SUMMARY
Mycobacterium tuberculosis (Mtb), the etiological agent of tuberculosis (TB), is recognized as a global health emergency as promoted by the World Health Organization. Over 1 million deaths per year, along with the emergence of multi- and extensively-drug resistant strains of Mtb, have triggered intensive research into the pathogenicity and biochemistry of this microorganism, guiding the development of anti-TB chemotherapeutic agents. The essential mycobacterial cell wall, sharing some common features with all bacteria, represents an apparent ‘Achilles heel’ that has been targeted by TB chemotherapy since the advent of TB treatment. This complex structure composed of three distinct layers, peptidoglycan, arabinogalactan and mycolic acids, is vital in supporting cell growth, virulence and providing a barrier to antibiotics. The fundamental nature of cell wall synthesis and assembly has rendered the mycobacterial cell wall as the most widely exploited target of anti-TB drugs. This review provides an overview of the biosynthesis of the prominent cell wall components, highlighting the inhibitory mechanisms of existing clinical drugs and illustrating the potential of other unexploited enzymes as future drug targets.
Key words: tuberculosis, cell wall, peptidoglycan, arabinogalactan, mycolic acids, antibiotics
INTRODUCTION
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), is regarded as the world's most successful pathogen (Hingley-Wilson et al. 2003). Responsible for an estimated 1·4 million deaths and 10·4 million new cases of TB, including 480 000 new cases of multi-drug resistant (MDR)-TB in 2015 (World Health Organization, 2016), Mtb remains a global health emergency as declared by the World Health Organization (WHO) (World Health Organization, 2014). New chemotherapeutic agents to complement or replace existing front-line treatment regimens are urgently required to reduce treatment time (currently 6-month course) and to combat the increasing threat by this microorganism.
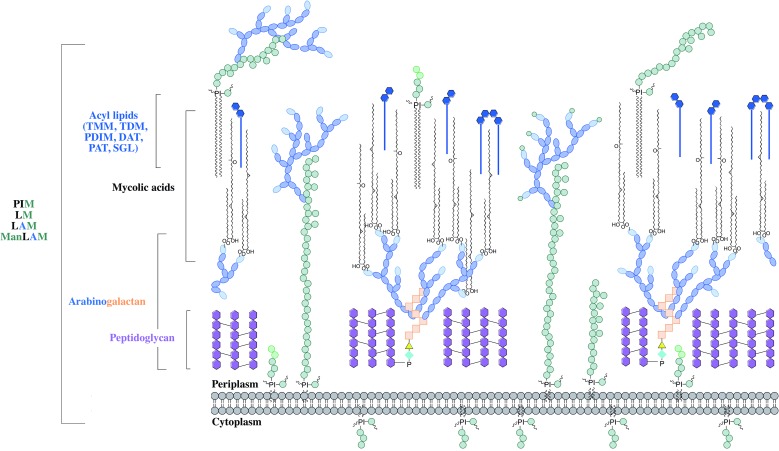
The distinguishing feature of mycobacteria, the complex cell wall, is a well-recognized drug target. The cell wall is common to all bacteria, both Gram-positive and Gram-negative, but can have vast differences in terms of the biochemical and structural features. Over the past decade, extensive research into cell wall assembly, aided by whole-genome sequencing, has led to an increased understanding of mycobacterial cell wall biosynthesis. This has promoted further exploration into the discovery and development of chemotherapeutic agents (from an enzymatic and phenotypic perspective) directed against the synthesis of this unique macromolecule structure in Mtb. The Mtb cell envelope is an expansive structure and is summarized in Fig. 1. The inner membrane phospholipid bilayer contains glycolipids that extend into the periplasmic space. The essential core cell wall structure is composed of three main components: a cross-linked polymer of peptidoglycan, a highly branched arabinogalactan polysaccharide, and long-chain mycolic acids. Intercalated into the mycolate layer are solvent-extractable lipids including non-covalently linked glycophospholipids and inert waxes, forming the outer membrane. The capsule forms the outermost layer and is mainly composed of proteins and polysaccharides. The lipid- and carbohydrate-rich layers of the cell wall serve not only as a permeability barrier, providing protection against hydrophilic compounds, but also are critical in pathogenesis and survival. It is these traits that make the biosynthesis and assembly of the cell wall components attractive drug targets. This review focuses on the synthesis of the key cell wall components, highlighting previously validated targets and the ongoing drug discovery efforts to inhibit other essential enzymes in mycobacterial cell wall biosynthesis.
Fig. 1.
The mycobacterial cell wall. A schematic representation of the mycobacterial cell wall, depicting the prominent features, including the glycolipids (PIMs, phosphatidyl-myo-inositol mannosides; LM, lipomannan; LAM, lipoarabinomannan; ManLAM, mannosylated lipoarabinomannan), peptidoglycan, arabinogalactan and mycolic acids. Intercalated into the mycolate layer are the acyl lipids (including TMM, trehalose monomycolate; TDM, trehalose dimycolate; DAT, diacyltrehalose; PAT, polyacyltrehalose; PDIM, phthiocerol dimycocerosate; SGL, sulfoglycolipid). The capsular material is not illustrated.
PEPTIDOGLYCAN
Peptidoglycan is a major component of the cell wall of both Gram-positive and Gram-negative bacteria (Vollmer et al. 2008). It is a polymer of alternating N-acetylglucosamine and N-acetylmuramic acid residues via β(1 → 4) linkages with side chains of amino acids cross-linked by transpeptide bridges (Brennan and Nikaido, 1995). Mycobacterial peptidoglycan has a number of unique features that diversifies the cell wall from the typical structure including N-glycolyl- and N-acetyl-muramic acid residues (Mahapatra et al. 2005a), amidation of the carboxylic acids in the peptide stems (Mahapatra et al. 2005b) and additional glycine or serine residues (Vollmer et al. 2008). The function of peptidoglycan is not only to provide shape and rigidity, but it is responsible for counteracting turgor pressure and hence it is essential for growth and survival (Vollmer et al. 2008). Peptidoglycan is unique to bacterial cells, and it is this property that has led to numerous enzymes involved in its synthesis to be targeted by potent antibiotics, with others representing attractive targets in the development of future antibiotics.
PEPTIDOGLYCAN BIOSYNTHESIS
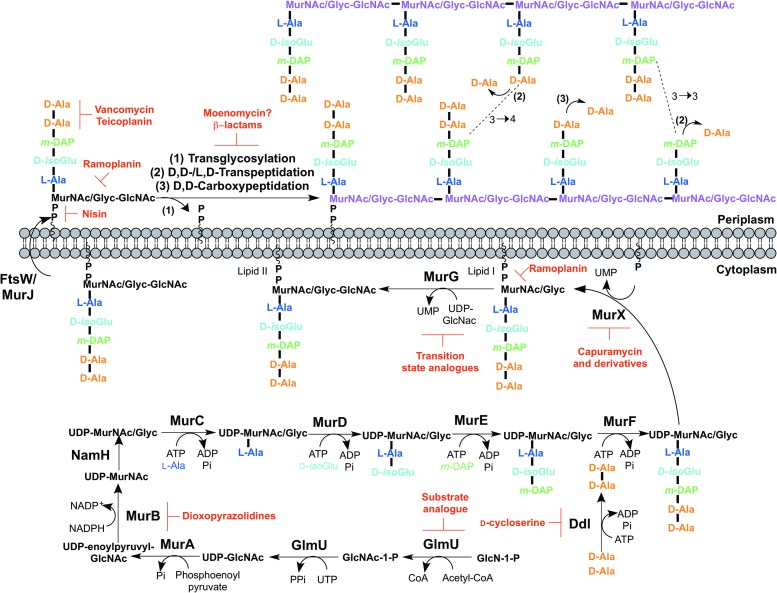
The biosynthesis of peptidoglycan is summarized in Fig. 2. The first committed step is the generation of uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc). This is catalysed by the acetyltransferase and uridyltransferase activities of GlmU (Zhang et al. 2009), where first the acetyl group from acetyl-CoA is transferred to glucosamine-1-phosphate (GlcN-1-P) to produce N-acetylglucosamine-1-phosphate (GlcNAc-1-P). Secondly, uridine-5′-monophosphate from UTP is transferred to GlcNAc-1-P to yield UDP-GlcNAc (Zhang et al. 2009). The abundance of GlcNAc-1-P in eukaryotes (Mio et al. 1998) and the functional similarity of the GlmU uridyltransferase with human enzymes (Peneff et al. 2001) makes this domain an unsuitable drug target (Rani and Khan, 2016). However, the absence of GlcN-1-P from humans makes the acetyltransferase domain a potential target (Mio et al. 1998). Efforts to identify inhibitors of this domain are underway (Tran et al. 2013). A substrate analogue of GlcN-1-P has been designed and exhibits inhibitory effect against GlmU, providing a candidate for further optimization (Li et al. 2011).
Fig. 2.
Inhibitors targeting peptidoglycan biosynthesis. The roles of the key enzymes involved in peptidoglycan biosynthesis are illustrated. Reported inhibitors are shown in red.
The next step involves the generation of the UDP-N-acetylmuramic acid (UDP-MurNAc)-pentapeptide, which is synthesized in a sequential pathway catalysed by the Mur ligases A–F (Barreteau et al. 2008), whereby most of the Mtb genes have been found through homology. MurA, a UDP-N-acetylglucosamine 1-carboxyvinyltransferase, and MurB, a UDP-N-acetylenolpyruvoylglucosamine reductase, are involved in generating UDP-MurNAc from UDP-GlcNAc, by first the addition of the enoylpyruvyl moiety of PEP, followed by reduction to a lactoyl ether moiety via NADPH. At this point, NamH, a UDP-N-acetylmuramic acid hydroxylase, hydroxylates UDP-MurNAc to UDP-N-glycolylmuramic acid (UDP-MurNGlyc), providing both types of UDP-muramyl substrates; Mtb cell walls are dominated by the latter (Mahapatra et al. 2005a). This structural modification is unique to mycobacteria (and closely related genera) and is considered to increase the intrinsic strength of peptidoglycan, by potentially alleviating susceptibility to lysozyme and providing sites for additional hydrogen bonding (Raymond et al. 2005). Inhibitors of Mtb MurA and MurB are yet to be discovered. Whilst the natural product, broad spectrum antibiotic, fosfomycin, targets Gram-negative MurA, the critical residue for inhibition is absent in Mtb, providing intrinsic resistance against this antibiotic (Kim et al. 1996). Consequently, an inhibitor with a new mode of action is required to target Mtb MurA. A limited number of inhibitors have been reported against MurB. Molecular dynamics and docking studies of existing MurB inhibitors (3,5-dioxopyrazolidine derivatives) onto the Mtb MurB structure reveal the potential potent activity of these compounds, which can be used to guide future structure-based drug design (Kumar et al. 2011). Inhibitors of NamH have not been documented; namH is not essential in Mycobacterium smegmatis, and therefore is not condusive to a characteristic target property. However, gene deletion results in a strain hypersusceptible to β-lactam antibiotics and lysozyme and therefore inhibitors of NamH could potentiate the effect of β-lactams (Raymond et al. 2005).
The pentapeptide chain is incorporated onto the UDP-MurNAc/Glyc substrates by the successive addition of amino acid residues L-alanine, D-isoglutamate, meso-diaminopimelate (m-DAP) and D-alanyl-D-alanine [generated by the D-Ala: D-Ala ligase (Ddl)] by the ATP-dependent Mur ligases C-F respectively (Munshi et al. 2013). This results in the muramyl-pentapeptide product, UDP-MurNAc/Glyc-L-Ala-D-isoGlu-m-DAP-D-Ala-D-Ala, also known as Park's nucleotide (Kurosu et al. 2007). Despite the different amino acid specificities, the four ligases share common properties: the reaction mechanism; six invariant ‘Mur’ residues; an ATP-binding consensus; three-dimensional structural domains (Barreteau et al. 2008). Due to these similarities, it is plausible that a single inhibitor could target more than one Mur ligase and such inhibitors have been reported in the literature (Tomasic et al. 2010). Numerous small molecule inhibitors of the Mur ligases have been discovered and are the subject of an extensive review (Hrast et al. 2014). In most cases, the inhibitors were identified from high-throughput screening (HTS) campaigns of compound libraries employing in vitro kinetic assays. These types of in vitro screening methods are limited in use against Mtb Mur ligases given that only MurC and MurE have been biochemically characterized (Mahapatra et al. 2000; Li et al. 2011). This dictates the next rational step towards the target-based discovery of Mur ligase inhibitors. Ddl is the target of D-cycloserine (Bruning et al. 2011), a second-line drug used in the treatment of TB, and is at the cornerstone of treatment for MDR and extensively drug resistant (XDR)-TB. D-cycloserine acts as a structural analogue of D-Ala, inhibiting the binding of either D-Ala to Ddl (Prosser and de Carvalho, 2013a, b).
The first membrane-anchored peptidoglycan precursor is generated by the translocation of Park's nucleotide to decaprenyl phosphate (C50-P), catalysed by MurX (also known as MraY), forming Lipid I (Kurosu et al. 2007). There are a number of nucleoside-based complex natural products that inhibit MurX, including muraymycin, liposidomycin, caprazamycin and capuramycin (Dini, 2005). Capuramycin and derivatives exhibit killing in vitro and in vivo and more significantly, analogues of capuramycin have been shown to kill non-replicating Mtb, a feature not common to the majority of cell wall biosynthesis inhibitors (Koga et al. 2004; Reddy et al. 2008; Nikonenko et al. 2009; Siricilla et al. 2015). Significantly, the analogue SQ641 is in preclinical development (http://www.newtbdrugs.org).
The final intracellular step of peptidoglycan synthesis is performed by the glycosyltransferase, MurG. A β(1 → 4) linkage between GlcNAc (from UDP-GlcNAc) and MurNAc/Glyc of Lipid I is formed, leading to the generation of Lipid II, the monomeric building block of peptidoglycan (Mengin-Lecreulx et al. 1991). A library of transition state mimics have been designed for Escherichia coli MurG, and tested against Mtb MurG with partial success, one being the first inhibitor identified against the Mtb enzyme (Trunkfield et al. 2010).
The enzyme catalysing the translocation of Lipid II across the plasma membrane has been the subject of much debate. To date, there is evidence for two different enzymes with ‘flippase’ activity: MurJ and FtsW (Ruiz, 2008, 2015; Mohammadi et al. 2011, 2014; Sham et al. 2014). Further biochemical characterization is required to confirm the identification of the ‘flippase’. Inhibitors against this enzyme would be expected to exhibit broad-spectrum activity, targeting a vital activity in all bacteria.
