Abstract
Aggregatibacter actinomycetemcomitans is a Gram-negative organism, strongly associated with aggressive forms of periodontitis. An important virulence property of A. actinomycetemcomitans is its ability to form tenacious biofilms that can attach to abiotic as well as biotic surfaces. The histone-like (H-NS) family of nucleoid-structuring proteins act as transcriptional silencers in many Gram-negative bacteria. To evaluate the role of H-NS in A. actinomycetemcomitans, hns mutant derivatives of serotype a strain D7S were generated. Characteristics of the hns mutant phenotype included shorter and fewer pili, and substantially lower monospecies biofilm formation relative to the wild type. Furthermore, the D7S hns mutant exhibited significantly reduced growth within a seven-species oral biofilm model. However, no apparent difference was observed regarding the numbers and proportions of the remaining six species regardless of being co-cultivated with D7S hns or its parental strain. Proteomics analysis of the strains grown in monocultures confirmed the role of H-NS as a repressor of gene expression in A. actinomycetemcomitans. Interestingly, proteomics analysis of the multispecies biofilms indicated that the A. actinomycetemcomitans wild type and hns mutant imposed different regulatory effects on the pattern of protein expression in the other species, i.e., mainly Streptococcus spp., Fusobacterium nucleatum, and Veillonella dispar. Gene ontology analysis revealed that a large portion of the differentially regulated proteins was related to translational activity. Taken together, our data suggest that, apart from being a negative regulator of protein expression in A. actinomycetemcomitans, H-NS promotes biofilm formation and may be an important factor for survival of this species within a multispecies biofilm.
Periodontal disease: Protein promoting biofilm formation
A member of a specific group of gene-regulating proteins promotes biofilm formation by a bacterium associated with aggressive forms of gum disease. Forming biofilms helps the bacterium to cause persistent infections. Researchers at Karolinska Institutet and Umeå University (Sweden), and University of Zürich (Switzerland), led by Jan Oscarsson at Umeå University, investigated the role of the “histone-like” protein H-NS in Aggregatibacter actinomycetemcomitans infections. These proteins are known to suppress the activity of specific genes in many bacteria, a property confirmed in this research. By studying mutant bacterial strains deficient in H-NS protein, the researchers demonstrated that this protein promotes the formation of biofilms by the bacteria. The results suggest that H-NS plays a significant role in allowing Aggregatibacter actinomycetemcomitans to thrive in biofilms containing mixed populations of bacteria. This effect appears to involve activating production of hair-like appendages called pili on the bacterial surface.
Introduction
Aggregatibacter actinomycetemcomitans is a Gram-negative bacterium from the Pasteurellaceae family. Colonization by A. actinomycetemcomitans is strongly associated with aggressive forms of periodontitis in adolescents and young adults.1 The microorganism is also a pathogen, associated with non-oral infections, such as endocarditis,2 and is a candidate bacterial trigger of anti-citrulline autoimmunity in rheumatoid arthritis.3 Some of the virulence factors of A. actinomycetemcomitans involved in oral colonization and induction of periodontal inflammation have been thoroughly studied and well characterized, including, e.g., leukotoxin (LtxA) and cytolethal distending toxin (for a recent review see ref.4). An important virulence property of A. actinomycetemcomitans is its ability to form tenacious biofilms that can attach to abiotic as well as biotic surfaces.5 Adherence and biofilm growth in A. actinomycetemcomitans is mediated by the tight-adherence (tad) gene locus, which consists of 14 genes (flp-1, flp-2, tadV, rcpCAB, tadZABCDEFG),6,7 and also depends on the production of an extracellular carbohydrate polymer of β (1,6)-linked N-acetyl-d-glucosamine.8
The histone-like (H-NS) family of DNA-binding, nucleoid-structuring proteins is widespread in Gram-negative bacteria.9 H-NS appears to act primarily as a global silencer of AT-rich DNA acquired by horizontal gene transfer,10 but evidence has also been presented that it stimulates translation of genes with suboptimal ribosome-binding sequences.11 H-NS has been demonstrated to globally control the expression of ~5% of all genes in Escherichia coli, many of which are involved in transcription and translation and in the production of cell envelope components required for adjustment to varying environments.12 However, in other species, the number of genes regulated by H-NS appears to be more restricted.9 H-NS plays a central role in the regulation of virulence-associated genes in several pathogens, and hns mutants have been demonstrated to exhibit reduced virulence in various in vitro and in vivo models.13,14 For example, in Actinobacillus pleuropneumoniae and Vibrio cholerae, H-NS acts as a repressor of exopolysaccharide biosynthesis genes, suppressing biofilm formation.15,16 On the other hand, in E. coli, deletion of the hns gene decreased biofilm formation,17 whereas accordingly it was increased upon expression of the hns gene in trans.18
A. actinomycetemcomitans encodes an H-NS protein, which is 132 amino acids, and exhibits ~50% amino acid identity to the 15.5 kDa E. coli protein. The H-NS protein of A. actinomycetemcomitans is well conserved among strains, i.e., identical proteins are encoded by all 38 genome-sequenced strains available in the National Center for Biotechnology (NCBI) database, representing all serotypes (a–g). To date, the function of H-NS in A. actinomycetemcomitans has not been described. Hence, we sought to investigate if H-NS may act as a global regulator in A. actinomycetemcomitans, and to examine the influence of H-NS on biofilm formation. It was recently demonstrated that presence of A. actinomycetemcomitans in an in vitro multispecies oral biofilm mimicking subgingival dental plaque had a regulatory effect on the other species, i.e., their overall protein expression profiles were altered.19 This prompted us in the present work to assess whether an A. actinomycetemcomitans wild type and hns mutant might induce different patterns of altered protein expression in the multispecies oral biofilm.
Results and discussion
Inactivation of the A. actinomycetemcomitans hns gene results in impaired biofilm growth in monospecies biofilm
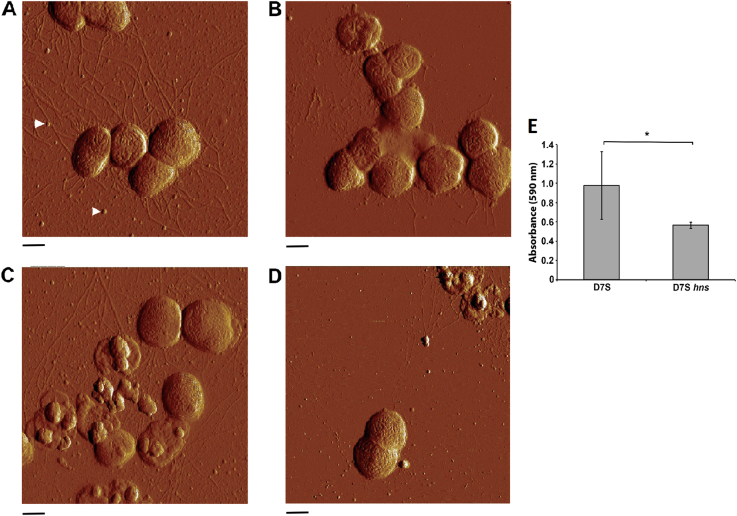
To investigate if lack of hns expression in A. actinomycetemcomitans might result in apparent phenotypical differences regarding gene expression with emphasis on virulence, hns mutants were generated in strains D7S and D7SS, as described in the Methods section. The abolished H-NS production in the mutants was confirmed using western blot, and a polyclonal antiserum made against E. coli H-NS (Supplementary Fig. 1). To investigate if inactivation of hns caused a visible effect on the cell morphology, atomic force microscopy (AFM) was conducted, assessing strains D7S and D7SS, and their corresponding hns mutants, cultivated at 30 and 37 °C, respectively. According to our results (Fig. 1a–d), at both temperatures the D7S hns mutant appeared less piliated, exhibiting pili that were shorter than those of the parental strain. However, no apparent dissimilarity was seen comparing the smooth-colony type derivative, D7SS, and its hns mutant (data not shown). Moreover, in contrast to findings with E. coli,20 AFM revealed no substantial difference in the amount of outer membrane vesicles (OMVs) released by the wild type and the hns mutant in either D7S or D7SS (Fig. 1a–d, and data not shown). To test whether the reduced piliation in the D7S hns mutant might be associated with impaired biofilm growth, monocultures of the strains cultivated at 37 °C were stained with crystal violet (Fig. 1e). This revealed decreased (reduced to approximately 58%; p < 0.05) biofilm formation in the D7S hns mutant relative to the wild type. Together, these observations support the notion that hns may act as an activator of pili production in A. actinomycetemcomitans, promoting biofilm growth.
