard, and the major CYP2J2 product of Δ9-THC. (C–F) Spectra of the (C) Δ8-THC-OH product, (D) CBD-OH product, (E) CBG-OH product, and (F) CBC-OH product. Corresponding standard spectra for (C–F) are shown as grey dashes and grey font. Elution times of products are given. Masses that indicate 1’/1”-OH as the product are labelled in red and underlined. Masses were considered significant if within 5 ppm of the predicted masses.
A resorcinolic ion (193.12 m/z) is produced upon the fragmentation of the Δ9-THC and Δ9-THC-11-OH standards (Figure 3A–B) [45]. The major Δ9-THC-OH product of the CYP2J2 metabolism shows a mass corresponding to mono-oxygenation of the resorcinolic ion followed by the elimination of a water molecule (209.12 m/z → 191.11 m/z) (Figure 3A–B). The Δ9-THC standard additionally fragments between Carbons 1’ and 2’ to produce an ion of 259.17 m/z. The CYP2J2 product shows a mass indicating mono-oxygenation (275.16 m/z) of this ion, which is not present in the Δ9-THC-11-OH standard, followed by the elimination of a water molecule (257.16 m/z). This identifies the CYP2J2 product as Δ9-THC-1’-OH. Furthermore, the 257.16 m/z peak is the largest peak in the CYP2J2 product spectrum, but not the Δ9-THC-11-OH standard. Fragmentation between Carbons 1’ and 2’ is likely facilitated by the elimination of the 1’-OH as a water molecule, thereby making it the most abundant ion.
The Δ8-THC, CBD, CBG, and CBC (Figures 3C–F) standards show similar fragmentation patterns as the Δ9-THC standard (Figure 3B). Likewise, the major mono-oxygenated products for these compounds show similar fragmentation patterns as the CYP2J2 product for Δ9-THC, as mentioned above (Figures 3C–F). Therefore, the metabolites for these compounds can be identified as Δ8-THC-1’-OH, CBD-1”-OH, CBG-1”-OH, and CBC-1”-OH. However, there are additional ions in the CBD sample that are reminiscent of ions in the Δ9-THC-11-OH standard, notably the resorcinolic 193.12 m/z ion (Figures 3A and 3D). Therefore, it is likely that CYP2J2 additionally produces CBD-7-OH, the analogous product compared to Δ9-THC-11-OH. The CBD-7-OH and CBD-1”-OH co-elute and could not be separated further.
CBN fragments differently when compared to the other pCBs due to the extended conjugation of the dibenzene moiety (Figure 4A). As a result, no ion corresponding to the separation of the resorcinol and terpenoid halves is observed in either the CBN standard or the product (Figure 4). There are two important ions of the product that are mono-oxygenated fragments of the parent compound (237.13 m/z → 253.12 m/z and 195.12 m/z → 211.11 m/z) (Figure 4B). These two fragments share the dibenzene moiety and Carbons 11 and 1’ (Figure 4). There are no other fragments of either the standard or the product that could distinguish between Carbons 11 and 1’. Therefore, the product is CBN-11-OH, CBN-1’-OH, or a combination of the two. The pentyl tail of CBN fragments to produce an ion of 71.09 m/z in the CBN standard, which is 69.07 m/z in the product (Figure 4B). This could be the result of a hydroxyl cleavage at the 1’ position and subsequent olefination of this fragment, which supports CBN-1’-OH as the identity of the product.
3.1.2 Formation of other pCB metabolites by CYP2J2
All pCBs were metabolized by CYP2J2 to produce ions suggestive of pCB carboxylation by CYP2J2. Δ9-THC-11-COOH was identified and compared to a standard, which is verified by a fragment resembling the carboxylated species without the pentyl side chain (Figure S1, Δ9-THC, Ion 9). Other carboxylated metabolites for each of the pCBs follow similar fragmentation, where an ion lacking an aliphatic carbon chain can be detected. Some pCBs (CBC, CBG, CBN) possess fragmentation suggestive of multiple carboxylation sites, due to the presence of representative ions being restricted to either positive or negative ion modes.
Each pCB was also metabolized by CYP2J2 into di-hydroxylated products in addition to the 1’/1” hydroxylation. Although MS analysis suggests their presence, the identities of these di-hydroxylated metabolites could not be determined from their MS/MS spectra due to lack of standards.
Overall, the relative abundances of these ions are much lower compared to the major hydroxylated products. Findings related to the formation of other metabolites are summarized in Figure 5 but are presented in greater detail in the Supplementary Material.
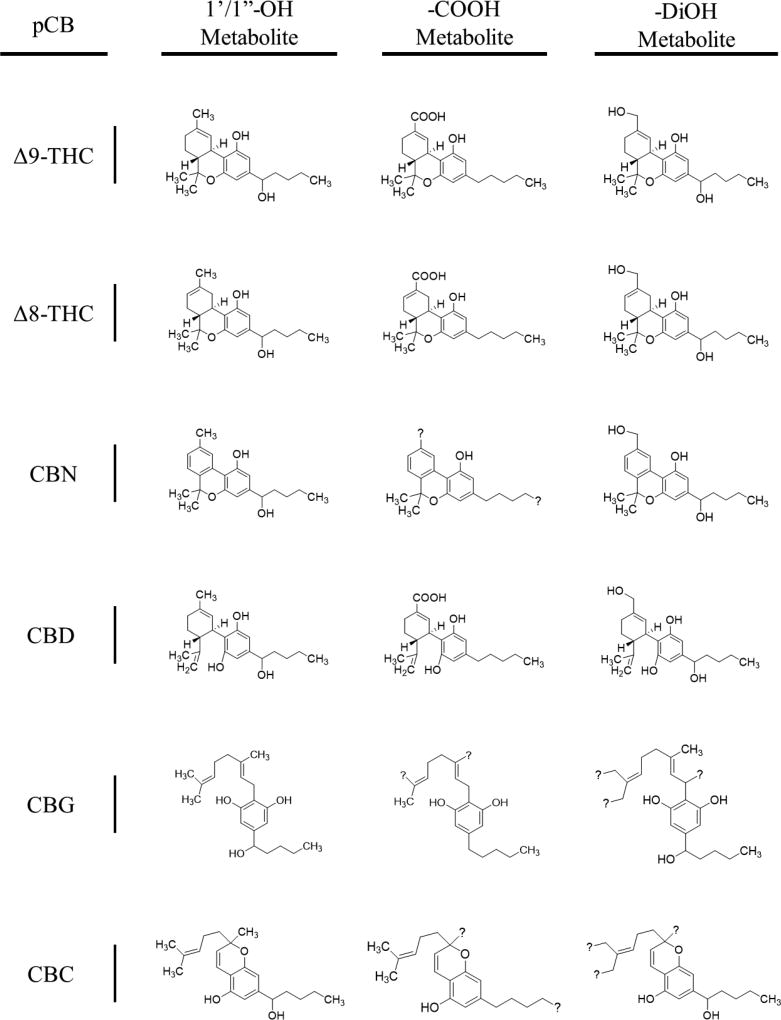
Figure 5. Major products of CYP2J2-pCB metabolism.
