Abstract
A current goal in molecular medicine is the development of new strategies to interfere with gene expression in living cells in the hope that novel therapies for human disease will result from these efforts. This review focuses on small-molecule or chemical approaches to manipulate gene expression by modulating either transcription of messenger RNA-coding genes or protein translation. The molecules under study include natural products, designed ligands, and compounds identified through functional screens of combinatorial libraries. The cellular targets for these molecules include DNA, messenger RNA, and the protein components of the transcription, RNA processing, and translational machinery. Studies with model systems have shown promise in the inhibition of both cellular and viral gene transcription and mRNA utilization. Moreover, strategies for both repression and activation of gene transcription have been described. These studies offer promise for treatment of diseases of pathogenic (viral, bacterial, etc.) and cellular origin (cancer, genetic diseases, etc.).
Keywords: Gene transcription, Molecular medicine, Novel therapies, Therapeutic strategies
CONSIDERABLE effort has been expended over the past decade to devise methods to interfere with gene expression in living cells in the hope that therapeutic strategies will come from these studies. These approaches include both interference with translation of messenger RNA into protein (with antisense oligonucleotides, peptide nucleic acids, ribozymes, deoxyribozymes) and the direct interference with gene transcription (with triple helix-forming oligonucleotides, peptide nucleic acids, decoys, and synthetic minor groove-binding ligands). Strategies for both repression and activation of gene transcription have been devised and will be considered below. We will concentrate in this review on small-molecule or chemical approaches to manipulate gene expression at both the level of transcription and translation, but we will begin by citing recent exciting studies on other related methods.
Recent advances for controlling translation with antisense oligonucleotides (15,24) or antisense peptide nucleic acids (PNA) (28,56) have been reviewed. A new modification of antisense technology is the development of circular antisense deoxyoligonucleotides that are resistant to exonuclease degradation. These molecules have been demonstrated to effectively downregulate the bcr-abl oncogene in a cell culture assay (61). Significantly, circular ODNs were shown to be more stable and more effective in down-regulation of protein levels than the corresponding linear molecules. A recent advance in ribozyme technology is the ability to tailor the activity of ribozymes to respond to small-molecule effectors, thus making ribozymes “allosteric molecular switches” (66). Additionally, translational control by deoxyribozymemediated cleavage of various mRNAs has been reported (5,62,67). Among a number of recent successful applications of this technology, a deoxyribozyme that targeted the translation initiation region of cMyc mRNA was found to cleave its target substrate in vitro and inhibit Myc-dependent smooth muscle cell proliferation (67). Deoxyribozymes offer the promise of greater stability over their ribozyme cousins.
Decoy molecules, corresponding to RNA domains that bind regulatory proteins, have also been used as inhibitors of RNA function in transcription, and circular versions of these molecules offer promise of greater stability over their linear counterparts (4). Other, nonchemical, approaches for direct inhibition and/or activation of gene transcription include designed or selected zinc finger peptides that recognize predetermined DNA sequences (1,12,41) and DNA-cleaving ribozymes (59). Triple helix-forming oligonucleotides (TFOs) (30), TFOs coupled with functional groups for DNA modification or cleavage (76), and PNAs (47,56) have been used to interfere with gene transcription both in vitro and in cell culture experiments. Transcription factor decoy molecules, whether made of DNA, RNA, or novel nucleic acid analogues, also offer promise for regulating gene activity. Recently, a PNA–DNA hybrid was shown to effectively compete for the transcription factor NF-κB in vitro (52), but the utility of this approach has not been investigated in cell culture. In vitro selection methods have also been employed to isolate a small RNA aptamer that binds the transcription factor NF-κB with high affinity (44). While this molecule effectively competes with NF-κB target site DNAs for transcription factor binding, expression of this RNA in cells did not lead to downregulation of NF-κB dependent gene expression. Thus, further improvements will be needed to render the decoy approach viable for intracellular gene regulation.
For any gene regulatory approach to be successful, several criteria must be met by the therapeutic agent. First, the agent must not posses any general cell toxicity and should not elicit an immune response. Second, the agent must be cell permeable or amenable to cell-type-specific delivery. In the case of the DNA-binding molecules, the therapeutic agent must transit to the nucleus and bind the target sequence with high affinity and specificity in the context of cellular chromatin. Third, binding of the agent to its DNA or RNA target sequence must interfere with gene transcription or translation. Each of the approaches listed above has its own unique advantages and limitations. For example, while nucleic acid-based approaches (antisense, decoy, and triple helix-forming oligonucleotides and ribozymes) have the potential for sequence selectivity and can effectively inhibit transcription or translation in vitro, these molecules suffer from poor cell permeability; other delivery systems, such as retroviral or adenovirus vectors, liposomes, or other delivery strategies must be used for effective gene inhibition. Similarly, zinc finger peptides must be introduced via a gene therapy approach with an appropriate viral expression vector because these peptides cannot directly enter cells (13). One additional problem with gene therapy approaches is that they must be performed on cells ex vivo and, once a “modified” cell population is established, these cells are then transferred back to the patient.
In contrast to gene therapy approaches, cell-permeable sequence-specific DNA- or RNA-binding ligands circumvent the problems associated with other forms of gene therapy and could compliment other therapeutic strategies. Below, we review several different classes of DNA and RNA ligands and inhibitors that have been studied over the past few years. Other approaches for controlling gene expression through protein-protein interactions, such as protein dimerizers that bring together molecules involved in transcriptional activation, have recently been reviewed (17,60) and will not be considered here.
DNA-BINDING AGENTS
Minor Groove-Binding and Intercalating Ligands
A recent review has summarized the chemistry and mode of binding of the various natural and synthetic ligands that bind to DNA in the minor groove (71). Here, we consider the use of these molecules, and other synthetic ligands, to inhibit protein–DNA interactions involved in transcription, and transcription by RNA polymerase II. Among these ligands, the natural product distamycin A (Fig. 1) and analogues of distamycin have received the most attention. Because distamycin binds in the minor groove of DNA preferentially to five-base pair A + T tracts, it seemed reasonable to suspect that distamycin might inhibit binding of the general eukaryotic transcription factor TATA box binding protein (TBP) to TATA elements. These DNA sequences (of the form 5′-TA TAAA-3′) are located approximately 25 bp upstream from the site of initiation of transcription of TATA-containing, messenger RNA-coding genes by RNA polymerase II. An early study from Beerman and collaborators (10) established that distamycin inhibited TBP binding, with 50% inhibition at 0.16 μM ligand concentration. Another minor groove-binding ligand, CC-1065 (Fig. 1), was found to be more effective than distamycin in inhibition of TBP binding (50% inhibition at ∼1 nM CC-1065). Unlike distamycin, CC-1065 binds covalently to DNA. Both ligands were able to dissociate preformed TBP-DNA complexes, and both small molecules could also inhibit the TBP-TFIIA-DNA complex (10,11). TFIIA stabilizes the TBP-DNA complex, and greater drug concentrations were needed to compete this ternary complex than the binary TBP-DNA complex. In a similar study, distamycin and its alkylating benzoyl mustard derivative, tallimustine (Fig. 1), were found to be effective in inhibiting TBP–DNA, TBP–TFIIA–DNA, and TBP–TFIIA–TFIIB–DNA complexes (2). Notably, both ligands were shown to inhibit basal transcription of a minimal TATA-containing promoter at the concentrations needed for inhibition of protein–DNA interactions.
Figure 1.
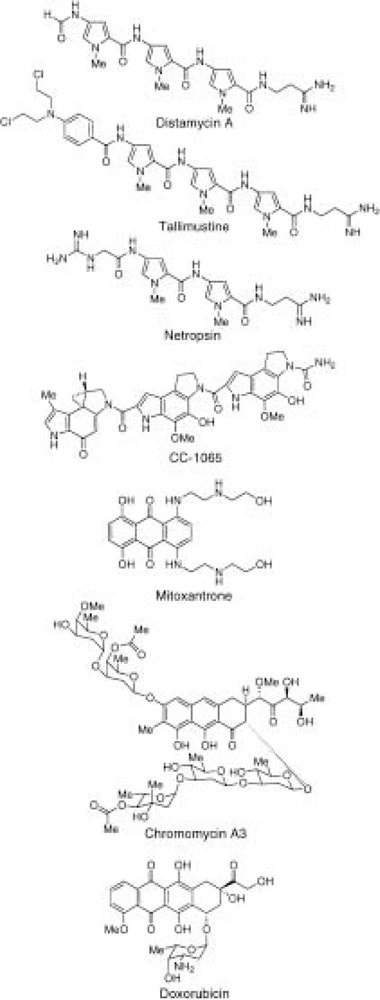
Structures of small molecules that bind DNA.
