ABSTRACT
The importance of mitochondria in energy metabolism, signal transduction and aging in post-mitotic tissues has been well established. Recently, the crucial role of mitochondrial-linked signaling in stem cell function has come to light and the importance of mitochondria in mediating stem cell activity is becoming increasingly recognized. Despite the fact that many stem cells exhibit low mitochondrial content and a reliance on mitochondrial-independent glycolytic metabolism for energy, accumulating evidence has implicated the importance of mitochondrial function in stem cell activation, fate decisions and defense against senescence. In this Review, we discuss the recent advances that link mitochondrial metabolism, homeostasis, stress responses, and dynamics to stem cell function, particularly in the context of disease and aging. This Review will also highlight some recent progress in mitochondrial therapeutics that may present attractive strategies for improving stem cell function as a basis for regenerative medicine and healthy aging.
KEY WORDS: Stem cell, Mitochondria, Cell fate, Aging
Summary: This Review looks beyond the role of mitochondria in cellular energy metabolism and highlights recent studies that implicate mitochondrial function in stem cell activation, fate decisions and defense against senescence.
Introduction
In adults, tissue-specific stem cells maintain tissue homeostasis and provide committed progenitors for regeneration after tissue injury. To maintain the stem cell pool and provide progenitors for tissue-specific cell differentiation, stem cells must undergo a process of self-renewal. The balance between the preservation of stem cells and tissue regeneration relies on the maintenance of cell fate decisions, which is under precise regulation by many different factors: growth factors, inflammatory mediators, the extracellular environment, cell-cell signaling, cellular metabolism and so on. It is now well known that an imbalance towards cell lineage commitment at the expense of self-renewal can have detrimental effects under certain circumstances, for example in disease or during aging.
Over time, stem cells progressively lose plasticity in response to chronological and replicative senescence. This has important consequences for the maintenance of tissue function; slower wound healing, muscle weakness, decreased immunity and hair graying or loss all appear with age as a result of changes in tissue-specific stem cell activity (Signer and Morrison, 2013; Van Zant and Liang, 2003). Despite a general decline in stem cell function during aging, the specific changes in the stem cell phenotype depend on their local tissue niche. During aging, tissues with low stem cell turnover rates often exhibit an accentuated depletion of the stem cell pool, as is commonly seen in skeletal muscle (Bengal et al., 2017), certain parts of the nervous system (Maslov et al., 2004) and hair follicles of epithelial tissue (Nishimura et al., 2005). In other tissues, stem cells can maintain their population, or even increase in numbers with aging, as seen for hematopoietic stem cells (HSCs) (Rossi et al., 2005). However, despite the expansion of HSCs during aging, there is still a decline in cell function (Morrison et al., 1996) and altered cell fate decisions (Rossi et al., 2005; Sudo et al., 2000). Similarly, an increase of cell number and decline in cell function is also seen in intestinal stem cell aging (Biteau et al., 2008; Choi et al., 2008; Martin et al., 1998). Understanding the diversity of stem cell responses to aging within different stem cell niches requires comprehensive mechanistic studies of the molecular events that influence stem cells during aging.
Mitochondria are bioenergetic organelles that produce ATP via oxidative phosphorylation (OXPHOS), a process driven by the formation of reducing equivalents NADH and FADH2. The importance of mitochondria in mediating stem cell activity has been largely neglected owing to the low abundance of mitochondria in many types of stem cells, and their widely recognized dependence on glycolysis for energy (Rafalski et al., 2012). Recent findings have revealed how mitochondria influence stem cell fate and function, whether it be in healthy tissue or during aging and disease (Ansó et al., 2017; Buck et al., 2016; Jin et al., 2018; Khacho et al., 2016; Mohrin et al., 2015; Zhang et al., 2016). Besides their known role in energy harvesting, mitochondria have a signaling function and various forms of stress in the mitochondria can generate retrograde signals, for example reactive oxygen species (ROS), that affect other cellular sites and are known to influence stem cell activity and function (Sun et al., 2016). Mitochondria also compartmentalize several key metabolic pathways, such as the tricarboxylic acid (TCA) cycle, fatty acid β-oxidation and the one-carbon cycle. Metabolites generated by these pathways can also act as retrograde signals, at least in post-mitotic tissues. Protein lysine modifications by malonylation, succinylation and glutarylation all utilize substrates from mitochondrial fatty acid and amino acid metabolism (Hirschey and Zhao, 2015). Interestingly, the sirtuin enzymes can catalyze the cleavage of the above-mentioned post-translational modifications (PTMs) using the mitochondrial metabolite NAD+ as a co-factor (Menzies et al., 2016). Furthermore, the TCA cycle intermediate α-ketoglutarate (α-KG) is a substrate for enzymes with DNA and histone demethylation activity (Teperino et al., 2010). Although most of these mitochondrial metabolism-mediated PTMs were discovered in post-mitotic adult tissue, similar cases have been described in stem cells during health, disease and aging (Chandel et al., 2016).
As mitochondria play an important role in regulating stem cell activity, it follows that a decline in stem cell mitochondrial function might underpin age-related deterioration in stem cell function and self-renewal in multiple different tissues (Ahlqvist et al., 2012; Ansó et al., 2017; Khacho et al., 2017; Rera et al., 2011; Stoll et al., 2011; Zhang et al., 2016) (Fig. 1). In this Review, we discuss the emerging role of mitochondrial energy metabolism, mitochondrial proteostasis, mitophagy and key mitochondrial signaling events in stem cell function and how this relates to tissue homeostasis in disease and aging.
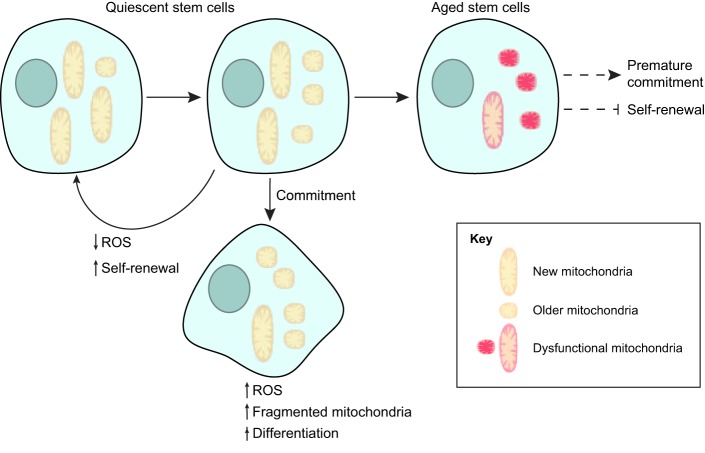
Fig. 1.
Mitochondrial influence on stem cell fate. Asymmetric division generates two daughter cells with different mitochondrial properties. The daughter cell that received a greater proportion of new mitochondria maintains its stem cell traits whereas the daughter cell that received more older mitochondria tends to differentiate. Other mitochondrial signaling mechanisms, such as ROS and mitochondrial dynamics, are also involved in the balance between stem cell self-renewal and commitment. Mitochondrial reduction and dysfunction can further lead to stem cell aging, which is typically characterized by a reduction in stem cell renewal and premature commitment.
Energy metabolism in stem cells
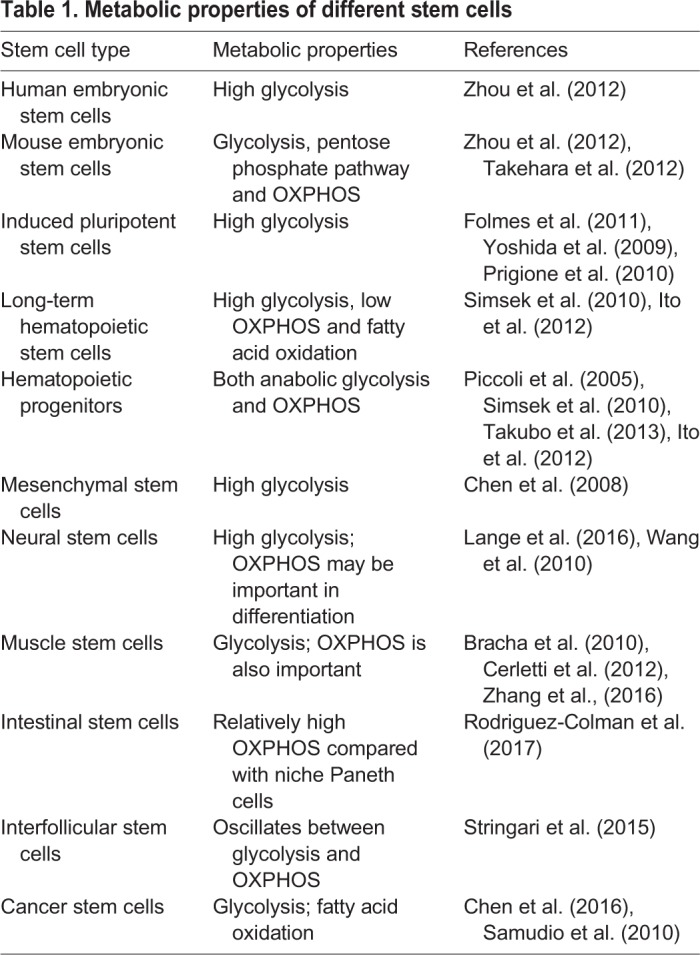
Different types of stem cells have different metabolic properties (Table 1); however, most reported data describe stem cells as glycolytic (reviewed by Rafalski et al., 2012). Human and murine HSCs (Miharada et al., 2011; Piccoli et al., 2005; Simsek et al., 2010; Takubo et al., 2013; Yu et al., 2013), murine neural stem cells (NSCs) (Lange et al., 2016; Wang et al., 2010), human embryonic stem cells (ESCs) (St John et al., 2005; Zhang et al., 2011) and human bone marrow-derived mesenchymal stem cells (MSCs) (Chen et al., 2008) are all highly dependent on glycolytic metabolism for stem cell maintenance and/or self-renewal. Consistent with a reliance on glycolysis and low rates of mitochondrial oxidative respiration, human HSCs (Piccoli et al., 2005), MSCs (Chen et al., 2008) and ESCs (Chung et al., 2007) have few mitochondria and an immature inner structure. Furthermore, stem cell self-renewal and/or multi- or pluripotency are promoted by hypoxia in murine HSCs, MSCs and NSCs and human ESCs (D'Ippolito et al., 2006; Ezashi et al., 2005; Jeong et al., 2007; Morrison et al., 2000).
Table 1.
Metabolic properties of different stem cells
Several explanations have been proposed to account for the relative importance of glycolysis over mitochondrial OXPHOS activity in stem cells. For instance, dependence on anaerobic metabolism may be a long-term evolutionary adaption of stem cells to their low oxygen niche. An example of this is the hypoxic bone marrow environment of HSCs (Miharada et al., 2011; Simsek et al., 2010), as well as the rates of glycolysis in cancer stem cells (CSCs) within solid tumors (Zu and Guppy, 2004). Another explanation might be the benefit of a glycolytic metabolism in providing intermediates necessary for supporting anabolic pathways that are essential for stem cell self-renewal and the generation of progeny (reviewed by Suda et al., 2011). Anaerobic metabolism can also help to avoid oxidative damage from mitochondria-generated ROS [reviewed by Suda et al. (2011) and further discussed in the next sections].
