Abstract
To assess the role of hepatocyte nuclear factor-3β (HNF-3β) in hepatocyte-specific gene transcription, we reported the characterization of the liver phenotype with transgenic mice in which the −3-kb transthyretin (TTR) promoter functioned to increase HNF-3β expression. During breeding of the TTR-HNF-3β transgenic mice we noticed that they displayed severe ataxia. In this study, we describe the analysis of our transgenic cerebellar phenotype and demonstrate that ectopic expression of HNF-3β disrupted cerebellar morphogenesis and caused reduction in cerebellar size. In postnatal cerebellum, the HNF-3β transgene expression pattern is colocalized to glial fibrillary acidic protein-positive cerebellar astrocytes and Bergmann glial cells. As a result of protracted expression, the transgenic cerebella are impaired in terms of astrocyte dispersal and formation of Bergmann glial cell processes. This caused a disruption in neuronal cell migration to the cortical laminar layers and Purkinje dendritic arbor maturation, thus leading to diminished foliation. Differential hybridization of cDNA arrays was used to identify altered expression of cerebellar genes, which is consistent with the observed defect in transgenic cerebellar morphogenesis and size as well as glial maturation. These include diminished expression of the brain lipid-binding protein, which is required for glial morphological differentiation, and the basic helix–loop–helix NeuroD/Beta2 and homeodomain Engrailed-2 transcription factors, which are required for normal cerebellar morphogenesis and foliation. Undetectable levels of ataxia telangiectasia (ATM), which is required for proper development of the Purkinje dendritic arbor, were found in postnatal transgenic cerebella. Furthermore, the transgenic cerebella displayed levels of insulin-like growth factor binding protein-1 elevated to 22 times greater than those measured for wild-type cerebella, an elevation consistent with the reduction in transgenic cerebellar size.
Keywords: Cerebellar expression, Transgenic mice, Winged helix domain, Granule cells, Purkinje cells Astrocytes, Neuronal migration, Transthyretin promoter, Reeler mice
DURING mouse embryogenesis, the cerebellum arises as a very complex outgrowth on the dorsal side of the metencephalon. Proliferating cerebellar neuroepithelial cells migrate from the ventricular zone to populate the deep cerebellar nuclei and Purkinje cell layers (29). Postnatal differentiation of Purkinje cells leads to the development of an extensive dendritic arbor in the molecular cell layer, which is located at the exterior of the adult cerebellum (64). The cerebellum also undergoes considerable postnatal granule neuron migration using a radial glial cell scaffolding (29). Granule cells migrate from the external granule cell layer through the molecular and Purkinje cell layers to the internal granule cell layer. In the molecular layer, parallel fiber axons elongate from granule neurons and synapse with Purkinje dendrites to establish neuronal communication required for cerebellar function. In the adult cerebellar cortex, bushy astrocytes and radial Bergmann glial cells are distributed throughout both the internal granule layer and Purkinje cells, respectively, and these glial processes interact extensively with their respective neuronal synapses (28). During cerebellar development, neuronal cell migration to the cortical laminar layers is guided by radial and Bergmann glial cell processes (29). The cell adhesion receptor systems involved in this neuronal cell migration include the netrin receptors [e.g., rostral cerebellar malformation (rcm)], erbB receptors (e.g., ErbB4), the integrin family (e.g., integrin α5), and the cadhedrin family (53). Recent studies using genetically marked precursor cells have demonstrated that radial glial cells may not only be important for neuronal guidance, but the proliferative radial glial cells can differentiate into neurons (48).
Cellular differentiation results in transcriptional induction of distinct sets of cell-specific genes whose expression is required for organ function. We have utilized the DNA regulatory regions of the transthy-retin (TTR) gene, which encodes the serum and cerebral spinal fluid carrier protein of thyroxine and vitamin A (21), as a model to understand hepatocyte-specific gene transcription (13,14). Functional analysis of the TTR and numerous other regulatory regions of liver-specific genes determined that hepatocyte-specific gene transcription is dependent on recognition of multiple DNA binding sites by distinct families of hepatocyte nuclear factors (HNF) as well as by widely distributed transcription factors (11,15). These studies also revealed that detectable promoter activity required combinatorial interactions among multiple HNF proteins and that this requirement plays an important role in maintaining cell-specific gene expression (13,14,22). The hepatocyte nuclear factor-3α (HNF-3α), -3β, and -3γ proteins were originally identified as mediating transcription of hepatocyte-specific genes (14,41,42) and sharing homology in the winged helix/fork head DNA binding domain (12). The HNF-3/fork head proteins are a growing family of transcription factors that play important roles in cellular proliferation and differentiation (16,39) and have recently been renamed as the Forkhead box (Fox) family (8).
The HNF-3β (also called Foxa2) protein is important not only for hepatocyte-specific gene expression, but also participates in gene regulation in epithelial cells of the esophagus, trachea, lung, stomach, intestine, and pancreas (37,46,54,75). HNF-3β expression initiates during gastrulation in the node, notochord mesoderm, lloorplate neuroepithelium, and in visceral and definitive endoderm (4,46,59,61). Consistent with this expression pattern, Hnf3β–/– mouse embryos die in utero and lack node and notochord, leading to defects in neurotube, somites, and gut endoderm formation (3,69). Moreover, ectopic expression of HNF-3β in the hindbrain/midbrain region of day 8.5 postcoitum (p.c.) mouse embryos mediates the conversion of the dorsal neurotube to lloorplate neuroepithelium. This resulted in severe defects in skull, midbrain, colliculi, and cerebellum formation in transgenic mouse embryos (62).
Because Hnf3β–/– embryos die in utero prior to liver formation, we previously assessed the role of HNF-3β in hepatocyte-specific gene regulation by increasing HNF-3β expression in transgenic mice using the −3-kb TTR promoter region (55). Our studies show that postnatal transgenic mice exhibit liver dysfunction leading to diminished postnatal growth and liver glycogen storage and elevated serum levels of bile acids and bilirubin. These defects are coincident with diminished postnatal expression of hepatocyte genes involved in glucose, bile acid, and bilirubin homeostasis. Furthermore, the TTR-HNF-3β transgenic mice display increased hepatic expression of IGFBP-1, which limits the biological effects of IGFs necessary for postnatal growth. In this study, we describe the characterization of a severe cerebellar phenotype in the TTR-HNF-3β transgenic mice resulting from ectopic expression of the HNF-3β transgene in the developing mouse cerebellum. The HNF-3β trans-gene expression pattern becomes restricted to astrocytes in postnatal cerebellum, which disrupts formation of the radial glial scaffolding and results in abnormal neuronal migration and organization in the transgenic cerebellar cortex. Differential hybridization of cDNA arrays was used to identify altered expression of cerebellar genes, which is consistent with the observed defect in transgenic cerebellar morphogenesis and size.
MATERIALS AND METHODS
Generation of Transgenic Mice
The transthyretin (TTR) minigene construct consists of the –3-kb TTR promoter region, the first and second TTR exons fused to the SV40 3′ end and poly (A) sequences (72). The creation of the TTR-HNF-3β transgenic founder CD-I T-60 and T-77 mouse lines has been described previously (55). Individual founder T-60 and T-77 mice were mated with CD-I wild-type mice to generate Fl transgenic mice, which were used for analysis of the cerebellar phenotype.
In Situ Hybridization and Immunohistochemical Staining
For paraffin wax microtome sections, dissected embryos or brains were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) at 4°C overnight, dehydrated, and embedded in paraffin. Sections of 7 μm were used for in situ hybridization or immunohistochemical staining. For immunohistochemistry, the paraffin wax was removed from sections with xylenes, rehydrated in ethanol, and then placed in PBS plus 0.25% Triton X-100 (PBT). We used the microwave-based antigen retrieval method to enhance antigenic reactivity of antibodies with paraformaldehyde-fixed dewaxed paraffin-embedded sections as described previously (75). For cryostat sections, brains were fixed overnight, washed three times in PBS at room temperature, and incubated overnight in 30% sucrose. Sections of 20 μm were cut in a cryostat and were collected either in PBS for free-floating immunostaining or on Superfrost plus slides (Fisher), air dried, and later rehydrated in Tris-buffed saline (TBS). The sections were blocked with 3% normal serum and incubated with primary antibody diluted in 1% normal serum in TBS overnight at 4°C [primary antibodies for HNF-3β (1:100); calbindin (Sigma, 1:200); GAD67 (Chemicon 1:2000), glial fibrillary acidic protein (GFAP) (Boerhinger Mannheim; 1:4)]. Sections were then washed in TBS three times and incubated with secondary antibody diluted in 1% normal serum in TBS for 1 h at room temperature. Primary antibodies were detected using secondary anti-mouse IgG coupled to FITC (1:100) or anti-rabbit IgG antibodies coupled to TRITC. Labeled sections were viewed and photographed using either a Zeiss microscope for visualization of horseradish peroxidase staining or a Zeiss or Leica laser scanning confocal microscope for double or single immunofluorescent labeling experiments.
The following cDNAs were used as templates for synthesizing [33P]UTP-labeled antisense RNA probes: hindIII digested rat HNF-3β cDNA (1.5 kb) pGEMl plasmid using T7 RNA polymerase; NotI digested mouse rcm cDNA (600 bp) pGEMl plasmid using sP6 RNA polymerase; EcoR1 digested rat reelin cDNA (760 bp) pGEMl plasmid using T7 RNA polymerase; mouse math-1 cDNA (1.4 kb) pBlue-script SK+ (pBS) plasmid using T3 RNA polymerase; an EcoRI linearized GABAA α1 receptor subunit cDNA β04 bp) pGEM-1 plasmid using SP6 RNA polymerase; and a BamHI linearized GABAA α6 receptor subunit cDNA β48 bp) pGEM-1 plasmid using SP6 RNA polymerase (74).
The following antisense RNase protection probes were isolated by RT-PCR of postnatal mouse brain RNA (primer sequences are written in the 5′ to 3′ direction): HindIII digested Integrin α5 template (caccagcagtcagagatggatac and gcccaacgtcttcttccgtct cag) was transcribed with T7 RNA polymerase. EcoR1 digested ErbB4 pBS template (ctgtgtgtgctgaa caatgtgatg and agacaaatgtttggggacactcag) was transcribed by T3 RNA polymerase. Nonmuscle myosin light chain 3 digested pBS template (cgaacaggtgatgg caagatcc and actcgggatacagaatgtctcagc) was transcribed by T7 RNA polymerase. An EcoRI-ClaI fragment close to the 3′ of the thyroid hormone binding protein p55 or protein disulfide isomerase (PDI) (60) cDNA was cloned into pBS vector. The construct was digested with EcoRI and transcribed by T7 RNA polymerase to generate antisense probe. All PCR-generated cDNA probes were verified by DNA sequencing. RNase protection assays and the synthesis of the insulin-like growth factor binding protein 1 (IGFBP-1) RNA probe was described previously (55). In situ hybridization of paraffin-embedded tissue sections was based on a protocol described previously (56). After hybridization, stringent washes, and autoradiography, dark field microscopy was used to visualize the hybridization signals of the expressing cells in the tissues.
Analysis of Cerebellar Postnatal Apoptosis
The number of cells undergoing apoptosis was measured in wild-type and transgenic cerebella of mice at postnatal day 2 (P2), P7, P14, P22, and P56 of age. Apoptosis was measured using the terminal DNA transferase mediated fluorescein-dUTP end labeling (TUNEL) assay with the ApoTag apopotosis kit (Intergen, Purchase, NY) following the manufacturer’s instructions. The apoptotic cells were counted in three 100× cerebellar micrographs, and mean and standard deviation were determined by using the Analysis ToolPak in Microsoft Excel 98.
