Abstract
Long noncoding RNAs (lncRNAs) have recently emerged as novel regulators of lineage commitment, differentiation, development, viability and disease progression. Few studies have examined their role in osteogenesis; however, given their critical and wide-ranging roles in other tissues, lncRNAs are most likely vital regulators of osteogenesis. In this study we extensively characterized lncRNA expression in mesenchymal cells during commitment and differentiation to the osteoblast lineage using a whole transcriptome sequencing approach (RNA-Seq). Using mouse primary mesenchymal stromal cells (mMSC), we identified 1438 annotated lncRNAs expressed during MSC differentiation, 462 of which are differentially expressed. We performed guilt-by-association analysis using lncRNA and mRNA expression profiles to identify lncRNAs influencing MSC commitment and differentiation. These findings open novel dimensions for exploring lncRNAs in regulating normal bone formation and in skeletal disorders.
Keywords: long noncoding RNA, mesenchymal stromal cells, osteoblast, RNA-Seq, gene expression, osteogenesis
INTRODUCTION
Osteogenesis is a complex biological process that requires mesenchymal stem cell differentiation through the osteoblast lineage together with the precise timing of developmental signaling cascades, expression of transcriptional regulators of genes producing bone matrix and promoting mineral deposition (1, 2). In mammals, while 75% of the genome is transcribed, only 2% encodes proteins. In addition to the messenger RNA (mRNA) transcribed genes, a large part of the osteoblast genome is represented by numerous classes of non-coding RNAs, among which are the well studied microRNAs that function in regulating protein translation (3). It is becoming increasingly apparent that the group of long noncoding RNAs (lncRNAs) contribute significantly to regulation of gene expression.
To date, 15,778 human and 11,975 mouse lncRNAs have been identified(4, 5); and only a small fraction of these are characterized. LncRNAs include enhancer RNAs, snoRNA hosts, intergenic transcripts and transcripts overlapping other transcripts in either sense or antisense orientations (6). All ncRNA transcripts longer than 200 nucleotides, with little or no coding potential, are considered lncRNAs. There is wide variability in lncRNA length, with some examples significantly longer than several kb. Like mRNAs, lncRNAs are often polyadenylated and transcribed by RNA polymerase II. They can have multiple isoforms and are regulated by transcription factors just like their protein coding counterparts. Generally, lncRNAs are expressed at lower levels than protein-coding transcripts, and their expression is more restricted in terms of tissue specificity (7). lncRNAs are nuclear and/or cytoplasmic and they have been found to act in a variety of biological processes, for example X-chromosome inactivation (8) and imprinting (9, 10). lncRNAs are often differentially expressed during development, indicating that they may influence cell fate (6, 11, 12). They have been demonstrated to be essential regulators of lineage commitment during adipogenesis (13), hematopoiesis (14), erythropoiesis (15), keratinocyte differentiation (16) and myogenesis (17), and are associated with numerous diseases including neurological, autoimmune, cardiovascular and cancer (6).
lncRNAs function in a variety of ways, including recruiting chromatin modifying enzymes to activate or repress transcription(18). Subsets of nuclear lncRNAs, known as enhancer RNAs (eRNAs), activate gene expression (19, 20). Cytoplasmic lncRNAs often have sequence complementarity with other transcripts, which is used to modulate translational control and stability of target RNAs. They can also act as competing endogenous RNAs (ceRNAs) or “miRNA sponges”, sequestering specific miRNAs, thereby protecting target mRNA from silencing (21–25). In addition, lncRNAs are multifunctional, and can interact with numerous components of the gene regulatory machinery (proteins, RNA, DNA). The identification and characterization of lncRNAs that are critical for osteogenesis will extend understanding of the cellular and molecular mechanisms that regulate bone formation as well as provide insight into the broader biological role of lncRNAs.
