Abstract
Background
The overall 5-year survival rate of lung cancer is about 15% even with therapeutic drugs like tyrosine kinase inhibitors. Ideal models are urgently needed for exploring mechanisms and finding new drugs. Patient-derived xenografts (PDX) models and primary cells are both used to screen therapeutic regimens for cancer. However, PDX models and primary cells from the same patient are difficult to establish. Their consistency to the original tumor tissue is not well studied.
Methods
31 lung cancer patient tissues were procured to establish the lung cancer PDX models and primary cell lines. Tumor growth measurements, histological and immunohistochemistry analysis, Western blotting, EGFR and K-RAS mutation detection and gefitinib sensitive assay were performed to evaluate the characteristic of established PDX models. Immunofluorescence analysis, anchorage-independent cell growth, Western blotting and gefitinib sensitive assay were performed to assay the characteristic of established primary cell lines. The whole-exome sequencing was used to compare the characteristic of the patient’s tumor tissue, established PDX and primary cell line.
Results
Twenty-one lung cancer PDX models (67.74%, 21/31) and ten primary cell lines (32.25%, 10/31) were established from patients’ tumor tissues. The histology and pathological immunohistochemistry of PDX xenografts are consistent with the patients’ tumor samples. Various signal pathways were activated in different PDX models (n = 5) and primary cell lines (n = 2). EGFR mutation PDX model and primary cell line (LG1) were sensitive to gefitinib treatment. The expression of CK8/18, TTF1 and NapsinA in LG1 and LG50 primary cells were also positive. And the activated signal pathways were activated in LG1 and LG50 primary cell lines. Furthermore, the gene mutation in PDX tumor tissues and primary cell line (LG50) was consistent with the mutation in LG50 patient’s tumor tissues.
Conclusion
These data suggested that established lung cancer PDX models and primary cell lines reserved mostly molecular characteristics of primary lung cancer and could provide a new tool to further understand the mechanisms and explore new therapeutic strategies.
Keywords: Lung cancer, Patient-derived xenograft, Signal pathway, Primary cell culture
Background
Lung cancer is the leading cause of cancer-related deaths worldwide [1–4]. Lung cancer treatments are dominated by chemotherapy, radiotherapy, surgery, even molecular targeted drugs [5, 6]. However, the overall 5-year survival rate of lung cancer is about 15% [7], even lower in small-cell lung carcinomas (SCLC) [5]. Molecular mechanism study of lung cancer is an immediate priority to find personal and targeted therapeutic strategies. In recent years, patient-derived xenografts have been established with the original molecular characteristics and heterogeneity of the cancer tissues [8–10]. These PDX models were even used to screen therapeutic regimens for breast cancer, gastric cancer and esophageal cancer [11–13]. Moreover, the primary cells are useful tools for mechanism study compared with stable cell lines, because of maintaining heterogeneity of tumors. The molecular changes in both PDX models and primary cell lines are valuable for mechanism research, new drug development and personalized treatments [14, 15].
Here, we established and characterized lung cancer patient-derived xenografts and established primary cell lines from patients’ tissues. We evaluated the pathological characteristic of the PDX models and primary cell lines. The differences between PDX models and the original tumors were also investigated. We found both the PDX models and primary cell lines mostly reserved molecular characteristics and heterogeneity of original cancer tissues, even drug sensitivity. Therefore, these PDX models and primary cell lines provide a platform for further understanding the molecular mechanisms and therapeutic screening, regimen for lung patient from cell to the animal level.
Methods
Patient tissue procurement
31 lung cancer patients had undergone the surgical procedures at the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China) and lung cancer tissues were obtained from August 2014 to October 2015. All patients did not receive both chemotherapy and radiotherapy and followed written informed consent before surgery. All research protocols were approved by the research ethics committee of Zhengzhou University. Tissue histology was analyzed and confirmed by two pathologists. Lung cancer specimens were harvested from the periphery of whole tumor tissues to maintain high activity and low necrotic part. All tissues were transported to the lab in transport media (FBS-free PRMI DMEM with penicillin and streptomycin).
Animals
6–8 weeks old female CB17/severe combined Immunodeficiency (SCID) mice of 18 ~ 20 g average body weight (Vital River, Beijing, China) were used in the studies. 4–5 mice were kept in a pathogen-free environment with light controlled rooms (12 h cycles) and provided with food and water adlibitum.
Patient tumor xenografts
Tissue samples were placed in Petri dishes containing PBS with penicillin and streptomycin. The tissue was divided into four parts (implanted into SCID mice, fixed in 10% formalin, in liquid nitrogen for protein extraction and for primary cells). The detailed method of implanting into SCID mice was described in our previous works [12]. In brief, the tumor tissues were implanted subcutaneously in mice. The tumor was passaged when the tumor size reached ~ 1500 mm3. PDX models were used for study when the tumors were passaged for three generations. All animal studies were performed according to guidelines approved by the Zhengzhou University Institutional Animal Care and Use Committee.
Tumor growth measurements
The tumors were measured with a vernier caliper twice a week. Tumor volume was calculated using the formula V = LD × (SD)2/2, where V was tumor volume, LD was longest tumor diameter, and SD was the shortest tumor diameter. Tumor growth curves were plotted as tumor volume.
Histological and immunohistochemistry analysis
One part of tumor tissue was embedded into paraffin for histopathologic examination and immunohistochemistry analysis. All the slides were stained with Harris Hematoxylin after dewaxing 5 μm thick sections with dimethylbenzene and were evaluated by two pathologists. Tissue sections were incubated with CK5/6, P63, P40, or CK8/18, TTF1, NapsinA antibodies (Abcam, England) overnight at 4 °C. HRP-IgG secondary antibody was used at 37 °C for 15 min and detected by the diaminobenzidine (DAB) reactions. All slides were observed and measured by Olympus microscope (Japan).
Western blotting
The PDX tissues were ground in liquid nitrogen and then were lysed. Next, protein was extracted by centrifugation for 30 min at 12,000 rpm. Protein (50 μg) was separated in 10% SDS-PAGE gel, and transferred to PVDF membrane at 90 V for 2 h. The membrane was blocked with 5% no-fat milk for 60 min. Later, membrane was incubated with mTOR, p-mTOR (Ser2481), STAT1, p-STAT1 (Tyr701), STAT3, p-STAT3 (Tyr705), AKT1, p-AKT (Ser473), ERK, p-ERK (Thr202/Tyr204) antibodies (Cell Signaling Technology, America) overnight at 4 °C. All antibodies were used at 1:1000. The STAT3 antibody was from mouse, other antibodies were from rabbit. The fluorescent secondary antibody was incubated at room temperature for 1.5 h. The PVDF membranes were scanned by Odyssey and analyzed by Image Studio Ver 2.1.
