ABSTRACT
Inflammation is the host self-protection mechanism to eliminate pathogen invasion. The excessive inflammatory response can result in uncontrolled inflammation, autoimmune diseases, or pathogen dissemination. Recent studies have widely shown that microRNAs (miRNAs) contribute to the regulation of inflammation in mammals by repressing gene expression at the posttranscriptional level. However, the miRNA-mediated mechanism in the inflammatory response in fish remains hazy. In the present study, the regulatory mechanism of the miR-216a-mediated inflammatory response in teleost fish was addressed. We found that the expression of miR-216a could be significantly upregulated in the miiuy croaker after challenge with Vibrio anguillarum and lipopolysaccharide. Bioinformatics predictions demonstrated a potential binding site of miR-216a in the 3′ untranslated region of the p65 gene, and the result was further confirmed by luciferase assay. Moreover, both the mRNA and protein levels of p65 in macrophages were downregulated by miR-216a. Deletion mutant analysis of the miR-216a promoter showed that the Ap1 and Sp1 transcription factor binding sites are indispensable for the transcription of miR-216a. Further study revealed that overexpression of miR-216a suppresses inflammatory cytokine expression and negatively regulates NF-κB signaling, which inhibit an excessive inflammatory response. The collective results indicate that miR-216a plays a role as a negative regulator involved in modulating the bacterium-induced inflammatory response.
KEYWORDS: microRNA-216a, inflammatory responses, p65, NF-κB, teleost fish
INTRODUCTION
Innate immunity is the initial defense against pathogen infections. Detection of the invading pathogen is an essential step of innate immune responses and is mediated by various germ line-encoded pattern recognition receptors (PRRs), which sense pathogen-associated molecular patterns (PAMPs) derived from bacteria, viruses, fungi, and protozoa (1, 2). As one of the most studied PRRs, Toll-like receptors (TLRs) are known to control multiple aspects of the innate immune response (2). TLR signals elicit innate immunity by initiating multiple intracellular signaling cascades that include variable adaptor proteins and transcriptional factors. Activation of NF-κB leads to the expression of a wide spectrum of inflammation-associated genes, including chemokines, cytokines, and adhesion molecules, which are closely related to eliminate the pathogen and which are important for the development of lasting protection against future infection (3). To maintain a balance between inflammatory pathology and protective immunity, the immune response and the inflammation that occurs upon infection need to be tightly regulated. Any dysregulation of this process results in uncontrolled inflammation, autoimmune diseases, or pathogen dissemination.
NF-κB is an important transcription factor which regulates the transcription and expression of multiple genes. The NF-κB family has five members, including p65 (RelA), NF-κB1 (p50/p105), NF-κB2 (p52/p100), RelB, and c-Rel (4). These members generally bind as a homodimer or heterodimer, with the heterodimeric p65/p50 complex appearing to be the most abundant form (4, 5). p65, a key component of the NF-κB family, is closely related to manipulation of the process of transcription of target genes through combination with p50. In mammals, when the signaling cascades are activated, the p65/p50 complex exposes the nuclear localization signal and translocates into the nucleus, thereby modulating downstream gene transcription (6, 7). In fish, the signaling cascades are conserved as well and include orthologous MyD88, SARM1, Tollip, IκB kinase (IKK) complex-associated protein (IKAP), IRAP, TRIF, and the central intermediator interferon regulatory factors, which lead to the translocation of NF-κB to the nucleus and the secretion of inflammatory cytokines (8). Growing evidence has shown that many molecules are positive or negative regulators involved in the process of NF-κB activation. For instance, RIG-I can bind with the 3′ untranslated region (3′ UTR) of NF-κB1 to enhance its translation by recruiting ribosomal protein Rpl8/13 and rRNA (9). The V protein of measles virus interacts with p65, the subunit of NF-κB, to negatively regulate NF-κB nuclear accumulation (10), and the methyltransferase SETD6 has also been reported to methylate p65 to inhibit NF-κB-dependent transcription (11). In mammals, IKK activity is critical for activating NF-κB signaling. However, evidence has also pointed out that zebrafish IKK1, unlike mammalian IKK1, can function as a repressor of NF-κB (12).
MicroRNAs (miRNAs) are a class of short (19- to 24-nucleotide), endogenous, noncoding RNAs that regulate gene expression at the posttranscriptional level via an RNA interference mechanism (13). Currently, thousands of miRNAs have been identified in mammals, and as many as 60% of all mRNAs are reported to be regulated by miRNAs (14, 15). miRNAs play important roles in many fundamental biological processes, such as immune regulation and inflammatory responses (16). NF-κB is well-known to play a critical role in inflammation responses, and a series of miRNAs has been shown to be involved in the regulation of NF-κB activation. For example, miRNA-146 (miR-146), one of the most studied miRNAs, inhibits NF-κB activity by targeting and regulating IRAK1 and TRAF6 upon lipopolysaccharide (LPS) stimulation (17). In contrast, miR-26b enhances NF-κB activity and thus upregulates cytokine production in alveolar macrophages challenged with LPS through the negative regulation of PTEN (18). miR-301a increases NF-κB activity by inhibition of NF-κB repressing factor (NκRF), which in turn promotes the transcription of miR-301a (19). Nevertheless, the function of miRNAs in regulating NF-κB signaling against varied pathogens or stimuli in fish remains largely unknown.
The miiuy croaker (Miichthys miiuy), a member of the Sciaenidae family, is an economically important marine fish in China. The species has been studied in-depth at the transcriptome (20), whole-genome (21), and functional gene and immune pathway regulation (22–25) levels, which makes the miiuy croaker an excellent model for studying the immune response of fish. However, with the increasing industrial farming of the miiuy croaker, the fish has been challenged by many more diseases caused by bacteria, parasites, and viruses than ever, resulting in severe infections and economic losses. Among these infectious pathogens is Vibrio anguillarum, a Gram-negative bacterium that can cause severe epidemic vibriosis, resulting in high mortality and enormous economic losses. V. anguillarum grows rapidly at temperatures of between 25°C and 30°C, forming cream-colored and round colonies (26). Typical external clinical signs of infection with the bacterium include weight loss and red spots on the ventral and lateral areas of the fish, and the pathogen is found in high concentrations in the blood and hematopoietic tissues, such as the spleen and kidney (26, 27).
In the current study, we report that miR-216a acts as a novel negative regulator involved in the inflammation reaction through repression of NF-κB signaling. Previous studies have shown that miR-216a functions as a tumor suppressor by regulating pancreatic cancer cells by targeting and regulating the JAK/STAT pathway (28). Herein, we demonstrate that miR-216a, which is upregulated by V. anguillarum and LPS, could subsequently repress the production of inflammatory cytokines by targeting and regulating the NF-κB subunit p65. These findings suggest that the tight regulation of miR-216a is critical for inhibition of an excessive inflammation reaction and provide a new miRNA-mediated mechanism of regulation of the response against pathogen infection in fish species.
RESULTS
V. anguillarum and LPS enhance miR-216a expression.
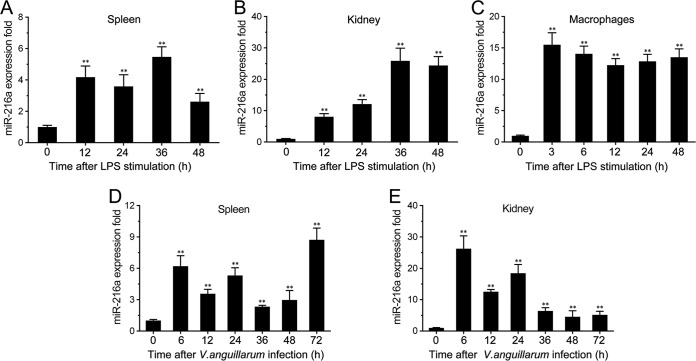
To explore the miRNAs potentially involved in the response to bacterial stimuli, the miRNA expression pattern in LPS-stimulated miiuy croaker spleen tissues was tested. Deep sequencing analysis revealed that a series of miRNAs presented differential expression, and we focused on miR-216a, whose expression was significantly upregulated following LPS stimulation (see Table S2 in the supplemental material). Further validation of the miR-216a expression pattern was analyzed in vivo and in vitro using quantitative real-time PCR. As shown in Fig. 1A and B, miR-216a expression was upregulated in both LPS-stimulated spleen and kidney tissues, and the highest level of miR-216a was expressed at 36 h poststimulation in both spleen and kidney tissues. Moreover, miR-216a expression levels were also examined in miiuy croaker macrophages, and the expression of miR-216a presented a significant increase and reached a peak at 3 h after stimulation with LPS (Fig. 1C). To further validate the findings, we determined the profiles of miR-216a under infection with the Gram-negative bacterium V. anguillarum. V. anguillarum is a Gram-negative, comma-shaped, rod bacterium which can induce a deadly vibriosis and affect various fish species (26, 27). The results revealed that miR-216a was highly expressed in both spleen and kidney tissues upon V. anguillarum infection (Fig. 1D and E). As shown in Fig. 1D, the expression of miR-216a in spleen tissue reached a peak at 72 h. In comparison, miR-216a expression levels in kidney tissue reached a peak at the early stage of infection (Fig. 1E). Together, these data indicate that miR-216a expression can be upregulated by LPS and V. anguillarum and that miR-216a may participate in the regulation of the immune response upon bacterial infection.
