Abstract
Although most activating mutations of epidermal growth factor receptor (EGFR)-mutant non–small cell lung cancers (NSCLCs) are sensitive to available EGFR tyrosine kinase inhibitors (TKIs), a subset with alterations in exon 20 of EGFR and HER2 are intrinsically resistant and lack an effective therapy. We used in silico, in vitro, and in vivo testing to model structural alterations induced by exon 20 mutations and to identify effective inhibitors. 3D modeling indicated alterations restricted the size of the drug-binding pocket, limiting the binding of large, rigid inhibitors. We found that poziotinib, owing to its small size and flexibility, can circumvent these steric changes and is a potent inhibitor of the most common EGFR and HER2 exon 20 mutants. Poziotinib demonstrated greater activity than approved EGFR TKIs in vitro and in patient-derived xenograft models of EGFR or HER2 exon 20 mutant NSCLC and in genetically engineered mouse models of NSCLC. In a phase 2 trial, the first 11 patients with NSCLC with EGFR exon 20 mutations receiving poziotinib had a confirmed objective response rate of 64%. These data identify poziotinib as a potent, clinically active inhibitor of EGFR and HER2 exon 20 mutations and illuminate the molecular features of TKIs that may circumvent steric changes induced by these mutations.
Approximately 10–15% of NSCLCs harbor activating mutations in EGFR. For the majority of patients whose tumors have ‘classical’ sensitizing mutations (including deletions in exon 19 and the mutation encoding p.L858R), TKIs, such as gefitinib and erlotinib, provide dramatic clinical benefit; approximately 70% of patients treated with TKIs experience an objective response (OR), improved progression free-survival (PFS), and improved quality of life compared to chemotherapy alone1,2,3,4,5,6,7,8,9,10,11,12,13,14. However, approximately 10–12% of EGFR-mutant NSCLC tumors have an in-frame insertion within exon 20 of EGFR6,15,16,17 and are generally resistant to EGFR TKIs. Historical data for patients with EGFR exon 20 insertion mutations have shown that overall response rates are approximately 3–8% to first-line therapy with erlotinib, gefitinib, or afatinib16,18. In addition, 90% of HER2 mutations in NSCLC are exon 20 mutations, and approximately 3% of patients with NSCLC harbor HER2 mutations19,20. Together, EGFR and HER2 exon 20 mutations are found in approximately 4% of all patients with NSCLC19. The data thus far suggest that TKIs targeting HER2 (afatinib, lapatinib, neratinib, dacomitinib) have limited activity in patients with HER2-mutant tumors, with objective response rates (ORRs) of below 40% reported by many studies19,20,21,22,23,25, although some preclinical activity was observed in mouse models bearing mutated HER2 that were treated with afatinib26.
Exon 20 of EGFR and HER2 contains two major regions, the α-C helix (residues 762–766 in EGFR and 770–774 in HER2) and the loop following the α-C helix (residues 767–774 in EGFR and 775–783 in HER2)16,19,20,27. Crystallography of the EGFR exon 20 insertion D770insNPG has revealed a stabilized and rigid active conformation inducing resistance to first-generation TKIs in the insertions after residue 764. However, modeling of EGFR A763insFQEA demonstrated that insertions before residue 764 do not exhibit this effect and do not induce drug resistance16. Moreover, in a patient-derived xenograft (PDX) model of NSCLC driven by an EGFR exon 20 mutation in which the insertions are in the loop after the α-C helix (EGFR H773insNPH), the third-generation EGFR TKIs osimertinib (AZD9291) and rociletinib (CO-1696) were found to have minimal activity28. In a recent study of rare EGFR and HER2 exon 20 mutations, the authors found a heterogeneous response to covalent quinazoline-based second-generation inhibitors, such as dacomitinib and afatinib; however, the concentrations that were required to target more common exon 20 insertion mutations were above what are clinically achievable24. Therefore, there is a substantial clinical need to identify new therapies to overcome the innate drug resistance of NSCLC tumors harboring exon 20 insertions in EGFR and HER2.
Results
EGFR and HER2 exon 20 insertion mutations are resistant to reversible and irreversible EGFR TKIs
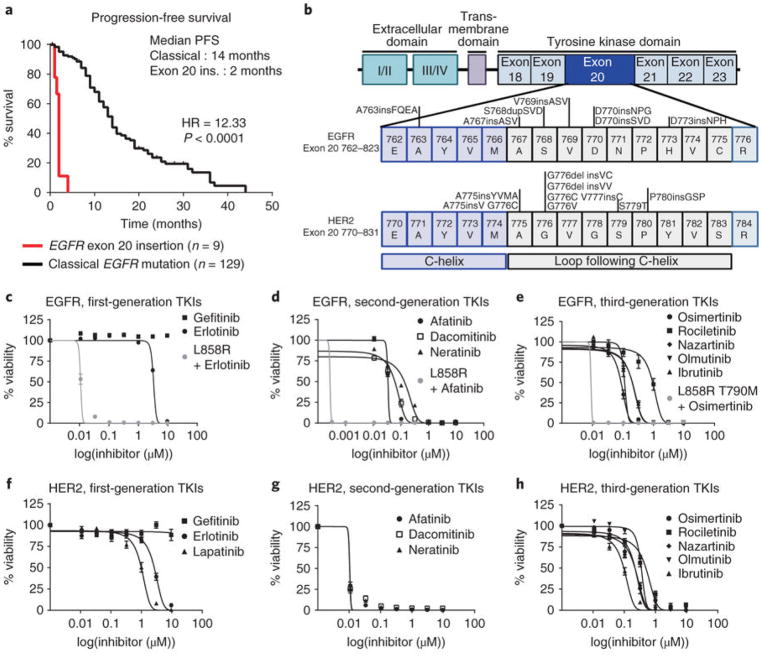
We investigated clinical responses to TKIs in patients with tumors harboring EGFR exon 20 insertion mutations in our clinical database. Among 280 patients with EGFR-mutant NSCLC, we identified 129 patients with classical EGFR mutations (exon 19 deletions and the mutations encoding p.L858R and p.L861Q) and 9 patients with EGFR exon 20 insertion mutations that received single-agent treatment with erlotinib, gefitinib, or afatinib. Patients with NSCLC harboring classical EGFR mutations had a median PFS of 14 months, whereas patients with EGFR exon 20 insertion mutations had a median PFS of only 2 months (P < 0.0001, log-rank test; Fig. 1a). Of the nine patients with an EGFR exon 20 insertion, OR was observed in only one patient harboring a deletion–insertion mutation (S768delinsIL) who received afatinib (Supplementary Fig. 1a). These clinical data as well as results from prior studies16,18 demonstrate the limited activity of the available EGFR TKIs in NSCLC driven by an EGFR exon 20 insertion and validate the need for alternative treatment strategies for these specific tumors.
Fig. 1. Exon 20 insertion mutations induce de novo resistance to covalent and noncovalent TKIs.
a, PFS of patients with classical EGFR mutations and exon 20 insertion mutations in EGFR demonstrating resistance to first-line therapy (log-rank P < 1.0 × 10−9). b, Schematic of EGFR and HER2 exon 20 insertions generated in a stable Ba/F3 model. c–h, Averaged dose response curves of cell viability of Ba/F3 cell lines expressing six different EGFR (c–e) and six different HER2 (f–h) exon 20 insertion mutations indicated in bold in b treated with first-, second-, or third-generation TKIs for 72 h. In c–h, the mean ± s.e.m. of the six cell lines is plotted for each concentration (n = 3 biologically independent experiments). i, 3D modeling of EGFR D770insNPG (green) and EGFR T790M (yellow). The NPG insertion is highlighted in pink; the P-loop is highlighted in red. Shifts of the P-loop (red arrow) and the α-C helix (blue arrow) into the binding pocket result in steric hindrance, reducing the size of the binding pocket. j, 3D modeling of HER2 A775insYVMA (blue) and HER2-WT (yellow). The YVMA insertion is highlighted in pink, and the P-loop is highlighted in red. Overall shifts of the P-loop (red arrow) and the α-C helix (blue arrow) into the binding pocket result in an overall reduction in the size of the binding pocket.