EGFR and K-RAS mutation detection
Lung cancer patients’ tissues and xenograft tissues from PDX models were pathologically reviewed to ensure that tumor cells were more than 80% and that no significant tumor necrosis had occurred. Genomic DNA was extracted from each sample using Puregene Cell and Tissue Kit (QIAGEN, Cat#158388, Germany). The quantity and quality of DNA samples were measured by Nanodrop ND-1000 UV/VIS spectrophotometer (Thermo Scientific, USA). DNA fragment integrity was confirmed by electrophoresis using 1% agarose gel. The concentration of DNA samples was normalized to 20 ng/μl and stored at − 20 °C. ‘Hotspot’ mutations in epidermal growth factor receptor (EGFR) (exons 18, 19, 20, 21) and K-RAS (exons 2 and 3) were screened by the mutant-enriched liquid chip polymerase chain reaction method.
Gefitinib treatment for PDX and primary cell lines
After establishing PDX models, we chose LG1 and LG50 PDX xenografts for gefitinib treatment. Mice were divided into two groups (10 mice per group) which were vehicle control and gefitinib treatment group. Once the tumor volumes reached approximately 25 mm3, mice were treated by oral gavages with vehicle control (0.9% NaCl) and gefitinib (100 mg/kg). Body weight and tumor size measurements were performed twice a week.
LG1 and LG50 primary cells (1 × 103 per well) for proliferation were seeded into 96-well plates. After overnight incubation, cells were treated with different concentrations of geftinib (0, 0.25, 0.5 and 1 μM) and incubated for 24, 48, 72, or 96 h. CCK8 Solution (10 μl, Dojindo, Japan) was then added and cells were incubated for another 1.5–2 h. Absorbance was measured at 450 nm.
Establishment of primary lung cancer cell lines from the patients’ lung cancer tissues
The fresh lung cancer tissues were minced into small pieces less than 1 mm3 using sterile eye scissors, followed by extensive washing in RPMI-DMEM medium and centrifuging at 300g for 5 min. Next, the tissues were resuspended in RPMI-DMEM medium containing collagenase II (Invitrogen, USA) at the concentration of 200 U/ml and digested for 2–4 h at 37 °C. The enzymatic digestion was stopped when most of the tissues became cell suspensions. Following washing in RPMI-DMEM and centrifuging at 300g for 5 min, cells were transferred into standard tissue culture coated flasks (Corning Life Sciences, USA) and cultured in the Defined Keratinocyte-Serum Free Medium (DK-SFM) supplemented with l-glutamine (Invitrogen, USA), EGF 20 ng/ml, basic-FGF 10 ng/ml (PeproTech Inc., USA), 2% B27 (Invitrogen, USA), penicillin and streptomycin, and amphotericin B (0.25 mg/ml; Invitrogen, USA). All primary cells were cultured at 37 °C in a humidified incubator with 5% CO2. Culture medium was changed every 2–3 days. Cells were passaged after 80–90% confluence.
Immunofluorescence analysis
2.5 × 105 primary cells were seeded in 24-well plate which was placed a sterilized glass slide in every well, incubated for 24 h, and fixed with 4% paraformaldehyde for 30 min. CK8/18, TTF1, NapsinA antibodies (Rat anti-human, 1:50; Santa Cruz Biotechnology) was incubated at 4 °C overnight, and then FITC-conjugated rabbit IgG antibody was used. Cells with green fluorescent signals in the nucleus and cytoplasm were counted as positive expression cells. The In Cell 6000 Analyzer (GE) was used for detecting the fluorescence.
Anchorage-independent cell growth
A total of 8000 primary cells were suspended in 0.33% basal medium eagle agar supplemented with 10% FBS. The cells were maintained at 37 °C, 5% CO2 for 2–3 weeks, and cell colony numbers were counted by In Cell 6000 Analyzer (GE).
Whole-exome sequencing
We used the Illumina HiSeq to perform whole-exome sequencing of LG50 patient tissue, LG50 PDX xenografts, and LG50 primary cell line. All genomic variations including SNPs and In-Dels were detected by the software, such as HaplotypeCaller of GATK (v3.3.0). After that, the hard-filtering method was applied to get high-confident variant calls. Then the SnpEff tool (http://snpeff.sourceforge.net/SnpEff_manual.html) was applied to perform a series of annotations for variants. The final variants and annotation results were used in the advanced downstream analysis.
Statistical analysis
All statistical analyses were performed using SPSS statistical software 17.0. Experimental values were reported as mean ± standard deviation. One-way analysis of variance was used for statistical analysis. p < 0.05 was considered as statistically significant.
Results
Clinical characteristics of the patients
In this study, 31 lung cancer patients underwent surgical resection. These patients consisted of twenty-six men and five women ranging from 40 to 75 years in age (60.5 ± 10.0 years). All the patients did not have any apparent distant metastases before surgery and had not been previously treated. In these samples, there are 10 adenocarcinoma samples, 1 adenosquamous carcinoma, 1 mucoepidermoid carcinoma, 1 neuroendocrine and SCLC, 1 non-small cell lung carcinomas (NSCLC), 1 SCLC, 16 squamous carcinomas. There are 2 well-differentiated samples, 4 moderately-well, 12 moderate, 7 moderately-poor and 6 poor differentiated samples. According to lymph node metastasis, 8 of 11 samples with lymph node metastasis had been established, and 13 of 20 samples without lymph node metastasis had been established. All patients’ clinical data were shown in Table 1.
Table 1.