Several studies have documented that distamycin and other minor groove-binding ligands also inhibit the DNA-binding activity of various other eukaryotic transcription factors that bind A + T-rich regions of DNA. For example, distamycin was found to inhibit two transcription factors, SRF and MEF2, that are required for transcription of muscle-specific genes, such as cardiac and skeletal α-actin, and distamycin was shown to downregulate these genes in a myogenic cell line and SRF- and MEF2-dependent transcription in transient transfection experiments (68). Conversely, Sp1 and MyoD, which bind G + C-rich sequences, were not affected by distamycin. Distamycin and its two-pyrrole-ring analogue, netropsin (Fig. 1), have also been found to inhibit binding of the transcription factor E2F1 to its cognate site in the hamster dihydrofolate reductase (DHFR) promoter, with the effect that distamycin can also inhibit DHFR RNA synthesis by RNA polymerase II in a HeLa cell nuclear extract (8).
The microgonotropen (MGT) class of synthetic agents consist of an A + T-selective minor groove-binding tripyrrole (similar to distamycin) and polyamine chains attached to the central pyrrole that extend the ligand contact to the major groove (7,9). In in vitro studies, the MGT inhibitors were found to be highly efficient at inhibition of the DNA-binding activity of transcription factor E2F1, with 50% inhibition at ∼1 nM compound.
Various other minor groove-binding compounds and DNA intercalators have been tested for their effects on protein–DNA interactions: the G + C-binding drugs doxorubicin, hedamycin, mitoxantrone, nogalamycin, and chromomycin A3 (Fig. 1) have been shown to inhibit binding of the zinc finger proteins Sp1, EGR-1, WT1, and NIL2A (3,8,11,70); distamycin, however, failed to inhibit these major groove-binding zinc finger proteins (8). Sp1-dependent transcription from the DHFR promoter is also inhibited by these GC-binding drugs (8). Adriamycin has been shown to inhibit members of the octamer-binding transcription factors, Oct-1, -3, and -5, and transcription by E. coli RNA polymerase (16) and doxorubicin has been shown to interfere with muscle-specific gene expression in cell culture experiments (43). Interestingly, in these latter experiments, doxorubicin inhibited MyoD-dependent gene expression; however, the target of doxorubicin was shown to be the promoter element of the MyoD regulatory protein Id, rather than MyoD itself. While these natural and synthetic small molecules are effective inhibitors of protein–DNA complexes, generally at micromolar concentrations of drug, these compound suffer from low affinities in comparison to their DNA-binding protein targets, and poor sequence selectivities. Thus, a major goal has been the development of new generations of synthetic, cell-permeable molecules that can bind to a wide range of predetermined DNA sequences with affinities similar to transcription factors.
Calicheamicin Oligosaccharides
The calicheamicin oligosaccharides are bacterial antibiotics that are members of the enediyne class of minor groove-binding, DNA-cleaving agents [(55,65) and references therein]. Calicheamicin γ1 I (CLM) (Fig. 2) has been shown to bind duplex DNA through its aryltetrasaccharide motif while the enediyne portion of the molecule contributes to both binding affinity and sequence preference. DNA cleavage studies with CLM indicate that this compound prefers to bind the four-base pair, homopyrimide sequences 5′-TCCT-3′, 5′-TCTC-3′, and 5′-TTTT-3′. Because CLM is amenable to total chemical synthesis (55,65), novel derivatives, such as head-to-head dimers (46) and the methylglycoside of the carbohydrate moiety of CLM (CLM-MG, (34)), have been synthesized (Fig. 2). Compounds lacking the enediyne moiety bind DNA with the same sequence preference as the parent compound but do not cleave DNA (34).
Figure 2.
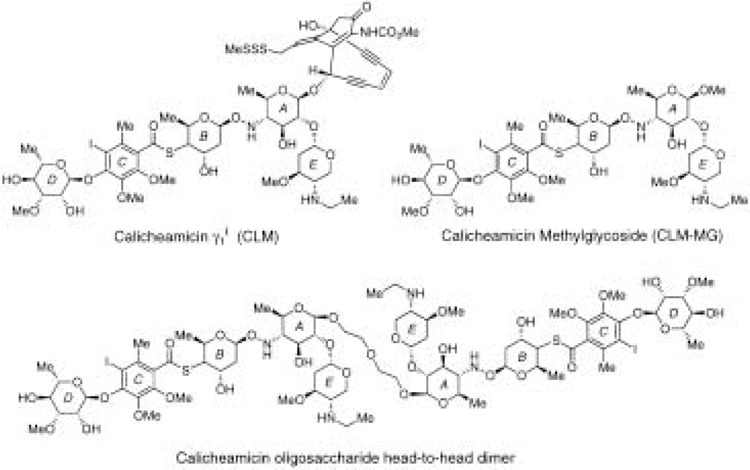
Structures of calicheamicin γ1 I (CLM), the methylglycoside of the carbohydrate moiety of CLM (CLM-MG), and the head-to-head dimer of the CLM oligosaccharide.
These derivatives and the parent compound have been used to block DNA binding by both eukaryotic and prokaryotic transcription factors (34,46,64). The DNA-binding proteins investigated contained a variety of protein structural motifs utilized for DNA binding. As expected for a minor groove-binding agent, proteins that make minor groove contacts are more easily inhibited by CLM and its derivatives than proteins that exclusively contact the major groove (34,64). Nonetheless, the bZip protein AP1, which contacts the major groove of DNA, can be inhibited when the preferred binding site for CLM is present near the AP1 recognition site (46). CLM-MG has also been shown to displace a preformed protein–DNA complex (34). CLM and its derivatives have been shown to block RNA polymerase II transcription in vitro (46) and in cell culture experiments (34). This latter finding indicates that the calicheamicins are sufficiently hydrophobic to pass through cell membranes; however, due to the limited sequence preference (4 bp) and relatively low binding affinities of the calicheamicins [micromolar or higher concentrations required for inhibition of protein–DNA interactions and transcription (34,46)], new derivatives that possess higher degrees of DNA sequence specificity and affinity are clearly needed.
Pyrrole-Imidazole Polyamides
The pyrrole-imidazole polyamides are the only available class of synthetic small molecules that can be designed to bind predetermined DNA sequences (69,73). These molecules, containing N-methylpyrrole (Py) and N-methylimidazole (Im) amino acids, bind their target sequences with exceptionally high affinities, comparable to the binding affinities of natural DNA-binding transcriptional regulatory proteins (with dissociation constants in the picomolar to nanomolar range) (69). The development of the Py-Im polyamides and the structural basis for DNA recognition by polyamides has recently been reviewed (18). Briefly, base-sequence specificity depends on side-by-side pairing of Py and Im amino acids in the minor groove of DNA. A pairing of an Im opposite a Py targets a G•C base pair, whereas a Py opposite an Im targets a C•G base pair (Fig. 3). The Py/Py pair is degenerate and targets both A•T and T•A base pairs. Recent studies have shown that a 3-hydroxypyrrole/Py pair can discriminate between T•A and A•T base pairs and specifically recognizes T•A (73). The generality of these pairing rules has been demonstrated by targeting a variety of sequences 6–16 base pairs in length [for examples, see (18) and references therein] and is supported by direct NMR structural studies (25) and by X-ray crystallography (36,37,39).
Figure 3.
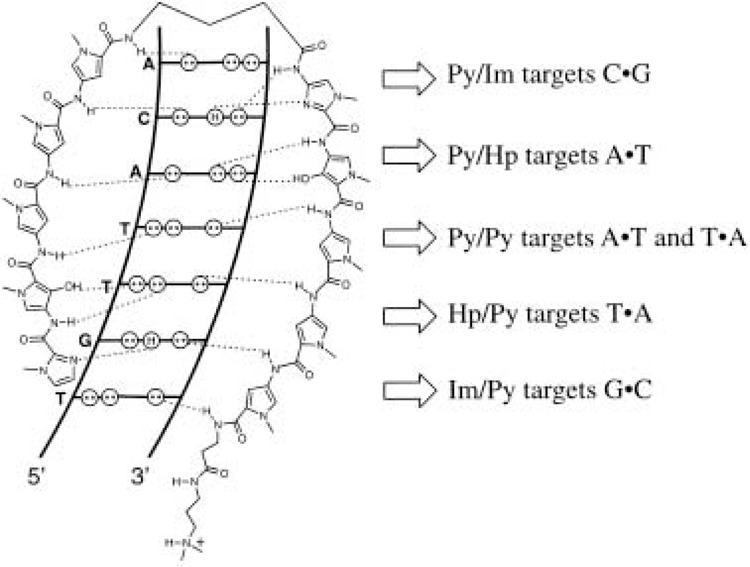
Binding model for the complex formed between ImHpPyPyPy-y-ImHpPyPyPy-P-Dp and a 5′-TGTTACA-3′ sequence. Circles with dots represent the lone pairs of N(3) of purines and O(2) of pyrimidines, and circles containing an H represent the 2-amino group of guanine. Hydrogen bonds are shown as dashed lines.