Fig. 1.
Assessment of the influence of H-NS on A. actinomycetemcomitans cell morphology and monoculture biofilm formation. Atomic force microscopy was used to analyze the following A. actinomycetemcomitans strains, cultivated on agar at the indicated temperatures: D7S 30 °C (a), D7S hns 30 °C (b), D7S 37 °C (c), and D7S hns 37 °C (d). Arrows indicate examples of the released OMVs. Scale bars = 500 nm. Crystal violet was used to quantify the biofilm formation of the strains D7S and D7S hns, respectively, after cultivation in 24-well cell culture plates for 3 days at 37 °C (e). Shown are the means of OD 590 nm ± standard deviation (SD) for six experiments; *p = 0.0169 for D7S vs. D7S hns
Inactivation of the hns gene results in impaired growth of A. actinomycetemcomitans in a multispecies oral biofilm, without altering the proportions of the other species
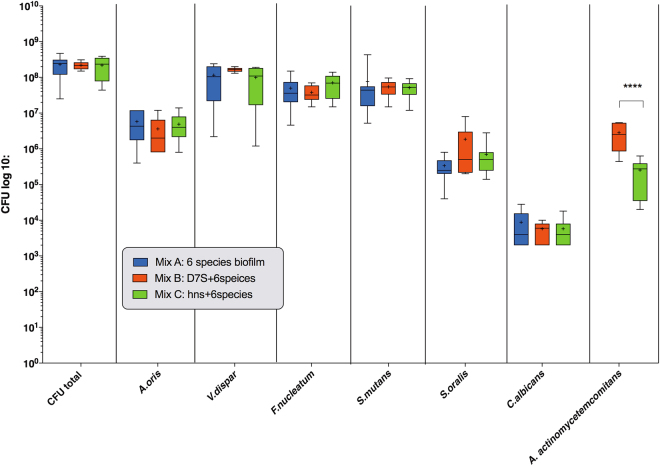
We next investigated the influence of the hns mutation on the growth in a multispecies (n = 6) biofilm setting. After anaerobic cultivation for 64 h at 37 °C, the numbers of the individual oral microbial species within the biofilms were estimated by colony-forming unit (CFU) counting (Fig. 2). Comparative investigation of strain D7S and its hns mutant within the biofilm context, respectively, revealed that there were significantly lower numbers of the hns mutant, i.e., more than 11 times reduced relative to the wild type (p < 0.0001) (Fig. 2). This is consistent with the reduced biofilm formation of the hns mutant observed in monocultures (Fig. 1). On the other hand, neither D7S nor D7S hns had a significant impact on the growth of the other six species within the biofilm, as compared to when the biofilm was cultivated without an A. actinomycetemcomitans strain (Fig. 2).
Fig. 2.
Analysis of the quantitative composition of the multispecies oral biofilms. Quantification was performed used CFU counting for each species per biofilm disc as described in the Methods. As indicated, control biofilms did not include an A. actinomycetemcomitans strain (Mix A), whereas experimental biofilms either contained strain D7S (mix B) or its hns mutant (Mix C). The data are expressed as the bacterial mean counts ± SD from 10 biological replicates. ANOVA test: ****p < 0.0001
Inactivation of the A. actinomycetemcomitans hns gene does not alter the structure of the multispecies oral biofilm
A further investigation of the structure of the multispecies biofilm by means of confocal laser scanning microscopy (CLSM) corroborated the CFU results. This revealed the localization of the A. actinomycetemcomitans cells along with the other species. Evidently, strain D7S cells appeared to be more abundant in the biofilms as compared to cells of the D7S hns mutant (Supplementary Fig. 2). On the other hand, CLSM revealed no apparent alterations regarding the general structural conformation of the whole biofilms.
Inactivation of the hns gene alters the proteome of A. actinomycetemcomitans strain D7S
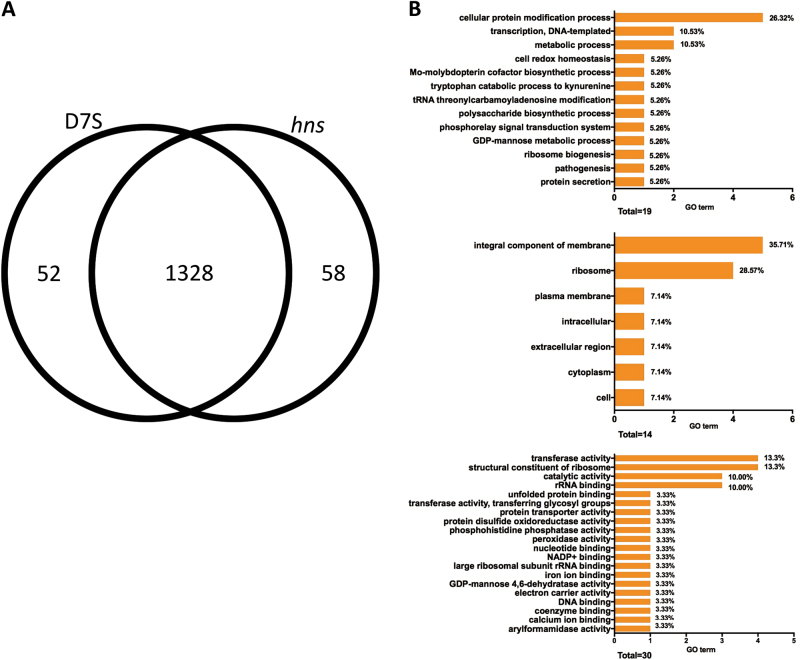
To investigate if H-NS may act as a regulator of multiple genes in A. actinomycetemcomitans, we investigated the influence of the hns mutation on the whole proteome of strain D7S. For this, whole cell lysates from the agar monocultures were collected and processed for quantitative proteomics analysis. For each strain, three independent protein preparations were analyzed. For identification, proteins with at least two unique peptides were accepted. A total of 1438 proteins with corresponding false discovery rate (FDR) of 3% at the protein level and 0.18% at the peptide level were identified (Supplementary Table 1). The majority of proteins were identical in both variants (1328 out of 1438). In all, 58 proteins were exclusive to the wild type D7S, whereas 52 were found only in the hns mutant strain (Fig. 3a). Quantitative differences in protein expression were assessed by the Progenesis software, as detailed in the Methods section. Heat maps were used to assess the correlation between samples (Supplementary Fig. 3A). It was found that the D7S and D7S hns samples were clustered in two separate groups. The quantification by the Progenesis software showed that 31 proteins were differently expressed among these two strains with analysis of variance (ANOVA) p value <0.05 and log2 fold ≥1 (Supplementary Table 2). Thus, under these conditions, the H-NS-regulated proteome constituted a relatively small proportion of the protein-encoding genes (i.e., 2581 in D7S).21 The majority of these 31 proteins, i.e. 29, were upregulated in the hns mutant, which is consistent with the notion that H-NS is mainly acting as a repressor of gene expression in A. actinomycetemcomitans. The two proteins that showed significantly higher levels in D7S relative to the hns mutant strain were HK1651_07880 (i.e., H-NS), and ABC transporter ATP-binding protein.