Question marks (?) are positioned to suggest multiple species derived from the fragmentation pathways for each metabolite (reference Supplementary Materials). Metabolites are as follows (left to right, row by row): Δ9-THC-1’-OH, Δ9-THC-11-COOH, Δ9-THC-DiOH; Δ8-THC-1’-OH, Δ8-THC-11-COOH, Δ8-THC-DiOH; CBN-1’-OH, CBN-5”/11-COOH, CBN-DiOH; CBD-1’-OH, CBD-7-COOH, CBD-DiOH; CBG-1’-OH, CBG-8’/9’/10’-COOH, CBG-DiOH; CBC-1’-OH, CBC-5”/9’-COOH, CBC-DiOH. Structures of -DiOH products are representative as the position of the second OH could not be determined.
3.2 Kinetics of pCB metabolism by CYP2J2
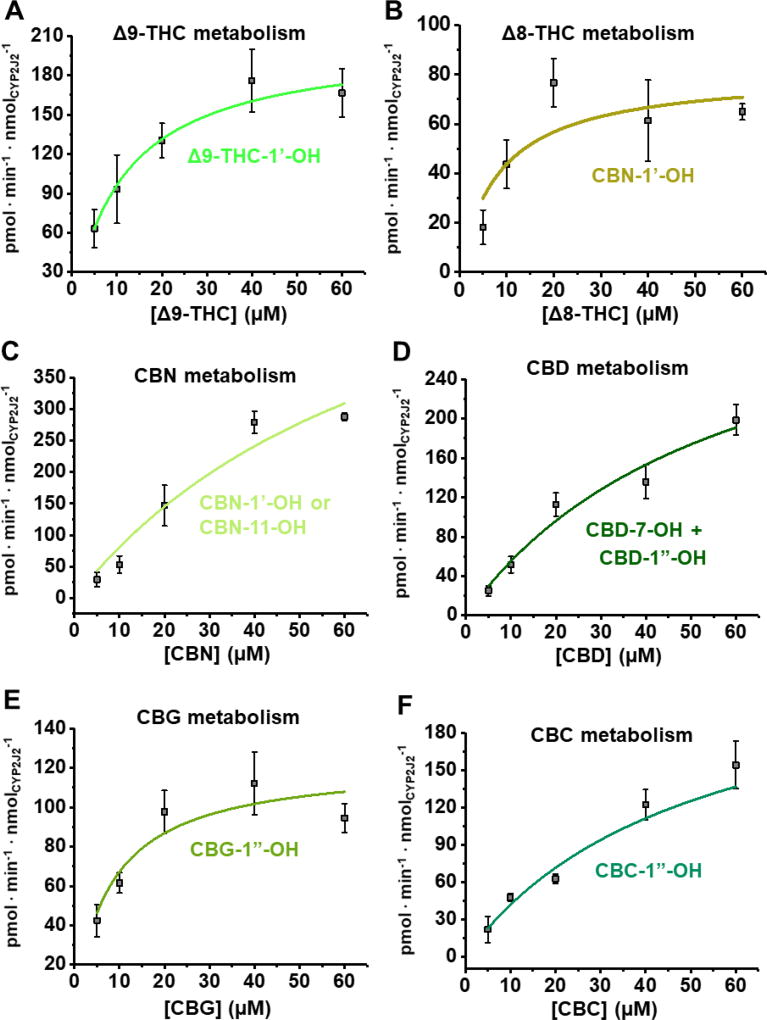
We next determined the kinetics of metabolism for the pCBs. To do this, we developed an ultraviolet/visible wavelength (UV-Vis) HPLC quantification method based on the absorbance of the resorcinol moiety (λ = 278–283 nm) as described in the Materials and Methods section. The initial mono-oxygenated products were analyzed to determine the steady-state kinetics (Figure 6, Table 1). These correspond to Δ9-THC-1’-OH; Δ8-THC-1’-OH; CBN-1’-OH or CBN-11-OH; CBD-1”-OH and CBD-7-OH; CBG-1”-OH; and CBC-1”-OH. The other metabolites were below the detection limit.
Figure 6. Kinetics of pCB metabolism.
Mono-oxygenated products identified by LC-MS/MS analysis for the metabolism of (A) Δ9-THC, (B) Δ8-THC, (C) CBN, (D), CBD, (E) CBG, and (F) CBC were quantified using UV-HPLC as stated in the Materials and Methods section. Data fit to the Michaelis-Menten equation. Error represents the SEM of 3 experiments.
Table 1.