Several studies have now documented that the Py-Im polyamides effectively block eukaryotic transcription factors from binding to their target DNA sites and can also inhibit transcription by RNA polymerases II and III, both in vitro and in cell culture experiments. An early study showed that a polyamide could co-occupy a DNA fragment along with the major groove-binding transcription factor GCN4 (57). Subsequent studies have shown that polyamides are potent inhibitors of protein–DNA interactions when the DNA-binding protein either contacts the minor groove exclusively or makes contacts with both the major and minor grooves. Table 1 lists the various transcription factors that have been inhibited by polyamides, the structural motifs utilized by these proteins for DNA recognition, and the genes regulated by these proteins.
TABLE 1.
PROTEIN TARGETS FOR POLYAMIDE INHIBITORS
| Protein | DNA-Binding Motif | Gene Target |
|---|---|---|
| TFIIIA | zinc finger | 5S ribosomal RNA |
| TBP/TFIID | minor groove/saddle | mRNA-coding genes |
| LEF-1 | HMG box | HIV-1 |
| Ets-1 | winged-helix–turn–helix | HIV-1 |
| IE-86 | minor groove | CMV MIEP |
| Estrogen receptor | C4 zinc finger | estrogen-responsive genes |
| Deadpan | basic helix–loop–helix | Drosophila neural genes |
| GCN4 | bZip | yeast mRNA-coding genes |
| Tax | minor groove | HTLV-I |
We showed that a high-affinity polyamide, which was designed to bind within the intragenic promoter element of the Xenopus 5S RNA gene, inhibits TFIIIA binding and 5S RNA transcription by RNA polymerase III (29,53). TFIIIA is the prototype zinc finger protein, and the binding site for the inhibitory polyamide was chosen to coincide with a zinc finger–minor groove interaction (14) (Fig. 4). The inhibitory polyamide selectively disrupts transcription complexes on the chromosomal 5S RNA genes in Xenopus kidney-derived fibroblasts grown in culture (29). This latter result demonstrated that Py-Im polyamides are cell permeable and can inhibit transcription of target genes in living cells.
Figure 4.
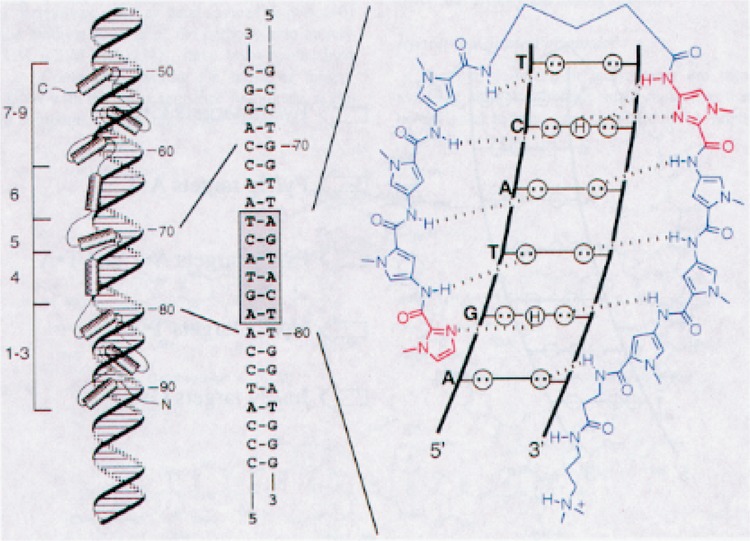
Left: model of the nine-zinc finger protein TFIIIA with the 5S RNA gene internal control region (ICR). Middle: sequence of the ICR recognized by zinc finger 4 in the minor groove. Right: complex of a polyamide with its target site, 5′-AGTACT-3′. Circles with dots represent the lone pairs of N(3) of purines and O(2) of pyrimidines, and circles containing an H represent the 2-amino group of guanine. Hydrogen bonds are shown as dashed lines.
Our studies have been extended to both cellular and viral transcription factors utilized by genes transcribed by RNA polymerase II (19–21,45). Polyamides are effective inhibitors of tissue-specific and general transcription factors (19,20) as well as viral repressors (21) and transactivators (45). Because batteries of genes utilize common general and tissue-specific transcription factors, polyamides were synthesized to bind sequences adjacent to the binding sites for required transcription factors (19). The central transcription factor utilized by most eukaryotic genes is the TATA box-binding protein, TBP (33). Because TBP binds DNA in the minor groove, and protein binding is accompanied by a large distortion of the DNA helix (bending and unwinding) (40,42), we reasoned that a polyamide might lock the DNA into a B-type conformation (36) and prevent TBP binding. A polyamide was synthesized to target the sequences adjacent to the TATA box element of the HIV-1 promoter (Fig. 5): this polyamide inhibits TBP binding and basal transcription by RNA polymerase II in vitro. This approach, of targeting TATA-adjacent sequences, might prove effective for inhibition of TATA-dependent basal transcription from various mRNA-coding genes.
Figure 5.
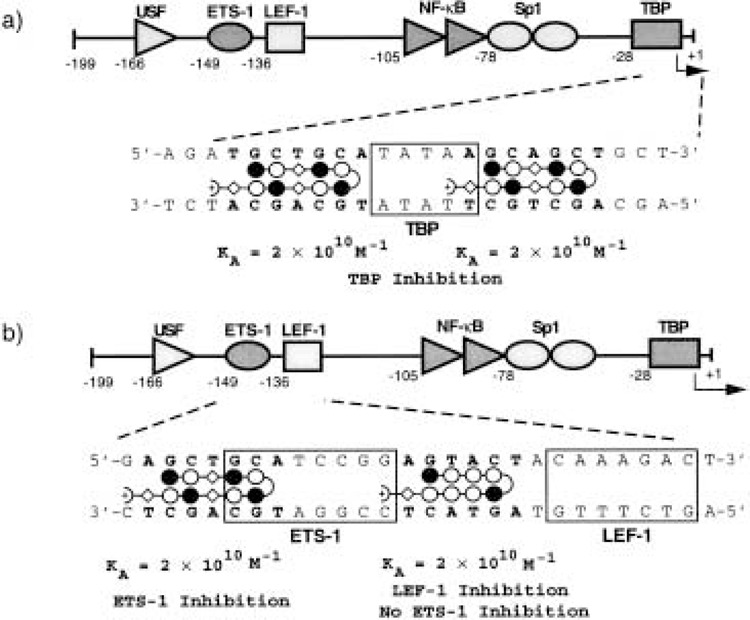
Schematic of the HIV-1 enhancer and promoter (nucleotide positions −199 to +1) showing binding sites for polyamides and the transcription factors upstream stimulatory factor (USF), Ets-1, LEF-1, NF-kB, Sp1, and TFIID (TBP). For polyamide binding models: shaded and unshaded circles, Im and Py rings, respectively; curved lines, γ-aminobutyric acid (γ); diamonds, β-alanine (β); and Dp, dimethylamino-propylamide. (a) A polyamide binds to sequences adjacent to the TATA box element of the HIV-1 promoter and inhibits TBP binding. (b) Polyamides bind adjacent to the binding sites for LEF-1 and Ets-1 and inhibit binding of these transcription factors.
A polyamide has also been synthesized to bind adjacent to the binding site for the lymphoid enhancer factor LEF-1 in the HIV-1 proviral enhancer (19), and was shown to inhibit LEF-1 DNA binding (Fig. 5b). LEF-1 recognizes a specific nucleotide sequence through its HMG domain and proteins containing HMG domains bind DNA in the minor groove and bend and unwind the double helix (27,48) in a manner similar to TBP. Thus, polyamide inhibition of LEF-1 binding might be via the same mechanism as proposed above for inhibition of TBP binding. The LEF-inhibitory polyamide was shown to be effective in inhibition of LEF-1-activated transcription in vitro from the HIV-1 promoter (19).