Summary of the clinical data of lung cancer patients for PDX models
| # | Number | Age | Sex | Pattern of organization | Tumor differentiation | TNM Staging | Lymph Node Metastasis | Engrafted model | Primary cell lines | EGFR mutation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | LG1 | 45 | Female | Adenocarcinoma | Poor | T2bN0M0 | Y | Y | Y | L858R |
| 2 | LG2 | 69 | Female | Adenocarcinoma | Moderate | T1aN0M0 | N | Y | N | Exon19 delete |
| 3 | LG3 | 74 | Male | Adenocarcinoma | Moderately-poor | T1bN0M0 | N | Y | N | Wild type |
| 4 | LG6 | 60 | Male | Squamous carcinoma | Moderately-poor | T2aN0M0 | N | Y | Y | Wild type |
| 5 | LG7 | 43 | Male | Mucoepidermoid carcinoma | Moderately-poor | T2bN0M0 | Y | Y | Y | Wild type |
| 6 | LG10 | 62 | Female | Adenocarcinoma | Moderate | T1N0M0 | N | N | N | Wild type |
| 7 | LG12 | 75 | Male | Squamous carcinoma | Moderately-poor | T2bN0M0 | N | Y | Y | Wild type |
| 8 | LG14 | 77 | Male | NE and SCLC | Well | T2N1M0 | Y | Y | N | Wild type |
| 9 | LG17 | 72 | Male | Adenocarcinoma | Moderately-good | T2N0M0 | N | Y | N | Wild type |
| 10 | LG19 | 53 | Male | Squamous carcinoma | Moderate | T1N0M0 | N | N | Y | Wild type |
| 11 | LG21 | 67 | Male | Adenocarcinoma | Moderate | T2N0M0 | N | N | N | Wild type |
| 12 | LG22 | 55 | Male | Squamous carcinoma | Moderate | T2aN0M0 | N | Y | N | L858R |
| 13 | LG24 | 40 | Male | SCLC | Moderately-poor | T2N0M0 | N | Y | N | Wild type |
| 14 | LG26 | 50 | Male | Squamous carcinoma | Moderately-poor | T2bN0M0 | N | Y | N | Wild type K-RAS |
| 15 | LG28 | 60 | Male | Squamous carcinoma | Moderate | T2bN1M0 | Y | Y | N | colon12 GGT > CGT |
| 16 | LG29 | 58 | Male | Squamous carcinoma | Moderately-good | T2bN0M0 | N | Y | N | Wild type |
| 17 | LG33 | 58 | Male | Squamous carcinoma | Moderate | T3N1M0 | N | Y | N | Wild type |
| 18 | LG37 | 64 | Male | Squamous carcinoma | Poor | T4N1M0 | Y | Y | Y | Wild type |
| 19 | LG38 | 46 | Male | Squamous carcinoma | Moderately-good | T2N0M0 | N | Y | N | Wild type |
| 20 | LG42 | 67 | Male | Squamous carcinoma | Moderately-good | T2N1M0 | Y | N | N | Wild type |
| 21 | LG44 | 58 | Male | NSCLC | Poor | T2aN0M0 | N | Y | N | Wild type |
| 22 | LG46 | 56 | Female | Squamous carcinoma | Poor | T2aN1M0 | Y | Y | N | Wild type |
| 23 | LG48 | 50 | Male | Adenocarcinoma | Moderate | T2N1M0 | Y | N | Y | Wild type |
| 24 | LG50 | 75 | Female | Adenocarcinoma | Moderate | T2N0M0 | N | Y | Y | Wild type |
| 25 | LG52 | 56 | Male | Squamous carcinoma | Moderate | T1N1M0 | Y | N | N | Wild type |
| 26 | LG53 | 65 | Male | Adenosquamous carcinoma | Poor | T2aN1M0 | Y | Y | N | Wild type |
| 27 | LG54 | 70 | Male | Adenocarcinoma | Moderate | T1N0M0 | N | N | N | Wild type |
| 28 | LG55 | 64 | Male | Adenocarcinoma | Moderate | T2aN0M0 | N | N | Y | Wild type |
| 29 | LG59 | 68 | Male | Squamous carcinoma | Well | T1N0M0 | N | N | N | Wild type |
| 30 | LG61 | 59 | Male | Squamous carcinoma | Moderately-poor | T2N2M0 | Y | Y | Y | Wild type |
| 31 | LG62 | 67 | Male | Adenocarcinoma | Poor | T2bN0M0 | N | N | N | Wild type |
Y yes; N no
Growth of lung cancer xenografts in SCID mice
31 tumor specimens were transplanted to SCID mice and 21 xenografts were established (Table 1). The successful growth rate is 67.7%. The pathological types included SCLC, adenocarcinomas and squamous carcinomas. The generation harboring the patient-derived tumor tissue was termed P0, with subsequent generations numbered consecutively (P1, P2, P3 and so on). The growth curves of three passages (P1, P2, and P3) of LG50 and LG33 (patient ID) were plotted using the data obtained each week (Fig. 1). The latency time before passaging of P1 for LG50 and LG33 was 168 and 154 days, respectively, the latency time of P2 was 70 and 77 days, and P3 was 35 and 56 days. It was noticed that the latency time of each passage became shorter and it decreased to 1–3 months in subsequent passages after P3.
Fig. 1.
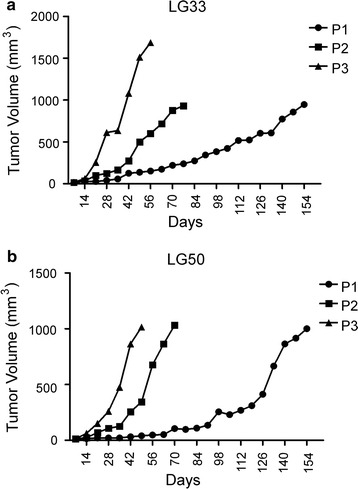
Growth curves of lung cancer PDXs in SCID mice. a The generation harboring the patient-derived tumor tissue was termed P0, with subsequent generations numbered consecutively (P1, P2, P3 and so on). The growth of PDX xenografts reaches a steady state level in third and more passages. Growth curves of three passage xenografts (1st, 2nd and 3rd passage) plotted as tumor volume in PDX xnografts LG33. b The tumor volume of 1st, 2nd and 3rd passage was measured in lung adenocarcinoma LG50
Histology and immunohistochemistry comparison between xenografts and patients’ tumors
Next, LG33 with squamous carcinoma and LG50 with adenocarcinoma were used as representative to compare histology and immunohistochemistry of the primary tumor tissue with P3 PDX tumor tissues. The HE staining results indicated that the morphology was similar between primary tumor tissues and PDX tumor tissues (Fig. 2a, b). P40, P63 and CK5/6 were specific clinical diagnosis indexes for squamous carcinoma. CK8/18, TTF1 and NapsinA were specific clinical diagnosis indexes for adenocarcinoma. Our results indicated CK5/6, p40, and p63 were positive in both P0 and P3 of lung squamous carcinoma LG33 (Fig. 2a) and CK8/18, NapsinA, and TTF-1 were positive in both P0 and P3 of lung adenocarcinoma LG50 (Fig. 2b). These results indicated that the original tumors’ characteristics were maintained in established PDX models.
Fig. 2.
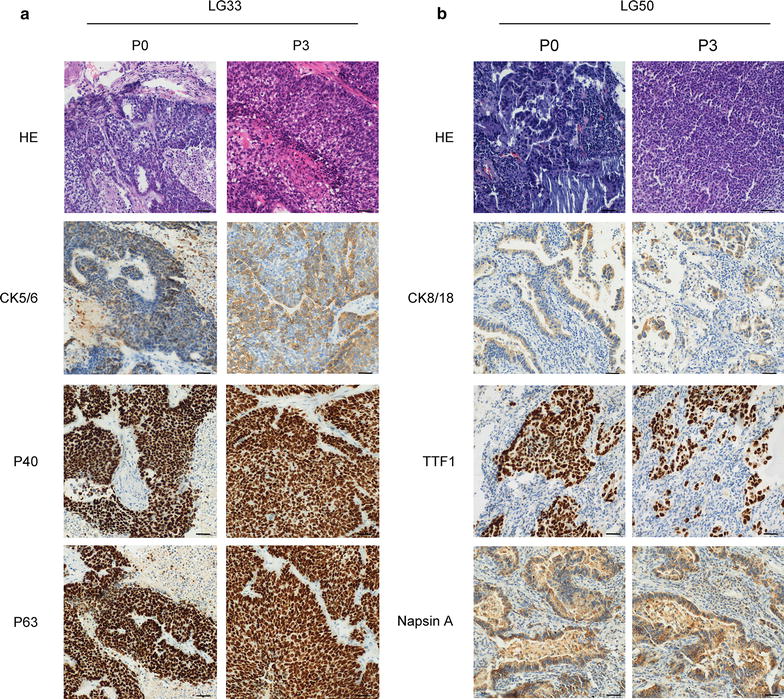
Histologic and immunohistochemistric features of patients’ lung cancers and their corresponding xenograft. a Histologic sections (H&E) and CK5/6, p40, p63 expression in LG33 lung cancer patient’s tissues and the third passage xenografts (Bar: 50 μm). Tumor tissue slides from each group were prepared with paraffin sections after fixation with formalin and then stained with HE and the indicated antibodies. P40, P63 and CK5/6 were specific clinical diagnosis indexes for squamous carcinoma. Expression of CK5/6, p40 and p63 were visualized by microscopy in LG33. b Histologic sections (H&E) and CK8/18, TTF1, NapsinA expression in LG50 lung cancer patient’s tissues and the third passage xenografts (Bar: 50 μm). CK8/18, TTF1 and NapsinA were specific clinical diagnosis indexes for adenocarcinoma. Expression of CK8/18, TTF1 and NapsinA were visualized by microscopy in LG50
Activated signal transduction pathways in the established lung cancer PDXs
mTOR, p70S6K, Akt, STAT1, STAT3, and ERK, as well as their corresponding phosphorylated forms, were examined in PDX models by Western blotting. LG1, LG14, LG17, LG22 and LG50 were representative as PDX models. Among these models, the level of p-mTOR, p-AKT473 and p-p70S6K (Thr389) was lowest in LG22. p-p70S6K (Ser424) in LG17 (Fig. 3a). p-STAT1 was lowest in LG1 and LG14. However, p-STAT3 was lower in LG1 and LG22 (Fig. 3b). Moreover, p-ERK was lowest in LG14 (Fig. 3c).