Kinetic parameters of experiments.
| Metabolism | AEA Metabolism Inhibition | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Metabolite(s) | Vmaxa | Km (µM) | kcat · Km−1b | Ki (µM) | Type | |
|
| ||||||
| AEA | EET-EAs | 135 ± 14 | 30.9 ± 7.3 | 4.47 | --- | --- |
|
| ||||||
| Δ9-THC | -1’-OH | 211 ± 28 | 11.9 ± 4.8 | 18.7 | 9.86 ± 0.45 | Noncompetitive |
| Δ8-THC | -1’-OH | 80.9 ± 14.3 | 8.52 ± 5.30 | 9.50 | 20.2 ± 1.9 | Noncompetitive |
| CBN | -1’-OH or -11-OH | 710 ± 248 | 77.8 ± 42.2 | 9.13 | 19.4 ± 2.0 | Noncompetitive |
| CBD | -7-OH + -1”-OH | 372 ± 100 | 57.2 ± 26.5 | 6.50 | 16.9 ± 1.1 | Noncompetitive |
| CBG | -1”-OH | 123 ± 14 | 8.27 ± 3.24 | 14.9 | 10.8 ± 1.4 | Competitive |
| CBC | -1”-OH | 371 ± 134 | 84.2 ± 46.2 | 4.41 | 21.1 ± 1.4 | Noncompetitive |
(pmol · min−1 · nmolCYP2J2−1)
(nM−1 · min−1)
The pCBs were effectively metabolized with differing kinetic characteristics. The Vmax values ranged from 80.9 to 710 pmol · min−1 · nmolCYP2J2−1 and the Km values ranged from 8.27 to 84.2 µM. Δ9-THC displayed the greatest catalytic efficiency (18.9 min−1 · nM−1). This value is twice that of the other dibenzopyran cannabinoids, Δ8-THC and CBN, which had catalytic efficiencies of 9.50 and 9.13 min−1 · nM−1, respectively. CBC, CBD, and CBG varied in their efficiencies from 4.41 to 14.9 min−1 · nM−1. Compared to the kinetics we had previously determined for AA, the canonical endogenous substrate of CYP2J2, these pCBs are metabolized more efficiently [46].
3.3 Binding of pCBs to CYP2J2-Nanodiscs (CYP2J2-NDs)
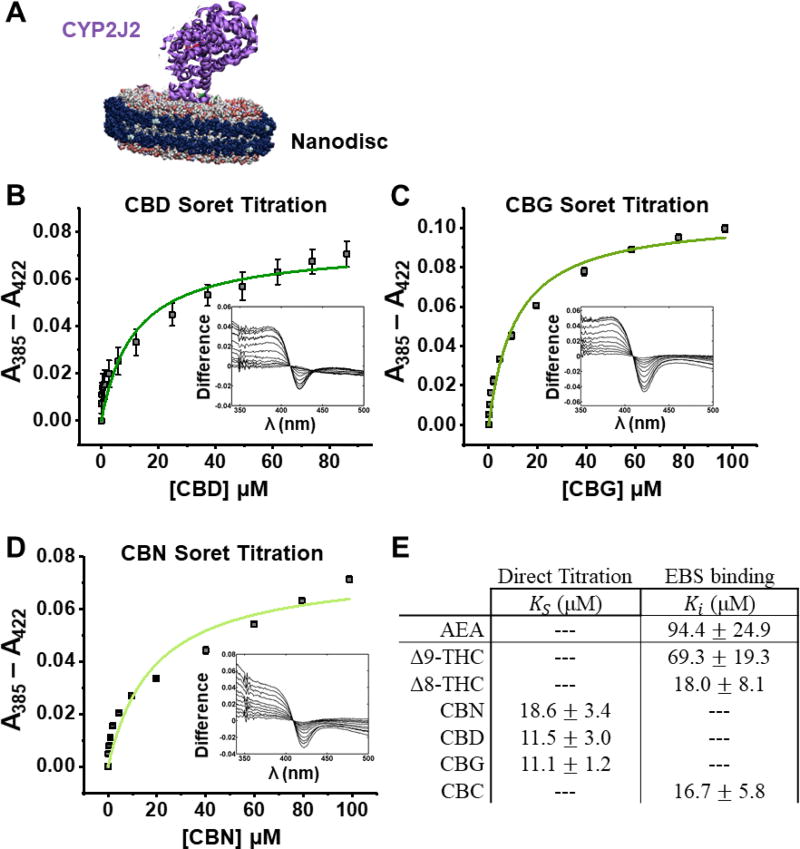
We next determined the binding of the pCBs and AEA to CYP2J2 using UV-Vis Soret titration binding. To produce accurate binding spectra, we used CYP2J2-NDs, which solubilize CYP2J2 in a membrane mimic (Figure 7A). Substrates binding to CYPs typically displace a water molecule from the 6th coordination position of the iron-heme, producing a high-spin iron. This results in a characteristic shift in the Soret absorbance from ~417 nm to ~390 nm. The pCBs were titrated into CYP2J2-NDs and the spin-state equilibrium constants (KS) were determined, which is used as an approximation of the KD for CYPs. CBD, CBG, and CBN produced a measurable Soret shift upon binding (Figure 7B–E). At saturating concentrations, CBD, CBG, and CBN produced 32%, 48%, and 38% high-spin shift, respectively. Among these, CBD and CBG showed the tightest binding (KS of 11.5 ± 3.0 µM and 11.1 ± 1.2 µM for CBD and CBG, respectively). The binding of Δ9-THC, Δ8-THC, CBC, and AEA did not produce a significant change in the high-spin content. Therefore, to measure their relative binding affinity, we measured their inhibition of ebastine binding to CYP2J2, a technique we had previously used to determine the binding constant of AA [46]. All the pCBs displayed binding affinities that are greater than AEA, with most between 10 and 20 µM, and Δ9-THC being the weakest (Figure 7E).
Figure 7. Soret binding titration.
(A) CYP2J2 in Nanodiscs. (B) CBD, (C) CBG, and (D) CBN produced a measurable Soret shift with 32%, 48%, and 38% high-spin shift, respectively. (E) Binding parameters as determined directly from the Soret titration or through inhibition of ebastine (EBS) binding. Data displayed one-site binding and data shown are the mean ± SEM of 3 titrations.