Fig. 3.
H-NS-dependent proteome of A. actinomycetemcomitans strain D7S. Venn diagram indicating the numbers of identified proteins in A. actinomycetemcomitans strain D7S and D7S hns monocultures grown on agar (a). Gene ontology (GO) analysis of the 29 proteins found to be repressed by H-NS (b). The numbers and proportions of GO terms from each of the three categories, biological process, cellular component, and molecular function, are shown. Sub-domains with their GO proportion less than 2% were classified as “other”
To determine putative cellular functions repressed by H-NS, the 29 proteins were grouped according to their Gene Ontology (GO) terms for their predicted biological process, cellular component (i.e., subcellular localization), and molecular function, respectively (Fig. 3b). A majority of these proteins were integral components of membrane (35.71%) or structural constituent of ribosome (28.57%), which is in line with findings in other species, e.g., E. coli.12 Concomitantly, the two most predominant molecular functions of these proteins were structural constituent of ribosome (13.33%) and transferase activity (13.33%). Regarding the biological processes repressed by H-NS, the majority of the proteins were represented in cellular protein modification processes (26.32%), metabolic process (10.53%), and transcription, DNA-templated (10.53%). Of the known virulence factors, we observed that two proteins encoded by the leukotoxin (ltxCABD) gene locus, i.e., leukotoxin (LtxA), and LtxD, were among the proteins upregulated in the hns mutant. This is consistent with findings with the homologous hemolysin operons hlyCABD and ehxCABD in E. coli, and rtxACBD in V. cholerae, which are also repressed by H-NS.13,22,23 Functional homologs to some of the other upregulated proteins in D7S hns have been shown earlier to promote virulence, including biofilm formation in other species, e.g., glycosyl transferases,24 galactose metabolism pathway proteins,25 and prepilin peptidase.26 Moreover, TorR (also known as ArcA) is a two-component system response regulator and a global regulator of virulence factor expression.27 Albeit implied by AFM (Fig. 1a–d), we detected no significant regulation of any component encoded by the tad gene locus. For Flp-1, the major fimbrial subunit, this may be a result of low detection efficiency in liquid chromatography– tandem mass spectrometry (LC-MS/MS) due to its small molecular size (8 kDa) or to limited sensitivity for low abundant proteins. Our data also identified several unknown proteins repressed by H-NS, which might represent factors affecting virulence and biofilm development. Among these, the protein exhibiting the highest level of upregulation in the D7S hns mutant versus the wild type was hypothetical protein ACT75_01010. Whether they may play a role in A. actinomycetemcomitans virulence and biofilm formation will be subject to future studies.
A. actinomycetemcomitans D7S and D7S hns induce different patterns of protein expression in the microbial species of the oral biofilm
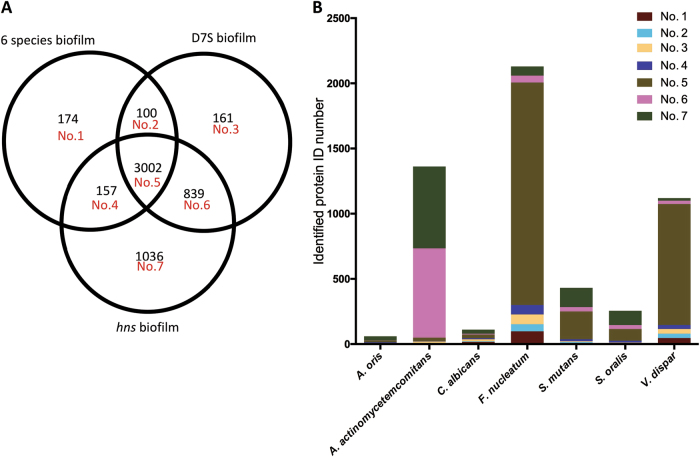
To evaluate if D7S and D7S hns may differentially induce proteomic changes in multispecies oral biofilms, lysates from the D7S (n = 10), the D7S hns (n = 10), and the control biofilm (A. actinomycetemcomitans excluded; n = 9) were collected and processed for proteomics analysis. From these analyses, a total of 5469 proteins with FDR of 2.8% on protein level and 0.68% on peptide level were identified (Supplementary Table 3). Comparisons of the numbers of identified proteins in the different biofilms and for each species in the biofilms are shown in Fig. 4a, b, respectively. According to our data, 3002 proteins were identified in all three biofilms, and a relatively large number of the 1036 proteins were uniquely identified in the D7S hns biofilm. Moreover, there was an overlap of 839 proteins between the D7S and D7S hns biofilms. For F. nucleatum and V. dispar, most of their identified proteins (1706 of 2130 F. nucleatum proteins, and 927 of 1120 V. dispar proteins) were identified in all three biofilms, whereas for A. actinomycetemcomitans (627 of 1362 proteins), S. mutans (147 of 431 proteins), and S. oralis (109 of 225 proteins), a relatively large fraction of the proteins was merely identified in the D7S hns biofilm.
Fig. 4.
Qualitative analysis of the differential effect of A. actinomycetemcomitans strain D7S and D7S hns on protein expression in a multispecies oral biofilm. Venn diagram indicating the numbers of identified proteins in the three different biofilms using LC-MS/MS (a). Numbers of identified proteins per species in the biofilms are indicated in the bar chart (b) with corresponding domain numbers from the Venn diagram
According to the label-free quantification results, the presence of an A. actinomycetemcomitans strain resulted in altered protein expression in the multispecies biofilm compared to when it was absent. Interestingly, when biofilms were clustered based on the overall protein expressions, D7S biofilms formed one group, whereas the D7S hns and the 6-species control biofilms (control biofilm without D7S) were clustered together in another group based on our algorithm (Supplementary Fig. 3). Considering the tendency of Progenesis to aggressively match features,28 A. actinomycetemcomitans proteins were not included in these analyses when comparisons were made against the control, i.e., either of the A. actinomycetemcomitans-containing biofilms versus the 6-species biofilm. The total numbers of proteins identified as regulated between the different biofilms were 872 in the D7S wild type versus the control biofilm without D7S (Supplementary Table 4), 214 in the D7S hns versus the control biofilm without D7S (Supplementary Table 5), and 610 in the D7S versus the D7S hns biofilm (Supplementary Table 6). Comparing the pattern of regulated non-A. actinomycetemcomitans (non-Aa) proteins in the D7S versus the 6-species control biofilm (Fig. 5a) revealed that 16 were upregulated in the D7S biofilm, whereas a large majority, i.e., 411, were downregulated. The downregulated proteins mainly originated from F. nucleatum (n = 274) and V. dispar (n = 132). In contrast, when comparing the D7S hns with the control biofilm without D7S (Fig. 5b), the total number of regulated non-Aa proteins was much lower, and the majority (n = 104) were up- rather than downregulated (n = 8). These regulated proteins mainly originated from Streptococcus spp. (11 S. mutans and 83 S. oralis proteins, all being upregulated), F. nucleatum (8 upregulated and 1 downregulated), and V. dispar (5, all downregulated) in the D7S relative to the 6-species biofilm.
