SUMMARY
Microglia are an exquisitely tiled and self-contained population in the CNS that do not receive contributions from circulating monocytes in the periphery. While microglia are long-lived cells, the extent to which their cell bodies are fixed and the molecular mechanisms by which the microglial landscape is regulated have not been determined. Using chronic in vivo two-photon imaging to follow the microglial population in young adult mice, we document a daily rearrangement of the microglial landscape. Furthermore, we show that the microglial landscape can be modulated by severe seizures, acute injury, and sensory deprivation. Finally, we demonstrate a critical role for microglial P2Y12Rs in regulating the microglial landscape through cellular translocation independent of proliferation. These findings suggest that microglial patrol the CNS through both process motility and soma translocation.
In Brief
Using a chronic in vivo imaging approach, Eyo et al. show that the physical positions of brain microglia change daily and that these changes increase following certain experimental manipulations. The mechanism underlying these changes involves cell translocation controlled by microglial-specific P2Y12 receptors.
INTRODUCTION
Microglia, the tissue-resident macrophages of the CNS, are extremely dynamic and exhibit elaborate remodeling of their processes (Davalos et al., 2005; Nimmerjahn et al., 2005), through which they make repeated contacts with other CNS elements (Tremblay et al., 2010; Wake et al., 2009). Roles are now recognized for microglia in early CNS development where they clear apoptotic cells (Marín-Teva et al., 2004), facilitate brain wiring (Squarzoni et al., 2014), and regulate synaptic development (Paolicelli et al., 2011; Schafer et al., 2012). In adults, microglia participate in mechanisms underlying learning and behavior (Chen et al., 2010; Parkhurst et al., 2013).
Interest in understanding microglial homeostatic maintenance has revealed that they are an autonomous self-renewing population (Ajami et al., 2011) lacking long-term contributions from peripheral cells. In both the rodent and human brain, microglial numbers are maintained throughout adult life (Askew et al., 2017; Davies et al., 2017). Indeed, it is assumed that, as somatically stationary but motile cells, the microglial landscape is largely fixed. However, the mechanisms by which the microglial landscape is organized and maintained are not entirely clear. To address this, we used chronic two-photon in vivo imaging in the intact brain of double-transgenic CX3CR1GFP/+:Thy1YFP/+ mice to visualize microglia (CX3CR1-GFP) and dendrites (Thy1-yellow fluorescent protein [YFP]), which were used as landmarks for repeated positional imaging (Eyo et al., 2014; Feng et al., 2000; Jung et al., 2000). Specifically, we studied the daily changes in the microglial landscape, modulated these changes experimentally, and determined the mechanistic processes as well as a molecular factor underlying such changes.
RESULTS
Daily Microglial Landscape Rearrangement Occurs via Translocation in the Healthy Brain
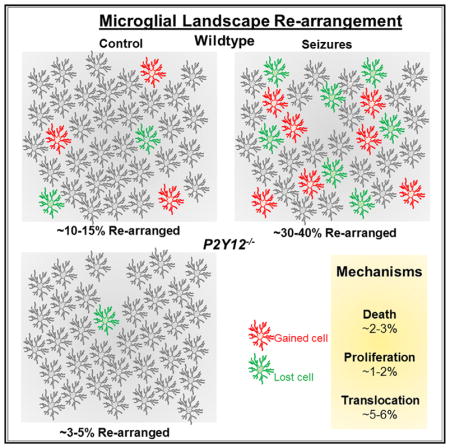
Two to four weeks after window implantation, microglia underwent a round of activation and returned to a ramified morphology (Figure S1A). To re-locate specific fields of view (FOVs), blood vessels and dendrites were used as gross and fine landmarks, respectively (Figure S1B). In the limb/trunk region of the primary somatosensory cortex, microglia exhibited a rearrangement of their landscape: some cells emerged (‘‘gained’’) into the FOV at a position where no cells resided previously, while others were missing (‘‘lost’’) from the FOV at positions where they resided previously (Figure 1A). Approximately 10%–15% of microglia were rearranged (Figure S2A), regardless of the frequency of daily observation (Figure S2B) or gender (Figure S2C). Similar degrees of rearrangement occurred in both the superficial layer I (depth, 50–120 μm) and deeper layer II/III (depth, 180–240 μm) of the cortex (Figure 1B), though rearrangement differed between the limb/trunk and whisker barrel areas (Figure 1C).
Figure 1. Microglial Landscape Rearrangement via Cell Translocation.
(A) Microglia in the healthy cortex during chronic in vivo two-photon imaging showing ‘‘lost’’ (red) and ‘‘gained’’ (green) cells. BV, blood vessels.
