Abstract
Peroxisome proliferators, which function as peroxisome proliferator-activated receptor-α (PPARα) agonists, are a group of structurally diverse nongenotoxic hepatocarcinogens including the fibrate class of hypolipidemic drugs that induce peroxisome proliferation in liver parenchymal cells. Sustained activation of PPARα by these agents leads to the development of liver tumors in rats and mice. To understand the molecular mechanisms responsible for the pleiotropic effects of these agents, we have utilized the cDNA microarray to generate a molecular portrait of gene expression in the liver of mice treated for 2 weeks with Wy-14,643, a potent peroxisome proliferator. PPARα activation resulted in the stimulation of expression (fourfold or greater) of 36 genes and decreased the expression (fourfold or more decrease) of 671 genes. Enhanced expression of several genes involved in lipid and glucose metabolism and many other genes associated with peroxisome biogenesis, cell surface function, transcription, cell cycle, and apoptosis has been observed. These include: CYP2B9, CYP2B10, monoglyceride lipase, pyruvate dehydrogenase-kinase-4, cell death-inducing DNA-fragmentation factor-α, peroxisomal biogenesis factor 11p, as well as several cell recognition surface proteins including annexin A2, CD24, CD39, lymphocyte antigen 6, and retinoic acid early transcript-γ, among others. Northern blotting of total RNA extracted from the livers of PPARα−/− mice and from mice lacking both PPARα and peroxisomal fatty acyl-CoA oxidase (AOX), that were fed control and Wy-14,643-containing diets for 2 weeks, as well as time course of induction following a single dose of Wy-14,643, revealed that upregulation of genes identified by microarray procedure is dependent upon peroxisome proliferation vis-á-vis PPARα. However, cell death-inducing DNA-fragmentation factor-α mRNA, which is increased in the livers of wild-type mice treated with peroxisome proliferators, was not enhanced in AOX mice with spontaneous peroxisome proliferation. These observations indicate that the activation of PPARα leads to increased and decreased expression of many genes not associated with peroxisomes, and that delayed onset of enhanced expression of some genes may be the result of metabolic events occurring secondary to PPARα activation and alterations in lipid metabolism.
Keywords: Peroxisome proliferator-activated receptor-α (PPARα), Peroxisome proliferator, Liver DNA microarray analysis
PEROXISOMES, cytoplasmic organelles of about 0.5 μm in diameter, surrounded by a membrane, are present in virtually all eukaryotic cells (18,58). These organelles contain more than 60 proteins and participate in several metabolic functions, including simple respiration characterized by H2O2 production and H2O2 degradation, β-oxidative chain shortening of long chain and very long chain fatty acids, metabolism of glyoxalate, degradation of uric acid, and synthesis of ether-phospholipids, cholesterol, and bile acids, among others (45,58). The importance of peroxisomes in biology is further underscored by: i) the identification of several genetic diseases affecting peroxisome biogenesis that lead to disturbances in lipid metabolism, as most of them are lethal during childhood (71), and ii) by the association that sustained increases in peroxisome population in hepatic parenchymal cells, when induced by several structurally diverse nonmutagenic chemicals designated as peroxisome proliferators, lead to the development of liver tumors in rats and mice (55,56,57,72). Peroxisome proliferators form a broad spectrum of compounds of industrial, pharmaceutical, and agricultural importance and include certain phthalate ester plasticizers and herbicides, leukotriene D4 receptor antagonists, and lipid-lowering drugs, such as clofibrate, ciprofibrate, fenofibrate, gemfibrozil, and Wy-14,643, among others (59,72). Despite their structural diversity, peroxisome proliferators induce qualitatively predictable immediate and delayed pleiotropic responses including the development of hepatocellular carcinomas, but the implications to human health of exposure to these agents is an unresolved matter of considerable debate (2,9). Peroxisome proliferator-induced pleiotropic responses are mediated by peroxisome proliferator-activated receptor-a (PPARα) (38), a member of the family of ligand-dependent nuclear transcription factors that regulate the expression of genes associated with lipid metabolism and adipocyte differentiation (19). In the human, mouse, rat, Xenopus, and other species, the PPAR subfamily of nuclear receptors has three isotypes (α, β or 8, and γ) (21,79), which exhibit distinct patterns of tissue distribution and differ considerably in their ligand binding domains and ligand specificities, suggesting that they perform different functions in different cell types and activate different target genes (6,39). The PPARs form heterodimers with another nuclear receptor, the 9-cis-retinoic acid receptor (RXR), and the PPAR/RXR heterodimers bind to DNA sequences, termed PPAR response elements (PPREs), containing direct repeats (DR) of the hexanucleotide sequence AGGTCA separated by one nucleotide, known as a DR-1 response element, present in the 5′-flanking region of target genes (42).
Of the three PPARs, the PPARα isotype is solely responsible for the peroxisome proliferator-induced pleiotropic responses, including hepatic peroxisome proliferation and the development of liver tumors (29,43,57,60). Mice lacking PPARα (PPARα−/−) are refractory to the induction of peroxisome proliferation in liver cells and the PPARα-regulated changes in gene expression by synthetic as well as natural PPARα ligands (33,43). The PPARα-dependent induction of peroxisome proliferation in liver is associated with transcriptional activation of genes encoding for the classical peroxisomal straight chain fatty acid β-oxidation system (3,78), microsomal cytochrome P450 4A isoforms, CYP4A1, and CYP4A3 (40), and some of the genes involved in the mitochondrial P-oxidation (30), among others. The classical peroxisomal β-oxidation system, induced by synthetic peroxisome proliferators, utilizes straight chain fatty acyl-CoAs as substrates, and it consists of three enzymes, namely fatty acyl-CoA oxidase (AOX), enoyl-CoA hydratase/l-3-hydroxyacyl-CoA dehydrogenase (l-bifunctional enzyme (L-PBE), and 3-ketoacyl-CoA thiolase (PTL) (58). Mice deficient in AOX (AOX−/−), the first and rate-limiting enzyme of this inducible peroxisomal β-oxidation pathway, exhibit sustained activation of PPARα, resulting in profound spontaneous peroxisome proliferation in liver cells as well as induction of genes that are regulated by PPARα. As a result of sustained spontaneous hyper-activation of PPARα by its natural ligands, these animals eventually develop liver tumors as do wild-type animals chronically exposed to synthetic peroxisome proliferators (23,24,53,76). These observations imply that AOX is responsible for the metabolic processing of natural PPARα ligands and the unmetabolized substrates of AOX function as endogenous ligands for PPARα, leading to an enzyme receptor cross-talk, because AOX is transcriptionally regulated by this receptor (24,33). The magnitude of peroxisome proliferation occurring spontaneously in the liver of adult AOX mice as a result of activation of PPARα by unmetabolized AOX substrates is comparable with that induced in the liver of wild-type mice exposed to exogenous peroxisome proliferators (23,24,33).
In addition to the key enzymes of the peroxisomal, mitochondrial, and microsomal fatty acid oxidation systems that are markedly upregulated in livers with peroxisome proliferation induced by synthetic and natural PPARα ligands, certain other genes have recently been identified as PPARα targets (35,67). To understand the physiological significance of peroxisome proliferation and the molecular basis of peroxisome proliferation-associated immediate and delayed pleiotropic responses, it is important to identify the full spectrum of target genes that become activated in response to PPARα ligands in the liver. In an effort to identify novel genes expressed in livers with sustained activation of PPARα, we recently used a gene expression screen and identified Ly-6D (lymphocyte antigen 6 complex, locus D), which belongs to a distinctive family of low molecular weight phos-phatidyl inositol anchored cell surface glycoproteins (31), as upregulated in mouse liver with peroxisome proliferation in a PPARα-dependent manner (15). The identification and characterization of differentially expressed genes by cDNA subtraction and other time-consuming methods are not optimally suited for efficient and global identification of changes in gene expression profiles. In an effort to gain appreciation of the extent of changes in gene expression profiles and to identify novel PPARα target genes in the liver, we used the emerging high-throughput cDNA mi-croarray technology to track changes in gene expression levels in mice treated for 2 weeks with Wy-14,643, a synthetic PPARα ligand (11,59). We then determined the expression of the candidate target genes by Northern blotting in livers of AOX−/− mice, and also in wild-type, PPARα−/−, and PPARα−/− AOX−/− double nulls treated with peroxisome proliferators, to ascertain if the observed induction of the novel genes is PPARα dependent. Using this strategy, we have identified several novel PPARα target genes. These include signaling molecules, transcription factors, membrane-associated proteins involved in adhesion, and others. These results not only point to the profound and defined changes in gene expression in livers exposed to both synthetic and biological PPARα ligands, but also provide a baseline for identifying sequential alterations in global gene expression profiles in animals with sustained activation of PPARα and their possible role in hepatocarcinogenesis.