Mouse cDNA Array Analysis
Radioactively labeled cDNA was synthesized to either wild-type or transgenic mouse cerebellar RNA (P21) and used to hybridize to either the Atlas mouse 1.2 or mouse stress cDNA expression arrays (Clontech, Palo Alto, CA) as described previously (73). For comparison, we hybridized the mouse stress array with cDNA probe from P21 wild-type liver RNA. Total RNA from wild-type or TTR-HNF-3β transgenic cerebellum or from wild-type liver was used to prepare labeled cDNA probe with [32P]dCTP and hybridized to membranes following protocols supplied by the manufacturer (Clontech, Palo Alto, CA). The arrays were then exposed to the phosphorimaging screens for 1 or 2 days and scanned with a storm 840 Phosphorlmager. Following subtraction of background, hybridization signals were normalized to the cytoplasmic β-actin (M12481) and 40S ribosomal protein S29 (L31609) controls. Normalized expression levels from transgenic and wild-type cerebellum cDNA hybridizations were determined by using the Atlaslmage 1.5 program (Clontech).
RESULTS
Ectopic Expression of HNF-3β in the Developing Cerebellum Inhibits Neuronal Cell Migration and Induces Ataxia
In order to assess the role of HNF-3β in regulating hepatocyte genes in vivo, we previously reported on the use of the −3-kb TTR promoter construct (see Fig. 1A) to increase hepatocyte expression of HNF-3β in the developing liver (55). Two transgenic lines (T-77 and T-60) were developed and characterized for defects in liver function and corresponding changes in liver gene transcription (55). The T-77 transgenic mouse line displayed severe liver dysfunction, leading to an increase in serum levels of bile acids and bilirubin, a complete absence of hepatic glycogen storage, and an aberrant increase in hepatic expression of the IGFBP-1 gene (55). In contrast, T-60 transgenic mice lost postnatal hepatic expression of the HNF-3β transgene and displayed only a transient depletion of hepatic glycogen storage and possessed normal serum levels of bilirubin and bile acids.
Figure 1.
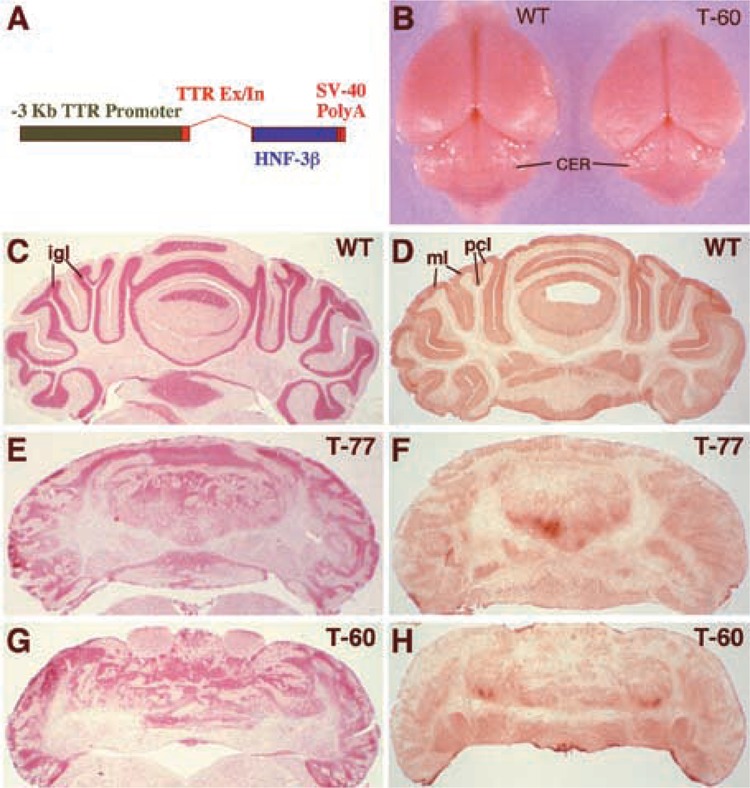
Ectopic expression of HNF-3β in the cerebellum elicits defects in cerebellar size and cellular organization. (A) Diagram representation of the mouse −3-kb transthyretin (TTR) promoter-HNF-3β transgene construct. Transgenic mice were created with the –3-kb TTR promoter region (green box) driving expression of the rat HNF-3β cDNA (blue box), which was cloned into the TTR second exon (red box) (55,72). The creation and characterization of the liver phenotype displayed by the T-60 and T-77 transgenic lines was described previously (55). (B) Cerebellar morphogenesis is disrupted in the TTR-HNF-3β transgenic mice. (C–H) Histological analysis of transgenic cerebellum reveals disruption of the internal granule, Purkinje cell, and molecular layers. Cryostat sections of adult cerebellum sections were stained either with neutral red (C, E, G), which stains all cell bodies including granule neurons and Purkinje cells, or with glutamic acid decarboxyl-ase67 (GAD67) antibody (D, F, H), which stains GABAergic cells. This includes Purkinje cell dendrites and somata organized in the molecular layer and Purkinje cell layers, respectively. Wild-type cerebellum (C, D) shows distinct cellular organization that is lacking in the cerebella of transgenic lines T-77 (E, F) and T-60 (G, H). Abbreviations: igl, internal granule cell layer; pel, Purkinje cell layer; ml, molecular layer.
Surprisingly, we noticed that the Fl heterozygous progeny of both the T-60 and T-77 transgenic mouse lines displayed severe ataxia (a complete absence of motor coordination). The established T-60 and T-77 transgenic lines were used for further characterization of the ataxia phenotype, which was associated with disruption in cerebellar morphogenesis (Fig. 1B) due to ectopic expression of the HNF-3β transgene in the developing cerebellum (see below). This defect was not due to liver dysfunction because the T-60 mice possessed normal liver function with only a transient decrease in hepatic glycogen levels (55). To further characterize the cerebellar defect, distinct cellular layers of the adult cerebellum were visualized by staining the tissue with either neutral red (cell body stain, Fig. 1C, E, G) or glutamate decarboxylase 67 kDa antibody (GAD67, Fig. 1D, F, H), the latter of which stains γ-aminobutyric acid (GABA) containing Purkinje cells and their dendritic arbors (70). In wild-type cerebellum, the neutral red stained the cell bodies of the internal granule cell layer (igl) and the Purkinje cell layer (pcl), highlighting the convoluted wild-type cerebellar cortex (Fig. 1C). Moreover, immunohistochemical staining for GAD67 protein detected the Purkinje cell bodies and the Purkinje cell dendritic arbors in the molecular layer (ml), which synapses with the granule cell parallel fibers as well as a small population of basket and Golgi type II GABAergic interneurons scattered throughout the cerebellum (Fig. 1D). In contrast, histological staining demonstrated that the TTR-HNF-3β transgenic cerebella (T-60 and T-77 lines) displayed a complete disorganization of the molecular, Purkinje cell, and internal granule layers, suggesting defective neuronal migration during cerebellar morphogenesis (Fig. 1E–H). As a result, the tangential growth of the cerebellar cortex was reduced and there was almost no foliation in the transgenic cerebellum (Fig. 1), which is likely due to the inhibition of Purkinje and granule cell migration that is necessary for cerebellar cortex foliation (9).
We next used in situ hybridization to determine the temporal and spatial expression patterns of the HNF-3β transgene in the developing cerebellum. Expression of the HNF-3β transgene was first detected in the proliferating neuroepithelial cells of the cerebellar ventricular zone and in the precerebellar neuroepithelium of 12.5 day p.c. embryos (Fig. 2A–D). Throughout embryonic cerebellar development, transgene expression was confined to the neuroepithelial cells of the cerebellar ventricular zone as well as the cerebellar parenchyma cells that migrated from the ventricular zone (Fig. 2E–H). By contrast, no HNF-3β hybridization signals were detected in non-transgenic cerebella (Fig. 2I–J) or in the external granule cell layer (egl) of the transgenic cerebellum (Fig. 2E–L). Defects in cerebellar morphogenesis were apparent in both transgenic lines by 4 days postnatal (P4), at which time it was noted that the transgenic cerebellum possessed a limited number of folia and appeared to retain an embryonic morphology (Fig. 2J–K). Only the T-60 transgenic mouse brain displayed consistent choroid plexus expression of the HNF-3β transgene, demonstrating that the cerebellar phenotype is not the result of ectopic HNF-3β expression in the choroid plexus (Fig. 2E–H).
Figure 2.
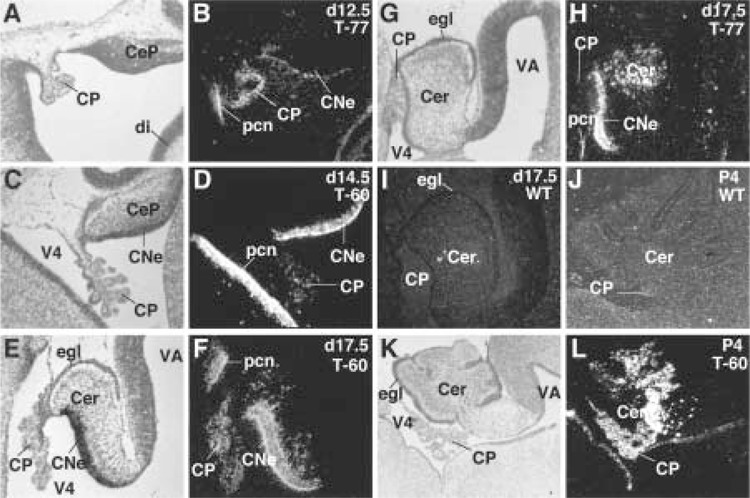
HNF-3β transgene expression in embryonic and postnatal mouse cerebellum. In situ hybridization with [33P]UTP antisense HNF-3β RNA probe was used to examine the expression pattern of the HNF-3β transgene in developing mouse cerebellum. HNF-3β transgene is expressed in the cerebellar and precerebellar neuroepithelium of 12.5 day (A, B), 14.5 day (C, D), and 17 day (E–H) postcoitum (p.c.) transgenic mouse embryos, which are absent in the wild-type cerebellum (I, J). HNF-3β is also observed in the transgenic cerebellar parenchyma at day 17.5 (E–H) and 4 days postnatal (P4) (K, L), which lacks foliation compared to the wild-type P4 cerebellum (J). Consistent choroid plexus expression is only observed in the T-60 transgenic mouse brain. Abbreviations: CNe, cerebellar neuroepithelium; CeP, cerebellar primordium; Cer, cerebellar parenchyma; CP, choroid plexus; egl, external granule cell layer; V4, fourth ventricle; pen, precerebellar neuroepithelium; VA, aqueduct.
