Abstract
Chronic myelogenous leukemia (CML) stem cells (LSCs) are responsible for initiating and maintaining clonal hematopoiesis. These cells persist in the bone marrow (BM) despite effective inhibition of BCR-ABL kinase activity by tyrosine kinase inhibitors (TKIs). Here, we show that although miR-126 supports the quiescence, self-renewal and engraftment capacity of CML LSCs, miR-126 levels are lower in CML LSCs as compared to normal long-term hematopoietic stem cells (LT-HSCs). Down-regulation of miR-126 levels in CML LSCs is due to phosphorylation of SPRED1 by BCR-ABL, leading to inhibition of the RAN/EXP-5/RCC1 complex that mediates miRNA maturation. Endothelial cells (ECs) in the BM supply miR-126 to CML LSCs to support quiescence and leukemia growth, as shown using CML mouse models with conditional miR-126 knock-out (KO) in ECs and/or LSCs. Inhibition of BCR-ABL by TKI treatment causes an undesired increase in endogenous miR-126 levels, thereby enhancing LSC quiescence and persistence. miR-126 KO in LSCs and/or ECs, or treatment with a CpG-miR-126 inhibitor targeting miR-126 in both LSCs and ECs, enhances the in vivo anti-leukemic effects of TKI treatment and strongly diminishes LSC leukemia-initiating capacity, providing a new strategy for the elimination of LSCs in CML.
Keywords: microRNA, BM niche, CML, LSC, chemoresistance
Introduction
Chronic myelogenous leukemia (CML) is a clonal myeloproliferative disorder characterized at the cytogenetic level by the translocation of chromosomes 9q34 and 22q111. This translocation creates a fusion gene, BCR-ABL, which encodes a constitutively activated tyrosine kinase responsible for transforming normal hematopoietic stem cells (HSCs) into leukemia stem cells (LSCs). LSCs are characterized by growth factor-independent proliferation and enhanced survival, resulting in uncontrolled myeloproliferation that eventually evolves into fatal blast crisis if left untreated. CML LSCs are at the apex of malignant clonal hematopoiesis and initiate and maintain leukemia growth. In CML, LSC activity is restricted to the LT-HSC-enriched Lin−CD34+CD38−CD90+ cell population in humans, and the Lin−Sca-1+c-Kit+Flt3−CD150+CD48− cell population in mice2. CML LSCs are thought to reside in a leukemia niche that may be anatomically and functionally different from that of normal HSCs.
Currently, oral tyrosine kinase inhibitors (TKI) are used as the first-line treatment to induce long-term disease remission in CML patients. Although most patients treated with TKI monotherapy achieve major clinical and molecular responses, cells from the original BCR-ABL clone frequently persist, likely due to the failure of these agents to eliminate CML LSC3, and treatment discontinuation frequently results in disease relapse. Thus, the identification of mechanisms that support CML LSC persistence is clinically relevant as it may enable the design of new targeting strategies aimed at complete disease elimination, allowing for discontinuation of life-long TKI therapy.
miR-126-3p (miR-126) is a microRNA (miRNA) that is highly expressed in normal HSCs and hematopoietic progenitor cells (HPCs) and restrains cell-cycle progression during hematopoiesis4. Our group and others have shown that increased miR-126 levels are associated with an increased frequency of quiescent LSCs and a worse outcome in acute myeloid leukemia (AML)5–8. Here we show that miR-126 biogenesis in CML LSCs is down-regulated through a BCR-ABL-dependent mechanism, a finding which is seemingly inconsistent with a pro-leukemic role for miR-126. However, miR-126 is also highly expressed in endothelial cells (ECs)9. Anatomical and functional connections between the endothelium and normal HSCs regulate normal hematopoiesis10. We hypothesized that miR-126 may mediate a functional interplay between ECs and LSCs in the leukemia BM niche that regulates CML progression. Consistent with this hypothesis, we found that ECs supply miR-126 to CML LSCs to modulate their quiescence and self-renewal.
Results
Higher miR-126 levels are associated with human and mouse CML LSCs
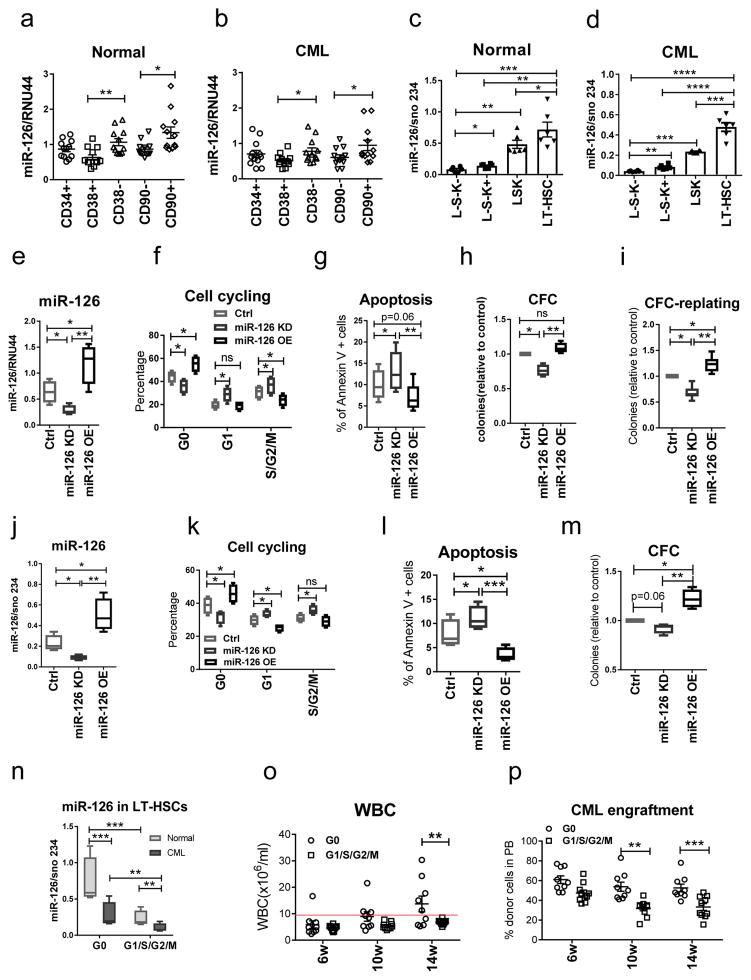
miR-126 has been shown to contribute to leukemogenesis in acute leukemia6,11,12. To determine miR-126 expression in CML cell subpopulations, we sorted immunophenotypically defined subsets of HPCs [Lin−CD34+(CD34+) and Lin−CD34+CD38+ (CD38+)], HSCs [Lin−CD34+CD38− (CD38−) and Lin−CD34+CD38−CD90− (CD90−)] and LT-HSCs [Lin−CD34+CD38−CD90+ (CD90+)] from peripheral blood (PB) and BM samples of normal donors (n=12) and newly diagnosed chronic phase (CP) CML patients (n=12). LT-HSCs in both normal and CML samples showed the highest expression of miR-126 (Fig. 1a, b). Similar results were obtained in wild-type (WT) B6 and inducible SCLtTA/BCR-ABL transgenic B6 mice, a well established CML mouse model13. We isolated Lin−Sca-1−c-Kit− (L−S−K−), Lin−Sca-1−c-Kit+ (L−S−K+) [including common myeloid progenitors (CMP), granulocyte-macrophage progenitors (GMP) and megakaryocyte-erythrocyte progenitors (MEP)], Lin−Sca-1+c-Kit+ (LSK) and LSK Flt3−CD150+CD48− (LT-HSC) cells from the BM of WT mice and CML mice after BCR-ABL induction by tetracycline withdrawal (Supplementary Fig. 1a). As in the human samples, mouse normal and CML LT-HSCs showed the highest expression of miR-126 (Fig. 1c, d).
Figure 1. Human and mouse CML LSCs express the highest levels of miR-126 among CML subpopulations.