Only a few lncRNAs functioning in osteoblasts and bone tissue are known and a more in depth description of them can be found in these reviews (26–28). The first well-characterized lncRNA to effect dental and craniofacial development was Msx1-as. Msx1-as is an important osteogenic regulator that negatively affects the expression of the homeobox protein Msx1 and its inhibition of Dlx5 (29–31). Recently, the lncRNA H19, which is upregulated during osteogenic differentiation, was found to act as a competing endogenous RNA for miR-141 and miR-22, thereby were regulating the Wnt/β-catenin pathway and osteogenesis (25). Another lncRNA DANCR (differentiation antagonizing non-coding RNA), has been shown to effect osteoblast differentiation by its association with enhancer of zeste homolog 2 (EZH2) (32). It has been demonstrated that knockout mice of the lncRNA Dnm3os die shortly after birth and demonstrate skeletal abnormalities, including craniofacial hypoplasia, defects in dorsal neural arches and spinous processes of the vertebrae, and osteopenia (33). Targeted disruption of Hotair results in malformation of metacarpal-carpal bones and homeotic transformation of the spine (34).
We hypothesize that multiple lncRNAs are instrumental in osteogenesis and have embarked on identifying lncRNAs with critical functions for osteoblastogenesis. Zuo et al. examined the lncRNA profile of early differentiated C3H10T1/2 MSCs by lncRNA array (35) and recently Huang et al. have examined the expression of lncRNA in differentiating human adipose stem cells (36). Herein, we profile the complete transcriptome of mouse MSCs from an undifferentiated cell, to commitment and ECM deposition and finally mineralization by complete genome sequencing (RNA-Seq).
MATERIALS AND METHODS
Ethics Statement
All animal work was reviewed and approved by UVM Institutional Animal Care and Use Committee (IACUC).
Tissue Culture and Osteogenic Differentiation
Primary mouse bone marrow MSCs were isolated from femurs and tibias of 6–8 week old SMAA-mCherry mice, cultured and sorted as previously described (37, 38). mMSCs of passage 6 to 10 were used for all the experiments in this study. To induce osteogenic differentiation of mouse MSCs, complete alpha-MEM was supplemented with 280 μM ascorbic acid and 10 mM beta-glycerophosphate (Sigma Aldrich, St. Louis, MO, USA). Cells were maintained at 37°C in a humidified 5% CO2 environment and media replaced every 2 to 3 days for the duration of all experiments.
RNA Isolation and Library Preparation
Total RNA was isolated at 4 stages of differentiation (days 0, 7, 14 and 21) using Trizol reagent (Invitrogen) according to the manufacturers’ specifications, or using the Direct-zol RNA Miniprep kit (Zymo Research), with DNaseI treatment. RNA integrity was assessed by Bioanalyzer (Agilent). RNA-Seq libraries were built with the TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold (Illumina) according to manufacturer’s protocol. Library quality was assessed by Bioanalyzer using DNA HS chip and quantified by Kapa Library Quantification Kit for Illumina Platforms (Kapa Biosystems). Three biologically independent RNA-Seq libraries were prepared for each time point during mMSC differentiation. All RNA-Seq libraries were single-end sequenced (SE100) on a HiSeq-1500. Base calls and sequence reads were generated by bcl2fastq software (version 1.8.4, Illumina).
RNA-Seq Analysis
Data analysis was conducted using the Galaxy platform (39) and RStudio (40). Quality control analysis of fastq raw data was performed using FastQC (41). Reads were aligned to reference genome mm10 using STAR (42) and expression counts were determined by HTSeq (43) with Gencode vM10 gene annotation (5). Genes with > 0.2FPKM were considered to be expressed. Differential expression analysis was performed with DESeq2 (44). For differential gene expression analyses, the cutoff for significant fold change was >2, adjusted p-value <0.05. Cells at all differentiation stages (days 7, 14 and 21) were compared with undifferentiated cells (day 0) as well as to each other.
Heatmaps
The Gencode annotation file was used to identify the protein coding genes (mRNAs) and long noncoding RNA groups. The lncRNA group included the following categories: 3prime overlapping ncRNA, antisense, bidirectional promoter lncRNA, lincRNA (long intergenic noncoding RNA), processed transcript, sense intronic, sense overlapping, TEC (to be experimentally confirmed). Other noncoding RNAs (ie: miRNA, ribosomal RNA, snoRNAs, tsRNA etc) were removed from analysis. Clustering analysis of row normalized differentially expressed gene count data was performed for each group using k-means with 6 groups.
Gene Ontology Analysis
mRNA groups with similar expression patterns were merged and Gene Ontology (GO) annotation analyses of gene sets were performed. GO analysis was derived from Gene Ontology (www.geneontology.org) (45, 46).