MATERIALS AND METHODS
Animals and Treatment With a Peroxisome Proliferator
Wild-type (C57BL/6J), AOX−/− null mice (23), PPARα −/− null mice (43), and PPARα−/−AOX−/− double null (DKO) mice (33), 3–4 months of age, were used in these studies. They were fed a powdered diet with or without a peroxisome proliferator [Wy-14,643 (0.125%, w/w), nafenopin (0.125%, w/w), or ciprofibrate (0.025%, w/w)] for 2 weeks. For time course of induction, single dose of Wy-14,643 (250 mg/kg body weight) was given by gavage to wild-type mice and they were killed 0, 2, 4, 8, or 16 h after dosing. Control mice received 0.15 ml of dimethyl sulfoxide by gavage, which was used as solvent for the drugs. All animal procedures used in this study were reviewed and approved by the Institutional Review Board for Animal Research of the Northwestern University.
Microarray Analysis
Poly(A)+ selected RNA was isolated using the Oligotex kit (Qiagen, Valencia, CA) from wild-type untreated mice, and wild-type mice treated with Wy-14,643 (0.125%, w/w) for 2 weeks. Poly(A)+ RNA was labeled with Cy3 and Cy5 fluorescent dyes for microarray hybridization on UniGene mouse cDNA microarray (Incyte Genomics, St Louis, MO), as described elsewhere (77). cDNA mouse clones, corresponding to spotted cDNAs on the microarray, were obtained from Incyte Genomics (St. Louis, MO), amplified and sequenced to verify the correct identity of each clone. The data were analyzed using Incyte GEMTools vision 2.4.2a software (Incyte). A total of 6913 genes passed the quality control criteria and of these 713 showed fourfold or greater differential expression, whereas 1656 genes showed twofold or greater differential expression.
Northern Hybridization
Total RNA was isolated from the liver using Trizol reagent (Life Technologies, Carlsbad, CA). After glyoxylation, samples were electrophoresed on 0.8% agarose gel, transferred to nylon membranes, and hybridized at 42°C in 50% formamide hybridization solution using 32P-labeled cDNA probes as described previously (23).
RESULTS AND DISCUSSION
Microarray Analysis of Overall Gene Expression in Wy-14,643-Treated Mouse Liver
Wy-14,643 is a potent hypolipidemic chemical and a hepatic peroxisome proliferator (11,56). It exerts its pleiotropic effects by functioning as a PPARα activating ligand in the liver (38). As a peroxisome proliferator, Wy-14,643 is a member of the carcinogenic nongenotoxic molecules that induce hepatocellular carcinomas in rodents (60). Elucidation of the expression patterns of some genes in livers with peroxisome proliferation and the fact that their expression is PPARα dependent have established a functional role for PPARα in lipid metabolism, atherosclerosis, inflammation, and hepatocarcinogenesis (20,32,57,61,67). Peroxisome proliferation with associated induction of H2O2-generating enzymes and the resulting sustained oxidative stress, as well as subtle increases in cell proliferation in liver, are tightly linked to the development of liver tumors in rats and mice (54,76). While the evidence is overwhelming that downstream events of PPARα activation lead to the development of liver cancer in rodents, the relative roles of the possible, nonexclusive, downstream mechanisms—oxidative stress, cell proliferation, and apoptosis—in the initiation of cancer remain unresolved and controversial given the nongenotoxic nature of these PPARα ligands (29,57). Furthermore, lack of definitive information on the peroxisome proliferator-induced immediate pleiotropic responses, namely the induction of peroxisome proliferation and associated enzymes in human liver following exposure to pharmacologically effective dose levels of the fibrate class of lipid-lowering drugs or exposure to environmental PPARα ligands, is viewed as evidence that the carcinogenic risk for humans to chronic exposure to these potent PPARα ligands may be negligible (2,9,51). The conclusion that humans are insensitive to peroxisome proliferator-induced changes in liver is based on the limited information regarding changes in peroxisome volume densities and expression of PPARα target genes involved in the control of lipid metabolism. To identify genes that are regulated by PPARα ligands, we used the cDNA arrays to survey the expression pattern of a large number of genes in Wy-14,643-treated mouse liver. The microarray used in this study consisted of a total of 7483 unique genes/EST clusters, of which 6499 were annotated (Incyte Genomics). Figure 1 plots the differential mRNA expression with points lying above or below the upper and lower boundaries representing expression in Wy-14,643-treated mouse liver either two- or fourfold higher, or two- or fourfold lower than wild-type untreated control. When twofold change is used as the cut off for either up- or down-regulation, we found that 1656 genes were affected of which 246 were upregulated and 1410 were down-regulated in Wy-14,643-treated livers (Fig. 1A). When fourfold deviation is used, a total of 36 genes were upregulated fourfold or greater, and a total of 671 genes were downregulated fourfold or greater in mice treated with Wy-14,643 for 2 weeks (Fig. 1B). Table 1 lists the transcripts displaying a fourfold or more increase. This group includes nine genes that have previously been shown to be induced in livers with peroxisome proliferation and identified as PPARα regulated: CYP4A1, CYP4A3 (40), AOX (58), acyl-CoA thioesterase (ACTE) (35), CD36 (13), 3-hydroxy-3-methylglutaryl-coenzyme A synthase-2 (HMG-CoA synthase) (62), peroxisomal membrane protein 70 (PMP70) (36), long chain acyl-CoA synthetase (fatty acid transporter 2) (47), and cytosolic epoxide hydrolase (49). The other 27 genes upregulated, fourfold or greater, appear to be novel findings in the Wy-14,643 mouse liver and include some genes with known function and many others whose function is unclear (Table 1). While this microarray screen confirms the upregulation of previously known PPARα-regulated genes present on the array, validating the application of the procedure, this procedure also highlights the limitation because some of the other known, and possibly many unknown, PPARα upregulated genes are not on the microarray. Therefore, the finding of only 27 additional genes as novel most highly upregulated PPARα downstream genes is considered an underestimate. Because a large number of transcripts displayed a fourfold or greater decrease, we chose to list only some of the genes reflecting a fourfold or more decrease (Table 2).
Figure 1.
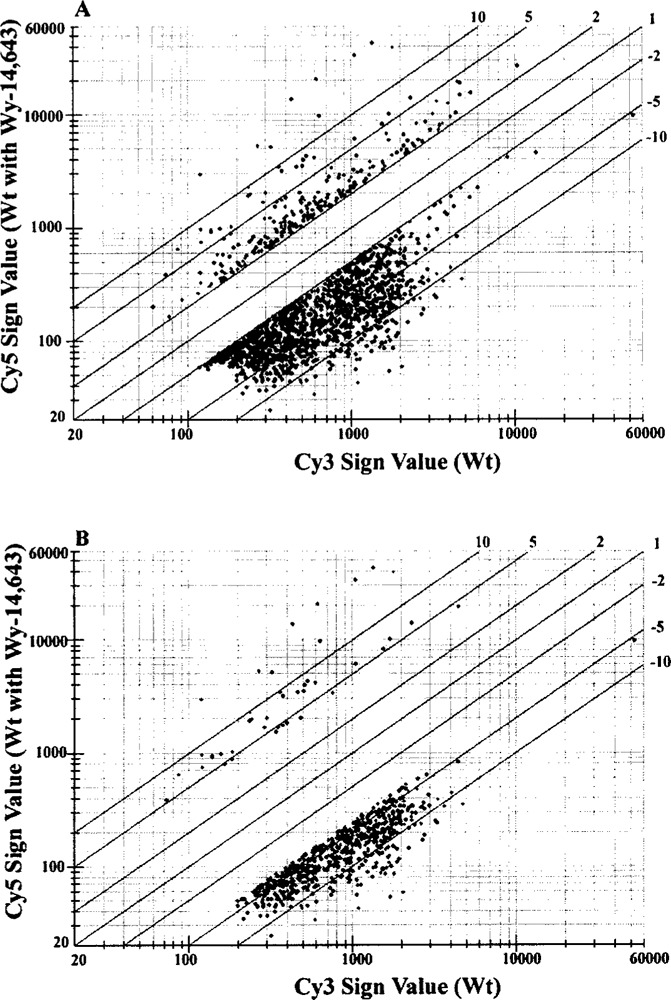
Representative scatter plots of cDNA microarray analysis. Mouse liver treated with Wy-14,643 (0.125%, w/w) for 2 weeks (labeled with Cy5) and liver of untreated wild-type mice (labeled with Cy3) were hybridized to UniGene mouse cDNA microarray (Incyte Genomics). (A) All genes whose expression was changed twofold. (B) All genes whose expression was changed fourfold.
TABLE 1.