As an initial step in screening for drugs targeting EGFR or HER2 exon 20 mutants, we stably expressed 7 EGFR and 11 HER2 mutations in Ba/F3 cells. The locations of the EGFR and HER2 exon 20 mutations are summarized in Fig. 1b. To assess which exon 20 mutations of EGFR and HER2 were activating, Ba/F3 cell lines were screened for ability to proliferate independent of IL-3 supplmentation. We found that all EGFR exon 20 insertions tested were activating mutations (Supplementary Fig. 1b), and six HER2 exon 20 mutations and HER2 L755P, which is located in exon 19, were activating mutations (Supplementary Fig. 1c). Next, we tested the sensitivity of the exon 20 insertions to EGFR and HER2 TKIs that have undergone clinical evaluation, including reversible (first-generation), irreversible (second-generation) and irreversible mutant-specific (third-generation) TKIs, and then we compared this sensitivity to that of EGFR L858R, a classical sensitizing mutation. With the exception of EGFR A763insFQEA, EGFR exon 20 insertions (n = 6) were resistant to first-generation (Fig. 1c, half-maximal inhibitory concentration (IC50) = 3.3–10 μM), second-generation (Fig. 1d, IC50 = 40–135 nM), and third-generation (Fig. 1e, IC50 = 103–850 nM) EGFR TKIs (Supplementary Fig. 2 and Supplementary Table 1). In addition, HER2 exon 20 mutants (n = 6) were resistant to first-generation (Fig. 1f, IC50 = 1.2–13 μM), and third-generation (Fig. 1h, IC50 = 114–505 nM) TKIs. Second-generation TKIs did exhibit some activity against Ba/F3 HER2 exon 20 mutated cell lines (Fig. 1g, Supplementary Fig. 3, and Supplementary Table 1, IC50 = 10–12 nM). In accordance with results from our drug screening, western blotting of Ba/F3 cells bearing an EGFR exon 20 insertion mutation demonstrated that erlotinib and osimertinib did not substantially inhibit phosphorylated EGFR (p-EGFR), with the exception of the EGFR A763insFQEA mutant cell line, in which there was partial inhibition at low doses of drug. Furthermore, western blotting of Ba/F3 cell lines expressing HER2 exon 20 insertion mutations treated with erlotinib and osimertinib only show considerably inhibited p-HER2 at a concentration of 500 nM (Supplementary Fig. 4a–d).
EGFR and HER2 exon 20 insertion mutations cause steric hindrance of the drug-binding pocket
To investigate why exon 20 insertions are resistant to first- and third-generation EGFR TKIs, we performed 3D modeling of the solved crystal structures of EGFR D770insNPG with EGFR T790M and wild-type (WT) EGFR to visualize changes within the drug-binding pocket. Our modeling suggested that EGFR exon 20 insertion mutants (Supplementary Fig. 5a) are similar to EGFR T790M in the alignment of the gatekeeper residue Thr790, which results in increased affinity to ATP and a reduced binding of first-generation inhibitors, rendering these mutations resistant to noncovalent inhibitors. In addition, HER2 exon 20 insertions (Supplementary Fig. 5b) induce a constitutively active conformation, preventing the binding of the noncovalent HER2 inhibitor lapatinib, which binds to HER2 in the inactive conformation. Moreover, EGFR and HER2 exon 20 insertions have a dramatic effect on the drug-binding pocket. In silico modeling of EGFR (Fig. 1i) and HER2 (Fig. 1j) exon 20 insertions revealed a notable shift of the α-C helix into the drug-binding pocket (Fig. 1j, blue arrow) due to the insertions at the C-terminal end of the α-C helix (Fig. 1j, pink), forcing a rigid placement of the α-C helix in the inward, activated position. In addition, 3D modeling suggested a considerable shift of the phosphate-binding loop (P-loop) into the drug-binding pocket (Fig. 1i,j and Supplementary Fig. 5b,c, red arrows) of both receptors. Together, these shifts result in steric hindrance of the drug-binding pocket from two directions in both EGFR and HER2 exon 20 mutants. Consistent with results from our aforementioned in vitro testing, 3D modeling supported the observation that afatinib inhibits exon 20 insertions more effectively than osimertinib. Osimertinib has a large terminal 1-methylindole group connected directly to a rigid pyrimidine core. This large inflexible group reduces the ability of osimertinib to reach the Cys797 residue as effectively as afatinib in EGFR exon 20 insertion (Supplementary Fig. 5c). In contrast, afatinib has a smaller 1-chloro-2-flurobenzene ring terminal group indirectly linked to a quinazoline core via a secondary amine group, enabling afatinib to fit into the sterically hindered binding pocket. Similarly, steric hindrance prevents binding of osimertinib to HER2 A775insYVMA (Supplementary Fig. 5d). Taken together, our in vitro data and in silico modeling indicate that small, flexible quinazoline derivatives may be capable of targeting EGFR and HER2 exon 20 insertion mutants.
Poziotinib is a potent inhibitor of EGFR and HER2 exon 20 insertion mutants in vitro
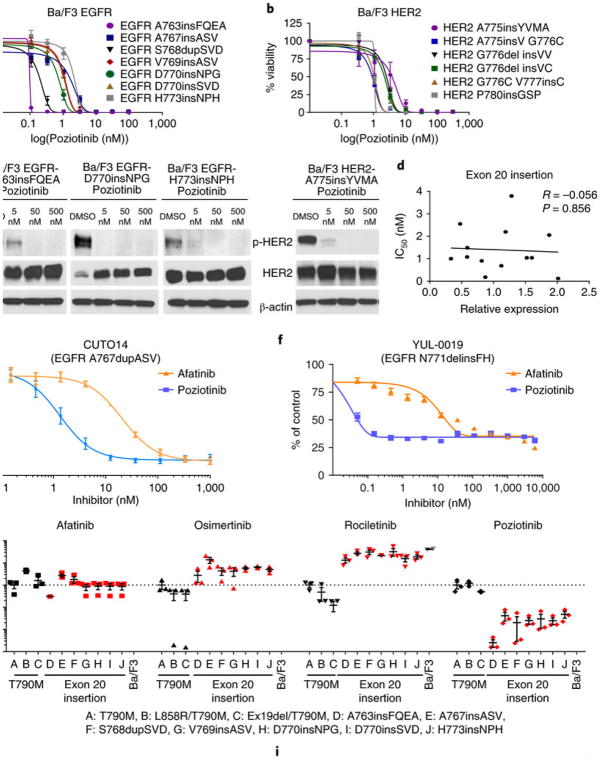
We next sought to identify TKIs with enhanced activity against exon 20 insertions. Previous studies of EGFR exon 20 insertions suggested that the drug-binding pocket of exon 20 may be altered, affecting drug binding17. In a study by Cha et al.29, the authors demonstrate that poziotinib (HM781-36B) covalently binds and inhibits mutant EGFR and HER2 kinases and induces cell death in EGFR and HER2 mutated NSCLC cell lines in vitro. Poziotinib, like afatinib, also contains a small terminal group and a flexible quinazoline core. In addition, poziotinib has smaller substituent groups linking the Michael acceptor group to the quinazoline core, can rotate freely around the amine and ether groups, and has increased halogenation of the terminal benzene ring compared to afatinib. The electron-rich moiety within the terminal group also interacts with basic residues of EGFR, such as Lys745, to further stabilize its binding (Supplementary Fig. 6a). On the basis of these characteristics, we hypothesized that poziotinib may effectively bind and inhibit EGFR and HER2 exon 20 insertion mutants. Therefore, we tested poziotinib in vitro and found that it potently inhibited the growth of Ba/F3 cell lines with an EGFR exon 20 mutation (Fig. 2a) or a HER2 exon 20 mutation (Fig. 2b). Poziotinib had an average IC50 value of 1.0 nM in Ba/F3 cell lines with an EGFR exon 20 mutation, making poziotinib approximately 100 times more potent than osimertinib and 40 times more potent than afatinib in vitro. Moreover, poziotinib had an average IC50 value of 1.9 nM in Ba/F3 cell lines with a HER2 exon 20 mutation, making poziotinib 200 times more potent than osimertinib and 6 times more potent than afatinib in vitro. These results were validated through western blotting, in which poziotinib inhibited phosphorylation of EGFR and HER2 at concentrations as low as 5 nM (Fig. 2c and Supplementary Fig. 7a). Furthermore, to validate that poziotinib sensitivity was not due to the level of expression of EGFR or HER2 mutants, expression of each mutant was determined using ELISA and then plotted against IC50 values (Fig. 2d). No correlation was found between IC50 and expression (R = –0.056, P = 0.856). In addition, poziotinib effectively inhibited growth of the human cell line H1781, which harbors a HER2 exon 20 insertion mutation, and of patient-derived cell lines CUTO14 (EGFR A767dupASV) and YUL-0019 (EGFR N771delinsFH), which had an average IC50 value of 7.7 nM, 1.84 nM, and 0.30 nM, respectively. Poziotinib was 2.7 times more potent than afatinib in the H1781 cell line, 15 times more potent than afatinib in the CUT014 cell line, and more than 100 times more potent than afatinib in the YUL-0019 cell line (Fig. 2e,f and Supplementary Fig. 7b). Western blotting of the CUT014 cell line revealed that there was considerable inhibition of p-EGFR following treatment with 10 nM poziotinib, but p-EGFR was not substantially inhibited by afatinib until a treatment concentration of 1,000 nM was reached (Supplementary Fig. 7c,d).