The DNA-binding activity of the winged-helix–turn–helix transcription factor Ets-1 can also be inhibited by a polyamide that binds adjacent to the Ets-1 site in the HIV-1 proviral enhancer (19). Winged-helix–turn–helix factors interact with their cognate DNA elements through a recognition helix bound in the major groove with additional phosphate contacts on either side of this major groove interaction. The inhibitory polyamide likely interferes with phosphate contacts made by Ets-1 across one adjacent minor groove (Fig. 5b). This polyamide also inhibits the cooperative interaction of Ets-1 and NF-κB on the HIV-1 enhancer (20). Another important family of transcriptional regulators is the basic helix–loop–helix family. DNA recognition by this family of proteins is through basic region–major groove interactions; however, in a recent study, we showed that a polyamide located immediately adjacent to this major groove site could inhibit a specific DNA contact made by a lysine residue in the loop region of the protein (74). This lysine residue likely stretches across the minor groove to make a DNA phosphate contact (22). These findings broaden the application range of the polyamides to proteins that recognize specific DNA sequences in the major groove, as long as these proteins also make additional contacts outside of the major groove. Numerous DNA-binding transcription factors fall into this category [for a recent review, see (75)].
To check for the biological consequences of inhibition of protein–DNA interactions on the HIV-1 promoter and enhancer, HIV-1 replication was monitored in a cell culture assay with normal human donor lymphoid cells. A combination of the polyamides that target the TATA element, LEF-1 and Ets-1, were found to inhibit HIV-1 replication in this system, while mismatch polyamides had no effect (19). The polyamides had no cytotoxic effect on these primary lymphoid cells and actually protected the cells from the cytopathic effect of HIV-1 infection. Inhibition of virus replication with the polyamides was likely due to interference with the DNA-binding activities of the cellular transcription factors utilized by HIV-1, but it is possible that inhibition of cellular genes involved in T-cell activation could have had an indirect effect on HIV-1 replication. To assess this possibility, we performed an RNAase protection assay for transcripts of a battery of cytokine and growth factor genes that are actively transcribed in human lymphoid cells. Some of these genes had single base mismatches from the HIV-1 TATA-flanking sequence; however, the transcript levels for none of these genes were affected by the polyamides that were inhibitory to HIV-1 replication. This lack of inhibition of nontarget gene transcription suggests that the polyamides reduce virus replication in cells by a direct effect on HIV-1 RNA transcription.
Additional studies have demonstrated that polyamides can be used to activate gene expression by interfering with the DNA-binding activity of a viral repressor protein, the human cytomegalovirus IE86 protein (21) (Fig. 6). IE86 represses transcription of the CMV major immediate early promoter through its cognate cis recognition sequence located between the TATA box and the transcription initiation site. IE86-mediated transcriptional repression in vitro was reversed by a match polyamide but not by a mismatch polyamide. In another study, the DNA-binding activity of a viral transactivator, the human T-cell leukemia virus Tax protein, was inhibited by a polyamide, leading to downregulation of HTLV-I transcription (45). Thus, polyamides can be designed to interfere with both cellular and viral transcription factors.
Figure 6.
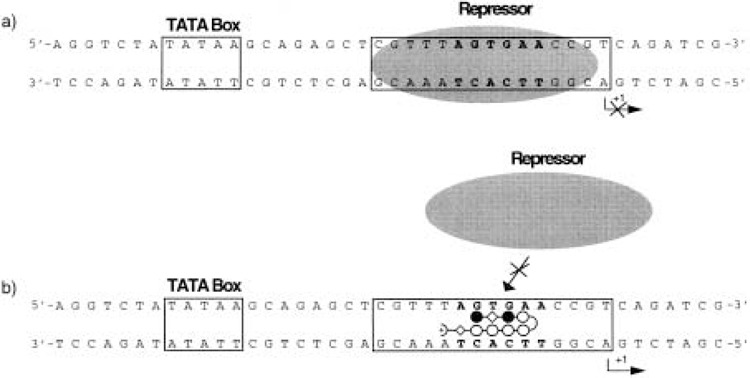
Sequence of the human cytomegalovirus major immediate early promoter region from position —34 to position +8. The TATA box and the repressor binding site (located at —14 to +1 relative to the start site of transcription) are boxed. (a) Without inhibition, repressor binds and blocks transcription. (b) Polyamide binding inhibits repressor binding, and transcription occurs. The polyamide is schematically represented at its DNA-binding site. The black and white circles represent imidazole and pyrrole rings, respectively; the hairpin junction (curved line) is formed with γ-aminobutyric acid, and the diamond represents β-alanine.
In each of the studies mentioned above, the DNA binding proteins either make direct contacts with the minor groove or phosphate contacts across the adjacent minor groove from a major groove interaction, allowing for polyamide inhibition of protein binding (Table 1). In another approach, the Dervan laboratory has synthesized a polyamide conjugated to the tripeptide Arg-Pro-Arg at the carboxy-terminus of the DNA ligand (6). This minor groove-binding polyamide delivers a positive residue to the DNA backbone and was shown to inhibit binding of the bZip transcription factor GCN4 in the adjacent major groove (Fig. 7).
Figure 7.
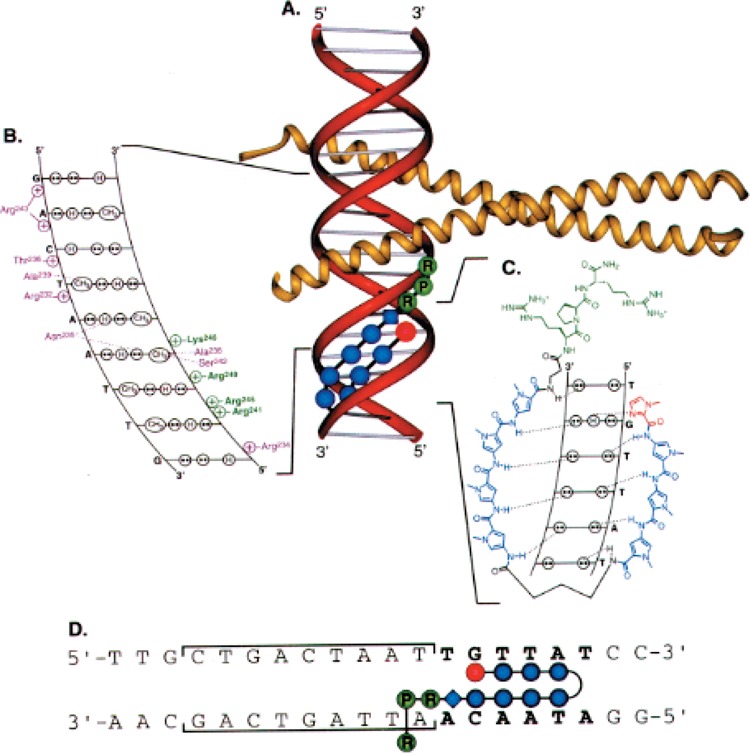
A schematic model of Arg-Pro-Arg (RPR) polyamides targeted to the major groove transcription factor GCN4. (A) The a-helical GCN4 dimer (yellow) is shown binding to adjacent major grooves. The Arg-Pro-Arg-hairpin polyamide is shown as red, blue, and green balls, which represent imidazole, pyrrole, and Arg-Pro-Arg amino acids, respectively. The blue diamond represents β-alanine. γ-Aminobutyric acid is designated as a curved line. (B) The contacts between one GCN4 monomer and the major groove of one half site of 5′-CTGACTAAT-3′ are depicted. Circles with two dots represent the lone pairs of the N7 of purines, the O4 or thymine and the O6 of guanine. Circles containing an H represent the N6 and N4 hydrogens of the exocyclic amines of adenine and cytosine, respectively. The C5 methyl group of thymine is depicted as a circle with CH3 inside. Protein side chains that make hydrogen bonds or van der Waals contacts to the bases are shown in purple and connected to the DNA via a dotted line. Green and purple plus signs represent protein residues that electrostatically contact the phosphate backbone. The residues that are predicted to be disrupted by an Arg-Pro-Arg polyamide are shown in green. (C) The hydrogen-bonding model of the eight-ring hairpin polyamide ImPyPyPy-y-PyPyPyPy-P-RPR bound to the minor groove of 5′-TGTTAT-3′. Circles with two dots represent the lone pairs of N3 of purines and O2 of pyrimidines. Circles containing an H represent the N2 hydrogens of guanines. Putative hydrogen bonds are illustrated by dotted lines. Py and Im rings are represented as blue and red rings, respectively. The Arg-Pro-Arg moiety is green. (D) The model of the polyamide binding its target site (bold) adjacent to the GCN4 binding site (brackets). Polyamide residues are as in (A).