Recent studies have demonstrated that stem cells also utilize mitochondrial fatty acid oxidation (FAO) in addition to glycolysis during self-renewal (Ito et al., 2012). Inhibition of FAO in HSCs results in the loss of the asymmetric division of HSC daughter cells, which is an essential process for maintaining the ‘reserve’ stem cell pool during the simultaneous expansion of stem cell differentiation (Ito et al., 2012). Indeed, loss of the mitochondrial FAO activator peroxisome proliferator-activated receptor δ (PPAR-δ) induces a loss of HSC maintenance, whereas treatment with PPAR-δ agonists improves HSC maintenance by promoting asymmetric HSC division (Ito et al., 2012). Lipid metabolism is also important for maintenance of NSC proliferation (Knobloch et al., 2013). Conditional deletion of fatty acid synthase (FASN), the key enzyme of de novo lipogenesis, in mouse NSCs impairs adult neurogenesis (Knobloch et al., 2013). Similar to HSCs and NSCs, cancer cells are generally considered to be glycolytic, a result of the Warburg effect; however, glioma stem cells have been reported to contain higher levels of ATP and rely mainly on OXPHOS as an energy source (Vlashi et al., 2011). Moreover, several types of tumor-initiating stem cells exhibit mitochondrial FAO as a mechanism for self-renewal and resistance to chemotherapy (Chen et al., 2016; Samudio et al., 2010). Thus, the combination of mitochondrial FAO and glycolysis might play a role in self-preservation in some types of CSCs. Related to this, intestinal stem cells (ISCs) exhibit an interesting phenomenon whereby their proper function depends both on their own mitochondrial activity, and on Paneth cells in their surrounding niche that are reliant on glycolysis (Rodríguez-Colman et al., 2017).
Consistent with the importance of mitochondrial OXPHOS activity in stem cell function and maintenance, the clearance of ‘older’ mitochondria away from stem cells during asymmetric cell division seems to be essential for retaining stemness in mammary stem-like cells (Katajisto et al., 2015) (Fig. 1). Calorie restriction (CR), which is known to improve mitochondrial function in post-mitotic tissues, increases the abundance of muscle stem cells (MuSCs) (Cerletti et al., 2012) and improves the self-renewal of many stem cell populations, such as germline stem cells (GSCs) in flies (Mair et al., 2010) and HSCs (Chen et al., 2003; Cheng et al., 2014) and ISCs (Igarashi and Guarente, 2016; Yilmaz et al., 2012) in mice. Conversely, caloric excess reduces mitochondrial function (Bournat and Brown, 2010) and impairs stem cell function: in mouse models of high fat feeding or obesity and type 2 diabetes (ob/ob and db/db mice, respectively) muscle regeneration is blunted with a reduction in injury-induced MuSC proliferation (Hu et al., 2010; Nguyen et al., 2011). Similarly, a high fat diet dysregulates ISCs and their daughter cells, resulting in an increased incidence of intestinal tumors (Beyaz et al., 2016).
Interestingly, mouse and human ESCs have different metabolic properties (reviewed by Mathieu and Ruohola-Baker, 2017). In mice, despite the more immature appearance of mitochondria and lower mitochondrial content, basal and maximal mitochondrial respiration are substantially higher in ESCs compared with the more differentiated (primed) epiblast stem cells (EpiSCs), which are derived from a post-implantation epiblast at a later stage of development (Zhou et al., 2012). Conventional human ESCs (hESCs) do not appear to be naïve like mouse ESCs (mESCs) but more similar to primed mouse EpiSCs with regards to their gene expression profile and epigenetic state. In addition, hESCs are also more metabolically similar to rodent EpiSCs as they display a higher rate of glycolysis than do mouse ESCs (Sperber et al., 2015; Zhou et al., 2012). Ectopic expression of HIF1α or exposure to hypoxia can promote the conversion of mESCs to the primed state by favoring glycolysis, thereby suggesting an important role for mitochondrial metabolism in the maintenance of mESCs (Zhou et al., 2012). Indeed, upregulated mitochondrial transcripts and increased mitochondrial oxidative metabolism by STAT3 activation supports the enhanced proliferation of mESCs and the reprogramming of EpiSCs back to a naïve pluripotent state (Carbognin et al., 2016). In the human context, conventional, primed ESCs can transition to a more naïve state in vitro by treatment with histone deacetylase (HDAC) inhibitors (Ware et al., 2014). The fact that HDACs are largely NAD+ dependent (further discussed below) supports the role of metabolism in stem cell maintenance.
In addition to its role in stem cell self-renewal, metabolism is also an important regulator of stem cell identity and fate decisions. For instance, several glycolytic adult stem cells require OXPHOS activity for differentiation, including NSCs (Zheng et al., 2016), MSCs (Tang et al., 2016; Tormos et al., 2011; Zhang et al., 2013), HSCs (Inoue et al., 2010) and ESCs (Yanes et al., 2010). The reverse transition, from OXPHOS to glycolysis, is required for the induction of pluripotency from somatic cells (Folmes et al., 2012), which is consistent with the fact that induced pluripotent stem cells (iPSCs) generally exhibit an immature mitochondrial morphology and reliance on glycolytic metabolism (Prigione et al., 2010). Interestingly, it was later reported that the reprogramming of human and mouse iPSCs from fibroblasts requires a transient increase of OXPHOS (Kida et al., 2015; Prigione et al., 2014). The switch between glycolysis and OXPHOS appears to also causally affect HSC fate decisions, as electron transport chain (ETC) uncoupling facilitates the self-renewal of cultured HSCs, even under differentiation-inducing conditions (Vannini et al., 2016). NSCs exhibit changes in mitochondrial morphology and metabolic properties throughout various stages of differentiation in vivo and, in general, activated proliferating NSCs and committed neural progenitors rely more on OXPHOS (Khacho et al., 2016). Intriguingly, mouse epidermal stem cells undergo diurnal metabolic oscillations such that a higher NADH/NAD+ ratio matched by an increased glycolysis/OXPHOS ratio is observed during the night compared with the daytime. Moreover, this metabolic oscillation is coupled to the stem cell cycle (Stringari et al., 2015). It is not yet known whether the change in stem cell metabolism in circadian phase also occurs for other stem cell types.
Mitochondrial metabolites and stem cell fate decisions
The generation of mitochondrial metabolites represents a possible means by which mitochondria could regulate stem cell activity. Key enzymes that regulate chromatin (both DNA and histones) and protein modifications (i.e. acetylation and methylation) rely on mitochondrial metabolic intermediates as co-factors (Matilainen et al., 2017; Menzies et al., 2016). Hence, mitochondrial metabolism is inextricably coupled to gene expression and protein activity. Despite the fact that many of these findings were made using post-mitotic tissues, there are several emerging lines of evidence that suggest similar mechanisms may be at play in regulating stem cell fate decisions.
Several metabolites in the TCA cycle participate in additional pathways that regulate stem cell function and fate decisions (Fig. 2). Export of citrate from mitochondria to the cytoplasm provides acetyl coenzyme A (acetyl-CoA) for nucleo-cytoplasmic acetylation reactions (Wellen et al., 2009). In addition to acetylation, other protein PTMs, such as malonylation, succinylation and glutarylation, also use mitochondrial intermediates, including malonyl-CoA, succinyl-CoA and glutaryl-CoA, respectively, for the regulation of protein function (Matilainen et al., 2017; Sabari et al., 2017). α-KG is not only a substrate for the family of ten-eleven translocation (TET) dioxygenases, enzymes with DNA demethylation function, but also for the Jumonji-C (JMJC) domain-containing histone demethylase (JHDMs) family of proteins that catalyze histone demethylation (Su et al., 2016; Teperino et al., 2010). Furthermore, S-adenosyl-methionine (SAM) is a co-factor for histone methyltransferases connecting histone methylation to one-carbon metabolism (Mentch et al., 2015) and threonine metabolism (Shyh-Chang et al., 2013).
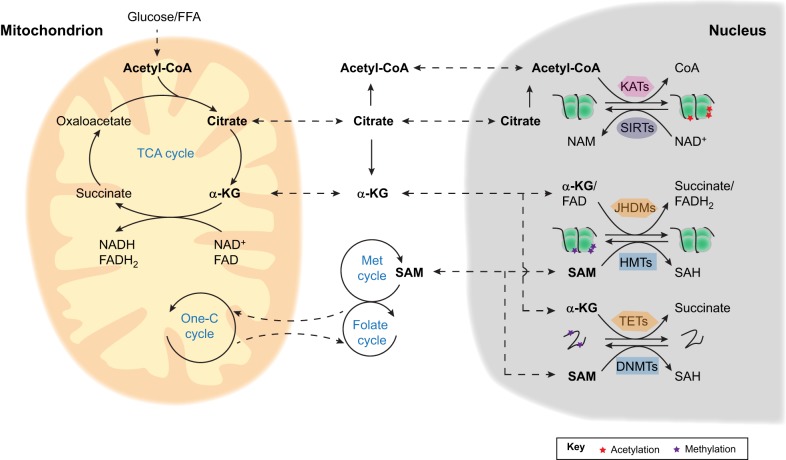
Fig. 2.
Mitochondrial metabolism and epigenetic regulation in stem cells. Mitochondrial TCA cycle intermediate metabolites, such as acetyl-CoA, citrate and α-KG, can be transported to the cytosol and nucleus where they may potentially control stem cell fate via the epigenetic regulation of histones and DNA. Acetyl-CoA is a co-factor of lysine acetyltransferases (KATs), which reverse the activity of the NAD+-dependent sirtuins (SIRTs) by catalyzing the acetylation of histones and other proteins. α-KG is the substrate for both histone (JHDMs) and DNA (TETs) demethylases. Mitochondrial one-carbon (One-C) metabolism coupled with cytosolic folate and methionine (Met) cycles generates SAM, which serves as the co-factor of histone (HMTs) and DNA (DNMTs) methyltransferases. FFA, free fatty acid; NAM, nicotinamide; SAH, S-adenosylhomocysteine.
Acetylation and deacetylation
Both histones and other proteins, such as the transcriptional factors/co-factors peroxisome proliferator-activated receptor-γ coactivator 1α (PPARGC1α), forkhead box protein O 1 (FOXO1) and tumor protein p53 (TP53/TRP53), can be acetylated on the ε-amino groups of site-specific lysine residues through N-ε-acetylation. This is a reversible PTM reaction catalyzed by lysine acetyltransferases (KATs) and lysine deacetylases (KDACs, such as sirtuins). KATs and sirtuins are highly sensitive to the concentration of acetyl-CoA and NAD+ as substrates that tightly connect energy metabolism to gene expression (Box 1 and reviewed by Menzies et al., 2016). Recently, control of such PTMs has been found to influence stem cell activity and senescence across a range of different stem cell types.
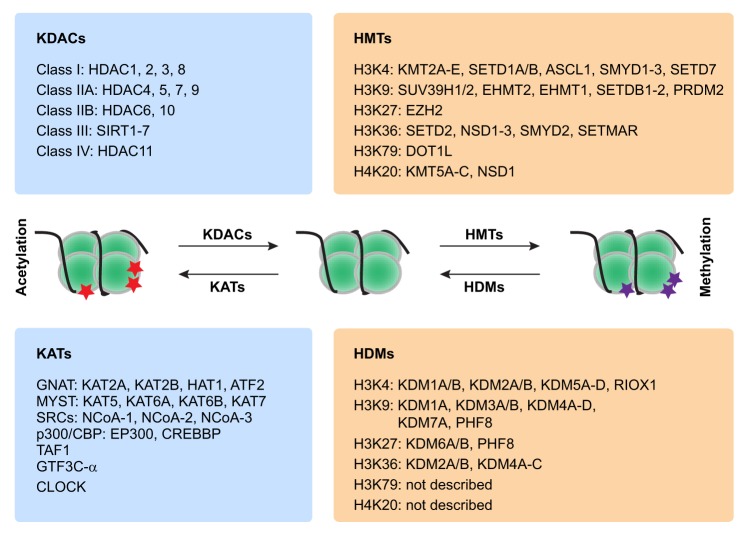
Box 1. Principal enzymes involved in protein acetylation and methylation.
