Allergic airway inflammation, a cardinal pathological feature of asthma is driven by production of IL-4, IL-5, and IL-13. Together, these mediators are referred to as ‘Th2 cytokines’ because a major producer of them is the differentiated CD4+ T-helper 2 (Th2) cell. IL-4 drives allergic sensitization, IgE production, and Th2 cell differentiation. IL-5 is critical in eosinophil accumulation and activation in the lung. IL-13 has pleiotropic effects in tissue remodeling and the development and progression of airway hyper-responsiveness. Because of these major effects, Th2 cytokines were identified as potential therapeutic targets for the treatment of asthma with a great hope in the field for their success (1). Unfortunately, the anti-Th2 cytokine therapeutics had generally poor results, although recent advances in identifying specific patients with endotypes responsive to these therapeutics have helped efficacy (2). These drugs likely struggled in the clinic because (in addition to their unique effects) the Th2 cytokines have numerous overlapping functions. Would it be possible therefore that upstream therapeutic targets like transcription factors that regulate multiple Th2 cytokine production would work better (3, 4)? To explore such option, it is imperative that we improve our understanding of the mechanisms that control the transcription of Th2 cytokine genes, especially during allergic airway inflammation.
The genes for IL-4, IL-13, and IL-5 are clustered in a 120-kb region on chromosome 11 in mice and a 160-kb region on chromosome 5 in humans. Th2 cytokines are expressed in a T-cell lineage-specific fashion and their transcription is therefore coordinately regulated by a complex network of transcription factors and other regulators that bind to gene promoters to activate and/or repress them. This process is accompanied by changes in chromatin structure (5). Indeed, the Th2 cytokine locus has been intensively studied as a model system of chromatin conformation of immune genes. How DNA is organized into three-dimensional structures in the nucleus and what are the functional consequences have been among the oldest questions in cell and molecular biology (6).
In this issue of Allergy, Hwang et al. (7) describe a specific region of DNA critical for transcription factor binding and complex formation that allows for efficient transcription of the Th2 cytokine genes. Transcription activators interact with the core transcription machinery through binding to enhancers while repressors work by recruiting corepressor complexes leading to chromatin condensation of enhancer regions. Repressors and activators may also function by competing with each other for occupation of shared DNA-binding sequences (8). Whether a transcription factor is activated or not is determined by its localization and ability to bind DNA. The initiation, termination, and regulation of transcription are influenced by epigenetic and metabolic processes (9, 10) and require ‘DNA looping’, an important mechanism to bring the promoters, enhancers, transcription factors, and RNA polymerases together (11). The transcription factors that can induce signature cytokines for distinct effector CD4 T-cell lineages and are both necessary and sufficient to elicit this process are called ‘master regulators’ (9).
Th2 cytokine genes are expressed preferentially from one chromosome rather than at random suggesting coordinated expression. How do master regulators act to activate the Th2 locus in a coordinated fashion? A locus control region (LCR) is a cis-acting element that confers tissue-specific high-level expression to linked genes. To identify the LCR, investigators deleted the conserved noncoding sequence-1 (CNS-1) in the Th2 cytokine locus and found that this reduced Th2 cell frequency (12) but did affect copy number-dependent expression of a linked IL-4 promoter reporter in transgenic mice. This suggested that CNS-1 is not an LCR but rather an important enhancer. Another study demonstrated that none of the previously described DNase I hypersensitive sites or conserved sequences in the Th2 cytokine locus had LCR activity, either individually or in combination as a minilocus. However, by evaluating copy number dependency in mice containing transgenes encoding the whole Th2 cytokine locus, Lee et al. (13, 14) found the LCR, located within a 25-kb region containing the 3′ portion of the RAD50 gene. The Rad50 gene separates the IL-4 and IL-13 genes from the IL-5 gene (15–17) (Fig. 1). There are four sites within the Th2 LCR that are ‘DNase I hypersensitive’, [i.e. exposed chromatin, highly susceptible to binding by proteins like DNase I or transcription factors (15)], implying their importance in transcription. The DNase I hypersensitive sites of the Rad50 gene are RHS4, RHS5, RHS6, and RHS7 (16, 18). Indeed, it has been demonstrated that these sites coordinate transcription of the Th2 cytokine genes, although the importance of RHS4 has not been fully established (10, 17, 19). RHS6 is thought to be the most important of the four sites for Th2 cytokine transcription as it is essential for the expression of each of the IL-4, IL-5, and IL-13 genes (17).
Figure 1.