Small-Molecule Activators of Transcription
It is well established that eukaryotic transcription factors contain modular activation and/or repression domains that can be artificially tethered to selected DNA-binding domains [see, e.g., (54)]. One way to target such activation and/or repression domains to a particular promoter or mRNA coding sequence is to chemically link these small protein motifs to a sequence-specific Py-Im polyamide. Activation of a reporter gene in vitro has been achieved with such an approach by tethering a dimerization element and acidic helical peptide to a hairpin polyamide (50). In these experiments, the coiled-coil dimerization element from the yeast transcription factor GCN4 and a synthetic acidic helical (AH) activation domain were chemically coupled to a nanomolar-binding Py-Im polyamide (Fig. 8). When this 4.2-kDa synthetic transcriptional activator was bound to a DNA template harboring three palindromic binding sites for the polyamide, a 13-fold activation of transcription was found in a yeast nuclear extract. Conjugates lacking the AH domain or those containing a mismatch Py-Im polyamide segment failed to activate transcription above basal levels. Other conjugates, in which the GCN4 coiled-coil was replaced with flexible straight-chain linkers, also activated transcription. Because the Py-Im polyamides can be synthesized to target a wide range of predetermined DNA sequences, this novel class of artificial transcription factors represents a key step toward upregulation of physiologically relevant genes by small molecules.
Figure 8.
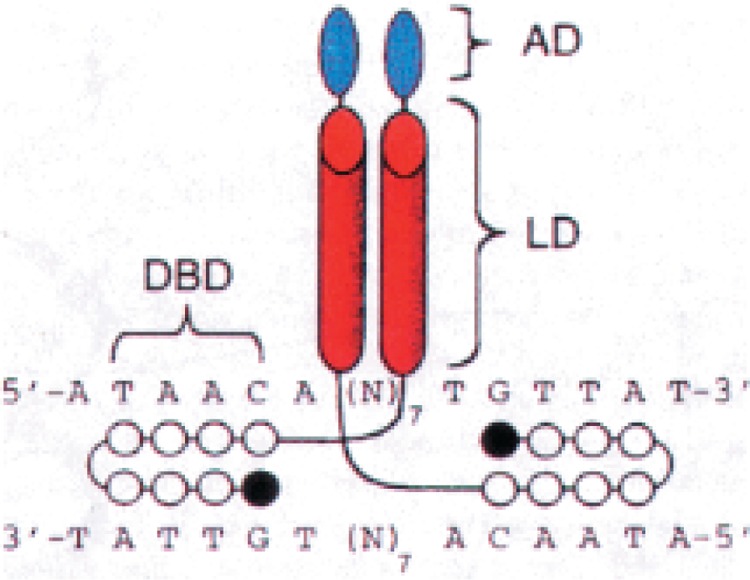
A synthetic activator consisting of an activation domain (AD), a dimerization or linker domain (LD), and a DNA-binding domain (DBD) complexed with the cognate palindromic DNA site of the hairpin polyamide-peptide conjugate. The black and white circles represent imidazole and pyrrole rings, respectively, and the hairpin junction (curved line) is formed with γ-aminobutyric acid.
Metallointercalators
Barton and coworkers have developed a novel series of metallointercalators that bind DNA in the major groove [(32,58) and references therein]. These are phenanthrenequinone diimine (phi) complexes of rhodium (III), such as Rh(phen)2phi3+, Δ-α-[Rh(R,R)-Me2trien]phi3+ (where Me2trien = 2,9-diamino-4,7-diazadecane), and Rh(MGP)2phi5+ (where MGP = 4-guanidylmethyl-1,10-phenanthroline) (Fig. 9). The phi ligand is an extended aromatic heterocycle that intercalates between DNA base pairs on the major groove side of the DNA helix. The other ligands of these phi complexes can be modified to provide sequence specificity in DNA recognition through base-specific interactions in the major groove. For example, Δ-α-[Rh(R,R)-Me2trien]phi3+ has been shown to recognize the sequence 5′-TGCA-3′ through hydrogen-bonding interactions between the axial amines of the ligand and guanines and by van der Waals contacts between methyl groups on the ligand and thymine methyls. Additionally, (Λ1-Rh(MGP)2phi5+ has been shown to bind preferentially to the sequence 5′-CATATG-3′ by a combination of direct readout and shape selection. Recently, the crystal structure of a rhodium intercalator bound to DNA has been solved at high resolution (38). As expected, the rhodium complex intercalates via the major groove. Specific contacts are formed with the edges of the bases at the target site and the phi ligand is deeply inserted into the DNA base pair stack. Although there is no change in sugar pucker from B-type DNA, the helical rise per residue doubles at the site of intercalation. Thus, the authors of this study conclude that the intercalator can be viewed as an additional base pair with specific functional groups positioned in the major groove.
Figure 9.
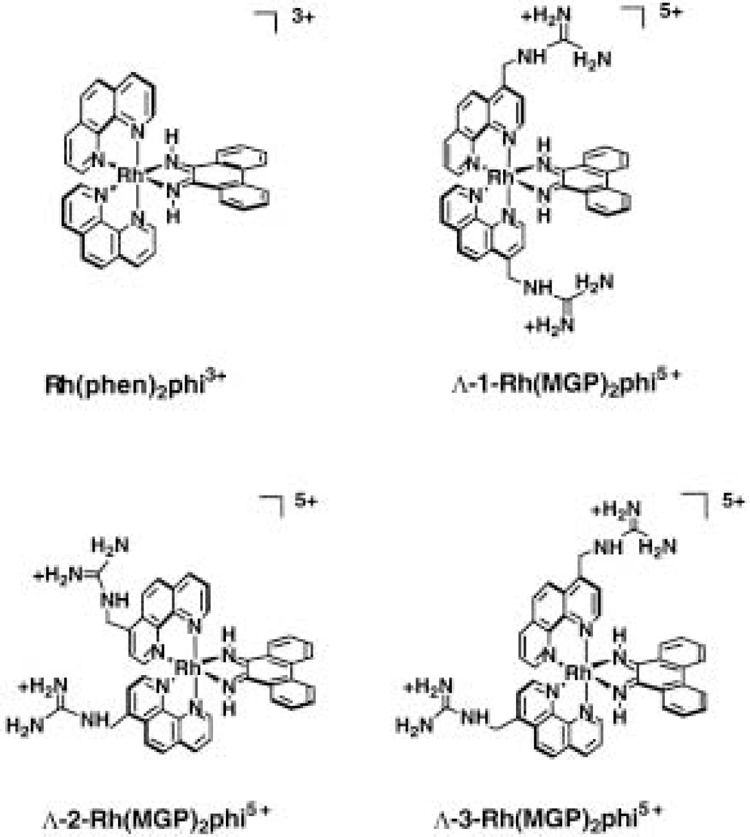
Structures of metallointercalators that bind DNA in the major groove.
When the binding site for Λ-1-Rh(MGP)2phi5+ was placed within the recognition element for the bZip transcription factor yAP-1, the yeast homologue of mammalian AP-1, this ligand was found to inhibit protein binding, with 50% inhibition at ∼100 nM ligand concentration. Inhibition was shown to be dependent on the presence of the target site in the DNA sequence and on the geometry of the ligand. Geometric isomers of Λ-1-Rh(MGP)2phi5+ showed no specific binding to the target site and failed to inhibit protein–DNA interactions. Additional studies in the Barton laboratory have established that peptide conjugates of rhodium complexes can be used to fine tune the sequence specificity of DNA binding [(32) and references therein]. For example, coupling of the 13 amino-acid recognition helix of a bacterial repressor protein to a relatively sequence-neutral metallointercalator partially conferred the sequence specificity of the parent protein on the metallointercalator. As for the parent protein, amino acid substitutions in the peptide alter the sequence preference of the conjugate (32). Thus, the metallointercalators and their peptide conjugates offer promise toward the development of highly specific inhibitors of major groove-binding proteins and transcription of genes that are regulated by such factors.
Combinatorial Approaches
Rebek and coworkers have described the synthesis and screening of small-molecule libraries for identification of novel compounds that bind DNA and inhibit transcription factor–DNA interactions (63). Tetraurea-based libraries were synthesized by solution methods and were derived by coupling modified amino acids to the tetraisocyanate of xanthene. Because two families of known DNA intercalators (the acridines and actinomycins) contain core structures resembling xanthene, it seemed likely that the xanthine core might intercalate between DNA base pairs and the linked amino acid side chains might provide base-specific interactions. Using eight amino acid derivatives per library, each library contained 2080 components and a total of five independent libraries, with different amino acid mixtures, were screened. A gel mobility shift assay used to monitor the effects of the library components on specific transcription factor–DNA interactions and deconvolution methods were employed to identify two active compounds (Fig. 10), each of which inhibited complex formation at 5–10 μM concentration. Two sea urchin-derived transcription factors were analyzed, SpP3A2 and the zinc finger transcription factor SpZ12-1. Surprisingly, both active compounds inhibited both transcription factors even though different target DNA sequences were bound by these two factors. This suggests that binding by the compounds was not DNA sequence specific and the precise mechanism of inhibition remains to be determined. Nonetheless, combinatorial synthesis approaches such as this offer the possibility for identification of new transcription antagonists.