Fig. 3.
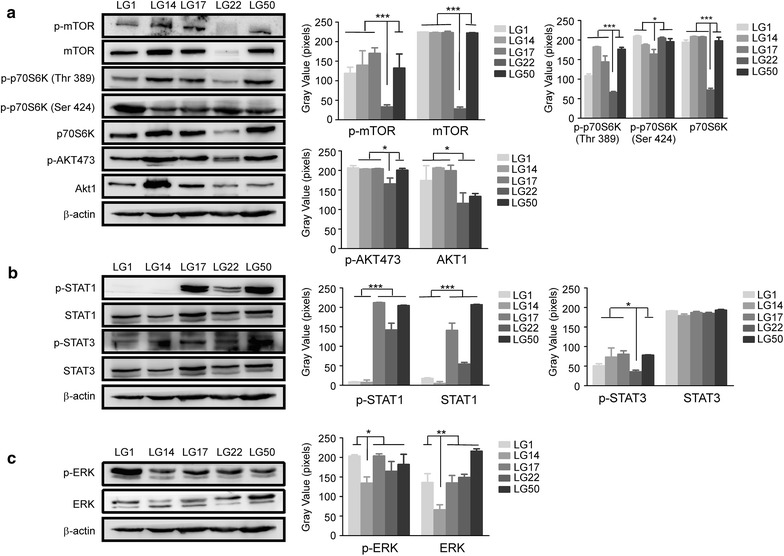
Activated signal transduction pathways in the established lung cancer PDXs. a AKT-mTOR axis pathway was activated in the different PDXs (LG1, LG14, LG17, LG22 and LG50). Protein levels of p-mTOR and mTOR, p-p70S6K (Thr389), p-p70S6K (Ser424) and p70S6K, p-AKT473 and AKT1 were visualized by Western blotting. β-actin was used to verify equal protein loading. b STAT1 and STAT3 pathway were activated in the different PDXs (LG1, LG14, LG17, LG22 and LG50). Protein levels of p-STAT1 and STAT1, p-STAT3 and STAT3 were visualized by Western blotting. β-actin was used to verify equal protein loading. c ERK pathway were activated in the different PDXs (LG1, LG14, LG17, LG22 and LG50). Protein levels of p-ERK and ERK was visualized by Western blotting. β-actin was used to verify equal protein loading. Each experiment was repeated three times. According to the results of Western blotting, we used image J to determine the gray value of each stripe, made histograms by the gray value, the asterisks (*,**, ***) indicate a significant (p < 0.05, 0.01, p < 0.001, respectively)
EGFR and K-Ras mutation and gefitinib sensitive assay
We sequenced EGFR and K-Ras gene locus of established PDX models. We found 2 EGFR L858R mutation, 1 EGFR Exon19 deletion mutation and 1 K-Ras codon12 GGT > CGT mutation (Table 1). Next, we chose LG1 xenografts with EGFR L858R and LG50 xenografts with wild-type EGFR to do the gefitinib treatment sensitive assay. We found that LG1 xenografts quickly shrunk after injecting gefitinib 100 mg/kg once daily for a week. By comparison, LG50 grew normally. This indicated LG1 was sensitive to gefitinib treatment (Fig. 4a), and LG50 was resistant (Fig. 4b). To assess the gefitinib sensitivity, LG1 primary cells and LG50 primary cells were treated by different doses of gefitinib (0, 0.25, 0.5 and 1 μM). Our results found that gefitinib suppressed the LG1 cell proliferation and had no effection on the LG50 primary cell proliferation by CCK8 assay (Fig. 4c). These indicated that EGFR mutant model and primary cells were sensitive to gefitinib.
Fig. 4.
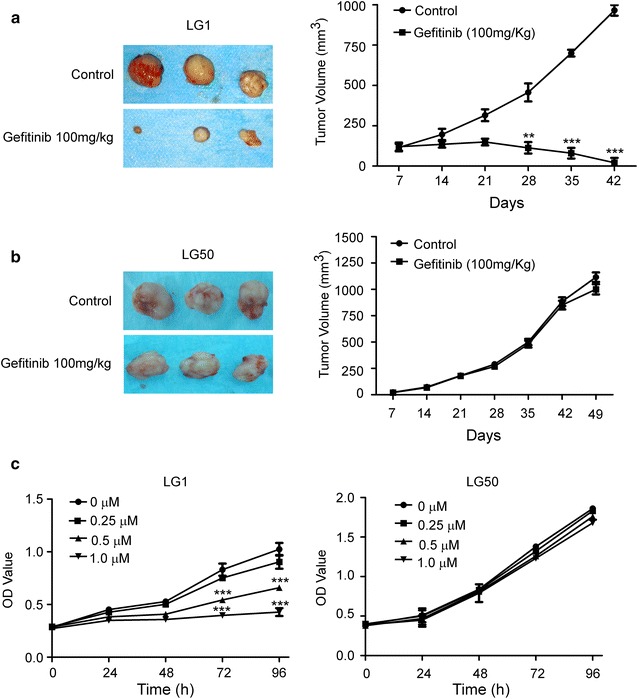
Gefitinib inhibits tumor cell growth in EGFR mutant PDX model and its corresponding primary cell. a Gefitinib (100 mg/kg) significantly suppresses tumor growth in an EGFR mutant PDX LG1. Mice were divided into two different groups as described in Materials and Methods. The asterisks (**, ***) indicate a significant (p < 0.01, p < 0.001, respectively). b Gefitinib (100 mg/kg) have no effective on tumor growth in EGFR wild type PDX LG50. Mice were divided into two different groups as described in Materials and Methods. There is no statistical significance in tumor volume compared to the vehicle-treated control (p > 0.05). c Gefitnib suppresses LG1 primary cell proliferation and have no effection on the LG50 primary cell proliferation. LG1 primary cells and LG50 primary cells were treated with different doses of gefitinib (0, 0.25, 0.5 and 1 μM), and proliferation was measured by CCK8 assay Significant differences were evaluated using a 2-way ANOVA to determine significant time and dose effects. The asterisks (**, ***) indicate a significant (p < 0.01, p < 0.001, respectively). There was no significant differences in the LG50 cell proliferation. The morphological characteristic was obseved by microscopes (Bar:100 μm)
Immunofluorescence analysis of primary cells
10 primary cell lines were successfully established. LG1, LG6, LG7 and LG50 was respective as these primary cell lines. The primary cells had the cellular atypia and pleomorphism (Fig. 5). Next, we evulate whether the cells have the primary characteristic of tumor cells. LG1 and LG50 as adenocarcinoma were tested by the clinical indexes (CK8/18, NapsinA, and TTF1). Immunofluorescence assay indicated the expresion of these indexes were positive in both LG1 and LG50 primary cells (Fig. 6).