Protein acetylation and methylation are two very common post-transcriptional protein modifications involved in the epigenetic regulation of stem cells (reviewed by Aloia et al., 2013; Oh et al., 2014; Portela and Esteller, 2010). The main enzymes in the class of lysine acetyltransferases (KATs) and lysine deacetylases (KDACs), which catalyze histone acetylation (red stars) and deacetylation, respectively, are given on the left. The main enzymes in the class of histone methytransferases (HMTs) and histone demethylases (HDMs), which catalyze histone methylation (purple stars) and demethylation, respectively, are given on the right. SRCs, steroid receptor co-activators.
In MuSCs, NAD+ levels have been linked to myogenic differentiation. Decreased levels of cellular NAD+, a co-factor for the sirtuin deacetylases (Cantó et al., 2015), leads to elevated H4K16 acetylation (H4K16ac), an activating modification that induces myogenic gene expression (Ryall et al., 2015). In aged mice, a decline in NAD+ levels in MuSCs leads to cell senescence, and boosting NAD+ levels with the dietary precursor nicotinamide riboside alleviates MuSCs senescence and extends mouse lifespan (Zhang et al., 2016). These effects are mediated, at least in part, by the sirtuin family of enzymes. NAD+ levels have also been shown to be limiting during aging in NSCs and can drive the decline in NSC oligodendrogenesis after cuprizone-induced brain demyelination (Stein and Imai, 2014). In HSCs, the level of H4K16ac decreases with age (Florian et al., 2012) and inhibition of the RhoGTPase CDC42 restores H4K16ac levels and reverses phenotypes of HSC aging in transplantation assays (Florian et al., 2012). It is less likely that the decrease of H4K16ac levels in aged HSCs is due to the activation of the NAD+-sirtuin signaling axis as the expression of two NAD+-dependent sirtuins, SIRT3 and SIRT7, have been reported to decline with age and lead to HSC dysfunction (Brown et al., 2013; Mohrin et al., 2015). Protein acetylation has been shown to modulate ESC function too. KDAC inhibitors, such as butyrate, support the extensive self-renewal of mouse and human ESCs and promotes their convergence toward an earlier developmental (more naïve) stage (Ware et al., 2014, 2009).
The evidence linking KAT activity or acetyl-CoA levels to stem cell function is mostly indirect. Recently, in Drosophila, acetyl-CoA levels were found to increase during aging. This increase is accompanied by an elevation in global histone acetylation, notably the acetylation of lysine 12 on histone H4 (H4K12ac). Consistent with this notion, a mutation in the gene encoding the H4K12-specific acetyltransferase Chameau extends lifespan in Drosophila (Peleg et al., 2016). Whether the effect of acetyl-CoA on Drosophila aging is related to stem cell function has yet to be investigated. In human and mouse ESCs, glycolysis-generated acetyl-coA has been reported to promote histone acetylation during pluripotency (Moussaieff et al., 2015). Inhibition of acetyl-coA production by blocking glycolysis causes a significant loss in pluripotency markers in human and mouse ESCs. It would be interesting to investigate whether cellular acetyl-CoA levels are also coupled with stem cell function in mammals in vivo.
Methylation and demethylation
The methylation of nucleic acids and proteins can significantly change their stability and transcription/translation efficiency and activity. Protein methylation can occur on both lysines and arginines and generate either mono- or di-methylated forms, and lysine can also be tri-methylated. The most well-studied protein methylation is histone N-terminal tail methylation, which can either negatively or positively affect gene transcriptional efficiency (Booth and Brunet, 2016; Teperino et al., 2010). DNA methylation and demethylation PTMs are catalyzed reversibly by DNA methyltransferases (DNMTs) and TET enzymes, respectively. Reversible histone methylation can be catalyzed by histone methyltransferases (HMTs) and histone demethylases (HDMs, such as JHDMs). Importantly, both DNA and histone methyltransferases use SAM as a substrate, whereas demethylases use α-KG (Fig. 2, Box 1). Therefore, these modifications are strictly dependent on the availability of mitochondrial metabolites (reviewed by Matilainen et al., 2017).
Both DNA and histone methylation regulate stem cell function. For instance, deficiency in DNA methyltransferase 3A and/or 3B (DNMT3A/B) impairs the differentiation potential of HSCs (Challen et al., 2014; Mayle et al., 2015). Conversely, reducing the function of the DNA demethylase TET2 by ascorbate depletion in mice increases HSC frequency and function, which leads to the acceleration of leukemogenesis (Agathocleous et al., 2017). TET2 restoration, or vitamin C treatment, promotes HSC DNA demethylation, promotes differentiation, and blocks leukemia progression (Cimmino et al., 2017). Maintenance of the euchromatic transcriptional state by inhibition of the euchromatic histone-lysine N-methyltransferase 2 (EHMT2, also known as G9A), delays HSC differentiation (Ugarte et al., 2015). However, in MuSCs, an H4K20 dimethyltransferase, lysine methyltransferase 5B (KMT5B), maintains stem cell quiescence by promoting the formation of facultative heterochromatin. Deletion of KMT5B induces transcriptional activation, further resulting in abnormal MuSC activation and differentiation. This leads to stem cell depletion and impaired long-term muscle regeneration (Boonsanay et al., 2016). Histone methylation also appears to be involved in stem cell aging. H3K4me3 (histone trimethylation) is a marker of gene activation and increases across HSC identity and self-renewal genes with age, which might explain the observed increase of HSC numbers in mouse aging (Sun et al., 2014). In contrast, H3K4me3 in MuSCs decreases with age, as the repressive histone methylation H3K27me3 increases (Liu et al., 2013).
Metabolites that modulate HMT and HDM activity can also impact on stem cell function and fate decisions. Reductions in α-KG levels in response to the knockdown of phosphoserine aminotransferase 1 (PSAT1) impairs mESC self-renewal and induces differentiation, mediated by lower DNA 5′-hydroxymethylcytosine levels and increased histone methylation (Hwang et al., 2016). In fact, there is evidence to suggest that pluripotency in mESCs might be regulated by the ratio of α-KG to succinate by virtue of its effect on H3K27me3 levels (Carey et al., 2015). Both α-KG and succinate are metabolites generated as a result of TCA metabolism in the mitochondrial matrix (Fig. 2). Similarly, regulation of DNA and histone methylation by SAM is also important in maintaining hESC and iPSC function. Methionine deprivation rapidly decreases intracellular SAM, preventing hESC and iPSC self-renewal and increasing overall differentiation potency, and eventually leading to cell apoptosis (Shiraki et al., 2014). Depletion of intracellular SAM is also seen after threonine deprivation in mouse ESCs, where it leads to slowed stem cell growth and increased differentiation (Fig. 2) (Shyh-Chang et al., 2013).
The mitochondrial unfolded protein response and stem cell aging
Maintenance of mitochondrial function relies on the coordinated expression of mitochondrial- and nuclear-encoded mitochondrial proteins. An altered balance between protein expressions from these genomes can result in proteotoxic stress and the subsequent accumulation of misfolded proteins within the mitochondria. When this occurs, a mitochondrial unfolded protein response, termed the UPRmt, is triggered (reviewed by Jovaisaite et al., 2014). This response likely synchronizes mitochondrial function to cellular homeostasis through the activation of a mitochondrion-to-nucleus retrograde signaling pathway, such as through prohibitin 1 (PHB) and 2 (PHB2) and/or c-Jun N-terminal protein kinase (JNK2; MAPK9) and the protein kinase RNA-activated (PKR; EIF2AK2)-activating transcription factor 4 (ATF4) pathway (Quirós et al., 2017). Overall, the UPRmt increases the expression of mitochondrial chaperones and proteases, and allows for a compensatory restoration of mitochondrial function following cellular stress (Jensen and Jasper, 2014; Jovaisaite et al., 2014; Lin and Haynes, 2016).
Activation of the UPRmt response has been shown to have beneficial effects in multiple species (Jovaisaite and Auwerx, 2015). For instance, disrupting subunits of the mitochondrial ETC, which stimulates the imbalance of the mito- and nuclear-encoded OXPHOS proteins and the induction of UPRmt, leads to overall lifespan extension in yeast, worms, flies and mice (Durieux et al., 2011; Houtkooper et al., 2013; Lee et al., 2003; Liu et al., 2005; Owusu-Ansah et al., 2013). Recently, the UPRmt pathway was shown to regulate stem cell function, and the lifespan-extending effects of UPRmt stimulators such as nicotinamide riboside was linked in part to the improvement of stem cell function in aged mice (Zhang et al., 2016).
Mitochondrial stress, which can be stimulated by treating NSCs with the bile acid tauro-ursodeoxycholic acid (TUDCA), generates a mitochondrial ROS-dependent retrograde signal that modulates NSC proliferation and differentiation (Xavier et al., 2014). However, because of the implication of TUDCA in endoplasmic reticulum (ER) stress (Malo et al., 2010), it was unclear until recently whether a mitochondrial-specific retrograde signal exists in stem cells and, if so, whether it regulates stem cell function. Supplementing mouse chow with nicotinamide riboside has been shown to induce the UPRmt to prevent MuSC, NSC and melanocyte stem cell (McSC) senescence (Zhang et al., 2016). In MuSCs of aged mice, nicotinamide riboside rescues cellular NAD+ depletion and thereby activates a SIRT1-dependent UPRmt signal to improve mitochondrial function and attenuate senescence (Zhang et al., 2016). Similarly, increased levels of NAD+ achieved via the overexpression of NAMPT, the rate-limiting NAD+ salvage enzyme, reduced cell senescence in aged MSCs (Ma et al., 2017). In HSCs, SIRT7 has been shown to mediate a regulatory branch of the UPRmt and to maintain HSC function in aging (Mohrin et al., 2015). Therefore, some level of UPRmt signaling may help maintain stem cell function during aging, possibly by preventing senescence. However, constitutive activation of the UPRmt, with tissue-specific loss of the mitochondrial chaperone protein HSP60 (HSPD1), can also have detrimental effects, and lead to the loss of stemness in ISCs (Berger et al., 2016). Further studies will be needed to address how the relative level of UPRmt signaling affects stem cells and the extent to which this is context dependent (Fig. 3).
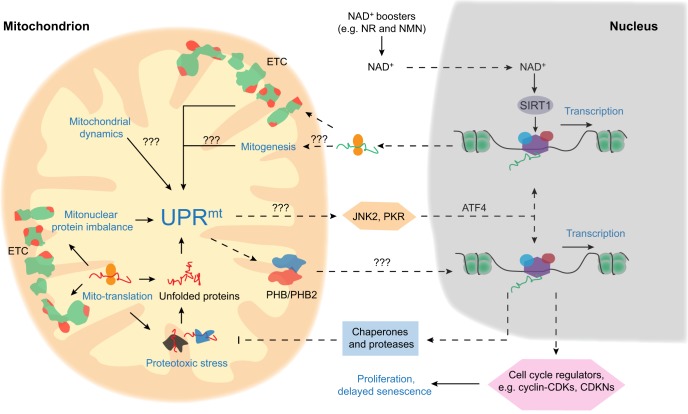
Fig. 3.