3.4 Inhibition of AEA metabolism by pCBs
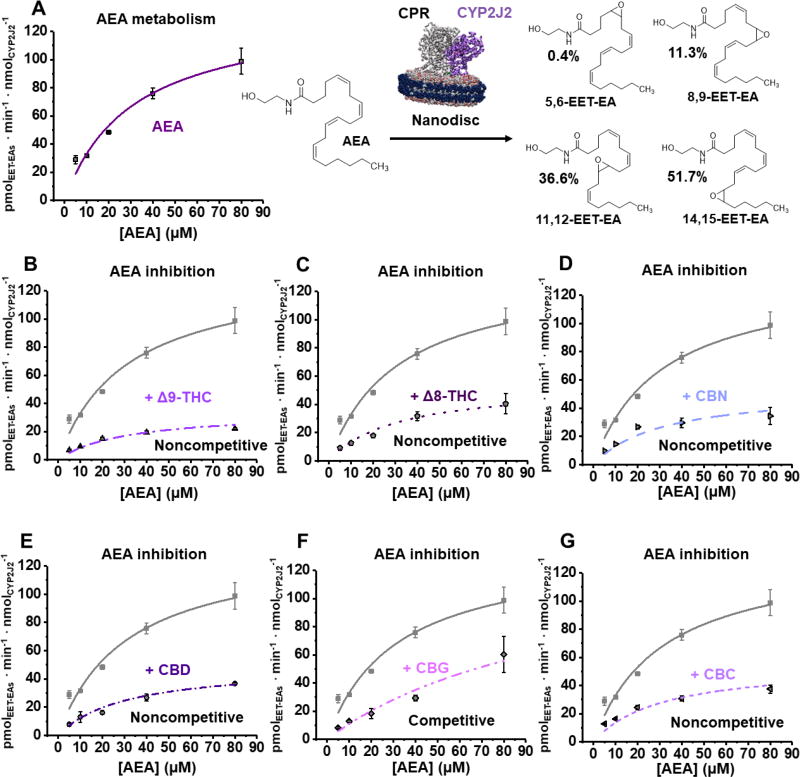
Having established that the pCBs are substrates of CYP2J2, we next determined the inhibition of the CYP2J2-mediated AEA metabolism by the pCBs using a CYP2J2-ND/CPR system (Figure 8A). AEA is metabolized by CYP2J2 with a Vmax of 135 ± 14.0 pmolEET-EAs · min−1 · nmolCYP2J2−1 and a Km of 30.9 ± 7.3 µM (Figure 8A, Table 1), which is a greater catalytic efficiency compared to AA,[46] but less than that of most of the pCBs. The predominant metabolite is 14,15-EET-EA (51.7% of the total) with 5,6-EET-EA being the least (0.4%) (Figure 8A). The metabolism of AEA was repeated in the presence of 30 µM of each pCB in separate experiments (Figure 8B–G, Table 1). CBG inhibited AEA metabolism competitively with a Ki of 10.8 ± 1.4 µM. The rest of the pCBs noncompetitively inhibited AEA metabolism. The apparent Vmax of the AEA metabolism was reduced to 20–45% of the uninhibited enzyme in the presence of Δ9-THC, Δ8-THC, CBN, CBD, and CBC (Figure 8B–G). Of these, Δ9-THC inhibited most strongly with a Ki of 9.86 ± 0.45 µM. The next strongest was CBD with a Ki that is almost double that of Δ9-THC (16.9 ± 1.1 µM for CBD). CBN, CBC, and Δ8-THC inhibited with Ki values around 20 µM. For all these pCBs, the Ki values are in close approximation to their determined binding affinities, except for Δ9-THC, which is comparatively stronger (Table 1).
Figure 8. Anandamide (AEA) metabolism inhibition by pCBs.
(A) Kinetics of AEA metabolism. A schematic of CYP2J2-Nanodiscs and CPR metabolizing AEA is given. The total amount of EET-EAs are plotted. The percent of each regioisomer to the total amount of EET-EAs is given to the right. (B–G) Inhibition of AEA metabolism by the indicated pCB. AEA data from Panel A is shown as grey for comparison and the model of inhibition is given. 30 µM of each pCB was used to determine inhibition. Error represents the SEM of 3 experiments.
4. DISCUSSION
Phytocannabinoids are known for their psychotropic effects. However, their role in overall human physiology remains to be elucidated. For instance, pCBs have strong effect on the human cardiovascular physiology, including inflammation; additionally, the principal component of cannabis, Δ9-THC, induces tachycardia in humans. Previous studies have produced variable results regarding their effects on cardiovascular health, which is partly due to the dearth of information regarding most of the pCBs, their metabolites, and their modulation of the endocannabinoid system.
Herein, we have performed direct in vitro assays to determine the effects of pCBs on a human cardioprotective enzyme, CYP2J2. Given that CYP2J2 is known to metabolize drugs and convert eCBs, such as AEA, to cardioprotective epoxides makes this enzyme a prime target for investigating the impact of pCBs on the metabolism of AEA. We have determined that the 6 most common pCBs, Δ9-THC, Δ8-THC, CBN, CBD, CBG, and CBC, are mostly converted to 1’/1”-OH metabolites by CYP2J2. We further determine that these pCBs demonstrate greater catalytic efficiencies compared to typical endogenous CYP2J2 substrates. Finally, we determined that these pCBs potently inhibit AEA metabolism. While CBG inhibits competitively, the other 5 pCBs inhibit via a noncompetitive model, thereby shutting down AEA metabolism through CYP2J2. Of these pCBs, Δ9-THC is the strongest inhibitor of AEA metabolism.