Fig. 5.
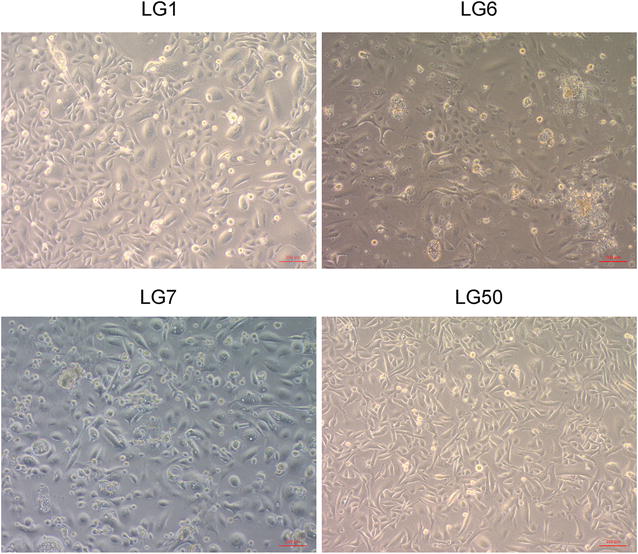
Morphological characteristic of LG1, LG6, LG7 and LG50 lung cancer primary cells. Primary cell line was established from the fresh lung cancer tissues LG1, LG6. LG7 and LG50
Fig. 6.
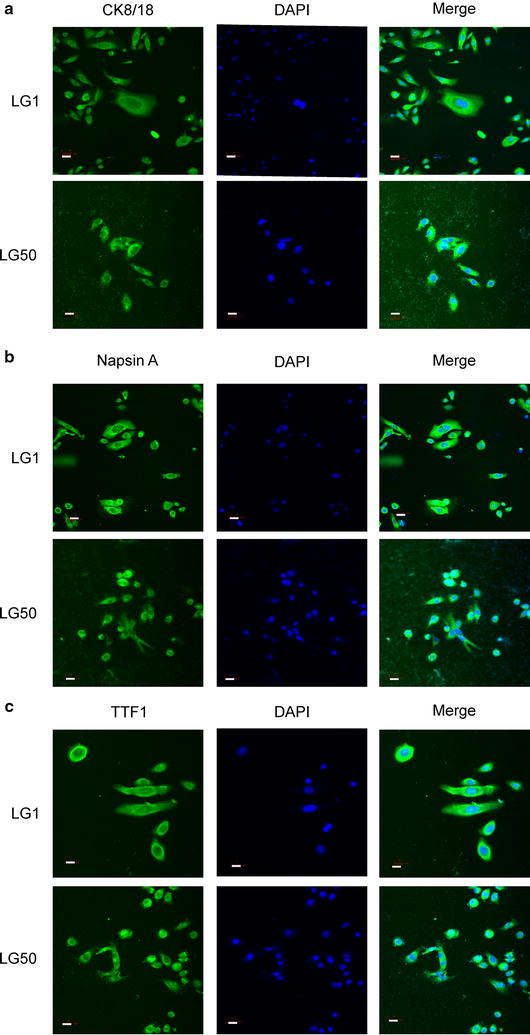
Immunofluorescence analysis of primary cells in LG1 and LG50. a CK8/18 was positive in LG1 and LG50 primary cells. LG1 and LG50 primary cells were fixed and subjected to immunofluorescence analysis. Sections were stained as described in “Methods” section using anti-CK8/18 followed by FITC-conjugated anti-rabbit IgG. Nuclei were counterstained with DAPI (Bar:15 μm). b Napsin A was positive in LG1 and LG50 primary cells. LG1 and LG50 primary cells were fixed and subjected to immunofluorescence analysis. Sections were stained as described in “Methods” section using anti-NapsinA followed by FITC-conjugated anti-rabbit IgG. Nuclei were counterstained with DAPI (Bar:15 μm). c TTF1 was positive in LG1 and LG50 primary cells. LG1 and LG50 primary cells were fixed and subjected to immunofluorescence analysis. Sections were stained as described in “Methods” section using anti-TTF1 followed by FITC-conjugated anti-rabbit IgG. Nuclei were counterstained with DAPI (Bar:15 μm)
Anchorage-independent growth and activated signal transduction pathway in established primary cells
We evaluated the characteristics of these primary tumor cells by anchorage-independent growth assay and found LG1 and LG50 primary cells both grow independently (Fig. 7a). The clone number of LG1 and LG50 was 2632.81 ± 275.89 and 2794.2 ± 93.54 every 8000 cells, respectively. The expressions of mTOR, p70S6K, AKT, and ERK, as well as their phosphorylation forms, were examined in both LG1 and LG50 primary cells. The level of p-mTOR, p-AKT473, and p-p70S6K (Thr389) was different between the LG1 and LG50 primary cell lines (Fig. 7b). These indicated different signal pathways were activated in different lung cancer; even they had the same pathology.
Fig. 7.