GENES CONSISTENTLY UPREGULATED GREATER THAN FOURFOLD IN MOUSE LIVER TREATED WITH Wy-14,643 FOR 2 WEEKS
| Genebank Acc. | Fold Change | Gene Description | Chrom. Mapping | Putative Function |
|---|---|---|---|---|
| AI893661 | 33.4 | CYP 450, 4A3 | 4 | ω-oxidation |
| AA097980 | 31.3 | CYP 450, 4A1 | 4 | ω-oxidation |
| W41358 | 24.7 | cell death-inducing DNA fragmentation factor α | 18 | apoptosis |
| AA458178 | 19.7 | CD36 antigen | 5 | lipid metabolism |
| AA288170 | 16.0 | long chain acyl-CoA thioesterase, peroxisomal | 12 | fatty acid metabolism |
| AA261287 | 15.1 | acyl-CoA oxidase, peroxisomal | 6 | fatty acid metabolism |
| AI451859 | 9.5 | retinoic acid early transcript γ (RAEγ) | — | cell surface protein |
| AA269533 | 9.5 | CYP 450, 2B9, phenobarbitol inducible | 7 | metabolism |
| AI390110 | 8.4 | monoglyceride lipase | 6 | lipid metabolism |
| AI325330 | 8.1 | CYP 450, 2B10, phenobarbitol inducible | 7 | metabolism |
| AA108340 | 8.1 | gene rich cluster, C3f | 6 | unknown |
| AI892192 | 8.0 | pre-B cell leukemia transcription factor 1 | 1 | transcription |
| AA163336 | 7.9 | lymphocyte antigen 6 complex | 15 | cell surface protein |
| AA238875 | 7.7 | alcohol dehydrogenase 3 | 11 | metabolism |
| AI322278 | 7.4 | pyruvate dehydrogenase kinase 4 | 6 | gluconeogenesis |
| AA437485 | 7.3 | ATP binding cassette, subfamily D (PMP70=ABD3) | 3 | peroxisomal biogenesis |
| AA286024 | 7.0 | similar to hypothetical 47.9 kDA protein | — | unknown |
| AA237383 | 6.7 | peroxisomal biogenesis factor 11p (Pex11P) | 3 | peroxisomal biogenesis |
| AA466094 | 6.5 | fat-specific gene 27 | — | unknown |
| W98974 | 6.2 | CD24 antigen | 10 | cell surface protein |
| AA108401 | 6.0 | fatty acid transporter 2 (VLFAacyl-CoA synthetase) | 2 | fatty acid metabolism |
| AA030116 | 5.9 | 3-hydroxy-3-methylglutaryl-CoA synthase 2 | 3 | ketone body synthesis |
| W09719 | 5.7 | Riken cDNA library clone | — | unknown |
| W29607 | 5.7 | dienoyl-CoA isomerase, peroxisomal/mitochondrial | 7 | fatty acid metabolism |
| W89518 | 5.5 | annexin A2 | 9 | cell surface protein |
| AA124202 | 5.3 | epoxide hydrolase 2, cytosolic | 14 | fatty acid metabolism |
| AI894027 | 5.2 | fatty acid binding protein 7, brain | 10 | fatty acid transport |
| AA097860 | 4.8 | CDC-like kinase 2 | 3 | cell cycle |
| W34507 | 4.7 | repeat element-containing transcript 3 | 6 | unknown |
| W97158 | 4.6 | retinoid binding protein 7 (Rbp7) | 5 | unknown |
| AA120757 | 4.4 | CD39 antigen | 12 | cell surface protein |
| AA087542 | 4.2 | similar to hypothetical protein KIAA0281 | — | unknown |
| AA030780 | 4.2 | A3,A2-Enoyl-CoA- isomerase, peroxisomal | — | fatty acid metabolism |
| W34612 | 4.0 | tissue transglutaminase | 2 | metabolism |
| AA276752 | 4.0 | similar to dJ483K16.1 | 9 | unknown |
| W83771 | 4.0 | similar to hypothetical protein C22G7.01C | — | unknown |
Mouse chromosome location of the gene, genebank accession number, and putative function are indicated. —: not known.
TABLE 2.
SELECTION OF GENES CONSISTENTLY DOWNREGULATED GREATER THAN FOURFOLD IN MOUSE LIVER TREATED WITH Wy-14, 643 FOR 2 WEEKS
| Genebank Acc. | Fold Change | Gene Description | Chrom. Mapping | Putative Function |
|---|---|---|---|---|
| AA546085 | −19.1 | antigen-containing epitope to antibody MMS-85/12 | — | unknown |
| AA546673 | −17.0 | MutS homolog 2 (E. coli) | 17 | cell cycle, DNA repair |
| AA387579 | −16.2 | pannexin 1 | — | cell–cell interaction |
| AI509029 | −16.2 | sine oculis-related homeobox 3 homolog | 17 | development |
| AA260001 | −14.7 | Wee 1 homolog (S. pombe) | — | cell cycle |
| AA277571 | −14.0 | polymeric immunoglobin receptor | 1 | apoptosis |
| AA-023077 | −14.0 | hydroxysteroid 11-β dehydrogenase 1 | — | steroid metabolism |
| AA517145 | −13.0 | L-threonine 3-dehydrogenase mRNA | — | metabolism |
| AA207872 | −12.8 | renin 1 | 1 | blood pressure control |
| AA538499 | −12.6 | phosphatidylinositol-4-phosphate 5-kinase, type II, α | — | signal transduction |
| AA268683 | −12.1 | kinesin-associated protein 3 | — | cytoskeletal |
| AA517717 | −11.6 | ankyrin repeat hooked to zinc finger motif | — | transcription |
| AA253725 | −11.4 | neurofilament-L | 14 | cytoskeletal |
| AA432951 | −11.3 | myoblastosis oncogene-like 2 | 2 | cell cycle |
| AA009039 | −10.6 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase | 11 | metabolism |
| AA387076 | −10.6 | testis-specific gene A 12 | 10 | unknown |
| AA426866 | −9.8 | serine/threonine kinase 18 | 13 | signal transduction |
| AA030546 | −9.6 | insulin-like growth factor 2 | 7 | cell growth |
| W45834 | −9.5 | proteoglycan 3 | 2 | extracellular matrix |
| AA020100 | −9.5 | tissue factor pathway inhibitor | — | blood coagulation |
| AA426845 | −8.5 | SRY-box containing gene 15 | 11 | testis determination factor |
| AI327515 | −8.4 | excision repair 1 | 7 | transcription/replication |
| AA422377 | −8.4 | growth differentiation factor 9 | 11 | cell growth |
| AA545607 | −8.4 | metal response element binding transcription factor 2 | — | transcription |
| AA162875 | −7.5 | CD53 antigen | 3 | apoptosis |
| AA270669 | −7.3 | laminin, alpha 4 | 10 | cytoskeletal |
| AA387258 | −7.2 | myosin IB | 1 | cytoskeletal |
| AA542013 | −6.8 | fibroblast growth factor receptor 1 | 8 | cell growth |
| AA473938 | −6.7 | CCAAT/enhancer binding protein alpha (C/EBP) | 17 | transcription |
| AA414632 | −6.7 | cyclin-dependent kinase like 2 | 5 | cell cycle |
| W55655 | −6.6 | myeloperoxidase | 11 | inflammation |
| W83974 | −6.3 | carboxypeptidase E | 8 | metabolism |
| AA285921 | −5.5 | major urinary protein 2 | — | pheromone |
| W89253 | −4.9 | insulin-like growth factor binding protein 5 | 1 | cell growth |
| AA274685 | −4.6 | hydroxysteroid dehydrogenase Δ5,3-β | 3 | steroid metabolism |
| AI326579 | −4.0 | nuclear receptor coactivator 1 | 12 | transcription |
Mouse chromosome location of the gene, Genebank accession number, and putative function are indicated. —: not known.
Confirmation of Inducibility of Genes Detected by cDNA Array
We validated the upregulation of some of the known PPARα responsive genes and many other genes that are hitherto not known to be regulated by PPARα (Table 1) at the mRNA level by using Northern analysis, and by in situ hybridization in some cases (data not shown). For Northern analysis we isolated total RNA from the livers of wild-type, AOX−/−, PPARα−/−, and PPARα−/− AOX−/− double null (DKO) mice maintained on normal diet or on Wy-14,643-containing diet for 2 weeks (Fig. 2). As expected, CYP4A1, CYP4A3, AOX, L-PBE, PTL, macrophage scavenger receptor CD36, ACTE, and Ly6-D mRNA levels were upregulated in the livers of wild-type mice fed Wy-14,643, a synthetic PPARα ligand. Of the 27 novel genes identified as upregulated in the Wy-14,643-treated livers, we validated the increases in the levels of mRNAs of seven genes by Northern analysis (Fig. 2). These include cell death-inducing DNA fragmentation factor a (CIDE) (37), Riken cDNA clone W09719 (8), pyruvate dehydrogenase kinase protein (PDK-4) (73), monoglyceride lipase (mLipase) (41), retinoic acid early transcript γ(RAEγ) (80), gene rich cluster C3f (1), Rbp7 (a member of the cellular retinoid-binding protein family) (14), CD39 (22), and CD24 (74). The other 20 novel genes found to be upregulated by microarray technique have not been validated by Northern blotting or by quantitative reverse transcriptase PCR, but the intensity of signals in Northern blots of genes evaluated thus far confirms the dynamic range of fold increases seen in microarray results, suggesting the reliability of changes in the upregulation of genes observed in the microarray. All these genes were also upregulated in AOX−/− mouse liver with spontaneous peroxisome proliferation, with the exception of AOX (because of null mutation of this gene in this mouse) and CIDE (Fig. 2). We also assessed the inducibility of these genes by two other synthetic peroxisome proliferators, namely ciprofibrate and nafenopin. Wild-type mice fed a powdered diet containing ciprofibrate (0.025%, w/w) or nafenopin (0.125%, w/ w), ad libitum for 2 weeks (Fig. 3), revealed increases in mRNA levels similar to those noted in the liver of Wy-14,643-fed mice. These observations support the view that the expression of genes identified as upregulated in the microarray parallels peroxisome proliferation in liver.