FIG 1.
Expression profiles of miR-216a detected by quantitative reverse transcription-PCR. (A to C) Expression profiles of miR-216a in LPS-stimulated miiuy croaker spleen (A), kidney (B), and macrophages (C); (D and E) expression profiles of miR-216a in spleen (D) and kidney (E) samples after V. anguillarum infection. The data were normalized to those for 5.8S rRNA. Results are standardized to a value of 1 for the control samples. Data represent the means ± SEs from three independent experiments performed in triplicate. **, P < 0.01 versus 0 h.
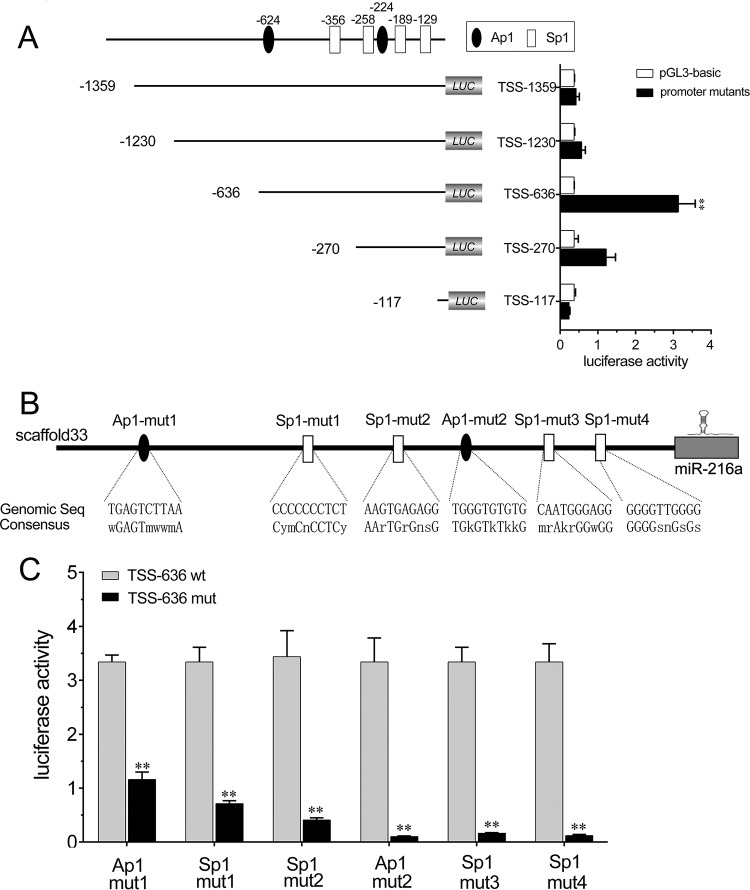
The Ap1 and Sp1 sites are significant for the transcriptional activity of the miR-216a promoter.
To investigate the transcriptional regulation of miR-216a, we first analyzed the genomic information for miR-216a from the miiuy croaker whole genome (scaffold 33) (21), and a schematic representation of the miR-216a promoter was predicted. To determine the fully intact promoter of miR-216a, deletion mutation assays were conducted. A set of luciferase reporters of different lengths was constructed from the sequence upstream of the putative transcription start site (TSS) of miR-216a and then transfected into HEK293 cells to determine the basal promoter activity. As shown in Fig. 2A, the highest luciferase activity was produced by the luciferase reporter TSS-636, which led to an 8.4-fold induction of luciferase activity compared with that achieved with the pGL3-basic empty plasmid. The results indicate that TSS-636 possesses fully intact promoter activity. However, transfection of the TSS-1359, TSS-1230, or TSS-117 reporter plasmid resulted in less basal promoter activity, which indicated that the region from positions −636 to −117 is required for intact promoter activity, the region from positions −1359 to −636 may contain a negative regulatory element(s), and multiple cis-acting elements in the promoter region coordinate to regulate the initiation activity. To investigate the transcription factors involved in the transcription of miR-216a, the transcription factors in the TSS-636 luciferase reporter plasmid involved in the transcription of miR-216a were analyzed with Alibaba (v2.1) software. The Ap1 and Sp1 sites were predicted to be separately located in the promoter region. We then constructed six mutations that mutated either the Ap1 site or the Sp1 site in the TSS-636 plasmid to explore the essential transcription factor binding sites (Fig. 2B). As shown in Fig. 2C, mutation of any of the Ap1 or Sp1 binding sites in the miR-216a promoter resulted in the significant attenuation of promoter activity. These results indicate that the Ap1 and Sp1 sites are each essential for the transcription of miR-216a and that mutation of either of them attenuates the promoter activity. Taken together, these data suggest that the region from positions −636 to −117 is required for basal miR-216a promoter activity and that the Ap1 and Sp1 transcription factor binding sites in the promoter of miR-216a are required for the transcription of miR-216a.
FIG 2.
Ap1 and Sp1 sites are required for the transcription of miR-216a. (A) Schematic representation of the region encoding miiuy croaker miR-216a. (Top) The putative transcription start sites (TSS) of miR-216a are indicated. (Bottom left) miR-216a promoter reporter mutants containing various lengths of the miR-216a promoter region used to transfect HEK293 cells. (Bottom right) At 24 h posttransfection, luciferase activity was measured, and the luciferase activity of the mock-transfected empty vector pGL3-basic group was regarded as the control. (B) Schematic representation of the binding sites for Ap1 and Sp1. Seq, sequence. (C) HEK293 cells were transfected with the wild-type miR-216a promoter reporters (TSS-636 wt) or the mutated miR-216a promoter reporters (TSS-636 mut). At 24 h posttransfection, the luciferase activity was measured. All data are representative of those from at least three independent experiments. **, P < 0.01 versus the controls.
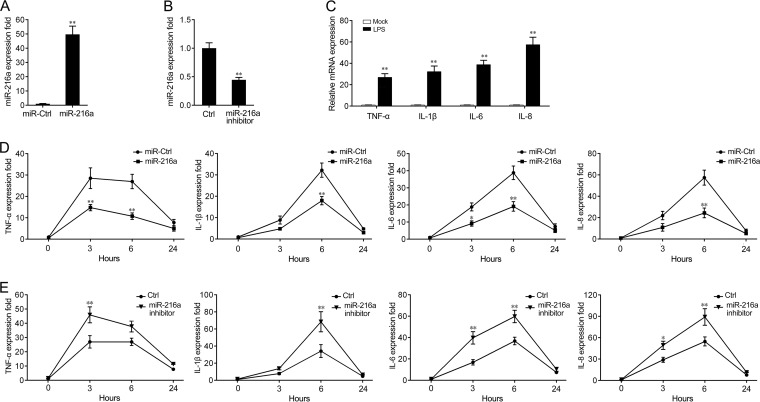
miR-216a suppresses the expression of LPS-induced inflammatory cytokines.