The HNF-3β Transgene Is Expressed in the GFAP-Positive Cerebellar Astrocytes and Bergmann Glial Cells
To identify cerebellar cells that express the HNF-3β transgene, we performed indirect double immunofluorescence labeling of postnatal and adult cerebellar sections. We distinguished the different cell types in the postnatal cerebellum using antisera generated against different cell-specific marker proteins, which were detected by immunofluorescence using a FITC (green)-conjugated secondary antibody. Astrocytes and activated Bergmann radial glial cells were detected with the glial fibrillary acidic protein (GFAP) antibody β1) (Fig. 3A–D), and Purkinje cells were detected with the calbindin D-28K antibody (10) (Fig. 3F–G). Nuclear HNF-3β staining was performed with an affinity-purified rabbit antibody generated against the HNF-3β amino-terminus β5) and detected with TRITC (red)-conjugated secondary antibody (Fig. 3B). Double immunofluorescent staining of adult transgenic cerebellum demonstrates that the HNF-3β transgene protein is colocalized with GFAP-positive astrocytes and Bergmann glial cells (Fig. 3C, D), which orchestrate neuronal cell migration during prenatal and postnatal cerebellar development (18,29). Moreover, the HNF-3β transgene protein is expressed throughout the P22 transgenic cerebellum (Fig. 3E), but no endogenous HNF-3β protein is detected in wild-type cerebellum (Fig. 3F). Calbindin D-28K immunostaining of wild-type cerebella highlights the precise alignment of the Purkinje neurons (Fig. 3F), which is completely disrupted in the transgenic cerebella (Fig. 3G). Moreover, HNF-3β staining does not colocalize with calbindin D-28K Purkinje cells in transgenic cerebella (Fig. 3G). In situ hybridization data demonstrate that the external granule cell layer is negative for HNF-3β transgene expression (Fig. 2E–H), suggesting that the granule cells do not ectopically express the HNF-3β gene.
Figure 3.
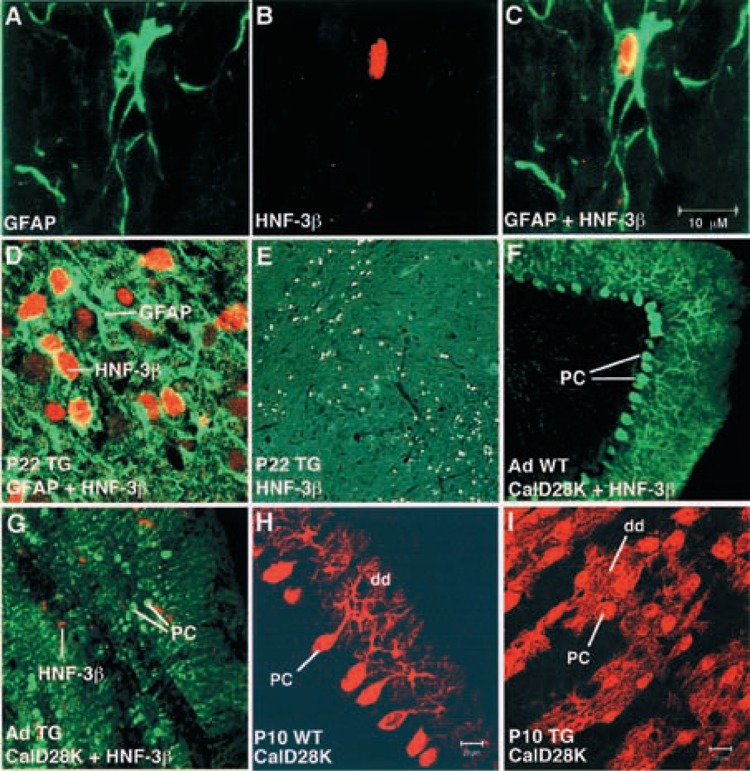
Immunohistochemistry colocalization of the HNF-3β transgene protein with GFAP-positive astrocytes and Bergmann glial cells of adult cerebellum using laser scanning confocal microscopy. HNF-3β transgene-expressing cells are labeled with HNF-3β antibody β5) and detected by immunofluorescence with TRITC (red)-conjugated secondary antibody. Astrocytes and Bergmann glial cells are labeled with glial fibrillary acidic protein (GFAP) antibody (31) and Purkinje cells are labeled with Calbindin D28K (CalD28K) antibody (10), and both are detected by immunofluorescence with FITC (green)-conjugated secondary antibody. (A) Detection of GFAP-positive astrocytes in TTR-HNF-3β transgenic cerebellum. (B) Detection of HNF-3β-positive nuclei in TTR-HNF-3β transgenic cerebellum. (C) Overlay of (A) and (B) showing that HNF-3β transgene protein expression fills the somata of GFAP-positive astrocytes in TTR-HNF-3β transgenic cerebellum. (D) Colocalization of HNF-3β transgene protein with GFAP-positive astrocytes in a region of transgenic cerebellum containing a cluster of astrocytes. Note that similar colocalization results were obtained for both T-60 and T-77 transgenic mouse lines. (E) Low-power magnification (10×) depicting HNF-3β immunohistochemical staining of transgenic cerebellum. (F) Precise organization of Purkinje cells in wild-type cerebellum as detected by immunofluorescence with Calbindin D28K antibody. Note that no HNF-3β protein expression is detected in wild-type cerebellum. (G) HNF-3β transgene protein is not colocalized with Calbindin D28K-positive Purkinje cells and disorganization of Purkinje cells in the transgenic cerebellum. (H) Postnatal wild-type cerebella display normal maturation of dendritic arbor. (I) Postnatal transgenic cerebella possess diminished Purkinje cell dendritic arbor. Immunofluorescence staining of wild-type (H) and transgenic (I) P10 cerebellum with CalD28K antibody was detected by TRITC (red)-conjugated secondary antibody using confocal microscopy. Abbreviations: CalD28K, Calbindin D28K antibody; GDAP, glial fibrillary acidic protein antibody; pc, Purkinje cell; dd, Purkinje cell dendrites.
Defective Astrocyte Organization in Transgenic Cerebellum Is Associated With Inappropriate Migration of Purkinje and Granule Cells
Immunohistochemical staining of cerebellar Purkinje cells with calbindin D-28K antibody (Fig. 4I–L) and hematoxylin/eosin staining (Fig. 4A–D) demonstrated that transgenic cerebella displayed abnormal Purkinje and granule cell organization. Wild-type P7 cerebellar cortex possessed distinct cellular layers, including the external granule cell layer (egl), molecular layer (ml), and internal granule layer (igl) (Fig. 4A, C). Calbindin D-28K antibody staining of wild-type cerebella highlighted precisely aligned cell bodies in the Purkinje cell layer and labeled the branching Purkinje cell dendrites facing toward the cerebellar surface in the molecular layer (Fig. 4I–K and Fig. 3H–I). In contrast, the transgenic cerebellar cortex lacked clearly defined laminar cell layers (Fig. 4B, D), the Purkinje cell bodies were randomly oriented and they displayed immature dendritic arbor (Fig. 4J–L and Fig. 3H–I). Immunostaining of wild-type cerebella with GFAP antibody detected the bushy astrocytes in the internal granule layer and the radial Bergmann glial cell bodies in the Purkinje cell layer, from which they extend radial processes into the molecular layer (Fig. 4E, G). In contrast, the bushy astrocytes and Bergmann glial cells were disorganized in the transgenic cerebella and the Bergmann glial radial processes failed to mature (Fig. 4E–H). Taken together, these data suggest that HNF-3β transgene expression disrupted astrocyte cytoarchitecture and function, which is critical for neuronal migration and terminal differentiation.
Figure 4.
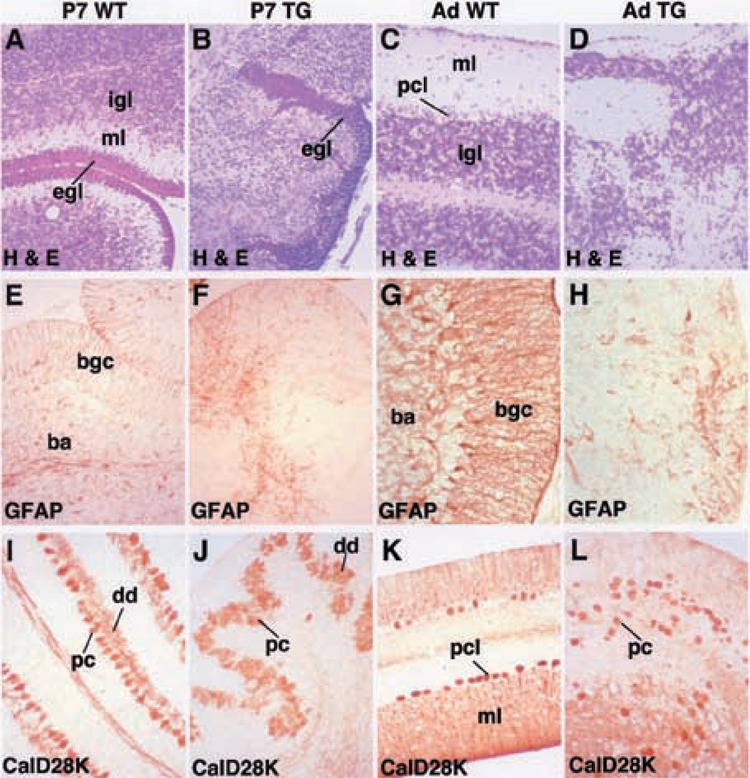
The TTR-HNF-3β transgenic cerebellum exhibits disorganization and maturation of astrocytes, Purkinje cells, and granule cells. Postnatal day 7 (P7; left two columns) or adult cerebella (Ad; right two columns) from either wild-type (WT; A, E, I, C, G, K) or transgenic (TG; B, F, J, D, H, L) mice were stained with either hematoxylin/eosin (H&E; A–D), which stains all the cell bodies, with glial fibrillary acidic protein (GFAP; E–H) antibody used to label astrocytes, or with Calbindin D28K (Cald28k; I–L) antibody to label Purkinje cells. (A–D) Hematoxylin/eosin histological staining of wild-type and transgenic cerebellum. Wild-type cerebellum contains laminar structures including molecular layer (ml), Purkinje cell layer (pel), and internal granule layer (igl), which are absent in transgenic cerebellum. In P7 cerebellum the transient external granule cell layer (egl) is present in both wild-type and transgenic cerebella. (E–H) Immunohistochemical staining of astrocytes with GFAP antibody in transgenic and wild-type cerebella. Altered cellular organization of GFAP-positive bushy astrocytes (ba) and Bergmann glial cells (bgc) in P7 transgenic cerebellum (E, F). In adult transgenic cerebellum, the Bergmann glial cells fail to align in the Purkinje cell layer and extend their processes to the cerebellar surface and bushy astrocytes are not found in the internal granule layer (G, H). (I–L) Immunohistochemical staining of Purkinje cell somata and dendrites with Cald28k antibody in transgenic and wild-type cerebella. Alignment of Purkinje cells is disrupted in both P7 and adult transgenic cerebella and the Purkinje cell dendrites (dd) are stunted.
Induced Expression of IGFBP-1 Expression Correlates With Diminished Size of Transgenic Cerebellum
During the breeding process, we noted that both TTR-HNF-3β transgenic mouse lines weighed significantly less than their postnatal wild-type litter mates (55). RNase protection assays revealed an aberrant 20-fold increase in postnatal hepatic expression of insulin-like growth factor binding protein-1 (IGFBP-1) (55), which limits the biological availability of IGFs required for postnatal growth (26,52). Because there was a significant reduction in transgenic cerebellar size, we examined IGFBP-1 mRNA levels in P8 cerebellum of wild-type and T-77 transgenic mice using RNase protection assays (55). While IGFBP-1 levels were almost undetectable in wild-type P8 cerebella, the T-77 transgenic cerebella displayed a 22-fold increase in IGFBP-1 expression (Fig. 5A), which is similar to that observed with T-77 postnatal liver (55). This result suggests that increased transgenic cerebellar expression of IGFBP-1 may sequester the biological activity of the IGF proteins, which are required for postnatal growth. Consistent with the smaller size of the TTR-HNF-3β cerebellum, transgenic mice studies demonstrated that increased expression of IGFBP-1 in the developing brain results in significant reductions in postnatal brain growth (20).