Figure 2.
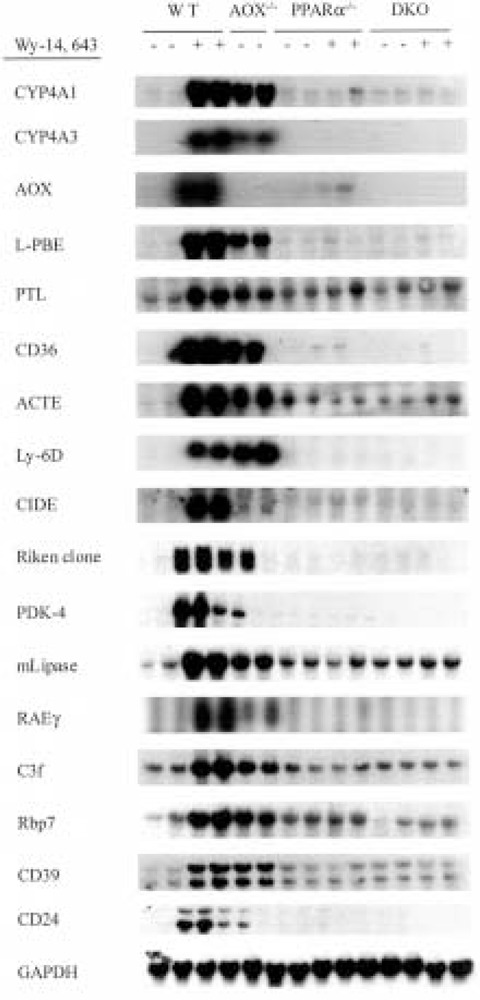
Confirmation of the microarray results by Northern blot analysis with total RNA isolated from livers of wild-type and different knockout mice. Total RNA (20 μg) from wild-type mice (lanes 1–4), AOX−/− mice (lanes 5 and 6), PPARα−/− (lanes 7–10), and PPARα−/− AOX−/− double null mice (lanes 11–14) untreated (–) or treated with Wy-14,643 (+) was hybridized with 32P-labeled cDNA probes as indicated. GAPDH was used as loading indicator.
Figure 3.
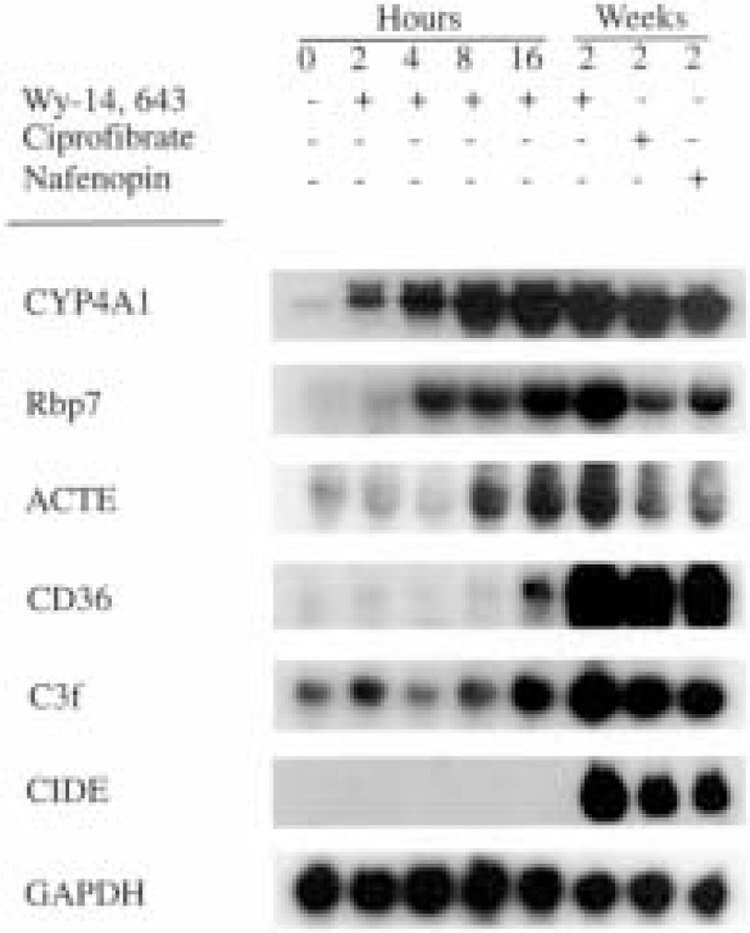
Time course of induction. Northern blot analysis of total RNA (20 μg) isolated from livers of wild-type mice administered Wy-14,643 in a single intragastric dose (250 mg/kg body weight). Animals were killed at 0 h (lane 1), 2 h (lane 2), 4 h (lane 3), 8 h (lane 4), and 16 h (lane 5) after gavage. Lanes 6–8 represent RNAs isolated from the liver of wild-type animals fed with Wy-14,643 (0.125%, w/w), ciprofibrate (0.025%, w/w), and nafenopin (0.125%, w/w) for 2 weeks. Northern blots were hybridized with 32P-labeled cDNA probes as indicated. GAPDH was used as loading indicator.
PPARα Dependency of the Inducible Genes
Northern blot data presented in Figures 1 and 3 strongly suggest that the upregulation of genes in livers with peroxisome proliferation is under the control of PPARα. To assess the role of PPARα in the induction of these genes, we analyzed total RNA extracted from the livers of PPARα−/− and DKO mice fed control and Wy-14,643-containing diets for 2 weeks by Northern blotting for the expression of selected upregulated genes. As illustrated in Figure 2, mRNA levels of all genes evaluated by Northern blotting showed no increases in the livers of PPARα−/− and DKO mice fed Wy-14,643 for 2 weeks. These data imply that the upregulation of genes identified by microarray procedure is dependent upon peroxisome proliferation vis-à-vis PPARα activation, with the exception of CIDE mRNA, which is increased in the livers of wild-type mice treated with peroxisome proliferators but not prominently in AOX−/− mouse liver with spontaneous peroxisome proliferation (Figs. 2 and 3). To further examine the role of PPARα, we have evaluated the time course of induction of some of these genes by Northern blotting of total RNA extracted from the livers of wild-type mice that were killed at 2, 4, 8, and 16 h after they were given a single intragastric dose of Wy-14,643 (Fig. 3). Time-dependent increases in mRNA levels of CYP4A1, Rbp7, ACTE, C3f, and CD36 were evident and the apparent lag in the induction of some genes when compared to CYP4A1 most likely reflects the relatively marginal basal levels of their expression. No increase in CIDE mRNA level was detected even at 16 h after dosing (Fig. 3). The failure to observe an increase by 16 h after Wy-14,643 administration and the relative noninduction of this gene in AOX livers strongly suggest that CIDE gene may not be directly activated by PPARα dependence and may reflect delayed/secondary alterations of these synthetic peroxisome proliferators (Fig. 4).
Figure 4.
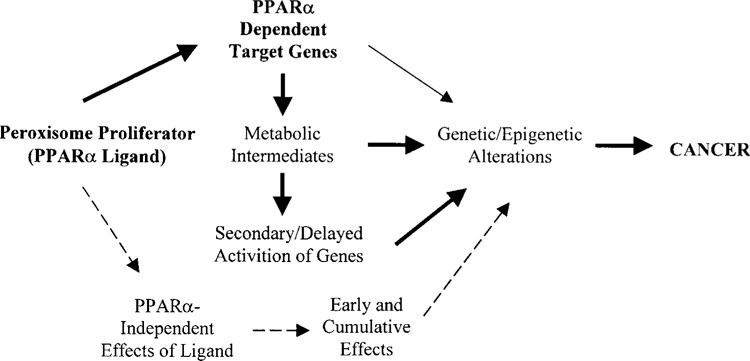
Model of gene expression alterations occurring in the liver following exposure to a peroxisome proliferator (PPARα). PPARα-mediated and PPARα-independent effects of ligand can be acute and sustained as long as the ligand is present. Delayed onset of gene expression patterns may be secondary to sustained PPARα activation and the resulting metabolic intermediates (e.g., reactive action intermediates). Sequential determination of gene expression profiles during chronic exposure to peroxisome proliferators should provide clues regarding the primary and secondary genetic alterations.