Δ8/Δ 9-THC-11-OH are the best studied metabolites of the THCs as these metabolites are psychotropic and are the most abundant metabolite formed by CYPs in liver microsomes.[47–49] Other CYP-mediated oxidations of THCs the liver involve C-ring oxidations and acyl hydroxylation, namely 3’-OH (CYP1A1) and 4’-OH (CYP2C11) [50–52]. Other organs, such as the brain and lung, produce primarily hydroxylated metabolites on the acyl chain, notably -4’-OH and -5’-OH [53, 54]. However, despite being found as a metabolite of pCBs, the 1’/1” hydroxylation was found to be a minor metabolite of other CYPs that have been tested, such as CYP1A1/2 [48]. Likewise, CBD is hydroxylated on both the C ring and pentyl side chain, and CBD-1”-OH has been primarily shown to be produced by CYP1A1/2, albeit it is still not the major metabolite of these CYP enzymes [8]. The primarily 1’/1” hydroxylation is therefore unique to CYP2J2 and may be used as a probe regarding its activity. In smaller quantities, we also propose that CYP2J2 generates Δ8/Δ9-THC-11-COOH, CBN-11/5”-COOH, CBD-7-COOH, CBG-8’/9’/10’-COOH, and CBC-9’/5”-COOH. Throughout the literature, THC and CBD are the best studied cannabinoids with respect to CYP metabolism. Herein, we measure the CYP2J2-mediated metabolism of CBC, CBG, and CBN that are not extensively studied but nonetheless very important constituents of cannabis.
All the pCBs inhibited CYP2J2-mediated AEA metabolism. Interestingly, two modes of AEA inhibition by the pCBs were observed. CBG inhibited AEA metabolism competitively. Compared to the other pCBs, CBG has a structure that is the most linear and most resembles the structure of AEA. We have previously determined that the competitive nature of PUFAs arises from their flexibility, which allows them to occupy most of the active site volume [46]. Likewise, CBG and AEA may be competitive for similar reasons.
The other pCBs inhibit AEA metabolism in a noncompetitive manner. Classically, noncompetitive inhibition arises from the binding of the inhibitor to an allosteric site, preventing the metabolism of a substrate when both are bound. CYP2C9’s metabolism of diclofenac was shown to be noncompetitively inhibited by 6-hydroxyflavone in an atypical manner. 6-hydroxyflavone was shown to bind into a site that prevents full access of diclofenac to the heme [55]. Since these pCBs are also substrates of CYP2J2, they must bind near the heme. Therefore, in addition to the classical mode of noncompetitive inhibition, the pCBs may be binding the active site along with AEA and preventing AEA from being metabolized. We had previously demonstrated that 7-deoxydoxorubicin aglycone concurrently binds the active site with AA and alters AA metabolism by changing the binding conformation of AA [44]. This is mediated in part to the polycyclic and planar structure of 7-deoxydoxorubicin aglycone. The rest of the pCBs other than CBG likewise have polycyclic structures, giving them less flexibility.
Another intriguing characteristic of the AEA inhibition by these pCBs is the differences in the inhibitory strength concerning the dibenzopyran varieties, Δ9-THC, Δ8-THC, and CBN. Among these, Δ9-THC has a Ki value for the inhibition of AEA that is twofold tighter than Δ8-THC or CBN. Coincidentally, the catalytic efficiency of the Δ9-THC metabolism was determined to be twice that of the metabolism of Δ8-THC or CBN. These data demonstrate that the positioning and frequency of double bonds in the C-ring of the dibenzopyran pCBs governs the metabolism of these pCBs and the inhibition of AEA metabolism, with the Δ9 position being crucial.
All the pCBs tested herein potently inhibit AEA metabolism by CYP2J2. These data have intriguing physiological implications, particularly in relation to trafficking and molecular recognition of cannabinoid-like ligands within the ECS. The pCBs have been shown to stall the catabolism of AEA by inhibiting fatty-acid-binding proteins that aid in the delivery of AEA to fatty acid amide hydrolase (FAAH) [56], and FAAH has been shown to be weakly inhibited by CBD [57]. As a result, the extended lifetime of AEA [57] results in greater targeting of CBR1, which is further amplified by the activation of CBR1 by select pCBs themselves. Through noncompetitive inhibition, our data shows that in addition to pCB signaling through CBR1 receptors, they also prevent CYP2J2-mediated AEA metabolism. As EET-EAs are CBR2-preferring agonists [30, 42, 58], pCBs prevent the formation of CBR2-preferring ligands in addition to increasing actions at CBR1. Interestingly, a study performed on Rhesus monkeys shows that Δ8-THC-1’-OH is the least effective at inducing behavioral changes associated with cannabis use [59]. As CBR1 is responsible for the psychotropic effects of cannabinoids, this presumes that Δ8-THC-1’-OH is the weakest agonist of CBR1. Such crosstalk is not only mediated by CYP metabolism, but also by CBR signaling. It has been shown previously that selective blockade of CBR1 receptor and activation of CBR2 is ideal for the treatment and management of cardiovascular disease [60]. Therefore, the inhibited formation of CBR2-preferring EET-EAs and stronger CBR1 activation by select pCBs may explain the increase risk of myocardial infarction after the first hour of cannabis use. CYP2J2 then clears the pCBs by converting them to weaker CBR1 agonists and resumes normal EET-EA metabolism after the first hour and the cardiotoxic risk is lowered. Taken together, this study indicates that pCBs will elicit complex cardiovascular physiology that will be partially mediated through inhibition of human cardiac CYP2J2.
Supplementary Material
Acknowledgments
We would like to thank Dr. Lucas Li of the Roy J. Carver Metabolomics Center (UIUC) for performing the LC-MS/MS analyses.
Funding Sources. Partially supported by American Heart Association Scientist Development Grant 15SDG25760064 (A.D.), National Institutes of Health (NIH) Grant R01 GM1155884 (A.D.) and NIH Grant R03 DA042365.
ABBREVIATIONS
- Δ8-THC
Δ8-tetrahydracannabinol
- Δ9-THC
Δ9-tetrahydracannabinol
- AEA
anandamide
- AA
arachidonic acid
- CBC
cannabichromene
- CBD
cannabidiol
- CBG
cannabigerol
- CBR
cannabinoid receptor
- CBN
cannabinol
- -COOH
carboxy
- CYP
cytochrome P450
- CPR
cytochrome P450 reductase
- eCB
endocannabinoid
- ECS
endocannabinoid system
- EET
epoxyeicosatrienoic acid
- EET-EA
epoxyeicosatrienoyl ethanolamide
- -OH
hydroxy
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- ND
Nanodisc
- pCB
phytocannabinoid
- PUFA
polyunsaturated fatty acid
Footnotes
AUTHOR CONTRIBUTIONS
WRA designed and performed the experiments, made figures, and analyzed the data and wrote the manuscript. ATW performed experiments, made figures, analyzed data, and wrote the manuscript. AD designed the experiments and wrote the manuscript. All authors analyzed the results and approved the final version of the manuscript.
