Summary
The gut microbiome is comprised of microbes from multiple kingdoms, including bacteria but also fungi, viruses and perhaps other agents. In this issue of Cell Host & Microbe, Jiang et al. (Jiang et. al., 2017) reveal that fungal monocolonization after antibiotic-mediated depletion of intestinal bacteria prevents colitis, thus highlighting beneficial roles of fungi.
Preview
The bacterial microbiome, predominantly the bacterial communities in the gut, has captured the attention of the scientific community for decades. Gut commensal bacteria are extremely beneficial to human health, facilitating nutrient metabolism and colonization resistance, promoting epithelial cell integrity and immune system development, and positively influencing immune responses to pathogens in extra-intestinal organs such as the lung (Jandhyala et al., 2015). However, bacteria are not the only microorganisms inhabiting mucosal surfaces; fungi and viruses occupy the same niches and have also been demonstrated to be crucial in maintaining intestinal homeostasis and promoting systemic immunity. Despite their importance, there is a paucity of scientific literature addressing the interactions of this kingdom of microorganisms with the other constituents of the microbiome, and their contribution to health and disease. In fact, conducting a PubMed search using either mycobiome (fungal microbiome) or microbiome as input yields 142 and 37,452 results, respectively. In other words, less than 0.4% of the microbiome literature accounts for the presence of fungi in commensal microbial communities. But recent studies in mice and humans have highlighted the key role of fungi in intestinal homeostasis and systemic immunity (Iliev et al., 2012; Iliev and Leonardi, 2017; Sokol et al., 2013; Tang et al., 2015; Wang et al., 2016), making it difficult to ignore the contribution of fungi to the global effects of the microbiome in health and disease.
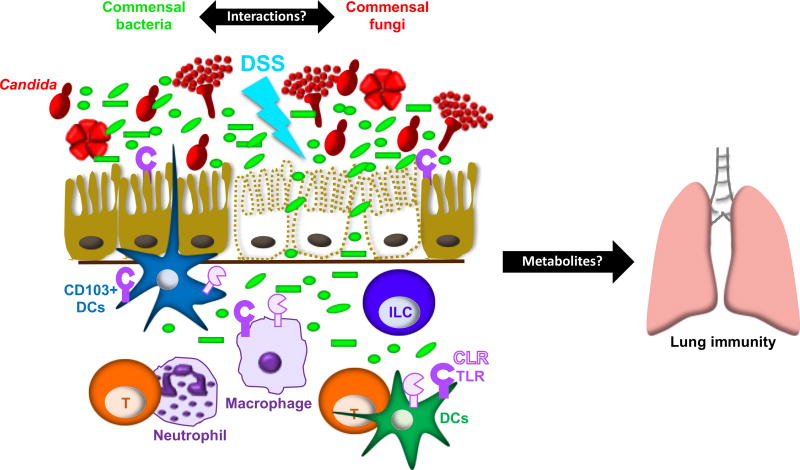
In this issue, Jiang et.al. (Jiang et. al., 2017) demonstrate that in mice treated with broad-spectrum antibiotics, and thus depleted of commensal bacteria, monocolonization with either Candida albicans or Saccharomyces cerevisiae, protects mice from dextran sulfate sodium (DSS)-induced colitis. Accordingly, antifungal treatment of C. albicans-colonized animals results in exacerbated colitis. Therefore, fungi replaced bacteria in averting colitis induced by epithelial cell damage (Figure).
Figure 1.
The Intestinal Mycobiome and Resistance to Colitis and Influenza A Infection Bacterial and fungal communities cohabit mucosal surfaces, but little is known about the interactions between these microbial communities and how such interactions affect local and systemic immunity. In mice treated with broad-spectrum antibiotics, monocolonization with Candida albicans or Saccharomyces cerevisiae (not depicted) replaced the protective effect of bacteria against DSS-induced colitis (Jiang et al., 2017). This protective effect required Candida cell wall mannan and host TLR4. Colonization with C. albicans also protected against influenza infection. Additional microbial products and/or host factors directly responsible for C. albicans’ protective effect remain to be elucidated. Prior studies have shown that the absence of pattern recognition pathways crucial in antifungal immunity (dectin 1, dectin 3, and Card 9) results in exacerbated colitis. In some instances, aggravated disease is a result of mouse genotype and, in other instances, alterations in the microbiota. Regardless of the cause of exacerbated disease, it is clear that fungi are a key component of intestinal health and thus should be included in efforts to fully understand the microbiome.
These findings support recent studies demonstrating that the inability to recognize molecular patterns in the fungal cell wall exacerbates DSS-induced colitis. In fact, Dectin 1−/− (Iliev et al., 2012), Dectin 3−/− (Wang et al., 2016) and Card 9−/− mice (Lamas et al., 2016; Sokol et al., 2013), which lack pattern recognition pathways essential for sensing carbohydrates often found in the fungal cell wall, exhibit increased susceptibility to colitis. In considering these studies, it is important to distinguish between mouse genotype, and variations in the microbial communities across mouse strains and laboratories. For example, one study showed that Dectin 1−/− mice are more susceptible to DSS-induced colitis due to penetration of the fungus C. tropicalis into the lamina propria (Iliev et al., 2012). There were no differences in the bacterial communities between wild-type and Dectin 1−/− mice in that study. In another study, Dectin 1−/− mice (Tang et al., 2015) were shown to be protected from colitis due to the overgrowth of Lactobacilli, which promoted the expansion of regulatory T cells and consequently, decreased inflammation. A difference between these two studies is that in the former, animals were colonized by fungal commensals whereas in the latter, mice were not initially colonized by fungi. Regardless of the discrepancies in the microbial communities between laboratories, and whether or not disease phenotypes are due to mouse genotype or the microbiota, it is clear that the mycobiota is a key component of intestinal homeostasis, and microbiome analyses that ignore fungal communities are thus incomplete.