Fig. 5.
Quantitative analysis of the differential effect of A. actinomycetemcomitans strain D7S and D7S hns on protein expression in a multispecies oral biofilm. Regulation trends of label-free quantified proteins from each species are shown in the figure based on whether they were upregulated (brown) or downregulated (blue) in a D7S compared with 6 species biofilms, b D7S hns compared with 6 species biofilms, and c D7S compared with D7S hns biofilms
Our results are consistent with the notion that the A. actinomycetemcomitans strain D7S and its hns mutant promote distinctly different proteomic changes in the multispecies oral biofilms, i.e., as judged by the numbers of enhanced protein expression, the regulatory impact on the other biofilm species appeared to be largely compromised by the hns deletion. For example, the relatively large downregulation of proteins in F. nucleatum and V. dispar was observed only when D7S was present in the multispecies biofilm, whereas D7S hns instead caused a significant alteration of proteome (i.e., upregulation) in S. oralis.
For the species exhibiting different patterns of expressed proteins when co-incubated with D7S and D7S hns, respectively, several potential relationships with A. actinomycetemcomitans have been described. These include evidence that F. nucleatum co-aggregates with A. actinomycetemcomitans among a large number of other oral microorganisms.29 Results obtained with multispecies biofilm models suggest a potential mutualistic relationship, i.e., that A. actinomycetemcomitans and F. nucleatum may support the growth of each other in the oral cavity.30 Moreover, F. nucleatum was found to enhance the attachment and invasion of A. actinomycetemcomitans to epithelial cells.31 As early colonizers of the oral biofilm, Streptococcus spp. were shown to localize on the tooth surface, optimizing the microenvironment for later colonizers.32 Autoinducer-2, a member of a family of signaling molecules for quorum sensing, is expressed by both Streptococcus spp.33 and A. actinomycetemcomitans,34 suggesting that interspecies signaling might occur between A. actinomycetemcomitans and S. oralis. In addition, A. actinomycetemcomitans can digest lactate secreted by Streptococcus spp.35 However, in spite of these potential relationships, very few S. oralis proteins were regulated in the biofilm when the A. actinomycetemcomitans wild-type strain was present. This is consistent with our previous findings with the multispecies biofilm model using an hns+ A. actinomycetemcomitans strain,19 suggesting that the regulatory effect on S. oralis is mediated by factors normally suppressed by H-NS. The large amount of S. oralis proteins that were uniquely identified (Fig. 4b) is also consistent with this hypothesis. Finally, comparing the D7S with the D7S hns biofilm, we observed that all 442 regulated A. actinomycetemcomitans proteins exhibited higher levels in the D7S biofilm (Fig. 5c). This, which would argue against H-NS acting as a repressor in the multispecies biofilm, may be at least partly a result of the impaired growth (i.e., lower abundance) of D7S hns in the biofilm (Figs. 1b and 2). In contrast, F. nucleatum proteins (n = 78) and S. oralis (n = 78) constituted a large majority of the 164 that were downregulated in the D7S versus the D7S hns biofilm.
Different patterns of biological pathways expressed in the multispecies biofilm containing D7S and D7S hns, respectively
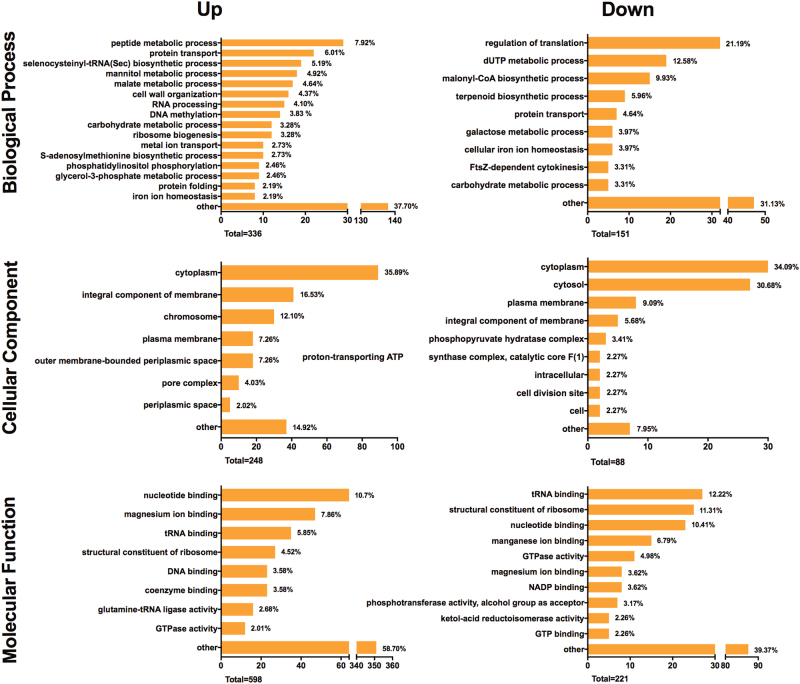
As D7S and D7S hns differentially induced proteomic changes in the species of the multispecies biofilm, we aimed to assess an overview of the potential functional differences of the protein profiles expressed in the two biofilms (i.e., the D7S versus the D7S hns biofilm). To this end, all individual proteins, including also A. actinomycetemcomitans proteins, were grouped according to their GO terms for their predicted biological process, cellular component (i.e., subcellular localization), and molecular function, respectively. The GO terms of all the regulated proteins were collectively pooled to decipher the patterns of biological pathways expressed in the multispecies biofilm containing either strain D7S or D7S hns (Fig. 6). In brief, 366, 248, and 598 GO terms from the biological process, cellular component, and molecular function category, respectively, were generated from the proteins that were upregulated in the D7S biofilm. The enriched GO terms from the downregulated proteins were 151, 88, and 221 from these three domains. In general, both the up- and downregulated proteins in the D7S versus the D7S hns biofilm have diverse functions with 31.13 and 58.70% GO terms enriched in the “other” category in biological process and molecular function domains. This is similar to observations with E. coli that H-NS represses a large number of poorly characterized, unrelated genes.12 Interestingly, the proteins expressed at higher levels in the D7S hns biofilm contained relatively large proportions of GO terms associated with the biological process “regulation of translation” (21.19%), and with the molecular functions “tRNA binding” (12.22%) and “structural constituent of ribosome” (11.31%). Thus, our data suggest that mutational loss of A. actinomycetemcomitans H-NS had a significant impact on the overall translational activity of the biofilm. This would be in accordance with observations that H-NS controls multiple genes involved in transcription and translation in Gram-negative bacteria,12 albeit putative indirect mechanism(s) behind the regulatory effects onto the other species remain to be characterized. As cell–cell physical interactions via pili can moderate bacterial swarming behavior,36 it cannot be excluded that pili could be involved in inter-bacterial interactions, impacting on regulatory pathways controlling protein expression.