Guilt-by-association Analysis
We generated a correlation matrix between lncRNAs and mRNAs by computing the Spearman correlation coefficient between all pairs of lncRNAs-mRNAs across our time course. We applied k-means clustering (k = 60 for mRNA and k = 50 for lncRNAs) to find groups of lncRNAs whose expression patterns correlate closely with groups of mRNAs. Clusters with ρ > 0.7 or ρ < −.7 were selected for further analysis by performing gene ontology analysis as described above.
RESULTS AND DISCUSSION
mMSCs were isolated from bone marrow of 8-week transgenic male mice and differentiated over 21 days. RNA was isolated 4 stages of differentiation: undifferentiated (day0), commitment (day7), extracellular matrix (ECM) deposition (day14) and mineralization (day21). Expression patterns of known osteoblast marker genes confirmed data quality at the differentiation time points. From this analysis we identified 12,391 mRNAs and 1,438 annotated lncRNAs expressed during mMSC osteoblast differentiation. Differential expression of mRNAs and lncRNAs in mMSCs was analyzed and 3,100 mRNAs and 462 lncRNAs were identified as being differentially expressed (fold-change >2-fold; FDR adjusted p-value <0.05). Clustering of the differentially expressed mRNAs showed distinct profiles across development (Figure 1a). Cluster 1 consisted of mRNAs that exhibit increasing expression with differentiation. An examination of mRNAs in this cluster revealed several known osteoblast marker genes (ie: Ibsp, Bglap, Bmp2) and GO analysis identified this cluster to be enriched in genes associated with ossification, skeletal development and mineralization (Table 1).
Figure 1. Gene expression profiling in differentiating mouse MSCs.
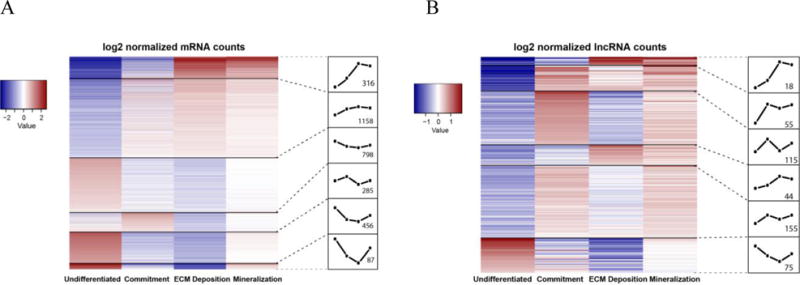
(a) Heatmap of differentially expressed mRNAs in mMSCs at 4 time points (undifferentiated, commitment, ECM deposition, mineralization). (b) Heatmap of differentially expressed lncRNAs in the same mMSCs at 4 stages of osteogenic differentiation. The number of genes in each cluster is noted in the respective expression profile box.
Table 1.
Gene ontology of the mRNAs in cluster 1 of Figure 1a is shown with the associated p-value for the enrichment of each Gene Ontology term
| GO Biological Process | p-value |
|---|---|
| regulation of ossification (GO:0030278) | 5.18E-08 |
| regulation of biomineral tissue development (GO:0070167) | 1.95E-07 |
| regulation of bone mineralization (GO:0030500) | 2.01E-06 |
| biomineral tissue development (GO:0031214) | 3.70E-05 |
| ossification (GO:0001503) | 4.55E-05 |
| regulation of developmental process (GO:0050793) | 2.99E-03 |
| regulation of pathway-restricted SMAD protein phosphorylation (GO:0060393) | 8.74E-03 |
| tissue development (GO:0009888) | 9.70E-03 |
| regulation of multicellular organismal process (GO:0051239) | 1.01E-02 |
| system development (GO:0048731) | 1.87E-02 |
| single-multicellular organism process (GO:0044707) | 2.25E-02 |
| regulation of multicellular organismal development (GO:2000026) | 2.50E-02 |
| animal organ development (GO:0048513) | 4.68E-02 |
By comparing the expression profiles in both the mRNA and lncRNA heatmaps, it is apparent that the expression of lncRNAs in cluster 1 (Figure 1b) may closely mimic mRNAs and may yield a clue as to which lncRNAs would be of interest to study in osteogenesis. As there is currently no way to postulate potential function of lncRNAs by sequence, researchers have been using this type of bioinformatics “guilt-by-association” approach. Using this approach, it assumed that lncRNAs and mRNAs that are tightly co-expressed are co-regulated and families of lncRNAs can be identified (12, 47). We therefore performed a guilt-by-association analysis using much smaller gene clusters, to identify groups of lncRNAs and mRNAs that are co-expressed and have highly correlated expression patterns (Figure 2a). We identified cluster 18 as highly associated with osteogenesis and mineralized tissues (Figure 2b) so we focused on lncRNAs that had expression patterns highly correlative or negatively correlated to it. We identified 2 clusters of lncRNAs whose patterns of expression highly correlated with osteogenic mRNAs (Figure 2c,d) and 1 cluster that highly anti-correlation with lncRNAs and mRNAs showing diametrically opposed expression patterns (Figure 2e).