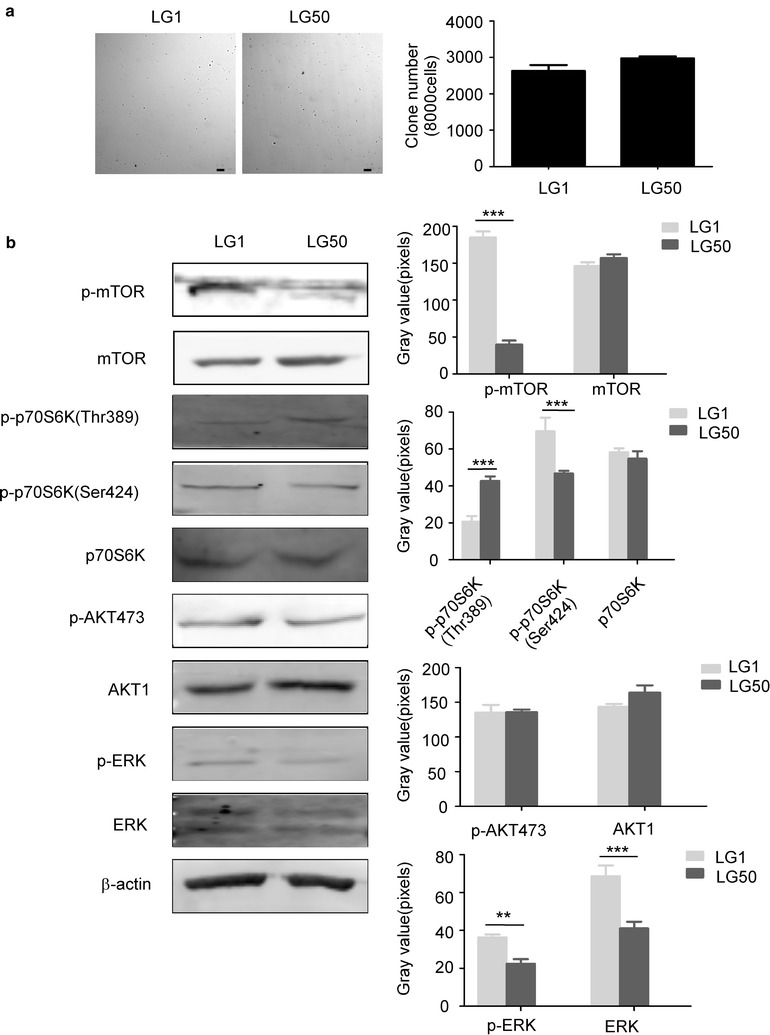
Anchorage-independent growth assay and activated signal transduction pathways in LG1 and LG50 primary cells. a Representative images from the anchorage-independent growth assay for LG1 (left) and LG50 (right) (Bar:100 μm). Data are presented as mean values ± S.D. from triplicate experiments. b Activated signal transduction pathways in LG1 and LG50 primary cells. The levels of phosphorylated and total proteins of AKT-mTOR axis, ERK in LG1 and LG50 primary cells were visualized by Western blotting. β-actin was used to verify equal protein loading. Each experiment was repeated three times. According to the results of Western blotting, we used image J to determine the gray value of each stripe, made histograms by the gray value, and analyzed by SPSS 17.0. The asterisks (**, ***) indicate a significant (p < 0.01, p < 0.001, respectively)
Whole-exome sequencing
To evaluate the genetic variations between the patient’s tumors and the established models, single nucleotide polymorphisms (SNPs) and INsertion/DELetion (InDel) were analyzed among LG50 patient tissue, LG50 primary cells, and PDX tumor tissue. The summary statistics of SNPs was shown in Table 2. There were 102,402, 94,847, 95,908 SNPs in the patient tissue, primary cell, and PDX tissue respectively. 98.23, 98.31, 94.59% were represented in dbSNP, while 95.81, 95.81, 92.17% were annotated in the 1000 genomes project database, respectively. In these genes, there are similar synonymous, missense, stop loss, stop-gain, start loss in three samples in Table 3. Moreover, the summary statistics of InDels was shown in Table 4. There were 15,964, 14,176, 14,958 InDels in three samples, respectively. In these variants, 80.62, 82.15, 78% were represented in dbSNP and 60.42, 60.77, 57.28% were annotated in the 1000 genomes project database. The number of novel InDels was 2728, 2223 and 2955, respectively. In three InDels groups in Table 5, 250, 252, 264 were frameshift, 7, 7, 9 were stop loss, 2 were all start loss and 59, 55, 58 were splice site. The length distribution of the InDels in coding sequence region (CDS) were also plotted in Fig. 8.
Table 2.
Summary statistics for identified SNPs for LG50 tumor tissue, primary cell line and PDX xenografts
| Samples | Total SNPs | Fraction of SNPs in dbSNP (%) | Fraction of SNPs in 1000 genomes (%) | Novel | Homozygous | Heterozygous | Intron | 5′ UTRs | 3′ UTRs | Upstream | Down stream | Intergenic | Ti/Tv |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient tumor tissue | 102,402 | 98.23 | 95.81 | 1473 | 1473 | 54,009 | 69,062 | 1567 | 3298 | 2077 | 1538 | 2330 | 2.34 |
| Primary cell line | 94,847 | 98.23 | 95.81 | 1473 | 44,541 | 50,306 | 62,661 | 1426 | 2974 | 1793 | 1377 | 2188 | 2.35 |
| PDX tumor tissue | 95,908 | 94.59 | 92.17 | 1473 | 44,875 | 51,033 | 61,376 | 1468 | 3130 | 1778 | 1359 | 2170 | 2.33 |
Table 3.
Functional categories for coding SNPs for LG50 tumor tissue, primary cell line and PDX xenografts
| Samples | Synonymous | Missense | Stopgain | Stoploss | Startloss | Splicing |
|---|---|---|---|---|---|---|
| Patient tumor tissue | 10,587 | 9794 | 79 | 33 | 16 | 80 |
| Primary cell line | 10,536 | 9774 | 80 | 32 | 17 | 79 |
| PDX tumor tissue | 12,431 | 10,036 | 82 | 31 | 15 | 89 |
Table 4.
Summary statistics for identified InDels for LG50 tumor tissue, primary cell line and PDX xenografts
| Samples | Total InDels | Fraction of InDels in dbSNP (%) | Fraction of InDels in 1000 genomes (%) | Novel | Homozygous | Heterozygous | Intron | 5′ UTRs | 3′ UTRs | Upstream | Down stream | Intergenic |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient tumor tissue | 15,964 | 80.62 | 60.42 | 2728 | 6044 | 9920 | 13,059 | 234 | 664 | 340 | 296 | 339 |
| Primary cell line | 14,176 | 82.15 | 60.77 | 2223 | 5567 | 8609 | 11,616 | 195 | 561 | 264 | 242 | 309 |
| PDX tumor tissue | 14,958 | 78.09 | 57.28 | 2223 | 5379 | 9579 | 12,205 | 214 | 616 | 273 | 259 | 328 |
Table 5.
Functional categories for coding InDels for LG50 tumor tissue, primary cell line and PDX xenografts
| Samples | Frameshift | Non-frameshift insertion | Non-frameshift deletion | Stoploss | Startloss | Splicing |
|---|---|---|---|---|---|---|
| Patient tumor tissue | 250 | 122 | 142 | 7 | 2 | 59 |
| Primary cell line | 252 | 111 | 144 | 9 | 2 | 55 |
| PDX tumor tissue | 264 | 117 | 144 | 9 | 2 | 58 |
Fig. 8.