Figure 10.
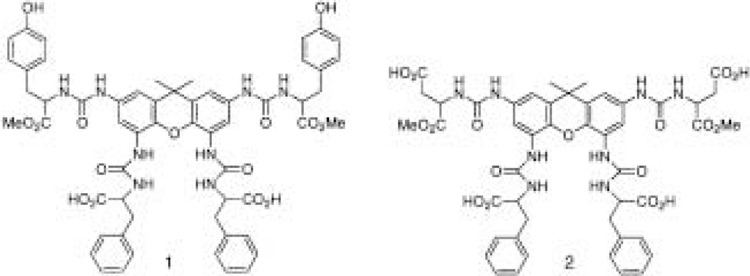
Structures of two xanthene tetraureas active in binding to DNA.
SMALL MOLECULE RNA LIGANDS
Transcription and RNA Transport Inhibitors
Synthetic ligands have been developed to bind specific RNAs in order to interfere with gene expression at the levels of transcription, RNA transport, and translation. Considerable effort has been expended to develop ligands for the HIV-1 transactivation response region (TAR) and the Rev response element (RRE). TAR is the binding site for the viral Tat protein, and is located near the 5′ end of the nascent viral RNA. Tat is also a protein partner of the host cell transcription elongation factor b kinase (TEFb, composed of cyclin dependent kinase 9 and cyclin T). Once recruited to the Tat–TAR complex, TEFb phosphorylates the carboxy-terminal domain of the largest subunit of host cell RNA polymerase II, leading to high levels of transcription. In one approach, a small circular RNA decoy containing the stem, bulge, and loop of HIV-1 TAR was used to effectively compete with Tat and its cellular cofactors in cell-free extracts (4). Additionally, small-molecule inhibitors of this protein–RNA interaction have been selected from compound libraries (51) and synthesized based on structural characterization of the protein-RNA complex (26,31). These molecules have been shown to interfere with Tat-TAR interactions in vitro and, in one instance, in both Tat-activated transcription and HIV-1 replication in lymphoid cells (51). In another approach, TAR-binding synthetic tripeptides were isolated from an encoded combinatorial library of ∼24,000 members consisting of both d- and l-amino acids (35). The most effective tripeptide, NH2-(l)Lys-(d)Lys-(l)Asn-OH, inhibited Tat transactivation in human cells with an IC50 of ∼50 nM.
Another HIV-1 RNA target of interest is the Rev response element or RRE, the target of the viral Rev protein. Rev is important for nuclear-cytoplasmic transit of viral RNA for synthesis of HIV-1 structural proteins. Green and coworkers have identified small-molecule inhibitors of the Rev–RRE interaction that are active inhibitors of virus replication, including both aminoglycosides such as neomycin B (77) and novel aromatic heterocyclic compounds (78). Although these studies have so far focused on HIV-1, small-molecule approaches offer the promise for inhibition of a variety of protein–RNA interactions involved in transcription, RNA processing, and RNA transport.
Small-Molecule Inhibitors of Translation
Based on the finding that aminoglycosides can bind the HIV-1 RRE, Werstuck and Green (72) have devised a novel strategy for regulation of translation with small molecules in living cells. These authors selected short RNA aptamers that specifically bind to either aminoglycoside or Hoechst minor groove-binding ligands. These RNA aptamers were selected in vitro from randomized pools of RNAs. When these sequences were inserted into the 5′-untranslated region of reporter mRNAs, these ligands specifically inhibited translation of the RNAs harboring their cognate binding sites. These experiments were performed with bacterial cells (for the aminoglycosides tobramycin and kanamycin) and with mammalian cells in culture (for the Hoechst dyes H33258 and H33342). For this strategy to be practical in human medicine, small molecules will need to be selected from compound libraries or rationally synthesized to bind to specific RNA sequences in the messenger RNAs be downregulated. Nonetheless, the success of this aptamer study (72) indicates that small-molecule regulation of translation is also a feasible approach to manipulate gene expression.
BLOCKING TRANSCRIPTION WITH PROTEIN PHOSPHORYLATION INHIBITORS
Flores and colleagues (23,49) have identified a series of compounds that inhibit the TEFb protein kinase activity that is required for Tat activation of HIV-1 transcription (see above). These inhibitor studies establish the role of this kinase in Tat-activated transcription and show a specific effect of these inhibitors on both HIV-1 transcription and virus replication (23). These compounds were identified from an in vitro screen of a library >100,000 compounds for inhibition of Tat-dependent HIV-1 transcription, and included the well-nown inhibitor of RNA polymerase II transcription, 5,6-dichloro-1-β-d-ribofura-nosylbenzimidazole (DRB) and related nucleosides, benzimidazoles, isoquinoline sulfonamides, flavonoids, and other novel compounds (49). Although these compounds were isolated based on an in vitro assay, the ability of each compound to inhibit Tat activation in a cell culture assay was directly proportional to its in vitro activity. Strikingly, these compounds had no effect on the transcription of control genes, either in vitro or in transient transfection assays. Moreover, DRB, a benzimidazole and an isoxazole were also potent inhibitors of HIV-1 replication (23). Thus, small-molecule inhibitors of enzymes required for postsynthetic modifications of transcription factors and RNA polymerase might prove to be valuable reagents for regulation of specific gene expression.
ACKNOWLEDGMENTS
Research in the J.M.G. and P.B.D. laboratories is supported by grants from the National Institutes of General Medical Sciences. J.M.T. thanks the Howard Hughes Medical Institute for a predoctoral fellowship. We thank Dr. J. K. Barton for providing Figure 9.
REFERENCES
- 1. Beerli R. R.; Segal D. J.; Dreier B.; Barbas C. F. 3rd. Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. USA 95:14628–14633; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bellorini M.; Moncollin V.; D’Incalci M.; Mongelli N.; Mantovani R. Distamycin A and tallimustine inhibit TBP binding and basal in vitro transcription. Nucleic Acids Res. 23:1657–1673; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bianchi N.; Passadore M.; Rutigliano C.; Feriotto G.; Mischiati C.; Gambari R. Targeting of the Sp1 binding sites of HIV-1 long terminal repeat with chro-momycin. Disruption of nuclear factor.DNA complexes and inhibition of in vitro transcription. Biochem. Pharmacol. 52:1489–1498; 1996. [DOI] [PubMed] [Google Scholar]
- 4. Bohjanen P. R.; Liu Y.; Garcia-Blanco M. A. TAR RNA decoys inhibit tat-activated HIV-1 transcription after preinitiation complex formation. Nucleic Acids Res. 25:4481–4486; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breaker R. R. Catalytic DNA: In training and seeking employment. Nat. Biotechnol. 17:422–423; 1999. [DOI] [PubMed] [Google Scholar]
- 6. Bremer R. E.; Baird E. E.; Dervan P. B. Inhibition of major-groove-binding proteins by pyrrole-imidazole polyamides with an Arg-Pro-Arg positive patch. Chem. Biol. 5:119–133; 1998. [DOI] [PubMed] [Google Scholar]
- 7. Bruice T. C.; Sengupta D.; Blasko A.; Chiang S. Y.; Beerman T. A. A microgonotropen branched decaaza decabutylamine and its DNA and DNA/transcription factor interactions. Bioorg. Med. Chem. 5:685–692; 1997. [DOI] [PubMed] [Google Scholar]
- 8. Chiang S. Y.; Azizkhan J. C.; Beerman T. A. A comparison of DNA-binding drugs as inhibitors of E2F1-and Sp1-DNA complexes and associated gene expression. Biochemistry 37:3109–3115; 1998. [DOI] [PubMed] [Google Scholar]