Functional Relevance of Inducible Genes
Energy Metabolism
Microarray data showed that the expression of several genes involved in different steps of lipid and glucose metabolism is increased by treatment with Wy-14,643 (Table 1). Sixteen of the 36 genes upregulated fourfold or greater in the liver of Wy-14,643-treated mouse participate in lipid metabolism. Nine of these 16 genes involved in lipid metabolism have been described previously to be associated with PPARα-dependent peroxisome proliferation and include AOX (23), L-PBE (3), ACTE (35), CYP4A1 and CYP4A3 (40), HMG-CoA synthase (62), very long chain fatty acyl-CoA synthetase (47), CD36 (25), and epoxide hydrolase (49). It is pertinent to note that increases in the expression of 7 of these 16 genes have been identified for the first time to be associated with peroxisome proliferation.
The fatty acid ω-oxidation is increased in Wy-14,643-treated liver, as evidenced by the induction of CYP4A1 and CYP4A3 mRNAs. Beside these two proteins, two novel cytochrome family members, CYP2B9 (9.5-fold), and a CYP2B10 (8.1-fold) are also induced by Wy-14,643 treatment. CYP2B10 presents a high homology with NADPH cytochrome P450 oxidoreductase (16) and has been associated with regenerative changes in hepatocyte morphology (52). This is in line with increases in the classical peroxisomal β-oxidation system, as demonstrated by sixfold induction of very long chain acyl-CoA synthetase (fatty acid transporter 2) involved in the activation of fatty acids to their CoA esters and present on peroxisomal membrane (50), and 15-fold increase in AOX mRNA, encoding the first and rate-limiting enzyme of peroxisomal β-oxidation system (58). Increase in Δ3, Δ2-enoyl-CoA isomerase points to an increase in the peroxisomal degradation of polyunsaturated fatty acids. Fatty acid translocase (FAT/CD36), a scavenger receptor that is known to interact with a large variety of ligands, including oxidized low-density lipoproteins and also to function as a putative long chain fatty acid transporter (25), is also upregulated ∼ 20-fold following treatment with Wy-14,643. Activation of PPARα promotes foam cell formation by increasing CD36 expression in macrophages (13), and an increase in CD36 expression in liver possibly reflects enhanced hepatic fatty acid uptake of triglyceride-rich very low-density lipoproteins following exposure to hypolipidemic drug Wy-14,643. Combined action of two lipases, namely hormone-lipoprotein lipase and mLipase, is required for the hydrolysis of triglycerides (41). Enhanced expression of mLipase in the liver of Wy-14,643-treated mouse observed in this study suggests that this enzyme catalyzes the degradation of monoglycerides to fatty acids and glycerol and fatty acids liberated then serve as substrates for the β-oxidation in peroxisomes and mitochondria and ω-oxidation in microsomes (58). Consequence of the induction of β-oxidation is an increased intracellular pool of acetyl-CoA, which can be metabolized by 3-HMG-CoA synthase (48). A nearly sixfold increase in 3-HMG-CoA synthase and ∼ 16-fold increase in ACTE have been found in Wy-14,643-treated livers. ACTE cleaves CoA thioester bonds of acteyl-CoA, regenerating cellular CoA and releasing acetyl molecules, which are then condensed by 3-HMG-CoA synthase (47). Increases in these two enzymes can account for the clearance of acetyl-CoA, the end product of fatty acid oxidation, as acetoacetate via the ketone body pathway.
There is an increasing recognition that PPARα regulates glucose metabolism (32,63). Wy-14,643 increased the expression of PDK4 in liver by 7.4-fold. This enzyme phosphorylates and inactivates the pyruvate dehydrogenase complex, which is essential to initiate the catalysis of the irreversible oxidative glucose metabolism. Increases in hepatic PDK4 would result in decreases in glucose oxidation in this organ with PPARα-mediated peroxisome proliferation. This is in contrast to the decreased expression of PDK4 in skeletal muscle following activation of PPARγ by its agonists (73). These observations suggest that activation of PPARα in liver and of PPARγ in skeletal muscle by their respective ligands would result in opposite effects in these target tissues. Consistent with these observations is the marked downregulation of 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD) gene expression in the liver of mice treated for 2 weeks with Wy-14,643 (Table 2). Downregulation of 11β-HSD would lead to decreased levels of functionally active cortisol in liver, resulting in decreased hepatic gluconeogenesis and reduced serum glucose (5,34,66). The hepatic glycogen content in AOX−/− mice that exhibit spontaneous PPARα activation and in wild-type mice fed a PPARα ligand is lower than that seen in wild-type mice, suggesting that PPARα activation leads to a reduction in hepatic glycogen content (32). The reduction in 11β-HSD level observed in this study is consistent with these observations.
Peroxisome Biogenesis
Besides the genes involved in lipid and glucose metabolism, peroxisome proliferators are also known to induce several membrane proteins, which participate in peroxisome biogenesis. One of these is PMP70, a member of the ATP-binding cassette family (36). We noted increases in the mRNA levels of PMP70 (∼7.3-fold increase) and peroxisomal biogenesis factor 11β (PEX11β) (∼6.7-fold increase) in this study. PEX11 encodes the peroxisomal membrane protein Pex11p, which appears to be required for medium chain fatty acid β -oxidation in peroxisomes and also in the fission of larger peroxisomes into smaller ones (70). It appears that Pex11α is involved either directly or indirectly in directing the entry of medium chain fatty acids into the peroxiosme matrix (70). While Pex11α isoform regulates peroxisome abundance in response to extracellular stimuli, PEX11β has been to be sufficient to induce peroxisome proliferation in the absence of extracellular stimuli (64).
Cell Surface Marker Proteins
Phenotypic changes in cells often parallel altered expression patterns of various genes, some of them expressed only under special conditions like proliferation, differentiation, or neoplastic conversion. The identification of such genes and evaluation of possible consequences can serve as a baseline for defining the phenotype of cells. cDNA microarry data revealed for the first time that several mRNAs encoding liver cell surface marker proteins such as annexin A2 (26), CD24 (74), CD39 (22), Ly-6D (15), and RAEγ (80) are markedly increased in liver with peroxisome proliferation (Table 1). Annexin A2 is a Ca2+/phospholipid-binding membrane-associated protein, which is a tyrosine-kinase substrate (26). It has been suggested that overex-pression of annexin plays a role in metastatic progression and is associated with stress response and cellular proliferation (68). CD24, also called heat-stable antigen, is a differentiation marker generally expressed in hematopoietic tissues, and has recently been shown to be expressed in human breast carcinoma (27,74). CD39 (ecto-ATP diphosphohydrolase), which hydrolyzes the extracellular ATP and ADP, is of critical importance in homotypic adhesion and platelet aggregation and is overexpressed in differentiated human melanomas (22). Lymphocyte antigen 6 complex, locus D (Ly-6D; mouse ThB) is a glycosylphosphatidylinositol (GPI)-anchored cell surface glycoprotein that is highly expressed in tumor cells of high malignancy (75). The human homolog of Ly-6D, designated as E48, is expressed in squamous carcinoma and plays a role in keratinocyte cell-cell adhesion (7). RAEγ, also a GPI-anchored cell surface protein, which shares weak homology to the major histocompatibility complex class I, is detected in early embryos and not in adult mouse tissues (10). RAEγ is inducible in F9 mouse embryonal carcinoma cells by retinoic acid (80). We found an increase in RAEγ expression in Wy-14,643-treated liver. Finally, CD36, a transmembrane glycoprotein involved in lipid metabolism, which is also increased in Wy-14,643-treated mouse liver, is known to interact with integrins in human melanoma cells (69). These studies clearly establish that sustained increases in peroxisome volume density in liver are associated with upregulation of several cell surface marker proteins. Although enhanced expression of some of these genes was not observed in the livers of Wy-14,643-treated PPARα−/− mice, implying an obligatory role of PPARα. Additional studies are needed to ascertain if these alterations are mediated by PPARα, or occur secondarily in livers with sustained peroxisome proliferation and establish the role of these changes, if any, in liver tumorigenesis.
Transcription, Cell Cycle, and Apoptosis
Peroxisome proliferators are known to cause increased DNA synthesis and have been demonstrated to suppress apoptosis in hepatocytes (4,28,65). The cell death-inducing fragmentation factor (CIDE) is induced in Wy-14,643-treated mouse liver but not in AOX null mouse liver, suggesting that its induction may not be dependent upon PPARα (Fig. 2). Microarray data also show that fat-specific gene 27 (FSP27), which is highly homologous to CIDE-A, is also upregulated (Table 1). CIDE is a member of family proteins that activate apoptosis (37). CIDE-A, CIDE-B, and FSP27 proteins share homology at their N-terminal regions and control cell terminal differentiation and programmed cell death (17,44). The other novel genes found to be upregulated in Wy-14,643-treated livers include C3f gene belonging to the gene-rich cluster (1), Rbp7 (14), and an unknown gene from the Riken cDNA library (8). Rbp7 is a member of cellular retinoid binding protein family, and is highly expressed in white adipose tissue and mammary gland (14). Increases in both RAEy and Rbp7 expression suggest that alterations in retinoid metabolism occur in the liver of mice treated with PPARα ligand. This is of importance, because PPARα heterodimerizes with RXR, a 9-cis-retinoic acid-activated nuclear receptor. The expression of both RAEγ and Rbp7 appears to be under the control of PPARα as evidenced by the lack of enhanced expression in Wy-14, 643-treated PPARα−/− and double nulls.
