Structural variation is often regarded as a genetic abnormality with negative consequences. This is perhaps not surprising, since some of the best-documented instances of structural variation were detected through analysis of aberrant human cells1,2 and tumors3. However, recent analyses show that structural variation is a normal aspect of natural genetic variation. For example, a recent analysis of 2504 human genome sequences finds that structural variation is common between human individuals and population groups4. While it is still unclear to what extent such structural variation helped shape human evolution, experiments with microbes clearly indicate that large-scale rearrangements are an important driver of genetic adaptation to new environments (reviewed in ref. 5).
Interestingly, little is known about how structural variation can lead to phenotypic changes, and few studies explore the possibilities to exploit structural variation. This can be partly attributed to the use of short-read sequencing technologies, which are great at detecting SNPs and small insertions and deletions, but only offer a limited capacity to detect structural variation. Secondly, it is difficult to induce and control structural variations experimentally, impeding further investigation and exploitation.
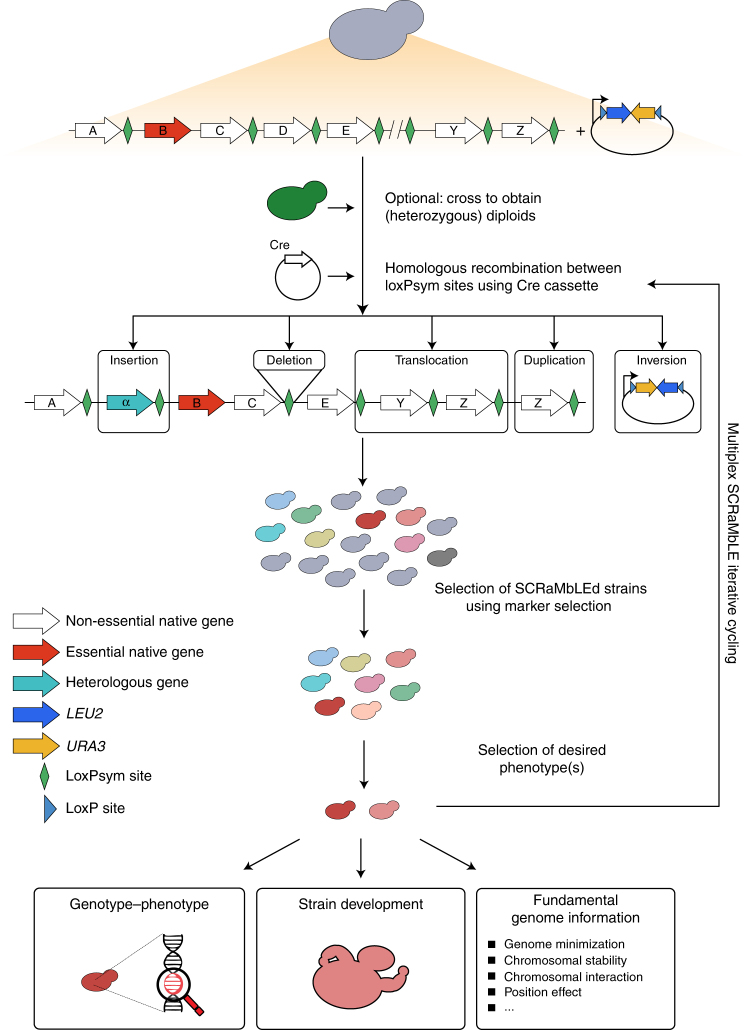
Advanced synthetic biology allows inducing structural variation in genomes. The technology, appropriately called “Synthetic Chromosome Recombination and Modification by LoxP-mediated Evolution” (SCRaMbLE), makes it possible to rapidly induce recombination events that lead to extensive rearrangements between specific sites. SCRaMbLEing genomes does not only allow investigating if and how gross rearrangements lead to phenotypic changes, but also opens new avenues to create superior (micro)organisms (Fig. 1).
Fig. 1.
Overview of the SCRaMbLE technology and its relevance for biotechnology
SCRaMbLE was first introduced as part of the Sc2.0-project, where an international team of researchers aims at synthesizing the entire Saccharomyces cerevisiae genome. While the Sc2.0 project largely kept the natural genomic DNA sequence, a few notable changes were introduced. One of these changes was the insertion of site-specific recombination sites, called loxPsym sites, 3 bp downstream of the stop codon of every non-essential gene, as well as near chromosomal landmarks like the centromere and telomeres. Like the natural loxP sites derived from bacteriophage P1, loxPsym sites promote direct recombination by the Cre recombinase6,7. However, in contrast to the natural sites, loxPsym sites facilitate recombination in either direction, which vastly expands the spectrum of potential rearrangements8. To ensure fast (de)activation and low leaky expression of the recombinase enzyme, several modified Cre expression vectors were developed9. Together, the combination of strategically placed recombination sites and tightly controlled Cre activity allows inducing a temporary burst in structural variation.
The first demonstration of SCRaMbLE was published in 20119. Activating the recombinase in strains carrying a partly synthetic chromosome with 43 loxPsym sites resulted in mutant lines that showed a large variation in growth rate. Deep sequencing of 64 SCRaMbLEd mutants of a strain containing a circular synIXR chromosome revealed a unique pattern of on average 6.2 complex recombination events in each mutant10. Importantly, recombination events were limited to loxPsym sites, with the rest of the genome left untouched.
SCRaMbLE allows investigating how structural variation can lead to phenotypic changes. One way to do this will be to grow pools of variants in different stressful environments. Variants showing improved fitness can easily be enriched by prolonged exposure to the stress, and long-read whole-genome sequencing allows determining the structure of the recombined genomes. This enables researchers to study the phenotypic effect of genetic perturbations that are difficult to obtain using “traditional” genetic engineering strategies (inversions, duplications, translocations, etc.), and will thus further expand our knowledge of complex phenotypes.