Following translocation across the plasma membrane, Lipid II is polymerized by the monofunctional and bifunctional Penicillin-binding proteins (PBPs) (Sauvage et al. 2008). Bifunctional PBPs (PonA1/PBP1 and PonA2/PBP2) possess transglycosylase and transpeptidase domains. The former domain is responsible for linking the disaccharide building blocks of Lipid II to the pre-existing glycan chains (with the concomitant release of decaprenyl pyrophosphate), whereas the latter domain catalyses the formation of the classical (3 → 4) cross-links, between m-DAP and D-Ala of the adjacent pentapeptide chains, with the cleavage of the terminal D-Ala. D,D-transpeptidation and D,D-carboxypeptidation is performed by the monofunctional PBPs, both resulting in the cleavage of the terminal D-Ala of the peptide stem (Goffin and Ghuysen, 2002). Only 20% of the cross-links in Mtb peptidoglycan are (3 → 4) (Kumar et al. 2012). The majority are (3 → 3) links between two tetrapeptide stems, with the release of the fourth position D-Ala (Lavollay et al. 2008). This reaction is catalysed by the L,D-transpeptidases, with D,D-carboxypeptidation as a prerequisite activity. The L,D-transpeptidases are structurally unrelated to PBPs, with different active site residues (cysteine and serine, respectively) (Mainardi et al. 2005; Biarrotte-Sorin et al. 2006). The β-lactam antibiotics have been used in the treatment of bacterial infections for nearly a century, and gave rise to the discovery of their target, the PBPs. The L,D-transpeptidases are resistant to most β-lactam antibiotics, except the carbapenems (Dubee et al. 2012). Until recently, β-lactams were not considered for use in the treatment of TB, due to the expression of a broad-spectrum β-lactamase, BlaC. However, it has been shown that BlaC is irreversibly inactivated by clavulanic acid, yet hydrolyses carbapenems at a low rate (Hugonnet et al. 2009). Combined treatment of the β-lactam with the β-lactamase inhibitor has been shown to be bactericidal against both replicating and non-replicating forms of Mtb, and combinations are now being explored in clinical trials (Hugonnet et al. 2009; Rullas et al. 2015). A well-documented inhibitor of the transglycosylase of PBPs, moenomycin (van Heijenoort et al. 1987), a natural product glycolipid, is yet to have proven efficacy against Mtb.
The inhibitors discussed thus far directly target the enzymes involved in peptidoglycan biosynthesis. There are, however, other antibiotics that act on the peptidoglycan precursors. For example, the glycopeptides, vancomycin and teicoplanin, bind to the D-Ala-D-Ala terminus of the pentapeptide stem, preventing polymerization reactions (Reynolds, 1989). Members of the lantibiotic family of antibiotics, such as nisin, interact with the pyrophosphate moiety of Lipid II, forming a pore in the cytoplasmic membrane, but also inhibiting peptidoglycan biosynthesis (Wiedemann et al. 2001). The lipoglycodepsipeptide ramoplanin inhibits the action of MurG by binding to Lipid I. Ramoplanin also binds to Lipid II, preventing its polymerization (Lo et al. 2000).
ARABINOGALACTAN
The major cell wall polysaccharide, arabinogalactan (Fig. 1), as the name suggests, is composed of galactose and arabinose sugar residues, in the furanose (f) ring form (Galf) (McNeil et al. 1987). Arabinogalactan is attached to peptidoglycan via a single linker unit (McNeil et al. 1990). The galactan component is a linear chain of approximately 30 alternating 5- and 6-linked β-D-Galf residues (Daffe et al. 1990). Three highly branched arabinan chains, consisting of approximately 30 Araf residues, are attached to the galactan chain (Besra et al. 1995). The non-reducing termini of the arabinan chains act as an attachment site for mycolic acids, succinyl and galactosamine (D-GalN) moieties (Draper et al. 1997; Bhamidi et al. 2008).
ARABINOGALACTAN BIOSYNTHESIS
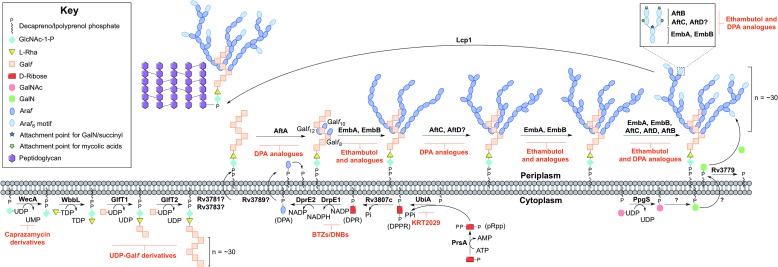
Arabinogalactan biosynthesis is illustrated in Fig. 3. The first committed step begins in the cytoplasm and proceeds by the formation of the linker unit connecting peptidoglycan to arabinogalactan, which is initiated by WecA, a GlcNAc-1-P transferase (Jin et al. 2010). This enzyme catalyses the transfer of GlcNAc-1-P to C50-P. WbbL, a rhamnosyltransferase catalyses the transfer of L-rhamnose (L-Rha) from dTDP-L-Rha to position 3 of C50-P-P-GlcNAc to form C50-P-P-GlcNAc-L-Rha, completing the linker unit (McNeil et al. 1990; Mills et al. 2004). WecA has been identified as the target of caprazamycin derivatives, such as CPZEN-45, with the original nucleoside antibiotic shown to target MraY (Ishizaki et al. 2013). Recently, a fluorescence-based assay for WecA activity has been developed and used to screen compound libraries with some success (Mitachi et al. 2016). Inhibitors targeting WbbL have yet to be identified. This essential enzyme, present in all mycobacteria, is recognized as a promising target and efforts are underway to characterize the enzyme via the establishment of a microtiter plate-based assay for its activity, which could be exploited in inhibitor library screening (Grzegorzewicz et al. 2008).
Fig. 3.
Inhibitors targeting arabinogalactan biosynthesis. The current understanding of the roles of enzymes involved in arabinogalactan biosynthesis. Reported inhibitors are shown in red.
The linker unit provides an attachment point for the polymerization of the galactan chain. This process also occurs in the cytoplasm. The bifunctional galactofuranosyltransferases (GlfT1 and GlfT2) (Alderwick et al. 2008) are responsible for the synthesis of the linear galactan chain. Initially, GlfT1 transfers Galf from UDP-Galf to the C-4 position of L-Rha, and then adds a second Galf residue to the C-5 position of the primary Galf, generating C50-P-P-GlcNAc-L-Rha-Galf2 (Mikusova et al. 2006; Alderwick et al. 2008; Belanova et al. 2008). GlfT2 sequentially transfers Galf residues to the growing galactan chain with alternating β(1 → 5) and β(1 → 6) glycosidic linkages (Kremer et al. 2001a; Rose et al. 2006). The galactan chains contain ~30 Galf residues in vivo, forming C50-P-P-GlcNAc-L-Rha-Galf30 (Daffe et al. 1990), but the chain length determination mechanism is yet to be fully understood. GlfT1 and GlfT2 are suitable targets, as rationalized by an in silico target identification program (Raman et al. 2008). UDP-Galf derivatives, with modifications to the C-5 and C-6 positions have been investigated as suitable inhibitors of these enzymes, whereby they cause premature galactan chain termination (Peltier et al. 2010).
The remainder of arabinogalactan synthesis occurs on the outside of the cell. Although the transport mechanism of this cell wall polysaccharide is not fully understood, Rv3781 and Rv3783, encoding an ABC transporter, are potential ‘flippase’ candidates (Dianiskova et al. 2011). Araf residues are transferred directly onto C50-P-P-GlcNAc-L-Rha-Galf30 from the lipid donor decaprenylphosphoryl-D-arabinose (DPA) (Wolucka et al. 1994). DPA is synthesized through a series of cytoplasmic steps, and originates exclusively from phospho-α-D-ribosyl-1-pyrophosphate (pRpp), prior to reorientation to the extracellular face of the plasma membrane. The pRpp synthetase, PrsA, catalyses the transfer of pyrophosphate from ATP to C-1 of ribose-5-phosphate, forming pRpp (Alderwick et al. 2011b). A decaprenyl moiety is added, catalysed by UbiA (decaprenol-1-phosphate 5-phosphoribosyltransferase), forming decaprenol-1-monophosphate 5-phosphoribose (Alderwick et al. 2005; Huang et al. 2005, 2008). Rv3807c encodes a putative phospholipid phosphatase, which catalyses C-5 dephosphorylation, generating decaprenol-1-phosphoribose (DPR) (Jiang et al. 2011). Finally, DPA is generated by an epimerization reaction of the ribose C-2 hydroxyl, catalysed by a two-step oxidation/reduction activity of the decaprenylphosphoribose-2′-epimerase consisting of subunits DprE1 and DprE2 (Mikusova et al. 2005).
The DPA synthetic pathway is a validated drug target. The nitro-benzothiazinones (BTZs) and the structurally related dinitrobenzamides target DprE1 and are effective against MDR and XDR strains of Mtb with low toxicity (Christophe et al. 2009; Batt et al. 2012; Makarov et al. 2014, 2015). The success of these compounds has led to the study of the other enzymes as potential drug targets. Conditional knockdown mutants of dprE1, dprE2, ubiA, prsA and Rv3807c have proven the essentiality of all except Rv3807c, and a target-based whole-cell screen has been developed using these strains of reduced expression levels to identify enzyme-specific inhibitors. Inhibitors targeting a particular enzyme cause increased sensitivity and this was confirmed with BTZ and KRT2029 targeting DprE1 and UbiA, respectively, and can be the subject of future medicinal chemistry efforts (Kolly et al. 2014).
The mechanism of DPA reorientation into the periplasm is unknown. The ‘flippase’ was recently considered to be Rv3789, but there is evidence that this protein plays a different role: to act as an anchor protein to recruit AftA (Kolly et al. 2015). AftA is the first arabinofuranosyltransferase (AraT), of a predicted six, to commence the addition of arabinose from DPA onto the galactan chain (Alderwick et al. 2006). AftA transfers a single Araf residue onto C-5 of β(1 → 6) Galf residues 8, 10 and 12 of C50-P-P-GlcNAc-L-Rha-Galf30 (Alderwick et al. 2005). EmbA and EmbB, so called because their discovery was based on the mode of action elucidation of ethambutol (EMB), catalyse the addition of further α(1 → 5) Araf polymerization (Alderwick et al. 2005). AftC introduces α(1 → 3) branching (Birch et al. 2008), with AftD having an equivalent role (Skovierova et al. 2009). The structure terminates in a well-defined hexa-arabinofuranosyl (Araf6) structural motif: [β-D-Araf-(1 → 2)-α-D-Araf]2-3,5-α-D-Araf-(1 → 5)-α-D-Araf. This motif is generated by EmbA, EmbB, AftC, AftD and AftB (Escuyer et al. 2001; Alderwick et al. 2005; Birch et al. 2008, 2010; Skovierova et al. 2009). AftB catalyses the transfer of the terminal β(1 → 2) Araf residues (Seidel et al. 2007). C-5 of the terminal β-D-Araf and the penultimate 2-α-D-Araf of this motif act as anchoring points for mycolic acids (McNeil et al. 1991).
The Emb arabinosyltransferases are inhibited by EMB, a well-recognized anti-TB drug, which is employed in the short-course treatment strategy of TB. Efforts are focused on investigating EMB analogues, such as SQ109 (Jia et al. 2005a, b, c; Sacksteder et al. 2012) and SQ775 (Bogatcheva et al. 2006), for future lead drug development. Interestingly, the other AraTs are not inhibited by EMB (Alderwick et al. 2006; Seidel et al. 2007; Birch et al. 2008) and screening for inhibitors against these enzymes is hindered due to the nature of the protein and substrate (membrane bound). However, there have been reports on the development of DPA analogues for the inhibition of arabinogalactan biosynthesis (Pathak et al. 2001; Owen et al. 2007). A recent study employing a cell free assay approach with membrane preparations has determined that various DPA analogues are able to limit the incorporation of a radiolabelled DP[14C]A (Zhang et al. 2011).
The primary structure of arabinogalactan is completed by the transfer of succinyl and D-GalN residues to the inner arabinan units. PpgS, polyprenyl-phospho-N-acetylgalactosaminyl synthase, catalyses the formation of polyprenol-P-D-GalNAc from polyprenyl-P and UDP-GalNAc, which is then translocated across the membrane (Skovierova et al. 2010; Rana et al. 2012). The deacylation to polyprenol-P-D-GalN occurs in an undetermined location and by an unknown mechanism. The glycosyltransferase, Rv3779, transfers D-GalN to arabinogalactan at the C-2 position of 3,5-branched Araf residue (Scherman et al. 2009; Skovierova et al. 2010; Peng et al. 2012; Rana et al. 2012). Succinylated Araf residues have also been detected at this position of non-mycolated arabinan chains (Bhamidi et al. 2008), but the enzyme responsible is currently unknown. A comprehensive mechanistic and functional understanding of these enzymes is required for evaluation as suitable drug targets and to date, there are no identified inhibitors against these processes. The final stage is the attachment of the arabinogalactan macromolecule to peptidoglycan. The enzyme responsible for this essential ligation has recently been elucidated to be Lcp1 (Harrison et al. 2016).
PHOSPHATIDYL-MYO-INOSITOL MANNOSIDES, LIPOMANNAN AND LIPOARABINOMANNAN
The glycolipids, phosphatidyl-myo-inositol mannosides (PIMs), and the related lipoglycans, lipomannan (LM) and lipoarabinomannan (LAM), are non-covalently anchored into the inner and outer membranes of the cell wall via the phosphatidyl-myo-inositol unit (Ortalo-Magne et al. 1996) (Fig. 1). The core structure of PIM consists of an acylated sn-glycerol-3-phospho-(1-D-myo-inositol), the phosphatidyl inositol (PI) unit. Glycosylation with mannopyranose (Manp) residues at the O-2 and O-6 positions of myo-inositol, results in the mannosyl phosphate inositol (MPI) anchor (Ballou et al. 1963; Ballou and Lee, 1964; Nigou et al. 2004). The MPI structure is highly diverse, with variations in the type (commonly palmitic and tuberculostearic chains (Pitarque et al. 2005)), number and location of acyl chains. The most prevalent forms of PIMs in mycobacteria are tri- and tetra-acylated phospho-myo-inositol di/hexamannosides (Ac1PIM2, Ac1PIM6, Ac2PIM2, Ac2PIM6), where in the hexamannosides, there is one Manp unit on the O-2 and five Manp units on the O-6 position of myo-inositol (Gilleron et al. 2001). Extensions of mannan and arabinomannan chains on the MPI anchor form LM and LAM, respectively. In both LM and LAM, the mannan chain consists of approximately 21–34 α(1 → 6) linked Manp units, decorated with single α(1 → 2)-Manp residues (Kaur et al. 2008). In LAM, the mannan chain is glycosylated through an α(1 → 2) linkage with ~50–80 Araf residues (Khoo et al. 1996).