UPRmt and stem cell regulation. Mitochondrial electron transport chain proteins are encoded by both the nuclear (green) and mitochondrial (red) genomes. Mitonuclear protein imbalance and proteotoxic stress, mitochondrial dynamics, and mitogenesis can all lead to mitochondrial stress, which activates the mitochondrial unfolded protein response (UPRmt). Retrograde UPRmt signaling, through prohibitin 1 and 2 (PHB and PHB2) and/or c-Jun N-terminal protein kinase (JNK2) and the protein kinase RNA-activated (PKR)-activating transcription factor 4 (ATF4) pathway (Quiros et al., 2017) can specifically regulate nuclear gene expression to generate more chaperones and proteases to alleviate proteotoxic stress. UPRmt-induced gene expression changes also lead to stem cell proliferation and a delayed senescence response. Pathways with the question marks are speculative and require further experimental confirmation. Solid and dashed arrows indicate direct and indirect pathway/links, respectively. NR, nicotinamide riboside; NMN, nicotinamide mononucleotide.
Mitochondrial DNA mutations and stem cell function
The mitochondrial genome encodes 37 genes, including 13 of the OXPHOS subunits, two rRNAs and 22 tRNAs. The accumulation of mitochondrial mutations has been observed in both rodent and human tissues in vivo during aging (Cortopassi and Arnheim, 1990; Pikó et al., 1988), as well as in in vitro cell culture (Coller et al., 2005). Mitochondrial DNA (mtDNA) integrity plays an important role in stem cell fate decisions; NSCs isolated from mice deficient for 8-oxoguanine DNA glycosylase (OGG1; the enzyme essential for mtDNA damage repair) accumulate mtDNA damage and shift their differentiation trajectory toward an astrocytic lineage at the expense of neurogenesis (Wang et al., 2011). mtDNA defects have also been observed in human stem cell aging (McDonald et al., 2008; Taylor et al., 2003) and age-associated mitochondrial DNA mutations have been reported to lead to abnormal cell proliferation and apoptosis in human colonic crypts (Taylor et al., 2003). The frequency of mtDNA defects is also greater in iPSCs generated from aged compared with young people (Kang et al., 2016). However, owing to limitations in the availability of human tissue and a lack of robust stem cell markers, the direct link between mtDNA mutations and stem cell function in vivo remains to be fully elucidated.
Clear evidence for a causal relationship between mtDNA mutations and aging is provided by the nuclear-encoded mtDNA polymerase γ (POLG) knockout mouse model in which spontaneous mutations or replication errors lead to the accumulation of mtDNA mutations, severe OXPHOS deficiency and premature aging (Kujoth et al., 2005; Trifunovic et al., 2004). Similar to the situation in physiological aging, stem cell deficiencies were observed in multiple tissues of the POLG mice. mtDNA mutations inhibit early differentiation of HSCs (Norddahl et al., 2011), resulting in abnormal myeloid lineages in the mutant mice (Ahlqvist et al., 2012; Chen et al., 2009). POLG mice have fewer quiescent NSCs (Ahlqvist et al., 2012), as well as increased levels of apoptosis, decreased cell proliferation and impaired SC-derived intestinal organoid formation (Fox et al., 2012). Moreover, POLG-mutant cells have a markedly impaired capacity for reprogramming during the generation of iPSCs (Hämäläinen et al., 2015). Although these studies link mtDNA mutations to stem cell dysfunction and aging, the stem cell defects in POLG mice do not appear to recapitulate accurately the pace of physiological aging, as the level of mtDNA mutations seen in these models is much higher than that seen during the normal aging process (Norddahl et al., 2011). Therefore, although the levels of mtDNA mutations increase in aged stem cells, it remains unclear whether this increase in mtDNA mutations plays a fundamental role in stem cell aging.
Influence of mitochondrial ROS on stem cell fate
Mitochondrial ROS are primarily generated from electron leakage from the mitochondrial OXPHOS complexes I and III (Box 2), which damages all components of the cell, including DNA, lipids and proteins. Dysfunction of the mitochondrial respiratory chain and inefficient OXPHOS may lead to more electron leakage and further increases in ROS generation, resulting in a detrimental cycle that causes eventual, irreversible damage to cells and contributes to aging (Harman, 1972).
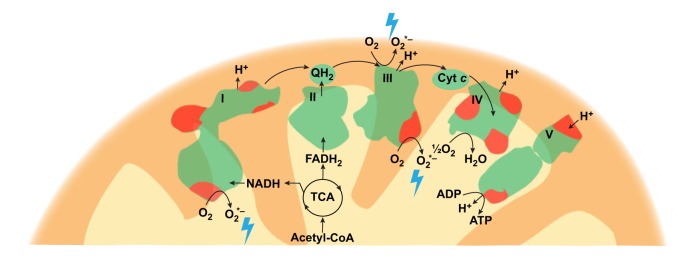
Box 2. Reactive oxygen species generation.
The mitochondrial ETC consists of five respiratory complexes (I-V), which are composed of proteins encoded by both nuclear (green) and mitochondrial (red) genomes. Mitochondrial ROS (blue lightning bolt) are mainly generated from complex I and III of the ETC. Cyt c, cytochrome c; QH2, ubiquinol.
One of the first observations that suggested a role for ROS in cell aging was that the replicative lifespans of in vitro cultured cells were significantly extended by culturing the cells in a low-oxygen environment, which might be due to less ROS generation (Packer and Fuehr, 1977; Parrinello et al., 2003). Mice with mutations in the DNA damage sensor ataxia telangiectasia (ATM) show a defect in HSC function that is associated with elevated ROS production, and treatment with the anti-oxidant N-acetyl-L-cysteine (NAC) restored HSC function in these mice (Ito et al., 2004). A similar increase in ROS production and DNA damage was seen in Bmi1-deficient mice, which were also rescued by NAC treatment (Liu et al., 2009). Furthermore, FOXO proteins, which are key mediators in ROS signaling, may play essential roles in the response to physiological oxidative stress and thereby preserve HSC function by upregulating antioxidant gene expression (Tothova et al., 2007).
In POLG mice, the deficiencies of NSC and HSC function were also rescued by supplementation with NAC (Ahlqvist et al., 2012). NAC also restored the ROS-mediated deficiency of POLG-mutant cells to be reprogrammed into iPSCs (Hämäläinen et al., 2015), again suggesting the effects of ROS signaling in mediating the balance between quiescent and active status of stem cells. Some studies suggest that mtDNA mutations might cause slight alterations in redox status, thus affecting stem cell fate decisions (Ahlqvist et al., 2012). Indeed, physiological ROS appears to be important for mediating NSC fate decisions (reviewed by Bigarella et al., 2014). In NSCs, mitochondrial fusion and fission (further discussed below) triggers ROS signaling to moderate self-renewal and differentiation via NRF2 (also known as nuclear factor, erythroid 2 like 2, NFE2L2)-mediated mito-nuclear retrograde signaling (Khacho et al., 2016).
Similar to the role of the mtDNA mutations as previously discussed, there is much controversy regarding the effect of mitochondrial ROS on stem cell dysfunction and aging due to a number of observations. First, long-lived species do not always demonstrate lower levels of ROS and the accompanying oxidative damage (Andziak et al., 2006; Chen et al., 2007). For example, a rise in ROS increases the lifespan of worms (Yang and Hekimi, 2010; Zarse et al., 2012). Second, it is known that the widely used anti-oxidative reagent NAC affects physiological processes beyond ROS scavenging. Therefore, NAC treatment could benefit stem cell phenotypes in mtDNA mutant mice by affecting mechanisms beyond redox status. Finally, there are an increasing number of examples in which ROS appears to play a positive and necessary signaling role in stem cell biology (Bigarella et al., 2014; Hameed et al., 2015; Maryanovich et al., 2015). Altogether, the effect and detailed mechanism of ROS in stem cell function still needs further investigation.
Role of mitophagy and mitochondrial dynamics on stem cell fate
Mitochondria have been shown to be asymmetrically segregated during stem cell division (Katajisto et al., 2015). This asymmetric allocation of mitochondria appears to be a unique feature of adult stem cells that enables the exclusion of older mitochondria. Beyond cell division, mitochondria in quiescent stem cells are subject to mitochondrial dynamics, involving fusion/fission and mitochondria-specific autophagy, termed mitophagy. Recently, mitochondrial dynamics has been linked to stem cell function and aging.
In general, compared with post-mitotic cells, mitochondrial networks are fragmented in stem cells of various types, including ESCs (Folmes et al., 2011; Zhou et al., 2012). This normally suggests a high level of mitochondrial fission and a low level of mitochondrial fusion events. Indeed, DRP1 (dynamin-related protein 1; also known as DNM1L)-dependent mitochondrial fission has been shown to be necessary for cell reprogramming (Prieto et al., 2016). However, two studies also illustrate the potential importance of mitochondrial fusion proteins in maintaining stem cell function. In HSCs, mitofusin 2 (MFN2), a protein involved in promoting mitochondrial fusion and mitochondria-ER tethering, increases the buffering capacity for intracellular Ca2+ and thereby inhibits the translocation and transcriptional activity of the nuclear factor of activated T cells (NFAT) (Luchsinger et al., 2016). The inhibition of NFAT is essential for the maintenance of lymphoid potential of HSCs. In NSCs, mitochondrial dynamics (fusion and fission) regulates NSC identity, proliferation, and fate decisions by orchestrating nuclear transcriptional programs. NSC-specific deletion of either OPA1 or MFN1/2 induces ROS and NRF2-dependent retrograde signaling, which suppresses NSC self-renewal and promotes differentiation, leading to age-dependent NSC depletion, defects in neurogenesis, and cognitive impairment (Khacho et al., 2016) (Fig. 1).
When mitochondrial damage or unfolded protein accumulates in the mitochondria, the UPRmt activates mitochondrial chaperones and proteases to help protein folding and cleavage. Mitochondrial fusion/fission might then be activated to dilute or separate damaged proteins for further degradation. Simultaneously, the clearance of mitochondria through mitophagy may help to dispose of damaged proteins. In fact, evidence for the importance of autophagy/mitophagy for the maintenance of stem cell homeostasis is accumulating. For instance, autophagy levels are higher in HSCs and skin stem cells compared with the surrounding cells (Salemi et al., 2012). In the dentate gyrus of the mouse brain, areas enriched for NSCs appear to have higher rate of autophagy (Sun et al., 2015). Moreover, several recent studies suggest a causative link between mitophagy and the prevention of stem cell aging (García-Prat et al., 2016; Ho et al., 2017; Vazquez-Martin et al., 2016). Failure of autophagy in aged MuSCs, or the genetic impairment of autophagy in young MuSCs, causes entry into senescence by loss of proteostasis, increased mitochondrial dysfunction and oxidative stress, resulting in a decline in the function and number of MuSCs (García-Prat et al., 2016). However, MuSC senescence can be reversed through the re-establishment of autophagy (García-Prat et al., 2016). Loss of autophagy in HSCs causes the accumulation of mitochondria and an activated metabolic state that drives accelerated myeloid differentiation and impairs HSC self-renewal activity and regenerative potential (Ho et al., 2017; Mortensen et al., 2011). In agreement with this, lowering the mitochondrial activity by uncoupling the ETC stimulates autophagy and drives self-renewal of HSCs (Vannini et al., 2016). Furthermore, autophagy maintains HSC function in aged mice (Ho et al., 2017), and is also likely to be involved in the metabolic switch during iPSC reprogramming (Ma et al., 2015).
Interestingly, SIRT1, FOXO and mTORC1 signaling have all been implicated in the regulation of autophagy during stem cell activation, reprogramming and survival under stress (Tang and Rando, 2014; Warr et al., 2013; Wu et al., 2015). Whether these proteins and signaling pathways are also linked to mitophagy, and play a similar role to autophagy, in stem cells still needs confirmation, yet mitophagy and mitochondrial metabolic pathways might be tightly conjugated and are likely to regulate stem cell function and aging in a coordinated manner.