Figure 5.
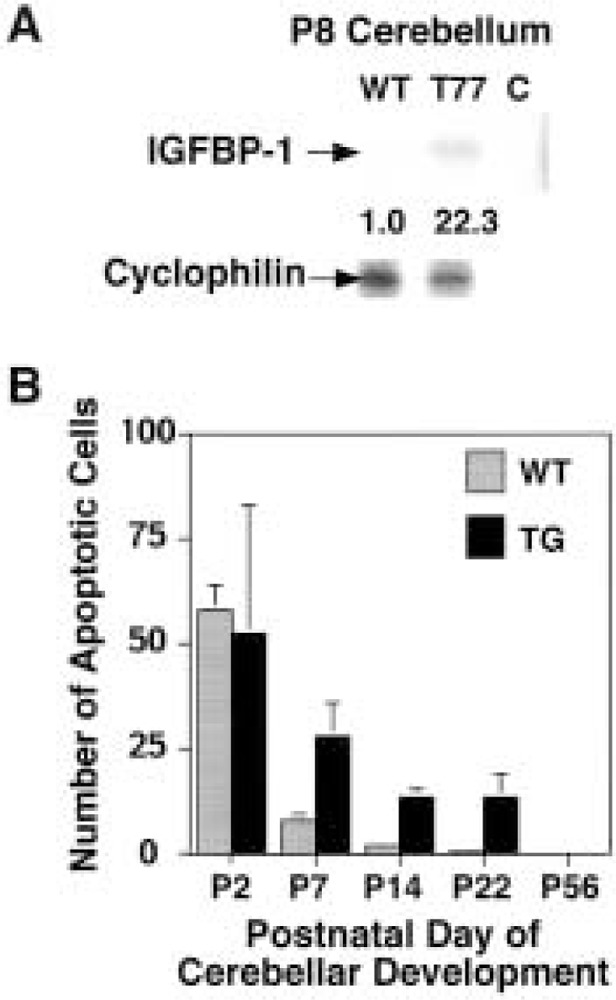
Aberrant expression of IGFBP-1 in TTR-HNF-3β transgenic cerebellum correlates with diminished cerebellar size. (A) RNase protection assay demonstrates aberrant expression of IGFBP-1 in transgenic cerebellum. Total RNA was prepared from postnatal day 8 (P8) wild-type and T-77 transgenic mouse cerebella, and RNase protection assays were used to analyze for expression of insulin-like growth factor binding protein-1 (IGFBP-1) and cyclophilin. Cyclophilin RNase protected band was used as a normalization internal control, and shown are representative RNase protection assays displaying aberrant mRNA levels of IGFBP-1 in transgenic cerebellum. (B) Tunnel assay detects prolonged apoptosis in postnatal transgenic cerebella. We used the TUNEL assay to stain P2, P7, P14, P22, and P56 cerebella from either transgenic or wild-type mice (see Materials and Methods). Graphically shown are the mean and SD of the number of apoptotic cells from three distinct cerebellar micrographs from either transgenic (TG) or wild-type (WT) mice. Transgenic cerebella (P7, P14, and P22) displayed significant increase in apoptosis compared with wild-type cerebella, whereas no significant difference was observed at P2. Note that no apoptosis was detected in transgenic cerebellum from P56 mice.
Because appropriate neuronal cell migration and alignment is required for proper formation of synapses, the transgenic cerebella can be expected to display diminished synaptogenesis, leading to an increase in postnatal apoptosis (71). We therefore used the TUNEL assay to measure cerebellar apoptosis in postnatal wild-type and transgenic mice (see Materials and Methods). Although P2 wild-type and transgenic cerebella demonstrated no significant difference in apoptosis, transgenic cerebella exhibited an aberrant increase in apoptosis between P7 and P22, which subsided by P56 (Fig. 5B). These data indicate that an increase in postnatal apoptosis as well as induced expression of IGFBP-1 is consistent with reduction in transgenic cerebellar size.
Continued Expression of Differentiated Marker Genes in Adult Transgenic Cerebella
We next examined adult wild-type and transgenic cerebella for expression patterns of genes associated with neuronal differentiation (Fig. 6). In wild-type cerebella expression of the GABAA α6 receptor subunit (74) is a marker for mature granule neurons (Fig. 6C), while the GABAA α1 receptor subunit (43) is a marker gene for mature Purkinje cells, granule cells, and deep cerebellar nuclei (Fig. 6E). In situ hybridization of these probes with adult transgenic cerebella highlights abnormal neuronal organization and suggests that their expression does not require appropriate neuronal migration (Fig. 6D, F). Likewise, transgenic cerebella exhibit a diffuse hybridization pattern with either nonmuscle myosin light chain 3 (Fig. 6H) or thyroid hormone binding protein p55 (60), which are differentiated markers for cerebellar glial cells and neurons (Fig. 6G) and glial cells (Fig. 6I), respectively. Taken together, these data suggest that disruption of transgenic cerebellar morphogenesis still allows expression of cell-specific marker genes, but the glial and neurons fail to terminally differentiate and develop their cellular processes.
Figure 6.
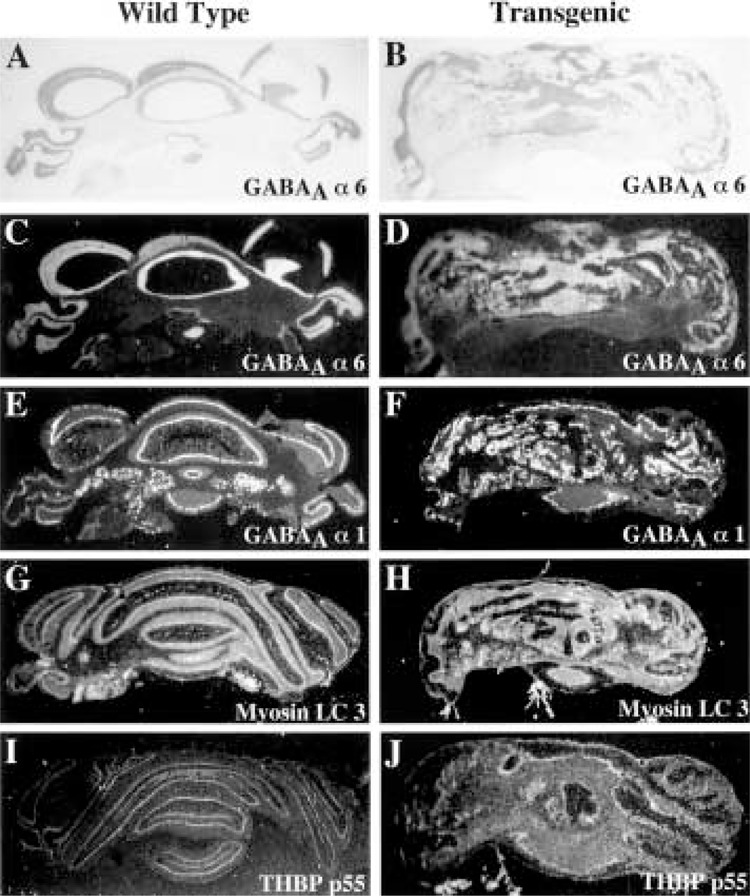
Continued expression of differentiated marker genes in adult TTR-HNF-3β transgenic cerebellum. In situ hybridization of wild-type (left panels) and transgenic (right panels) adult cerebellum with 33P-labeled antisense RNA probes made to various differentiated marker cDNAs. Shown are the bright field microscopy (A, B) and the dark field illumination depicting the hybridization signals (C–J). In situ hybridization of wild-type or transgenic cerebellum with antisense RNA probe made from the GABAA α6 receptor subunit cDNA (C, D), which labels granule neurons (74); the GABAA α1 receptor subunit cDNA (E, F), which labels mature Purkinje and granule cells and deep cerebellar nuclei (43); the nonmuscle myosin light chain 3 (Myosin LC 3) cDNA (G, H), which labels glial cells and neurons (23); and the thyroid hormone binding protein p55 (THBP p55) cDNA (I, J), which is identical to protein disulfide isomerase and is expressed in mature glial cells (60).
Aberrant Expression of Integrin α5 and ErbB4 Receptor Genes in Adult Transgenic Astrocytes
We next determined the expression of astrocyte genes important for neuronal cell migration in adult wild-type (Fig. 7, left panel) and transgenic (Fig. 7, right panel) cerebella by in situ hybridization. The transmembrane protein integrin α5 is transiently expressed in radial glial cells during cerebellar morphogenesis, suggesting its involvement in glial-guided neuronal migration β2). Likewise, Bergmann glial cells transiently express the ErbB4 receptor, which mediates granule cell migration through interaction with the neuregulin protein (49,58). While expression of neither the integrin α5 subunit nor ErbB4 receptor is detectable in wild-type adult cerebellum (Fig. 7C, G), transgenic glial cells display persistent expression of these genes (Fig. 7D, H). These studies demonstrate that the adult transgenic cerebellum continues to express genes involved in neuronal guidance, whose expression is normally restricted to the period of cerebellar morphogenesis.
Figure 7.
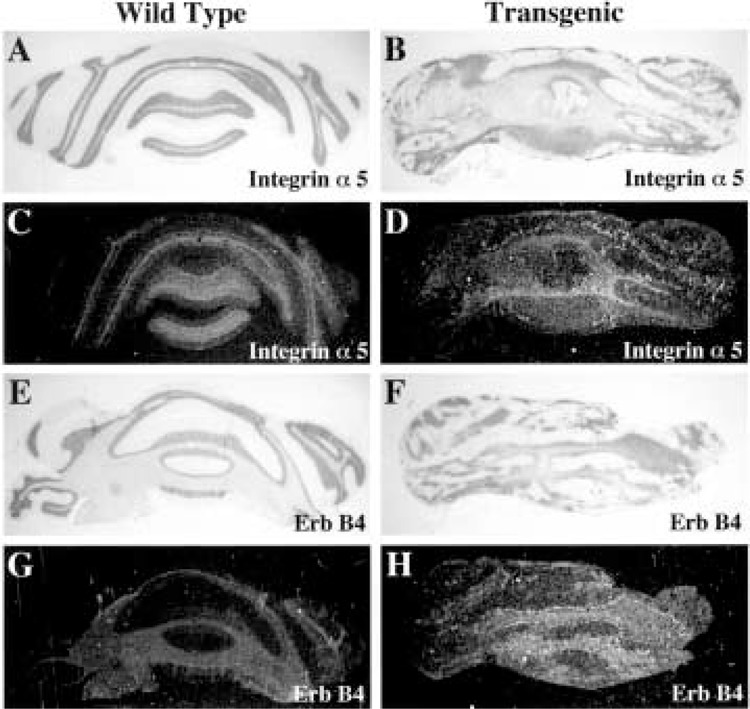
Aberrant expression of neuronal migration genes in adult TTR-HNF-3β transgenic cerebellum. In situ hybridization of wild-type (left panels) and transgenic (right panels) adult cerebellum with 33P-labeled antisense RNA probes made to neuronal cell migration cDNAs, which are exclusively expressed during cerebellar morphogenesis. Shown are the bright field microscopy (A, B, E, F) and the dark field illumination depicting the hybridization signals (C, D, G, H). In situ hybridization of wild-type or transgenic cerebellum with antisense RNA probe made from the integrin α5 subunit cDNA (C, D) and the ErbB4 receptor cDNA (G, H). The integrin α5 subunit is transiently expressed in radial glial cells in developing cerebella, suggesting an involvement in glial-guided neuronal migration β2), and the ErbB4 receptor cDNA is transiently expressed in Bergmann glial cells during granule cell migration and is the ligand for granule cell neuregulin (58).