However, there are still some pitfalls. Firstly, rearrangements are difficult to re-engineer one by one, making it difficult to distinguish the rearrangements that are associated with the change in fitness from other, neutral or slightly detrimental rearrangements. A second, and perhaps more important pitfall is that the rearrangements obtained through SCRaMbLE only occur between loxPsym sites and may therefore not be a perfect representation of naturally occurring events. Moreover, it is also still unclear if and how the rearrangements are influenced by the relative positioning of the loxPsym sites. Thirdly, deleting or amplifying certain genes can be lethal, causing recombination biases that can stretch over several kilobases. Lastly, SCRaMbLE is limited to strains harbouring the loxPsym sites, which is currently only the S288c lab strain.
It is easy to imagine how SCRaMbLE could be exploited to fine-tune, prototype or even discover new complex pathways for many biotechnological applications. For example, while the production of artemisinic acid in yeast is arguably the greatest achievement in fungal synthetic biology thus far, it was estimated to have required roughly 100 postdoc years11,12. Theoretically, engineering such complex pathways could be sped up drastically with clever design of loxPsym-containing neochromosomes, or recombination-mediated integration of LoxPsym-flanked DNA containing required genes. Since gene expression depends on the chromosomal location13, this approach can be applied for combinatorial optimization of pathway gene expression levels. Moreover, since the loxP-mediated recombination system is not specific for S. cerevisiae, the general concept of SCRaMbLE can be extrapolated to other industrially relevant yeasts and bacteria14,15. As the cost of synthetic DNA keeps falling, and the availability of CRISPR-Cas9-based strategies for rapid and efficient integration of heterologous DNA, it seems likely that other industrially relevant organisms will be equipped with recombination sites to perform SCRaMbLE.
Taken together, it is clear that SCRaMbLE is a promising new way to obtain superior microbes for industrial applications. Large pools of genetic variants differing in gene content and genome structure can be obtained in just a matter of days and, thus, parallel optimization of complex multigenic traits can be performed. The technology complements the existing toolbox, from classic breeding, mutagenesis and genome shuffling over directed evolution to genetic engineering16. Importantly, the technology is orthogonal to these existing methods, as the type of changes that SCRaMbLE induces is fundamentally different. Hence, it seems interesting to combine SCRaMbLE with other techniques.
That said, there are also limitations. Firstly, the dependence on loxP or loxPsym sites implies that recombination is not completely random and that the recombination sites need to be carefully chosen. Moreover, as SCRaMbLE results in extremely large pools of genetically diverse mutants, its applicability for industrial strain development depends on whether the phenotype of interest is easily selectable for. Therefore, finding clever strategies to detect improved variants, even for “difficult” phenotypes like production of specific metabolites, is vital. Lastly, it is important to realize that structural variation is sometimes seen as a “quick and dirty” way to rapidly optimize a phenotype. Other mutation events, including smaller rearrangements and changes in the DNA sequence may be needed to further optimize the phenotype under selection without scarifying other desirable traits. Still, SCRaMbLE is clearly more than a gimmick, and it will be exciting to see how the technology will help us to develop novel drugs, biomaterials or chemicals, and reach higher productivity, quality and sustainability in various industries.
Acknowledgements
This work was supported by a FWO grant to J.S. Research in the laboratory of K.J.V. is supported by VIB, AB-InBev Ballet Latour Fund, FWO, VLAIO, and a European Research Council (ERC) Consolidator Grant CoG682009.
Author contributions
J.S., A.G., and K.J.V. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Jan Steensels, Anton Gorkovskiy.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wyandt, H. E. & Tonk, V. S. Atlas of Human Chromosome Heteromorphisms (Springer, New York, 2004).
- 2.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet. 2013;14:125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 3.Macintyre G, Ylstra B, Brenton JD. Sequencing structural variants in cancer for precision therapeutics. Trends Genet. 2016;32:530–542. doi: 10.1016/j.tig.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voordeckers K, Verstrepen KJ. Experimental evolution of the model eukaryote Saccharomyces cerevisiae yields insight into the molecular mechanisms underlying adaptation. Curr. Opin. Microbiol. 2015;28:1–9. doi: 10.1016/j.mib.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 6.Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:2087–2096. doi: 10.1128/MCB.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination: I. Recombination between loxP sites. J. Mol. Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 8.Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in PI site-specific recombination. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dymond JS, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen Y, et al. SCRaMbLE generates designed combinatorial stochastic diversity in synthetic chromosomes. Genome Res. 2016;26:36–49. doi: 10.1101/gr.193433.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ro DK, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 12.Paddon CJ, Keasling JD. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat. Rev. Microbiol. 2014;12:355–367. doi: 10.1038/nrmicro3240. [DOI] [PubMed] [Google Scholar]
- 13.Thompson A, Gasson MJ. Location effects of a reporter gene on expression levels and on native protein synthesis in Lactococcus lactis and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2001;67:3434–3439. doi: 10.1128/AEM.67.8.3434-3439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xin Y, Guo T, Mu Y, Kong J. Coupling the recombineering to Cre-lox system enables simplified large-scale genome deletion in Lactobacillus casei. Microb. Cell Fact. 2018;17:21. doi: 10.1186/s12934-018-0872-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marx H, Mattanovich D, Sauer M. Overexpression of the riboflavin biosynthetic pathway in Pichia pastoris. Microb. Cell Fact. 2008;7:23. doi: 10.1186/1475-2859-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steensels J, Snoek T, Meersman E, Nicolino MP, Voordeckers K, Verstrepen KJ. Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol. Rev. 2014;38:947–995. doi: 10.1111/1574-6976.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]