Abstract
Migraineurs show an increased load of white matter hyperintensities (WMHs) and more rapid deep WMH progression. Previous methods for WMH segmentation have limited efficacy to detect small deep WMHs. We developed a new fully automated detection pipeline, DEWS (DEep White matter hyperintensity Segmentation framework), for small and superficially-located deep WMHs. A total of 148 non-elderly subjects with migraine were included in this study. The pipeline consists of three components: 1) white matter (WM) extraction, 2) WMH detection, and 3) false positive reduction. In WM extraction, we adjusted the WM mask to re-assign misclassified WMHs back to WM using many sequential low-level image processing steps. In WMH detection, the potential WMH clusters were detected using an intensity based threshold and region growing approach. For false positive reduction, the detected WMH clusters were classified into final WMHs and non-WMHs using the random forest (RF) classifier. Size, texture, and multi-scale deep features were used to train the RF classifier. DEWS successfully detected small deep WMHs with a high positive predictive value (PPV) of 0.98 and true positive rate (TPR) of 0.70 in the training and test sets. Similar performance of PPV (0.96) and TPR (0.68) was attained in the validation set. DEWS showed a superior performance in comparison with other methods. Our proposed pipeline is freely available online to help the research community in quantifying deep WMHs in non-elderly adults.
Keywords: Deep white matter hyperintensity, Automated detection, Migraine
Highlights
-
•
A fully automated deep white matter hyperintensity detection algorithm was developed.
-
•
An exquisite white matter mask region was constructed.
-
•
Size, texture, and multi-scale deep features were used for false positive reduction.
-
•
DEWS detected white matter hyperintensities with a high positive predictive value.
1. Introduction
Migraine is neurological disorder affecting ~20% of people worldwide. While it is believed that migraine is a benign disease, the risk of stroke, cardiovascular diseases, and death is increased in migraineurs (Kurth et al., 2016). Migraineurs show an increased load of white matter hyperintensities (WMHs) and more rapid WMH progression than migraine-free controls (Erdélyi-Bõtor et al., 2015; Kruit et al., 2004; Palm-Meinders et al., 2012). In addition, common psychiatric comorbidities of migraine such as depression and increased suicidality are also associated with increased WMH load (Herrmann et al., 2008; Serafini et al., 2011). A vascular hypothesis is commonly proposed as a possible pathophysiology underlying deep WMH development, while the development of periventricular WMH is currently more debated (Fazekas et al., 1993; Fernando et al., 2006).
WMHs have been linked to several neurological disorders such as vascular cognitive impairment. WMHs are also prevalent in the healthy population, which has led to debate on the clinical importance of WMHs in asymptomatic subjects (Mineura et al., 1995). However, recent studies have shown that WMHs are associated with an increased risk of cognitive decline, incident dementia, ischemic stroke, and death in asymptomatic healthy subjects (Debette and Markus, 2010; Murray et al., 2010; Vermeer et al., 2003). While the causative role of WMH for these conditions is still considered controversial, these findings may indicate that WMH can be a marker of brain damage, warranting more research on their development in earlier life.
As first suggested by Fazekas et al., WMHs have been classified into periventricular and deep WMHs (Fazekas et al., 1987). Risk factors and clinical implications differ between the two types of WMHs (Griffanti et al., 2017; Kim et al., 2008). In the CAMERA-2 study, women with migraine had a higher incidence and progression of deep WMHs, while such an association was not found for periventricular WMHs (Palm-Meinders et al., 2012). Longitudinal studies demonstrated that the progression of periventricular WMH is associated with a decline in cognitive function and cerebral blood flow, while no such association was found with deep WMHs (Seo et al., 2012; ten Dam et al., 2007; Van Dijk et al., 2008). Different pathogeneses may be involved in the development of periventricular and deep WMHs (Kim et al., 2008). Autopsy studies suggested that deep WMHs were of hypoxic/ischemic origin, while periventricular WMHs seldom showed markers of ischemia. Periventricular WMHs are strongly related to advanced age and arterial hypertension, but this association is weaker for deep WMHs (Griffanti et al., 2017). Taken together, deep WMHs might be more relevant to migraine and its ischemic complications than periventricular WMHs.
Currently available methods for automated quantification of WMH are less robust in the segmentation of small, juxtacortical deep WMHs (Griffanti et al., 2017). In previous studies on WMH segmentation, only elderly subjects with a high load of both periventricular and deep WMHs were recruited (Griffanti et al., 2016; Jeon et al., 2011; Klöppel et al., 2011; Yoshita et al., 2006). However, in young healthy subjects, WMHs are often discrete, small-sized, and located in the deep white matter (Hopkins et al., 2006). Therefore, accuracy of the detection of small, superficially-located WMHs has not been adequately evaluated in the literature. Furthermore, a simple intensity-based thresholding technique has been widely used to detect WMHs in previous studies (Hulsey et al., 2012; Ithapu et al., 2014; Jeon et al., 2011; Klöppel et al., 2011). However, this technique is not optimal for detection of small or low-intensity WMHs because lowering the threshold of WMH segmentation increases the rate of false-positives. In addition, this technique might underestimate superficially-located deep WMHs due to the similar intensities between gray matter (GM) and WMHs (Jeon et al., 2011). However, when examining WMHs among young healthy individuals, it is crucial to detect small, relatively low-intensity, and superficially-located deep WMHs, which have been difficult to identify to date. To overcome the limitations of previous detection methods, several characteristics of deep WMHs, such as intensity value, shape, and location should be considered.
In the current study, we developed a new, fully-automated, machine learning-based pipeline for detecting deep WMHs, DEWS (DEep White matter hyperintensity Segmentation framework), using non-elderly migraineurs. For accurate detection of small, superficially-located deep WMHs, we established a new procedure for WM mask extraction and a classification model based on size, texture and multi-scale deep features as well as intensity threshold information.
2. Materials and methods
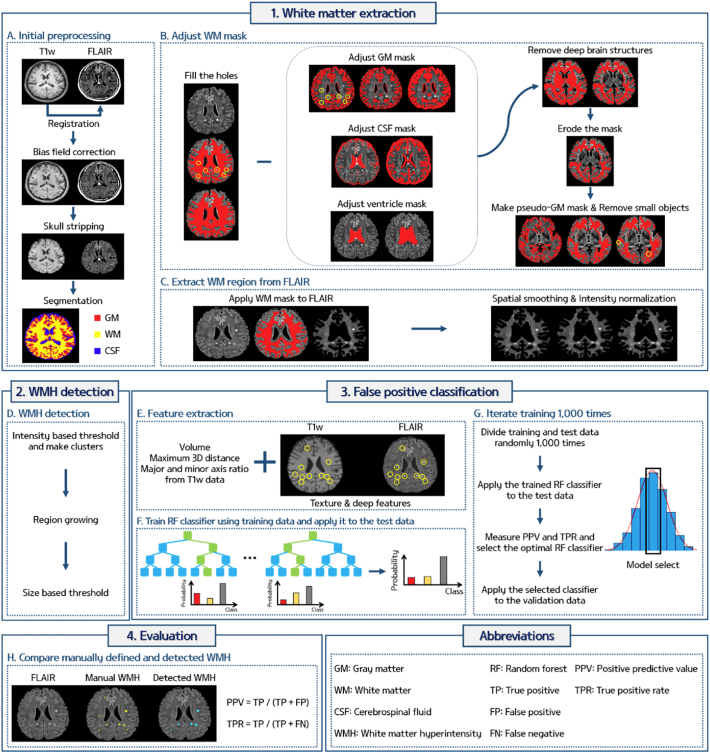
The proposed pipeline of this study consisted of three components: 1) WM extraction, 2) WMH detection, and 3) false positive (FP) reduction. The overall scheme of our pipeline is given in Fig. 1.