Cellular Expression Patterns of Math-1, Reelin, and rcm in Neonatal Transgenic Cerebellum
In situ hybridization of P4 transgenic and wild-type cerebella was used to determine the expression patterns of genes involved in neuronal guidance (Fig. 8). The basic helix-loop-helix Math-1 gene is transiently expressed in developing granule neurons (2,7) and stains the external granule cell layer of wild-type and transgenic cerebellum (Fig. 8C–D). We also examined the expression of two neuronal guidance genes, which are dysfunctional in naturally occurring cerebellar mouse mutants and whose precise expression pattern is critical for proper neuronal migration. These are the rostral cerebellar malformation (rcm) gene, which encodes the receptor for the neural guidance netrin protein (1), and the reelin gene, which is an extracellular matrix protein that dictates migration of neurons in the laminated structures including cerebellum (17). As expected, the Rcm probe labeled both the granule and Purkinje cells (Fig. 8E) and the Reelin probe hybridized to the external and internal granule cell layers (Fig. 8G) in wild-type cerebellum (50,63). In contrast, the expression patterns of the rcm and reelin genes were severely altered in the TTR-HNF-3β transgenic cerebella because of disruption in neuronal cell migration and laminar organization (Fig. 8F, H).
Figure 8.
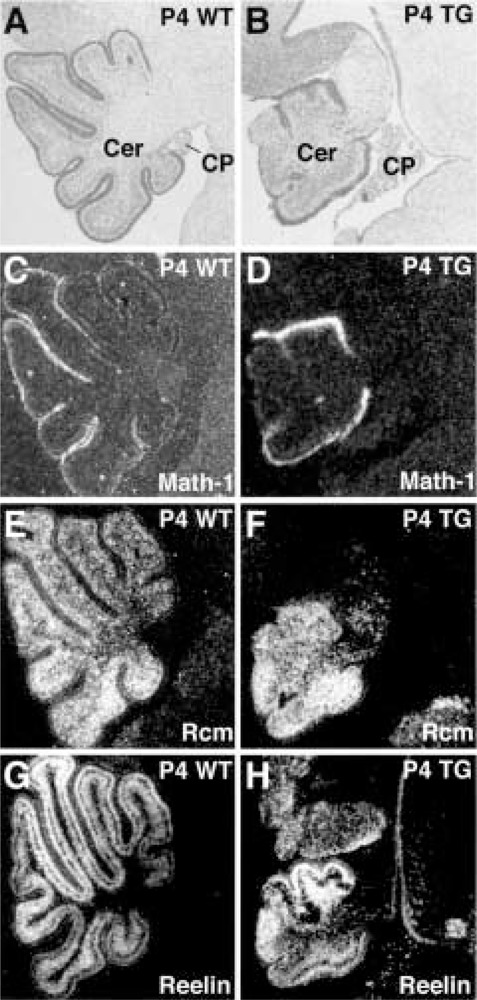
Continued expression of reelin and rcm in neonatal TTR-HNF-3β transgenic cerebellum. In situ hybridization of wild-type (left panels) and transgenic (right panels) P4 cerebellum with 33P-labeled antisense RNA probes made to various cerebellar marker cDNAs and hybridization signals are shown by dark field illumination (C–H). (A, B) Bright field micrographs of wild-type (A) and transgenic (B) P4 mouse cerebellum demonstrating lack of foliation in transgenic cerebellum stained with hematoxylin/eosin. In situ hybridization of wild-type or transgenic cerebellum with antisense RNA probe made from the Math-1 cDNA (C, D), which labels the external granule cell layer (2,7); the rostral cerebellar malformation (Rcm) cDNA (E, F), which labels both the granule and Purkinje cells (1); and Reelin cDNA (G, H), which labels the external and internal granule cell layers (50,63).
Analysis of cDNA Expression Array Identified Altered Expression of Genes That Are Consistent With the Cerebellar Phenotype
Differential hybridization of cDNA arrays was used to identify cerebellar genes whose altered expression is consistent with the transgenic cerebellar phenotype. Radioactive cDNA prepared from either P21 transgenic or wild-type cerebella were hybridized to either the mouse Atlas 1.2 or stress Expression cDNA Array Blots and phosphorimager files were analyzed by the AtlasImage 1.5 program (Clontech; see Materials and Methods). Analysis of the Atlas 1.2 Expression cDNA Array Blot confirms the large stimulation in transgenic cerebellar levels of IGFBP-1 and a smaller increase in IGFBP-2 (Table 1, genes 1–2), which is consistent with reduction in transgenic cerebellar size (19). Coincident with an increase in postnatal apoptosis in transgenic cerebella, we observed an increase in the tumor suppressor p53 and a p53 regulated gene (Table 1, genes 3–4) (68). Transgenic cerebella also display diminished expression of genes, which is consistent with the observed defect in transgenic cerebellar morphogenesis, foliation, and glial maturation (see Discussion section). These includes the basic helix–loop–helix NeuroD and the homeodomain engrailed-2 transcription factors (36,44,45), the brain lipid-binding protein (BLBP) gene (25), and undetectable levels of ataxia telangiectasia (ATM) (8) (Table 1, genes 5–7 and Table 2, gene 10). Moreover, we observed altered expression of genes involved in cellular proliferation, oxidative stress (Table 1, genes 8–13), cell adhesion and migration (Table 1, genes 14–19), ion channels, signal transduction pathways (Table 1, genes 20–22), and mediating an adaptive response to stress (Table 2).
TABLE 1.
ANALYSIS OF MOUSE ATLAS 1.2 cDNA EXPRESSION ARRAY BETWEEN WT AND TG CEREBELLUM
| Gene Name | GenBank | WT | TG | TG/WT |
|---|---|---|---|---|
| 1. Insulin-like growth factor binding protein-1 (IGFBP-1) | X81579 | 5 | 89 | 17.8 |
| 2. IGFBP-2 | X81580 | 58 | 155 | 2.7 |
| 3. Cellular tumor suppressor p53 (TRP53; TP53) | K01700 | 12 | 35 | 2.9 |
| 4. Etoposide induced p53 responsive (EI24) mRNA | U41751 | 11 | 41 | 3.7 |
| 5. Neurogenic differentiation factor (NeuroD; Beta2) | U28068 | 97 | 39 | 0.4 |
| 6. Engrailed-2 protein (En-2) homolog | L12705 | 55 | 30 | 0.5 |
| 7. Brain lipid-binding protein (BLBP) | S69799 | 133 | 60 | 0.5 |
| 8. p27kipl; G1 cyclin-Cdk protein kinase inhibitor | U10440 | 51 | 29 | 0.6 |
| 9. Proliferation-associated protein 1 (PLFAP) | U43918 | 12 | 56 | 4.7 |
| 10. Tyro3 growth factor receptor tyrosine kinase | U18342 | 42 | 83 | 2.0 |
| 11. Glutathione reductase | X76341 | 29 | 91 | 3.1 |
| 12. Microsomal glutathione S-transferase (GST12) | J03752 | 28 | 99 | 3.5 |
| 13. Oxidative stress-induced protein mRNA | U40930 | 24 | 62 | 2.6 |
| 14. Fibronectin 1 (FN1) | X82402 | 75 | 215 | 2.9 |
| 15. Chondroitin sulfate proteoglycan 3 | X84727 | 22 | 71 | 3.2 |
| 16. Plasminogen | J04766 | 55 | 217 | 3.9 |
| 17. Dystroglycan 1 | U43512 | 47 | 92 | 2.0 |
| 18. Neurofibromatosis 1 | X54924 | 60 | 140 | 2.3 |
| 19. Lamimin receptor 1 (LAMR1) | J02870 | 102 | 39 | 0.4 |
| 20. Voltage-gated sodium channel | L36179 | 52 | 22 | 0.4 |
| 21. Potassium large conductance calcium-activated channel | L16912 | 26.0 | 57.0 | 2.2 |
| 22. Calcium/calmodulin-dependent protein kinase IV | X58995 | 356 | 134 | 0.4 |
Radioactive cDNA probes were made to postnatal day 21 (P21) RNA isolated from either wild-type (WT) or T-77 transgenic cerebella (TG; TTR-HNF-3β) and hybridized to the Mouse Atlas cDNA Expression Array Blot (Clontech, Palo Alto, CA), rinsed, and then exposed for phosphorimager analysis. Expression levels of cDNA array were determined by using Atlaslmage program version 1.5 (Clontech) following normalization to β-actin and ribosomal S29 protein (dividing by the average of these control genes) as well as subtraction of background signal. All signals were at least 200% of background levels. Genes in regular type face display increased expression and genes in italic type face exhibit diminished expression in transgenic cerebellum. Abbreviations: Genbank, Genbank Accession Number; TG/WT, ratio of TG signals divided by WT levels.
TABLE 2.
ANALYSIS OF STRESS BLOT cDNA EXPRESSION ARRAY BETWEEN WT AND TG CEREBELLUM
| Gene Name | GenBank | Cerebellum | Liver WT | |
|---|---|---|---|---|
| WT | TG | |||
| 1. XPAC; xeroderma pigmentosum group A correcting protein | X74351 | 27.4 | ND | 13.3 |
| 2. MDR1; P-glycoprotein | M14757 | 36.3 | ND | 15.9 |
| 3. Peroxisome proliferator-activated receptor γ (PPARγ) | U01664 | 35.0 | ND | 22.5 |
| 4. Dioxin-inducible cytochrome P450 | M10021 | 34.6 | ND | 16.9 |
| 5. Cytochrome P450 IIIA (CYP3A13) | X63023 | 27.0 | ND | 36.2 |
| 6. DNA-(apurinic/apyrimidinic) endonuclease (APE/REF-1) | U12273 | 31.8 | ND | ND |
| 7. DNA ligase I | U04674 | 28.5 | ND | ND |
| 8. Proliferation cell nuclear antigen (PCNA) | X53068 | 26.6 | ND | ND |
| 9. Meiotic recombination protein DMC1/LIM15 homolog | D64107 | 29.1 | ND | ND |
| 10. Atm; ataxia telangiectasia | U43678 | 31.4 | ND | ND |
| 11. d-amino acid xxidase | M32299 | 22.1 | ND | ND |
| 12. DNA-repair protein complementing XP-C cells (p125) | U27398 | ND | 25.2 | 20.7 |
| 13. ATP-dependent DNA helicase II; Ku autoantigen protein | X66323 | ND | 23.5 | 15.4 |
| 14. Heme oxygenase 1 | M33203 | ND | 22.9 | 46.2 |
| 15. MAP kinase-activated protein kinase | X76850 | ND | 22.2 | 31.8 |
| 16. Pancreatitis-associated protein 1 precursor | D63359 | ND | 22.1 | 16.5 |
| 17. Cyclooxygenase-1 (COX-1) | M34141 | ND | 27.8 | 29.7 |
| 18. PPAR α | X57638 | ND | 23.8 | 109.7 |
| 19. PPAR β | L28116 | ND | 14.8 | 14.0 |
| 20. Cytochrome P450 16α (testosterone 16-α hydroxylase) | M21855 | ND | 32.4 | 237.2 |
| 21. Cytochrome P450 7B1 (CYP7B1) | U36993 | ND | 25.3 | 26.7 |
| 22. Cytochrome P450 4A12 (CYP4A12) | X71479 | ND | 17.9 | 66.8 |
| 23. Serum paraoxonase/arylesterase 3 | L76193 | ND | 20.3 | 45.2 |
Radioactive cDNA probes were made to postnatal day 21 (P21) RNA isolated from either wild-type (WT) or T-77 transgenic cerebella (TG; TTR-HNF-3β) and hybridized to the Mouse Stress cDNA Expression Array Blot (Clontech, Palo Alto, CA), rinsed, and then exposed for phosphorimager analysis. We also included cDNA synthesized from P21 wild-type mouse liver for hybridization comparison. Expression levels of cDNA array were determined by using Atlaslmage program version 1.5 (Clontech) following normalization to β-actin and subtraction of background signal of the array blot. Genes in regular type face represent genes that are either activated or repressed in transgenic cerebellum similar to their expression pattern in wild-type mouse liver. Genes in italic type face represent those that are repressed in transgenic liver. Abbreviations: Genbank, Genbank Accession Number; ND, not detectable.