In mycobacteria, PI and PIMs contribute up to 56% of all phospholipids in the cell wall and 37% in the cytoplasmic membrane (Goren, 1984). These significant quantities indicate their importance. Not only are they structural components, they also have roles in cell wall integrity, permeability and control of septation and division (Parish et al. 1997; Patterson et al. 2003; Fukuda et al. 2013). LM and LAM are involved in Mtb pathogenicity, with evidence to suggest they are modulators of host–pathogen interactions (Schlesinger et al. 1994; Nigou et al. 2002; Maeda et al. 2003). These features of PIMs, LM and LAM make them suitable targets in anti-TB drug discovery.
BIOSYNTHESIS OF PHOSPHATIDYL-MYO-INOSITOL MANNOSIDES, LIPOMANNAN AND LIPOARABINOMANNAN
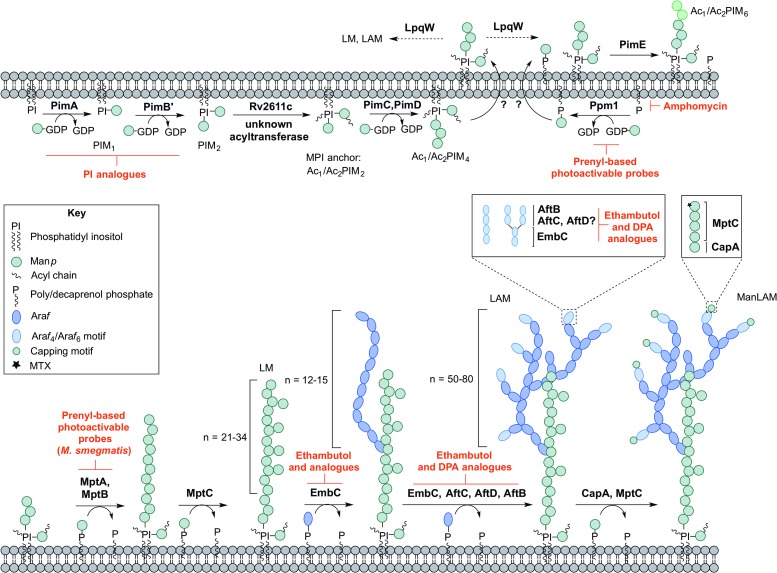
PIM biosynthesis begins in the cytoplasm (Fig. 4). The α-mannopyranosyl transferase (ManpT), PimA, of the GT-A/B superfamily, transfers Manp from the donor GDP-Manp to position O-2 of the myo-inositol ring to form PIM1 (Kordulakova et al. 2002; Guerin et al. 2007). A second Manp residue is transferred to position O-6 of the myo-inositol ring by PimB’ to form PIM2 (Guerin et al. 2009). Acylation of the Manp residue of PIM1 is performed by the acyltransferase Rv2611c before or after the addition of the second Manp residue (Kordulakova et al. 2003). The acylation of the C-3 position of the myo-inositol ring is performed by an unknown acyltransferase. This finishes the synthesis of the MPI anchor. Mannosylation of Ac1/Ac2PIM2 to Ac1/Ac2PIM3 is performed by a ManpT, designated PimC, but this enzyme is yet to be confirmed in Mtb H37Rv (Kremer et al. 2002b). It is suspected that the subsequent addition of Manp to the non-reducing end of Ac1/Ac2PIM3 is performed by the unidentified PimC or PimD forming Ac1/Ac2PIM4. The ManpTs have been the subject of target-based screening programs. More specifically, in vitro PimA activity was screened with approximately 350 compounds. Several hit molecules exhibited significant inhibition, but the compounds did not exhibit in vivo activity in Mtb (Sipos et al. 2015). Substrate analogues of PimA and PimB’, galactose-derived phosphonate analogs of PI, have also been developed, which show enzyme inhibition in a cell-free system (Dinev et al. 2007).
Fig. 4.
Inhibitors targeting the biosynthesis of phosphatidyl-myo-inositol mannosides, lipomannan and lipoarabinomannan. The current understanding of the biosynthesis of PIMs, LM, LAM and ManLAM. Reported inhibitors are shown in red.
The biosynthesis of Ac1/Ac2PIM4 marks the transition towards the synthesis of higher order PIMs, LM and LAM (Fig. 4). It is predicted that the synthesis of Ac1/Ac2PIM4 occurs on the cytoplasmic side of the membrane, and at this point, is flipped across the membrane by an unidentified translocase, with the remainder of the steps thought to occur in the periplasmic space. The integral membrane ManpTs (of the GT-C glycosyltransferase superfamily) are reliant on polyprenyl-phosphate-based mannose donors (PPM) rather than the nucleotide-based sugars (Berg et al. 2007). The polyprenol monophosphomannose synthase, Ppm1, catalyses the synthesis of PPM from GDP-Manp and polyprenol phosphates (Gurcha et al. 2002).
PimE catalyses the transfer of an α(1 → 2)-linked Manp residue onto Ac1/Ac2PIM4, generating Ac1/Ac2PIM5 (Morita et al. 2006). The transfer of the last Manp residue is either performed by PimE or by an unidentified GT-C glycosyltransferase forming Ac1/Ac2PIM6 (Morita et al. 2006). The distal 2-linked Manp residues are not present in the mannan core of LM or LAM; Ac1/Ac2PIM4 is the likely precursor for the extension of the mannan chain. Recent evidence suggests that the putative lipoprotein LpqW channels intermediates such as Ac1/Ac2PIM4 towards either PimE (to form the polar lipids) or to LM and LAM synthesis (Crellin et al. 2008). The mannosyltransferases, MptA and MptB (Mishra et al. 2007, 2008), are responsible for the α(1 → 6)-linked mannan core of LM and LAM. MptC catalyses the transfer of the monomannose side chains via α(1 → 2) linkages, forming mature LM (Kaur et al. 2008; Mishra et al. 2011). Modification of LM leads to LAM. Approximately 50–80 Araf residues are added using DPA as the donor, comparable to that of the arabinogalactan domain. An unidentified ArafT primes the mannan chain, which is further elongated by EmbC, adding 12–16 Araf residues with α(1 → 5) linkages (Shi et al. 2006; Alderwick et al. 2011a). AftC, the same enzyme involved in arabinogalactan synthesis, integrates α(1 → 3) Araf branches (Birch et al. 2008). It has also been speculated that AftD introduces α(1 → 3) Araf, but its function is yet to be confirmed (Skovierova et al. 2009). The arabinan domain is terminated by β(1 → 2) Araf linkages, predicted to be performed by AftB, resulting in branched hexa-arabinoside or linear tetra-arabinoside motifs. Further structural heterogeneity is introduced by capping motifs. These moieties consist of a number of α(1 → 2)-linked Manp residues, producing mannosylated LAM (ManLAM) (Kaur et al. 2008). Using PPM, the α(1 → 5) ManpT, CapA, attaches the first Manp residue (Dinadayala et al. 2006). MptC catalyses the addition of subsequent α(1 → 2) Manp residues (Kaur et al. 2008), which can be decorated with an α(1 → 4)-linked 5-deoxy-5-methyl-thio-xylofuranose (MTX) residue (Ludwiczak et al. 2002; Turnbull et al. 2004). The enzymes involved in the addition of MTX and succinyl residues to LAM are still to be determined.
The essentiality of PPM in lipoglycan biosynthesis makes Ppm1 an attractive drug target. Amphomycin, a lipopeptide antibiotic, inhibits the synthesis of PPM by sequestering the polyprenol phosphates, and consequently inhibits the extracellular ManpTs (Banerjee et al. 1981; Besra et al. 1997). Guy et al. (2004) designed a variety of prenyl-based photoactivable probes. Upon photoactivation, a number of the probes exhibited inhibitory activity against Mtb Ppm1 and M. smegmatis α(1 → 6) ManpTs (Guy et al. 2004). Substrate analogues of the ManpTs have been designed to investigate enzyme–substrate interactions and mechanisms of action (Brown et al. 2001; Tam and Lowary, 2010). These types of studies will provide an invaluable insight into the interactions involved and for the future design of inhibitors.
MYCOLIC ACIDS
The final distinctive component of the mycobacterial cell wall is the unique fatty acids, termed the mycolic acids (Fig. 1). These unique long chain α-alkyl-β-hydroxy fatty acids (comprised a meromycolate chain of C42–C62 and a long saturated α-chain C24–C26) are attached to the arabinogalactan layer, but also make up other outer cell envelope lipids such as trehalose mono/di-mycolates and glucose monomycolate. There are three subclasses of mycolic acids: α-mycolates, containing cyclopropane rings in the cis-configuration; methoxy-mycolates and keto-mycolates containing methoxy or ketone groups, respectively, and have cyclopropane rings in the cis- or trans- configuration (Brennan and Nikaido, 1995; Watanabe et al. 2001, 2002). Mycolic acids contribute to the permeability of the cell wall, and as such are essential for cell viability, and are also essential in virulence, making the biosynthesis of mycolates suitable drug targets (Liu et al. 1996).
MYCOLIC ACID BIOSYNTHESIS
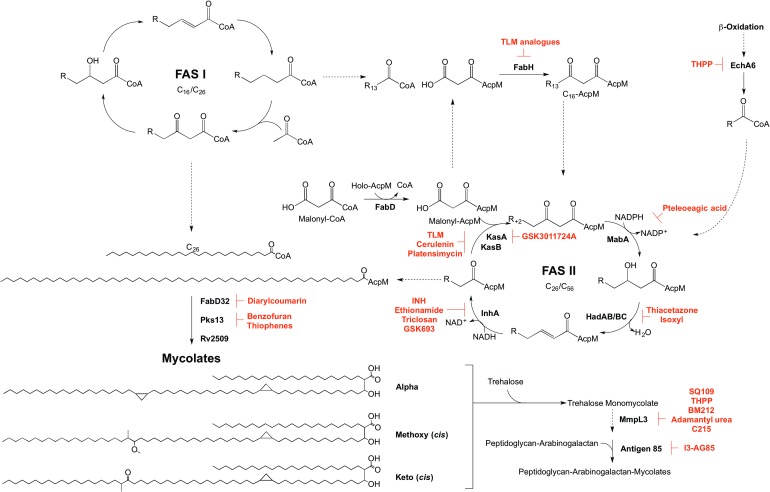
Mycolic acid biosynthesis occurs in the cytoplasm, involving two distinct pathways, termed fatty acid synthase types I and II (FAS I and FAS II) (Fig. 5). FAS I (Rv2524c), a multifunctional polypeptide, generates short-chain fatty acyl-CoA esters that can either form the saturated α-branch (C24), or be extended by FAS II to form the meromycolate chain (Cole et al. 1998). Elongation of the fatty acids is dependent on the availability of holo-AcpM, an acyl carrier protein, and malonyl-CoA. FabD, the malonyl:AcpM transacylase generates malonyl-AcpM (Kremer et al. 2001b). C14-CoA primers from FAS I are condensed with malonyl-AcpM, catalysed by FabH (β-ketoacyl ACP synthase) (Choi et al. 2000), forming a pivotal link between the FAS I and FAS II pathways. The C16-AcpM formed is channeled to the FAS II pathway (Bhatt et al. 2007), where it undergoes a round of keto-reduction, dehydration and enoyl-reduction, catalysed by: MabA, a β-ketoacyl-AcpM reductase (Marrakchi et al. 2002); HadAB/BC, a β-hydroxyacyl-AcpM hydratase (Sacco et al. 2007); InhA, an enoyl-AcpM reductase (Banerjee et al. 1994). Successive cycles ensue, whereby the condensation reaction of FabH is replaced by the activities of KasA and KasB, β-ketoacyl synthases (Schaeffer et al. 2001; Kremer et al. 2002a). The AcpM-bound acyl chain extends by two carbon units in each cycle, forming a saturated long-chain meromycolate of C42–C62, which is subject to modifications such as cis-/trans-cyclopropanation, and the addition of methoxy and keto groups (Dubnau et al. 2000; Glickman et al. 2000; Glickman, 2003; Barkan et al. 2010). FabD32, a fatty acyl-AMP ligase, activates the meromycolate chain (Trivedi et al. 2004) and the subsequent meromycolyl-AMP is linked with the α-alkyl-CoA ester, catalysed by Pks13, to generate a α-alkyl-β-keto-mycolic acid (Gande et al. 2004; Portevin et al. 2004). Finally a reduction step, catalysed by Rv2509, generates a mature mycolate (Bhatt et al. 2008). Transport of the mycolates to either the cell envelope or for attachment to arabinogalactan remains to be elucidated. It is considered that the mycolates are transported in the form of trehalose monomycolate (TMM). In the generation of TMM, Takayama et al. (2005) propose that a mycolyltransferase transfers the mycolyl group from mycolyl-Pks13 to D-mannopyranosyl-1-phosphoheptaprenol (Besra et al. 1994). The mycolyl group of mycolyl-D-mannopyranosyl-1-phosphoheptaprenol is transferred to trehalose-6-phosphate by a second mycolyltransferase, forming TMM-phosphate. The phosphate moiety is removed by a trehalose-6-phosphate phosphatase, and the TMM is immediately translocated outside of the cell using a resistance-nodulation-division (RND) family of efflux pumps, termed mycobacterial membrane proteins large (MmpL), limiting TMM accumulation in the cytoplasm (Takayama et al. 2005; Grzegorzewicz et al. 2012; Varela et al. 2012). Finally, the mycolyltransferase Antigen 85 complex, formed of Ag85A, Ag85B and Ag85C, attaches the mycolic acid moiety from TMM to arabinogalactan (Jackson et al. 1999). This complex also catalyses the formation of trehalose dimycolate, TDM, from two TMM molecules with the release of trehalose (Takayama et al. 2005). TDM, or ‘cord factor’, is implicated in the pathogenicity of Mtb.
Fig. 5.
Inhibitors targeting mycolic acid biosynthesis. The enzymes involved in the mycolic acid biosynthetic pathway are presented. Reported inhibitors are shown in red. ‘R’ represents an acyl chain of varying carbon units in length.
The enzymes involved in mycolic acid biosynthesis are the targets of numerous inhibitors. In 1952, shortly after its discovery, isoniazid (INH) was administered as a front-line and essential antibiotic in the treatment of TB (Medical Research Council, 1952) and has only recently had the mode of action elucidated. Initially thought to target KatG due to mutations in the corresponding gene in resistant isolates (Zhang and Young, 1994; Rouse and Morris, 1995), INH was later revealed to be a pro-drug, with the true target being InhA (Banerjee et al. 1994; Larsen et al. 2002). Ethionamide, a structural analogue of INH, also requires cellular activation via EthA, before targeting InhA (Banerjee et al. 1994). Direct inhibitors of InhA that do not require activation are now being searched for (Lu et al. 2010; Vilcheze et al. 2011; Pan and Tonge, 2012; Encinas et al. 2014; Manjunatha et al. 2015; Sink et al. 2015; Martinez-Hoyos et al. 2016). One such molecule is the broad-spectrum antibiotic triclosan, which has not been adopted in TB treatment due to its sub-optimal bioavailability (Wang et al. 2004). In the last year, GlaxoSmithKline have published a set of thiadiazole compounds, which directly target InhA, with GSK693 demonstrating in vivo efficacy comparable to INH (Martinez-Hoyos et al. 2016). Therefore, old drug targets should not be discounted in the search for new anti-tubercular agents.