Conclusions and perspective
In many early studies, the evidence for an association between mitochondria and stem cell function and especially aging was mostly correlative, such as the increase of mtDNA mutations, ROS levels and the decline in stem cell function with aging. Increasingly, however, causative connections are being established. Mitochondria regulate stem cell function and fate decision through different strategies: mitochondrial metabolism generates metabolites that serve as DNA and protein modification substrates, such as NAD+, acetyl-CoA, citrate, α-KG and fatty acids. Through epigenetic modulation of gene expression, the availability of these key mitochondrial metabolites directly regulates stem cell fate decisions and aging. Moreover, UPRmt, mitochondrial dynamics and stress can generate retrograde signals from the mitochondria to control nuclear gene expression. With stem cell mitochondria-nuclear cross-talk, mitochondria not only exert a strict quality control but may also impact gene expression patterns in order to alter cell fate decisions and physiological functions during aging.
Despite much progress, this field of research is still in its infancy, and there are many research questions that should now be answered. Importantly, we know that metabolism and mitochondrial function are different amongst stem cell types and can change with stem cell dynamics. However, it is still not clear to what extent mitochondria determine stem cell properties and fate decisions. Also, the mechanisms that underpin the heterogeneity of metabolic and mitochondrial properties in different stem cell types remains undefined. Another interesting topic is stem cell aging, as it represents not only a fundamental scientific issue but is also directly linked to several age-related diseases. We know that mitochondrial function declines in several types of adult stem cells, but it is still not known whether all types of stem cells, or indeed each individual stem cell, have similar mitochondrial and metabolic changes upon aging. Also, it is important to unveil the key signaling pathways that control mitochondrial dysfunction and mediate downstream cellular senescence. One of the interesting related topics to explore includes the identification of key regulators of UPRmt, mitophagy and mitochondrial dynamics in different stem cells.
Further genetic and molecular studies are required to identify the key differences in how mitochondria influence stem cell function across a range of different tissue contexts. The application of a set of new technologies to stem cell research will help to address this. For instance, the simultaneous analysis of the genome, transcriptome, proteome and metabolome to connect multiple layers of information has been successfully applied to studies of genetic reference populations (Williams et al., 2016; Wu et al., 2014). Such multilayered studies would also be of value to define the regulatory networks of stem cell function and fate decision. Another powerful approach to investigating cell diversity among stem cells would be to make use of emerging single-cell technologies, such as single-cell RNA sequencing. This method may be specifically relevant because of its high sensitivity and the limited availability of stem cells. Finally, the newer precision gene editing methods will also aid exploration of the detailed mechanisms of stem cell regulation.
Modulation of mitochondrial function or improving mitochondrial quality control might be effective strategies to control stem cell amplification, to stimulate stem cell activation for regenerative medicine and for combating stem cell dysfunction during aging (Box 3). Application of such ‘mitochondrial medicine’ (Andreux et al., 2013; Sorrentino et al., 2017) may not only provide a means of slowing down aging at the stem cell level but also at the organismal level by extending lifespan (Rera et al., 2011; Zhang et al., 2016). This is an exciting prospect, and one that puts the interplay between mitochondria and stem cells at the center stage of the aging field.
Box 3. Mitochondrial therapeutics in stem cells.
Targeting mitochondrial function represents a possible treatment strategy for some inherited genetic diseases, metabolic syndromes and neurodegenerative diseases. There are several ways to improve mitochondrial function, for example by targeting sirtuins, AMP-activated protein kinase (AMPK), mTOR, NAD+ metabolism, nuclear receptors (such as PPARs and estrogen-related receptors), transcriptional factors/co-factors (such as PPARGC1α, FOXO, NCORs), along with activators of UPRmt and mitochondrial fusion/fission or mitophagy (reviewed by Sorrentino et al., 2017). Among these, however, there are very few descriptions of how these treatment strategies affect stem cells specifically. It was recently shown that PPARδ agonists improve hematopoietic stem cell function by improving fatty acid oxidation (Ito et al., 2012). This finding highlights the therapeutic potential of PPARδ agonists in improving the efficacy of bone marrow transplantation and the treatment of hematological malignancies. NAD+ boosters, such as nicotinamide riboside and nicotinamide mononucleotide, have been shown to improve mitochondrial function in muscle and neural stem cells in aging (Stein and Imai, 2014; Zhang et al., 2016), and thus may be considered for the prevention of muscle weakness and neural degeneration in aging. Improving mitochondrial function by activating autophagy is another possibility for ameliorating stem cell function. Treatment with the autophagy activator bafilomycin has been shown to improve aged muscle stem cell function (Garcia-Prat et al., 2016). An important limitation of these findings is that they are all based on rodent studies: whether such treatments have a similar effect on human cells remains to be seen. Although mitochondrial therapeutics in stem cells is a relatively recent avenue for exploration, progress so far has been good and the field has gained considerable traction.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
H.Z. is the recipient of a fellowship from the CARIGEST SA and his research is supported by Sun Yat-sen University. K.J.M. is supported by the Canadian Institutes of Health Research and Kidney Foundation of Canada. The research of J.A. is supported by the École Polytechnique Fédérale de Lausanne, the National Institutes of Health (R01AG043930), SystemsX (SySX.ch 2013/153), the Velux Stiftung (Switzerland), and the Swiss National Science Foundation (Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung; 31003A-140780). Deposited in PMC for release after 12 months.
References
- Agathocleous M., Meacham C. E., Burgess R. J., Piskounova E., Zhao Z., Crane G. M., Cowin B. L., Bruner E., Murphy M. M., Chen W. et al. (2017). Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. 549, 476-481. 10.1038/nature23876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlqvist K. J., Hämäläinen R. H., Yatsuga S., Uutela M., Terzioglu M., Götz A., Forsstrom S., Salven P., Angers-Loustau A., Kopra O. H. et al. (2012). Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. 15, 100-109. 10.1016/j.cmet.2011.11.012 [DOI] [PubMed] [Google Scholar]
- Aloia L., Di Stefano B. and Di Croce L. (2013). Polycomb complexes in stem cells and embryonic development. 140, 2525-2534. 10.1242/dev.091553 [DOI] [PubMed] [Google Scholar]
- Andreux P. A., Houtkooper R. H. and Auwerx J. (2013). Pharmacological approaches to restore mitochondrial function. 12, 465-483. 10.1038/nrd4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andziak B., O'Connor T. P., Qi W., DeWaal E. M., Pierce A., Chaudhuri A. R., Van Remmen H. and Buffenstein R. (2006). High oxidative damage levels in the longest-living rodent, the naked mole-rat. 5, 463-471. 10.1111/j.1474-9726.2006.00237.x [DOI] [PubMed] [Google Scholar]
- Ansó E., Weinberg S. E., Diebold L. P., Thompson B. J., Malinge S., Schumacker P. T., Liu X., Zhang Y., Shao Z., Steadman M. et al. (2017). The mitochondrial respiratory chain is essential for haematopoietic stem cell function. 19, 614-625. 10.1038/ncb3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengal E., Perdiguero E., Serrano A. L. and Muñoz-Cánoves P. (2017). Rejuvenating stem cells to restore muscle regeneration in aging. 6, 76 10.12688/f1000research.9846.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger E., Rath E., Yuan D., Waldschmitt N., Khaloian S., Allgäuer M., Staszewski O., Lobner E. M., Schöttl T., Giesbertz P. et al. (2016). Mitochondrial function controls intestinal epithelial stemness and proliferation. 7, 13171 10.1038/ncomms13171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyaz S., Mana M. D., Roper J., Kedrin D., Saadatpour A., Hong S.-J., Bauer-Rowe K. E., Xifaras M. E., Akkad A., Arias E. et al. (2016). High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. 531, 53-58. 10.1038/nature17173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigarella C. L., Liang R. and Ghaffari S. (2014). Stem cells and the impact of ROS signaling. 141, 4206-4218. 10.1242/dev.107086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Hochmuth C. E. and Jasper H. (2008). JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. 3, 442-455. 10.1016/j.stem.2008.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonsanay V., Zhang T., Georgieva A., Kostin S., Qi H., Yuan X., Zhou Y. and Braun T. (2016). Regulation of skeletal muscle stem cell quiescence by Suv4-20h1-dependent facultative heterochromatin formation. 18, 229-242. 10.1016/j.stem.2015.11.002 [DOI] [PubMed] [Google Scholar]
- Booth L. N. and Brunet A. (2016). The aging epigenome. 62, 728-744. 10.1016/j.molcel.2016.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournat J. C. and Brown C. W. (2010). Mitochondrial dysfunction in obesity. 17, 446-452. 10.1097/MED.0b013e32833c3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha A. L., Ramanathan A., Huang S., Ingber D. E. and Schreiber S. L. (2010). Carbon metabolism-mediated myogenic differentiation. 6, 202-204. 10.1038/nchembio.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K., Xie S., Qiu X., Mohrin M., Shin J., Liu Y., Zhang D., Scadden D. T. and Chen D. (2013). SIRT3 reverses aging-associated degeneration. 3, 319-327. 10.1016/j.celrep.2013.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M. D., O'Sullivan D., Klein Geltink R. I., Curtis J. D., Chang C.-H., Sanin D. E., Qiu J., Kretz O., Braas D., van der Windt G. J. et al. (2016). Mitochondrial dynamics controls T cell fate through metabolic programming. 166, 63-76. 10.1016/j.cell.2016.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C., Menzies K. J. and Auwerx J. (2015). NAD(+) metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. 22, 31-53. 10.1016/j.cmet.2015.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbognin E., Betto R. M., Soriano M. E., Smith A. G. and Martello G. (2016). Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. 35, 618-634. 10.15252/embj.201592629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B. W., Finley L. W. S., Cross J. R., Allis C. D. and Thompson C. B. (2015). Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. 518, 413-416. 10.1038/nature13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerletti M., Jang Y. C., Finley L. W. S., Haigis M. C. and Wagers A. J. (2012). Short-term calorie restriction enhances skeletal muscle stem cell function. 10, 515-519. 10.1016/j.stem.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen G. A., Sun D., Mayle A., Jeong M., Luo M., Rodriguez B., Mallaney C., Celik H., Yang L., Xia Z. et al. (2014). Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. 15, 350-364. 10.1016/j.stem.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel N. S., Jasper H., Ho T. T. and Passegué E. (2016). Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. 18, 823-832. 10.1038/ncb3385 [DOI] [PubMed] [Google Scholar]