Fig. 2. Poziotinib potently inhibits EGFR and HER2 exon 20 insertion mutants.
a,b, Dose-response curves showing the cell viability of Ba/F3 cell lines expressing EGFR (a) and HER2 (b) exon 20 insertion mutations that were treated with poziotinib for 72 h. The mean ± s.e.m. of each individual cell line is plotted for each concentration (n = 3 biologically independent experiments). c, Western blotting showing inhibition of p-EGFR and p-HER2 in Ba/F3 cell lines after 2 h of poziotinib treatment (n = 2 biologically independent experiments). β-actin was used as a loading control. Uncropped blots are available in Supplementary Fig. 10. d, Correlation of expression levels of Ba/F3 exon 20 insertions compared to sensitivity (n = 2 biologically independent experiments). The Pearson correlation coefficient of 13 biologically independent samples and the P value were determined using GraphPad Prism. e,f, Dose-response curves showing the cell viability of the patient-derived cell lines CUTO14 expressing EGFR A767dupASV (e) and YUL-0019 expressing EGFR N771delinsFH (f) that were treated with poziotinib or afatinib for 72 h (n = 3 biologically independent experiments). The mean ± s.e.m. of the experimental replicates is plotted for each concentration. g, IC50 values of EGFR-mutant Ba/F3 cells normalized to the IC50 values of Ba/F3 EGFR-T790M cell line after incubation with afatinib, osimertinib, rociletinib, or poziotinib for 72 h (n = 3 biologically independent experiments). Dot plots are representative of mean ± s.e.m. Values greater than 1 are indicative of less potent inhibition compared to T790M, whereas values less than one indicate more potent inhibition of exon 20 insertions compared to T790M. h, EGFR-T790M (yellow) with osimertinib (blue) has a very large binding pocket (h) compared to EGFR D770insNPG (green) with poziotinib (orange) (i). Steric hindrance induced by the NPG insertion is shown in red. Blue also indicates changes occurring in the drug binding pocket that compensate for the steric hindrance caused by exon 20 insertions.
Poziotinib is a relatively selective inhibitor of EGFR exon 20 mutants over EGFR T790M mutants
To determine the specificity of poziotinib to inhibit EGFR exon 20 mutants compared to EGFR T790M mutants, we compared the IC50 values of afatinib, osimertinib, rociletinib, and poziotinib in Ba/F3 cell lines with exon 20 mutants to the IC50 values of these drugs in Ba/F3 cell lines harboring the EGFR T790M mutant. IC50 values are displayed normalized to that of the EGFR T790M mutant, and values <1 indicate relative specificity for the exon 20 insertion mutant as compared to EGFR T790M (Fig. 2g). When compared to EGFR T790M mutants, EGFR exon 20 insertions were 65 times more sensitive to poziotinib. Moreover, EGFR exon 20 insertion mutants were 1.4 times more resistant to afatinib, 5.6 times more resistant to osimertinib, and 24 times more resistant to rociletinib than the EGFR T790M mutant (Fig. 2g). Western blotting of cells from the EGFR-WT NSCLC cell line H292 that were treated with erlotinib, afatinib, poziotinib, or osimertinib showed similarly decreased p-EGFR and total EGFR levels in afatinib- and poziotinib-treated samples, but no or little reduction in p-EGFR and total EGFR levels in erlotinib- and osimertinib-treated samples (Supplementary Fig. 7e–g), suggesting that poziotinib activity against EGFR-WT is similar to that of afatinib.
Poziotinib overcomes steric hindrance and binds tightly in the drug-binding pocket
To examine why poziotinib, but not third-generation TKIs, such as osimertinib, selectively and potently inhibits exon 20 mutants, 3D modeling was performed to determine how changes in the drug-binding pocket affect drug binding. Although osimertinib fits into the drug-binding pocket of EGFR T790M and EGFR L858R/T790M mutant receptors (Fig. 2h and Supplementary Fig. 6b), in exon 20 mutants, large changes within the drug-binding pocket sterically hinder the binding of third-generation inhibitors (Fig. 2i, denoted in red). However, poziotinib is smaller and has greater flexibility, allowing it to fit into the sterically hindered drug- binding pocket of exon 20 insertion mutants (Fig. 2i). 3D modeling of EGFR D770insNPG with poziotinib and afatinib suggests that the shifted P-loop (red) into the drug-binding pocket causes poziotinib (orange) to bind more tightly into the drug-binding pocket than afatinib (Supplementary Fig. 6c, blue). Calculations of structural modeling indicate that the free energy of binding (London ΔG) for poziotinib is lower than afatinib, indicating stronger binding affinity of poziotinib (Supplementary Fig. 6c). 3D modeling of HER2-WT with osimertinib (Supplementary Fig. 6d) revealed that the binding pocket of HER2-WT is larger than the binding pocket of HER2 A775insYVMA (Supplementary Fig. 6e). Thus, poziotinib tightly binds deep into the sterically hindered drug-binding pocket of HER2 A775insYVMA, overcoming structural changes induced by exon 20 insertions.