Fig. 1.
The overall scheme of the pipeline for automated deep WMH detection.
2.1. Participants and imaging data
We prospectively collected magnetic resonance imaging (MRI) data of new patients diagnosed with migraine at the Samsung Medical Center headache clinic from January 2015 to January 2017. The diagnosis of migraine was confirmed by two headache specialists (MJL and C-SC) based on the International Classification of Headache Disorders-3rd edition beta version (ICHD-3 beta) (Headache Classification Committee of the International Headache Society [IHS], 2013). We included patients with 1.1 migraine without aura, 1.2.1 migraine with typical aura, and 1.3 chronic migraine. A total of 233 non-elderly patients aged ≤ 65 who voluntarily underwent brain MRI during the study period were considered eligible for the analysis. After reviewing all MRI data, we excluded 67 subjects with motion-related artifacts and 18 subjects who did not have deep WMHs in their MRI scan. Finally, 148 subjects were enrolled in the study. This study was approved by the Institutional Review Board (IRB) of Samsung Medical Center. Written consent was waived by the IRB.
The T1-weighted and fluid attenuated inversion recovery (FLAIR) MRI scans were acquired using a 3 Tesla MR scanner (Achieva, Philips Medical Systems, Best, Netherlands). The imaging parameters of T1-weighted data were as follows: repetition time (TR) = 9.9 ms; echo time (TE) = 4.6 ms; field of view (FOV) = 240 × 240 mm2; acquisition matrix = 480 × 480 pixels; and slice thickness = 1 mm with 360 slices. The imaging parameters of the FLAIR data were as follows: TR = 11,000 ms; TE = 125 ms; inversion time = 2800 ms; FOV = 240 × 240 mm2; acquisition matrix = 512 × 512 pixels; and slice thickness = 2 mm with 80 slices. The same MRI scanner and protocol were applied for all subjects during the study period.
2.2. Manual annotations of WMHs
The manual annotations of deep WMHs were drawn on the 2D slice of FLAIR images by two investigators (MJL, a neurologist with 8 years of experience in clinical neurology, and JC with 11 years of experience in neuroradiology) who were blinded to the clinical information. WMHs were defined as a round- or oval-shaped FLAIR hyperintensity with a variable size in the U-fiber or subcortical WM, which can be discrete or confluent and showed T1 iso- or hypo-intensity (Wardlaw et al., 2013). WMHs were carefully differentiated from subcortical infarctions, perivascular spaces, and artifacts (Kwee and Kwee, 2007; Wardlaw et al., 2013). Periventricular WMHs and lacunes in deep nuclei were excluded from the manual annotations. Periventricular WMH was defined as hyperintensities along the walls of ventricles with an appearance of small caps, thin rims, or confluent lesions (Fazekas et al., 1987; van den Heuvel et al., 2006). The intra-class correlation coefficient between the two raters was 0.994 (95% confidence interval between 0.968 and 0.999) for the number of WMHs for each subject.
2.3. WM extraction
The overall processing was performed using AFNI, FSL, and MATLAB (Cox, 1996; Jenkinson et al., 2012). The T1-weighted and FLAIR data were reoriented to the right-posterior-inferior (RPI) direction and the T1-weighted data were registered onto the FLAIR data using rigid body transformation. The magnetic field bias for both the T1-weighted and FLAIR data was corrected and the skull was removed (Fig. 1A). The T1-weighted data were segmented into GM, WM, and cerebrospinal fluid (CSF) using FSL (Fig. 1A). However, due to the similar intensities between WMH and GM, some voxels of the WMH were misclassified to GM. The following steps were performed to adjust the WM mask to include WMH voxels. The segmented WM mask was dilated and eroded in the axial plane (both x and y directions) with disk size of 5 voxels to fill the holes (shown in yellow circles) in the WM mask which were due to the misclassified WMH voxels (Fig. 1B). The segmented GM mask was adjusted by multiplying the GM partial volume effect (PVE) mask with the complement (i.e., logical negative) of the WM mask of the previous step. The adjusted GM mask was dilated in the axial plane with a disk size of 2 voxels (Fig. 1B). The segmented CSF mask was skeletonized and dilated in the axial direction with a disk size of 6. The ventricle mask was extracted from the segmented CSF mask using the region growing method in each slice. The ventricle mask was dilated in all three directions with a sphere radius of 5 voxels to remove potential periventricular WMHs and MRI induced artifacts near the ventricle which could be misjudged as periventricular WMHs (Fig. 1B). The deep brain structures of the hippocampus, amygdala, caudate, putamen, pallidum, thalamus, hypothalamus, nucleus accumbens, mammillary body, subthalamic nuclei, substantia nigra, and red nucleus that were specified by the automated anatomical labeling (AAL) atlas were registered onto the FLAIR space (Fig. 1B). The adjusted GM, CSF, ventricle, and deep brain structure masks were removed from the WM mask. The adjusted WM mask was eroded in the axial direction with a disk size of 1 (Fig. 1B). The adjusted WM mask might still contain non-WM regions near the boundary between the GM and WM, and thus additional processing was performed. The pseudo-GM mask was constructed by removing small objects (smaller than 500 voxels) within the WM from the GM PVE mask, and the pseudo-GM mask was removed from the adjusted WM mask (Fig. 1B). After these steps, the WM mask was finalized and applied to the FLAIR image to obtain only the WM region (Fig. 1C). The WM region of the FLAIR image was spatially smoothed with a full width at half maximum (FWHM) of 0.7 mm and intensity normalization was performed with a mean value of 500 (Fig. 1C). This was considered as the final WM region which contained deep WMHs.
2.4. WMH detection
The detection of deep WMHs was performed (Fig. 1D) only within the WM region specified from the previous steps. Potential deep WMH voxels were detected by applying a threshold of 2.9 times the mean intensity of FLAIR to WM FLAIR voxels. The thresholded voxels were clustered and clusters smaller than 4 voxels (<1 mm) were removed (Ghafoorian et al., 2016). We applied region growing for the remaining clusters that were <20 voxels in size. Region growing terminated when the growing boundary met a voxel with an intensity value lower than 2.8 times the mean FLAIR intensity. A maximum of 5 mm in Euclidean distance was allowed for region growing. When there were two or more contiguous clusters, the clusters were merged into a single cluster. Finally, clusters with an effective diameter smaller than 1 mm, which was defined as the major axis of the cluster in a 2D slice, were removed. The WMH detection using the Gaussian mixture model (GMM) distribution clustering algorithm was also performed (Biernacki et al., 2000; Zhuang et al., 1996), and detailed methods and relevant results were reported in the Supplementary material.