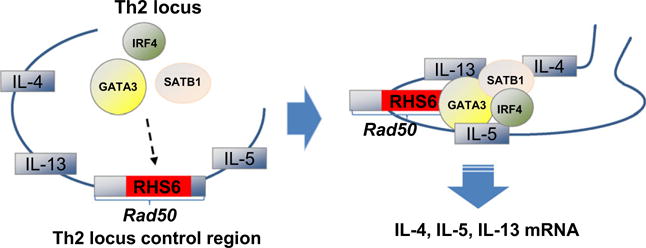
Activation of the Th2 cytokine gene locus. Gene activation of Th2 cytokines (IL-4, IL-5, and IL-13) is tightly controlled by transcription factors and DNA regulatory elements. RHS6 is a regulatory element of the Rad50 gene which is located between the IL-4/IL-13 and IL-5 genes. RHS6 is essential for the formation of a transcription factor complex consisting of GATA3, SATB1, and IRF4 in Th2 cells during allergic airway inflammation. The formation of this transcription factor complex is necessary for the efficient production of IL-4, IL-5, and IL-13 mRNA.
Hwang et al. (7) here hypothesized that this regulatory element was critical to Th2 cytokine production during allergic airway inflammation. Using a model of ovalbumin-induced airway inflammation in mice, they established that RHS6 was required for the production of IL-4, IL-5, and IL-13 from Th2 cells and this effect was specific for Th2-type inflammation as a Th1/Th17-mediated lung disease model was unchanged in RHS6−/− mice. Because DNase I hypersensitive regions are typically important sites for transcription factor binding, Hwang et al. also hypothesized that transcription factors bound to RHS6 to coordinate transcription during allergic airway inflammation. Indeed, RHS6 contained a number of sites for transcription factor binding, including two direct sites for GATA3. While several transcription factors are implicated in the regulation of the Th2 cytokine genes, GATA3 is the master regulator of Th2 cell differentiation and is essential for expression of IL-5 and IL-13, but not IL-4 (20). The authors found that large protein complexes were formed at the RHS6 DNA subregion, suggesting that additional transcription factors may also participate in Th2 cytokine gene activation.
To investigate what other transcription factors might be found to be bound to RHS6, the DNA subregions were isolated and mass spectrometry was used (7). Although many proteins were identified, SATB1 and IRF4 were assessed further because these transcription factors perform similar functions to GATA3. Notably, they were found in all of the isolated subregions of RHS6. IRF4 is required for Th2 cell differentiation, binds to the IL-4 promoter to induce its expression, and acts upstream and downstream of GATA3 (20). SATB1 regulates GATA3, coordinating Th2 development (21). In the absence of RHS6, GATA3, IRF4, and SATB1 binding was reduced at all sites in the Th2 cytokine LCR. These data highlight the importance of RHS6 as the critical regulatory element for effective Th2 cytokine transcription, as suggested by others (17). The authors also found that RHS6 was required for the formation of a physical protein complex comprised of GATA3, SATB1, and IRF4, among others. Transcription of IL-4, IL-5, and IL-13 was dependent on formation of this complex at RHS6. In the future, it would be interesting to understand what other transcription factors are present in this protein complex and how they function to mediate Th2 cytokine transcription. In addition, in NK/NKT cells Hwang et al. demonstrated that RHS6’s role in mediating Th2 cytokine transcription is conserved among cell types. Because RHS6 is located between the IL-4/IL-13 and IL-5 genes, it is intriguing that RHS6 can mediate the transcription of up- and downstream targets. This fact brings up the importance of the three-dimensional structure of chromatin and suggests that regulatory elements throughout the genome can have profound effects on genes both up- and downstream.
The present study by Hwang et al. revealed transcriptional insight into Th2-driven allergic airways disease. It was established that GATA3, IRF4, and SATB1 physically complex at RHS6 in the Rad50 gene of the Th2 cytokine locus during allergic airway inflammation. Presence of RHS6 was essential for the transcription factor complex formation, expression of Th2 cytokine genes, and allergic airway inflammation. There are a number of outstanding questions that remain for future investigations. The pathogenic significance of RHS6, for example, must be confirmed in CD4+ cells from asthmatic patients as they undergo Th2 differentiation. Group 2 innate lymphoid cells have recently been implicated in the pathogenesis of asthma. These cells preferentially express IL-5 and IL-13 upon activation (22). It would be interesting to investigate whether RHS6 also coordinates cytokine transcription in these cells. A number of genomewide association studies have revealed that single nucleotide polymorphisms in the Rad50 gene are associated with asthma and allergy (23–25), but the involvement of the RHS6 region remain unclear in these. Importantly, the data presented here suggest that Th2 cytokine production is controlled by transcription factor binding to RHS6. Impeding the physical association of the GATA3, SATB1, and IRF4 complex by targeting RHS6 presents an attractive, novel therapeutic possibility to reduce Th2 cytokine production during chronic airway inflammation. Further studies are warranted on targeting this and other mechanisms regulating coordinated Th2 cytokine transcription so that these potential therapeutics could become reality.
Acknowledgments
This work was supported by R21Al116121 and R01AI072197 (AH); T32 ES007059 (CHF).
Footnotes
Author contributions
Cameron H Flayer has provided the first draft of the manuscript and the illustration. Angela Haczku edited and finalized the manuscript and the figure.
Conflicts of interest
The authors declare that they have no conflicts of interest.