The β-ketoacyl synthases, KasA and KasB, are the targets of the natural products cerulenin (Parrish et al. 1999; Schaeffer et al. 2001; Kremer et al. 2002a), platensimycin (Brown et al. 2009), and thiolactomycin (TLM) (Kremer et al. 2000; Schaeffer et al. 2001). There has been significant interest in TLM due to its broad-spectrum activity and numerous analogues have been synthesized to improve on potency and pharmacokinetic properties (Kremer et al. 2000; Senior et al. 2003, 2004; Kim et al. 2006). The biphenyl-based 5-substituents of TLM also exhibit in vitro activity against FabH, but with no whole-cell activity (Senior et al. 2003, 2004). The 2-tosylnaphthalene-1,4-diol pharmacophore of TLM also has in vitro activity against FabH, however, whole-cell data are yet to be published (Alhamadsheh et al. 2008). Recently, a new anti-TB compound, an indazole sulfonamide GSK3011724A, was discovered from a phenotypic whole-cell HTS (Abrahams et al. 2016). The compound was shown to target KasA specifically, with no discernable target engagement with KasB or FabH, and is currently the focus of medicinal chemistry optimization (Abrahams et al. 2016).
Due to the success of InhA as a chemotherapeutic target, there is a mounting interest in the other enzymes involved in mycolic acid biosynthesis from a drug target perspective that could bypass INH resistance in MDR and XDR-TB. Formerly used in the treatment of TB, the thiocarbamide-containing drugs, thiacetazone and isoxyl, were shown to target mycolic acid biosynthesis and the inhibition mechanism has recently been elucidated. Following activation by EthA, both drugs target the HadA subunit of the HadABC dehydratase, forming a covalent interaction with the active site cysteine (Grzegorzewicz et al. 2015). It has also been shown that thiacetazone inhibits cyclopropanation of mycolic acids (Alahari et al. 2007). MabA has been the subject of a molecular docking study. Comparable with the control inhibitory substrate isonicotinic-acyl-NADH, pteleoellagic acid had a high docking score with in vivo activity to be confirmed (Shilpi et al. 2015). Through a target-based screening approach linked with whole-genome sequencing of resistant mutants, a benzofuran has been shown to target Pks13 (Ioerger et al. 2013). Additionally, Pks13 is the target of thiophene compounds (Wilson et al. 2013) including 2-aminothiophenes (Thanna et al. 2016). From a GFP reporter-based whole-cell HTS, a diarylcoumarin exhibited potent activity against Mtb and this structural class was shown to target FadD32 by inhibiting the acyl–acyl carrier protein synthetase activity (Stanley et al. 2013). The homologue of the Rv2509 reductase in M. smegmatis is non-essential but loss of function increases susceptibility to lipophilic antibiotics such as rifampicin. Targeting this ‘secondary’ drug target in Mtb could increase the susceptibility of the bacilli to antibiotics (Bhatt et al. 2008). The Antigen 85 complex has been the focus of a number of inhibitor-based screening studies (Belisle et al. 1997; Gobec et al. 2004; Sanki et al. 2008, 2009; Elamin et al. 2009; Barry et al. 2011). Recently, an inhibitor from a compound library was shown to bind to Antigen 85C, and derivatives of this compound have been synthesized, with 2-amino-6-propyl-4,5,6,7-tetrahydro-1-benzothiphene-3-carbonitrile (I3-AG85) exhibiting the lowest MIC in Mtb and drug-resistant strains (Warrier et al. 2012).
In the target identification of new anti-tubercular compounds, some targets can be regarded as promiscuous, inhibited by multiple different chemical scaffolds, exemplified by MmpL3 (Grzegorzewicz et al. 2012; La Rosa et al. 2012; Stanley et al. 2012; Tahlan et al. 2012; Lun et al. 2013; Remuinan et al. 2013), a predicted TMM transporter. Through the generation and sequencing of spontaneous resistant mutants, a number of inhibitors with diverse chemical structures have been shown to target MmpL3 (Grzegorzewicz et al. 2012; La Rosa et al. 2012; Stanley et al. 2012; Tahlan et al. 2012; Lun et al. 2013; Remuinan et al. 2013). However, a recent chemoproteomics approach determined that one of the proposed inhibitor classes of MmpL3, the tetrahydropyrazo[1,5-a]pyrimidine-3-carboxamides (THPPs), has a novel alternative target, EchA6 (Cox et al. 2016). Sequence analysis predicted EchA6 to be an enoyl-CoA hydratase, but it lacks the residues required for catalytic activity. Through an extensive biochemical investigation, Cox et al. (2016) predicted that EchA6 shuttles fatty acyl-CoA esters from the β-oxidation pathway into FAS II, ready for the condensation activities of KasA or KasB with malonyl-AcpM. This research demonstrates that target identification of inhibitory compounds can unveil not only a new biological pathway, but also an untapped area for drug targets.
DRUG DISCOVERY EFFORTS
The strategies involved in drug discovery are forever evolving. Traditional enzyme screening campaigns and medicinal chemistry focused on ligand-based inhibitor designs (such as substrate or transition state analogues) that once dominated drug discovery are being superseded by phenotypic HTS. The former approach often relies on the X-ray crystal structure of the enzyme or biochemical understanding, and successful inhibitors from these screens are further challenged by target engagement in vivo. Over recent years, HTS has become the lead approach in drug discovery. HTS employs extensive compound libraries of diverse chemical structures, and as a consequence, these methods can identify a multitude of inhibitors with novel chemical scaffolds. Phenotypic HTS can reveal anti-TB agents with whole-cell activity and unknown modes of action, having the potential to unveil new biochemical pathways (Abrahams et al. 2012, 2016; Gurcha et al. 2014; Mugumbate et al. 2015). Alternatively, phenotypic HTS can be target-based, focusing on enzymes or pathways such as those involved in cell wall biosynthesis. This can be a very effective way to identify novel anti-TB compounds with known modes of action, but is limited by the specified target (Batt et al. 2015; Martinez-Hoyos et al. 2016). Target assignment is a fundamental step in in the drug discovery pipeline. Without knowledge of the physiological target, efforts can be wasted on developing compounds against an unsuitable target, such as those homologous in humans. Establishing the mode of action of an inhibitor is a prerequisite for facilitating medicinal chemistry efforts to convert compounds into potential drug candidates.
Concluding remarks
The essential mycobacterial cell wall, responsible for structural integrity, permeability and pathogenicity, is an attractive drug target, both structurally and biosynthetically. Recent advancements in biochemical and omics-based techniques have led to the discovery and mechanistic understanding of enzymes involved in mycobacterial cell wall synthesis and assembly. Although a number of key enzymes are yet to be established, there are a plethora of suitable targets, exploited not only in current treatment programmes but also for anti-TB drug discovery. In the current TB treatment regimen, two of the front-line drugs, INH and EMB, target mycolic acid and arabinogalactan biosynthesis, respectively, with the second-line drugs such as ethionamide and D-cycloserine also targeting cell wall production. The proven success of these drugs validates the future development of inhibitors targeting the unique mycobacterial cell wall, which remains a source of unexploited clinically relevant drug targets. The continued progression in drug discovery approaches and the optimization of biochemical techniques, will enable the rapid identification of anti-TB agents, many of which are likely to target the biosynthesis of the so-called ‘Achilles heel’ of Mtb.
ACKNOWLEDGEMENTS
The authors would like to thank Jonathan Cox for his technical support and advice.
FINANCIAL SUPPORT
G.S.B. acknowledges support in the form of a Personal Research Chair from Mr James Bardrick, a Royal Society Wolfson Research Merit Award, the Medical Research Council (MR/K012118/1) and the Wellcome Trust (081569/Z/06/Z).
REFERENCES
- Abrahams K. A., Cox J. A., Spivey V. L., Loman N. J., Pallen M. J., Constantinidou C., Fernandez R., Alemparte C., Remuinan M. J., Barros D., Ballell L. and Besra G. S. (2012). Identification of novel imidazo[1,2-a]pyridine inhibitors targeting M. tuberculosis QcrB. PLoS ONE 7, e52951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams K. A., Chung C. W., Ghidelli-Disse S., Rullas J., Rebollo-Lopez M. J., Gurcha S. S., Cox J. A., Mendoza A., Jimenez-Navarro E., Martinez-Martinez M. S., Neu M., Shillings A., Homes P., Argyrou A., Casanueva R., Loman N. J., Moynihan P. J., Lelievre J., Selenski C., Axtman M., Kremer L., Bantscheff M., Angulo-Barturen I., Izquierdo M. C., Cammack N. C., Drewes G., Ballell L., Barros D., Besra G. S. and Bates R. H. (2016). Identification of KasA as the cellular target of an anti-tubercular scaffold. Nature Communications 7, 12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alahari A., Trivelli X., Guerardel Y., Dover L. G., Besra G. S., Sacchettini J. C., Reynolds R. C., Coxon G. D. and Kremer L. (2007). Thiacetazone, an antitubercular drug that inhibits cyclopropanation of cell wall mycolic acids in mycobacteria. PLoS ONE 2, e1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderwick L. J., Radmacher E., Seidel M., Gande R., Hitchen P. G., Morris H. R., Dell A., Sahm H., Eggeling L. and Besra G. S. (2005). Deletion of Cg-emb in corynebacterianeae leads to a novel truncated cell wall arabinogalactan, whereas inactivation of Cg-ubiA results in an arabinan-deficient mutant with a cell wall galactan core. Journal of Biological Chemistry 280, 32362–32371. [DOI] [PubMed] [Google Scholar]
- Alderwick L. J., Seidel M., Sahm H., Besra G. S. and Eggeling L. (2006). Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. Journal of Biological Chemistry 281, 15653–15661. [DOI] [PubMed] [Google Scholar]
- Alderwick L. J., Dover L. G., Veerapen N., Gurcha S. S., Kremer L., Roper D. L., Pathak A. K., Reynolds R. C. and Besra G. S. (2008). Expression, purification and characterisation of soluble GlfT and the identification of a novel galactofuranosyltransferase Rv3782 involved in priming GlfT-mediated galactan polymerisation in Mycobacterium tuberculosis. Protein Expression and Purification 58, 332–341. [DOI] [PubMed] [Google Scholar]
- Alderwick L. J., Lloyd G. S., Ghadbane H., May J. W., Bhatt A., Eggeling L., Futterer K. and Besra G. S. (2011a). The C-terminal domain of the Arabinosyltransferase Mycobacterium tuberculosis EmbC is a lectin-like carbohydrate binding module. PLoS Pathogens 7, e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderwick L. J., Lloyd G. S., Lloyd A. J., Lovering A. L., Eggeling L. and Besra G. S. (2011b). Biochemical characterization of the Mycobacterium tuberculosis phosphoribosyl-1-pyrophosphate synthetase. Glycobiology 21, 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhamadsheh M. M., Waters N. C., Sachdeva S., Lee P. and Reynolds K. A. (2008). Synthesis and biological evaluation of novel sulfonyl-naphthalene-1,4-diols as FabH inhibitors. Bioorganic and Medicinal Chemistry Letters 18, 6402–6405. [DOI] [PubMed] [Google Scholar]
- Ballou C. E. and Lee Y. C. (1964). The structure of a myoinositol mannoside from Mycobacterium tuberculosis glycolipid. Biochemistry 3, 682–685. [DOI] [PubMed] [Google Scholar]
- Ballou C. E., Vilkas E. and Lederer E. (1963). Structural studies on the myo-inositol phospholipids of Mycobacterium tuberculosis (var. bovis, strain BCG). Journal of Biological Chemistry 238, 69–76. [PubMed] [Google Scholar]
- Banerjee A., Dubnau E., Quemard A., Balasubramanian V., Um K. S., Wilson T., Collins D., de Lisle G. and Jacobs W. R. Jr. (1994). inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263, 227–230. [DOI] [PubMed] [Google Scholar]
- Banerjee D. K., Scher M. G. and Waechter C. J. (1981). Amphomycin: effect of the lipopeptide antibiotic on the glycosylation and extraction of dolichyl monophosphate in calf brain membranes. Biochemistry 20, 1561–1568. [DOI] [PubMed] [Google Scholar]
- Barkan D., Rao V., Sukenick G. D. and Glickman M. S. (2010). Redundant function of cmaA2 and mmaA2 in Mycobacterium tuberculosis cis cyclopropanation of oxygenated mycolates. Journal of Bacteriology 192, 3661–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreteau H., Kovac A., Boniface A., Sova M., Gobec S. and Blanot D. (2008). Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiology Reviews 32, 168–207. [DOI] [PubMed] [Google Scholar]
- Barry C. S., Backus K. M., Barry C. E. III and Davis B. G. (2011). ESI-MS assay of M. tuberculosis cell wall antigen 85 enzymes permits substrate profiling and design of a mechanism-based inhibitor. Journal of the American Chemical Society 133, 13232–13235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batt S. M., Jabeen T., Bhowruth V., Quill L., Lund P. A., Eggeling L., Alderwick L. J., Futterer K. and Besra G. S. (2012). Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors. Proceedings of the National Academy of Sciences of the United States of America 109, 11354–11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batt S. M., Izquierdo M. C., Pichel J. C., Stubbs C. J., Del Peral L. V.-G., Perez-Herran E., Dhar N., Mouzon B., Rees M., Hutchinson J. P., Young R. J., McKinney J. D., Barros-Aguirre D., Ballell Pages L., Besra G. S. and Argyrou A. (2015). Whole cell target engagement identifies novel inhibitors of Mycobacterium tuberculosis decaprenylphosphoryl-β-D-ribose oxidase. ACS Infectious Diseases 1, 615–626. [DOI] [PubMed] [Google Scholar]
- Belanova M., Dianiskova P., Brennan P. J., Completo G. C., Rose N. L., Lowary T. L. and Mikusova K. (2008). Galactosyl transferases in mycobacterial cell wall synthesis. Journal of Bacteriology 190, 1141–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle J. T., Vissa V. D., Sievert T., Takayama K., Brennan P. J. and Besra G. S. (1997). Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276, 1420–1422. [DOI] [PubMed] [Google Scholar]