Poziotinib inhibits EGFR- and HER2-mutant NSCLC in vivo more potently than afatinib
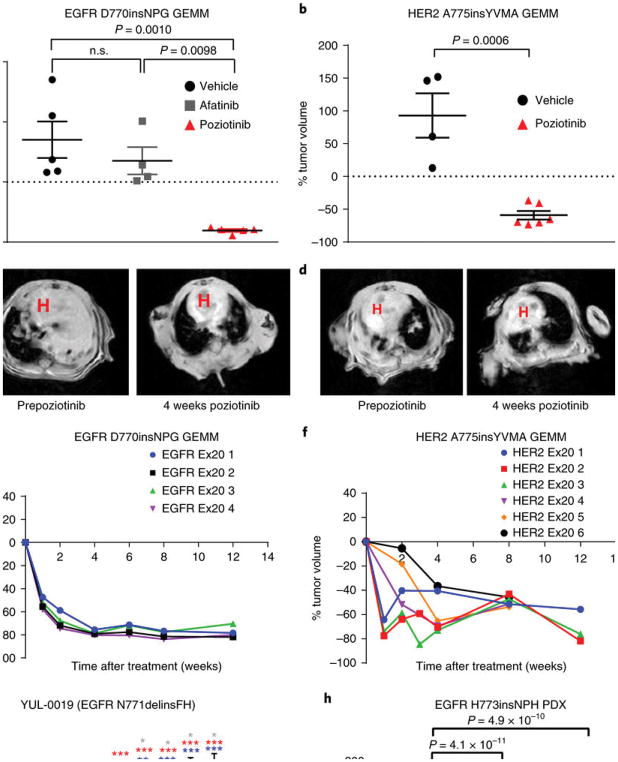
We next tested the in vivo efficacy of poziotinib using genetically engineered mouse models (GEMMs) of NSCLC driven by an EGFR or HER2 exon 20 insertion. Lung tumors were induced in mice harboring EGFR D770insNPG30 as previously described, and once the mice had obvious lung tumor formation as determined through magnetic resonance imaging (MRI), mice with equal tumor volume received poziotinib (10 mg per kg body weight (mg/kg)), afatinib (20 mg/kg), or vehicle daily for 4 weeks. Tumor volume was determined through MRI, and poziotinib reduced tumor volume by 80% in GEMMs with an EGFR exon 20 mutation (Fig. 3a,c), whereas mice treated with a clinically relevant dose of afatinib31,32 had a 35% increase in tumor volume and were not significantly different from mice treated with vehicle control. In addition, lung tumors were induced in mice bearing a HER2 A775insYVMA mutation26 as previously described. Mice received poziotinib (10 mg/kg) or vehicle daily for 4 weeks. Tumor volume was reduced by 60% in GEMMs bearing a HER2 exon 20 insertion (Fig. 3b,d); this is a higher level of tumor reduction than the 37% previously observed for afatinib in the identical GEMM26. Representative MRI images of tumors before and after poziotinib are shown for both EGFR- and HER2-mutant GEMMs (Fig. 3c,d). In both EGFR- and HER2-mutant GEMMs, mice treated with 10 mg poziotinib per kg body weight demonstrated durable regression without signs of progression at 12 weeks (Fig. 3e,f). Next, we tested the efficacy of poziotinib versus afatinib in a xenograft model of a patient-derived cell line harboring an EGFR N771delinsFH mutation. YUL-0019 cells were injected into the flanks of nude mice, and mice were randomized to receive poziotinib (10 mg/kg or 5 mg/kg), afatinib (20 mg/kg), or vehicle daily for 10 d. Poziotinib-treated mice displayed significant reduction in tumor burden by 50% and 56% for the 5 mg/kg and 10 mg/kg doses, respectively (Fig. 3g and Supplementary Table 2). In contrast, mice that received afatinib showed no significant reduction in tumor burden; however, tumor volume did not increase, unlike tumors in mice treated with vehicle control (Fig. 3g). Lastly, poziotinib treatment (5 mg/kg or 10 mg/kg) completely reduced tumors by >85% in eight out of nine mice in 14 d in the PDX model LU0387 bearing an EGFR H773insNPH exon 20 insertion (Fig. 3h).
Fig. 3. Poziotinib reduces tumor burden in mouse models bearing EGFR or HER2 exon 20 insertion mutants.
a,b, Dot plots showing tumor volume of mice bearing EGFR D770insNPG (a) or HER2 A775insYVMA (b) that were treated daily with vehicle (EGFR, n = 5 mice; HER2, n = 4 mice), 20 mg/kg of afatinib daily (EGFR n = 4 mice), or 10 mg/kg of poziotinib daily (EGFR, n = 5 mice; HER2, n = 6 mice) for 4 weeks. The mean ± s.e.m. of percent change in tumor volume after 4 weeks of treatment is plotted. A two-sided Student’s t-test was used to calculate the P values; n.s., nonsignificant. c,d, Representative MRI images of EGFR-mutant (c) and HER2-mutant (d) GEMMs before and after 4 weeks of poziotinib treatment showing robust tumor regression. The red H marks the heart. e,f, Plots of tumor volume of each mouse (numbered Ex20 1–6) bearing EGFR D770insNPG (e; n = 4 biologically independent mice) or HER2 A775insYVMA (f; n = 6 biologically independent mice) treated with 10 mg/kg of poziotinib 5 d per week for 12 weeks showing that mice continue to respond to poziotinib treatment. g, Tumor area in nude mice in which cells from the YUL-0019 line were grown. Mice were treated with vehicle control (n = 6 biologically independent mice), 20 mg/kg afatinib (n = 6 biologically independent mice), 5 mg/kg poziotinib (n = 4 biologically independent mice), or 10 mg/kg poziotinib (n = 3 biologically independent mice). Mean ± s.e.m. of tumor area is plotted for each measurement. A multiple-comparisons test using the Holm–Sidak method was used to determine statistical significance between groups, and P values can be found in Supplementary Table 3. *P < 0.05, **P < 0.01, ***P < 0.001. h, Dot plots of tumor burden in mice bearing EGFR H773insNPH PDXs were treated with vehicle control (n = 6 biologically independent mice), 5 mg/kg poziotinib (n = 6 biologically independent mice), or 10 mg/kg poziotinib (n = 3 biologically independent mice). The mean ± s.e.m. of percent change in tumor volume after 4 weeks of treatment is plotted. A one-way ANOVA analysis was used in combination with Tukey’s test to determine statistical significance.
C797S and epithelial-to-mesenchymal transition (EMT) are possible acquired resistance mechanisms of poziotinib
To determine whether poziotinib, like other irreversible inhibitors, binds covalently at Cys797, we generated Ba/F3 cell lines with the C797S mutation, which was observed in ~30% of patients with osimertinib resistance33,34. We found that the C797S mutation induced resistance to poziotinib, which had an IC50 value of >10 μM in cell lines with this mutant (Supplementary Fig. 8a). In addition, EGFR-mutant NSCLC cell lines HCC4006 and HCC827, having undergone EMT owing to erlotinib resistance (ER)35, were also resistant to poziotinib (average IC50 value of seven ER cell lines = 8.6 μM; Supplementary Fig. 8b). Together, these experiments suggest that poziotinib may be susceptible to similar mechanisms of acquired resistance as other third-generation TKIs. Further studies to understand poziotinib resistance are currently being performed.
Poziotinib is a clinically active inhibitor of EGFR and HER2 exon 20 mutant NSCLC
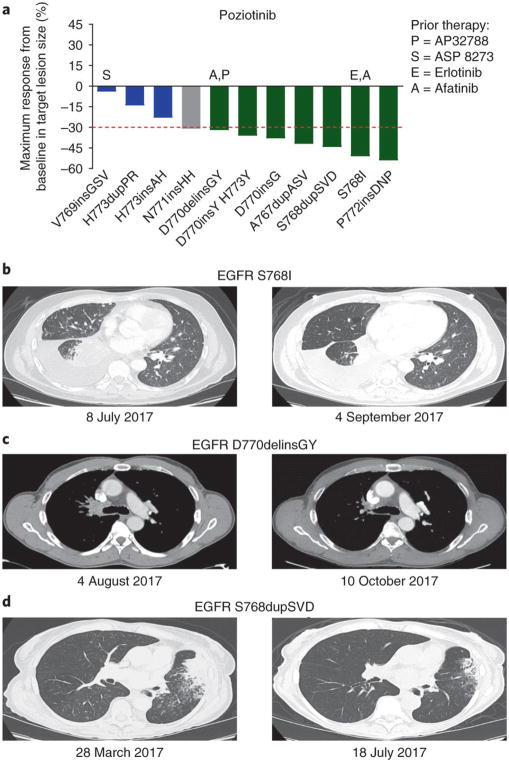
On the basis of these preclinical data, a 65-year-old female never-smoker with stage IV adenocarcinoma of lung who harbored HER2 exon 20 insertion A771insAYVM was placed on 16 mg poziotinib daily under a compassionate-use protocol (CIND16-0055). Positron emission tomography–computed tomography (PET–CT) imaging was obtained after 4 weeks of poziotinib therapy and was compared to baseline PET–CT imaging that was obtained 1 d before poziotinib commencement. The patient experienced a considerable radiological response with reduction in fluorodeoxyglucose (FDG) avidity in the right humerus, left seventh rib, right sacrum, and a right lower lobe nodule among others (Supplementary Fig. 9a). Furthermore, reduction in tumor size corresponded to a drop in HER2 A771insAYVM circulating free DNA (cfDNA) from 2.4% to undetectable levels (<0.3%) (Supplementary Fig. 9b). We next investigated the clinical activity of poziotinib in a phase 2 study in patients with NSCLC with EGFR exon 20 mutations (NCT03066206). The starting dose was 16 mg daily orally. The majority (55%) of patients received a dose reduction, with the two most common adverse events being known EGFR-inhibitor-related toxicities: skin rash and diarrhea. Of the first 11 patients, 7 of 11 (64%) had a confirmed objective partial radiological response based on response evaluation criteria in solid tumors (RECIST) that was due to poziotinib (Fig. 4a). Previous treatments, dose reductions, and patient characteristics can be found in Supplementary Table 3. Of note, two patients with EGFR exon 20 insertion mutations were pretreated with EGFR TKIs (erlotinib, afatinib, and/or AP32788) and displayed radiological partial responses (Fig. 4b,c). Although the clinical data is not yet mature, as of January 2018, 5 of 11 patients have progressed, and the median PFS has not yet been reached (median follow-up, 6.6 months). Representative computed tomography (CT) scans of patients with the common EGFR exon 20 insertion mutation S768dupSVD illustrate a confirmed robust radiological response after 4 months of poziotinib treatment (Fig. 4d). More detailed clinical data from the full cohort of patients will be reported separately when available.