2.5. False positive reduction
2.5.1. Feature extraction
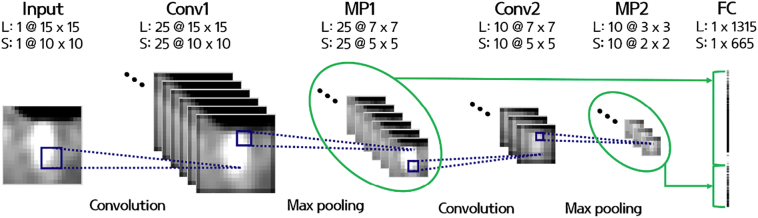
After the initial detection of potential deep WMH clusters, FP voxels still existed in the detected clusters. Reduction of FP voxels is one of the most challenging issues in WMH detection (Ghafoorian et al., 2016; Ithapu et al., 2014; Jeon et al., 2011). We used the random forest (RF) classifier, a supervised machine learning technique, based on manual annotation to distinguish WMHs from non-WMH clusters using volume, maximum 3D distance, the ratio between the major and minor axis, texture, and multi-scale deep features (Table 1 and Fig. 1E). The texture features consisted of 19 first order statistical based features that were calculated using the voxel intensities of the clusters. The features were max, min, median, mean, variance, energy, SD, skewness, kurtosis, root mean square, range, inter quartile range, entropy, uniformity, and percentiles of 2.5, 25, 50, 75, and 97.5 (Table 1). The multi-scale deep features were computed by constructing a network architecture commonly found in the convolutional neural network (CNN) (Fig. 2). The architecture consisted of two convolutional, two max pooling, and one fully-connected layer. The architecture operated on image patches of two different scales (15 × 15 and 10 × 10) that covered potential deep WMH clusters. In the first convolutional layer, 25 2D filters of average, disk, Gaussian, log of Gaussian, Laplacian, Prewitt, Sobel, and motion filters with different hyper-parameters (Table 1) were applied and in the next max pooling layer, the patches were down-sampled with a kernel size of two. In the second convolutional layer, 10 3D filters of average, ellipsoid, Gaussian, and log of Gaussian with different hyper-parameters (Table 1) were used and then subsequently max pooling was applied with a size of two. The output image patches of the first and second max pooling layers (7 × 7 × 25 and 3 × 3 × 10 for large patches and 5 × 5 × 25 and 2 × 2 × 10 for small patches) were vectorized and concatenated in the fully connected layer. Finally, 1980 multi-scale deep features were computed. The texture and multi-scale deep features were computed from both the T1-weighted and FLAIR images.
Table 1.
Features and hyper-parameters used to train the random forest model.
| Classes | Features or filters | Hyper-parameters | Modality |
|---|---|---|---|
| Size | Volume | – | T1w |
| Maximum 3D distance | – | ||
| Major and minor axis ratio | – | ||
| Texture | Max | – | T1w & FLAIR |
| Min | – | ||
| Median | – | ||
| Mean | – | ||
| Variance | – | ||
| Energy | – | ||
| Standard deviation | – | ||
| Skewness | – | ||
| Kurtosis | – | ||
| Root mean square | – | ||
| Range | – | ||
| Inter quartile range | 0.25–0.75 | ||
| Entropy | – | ||
| Uniformity | – | ||
| Percentile | 2.5, 25, 50, 75, 97.5 | ||
| Multi-scale deep | 2D Average | Kernel size = 3 | T1w & FLAIR |
| 2D Disk | Radius = 1 | ||
| 2D Gaussian | Kernel size = 3 σ = 0.5, 1, 1.5, 2 |
||
| 2D Log of Gaussian | Kernel size = 3 σ = 0.5, 1, 1.5, 2 |
||
| 2D Laplacian | σ = 0, 0.25, 0.5, 0.75, 1 | ||
| 2D Prewitt | Direction = 0, 90, 180, 270° | ||
| 2D Sobel | Direction = 0, 90, 180, 270° | ||
| 2D Motion | Length = 3, angle = 25, 50° | ||
| 3D Average | Kernel size = 3 | ||
| 3D Ellipsoid | Kernel size = 3 | ||
| 3D Gaussian | Kernel size = 3 σ = 0.5, 1, 1.5, 2 |
||
| 3D Log of Gaussian | Kernel size = 3 σ = 0.5, 1, 1.5, 2 |
Fig. 2.
The architecture for computing multi-scale deep features. L, large patch; S, small patch; Conv1, convolutional layer 1; MP1, max pooling layer 1; Conv2, convolutional layer 2; MP2, max pooling layer 2; FC, fully-connected layer.
2.5.2. RF classifier
Among 148 subjects, 128 subjects were used to train and test the RF classifier and the remaining 20 subjects were used for validation. The 128 subjects were randomly divided into the training (n = 102) and test (n = 26) set 1000 times. The RF classifier was constructed for each iteration using the features extracted from the training data. The trained RF classifier was tested using the test data. The number of true positives (TP), FP, and false negatives (FN) were counted by comparing the detected and manually drawn WMH clusters. The quality of the RF classifier was assessed by calculating the positive predictive value [PPV = TP/(TP + FP)] and true positive rate [TPR = TP/(TP + FN)]. Histograms of the PPV and TPR were constructed and the model that appeared most frequently (i.e., mode of the histogram) was selected as the optimal RF classifier. The selected RF classifier was applied to the independent validation dataset (Fig. 1G).
2.6. Evaluation
The quality of our algorithm was assessed by comparing the detected deep WMH clusters and manual annotations. We compared the locations of the detected and manually drawn deep WMH clusters. If the detected and manual clusters overlapped spatially, we considered our algorithm to have successfully detected the WMH cluster. After determining TP, FP, and FN, FP were reviewed by investigators and re-classified into either FP or TP. This two-tiered approach was adopted to minimize FP which were initially missed in the manual annotation and were only detected by our automated method. The PPV [=TP/(TP + FP)] and TPR [=TP/(TP + FN)] were calculated to assess the quality of the results (Fig. 1H).