- Chen J., Astle C. M. and Harrison D. E. (2003). Hematopoietic senescence is postponed and hematopoietic stem cell function is enhanced by dietary restriction. 31, 1097-1103. 10.1016/S0301-472X(03)00238-8 [DOI] [PubMed] [Google Scholar]
- Chen J.-H., Hales C. N. and Ozanne S. E. (2007). DNA damage, cellular senescence and organismal ageing: causal or correlative? 35, 7417-7428. 10.1093/nar/gkm681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-T., Shih Y.-R. V., Kuo T. K., Lee O. K. and Wei Y.-H. (2008). Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. 26, 960-968. 10.1634/stemcells.2007-0509 [DOI] [PubMed] [Google Scholar]
- Chen M. L., Logan T. D., Hochberg M. L., Shelat S. G., Yu X., Wilding G. E., Tan W., Kujoth G. C., Prolla T. A., Selak M. A. et al. (2009). Erythroid dysplasia, megaloblastic anemia, and impaired lymphopoiesis arising from mitochondrial dysfunction. 114, 4045-4053. 10.1182/blood-2008-08-169474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-L., Uthaya Kumar D. B., Punj V., Xu J., Sher L., Tahara S. M., Hess S. and Machida K. (2016). NANOG metabolically reprograms tumor-initiating stem-like cells through tumorigenic changes in oxidative phosphorylation and fatty acid metabolism. 23, 206-219. 10.1016/j.cmet.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.-W., Adams G. B., Perin L., Wei M., Zhou X., Lam B. S., Da Sacco S., Mirisola M., Quinn D. I., Dorff T. B. et al. (2014). Prolonged fasting reduces IGF-1/PKA to promote hematopoietic-stem-cell-based regeneration and reverse immunosuppression. 14, 810-823. 10.1016/j.stem.2014.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N.-H., Kim J.-G., Yang D.-J., Kim Y.-S. and Yoo M.-A. (2008). Age-related changes in Drosophila midgut are associated with PVF2, a PDGF/VEGF-like growth factor. 7, 318-334. 10.1111/j.1474-9726.2008.00380.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Dzeja P. P., Faustino R. S., Perez-Terzic C., Behfar A. and Terzic A. (2007). Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. 4 Suppl. 1, S60-S67. 10.1038/ncpcardio0766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L., Dolgalev I., Wang Y., Yoshimi A., Martin G. H., Wang J., Ng V., Xia B., Witkowski M. T., Mitchell-Flack M. et al. (2017). Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. 170, 1079-1095 e1020. 10.1016/j.cell.2017.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller H. A., Khrapko K., Herrero-Jimenez P., Vatland J. A., Li-Sucholeiki X.-C. and Thilly W. G. (2005). Clustering of mutant mitochondrial DNA copies suggests stem cells are common in human bronchial epithelium. 578, 256-271. 10.1016/j.mrfmmm.2005.05.014 [DOI] [PubMed] [Google Scholar]
- Cortopassi G. A. and Arnheim N. (1990). Detection of a specific mitochondrial DNA deletion in tissues of older humans. 18, 6927-6933. 10.1093/nar/18.23.6927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ippolito G., Diabira S., Howard G. A., Roos B. A. and Schiller P. C. (2006). Low oxygen tension inhibits osteogenic differentiation and enhances stemness of human MIAMI cells. 39, 513-522. 10.1016/j.bone.2006.02.061 [DOI] [PubMed] [Google Scholar]
- Durieux J., Wolff S. and Dillin A. (2011). The cell-non-autonomous nature of electron transport chain-mediated longevity. 144, 79-91. 10.1016/j.cell.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezashi T., Das P. and Roberts R. M. (2005). Low O2 tensions and the prevention of differentiation of hES cells. 102, 4783-4788. 10.1073/pnas.0501283102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian M. C., Dörr K., Niebel A., Daria D., Schrezenmeier H., Rojewski M., Filippi M.-D., Hasenberg A., Gunzer M., Scharffetter-Kochanek K. et al. (2012). Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. 10, 520-530. 10.1016/j.stem.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes C. D. L., Nelson T. J., Martinez-Fernandez A., Arrell D. K., Lindor J. Z., Dzeja P. P., Ikeda Y., Perez-Terzic C. and Terzic A. (2011). Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. 14, 264-271. 10.1016/j.cmet.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes C. D. L., Nelson T. J., Dzeja P. P. and Terzic A. (2012). Energy metabolism plasticity enables stemness programs. 1254, 82-89. 10.1111/j.1749-6632.2012.06487.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox R. G., Magness S., Kujoth G. C., Prolla T. A. and Maeda N. (2012). Mitochondrial DNA polymerase editing mutation, PolgD257A, disturbs stem-progenitor cell cycling in the small intestine and restricts excess fat absorption. 302, G914-G924. 10.1152/ajpgi.00402.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Prat L., Martínez-Vicente M., Perdiguero E., Ortet L., Rodríguez-Ubreva J., Rebollo E., Ruiz-Bonilla V., Gutarra S., Ballestar E., Serrano A. L. et al. (2016). Autophagy maintains stemness by preventing senescence. 529, 37-42. 10.1038/nature16187 [DOI] [PubMed] [Google Scholar]
- Hämäläinen R. H., Ahlqvist K. J., Ellonen P., Lepistö M., Logan A., Otonkoski T., Murphy M. P. and Suomalainen A. (2015). mtDNA mutagenesis disrupts pluripotent stem cell function by altering redox signaling. 11, 1614-1624. 10.1016/j.celrep.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed L. S., Berg D. A., Belnoue L., Jensen L. D., Cao Y. and Simon A. (2015). Environmental changes in oxygen tension reveal ROS-dependent neurogenesis and regeneration in the adult newt brain. 4, e08422 10.7554/eLife.08422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. (1972). The biologic clock: the mitochondria? 20, 145-147. 10.1111/j.1532-5415.1972.tb00787.x [DOI] [PubMed] [Google Scholar]
- Hirschey M. D. and Zhao Y. (2015). Metabolic regulation by lysine malonylation, succinylation, and glutarylation. 14, 2308-2315. 10.1074/mcp.R114.046664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T. T., Warr M. R., Adelman E. R., Lansinger O. M., Flach J., Verovskaya E. V., Figueroa M. E. and Passegué E. (2017). Autophagy maintains the metabolism and function of young and old stem cells. 543, 205-210. 10.1038/nature21388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R. H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R. W. and Auwerx J. (2013). Mitonuclear protein imbalance as a conserved longevity mechanism. 497, 451-457. 10.1038/nature12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Wang H., Lee I. H., Modi S., Wang X., Du J. and Mitch W. E. (2010). PTEN inhibition improves muscle regeneration in mice fed a high-fat diet. 59, 1312-1320. 10.2337/db09-1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I.-Y., Kwak S., Lee S., Kim H., Lee S. E., Kim J.-H., Kim Y. A., Jeon Y. K., Chung D. H., Jin X. et al. (2016). Psat1-dependent fluctuations in alpha-ketoglutarate affect the timing of ESC differentiation. 24, 494-501. 10.1016/j.cmet.2016.06.014 [DOI] [PubMed] [Google Scholar]
- Igarashi M. and Guarente L. (2016). mTORC1 and SIRT1 cooperate to foster expansion of gut adult stem cells during calorie restriction. 166, 436-450. 10.1016/j.cell.2016.05.044 [DOI] [PubMed] [Google Scholar]
- Inoue S.-I., Noda S., Kashima K., Nakada K., Hayashi J.-I. and Miyoshi H. (2010). Mitochondrial respiration defects modulate differentiation but not proliferation of hematopoietic stem and progenitor cells. 584, 3402-3409. 10.1016/j.febslet.2010.06.036 [DOI] [PubMed] [Google Scholar]
- Ito K., Hirao A., Arai F., Matsuoka S., Takubo K., Hamaguchi I., Nomiyama K., Hosokawa K., Sakurada K., Nakagata N. et al. (2004). Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. 431, 997-1002. 10.1038/nature02989 [DOI] [PubMed] [Google Scholar]
- Ito K., Carracedo A., Weiss D., Arai F., Ala U., Avigan D. E., Schafer Z. T., Evans R. M., Suda T., Lee C.-H. et al. (2012). A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. 18, 1350-1358. 10.1038/nm.2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. B. and Jasper H. (2014). Mitochondrial proteostasis in the control of aging and longevity. 20, 214-225. 10.1016/j.cmet.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C.-H., Lee H.-J., Cha J.-H., Kim J. H., Kim K. R., Kim J.-H., Yoon D.-K. and Kim K.-W. (2007). Hypoxia-inducible factor-1 alpha inhibits self-renewal of mouse embryonic stem cells in Vitro via negative regulation of the leukemia inhibitory factor-STAT3 pathway. 282, 13672-13679. 10.1074/jbc.M700534200 [DOI] [PubMed] [Google Scholar]
- Jin G., Xu C., Zhang X., Long J., Rezaeian A. H., Liu C., Furth M. E., Kridel S., Pasche B., Bian X.-W. et al. (2018). Atad3a suppresses Pink1-dependent mitophagy to maintain homeostasis of hematopoietic progenitor cells. 19, 29-40. 10.1038/s41590-017-0002-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovaisaite V. and Auwerx J. (2015). The mitochondrial unfolded protein response-synchronizing genomes. 33, 74-81. 10.1016/j.ceb.2014.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovaisaite V., Mouchiroud L. and Auwerx J. (2014). The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. 217, 137-143. 10.1242/jeb.090738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang E., Wang X., Tippner-Hedges R., Ma H., Folmes C. D., Gutierrez N. M., Lee Y., Van Dyken C., Ahmed R., Li Y. et al. (2016). Age-related accumulation of somatic mitochondrial DNA mutations in adult-derived human iPSCs. 18, 625-636. 10.1016/j.stem.2016.02.005 [DOI] [PubMed] [Google Scholar]
- Katajisto P., Dohla J., Chaffer C. L., Pentinmikko N., Marjanovic N., Iqbal S., Zoncu R., Chen W., Weinberg R. A. and Sabatini D. M. (2015). Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. 348, 340-343. 10.1126/science.1260384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khacho M., Clark A., Svoboda D. S., Azzi J., MacLaurin J. G., Meghaizel C., Sesaki H., Lagace D. C., Germain M., Harper M.-E. et al. (2016). Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. 19, 232-247. 10.1016/j.stem.2016.04.015 [DOI] [PubMed] [Google Scholar]
- Khacho M., Clark A., Svoboda D. S., MacLaurin J. G., Lagace D. C., Park D. S. and Slack R. S. (2017). Mitochondrial dysfunction underlies cognitive defects as a result of neural stem cell depletion and impaired neurogenesis. 26, 3327-3341. 10.1093/hmg/ddx217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y. S., Kawamura T., Wei Z., Sogo T., Jacinto S., Shigeno A., Kushige H., Yoshihara E., Liddle C., Ecker J. R. et al. (2015). ERRs mediate a metabolic switch required for somatic cell reprogramming to pluripotency. 16, 547-555. 10.1016/j.stem.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch M., Braun S. M., Zurkirchen L., von Schoultz C., Zamboni N., Araúzo-Bravo M. J., Kovacs W. J., Karalay O., Suter U., Machado R. A. et al. (2013). Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. 493, 226-230. 10.1038/nature11689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth G. C., Hiona A., Pugh T. D., Someya S., Panzer K., Wohlgemuth S. E., Hofer T., Seo A. Y., Sullivan R., Jobling W. A. et al. (2005). Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. 309, 481-484. 10.1126/science.1112125 [DOI] [PubMed] [Google Scholar]
- Lange C., Turrero Garcia M., Decimo I., Bifari F., Eelen G., Quaegebeur A., Boon R., Zhao H., Boeckx B., Chang J. et al. (2016). Relief of hypoxia by angiogenesis promotes neural stem cell differentiation by targeting glycolysis. 35, 924-941. 10.15252/embj.201592372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. S., Lee R. Y. N., Fraser A. G., Kamath R. S., Ahringer J. and Ruvkun G. (2003). A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. 33, 40-48. 10.1038/ng1056 [DOI] [PubMed] [Google Scholar]