(B) Percentage of rearranged microglia in superficial (50–120 μm) and deeper (180–240 μm) cortex (n = 12 FOVs each for superficial and deeper regions from 4 mice).
(C) Percentage of rearranged microglia in the somatosensory limb/trunk and barrel cortices (n = 12 FOVs from 4 mice for each cortical region).
(D) Cortical and dentate gyrus (DG) microglia and BrdU with white (BrdU-only cells) and yellow (BrdU/Iba1 cells) arrows.
(E and F) BrdU/Iba1 double-immunoreactive quantification in the (E) wild-type cortex and DG and in the (F) wild-type and Trem2−/− cortex; n = 4 for (E) and 4 mice each for (F).
(G) Microglia per FOV in wild-type, CX3CR1−/−, and Trem2−/− mice (n = 3 wild-type, 3 CX3CR1−/−, and 4 Trem2−/− mice).
(H) Percentage of rearranged microglia in wild-type, CX3CR1−/−, and Trem2−/− mice (n = 16 FOVs from 5 wild-type mice, 15 FOVs from 5 CX3CR1−/− mice, and 11 FOVs from 4 Trem2−/− mice).
(I) Percentage of daily ‘‘gained’’ or ‘‘lost’’ cells in wild-type, CX3CR1−/−, and Trem2−/− mice (n = 4 wild-type, 3 CX3CR1−/−, and 4 Trem2−/− mice).
(J) Static (circles) and translocated (arrows: white for starting and yellow for translocated position) microglia from daily imaging.
*p < 0.05; **p < 0.01; and ***p < 0.001. Data are presented as mean ± SEM.
Since coupled proliferation and death (Askew et al., 2017) or microglial translocation could explain these findings, we assessed proliferation by bromodeoxyuridine (BrdU) injection. BrdU rarely co-localized with cortical microglia, though robust co-localization was detected in the dentate gyrus (DG) as expected (Askew et al., 2017; Tay et al., 2017) (Figures 1D and 1E). Similar minimal co-localization in the cortex was observed for microglia with Ki67 (Figure S3). We investigated contributions by receptors implicated in microglial proliferation and translocation/chemotaxis: Trem2 (Zhang et al., 2014; Zheng et al., 2017) and CX3CR1 (Cardona et al., 2006; Liang et al., 2009), respectively. Trem2−/− microglia showed a 59% reduction in cortical (Figure 1G) and an 87% reduction in DG (data not shown) proliferation. Trem2−/−, but not CX3CR1−/−, mice had reduced microglial numbers in 2-month-old mice (Figure 1G). However, neither genotype differed from wild-type in microglial rearrangement (Figure 1H) or ‘‘gained’’ and ‘‘lost’’ cells (Figure 1I). This indicates that the mechanism underlying microglial rearrangement couples ‘‘gained’’ and ‘‘lost’’ cells independent of proliferation. Carefully monitoring microglia daily, we noted both static and, presumably, translocating cells (Figure 1J).
Acute Experimental Modulation of the Microglial Landscape
Next, we sought to determine whether the observed daily rearrangement could be experimentally altered. We first tested the possibility that systemic activation of microglia could alter their rearrangement via intraperitoneal (i.p.) lipopysaccharide (LPS) (2 mg/kg) treatment. However, cortical microglia did not alter their daily rearrangement within 24 hr of LPS treatment (Figures S2D and S2E). Then, we turned our attention to systemic brain hyperactivity to modulate the microglial landscape. Our previous work showed that i.p. KA-induced seizures altered microglial morphologies (Eyo et al., 2014, 2017). Therefore, we used this treatment paradigm to experimentally alter brain activity and found that severe (stage-5), but not mild (stage-3), seizures transiently increased microglial rearrangement especially within the first 24 hr (Figures 2A–2C). By the third day of seizures, basal levels were restored and maintained thereafter for at least a month (Figures S2F and S2G). Similarly, i.p. pilocarpine-induced seizures increased microglial rearrangement within the first 24 hr of seizures (Figures 2D and 2E). Under these conditions, microglial translocations could also be detected (Figure 2F). Together, these results indicate that global increases in neuronal activity resulting from seizures increased microglial somatic rearrangements.
Figure 2. Global Experimental Modulation of the Microglial Landscape.
(A) Representative images of microglia in a field of view showing lost cells (yellow) and gained cells (red) under control conditions and then 24 hr after a severe kainic-acid-induced seizure.
(B) Percentage of rearranged microglia before and after severe kainic-acid-induced seizures (n = 19 FOVs from 6 mice).