2.7. Comparison with other methods
We compared the accuracy of our DEWS with the Wisconsin WMH Segmentation Toolbox (W2MHS) (Ithapu et al., 2014; https://www.nitrc.org/projects/w2mhs/), Brain Intensity AbNormality Classification Algorithm (BIANCA) (Griffanti et al., 2016; https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/BIANCA/), and Lesion-TOADS framework of MIPAV software (Shiee et al., 2010). We chose the W2MHS, BIANCA, and Lesion-TOADS to compare with our DEWS since they were open source software algorithms and have been used as reference algorithms in other studies (Baggio et al., 2015; Harrison et al., 2015; Paternicò et al., 2016). A brief overview of the W2MHS is as follows: The FLAIR data was registered onto the T1-weighted data and corrected for magnetic field bias. The PVE masks of GM, WM, and CSF were generated based on the T1-weighted data and were applied to FLAIR to obtain the actual tissue maps of FLAIR. The ventricle mask was extracted and dilated in all three directions to detect periventricular and deep WMHs separately. The modified WM mask was used for detecting WMHs. The intensity threshold of 0.6 times the max intensity of WM voxels of FLAIR was applied to the WM FLAIR voxels and the pre-trained random forest model was used to reduce FP. Potential WMH voxels near the GM and CSF were removed. BIANCA is a supervised machine learning algorithm which requires manually annotated WMH images. It detects WMHs based on the k-nearest neighbor algorithm using information from the voxel- and patch-based intensity values, and prior spatial coordinates of the Montreal Neurological Institute (MNI) template. The leave-one-out cross validation approach was applied to T1-weighted, FLAIR, and manually segmented WMH images. BIANCA produced a probability map of WMHs and the map was thresholded and binarized with a value of 0.5. Segmentation of WM lesions using Lesion-TOADS was performed based on the manually delineated topological atlas (Shiee et al., 2010). A fuzzy classification algorithm was iteratively performed and membership functions were used to classify between the lesion and non-lesion voxels. The false positives were removed based on the distance from the boundary of GM and ventricles. The detected WMH voxels were clustered and compared with our manual annotations as described in the “Evaluation” section.
2.8. Statistical analysis
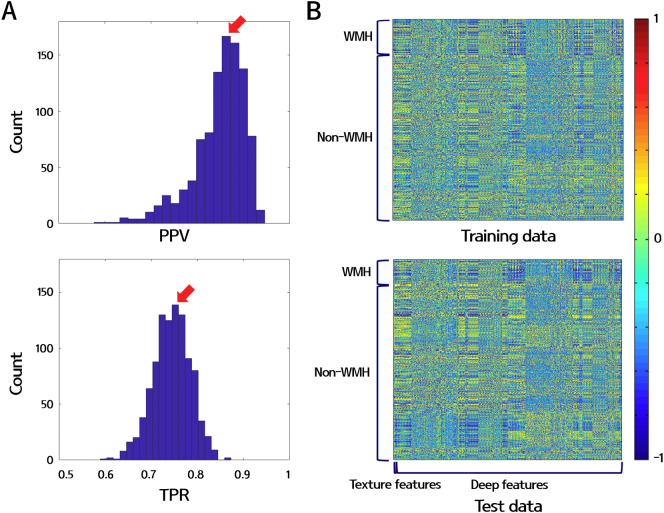
Statistical results of the demographics of the participants are presented as the mean (SD) or number (%). Descriptive analyses for clinical data were performed using Stata 15.0 (StataCorp. 2017. Stata Statistical Software: Release 15. College Station, TX: StataCorp LLC). The PPV [=TP/(TP + FP)] and TPR [=TP/(TP + FN)] were calculated using MATLAB 2016a (Mathworks Inc., Natick, MA, USA). The optimal RF classifier model was selected by constructing histograms of PPV and TPR while iterating 1000 times with different training and test sets (Fig. 3A). The RF model that appeared most frequently in terms of PPV and TPR was selected (red arrows in Fig. 3A). The features used for the RF classifier was normalized to z-scores. The heatmap of z-normalized features for the training and test sets that were used to construct the selected RF model is shown in Fig. 3B.
Fig. 3.
(A) Histograms of PPV (upper) and TPR (bottom) calculated from the 1000 iterations of the test set. The red arrows indicate the peak point of PPV and TPR. (B) Heatmaps of the potential WMH features of the training (upper) and test (bottom) data. The volume, maximum 3D distance, ratio between the major and minor axis, 38 textures, and 3960 deep features were plotted (horizontal axis) for 102 training and 26 test subjects (vertical axis). All features were z-normalized.
3. Results
3.1. Demographics of the participants
Demographics and characteristics of our study subjects are summarized in Table 2. The mean age was 44.4 (SD 12.40) years, with a range of 14 to 65 years. 60 (40.5%) subjects were below the age of 50. 70 (47.3%) had episodic migraine (median monthly headache days, 6; interquartile range, 3–10), while 31 (20.9%) had chronic migraine (median monthly headache days, 24; interquartile range 18–30). The prevalence of hypertension, diabetes, dyslipidemia, stroke, and smoking were 11.5%, 0.7%, 9.5%, 0%, and 2.7%, respectively (Table 2).
Table 2.
Demographics of the study subjects. Data are presented as the median (interquartile range) or number (percentage).
| Information | Subjects with migraine (n = 148) | |
|---|---|---|
| Age, years | 44.4 (12.40) | |
| Females | 82 (55.4%) | |
| Disease duration, years | 10 (4–20) | |
| Diagnosis | Migraine without aura | 88 (59.5%) |
| Migraine with aura | 13 (8.8%) | |
| Chronic migraine | 30 (20.3%) | |
| Monthly headache days | 10 (4–27) | |
| Hypertension | 17 (11.5%) | |
| Diabetes | 1 (0.7%) | |
| Dyslipidemia | 14 (9.5%) | |
| Stroke | 0 (0.0%) | |
| Smoking | 4 (2.7%) | |
3.2. WMH characteristics
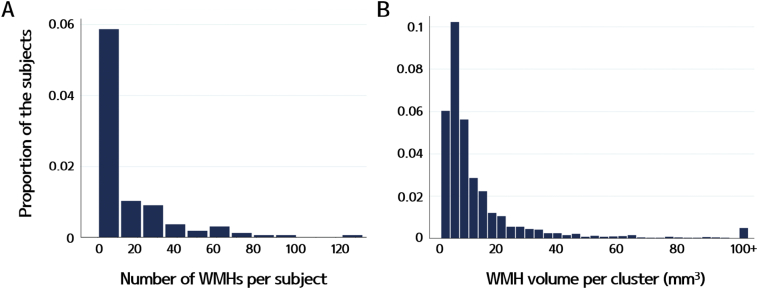
Our subjects had a median of 6.5 WMHs (interquartile range, 4–20) per subject. The volume of each WMH was small, with a median estimated volume of 8.0 mm3 (interquartile range, 5.3–13.7) per cluster. The total WMH volume was a median of 53.0 mm3 (interquartile range, 19.2–226.6) per subject. Fig. 4 shows the distribution of the number of WMHs per subject and WMH volumes per cluster.
Fig. 4.
Histograms of (A) the number of WMHs per subject and (B) WMH volumes per cluster. Most of our subjects had <10 WMHs, and the volume of each WMH was small for most subjects.