- Lin Y.-F. and Haynes C. M. (2016). Metabolism and the UPR(mt). 61, 677-682. 10.1016/j.molcel.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao L., Chen J., Song S., Lee I. H., Quijano C., Liu H., Keyvanfar K., Chen H., Cao L.-Y. et al. (2009). Bmi1 regulates mitochondrial function and the DNA damage response pathway. 459, 387-392. 10.1038/nature08040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Jiang N., Hughes B., Bigras E., Shoubridge E. and Hekimi S. (2005). Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. 19, 2424-2434. 10.1101/gad.1352905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Cheung T. H., Charville G. W., Hurgo B. M. C., Leavitt T., Shih J., Brunet A. and Rando T. A. (2013). Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. 4, 189-204. 10.1016/j.celrep.2013.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchsinger L. L., de Almeida M. J., Corrigan D. J., Mumau M. and Snoeck H.-W. (2016). Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. 529, 528-531. 10.1038/nature16500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Li J., Xu Y., Yu C., Xu T., Wang H., Liu K., Cao N., Nie B.-M., Zhu S.-Y. et al. (2015). Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. 17, 1379-1387. 10.1038/ncb3256 [DOI] [PubMed] [Google Scholar]
- Ma C., Pi C., Yang Y., Lin L., Shi Y., Li Y., Li Y. and He X. (2017). Nampt expression decreases age-related senescence in rat bone marrow mesenchymal stem cells by targeting sirt1. 12, e0170930 10.1371/journal.pone.0170930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W., McLeod C. J., Wang L. and Jones D. L. (2010). Dietary restriction enhances germline stem cell maintenance. 9, 916-918. 10.1111/j.1474-9726.2010.00602.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo A., Krüger B., Seyhun E., Schäfer C., Hoffmann R. T., Göke B. and Kubisch C. H. (2010). Tauroursodeoxycholic acid reduces endoplasmic reticulum stress, trypsin activation, and acinar cell apoptosis while increasing secretion in rat pancreatic acini. 299, G877-G886. 10.1152/ajpgi.00423.2009 [DOI] [PubMed] [Google Scholar]
- Martin K., Kirkwood T. B. L. and Potten C. S. (1998). Age changes in stem cells of murine small intestinal crypts. 241, 316-323. 10.1006/excr.1998.4001 [DOI] [PubMed] [Google Scholar]
- Maryanovich M., Zaltsman Y., Ruggiero A., Goldman A., Shachnai L., Zaidman S. L., Porat Z., Golan K., Lapidot T. and Gross A. (2015). An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. 6, 7901 10.1038/ncomms8901 [DOI] [PubMed] [Google Scholar]
- Maslov A. Y., Barone T. A., Plunkett R. J. and Pruitt S. C. (2004). Neural stem cell detection, characterization, and age-related changes in the subventricular zone of mice. 24, 1726-1733. 10.1523/JNEUROSCI.4608-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J. and Ruohola-Baker H. (2017). Metabolic remodeling during the loss and acquisition of pluripotency. 144, 541-551. 10.1242/dev.128389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilainen O., Quirós P. M. and Auwerx J. (2017). Mitochondria and epigenetics - crosstalk in homeostasis and stress. 27, 453-463. 10.1016/j.tcb.2017.02.004 [DOI] [PubMed] [Google Scholar]
- Mayle A., Yang L., Rodriguez B., Zhou T., Chang E., Curry C. V., Challen G. A., Li W., Wheeler D., Rebel V. I. et al. (2015). Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. 125, 629-638. 10.1182/blood-2014-08-594648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald S. A. C., Greaves L. C., Gutierrez-Gonzalez L., Rodriguez-Justo M., Deheragoda M., Leedham S. J., Taylor R. W., Lee C. Y., Preston S. L., Lovell M. et al. (2008). Mechanisms of field cancerization in the human stomach: the expansion and spread of mutated gastric stem cells. 134, 500-510. 10.1053/j.gastro.2007.11.035 [DOI] [PubMed] [Google Scholar]
- Mentch S. J., Mehrmohamadi M., Huang L., Liu X., Gupta D., Mattocks D., Gómez Padilla P., Ables G., Bamman M. M., Thalacker-Mercer A. E. et al. (2015). Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. 22, 861-873. 10.1016/j.cmet.2015.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies K. J., Zhang H., Katsyuba E. and Auwerx J. (2016). Protein acetylation in metabolism - metabolites and cofactors. 12, 43-60. 10.1038/nrendo.2015.181 [DOI] [PubMed] [Google Scholar]
- Miharada K., Karlsson G., Rehn M., Rörby E., Siva K., Cammenga J. and Karlsson S. (2011). Cripto regulates hematopoietic stem cells as a hypoxic-niche-related factor through cell surface receptor GRP78. 9, 330-344. 10.1016/j.stem.2011.07.016 [DOI] [PubMed] [Google Scholar]
- Mohrin M., Shin J., Liu Y., Brown K., Luo H., Xi Y., Haynes C. M. and Chen D. (2015). Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. 347, 1374-1377. 10.1126/science.aaa2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S. J., Wandycz A. M., Akashi K., Globerson A. and Weissman I. L. (1996). The aging of hematopoietic stem cells. 2, 1011-1016. 10.1038/nm0996-1011 [DOI] [PubMed] [Google Scholar]
- Morrison S. J., Csete M., Groves A. K., Melega W., Wold B. and Anderson D. J. (2000). Culture in reduced levels of oxygen promotes clonogenic sympathoadrenal differentiation by isolated neural crest stem cells. 20, 7370-7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M., Watson A. S. and Simon A. K. (2011). Lack of autophagy in the hematopoietic system leads to loss of hematopoietic stem cell function and dysregulated myeloid proliferation. 7, 1069-1070. 10.4161/auto.7.9.15886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A., Rouleau M., Kitsberg D., Cohen M., Levy G., Barasch D., Nemirovski A., Shen-Orr S., Laevsky I., Amit M. et al. (2015). Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. 21, 392-402. 10.1016/j.cmet.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Nguyen M.-H., Cheng M. and Koh T. J. (2011). Impaired muscle regeneration in ob/ob and db/db mice. 11, 1525-1535. 10.1100/tsw.2011.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura E. K., Granter S. R. and Fisher D. E. (2005). Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. 307, 720-724. 10.1126/science.1099593 [DOI] [PubMed] [Google Scholar]
- Norddahl G. L., Pronk C. J., Wahlestedt M., Sten G., Nygren J. M., Ugale A., Sigvardsson M. and Bryder D. (2011). Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. 8, 499-510. 10.1016/j.stem.2011.03.009 [DOI] [PubMed] [Google Scholar]
- Oh J., Lee Y. D. and Wagers A. J. (2014). Stem cell aging: mechanisms, regulators and therapeutic opportunities. 20, 870-880. 10.1038/nm.3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E., Song W. and Perrimon N. (2013). Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. 155, 699-712. 10.1016/j.cell.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer L. and Fuehr K. (1977). Low oxygen concentration extends the lifespan of cultured human diploid cells. 267, 423-425. 10.1038/267423a0 [DOI] [PubMed] [Google Scholar]
- Parrinello S., Samper E., Krtolica A., Goldstein J., Melov S. and Campisi J. (2003). Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. 5, 741-747. 10.1038/ncb1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S., Feller C., Forne I., Schiller E., Sévin D. C., Schauer T., Regnard C., Straub T., Prestel M., Klima C. et al. (2016). Life span extension by targeting a link between metabolism and histone acetylation in Drosophila. 17, 455-469. 10.15252/embr.201541132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli C., Ria R., Scrima R., Cela O., D'Aprile A., Boffoli D., Falzetti F., Tabilio A. and Capitanio N. (2005). Characterization of mitochondrial and extra-mitochondrial oxygen consuming reactions in human hematopoietic stem cells. Novel evidence of the occurrence of NAD(P)H oxidase activity. 280, 26467-26476. 10.1074/jbc.M500047200 [DOI] [PubMed] [Google Scholar]
- Pikó L., Hougham A. J. and Bulpitt K. J. (1988). Studies of sequence heterogeneity of mitochondrial DNA from rat and mouse tissues: evidence for an increased frequency of deletions/additions with aging. 43, 279-293. 10.1016/0047-6374(88)90037-1 [DOI] [PubMed] [Google Scholar]
- Portela A. and Esteller M. (2010). Epigenetic modifications and human disease. 28, 1057-1068. 10.1038/nbt.1685 [DOI] [PubMed] [Google Scholar]
- Prieto J., León M., Ponsoda X., Sendra R., Bort R., Ferrer-Lorente R., Raya A., López-Garcia C. and Torres J. (2016). Early ERK1/2 activation promotes DRP1-dependent mitochondrial fission necessary for cell reprogramming. 7, 11124 10.1038/ncomms11124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigione A., Fauler B., Lurz R., Lehrach H. and Adjaye J. (2010). The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. 28, 721-733. 10.1002/stem.404 [DOI] [PubMed] [Google Scholar]
- Prigione A., Rohwer N., Hoffmann S., Mlody B., Drews K., Bukowiecki R., Blümlein K., Wanker E. E., Ralser M., Cramer T. et al. (2014). HIF1alpha modulates cell fate reprogramming through early glycolytic shift and upregulation of PDK1-3 and PKM2. 32, 364-376. 10.1002/stem.1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós P. M., Prado M. A., Zamboni N., D'Amico D., Williams R. W., Finley D., Gygi S. P. and Auwerx J. (2017). Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. 216, 2027-2045. 10.1083/jcb.201702058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski V. A., Mancini E. and Brunet A. (2012). Energy metabolism and energy-sensing pathways in mammalian embryonic and adult stem cell fate. 125, 5597-5608. 10.1242/jcs.114827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M., Bahadorani S., Cho J., Koehler C. L., Ulgherait M., Hur J. H., Ansari W. S., Lo T. Jr, Jones D. L. and Walker D. W. (2011). Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. 14, 623-634. 10.1016/j.cmet.2011.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Colman M. J., Schewe M., Meerlo M., Stigter E., Gerrits J., Pras-Raves M., Sacchetti A., Hornsveld M., Oost K. C., et al. (2017). Interplay between metabolic identities in the intestinal crypt supports stem cell function. 543, 424-427. 10.1038/nature21673 [DOI] [PubMed] [Google Scholar]
- Rossi D. J., Bryder D., Zahn J. M., Ahlenius H., Sonu R., Wagers A. J. and Weissman I. L. (2005). Cell intrinsic alterations underlie hematopoietic stem cell aging. 102, 9194-9199. 10.1073/pnas.0503280102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryall J. G., Dell'Orso S., Derfoul A., Juan A., Zare H., Feng X., Clermont D., Koulnis M., Gutierrez-Cruz G., Fulco M. et al. (2015). The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. 16, 171-183. 10.1016/j.stem.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari B. R., Zhang D., Allis C. D. and Zhao Y. (2017). Metabolic regulation of gene expression through histone acylations. 18, 90-101. 10.1038/nrm.2016.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi S., Yousefi S., Constantinescu M. A., Fey M. F. and Simon H.-U. (2012). Autophagy is required for self-renewal and differentiation of adult human stem cells. 22, 432-435. 10.1038/cr.2011.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samudio I., Harmancey R., Fiegl M., Kantarjian H., Konopleva M., Korchin B., Kaluarachchi K., Bornmann W., Duvvuri S., Taegtmeyer H. et al. (2010). Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. 120, 142-156. 10.1172/JCI38942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki N., Shiraki Y., Tsuyama T., Obata F., Miura M., Nagae G., Aburatani H., Kume K., Endo F. and Kume S. (2014). Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. 19, 780-794. 10.1016/j.cmet.2014.03.017 [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N., Locasale J. W., Lyssiotis C. A., Zheng Y., Teo R. Y., Ratanasirintrawoot S., Zhang J., Onder T., Unternaehrer J. J., Zhu H. et al. (2013). Influence of threonine metabolism on S-adenosylmethionine and histone methylation. 339, 222-226. 10.1126/science.1226603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer R. A. J. and Morrison S. J. (2013). Mechanisms that regulate stem cell aging and life span. 12, 152-165. 10.1016/j.stem.2013.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek T., Kocabas F., Zheng J., Deberardinis R. J., Mahmoud A. I., Olson E. N., Schneider J. W., Zhang C. C. and Sadek H. A. (2010). The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. 7, 380-390. 10.1016/j.stem.2010.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino V., Menzies K. J. and Auwerx J. (2017). repairing mitochondrial dysfunction in disease. 58, 353-389. https://doi.org/10.1146/annurev-pharmtox-010716-104908 [DOI] [PubMed] [Google Scholar]