(a,b) miR-126 expression, as assessed by QPCR, in HPCs [Lin−CD34+(CD34+) and Lin−CD34+CD38+ (CD38+)], HSCs [Lin−CD34+CD38− (CD38−) and Lin−CD34+CD38−CD90− (CD90−)] and LT-HSCs [Lin−CD34+CD38−CD90+ (CD90+)] from blood and BM samples from normal donors (n=12 biologically independent samples) (a) and newly diagnosed CP CML patients (n=12 biologically independent samples) (b). (c–d) miR-126 expression, as assessed by QPCR, in the indicated BM subpopulations from normal (c) and CML (d) mice (n=6). (e–i) miR-126 expression (e), cell cycle analysis (f), apoptosis (g), CFCs (h) and CFC replating efficiency (i) of CML Lin−CD34+CD38− cells transduced with anti-miR-126 (KD), miR-126 precursor (OE) or control (Ctrl) lentiviruses (n=4 biologically independent samples). (j–m) miR-126 expression (j), cell cycle analysis (k), apoptosis (l), and CFCs (m) of LT-HSCs from induced SCLtTA/BCR-ABL mice after transduction with miR-126 KD, miR-126 OE, or control lentiviruses (n=4 independent experiments). (n) miR-126 expression, as assessed by QPCR, in quiescent Hoechst−Pyronin− (G0) LT-HSCs and proliferating Hoechst+/−Pyronin+ (G1/S/G2/M) LT-HSCs from normal or SCLtTA/BCR-ABL mice (n=4 independent samples). (o,p) White blood cell (WBC) counts (o) and donor CML cell engraftment in PB (p) of mice transplanted with G0 or G1/S/G2/M LT-HSCs from CML mice (n=10). Comparison between groups was performed by two-tailed, paired Student’s t-test. P values ≤0.05 were considered significant. Results shown represent mean ± SEM. *p ≤ 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To test the effects of miR-126 on quiescence of CML LSCs, we knocked down miR-126 expression in human CML Lin−CD34+CD38− cells (HSCs) and mouse CML LT-HSCs using GFP-expressing miRZip anti-miR-126 (miR-126 KD) or miR-126 precursor (overexpression, OE) lentiviral vectors. After transduction, GFP+ cells were selected and cultured for 72 hours (h). miR-126 KD increased cell cycling and apoptosis, decreased the number of colony forming cells (CFCs) and CFC replating efficiency of both human CML HSCs (Fig. 1e–i) and mouse CML LT-HSCs (Fig. 1j–m); conversely, miR-126 OE decreased cell cycling and apoptosis and increased CFC replating efficiency. We validated these results in vivo, first by showing that the quiescent Hoechst−Pyronin− (G0) fraction of CML LT-HSCs from induced SCLtTA/BCR-ABL mice (CD45.2) expressed significantly higher miR-126 levels than the proliferating Hoechst+/−Pyronin+ (G1/G2/S/M) fraction of CML LT-HSCs (p= 0.0019; Fig. 1n). The association of miR-126 with LSC activity was then demonstrated by showing that quiescent CML LT-HSCs had a significantly higher rate of long-term engraftment and leukemogenic capacity than did proliferating CML LT-HSCs after transplantation into CD45.1 congenic recipient mice (Fig. 1o–p).
BCR-ABL down-regulates miR-126 expression in CML cells
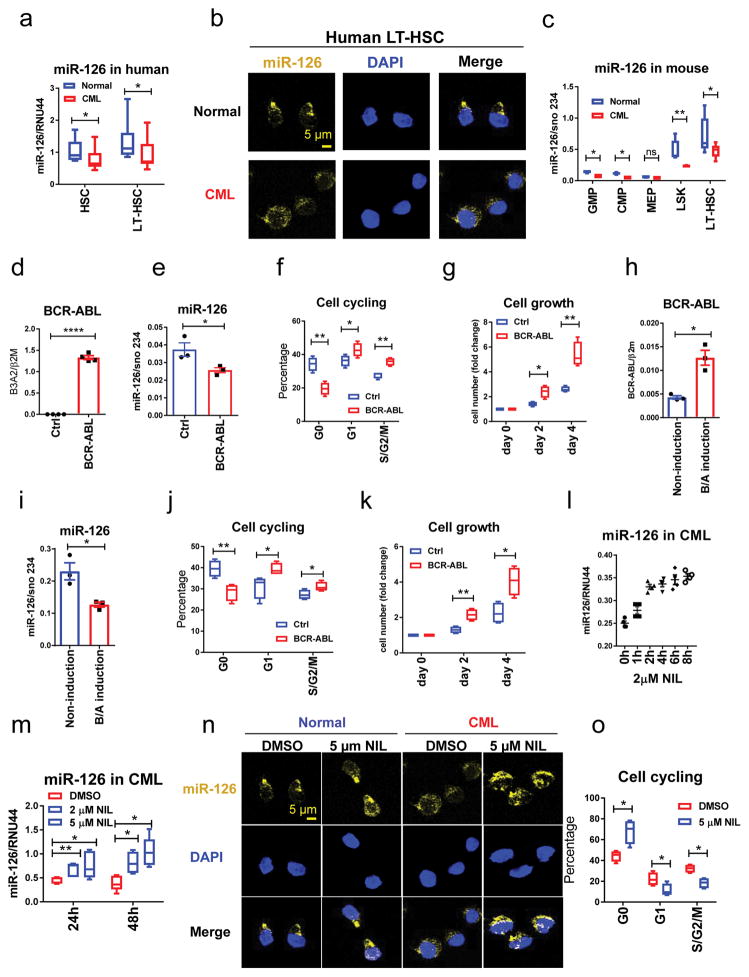
Although miR-126 has similar patterns of expression and function in CML as in normal hematopoiesis4, we noted that human CML Lin−CD34+CD38− (HSCs) and Lin−CD34+CD38−CD90+ cells (LT-HSCs) had significantly lower miR-126 levels than their normal counterparts (Fig. 2a–b); similar differences were also observed in mouse samples (Fig. 2c). Consistent with this finding, CML HSCs and LT-HSCs expressed higher levels of PIK3R2 and SPRED1, two validated targets of miR-1264,7,9,14,15, as compared to their normal counterparts (Supplementary Fig. 1b, c).
Figure 2. BCR-ABL down-regulates miR-126 expression in CML cells.