As our results have demonstrated that miR-216a could be upregulated by Gram-negative bacteria, we thus further investigated the role of miR-216a in the regulation of the immune response during pathogen infection. First, the effects of synthetic miR-216a mimics and the miR-216a inhibitor on the expression of miR-216a in miiuy croaker macrophages were examined. miRNA mimics are synthetic double-stranded RNAs that stimulate naturally occurring mature miRNAs, whereas the miRNA inhibitor is a synthetic single-stranded RNA that sequesters and inhibits intracellular miRNAs. Miiuy croaker macrophages were transfected with miR-216a mimics or control mimics and the miR-216a inhibitor or the control inhibitor. As expected, the miR-216a mimics significantly increased the level of miR-216a expression, whereas the miR-216a inhibitor decreased the level of miR-216a expression (Fig. 3A and B). To explore whether LPS stimulates the expression of inflammatory cytokines in miiuy croaker macrophages, cells were stimulated with purified LPS, and then some of the inflammatory cytokines were monitored. The results demonstrated that the mRNA levels of expression of certain inflammatory cytokines, including tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β), IL-6, and IL-8, were significantly upregulated in miiuy croaker macrophages after stimulation with LPS (Fig. 3C). Afterwards, the effects of miR-216a and LPS stimulation on the levels of mRNA expression of inflammatory cytokines were investigated. To this end, we transfected miiuy croaker macrophages with miR-216a mimics or control mimics and then challenged the macrophages with LPS for 3 h, 6 h, and 24 h. As shown in Fig. 3D, transfection with the miR-216a mimics inhibited the mRNA expression of LPS-induced TNF-α, IL-1β, IL-6, and IL-8. The results also demonstrated that the miR-216a mimics exerted a time-dependent effect on the expression of the inflammatory cytokines, in which miR-216a markedly inhibited the expression levels of the inflammatory cytokines at 6 h poststimulation (Fig. 3D). Accordingly, inhibition of miR-216a significantly increased the mRNA expression of the LPS-induced inflammatory cytokines (Fig. 3E). Taken together, these data indicate that miR-216a can downregulate the mRNA expression of inflammatory cytokines upon LPS stimulation and that it may act as a negative regulator involved in LPS-induced immune responses.
FIG 3.
miR-216a suppresses the mRNA expression of LPS-induced TNF-α, IL-1β, IL-6, and IL-8. (A and B) Miiuy croaker macrophages were transfected with control mimics (miR-Ctrl) or miR-216a mimics (miR-216a) (A) and the miR-216 inhibitor or control inhibitor (Ctrl) (B) at a final concentration of 100 nM. At 24 h posttransfection, miR-216a expression was measured by quantitative reverse transcription-PCR, and the level of expression was normalized to that of 5.8S rRNA. (C) After 3 h of LPS stimulation of miiuy croaker macrophages, the levels of expression of TNF-α, IL-1β, IL-6, and IL-8 were determined by qPCR and normalized to the level of expression of β-actin in each sample. (D and E) Miiuy croaker macrophages were transfected with miR-216a mimics (miR-216a) or control mimics (miR-Ctrl) (D) and the miR-216a inhibitor or the control inhibitor (Ctrl) (E). After 24 h, the cells were stimulated with LPS for 3 h, 6 h, and 24 h, and the expression levels of TNF-α, IL-1β, IL-6, and IL-8 were analyzed by quantitative reverse transcription-PCR. Results are standardized to those for the control cells, for which the results were given a value of 1. Data are presented as the means ± SEs from three independent experiments performed in triplicate. **, P < 0.01 versus the controls; *, P < 0.05 versus the controls.
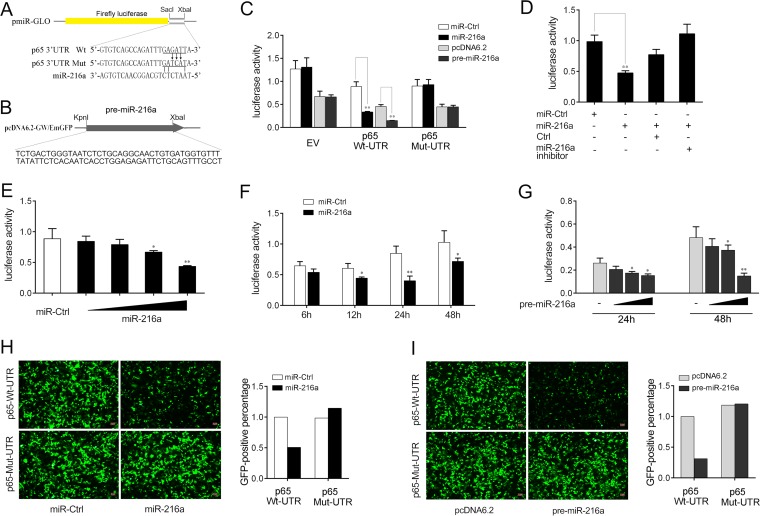
miR-216a targets p65.
We next investigated the possible targets through which miR-216a acts to modulate inflammatory cytokine expression. Computational predictions made by use of the TargetScan, miRanda, and miRInspector algorithms were used to search for potential miR-216a targets. We found putative miR-216a binding sites in the 3′ UTR of the miiuy croaker p65 gene (Fig. 4A). To obtain direct evidence that miR-216a targets the 3′ UTR of the p65 gene, luciferase reporter constructs were generated by cloning either the wild-type 3′ UTR or the mutated 3′ UTR of p65 into the pmir-GLO vector (Fig. 4A). In addition, the pre-miR-216a plasmid was constructed by cloning the miiuy croaker pre-miR-216a sequence into the pcDNA6.2-GW/EmGFP vector (Fig. 4B). Then, HEK293 cells were transfected with the reporter plasmids together with the miR-216a mimics, the pre-miR-216a plasmid, or their controls. The results showed that the miR-216a mimics, as well as the pre-miR-216a plasmid, markedly decreased the luciferase activity in cells transfected with the p65 3′ UTR plasmid compared with that in cells transfected with the pmir-GLO empty plasmid. However, no effect on luciferase activity was observed in cells transfected with the mutated 3′ UTR p65 plasmid (Fig. 4C). Furthermore, we also observed that the introduction of the corresponding base pair exchanges in the miRNA seed region could restore the repression of the mutated reporter (Fig. S1).
FIG 4.
miR-216a targets the 3′ UTR of the miiuy croaker p65 gene. (A) Schematic diagram of the predicted target sites of miR-216a in the 3′ UTR of p65. (B) Construction of the pre-miR-216a plasmid. (C) HEK293 cells were cotransfected with the wild-type p65 3′ UTR (Wt-UTR), the mutant p65 3′ UTR (Mut-UTR), or the empty pmir-GLO vector (EV) together with miR-216a or control mimics (miR-Ctrl) and the pre-miR-216a plasmid or pcDNA6.2-GW/EmGFP. The control mimics and pcDNA6.2-GW/EmGFP were used to control for the same amount of molecules for the transfections. The luciferase activity was determined and normalized to the Renilla luciferase activity. (D) HEK293 cells were cotransfected with the p65 3′ UTR (wild type) together with control mimics (miR-Ctrl), miR-216a, the control inhibitor (Ctrl), or the miR-216a inhibitor for 24 h, and the luciferase activity was determined. For each transfection, the total amount of oligonucleotides was 100 nM. (E) miR-216a (0, 10, 40, 70, and 100 nM) together with control mimics (miR-Ctrl; 100, 90, 60, 30, and 0 nM) was cotransfected into HEK293 cells for 24 h, and then the luciferase activity was determined. (F) Time gradient for transfection of miR-216a. (G) HEK293 cells were transfected with the p65 3′ UTR (wild type) together with a concentration gradient of the pre-miR-216a plasmid. After 24 h or 48 h, the luciferase activity was determined and pcDNA6.2-GW/EmGFP was used as a control. Luciferase activity was normalized to the Renilla luciferase activity. (H and I) The p65 3′ UTR or its mutant was inserted into the mVenus-C1 vector (Addgene), which included the sequence of enhanced green fluorescent protein (GFP). HEK293 cells were cotransfected with the wild-type mVenus-p65 3′ UTR (Wt-UTR) or the mutant mVenus-p65 3′ UTR (Mut-UTR) together with miR-216a or control mimics (H) and pre-miR-216a or pcDNA6.2-GW/EmGFP (I). At 48 h posttransfection, the fluorescence intensity was evaluated. Data are presented as the means ± SEs from at least three independent experiments performed in triplicate. **, P < 0.01 versus the controls; *, P < 0.05 versus the controls.