DISCUSSION
Mouse genetic studies demonstrate that HNF-3β is required for formation of the notochord and floor-plate neuroepithelium, which provide inductive signaling for patterning of the neurotube (3,69). Misexpression of the HNF-3β transgene in the dorsal midbrain/hindbrain region of the mouse embryo (day 8.5 p.c.) demonstrated that early expression of HNF-3β alters specification of the dorsal neuroepithelial cells (62). In the most severe phenotype, the transgenic mouse embryos are inhibited in formation of the cerebellum, skull, midbrain, and colliculi. In this study, the TTR promoter elicited ectopic HNF-3β expression in the ventricular neuroepithelial cells of the developing cerebellum from day 12.5 p.c, which allowed formation of the cerebellum, but its expression inhibited cerebellar morphogenesis. In both the T-60 and T-77 transgenic lines, ectopic HNF-3β expression was restricted to cerebellar astrocytes, which disrupted formation of the radial glial scaffolding, leading to abnormal neuronal migration to the laminar cell layers. Recent studies using genetically marked precursor cells have demonstrated that radial glial cells may not only be important for neuronal guidance, but the proliferative radial glial cells can differentiate into neurons (48). This cell fate study therefore implies that inhibition of radial glial cell maturation in the transgenic cerebella may also contribute to defective development of cerebellar neurons. Furthermore, we demonstrate that although transgenic glial, granule cell, and Purkinje cell neurons did express appropriate cell-specific marker genes, they failed to terminally differentiate and develop their normal cellular processes. This cerebellar defect was not due to liver dysfunction because the T-60 mice possessed normal liver function with only a transient decrease in hepatic glycogen levels (55). Moreover, ectopic cerebellar expression of the FoxMlB (HFH-11B) transcription factor elicited normal cerebellar development (data not shown) in the TTR-HFH-11B transgenic mice (73), suggesting that the observed cerebellar phenotype is specific to the HNF-3β transgene.
HNF-3β Transgene Expression in Astrocytes and Bergmann Glial Cells Causes Diminished BLBP Expression, Which Is Required for Glial Morphological Differentiation
Ectopic HNF-3β transgene expression in postnatal and adult astrocytes and Bergmann glial cells is associated with abnormal arrangement of both bushy astrocytes in the granule cell layer and alignment of Bergmann glial cell bodies in the Purkinje cell layer (Figs. 3 and 4). Furthermore, Bergmann glial cells failed to assemble into radial glial scaffolding and did not extend their processes to the transgenic cerebellar surface. Because cerebellar astrocytes play an essential role in guiding neuronal migration (18,29), we propose that disorganization of transgenic cerebellar astrocytes results in defective neuronal cell migration to the molecular, Purkinje cell, and internal granule cell layers. Our cDNA array analysis revealed that transgenic cerebella exhibited diminished expression of the brain lipid-binding protein (BLBP) gene (Table 1, gene 7), whose expression is required for maintaining glial differentiation in vitro (25). Antibodies against BLBP were shown to block glial differentiation in response to neurons in mixed primary cultures, but did not disrupt glial-neuronal adhesion (25). Disruption of Bergmann glial processes and their assembly into radial glial scaffolding is associated with diminished transgenic cerebellar expression of BLBP.
Disruption in Bergmann Glial Cell Maturation Inhibits Purkinje Cell Migration and Differentiation
Cerebellar development involves periods of cell proliferation, followed by migration to appropriate cell layers and, ultimately, cells undergo terminal differentiation. The different cell types, including astrocytes, Purkinje cells, and granule cells, were present in the transgenic cerebellum but they were randomly dispersed in the cerebellar cortex, suggesting a defect in neuronal migration to their respective cell layers (Fig. 4). Disruption of cerebellar neuronal cell migration is likely to inhibit molecular layer synaptogenesis between the granule cell parallel fiber axons and the Purkinje cell dendrites. Consistent with these findings, maturation of the Purkinje cell dendritic arbor was reduced in the transgenic cerebellum (Fig. 4), which requires formation of parallel fiber synapses in the molecular layer (27). Impaired migration of neuronal cells is likely contributing to limited foliation of the transgenic cerebella, which requires proper Purkinje and granule cell organization and maturation (27).
In our current study, ectopic expression of HNF-3β in the Bergmann glial cells and astrocytes is paralleled by protracted expression of integrin α5 and ErbB4 receptor genes, which are transiently expressed during radial glial-guided neuronal migration during cerebellar morphogenesis (32,58). The abnormally protracted expression of genes associated with the neuronal migratory period of cerebellar formation supports the hypothesis that transgenic cerebella are maintained in a developmental stage of morphogenesis. Alternatively, increased astrocyte expression of integrin α5 β3 receptor is also found following stroke injury, suggesting the possibility that its elevated expression in transgenic cerebella is a response to glial cell injury or a failure in glial cell maturation (24). Moreover, we observed altered expression of extracellular matrix and cell migration genes (Table 1, genes 14–19), which may be a response to the defect in neuronal migration of the transgenic cerebella.
Diminished Expression of NeuroD, Engrailed-2, and ATM Correlates With Defects in TTR-HNF-3β Cerebellar Morphogenesis, Neuronal Cell Differentiation, and Foliation
Analysis of cDNA expression arrays determined that transgenic cerebella display diminished expression of the basic helix–loop–helix NeuroD and the homeodomain engrailed-2 (En-2) transcription factors (44,45), which are required for proper cerebellar morphogenesis and foliation. Although En-2–/– mice do not exhibit ataxia, they displayed reduction in cerebellar size and a distinct patterning defect of the cerebellar folia (36,44). Reduced expression of En-2 in the TTR-HNF-3β transgenic cerebella may contribute to the observed foliation defects. It is interesting to note that ectopic expression of HNF-3β in the dorsal neurotube of transgenic mice also elicited diminished expression of the En-2 gene in the roof plate of the developing transgenic neurotube (62). Using the insulin-NeuroD transgene to rescue the neonatal diabetes phenotype of NeuroD/Beta2–/– mice, a neuronal deficit in the cerebellar and hippocampus granule cell layers was observed (45). NeuroD/Beta2-deficient cerebella exhibit defects in granule cell differentiation, which is required for their survival. Diminished transgenic cerebellar levels of NeuroD/Beta2 may contribute to defective granule cell differentiation and an increase in postnatal apoptosis.
Undetectable levels of ataxia telangiectasia (ATM), which is required for proper development of the Purkinje dendritic arbor, were found in postnatal transgenic cerebella. This is consistent with the phenotype of ATM-deficient mice, which display ataxia resulting from neuronal degeneration and inhibition of Purkinje cell dendritic arbor maturation (8). Furthermore, we observed repressed levels of the d-amino acid oxidase (DAO) gene, which is expressed in astrocytes and Bergmann glial cells β4). This defect may influence neuronal transmission because DAO is required for degradation of d-amino acids, which may lead to the prolonged stimulation of the N-methyl-d-aspartate (NMDA) receptor (47). Interestingly, treatment of chick embryos with the NMDA receptor antagonist, NPC 12626, resulted in a 19% reduction in Purkinje cell dendritic tree area and a 13% reduction in the number of dendritic branch points (67). The reduction of DAO expression in transgenic cerebellum may therefore exacerbate the defect in Purkinje cell maturation. Furthermore, calcium/calmodulin-dependent protein kinase IV (CaMKIV)-deficient mice exhibited impaired neuronal cAMP-responsive element binding protein (CREB) phosphorylation and CREB transcriptional activation and exhibited defects in cerebellar Purkinje neuronal function (33). A decrease in CaMKIV expression (Table 1, gene 22) may reflect the incomplete maturation of the Purkinje dendritic arbor and diminished neuronal signal transduction in the transgenic cerebellum. Moreover, altered expression of sodium- and calcium-activated channels in transgenic cerebellum may influence neuronal transmission and cerebellar function (40).
Induction of IGFBP-1 Expression Correlates With Reduction in Transgenic Cerebellar Size
We previously reported that the reduction in postnatal growth of the TTR-HNF-3β transgenic mice is associated with a 20-fold stimulation in hepatic expression of the IGFBP-1 gene compared with wild-type littermates (55). Increased hepatic expression of IGFBP-1 in transgenic mice causes significant reductions in postnatal growth, confirming its role in regulating IGF-1 biological activity (26,52). Likewise, we observed a 22-fold induction of cerebellar IGFBP-1 levels as well as a smaller increase in IGFBP-2 expression (Fig. 5, Table 1), which function to limit the local biological activity of IGF growth factors in the brain (19). Consistent with the smaller size of the TTR-HNF-3β cerebellum, transgenic mice that ectopically express IGFBP-1 in the developing brain display a 60% reduction in postnatal brain growth (20). Methylation interference and cotransfection studies demonstrated that the HNF-3β protein specifically binds to and activates expression of the IGFBP-1 promoter region (55,66). Taken together, these data suggest that HNF-3β is also able to activate in vivo transcription of the IGFBP-1 gene in either hepatocytes or glial cells.
We demonstrated that the transgenic cerebella exhibited an increase in postnatal apoptosis compared with wild-type littermates. Analysis of the cDNA arrays indicates that the transgenic cerebella exhibit altered expression of genes involved in DNA repair, oxidative stress, and proliferation, which may increase the incidence of apoptosis (Tables 1 and 2). We observed an increase in transgenic cerebellar levels of the tumor suppressor p53 gene (Table 1), and recent studies suggest that an increase in oxidative stress functions to inhibit Mdm2-mediated degradation of the p53 protein (5). We speculate that increased expression of oxidative stress genes in transgenic cerebella may also function to stabilize p53 protein leading to the observed increase in postnatal apoptosis. Furthermore, we observed undetectable levels of PPARγ, which is critical to inhibit inflammatory mediated neuronal apoptosis (30), in transgenic cerebella (Table 2). Moreover, transgenic cerebella display increased levels of heme oxygenase 1, ATP-dependent DNA helicase II, and lupus Ku autoantigen protein (Tables 1 and 2), all of which are known to contribute to neuronal degeneration and apoptosis (6,51,65).