Fig. 6.
Functional classification of proteins differentially regulated in the multispecies oral biofilms containing A. actinomycetemcomitans strain D7S and D7S hns, respectively. All up- or downregulated bacterial protein functions in the D7S relative to the D7S hns biofilms were annotated by enrichment of Gene Ontology (GO) terms, and displayed as proportions (%) of the total numbers of regulated (up/down) GO terms. Indicated is the numbers of GO terms for biological process, cellular component, and molecular function, respectively. Sub-domains with their GO proportion less than 2% were classified as “other”
Concluding remarks
In the present work we have used quantitative proteomics to investigate the role of H-NS in gene expression in the A. actinomycetemcomitans model strain D7S, cultured in both mono- and multispecies biofilm settings. We employed the “supragingival” variant of the in vitro multispecies biofilm model, as in the clinical settlement A. actinomycetemcomitans is more likely to establish first in supragingival plaque, before expanding subgingivally. We demonstrated that H-NS acted as a repressor of gene expression in A. actinomyctemcomitans, which is consistent with its role in several other Gram-negative bacteria. Our present study also underscores the importance of this global gene regulator for the behavior of this organism in a multiple species bacterial community. It will be of interest to elucidate the mechanism(s) of how H-NS promotes A. actinomycetemcomitans biofilm formation and contributes to the survival of this species within the multispecies biofilm, which was not revealed from our present proteomics data. Our results are consistent with the notion that H-NS caused qualitative and quantitative proteomic alterations in the multispecies biofilm, and therefore likely contributed to the ecological pressure exerted by A. actinomycetemcomitans onto the other species. Although GO terms associated with translational activity were highly abundant among the proteins upregulated in the hns mutant multispecies biofilm, we also concluded that there was a large amount of H-NS-regulated proteins corresponding to various unrelated functions (i.e., GO terms classified as “other”). These poorly characterized genes may frequently have been acquired by horizontal gene transfer,37 and it cannot be excluded that H-NS may play a role in silencing such genes in A. actinomycetemcomitans, and/or in facilitating their sequence diversification as has been demonstrated in other bacteria.38
Methods
Bacterial strains and growth conditions
A. actinomycetemcomitans wild-type fimbriated strain D7S (serotype a) was originally isolated from a patient with aggressive periodontal disease, and D7SS is a smooth colony derivative of D7S.39 Mutant derivatives of D7S and D7SS with allelic replacement of the hns gene, i.e., D7S hns::kan [Kanr] and D7SS hns::kan [Kanr], were generated in the present work. A. actinomycetemcomitans strains were routinely cultivated in air supplemented with 5% CO2 at 37 °C for 3 days unless otherwise stated. For this we used blood agar plates (5% defibrinated horse blood, 5 mg hemin/l, 10 mg Vitamin K/l, Columbia agar base), on tryptic soy agar plates or tryptic soy broth (TSB), supplemented with 0.6% yeast extract, and 0.8% glucose (Difco). Alternatively, for transformation assays, the strains were grown on trypticase soy broth supplemented with 0.1% yeast extract, 5% heat-inactivated horse serum, and 1.5% agar (sTSB agar), and when needed, supplemented with 100 μg/ml (final concentration) kanamycin. For biofilm growth of A. actinomycetemcomitans strains, 2 × 108 bacterial cells were inoculated in 2 ml tryptic soy broth (Difco) in 24-well cell culture plates (Nunc), which were incubated in static culture in air supplemented with 5% CO2, at 37 °C for 3 days. Biofilms were stained with crystal violet as previously described40 and the amount of bound dye, which is proportional to the biofilm mass, was quantitated by measuring its absorbance at 590 nm. Escherichia coli laboratory strain DH5α41 was used for maintenance of plasmids, and E. coli strains JGJ102 (hns+) and JGJ103 (hns)42 were used as controls in western blot. E. coli strains were cultured aerobically at 37 °C in Luria-Bertani (LB) broth or on LB broth solidified with 1.5% (w/v) agar.
Generation of strain D7S and D7SS hns allelic replacement mutants
A PCR-based approach following standard cloning procedures43 was used to construct hns gene replacement mutants in A. actinomycetemcomitans strains D7SS and D7S. Strain D7S-1 complete genome (GenBank accession CP003496) was used as reference in oligonucleotide synthesis. In brief, PCR fragments flanking the gene locus encoding the H-NS protein (GenBank AFI86019) were amplified using primers H1 (5’-CGCCTTGTAGAAAATCCACGCC-3’) with H2 (5’-CAATCCAAGCAGAATTCGATAAAGGTAAG-3’), and H3 (5’-TACGCAAGCTACGAATTCTCGTTAATATT-3’) with H4 (5’-CTTACACCACCGGTGACTAAAGATAC-3’). The PCR primers H2 and H3 introduced an EcoRI restriction site (underlined sequences), allowing ligation of the PCR fragments to flank the kanamycin resistance gene from pUC4K.44 Ligation products were then used to transform D7SS on agar plates using procedures described earlier.39 The hns::Kan allele of D7SS hns was transferred to D7S, using natural transformation,39 generating D7S hns. Confirmation of allelic replacements and the orientation of the inserted resistance cassette were done by DNA sequencing and PCR. For this we used primer H1 and H4, respectively, in combination with a primer specific for the kanamycin determinant (H7: 5’-GATTTATTCAACAAAGCCGCCGTCC-3’).
Western blot
Standard procedures for sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blot analysis were used.43 For immune detection, we used a rabbit polyclonal antiserum specific for E. coli H-NS45 (final dilution 1:10,000). As secondary antibody, anti-rabbit horseradish peroxidase-conjugate was used (Jackson ImmunoResearch, Newmarket, UK) (1:10,000). Immunoreactive bands were visualized using Clarity™ Western ECL Substrate (Bio-Rad) and the ChemiDoc™ XRS+ System (Bio-Rad).
Atomic force microscopy
For AFM, bacterial cells were diluted with ultrapure water (Millipore) and placed onto a freshly cleaved mica surface. Samples were incubated for 5 min at room temperature, washed with ultrapure water, and then placed in a desiccator for ~2 h in order to dry. The samples were finally magnified through a Nanoscope V Atomic Force Microscope (Bruker AXS GmbH, Karlsruhe, Germany), using tapping mode. Final images were plane fitted in both the x- and y-axes and are presented in amplitude mode.
Multispecies biofilm formation and harvesting
In addition to A. actinomycetemcomitans, the following six oral microbial strains were used in this study: Actinomyces oris (OMZ 745), Candida albicans (OMZ 110), Fusobacterium nucleatum subsp. nucleatum KP-F2 (OMZ 598), Streptococcus oralis SK248 (OMZ 607), Streptococcus mutans UA159 (OMZ 918), and Veillonella dispar ATCC 17748T (OMZ 493). A multiple-species biofilm with the latter 6 species (A. actinomycetemcomitans excluded) was cultivated as previously reported,46 and is in the present work referred to as “control biofilm” in experiments including A. actinomycetemcomitans. Two modified 7-species biofilms, namely the D7S biofilm and the D7S hns-deficient biofilm, were also developed in parallel. Briefly, 200 μl of each species with similar densities (OD550 = 1.0 ± 0.05) were loaded on the hydroxyapatite dishes and anaerobically incubated for 64 h. During the incubation, the cultivated medium was replenished at 16 h and 40 h. The biofilm dishes were dip-washed in 0.9% w/v NaCl at 16 h, 20 h, 40 h, 44 h, 48 h, and 64 h. After being developed, biofilms were then either suspended in 0.9% w/v NaCl for CFU count as well as proteomic analysis, or fixed in 4% paraformaldehyde for image analysis.