To further verify this specific binding, HEK293 cells were transfected with miR-216a mimics, the miR-216a inhibitor, or their controls. As shown in Fig. 4D, the inhibition of luciferase activity by the miR-216a mimics was attenuated after cotransfection with the miR-216a inhibitor. Furthermore, concentration and time gradient experiments were conducted with the miR-216a mimics, and the results revealed that the miR-216a mimics diminished luciferase activity in dose-dependent and time-dependent manners (Fig. 4E and F). Similar results were obtained by transfection of the pre-miR-216a plasmid. As shown in Fig. 4G, the pre-miR-216a plasmid also showed a dose-dependent effect on luciferase activity at both 24 h and 48 h posttransfection, and significant inhibition was shown at 48 h posttransfection. Additionally, the miR-216a mimics and the pre-miR-216a plasmid could also downregulate green fluorescent protein (GFP) gene expression when the 3′ UTR of p65 was cloned into the 3′ UTR region of the GFP vector, whereas no change in fluorescence intensity was observed in cells transfected with the mutant p65 3′ UTR (Fig. 4H and I). Collectively, these data demonstrate that miR-216a specifically binds to the 3′ UTR of the miiuy croaker p65 gene.
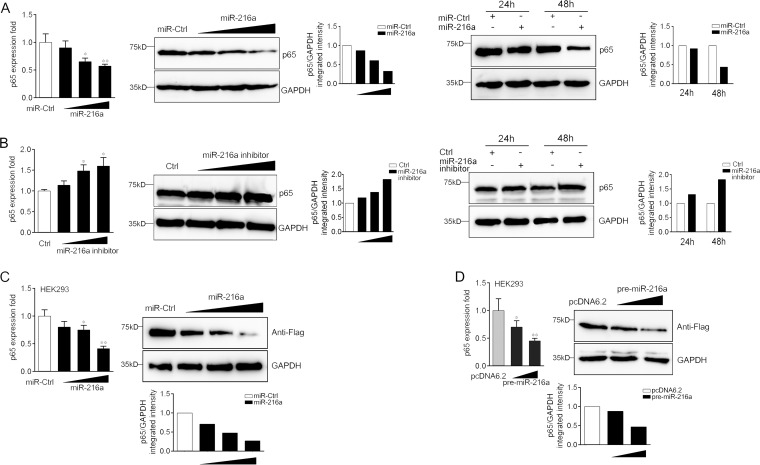
miR-216a modulates the expression of p65 at the posttranscriptional level.
To further validate the function of miR-216a in the regulation of p65 expression, the expression levels of endogenous p65 in miiuy croaker macrophages that had been treated with the miR-216a mimics or the miR-216a inhibitor were examined. As shown in Fig. 5A and B, transfection of the miR-216a mimics significantly suppressed p65 mRNA and protein levels in dose-dependent and time-dependent manners (Fig. 5A), whereas the miR-216a inhibitor obviously increased the level of p65 expression (Fig. 5B). The results indicate that miR-216a can decrease the level of p65 expression at both the mRNA and the protein levels. In addition, a p65 expression plasmid was constructed and transfected into HEK293 cells to further examine the regulation mechanism. To construct the p65 expression plasmid, the full-length coding DNA sequence (CDS) region and the 3′ UTR of the miiuy croaker p65 gene were amplified and cloned into the pcDNA3.1 vector with a Flag tag. HEK293 cells were transfected with the p65 expression plasmid together with the miR-216a mimics, the pre-miR-216a plasmid, or their controls. As expected, the results showed that the miR-216a mimics could significantly suppress the expression of p65 at both the protein and the mRNA levels in a dose-dependent manner (Fig. 5C). A consistent effect was shown by the pre-miR-216a plasmid (Fig. 5D). These data suggest that p65 is a direct target of miR-216a and that miR-216a regulates p65 expression at both the mRNA and the protein levels.
FIG 5.
miR-216a suppresses the expression of p65 at both the mRNA and the protein levels. (A) (Left) Miiuy croaker macrophages were transfected with miR-216a (0, 30, 60, and 90 nM) or control mimics (miR-Ctrl; 90, 60, 30, and 0 nM), and the mRNA and protein levels of p65 were determined by quantitative reverse transcription-PCR and Western blotting, respectively. (Right) The time gradient experiment was also conducted to measure the effect of miR-216a on p65 protein levels. (B) (Left) Miiuy croaker macrophages were transfected with the miR-216a inhibitor (0, 30, 60, and 90 nM) or the control inhibitor (90, 60, 30, and 0 nM), and the mRNA and protein levels of p65 were determined by quantitative reverse transcription-PCR and Western blotting, respectively. (Right) The time gradient experiment was also conducted to measure the effect of the miR-216a inhibitor on p65 protein levels. (C and D) (Left) HEK293 cells were cotransfected with the p65 expression plasmid, which contains the full-length CDS region and the 3′ UTR of miiuy croaker p65, along with control mimics (miR-Ctrl) or miR-216a (C) and the pre-miR-216a plasmid or pcDNA6.2-GW/EmGFP (D) in a concentration gradient manner. The control mimics (C) and pcDNA6.2-GW/EmGFP (D) were used to control for the same amount of molecules for the transfections. (Right) After 48 h, p65 mRNA and protein levels were determined by quantitative reverse transcription-PCR and Western blotting, respectively. Data are presented as the means ± SEs from three independent experiments performed in triplicate. **, P < 0.01 versus the controls; *, P < 0.05 versus the controls. Wt-UTR, wild-type 3′ UTR; Mut-UTR, mutated 3′ UTR.
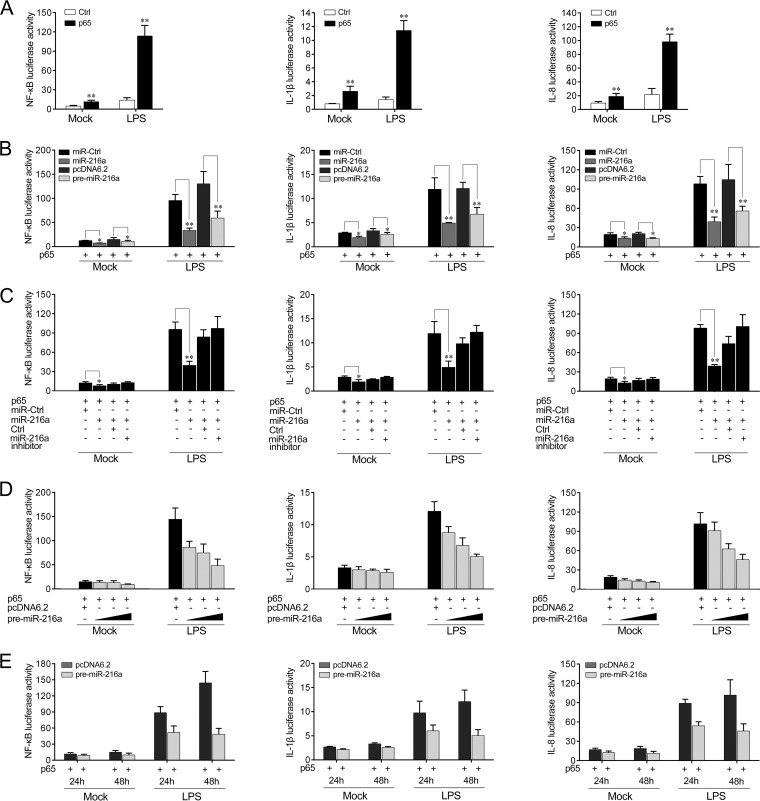
miR-216a modulates the NF-κB signaling pathway.
Previous studies have indicated that activation of the NF-κB pathway leads to the transcription of a variety of genes, such as those for cytokines, chemokines, and adhesion molecules (29). Given that miR-216a modulates the expression of inflammatory cytokines, we thus examined whether miR-216a affects NF-κB signaling. We first transfected HeLa cells with the miiuy croaker p65 plasmid in combination with the NF-κB, IL-1β, or IL-8 reporter gene for 24 h and then stimulated the cells with LPS for 6 h. As shown by the results of dual-luciferase reporter assays (Fig. 6A), the miiuy croaker p65 gene was sufficient to activate the NF-κB reporter gene, as well as the IL-1β and IL-8 reporter genes, in cells stimulated with LPS, indicating the regulatory role of the miiuy croaker p65 gene upon LPS stimulation. Because we found that miR-216a targets and negatively regulates p65 expression, we then examined whether miR-216a modulates NF-κB signaling. To this end, we transfected HeLa cells with the p65 expression plasmid, which contained the full-length CDS region and the 3′ UTR of the miiuy croaker p65 gene together with the miR-216a mimics, the pre-miR-216a plasmid, or their controls, and then stimulated the cells with LPS. The results revealed that both miR-216a mimics and the pre-miR-216a plasmid participated in the downregulation of the NF-κB, IL-1β, and IL-8 reporter genes (Fig. 6B). The regulation of inhibition by miR-216a was shown to be more significant in cells treated with LPS. However, as shown in Fig. 6C, the inhibition effect could be attenuated when the cells were cotransfected with the miR-216a inhibitor. Additionally, concentration and time gradient experiments of the pre-miR-216a plasmid were carried out. As shown in Fig. 6D and E, the pre-miR-216a plasmid showed a dose-dependent effect on the inhibition of the NF-κB, IL-1β, and IL-8 reporter genes (Fig. 6D), and the downregulation effect was shown to be more significant at 48 h posttransfection (Fig. 6E). Collectively, these data sufficiently indicate that miR-216a participates in negatively regulating NF-κB signaling upon LPS stimulation.