One of the most remarkable findings of the current study reported here is that the protective effects of fungal colonization were not restricted to the intestine, as depletion of C. albicans via the antifungal fluconazole resulted in decreased survival following respiratory infection with influenza virus. A prior study showed that oral treatment with antifungal drugs resulted in alterations of both fungal and bacterial communities, and increased susceptibility to allergic airway disease (Wheeler et al., 2016). What host and microbial elements establish communication between the gut and the lung? It has been shown that migrating immune cells, and bacterial ligands and metabolites such as lipopolysaccharide (LPS), polysaccharide A (PSA), and short chain fatty acids (SCFA), play an important role in facilitating communication in the “gut-lung axis (Budden et al., 2017).” Mechanistically, bacterial metabolites promote production of the immunosuppressive cytokine IL-10 and regulatory T cell responses that alleviate allergic airway disease. Ligation of nucleotide-binding oligomerization domain (NOD)-like receptors and Toll-like receptors (TLR) by gut bacteria promotes innate and adaptive immunity against pulmonary bacterial and viral infections (Budden et al., 2017). These observations prompt questions such as: “How do fungi that inhabit the intestine affect bacterial metabolism?”; and “What are the fungal structural components and/or metabolites that themselves serve as liaisons of communication between the gut and the lung?”
Jiang, et.al. took a step in this direction by identifying mannan as the fungal cell wall component responsible for the protective effect of C. albicans colonization against colitis and influenza infection. Interestingly, mannan administration resulted in complete protection against colitis, but only partial protection against influenza A, suggesting a requirement for other fungal products in “positively calibrating” systemic immunity. In addition, the authors identified TLR4 as the pattern recognition receptor (PRR) mediating C. albicans’ protective effect against colitis. These findings pose various questions that might be tackled in future studies. For example, (i) Does TLR4 directly recognize mannan in vivo? (ii) Is TLR4 engagement in the gut a requirement for the establishment of effective lung immunity? (iii) Do PRRs cooperate in the recognition of C. albicans in the gut to establish lung antiviral immunity? Unraveling the mechanisms by which fungal structural elements and, perhaps, metabolites confer protective immunity both locally and in extra-intestinal sites such as the lung is of paramount importance, particularly in the context of broad spectrum antibiotic treatment.
The current study represents an important advance in our understanding of the contribution of fungi to gut health and to the establishment of effective antimicrobial immunity in distal organs such as the lung. Moreover, the findings highlight the importance of conducting comprehensive microbiome studies that reflect the diversity of the microbial communities that inhabit mucosal surfaces. Ignoring fungi in microbiome studies robs the scientific community of the opportunity to discover microbial compounds that aid in establishing beneficial lines of communication between the gut and other peripheral organs, and yields an incomplete picture of the contribution of microbes to health and disease.
Acknowledgments
The authors are supported by Burroughs Wellcome Fund grant 1016190 (NHS), NIAID T32 AI055397 (BSK), and R01 grants AI035681 and AI040996 (BSK).
References
- Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, Hansbro PM. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol. 2017;17:635–646. doi: 10.1038/nri.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang TT, Shao Tzu-Yu, Gladys Ang WX, Kinder Jeremy M, Turner Lucien, Pham Giang, Whitt Jordan, Theresa Alenghat, Way Sing Sing. Commensal fungi replace the protective benefits of intestinal bacteria. Cell Host Microbe. 2017 doi: 10.1016/j.chom.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Conway KL, Zhang M, Choi M, Morin B, Cao Z, Villablanca EJ, Li C, Wijmenga C, Yun SH, et al. Card9 mediates intestinal epithelial cell restitution, T-helper 17 responses, and control of bacterial infection in mice. Gastroenterology. 2013;145:591–601. e593. doi: 10.1053/j.gastro.2013.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Kamiya T, Liu Y, Kadoki M, Kakuta S, Oshima K, Hattori M, Takeshita K, Kanai T, Saijo S, et al. Inhibition of Dectin-1 Signaling Ameliorates Colitis by Inducing Lactobacillus-Mediated Regulatory T Cell Expansion in the Intestine. Cell Host Microbe. 2015;18:183–197. doi: 10.1016/j.chom.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Wang T, Pan D, Zhou Z, You Y, Jiang C, Zhao X, Lin X. Dectin-3 Deficiency Promotes Colitis Development due to Impaired Antifungal Innate Immune Responses in the Gut. PLoS pathogens. 2016;12:e1005662. doi: 10.1371/journal.ppat.1005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, Brown J, Funari VA, Wang HL, Crother TR, et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe. 2016 doi: 10.1016/j.chom.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]