Figure 2. Guilt-by-association Analysis.
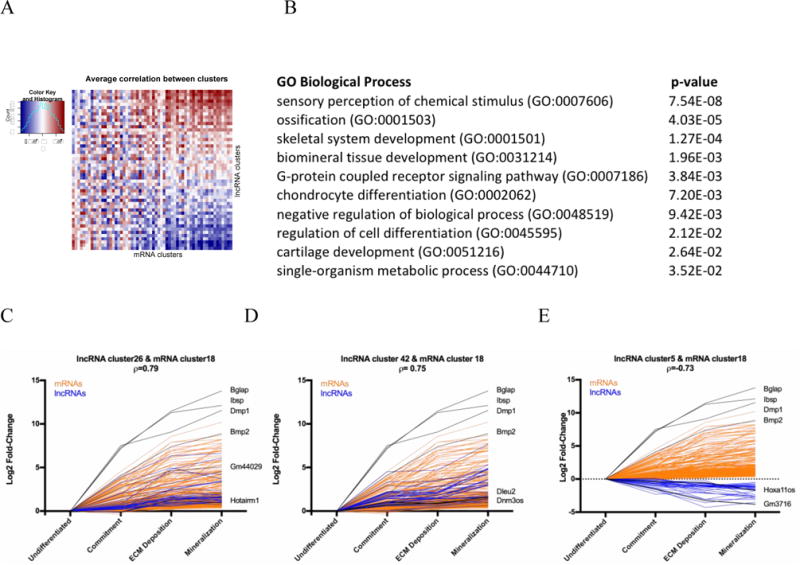
A) Association matrix of mRNAs and lncRNAs. 60 mRNA expression profile clusters (columns) and 50 lncRNA expression profile clusters (rows) are shown as positively (blue), negatively (red) or not associated (white). B) Gene ontology of the mRNAs in cluster 18 with the associated p-value. C,D) Log2 fold-change expression of mRNAs (orange) and lncRNAs (blue) across development are depicted. mRNAs in cluster18 show a highly correlative pattern of expression with lncRNA clusters 26 and 42, and a negatively correlative expression pattern with lncRNA cluster 5 (E).
Examination of the lncRNAs in lncRNA cluster 42, we identified Dnm3os as a potential osteogenic lncRNA. Dnm3os has been found to be essential for normal growth and skeletal development (33) and thereby validates the guilt-by-association strategy to identify lncRNAs that may be essential for osteogenesis. Dleu2 is also identified in this cluster and this lncRNA was identified as being associated with bone mineral density variation in postmenopausal women (48). In examining the negatively correlated lncRNA cluster 5, we found Hoxa11os. While little is known about the function of Hoxa11os in mice, its human homologue, HOXA11AS is associated with cell proliferation and therefore a decrease in its expression upon differentiation would be expected.
We have acquired the first comprehensive transcriptome from differentiating mouse MSCs across 4 stages of osteogenesis. Although lncRNAs are transcribed, as are mRNAs that are encoded into protein, the lncRNAs cannot be functionally annotated as readily. Their functional diversity and the significant roles they play in gene regulation and chromatin interaction challenges us to determine the secrets they hold in establishing cell phenotypes. The distribution of distinct lncRNAs at different stages of differentiation and the examples we have provided through lncRNA guilt-by-association promises to be a novel dimension in understanding the epigenetic mechanism regulating normal bone formation and their contributions to disease states of the skeleton.