FIG 6.
miR-216a inhibits NF-κB signaling upon LPS stimulation. (A) HeLa cells were cotransfected with the p65 expression plasmid or the pcDNA3.1 vector as a control (Ctrl) together with the pRL-TK Renilla luciferase plasmid and the luciferase reporter gene NF-κB, IL-1β, or IL-8, for 24 h and then stimulated with LPS (2 μg/ml) for 6 h. The luciferase activity was measured, and the pcDNA3.1 empty vector was used to control for the same amount of molecules for the transfections. (B) HeLa cells were cotransfected with control mimics (miR-Ctrl), miR-216a, pcDNA6.2-GW/EmGFP, or the pre-miR-216a plasmid together with the NF-κB, IL-1β, or IL-8 luciferase reporter and the p65 expression plasmid. Then the cells were stimulated with LPS (2 μg/ml) for another 6 h and the luciferase activity was measured. The control mimics and the pcDNA6.2-GW/EmGFP plasmid were used to control for the same amount of molecules for transfections. (C) HeLa cells were transfected with control mimics, miR-216a, the control inhibitor (Ctrl), or the miR-216a inhibitor together with the NF-κB, IL-1β, or IL-8 luciferase reporter and the p65 expression plasmid. Then the cells were stimulated with LPS (2 μg/ml) for another 6 h and the luciferase activity was measured. For each transfection, the total amount of oligonucleotides was 100 nM. (D) HeLa cells were transfected with the pre-miR-216a plasmid in a concentration gradient together with the NF-κB, IL-1β, or IL-8 luciferase reporter for 48 h and stimulated with LPS (2 μg/ml) for another 6 h. Afterwards, the luciferase activity was measured, and the pcDNA6.2-GW/EmGFP plasmid was used to control for the same amount of molecules for the transfections. (E) The time gradient experiment with the pre-miR-216a plasmid was conducted in HeLa cells treated with or without LPS. Luciferase activity was normalized to the Renilla luciferase activity. All data are representative of those from at least three independent experiments. **, P < 0.01 versus the controls; *, P < 0.05 versus the controls.
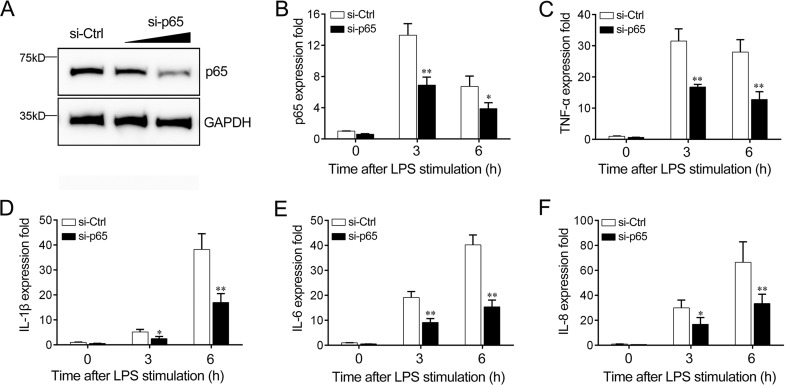
Knockdown of p65 inhibits the LPS-induced inflammatory response.
To confirm the role of p65 in the regulation of LPS-induced inflammatory cytokine expression, we transfected miiuy croaker macrophages with p65-specific small interfering RNA (siRNA) (si-p65) to knock down the expression of p65. The significant inhibition of p65 at both the protein and the mRNA levels was found (Fig. 7A and B). Afterwards, we silenced p65 and tested the expression of the inflammatory cytokine in LPS-challenged miiuy croaker macrophages. As shown in Fig. 7C to F, the knockdown of p65 markedly reduced LPS-triggered TNF-α, IL-1β, IL-6, and IL-8 expression, suggesting that p65 in miiuy croaker macrophages play an essential role in regulating the effect of LPS on the production of inflammatory cytokines.
FIG 7.
Expression levels of p65 and inflammatory cytokines after p65 interference. (A and B) Miiuy croaker macrophages were transfected with control siRNA (si-Ctrl) or p65-specific siRNA (si-p65). After 48 h, p65 protein (A) and mRNA (B) levels were determined by Western blotting and quantitative reverse transcription-PCR, respectively. (C to F) At 48 h after transfection with si-Ctrl or si-p65, miiuy croaker macrophages were stimulated with LPS for 3 h or 6 h. The expression levels of TNF-α (C), IL-1β (D), IL-6 (E), and IL-8 (F) were determined and normalized to the level of β-actin expression. All data are representative of those from at least three independent experiments. **, P < 0.01 versus the controls; *, P < 0.05 versus the controls.
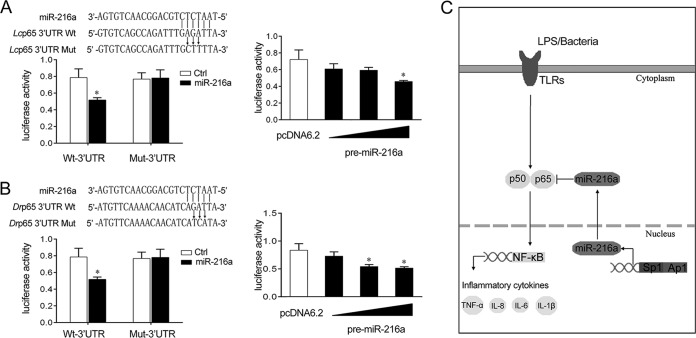
miR-216a regulation of the p65 gene is widely found in teleost fish.
To address the generality of the finding that miR-216a targets and regulates p65 in other fish species, we explored the results in other model animals, including the large yellow croaker and zebrafish. As shown in Fig. 8A, the results indicated that miR-216a mimics, as well as the pre-miR-216a plasmid, showed an inhibitory effect on luciferase activity in the large yellow croaker. A similar regulation mechanism was also observed in the zebrafish (Fig. 8B), suggesting the generality of our findings. Collectively, as shown in Fig. 8C, the transcription factors Ap1 and Sp1 are significant for the transcription of miR-216a. Upon bacterial infection, miR-216a inhibits the expression of inflammatory cytokines by targeting p65 and modulating NF-κB signaling, thereby inhibiting the excessive inflammatory response.
FIG 8.
Proposed model for the mechanism regulated by miR-216a in fish species. (A) (Left top) Schematic diagram of the predicted target sites of miR-216a in the 3′ UTR of Larimichthys crocea p65 (Lcp65). (Left bottom) HEK293 cells were transfected with miR-216a or control mimics (miR-Ctrl) along with the wild-type L. crocea p65 3′ UTR (Wt-UTR) or the mutant L. crocea p65 3′ UTR (Mut-3′ UTR) for 24 h, and the luciferase activity was determined. (Right) HEK293 cells were transfected with the pre-miR-216a plasmid or pcDNA6.2-GW/EmGFP together with the wild-type L. crocea p65 3′ UTR. After 48 h posttransfection, the luciferase activity was determined. (B) (Left top) Schematic diagram of the predicted target sites of miR-216a in the 3′ UTR of Danio rerio p65 (Drp65). (Left bottom) HEK293 cells were transfected with miR-216a or control mimics along with the wild-type D. rerio p65 3′ UTR (Wt-UTR) or the mutant D. rerio p65 3′ UTR (Mut-3′ UTR) for 24 h, and the luciferase activity was determined. (Right) HEK293 cells were transfected with the pre-miR-216a plasmid or pcDNA6.2-GW/EmGFP together with the wild-type D. rerio p65 3′ UTR. After 48 h posttransfection, the luciferase activity was determined. Data are presented as the means ± SEs from three independent experiments performed in triplicate. *, P < 0.05 versus the controls. (C) Proposed model for the mechanism by which induced miR-216a negatively regulates inflammatory cytokine expression by targeting p65 and inhibiting NF-κB signaling, thereby inhibiting the inflammatory response.