3.3. Detection results of deep WMH clusters
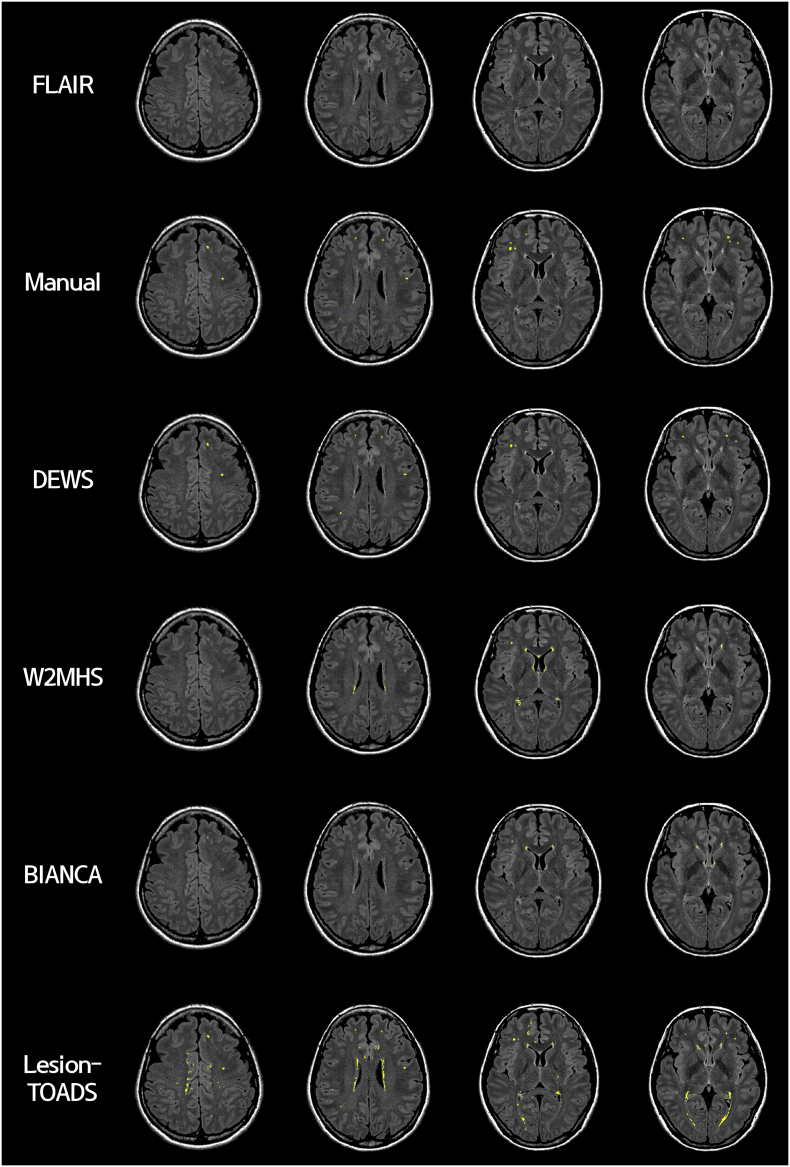
Deep WMH clusters were detected using T1-weighted and FLAIR data for 148 participants and compared with manual annotations. DEWS yielded good results with a mean PPV of 0.98 (SD 0.12) and mean TPR of 0.70 (SD 0.24) for the training and test sets. In the validation set, a mean PPV of 0.96 (SD 0.12) and mean TPR of 0.68 (SD 0.18) were observed. Results from a representative participant are reported in Fig. 5.
Fig. 5.
Results from a representative case. Images are presented in the following order: 1st row, the FLAIR image corrected for magnetic field bias; 2nd row, manually drawn deep WMH clusters (in yellow); 3rd ~ 6th rows, detected deep WMH clusters using our pipeline (DEWS), W2MHS, BIANCA, and Lesion-TOADS, respectively.
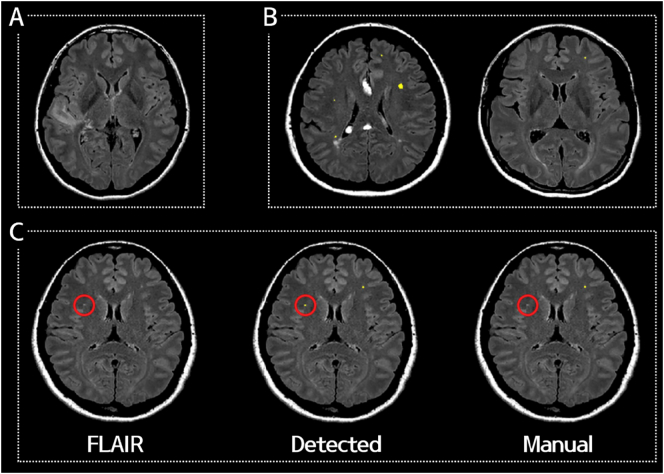
Of the 32 clusters which were initially classified as FP, 18 were re-classified into TP (6 missed WMH in the manual annotation and 12 small WMHs which were present in the manual drawing but excluded during the validation procedure due to the small size, i.e. a maximal diameter of <1 mm). Only 14 were determined to be real FP (1 periventricular WMHs, 9 cortices, and 4 artifacts). Otherwise, periventricular WMHs and silent infarctions were successfully removed. Representative images are shown in Fig. 6.
Fig. 6.
Representative cases that successfully removed (A) a silent infarction and (B) periventricular WMHs. (C) A representative case of a detected WMH cluster that was missed in the manual annotation (red circle).
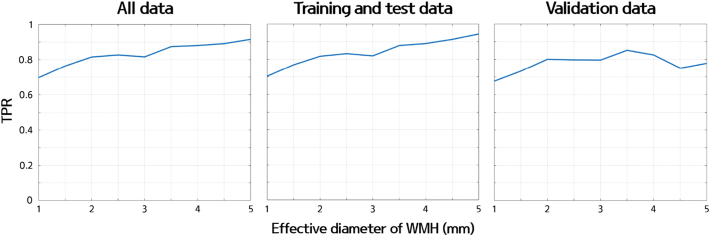
We also explored the change in sensitivity with respect to the size of WMHs to gauge the lower limit of our algorithm. We applied different thresholds from 1 mm to 5 mm with an interval of 0.5 mm and computed the TPR of our pipeline to detect manually drawn WMHs with larger diameters than the predefined threshold. The TPR of all data (n = 148) was 0.70 for 1 mm, 0.76 for 1.5 mm, 0.82 for 2 mm, 0.83 for 2.5 mm, 0.82 for 3 mm, 0.87 for 3.5 mm, 0.88 for 4 mm, 0.89 for 4.5 mm, and 0.92 for 5 mm (Fig. 7). The TPR of the training, test, and validation data is also plotted in Fig. 7. The TPR showed an increasing trend as the size threshold of the effective diameter increased. The TPR plot showed a general increasing trend but was not monotonically increasing. If we increased the size threshold, there were fewer WMH clusters and thus the sample size from which the TPR was computed decreased. With the decreased sample size, the value of the TPR became unstable, which might lead to the non-monotonic shape of the TPR curve.
Fig. 7.
The graph of the TPR with respect to the effective diameter of WMH clusters for all data (left), training and test data (middle), and validation data (right).