Comparison of the TTR-HNF-3β Cerebellar Phenotype to Other Naturally Occurring Mouse Models of Ataxia
Insight into the mechanisms regulating cerebellar development has been provided by the molecular analysis of neurological mutant mice with defects in the patterning and histogenesis of the cerebellar cortex (57). Mutations in the rcm gene, which encodes the receptor for the neural guidance netrin protein, result in disorganization of Purkinje and granule cells in the rostral portion of the cerebellum (1). The cerebella of rcm mutant mice are smaller and possess fewer folia than wild-type. The naturally occurring reeler mouse is mutated in the reelin gene display severe ataxia and possesses cerebellar defects, which are similar to those observed with TTR-HNF-3β transgenic mice. Mutations in the reelin gene allow neurons to migrate out of the proliferative zones during cerebellar development, but they do not align correctly into distinct cellular layers and therefore neuronal differentiation occurs in the wrong location (17). Cerebella of reeler mice possess defects in laminar structure and exhibit a significant reduction in the number of cerebellar folia. In a similar manner, TTR-HNF-3β transgenic cerebella continued to express differentiated marker genes for granule and Purkinje neurons including the GABAA α6 and GABAA α1 receptor subunits, suggesting that appropriate neuronal cell migration is not necessary for expression of neuronal differentiation genes (Fig. 6). Likewise, the expression pattern of these GABAA receptor subunit genes was completely altered in the transgenic and reeler cerebella due to the lack of laminar structure. Because our transgenic cerebella continued to express the reelin and rcm genes (Fig. 8), however, it appears that the morphological defects observed in the TTR-HNF-3β transgenic cerebella are caused by mechanisms that differ from these naturally occurring reeler (17) and rcm (1) mouse strains. The defective neuronal cell migration in the TTR-HNF-3β transgenic cerebellum is likely the result of abnormal radial glial and astrocyte organization (18). These transgenic mice will therefore provide us with an experimental cerebellar developmental system in which to identify neuronal genes whose expression is influenced by radial glial-specific interactions during neuronal cell migration.
In summary, we describe the characterization of a severe cerebellar phenotype in the TTR-HNF-3β transgenic mice resulting from ectopic expression of the HNF-3β transgene in the developing mouse cerebellum. The HNF-3β transgene expression pattern becomes restricted to astrocytes in postnatal cerebellum, which disrupts formation of the radial glial scaffolding, resulting in abnormal neuronal migration and organization of the transgenic cerebellar cortex. The transgenic cerebella also display defects in maturation of Purkinje dendritic arbor and extension of the radial glial processes, thus leading to diminished foliation. Differential hybridization of cDNA arrays identified altered expression of cerebellar genes, which is consistent with the observed defect in transgenic cerebellar morphogenesis and size as well as glial maturation. These include diminished expression of the BLBP, which is required for glial morphological differentiation, and the transcription factors NeuroD/Beta2 and Engrailed-2, which are required for normal cerebellar morphogenesis and foliation. Undetectable levels of ATM were found in postnatal transgenic cerebella, which is required for proper development of the Purkinje dendritic arbor. Furthermore, the transgenic cerebella displayed aberrant increased levels of IGFBP-1, which is associated with a reduction in transgenic cerebellar size.
ACKNOWLEDGMENTS
We thank P. Raychaudhuri, V. V. Kalinichenko, Y. Tan, D. R. Grayson, and F. Rausa for critically reading the manuscript, and we thank D. R. Grayson, A. Guidotti, and E. Costa for helpful discussions. We are also grateful to the following investigators for providing various cDNA clones: D. R. Grayson for the rat GABAA α6 and the GABAA α1 receptor sub-unit cDNAs; B. Knowles for the mouse Rcm cDNA; A. Guidotti for the rat Reelin cDNA; M. Takeichi for the N-Cadherin cDNA; and R. Kageyama for the Math-1 cDNA. The University of Illinois at Chicago Research Resource Center provided the use of the confocal microscope. This work was supported by Public Health Service grant ROI GM43241 from the National Institute of General Medical Sciences (R.H.C.).
REFERENCES
- 1. Ackerman S. L.; Kozak L. P.; Przyborski S. A.; Rund L. A.; Boyer B. B.; Knowles B. B. The mouse rostral cerebellar malformation gene encodes an UNC-5-like protein. Nature 386:838–842; 1997. [DOI] [PubMed] [Google Scholar]
- 2. Akazawa C.; Ishibashi M.; Shimizu C.; Nakanishi S.; Kageyama R. A mammalian helix–loop–helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J. Biol. Chem. 270:8730–8738; 1995. [DOI] [PubMed] [Google Scholar]
- 3. Ang S. L.; Rossant J. HNF-3b is essential for node and notochord formation in mouse development. Cell 78:561–574; 1994. [DOI] [PubMed] [Google Scholar]
- 4. Ang S. L.; Wierda A.; Wong D.; Stevens K. A.; Cascio S.; Rossant J.; Zaret K. S. The formation and maintenance of the definitive endoderm lineage in the mouse: Involvement of HNF3/forkhead proteins. Development 119:1301–1315; 1993. [DOI] [PubMed] [Google Scholar]
- 5. Ashcroft M.; Taya Y.; Vousden K. H. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 20:3224–3233; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bakalkin G.; Yakovleva T.; Hurd Y. L.; Nussenzweig A.; Li G. C.; Terenius L. Autoantigen Ku in the brain. Developmentally regulated expression and subcellular localization. Neuroreport 9:2147–2151; 1998. [DOI] [PubMed] [Google Scholar]
- 7. Ben-Arie N.; Bellen H. J.; Armstrong D. L.; McCall A. E.; Gordadze P. R.; Guo Q.; Matzuk M. M.; Zoghbi H. Y. Mathl is essential for genesis of cerebellar granule neurons. Nature 390:169–172; 1997. [DOI] [PubMed] [Google Scholar]
- 8. Borghesani P. R.; Alt F. W.; Bottaro A.; Davidson L.; Aksoy S.; Rathbun G. A.; Roberts T. M.; Swat W.; Segal R. A.; Gu Y. Abnormal development of Purkinje cells and lymphocytes in Atm mutant mice. Proc. Natl. Acad. Sci. USA 97:3336–3341; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caviness V. S. Jr.; Rakic P. Mechanisms of cortical development: A view from mutations in mice. Annu. Rev. Neurosci. 1:297–326; 1978. [DOI] [PubMed] [Google Scholar]
- 10. Celio M. R. Calbindin D-28k and parvalbumin in the rat nervous system. Neuroscience 35:375–475; 1990. [DOI] [PubMed] [Google Scholar]
- 11. Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 10:267–282; 1996. [PubMed] [Google Scholar]
- 12. Clark K. L.; Halay E. D.; Lai E.; Burley S. K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364:412–420; 1993. [DOI] [PubMed] [Google Scholar]
- 13. Costa R. H.; Grayson D. R. Site-directed mutagenesis of hepatocyte nuclear factor (HNF) binding sites in the mouse transthyretin (TTR) promoter reveal synergistic interactions with its enhancer region. Nucleic Acids Res. 19:4139–4145; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Costa R. H.; Grayson D. R.; Darnell J. E. Jr. Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and al-antitrypsin genes. Mol. Cell. Biol. 9:1415–1425; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Costa R. H.; Holterman A.-X.; Rausa F. M.; Adami G. Gene regulation and in vivo function of liver transcription factors. In: Arias I. M.; Boyer J. L.; Chisari F. V.; Fausto N.; Schachter D.; Shafritz D., eds. The liver: Biology and pathobiology. Philadelphia, PA: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 16. Costa R. H.; Kalinichenko V. V.; Lim L. Transcription factors in mouse lung development and function. Am. J. Physiol. Lung Cell. Mol. Physiol. 280:L823–L838; 2000. [DOI] [PubMed] [Google Scholar]
- 17. D’Arcangelo G.; Miao G. G.; Chen S. C.; Soares H. D.; Morgan J. I.; Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 374:719–723; 1995. [DOI] [PubMed] [Google Scholar]
- 18. Delaney C. L.; Brenner M.; Messing A. Conditional ablation of cerebellar astrocytes in postnatal transgenic mice. J. Neurosci. 16:6908–6918; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. D’Ercole A. J.; Ye P.; Calikoglu A. S.; Gutierrez-Ospina G. The role of the insulin-like growth factors in the central nervous system. Mol. Neurobiol. 13: 227–255; 1996. [DOI] [PubMed] [Google Scholar]
- 20. D’Ercole A. J.; Ye P.; Dai Z. Human insulin-like growth factor binding protein-1 (hIGFBP-1) transgenic mice: Insights into hIGFBP-1 regulation and actions. Prog. Growth Factor Res. 6:417–423; 1995. [DOI] [PubMed] [Google Scholar]
- 21. Dickson P. W.; Howlett G. J.; Schreiber G. Rat transthyretin (prealbumin). Molecular cloning, nucleotide sequence, and gene expression in liver and brain. J. Biol. Chem. 260:8214–8219; 1985. [PubMed] [Google Scholar]
- 22. DiPersio C. M.; Jackson D. A.; Zaret K. S. The extracellular matrix coordinately modulates liver transcription factors and hepatocyte morphology. Mol. Cell. Biol. 11:4405–4414; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edelman A. M.; Higgins D. M.; Bowman C. L.; Haber S. N.; Rabin R. A.; Cho-Lee J. Myosin light chain kinase is expressed in neurons and glia: Immunoblotting and immunocytochemical studies. Brain Res. Mol. Brain Res. 14:27–34; 1992. [DOI] [PubMed] [Google Scholar]
- 24. Ellison J. A.; Velier J. J.; Spera P.; Jonak Z. L.; Wang X.; Barone F. C.; Feuerstein G. Z. Osteopontin and its integrin receptor alpha(v)beta3 are upregulated during formation of the glial scar after focal stroke. Stroke 29:1698–1706; 1998. [DOI] [PubMed] [Google Scholar]
- 25. Feng L.; Hatten M. E.; Heintz N. Brain lipid-binding protein (BLBP): A novel signaling system in the developing mammalian CNS. Neuron 12:895–908; 1994. [DOI] [PubMed] [Google Scholar]
- 26. Gay E.; Seurin D.; Babajko S.; Doublier S.; Cazillis M.; Binoux M. Liver-specific expression of human insulin-like growth factor binding protein-1 in transgenic mice: Repercussions on reproduction, ante- and perinatal mortality and postnatal growth. Endocrinology 138:2937–2947; 1997. [DOI] [PubMed] [Google Scholar]
- 27. Goffinet A. M. Determinants of nerve cell patterns during development: A review. Eur. J. Morphol. 28:149–168; 1990. [PubMed] [Google Scholar]
- 28. Grosche J.; Matyash V.; Moller T.; Verkhratsky A.; Reichenbach A.; Kettenmann H. Microdomains for neuron–glia interaction: Parallel fiber signaling to Bergmann glial cells. Nat. Neurosci. 2:139–143; 1999. [DOI] [PubMed] [Google Scholar]
- 29. Hatten M. E. Central nervous system neuronal migration. Annu. Rev. Neurosci. 22:511–539; 1999. [DOI] [PubMed] [Google Scholar]