DISCUSSION
Innate immune and inflammatory responses are the normal self-protection mechanism to eliminate pathogens and resist microbial invaders, and the regulatory mechanisms in these processes are largely dependent on the NF-κB pathway (1, 2, 4). The excessive immune response and prolonged inflammation can cause a variety of pathologies, leading to either cell lesions or tissue damage. Hence, various layers of negative mechanisms and regulators participate in controlling the homeostasis of the immune system and avoiding excessive inflammation. Because of the complexity of their living environment, fish species are constantly threatened by pathogenic microorganisms, of which bacteria are an important group. Diseases caused by bacterial infections in fish species lead to high mortality and severe economic losses because of their wide prevalence and high incidence and the great harm that they cause. Thus, the regulatory mechanisms in response to bacterial infection are particularly important and prominent. In the present study, the underlying miRNA-mediated mechanisms for avoiding excessive inflammation in teleost fish upon Gram-negative bacterial infection were addressed. miR-216a has been documented to be a negative regulator involved in the inflammatory response in the miiuy croaker. Specifically, we found that the miR-216a expression level could be markedly increased in the miiuy croaker when it was challenged with the Gram-negative bacterium V. anguillarum and LPS. Upregulated miR-216a could modulate the expression of LPS-induced inflammatory cytokines, including TNF-α, IL-1β, IL-6, and IL-8. Further findings suggest that miR-216a targets p65 and negatively regulates inflammatory cytokine expression via NF-κB signaling, thereby inhibiting excessive and prolonged inflammation. These findings therefore provide an opportunity to obtain further understanding of the impact of miRNAs associated with inflammatory responses against bacterial stimuli.
NF-κB is a transcription factor that controls the transcription and expression of a variety of genes. The activation of NF-κB is essential for regulating cell activation, cell proliferation, inflammation, and the immune response (30). p65 is an important subunit of NF-κB which controls gene transcription in combination with the p50 subunit of NF-κB. At steady state, NF-κB is associated with members of the inhibitor of κB (IκB) protein family and is sequestered in the cytoplasm, in which it has no known activity. However, when the signaling pathway is activated, signals through PRRs lead to the phosphorylation of IκB proteins and, ultimately, their degradation. The p65/p50 complex then translocates into the nucleus, binds to κB sites in the promoters/enhancers of the target genes, and thus regulates the transcription of inflammatory cytokines (4, 5, 30). Because these responses are vital for eliminating invading pathogens, NF-κB activity must be tightly regulated to avoid an excessive immune response, and many regulators have been reported to be involved in the process. For example, as a cytoplasmic LIM protein, PDLIM1 binds to and sequesters the NF-κB subunit p65, thereby inhibiting NF-κB-mediated inflammatory signaling (31), and casein kinase 1γ1 (CK1γ1) phosphorylates p65 and promotes its degradation to inhibit the RIG-I/TLR signaling pathway (32). Besides, RIG-I functions as a positive regulator of NF-κB signaling by interacting with NF-κB1 and enhancing its translation (6).
Evidence is rapidly growing that noncoding small RNAs, such as miRNAs, play crucial roles in the initiation and progression of immune responses and pathological inflammation. Many studies have reported that miRNAs participate in the regulation of the NF-κB signaling network (33). Some miRNAs contribute to NF-κB activation in response to pathogen infection. For example, miR-301a can downregulate NF-κB-repressing factor (NκRF) and, consequently, elevate NF-κB activation (19), and miR-381 targets IκBα, which is an inhibitor of NF-κB, and increases the activity of NF-κB, thereby increasing the levels of expression of certain inflammatory cytokines (34). In contrast, many miRNAs are involved in the negative regulation of NF-κB signaling. For instance, miR-146 can inhibit the expression of TRAF6, as well as that of IRAK1, by targeting their 3′ UTRs to downregulate NF-κB activity (17); miR-155 can also limit NF-κB activation by inhibiting both IKKβ and IKKε, leading to a negative regulatory feedback loop (35). Although these reports have documented the molecular mechanisms by which endogenous cellular miRNAs regulate NF-κB signaling in mammals, the underlying mechanisms of miRNAs in fish species are just beginning to be explored. Recently, our studies have reported that fish miR-3570 (24) and miR-214 (36) participate in the regulation of NF-κB signaling by modulating the adaptor protein MyD88 at both the mRNA and the protein levels. Additionally, reports have also indicated that miR-3570 can target and regulate MAVS expression to downregulate NF-κB signaling (22). In this study, endogenous miR-216a in miiuy croaker macrophages was found to negatively regulate NF-κB signaling. The finding that the miR-216a-mediated regulation mechanism occurs by targeting a subunit of NF-κB p65 may provide an indication that it has more immediate effects on the NF-κB signaling pathway.
miR-216a is highly expressed in the mammalian pancreas (37). miR-216a has been shown to function as a tumor suppressor in modulating pancreatic cancer cells by targeting and regulating the JAK/STAT pathway (28). miR-216a is also regarded as a serum biomarker of exocrine pancreas injury during acute pancreatitis (38). PTEN (39) and Smad7 (40) have been identified to be the targets of miR-216a which can regulate the expression of PTEN and Smad7 and affect the expression of its downstream genes. Despite all of these reports, there are very few functional reports of how miR-216a is associated with the inflammatory response. We demonstrated that miR-216a regulates inflammation by suppressing the expression of TNF-α, IL-1β, IL-6, and IL-8. Because of the importance of miRNA activity in gene regulation, interest has focused on the mechanisms by which miRNAs are governed. In the study described here, we investigated the transcriptional regulation of miR-216a and demonstrated that the two Ap1 binding sites and four Sp1 binding sites in the promoter of miR-216a are indispensable for the transcription of miR-216a. Ap1 is a cellular transcription factor complex involved in several cellular functions, including immune responses. Reports have indicated that Ap1 can be induced during infections and that it can also get itself activated by proinflammatory cytokines, such as TNF-α and IL-1 (41). With regard to Sp1, it is a ubiquitous transcription factor that has been involved in regulating many genes by binding the promoter or by acting as a coregulator interacting with other transcription factors (42). In response to LPS, Sp1 has been demonstrated to be activated through the p38/mitogen-activated protein kinase pathway. Considering that our data show that both the Ap1 and the Sp1 binding sites are indispensable for the transcription of miR-216a and the fact that Ap1 gets itself activated by proinflammatory cytokines, a classical negative-feedback loop may develop to prevent host inflammation during bacterial induction. Further studies are needed to test this possibility.
In conclusion, our data demonstrate that the host miRNA miR-216a is associated with the immune response to Gram-negative bacteria and that it acts as a negative regulator of inflammatory cytokine expression in fish. The underlying regulatory mechanism of miR-216a is through targeting and downregulating an important subunit of NF-κB p65 and negatively modulating NF-κB signaling, which ultimately lead to the inhibition of an excessive immune response. These findings suggest a novel miRNA-mediated mechanism for avoiding excessive inflammation and immune escape in fish, which provides new insights into the regulatory mechanism of the NF-κB signaling pathway.
MATERIALS AND METHODS
Animals and challenge.
Healthy miiuy croakers (weight, ∼750 g) were obtained from Zhoushan Fisheries Research Institute, Zhejiang Province, China. The fish were acclimated in aerated seawater tanks at 25°C and grown for at least a week for acclimation before the experiments. Bacterial challenge was performed as described previously (25, 43). Briefly, healthy miiuy croakers were challenged with 1 ml V. anguillarum (1.5 × 108 CFU/ml) or 1 ml suspension of ultrapure LPS (1 mg/ml) through the intraperitoneal route, and individuals challenged with 1 ml physiological water served as a comparison group. The fish were sacrificed, and the immune tissues (spleen and kidney) were collected for RNA extraction. All animal experimental procedures were performed in accordance with the National Research Council's Guide for the Care and Use of Laboratory Animals (44), and the experimental protocol was reviewed and approved by the Research Ethics Committee of the College of Marine Science, Zhejiang Ocean University.
Cell culture and treatment.