Fig. 4. Poziotinib inhibits EGFR exon 20 insertion mutants in patients with NSCLC.
a, Waterfall plot of the first 11 patient responses on clinical trial NCT03066206. Objective partial responses are shown in green (n = 7), stable disease is shown in blue (n = 3), and unconfirmed response is shown in gray (n = 1). b, CT scan of a patient with EGFR S768I mutant NSCLC 1 d before poziotinib treatment and after 8 weeks of poziotinib therapy. The patient had previously been treated with both erlotinib and afatinib with progression and had a 50% reduction in the volume of target lesions after 4 weeks of poziotinib therapy. c, CT scans of a patient with EGFR D770delinsGY 1 d before and after 8 weeks of poziotinib treatment. Patient had been previously treated with afatinib and AP32788 with no response but had a 32% reduction in the volume of target lesions with poziotinib treatment. d, CT scans of a patient with EGFR S768dupSVD 1 d before and after 16 weeks of poziotinib treatment. The patient had a confirmed objective partial response as seen in the second scan.
Discussion
Each year in the United States alone, approximately 4% of patients diagnosed with NSCLC, which is approximately 7,000 people, bear EGFR and/or HER2 exon 20 insertion mutations36. Previously, patients with EGFR and/or HER2 exon 20 mutations have been excluded from clinical trials owing to lack of response to targeted therapy, demonstrating the urgent need for new approaches to treat these patients15. Here, we report that exon 20 mutants exhibit de novo resistance both clinically and preclinically to first-, second-, and third-generation TKIs. Using 3D modeling of EGFR D770insNPG and HER2 A775insYVMA, we identified poziotinib as having structural features that could overcome changes within the drug-binding pocket induced by insertions in exon 20. Moreover, the predicted activity of poziotinib was confirmed using in vitro and in vivo models demonstrating the potent antitumor activity of poziotinib in cells with these mutations. Finally, we provide clinical evidence of the activity of poziotinib in 11 patients with EGFR exon 20 mutated NSCLC, with a confirmed ORR of 64%.
Our observations indicate that poziotinib inhibits the most common and complex EGFR and HER2 exon 20 insertion mutations at concentrations well below clinically achievable levels. Although many patients required dose reduction, results from phase 1 and 2 clinical testing of poziotinib demonstrate that 16 mg/day doses are tolerable and that a plasma concentration of 150 nM can be achieved37,38,39,40. As determined on the basis of our data using cell lines bearing EGFR or HER2 exon 20 mutations, this plasma level is more than 125 times the average IC50 value calculated in vitro. Although plasma protein-binding to poziotinib may reduce poziotinib activity, this plasma concentration is well above the required concentration for inhibition of mutant EGFR exon 20. By contrast, other first-, second-, and third-generation TKIs had IC50 values that were above the clinically achievable plasma concentrations previously observed in patients31,41,42,43,44,45. In concordance with this, clinical testing of afatinib, neratinib, and dacomitinib achieved overall response rates of 8.7%, 9.5%, and 12%, respectively, for patients harboring EGFR or HER2 exon 20 mutations25,42,46,47. Here, we report that in vitro, poziotinib is approximately 40 times more potent than afatinib and 65 times more potent than dacomitinib in cell lines bearing EGFR exon 20 mutants. Moreover, poziotinib is six times more potent that afatinib and dacomitinib in cell lines with HER2 exon 20 mutants in vitro. Lastly, in GEMM and xenograft models, poziotinib resulted in 80% tumor reduction in 4 weeks and 50% tumor reduction in 10 d, whereas afatinib did not reduce tumor burden in either model. Taken together, these data indicate that although poziotinib shares a similar quinazoline backbone with afatinib and dacomitinib, additional features of the kinase inhibitor result in increased activity and relative specificity for EGFR exon 20 mutants compared with the more common T790M mutant.
Our 3D modeling suggests that the smaller size, increased halogenation, and flexibility of poziotinib give the inhibitor a competitive advantage in the sterically hindered drug-binding pocket of exon 20 mutant EGFR and HER2. Furthermore, our data suggest that the size of the insertion may affect drug sensitivity. In our patient set, the one responder to afatinib harbored an S768delinsIL mutation, which produced a net gain of only one amino acid. Furthermore, our patient-derived cell line, YUL-0019 (N771delinsFH), which had a net gain of only one amino acid, was more sensitive to quinazoline-based pan-HER inhibitors than cell lines with larger EGFR exon 20 insertions. In previously published clinical studies, patients with only a single amino acid insertion responded better to already-established therapies, such as erlotinib plus cetuximab or irreversible inhibitors, compared to patients with larger insertions comprised of three amino acids48,49,50. Further studies are needed to determine the precise effect of insertion size on specific drug combinations and sensitivity.
In a recent phase 2 study of EGFR-mutated NSCLC with acquired resistance to erlotinib or gefitinib, in which 49% of patients had a T790M mutation, there was an 8% ORR to poziotinib39. However, 46% of patients had a minor response to poziotinib39. In our study, we found that poziotinib was 65 times more potent in inhibiting cell lines with EGFR exon 20 insertions than EGFR T790M mutant cell lines, indicating poziotinib selectively inhibits exon 20 insertion mutations compared to T790M mutations. This finding suggests that patients with EGFR exon 20 insertion mutations may receive greater clinical benefit from poziotinib treatment compared to patients with EGFR T790M mutations. This hypothesis is supported by the 64% ORR observed in the 11 patients currently enrolled in clinical trial NCT03066206 and described in this study. Taken together, these clinical and preclinical findings demonstrate that poziotinib has potent antitumor activity against NSCLC with an EGFR and/or HER2 exon 20 mutation and support further clinical testing of poziotinib in patients with NSCLC with exon 20 mutations in EGFR or HER2 to determine overall survival, disease control rate, duration of results, safety, and toxicity.
Lastly, overall clinical benefit and the mechanisms underlying resistance to poziotinib remain to be determined. Our data indicate that in the setting of classical EGFR activating mutations (exon 19 deletions or the mutation encoding p.L858R), the secondary mutation encoding p.C797S renders tumor cells resistant to poziotinib. It is currently unknown whether C797S or other secondary mutations occur in patients with NSCLC harboring EGFR exon 20 insertion mutations with acquired resistance to poziotinib. Furthermore, a recent phase 2 study of poziotinib in patients with NSCLC bearing sensitizing mutations in EGFR (exon 19 deletions or the mutation encoding p.L858R) found that MET amplification, PIK3CA mutations, and EGFR T790M mutations were mechanisms underlying resistance39. Ongoing and future clinical studies will be needed to evaluate these questions and aid in the design of more effective therapeutic regimens for NSCLC and other cancer types bearing EGFR and HER2 exon 20 mutations.