- Berg S., Kaur D., Jackson M. and Brennan P. J. (2007). The glycosyltransferases of Mycobacterium tuberculosis – roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology 17, 35–56R. [DOI] [PubMed] [Google Scholar]
- Besra G. S., Sievert T., Lee R. E., Slayden R. A., Brennan P. J. and Takayama K. (1994). Identification of the apparent carrier in mycolic acid synthesis. Proceedings of the National Academy of Sciences of the United States of America 91, 12735–12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besra G. S., Khoo K. H., McNeil M. R., Dell A., Morris H. R. and Brennan P. J. (1995). A new interpretation of the structure of the mycolyl-arabinogalactan complex of Mycobacterium tuberculosis as revealed through characterization of oligoglycosylalditol fragments by fast-atom bombardment mass spectrometry and 1H nuclear magnetic resonance spectroscopy. Biochemistry 34, 4257–4266. [DOI] [PubMed] [Google Scholar]
- Besra G. S., Morehouse C. B., Rittner C. M., Waechter C. J. and Brennan P. J. (1997). Biosynthesis of mycobacterial lipoarabinomannan. Journal of Biological Chemistry 272, 18460–18466. [DOI] [PubMed] [Google Scholar]
- Bhamidi S., Scherman M. S., Rithner C. D., Prenni J. E., Chatterjee D., Khoo K. H. and McNeil M. R. (2008). The identification and location of succinyl residues and the characterization of the interior arabinan region allow for a model of the complete primary structure of Mycobacterium tuberculosis mycolyl arabinogalactan. Journal of Biological Chemistry 283, 12992–13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt A., Molle V., Besra G. S., Jacobs W. R. Jr. and Kremer L. (2007). The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. Molecular Microbiology 64, 1442–1454. [DOI] [PubMed] [Google Scholar]
- Bhatt A., Brown A. K., Singh A., Minnikin D. E. and Besra G. S. (2008). Loss of a mycobacterial gene encoding a reductase leads to an altered cell wall containing beta-oxo-mycolic acid analogs and accumulation of ketones. Chemistry and Biology 15, 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biarrotte-Sorin S., Hugonnet J. E., Delfosse V., Mainardi J. L., Gutmann L., Arthur M. and Mayer C. (2006). Crystal structure of a novel beta-lactam-insensitive peptidoglycan transpeptidase. Journal of Molecular Biology 359, 533–538. [DOI] [PubMed] [Google Scholar]
- Birch H. L., Alderwick L. J., Bhatt A., Rittmann D., Krumbach K., Singh A., Bai Y., Lowary T. L., Eggeling L. and Besra G. S. (2008). Biosynthesis of mycobacterial arabinogalactan: identification of a novel alpha(1→3) arabinofuranosyltransferase. Molecular Microbiology 69, 1191–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch H. L., Alderwick L. J., Appelmelk B. J., Maaskant J., Bhatt A., Singh A., Nigou J., Eggeling L., Geurtsen J. and Besra G. S. (2010). A truncated lipoglycan from mycobacteria with altered immunological properties. Proceedings of the National Academy of Sciences of the United States of America 107, 2634–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogatcheva E., Hanrahan C., Nikonenko B., Samala R., Chen P., Gearhart J., Barbosa F., Einck L., Nacy C. A. and Protopopova M. (2006). Identification of new diamine scaffolds with activity against Mycobacterium tuberculosis. Journal of Medicinal Chemistry 49, 3045–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan P. J. and Nikaido H. (1995). The envelope of mycobacteria. Annual Review of Biochemistry 64, 29–63. [DOI] [PubMed] [Google Scholar]
- Brown A. K., Taylor R. C., Bhatt A., Futterer K. and Besra G. S. (2009). Platensimycin activity against mycobacterial beta-ketoacyl-ACP synthases. PLoS ONE 4, e6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. R., Field R. A., Barker A., Guy M., Grewal R., Khoo K. H., Brennan P. J., Besra G. S. and Chatterjec D. (2001). Synthetic mannosides act as acceptors for mycobacterial alpha1-6 mannosyltransferase. Bioorganic and Medicinal Chemistry 9, 815–824. [DOI] [PubMed] [Google Scholar]
- Bruning J. B., Murillo A. C., Chacon O., Barletta R. G. and Sacchettini J. C. (2011). Structure of the Mycobacterium tuberculosis D-alanine:D-alanine ligase, a target of the antituberculosis drug D-cycloserine. Antimicrobial Agents and Chemotherapy 55, 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. H., Kremer L., Besra G. S. and Rock C. O. (2000). Identification and substrate specificity of beta-ketoacyl (acyl carrier protein) synthase III (mtFabH) from Mycobacterium tuberculosis. Journal of Biological Chemistry 275, 28201–28207. [DOI] [PubMed] [Google Scholar]
- Christophe T., Jackson M., Jeon H. K., Fenistein D., Contreras-Dominguez M., Kim J., Genovesio A., Carralot J. P., Ewann F., Kim E. H., Lee S. Y., Kang S., Seo M. J., Park E. J., Skovierova H., Pham H., Riccardi G., Nam J. Y., Marsollier L., Kempf M., Joly-Guillou M. L., Oh T., Shin W. K., No Z., Nehrbass U., Brosch R., Cole S. T. and Brodin P. (2009). High content screening identifies decaprenyl-phosphoribose 2′ epimerase as a target for intracellular antimycobacterial inhibitors. PLoS Pathogens 5, e1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E. III, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J. et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. [DOI] [PubMed] [Google Scholar]
- Cox J. A. G., Abrahams K. A., Alemparte C., Ghidelli-Disse S., Rullas J., Angulo-Barturen I., Singh A., Gurcha S. S., Nataraj V., Bethell S., Remuiñán M. J., Encinas L., Jervis P. J., Cammack N. C., Bhatt A., Kruse U., Bantscheff M., Futterer K., Barros D., Ballell L., Drewes G. and Besra G. S. (2016). THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria. Nature Microbiology 1, 1–10. [DOI] [PubMed] [Google Scholar]
- Crellin P. K., Kovacevic S., Martin K. L., Brammananth R., Morita Y. S., Billman-Jacobe H., McConville M. J. and Coppel R. L. (2008). Mutations in pimE restore lipoarabinomannan synthesis and growth in a Mycobacterium smegmatis lpqW mutant. Journal of Bacteriology 190, 3690–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffe M., Brennan P. J. and McNeil M. (1990). Predominant structural features of the cell wall arabinogalactan of Mycobacterium tuberculosis as revealed through characterization of oligoglycosyl alditol fragments by gas chromatography/mass spectrometry and by 1H and 13C NMR analyses. Journal of Biological Chemistry 265, 6734–6743. [PubMed] [Google Scholar]
- Dianiskova P., Kordulakova J., Skovierova H., Kaur D., Jackson M., Brennan P. J. and Mikusova K. (2011). Investigation of ABC transporter from mycobacterial arabinogalactan biosynthetic cluster. General Physiology and Biophysics 30, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinadayala P., Kaur D., Berg S., Amin A. G., Vissa V. D., Chatterjee D., Brennan P. J. and Crick D. C. (2006). Genetic basis for the synthesis of the immunomodulatory mannose caps of lipoarabinomannan in Mycobacterium tuberculosis. Journal of Biological Chemistry 281, 20027–20035. [DOI] [PubMed] [Google Scholar]
- Dinev Z., Gannon C. T., Egan C., Watt J. A., McConville M. J. and Williams S. J. (2007). Galactose-derived phosphonate analogues as potential inhibitors of phosphatidylinositol biosynthesis in mycobacteria. Organic & Biomolecular Chemistry 5, 952–959. [DOI] [PubMed] [Google Scholar]
- Dini C. (2005). MraY Inhibitors as novel antibacterial agents. Current Topics in Medicinal Chemistry 5, 1221–1236. [DOI] [PubMed] [Google Scholar]
- Draper P., Khoo K. H., Chatterjee D., Dell A. and Morris H. R. (1997). Galactosamine in walls of slow-growing mycobacteria. Biochemical Journal 327 (Pt 2), 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubee V., Triboulet S., Mainardi J. L., Etheve-Quelquejeu M., Gutmann L., Marie A., Dubost L., Hugonnet J. E. and Arthur M. (2012). Inactivation of Mycobacterium tuberculosis l,d-transpeptidase LdtMt(1) by carbapenems and cephalosporins. Antimicrobial Agents and Chemotherapy 56, 4189–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau E., Chan J., Raynaud C., Mohan V. P., Laneelle M. A., Yu K., Quemard A., Smith I. and Daffe M. (2000). Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Molecular Microbiology 36, 630–637. [DOI] [PubMed] [Google Scholar]
- Elamin A. A., Stehr M., Oehlmann W. and Singh M. (2009). The mycolyltransferase 85A, a putative drug target of Mycobacterium tuberculosis: development of a novel assay and quantification of glycolipid-status of the mycobacterial cell wall. Journal of Microbiological Methods 79, 358–363. [DOI] [PubMed] [Google Scholar]
- Encinas L., O'Keefe H., Neu M., Remuinan M. J., Patel A. M., Guardia A., Davie C. P., Perez-Macias N., Yang H., Convery M. A., Messer J. A., Perez-Herran E., Centrella P. A., Alvarez-Gomez D., Clark M. A., Huss S., O'Donovan G. K., Ortega-Muro F., McDowell W., Castaneda P., Arico-Muendel C. C., Pajk S., Rullas J., Angulo-Barturen I., Alvarez-Ruiz E., Mendoza-Losana A., Ballell Pages L., Castro-Pichel J. and Evindar G. (2014). Encoded library technology as a source of hits for the discovery and lead optimization of a potent and selective class of bactericidal direct inhibitors of Mycobacterium tuberculosis InhA. Journal of Medicinal Chemistry 57, 1276–1288. [DOI] [PubMed] [Google Scholar]
- Escuyer V. E., Lety M. A., Torrelles J. B., Khoo K. H., Tang J. B., Rithner C. D., Frehel C., McNeil M. R., Brennan P. J. and Chatterjee D. (2001). The role of the embA and embB gene products in the biosynthesis of the terminal hexaarabinofuranosyl motif of Mycobacterium smegmatis arabinogalactan. Journal of Biological Chemistry 276, 48854–48862. [DOI] [PubMed] [Google Scholar]
- Fukuda T., Matsumura T., Ato M., Hamasaki M., Nishiuchi Y., Murakami Y., Maeda Y., Yoshimori T., Matsumoto S., Kobayashi K., Kinoshita T. and Morita Y. S. (2013). Critical roles for lipomannan and lipoarabinomannan in cell wall integrity of mycobacteria and pathogenesis of tuberculosis. American Society for Microbiology 4, e00472–00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gande R., Gibson K. J., Brown A. K., Krumbach K., Dover L. G., Sahm H., Shioyama S., Oikawa T., Besra G. S. and Eggeling L. (2004). Acyl-CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg-pks), are key to mycolic acid biosynthesis in Corynebacterianeae such as Corynebacterium glutamicum and Mycobacterium tuberculosis. Journal of Biological Chemistry 279, 44847–44857. [DOI] [PubMed] [Google Scholar]
- Gilleron M., Ronet C., Mempel M., Monsarrat B., Gachelin G. and Puzo G. (2001). Acylation state of the phosphatidylinositol mannosides from Mycobacterium bovis bacillus Calmette Guerin and ability to induce granuloma and recruit natural killer T cells. Journal of Biological Chemistry 276, 34896–34904. [DOI] [PubMed] [Google Scholar]
- Glickman M. S. (2003). The mmaA2 gene of Mycobacterium tuberculosis encodes the distal cyclopropane synthase of the alpha-mycolic acid. Journal of Biological Chemistry 278, 7844–7849. [DOI] [PubMed] [Google Scholar]
- Glickman M. S., Cox J. S. and Jacobs W. R. Jr. (2000). A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Molecular Cell 5, 717–727. [DOI] [PubMed] [Google Scholar]
- Gobec S., Plantan I., Mravljak J., Wilson R. A., Besra G. S. and Kikelj D. (2004). Phosphonate inhibitors of antigen 85C, a crucial enzyme involved in the biosynthesis of the Mycobacterium tuberculosis cell wall. Bioorganic and Medicinal Chemistry Letters 14, 3559–3562. [DOI] [PubMed] [Google Scholar]
- Goffin C. and Ghuysen J. M. (2002). Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiology and Molecular Biology Reviews 66, 702–738 (table of contents). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren M. B. (1984). Biosynthesis and structures of phospholipids and sulphatides. In The Mycobacteria. A Sourcebook (ed. Kubica G. P. and Wayne L. G.), pp. 379–415. Marcel Dekker Inc., New York. [Google Scholar]
- Grzegorzewicz A. E., Ma Y., Jones V., Crick D., Liav A. and McNeil M. R. (2008). Development of a microtitre plate-based assay for lipid-linked glycosyltransferase products using the mycobacterial cell wall rhamnosyltransferase WbbL. Microbiology 154, 3724–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzegorzewicz A. E., Pham H., Gundi V. A., Scherman M. S., North E. J., Hess T., Jones V., Gruppo V., Born S. E., Kordulakova J., Chavadi S. S., Morisseau C., Lenaerts A. J., Lee R. E., McNeil M. R. and Jackson M. (2012). Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nature Chemical Biology 8, 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzegorzewicz A. E., Eynard N., Quemard A., North E. J., Margolis A., Lindenberger J. J., Jones V., Kordulakova J., Brennan P. J., Lee R. E., Ronning D. R., McNeil M. R. and Jackson M. (2015). Covalent modification of the Mycobacterium tuberculosis FAS-II dehydratase by Isoxyl and Thiacetazone. ACS Infectious Diseases 1, 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin M. E., Kordulakova J., Schaeffer F., Svetlikova Z., Buschiazzo A., Giganti D., Gicquel B., Mikusova K., Jackson M. and Alzari P. M. (2007). Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. Journal of Biological Chemistry 282, 20705–20714. [DOI] [PubMed] [Google Scholar]
- Guerin M. E., Kaur D., Somashekar B. S., Gibbs S., Gest P., Chatterjee D., Brennan P. J. and Jackson M. (2009). New insights into the early steps of phosphatidylinositol mannoside biosynthesis in mycobacteria: PimB’ is an essential enzyme of Mycobacterium smegmatis. Journal of Biological Chemistry 284, 25687–25696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurcha S. S., Baulard A. R., Kremer L., Locht C., Moody D. B., Muhlecker W., Costello C. E., Crick D. C., Brennan P. J. and Besra G. S. (2002). Ppm1, a novel polyprenol monophosphomannose synthase from Mycobacterium tuberculosis. Biochemical Journal 365, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurcha S. S., Usha V., Cox J. A., Futterer K., Abrahams K. A., Bhatt A., Alderwick L. J., Reynolds R. C., Loman N. J., Nataraj V., Alemparte C., Barros D., Lloyd A. J., Ballell L., Hobrath J. V. and Besra G. S. (2014). Biochemical and structural characterization of mycobacterial aspartyl-tRNA synthetase AspS, a promising TB drug target. PLoS ONE 9, e113568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy M. R., Illarionov P. A., Gurcha S. S., Dover L. G., Gibson K. J., Smith P. W., Minnikin D. E. and Besra G. S. (2004). Novel prenyl-linked benzophenone substrate analogues of mycobacterial mannosyltransferases. Biochemical Journal 382, 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J., Lloyd G., Joe M., Lowary T. L., Reynolds E., Walters-Morgan H., Bhatt A., Lovering A., Besra G. S. and Alderwick L. J. (2016). Lcp1 is a phosphotransferase responsible for ligating arabinogalactan to peptidoglycan in Mycobacterium tuberculosis. American Society for Microbiology 7, e00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingley-Wilson S. M., Sambandamurthy V. K. and Jacobs W. R. Jr. (2003). Survival perspectives from the world's most successful pathogen, Mycobacterium tuberculosis. Nature Immunology 4, 949–955. [DOI] [PubMed] [Google Scholar]