HEK293 and HeLa cells were cultured in Dulbecco modified Eagle medium (HyClone) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C in 5% CO2. Miiuy croaker macrophages were isolated from head kidney tissues as described in our previous studies (43). In brief, tissues were aseptically pushed through a 100-μm-mesh-size nylon mesh to give a cell suspension that was then loaded onto a 34%/51% Percoll (Pharmacia, USA) density gradient to obtain macrophages. The cells were cultured in L-15 medium (HyClone) supplemented with 20% FBS and seeded into six-well plates at 26°C in 4% CO2. The macrophages were allowed to adhere for 12 h and then stimulated with ultrapure LPS, which was purified by ion-exchange chromatography (10 μg/ml), and harvested at different times for RNA extraction. Cell viability was determined by a trypan blue dye exclusion assay.
Plasmid construction.
In order to construct the p65 3′ UTR reporter vector, the full-length 3′ UTR of the miiuy croaker p65 gene was amplified using PCR and cloned into the pmir-GLO luciferase reporter vector (Promega) using the SacI and XbaI restriction sites. Similarly, the 3′ UTRs of the zebrafish (Danio rerio) p65 gene and the large yellow croaker (Larimichthys crocea) p65 gene were amplified and inserted into the SacI and XbaI restriction sites of the pmir-GLO vector. The mutant type of the p65 3′ UTR reporter vector was constructed by using a Mut Express II fast mutagenesis kit (v2; Vazyme) with specific primers (see Table S1 in the supplemental material). Moreover, the miiuy croaker p65 3′ UTR or its mutant was inserted into the mVenus-C1 vector (Addgene), which included the sequence of enhanced green fluorescent protein (GFP). In addition, to construct the pre-miRNA vector, the pre-miR-216a sequence was PCR amplified and then cloned into the pcDNA6.2-GW/EmGFP vector (Invitrogen). The luciferase reporter plasmids TSS-1359, TSS-1230, TSS-636, TSS-270, and TSS-117, which contained the 1,359-, 1,230-, 636-, 270-, and 117-bp proximal promoter sequences of miiuy croaker miR-216a, respectively, were constructed by PCR amplification using genomic DNA of the miiuy croaker as the template and subsequent cloning into the pGL3-basic vector (Promega). miR-216a promoter reporter constructs containing mutations for Sp1 and Ap1 were constructed with specific primers (Table S1). To construct the p65 expression vector, the full length of the CDS region and the 3′ UTR of the miiuy croaker p65 gene were amplified with specific primer pairs with the Flag tag and cloned into the pcDNA3.1 vector (Invitrogen) using the BamHI and XbaI restriction sites. The total plasmid sequences were verified by Sanger sequencing and extracted through the use of an endotoxin-free plasmid DNA miniprep kit (Tiangen).
miRNA mimics and inhibitor.
miR-216a mimics and the miR-216a inhibitor were commercially synthesized by GenePharma (Shanghai, China). The primer sequences were as follows: for the miR-216a mimics, 5′-UAAUCUCUGCAGGCAACUGUGA-3′ (sense) and 5′-ACAGUUGCCUGCAGAGAUUAUU-3′ (antisense); for the control mimics, 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (antisense); for the miR-216a inhibitor, 5′-UCACAGUUGCCUGCAGAGAUUA-3′ (chemically modified by 2′-Ome); and for the control inhibitor, 5′-CAGUACUUUUGUGUAGUACAA-3′. Miiuy croaker macrophages were seeded for up to 12 h and then transfected with 100 nM miR-216a mimics and the miR-216a inhibitor using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocols. At 24 h posttransfection, the cells were stimulated with ultrapure LPS (10 μg/ml). The cells were collected after LPS induction for 3 h, 6 h, and 24 h.
RNA interference.
The p65-specific siRNA (si-p65) sequences were 5′-GGAGGAGUUUGACCUGAAUTT-3′ (sense) and 5′-AUUCAGGUCAAACUCCUCCTT-3′ (antisense). The scrambled control RNA (si-Ctrl) sequences were 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (antisense). Miiuy croaker macrophages were transfected with each siRNA by using the Lipofectamine 2000 reagent for up to 48 h and then stimulated with ultrapure LPS (10 μg/ml) for 3 h and 6 h.
qPCR.
Total RNA was extracted with the TRIzol reagent (Invitrogen) according to the instructions of the manufacturer. For mRNA analysis, reverse transcription was performed using a FastQuant reverse transcription kit (Tiangen). To quantify mature miR-216a expression, the small RNA was extracted by using an miRcute miRNA isolation kit (Tiangen), and an miRcute miRNA FirstStrand cDNA synthesis kit (Tiangen) was applied for reverse transcription of miRNAs. All gene transcripts were measured by quantitative real-time PCR (qPCR) using a 7500 real-time PCR system (Applied Biosystems) and SYBR Premix Ex Taq polymerase (TaKaRa) (25). Then, primers specific for the genes were designed, and β-actin and 5.8S rRNA were applied as internal controls (Table S1). The fold change in expression was calculated using the 2−ΔΔCT threshold cycle (CT) method of relative quantification. All experiments were conducted in triplicate.
Computational prediction of miR-216a targets.
To further analyze the functions of miR-216a, we used three computational approaches with the TargetScan (45), miRanda (46), and miRInspector (47) algorithms to predict the targets of miR-216a. Predictions were ranked on the basis of the predicted efficacy of targeting, as calculated using the context and the scores for the sites.
Dual-luciferase reporter assays.
For miRNA target identification, HEK293 cells were transfected with the p65 3′ UTR reporter vector together with the miR-216a mimics, the miR-216a inhibitor, the pre-miR-216a plasmid, or their controls. Reporter luciferase activities were measured using a dual-luciferase reporter assay system (Promega). To determine the functional regulation of p65 during LPS stimulation, HeLa cells were transfected with the p65 expression plasmid together with a Renilla luciferase plasmid and NF-κB, IL-8, and IL-1β luciferase reporters and then stimulated with ultrapure LPS (2 μg/ml). After they were treated with LPS for 6 h, the cells were collected and assayed for reporter activity using the dual-luciferase reporter assay system. In addition, miR-216a promoter luciferase constructs or constructs with mutations in the miR-216a promoter were transfected into HEK293 cells to measure luciferase activity. Cotransfection was performed with the constructed plasmids, the miRNA mimics, or the inhibitor using the Lipofectamine 2000 reagent. The luciferase activity values were achieved by comparison with the value for the Renilla luciferase control. For each experiment, three independent experiments were conducted, and each experiment was done in triplicate.
Western blotting.
Cellular lysates were generated by using 1× SDS-PAGE loading buffer. Proteins were extracted from the cells, the concentrations were measured with a bicinchoninic acid protein assay kit (Vazyme), and then the proteins were subjected to SDS-PAGE (10%) and transferred to polyvinylidene difluoride membranes (Millipore) by semidry blotting (Bio-Rad Trans Blot Turbo system). The membranes were blocked with 5% bovine serum albumin. The proteins were blotted with different antibodies. The antibody against p65 was diluted 1:1,000 (Cell Signaling Technology), anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was diluted 1:2,000 (Sigma-Aldrich), and horseradish peroxidase-conjugated anti-rabbit IgG or anti-mouse IgG (Abbkine) was diluted 1:5,000. The results are the representative of those from three independent experiments. The immunoreactive proteins were detected by using a WesternBright enhanced chemiluminescence assay (Advansta). Digital imaging was performed with a cold charge-coupled-device camera.
Statistical analysis.
For all the experiments, at least three independent experiments with three technical replicates for each experiment were performed. The relative gene expression data were acquired using the 2−ΔΔCT method, and comparisons between groups were analyzed by one-way analysis of variance (ANOVA) followed by Duncan's multiple-comparison test (48). Results are expressed as the mean ± standard error (SE), and differences between means with P values of <0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by the National Natural Science Foundation of China (31370049 and 31672682).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00256-18.