The authors claim no conflict of interest.
References
- 1.Mendizabal VE, Adler-Graschinsky E. Cannabinoids as therapeutic agents in cardiovascular disease: a tale of passions and illusions. Brit J Pharmacol. 2007;151:427–440. doi: 10.1038/sj.bjp.0707261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Randall MD, Kendall DA, O'Sullivan S. The complexities of the cardiovascular actions of cannabinoids. Br J Pharmacol. 2004;142:20–26. doi: 10.1038/sj.bjp.0705725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fredericks AB, Benowitz NL, Savanapridi CY. The cardiovascular and autonomic effects of repeated administration of delta-9-tetrahydrocannabinol to rhesus monkeys. J Pharmacol Exp Ther. 1981;216:247–253. [PubMed] [Google Scholar]
- 4.Dewey WL. Cannabinoid pharmacology. Pharmacol Rev. 1986;38:151–178. [PubMed] [Google Scholar]
- 5.Adams MD, Chait LD, Earnhardt JT. Tolerance to the cardiovascular effects of delta9-tetrahydrocannabinol in the rat. Br J Pharmacol. 1976;56:43–48. doi: 10.1111/j.1476-5381.1976.tb06957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones RT. Cardiovascular system effects of marijuana. J Clin Pharmacol. 2002;42:58s–63s. doi: 10.1002/j.1552-4604.2002.tb06004.x. [DOI] [PubMed] [Google Scholar]
- 7.Guengerich FP, Wu ZL, Bartleson CJ. Function of human cytochrome P450s: characterization of the orphans. Biochem Biophys Res Commun. 2005;338:465–469. doi: 10.1016/j.bbrc.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 8.Jiang R, Yamaori S, Takeda S, Yamamoto I, Watanabe K. Identification of cytochrome P450 enzymes responsible for metabolism of cannabidiol by human liver microsomes. Life Sci. 2011;89:165–170. doi: 10.1016/j.lfs.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 9.Yamaori S, Okushima Y, Masuda K, Kushihara M, Katsu T, Narimatsu S, Yamamoto I, Watanabe K. Structural requirements for potent direct inhibition of human cytochrome P450 1A1 by cannabidiol: role of pentylresorcinol moiety. Biol Pharm Bull. 2013;36:1197–1203. doi: 10.1248/bpb.b13-00183. [DOI] [PubMed] [Google Scholar]
- 10.Yamaori S, Kushihara M, Yamamoto I, Watanabe K. Characterization of major phytocannabinoids, cannabidiol and cannabinol, as isoform-selective and potent inhibitors of human CYP1 enzymes. Biochem Pharmacol. 2010;79:1691–1698. doi: 10.1016/j.bcp.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 11.Yamaori S, Maeda C, Yamamoto I, Watanabe K. Differential inhibition of human cytochrome P450 2A6 and 2B6 by major phytocannabinoids. Forensic Toxicol. 2011;29:117–124. [Google Scholar]
- 12.Bornheim LM, Everhart ET, Li JM, Correia MA. Induction and Genetic-Regulation of Mouse Hepatic Cytochrome-P450 by Cannabidiol. Biochem Pharmacol. 1994;48:161–171. doi: 10.1016/0006-2952(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 13.Bland TM, Haining RL, Tracy TS, Callery PS. CYP2C-catalyzed delta9-tetrahydrocannabinol metabolism: kinetics, pharmacogenetics and interaction with phenytoin. Biochem Pharmacol. 2005;70:1096–1103. doi: 10.1016/j.bcp.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Jaeger W, Benet LZ, Bornheim LM. Inhibition of cyclosporine and tetrahydrocannabinol metabolism by cannabidiol in mouse and human microsomes. Xenobiotica. 1996;26:275–284. doi: 10.3109/00498259609046707. [DOI] [PubMed] [Google Scholar]
- 15.Yamaori S, Koeda K, Kushihara M, Hada Y, Yamamoto I, Watanabe K. Comparison in the in vitro inhibitory effects of major phytocannabinoids and polycyclic aromatic hydrocarbons contained in marijuana smoke on cytochrome P450 2C9 activity. Drug Metab Pharmacokinet. 2012;27:294–300. doi: 10.2133/dmpk.dmpk-11-rg-107. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto I, Watanabe K, Narimatsu S, Yoshimura H. Recent advances in the metabolism of cannabinoids. Int J Biochem Cell Biol. 1995;27:741–746. doi: 10.1016/1357-2725(95)00043-o. [DOI] [PubMed] [Google Scholar]
- 17.Jiang RR, Yamaori S, Okamoto Y, Yamamoto I, Watanabe K. Cannabidiol Is a Potent Inhibitor of the Catalytic Activity of Cytochrome P450 2C19. Drug Metab Pharmacok. 2013;28:332–338. doi: 10.2133/dmpk.dmpk-12-rg-129. [DOI] [PubMed] [Google Scholar]
- 18.Yamaori S, Okamoto Y, Yamamoto I, Watanabe K. Cannabidiol, a Major Phytocannabinoid, As a Potent Atypical Inhibitor for CYP2D6. Drug Metabolism and Disposition. 2011;39:2049–2056. doi: 10.1124/dmd.111.041384. [DOI] [PubMed] [Google Scholar]
- 19.Gaston TE, Friedman D. Pharmacology of cannabinoids in the treatment of epilepsy. Epilepsy Behav. 2017;70:313–318. doi: 10.1016/j.yebeh.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Bornheim LM, Grillo MP. Characterization of cytochrome P450 3A inactivation by cannabidiol: possible involvement of cannabidiol-hydroxyquinone as a P450 inactivator. Chem Res Toxicol. 1998;11:1209–1216. doi: 10.1021/tx9800598. [DOI] [PubMed] [Google Scholar]