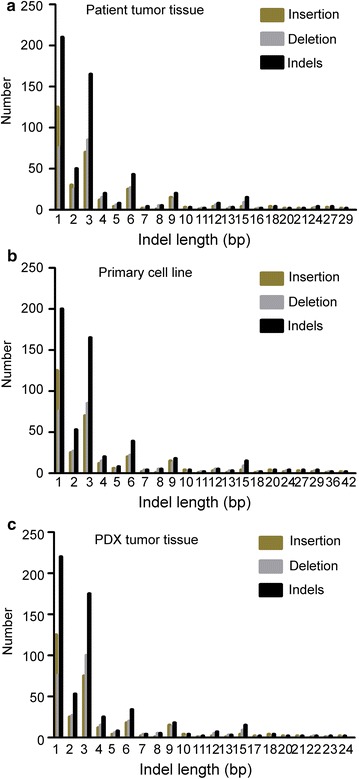
The distribution of lengths of coding InDel variants (CDS) in LG50 patient tumor tissue, primary cell line and PDX tumor tissue. a The distribution of lengths of CDS was in LG50 patient tumor tissue. DNA was extracted from LG50 patient tumor tissue and was performed the whole-exome sequencing by Illumina HiSeq. There were InDels in this sample. The length distribution of the InDels, Deletion and Insertion in CDS were also plotted. b The distribution of lengths of CDS was in LG50 primary cells. DNA was extracted from LG50 primary cell line and was performed whole-exome sequencing by Illumina HiSeq. There were InDels in this sample. The length distribution of the InDels, Deletion and Insertion in CDS were also plotted. c The distribution of CDS was in LG50 PDX tumor tissues. DNA was extracted from LG50 PDX tumor tissue and was performed whole-exome sequencing by Illumina HiSeq. There were InDels in this sample. The length distribution of the InDels, deletion and insertion in CDS were also plotted
Discussion
Identification and characterization of tumor molecular changes and genetics are pivotal for screening targeted drugs and precise treatments [8, 16, 17]. PDXs maintain the histology, as well as the molecular and genetic characteristics of the original tumor. Therefore, PDX models have advantages over conventional cell-line-derived xenografts and other models [10]. Here, we established 21 lung cancer PDX models from 31 lung cancer patients, and the success rate is 67.7% which is higher than others [8, 18]. Monitoring of tumor growth and location was important for evaluating the PDX models [19]. One defining feature of PDX model is its lag time before exponential growth that can be stable or variable during subsequent passages of the tumor. PDX tumor reached a steady state level in third and more passages, probably due to the progressive substitution of the original human tumor stroma with SCID mice stroma. In our PDX models, the first passage reached 1500 mm3 on day 168 and 154 for LG50 and LG33, respectively.
Different pathological type of cancer have different indexes in clinical. P40, P63 and CK5/6 were specific clinical diagnosis indexes for squamous carcinoma. CK8/18, TTF1 and NapsinA were specific clinical diagnosis indexes for adenocarcinoma. Our results found lung squamous carcinoma indexes and lung adenocarcinoma indexes still maintained in the third passage of the PDX models. These data indicated that our PDX models reflected the characteristics of patients’ samples. Different signaling pathways such as AKT/mTOR pathway, STAT1, and STAT3 pathway, ERK pathway played a critical role in the progression of lung cancer [20–22]. Targeting mTOR and AKT is a promising way to personalized treatment of lung cancer [23, 24]. AKT/mTOR pathways were strongly activated in established PDX models and primary cell lines (Figs. 3 and 7). Even in the same pathological type of lung cancer, the different pathways were activated. These indicated that our models provided a platform to screen the individual pathway inhibitors in the future.
Genetic and epigenetic abnormalities of primary cancer influence the processes of invasion, metastasis and drug resistance [6, 25, 26]. EGFR plays an essential role in regulating cell proliferation and apoptosis of lung adenocarcinoma. EGFR mutation and K-Ras mutation are regarded as mutation initiator in lung cancer patients [27, 28]. Mutation of EGFR in 17% of NSCLC patients is more frequent than in 5% of SCLC [20]. In the established PDX models, we found 4 EGFR mutation and 1 K-Ras mutation. We assessed the EGFR inhibitor sensitivity of these models. PDX model with EGFR L858R mutation was sensitive to gefitinib. However, PDX model with EGFR wild-type was resistant to gefitinib (Fig. 5). These results indicated PDX models are suitable tools for studying molecular diversity, drug screening and precise therapies.
The primary cell lines have advantages over conventional cell lines which may lose the diversity of tumor cells. Therefore, we established the primary cell lines from the patients’ lung cancer tissues. We checked the molecular characteristics of these primary cell lines. CK8/18, TTF1, and NapsinA were mostly located in the cytoplasm and cell nucleus of the LG1 and LG50 primary cells (Fig. 6). The LG1 and LG50 primary cells have a colony formation by anchorage-independent growth assay. Activated pathways in primary cells were also different (Fig. 7). It shows that primary cell lines maintain the tumor’s heteromorphism. Whole-exome sequencing is an important method to understand the diversity of tumor [29, 30]. Whole-exome sequencing results found that the established primary cell line and the PDX model remained more than 90% gene characteristics of the patient’s tumor tissues in LG50 (Tables 2, 3, 4, 5 and Fig. 8). Even through we indicated the established primary cell line and the PDX model remained more than 90% gene characteristics of the patient’s tumor tissues, more samples are needed to verify this result. We will continue amplify the library and put into clinical test.
Conclusions
We established lung cancer PDX models and paired primary cell lines. The established PDX models and primary cell lines have uttermostly reserved molecular characteristics, heterogeneity, and drug sensitivity of patients’ tumor. These PDX models and primary cell lines provide a useful tool for understanding the molecular mechanisms and screening new compounds for lung cancer therapy.
Authors’ contributions
YJ, JZ, YZ, KL, TL, XL and XC conducted experiments; XY, YW and SZ collected the samples, KL ZD and YJ designed the study, analyzed data and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Yihui Ma, The First Affiliated Hospital of Zhengzhou University, Zhengzhou University for pathology. We also thank Dr. Linlin Li, Public Health College, Zhengzhou University for statistical analysis.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved and supervised by the research ethics committee of Zhengzhou University, Zhengzhou, China. All animal experiments were performed in accordance with the animal experimental guidelines of Zhengzhou University.
Funding
This work was supported in by Natural Science Foundation of China (Nos. 81372269, 81472324, 81572812), Science Foundation of the Henan Province of China (Nos. 13HASTIT022, 17A310007) and China Scholarship Council.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- TKIs
tyrosine kinase inhibitors
- PDX
patient-derived xenografts
- SCLC
small-cell lung carcinomas
- SCID
severe combined immunodeficiency
- NSCLC
non-small cell lung carcinomas
- SNPs
single nucleotide polymorphisms
- InDel
INsertion/DELetion
- CDS
coding sequence region
Footnotes
Yanan Jiang, Jimin Zhao, Yi Zhang and Ke Li contributed equally to this work
Contributor Information
Yanan Jiang, Email: jyn163@hotmail.com.
Jimin Zhao, Email: zjm.0427@163.com.
Yi Zhang, Email: zhangyimed@163.com.
Ke Li, Email: LiK0703@163.com.
Tiepeng Li, Email: claylee5405@163.com.
Xinhuan Chen, Email: chen_xinhuan@126.com.
Simin Zhao, Email: szhao@hi.umn.edu.
Song Zhao, Email: zhaosong@zzu.edu.cn.
Kangdong Liu, Phone: 86-371-6778-1952, Email: kangdongliu@126.com.