(a–b) miR-126 expression in normal and CML cell populations from human samples, as assessed by QPCR (n=10 biologically independent samples) (a) and miRNA staining (b) using 3 independent samples with similar results. (c) miR-126 expression, as assessed by QPCR, in the indicated BM cell populations from normal and CML mice (n=6). (d,e) BCR-ABL (d) and miR-126 (e) expression in normal mouse BM LSK cells transduced with BCR-ABL or control (Ctrl) retroviruses, as assessed by QPCR at 24 h after transduction (n=4 independent experiments). (f,g) Cell cycle (f) and cell growth (g) analyses (n=4 independent experiments) of the cells from d, e at 48 h after transduction. (h,i) BCR-ABL (h) and miR-126 (i) expression, as assessed by QPCR, in LT-HSCs from non-induced CML mice (tet off) cultured for 24 h in the presence (non-induction) or absence (BCR-ABL (B/A) induction) of tetracycline (2 μg/ml) (n=3 independent animals in each group). (j,k) Cell cycle (j) and cell growth (k) analyses (n=4 independent experiments in each group) of the cells from h,i at 48 h after BCR-ABL induction. (l) miR-126 expression, as assessed by QPCR, in human CML Lin−CD34+CD38− cells treated with NIL (2 μM) for the indicated periods of time (n=4 independent experiments). Mean±SEM was shown. (m) miR-126 expression, as assessed by QPCR, in CML Lin−CD34+CD38− cells treated with DMSO (Ctrl), 2 μM or 5 μM NIL (n=5 independent samples). (n) miR-126 expression, as assessed by miRNA staining, in normal and CML Lin−CD34+CD38− cells treated with DMSO or NIL (5 μM ) The experiments were repeated using 4 independent samples with similar results. (o) Cell cycle analysis of human CML Lin−CD34+CD38− cells treated with DMSO (Ctrl) or 5 μM NIL (n=4). Comparison between groups was performed by two-tailed, unpaired Student’s t-test. P values ≤0.05 were considered significant. Results shown represent mean ± SEM. *p ≤ 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
This differential expression of miR-126 observed led us to postulate that BCR-ABL itself might be involved in lowering miR-126 levels in CML cells. To test this hypothesis, we transduced normal mouse BM LSK cells with retroviral BCR-ABL or control vectors, and selected GFP+ cells and cultured them for 72 h. Upon BCR-ABL induction (Fig. 2d), miR-126 expression decreased (Fig. 2e) and PIK3R2 and SPRED1 expression increased compared to the control cells (Supplementary Fig. 1d, e); this was associated with an increase in cell cycling (Fig. 2f) and cell growth (Fig. 2g). To further validate this finding, we sorted LT-HSCs from non-induced BCR-ABL transgenic mice and cultured them with or without tetracycline. Upon tetracycline withdrawal and BCR-ABL induction (Fig. 2h), we observed reduced miR-126 levels (Fig. 2i) and increased cell cycling (Fig. 2j) and cell growth (Fig. 2k) as compared with non-induced controls. Conversely, BCR-ABL inhibition by nilotinib (NIL), a first-line TKI for CML treatment, led to increased miR-126 expression (Fig. 2l–n), decreased PIK3R2 and SPRED1 levels (Supplementary Fig. 1f, g) and an increased fraction of quiescent cells in human CML HSCs (Fig. 2o), but not in normal HSCs (Supplementary Fig. 1h), as compared with vehicle alone. NIL treatment also resulted in increased miR-126 expression in human BCR-ABL+ K562 cells (Supplementary Fig. 1i).
BCR-ABL deregulates miR-126 biogenesis
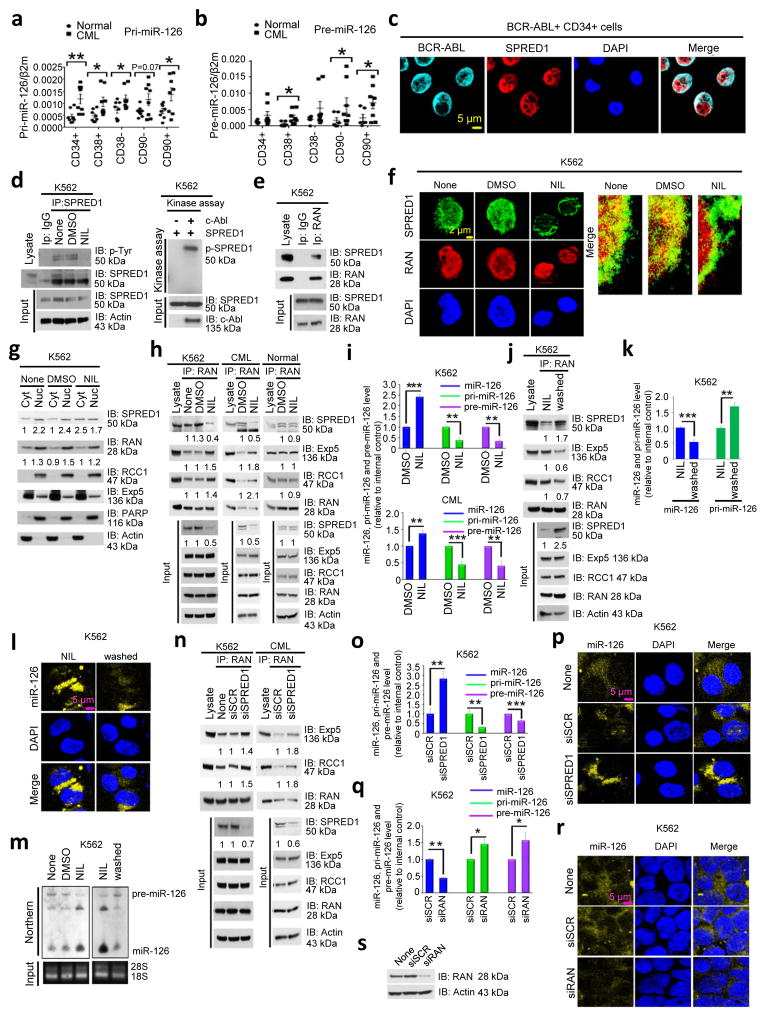
Whereas CML cells harbored lower levels of mature miR-126 than their normal counterparts, we noted that BCR-ABL-positive cells had higher levels of primary (pri-) and precursor (pre-) miR-126 than their normal counterparts (Fig. 3a, b). This result led us to hypothesize that BCR-ABL might interfere with miR-126 biogenesis.
Figure 3. BCR-ABL deregulates miR-126 biogenesis.
(a,b) pri-miR-126 (n=8 biologically independent samples) (a) and pre-miR-126 (n=8 biologically independent samples) (b) expression levels, as assessed by QPCR, in the indicated human normal and CML cell populations. (c) BCR-ABL and SPRED1 staining in CML CD34+ cells by immunofluorescence (IF). (d) Immunoprecipitation (IP) with anti-SPRED1 followed by immunoblotting (IB) with anti-SPRED1 and anti-phosphotyrosine (p-Tyr) antibodies (left) and an in vitro kinase assay (right), as performed by IP with anti-c-Abl or anti-normal mouse IgG as control and immunoblotting with anti-SPRED1, in lysates of K562 cells treated with none, DMSO (vehicle) or NIL. (e) IP with anti-RAN followed by IB with anti-SPRED1 and anti-RAN antibodies in lysates of K562 cells. (f) SPRED1 and RAN staining by IF in K562 cells treated with none, DMSO or NIL. (g) SPRED1, RAN, RCC1 and Exp-5 expression in cytoplasmic (Cyt) and nuclear (Nu) fractions from K562 cells, treated with DMSO or NIL, as assessed by IB. Densitometric quantification of selected bands is shown (normalized to the actin loading control for total and Cyt lysates or to the PARP loading control for Nu lysates). (h) IP with anti-RAN followed by IB with anti-SPRED1, RAN, Exp-5 and RCC1 antibodies in lysates of K562 cells, CML CD34+ cells, and normal CD34+ cells treated with DMSO or NIL. Densitometric quantification of selected bands is shown (normalized to the actin loading control). (i) Mature, pri- and pre-miR-126 expression, as assessed by QPCR, in K562 and CML CD34+ cells treated with DMSO or NIL (n=3 independent experiments for K562 and 3 independent samples for CML cells). (j) IP with anti-RAN followed by IB with anti-SPRED1, Exp-5, RCC1 and RAN antibodies in lysates of K562 cells without or with washing-off of NIL. Densitometric quantification of selected bands is shown (normalized to the actin loading control). (k–m) Mature and pri-miR-126 expression