- 30. Heneka M. T.; Klockgether T.; Feinstein D. L. Peroxisome proliferator-activated receptor-gamma ligands reduce neuronal inducible nitric oxide synthase expression and cell death in vivo. J. Neurosci. 20:6862–6867; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hewicker-Trautwein M.; Trautwein G. An immunohistochemical study of the fetal sheep neocortex and cerebellum with antibodies against nervous system-specific proteins. J. Comp. Pathol. 109:409–421; 1993. [DOI] [PubMed] [Google Scholar]
- 32. Hirsch E.; Gullberg D.; Balzac F.; Altruda F.; Silengo L.; Tarone G. αv integrin subunit is predominantly located in nervous tissue and skeletal muscle during mouse development. Dev. Dyn. 201:108–120; 1994. [DOI] [PubMed] [Google Scholar]
- 33. Ho N.; Liauw J. A.; Blaeser F.; Wei F.; Hanissian S.; Muglia L. M.; Wozniak D. F.; Nardi A.; Arvin K. L.; Holtzman D. M.; Linden D. J.; Zhuo M.; Muglia L. J.; Chatila T. A. Impaired synaptic plasticity and cAMP response element-binding protein activation in Ca2+/calmodulin-dependent protein kinase type IV/Gr-deficient mice. J. Neurosci. 20:6459–6472; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Horiike K.; Tojo H.; Arai R.; Yamano T.; Nozaki M.; Maeda T. Localization of D-amino acid oxidase in Bergmann glial cells and astrocytes of rat cerebellum. Brain Res. Bull. 19:587–596; 1987. [DOI] [PubMed] [Google Scholar]
- 35. Jacob A.; Budhiraja S.; Qian X.; Clevidence D.; Costa R. H.; Reichel R. R. Retinoic acid-mediated activation of HNF-3αa during EC stem cell differentiation. Nucleic Acids Res. 22:2126–2133; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Joyner A. L.; Herrup K.; Auerbach B. A.; Davis C. A.; Rossant J. Subtle cerebellar phenotype in mice homozygous for a targeted deletion of the En-2 homeobox. Science 251:1239–1243; 1991. [DOI] [PubMed] [Google Scholar]
- 37. Kaestner K. H.; Hiemisch H.; Luckow B.; Schutz G. The HNF-3 gene family of transcription factors in mice: Gene structure, cDNA sequence, and mRNA distribution. Genomics 20:377–385; 1994. [DOI] [PubMed] [Google Scholar]
- 38. Kaestner K. H.; Knochel W.; Martinez D. E. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 14:142–146; 2000. [PubMed] [Google Scholar]
- 39. Kaufmann E.; Knochel W. Five years on the wings of fork head. Mech. Dev. 57:3–20; 1996. [DOI] [PubMed] [Google Scholar]
- 40. Kohrman D. C.; Smith M. R.; Goldin A. L.; Harris J.; Meisler M. H. A missense mutation in the sodium channel Scn8a is responsible for cerebellar ataxia in the mouse mutant jolting. J. Neurosci. 16:5993–5999; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lai E.; Prezioso V. R.; Smith E.; Litvin O.; Costa R. H.; Darnell J. E. Jr. HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 4:1427–1436; 1990. [DOI] [PubMed] [Google Scholar]
- 42. Lai E.; Prezioso V. R.; Tao W. F.; Chen W. S.; Darnell J. E. Jr. Hepatocyte nuclear factor 3α belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 5:416–427; 1991. [DOI] [PubMed] [Google Scholar]
- 43. Laurie D. J.; Wisden W.; Seeburg P. H. The distribution of thirteen GABAA receptor subunit mRNAs in the rat brain. III. Embryonic and postnatal development. J. Neurosci. 12:4151–4172; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Millen K. J.; Wurst W.; Herrup K.; Joyner A. L. Abnormal embryonic cerebellar development and patterning of postnatal foliation in two mouse Engrailed-2 mutants. Development 120:695–706; 1994. [DOI] [PubMed] [Google Scholar]
- 45. Miyata T.; Maeda T.; Lee J. E. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 13:1647–1652; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Monaghan A. P.; Kaestner K. H.; Grau E.; Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 α, β and γ genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development 119:567–578; 1993. [DOI] [PubMed] [Google Scholar]
- 47. Mothet J. P.; Parent A. T.; Wolosker H.; Brady R. O. Jr.; Linden D. J.; Ferris C. D.; Rogawski M. A.; Snyder S. H. D-serine is an endogenous ligand for the glycine site of the N-methyl-D-aspartate receptor. Proc. Natl. Acad. Sci. USA 97:4926–4931; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Noctor S. C.; Flint A. C.; Weissman T. A.; Dammerman R. S.; Kriegstein A. R. Neurons derived from radial glial cells establish radial units in neocortex. Nature 409:714–720; 2001. [DOI] [PubMed] [Google Scholar]
- 49. Ozaki M.; Kishigami S.; Yano R. Expression of receptors for neuregulins, ErbB2, ErbB3 and ErbB4, in developing mouse cerebellum. Neurosci. Res. 30:351–354; 1998. [DOI] [PubMed] [Google Scholar]
- 50. Pesold C.; Impagnatiello F.; Pisu M. G.; Uzunov D. P.; Costa E.; Guidotti A.; Caruncho H. J. Reelin is preferentially expressed in neurons synthesizing gamma-aminobutyric acid in cortex and hippocampus of adult rats. Proc. Natl. Acad. Sci. USA 95:3221–3226; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Premkumar D. R.; Smith M. A.; Richey P. L.; Petersen R. B.; Castellani R.; Kutty R. K.; Wiggert B.; Perry G.; Kalaria R. N. Induction of heme oxygenase-1 mRNA and protein in neocortex and cerebral vessels in Alzheimer’s disease. J. Neurochem. 65:1399–1402; 1995. [DOI] [PubMed] [Google Scholar]
- 52. Rajkumar K.; Barron D.; Lewitt M. S.; Murphy L. J. Growth retardation and hyperglycemia in insulin-like growth factor binding protein-1 transgenic mice. Endocrinology 136:4029–4034; 1995. [DOI] [PubMed] [Google Scholar]
- 53. Rakic P.; Cameron R. S.; Komuro H. Recognition, adhesion, transmembrane signaling and cell motility in guided neuronal migration. Curr. Opin. Neurobiol. 4:63–69; 1994. [DOI] [PubMed] [Google Scholar]
- 54. Rausa F.; Samadani U.; Ye H.; Lim L.; Fletcher C. F.; Jenkins N. A.; Copeland N. G.; Costa R. H. The cut-homeodomain transcriptional activator HNF-6 is coexpressed with its target gene HNF-3β in the developing murine liver and pancreas. Dev. Biol. 192:228–246; 1997. [DOI] [PubMed] [Google Scholar]
- 55. Rausa F. M.; Tan Y.; Zhou H.; Yoo K.; Stolz D. B.; Watkins S.; Franks R. R.; Unterman T. G.; Costa R. H. Elevated levels of HNF-3β in mouse hepatocytes influence expression of genes involved in bile acid and glucose homeostasis. Mol. Cell. Biol. 20:8264–8282; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rausa F. M.; Ye H.; Lim L.; Duncan S. A.; Costa R. H. In situ hybridization with 33P-labeled RNA probes for determination of cellular expression patterns of liver transcription factors in mouse embryos. Methods 16:29–41; 1998. [DOI] [PubMed] [Google Scholar]
- 57. Rice D. S.; Curran T. Mutant mice with scrambled brains: Understanding the signaling pathways that control cell positioning in the CNS. Genes Dev. 13:2758–2773; 1999. [DOI] [PubMed] [Google Scholar]
- 58. Rio C.; Rieff H. I.; Qi P.; Corfas G. Neuregulin and erbB receptors play a critical role in neuronal migration. Neuron 19:39–50; 1997. [DOI] [PubMed] [Google Scholar]
- 59. Ruiz i Altaba A.; Prezioso V. R.; Darnell J. E.; Jessell T. M. Sequential expression of HNF-3β and HNF-3α by embryonic organizing centers: The dorsal lip/node, notochord and floor plate. Mech. Dev. 44:91–108; 1993. [DOI] [PubMed] [Google Scholar]
- 60. Safran M.; Leonard J. L. Characterization of a N-bromoacetyl-L-thyroxine affinity-labeled 55-kilodalton protein as protein disulfide isomerase in cultured glial cells. Endocrinology 129:2011–2016; 1991. [DOI] [PubMed] [Google Scholar]
- 61. Sasaki H.; Hogan B. L. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development 118:47–59; 1993. [DOI] [PubMed] [Google Scholar]
- 62. Sasaki H.; Hogan B. L. HNF-3b as a regulator of floor plate development. Cell 76:103–115; 1994. [DOI] [PubMed] [Google Scholar]
- 63. Schiffmann S. N.; Bernier B.; Goffinet A. M. Reelin mRNA expression during mouse brain development. Eur. J. Neurosci. 9:1055–1071; 1997. [DOI] [PubMed] [Google Scholar]
- 64. Takacs J.; Hamori J. Developmental dynamics of Purkinje cells and dendritic spines in rat cerebellar cortex. J. Neurosci. Res. 38:515–530; 1994. [DOI] [PubMed] [Google Scholar]
- 65. Takeda A.; Perry G.; Abraham N. G.; Dwyer B. E.; Kutty R. K.; Laitinen J. T.; Petersen R. B.; Smith M. A. Overexpression of heme oxygenase in neuronal cells, the possible interaction with Tau. J. Biol. Chem. 275:5395–5399; 2000. [DOI] [PubMed] [Google Scholar]
- 66. Unterman T. G.; Fareeduddin A.; Harris M. A.; Goswami R. G.; Porcella A.; Costa R. H.; Lacson R. G. Hepatocyte nuclear factor-3 (HNF-3) binds to the insulin response sequence in the IGF binding protein-1 (IGFBP-1) promoter and enhances promoter function. Biochem. Biophys. Res. Commun. 203:1835–1841; 1994. [DOI] [PubMed] [Google Scholar]
- 67. Vogel M. W.; Prittie J. Purkinje cell dendritic arbors in chick embryos following chronic treatment with an N-methyl-D-aspartate receptor antagonist. J. Neurobiol. 26:537–552; 1995. [DOI] [PubMed] [Google Scholar]
- 68. Vousden K. H. p53. Death star. Cell 103:691–694; 2000. [DOI] [PubMed] [Google Scholar]
- 69. Weinstein D. C.; Ruiz i Altaba A.; Chen W. S.; Hoodless P.; Prezioso V. R.; Jessell T. M.; Darnell J. Jr. The winged-helix transcription factor HNF-3β is required for notochord development in the mouse embryo. Cell 78:575–588; 1994. [DOI] [PubMed] [Google Scholar]
- 70. Willcutts M. D.; Morrison-Bogorad M. Quantitative in situ hybridization analysis of glutamic acid decarboxylase messenger RNA in developing rat cerebellum. Brain Res. Dev. Brain Res. 63:253–264; 1991. [DOI] [PubMed] [Google Scholar]
- 71. Wood K. A.; Dipasquale B.; Youle R. J. In situ labeling of granule cells for apoptosis-associated DNA fragmentation reveals different mechanisms of cell loss in developing cerebellum. Neuron 11:621–632; 1993. [DOI] [PubMed] [Google Scholar]
- 72. Wu H.; Wade M.; Krall L.; Grisham J.; Xiong Y.; Van Dyke T. Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development and regeneration. Genes Dev. 10:245–260; 1996. [DOI] [PubMed] [Google Scholar]
- 73. Ye H.; Holterman A.; Yoo K. W.; Franks R. R.; Costa R. H. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S-phase. Mol. Cell. Biol. 19:8570–8580; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zheng T.; Santi M. R.; Bovolin P.; Marlier L. N.; Grayson D. R. Developmental expression of the a6 GABAA receptor subunit mRNA occurs only after cerebellar granule cell migration. Brain Res. Dev. Brain Res. 75:91–103; 1993. [DOI] [PubMed] [Google Scholar]
- 75. Zhou L.; Lim L.; Costa R. H.; Whitsett J. A. Thyroid transcription factor-1, hepatocyte nuclear factor-3b, surfactant protein B, C, and Clara cell secretory protein in developing mouse lung. J. Histochem. Cytochem. 44:1183–1193; 1996. [DOI] [PubMed] [Google Scholar]