Image analysis with CLSM
The paraformaldehyde-fixed biofilms were stained by fluorescence in situ hybridization and subjected to CLSM for imagine analysis. For this we used Act639, cy3-labeled 16S rRNA oligonucleotide probe of A. actinomycetemcomitans (5’-CTCCAGACCCCCAGTATG-3’; formamide concentration: 40%, and NaCl concentration in wash buffer: 46 mM)47 and FUS664, cy5-labeled 16S rRNA oligonucleotide probe of F. nucleatum (5’-CTTGTAGTTCCG C/T ACCTC-3’; formamide concentration: 40%, NaCl concentration in wash buffer: 46 mM).48 YoPro-1 iodide and Sytox Green (1:1 v/v) (Thermo Fisher) were used to counterstain the biofilm following the protocol previously reported.49 All images were captured with a 63× objective (glycerol immersion, NA 1.3, Leica Microsystems) on a Leica sp5 confocal microscope (Leica Microsystems). The filters on microscope were set to 500–540 nm, 570–630 nm, and 660–710 for the detection of colors from YoPro-1 iodide and Sytox Green mixture, Cy3 and Cy5, respectively. The captured images were processed using Imaris software (version 7.4.0, Bitplane) to reconstruct the biofilm.
CFU count on selective plates
The CFU counts were performed to quantify the numbers of individual species in different biofilm models. Biofilm suspensions were diluted into seven 10-fold serial dilutions in 0.9% w/v NaCl to obtain at least one plate containing 20–200 CFUs. Briefly, 50 μl of each diluted suspension was plated using a EDDY Jet Auto Spiral Diluter (IUL instruments). Difco™ mitis salivarius agar plates (Becton, Dickinson and Company) supplemented with 0.001% w/v sodium tellurite (BDH Chemicals Ltd) were used to select S. oralis and S. mutans. Fastidious anaerobe agar plates (Neogen) containing 1 mg/l erythromycin (Sigma-Aldrich), 4 mg/l vancomycin (Sigma-Aldrich), and 1 mg/l norfloxacin were used to selectively grow F. nucleatum. Biggy agar plates (Difco) were used to selectively grow C. albicans. Columbia blood agar plates (Oxoid) supplemented with 5% whole human blood were used to selectively cultivate the remaining species. Ten biological replicates were performed for counting purposes.
Bacterial and biofilm protein extraction
Bacterial lysates for LC-MS/MS were obtained as follows. Multispecies biofilm pellets for control (n = 9), D7S wild-type (n = 10) and D7S hns mutant (n = 10) were collected and lysed from suspensions as previously described.19 Three biological replicates of A. actinomycetemcomitans strain D7S and A. actinomycetemcomitans strain D7S hns grown on blood agar as monospecies biofilm were also processed using the same method. Briefly, sample preparations were randomized and the samples were suspended with 30 μl lysis buffer consisting 4% w/v sodium dodecyl sulfate (SDS), 0.1 mM dithiothreitol, and 100 mM Tris-HCl pH 8.2, heat shocked at 95 °C for 5 min, and then sonicated (UTR2000, Hielscher) 3× 3 min with 0.5 cycle for intervals at 65 % ultrasonic amplitude. The protein concentrations of lysed mixtures were evaluated using Qubit Protein Assay Kit (Life Technologies).
Filter-aided sample preparation digestion and C18 clean-up
Microcon YM-30 centrifugal filter unit (Millipore) was used to lyse the samples and remove SDS contamination following the protocol described previously.19 Briefly, 200 μl of urea buffer containing 8 M urea, 0.1 mM dithiothreitol, and 100 mM Tris/HCl buffer (pH 8.2) were mixed with 20 μg of each lysed sample and loaded in a filter unite. Each of these mixtures was denatured with additional 200 μl of urea buffer, alkylated with 100 μl of 0.05 M iodoacetamide, and washed three times with 100 μl 0.5 M NaCl. Reagents were then completely removed by centrifugation at 14,000 × g for 20 min (or 17 min, for removal of NaCl) at 35 °C. The samples were digested overnight by trypsin (Sigma-Aldrich) in enzyme/protein ratio = 1:50 w/w at room temperature and desalted with StageTips, C18 material, 200 µl tip (Thermo Scientific). The samples were then concentrated using a Speedvac (Thermo Savant SPD121P, Thermo Scientific) and stored at –20 °C until further use.
LC-MS/MS analysis
Each sample was divided into two technical replicates. The desalted samples were reconstituted with 3% acetonitrile in 0.1% formic acid, and a pooled sample of all samples was prepared to serve as an alignment reference in the quantification analyses stage. Randomization for sample run order was applied and the samples were individually analyzed in a Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific), coupled with EASY-Spray™ LC Columns (Thermo Scientific) column and emitter for chromatographic separation, and a linear gradient of acetonitrile/water (2 to 35% acetonitrile in 80 min, containing 0.1% formic acid) with a flow rate of 300 nl/min, and an automatic switching between MS and MS/MS scans using a top-12 method. MS spectra were acquired for a mass range of 300–1700 m/z in profile mode at a resolution of 60,000 at m/z 400. The high collusion-induced dissociation fragmentation was performed on 28 normalized collision energy at high resolution.
Protein identification and label-free quantification
All raw files from LC-MS/MS were searched with Mascot (version 2.5.1) against a database containing 1,535,919 sequences; 551,265,940 residues of human, bacterial, and fungal proteins including those from A. oris, Aggregatibacter aphrophilus, A. actinomycetemcomitans, C. albicans, F. nucleatum, S. mutans, Streptococcus oralis, and V. dispar. All sequences were downloaded from NCBI on 27 May 2016, and concatenated to 261 sequences known as MS contaminants and reversed (decoyed) to generate the search database. The following parameters were set for the database search: tryptic digests, max two missed cleavages for each peptide, iodoacetamide derivative as a fixed modification on cysteine, acetylation on the protein N-term, deamidation on asparagine to glutamine, and oxidation on methionine residues as variable modification. Peptide tolerance was set to ±10 ppm, and MS/MS tolerance to ±0.6 Da. Mascot search results were imported into Scaffold (version 4.2.1, Proteome software) for validation of the MS/MS-based peptide and protein identifications. The following protein identification thresholds were set for the Scaffold research: 3.0% FDR at the protein level, at least 2 minimal peptides, and 1.0% FDR at the peptide level.