- 9. Chiang S. Y.; Bruice T. C.; Azizkhan J. C.; Gawron L.; Beerman T. A. Targeting E2F1-DNA complexes with microgonotropen DNA binding agents. Proc. Natl. Acad. Sci. USA 94:2811–2816; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiang S. Y.; Welch J.; Rauscher F. J. 3rd; Beerman T. A. Effects of minor groove binding drugs on the interaction of TATA box binding protein and TFIIA with DNA. Biochemistry 33:7033–7040; 1994. [DOI] [PubMed] [Google Scholar]
- 11. Chiang S. Y.; Welch J. J.; Rauscher F. J. 3rd; Beerman T. A. Effect of DNA-binding drugs on early growth response factor-1 and TATA box-binding protein complex formation with the herpes simplex virus latency promoter. J. Biol. Chem. 271:23999–234004; 1996. [DOI] [PubMed] [Google Scholar]
- 12. Choo Y.; Castellanos A.; Garcia-Hernandez B.; Sanchez-Garcia I.; Klug A. Promoter-specific activation of gene expression directed by bacteriophage-selected zinc fingers. J. Mol. Biol. 273:525–532; 1997. [DOI] [PubMed] [Google Scholar]
- 13. Choo Y.; Sanchez-Garcia I.; Klug A. In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature 372:642–645; 1994. [DOI] [PubMed] [Google Scholar]
- 14. Clemens K. R.; Liao X. B.; Wolf V.; Wright P. E.; Gottesfeld J. M. Definition of the binding sites of individual zinc fingers in the transcription factor IIIA-5 S RNA gene complex. Proc. Natl. Acad. Sci. USA 89: 10822–10826; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cook P. D. Making drugs out of oligonucleotides: A brief review and perspective. Nucleosides Nucleotides 18:1141–1162; 1999. [DOI] [PubMed] [Google Scholar]
- 16. Cutts S. M.; Parsons P. G.; Sturm R. A.; Phillips D. R. Adriamycin-induced DNA adducts inhibit the DNA interactions of transcription factors and RNA polymerase. J. Biol. Chem. 271:5422–5429; 1996. [DOI] [PubMed] [Google Scholar]
- 17. Denison C.; Kodadek T. Small-molecule-based strategies for controlling gene expression. Chem. Biol. 5: 129–145; 1998. [DOI] [PubMed] [Google Scholar]
- 18. Dervan P. B.; Burli R. W. Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol. 3: 688–693; 1999. [DOI] [PubMed] [Google Scholar]
- 19. Dickinson L. A.; Gulizia R. J.; Trauger J. W.; Baird E. E.; Mosier D. E.; Gottesfeld J. M.; Dervan P. B. Inhibition of RNA polymerase II transcription in human cells by synthetic DNA-binding ligands. Proc. Natl. Acad. Sci. USA 95:12890–12895; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dickinson L. A.; Trauger J. W.; Baird E. E.; Dervan P. B.; Graves B. J.; Gottesfeld J. M. Inhibition of Ets-1 DNA binding and ternary complex formation between Ets-1, NF-kB, and DNA by a designed DNA-binding ligand. J. Biol. Chem. 274:12765–12773; 1999. [DOI] [PubMed] [Google Scholar]
- 21. Dickinson L. A.; Trauger J. W.; Baird E. E.; Ghazal P.; Dervan P. B.; Gottesfeld J. M. Anti-repression of RNA polymerase II transcription by pyrrole-imidazole polyamides. Biochemistry 38:10801–10807; 1999. [DOI] [PubMed] [Google Scholar]
- 22. FerréD’Amaré A. R.; Prendergast G. C.; Ziff E. B.; Burley S. K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363:38–45; 1993. [DOI] [PubMed] [Google Scholar]
- 23. Flores O.; Lee G.; Kessler J.; Miller M.; Schlief W.; Tomassini J.; Hazuda D. Host-cell positive transcription elongation factor b kinase activity is essential and limiting for HIV type 1 replication. Proc. Natl. Acad. Sci. USA 96:7208–7213; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Galderisi U.; Cascino A.; Giordano A. Antisense oligonucleotides as therapeutic agents. J. Cell. Physiol. 181:251–257; 1999. [DOI] [PubMed] [Google Scholar]
- 25. Geierstanger B. H.; Mrksich M.; Dervan P. B.; Wemmer D. E. Design of a G. C-specific DNA minor groove-binding peptide. Science 266:646–650; 1994. [DOI] [PubMed] [Google Scholar]
- 26. Gelus N.; Hamy F.; Bailly C. Molecular basis of HIV-1 TAR RNA specific recognition by an acridine tat-antagonist. Bioorg. Med. Chem 7:1075–1079; 1999. [DOI] [PubMed] [Google Scholar]
- 27. Giese K.; Pagel J.; Grosschedl R. Functional analysis of DNA bending and unwinding by the high mobility group domain of LEF-1. Proc. Natl. Acad. Sci. USA 94:12845–12850; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Good L.; Nielsen P. E. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 16:355–358; 1998. [DOI] [PubMed] [Google Scholar]
- 29. Gottesfeld J. M.; Neely L.; Trauger J. W.; Baird E. E.; Dervan P. B. Regulation of gene expression by small molecules. Nature 387:202–205; 1997. [DOI] [PubMed] [Google Scholar]
- 30. Hamel Y.; Lacoste J.; Frayssinet C.; Sarasin A.; Garestier T.; Francois J. C.; Helene C. Inhibition of gene expression by anti-sense C-5 propyne oligonucleotides detected by a reporter enzyme. Biochem. J. 339: 547–553; 1999. [PMC free article] [PubMed] [Google Scholar]
- 31. Hamy F.; Brondani V.; Florsheimer A.; Stark W.; Blommers M. J.; Klimkait T. A new class of HIV-1 Tat antagonist acting through Tat-TAR inhibition. Biochemistry 37:5086–5095; 1998. [DOI] [PubMed] [Google Scholar]
- 32. Hastings C. A.; Barton J. K. Perturbing the DNA sequence selectivity of metallointercalator-peptide conjugates by single amino acid modification. Biochemistry 38:10042–10051; 1999. [DOI] [PubMed] [Google Scholar]
- 33. Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 7:1291–1308; 1993. [DOI] [PubMed] [Google Scholar]
- 34. Ho S. N.; Boyer S. H.; Schreiber S. L.; Danishefsky S. J.; Crabtree G. R. Specific inhibition of formation of transcription complexes by a calicheamicin oligosaccharide: A paradigm for the development of transcriptional antagonists. Proc. Natl. Acad. Sci. USA 91: 9203–9307; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hwang S.; Tamilarasu N.; Ryan K.; Huq I.; Richter S.; Still W. C.; Rana T. M. Inhibition of gene expression in human cells through small molecule-RNA interactions. Proc. Natl. Acad. Sci. USA 96:12997–13002; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kielkopf C. L.; Baird E. E.; Dervan P. B.; Rees D. C. Structural basis for GC recognition in the DNA minor groove. Nat. Struct. Biol. 5:104–109; 1998. [DOI] [PubMed] [Google Scholar]
- 37. Kielkopf C. L.; Bremer R. E.; White S.; Szewczyk J. W.; Turner J. M.; Baird E. E.; Dervan P. B.; Rees D. C. Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove. J. Mol. Biol. 295:557–567; 2000. [DOI] [PubMed] [Google Scholar]
- 38. Kielkopf C. L.; Erkkila K. E.; Hudson B. P.; Barton J. K.; Rees D. C. Structure of a photoactive rhodium complex intercalated into DNA. Nat. Struct. Biol. 7: 117–121; 2000. [DOI] [PubMed] [Google Scholar]
- 39. Kielkopf C. L.; White S.; Szewczyk J. W.; Turner J. M.; Baird E. E.; Dervan P. B.; Rees D. C. A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA. Science 282:111–115.; 1998. [DOI] [PubMed] [Google Scholar]
- 40. Kim J. L.; Nikolov D. B.; Burley S. K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 365:520–527; 1993. [DOI] [PubMed] [Google Scholar]
- 41. Kim J. S.; Pabo C. O. Transcriptional repression by zinc finger peptides. Exploring the potential for applications in gene therapy. J. Biol. Chem. 272:29795–29800; 1997. [DOI] [PubMed] [Google Scholar]
- 42. Kim Y.; Geiger J. H.; Hahn S.; Sigler P. B. Crystal structure of a yeast TBP/TATA-box complex. Nature 365:512–520; 1993. [DOI] [PubMed] [Google Scholar]
- 43. Kurabayashi M.; Jeyaseelan R.; Kedes L. Doxorubicin represses the function of the myogenic helix-loop-helix transcription factor MyoD. Involvement of Id gene induction. J. Biol. Chem. 269:6031–6039; 1994. [PubMed] [Google Scholar]