(C) Percentage of rearranged microglia before and after mild kainic-acid-induced seizures (n = 9 FOVs from 3 mice).
(D) Representative images of microglia in a field of view showing lost (yellow) and gained (red) cells under control conditions and then 24 hr after a pilocarpine-induced seizure.
(E) Percentage of rearranged microglia before and after pilocarpine-induced seizures (n = 9 FOVs from 3 mice).
(F) Static (circles) and translocated (arrows: white for starting position and yellow for translocated position) microglia before and 24 hr after kainic acid treatment.
***p < 0.001. Data are presented as mean ± SEM.
To complement this global approach, we tested the effect of more localized brain manipulations on the rearrangement of the microglial landscape. First, we performed a localized laser-induced tissue injury and monitored the microglial landscape at 6-hr intervals and report obvious somatic translocations within the first few days, with cells translocating the most within the first 12 hr of the injury (Figures 3A and 3B; Video S1). Then we monitored the microglial landscape in the barrel cortex following whisker trimming and also observed increased changes in the microglial landscape of whisker-trimmed mice (Figures 3C–3E). Thus, changes in brain activities globally and locally alter the microglial landscape.
Figure 3. Local Experimental Modulation of the Microglial Landscape.
(A) Representative images of microglia in a field of view following a localized laser-induced injury (asterisks) with images taken at 6- to 12-hr intervals following the injury. Microglia show soma translocations where new cells (red) emerge into the field of view or migrate toward the injury focus (yellow cells with dashed arrows). The majority of the translocation occurs within the first 24 hr (n = 3 FOVs each with or without laser-induced injuries from 2 mice).
(B) Quantification of cells that translocate toward the center of a non-injured and an injured field of view between 0 and 12 and between 12 and 24 hr of the injury.
(C) Representative images of microglia in a field of view showing lost (yellow) and gained (red) cells 24 hr after whisker trimming.
(D) Time course of the percentage of rearranged microglia before and following daily whisker trimming (n = 14 FOVs from 4 mice).
(E) Average percentage of rearranged microglia before and following daily whisker trimming (n = 14 FOVs from 4 mice).
**p < 0.01; ***p < 0.001. Data are presented as mean ± SEM.
P2Y12 Receptors Regulate the Microglial Landscape
Finally, we sought to determine a molecular regulator of the changing microglial landscape. Though we showed that Trem2 receptors regulate microglial proliferation (Figure 1G) and CX3CR1 receptors are known to regulate microglial process activity (Liang et al., 2009), we ruled out roles for these two receptors in the microglial landscape rearrangement (Figure 1H). Considering established roles for microglial P2Y12Rs in microglial process movements in situ (Haynes et al., 2006; Wu et al., 2007) and soma movements in vitro (Honda et al., 2001), we tested its involvement in regulating the daily microglial landscape.
P2Y12Rs are highly and uniquely expressed by brain microglia in the healthy brain (Hickman et al., 2013; Zhang et al., 2014). We found an upregulation of these receptors on microglia following KA-induced seizures and whisker trimming in the mouse cortex (data not shown). Remarkably, P2Y12−/− mice exhibited significantly reduced microglial rearrangement under basal conditions (Figures 4A and 4B), with a similar percent of ‘‘gained’’ and ‘‘lost’’ cells (Figure 4C) and no significant difference in microglial proliferation (Figure 4D). Consistently, following seizures and whisker trimming, there was a significant reduction in rearranged microglia in P2Y12−/− mice (Figures 4E and 4F; Figure S4). Translocation in wild-type microglia occurred in a fashion where days of movement were often interrupted by a day (or days) of stasis (Figure 4G; Video S2), but microglial translocation was rarely observed in P2Y12−/− mice (Figure 4G; Video S3).
Figure 4. P2Y12 Receptors Regulate the Microglial Landscape.
(A) Microglial rearrangement in naive wild-type and P2Y12−/− mice from the limb/trunk cortex (n = 25 FOVs from 7 wild-type and 24 FOVs from 7 P2Y12−/− mice).
(B) Microglial rearrangement in naive wild-type and P2Y12−/− mice from the barrel cortex (n = 3 mice each).
(C) Percentage of daily ‘‘gained’’ and ‘‘lost’’ microglia in naive wild-type and P2Y12−/− mice (n = 4 mice each).
(D) BrdU−/−Iba1 double-immunoreactive quantification in naive wild-type and P2Y12−/− cortex (n = 4 wild-type and 3 P2Y12−/− mice).