- Sperber H., Mathieu J., Wang Y., Ferreccio A., Hesson J., Xu Z., Fischer K. A., Devi A., Detraux D., Gu H. et al. (2015). The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. 17, 1523-1535. 10.1038/ncb3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L. R. and Imai S. (2014). Specific ablation of Nampt in adult neural stem cells recapitulates their functional defects during aging. 33, 1321-1340. 10.1002/embj.201386917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John J. C., Ramalho-Santos J., Gray H. L., Petrosko P., Rawe V. Y., Navara C. S., Simerly C. R. and Schatten G. P. (2005). The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. 7, 141-153. 10.1089/clo.2005.7.141 [DOI] [PubMed] [Google Scholar]
- Stoll E. A., Cheung W., Mikheev A. M., Sweet I. R., Bielas J. H., Zhang J., Rostomily R. C. and Horner P. J. (2011). Aging neural progenitor cells have decreased mitochondrial content and lower oxidative metabolism. 286, 38592-38601. 10.1074/jbc.M111.252171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringari C., Wang H., Geyfman M., Crosignani V., Kumar V., Takahashi J. S., Andersen B. and Gratton E. (2015). In vivo single-cell detection of metabolic oscillations in stem cells. 10, 1-7. 10.1016/j.celrep.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Wellen K. E. and Rabinowitz J. D. (2016). Metabolic control of methylation and acetylation. 30, 52-60. 10.1016/j.cbpa.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T., Takubo K. and Semenza G. L. (2011). Metabolic regulation of hematopoietic stem cells in the hypoxic niche. 9, 298-310. 10.1016/j.stem.2011.09.010 [DOI] [PubMed] [Google Scholar]
- Sudo K., Ema H., Morita Y. and Nakauchi H. (2000). Age-associated characteristics of murine hematopoietic stem cells. 192, 1273-1280. 10.1084/jem.192.9.1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Luo M., Jeong M., Rodriguez B., Xia Z., Hannah R., Wang H., Le T., Faull K. F., Chen R. et al. (2014). Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. 14, 673-688. 10.1016/j.stem.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Yun J., Liu J., Malide D., Liu C., Rovira I. I., Holmström K. M., Fergusson M. M., Yoo Y. H., Combs C. A. et al. (2015). Measuring in vivo mitophagy. 60, 685-696. 10.1016/j.molcel.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Youle R. J. and Finkel T. (2016). The mitochondrial basis of aging. 61, 654-666. 10.1016/j.molcel.2016.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehara T., Teramura T., Onodera Y., Hamanishi C. and Fukuda K. (2012). Reduced oxygen concentration enhances conversion of embryonic stem cells to epiblast stem cells. 21, 1239-1249. 10.1089/scd.2011.0322 [DOI] [PubMed] [Google Scholar]
- Takubo K., Nagamatsu G., Kobayashi C. I., Nakamura-Ishizu A., Kobayashi H., Ikeda E., Goda N., Rahimi Y., Johnson R. S., Soga T. et al. (2013). Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. 12, 49-61. 10.1016/j.stem.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A. H. and Rando T. A. (2014). Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. 33, 2782-2797. 10.15252/embj.201488278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Luo B., Deng Z., Wang B., Liu F., Li J., Shi W., Xie H., Hu X. and Li J. (2016). Mitochondrial aerobic respiration is activated during hair follicle stem cell differentiation, and its dysfunction retards hair regeneration. 4, e1821 10.7717/peerj.1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. W., Barron M. J., Borthwick G. M., Gospel A., Chinnery P. F., Samuels D. C., Taylor G. A., Plusa S. M., Needham S. J., Greaves L. C. et al. (2003). Mitochondrial DNA mutations in human colonic crypt stem cells. 112, 1351-1360. 10.1172/JCI19435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperino R., Schoonjans K. and Auwerx J. (2010). Histone methyl transferases and demethylases; can they link metabolism and transcription? 12, 321-327. 10.1016/j.cmet.2010.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormos K. V., Anso E., Hamanaka R. B., Eisenbart J., Joseph J., Kalyanaraman B. and Chandel N. S. (2011). Mitochondrial complex III ROS regulate adipocyte differentiation. 14, 537-544. 10.1016/j.cmet.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B. J., Lee B. H., Castrillon D. H., Cullen D. E., McDowell E. P., Lazo-Kallanian S., Williams I. R., Sears C. et al. (2007). FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. 128, 325-339. 10.1016/j.cell.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J. N., Rovio A. T., Bruder C. E., Bohlooly Y.-M., Gidlöf S., Oldfors A., Wibom R. et al. (2004). Premature ageing in mice expressing defective mitochondrial DNA polymerase. 429, 417-423. 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- Ugarte F., Sousae R., Cinquin B., Martin E. W., Krietsch J., Sanchez G., Inman M., Tsang H., Warr M., Passegué E. et al. (2015). Progressive chromatin condensation and H3K9 methylation regulate the differentiation of embryonic and hematopoietic stem cells. 5, 728-740. 10.1016/j.stemcr.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini N., Girotra M., Naveiras O., Nikitin G., Campos V., Giger S., Roch A., Auwerx J. and Lutolf M. P. (2016). Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. 7, 13125 10.1038/ncomms13125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zant G. and Liang Y. (2003). The role of stem cells in aging. 31, 659-672. 10.1016/S0301-472X(03)00088-2 [DOI] [PubMed] [Google Scholar]
- Vazquez-Martin A., Van den Haute C., Cufí S., Corominas-Faja B., Cuyàs E., Lopez-Bonet E., Rodriguez-Gallego E., Fernández-Arroyo S., Joven J., Baekelandt V. et al. (2016). Mitophagy-driven mitochondrial rejuvenation regulates stem cell fate. 8, 1330-1352. 10.18632/aging.100976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlashi E., Lagadec C., Vergnes L., Matsutani T., Masui K., Poulou M., Popescu R., Della Donna L., Evers P., Dekmezian C. et al. (2011). Metabolic state of glioma stem cells and nontumorigenic cells. 108, 16062-16067. 10.1073/pnas.1106704108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Osenbroch P., Skinnes R., Esbensen Y., Bjørås M. and Eide L. (2010). Mitochondrial DNA integrity is essential for mitochondrial maturation during differentiation of neural stem cells. 28, 2195-2204. 10.1002/stem.542 [DOI] [PubMed] [Google Scholar]
- Wang W., Esbensen Y., Kunke D., Suganthan R., Rachek L., Bjoras M. and Eide L. (2011). Mitochondrial DNA damage level determines neural stem cell differentiation fate. 31, 9746-9751. 10.1523/JNEUROSCI.0852-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware C. B., Wang L., Mecham B. H., Shen L., Nelson A. M., Bar M., Lamba D. A., Dauphin D. S., Buckingham B., Askari B. et al. (2009). Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. 4, 359-369. 10.1016/j.stem.2009.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware C. B., Nelson A. M., Mecham B., Hesson J., Zhou W., Jonlin E. C., Jimenez-Caliani A. J., Deng X., Cavanaugh C., Cook S. et al. (2014). Derivation of naive human embryonic stem cells. 111, 4484-4489. 10.1073/pnas.1319738111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr M. R., Binnewies M., Flach J., Reynaud D., Garg T., Malhotra R., Debnath J. and Passegué E. (2013). FOXO3A directs a protective autophagy program in haematopoietic stem cells. 494, 323-327. 10.1038/nature11895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R. and Thompson C. B. (2009). ATP-citrate lyase links cellular metabolism to histone acetylation. 324, 1076-1080. 10.1126/science.1164097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E. G., Wu Y., Jha P., Dubuis S., Blattmann P., Argmann C. A., Houten S. M., Amariuta T., Wolski W., Zamboni N. et al. (2016). Systems proteomics of liver mitochondria function. 352, aad0189 10.1126/science.aad0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Williams E. G., Dubuis S., Mottis A., Jovaisaite V., Houten S. M., Argmann C. A., Faridi P., Wolski W., Kutalik Z. et al. (2014). Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. 158, 1415-1430. 10.1016/j.cell.2014.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li Y., Zhang H., Huang Y., Zhao P., Tang Y., Qiu X., Ying Y., Li W., Ni S. et al. (2015). Autophagy and mTORC1 regulate the stochastic phase of somatic cell reprogramming. 17, 715-725. 10.1038/ncb3172 [DOI] [PubMed] [Google Scholar]
- Xavier J. M., Morgado A. L., Rodrigues C. M. P. and Sola S. (2014). Tauroursodeoxycholic acid increases neural stem cell pool and neuronal conversion by regulating mitochondria-cell cycle retrograde signaling. 13, 3576-3589. 10.4161/15384101.2014.962951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanes O., Clark J., Wong D. M., Patti G. J., Sánchez-Ruiz A., Benton H. P., Trauger S. A., Desponts C., Ding S. and Siuzdak G. (2010). Metabolic oxidation regulates embryonic stem cell differentiation. 6, 411-417. 10.1038/nchembio.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. and Hekimi S. (2010). A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. 8, e1000556 10.1371/journal.pbio.1000556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O. H., Katajisto P., Lamming D. W., Gultekin Y., Bauer-Rowe K. E., Sengupta S., Birsoy K., Dursun A., Yilmaz V. O., Selig M. et al. (2012). mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. 486, 490-495. 10.1038/nature11163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Takahashi K., Okita K., Ichisaka T. and Yamanaka S. (2009). Hypoxia enhances the generation of induced pluripotent stem cells. 5, 237-241. 10.1016/j.stem.2009.08.001 [DOI] [PubMed] [Google Scholar]
- Yu W.-M., Liu X., Shen J., Jovanovic O., Pohl E. E., Gerson S. L., Finkel T., Broxmeyer H. E. and Qu C. K. (2013). Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. 12, 62-74. 10.1016/j.stem.2012.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarse K., Schmeisser S., Groth M., Priebe S., Beuster G., Kuhlow D., Guthke R., Platzer M., Kahn C. R. and Ristow M. (2012). Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. 15, 451-465. 10.1016/j.cmet.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Khvorostov I., Hong J. S., Oktay Y., Vergnes L., Nuebel E., Wahjudi P. N., Setoguchi K., Wang G., Do A. et al. (2011). UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. 30, 4860-4873. 10.1038/emboj.2011.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Ryu D., Wu Y., Gariani K., Wang X., Luan P., D'Amico D., Ropelle E. R., Lutolf M. P., Aebersold R. et al. (2016). NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. 352, 1436-1443. 10.1126/science.aaf2693 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Marsboom G., Toth P. T. and Rehman J. (2013). Mitochondrial respiration regulates adipogenic differentiation of human mesenchymal stem cells. 8, e77077 10.1371/journal.pone.0077077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L., Ma L., Hamm M., Gage F. H. and Hunter T. (2016). Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. 5, e13374 10.7554/eLife.13374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., Blau C. A., Horwitz M. S., Hockenbery D., Ware C. et al. (2012). HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. 31, 2103-2116. 10.1038/emboj.2012.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu X. L. and Guppy M. (2004). Cancer metabolism: facts, fantasy, and fiction. 313, 459-465. 10.1016/j.bbrc.2003.11.136 [DOI] [PubMed] [Google Scholar]