as assessed by QPCR (n=3 independent experiments) (k), miR-126 staining (l), and mature and pre-miR-126 levels as assessed by Northern blotting (m) in K562 cells with or without washing-off of NIL. (n) IP with anti-RAN followed by IB with anti-Exp-5, RCC1 and RAN antibodies in lysates of K562 and CML CD34+ cells with control (siSCR) or SPRED1 (siSPRED1) knockdown. Densitometric quantification of selected bands is shown (normalized to the actin loading control. (o,p) Mature, pri- and pre-miR-126 expression, assessed by QPCR (n=3 independent experiments) (o) and miR-126 staining (p) in siSCR and siSPRED1 treated K562 cells. (q–s) Mature, pri- and pre-miR-126 expression, as assessed by QPCR (n=3 independent experiments) (q) and miR-126 staining (r) in K562 cells without (siSCR) or with RAN KD (siRAN), as assessed by IB with anti-RNA and anti-actin antibodies (s). All of the above IF, IP, IB and miRNA staining experiments including 3c–h, j, l–n, p, r, s, were repeated at least twice using independent samples, with similar results. Full-length gels and blots with molecular weight standards for 3d, e, g, h, j, m, n, q were provided in Supplementary Fig. 8–10. Comparison between groups was performed by two-tailed, unpaired Student’s t-test. P values ≤0.05 were considered significant. Results shown represent mean ± SEM. *p ≤ 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
miRNA nucleus-to-cytoplasm shuttling and maturation are mediated by a protein complex comprising the GTP-binding nuclear protein RAN (RAS-related nuclear protein, a member of the RAS superfamily), Exp-5 and RCC116. After activation via tyrosine phosphorylation, SPRED1 acts as a negative regulator of RAS superfamily proteins17. We therefore postulated that the BCR-ABL tyrosine kinase can phosphorylate SPRED1, allowing SPRED1 to bind RAN, and that this binding interferes with RAN/Exp-5/RCC1-mediated shuttling and maturation of miR-126. Using immunofluorescence (IF), immunoprecipitation (IP) and in vitro kinase activity assays to analyze BCR-ABL+ primary CD34+ and/or K562 cells, we showed that SPRED1 co-localized with BCR-ABL to the cytoplasm (Fig. 3c), was directly phosphorylated by BCR-ABL (Fig. 3d, Supplementary Fig. 8), and formed an intra/perinuclear protein complex with RAN (Fig. 3e, f, Supplementary Fig. 8). NIL treatment reversed these effects, resulting in SPRED1 de-phosphorylation (Fig. 3d, left panel, Supplementary Fig. 8), decreased binding and co-localization of SPRED1 with RAN (Fig. 3f), increased formation of RAN/Exp-5/RCC1 complex (Fig. 3g–h, Supplementary Fig. 8, 9), decreased pri- and pre-miR-126 levels, and increased mature miR-126 levels (Fig. 3i). Cell washing to remove NIL restored the binding of SPRED1 with RAN, decreased the binding of RAN with Exp-5/RCC1 (Fig. 3j, Supplementary Fig. 9), increased pri-miR-126 levels (Fig. 3k) and reduced mature miR-126 levels (Fig. 3k, l). Northern blotting confirmed that the ratio of pri- and pre-miR-126 to mature miR-126 levels decreased upon exposure of BCR-ABL+ cells to NIL and increased upon wash-off of NIL (Fig. 3m, Supplementary Fig. 10). SPRED1 knock-down (KD) by siRNA in BCR-ABL+ primary CD34+ and K562 cells enhanced formation of the RAN/Exp-5/RCC1 complex (Fig. 3n, Supplementary Fig. 9) and resulted in decreased pri- and pre-miR-126 levels (Fig. 3o) and increased mature miR-126 levels (Fig. 3o, p). Conversely, RAN KD by siRNA resulted in increased pri- and pre-miR-126 levels and reduced mature miR-126 levels (Fig. 3q–s, Supplementary Fig. 10). These results indicate that, in CML cells, BCR-ABL-induced SPRED1 phosphorylation interferes with RAN/Exp-5/RCC1-mediated miR-126 biogenesis and lowers mature miR-126 levels. Given that miR-126 suppresses SPRED1 expression, this BCR-ABL-mediated reduction in miR-126 biogenesis may cause a further increase in SPRED1 levels, such that higher SPRED1 levels result in lower mature miR-126 levels (Supplementary Fig. 1j) and that the endogenous levels of miR-126 are therefore controlled by its own target (SPRED1) in BCR-ABL+ cells.
Of note, BCR-ABL-dependent deregulation of miRNAs is unlikely to be restricted to down-regulation of miR-126. In fact, comparing LSK cells from non-induced versus induced CML mice, we found that a total of 33 miRNAs (including miR-126-3p and miR-126-5p, miR-125a-5p, 125b-5p, 181a-3p, 181b-5p, and 29b-3p) to be significantly decreased, and 75 miRNAs (including miR-142-3p and miR-142-5p, 146b-3p, 146b-5p, and 486-5p) to be significantly increased (GSE107431, Supplementary Fig. 1k), suggesting distinct BCR-ABL-dependent mechanisms causing miRNA deregulation in CML.
Endothelial cells in the BM niche supply miR-126 to CML LSCs
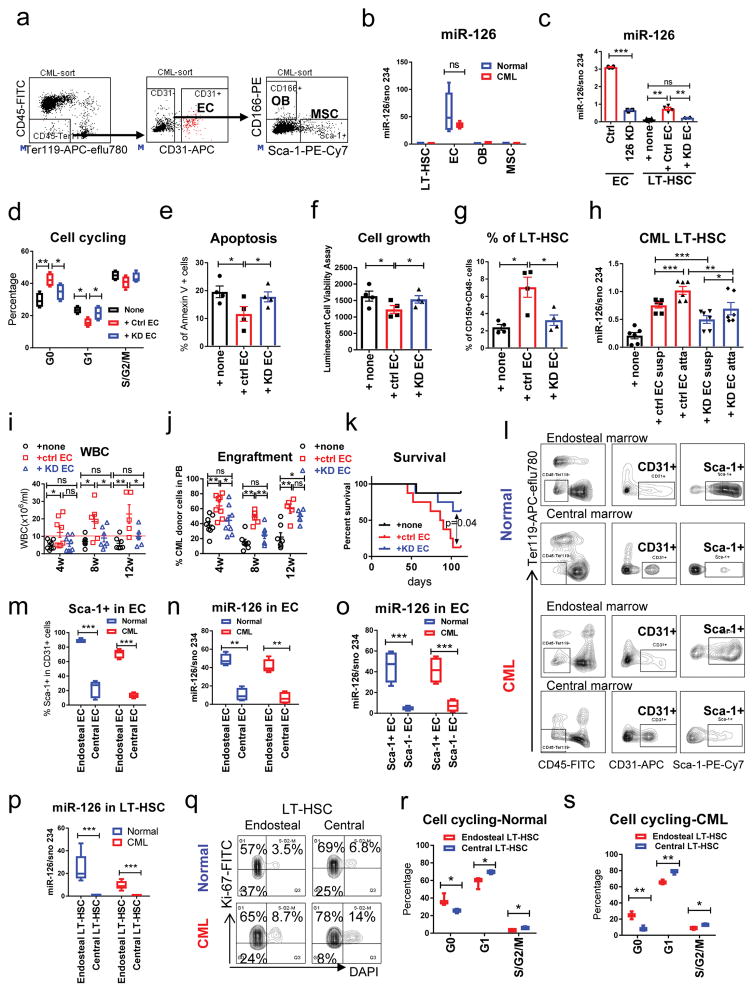
Given that miR-126 is necessary for the quiescence of LSCs and that BCR-ABL activation causes down-regulation of mature miR-126 levels in LSCs, we reasoned that an exogenous source of miR-126 might be needed for BCR-ABL+ LSCs to maintain quiescence and prevent clonal exhaustion. BCR-ABL+ LT-HSCs mainly reside in the BM niche, along with multiple regulatory non-hematopoietic cell types. miR-126 is one of the most abundantly expressed miRNAs in ECs and is involved in angiogenesis18,19. Consistent with this, we found that, in both normal and CML mice, BM ECs (CD45−Ter119−CD31+) expressed the highest levels of miR-126, as compared with LT-HSCs and other BM stromal cell populations, including osteoblasts (OB; CD45−Ter119−CD31−CD166+Sca-1−)20 and mesenchymal stem cells (MSC; CD45−Ter119−CD31−CD166−Sca-1+)21 (Fig. 4a, b). We therefore hypothesized that ECs supply miR-126 to CML LT-HSCs.
Figure 4. Endothelial cells in the niche supply miR-126 to normal and CML LT-HSCs.