3.4. Comparison with other methods
We applied three openly accessible software algorithms, the W2MHS, BIANCA, and Lesion-TOADS, to our dataset. The W2MHS had a mean PPV of 0.02 (SD 0.05) and mean TPR of 0.09 (SD 0.19) for all subjects. No WMH clusters were detected in the 69 subjects. BIANCA yielded a mean PPV of 0.02 (SD 0.04) and mean TPR of 0.02 (SD 0.04) for all subjects. Lesion-TOADS yielded a mean PPV of 0.02 (SD 0.03) and mean TPR of 0.76 (SD 0.28) for all subjects. A representative case is shown in Fig. 5. Both the W2MHS and BIANCA failed to detect small deep WMHs, thus yielding a high number of FP per subject (W2MHS: mean 40.82 with SD 67.45; BIANCA: mean 32.78 with SD 32.03). Lesion-TOADS showed better sensitivity in detecting small deep WMHs than W2MHS and BIANCA but yielded a large number of FP per subject (mean 1048.74 with SD 665.23). The number of FP was acceptable in our results using DEWS (mean 0.11 FP with SD 0.51).
3.5. Reproducible research and open software
The software for our proposed pipeline (DEWS) and limited anonymized imaging data are available at a software sharing site (https://github.com/bypark/DEWS).
4. Discussion
Non-elderly migraineurs showed small, discrete, and superficially-located deep WMHs, which have not been focused on in previous segmentation studies. Our algorithm successfully detected small deep WMHs with a higher PPV and TPR compared to previous methods. DEWS yielded few false positives and captured several deep WMHs that were missed by two investigators. It still showed moderate sensitivity on detecting deep WMHs (TPR = 0.7). Taken together, we recommend that the final results of DEWS should be monitored and validated by human experts at this stage. To the best of our knowledge, our proposed method is the first method developed for the research of deep WMHs in non-demented, non-elderly individuals.
We applied the W2MHS, BIANCA, and Lesion-TOADS software algorithms to our data from migraine patients for comparison. Our data contained WMHs that were mostly deep WMHs rather than periventricular WMHs. The W2MHS and BIANCA did not capture small deep WMHs and thus yielded low PPV and TPR. This might be due to several factors. First, the WMH detection procedure was performed on the T1-weighted space for W2MHS and MNI space for BIANCA rather than the FLAIR space. Registration errors of mapping FLAIR onto the T1-weighted or MNI space might negatively affect the locations and intensity values of WMHs especially for small deep WMHs. Our approach performed detection on the FLAIR space and thus registration errors had less of an effect. Second, these algorithms did not make fine adjustments to the WM mask and thus many WMHs were misclassified to GM. Third, in the W2MHS software, the threshold value for WMH detection was fixed at 0.6 times the maximum WM FLAIR intensity. Our dataset had MRI artifacts and the artifacts usually had high intensity values. Thus, adopting the threshold value using the max WM FLAIR intensity was not appropriate for our dataset. The Lesion-TOADS detected small deep WMHs but failed to remove a large number of FP. The FP reduction step in Lesion-TOADS was performed by considering the distance from the boundary of GM and ventricles, while our approach used size, texture, and multi-scale deep features. Our pipeline reduced a large number of FP compared to the Lesion-TOADS suggesting that the deep WMHs were better quantified using size, texture, and multi-scale deep features rather than only using location information.
We used numerous features to train the RF classifier including size, texture, and multi-scale deep features. We trained the RF classifiers with following different combinations of features: (1) size features only, (2) texture features only, (3) multi-scale deep features only, (4) size and texture features, (5) size and multi-scale deep features, (6) texture and multi-scale deep features, and (7) all features. Each RF classifier was constructed using the training data and tested using the test data for seven cases. We compared the PPV and TPR for each case to determine which case showed the best performance on classifying between WMHs and non-WMHs. The sum of PPV and TPR was the highest value when only multi-scale deep features or texture and multi-scale deep features were used and followed by using all features (Table S1). The differences in performance among using multi-scale deep features, texture and multi-scale deep features, and all features were small, but we believe using all features could be beneficial. Multi-scale deep features were inspired by CNN architecture which is very difficult to interpret. However, size and texture features, part of all features, are easy to interpret with proven performance and known biological implications. We wanted to keep those easily interpretable and robust features so that our model could be effectively applied to an independent data of possibly different WMH properties.
Multi-scale deep features were calculated by constructing an architecture inspired by the CNN. The CNN architecture allows extraction of various features that span many scales. These deep features might contain contrasting information to distinguish WMHs from non-WMHs as shown in the heatmap in Fig. 3B. Thus, these features might contribute positively toward the RF classifier. The CNN requires many hyper-parameters: the number of convolutional, pooling, and fully connected layers; the size and number of image patches; the type of pooling layer (e.g., max, average, or overlapping); and the number and types of filters. We chose these parameters in a typical fashion and did not attempt to optimize them as this was beyond the scope of our study.
The CNN is one of the powerful machine learning tools for segmenting WMHs (Ghafoorian et al., 2017a, Ghafoorian et al., 2017b). Studies have reported divergent CNN models including the number of convolutional, pooling, and fully connected layers, input patch size, number of filters, types of activation functions, and number of iteration steps to segment brain tumors, which implies that there is no gold standard CNN framework for brain tumor segmentation. The results of a previous study yielded a high Dice coefficient around 0.8 between the segmented and manually drawn images (Ghafoorian et al., 2017b). This high performance was achieved because the authors focused on segmenting big rather than small WMHs. Detecting small WMHs using the CNN remains a challenging task for future studies.
Our study has a few limitations. First, the Dice coefficient or Jaccard index were not used to report the validation results (Ithapu et al., 2014; Jeon et al., 2011). Because these indices are based on voxel-to-voxel comparisons and have high variability for small objects containing only a few voxels, they were considered inappropriate for the evaluation of small discrete WMHs (Ghafoorian et al., 2016). We calculated the PPV and TPR by counting the number of TP, FP, and FN to assess the quality of our proposed pipeline. We considered the detected cluster as TP if it overlapped with the manually annotated cluster. To overcome this limitation, all detected WMHs were reviewed and confirmed by clinical investigators (M.J.L. and J.C.). Second, DEWS required several small steps to specify the WM mask. Our imaging data contained many small deep WMHs which could easily be misclassified to GM during the tissue segmentation process. Thus, we had to adjust the WM mask in small heuristic increments so that misclassified WMHs were indeed included in the WM mask. There was no single step that could revert back the misclassified WMHs and many small adjustments were necessary as was done in previous studies (Ghafoorian et al., 2016; Ithapu et al., 2014; Jeon et al., 2011). Third, since our study is a single center study using a single clinical subset, external validation is warranted using data from other centers and subjects with clinical conditions other than migraine. Fourth, due to the limited sample size, we divided the whole sample into training (n = 102), test (n = 26), and validation (n = 20) sets. In future studies, we plan to collect larger cohort data to improve the reliability of the results. Fifth, there were possible confounding effects of risk factors and psychoactive treatments on WMHs. This issue might not be overwhelming as our goal was not to generate hypothesis on the pathogenesis of WMHs but to accurately detect WMHs.
5. Conclusions
Non-elderly migraineurs show discrete, small-sized, deep WMHs, which could not be detected by previous automated segmentation methods. DEWS, a fully automated detection pipeline dedicated for small and superficially-located deep WMHs, can aid further research on deep WMHs in migraineurs.