(E and F) Microglial rearrangement in wild-type and P2Y12−/− mice following (E) severe seizures and (F) whisker trimming. n = 11 FOVs from 3 wild-type and 20 FOVs from 3 P2Y12−/− mice for (E); and n = 14 FOVs from 4 wild-type and 4 P2Y12−/− mice for (F).
(G) Translocating cortical microglia from a naive wild-type (top) and a naive P2Y12−/− (bottom) cortex during daily imaging. Four different cells are tracked and are indicated with red, cyan, light purple, and yellow.
(H and I) Images showing (H) a dying microglia or (I) a proliferating microglia during repeated imaging.
(J) Quantification of the percentage of cells classified as dying, proliferating, and translocating from repeated imaging in the naive wild-type and P2Y12−/− cortex (n = 25 FOVs from 7 wild-type and 24 FOVs from 7 P2Y12−/− mice).
**p < 0.01; ***p < 0.001. Data are presented as mean ± SEM.
We classified cell behavior into (a) ‘‘death’’ (cells missing from the FOV compared to the previous day and that were not present in any other region of the FOV; Figure 4H; Video S4, left); (b) ‘‘proliferation’’ (cells emerging into the FOV compared to the previous day and that did not come from any other region of the FOV; Figure 4I; Video S4, right); and (c) ‘‘translocation,’’ as defined previously. Most cell changes belonged to the translocation category (5.8 ± 0.2%; n = 21 FOVs from 7 mice), while the rest were split between the ‘‘death’’ (2.1 ± 0.3%) and ‘‘proliferation’’ (1.3 ± 0.3%) categories. P2Y12−/− mice exhibited similar death (1.6 ± 0.2%) and proliferation (1.2 ± 0.3%) categories but showed significantly reduced translocating cells (1.0 ± 0.2%; n = 23 FOVs from 7 mice; Figure 4J). Therefore, we conclude that P2Y12 receptors regulate the microglial landscape predominantly through translocation mechanisms.
DISCUSSION
Microglia carry out elaborate surveillance of the brain by their never-resting process extensions and retractions (Davalos et al., 2005; Nimmerjahn et al., 2005). These processes interact with neuronal elements (Kato et al., 2016; Li et al., 2012), while microglial somata are assumed to be sessile. Evidence suggest a high capacity for soma translocation in murine microglia ex vivo (Eyo et al., 2016) and zebrafish microglia in vivo (Svahn et al., 2013) during early development. In addition, microglia in the adult Alzheimer’s brain exhibit locomotory capacities (Füger et al., 2017; Fuhrmann et al., 2010). Our findings suggest that a small population of microglia retain this ability in the healthy adult brain. Thus, microglial surveillance of the brain also (at least, for some microglia) includes cellular translocation.
Recently, it was suggested that the microglial landscape is altered predominantly by a coupling of proliferation and apoptosis (Askew et al., 2017), whereas other approaches suggested that cortical microglial proliferation is minimal (Tay et al., 2017). Our results indicate that proliferation and death are not the predominant mechanisms regulating the changing microglial landscape for three reasons. First, the degree of cortical proliferation is much too low, between 0.45% from our fixed tissue studies and 1.3% from our chronic in vivo imaging studies, when compared to the degree of daily re-arranged cells (10%–15%). Second, the percentage of daily translocating cells (5.8%), when doubled as ‘‘gained’’ and ‘‘lost’’ cells, would account for the 10%–15% daily rearrangement, with little contribution from proliferation or apoptosis. Finally, genetic ablation of Trem2, while reducing microglial proliferation (Zheng et al., 2017), did not significantly alter the basal daily re-arrangement. Our proliferation numbers from daily imaging (1.3%) is likely an overestimation, because some of the cells we identified as suddenly appearing in the FOV may have simply migrated into the FOV. Conversely, our proliferation quantification with BrdU (0.45%) is likely an underestimation, as we gave just one pulse of BrdU, which circulates for shorter than 24 hr and would miss some of the proliferating cells within that time period. Alternative to proliferation, we provide robust evidence of translocating microglia that accounts for the bulk of the daily changes in the microglial landscape.
Finally, we provide genetic evidence that, while proliferation and cell death are unaffected by a P2Y12R deficiency, translocation capacities are significantly reduced. P2Y12Rs are a molecular signature for brain microglia, as revealed by transcriptome studies (Hickman et al., 2013; Zhang et al., 2014). They function as the predominant receptor in microglial process chemotaxis toward injury (Haynes et al., 2006) and can heal the compromised blood-brain barrier by this chemotactic mechanism (Lou et al., 2016). Moreover, they mediate various microglial-to-neuronal physical interactions (Eyo et al., 2014, 2015, 2017) and are critical for synaptic plasticity in the visual system (Sipe et al., 2016). Since P2Y12Rs are required for microglial translocation, as shown in the present study, we would predict that they are important for adequate housekeeping surveillance functions of microglia. In summary, our findings demonstrate that, even in the homeostatic state, microglia should be better thought of as not merely stationary surveying cells but, more appropriately, as patrolling cells with migratory capacities.