Ziming Dong, Phone: 86-371-6773-9523, Email: dongzm@zzu.edu.cn.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, Salm M, Horswell S, Escudero M, Matthews N, Rowan A, Chambers T, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376(22):2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Swanton C, Govindan R. Clinical implications of genomic discoveries in lung cancer. N Engl J Med. 2016;374(19):1864–1873. doi: 10.1056/NEJMra1504688. [DOI] [PubMed] [Google Scholar]
- 5.Lemjabbar-Alaoui H, Hassan O, Yang YW, Buchanan P. Lung cancer: biology and treatment options. Biochem Biophys Acta. 2015;1856(2):189–210. doi: 10.1016/j.bbcan.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan W-L, Jain A, Takano A, Newell EW, Iyer NG, Lim W-T, Tan E-H, Zhai W, Hillmer AM, Tam W-L, Tan DSW. Novel therapeutic targets on the horizon for lung cancer. Lancet Oncol. 2016;17(8):e347–e362. doi: 10.1016/S1470-2045(16)30123-1. [DOI] [PubMed] [Google Scholar]
- 7.Anderson WC, Boyd MB, Aguilar J, Pickell B, Laysang A, Pysz MA, Bheddah S, Ramoth J, Slingerland BC, Dylla SJ, Rubio ER. Initiation and characterization of small cell lung cancer patient-derived xenografts from ultrasound-guided transbronchial needle aspirates. PLoS ONE. 2015;10(5):e0125255. doi: 10.1371/journal.pone.0125255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilie M, Nunes M, Blot L, Hofman V, Long-Mira E, Butori C, Selva E, Merino-Trigo A, Venissac N, Mouroux J, Vrignaud P, Hofman P. Setting up a wide panel of patient-derived tumor xenografts of non-small cell lung cancer by improving the preanalytical steps. Cancer Med. 2015;4(2):201–211. doi: 10.1002/cam4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao C, Wang L, Peng S, Cao M, Li H, Hu J, Huang X, Liu W, Zhang H, Wu S, Pataer A, Heymach JV, Eterovic AK, Zhang Q, Shaw KR, Chen K, et al. Gene mutations in primary tumors and corresponding patient-derived xenografts derived from non-small cell lung cancer. Cancer Lett. 2015;357(1):179–185. doi: 10.1016/j.canlet.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue T, Terada N, Kobayashi T, Ogawa O. Patient-derived xenografts as in vivo models for research in urological malignancies. Nat Rev Urol. 2017;14(5):267–283. doi: 10.1038/nrurol.2017.19. [DOI] [PubMed] [Google Scholar]
- 11.Holen I, Speirs V, Morrissey B, Blyth K. In vivo models in breast cancer research: progress, challenges and future directions. Dis Models Mech. 2017;10(4):359–371. doi: 10.1242/dmm.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y, Wu Q, Yang X, Zhao J, Jin Y, Li K, Ma Y, Chen X, Tian F, Zhao S, Xu J, Lu J, Yin X, Liu K, Dong Z. A method for establishing a patient-derived xenograft model to explore new therapeutic strategies for esophageal squamous cell carcinoma. Oncol Rep. 2016;35(2):785–792. doi: 10.3892/or.2015.4459. [DOI] [PubMed] [Google Scholar]
- 13.Owonikoko TK, Zhang G, Kim HS, Stinson RM, Bechara R, Zhang C, Chen Z, Saba NF, Pakkala S, Pillai R, Deng X, Sun SY, Rossi MR, Sica GL, Ramalingam SS, Khuri FR. Patient-derived xenografts faithfully replicated clinical outcome in a phase II co-clinical trial of arsenic trioxide in relapsed small cell lung cancer. J Transl Med. 2016;14(1):111–125. doi: 10.1186/s12967-016-0861-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Yao K, Shi C, Jiang Y, Liu K, Zhao S, Chen H, Reddy K, Zhang C, Chang X, Ryu J, Bode AM, Dong Z, Dong Z. 244-MPT overcomes gefitinib resistance in non-small cell lung cancer cells. Oncotarget. 2015;6(42):44274–44288. doi: 10.18632/oncotarget.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schild SE, Vokes EE. Pathways to improving combined modality therapy for stage III nonsmall-cell lung cancer. Ann Oncol. 2016;27(4):590–599. doi: 10.1093/annonc/mdv621. [DOI] [PubMed] [Google Scholar]
- 16.Urman A, Dean Hosgood H. Lung cancer risk, genetic variation, and air pollution. EBioMedicine. 2015;2(6):491–492. doi: 10.1016/j.ebiom.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Sun Y. Clinical experiences with molecular targeted therapy in lung cancer in China. Thorac Cancer. 2015;6(4):379–384. doi: 10.1111/1759-7714.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Y, Zhang P, An G, Zhang X, Zhang L, Si J, Zhang J, Yang Y. Induction of patient-derived xenograft formation and clinical significance of programmed cell death ligand 1 (PD-L1) in lung cancer patients. Med Sci Monit. 2016;22:4017–4025. doi: 10.12659/MSM.900661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siolas D, Hannon GJ. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 2013;73(17):5315–5319. doi: 10.1158/0008-5472.CAN-13-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch FR, Suda K, Wiens J, Bunn PA. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388(10048):1012–1024. doi: 10.1016/S0140-6736(16)31473-8. [DOI] [PubMed] [Google Scholar]
- 21.Siegfried JM, Farooqui M, Rothenberger NJ, Dacic S, Stabile LP. Interaction between the estrogen receptor and fibroblast growth factor receptor pathways in non-small cell lung cancer. Oncotarget. 2017;8(15):24063–24076. doi: 10.18632/oncotarget.16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka K, Kumano K, Ueno H. Intracellular signals of lung cancer cells as possible therapeutic targets. Cancer Sci. 2015;106(5):489–496. doi: 10.1111/cas.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yip PY. Phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin (PI3 K-Akt-mTOR) signaling pathway in non-small cell lung cancer. Transl Lung Cancer Res. 2015;4(2):165–176. doi: 10.3978/j.issn.2218-6751.2015.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fumarola C, Bonelli MA, Petronini PG, Alfieri RR. Targeting PI3K/AKT/mTOR pathway in non small cell lung cancer. Biochem Pharmacol. 2014;90(3):197–207. doi: 10.1016/j.bcp.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 26.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Wu Y-L, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 27.Castanon E, Rolfo C, Vinal D, Lopez I, Fusco JP, Santisteban M, Martin P, Zubiri L, Echeveste JI, Gil-Bazo I. Impact of epidermal growth factor receptor (EGFR) activating mutations and their targeted treatment in the prognosis of stage IV non-small cell lung cancer (NSCLC) patients harboring liver metastasis. J Transl Med. 2015;13:257–264. doi: 10.1186/s12967-015-0622-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan G, Zhang K, Ding J, Li J. Prognostic value of EGFR and KRAS in circulating tumor DNA in patients with advanced non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget. 2017;8(20):33922–33932. doi: 10.18632/oncotarget.15412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang G, Chen Y, Ju H, Bei F, Li J, Wang J, Sun J, Bu J. Carbamoyl phosphate synthetase 1 deficiency diagnosed by whole-exome sequencing. J Clin Lab Anal. 2018;32(2):e22241. doi: 10.1002/jcla.22241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gulati A, Somlo S. whole-exome sequencing: a state-of-the-art approach for defining (and exploring!) genetic landscapes in pediatric nephrology. Pediatr Nephrol. 2018;33(5):745–761. doi: 10.1007/s00467-017-3698-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