Label-free quantification was performed using the ProgenesisQI (for proteomics) software V4.0 (Nonlinear Dynamics, UK) as described previously.19 The comparison was made between the two monospecies A. actinomycetemcomitans biofilm lysates (i.e., D7S wild type versus D7S hns mutant) or between different types of multispecies biofilm lysates (i.e., 6-species biofilm, 7-species biofilm containing D7S wild-type strain, and 7-species biofilm containing D7S hns mutant strain). Briefly, raw files of each individual run were imported to Progenesis and aligned with a pool of all samples as align reference. Then, peak picking was applied to the aligned feature using default settings from Progenesis for feature detection, alignment, and quantification. Up to 6 of the best-ranked ms/ms spectra per aligned peptide ion were exported into a mascot generic file (mgf) using the top 200 peaks and de-isotoping as well as charge deconvolution. Mascot results were loaded into Scaffold. Using filter options of min1 pep, 10% protFDR, and 5% pepFDR, the spectrum report was reimported into ProgenesisQI software. Only unique peptides were included for quantification. For identification, we used all proteins identified with at least two features. Proteins were grouped with ProgenesisQI and only non-conflicting features were used for quantification. For protein quantification, the normalized abundance of all non-conflicting peptide ions from the same protein group were summed together individually for each sample. This generates the normalized quantitative protein abundance. For statistical testing, the parametric test (analysis of variance) on the transformed (hyperbolic arcsine transformation) normalized protein abundance was applied. A heat map was used to obtain a global visualization and assessment of protein expression in different biofilms and to remove obvious outliers (Supplementary Fig. 3). The cluster analysis and heat maps were generated using the R programming language (R Core Team) and additional packages such as quantable and gplots (CRAN). As there were no outliers in the mono-species biofilms, all three replicates were taken under consideration for quantification. In contrast, there were a few samples in the multispecies biofilms could not be associated with any obvious factor and were considered as outliers and removed. The final number of biological replicates used for protein quantification included the following groups: D7S biofilm (n = 6), D7S hns biofilm (n = 8), and 6-species biofilm (n = 7). Quantified proteins with significant raw p value (p < 0.05), at least two unique peptides, and absolute value of log2 fold-change ≥1 were considered as true regulated proteins.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE50 partner repository with the dataset identifier PXD008444.
Ontology analysis
GO terms from all regulated proteins for each comparison were put together to estimate the role of A. actinomycetemcomitans to entire biofilms or to their co-cultured species within the biofilm. The GO lists were generated with Uniprot (released on May 2017). For this, “Retrieve/ID Mapping” function was utilized with redundant terms removed based on analyses from REVIGO (released on May 2017) using the “small (0.5)” similarities. These GO lists were then manually summarized based on the defined classification of gene nomenclatures (molecular functional, biology process, and cellular component) on GO terms and presented in bar charts. The GO terms representing less than 2% of the whole GO in each domain were clustered into the category “other”.
Statistical analysis
Unpaired t-test or a one-way ANOVA was used to calculate the statistical significances of the microbiological data from biofilm growth and CFU counting. For the latter, Bonferroni post hoc test was used for comparisons between the individual groups (Prism v.6 software GraphPad). The data were considered significant at p < 0.05. The results are presented as means ± standard deviations.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD008444. The authors declare that all other data supporting the findings of this study are available within the article and its Supplementary Information, or upon request from the corresponding author.
Electronic supplementary material
Acknowledgements
The authors would like to thank Dr. Nathalie Selevsek and Dr. Peter Gehrig at the Functional Genomics Center Zürich for their support on the LC-MS/MS. We are also grateful to Manuela Flury, Elisabeth Granström, and Elpida Plattner for their excellent technical assistance. We thank Dr. Bernt Eric Uhlin for kindly providing the rabbit polyclonal antiserum specific for E. coli H-NS. This work was supported by the authors’ Institutional funds, by TUA grants from the County Council of Västerbotten, Sweden (to J.O.), and by funds from Insamlingsstiftelsen, Medical Faculty, Umeå University (to J.O.).
Author contributions
Conceived and designed the study: K.B., N.B., G.N.B., and J.O.; performed the laboratory experiments: K.B., T.T., B.T., and J.O.; performed the proteomic analysis: K.B., J.G., and W.E.W.; performed the bioinformatics analyses: K.B., J.G., W.E.W., and N.B.; wrote the paper: K.B., N.B., G.N.B., and J.O.; critically reviewed the paper: T.T. J.G., and W.E.W.
Competing interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies the paper on the npj Biofilms and Microbiomes website (10.1038/s41522-018-0055-4).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Henderson B, Ward JM, Ready D. Aggregatibacter (Actinobacillus) actinomycetemcomitans: a triple A* periodontopathogen? Periodontol 2000. 2010;54:78–105. doi: 10.1111/j.1600-0757.2009.00331.x. [DOI] [PubMed] [Google Scholar]
- 2.Figuero E, et al. Quantification of periodontal pathogens in vascular, blood, and subgingival samples from patients with peripheral arterial disease or abdominal aortic aneurysms. J. Periodontol. 2014;85:1182–1195. doi: 10.1902/jop.2014.130604. [DOI] [PubMed] [Google Scholar]
- 3.Konig MF, et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci. Transl. Med. 2016;8:369ra176. doi: 10.1126/scitranslmed.aaj1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbert BA, Novince CM, Kirkwood KL. Aggregatibacter actinomycetemcomitans, a potent immunoregulator of the periodontal host defense system and alveolar bone homeostasis. Mol. Oral. Microbiol. 2016;31:207–227. doi: 10.1111/omi.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fine DH, Furgang D, Kaplan J, Charlesworth J, Figurski DH. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral. Biol. 1999;44:1063–1076. doi: 10.1016/S0003-9969(99)00089-8. [DOI] [PubMed] [Google Scholar]
- 6.Kachlany SC, et al. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 2001;40:542–554. doi: 10.1046/j.1365-2958.2001.02422.x. [DOI] [PubMed] [Google Scholar]
- 7.Schreiner HC, et al. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc. Natl. Acad. Sci. USA. 2003;100:7295–7300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izano EA, et al. Poly-N-acetylglucosamine mediates biofilm formation and detergent resistance in Aggregatibacter actinomycetemcomitans. Microb. Pathog. 2008;44:52–60. doi: 10.1016/j.micpath.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tendeng C, Bertin PN. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol. 2003;11:511–518. doi: 10.1016/j.tim.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Singh K, Milstein JN, Navarre WW. Xenogeneic silencing and its impact on bacterial genomes. Annu. Rev. Microbiol. 2016;70:199–213. doi: 10.1146/annurev-micro-102215-095301. [DOI] [PubMed] [Google Scholar]