Methods
Patient population and statistical analyses
Patients with NSCLC with mutated EGFR who enrolled and consented in the prospectively collected MD Anderson Lung Cancer Moon Shot GEMINI database were identified. EGFR mutation status was determined as a part of routine clinical care using PCR-based next-generation sequencing. PFS was calculated using the Kaplan–Meier method. PFS was defined as time from commencement of treatment with an EGFR TKI to radiologic progression or death. Restaging scans were obtained at 6- to 8-week intervals during treatment and were retrospectively assessed according to RECIST, version 1.1 to determine the response rate in patients with NSCLC with an EGFR exon 20 insertion. All patients provided written informed consent for enrollment into the MD Anderson Lung Cancer Moon Shot GEMINI database and/or for treatment with poziotinib on either compassionate-use protocol (MD Anderson Cancer Center CIND16-0055) or clinical trial NCT03066206. The protocols are approved by both the MD Anderson Cancer Center institutional review board and the Food and Drug Administration.
Cell line generation and IL-3 deprivation
The Ba/F3 cell line was cultured in complete RPMI-1640 medium (R8758; Sigma Life Science) supplemented with l-glutamine, 10% heat-inactivated FBS (Gibco), 1% penicillin–streptomycin (Sigma Life Science), and 10 ng/ml mouse IL-3 (R&D systems) under sterile conditions. Stable cell lines were generated through retroviral transduction of the Ba/F3 cell line for 12 h. Retroviruses were generated through transfecting the pBABE-Puro-based vectors summarized in Supplementary Table 4 (Addgene and Bioinnovatise) into the Phoenix 293 T Ampho packaging cell line (Orbigen) using Lipofectamine 2000 (Invitrogen). 72 h after transduction, 2 μg/ml puromycin (Invitrogen) was added to the medium. After 5 d of selection, cells were stained with FITC-conjugated HER2 (Biolegend) or phycoerythrin (PE)-conjugated EGFR (Biolegend) and sorted via FACS. Cell lines were then grown in the absence of IL-3 for 15 d, and cell viability was determined every 3 d using the Cell Titer Glo assay (Progema). Resulting stable cell lines were maintained in the complete RPMI-1640 medium described above without IL-3. HCC827 and HCC4006 lung cancer cell lines were obtained from ATCC and maintained in 10% RPMI medium under sterile conditions. Cell line identity was confirmed through DNA fingerprinting via short tandem repeats using the PowerPlex 1.2 kit (Promega). Fingerprinting results were compared with reference fingerprints from ATCC. All cell lines were free of mycoplasma. Two erlotinib-resistant cell lines were generated as previously described35; briefly, we cultured HCC827 and HCC4006 (both with mutated EGFR) cells with increasing concentrations of erlotinib until resistant variants emerged.
Cell viability assay and IC50 estimation
Cell viability was determined using the Cell Titer Glo assay (Promega). Cells were collected from suspension medium, spun down at 300 g for 5 min and resuspended in fresh RPMI medium and counted using a Countess automated cell counter and trypan blue (Invitrogen). 1,500 cells per well were plated in 384-well plates (Greiner Bio-One) in technical triplicates on the same plate. Cells were treated with seven different concentrations of inhibitors in serial threefold-diluted TKIs or vehicle alone for a final volume of 40 μl per well. After 72 h, 11 μl of Cell Titer Glo was added to each well. Plates were shaken for 10 min, and bioluminescence was determined using a FLUOstar OPTIMA multimode microplate reader (BMG LABTECH). Bioluminescence values were normalized to DMSO-treated cells, and normalized values were plotted in GraphPad Prism using nonlinear regression fit to normalized data with a variable slope. IC50 values were calculated using GraphPad Prism at 50% inhibition. Each experiment was replicated three separate times to give biological replicates unless indicated otherwise.
Tyrosine kinase inhibitors
Lapatinib, afatinib, dacomitinib, osimertinib, rociletinib, nazartinib, ibrutinib, and poziotinib were purchased from Selleck Chemical. Erlotinib and gefitinib were obtained from the institutional pharmacy at The University of Texas MD Anderson Cancer Center. Olmutinib was provided by Boehringer-Ingelheim. All inhibitors were dissolved in DMSO at a concentration of 10 mM and stored at –80 °C.
3D modeling
We retrieved the structure of EGFR D770insNPG protein (Protein Data Bank: 4LRM) and used it as a template to build our molecular 3D structural model of EGFR D770insNPG. HER2 A775insYVMA was built using the previously published model in Shen et al52. The homology models were built using MODELLER 9v6 and were further energetically minimized using the Molecular Operating Environment software package (Chemical Computing Group, Montreal, Canada). Molecular docking of TKIs into exon 20 mutant EGFR and HER2 were performed using GOLD software with default parameters unless otherwise noted. No early termination was allowed in the docking process. Restraints were used to model the covalent bond formations between receptors and inhibitors. The flexibility of residues within the binding pocket was addressed using GOLD software. Figures demonstrating interactions between EGFR and/or HER2 and inhibitors were visualized using PyMOL.
Western blotting of Ba/F3 mutants
For western blotting, cells were washed in PBS and lysed in protein lysis buffer (ThermoFisher) and protease inhibitor cocktail tablets (Roche). Protein (30–40 μg) was loaded into gels purchased from BioRad. BioRad semidry transfer was used and then probed with antibodies against p-EGFR (no. 2234), EGFR (no. 4267), pHER2 (no. 2247), and HER2 (no. 4290) (all 1:1,000; Cell Signaling). Blots were probed with antibodies against β-actin (Sigma-Aldrich, no. A2228) or vinculin (Sigma-Aldrich, no. V4505) as a loading control and exposed using SuperSignal West Pico PLUS Chemiluminescent Substrate (ThermoFisher) and the ChemiDoc Touch Imaging System (Bio-Rad) or radiographic film. Representative images are shown of two separate protein isolations and blots run in duplicate. Quantification of western blotting was completed in Photoshop and calculated as (background mean intensity – sample mean intensity) × (number of pixels) = band intensity. Samples were normalized first to loading control (β-actin or vinculin) then to DMSO and were graphed in GraphPad Prism. Significance from DMSO was calculated in GraphPad Prism. Details regarding antibodies can be found the Nature Research Reporting Summary.
ELISA and correlation of Ba/F3 mutants
Protein was harvested from the parental Ba/F3 cell line and each of the Ba/F3 exon 20 mutants found to be activating mutations as described above. ELISA was performed following the manufacturer’s instructions for total EGFR (Cell signaling, no. 7250) and total HER2 (Cell Signaling, no. 7310). Relative expression determined through ELISA was plotted against IC50 values calculated as described above. Pearson correlations and P values were determined using GraphPad Prism.
Patient-derived cell line studies
CUTO14 cells were generated from the pleural effusion of a patient with lung adenocarcinoma following informed consent at the University of Colorado through use of previously described culture methods52. Cell lines were treated with the indicated doses of afatinib or poziotinib for 72 h, and cell viability was determined through MTS assay (Promega). IC50 values were calculated as previously described (n = 3 biological replicates). Western blotting with patient-derived cell lines was completed as previously described53 (n = 3 biological replicates). Cells were treated for 2 h with the indicated doses of afatinib or poziotinib. All antibodies were purchased from Cell Signaling Technology, with the exception of total EGFR (BD Transduction Laboratories) and GAPDH (Calbiochem). Catalog numbers and dilutions for antibodies can be found in the Life Sciences Reporting Summary.