- Hrast M., Sosic I., Sink R. and Gobec S. (2014). Inhibitors of the peptidoglycan biosynthesis enzymes MurA-F. Bioorganic Chemistry 55, 2–15. [DOI] [PubMed] [Google Scholar]
- Huang H., Scherman M. S., D'Haeze W., Vereecke D., Holsters M., Crick D. C. and McNeil M. R. (2005). Identification and active expression of the Mycobacterium tuberculosis gene encoding 5-phospho-{alpha}-d-ribose-1-diphosphate: decaprenyl-phosphate 5-phosphoribosyltransferase, the first enzyme committed to decaprenylphosphoryl-d-arabinose synthesis. Journal of Biological Chemistry 280, 24539–24543. [DOI] [PubMed] [Google Scholar]
- Huang H., Berg S., Spencer J. S., Vereecke D., D'Haeze W., Holsters M. and McNeil M. R. (2008). Identification of amino acids and domains required for catalytic activity of DPPR synthase, a cell wall biosynthetic enzyme of Mycobacterium tuberculosis. Microbiology 154, 736–743. [DOI] [PubMed] [Google Scholar]
- Hugonnet J. E., Tremblay L. W., Boshoff H. I., Barry C. E. III and Blanchard J. S. (2009). Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science 323, 1215–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger T. R., O'Malley T., Liao R., Guinn K. M., Hickey M. J., Mohaideen N., Murphy K. C., Boshoff H. I., Mizrahi V., Rubin E. J., Sassetti C. M., Barry C. E. III, Sherman D. R., Parish T. and Sacchettini J. C. (2013). Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PLoS ONE 8, e75245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki Y., Hayashi C., Inoue K., Igarashi M., Takahashi Y., Pujari V., Crick D. C., Brennan P. J. and Nomoto A. (2013). Inhibition of the first step in synthesis of the mycobacterial cell wall core, catalyzed by the GlcNAc-1-phosphate transferase WecA, by the novel caprazamycin derivative CPZEN-45. Journal of Biological Chemistry 288, 30309–30319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M., Raynaud C., Laneelle M. A., Guilhot C., Laurent-Winter C., Ensergueix D., Gicquel B. and Daffe M. (1999). Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Molecular Microbiology 31, 1573–1587. [DOI] [PubMed] [Google Scholar]
- Jia L., Coward L., Gorman G. S., Noker P. E. and Tomaszewski J. E. (2005a). Pharmacoproteomic effects of isoniazid, ethambutol, and N-geranyl-N’-(2-adamantyl)ethane-1,2-diamine (SQ109) on Mycobacterium tuberculosis H37Rv. Journal of Pharmacology and Experimental Therapeutics 315, 905–911. [DOI] [PubMed] [Google Scholar]
- Jia L., Tomaszewski J. E., Hanrahan C., Coward L., Noker P., Gorman G., Nikonenko B. and Protopopova M. (2005b). Pharmacodynamics and pharmacokinetics of SQ109, a new diamine-based antitubercular drug. British Journal of Pharmacology 144, 80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Tomaszewski J. E., Noker P. E., Gorman G. S., Glaze E. and Protopopova M. (2005c). Simultaneous estimation of pharmacokinetic properties in mice of three anti-tubercular ethambutol analogs obtained from combinatorial lead optimization. Journal of Pharmaceutical and Biomedical Analysis 37, 793–799. [DOI] [PubMed] [Google Scholar]
- Jiang T., He L., Zhan Y., Zang S., Ma Y., Zhao X., Zhang C. and Xin Y. (2011). The effect of MSMEG_6402 gene disruption on the cell wall structure of Mycobacterium smegmatis. Microbial Pathogenesis 51, 156–160. [DOI] [PubMed] [Google Scholar]
- Jin Y., Xin Y., Zhang W. and Ma Y. (2010). Mycobacterium tuberculosis Rv1302 and Mycobacterium smegmatis MSMEG_4947 have WecA function and MSMEG_4947 is required for the growth of M. smegmatis. FEMS Microbiology Letters 310, 54–61. [DOI] [PubMed] [Google Scholar]
- Kaur D., Obregon-Henao A., Pham H., Chatterjee D., Brennan P. J. and Jackson M. (2008). Lipoarabinomannan of Mycobacterium: mannose capping by a multifunctional terminal mannosyltransferase. Proceedings of the National Academy of Sciences of the United States of America 105, 17973–17977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo K. H., Douglas E., Azadi P., Inamine J. M., Besra G. S., Mikusova K., Brennan P. J. and Chatterjee D. (1996). Truncated structural variants of lipoarabinomannan in ethambutol drug-resistant strains of Mycobacterium smegmatis. Inhibition of arabinan biosynthesis by ethambutol. Journal of Biological Chemistry 271, 28682–28690. [DOI] [PubMed] [Google Scholar]
- Kim D. H., Lees W. J., Kempsell K. E., Lane W. S., Duncan K. and Walsh C. T. (1996). Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry 35, 4923–4928. [DOI] [PubMed] [Google Scholar]
- Kim P., Zhang Y. M., Shenoy G., Nguyen Q. A., Boshoff H. I., Manjunatha U. H., Goodwin M. B., Lonsdale J., Price A. C., Miller D. J., Duncan K., White S. W., Rock C. O., Barry C. E. III and Dowd C. S. (2006). Structure-activity relationships at the 5-position of thiolactomycin: an intact (5R)-isoprene unit is required for activity against the condensing enzymes from Mycobacterium tuberculosis and Escherichia coli. Journal of Medicinal Chemistry 49, 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T., Fukuoka T., Doi N., Harasaki T., Inoue H., Hotoda H., Kakuta M., Muramatsu Y., Yamamura N., Hoshi M. and Hirota T. (2004). Activity of capuramycin analogues against Mycobacterium tuberculosis, Mycobacterium avium and Mycobacterium intracellulare in vitro and in vivo. Journal of Antimicrobial Chemotherapy 54, 755–760. [DOI] [PubMed] [Google Scholar]
- Kolly G. S., Boldrin F., Sala C., Dhar N., Hartkoorn R. C., Ventura M., Serafini A., McKinney J. D., Manganelli R. and Cole S. T. (2014). Assessing the essentiality of the decaprenyl-phospho-d-arabinofuranose pathway in Mycobacterium tuberculosis using conditional mutants. Molecular Microbiology 92, 194–211. [DOI] [PubMed] [Google Scholar]
- Kolly G. S., Mukherjee R., Kilacskova E., Abriata L. A., Raccaud M., Blasko J., Sala C., Dal Peraro M., Mikusova K. and Cole S. T. (2015). GtrA protein Rv3789 is required for arabinosylation of arabinogalactan in Mycobacterium tuberculosis. Journal of Bacteriology 197, 3686–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordulakova J., Gilleron M., Mikusova K., Puzo G., Brennan P. J., Gicquel B. and Jackson M. (2002). Definition of the first mannosylation step in phosphatidylinositol mannoside synthesis. PimA is essential for growth of mycobacteria. Journal of Biological Chemistry 277, 31335–31344. [DOI] [PubMed] [Google Scholar]
- Kordulakova J., Gilleron M., Puzo G., Brennan P. J., Gicquel B., Mikusova K. and Jackson M. (2003). Identification of the required acyltransferase step in the biosynthesis of the phosphatidylinositol mannosides of mycobacterium species. Journal of Biological Chemistry 278, 36285–36295. [DOI] [PubMed] [Google Scholar]
- Kremer L., Douglas J. D., Baulard A. R., Morehouse C., Guy M. R., Alland D., Dover L. G., Lakey J. H., Jacobs W. R. Jr., Brennan P. J., Minnikin D. E. and Besra G. S. (2000). Thiolactomycin and related analogues as novel anti-mycobacterial agents targeting KasA and KasB condensing enzymes in Mycobacterium tuberculosis. Journal of Biological Chemistry 275, 16857–16864. [DOI] [PubMed] [Google Scholar]
- Kremer L., Dover L. G., Morehouse C., Hitchin P., Everett M., Morris H. R., Dell A., Brennan P. J., McNeil M. R., Flaherty C., Duncan K. and Besra G. S. (2001a). Galactan biosynthesis in Mycobacterium tuberculosis. Identification of a bifunctional UDP-galactofuranosyltransferase. Journal of Biological Chemistry 276, 26430–26440. [DOI] [PubMed] [Google Scholar]
- Kremer L., Nampoothiri K. M., Lesjean S., Dover L. G., Graham S., Betts J., Brennan P. J., Minnikin D. E., Locht C. and Besra G. S. (2001b). Biochemical characterization of acyl carrier protein (AcpM) and malonyl-CoA:AcpM transacylase (mtFabD), two major components of Mycobacterium tuberculosis fatty acid synthase II. Journal of Biological Chemistry 276, 27967–27974. [DOI] [PubMed] [Google Scholar]
- Kremer L., Dover L. G., Carrere S., Nampoothiri K. M., Lesjean S., Brown A. K., Brennan P. J., Minnikin D. E., Locht C. and Besra G. S. (2002a). Mycolic acid biosynthesis and enzymic characterization of the beta-ketoacyl-ACP synthase A-condensing enzyme from Mycobacterium tuberculosis. Biochemical Journal 364, 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer L., Gurcha S. S., Bifani P., Hitchen P. G., Baulard A., Morris H. R., Dell A., Brennan P. J. and Besra G. S. (2002b). Characterization of a putative alpha-mannosyltransferase involved in phosphatidylinositol trimannoside biosynthesis in Mycobacterium tuberculosis. Biochemical Journal 363, 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Arora K., Lloyd J. R., Lee I. Y., Nair V., Fischer E., Boshoff H. I. and Barry C. E. III (2012). Meropenem inhibits D,D-carboxypeptidase activity in Mycobacterium tuberculosis. Molecular Microbiology 86, 367–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Saravanan P., Arvind A. and Mohan C. G. (2011). Identification of hotspot regions of MurB oxidoreductase enzyme using homology modeling, molecular dynamics and molecular docking techniques. Journal of Molecular Modeling 17, 939–953. [DOI] [PubMed] [Google Scholar]
- Kurosu M., Mahapatra S., Narayanasamy P. and Crick D. C. (2007). Chemoenzymatic synthesis of Park's nucleotide: toward the development of high-throughput screening for MraY inhibitors. Tetrahedron Letters 48, 799–803. [Google Scholar]
- La Rosa V., Poce G., Canseco J. O., Buroni S., Pasca M. R., Biava M., Raju R. M., Porretta G. C., Alfonso S., Battilocchio C., Javid B., Sorrentino F., Ioerger T. R., Sacchettini J. C., Manetti F., Botta M., De Logu A., Rubin E. J. and De Rossi E. (2012). MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrobial Agents and Chemotherapy 56, 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. H., Vilcheze C., Kremer L., Besra G. S., Parsons L., Salfinger M., Heifets L., Hazbon M. H., Alland D., Sacchettini J. C. and Jacobs W. R. Jr. (2002). Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Molecular Microbiology 46, 453–466. [DOI] [PubMed] [Google Scholar]
- Lavollay M., Arthur M., Fourgeaud M., Dubost L., Marie A., Veziris N., Blanot D., Gutmann L. and Mainardi J. L. (2008). The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. Journal of Bacteriology 190, 4360–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhou Y., Ma Y. and Li X. (2011). Design and synthesis of novel cell wall inhibitors of Mycobacterium tuberculosis GlmM and GlmU. Carbohydrate Research 346, 1714–1720. [DOI] [PubMed] [Google Scholar]
- Liu J., Barry C. E. III, Besra G. S. and Nikaido H. (1996). Mycolic acid structure determines the fluidity of the mycobacterial cell wall. Journal of Biological Chemistry 271, 29545–29551. [DOI] [PubMed] [Google Scholar]
- Lo M. C., Men H., Branstrom A., Helm J., Yao N., Goldman R. and Walker S. (2000). A new mechanism of action proposed for ramoplanin. Journal of the American Chemical Society 122, 3540–3541. [Google Scholar]
- Lu X. Y., You Q. D. and Chen Y. D. (2010). Recent progress in the identification and development of InhA direct inhibitors of Mycobacterium tuberculosis. Mini Reviews in Medicinal Chemistry 10, 181–192. [DOI] [PubMed] [Google Scholar]
- Ludwiczak P., Gilleron M., Bordat Y., Martin C., Gicquel B. and Puzo G. (2002). Mycobacterium tuberculosis phoP mutant: lipoarabinomannan molecular structure. Microbiology 148, 3029–3037. [DOI] [PubMed] [Google Scholar]
- Lun S., Guo H., Onajole O. K., Pieroni M., Gunosewoyo H., Chen G., Tipparaju S. K., Ammerman N. C., Kozikowski A. P. and Bishai W. R. (2013). Indoleamides are active against drug-resistant Mycobacterium tuberculosis. Nature Communications 4, 2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N., Nigou J., Herrmann J. L., Jackson M., Amara A., Lagrange P. H., Puzo G., Gicquel B. and Neyrolles O. (2003). The cell surface receptor DC-SIGN discriminates between Mycobacterium species through selective recognition of the mannose caps on lipoarabinomannan. Journal of Biological Chemistry 278, 5513–5516. [DOI] [PubMed] [Google Scholar]
- Mahapatra S., Crick D. C. and Brennan P. J. (2000). Comparison of the UDP-N-acetylmuramate:L-alanine ligase enzymes from Mycobacterium tuberculosis and Mycobacterium leprae. Journal of Bacteriology 182, 6827–6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S., Scherman H., Brennan P. J. and Crick D. C. (2005a). N Glycolylation of the nucleotide precursors of peptidoglycan biosynthesis of Mycobacterium spp. is altered by drug treatment. Journal of Bacteriology 187, 2341–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S., Yagi T., Belisle J. T., Espinosa B. J., Hill P. J., McNeil M. R., Brennan P. J. and Crick D. C. (2005b). Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. Journal of Bacteriology 187, 2747–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainardi J. L., Fourgeaud M., Hugonnet J. E., Dubost L., Brouard J. P., Ouazzani J., Rice L. B., Gutmann L. and Arthur M. (2005). A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. Journal of Biological Chemistry 280, 38146–38152. [DOI] [PubMed] [Google Scholar]