(a) Gating strategy for the isolation of ECs (CD45−Ter119−CD31+), osteoblasts (OBs, CD45−Ter119−CD31−CD166+Sca-1−) and mesenchymal stem cells (MSCs, CD45−Ter119−CD31−CD166−Sca-1+). These experiments were repeated 5 times independently with similar results. (b) miR-126 expression, as assessed by QPCR in LT-HSCs, ECs, OBs and MSCs from normal and CML mice (n=5 biologically independent samples). (c) miR-126 expression in ECs from the BM of CML mice that were transduced with miR-126 KD (126 KD) or control (Ctrl) lentiviruses, or in CML LT-HSCs that were cultured alone (none) or co-cultured with the ECs that had been transduced with control (Ctrl EC) or miR-126 KD (KD EC) lentiviruses (n=4 biologically independent samples). (d–g) Cell cycle analysis (n=4 biologically independent samples) (d), apoptosis (n=4) (e), cell growth (n=4) (f) and the percentage of Flt3−CD150+CD48− LSK cells (n=4) (g) in CML LT-HSCs cultured alone or with control or miR-126 KD ECs. (h) miR-126 expression, as assessed by QPCR, in suspended (Susp) and EC-attached (Atta) sub-fractions of CML LT-HSCs (n=6 independent experiments). (i–k) WBC counts (i), CML donor cell engraftment in PB at the indicated times (j) and survival (k) after recipient mice were transplanted with CML LT-HSCs that had been cultured alone (none) or co-cultured with Ctrl or KD EC for 96 h (1,000 cells/mouse, n=8 in each group). Comparison between groups was performed by two-tailed, unpaired Student’s t-test. The log-rank test was used to assess significant differences between survival curves. (l,m) Representative flow cytometry plots of EC staining (l) and frequency of Sca-1+ cells (m) in endosteal or central ECs from normal and CML mice (n=4 independent samples). (n–o) miR-126 expression, as assessed by QPCR, in endosteal or central ECs (n=4 independent samples) (n) and in Sca-1+ or Sca-1− ECs (n=4 independent samples) (o) from normal and CML mice. (p) miR-126 expression, as assessed by QPCR, in endosteal or central LT-HSCs from normal and CML mice (n=6 independent samples). (q–s) Representative flow cytometry plots of Ki-67 and DAPi staining of endosteal or central LT-HSCs from normal and CML mice (q), cell cycle analysis of endosteal or central LT-HSCs from normal (n=3) (r) or CML (n=3) (s) mice. Comparison between groups was performed by two-tailed, paired Student’s t-test. P values ≤0.05 were considered significant. Results shown represent mean ± SEM. *p ≤ 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To test this hypothesis, we sorted ECs from endosteal and central marrow of SCLtTA/BCR-ABL mice and transduced these cells with lentiviral GFP-expressing miR-126 KD or control vectors (Fig. 4c). LT-HSCs from induced SCLtTA/BCR-ABL mice were then co-cultured with GFP+ control or miR-126 KD ECs or were cultured without ECs for 96 h and analyzed for cell cycle and cell growth. After collecting the LT-HSCs that were in suspension, we separately collected the EC-attached LT-HSCs by flushing them gently from the culture flask with PBS buffer. EC-attached LT-HSCs were stained with CD45 to exclude EC contamination. CML cells co-cultured with control ECs showed higher miR-126 levels (Fig. 4c), decreased cell cycling (Fig. 4d), decreased apoptosis (Fig. 4e), decreased cell growth (Fig. 4f), and increased frequency of LT-HSCs (Flt3−CD150+CD48− LSK) (Fig. 4g) as compared to CML cells co-cultured with miR-126 KD ECs or cultured alone. The highest miR-126 levels were found in the LT-HSCs that were attached to control ECs (Fig. 4h). Next, we transplanted CML LT-HSCs (CD45.2, 1,000 cells/mouse) co-cultured for 96h with control or miR-126 KD ECs, or cultured alone, into congenic CD45.1 recipient mice. CML LT-HSCs co-cultured with control ECs generated higher white blood cell (WBC) counts, higher CML engraftment levels and reduced survival (p=0.04) in recipient mice, as compared with CML LT-HSCs co-cultured with miR-126 KD ECs or cultured alone (Fig. 4i–k).
A recent report showed that in normal BM, Sca-1+ ECs from arterial blood vessels are associated with quiescent HSCs, whereas Sca-1− ECs from permeable sinusoidal blood vessels are associated with proliferative HSCs10. We hypothesized that a similar association between EC immunophenotypic subpopulations and adjacent LT-HSC cell-cycle status may exist in CML mice and, given the role of miR-126 in cell quiescence, correlate with miR-126 levels. Upon sorting ECs and LT-HSCs from endosteal and central marrow of CML and normal mice, we found that >70% of endosteal ECs were Sca-1+ and >80% of central ECs were Sca-1−; furthermore, Sca-1+ ECs expressed higher miR-126 levels than did Sca-1− ECs, and total endosteal ECs had higher miR-126 levels than did total central ECs (Fig. 4l–o). Accordingly, endosteal LT-HSCs had higher levels of miR-126 (Fig. 4p) and were more quiescent (Fig. 4q–s) as compared to central LT-HSCs. These results demonstrate a direct association between miR-126 levels in ECs and the cell cycle status of adjacent LT-HSCs, and support the concept that there is active trafficking of miR-126 from ECs to LT-HSCs in vivo.
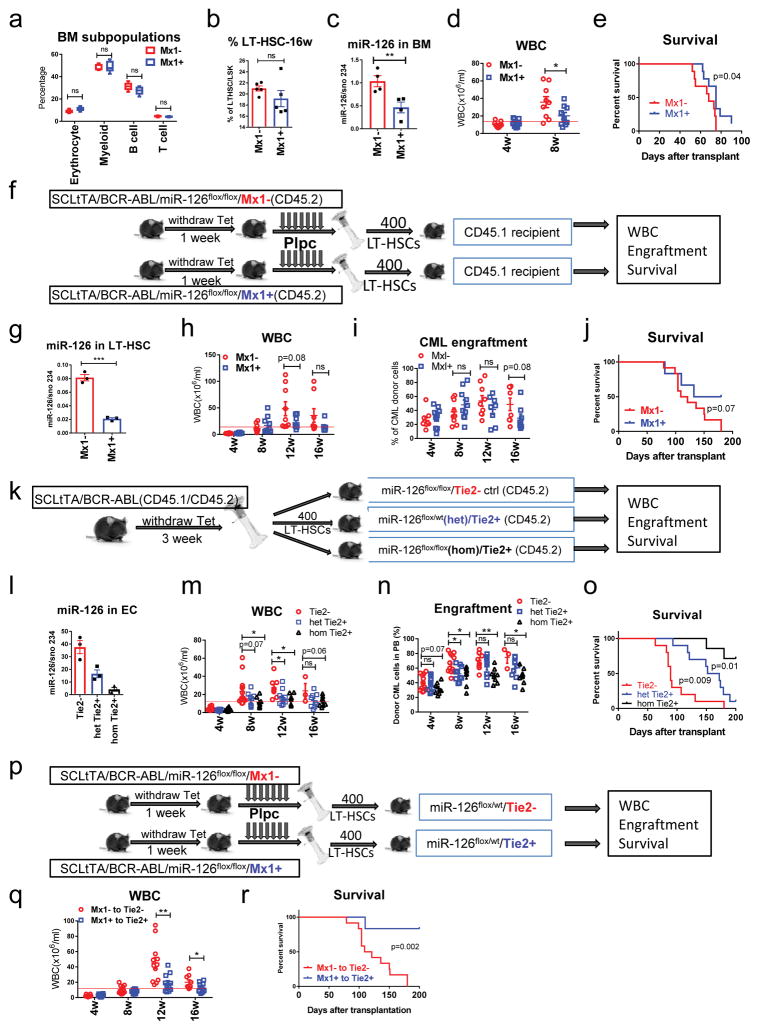
To test the functional role of EC-supplied miR-126 for leukemia growth, we generated CML or normal mice carrying miR-126 floxed alleles (flox) to enable conditional knock-out (c-KO) of miR-126 in LT-HSCs, in ECs, or in both. To generate these mice, we crossed miR-126flox/flox mice with Mx1-cre+ or Tie2-cre+ mice, followed by crossing with SCLtTA/BCR-ABL transgenic mice. These crosses led to the generation of the following strains: miR-126flox/flox/Mx1-cre, SCLtTA/BCR-ABL/miR-126flox/flox/Mx1-cre, miR-126flox/wt/Tie2-cre and miR-126flox/flox/Tie2-cre. After injecting miR-126flox/flox/Mx1-cre+ (Mx1+) mice with polyinosine-polycytosine (pIpC) to delete miR-126 in normal HSCs, we observed no significant changes in WBC counts in PB (data not shown) or BM mononuclear cell subpopulations (including LT-HSCs) (Fig. 5a–b) after 16 weeks of follow-up, as compared to control (Mx1-) mice. Using SCLtTA/BCR-ABL/miR-126flox/flox/Mx1+ mice, targeted miR-126 deletion in CML LSCs was obtained with tetracycline withdrawal to induce BCR-ABL expression and pIpC injection to induce cre expression. In these mice, miR-126 levels in BM cells were reduced by 60%, CML development was delayed (p=0.047) and survival was increased (p=0.04), as compared with similarly treated SCLtTA/BCR-ABL/miR-126flox/flox/Mx1- controls (Fig. 5c–e). To confirm that these results were not attributable to “leaky” miR-126 down-regulation in the non-hematopoietic compartment, we sorted CD45.2 CML LT-HSCs from BCR-ABL-induced and pIpC-injected SCLtTA/BCR-ABL/miR-126flox/flox/Mx1+ or Mx1- mice and transplanted these cells into CD45.1 congenic recipient mice (Fig. 5f). Recipients transplanted with Mx1+ (miR-126 KO) CML LT-HSCs showed a trend for decreased CML development and increased survival as compared with recipient mice transplanted with Mx1- control CML LT-HSCs (Fig. 5g–j).