The YUL-0019 cell line was established from malignant pericardial fluid obtained from a patient with advanced adenocarcinoma of the lung under an Institutional Review Board–approved protocol at Yale University. The cell line was cultured in RPMI + l-glutamine (Corning), supplemented with 10% heat-inactivated FBS (Atlanta Biologicals) and 1% penicillin–streptomycin (Corning). To confirm the presence of the EGFR mutation, RNA was extracted from cell pellet using the RNeasy mini kit (Qiagen no. 74104) according to the manufacturer’s instructions. cDNA was synthesized using the Superscript III First-Strand cDNA Synthesis Kit (Invitrogen no. 18080-051) and used as a template to amplify EGFR. PCR product was sequenced through Sanger sequencing using the following primers: EGFR-2080F: 5′-CTTACACCCAGTGGAGAAGC -3′ and EGFR-2507R 5′-ACCAAGCGACGGTCCTCCAA -3′. Forward and reverse sequence tracings were manually reviewed. The variant detected in the patient-derived cell line was a complex insertion in exon 20 of EGFR (N771delinsFH), leading to the replacement of amino acid asparagine at position 771 by two amino acids, phenylalanine and histidine. Cell viability and IC50 estimation was performed as described above.
Patient-derived xenograft and cell line studies
LU0387 PDX experiments were completed by Crown BioSciences. Briefly, fragments from tumors expressing EGFR H773insNPH were inoculated into 5- to 6-week old female nu/nu nude mice. When tumors reached a volume of 100–200 mm3 mice were randomized into three treatment groups: 5 mg/kg poziotinib, 10 mg/kg poziotinib, or vehicle control (20% PEG-400, 3% Tween-80 in dH2O). Tumor volumes and body weight were measured twice weekly. Mice treated with 5 mg/kg poziotinib received drug for 4–5 days, then were not treated for 4 d, and then received an additional 4 d of dosing. Mice were then observed for another 2 d without dosing. Mice treated with 10 mg/kg poziotinib received drug for 3–4 d, and then were observed for 10 d without dosing. Mice that were humanely euthanized for events unrelated to tumor burden were excluded from the final analysis.
YUL-0019 patient-derived cell line xenografts were created by injecting 5 × 106 cells in 50% matrigel into 5- to 6-week-old female nu/nu nude mice. When tumors reached 300 mm3, mice were randomized into four treatment groups: 20 mg/kg afatinib, 5 mg/kg poziotinib, 10 mg/kg poziotinib, or vehicle control (20% PEG-400, 3% Tween-80 in dH2O). Tumor volumes were measured daily. Mice received drug 5 days per week, but they received an additional period without treatment if body weight dropped by more than 10%. Mice humanely euthanized for events unrelated to tumor burden were excluded from final analysis. Experiments were completed in agreement with Good Animal Practices and with approval from the MD Anderson Cancer Center Institutional Animal Care and Use Committee (Houston, TX).
Genetically engineered mouse model studies
EGFR D770insNPG and HER2 A775insYVMA GEMMs were generated as previously described26,30. Mice were handled in accordance with Good Animal Practices as defined by the Office of Laboratory Animal Welfare and were done with approval from the Dana-Farber Cancer Institute Institutional Animal Care and Use Committee (Boston, MA). Mice were fed a continuous doxycycline diet from 6 weeks of age. Tumor volume was determined through MRI as previously described26,30. Mice with equal initial tumor volume were nonblindly randomized to vehicle, 20 mg/kg afatinib, or 10 mg/kg poziotinib daily upon obvious tumor formation as determined through MRI. Mice humanely euthanized for events unrelated to tumor burden were excluded from final analysis.
Preliminary results from patients receiving poziotinib
Patients provided written informed consent for treatment with poziotinib on either compassionate-use protocol (MD Anderson Cancer Center CIND16-0055) or clinical trial NCT03066206. The protocols are approved by both the MD Anderson Cancer Center institutional review board and the Food and Drug Administration.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software. Specifically, for patient PFS in retrospective analysis, the Kaplan–Meier method was used in conjunction with the log-rank test to determine statistical significance. For in vitro IC50 determinations, bioluminescence values were normalized to those of DMSO-treated cells, and these normalized values were plotted in GraphPad Prism using nonlinear regression fit to normalized data with a variable slope. IC50 values were calculated by interpolating the concentration of the compound at 50% inhibition using GraphPad Prism. Each experiment was replicated three times unless otherwise indicated. For correlations of protein expression and drug sensitivity, relative expression as determined through ELISA was plotted against IC50 values that were calculated as described above. Pearson correlation of the thirteen clones and P values were determined through GraphPad Prism. For in vivo GEMM studies, GraphPad prism was used to perform a two-sided Student’s t-test to determine the P value. For the 10-d PDX study, a multiple-comparisons test with the Holm–Sidak method was used to determine statistical significance between all groups and time points; P values can be found in Supplementary Table 3. Lastly, for the 14-d PDX study, a one-way ANOVA analysis was used in combination with a Tukey’s test to determine statistical significance at day 14 only. For all in vivo studies, mice humanely euthanized for events unrelated to tumor burden were excluded from the final analysis, and the exact number of mice per group is listed in figure legends.
Supplementary Material
Acknowledgments
The authors wish to thank the patients and their families for participation in this study and Spectrum Pharmaceuticals for providing the drug used in the compassionate-use protocol. The authors also wish to thank J.S. (a patient) for efforts in initiating the poziotinib trial. This work was supported by the Lung SPORE grant P50 CA070907 and P50 CA196530 (J.V.H.); the Lung Cancer Research Fund (Y.Y.E.); generous philanthropic contributions to the University of Texas MD Anderson Lung Cancer Moon Shots Program (J.V.H.); NIH R01 CA190628 (J.V.H.), NIH Cancer Center Support Grant P30 CA016672 (J.V.H.), the Rexanna Foundation for Fighting Lung Cancer (J.V.H.), the Exon 20 Group (J.V.H.), the University of Texas MD Anderson Cancer Center Bruton Endowed Chair in Tumor Biology (J.V.H.), the Stading Fund for EGFR inhibitor resistance (J.V.H.), the Hallman fund (J.V.H.), and the Fox Lung EGFR Inhibitor Fund (J.V.H.). Additional support was provided from the Christine J. Burge Endowment for Lung Cancer Research at the University of Colorado Cancer Center, the Burge family, the Miramont Cares foundation (R.C.D.), a fellowship from the Italian Association for Cancer Research (A.T.), and Cancer Prevention Research Institute of Texas DP150086, National Science Foundation CHE-1411859, and National Institute of General Medical Sciences GM070737 (S.Z.). The clinical study was supported by Spectrum Pharmaceuticals.
Footnotes
Contributions
J.P.R., Y.Y.E., and J.V.H. coordinated the study. J.P.R., Y.Y.E., Z.T., A.P., H.S., S.Z., S. Liu. S. Li, T.C., A.E-B., A.T.L., and J.V.H. designed and/or performed experiments. J.P.R., Y.Y.E., Z.T., B.W.C., S.Z., E.R., A.T., K.P., K.-K.W, R.C.D., and J.V.H. interpreted data. A.T., S.B.G., and K.P. validated and provided the YUL0019 cell line. J.R.B. referred two patients described in the study. V.P., C.L., M.A., Y.Y.E., and J.V.H. treated patients on the clinical study. J.P.R., M.B.N, Y.Y.E., and J.V.H wrote the manuscript. All authors edited and approved the manuscript.
Competing interests
J.P.R., M.B.N., and J.V.H. have filed patent applications under the Patent Cooperation Treaty and in Taiwan. J.V.H. has had grant or research support from AstraZeneca, Bayer, and GlaxoSmithKline and has served on advisory committees for AstraZeneca, Boehringer Ingelheim, Exelixis, Genentech, GSK, Lilly, Novartis, Spectrum, and Synta. R.C.D. has licensing fees, honorarium, and travel expenses from Ariad Pharmaceuticals, has a Sponsored Research Agreement from Threshold Pharmaceuticals, and has served as an advisory Board member for AstraZeneca.