EXPERIMENTAL PROCEDURES
Animals
Both male and female adult mice, 2 to 5 months of age, were used in accordance with institutional guidelines, as approved by the Institutional Animal and Care Committee (IACUC) animal care and use committee at the Mayo Clinic and Rutgers University. Heterozygous (CX3CR1GFP/+) GFP reporter mice expressing GFP under control of the fractalkine receptor (CX3CR1) promoter (Jung et al., 2000) and transgenic mice (Thy1YFP/+) expressing YFP (Feng et al., 2000) in a subset of pyramidal neurons under the control of the Thy1 promoter were purchased from the Jackson Laboratory. For some experiments, CX3CR1GFP/GFP mice were used as CX3CR1−/− mice. P2Y12−/− mice were originally donated by Dr. Michael Dailey at the University of Iowa (Iowa City, IA, USA) and have now been established in our lab. Trem2−/− mice were donated by Dr. Marco Colonna at Washington University (St. Louis, MO, USA).
Chronic Window Implantation
Mice were anesthetized with isoflurane (5% for induction; 1.5% for maintenance and surgery) combined with oxygen fitted into a custom-made stereo-taxic frame. Mice were maintained on a heating pad during surgery and then received a local subcutaneous injection of 15–20 mL 0.25% bupivicane and lubricant eye ointment (Artificial Tears, Henry Schein). The hair above the mouse head was shaved with a clipper (Wahl BravMini), after which the mouse was placed in a stereotactic frame (Kopf) cleaned with three alternating swabs of betadine and 75% alcohol. The skin above the head was then cut to expose the skull. The skull was cleaned using a cotton swab of 3%–5% hydrogen peroxide, and a dental drill (Osada Model EXL-M40) and drill bit (Fine Science Tools, 19008-07) were used to drill open a circular >3-mm-diameter window, which was carefully removed using sharp forceps. During drilling, bone debris was cleared away, and the skull was frequently irrigated with sterile saline. For the limb/trunk region of the somatosensory cortex, the skull was removed with the center at about −2.5 posterior and ±2 lateral to bregma, while for the barrel cortex, the skull was removed with the center at about −2.5 posterior and ±3 lateral to bregma. Additionally, once opened, the exposed brain surface was kept moist with sterile saline. A 3-mm glass coverslip previously sterilized in 75% ethanol was put inside the window and held in place with a pipette tip, while curing dental cement (Tetric EvoFlow) was applied around the glass coverslip and cured with a Kerr Demi Ultra LED Curing Light (Dental Health Products). The skull, excluding the region with the window, was then covered with IBond Total Etch glue (Heraeus) and cured with a curing light. Finally, a custom-made head plate was glued with another application of the dental cement and cured with a curing light to permanently attach the head plate. Mice were allowed to recover from anesthesia on a heating pad (~10 min) before they were returned to their home cage. Mice were allowed to recover from the surgery for 2 to 4 weeks. Mice that showed a loss in imaging window clarity before the 2- to 4-week period of observation were discarded from the study.
In Vivo Two-Photon Imaging
Single-transgenic heterozygous GFP reporter mice (Jung et al., 2000) or double-transgenic GFP reporter and YFP reporter mice were typically imaged using a two-photon microscope (Scientifica) with a Ti:Sapphire laser (Mai Tai; Spectra Physics) tuned to 900 nm with a 40× water-immersion lens (0.8 NA; Olympus). Fluorescence was detected using two photomultiplier tubes in whole-field detection mode and a 565-nm dichroic mirror with 525-/50-nm (green channel) and 620-/60-nm (red channel) emission filters. The laser power was maintained at 30–40 mW, and images were collected from 50 μm to 120 μm into the brain. For imaging microglial and neuronal YFP dynamics from each mouse, z stack images were collected at 1- to 2-μm intervals in several FOVs. For repeated imaging, blood vessels were used as gross landmarks, and dendrites were used as fine landmarks. To perform a general laser injury, we focused the laser 66× and parked it at 250 mW at 900 nm for 1–3 s.