Several genes that are upregulated between 2.5-fold- and 4-fold in Wy-14,643-treated mouse liver include phospholipase A2, uncoupling protein 2, uncoupling protein 3, carnitine palmitoyltransferase 1, proteasome 26S subunit, and HSP 74, among others (data not shown). UCP3 has been proposed to play a role as a regulator of lipid use and increases in the expression of UCP2 and UCP3 may reflect increased overall metabolism in livers with PPARα activation.
Downregulated Genes
Wy-14,643 treatment resulted in the downregulation of a large number of genes in liver (Table 2). These include: MutS homolog-MSH2, Wee 1 homolog, polymeric immunoglobulin receptor, 11β -HSD, pannexin 1, and others. It is reported that a basal expression of MutS homolog 2 is necessary for resting and differentiated cells, and that increased MSH2 protein expression is required when DNA replication is activated (46). This suggests that missing of cell cycle control by decreased expression of necessary proteins can cause alterations and destabilization of genome, leading to mutations that may be necessary for neoplastic transformation. Additional studies will be necessary to fully explore the molecular basis of these changes and the functional implication associated with sustained PPARα activation.
Perspective
We have obtained a portrait of gene expression, using cDNA microarray procedure, in the liver of the mouse treated for 2 weeks with Wy-14,43, a potent PPARα ligand. We found enhanced expression of several genes not previously known to be regulated by PPARα, in addition to many genes that are already known to be PPARα dependent. The novel genes identified in this study include CIDE, RAEγ, CYP2B9, CYP2B10, monoglyceride lipase, C3f, pre B-cell leukemia transcription factor 1, Ly-6D, alcohol dehydrogenase 3, PDK-4, Rbp7, annexin A2, and many others. Enhanced expression of several of these genes in Wy-14,643-treated mouse livers was confirmed by Northern analysis and an obligatory role for PPARα in the induction of these genes has been demonstrated, because an increase in these mRNAs was not obtained in PPARα−/− and PPARα−/− AOX−/− mice, but was noted in AOX−/− mice with spontaneous PPARα activation. Detailed studies are necessary to ascertain the presence of peroxisome proliferator-response elements (PPREs) in the promoter regions of these genes. Whether increases in mRNA levels translate into corresponding increases in protein level remains uncertain. Earlier studies have attempted to establish the protein expression patterns in liver of mice treated with peroxisome poliferators and these in most part were focused on peroxisomal β-oxidations enzymes (28,72). Recent developments in pro-teomic analysis should serve as a complementary approach to DNA microarray to analyze the expression profile of liver (12), at various intervals during acute and chronic exposure to PPARα ligands (Fig. 4). The availability of AOX null mice, which exhibit spontaneous peroxisome proliferation, should serve as parallel to compare the profiles resulting from the activation of PPARα by its natural and synthetic ligands.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Grant GM 23750 (to J.K.R), CA84472 (to M.S.R), VE505703 (to C.A.B.), and the Veterans Affairs Merit Review Grant (to A.V.Y). M.C.-M. was supported by a grant from the Association pour la Recherche Contre le Cancer. K.M. was supported by a fellowship of the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1. Ansari-Lari M. A.; Oeltjen J. C.; Schwartz S.; Zhang Z.; Muzny D. M.; Lu J.; Gorrell J. H.; Chinault A. C.; Belmont J. W.; Miller W.; Gibbs R. A. Comparative sequence analysis of gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 8:29–40; 1998. [PubMed] [Google Scholar]
- 2. Ashby J.; Brady A.; Elcombe C. R.; Elliot B. M.; Ishmael J.; Odum J.; Tugwood J. D.; Kettle S.; Purchase I. F. H. Mechanistically-based human hazard assessment of peroxisome proliferator-induced hepatocarcinogenesis. Hum. Exp. Toxicol. 13(Suppl. 2):S1–S117; 1994. [DOI] [PubMed] [Google Scholar]
- 3. Bardot O.; Aldridge T. C.; Latruffe N.; Green S. PPAR-RXR heterodimer activates a peroxisome proliferator response element upstream of the bifunctional enzyme gene. Biochem. Biophys. Res. Commun. 192: 37–45; 1993. [DOI] [PubMed] [Google Scholar]
- 4. Bayly A. C.; Roberts R. A.; Dive C. Suppression of liver cell apoptosis in vitro by the non-genotoxic hepatocarcinogen and peroxisome proliferator nafenopin. J. Cell Biol. 125:197–203; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berger J.; Tanen M.; Elbrecht A.; Hermanowski-Vosatka A.; Moller D. E.; Wright S. D.; Thieringer R. Peroxisome proliferator-activated receptor-γ ligands inhibit adipocyte 11 β-hydroxysteroid dehydrogenase type 1 expression and activity. J. Biol. Chem. 276: 12629–12635; 2001. [DOI] [PubMed] [Google Scholar]
- 6. Braissant O.; Foufelle F.; Scotto C.; Dauça M.; Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): Tissue distribution of PPAR-α, -β, and -γ in the adult rat. Endocrinology 137:354–366; 1996. [DOI] [PubMed] [Google Scholar]
- 7. Brakenhoff R. H.; Gerretsen M.; Knippels E. M.; van Dijk M.; van Essen H.; Weghuis D. O.; Sinke R. J.; Snow G. B.; van Dongen G. A. The human E48 antigen, highly homologous to the murine Ly-6 antigen ThB, is a GPI-anchored molecule apparently involved in keratinocyte cell-cell adhesion. J. Cell Biol. 129: 1677–1689; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carninci P.; Shibata Y.; Hayatsu N.; Sugahara Y.; Shibata K.; Itoh M.; Konno H.; Okazaki Y.; Muramatsu M.; Hayashizaki Y. Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 10:1617–1630, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cattley R. C.; DelUca J.; Elcombe C.; Fenner-Crisp P.; Lake B. G.; Marsman D. S.; Pastoor T. A.; Popp J. A.; Robinson D. E.; Schewtz B.; Tugwood J.; Wahli W. Do peroxisome proliferating compounds pose a hepatocarcinogenic hazard to humans? Regul. Toxicol. Pharmacol. 27:47–60; 1998. [DOI] [PubMed] [Google Scholar]
- 10. Cerwenka A.; Bakker B. H.; McClanahan T.; Wagner J.; Wu J.; Phillips J. H.; Lanier L. L. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity 12:721–727; 2000. [DOI] [PubMed] [Google Scholar]
- 11. Chatterjee B.; Demyan W. F.; Lalwani N. D.; Reddy J. K.; Roy A. K. Reversible alteration of hepatic messenger RNA species for peroxisomal and non-peroxisomal proteins induced by hypolipidemic drug Wy-14,643. Biochem. J. 241:879–883; 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chevalier S.; MacDonald N.; Tonge R.; Rayner S.; Rowlinson R.; Shaw J.; Young J.; Davison M.; Roberts R. A. Proteomic analysis of differential protein expression in primary hepatocytes induced by EGF, tumor necrosis factor α or the peroxisome proliferator nafenopin. Eur. J. Biochem. 2667:4624–4634; 2000. [DOI] [PubMed] [Google Scholar]
- 13. Chinetti G.; Lestavel S.; Bocher V.; Remaley A. T.; Neve B.; Torra I. P.; Teissier E.; Minnich A.; Jaye M.; Duverger N.; Brewer H. B.; Fruchart J. C.; Clavey V.; Staels B. PPAR-α and PPAR-γ activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat. Med. 7:53–57; 2001. [DOI] [PubMed] [Google Scholar]
- 14. Conforti L.; Tarlton A.; Mack T. G.; Mi W.; Buckmaster E. A.; Wagner D.; Perry V. H.; Coleman M. P. A Ufd2/D4Cole1e chimeric protein and overexpression of Rbp7 in the slow Wallerian degeneration (WldS) mouse. Proc. Natl. Acad. Sci. USA 97:11377–11382; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dadras S. S.; Cook W. S.; Yeldandi A. V.; Cao W.-Q.; Rao M. S.; Wang Z.; Reddy J. K. Peroxisome proliferator-activated receptor α-dependent induction of cell surface antigen Ly-6D gene in the mouse liver. Gene Expr. 9:173–181; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Damon M.; Fautrel A.; Marc N.; Guillouzo A.; Corcos L. Isolation of a new mouse cDNA clone: Hybrid form of cytochrome P450 2bl0 and NADPH-cytochrome P450 oxidoreductase. Biochem. Biophys. Res. Commun. 226:900–905; 1996. [DOI] [PubMed] [Google Scholar]