References
- 1.Boyman O, Kaegi C, Akdis M, Bavbek S, Bossios A, Chatzipetrou A, et al. EAACI IG Biologicals task force paper on the use of biologic agents in allergic disorders. Allergy. 2015;70:727–754. doi: 10.1111/all.12616. [DOI] [PubMed] [Google Scholar]
- 2.Chung KF. Asthma phenotyping: a necessity for improved therapeutic precision and new targeted therapies. J Intern Med. 2016;279:192–204. doi: 10.1111/joim.12382. [DOI] [PubMed] [Google Scholar]
- 3.Roth M, Black JL. Transcription factors in asthma: are transcription factors a new target for asthma therapy? Curr Drug Targets. 2006;7:589–595. doi: 10.2174/138945006776818638. [DOI] [PubMed] [Google Scholar]
- 4.Zhu J. Transcriptional regulation of Th2 cell differentiation. Immunol Cell Biol. 2010;88:244–249. doi: 10.1038/icb.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 6.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang SS, Jang SW, Lee KO, Kim HS, Lee GR. RHS6 coordinately regulates the Th2 cytokine genes by recruiting GATA3, SATB1, and IRF4. Allergy. 2017;72:772–782. doi: 10.1111/all.13078. [DOI] [PubMed] [Google Scholar]
- 8.Chatila TA, Li N, Garcia-Lloret M, Kim HJ, Nel AE. T-cell effector pathways in allergic diseases: transcriptional mechanisms and therapeutic targets. J Allergy Clin Immunol. 2008;121:812–823. doi: 10.1016/j.jaci.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 9.Wang C, Collins M, Kuchroo VK. Effector T cell differentiation: are master regulators of effector T cells still the masters? Curr Opin Immunol. 2015;37:6–10. doi: 10.1016/j.coi.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Mercer TR, Mattick JS. Understanding the regulatory and transcriptional complexity of the genome through structure. Genome Res. 2013;23:1081–1088. doi: 10.1101/gr.156612.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohrs M, Blankespoor CM, Wang ZE, Loots GG, Afzal V, Hadeiba H, et al. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat Immunol. 2001;2:842–847. doi: 10.1038/ni0901-842. [DOI] [PubMed] [Google Scholar]
- 13.Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 14.Lee GR, Fields PE, Flavell RA. Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity. 2001;14:447–459. doi: 10.1016/s1074-7613(01)00125-x. [DOI] [PubMed] [Google Scholar]
- 15.Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Koh BH, Hwang SS, Kim JY, Lee W, Kang M-J, Lee CG, et al. Th2 LCR is essential for regulation of Th2 cytokine genes and for pathogenesis of allergic asthma. Proc Natl Acad Sci USA. 2010;107:10614–10619. doi: 10.1073/pnas.1005383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams A, Lee GR, Spilianakis CG, Hwang SS, Eisenbarth SC, Flavell RA. Hypersensitive site 6 of the Th2 locus control region is essential for Th2 cytokine expression. Proc Natl Acad Sci USA. 2013;110:6955–6960. doi: 10.1073/pnas.1304720110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng WP. ‘All things considered’: transcriptional regulation of T helper type 2 cell differentiation from precursor to effector activation. Immunology. 2013;140:31–38. doi: 10.1111/imm.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim K, Kim N, Lee GR. Transcription factors Oct-1 and GATA-3 cooperatively regulate Th2 cytokine gene expression via the RHS5 within the Th2 locus control region. PLoS One. 2016;11:e0148576. doi: 10.1371/journal.pone.0148576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burute M, Gottimukkala K, Galande S. Chromatin organizer SATB1 is an important determinant of T-cell differentiation. Immunol Cell Biol. 2012;90:852–859. doi: 10.1038/icb.2012.28. [DOI] [PubMed] [Google Scholar]
- 22.Yang Q, Ge MQ, Kokalari B, Redai IG, Wang X, Kemeny DM, et al. Group 2 innate lymphoid cells mediate ozone-induced airway inflammation and hyperresponsiveness in mice. J Allergy Clin Immunol. 2016;137:571–578. doi: 10.1016/j.jaci.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Accordini S, Calciano L, Bombieri C, Malerba G, Belpinati F, Lo Presti AR, et al. An interleukin 13 polymorphism is associated with symptom severity in adult subjects with ever asthma. PLoS One. 2016;11:e0151292. doi: 10.1371/journal.pone.0151292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schieck M, Sharma V, Michel S, Toncheva AA, Worth L, Potaczek DP, et al. A polymorphism in the TH 2 locus control region is associated with changes in DNA methylation and gene expression. Allergy. 2014;69:1171–1180. doi: 10.1111/all.12450. [DOI] [PubMed] [Google Scholar]
- 25.Sharma V, Michel S, Gaertner V, Franke A, Vogelberg C, von Berg A, et al. Fine-mapping of IgE-associated loci 1q23, 5q31, and 12q13 using 1000 Genomes Project data. Allergy. 2014;69:1077–1084. doi: 10.1111/all.12431. [DOI] [PubMed] [Google Scholar]