Figure 5. Endothelial cells in the BM niche supply miR-126 to CML LT-HSCs.
(a, b) Frequency of BM mononuclear cell subpopulations (n=5 independent samples) (a) and LT-HSCs (n=5) (b) in miR-126flox/flox/Mx1-cre+ (Mx1+) and miR-126flox/flox/Mx1-cre- (Mx1-) mice at 16 weeks after pIpC injection. (c–e) miR-126 expression in CML BM cells (n=4 independent samples) (c), WBC counts (d) and survival (e) of SCLtTA/BCR-ABL/miR-126flox/flox/Mx1+ or Mx1- mice subjected to tetracycline withdrawal and pIpC injection (n=9). (f) CD45.2 CML LT-HSCs (400 cells/mouse) from BCR-ABL-induced and pIpC-injected SCLtTA/BCR-ABL/miR-126flox/flox/Mx1+ or Mx1− mice were transplanted into CD45.1 congenic recipient mice (n=10 in each group). (g–j) miR-126 expression, as assessed by QPCR, in donor CML LT-HSCs (n=3 independent samples) (g), WBC counts (h), CML cell engraftment in PB (i) and survival (j) of recipient mice from f (n=10 each). (k) CD45.1/CD45.2 CML LT-HSCs (400 cells/mouse) from induced CML mice were transplanted into CD45.2 congenic miR-126flox/flox/Tie2- (n=14), miR-126flox/wt(het)/Tie2+ (n=10) and miR-126flox/flox(hom)/Tie2+ recipient mice (n=8). (l–o) miR-126 expression, as assessed by QPCR, in ECs sorted from the recipient mice from k (n=3 independent samples) (l), WBC counts (m), CML cell engraftment in PB (n) and survival (o) of recipient mice from k. (p) CD45.2 CML LT-HSCs (400 cells/mouse) from BCR-ABL-induced and pIpC-injected SCLtTA/BCR-ABL/miR-126flox/flox/Mx1+ or Mx1− mice were transplanted into CD45.2 miR-126flox/wt/Tie2+ or Tie2- recipient mice (n=12 in each group), respectively. (q,r) WBC counts (q) and survival (r) of the recipient mice from p. Comparison between groups was performed by two-tailed, unpaired Student’s t-test. The log-rank test was used to assess significant differences between survival curves. P values ≤0.05 were considered significant. Results shown represent mean ± SEM. *p ≤ 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To assess the contribution of EC-derived miR-126 to leukemia growth, we sorted LT-HSCs from BCR-ABL-induced CD45.1/CD45.2 SCLtTA/BCR-ABL mice (used to track donor cells, and generated by crossing CD45.2 SCLtTA/BCR-ABL B6 mice with CD45.1 B6 mice) and transplanted these cells into CD45.2 congenic miR-126flox/flox/Tie2-cre- (Tie2-) (WT miR-126 allele in ECs), miR-126flox/wt(het)/Tie2-cre+ (Tie2+) (heterozygous miR-126 KO allele in ECs), or miR-126flox/flox(hom)/Tie2+ (homozygous miR-126 KO allele in ECs) recipient mice (Fig. 5k). Both het and hom Tie2+ recipient mice showed reduced CML cell engraftment, delayed CML development and significantly increased survival, as compared with Tie2- recipients at 16 weeks after transplantation (Fig. 5l–o; p=0.009 and 0.0003 for survival of het Tie2+ and hom Tie2+ mice respectively); a miR-126 dosage effect was evident, as 70% of hom Tie2+ mice versus 10% of het Tie2+ mice were alive at 28 weeks (Fig. 5o). Of note, no significant differences in donor cell output from transplanted normal LT-HSCs (CD45.1) were observed in PB and BM from CD45.2 miR-126flox/wt/Tie2+ recipients as compared with miR-126flox/wt/Tie2- recipients after 16 weeks of follow-up (data not shown).
Next, to assess the functional impact of concurrent miR-126 KO in both LT-HSCs and ECs, CML LT-HSCs from BCR-ABL-induced and pIpC-injected SCLtTA/BCR-ABL/miR-126flox/flox/Mx1+ or Mx1- mice were transplanted into miR-126flox/wt/Tie2+ or Tie2- recipient mice, respectively (Fig. 5p). Tie2+ mice transplanted with Mx1+ CML LT-HSCs showed a significantly delayed CML development (p=0.007) and prolonged survival (p=0.002) as compared with Tie2- mice transplanted with Mx1- CML LT-HSCs (Fig. 5q–r). At day 200, 83% of the mice with miR-126 KO in both ECs and LT-HSCs, but none of the controls with intact miR-126 in both ECs and LT-HSCs, were alive. Taken together, these results support a functional role of EC-derived miR-126 in sustaining leukemia growth in the CML LSC niche.
In assessing the relevance of these results to humans, we first showed that human umbilical vein EC (HUVECs) expressed significantly higher levels of miR-126 as compared with human CML CD34+ subpopulations (Supplementary Fig. 2a). We next knocked down miR-126 in HUVECs by transduction with lentiviral miR-126 KD or control vectors (Supplementary Fig. 2b, c) and then co-cultured CML Lin−CD34+CD38− cells (HSCs) with miR-126 KD or control HUVECs for 96 h. CML HSCs co-cultured for 96 h with control HUVECs had significantly higher miR-126 expression (Supplementary Fig. 2d, e), decreased cell cycle entry and apoptosis (Supplementary Fig. 2f–h) and an increased proportion of CD34+ cells (Supplementary Fig. 2i), as compared with CML cells co-cultured with KD HUVECs or cultured alone. Next, we transplanted CML CD34+ cells co-cultured with control or miR-126 KD HUVECs or cultured alone for 96 h into irradiated (300cGy) NSG-SGM3 (NOD-scid gamma IL3, GM-CSF, SCF triple transgenic) mice. At 16 weeks after transplantation, BCR-ABL expression in BM cells confirmed CML cell engraftment in the recipient mice (Supplementary Fig. 2j). Human CD45+ cell engraftment was significantly increased in mice receiving CML cells co-cultured with control HUVECs as compared with mice receiving CML cells co-cultured with KD HUVECs or cultured alone (Supplementary Fig. 2k–l).