- 17. Danesch U.; Hoeck W.; Ringold G. M. Cloning and transcriptional regulation of a novel adipocyte-specific gene, FSP27. CAAT-enhancer-binding protein (C/EBP) and C/EBP-like proteins interact with sequences required for differentiation-dependent expression. J. Biol. Chem. 267:7185–7193; 1992. [PubMed] [Google Scholar]
- 18. De Duve C.; Baudhuin P. Peroxisomes (microbodies and related particles). Physiol. Rev. 46:323–357; 1966. [DOI] [PubMed] [Google Scholar]
- 19. Desvergne B.; Wahli W. Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocr. Rev. 20:649–688; 1999. [DOI] [PubMed] [Google Scholar]
- 20. Devchand P. R.; Keller H.; Peters J. M.; Vazquez M.; Gonzalez F. J.; Wahli W. The PPARα-leukotriene B4 pathway to inflammation control. Nature 384:39–43; 1996. [DOI] [PubMed] [Google Scholar]
- 21. Dryer C.; Krey G.; Keller H.; Givel F.; Helftenbein G.; Wahli W. Control of the peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell 68:879–887; 1992. [DOI] [PubMed] [Google Scholar]
- 22. Dzhandzhugazyan K. N.; Kirkin A. F.; thor Straten P.; Zeuthen J. Ecto-ATP diphosphohydrolase/CD39 is overexpressed in differentiated human melanomas. FEBS Lett. 430:227–230; 1998. [DOI] [PubMed] [Google Scholar]
- 23. Fan C. Y.; Pan J.; Chu R.; Lee D.; Kluckman K. D.; Usuda N.; Singh I.; Yeldandi A. V.; Rao M. S.; Maeda N.; Reddy J. K. Hepatocellular and hepatic peroxisomal alterations in mice with a disrupted peroxisomal fatty acyl-coenzyme A oxidase gene. J. Biol. Chem. 271:24698–24710; 1996. [DOI] [PubMed] [Google Scholar]
- 24. Fan C. Y.; Pan J.; Usuda N.; Yeldandi A. V.; Rao M. S.; Reddy J. K. Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase. J. Biol. Chem. 273:15639–15645; 1998. [DOI] [PubMed] [Google Scholar]
- 25. Febbraio M.; Abumrad N. A.; Hajjar D. P.; Sharma K.; Cheng W.; Pearce S. F.; Silverstein R. L. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J. Biol. Chem. 274:19055–19062; 1999. [DOI] [PubMed] [Google Scholar]
- 26. Fey M. F.; Moffat G. J.; Vik D. P.; Meisenhelder J.; Saris C. J.; Hunter T.; Tack B. F. Complete structure of the murine p36 (annexin II) gene. Identification of mRNAs for both the murine and the human gene with alternatively spliced 5′ noncoding exons. Biochim. Biophys. Acta 1306:160–170; 1996. [DOI] [PubMed] [Google Scholar]
- 27. Fogel M.; Friederichs J.; Zeller Y.; Husar M.; Smirnov A.; Roitman L.; Altevogt P.; Sthoeger Z. M. CD24 is a marker for human breast carcinoma. Cancer Lett. 143:87–94; 1999. [DOI] [PubMed] [Google Scholar]
- 28. Giometti C. S.; Taylor J.; Gemmell M. A.; Tollaksen S. L.; Lalwani N. D.; Reddy J. K. A comparative study of the effects of clofibrate, ciprofibrate, WY-14,643, and di-(2-ethylhexyl)-phthalate on liver protein expression in mice. Appl. Theoret. Electrophor. 2:101–107; 1991. [PubMed] [Google Scholar]
- 29. Gonzalez F. J.; Peters J. M.; Cattley R. C. Mechansims of action of the nongenotoxic peroxisome proliferators: Role of the peroxisome proliferator-activated receptor. J. Natl. Cancer Inst. 90:1702–1709; 1998. [DOI] [PubMed] [Google Scholar]
- 30. Gulick T.; Cresci S.; Caira T.; Moore D. D.; Kelly D. P. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc. Natl. Acad. Sci. USA 91: 11012–11016; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gumley T. P.; McKenzie I. F. C.; Sandrin M. S. Tissue expression, structure and function of the murine Ly-6 family of molecules. Immunol. Cell Biol. 73: 277–296; 1995. [DOI] [PubMed] [Google Scholar]
- 32. Hashimoto T.; Cook W. S.; Qi C.; Yeldandi A. V.; Reddy J. K.; Rao M. S. Defect in peroxisome proliferator-activated receptor α inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J. Biol. Chem. 275:28918–28928; 2000. [DOI] [PubMed] [Google Scholar]
- 33. Hashimoto T.; Fujita T.; Usuda N.; Cook W.; Qi C.; Peters J. M.; Gonzalez F. J.; Yeldandi A. V.; Rao M. S.; Reddy J. K. Peroxisomal and mitochondrial fatty acid β-oxidation in mice nullizygous for both peroxisome proliferator-activated receptor α and peroxisomal fatty acyl-CoA oxidase. Genotype correlation with fatty liver phenotype. J. Biol. Chem. 274:19228–19236; 1999. [DOI] [PubMed] [Google Scholar]
- 34. Hermanowski-Vosatka A.; Gerhold D.; Mundt S. S.; Loving V. A.; Lu M.; Chen Y.; Elbrecht A.; Wu M.; Doebber T.; Kelly L.; Milot D.; Guo Q.; Wang P.-R.; Ippolito M.; Chao Y-S.; Wright S. D.; Thieringer R. PPAR agonists reduce 11-hydroxystreoid de-hydrogenase type 1 in the liver. Biochem. Biophys. Res. Commun. 279:330–336; 2000. [DOI] [PubMed] [Google Scholar]
- 35. Hunt M. C.; Nousiainen S. E.; Huttunen M. K.; Orii K. E.; Svensson L. T.; Alexson S. E. Peroxisome proliferator-induced long chain acyl-CoA thioesterases comprise a highly conserved novel multi-gene family involved in lipid metabolism. J. Biol. Chem. 274: 34317–34326; 1999. [DOI] [PubMed] [Google Scholar]
- 36. Imanaka T.; Aihara K.; Takano T.; Yamashita A.; Sato R.; Suzuki Y.; Yokota S.; Osumi T. Characterization of the 70-kDa peroxisomal membrane protein, an ATP binding cassette transporter. J. Biol. Chem. 274:11968–11976; 1999. [DOI] [PubMed] [Google Scholar]
- 37. Inohara N.; Koseki T.; Chen S.; Wu X.; Nunez G. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 17:2526–2533; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Issemann I.; Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 347:645–650; 1990. [DOI] [PubMed] [Google Scholar]
- 39. Jain S.; Pulikuri S.; Zhu Y.; Qi C.; Kanwar Y. S.; Yeldandi A. V.; Rao M. S.; Reddy J. K. Differential expression of the peroxisome proliferator-activated receptor γ (PPARγ) and its coactivators steroid receptor coactivator-1 and PPAR-binding protein PBP in the brown fat, urinary bladder, colon, and breast of the mouse. Am. J. Pathol. 153:349–354; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johnson E. F.; Palmer C. N. A.; Griffin K. J.; Hsu M.-H. Role of the peroxisome proliferator-activated receptor in cytochrome P450 4A gene regulation. FASEB J. 10:1241–1248; 1996. [DOI] [PubMed] [Google Scholar]
- 41. Karlsson M.; Contreras J. A.; Hellman U.; Tornqvist H.; Holm C. cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases J. Biol. Chem. 272: 27218–27223; 1997. [DOI] [PubMed] [Google Scholar]
- 42. Kliewer S. A.; Umesono K.; Noonan D. J.; Heyman R. A.; Evans R. M. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 358:771–774; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee S. S.-T.; Pineau T.; Drago J.; Lee E. J.; Owens J. W.; Kroetz D. L.; Fernandez-Salguero P. M.; Westphal H.; Gonzales F. J. Targeted disruption of the a isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 15:3012–3022; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lugovskoy A. A.; Zhou P.; Chou J. J.; McCarty J. S.; Li P.; Wagner G. Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis. Cell 99:747–755; 1999. [DOI] [PubMed] [Google Scholar]
- 45. Mannaerts G. P.; Van Veldhoven P. P.; Casteels M. Peroxisomal lipid degradation via β- and α-oxidation in mammals. Cell Biochem. Biophys. 32:73–87; 2000. [DOI] [PubMed] [Google Scholar]
- 46. Marra G.; Chang C. L.; Laghi L. A.; Chauhan D. P.; Young D.; Boland C. R. Expression of human MutS homolog 2 (hMSH2) protein in resting and proliferating cells. Oncogene 13:2189–2196; 1996. [PubMed] [Google Scholar]
- 47. Martin G.; Schoonjans K.; Lefebvre A.-M.; Staels B.; Auwerx J. Coordinate regulation of the expression of the fatty acid transport protein and acyl-CoA synthetase genes by PPARα and PPARγ activators. J. Biol. Chem. 272:28210–28217; 1997. [DOI] [PubMed] [Google Scholar]