Conflicts of interest
None.
Acknowledgements
This work was supported by the Institute for Basic Science (grant number IBS-R015-D1). This work was also supported by the NRF (National Research Foundation of Korea), grant numbers NRF-2016H1A2A1907833 (Mr. Park), NRF-2016R1A2B4008545 (Prof. Park), NRF-2017R1A2B2009086 (Dr. Chung), and NRF-2017R1A2B4007254 (Dr. Lee).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2018.02.033.
Appendix A. Supplementary data
Supplementary material
References
- Baggio H.-C., Segura B., Sala-Llonch R., Marti M.-J., Valldeoriola F., Compta Y., Tolosa E., Junqué C. Cognitive impairment and resting-state network connectivity in Parkinson's disease. Hum. Brain Mapp. 2015;36:199–212. doi: 10.1002/hbm.22622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernacki C., Celeux G., Govaert G. Assessing a mixture model for clustering with the integrated completed likelihood. IEEE Trans. Pattern Anal. Mach. Intell. 2000;22:719–725. [Google Scholar]
- Cox R.W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- ten Dam V.H., van den Heuvel D.M.J., de Craen A.J.M., Bollen E.L.E.M., Murray H.M., Westendorp R.G.J., Blauw G.J., van Buchem M.A. Decline in total cerebral blood flow is linked with increase in periventricular but not deep white matter hyperintensities. Radiology. 2007;243:198–203. doi: 10.1148/radiol.2431052111. [DOI] [PubMed] [Google Scholar]
- Debette S., Markus H.S. The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: systematic review and meta-analysis. BMJ. 2010;341:c3666. doi: 10.1136/bmj.c3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdélyi-Bõtor S., Aradi M., Kamson D.O., Kovács N., Perlaki G., Orsi G., Nagy S.A., Schwarcz A., Dõczi T., Komoly S., Deli G., Trauninger A., Pfund Z. Changes of migraine-related white matter hyperintensities after 3 years: a longitudinal MRI study. Headache. 2015;55:55–70. doi: 10.1111/head.12459. [DOI] [PubMed] [Google Scholar]
- Fazekas F., Chawluk J.B., Alavi A., Hurtig H.I., Zimmerman R.A. Mr signal abnormalities at 1.5-T in Alzheimer dementia and normal aging. Am. J. Roentgenol. 1987;149:351–356. doi: 10.2214/ajr.149.2.351. [DOI] [PubMed] [Google Scholar]
- Fazekas F., Kleinert R., Offenbacher H., Schmidt R., Kleinert G., Payer F., Radner H., Lechner H. Pathologic correlates of incidental-MRI white matter signal hyperintensities. Neurology. 1993;43:1683–1689. doi: 10.1212/wnl.43.9.1683. [DOI] [PubMed] [Google Scholar]
- Fernando M.S., Simpson J.E., Matthews F., Brayne C., Lewis C.E., Barber R., Kalaria R.N., Forster G., Esteves F., Wharton S.B., Shaw P.J., Brien J.T.O., Ince P.G. White matter lesions in an unselected cohort of the elderly. Stroke. 2006;37:1391–1398. doi: 10.1161/01.STR.0000221308.94473.14. [DOI] [PubMed] [Google Scholar]
- Ghafoorian M., Karssemeijer N., van Uden I.W.M., de Leeuw F.-E., Heskes T., Marchiori E., Platel B. Automated detection of white matter hyperintensities of all sizes in cerebral small vessel disease. Med. Phys. 2016;43:6246–6258. doi: 10.1118/1.4966029. [DOI] [PubMed] [Google Scholar]
- Ghafoorian M., Karssemeijer N., Heskes T., Bergkamp M., Wissink J., Obels J., Keizer K., de Leeuw F.E., van Ginneken B., Marchiori E., Platel B. Deep multi-scale location-aware 3D convolutional neural networks for automated detection of lacunes of presumed vascular origin. NeuroImage Clin. 2017;14:391–399. doi: 10.1016/j.nicl.2017.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafoorian M., Karssemeijer N., Heskes T., van Uden I.W.M., Sanchez C.I., Litjens G., de Leeuw F.-E., van Ginneken B., Marchiori E., Platel B. Location sensitive deep convolutional neural networks for segmentation of white matter Hyperintensities. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-05300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffanti L., Zamboni G., Khan A., Li L., Bonifacio G., Sundaresan V., Schulz U.G., Kuker W., Battaglini M., Rothwell P.M., Jenkinson M. BIANCA (brain intensity AbNormality classification algorithm): a new tool for automated segmentation of white matter hyperintensities. NeuroImage. 2016;141:191–205. doi: 10.1016/j.neuroimage.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffanti L., Jenkinson M., Suri S., Zsoldos E., Mahmood A., Filippini N., Sexton C.E., Topiwala A., Allan C., Kivimäki M., Singh-Manoux A., Ebmeier K.P., Mackay C.E., Zamboni G. Classification and characterization of periventricular and deep white matter hyperintensities on MRI: a study in older adults. NeuroImage. 2017 doi: 10.1016/j.neuroimage.2017.03.024. [DOI] [PubMed] [Google Scholar]
- Harrison D.M., Roy S., Oh J., Izbudak I., Pham D., Courtney S., Caffo B., Jones C.K., Van Zijl P., Calabresi P.A. Association of cortical lesion burden on 7-T magnetic resonance imaging with cognition and disability in multiple sclerosis. JAMA Neurol. 2015;72:1004–1012. doi: 10.1001/jamaneurol.2015.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headache Classification Committee of the International Headache Society (IHS) The international classification of headache disorders, 3rd edition (beta version) Cephalalgia. 2013;33:629–808. doi: 10.1177/0333102413485658. [DOI] [PubMed] [Google Scholar]
- Herrmann L.L., Le Masurier M., Ebmeier K.P. White matter hyperintensities in late life depression: a systematic review. J. Neurol. Neurosurg. Psychiatry. 2008;79:619–624. doi: 10.1136/jnnp.2007.124651. [DOI] [PubMed] [Google Scholar]
- van den Heuvel D.M.J., ten Dam V.H., de Craen A.J.M., Admiraal-Behloul F., Olofsen H., Bollen E.L.E.M., Jolles J., Murray H.M., Blauw G.J., Westendorp R.G.J., van Buchem M.A. Increase in periventricular white matter hyperintensities parallels decline in mental processing speed in a non- demented elderly population. J. Neurol. Neurosurg. Psychiatry. 2006;77:149–153. doi: 10.1136/jnnp.2005.070193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R.O., Beck C.J., Burnett D.L., Weaver L.K., Victoroff J., Bigler E.D. Prevalence of white matter hyperintensities in a young healthy population. J. Neuroimaging. 2006;16:243–251. doi: 10.1111/j.1552-6569.2006.00047.x. [DOI] [PubMed] [Google Scholar]