Acknowledgments
Studies reported here were supported by NIH/NIAMS R01AR039588 (GSS/JBL) and NIH/NIDCR R37 DE012528 (JBL). The next-generation sequencing was performed in the University of Vermont Cancer Center Advanced Genome Technologies Core and was supported by the University of Vermont Cancer Center, Lake Champlain Cancer Research Organization, and the University of Vermont College of Medicine.
Footnotes
Disclosures: The authors have nothing to disclose
References
- 1.Berendsen AD, Olsen BR. Bone development. Bone. 2015;80:14–8. doi: 10.1016/j.bone.2015.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Q, Shou P, Zheng C, Jiang M, Cao G, Yang Q, Cao J, Xie N, Velletri T, Zhang X, Xu C, Zhang L, Yang H, Hou J, Wang Y, Shi Y. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell death and differentiation. 2016;23(7):1128–39. doi: 10.1038/cdd.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang H, Hata A. The role of microRNAs in cell fate determination of mesenchymal stem cells: balancing adipogenesis and osteogenesis. BMB Rep. 2015;48(6):319–23. doi: 10.5483/BMBRep.2015.48.6.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22(9):1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mudge JM, Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm Genome. 2015;26(9–10):366–78. doi: 10.1007/s00335-015-9583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25(18):1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–42. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 9.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375(6526):34–9. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 10.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Molecular cell. 2008;32(2):232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 11.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22(9):1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, Yuan B, Kellis M, Lodish HF, Rinn JL. Long noncoding RNAs regulate adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(9):3387–92. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paralkar VR, Mishra T, Luan J, Yao Y, Kossenkov AV, Anderson SM, Dunagin M, Pimkin M, Gore M, Sun D, Konuthula N, Raj A, An X, Mohandas N, Bodine DM, Hardison RC, Weiss MJ. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123(12):1927–37. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez-Dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, van Oudenaarden A, Lodish HF. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123(4):570–81. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orom UA, Derrien T, Guigo R, Shiekhattar R. Long noncoding RNAs as enhancers of gene expression. Cold Spring Harbor symposia on quantitative biology. 2010;75:325–31. doi: 10.1101/sqb.2010.75.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neguembor MV, Jothi M, Gabellini D. Long noncoding RNAs, emerging players in muscle differentiation and disease. Skeletal muscle. 2014;4(1):8. doi: 10.1186/2044-5040-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidovich C, Cech TR. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA. 2015;21(12):2007–22. doi: 10.1261/rna.053918.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TK, Hemberg M, Gray JM. Enhancer RNAs: a class of long noncoding RNAs synthesized at enhancers. Cold Spring Harbor perspectives in biology. 2015;7(1):a018622. doi: 10.1101/cshperspect.a018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends in biochemical sciences. 2014;39(4):170–82. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang T, Zhou B, Shi L, Wang H, Chu Q, Xu F, Li Y, Chen R, Shen C, Schinckel AP. lncRNA AK017368 promotes proliferation and suppresses differentiation of myoblasts in skeletal muscle development by attenuating the function of miR-30c. FASEB J. 2017 doi: 10.1096/fj.201700560RR. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Sun X, Cai H, Sun Y, Plath M, Li C, Lan X, Lei C, Lin F, Bai Y, Chen H. Long non-coding RNA ADNCR suppresses adipogenic differentiation by targeting miR-204. Biochimica et biophysica acta. 2016;1859(7):871–82. doi: 10.1016/j.bbagrm.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Wu F, Song Y, Li X, Wu Q, Duan Y, Jin Z. Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Death Dis. 2016;7(8):e2327. doi: 10.1038/cddis.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang WC, Fu WM, Wang YB, Sun YX, Xu LL, Wong CW, Chan KM, Li G, Waye MM, Zhang JF. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Scientific reports. 