References
- 1.Bezjak A, et al. Symptom improvement in lung cancer patients treated with erlotinib: quality of life analysis of the National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol. 2006;24:3831–3837. doi: 10.1200/JCO.2006.05.8073. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Schmid-Bindert G, Zhou C. Erlotinib in the treatment of advanced non-small cell lung cancer: an update for clinicians. Ther Adv Med Oncol. 2012;4:19–29. doi: 10.1177/1758834011427927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wheatley-Price P, Ding K, Seymour L, Clark GM, Shepherd FA. Erlotinib for advanced non-small-cell lung cancer in the elderly: an analysis of the National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol. 2008;26:2350–2357. doi: 10.1200/JCO.2007.15.2280. [DOI] [PubMed] [Google Scholar]
- 5.Zhou C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 6.Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 2007;98:1817–1824. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu HA, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 10.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 11.Cappuzzo F, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 12.Costa DB, Kobayashi S, Tenen DG, Huberman MS. Pooled analysis of the prospective trials of gefitinib monotherapy for EGFR-mutant non-small cell lung cancers. Lung Cancer. 2007;58:95–103. doi: 10.1016/j.lungcan.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sequist LV, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 14.Maemondo M, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 15.Oxnard GR, et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol. 2013;8:179–184. doi: 10.1097/JTO.0b013e3182779d18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasuda H, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med. 2013;5:216ra177. doi: 10.1126/scitranslmed.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arcila ME, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–229. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol. 2012;13:e23–e31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 19.Arcila ME, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res. 2012;18:4910–4918. doi: 10.1158/1078-0432.CCR-12-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazières J, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol. 2013;31:1997–2003. doi: 10.1200/JCO.2012.45.6095. [DOI] [PubMed] [Google Scholar]
- 21.Costa DB, et al. Pulse afatinib for ERBB2 exon 20 insertion-mutated lung adenocarcinomas. J Thorac Oncol. 2016;11:918–923. doi: 10.1016/j.jtho.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Grève J, et al. Clinical activity of afatinib (BIBW 2992) in patients with lung adenocarcinoma with mutations in the kinase domain of HER2/neu. Lung Cancer. 2012;76:123–127. doi: 10.1016/j.lungcan.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Mazières J, et al. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2 targeted drugs: results from the EUHER2 cohort study. Ann Oncol. 2016;27:281–286. doi: 10.1093/annonc/mdv573. [DOI] [PubMed] [Google Scholar]
- 24.Kosaka T, et al. Response heterogeneity of EGFR and HER2 exon 20 insertions to covalent EGFR and HER2 inhibitors. Cancer Res. 2017;77:2712–2721. doi: 10.1158/0008-5472.CAN-16-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyman DM, et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature. 2018;554:189–194. doi: 10.1038/nature25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perera SA, et al. HER2YVMA drives rapid development of adenosquamous lung tumors in mice that are sensitive to BIBW2992 and rapamycin combination therapy. Proc Natl Acad Sci USA. 2009;106:474–479. doi: 10.1073/pnas.0808930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang SE, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Yang M, et al. NSCLC harboring EGFR exon-20 insertions after the regulatory C-helix of kinase domain responds poorly to known EGFR inhibitors. Int J Cancer. 2016;139:171–176. doi: 10.1002/ijc.30047. [DOI] [PubMed] [Google Scholar]
- 29.Cha MY. Antitumor activity of HM781-36B, a highly effective pan-HER inhibitor in erlotinib-resistant NSCLC and other EGFR-dependent cancer models. Int J Cancer. 2012;10:2445–2454. doi: 10.1002/ijc.26276. [DOI] [PubMed] [Google Scholar]
- 30.Cho J, et al. Cetuximab response of lung cancer-derived EGF receptor mutants is associated with asymmetric dimerization. Cancer Res. 2013;73:6770–6779. doi: 10.1158/0008-5472.CAN-13-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wind S, Schmid M, Erhardt J, Goeldner RG, Stopfer P. Pharmacokinetics of afatinib, a selective irreversible ErbB family blocker, in patients with advanced solid tumours. Clin Pharmacokinet. 2013;52:1101–1109. doi: 10.1007/s40262-013-0091-4. [DOI] [PubMed] [Google Scholar]
- 32.Li D, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–4711. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thress KS, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chabon JJ, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7:11815. doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nilsson MB. Stress hormones promote EGFR inhibitor resistance in NSCLC: implications for combinations with beta blockers. Sci Transl Med. 2017;9:eaao4307. doi: 10.1126/scitranslmed.aao4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 37.Kim D-W, et al. Phase I studies of HM781-36B, an irreversible pan-HER tyrosine kinase inhibitor (TKI) in patients with advanced solid tumor and the therapeutic potential in patients with advanced non-small cell lung cancer (NSCLC) J Thorac Oncol. 2013;8:S607. [Google Scholar]
- 38.Kim TM, et al. A phase I study of HM781–36B, a novel pan-HER inhibitor, in patients (pts) with advanced solid tumors. J Clin Oncol. 2012;30:3076. [Google Scholar]
- 39.Noh YH, et al. Population pharmacokinetics of HM781-36 (poziotinib), pan-human EGF receptor (HER) inhibitor, and its two metabolites in patients with advanced solid malignancies. Cancer Chemother Pharmacol. 2015;75:97–109. doi: 10.1007/s00280-014-2621-7. [DOI] [PubMed] [Google Scholar]
- 40.Han JY, et al. A phase II study of poziotinib in patients with epidermal growth factor receptor (EGFR)-mutant lung adenocarcinoma who have acquired resistance to egfr-tyrosine kinase inhibitors. Cancer Res Treat. 2017;49:10–19. doi: 10.4143/crt.2016.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Planchard D, et al. Osimertinib Western and Asian clinical pharmacokinetics in patients and healthy volunteers: implications for formulation, dose, and dosing frequency in pivotal clinical studies. Cancer Chemother Pharmacol. 2016;77:767–776. doi: 10.1007/s00280-016-2992-z. [DOI] [PubMed] [Google Scholar]
- 42.Jänne PA, et al. Phase I dose-escalation study of the pan-HER inhibitor, PF299804, in patients with advanced malignant solid tumors. Clin Cancer Res. 2011;17:1131–1139. doi: 10.1158/1078-0432.CCR-10-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kourie HR, Chaix M, Gombos A, Aftimos P, Awada A. Pharmacodynamics, pharmacokinetics and clinical efficacy of neratinib in HER2-positive breast cancer and breast cancer with HER2 mutations. Expert Opin Drug Metab Toxicol. 2016;12:947–957. doi: 10.1080/17425255.2016.1198317. [DOI] [PubMed] [Google Scholar]
- 44.Ito Y, et al. Safety, efficacy and pharmacokinetics of neratinib (HKI-272) in Japanese patients with advanced solid tumors: a Phase 1 dose-escalation study. Jpn J Clin Oncol. 2012;42:278–286. doi: 10.1093/jjco/hys012. [DOI] [PubMed] [Google Scholar]
- 45.Sequist LV, et al. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:3076–3083. doi: 10.1200/JCO.2009.27.9414. [DOI] [PubMed] [Google Scholar]
- 46.Yang JC, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015;16:830–838. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 47.Kris MG, et al. Targeting HER2 aberrations as actionable drivers in lung cancers: phase II trial of the pan-HER tyrosine kinase inhibitor dacomitinib in patients with HER2-mutant or amplified tumors. Ann Oncol. 2015;26:1421–1427. doi: 10.1093/annonc/mdv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsigelny IF, et al. Molecular determinants of drug-specific sensitivity for epidermal growth factor receptor (EGFR) exon 19 and 20 mutants in non-small cell lung cancer. Oncotarget. 2015;6:6029–6039. doi: 10.18632/oncotarget.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wheler J, et al. Revisiting clinical trials using EGFR inhibitor-based regimens in patients with advanced non-small cell lung cancer: a retrospective analysis of an MD Anderson Cancer Center phase I population. Oncotarget. 2013;4:772–784. doi: 10.18632/oncotarget.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wheler JJ, et al. Combining erlotinib and cetuximab is associated with activity in patients with non-small cell lung cancer (including squamous cell carcinomas) and wild-type EGFR or resistant mutations. Mol Cancer Ther. 2013;12:2167–2175. doi: 10.1158/1535-7163.MCT-12-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.