Experimental Manipulations
Seizure Induction
Mice with implanted windows were monitored for at least 5 days under basal conditions and received i.p. injections of either kainic acid at 22–24 mg/kg or pilocarpine at 260–280 mg/kg. For P2Y12-deficient mice, kainic acid was administered at 18–20 mg/kg to allow robust seizures without death, as these mice are more susceptible to kainic-acid-induced seizures (Eyo et al., 2014). Seizure behavior was monitored under a modified Racine scale as follows: (1) freezing behavior; (2) rigid posture with raised tail; (3) continuous head bobbing and forepaws shaking; (4) rearing, falling, and jumping; (5) continuous occurrence of level 4; and (6) loss of posture and generalized convulsion activity (Eyo et al., 2014). Mice that progressed to at least stage 3 were used for subsequent chronic daily imaging of mild (stages 3/4) to severe (stages 5/6) seizures.
Whisker Trimming
Mice with implanted windows were monitored for at least 5 days under basal conditions, and under anesthesia, all whiskers on the contralateral side to the implanted window were trimmed daily for 5 consecutive days with a clipper (Wahl BravMini).
LPS Treatment
Mice with implanted windows were monitored for at least 3 days under basal conditions and subsequently received i.p. injections of LPS at 2 mg/kg.
BrdU Labeling and Analysis
BrdU was used to label proliferating and recently post-mitotic cells in the brain. The BrdU solution was diluted in 1 M PBS just before use at a concentration of 10 mg/mL and intraperitoneally administered at 100 mg/kg in naive mice or 24 hr after mouse manipulation (seizures or whisker trimming). Mice were sacrificed 3 hr after the BrdU injection by perfusion, first with PBS and then with 4% paraformaldehyde (PFA). The brains were then incubated in 4% PFA overnight and transferred to 30% sucrose solution for at least 2 days. Brains were then cryosectioned to 15-μm thickness and attached to glass slides. For BrdU immunohistochemistry, the slides with brain sections were hydrated in Tris-buffered saline (TBS) for 10 s and transferred to a 50% form-amide in 2× saline sodium citrate (SSC) solution at 65°C for 2 hr. Slides with brain sections were then placed in a 2× SSC solution at room temperature for 15 min and were transferred to a 2 N HCl solution at 37°C for 20 min and then a 0.1 M borate buffer at room temperature for 10 min. Slides with brain sections were then washed three times in TBS (pH 7.6) in room temperature for 10 min each. Next, brain sections were blocked with 3% normal goat serum (NGS) in 1× TBS + 0.3% Triton-X at room temperature for 1 hr and then overnight at 4°C in the primary antibody Iba1 (Wako Pure Chemicals Industries, 1:500) or anti-BrdU (Sigma, 1: 500), washed 3 times in 1× TBS for 5 min each, and then incubated in the secondary antibody donkey anti-rabbit (1:500) or donkey anti-mouse (1:500) at room temperature for 2 hr. Slides with brain sections were then rinsed three times in 1× TBS for 5 min each and mounted for imaging on an EVOS fluorescence microscope. Images of Iba1- and BrdU-labeled tissues were collected at 10× magnification. Microglial proliferation was determined by assessing the colocalization of the BrdU signal with the Iba1 signal for microglia.
Landscape Rearrangement Analysis
For cell rearrangement analyses, microglia in FOVs from consecutive days within a volume of 60 × 330 × 330 μm between ~50 and 120 μm (or 180–240 μm) from the brain surface were compared. Individual cells were identified and marked with numbers from the previous day and transposed to the next day. Cell bodies were regarded as stable if they maintained their position within a 2-cell-body distance from the previous day (~10–15 μm). Cell bodies that were either absent or present at a distance of 2 cell bodies or more were marked as either ‘‘gained’’ if they were not there on the previous day or ‘‘lost’’ if they were there on the previous day but absent on the next day. The percent change of ‘‘rearranged’’ cells was determined as the number of both ‘‘gained’’ and ‘‘lost’’ cells divided by the total number of cells in each FOV × 100. Similar analysis, but specific for either ‘‘gained’’ or ‘‘lost’’ cells, was performed to determine the percent rearrangement of those cell groups (Figures 1I and 4C).
Statistical Analysis
Data were collected from at least 3 and up to 8 FOVs per mice and pooled together. Three to seven mice were used for each set of experiments. In our re-arrangement studies comparing changes in the microglial landscape before and after treatment, power analysis for paired t tests (α = 0.05, β = 0.2) was performed to establish the sufficiency of the sample size using GraphPad software. The power values achieved were as follows: (1) for an effect size of 30% change (from ~13% in control) in microglial rearrangement in the limb/trunk cortex, the power value was 0.90 for kainic acid (KA) treatment (n = 6 mice; combined SD: 17.09, severe seizures); (2) for an effect size of 20% change (from ~13% in control) in microglial rearrangement in the limb/trunk cortex, the power value was 0.80 for pilocarpine treatment (n = 3 mice, combined SD: 9.83); and (3) for an effect size of 14% (from ~7% in control) in microglial rearrangement in the barrel cortex, the power value was 0.80 for whisker trimming (n = 4 mice; combined SD: 8.58). Student’s t test was used for statistical analysis, Bonferroni correction was used for multiple comparisons, and significance was determined with a p value of 0.05 or less.