- Hulsey K.M., Gupta M., King K.S., Peshock R.M., Whittemore A.R., McColl R.W. Automated quantification of white matter disease extent at 3 T: comparison with volumetric readings. J. Magn. Reson. Imaging. 2012;36:305–311. doi: 10.1002/jmri.23659. [DOI] [PubMed] [Google Scholar]
- Ithapu V., Singh V., Lindner C., Austin B.P., Hinrichs C., Carlsson C.M., Bendlin B.B., Johnson S.C. Extracting and summarizing white matter hyperintensities using supervised segmentation methods in Alzheimer's disease risk and aging studies. Hum. Brain Mapp. 2014;35:4219–4235. doi: 10.1002/hbm.22472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C.F., Behrens T.E.J., Woolrich M.W., Smith S.M. Fsl. NeuroImage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Jeon S., Yoon U., Park J.S., Seo S.W., Kim J.H., Kim S.T., Kim S.I., Na D.L., Lee J.M. Fully automated pipeline for quantification and localization of white matter hyperintensity in brain magnetic resonance image. Int. J. Imaging Syst. Technol. 2011;21:193–200. [Google Scholar]
- Kim K.W., MacFall J.R., Payne M.E. Classification of white matter lesions on magnetic resonance imaging in elderly persons. Biol. Psychiatry. 2008;64:273–280. doi: 10.1016/j.biopsych.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S., Abdulkadir A., Hadjidemetriou S., Issleib S., Frings L., Thanh T.N., Mader I., Teipel S.J., Hüll M., Ronneberger O. A comparison of different automated methods for the detection of white matter lesions in MRI data. NeuroImage. 2011;57:416–422. doi: 10.1016/j.neuroimage.2011.04.053. [DOI] [PubMed] [Google Scholar]
- Kruit M.C., van Buchem M.A., Hofman P.A., Bakkers J.T.N., Terwindt G.M., Ferrari M.D., Launer L.J. Migraine as a risk factor for subclinical brain lesions. JAMA. 2004;291:427–434. doi: 10.1001/jama.291.4.427. [DOI] [PubMed] [Google Scholar]
- Kurth T., Winter A.C., Eliassen A.H., Dushkes R., Mukamal K.J., Rimm E.B., Willett W.C., Manson J.E., Rexrode K.M. Migraine and risk of cardiovascular disease in women: prospective cohort study. BMJ. 2016;353 doi: 10.1136/bmj.i2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee R.M., Kwee T.C. Virchow-Robin spaces at MR imaging. Radiographics. 2007;27:1071–1086. doi: 10.1148/rg.274065722. [DOI] [PubMed] [Google Scholar]
- Mineura K., Sasajima H., Kikuchi K., Kowada M., Tomura N., Monma K., Segawa Y. White matter hyperintensity in neurologically asymptomatic subjects. Acta Neurol. Scand. 1995;92:151–156. doi: 10.1111/j.1600-0404.1995.tb01030.x. [DOI] [PubMed] [Google Scholar]
- Murray M.E., Senjem M.L., Petersen R.C., John H., Preboske G.M., Weigand S.D., Knopman D.S., Ferman T.J., Dickson D.W., J. C.R., Jr. Functional impact of white matter hyperintensities in cognitively normal elderly. Arch. Neurol. 2010;67:1379–1385. doi: 10.1001/archneurol.2010.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm-Meinders I.H., Koppen H., Terwindt G.M., Launer L.J., Konishi J., Moonen J.M.E., Bakkers J.T.N., Hofman P.A.M., van Lew B., Middelkoop H.A.M., van Buchem M.A., Ferrari M.D., Kruit M.C. Structural brain changes in migraine. JAMA, J. Am. Med. Assoc. 2012;308:1889–1897. doi: 10.1001/jama.2012.14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternicò D., Premi E., Gazzina S., Cosseddu M., Alberici A., Archetti S., Cotelli M.S., Micheli A., Turla M., Gasparotti R., Padovani A., Borroni B. White matter hyperintensities characterize monogenic frontotemporal dementia with granulin mutations. Neurobiol. Aging. 2016;38:176–180. doi: 10.1016/j.neurobiolaging.2015.11.011. [DOI] [PubMed] [Google Scholar]
- Seo S.W., Lee J.M., Im K., Park J.S., Kim S.H., Kim S.T., Ahn H.J., Chin J., Cheong H.K., Weiner M.W., Na D.L. Cortical thinning related to periventricular and deep white matter hyperintensities. Neurobiol. Aging. 2012;33:1156–1167. doi: 10.1016/j.neurobiolaging.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Serafini G., Pompili M., Innamorati M., Fusar-poli P., Akiskal H.S., Rihmer Z., Lester D., Romano A., de Oliveira I.R., Strusi L., Ferracuti S., Girardi P., Tatarelli R. Affective temperamental pro fi les are associated with white matter hyperintensity and suicidal risk in patients with mood disorders. J. Affect. Disord. 2011;129:47–55. doi: 10.1016/j.jad.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Shiee N., Bazin P., Ozturk A., Reich D.S., Calabresi P.A., Pham D.L. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. NeuroImage. 2010;49:1524–1535. doi: 10.1016/j.neuroimage.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk E.J., Prins N.D., Vrooman H.A., Hofman A., Koudstaal P.J., Breteler M.M.B. Progression of cerebral small vessel disease in relation to risk factors and cognitive consequences: Rotterdam scan study. Stroke. 2008;39:2712–2719. doi: 10.1161/STROKEAHA.107.513176. [DOI] [PubMed] [Google Scholar]
- Vermeer S.E., Hollander M., Van Dijk E.J., Hofman A., Koudstaal P.J., Breteler M.M.B. Silent brain infarcts and white matter lesions increase stroke risk in the general population: the Rotterdam scan study. Stroke. 2003;34:1126–1129. doi: 10.1161/01.STR.0000068408.82115.D2. [DOI] [PubMed] [Google Scholar]
- Wardlaw J.M., Smith E.E., Biessels G.J., Cordonnier C., Fazekas F., Frayne R., Lindley R.I., O'Brien J.T., Barkhof F., Benavente O.R., Black S.E., Brayne C., Breteler M., Chabriat H., DeCarli C., de Leeuw F.E., Doubal F., Duering M., Fox N.C., Greenberg S., Hachinski V., Kilimann I., Mok V., van Oostenbrugge R., Pantoni L., Speck O., Stephan B.C.M., Teipel S., Viswanathan A., Werring D., Chen C., Smith C., van Buchem M., Norrving B., Gorelick P.B., Dichgans M. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 2013;12:822–838. doi: 10.1016/S1474-4422(13)70124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshita M., Fletcher E., Harvey D., Ortega M., Martinez O., Mungas D.M., Reed B.R., DeCarli C.S. Extent and distribution of white matter hyperintensities in normal aging, MCI, and AD. Neurology. 2006;67:2192–2198. doi: 10.1212/01.wnl.0000249119.95747.1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Huang Y., Palaniappan K., Zhao Y. Gaussian mixture density modeling, decomposition, and applications. IEEE Trans. Image Process. 1996;5:1293–1302. doi: 10.1109/83.535841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material