Extracellular vesicles (EVs) mediate miR-126 trafficking from ECs to LT-HSCs
To test whether intercellular miR-126 trafficking between ECs and HSCs involves extracellular vesicles (EVs), we isolated EVs from control and miR-126 KD HUVECs by differential ultracentrifugation. Using electron microscopy and nanoparticle tracking analysis by NanoSight, the size range of the EVs isolated from the supernatants of both control and miR-126 KD HUVECs was 40–150 nm (peaks at 105 and 125 nm, respectively, Supplementary Fig. 3a–b). By western blotting, EVs recovered from HUVECs showed expression of exosome-specific proteins (CD63, TSG101) and an EV-associated protein (HSP90)22,23 and lack of a non EV-associated protein (the mitochondrial protein cytochrome C) (Supplementary Fig. 3c, 10). Next, we fractionated EVs from HUVECs using magnetic beads coated with an anti-CD63 antibody and confirmed expression of exosome-specific proteins in the CD63+ EV fraction using antibodies to tetraspanins (CD63, CD9 and CD81) by cytofluorimetric analysis (Supplementary Fig. 3d). Significantly higher miR-126 levels were found in the CD63+ EV fraction as compared with the CD63− EV fraction, as assessed by QPCR (0.77 versus 0.19, Supplementary Fig. 3e). Notably, miR-126 levels were higher in EVs obtained from control HUVECs as compared to EVs from miR-126 KD HUVECs or EVs from human normal or CML Lin−CD34+CD38− HSC cells (Supplementary Fig. 3f).
To demonstrate intercellular trafficking of miR-126, we labeled miR-126 with a fluorescence probe in cultured HUVECs (Supplementary Fig. 3g) and subjected the supernatant from these cells to differential ultracentrifugation to collect EVs; treatment of BCR-ABL+ K562 cells with the collected HUVEC EVs resulted in the presence of a miR-126 fluorescence signal in the K562 cells, as assessed by confocal microscopy (Supplementary Fig. 3h). To corroborate these results, we cultured human CML HSCs for 96h with EVs (5×109 particles/ml as measured by NanoSight, equivalent to 5 μg/ml as measured by standard protein quantification methodology) isolated from control or KD HUVECs, or cultured the CML HSCs alone. CML HSCs co-cultured with control HUVEC-derived EVs had significantly increased miR-126 levels (Supplementary Fig. 3i) and reduced cell cycling and apoptosis rates (Supplementary Fig. 3j–m) as compared with CML HSCs co-cultured with KD HUVEC-derived EVs or cultured alone. Next, we co-cultured miR-126 KO CML LT-HSCs from tetracycline-off and pIpC-injected SCLtTA/BCR-ABL/miR-126flox/flox/Mx1+ mice with control or miR-126 KD HUVEC-derived EVs for 48 h, or cultured the CML LT-HSCs alone. miR-126 KO CML cells co-cultured with control HUVEC-derived EVs showed significantly increased miR-126 levels (Supplementary Fig. 3n) and reduced cell cycling and apoptosis rates (data not shown) as compared with miR-126 KO CML cells co-cultured with KD HUVEC-derived EVs or cultured alone.
Next, we transplanted primary CML CD34+ cells co-cultured with EVs (5×109 particles/ml) from control or miR-126 KD HUVECs for 96 h into NSG-SGM3 mice. Mice receiving cells co-cultured with control HUVEC-derived EVs showed enhanced human CD45+ (p=0.001; Supplementary Fig. 3o) and CD45+CD34+ (p=0.0008; Supplementary Fig. 3p) BM engraftment at 16 weeks, compared with mice receiving cells co-cultured with KD HUVEC-derived EVs. QPCR confirmed that the engrafted cells in both groups of mice were BCR-ABL+ (data not shown). We also sorted CML LT-HSCs from induced CD45.2 SCLtTA/BCR-ABL mice and co-cultured them with control or KD HUVEC-derived EVs for 96 h and then transplanted the cells into CD45.1 recipient mice. LT-HSCs co-cultured with control HUVEC-derived EVs showed increased miR-126 expression (p=0.009; Supplementary Fig. 3q), enhanced CML progression (p=0.002), an increased engraftment rate (p=0.06), and reduced survival (p=0.04) as compared with LT-HSCs co-cultured with KD HUVEC-derived EVs (Supplementary Fig. 3r–t). Altogether, these observations suggest that EV-mediated trafficking is responsible for the transfer of miR-126 from ECs to LT-HSCs.
Of note, although we could not completely exclude the possibility that decreased CML LT-HSC quiescence and engraftment capacity results from functional changes in ECs induced by miR-126 KD, rather than from a decrease in miR-126 trafficking from ECs to LT-HSCs, Kuo et al. previously showed that miR-126 KO mice have no substantial changes in BM EC structure9. Moreover, we did not observe detectable changes in the morphology and growth rate of miR-126 KD HUVECs or miR-126 KD mouse ECs compared with controls (data not shown).
miR-126 knockdown enhances TKI-mediated targeting of CML LSC
Given that BCR-ABL activity reduces endogenous levels of miR-126 in LT-HSCs and that pharmacologic inhibition of BCR-ABL by NIL increased miR-126 levels and the frequency of quiescent LT-HSCs (Fig. 2d–o), we postulated that miR-126 down-regulation may enhance the anti-leukemic activity of TKI treatment and eliminate CML LSCs. To test this hypothesis, we subjected human CML HSCs to miR-126 KD or OE by transduction of GFP-expressing lentiviruses (Supplementary Fig. 4a). We selected GFP+ cells and cultured them for 96h in the presence of NIL. Combined miR-126 KD and NIL treatment resulted in increased cell cycling and apoptosis (Supplementary Fig. 4b,c) and decreased cell growth, CFC and CFC replating efficiency (Supplementary Fig. 4d–f), as compared with only NIL-treated control cells. Conversely, combined miR-126 OE and NIL treatment resulted in decreased cell cycling and apoptosis and increased CFC and CFC replating efficiency (Supplementary Fig. 4b, c, e, f), as compared with only NIL-treated control cells. We obtained similar results for CML LT-HSCs from SCLtTA/BCR-ABL mice (Supplementary Fig. 4g–k).
To further test the concept that miR-126 KD can enhance the anti-leukemic activity of TKI treatment, we transduced human CML HSCs with miR-126 KD or control lentiviral vectors; we selected GFP+ cells and treated them with NIL for 96h and then transplanted them into irradiated NSG-SGM3 mice. Recipient mice receiving NIL-treated miR-126 KD cells showed reduced engraftment of human CD45+ cells in PB at 4 weeks (Fig. 6a) and in BM at 16 weeks (Fig. 6b and Supplementary Fig. 5a), as compared to controls. BCR-ABL levels were reduced in BM cells from NSG-SGM3 mice transplanted with miR-126 KD CML cells as compared with mice transplanted with control CML cells (Supplementary Fig. 5b).