- 21.Xu X, Zhang XA, Wang DW. The roles of CYP450 epoxygenases and metabolites, epoxyeicosatrienoic acids, in cardiovascular and malignant diseases. Adv Drug Deliv Rev. 2011;63:597–609. doi: 10.1016/j.addr.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Maccarrone M. Metabolism of the Endocannabinoid Anandamide: Open Questions after 25 Years. Front Mol Neurosci. 2017;10 doi: 10.3389/fnmol.2017.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zelasko S, Arnold WR, Das A. Endocannabinoid metabolism by cytochrome P450 monooxygenases. Prostag Oth Lipid M. 2015;116:112–123. doi: 10.1016/j.prostaglandins.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 24.McDougle DR, Kambalyal A, Meling DD, Das A. Endocannabinoids Anandamide and 2-Arachidonoylglycerol Are Substrates for Human CYP2J2 Epoxygenase. J Pharmacol Exp Ther. 2014;351:616–627. doi: 10.1124/jpet.114.216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evangelista EA, Kaspera R, Mokadam NA, Jones JP, 3rd, Totah RA. Activity, inhibition, and induction of cytochrome P450 2J2 in adult human primary cardiomyocytes. Drug Metab Dispos. 2013;41:2087–2094. doi: 10.1124/dmd.113.053389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaspera R, Kirby BJ, Sahele T, Collier AC, Kharasch ED, Unadkat JD, Totah RA. Investigating the contribution of CYP2J2 to ritonavir metabolism in vitro and in vivo. Biochem Pharmacol. 2014;91:109–118. doi: 10.1016/j.bcp.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee CA, Jones JP, 3rd, Katayama J, Kaspera R, Jiang Y, Freiwald S, Smith E, Walker GS, Totah RA. Identifying a selective substrate and inhibitor pair for the evaluation of CYP2J2 activity. Drug Metab Dispos. 2012;40:943–951. doi: 10.1124/dmd.111.043505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karkhanis A, Lam HY, Venkatesan G, Koh SK, Chai CL, Zhou L, Hong Y, Kojodjojo P, Chan EC. Multiple modes of inhibition of human cytochrome P450 2J2 by dronedarone, amiodarone and their active metabolites. Biochem Pharmacol. 2016;107:67–80. doi: 10.1016/j.bcp.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 29.Di Marzo V, Piscitelli F. The Endocannabinoid System and its Modulation by Phytocannabinoids. Neurotherapeutics. 2015;12:692–698. doi: 10.1007/s13311-015-0374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maccarrone M, Bab R, Biro T, Cabral GA, Dey SK, Di Marzo V, Konje JC, Kunos G, Mechoulam R, Pacher P, Sharkey KA, Zimmer A. Endocannabinoid signaling at the periphery: 50 years after THC. Trends Pharmacol Sci. 2015;36:277–296. doi: 10.1016/j.tips.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pacher P, Kunos G. Modulating the endocannabinoid system in human health and disease successes and failures. Febs J. 2013;280:1918–1943. doi: 10.1111/febs.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McPartland JM, Guy GW, Di Marzo V. Care and feeding of the endocannabinoid system: a systematic review of potential clinical interventions that upregulate the endocannabinoid system. PLoS One. 2014;9:e89566. doi: 10.1371/journal.pone.0089566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woods SC. Role of the endocannabinoid system in regulating cardiovascular and metabolic risk factors. Am J Med. 2007;120:S19–25. doi: 10.1016/j.amjmed.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Ashton JC, Smith PF. Cannabinoids and cardiovascular disease: the outlook for clinical treatments. Curr Vasc Pharmacol. 2007;5:175–185. doi: 10.2174/157016107781024109. [DOI] [PubMed] [Google Scholar]
- 35.O'Sullivan SE, Kendall PJ, Kendall DA. Endocannabinoids and the cardiovascular response to stress. J Psychopharmacol. 2012;26:71–82. doi: 10.1177/0269881111408457. [DOI] [PubMed] [Google Scholar]
- 36.Pacher P, Mukhopadhyay P, Mohanraj R, Godlewski G, Batkai S, Kunos G. Modulation of the endocannabinoid system in cardiovascular disease: therapeutic potential and limitations. Hypertension. 2008;52:601–607. doi: 10.1161/HYPERTENSIONAHA.105.063651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pacher P, Steffens S, Hasko G, Schindler TH, Kunos G. Cardiovascular effects of marijuana and synthetic cannabinoids: the good, the bad, and the ugly. Nat Rev Cardiol. 2017 doi: 10.1038/nrcardio.2017.130. [DOI] [PubMed] [Google Scholar]
- 38.Rezkalla S, Stankowski R, Kloner RA. Cardiovascular Effects of Marijuana. J Cardiovasc Pharmacol Ther. 2016;21:452–455. doi: 10.1177/1074248415627874. [DOI] [PubMed] [Google Scholar]
- 39.Montecucco F, Di Marzo V. At the heart of the matter: the endocannabinoid system in cardiovascular function and dysfunction. Trends Pharmacol Sci. 2012;33:331–340. doi: 10.1016/j.tips.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Steffens S, Pacher P. Targeting cannabinoid receptor CB2 in cardiovascular disorders: promises and controversies. Brit J Pharmacol. 2012;167:313–323. doi: 10.1111/j.1476-5381.2012.02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zubrzycki M, Liebold A, Janecka A, Zubrzycka M. A New Face of Endocannabinoids in Pharmacotherapy Part Ii. Role of Endocannabinoids in Inflammation-Derived Cardiovascular Diseases. J Physiol Pharmacol. 2014;65:183–191. [PubMed] [Google Scholar]