REFERENCES
- 1.Janeway AC Jr, Medzhitov R. 2002. Innate immune recognition. Annu Rev Immunol 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Kopp EB, Medzhitov R. 1999. The Toll-receptor family and control of innate immunity. Curr Opin Immunol 11:13–18. doi: 10.1016/S0952-7915(99)80003-X. [DOI] [PubMed] [Google Scholar]
- 4.Grilli M, Chiu JJ, Lenardo MJ. 1993. NF-kappaB and Rel: participants in a multiform transcriptional regulatory system. Int Rev Cytol 143:1–62. doi: 10.1016/S0074-7696(08)61873-2. [DOI] [PubMed] [Google Scholar]
- 5.Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S. 1995. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev 9:2723–2735. [DOI] [PubMed] [Google Scholar]
- 6.Hayden MS, Ghosh S. 2004. Signaling to NF-κB. Genes Dev 18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 7.Hayden MS, Ghosh S. 2012. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. 2003. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 9.Zhang HX, Liu ZX, Sun YP, Zhu J, Lu SY, Liu XS, Huang QH, Xie YY, Zhu HB, Dang SY, Chen HF, Zheng GY, Li YX, Kuang Y, Fei J, Chen SJ, Chen Z, Wang ZG. 2013. Rig-I regulates NF-κB activity through binding to Nf-κb1 3′-UTR mRNA. Proc Natl Acad Sci U S A 110:6459–6464. doi: 10.1073/pnas.1304432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuhmann KM, Pfaller CK, Conzelmann KK. 2011. The measles virus V protein binds to p65 (RelA) to suppress NF-κB activity. J Virol 85:3162–3171. doi: 10.1128/JVI.02342-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knippschild U, Gocht A, Wolff S, Huber N, Löhler J, Stöter M. 2005. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal 17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Correa RG, Matsui T, Tergaonkar V, Rodriguez-Esteban C, Izpisua-Belmonte JC, Verma IM. 2005. Zebrafish IκB kinase 1 negatively regulates NF-κB activity. Circ Res 15:1291–1295. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Kozomara A, Griffiths-Jones S. 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39:152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taganov KD, Boldin MP, Baltimore D. 2007. MicroRNAs and immunity: tiny players in a big field. Immunity 26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Gao M, Wang XH, Zhang X, Ha TZ, Liu L, Kalbfleisch JH, Gao X, Kao RL, Williams DL, Li CF. 2015. Attenuation of cardiac dysfunction in polymicrobial sepsis by microRNA-146a is mediated via targeting of IRAK1 and TRAF6 expression. J Immunol 195:672–682. doi: 10.4049/jimmunol.1403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Huang C, Guo Y, Hinsdale M, Lloyd P, Liu L. 2015. MicroRNA-26b modulates the NF-κB pathway in alveolar macrophages by regulating PTEN. J Immunol 195:5404–5414. doi: 10.4049/jimmunol.1402933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Z, Li Y, Takwi A, Li B, Zhang J, Conklin DJ, Young KH, Martin R, Li Y. 2011. miR-301a as an NF-κB activator in pancreatic cancer cells. EMBO J 30:57–67. doi: 10.1038/emboj.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu Q, Song W, Cui J, Xu T. 2017. Genome-guided transcriptome analysis of miiuy croaker provides insights into pattern recognition receptors and cytokines in response to Vibrio anguillarum. Dev Comp Immunol 73:72–78. doi: 10.1016/j.dci.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 21.Xu T, Xu G, Che R, Wang Y, Wang Y, Li J, Wang S, Shu C, Sun Y, Liu T, Liu J, Wang A, Han J, Chu Q, Yang Q. 2016. The genome of the miiuy croaker reveals well-developed innate immune and sensory systems. Sci Rep 6:21902. doi: 10.1038/srep21902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu T, Chu Q, Cui J, Bi D. 2018. Inducible microRNA-3570 feedback inhibits RIG-I-dependent innate immune response to rhabdovirus in teleost fish by targeting MAVS/IPS-1. J Virol 92:e01594. doi: 10.1128/JVI.01594-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, Chu Q, Cui J, Huo R, Xu T. 2017. IRF9 as a negative regulator involved in TRIF-mediated NF-κB pathway in a teleost fish, Miichthys miiuy. Mol Immunol 85:123–129. doi: 10.1016/j.molimm.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Chu Q, Sun Y, Cui J, Xu T. 2017. MicroRNA-3570 modulates the NF-κB pathway in teleost fish by targeting MyD88. J Immunol 198:3274–3282. doi: 10.4049/jimmunol.1602064. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Bi X, Chu Q, Xu T. 2016. Discovery of Toll-like receptor 13 exists in the teleost fish: miiuy croaker (Perciformes, Sciaenidae). Dev Comp Immunol 61:25–33. doi: 10.1016/j.dci.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Frans I, Michiels CW, Bossier P, Willems KA, Lievens B, Rediers H. 2011. Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J Fish Dis 34:643–661. doi: 10.1111/j.1365-2761.2011.01279.x. [DOI] [PubMed] [Google Scholar]
- 27.Grisez L, Chair M, Sorgeloos P, Ollevier F. 1996. Mode of infection and spread of Vibrio anguillarum in turbot Scophthalmus maximus larvae after oral challenge through live feed. Dis Aquat Org 26:181–187. doi: 10.3354/dao026181. [DOI] [Google Scholar]
- 28.Hou BH, Jian ZX, Cui P, Li SJ, Tian RQ, Ou JR. 2015. MiR-216a may inhibit pancreatic tumor growth by targeting JAK2. FEBS Lett 589:2224–2232. doi: 10.1016/j.febslet.2015.06.036. [DOI] [PubMed] [Google Scholar]
- 29.Dryden NH, Sperone A, Martin-Almedina S, Hannah RL, Birdsey GM, Khan ST, Layhadi JA, Mason JC, Haskard DO, Gottgens B, Randi AM. 2012. The transcription factor Erg controls endothelial cell quiescence by repressing activity of nuclear factor (NF)-κB p65. J Biol Chem 287:12331–12342. doi: 10.1074/jbc.M112.346791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker RG, Hayden MS, Ghosh S. 2011. NF-κB, inflammation, and metabolic disease. Cell Metab 13:11–22. doi: 10.1016/j.cmet.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono R, Kaisho T, Tanaka T. 2015. PDLIM1 inhibits NF-κB-mediated inflammatory signaling by sequestering the p65 subunit of NF-κB in the cytoplasm. Sci Rep 5:18327. doi: 10.1038/srep18327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Hu L, Tong X, Ye X. 2014. Casein kinase 1γ1 inhibits the RIG-I/TLR signaling pathway through phosphorylating p65 and promoting its degradation. J Immunol 192:1855–1861. doi: 10.4049/jimmunol.1302552. [DOI] [PubMed] [Google Scholar]
- 33.Ma X, Becker Buscaglia LE, Barker JR, Li Y. 2011. MicroRNAs in NF-κB signaling. J Mol Cell Biol 3:159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z, Dong D, Chen X, Huang H, Wen S. 2015. MicroRNA-381 negatively regulates TLR4 signaling in A549 cells in response to LPS stimulation. Biomed Res Int 2015:849475. doi: 10.1155/2015/849475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M. 2008. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res 36:6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu Q, Sun Y, Cui J, Xu T. 2017. Inducible microRNA-214 contributes to the suppression of NF-κB-mediated inflammatory response via targeting myd88 gene in fish. J Biol Chem 292:5282–5290. doi: 10.1074/jbc.M117.777078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shingara J, Keiger K, Shelton J, Laosinchai-Wolf W, Rowers P, Conrad R, Brown D, Labourier E. 2005. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA 11:1461–1470. doi: 10.1261/rna.2610405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Li T, Fan J, Wang S, Weng X, Han Q, Zhao RC. 2015. miR-216a rescues dexamethasone suppression of osteogenesis, promotes osteoblast differentiation and enhances bone formation, by regulating c-Cbl-mediated PI3K/AKT pathway. Cell Death Differ 22:1935–1945. doi: 10.1038/cdd.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato M, Putta S, Wang SM, Yuan H, Lanting L, Mair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, Rossi JJ, Natarajan R. 2009. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Ning X, Cui W, Bi M, Zhang D, Zhang J. 2015. Transforming growth factor (TGF)-beta-induced microRNA-216a promotes acute pancreatitis via Akt and TGF-beta pathway in mice. Dig Dis Sci 60:127–135. doi: 10.1007/s10620-014-3261-9. [DOI] [PubMed] [Google Scholar]
- 41.Shaulian E, Karin M. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 42.Kaczynski J, Cook T, Urrutia R. 2003. Sp1-and Krüppel-like transcription factors. Genome Biol 4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu Q, Xu T. 2016. miR-192 targeting IL-1RI regulates the immune response in miiuy croaker after pathogen infection in vitro and in vivo. Fish Shellfish Immunol 54:537–543. doi: 10.1016/j.fsi.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 44.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 45.Lewis BP, Shih IH. 2003. Prediction of mammalian microRNA targets. Cell 115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 46.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. 2004. Human microRNA targets. PLoS Biol 2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rusinov V, Baev V, Minkov IN, Tabler M. 2005. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res 33:696–700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.