- Makarov V., Lechartier B., Zhang M., Neres J., van der Sar A. M., Raadsen S. A., Hartkoorn R. C., Ryabova O. B., Vocat A., Decosterd L. A., Widmer N., Buclin T., Bitter W., Andries K., Pojer F., Dyson P. J. and Cole S. T. (2014). Towards a new combination therapy for tuberculosis with next generation benzothiazinones. EMBO Molecular Medicine 6, 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov V., Neres J., Hartkoorn R. C., Ryabova O. B., Kazakova E., Sarkan M., Huszar S., Piton J., Kolly G. S., Vocat A., Conroy T. M., Mikusova K. and Cole S. T. (2015). The 8-pyrrole-benzothiazinones Are Noncovalent Inhibitors of DprE1 from Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy 59, 4446–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunatha U. H., Rao S. P. S., Kondreddi R. R., Noble C. G., Camacho L. R., Tan B. H., Ng S. H., Ng P. S., Ma N. L., Lakshminarayana S. B., Herve M., Barnes S. W., Yu W., Kuhen K., Blasco F., Beer D., Walker J. R., Tonge P. J., Glynne R., Smith P. W. and Diagana T. T. (2015). Direct inhibitors of InhA are active against Mycobacterium tuberculosis. Science Translational Medicine 7, 269ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrakchi H., Ducasse S., Labesse G., Montrozier H., Margeat E., Emorine L., Charpentier X., Daffe M. and Quemard A. (2002). MabA (FabG1), a Mycobacterium tuberculosis protein involved in the long-chain fatty acid elongation system FAS-II. Microbiology 148, 951–960. [DOI] [PubMed] [Google Scholar]
- Martinez-Hoyos M., Perez-Herran E., Gulten G., Encinas L., Alvarez-Gomez D., Alvarez E., Ferrer-Bazaga S., Garcia-Perez A., Ortega F., Angulo-Barturen I., Rullas-Trincado J., Blanco Ruano D., Torres P., Castaneda P., Huss S., Fernandez Menendez R., Gonzalez Del Valle S., Ballell L., Barros D., Modha S., Dhar N., Signorino-Gelo F., McKinney J. D., Garcia-Bustos J. F., Lavandera J. L., Sacchettini J. C., Jimenez M. S., Martin-Casabona N., Castro-Pichel J. and Mendoza-Losana A. (2016). Antitubercular drugs for an old target: GSK693 as a promising InhA direct inhibitor. EBioMedicine 8, 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil M., Wallner S. J., Hunter S. W. and Brennan P. J. (1987). Demonstration that the galactosyl and arabinosyl residues in the cell-wall arabinogalactan of Mycobacterium leprae and Mycobacterium tuberculosis are furanoid. Carbohydrate Research 166, 299–308. [DOI] [PubMed] [Google Scholar]
- McNeil M., Daffe M. and Brennan P. J. (1990). Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. Journal of Biological Chemistry 265, 18200–18206. [PubMed] [Google Scholar]
- McNeil M., Daffe M. and Brennan P. J. (1991). Location of the mycolyl ester substituents in the cell walls of mycobacteria. Journal of Biological Chemistry 266, 13217–13223. [PubMed] [Google Scholar]
- Medical Research Council (1952). Treatment of pulmonary tuberculosis with isoniazid; an interim report to the Medical Research Council by their Tuberculosis Chemotherapy Trials Committee. British Medical Journal 2, 735–746. [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., Texier L., Rousseau M. and van Heijenoort J. (1991). The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine: N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. Journal of Bacteriology 173, 4625–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikusova K., Huang H., Yagi T., Holsters M., Vereecke D., D'Haeze W., Scherman M. S., Brennan P. J., McNeil M. R. and Crick D. C. (2005). Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. Journal of Bacteriology 187, 8020–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikusova K., Belanova M., Kordulakova J., Honda K., McNeil M. R., Mahapatra S., Crick D. C. and Brennan P. J. (2006). Identification of a novel galactosyl transferase involved in biosynthesis of the mycobacterial cell wall. Journal of Bacteriology 188, 6592–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills J. A., Motichka K., Jucker M., Wu H. P., Uhlik B. C., Stern R. J., Scherman M. S., Vissa V. D., Pan F., Kundu M., Ma Y. F. and McNeil M. (2004). Inactivation of the mycobacterial rhamnosyltransferase, which is needed for the formation of the arabinogalactan-peptidoglycan linker, leads to irreversible loss of viability. Journal of Biological Chemisrty 279, 43540–43546. [DOI] [PubMed] [Google Scholar]
- Mio T., Yabe T., Arisawa M. and Yamada-Okabe H. (1998). The eukaryotic UDP-N-acetylglucosamine pyrophosphorylases. Gene cloning, protein expression, and catalytic mechanism. Journal of Biological Chemistry 273, 14392–14397. [DOI] [PubMed] [Google Scholar]
- Mishra A. K., Alderwick L. J., Rittmann D., Tatituri R. V., Nigou J., Gilleron M., Eggeling L. and Besra G. S. (2007). Identification of an alpha(1→6) mannopyranosyltransferase (MptA), involved in Corynebacterium glutamicum lipomanann biosynthesis, and identification of its orthologue in Mycobacterium tuberculosis. Molecular Microbiology 65, 1503–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A. K., Alderwick L. J., Rittmann D., Wang C., Bhatt A., Jacobs W. R. Jr., Takayama K., Eggeling L. and Besra G. S. (2008). Identification of a novel alpha(1→6) mannopyranosyltransferase MptB from Corynebacterium glutamicum by deletion of a conserved gene, NCgl1505, affords a lipomannan- and lipoarabinomannan-deficient mutant. Molecular Microbiology 68, 1595–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A. K., Krumbach K., Rittmann D., Appelmelk B., Pathak V., Pathak A. K., Nigou J., Geurtsen J., Eggeling L. and Besra G. S. (2011). Lipoarabinomannan biosynthesis in Corynebacterineae: the interplay of two alpha(1→2)-mannopyranosyltransferases MptC and MptD in mannan branching. Molecular Microbiology 80, 1241–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitachi K., Siricilla S., Yang D., Kong Y., Skorupinska-Tudek K., Swiezewska E., Franzblau S. G. and Kurosu M. (2016). Fluorescence-based assay for polyprenyl phosphate-GlcNAc-1-phosphate transferase (WecA) and identification of novel antimycobacterial WecA inhibitors. Analytical Biochemistry 512, 78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T., van Dam V., Sijbrandi R., Vernet T., Zapun A., Bouhss A., Diepeveen-de Bruin M., Nguyen-Disteche M., de Kruijff B. and Breukink E. (2011). Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO Journal 30, 1425–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T., Sijbrandi R., Lutters M., Verheul J., Martin N. I., den Blaauwen T., de Kruijff B. and Breukink E. (2014). Specificity of the transport of lipid II by FtsW in Escherichia coli. Journal of Biological Chemistry 289, 14707–14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y. S., Sena C. B., Waller R. F., Kurokawa K., Sernee M. F., Nakatani F., Haites R. E., Billman-Jacobe H., McConville M. J., Maeda Y. and Kinoshita T. (2006). PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in mycobacteria. Journal of Biological Chemistry 281, 25143–25155. [DOI] [PubMed] [Google Scholar]
- Mugumbate G., Abrahams K. A., Cox J. A., Papadatos G., van Westen G., Lelievre J., Calus S. T., Loman N. J., Ballell L., Barros D., Overington J. P. and Besra G. S. (2015). Mycobacterial dihydrofolate reductase inhibitors identified using chemogenomic methods and in vitro validation. PloS ONE 10, e0121492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi T., Gupta A., Evangelopoulos D., Guzman J. D., Gibbons S., Keep N. H. and Bhakta S. (2013). Characterisation of ATP-dependent Mur ligases involved in the biogenesis of cell wall peptidoglycan in Mycobacterium tuberculosis. PLoS ONE 8, e60143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigou J., Gilleron M., Rojas M., Garcia L. F., Thurnher M. and Puzo G. (2002). Mycobacterial lipoarabinomannans: modulators of dendritic cell function and the apoptotic response. Microbes and Infection 4, 945–953. [DOI] [PubMed] [Google Scholar]
- Nigou J., Gilleron M., Brando T. and Puzo G. (2004). Structural analysis of mycobacterial lipoglycans. Applied Biochemistry and Biotechnology 118, 253–267. [DOI] [PubMed] [Google Scholar]
- Nikonenko B. V., Reddy V. M., Protopopova M., Bogatcheva E., Einck L. and Nacy C. A. (2009). Activity of SQ641, a capuramycin analog, in a murine model of tuberculosis. Antimicrobial Agents and Chemotherapy 53, 3138–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortalo-Magne A., Lemassu A., Laneelle M. A., Bardou F., Silve G., Gounon P., Marchal G. and Daffe M. (1996). Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. Journal of Bacteriology 178, 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D. J., Davis C. B., Hartnell R. D., Madge P. D., Thomson R. J., Chong A. K., Coppel R. L. and von Itzstein M. (2007). Synthesis and evaluation of galactofuranosyl N,N-dialkyl sulfenamides and sulfonamides as antimycobacterial agents. Bioorganic and Medicinal Chemistry Letters 17, 2274–2277. [DOI] [PubMed] [Google Scholar]
- Pan P. and Tonge P. J. (2012). Targeting InhA, the FASII enoyl-ACP reductase: SAR studies on novel inhibitor scaffolds. Current Topics in Medicinal Chemistry 12, 672–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish T., Liu J., Nikaido H. and Stoker N. G. (1997). A Mycobacterium smegmatis mutant with a defective inositol monophosphate phosphatase gene homolog has altered cell envelope permeability. Journal of Bacteriology 179, 7827–7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish N. M., Kuhajda F. P., Heine H. S., Bishai W. R. and Dick J. D. (1999). Antimycobacterial activity of cerulenin and its effects on lipid biosynthesis. Journal of Antimicrobial Chemotherapy 43, 219–226. [DOI] [PubMed] [Google Scholar]
- Pathak A. K., Pathak V., Khare N. K., Maddry J. A. and Reynolds R. C. (2001). Synthesis of disaccharides related to the mycobacterial arabinogalactan. Carbohydrate Letters 4, 117–122. [PubMed] [Google Scholar]
- Patterson J. H., Waller R. F., Jeevarajah D., Billman-Jacobe H. and McConville M. J. (2003). Mannose metabolism is required for mycobacterial growth. Biochemical Journal 372, 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier P., Belanova M., Dianiskova P., Zhou R., Zheng R. B., Pearcey J. A., Joe M., Brennan P. J., Nugier-Chauvin C., Ferrieres V., Lowary T. L., Daniellou R. and Mikusova K. (2010). Synthetic UDP-furanoses as potent inhibitors of mycobacterial galactan biogenesis. Chemistry and Biology 17, 1356–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peneff C., Ferrari P., Charrier V., Taburet Y., Monnier C., Zamboni V., Winter J., Harnois M., Fassy F. and Bourne Y. (2001). Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. EMBO Journal 20, 6191–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W., Zou L., Bhamidi S., McNeil M. R. and Lowary T. L. (2012). The galactosamine residue in mycobacterial arabinogalactan is alpha-linked. Journal of Organic Chemistry 77, 9826–9832. [DOI] [PubMed] [Google Scholar]
- Pitarque S., Herrmann J. L., Duteyrat J. L., Jackson M., Stewart G. R., Lecointe F., Payre B., Schwartz O., Young D. B., Marchal G., Lagrange P. H., Puzo G., Gicquel B., Nigou J. and Neyrolles O. (2005). Deciphering the molecular bases of Mycobacterium tuberculosis binding to the lectin DC-SIGN reveals an underestimated complexity. Biochemical Journal 392, 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portevin D., De Sousa-D'Auria C., Houssin C., Grimaldi C., Chami M., Daffe M. and Guilhot C. (2004). A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proceedings of the National Academy of Sciences of the United States of America 101, 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser G. A. and de Carvalho L. P. (2013a). Kinetic mechanism and inhibition of Mycobacterium tuberculosis D-alanine:D-alanine ligase by the antibiotic D-cycloserine. FEBS Journal 280, 1150–1166. [DOI] [PubMed] [Google Scholar]
- Prosser G. A. and de Carvalho L. P. (2013b). Metabolomics reveal d-alanine:d-alanine ligase as the target of d-cycloserine in Mycobacterium tuberculosis. ACS Medicinal Chemistry Letters 4, 1233–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman K., Yeturu K. and Chandra N. (2008). TargetTB: a target identification pipeline for Mycobacterium tuberculosis through an interactome, reactome and genome-scale structural analysis. BMC Systems Biology 2, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana A. K., Singh A., Gurcha S. S., Cox L. R., Bhatt A. and Besra G. S. (2012). Ppm1-encoded polyprenyl monophosphomannose synthase activity is essential for lipoglycan synthesis and survival in mycobacteria. PLoS ONE 7, e48211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani C. and Khan I. A. (2016). UDP-GlcNAc pathway: potential target for inhibitor discovery against M. tuberculosis. European Journal of Pharmaceutical Sciences 83, 62–70. [DOI] [PubMed] [Google Scholar]
- Raymond J. B., Mahapatra S., Crick D. C. and Pavelka M. S. Jr. (2005). Identification of the namH gene, encoding the hydroxylase responsible for the N-glycolylation of the mycobacterial peptidoglycan. Journal of Biological Chemistry 280, 326–333. [DOI] [PubMed] [Google Scholar]
- Reddy V. M., Einck L. and Nacy C. A. (2008). In vitro antimycobacterial activities of capuramycin analogues. Antimicrobial Agents and Chemotherapy 52, 719–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remuinan M. J., Perez-Herran E., Rullas J., Alemparte C., Martinez-Hoyos M., Dow D. J., Afari J., Mehta N., Esquivias J., Jimenez E., Ortega-Muro F., Fraile-Gabaldon M. T., Spivey V. L., Loman N. J., Pallen M. J., Constantinidou C., Minick D. J., Cacho M., Rebollo-Lopez M. J., Gonzalez C., Sousa V., Angulo-Barturen I., Mendoza-Losana A., Barros D., Besra G. S., Ballell L. and Cammack N. (2013). Tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide and N-benzyl-6′,7′-dihydrospiro[piperidine-4,4′-thieno[3,2-c]pyran] analogues with bactericidal efficacy against Mycobacterium tuberculosis targeting MmpL3. PLoS ONE 8, e60933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P. E. (1989). Structure, biochemistry and mechanism of action of glycopeptide antibiotics. European Journal of Clinical Microbiology and Infectious Diseases 8, 943–950. [DOI] [PubMed] [Google Scholar]
- Rose N. L., Completo G. C., Lin S. J., McNeil M., Palcic M. M. and Lowary T. L. (2006). Expression, purification, and characterization of a galactofuranosyltransferase involved in Mycobacterium tuberculosis arabinogalactan biosynthesis. Journal of the American Chemical Society 128, 6721–6729. [DOI] [PubMed] [Google Scholar]