- 48. Miziorko H. M.; Lane M. D. 3-Hydroxy-3-methylglutaryl-CoA synthase. Participation of acetyl-S-enzyme and enzyme-S-hydroxymethylgutaryl-SCoA intermediates in the reaction. J. Biol. Chem. 252:1414–1420; 1977. [PubMed] [Google Scholar]
- 49. Moody D. E.; Loury D. N.; Hammock B. D. Epoxide metabolism in the liver of mice treated with clofibrate (ethyl-a-)p-chlorophenoxyisobutyrate, a peroxisome proliferator. Toxicol. Appl. Pharmacol. 78:351–362; 1985. [DOI] [PubMed] [Google Scholar]
- 50. Motojima K.; Passilly P.; Peters J. M.; Gonzalez F. J.; Latruffe N. Expression of putative fatty acid transporter genes are regulated by peroxisome proliferator-activated receptor alpha and gamma activators in a tissue- and inducer-specific manner. J. Biol. Chem. 273:16710–16714; 1998. [DOI] [PubMed] [Google Scholar]
- 51. Palmer C. N. A.; Hsu M.-H.; Griffin K. J.; Raucy J. L.; Johnson E. F. Peroxisome proliferator activated receptor α expression in human liver. Mol. Pharmacol. 53:14–22; 1998. [PubMed] [Google Scholar]
- 52. Pellinen P.; Stenback F.; Kojo A.; Honkakoski P.; Gelboin H. V.; Pasanen M. Regenerative changes in hepatic morphology and enhanced expression of CYP2B10 and CYP3A during daily administration of cocaine. Hepatology 23:515–523; 1996. [DOI] [PubMed] [Google Scholar]
- 53. Qi C.; Zhu Y.; Reddy J. K. Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochem. Biophys. 32:187–204; 2000. [DOI] [PubMed] [Google Scholar]
- 54. Rao M. S.; Reddy J. K. Peroxisome proliferation and hepatocarcinogenesis. Carcinogenesis 8:631–636; 1987. [DOI] [PubMed] [Google Scholar]
- 55. Rao M. S.; Reddy J. K. Hepatocarcinogenesis of peroxisome proliferators. Ann. NY Acad. Sci. 804:573–587; 1996. [DOI] [PubMed] [Google Scholar]
- 56. Reddy J. K.; Azarnoff D. L.; Hignite C. E. Hypolipidaemic hepatic peroxisome proliferators form a novel class of chemical carcinogens. Nature 283:397–398; 1980. [DOI] [PubMed] [Google Scholar]
- 57. Reddy J. K.; Chu R. Peroxisome proliferator-induced pleiotropic responses: Pursuit of a phenomenon. Ann. NY Acad. Sci. 804:176–201; 1996. [DOI] [PubMed] [Google Scholar]
- 58. Reddy J. K.; Hashimoto T. Peroxisomal β-oxidation and PPARα: An adaptive metabolic system. Annu. Rev. Nutr. 21:193–230; 2001. [DOI] [PubMed] [Google Scholar]
- 59. Reddy J. K.; Krishnakantha T. P. Hepatic peroxisome proliferation: Induction by two novel compounds structurally unrelated to clofibrate. Science 190:787–789; 1975. [DOI] [PubMed] [Google Scholar]
- 60. Reddy J. K.; Lalwani N. D. Carcinogenesis by hepatic proxisome proliferators: Evaluation of the risk of hypolipidemic drugs and industrial plasticizers to human. Crit. Rev. Toxicol. 12:1–53; 1983. [DOI] [PubMed] [Google Scholar]
- 61. Reddy J. K.; Mannaerts G. P. Peroxisomal lipid metabolism. Annu. Rev. Nutr. 14:343–370; 1994. [DOI] [PubMed] [Google Scholar]
- 62. Rodriguez J. C.; Gil-Gomez G.; Hegardt F. G.; Haro D. Peroxisome proliferator-activated receptor mediates induction of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene by fatty acids. J. Biol. Chem. 269:18767–18772; 1994. [PubMed] [Google Scholar]
- 63. Schoonjans K.; Staels B.; Auwerx J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J. Lipid Res. 37:907–925; 1996. [PubMed] [Google Scholar]
- 64. Schrader M.; Reuber B. E.; Morrell J. C.; Jimenez-Sanchez G.; Obie C.; Stroh T. A.; Valle D.; Schroer T. A.; Gould S. J. Expression of PEX11β mediates peroxisome proliferation in the absence of extracellular stimuli. J. Biol. Chem. 273:29607–29614; 1998. [DOI] [PubMed] [Google Scholar]
- 65. Schulte-Hermann R.; Bursch W.; Kraupp-Grasl B.; Oberhammer F.; Wagner A.; Jirtle R. Cell proliferation and apoptosis in normal liver and preneoplastic liver foci. Environ. Health Perspect. 101(Suppl. 5):87–90; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Seckl J. R.; Walker B. R. 11β-hydroxysteroid dehydrogenase type 1—a tissue-sepecific amplifier of glucocorticoid action. Endocrinology 142:1371–1376; 2001. [DOI] [PubMed] [Google Scholar]
- 67. Staels B.; Auwerx J. Regulation of apo A-I gene expression by fibrates. Atherosclerosis 137(Suppl.):S19–23; 1998. [DOI] [PubMed] [Google Scholar]
- 68. Tanaka T.; Kondo S.; Iwasa Y.; Hiai H.; Toyokuni S. Expression of stress-response and cell proliferation genes in renal cell carcinoma induced by oxidative stress. Am. J. Pathol. 156:2149–2157; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thorne R. F.; Marshall J. F.; Shafren D. R.; Gibson P. G.; Hart I. R.; Burns G. F. The integrins alpha3beta1 and alpha6beta1 physically and functionally associate with CD36 in human melanoma cells. Requirement for the extracellular domain of CD36. J. Biol. Chem. 275:35264–35275; 2000. [DOI] [PubMed] [Google Scholar]
- 70. van Roermund C. W.; Tabak H. F.; van Den Berg M.; Wanders R. J.; Hettema E. H. Pex11p plays a primary role in medium-chain fatty acid oxidation, a process that affects peroxisome number and size in Saccharomyces cerevisiae . J. Cell Biol. 150:489–498; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wanders R. J. A. Peroxisomes, lipid metabolism and human disease. Cell Biochem. Biophys. 32:89–106; 2000. [DOI] [PubMed] [Google Scholar]
- 72. Watanabe T.; Lalwani N. D.; Reddy J. K. Specific changes in the protein composition of rat liver in response to the peroxisome proliferators ciprofibrate, clofibrate, Wy-14,643 and di-(2-ethylhexyl)phthalate. Biochem. J. 227:767–775; 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Way J. M.; Harrington W. W.; Brown K. K.; Gottschalk W. K.; Sundseth S. S.; Mansfield T. A.; Ramachandran R. K.; Willson T. M.; Kliewer S. A. Comprehensive messenger ribonucleic acid profiling reveals that peroxisome proliferator-activated receptor γ activation has coordinate effects on gene expression in multiple insulin-sensitive tissues. Endocrinology 142:1269–1277; 2001. [DOI] [PubMed] [Google Scholar]
- 74. Wenger R. H.; Ayane M.; Bose R.; Kohler G.; Nielsen P. J. The genes for a mouse hematopoietic differentiation marker called the heat-stable antigen. Eur. J. Immunol. 21:1039–1046; 1991. [DOI] [PubMed] [Google Scholar]
- 75. Witz I. P. Differential expression of genes by tumor cells of a low or a high malignancy phenotype: The case of murine and human Ly-6 proteins. J. Cell Bio-chem. 34(Suppl.):61–66; 2000. [PubMed] [Google Scholar]
- 76. Yeldandi A. V.; Rao M. S.; Reddy J. K. Hydrogen peroxide generation in peroxisome proliferator-induced oncogenesis. Mutat. Res. 448:159–177; 2000. [DOI] [PubMed] [Google Scholar]
- 77. Yue H.; Eastman P. S.; Wang B. B.; Minor J.; Doctolero M. H.; Nuttall R. L.; Stack R.; Becker J. W.; Montgomery J. R.; Vainer M.; Johnston R. An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res. 29:e41; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang B.; Marcus S. L.; Sajjadi F. G.; Alvares K.; Reddy J. K.; Subramani S.; Rachubinski R. A.; Capone J. P. Identification of a peroxisome proliferator-responsive element upstream of the gene encoding rat peroxisomal enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase. Proc. Natl. Acad. Sci. USA 89:7541–7545; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhu Y.; Qi C.; Korenberg J. R.; Chen X.-N.; Noya D.; Rao M. S.; Reddy J. K. Structural organization of mouse peroxisome proliferator-activated receptor γ (mPPARγ) gene: Alternative promoter use and different splicing yield two mPPARγ isoforms. Proc. Natl. Acad. Sci. USA 92:7921–7925; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zou Z.; Nomura M.; Takihara Y.; Yasunaga T.; Shimada K. Isolation and characterization of retinoic acid-inducible cDNA clones in F9 cells: A novel cDNA family encodes cell surface proteins sharing partial homology with MHC class I molecules. J. Biochem. 119:319–328; 1996. [DOI] [PubMed] [Google Scholar]