2016;6:20121. doi: 10.1038/srep20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huynh NP, Anderson BA, Guilak F, McAlinden A. Emerging roles for long noncoding RNAs in skeletal biology and disease. Connective tissue research. 2017;58(1):116–41. doi: 10.1080/03008207.2016.1194406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassan MQ, Tye CE, Stein GS, Lian JB. Non-coding RNAs: Epigenetic regulators of bone development and homeostasis. Bone. 2015;81:746–56. doi: 10.1016/j.bone.2015.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tye CE, Gordon JA, Martin-Buley LA, Stein JL, Lian JB, Stein GS. Could lncRNAs be the Missing Links in Control of Mesenchymal Stem Cell Differentiation? Journal of cellular physiology. 2015;230(3):526–34. doi: 10.1002/jcp.24834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blin-Wakkach C, Lezot F, Ghoul-Mazgar S, Hotton D, Monteiro S, Teillaud C, Pibouin L, Orestes-Cardoso S, Papagerakis P, Macdougall M, Robert B, Berdal A. Endogenous Msx1 antisense transcript: in vivo and in vitro evidences, structure, and potential involvement in skeleton development in mammals. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(13):7336–41. doi: 10.1073/pnas.131497098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berdal A, Lezot F, Pibouin L, Hotton D, Ghoul-Mazgar S, Teillaud C, Robert B, MacDougall M, Blin C. Msx1 homeogene antisense mRNA in mouse dental and bone cells. Connective tissue research. 2002;43(2–3):148–52. doi: 10.1080/03008200290000970. [DOI] [PubMed] [Google Scholar]
- 31.Babajko S, Petit S, Fernandes I, Meary F, LeBihan J, Pibouin L, Berdal A. Msx1 expression regulation by its own antisense RNA: consequence on tooth development and bone regeneration. Cells, tissues, organs. 2009;189(1–4):115–21. doi: 10.1159/000151748. [DOI] [PubMed] [Google Scholar]
- 32.Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochemical and biophysical research communications. 2013;432(4):612–7. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe T, Sato T, Amano T, Kawamura Y, Kawamura N, Kawaguchi H, Yamashita N, Kurihara H, Nakaoka T. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Developmental dynamics : an official publication of the American Association of Anatomists. 2008;237(12):3738–48. doi: 10.1002/dvdy.21787. [DOI] [PubMed] [Google Scholar]
- 34.Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, Helms JA, Chang HY. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell reports. 2013;5(1):3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuo C, Wang Z, Lu H, Dai Z, Liu X, Cui L. Expression profiling of lncRNAs in C3H10T1/2 mesenchymal stem cells undergoing early osteoblast differentiation. Molecular medicine reports. 2013;8(2):463–7. doi: 10.3892/mmr.2013.1540. [DOI] [PubMed] [Google Scholar]
- 36.Huang G, Kang Y, Huang Z, Zhang Z, Meng F, Chen W, Fu M, Liao W, Zhang Z. Identification and Characterization of Long Non-Coding RNAs in Osteogenic Differentiation of Human Adipose-Derived Stem Cells. Cell Physiol Biochem. 2017;42(3):1037–50. doi: 10.1159/000478751. [DOI] [PubMed] [Google Scholar]
- 37.Repic D, Torreggiani E, Franceschetti T, Matthews BG, Ivcevic S, Lichtler AC, Grcevic D, Kalajzic I. Utilization of transgenic models in the evaluation of osteogenic differentiation of embryonic stem cells. Connective tissue research. 2013;54(4–5):296–304. doi: 10.3109/03008207.2013.814646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H, Gordon JA, Whitfield TW, Tai PW, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Chromatin dynamics regulate mesenchymal stem cell lineage specification and differentiation to osteogenesis. Biochimica et biophysica acta. 2017;1860(4):438–49. doi: 10.1016/j.bbagrm.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, Gruning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic acids research. 2016;44(W1):W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.RStudio: Integrated Development for R. Boston, MA: RStudio Inc.; 2015. Version 0.98.978 ed. [Google Scholar]
- 41.Anders S. FastQC: a quality control tool for high throughput sequence data. 2010 [Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 42.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic acids research. 2015;43:D1049–56. doi: 10.1093/nar/gku1179. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reppe S, Refvem H, Gautvik VT, Olstad OK, Hovring PI, Reinholt FP, Holden M, Frigessi A, Jemtland R, Gautvik KM. Eight genes are highly associated with BMD variation in postmenopausal Caucasian women. Bone. 2010;46(3):604–12. doi: 10.1016/j.bone.2009.11.007. [DOI] [PubMed] [Google Scholar]