- Rouse D. A. and Morris S. L. (1995). Molecular mechanisms of isoniazid resistance in Mycobacterium tuberculosis and Mycobacterium bovis. Infection and Immunity 63, 1427–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N. (2008). Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 105, 15553–15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N. (2015). Lipid flippases for bacterial peptidoglycan biosynthesis. Lipid Insights 8, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rullas J., Dhar N., McKinney J. D., Garcia-Perez A., Lelievre J., Diacon A. H., Hugonnet J. E., Arthur M., Angulo-Barturen I., Barros-Aguirre D. and Ballell L. (2015). Combinations of beta-lactam antibiotics currently in clinical trials are efficacious in a DHP-I-deficient mouse model of tuberculosis infection. Antimicrobial Agents and Chemotherapy 59, 4997–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco E., Covarrubias A. S., O'Hare H. M., Carroll P., Eynard N., Jones T. A., Parish T., Daffe M., Backbro K. and Quemard A. (2007). The missing piece of the type II fatty acid synthase system from Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America 104, 14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacksteder K. A., Protopopova M., Barry C. E. III, Andries K. and Nacy C. A. (2012). Discovery and development of SQ109: a new antitubercular drug with a novel mechanism of action. Future Microbiology 7, 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanki A. K., Boucau J., Srivastava P., Adams S. S., Ronning D. R. and Sucheck S. J. (2008). Synthesis of methyl 5-S-alkyl-5-thio-D-arabinofuranosides and evaluation of their antimycobacterial activity. Bioorganic and Medicinal Chemistry 16, 5672–5682. [DOI] [PubMed] [Google Scholar]
- Sanki A. K., Boucau J., Ronning D. R. and Sucheck S. J. (2009). Antigen 85C-mediated acyl-transfer between synthetic acyl donors and fragments of the arabinan. Glycoconjugate Journal 26, 589–596. [DOI] [PubMed] [Google Scholar]
- Sauvage E., Kerff F., Terrak M., Ayala J. A. and Charlier P. (2008). The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiology Reviews 32, 234–258. [DOI] [PubMed] [Google Scholar]
- Schaeffer M. L., Agnihotri G., Volker C., Kallender H., Brennan P. J. and Lonsdale J. T. (2001). Purification and biochemical characterization of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthases KasA and KasB. Journal of Biological Chemistry 276, 47029–47037. [DOI] [PubMed] [Google Scholar]
- Scherman H., Kaur D., Pham H., Skovierova H., Jackson M. and Brennan P. J. (2009). Identification of a polyprenylphosphomannosyl synthase involved in the synthesis of mycobacterial mannosides. Journal of Bacteriology 191, 6769–6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger L. S., Hull S. R. and Kaufman T. M. (1994). Binding of the terminal mannosyl units of lipoarabinomannan from a virulent strain of Mycobacterium tuberculosis to human macrophages. Journal of Immunology 152, 4070–4079. [PubMed] [Google Scholar]
- Seidel M., Alderwick L. J., Birch H. L., Sahm H., Eggeling L. and Besra G. S. (2007). Identification of a novel arabinofuranosyltransferase AftB involved in a terminal step of cell wall arabinan biosynthesis in Corynebacterianeae, such as Corynebacterium glutamicum and Mycobacterium tuberculosis. Journal of Biological Chemistry 282, 14729–14740. [DOI] [PubMed] [Google Scholar]
- Senior S. J., Illarionov P. A., Gurcha S. S., Campbell I. B., Schaeffer M. L., Minnikin D. E. and Besra G. S. (2003). Biphenyl-based analogues of thiolactomycin, active against Mycobacterium tuberculosis mtFabH fatty acid condensing enzyme. Bioorganic and Medicinal Chemistry Letters 13, 3685–3688. [DOI] [PubMed] [Google Scholar]
- Senior S. J., Illarionov P. A., Gurcha S. S., Campbell I. B., Schaeffer M. L., Minnikin D. E. and Besra G. S. (2004). Acetylene-based analogues of thiolactomycin, active against Mycobacterium tuberculosis mtFabH fatty acid condensing enzyme. Bioorganic and Medicinal Chemistry Letters 14, 373–376. [DOI] [PubMed] [Google Scholar]
- Sham L. T., Butler E. K., Lebar M. D., Kahne D., Bernhardt T. G. and Ruiz N. (2014). Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345, 220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Berg S., Lee A., Spencer J. S., Zhang J., Vissa V., McNeil M. R., Khoo K. H. and Chatterjee D. (2006). The carboxy terminus of EmbC from Mycobacterium smegmatis mediates chain length extension of the arabinan in lipoarabinomannan. Journal of Biological Chemistry 281, 19512–19526. [DOI] [PubMed] [Google Scholar]
- Shilpi J. A., Ali M. T., Saha S., Hasan S., Gray A. I. and Seidel V. (2015). Molecular docking studies on InhA, MabA and PanK enzymes from Mycobacterium tuberculosis of ellagic acid derivatives from Ludwigia adscendens and Trewia nudiflora. In Silico Pharmacology 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sink R., Sosic I., Zivec M., Fernandez-Menendez R., Turk S., Pajk S., Alvarez-Gomez D., Lopez-Roman E. M., Gonzales-Cortez C., Rullas-Triconado J., Angulo-Barturen I., Barros D., Ballell-Pages L., Young R. J., Encinas L. and Gobec S. (2015). Design, synthesis, and evaluation of new thiadiazole-based direct inhibitors of enoyl acyl carrier protein reductase (InhA) for the treatment of tuberculosis. Journal of Medicinal Chemistry 58, 613–624. [DOI] [PubMed] [Google Scholar]
- Sipos A., Pato J., Szekely R., Hartkoorn R. C., Kekesi L., Orfi L., Szantai-Kis C., Mikusova K., Svetlikova Z., Kordulakova J., Nagaraja V., Godbole A. A., Bush N., Collin F., Maxwell A., Cole S. T. and Keri G. (2015). Lead selection and characterization of antitubercular compounds using the Nested Chemical Library. Tuberculosis (Edinburgh) 95 (Suppl. 1), S200–S206. [DOI] [PubMed] [Google Scholar]
- Siricilla S., Mitachi K., Wan B., Franzblau S. G. and Kurosu M. (2015). Discovery of a capuramycin analog that kills nonreplicating Mycobacterium tuberculosis and its synergistic effects with translocase I inhibitors. Journal of Antibiotics 68, 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovierova H., Larrouy-Maumus G., Zhang J., Kaur D., Barilone N., Kordulakova J., Gilleron M., Guadagnini S., Belanova M., Prevost M. C., Gicquel B., Puzo G., Chatterjee D., Brennan P. J., Nigou J. and Jackson M. (2009). AftD, a novel essential arabinofuranosyltransferase from mycobacteria. Glycobiology 19, 1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovierova H., Larrouy-Maumus G., Pham H., Belanova M., Barilone N., Dasgupta A., Mikusova K., Gicquel B., Gilleron M., Brennan P. J., Puzo G., Nigou J. and Jackson M. (2010). Biosynthetic origin of the galactosamine substituent of Arabinogalactan in Mycobacterium tuberculosis. Journal of Biological Chemistry 285, 41348–41355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S. A., Grant S. S., Kawate T., Iwase N., Shimizu M., Wivagg C., Silvis M., Kazyanskaya E., Aquadro J., Golas A., Fitzgerald M., Dai H., Zhang L. and Hung D. T. (2012). Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. ACS Chemical Biology 7, 1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S. A., Kawate T., Iwase N., Shimizu M., Clatworthy A. E., Kazyanskaya E., Sacchettini J. C., Ioerger T. R., Siddiqi N. A., Minami S., Aquadro J. A., Grant S. S., Rubin E. J. and Hung D. T. (2013). Diarylcoumarins inhibit mycolic acid biosynthesis and kill Mycobacterium tuberculosis by targeting FadD32. Proceedings of the National Academy of Sciences of the United States of America 110, 11565–11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahlan K., Wilson R., Kastrinsky D. B., Arora K., Nair V., Fischer E., Barnes S. W., Walker J. R., Alland D., Barry C. E. III and Boshoff H. I. (2012). SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy 56, 1797–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K., Wang C. and Besra G. S. (2005). Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clinical Microbiology Reviews 18, 81–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P. H. and Lowary T. L. (2010). Epimeric and amino disaccharide analogs as probes of an alpha-(1→6)-mannosyltransferase involved in mycobacterial lipoarabinomannan biosynthesis. Organic & Biomolecular Chemistry 8, 181–192. [DOI] [PubMed] [Google Scholar]
- Thanna S., Knudson S. E., Grzegorzewicz A., Kapil S., Goins C. M., Ronning D. R., Jackson M., Slayden R. A. and Sucheck S. J. (2016). Synthesis and evaluation of new 2-aminothiophenes against Mycobacterium tuberculosis. Organic & Biomolecular Chemistry 14, 6119–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasic T., Zidar N., Kovac A., Turk S., Simcic M., Blanot D., Muller-Premru M., Filipic M., Grdadolnik S. G., Zega A., Anderluh M., Gobec S., Kikelj D. and Peterlin Masic L. (2010). 5-Benzylidenethiazolidin-4-ones as multitarget inhibitors of bacterial Mur ligases. ChemMedChem 5, 286–295. [DOI] [PubMed] [Google Scholar]
- Tran A. T., Wen D., West N. P., Baker E. N., Britton W. J. and Payne R. J. (2013). Inhibition studies on Mycobacterium tuberculosis N-acetylglucosamine-1-phosphate uridyltransferase (GlmU). Organic & Biomolecular Chemistry 11, 8113–8126. [DOI] [PubMed] [Google Scholar]
- Trivedi O. A., Arora P., Sridharan V., Tickoo R., Mohanty D. and Gokhale R. S. (2004). Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature 428, 441–445. [DOI] [PubMed] [Google Scholar]
- Trunkfield A. E., Gurcha S. S., Besra G. S. and Bugg T. D. (2010). Inhibition of Escherichia coli glycosyltransferase MurG and Mycobacterium tuberculosis Gal transferase by uridine-linked transition state mimics. Bioorganic and Medicinal Chemistry 18, 2651–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull W. B., Shimizu K. H., Chatterjee D., Homans S. W. and Treumann A. (2004). Identification of the 5-methylthiopentosyl substituent in Mycobacterium tuberculosis lipoarabinomannan. Angewandte Chemie. International Edition In English 43, 3918–3922. [DOI] [PubMed] [Google Scholar]
- van Heijenoort Y., Leduc M., Singer H. and van Heijenoort J. (1987). Effects of moenomycin on Escherichia coli. Journal of General Microbiology 133, 667–674. [DOI] [PubMed] [Google Scholar]
- Varela C., Rittmann D., Singh A., Krumbach K., Bhatt K., Eggeling L., Besra G. S. and Bhatt A. (2012). MmpL genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria. Chemistry and Biology 19, 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcheze C., Baughn A. D., Tufariello J., Leung L. W., Kuo M., Basler C. F., Alland D., Sacchettini J. C., Freundlich J. S. and Jacobs W. R. Jr. (2011). Novel inhibitors of InhA efficiently kill Mycobacterium tuberculosis under aerobic and anaerobic conditions. Antimicrobial Agents and Chemotherapy 55, 3889–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W., Blanot D. and de Pedro M. A. (2008). Peptidoglycan structure and architecture. FEMS Microbiology Reviews 32, 149–167. [DOI] [PubMed] [Google Scholar]
- Wang L. Q., Falany C. N. and James M. O. (2004). Triclosan as a substrate and inhibitor of 3′-phosphoadenosine 5′-phosphosulfate-sulfotransferase and UDP-glucuronosyl transferase in human liver fractions. Drug Metabolism and Disposition: The Biological Fate of Chemicals 32, 1162–1169. [DOI] [PubMed] [Google Scholar]
- Warrier T., Tropis M., Werngren J., Diehl A., Gengenbacher M., Schlegel B., Schade M., Oschkinat H., Daffe M., Hoffner S., Eddine A. N. and Kaufmann S. H. (2012). Antigen 85C inhibition restricts Mycobacterium tuberculosis growth through disruption of cord factor biosynthesis. Antimicrobial Agents and Chemotherapy 56, 1735–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Aoyagi Y., Ridell M. and Minnikin D. E. (2001). Separation and characterization of individual mycolic acids in representative mycobacteria. Microbiology 147, 1825–1837. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Aoyagi Y., Mitome H., Fujita T., Naoki H., Ridell M. and Minnikin D. E. (2002). Location of functional groups in mycobacterial meromycolate chains; the recognition of new structural principles in mycolic acids. Microbiology 148, 1881–1902. [DOI] [PubMed] [Google Scholar]
- Wiedemann I., Breukink E., van Kraaij C., Kuipers O. P., Bierbaum G., de Kruijff B. and Sahl H. G. (2001). Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. Journal of Biological Chemistry 276, 1772–1779. [DOI] [PubMed] [Google Scholar]
- Wilson R., Kumar P., Parashar V., Vilcheze C., Veyron-Churlet R., Freundlich J. S., Barnes S. W., Walker J. R., Szymonifka M. J., Marchiano E., Shenai S., Colangeli R., Jacobs W. R. Jr., Neiditch M. B., Kremer L. and Alland D. (2013). Antituberculosis thiophenes define a requirement for Pks13 in mycolic acid biosynthesis. Nature Chemical Biology 9, 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolucka B. A., McNeil M. R., de Hoffmann E., Chojnacki T. and Brennan P. J. (1994). Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. Journal of Biological Chemistry 269, 23328–23335. [PubMed] [Google Scholar]
- World Health Organization (2014). Global Tuberculosis Report. World Health Organisation, Geneva, Switzerland. [Google Scholar]
- World Health Organization (2016). Global Tuberculosis Report. World Health Organisation, Geneva, Switzerland. [Google Scholar]
- Zhang J., Angala S. K., Pramanik P. K., Li K., Crick D. C., Liav A., Jozwiak A., Swiezewska E., Jackson M. and Chatterjee D. (2011). Reconstitution of functional mycobacterial arabinosyltransferase AftC proteoliposome and assessment of decaprenylphosphorylarabinose analogues as arabinofuranosyl donors. ACS Chemical Biology 6, 819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. and Young D. (1994). Molecular genetics of drug resistance in Mycobacterium tuberculosis. Journal of Antimicrobial Chemotherapy 34, 313–319. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Bulloch E. M., Bunker R. D., Baker E. N. and Squire C. J. (2009). Structure and function of GlmU from Mycobacterium tuberculosis. Acta Crystallographica. Section D: Biological Crystallography 65, 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]