- 44. Lebruska L. L.; Maher L. J. 3rd., Selection and characterization of an RNA decoy for transcription factor NF-kappa B. Biochemistry 38:3168–3174; 1999. [DOI] [PubMed] [Google Scholar]
- 45. Lenzmeier B. A.; Baird E. E.; Dervan P. B.; Nyborg J. K. The Tax protein–DNA interaction is essential for HTLV-I transactivation in vitro. J. Mol. Biol. 291: 731–744; 1999. [DOI] [PubMed] [Google Scholar]
- 46. Liu C.; Smith B. M.; Ajito K.; Komatsu H.; Gomez-Paloma L.; Li T.; Theodorakis E. A.; Nicolaou K. C.; Vogt P. K. Sequence-selective carbohydrate–DNA interaction: Dimeric and monomeric forms of the calicheamicin oligosaccharide interfere with transcription factor function. Proc. Natl. Acad. Sci. USA 93: 940–944; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lohse J.; Dahl O.; Nielsen P. E. Double duplex invasion by peptide nucleic acid: A general principle for sequence-specific targeting of double-stranded DNA. Proc. Natl. Acad. Sci. USA 96:11804–11808; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Love J. J.; Li X.; Case D. A.; Giese K.; Grosschedl R.; Wright P. E. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature 376:791–795; 1995. [DOI] [PubMed] [Google Scholar]
- 49. Mancebo H. S.; Lee G.; Flygare J.; Tomassini J.; Luu J.; Zhu Y.; Peng J.; Blau C.; Hazuda D.; Price D.; Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633–2644; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mapp A. K.; Ansari A. Z.; Ptashne M.; Dervan P. B. Activation of gene expression by small molecule transcription factors. Proc. Natl. Acad. Sci. USA 97: 3930–3935; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mei H. Y.; Cui M.; Heldsinger A.; Lemrow S. M.; Loo J. A.; Sannes-Lowery K. A.; Sharmeen L.; Czarnik A. W. Inhibitors of protein–RNA complexation that target the RNA: Specific recognition of human immunodeficiency virus type 1 TAR RNA by small organic molecules. Biochemistry 37:14204–14212; 1998. [DOI] [PubMed] [Google Scholar]
- 52. Mischiati C.; Borgatti M.; Bianchi N.; Rutigliano C.; Tomassetti M.; Feriotto G.; Gambari R. Interaction of the human NF-kappaB p52 transcription factor with DNA-PNA hybrids mimicking the NF-kappaB binding sites of the human immunodeficiency virus type 1 promoter. J. Biol. Chem. 274:33114–33122; 1999. [DOI] [PubMed] [Google Scholar]
- 53. Neely L.; Trauger J. W.; Baird E. E.; Dervan P. B.; Gottesfeld J. M. Importance of minor groove binding zinc fingers within the transcription factor IIIA DNA complex. J. Mol. Biol. 274:439–445; 1997. [DOI] [PubMed] [Google Scholar]
- 54. Nevado J.; Gaudreau L.; Adam M.; Ptashne M. Transcriptional activation by artificial recruitment in mammalian cells. Proc. Natl. Acad. Sci. USA 96: 2674–2677; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nicolaou K. C.; Pitsinos E. N.; Theodorakis E. A.; Saimoto H.; Wrasidlo W. Synthetic calicheamicin mimics with novel initiation mechanisms: DNA cleavage, cytotoxicity, and apoptosis. Chem. Biol. 1:57–66; 1994. [DOI] [PubMed] [Google Scholar]
- 56. Nielsen P. E. Peptide nucleic acids as therapeutic agents. Curr. Opin. Struct. Biol. 9:353–357; 1999. [DOI] [PubMed] [Google Scholar]
- 57. Oakley M. G.; Mrksich M.; Dervan P. B. Evidence that a minor groove-binding peptide and a major groove-binding protein can simultaneously occupy a common site on DNA. Biochemistry 31:10969–10975; 1992. [DOI] [PubMed] [Google Scholar]
- 58. Odom D. T.; Parker C. S.; Barton J. K. Site-specific inhibition of transcription factor binding to DNA by a metallointercalator. Biochemistry 38:5155–5163; 1999. [DOI] [PubMed] [Google Scholar]
- 59. Raillard S. A.; Joyce G. F. Targeting sites within HIV-1 cDNA with a DNA-cleaving ribozyme. Biochemistry 35:11693–11701; 1996. [DOI] [PubMed] [Google Scholar]
- 60. Rivera V. M. Controlling gene expression using synthetic ligands. Methods 14:421–429; 1998. [DOI] [PubMed] [Google Scholar]
- 61. Rowley P. T.; Kosciolek B. A.; Kool E. T. Circular antisense oligonucleotides inhibit growth of chronic myeloid leukemia cells. Mol. Med. 5:693–700; 1999. [PMC free article] [PubMed] [Google Scholar]
- 62. Santoro S. W.; Joyce G. F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA 94: 4262–4266; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shipps G. W. Jr.; Pryor K. E.; Xian J.; Skyler D. A.; Davidson E. H.; Rebek J. Jr. Synthesis and screening of small molecule libraries active in binding to DNA. Proc. Natl. Acad. Sci. USA 94:11833–11838; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sissi C.; Aiyar J.; Boyer S.; Depew K.; Danishefsky S.; Crothers D. M. Interaction of calicheamicin gam-ma1(I) and its related carbohydrates with DNA–protein complexes. Proc. Natl. Acad. Sci. USA 96:10643–10648; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smith A. L.; Nicolaou K. C. The enediyne antibiotics. J. Med. Chem. 39:2103–2117; 1996. [DOI] [PubMed] [Google Scholar]
- 66. Soukup G. A.; Breaker R. R. Nucleic acid molecular switches. Trends Biotechnol. 17:469–476; 1999. [DOI] [PubMed] [Google Scholar]
- 67. Sun L. Q.; Cairns M. J.; Gerlach W. L.; Witherington C.; Wang L.; King A. Suppression of smooth muscle cell proliferation by a c-myc RNA-cleaving deoxyribozyme. J. Biol. Chem. 274:17236–17241; 1999. [DOI] [PubMed] [Google Scholar]
- 68. Taylor A.; Webster K. A.; Gustafson T. A.; Kedes L. The anti-cancer agent distamycin A displaces essential transcription factors and selectively inhibits myogenic differentiation. Mol. Cell. Biochem. 169:61–72; 1997. [DOI] [PubMed] [Google Scholar]
- 69. Trauger J. W.; Baird E. E.; Dervan P. B. Subnanomolar sequence-specific recognition in the minor groove of DNA by designed ligands. Nature 382:559–561; 1996. [DOI] [PubMed] [Google Scholar]
- 70. Welch J. J.; Rauscher F. J. 3rd; Beerman T. A. Targeting DNA-binding drugs to sequence-specific transcription factor. DNA complexes. Differential effects of intercalating and minor groove binding drugs. J. Biol. Chem. 269:31051–31058; 1994. [PubMed] [Google Scholar]
- 71. Wemmer D. E.; Dervan P. B. Targeting the minor groove of DNA. Curr. Opin. Struct. Biol. 7:355–361; 1997. [DOI] [PubMed] [Google Scholar]
- 72. Werstuck G.; Green M. R. Controlling gene expression in living cells through small molecule-RNA interactions. Science 282:296–298; 1998. [DOI] [PubMed] [Google Scholar]
- 73. White S.; Szewczyk J. W.; Turner J. M.; Baird E. E.; Dervan P. B. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature 391:468–471; 1998. [DOI] [PubMed] [Google Scholar]
- 74. Winston R. L.; Ehley J. A.; Baird E. E.; Dervan P. B.; Gottesfeld J. M. Asymmetric DNA binding by a homodimeric bHLH protein. Biochemistry 39:9092–9098; 2000. [DOI] [PubMed] [Google Scholar]
- 75. Wolberger C. Multiprotein–DNA complexes in transcriptional regulation. Annu. Rev. Biophys. Biomol. Struct. 28:29–56; 1999. [DOI] [PubMed] [Google Scholar]
- 76. Zain R.; Marchand C.; Sun J.; Nguyen C. H.; Bisagni E.; Garestier T.; Helene C. Design of a triple-helix-specific cleaving reagent. Chem. Biol. 6:771–777; 1999. [DOI] [PubMed] [Google Scholar]
- 77. Zapp M. L.; Stern S.; Green M. R. Small molecules that selectively block RNA binding of HIV-1 Rev protein inhibit Rev function and viral production. Cell 74: 969–978; 1993. [DOI] [PubMed] [Google Scholar]
- 78. Zapp M. L.; Young D. W.; Kumar A.; Singh R.; Boykin D. W.; Wilson W. D.; Green M. R. Modulation of the Rev-RRE interaction by aromatic heterocyclic compounds. Bioorg. Med. Chem. 5:1149–1155; 1997. [DOI] [PubMed] [Google Scholar]