- 42.McDougle DR, Watson JE, Abdeen AA, Adili R, Caputo MP, Krapf JE, Johnson RW, Kilian KA, Holinstat M, Das A. Anti-inflammatory omega-3 endocannabinoid epoxides. Proc Natl Acad Sci U S A. 2017;114:E6034–E6043. doi: 10.1073/pnas.1610325114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bornheim LM, Kim KY, Chen B, Correia MA. The effect of cannabidiol on mouse hepatic microsomal cytochrome P450-dependent anandamide metabolism. Biochem Biophys Res Commun. 1993;197:740–746. doi: 10.1006/bbrc.1993.2541. [DOI] [PubMed] [Google Scholar]
- 44.Arnold WR, Baylon JL, Tajkhorshid E, Das A. Arachidonic Acid Metabolism by Human Cardiovascular CYP2J2 is Modulated by Doxorubicin. Biochemistry. 2017 doi: 10.1021/acs.biochem.7b01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harvey DJ. Mass-Spectrometry of the Cannabinoids and Their Metabolites. Mass Spectrom Rev. 1987;6:135–229. [Google Scholar]
- 46.Arnold WR, Baylon JL, Tajkhorshid E, Das A. Asymmetric Binding and Metabolism of Polyunsaturated Fatty Acids (PUFAs) by CYP2J2 Epoxygenase. Biochemistry. 2016;55:6969–6980. doi: 10.1021/acs.biochem.6b01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harvey DJ. Absorption, distribution, and biotransformation of the cannabinoids. Marihuana and Medicine. 1999:91–103. [Google Scholar]
- 48.Narimatsu S, Watanabe K, Matsunaga T, Yamamoto I, Imaoka S, Funae Y, Yoshimura H. Cytochrome P-450 isozymes in metabolic activation of delta 9-tetrahydrocannabinol by rat liver microsomes. Drug Metab Dispos. 1990;18:943–948. [PubMed] [Google Scholar]
- 49.Yamamoto I, Watanabe K, Matsunaga T, Kimura T, Funahashi T, Yoshimura H. Pharmacology and toxicology of major constituents of marijuana - On the metabolic activation of cannabinoids and its mechanism. J Toxicol-Toxin Rev. 2003;22:577–589. [Google Scholar]
- 50.Watanabe K, Matsunaga T, Narimatsu S, Yamamoto I, Imaoka S, Funae Y, Yoshimura H. Catalytic Activity of Cytochrome-P450 Isozymes Purified from Rat-Liver in Converting 11-Oxo-Delta-8-Tetrahydrocannabinol to Delta-8-Tetrahydrocannabinol-11-Oic Acid. Biochem Pharmacol. 1991;42:1255–1259. doi: 10.1016/0006-2952(91)90262-4. [DOI] [PubMed] [Google Scholar]
- 51.Matsunaga T, Shibayama K, Higuchi S, Tanaka H, Watanabe K, Yamamoto I. Characterization of microsomal alcohol oxygenase catalyzing the oxidation of 7-hydroxy-Delta(8)-tetrahydrocannabinol to 7-oxo-Delta(8)-tetrahydrocannabinol in rat liver. Biological & Pharmaceutical Bulletin. 2000;23:43–46. doi: 10.1248/bpb.23.43. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe K, Yamaori S, Funahashi T, Kimura T, Yamamoto I. Cytochrome P450 enzymes involved in the metabolism of tetrahydrocannabinols and cannabinol by human hepatic microsomes. Life Sci. 2007;80:1415–1419. doi: 10.1016/j.lfs.2006.12.032. [DOI] [PubMed] [Google Scholar]
- 53.Huestis MA. Human cannabinoid pharmacokinetics. Chem Biodivers. 2007;4:1770–1804. doi: 10.1002/cbdv.200790152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe K, Tanaka T, Yamamoto I, Yoshimura H. Brain Microsomal Oxidation of Delta-8-Tetrahydrocannabinol and Delta-9-Tetrahydrocannabinol. Biochem Bioph Res Co. 1988;157:75–80. doi: 10.1016/s0006-291x(88)80013-5. [DOI] [PubMed] [Google Scholar]
- 55.Si D, Wang Y, Zhou YH, Guo Y, Wang J, Zhou H, Li ZS, Fawcett JP. Mechanism of CYP2C9 inhibition by flavones and flavonols. Drug Metab Dispos. 2009;37:629–634. doi: 10.1124/dmd.108.023416. [DOI] [PubMed] [Google Scholar]
- 56.Elmes MW, Kaczocha M, Berger WT, Leung K, Ralph BP, Wang L, Sweeney JM, Miyauchi JT, Tsirka SE, Ojima I, Deutsch DG. Fatty acid-binding proteins (FABPs) are. intracellular carriers for Delta9-tetrahydrocannabinol (THC) and cannabidiol (CBD) J Biol Chem. 2015;290:8711–8721. doi: 10.1074/jbc.M114.618447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bisogno T, Hanus L, De Petrocellis L, Tchilibon S, Ponde DE, Brandi I, Moriello AS, Davis JB, Mechoulam R, Di Marzo V. Molecular targets for cannabidiol and its synthetic analogues: effect on vanilloid VR1 receptors and on the cellular uptake and enzymatic hydrolysis of anandamide. Br J Pharmacol. 2001;134:845–852. doi: 10.1038/sj.bjp.0704327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snider NT, Nast JA, Tesmer LA, Hollenberg PF. A Cytochrome P450-Derived Epoxygenated Metabolite of Anandamide Is a Potent Cannabinoid Receptor 2-Selective Agonist. Molecular Pharmacology. 2009;75:965–972. doi: 10.1124/mol.108.053439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohlsson A, Agurell S, Leander K, Dahmen J, Edery H, Porath G, Levy S, Mechoulam R. Synthesis and Psychotropic Activity of Side-Chain Hydroxylated Delta-6-Tetrahydrocannabinol Metabolites. Acta Pharm Suec. 1979;16:21–33. [PubMed] [Google Scholar]
- 60.Piomelli D. The molecular logic of endocannabinoid signalling. Nat Rev Neurosci. 2003;4:873–884. doi: 10.1038/nrn1247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.