Supplementary Material
Highlights.
The microglial landscape is rearranged daily in the healthy brain
The microglial landscape can be altered by manipulating brain activity
Daily microglial landscape rearrangement occurs predominantly by translocation
Rearrangement of the microglial landscape is controlled by the P2Y12 receptor
Acknowledgments
We thank all members of the Wu lab for critical discussions of the manuscript and especially Dr. Dale Bosco for help with editing the manuscript. This work is supported by grants from the NIH: R01NS088627 (to L.-J.W.), R21DE025689 (to L.-J.W.), K22NS104392 (to U.B.E.), and R01NS094450 (to D.J.M.).
Footnotes
AUTHOR CONTRIBUTIONS
U.B.E., P.X., and L.-J.W. designed the study and wrote the manuscript, with help from all authors. U.B.E. performed most of the experiments; M. Mo, M.-H.Y., and J.L. performed some experiments; U.B.E., J.L., M. Murugan, M.-H.Y., and R.Y. performed data analysis; D.J.M. provided training for the chronic window implantation studies and valuable discussions for the project.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Supplemental Information includes four figures and four videos and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.04.001.
References
- Ajami B, Bennett JL, Krieger C, McNagny KM, Rossi FM. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat Neurosci. 2011;14:1142–1149. doi: 10.1038/nn.2887. [DOI] [PubMed] [Google Scholar]
- Askew K, Li K, Olmos-Alonso A, Garcia-Moreno F, Liang Y, Richardson P, Tipton T, Chapman MA, Riecken K, Beccari S, et al. Coupled proliferation and apoptosis maintain the rapid turnover of microglia in the adult brain. Cell Rep. 2017;18:391–405. doi: 10.1016/j.celrep.2016.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona AE, Pioro EP, Sasse ME, Kostenko V, Cardona SM, Dijkstra IM, Huang D, Kidd G, Dombrowski S, Dutta R, et al. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. 2006;9:917–924. doi: 10.1038/nn1715. [DOI] [PubMed] [Google Scholar]
- Chen SK, Tvrdik P, Peden E, Cho S, Wu S, Spangrude G, Capecchi MR. Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell. 2010;141:775–785. doi: 10.1016/j.cell.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- Davies DS, Ma J, Jegathees T, Goldsbury C. Microglia show altered morphology and reduced arborization in human brain during aging and Alzheimer’s disease. Brain Pathol. 2017;27:795–808. doi: 10.1111/bpa.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyo UB, Peng J, Swiatkowski P, Mukherjee A, Bispo A, Wu LJ. Neuronal hyperactivity recruits microglial processes via neuronal NMDA receptors and microglial P2Y12 receptors after status epilepticus. J Neurosci. 2014;34:10528–10540. doi: 10.1523/JNEUROSCI.0416-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyo UB, Gu N, De S, Dong H, Richardson JR, Wu LJ. Modulation of microglial process convergence toward neuronal dendrites by extracellular calcium. J Neurosci. 2015;35:2417–2422. doi: 10.1523/JNEUROSCI.3279-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyo UB, Miner SA, Weiner JA, Dailey ME. Developmental changes in microglial mobilization are independent of apoptosis in the neonatal mouse hippocampus. Brain Behav Immun. 2016;55:49–59. doi: 10.1016/j.bbi.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyo UB, Peng J, Murugan M, Mo M, Lalani A, Xie P, Xu P, Margolis DJ, Wu LJ. Regulation of physical microglia-neuron interactions by fractalkine signaling after status epilepticus. eNeuro. 2017;3 doi: 10.1523/ENEURO.0209-16.2016. ENEURO.0209-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Füger P, Hefendehl JK, Veeraraghavalu K, Wendeln AC, Schlosser C, Obermüller U, Wegenast-Braun BM, Neher JJ, Martus P, Kohsaka S, et al. Microglia turnover with aging and in an Alzheimer’s model via long-term in vivo single-cell imaging. Nat Neurosci. 2017;20:1371–1376. doi: 10.1038/nn.4631. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M, Bittner T, Jung CK, Burgold S, Page RM, Mitteregger G, Haass C, LaFerla FM, Kretzschmar H, Herms J. Microglial Cx3cr1 knockout prevents neuron loss in a mouse model of Alzheimer’s disease. Nat Neurosci. 2010;13:411–413. doi: 10.1038/nn.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan WB, Julius D. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci. 2006;9:1512–1519. doi: 10.1038/nn1805. [DOI] [PubMed] [Google Scholar]
- Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Sasaki Y, Ohsawa K, Imai Y, Nakamura Y, Inoue K, Kohsaka S. Extracellular ATP or ADP induce chemotaxis of cultured microglia through Gi/o-coupled P2Y receptors. J Neurosci. 2001;21:1975–1982. doi: 10.1523/JNEUROSCI.21-06-01975.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G, Inada H, Wake H, Akiyoshi R, Miyamoto A, Eto K, Ishikawa T, Moorhouse AJ, Strassman AM, Nabekura J. Microglial contact prevents excess depolarization and rescues neurons from excitotoxicity. eNeuro. 2016;3 doi: 10.1523/ENEURO.0004-16.2016. ENEURO.0004-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Du XF, Liu CS, Wen ZL, Du JL. Reciprocal regulation between resting microglial dynamics and neuronal activity in vivo. Dev Cell. 2012;23:1189–1202. doi: 10.1016/j.devcel.2012.10.027. [DOI] [PubMed] [Google Scholar]
- Liang KJ, Lee JE, Wang YD, Ma W, Fontainhas AM, Fariss RN, Wong WT. Regulation of dynamic behavior of retinal microglia by CX3CR1 signaling. Invest Ophthalmol Vis Sci. 2009;50:4444–4451. doi: 10.1167/iovs.08-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou N, Takano T, Pei Y, Xavier AL, Goldman SA, Nedergaard M. Purinergic receptor P2RY12-dependent microglial closure of the injured blood-brain barrier. Proc Natl Acad Sci USA. 2016;113:1074–1079. doi: 10.1073/pnas.1520398113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Teva JL, Dusart I, Colin C, Gervais A, van Rooijen N, Mallat M. Microglia promote the death of developing Purkinje cells. Neuron. 2004;41:535–547. doi: 10.1016/s0896-6273(04)00069-8. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, 3rd, Lafaille JJ, Hempstead BL, Littman DR, Gan WB. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe GO, Lowery RL, Tremblay ME, Kelly EA, Lamantia CE, Majewska AK. Microglial P2Y12 is necessary for synaptic plasticity in mouse visual cortex. Nat Commun. 2016;7:10905. doi: 10.1038/ncomms10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarzoni P, Oller G, Hoeffel G, Pont-Lezica L, Rostaing P, Low D, Bessis A, Ginhoux F, Garel S. Microglia modulate wiring of the embryonic forebrain. Cell Rep. 2014;8:1271–1279. doi: 10.1016/j.celrep.2014.07.042. [DOI] [PubMed] [Google Scholar]
- Svahn AJ, Graeber MB, Ellett F, Lieschke GJ, Rinkwitz S, Bennett MR, Becker TS. Development of ramified microglia from early macrophages in the zebrafish optic tectum. Dev Neurobiol. 2013;73:60–71. doi: 10.1002/dneu.22039. [DOI] [PubMed] [Google Scholar]
- Tay TL, Mai D, Dautzenberg J, Fernández-Klett F, Lin G, Sagar Datta M, Drougard A, Stempfl T, Ardura-Fabregat A, et al. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat Neurosci. 2017;20:793–803. doi: 10.1038/nn.4547. [DOI] [PubMed] [Google Scholar]
- Tremblay ME, Lowery RL, Majewska AK. Microglial interactions with synapses are modulated by visual experience. PLoS Biol. 2010;8:e1000527. doi: 10.1371/journal.pbio.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake H, Moorhouse AJ, Jinno S, Kohsaka S, Nabekura J. Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J Neurosci. 2009;29:3974–3980. doi: 10.1523/JNEUROSCI.4363-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Vadakkan KI, Zhuo M. ATP-induced chemotaxis of microglial processes requires P2Y receptor-activated initiation of outward potassium currents. Glia. 2007;55:810–821. doi: 10.1002/glia.20500. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Jia L, Liu CC, Rong Z, Zhong L, Yang L, Chen XF, Fryer JD, Wang X, Zhang YW, et al. TREM2 promotes microglial survival by activating Wnt/β-catenin pathway. J Neurosci. 2017;37:1772–1784. doi: 10.1523/JNEUROSCI.2459-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