- 11.Park HS, Östberg Y, Johansson J, Wagner EG, Uhlin BE. Novel role for a bacterial nucleoid protein in translation of mRNAs with suboptimal ribosome-binding sites. Genes Dev. 2010;24:1345–1350. doi: 10.1101/gad.576310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hommais F, et al. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Ayala JC, Benitez JA, Silva AJ. RNA-seq analysis identifies new genes regulated by the histone-like nucleoid structuring protein (H-NS) affecting Vibrio cholerae virulence, stress response and chemotaxis. PLoS One. 2015;10:e0118295. doi: 10.1371/journal.pone.0118295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan B, et al. Global transcriptional regulation by H-NS and its biological influence on the virulence of Enterohemorrhagic Escherichia coli. Gene. 2016;588:115–123. doi: 10.1016/j.gene.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Ayala JC, Silva AJ, Benitez JA. The histone-like nucleoid structuring protein (H-NS) is a repressor of Vibrio cholerae exopolysaccharide biosynthesis (vps) genes. Appl. Environ. Microbiol. 2012;78:2482–2488. doi: 10.1128/AEM.07629-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosse JT, et al. Regulation of pga operon expression and biofilm formation in Actinobacillus pleuropneumoniae by sigmaE and H-NS. J. Bacteriol. 2010;192:2414–2423. doi: 10.1128/JB.01513-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belik AS, Tarasova NN, Khmel IA. [Regulation of biofilm formation in Escherichia coli K12: effect of mutations in HNS, StpA, lon, and rpoN genes] Mol. Gen. Mikrobiol. Virusol. 2008;4:3–5. [PubMed] [Google Scholar]
- 18.Hong SH, Wang X, Wood TK. Controlling biofilm formation, prophage excision and cell death by rewiring global regulator H-NS of Escherichia coli. Microb. Biotechnol. 2010;3:344–356. doi: 10.1111/j.1751-7915.2010.00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao K, Bostanci N, Selevsek N, Thurnheer T, Belibasakis GN. Quantitative proteomics reveal distinct protein regulations caused by Aggregatibacter actinomycetemcomitans within subgingival biofilms. PLoS One. 2015;10:e0119222. doi: 10.1371/journal.pone.0119222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouokam JC, et al. Active cytotoxic necrotizing factor 1 associated with outer membrane vesicles from uropathogenic Escherichia coli. Infect. Immun. 2006;74:2022–2030. doi: 10.1128/IAI.74.4.2022-2030.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kittichotirat W, Bumgarner RE, Asikainen S, Chen C. Identification of the pangenome and its components in 14 distinct Aggregatibacter actinomycetemcomitans strains by comparative genomic analysis. PLoS One. 2011;6:e22420. doi: 10.1371/journal.pone.0022420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juarez A, et al. Interaction of the nucleoid-associated proteins Hha and H-NS to modulate expression of the hemolysin operon in Escherichia coli. Adv. Exp. Med. Biol. 2000;485:127–131. doi: 10.1007/0-306-46840-9_17. [DOI] [PubMed] [Google Scholar]
- 23.Li H, Granat A, Stewart V, Gillespie JR. RpoS, H-NS, and DsrA influence EHEC hemolysin operon (ehxCABD) transcription in Escherichia coli O157:H7 strain EDL933. FEMS Microbiol. Lett. 2008;285:257–262. doi: 10.1111/j.1574-6968.2008.01240.x. [DOI] [PubMed] [Google Scholar]
- 24.Theilacker C, et al. Deletion of the glycosyltransferase bgsB of Enterococcus faecalis leads to a complete loss of glycolipids from the cell membrane and to impaired biofilm formation. BMC Microbiol. 2011;11:67. doi: 10.1186/1471-2180-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chai Y, Beauregard PB, Vlamakis H, Losick R, Kolter R. Galactose metabolism plays a crucial role in biofilm formation by Bacillus subtilis. mBio. 2012;3:e00184–e00112. doi: 10.1128/mBio.00184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Bentzmann S, Aurouze M, Ball G, Filloux A. FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J. Bacteriol. 2006;188:4851–4860. doi: 10.1128/JB.00345-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang F, et al. ArcA controls metabolism, chemotaxis, and motility contributing to the pathogenicity of avian pathogenic Escherichia coli. Infect. Immun. 2015;83:3545–3554. doi: 10.1128/IAI.00312-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisser H, et al. An automated pipeline for high-throughput label-free quantitative proteomics. J. Proteome Res. 2013;12:1628–1644. doi: 10.1021/pr300992u. [DOI] [PubMed] [Google Scholar]
- 29.Kolenbrander PE, Andersen RN, Moore LV. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect. Immun. 1989;57:3194–3203. doi: 10.1128/iai.57.10.3194-3203.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karched M, Bhardwaj RG, Asikainen SE. Coaggregation and biofilm growth of Granulicatella spp. with Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans. BMC Microbiol. 2015;15:114. doi: 10.1186/s12866-015-0439-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, et al. Coinfection with Fusobacterium nucleatum can enhance the attachment and invasion of Porphyromonas gingivalis or Aggregatibacter actinomycetemcomitans to human gingival epithelial cells. Arch. Oral. Biol. 2015;60:1387–1393. doi: 10.1016/j.archoralbio.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 32.Teles FR, et al. Early microbial succession in redeveloping dental biofilms in periodontal health and disease. J. Periodontal Res. 2012;47:95–104. doi: 10.1111/j.1600-0765.2011.01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rickard AH, et al. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 2006;60:1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x. [DOI] [PubMed] [Google Scholar]
- 34.Shao H, Lamont RJ, Demuth DR. Autoinducer 2 is required for biofilm growth of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect. Immun. 2007;75:4211–4218. doi: 10.1128/IAI.00402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown SA, Whiteley M. A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J. Bacteriol. 2007;189:6407–6414. doi: 10.1128/JB.00554-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anyan ME, et al. Type IV pili interactions promote intercellular association and moderate swarming of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2014;111:18013–18018. doi: 10.1073/pnas.1414661111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kittichotirat W, Bumgarner RE, Chen C. Evolutionary Divergence of Aggregatibacter actinomycetemcomitans. J. Dent. Res. 2016;95:94–101. doi: 10.1177/0022034515608163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higashi K, et al. H-NS facilitates sequence diversification of horizontally transferred DNAs during their integration in host chromosomes. PLoS Genet. 2016;12:e1005796. doi: 10.1371/journal.pgen.1005796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Goodman SD, Redfield RJ, Chen C. Natural transformation and DNA uptake signal sequences in Actinobacillus actinomycetemcomitans. J. Bacteriol. 2002;184:3442–3449. doi: 10.1128/JB.184.13.3442-3449.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaplan JB, Meyenhofer MF, Fine DH. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J. Bacteriol. 2003;185:1399–1404. doi: 10.1128/JB.185.4.1399-1404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 42.Johansson J, Dagberg B, Richet E, Uhlin BE. H-NS and StpA proteins stimulate expression of the maltose regulon in Escherichia coli. J. Bacteriol. 1998;180:6117–6125. doi: 10.1128/jb.180.23.6117-6125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook, J. E., Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, New York, 1989).
- 44.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 45.Johansson J, Uhlin BE. Differential protease-mediated turnover of H-NS and StpA revealed by a mutation altering protein stability and stationary-phase survival of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1999;96:10776–10781. doi: 10.1073/pnas.96.19.10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thurnheer T, van der Ploeg JR, Giertsen E, Guggenheim B. Effects of Streptococcus mutans gtfC deficiency on mixed oral biofilms in vitro. Caries Res. 2006;40:163–171. doi: 10.1159/000091065. [DOI] [PubMed] [Google Scholar]
- 47.Thurnheer T, Belibasakis GN. Integration of non-oral bacteria into in vitro oral biofilms. Virulence. 2015;6:258–264. doi: 10.4161/21505594.2014.967608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thurnheer T, Gmur R, Guggenheim B. Multiplex FISH analysis of a six-species bacterial biofilm. J. Microbiol. Methods. 2004;56:37–47. doi: 10.1016/j.mimet.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Bao K, et al. Role of Porphyromonas gingivalis gingipains in multi-species biofilm formation. BMC Microbiol. 2014;14:258. doi: 10.1186/s12866-014-0258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vizcaino JA, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD008444. The authors declare that all other data supporting the findings of this study are available within the article and its Supplementary Information, or upon request from the corresponding author.