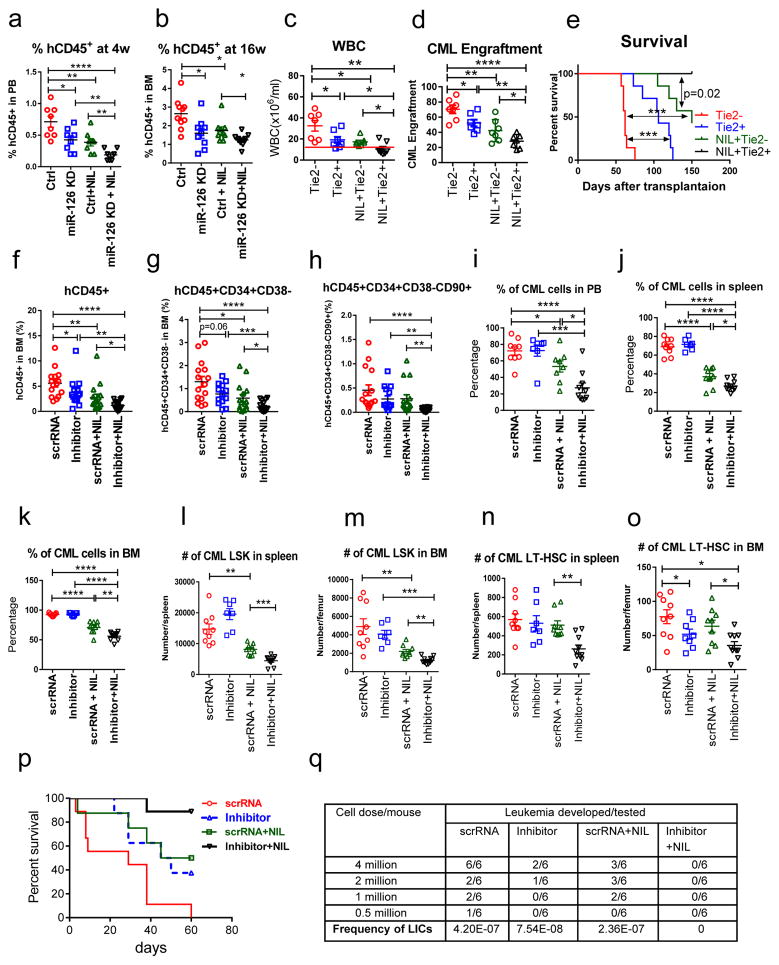
Figure 6. miR-126 knockdown by lentiviruses or CpG-miR-126 inhibitor in combination with nilotinib enhances the in vivo targeting of CML LSCs.
(a,b) Engraftment of human CD45+GFP+ cells in PB at 4 weeks (n=8 independent samples) (a) and in BM at 16 weeks (n=10 independent samples) (b) in NSG-SGM3 mice transplanted with human CML Lin−CD34+CD38− cells (5×105 cells/mouse, n=8–10 in each group), which were transduced with miR-126 KD or control lentiviruses and treated with NIL (5 μM) for 4 days. (c–e) WBC counts (c), CML donor cell engraftment (d) and survival (e) of CD45.2 miR-126flox/wt/Tie2+ or Tie2− mice transplanted with CD45.1/CD45.2 CML LT-HSCs and treated with or without NIL (n=7 in each group) for 3 weeks. The log-rank test was used to assess significant differences between survival curves. (f–h) Human CD45+ (f), CD45+CD34+CD38− HSC (g) and CD45+CD34+CD38−CD90+ LT-HSC (h) engraftment in BM of NSG-SGM3 mice transplanted with human CD34+ cells from CP CML patient samples and then treated with scrRNA (5mg/kg, i.v. 4 times a week, n=15), inhibitor (5mg/kg i.v. 4 times a week, n=14), scrRNA + NIL (50mg/kg, daily by gavage, n=15), or inhibitor + NIL (n=16) for 3 weeks (total 60 mice). (i–o) Percentage of donor CML cells in PB (i), spleen (j) and BM (k), numbers of donor CML LSK in spleen (l) and BM (m), and numbers of donor CML LT-HSC in spleen (n) and BM (o) of recipient mice transplanted with CML BM cells and then treated with scrRNA (n=9), inhibitor (n=7), scrRNA + NIL (n=8), or inhibitor + NIL (n=10) for 3 weeks (total 34 mice). (p) Survival of another cohort of CML mice treated with scrRNA (n=9), inhibitor (n=8), scrRNA + NIL (n=8), or inhibitor + NIL (n=9) for 3 weeks (total 34 mice). The log-rank test was used to assess significant differences between survival curves. (q) Frequency of leukemia initiating cells (LICs) in BM cells from treated leukemic mice, as assessed by leukemia development rate in secondary recipient mice transplanted with 4×106, 2×106, 1×106, and 5×105 BM cells/mouse from the treated mice (n=6 mice/dose/condition × 4 doses × 4 conditions = 96 mice) by L-Calc software. Comparison between groups was performed by two-tailed, unpaired Student’s t-test. P values ≤0.05 were considered significant. Results shown represent mean ± SEM. *p ≤ 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To test if miR-126 down-regulation in ECs also enhances the sensitivity of CML HSCs to TKI treatment, we cultured human CML HSCs alone or with HUVEC with or without miR-126 KD, and with or without NIL for 72 h. CML HSCs co-cultured with miR-126 KD HUVECs had significantly increased apoptosis and decreased cell growth and CFCs, as compared with CML HSCs co-cultured with control HUVECs with or without NIL (Supplementary Fig. 5c–e). Of note, CML HSCs co-cultured with KD HUVEC-derived EVs also showed significantly increased apoptosis and decreased cell growth as compared to CML HSCs co-cultured with control HUVEC-derived EVs, with or without NIL (Supplementary Fig. 5f, g). Next, we sorted CML LT-HSCs from induced CD45.1/CD45.2 SCLtTA/BCR-ABL mice and transplanted them into CD45.2 miR-126flox/wt/Tie2+ (heterozygous miR-126 KO allele in ECs) or Tie2- (WT miR-126 allele in ECs) mice. After confirming CML development, we treated the mice with NIL (50 mg/kg, daily by oral gavage) or vehicle for 3 weeks. Vehicle-treated Tie2+ mice showed delayed CML development (p=0.02) and increased survival (p=0.001) as compared with vehicle-treated Tie2- mice (Fig. 6c–e). Moreover, NIL-treated Tie2+ mice showed significantly reduced WBC counts in PB (p=0.03) and increased survival (p=0.02) as compared with NIL-treated Tie2- mice (Fig. 6c–e), and all NIL-treated Tie2+ mice were alive at day 150 (Fig. 6e). Altogether, these results support the hypothesis that miR-126 down-regulation in ECs and CML HSCs enhances the anti-leukemic activity of TKI treatment.
To sort out the mechanistic basis for the enhancement of TKI activity by miR-126 KD, we showed that, consistent with previous studies24,25, NIL treatment enhanced MAPK/ERK activation, increased BCL-2 expression and promoted survival in CML CD34+ cells (Supplementary Fig. 5h, 10). Thus, we reasoned that NIL could enhance MAPK/ERK activation through up-regulation of miR-126 and consequent down-regulation of SPRED1 (Supplementary Fig. 5h, i), a reported inhibitor of the MAPK/ERK pathway. In fact miR-126 KD in CML CD34+ cells resulted in SPRED1 up-regulation and decreased phospho-ERK (p-ERK) and BCL-2 levels (Supplementary Fig. 5j–k, 10), while SPRED1 KD increased p-ERK and BCL-2 levels and rescued NIL-induced apoptosis (Supplementary Fig. 5l–o, 10). Moreover, NIL-induced apoptosis was enhanced by the MEK inhibitor PD0325901 or BCL-2 KD (Supplementary Fig. 5p–s, 10). These results support a model in which NIL-induced up-regulation of miR-126 expression decreases SPRED1 expression and results in spurious activation of the MAPK/ERK pathway, and ultimate increase of BCL-2 levels. miR-126 KD counteracted these effects, thereby increasing the anti-leukemic activity of NIL.
Effective in vitro and in vivo uptake and gene silencing effects of the CpG-miR-126 inhibitor
In view of the enhancing effects of miR-126 KD on the anti-leukemic activity of NIL, we reasoned that miR-126 could represent a therapeutic target for eliminating LSC. Although microRNAs can be targeted with oligonucleotide therapeutics (ONTs), it remains challenging to achieve efficient and cell-selective delivery of ONTs in vivo. Thus, we designed a novel miR-126 inhibitor by linking an anti-miR-126 oligodeoxynucleotide (ODN) to a cytosine guanine dinucleotide (CpG) ODN, a ligand for the intracellular protein Toll-like receptor 9 (TLR9). To allow for systemic administration, we chemically modified the CpG-miR-126 inhibitor to resist serum nucleases, using phosphothioation and 2′OMe-modified nucleotides in the CpG ODN and anti-miR-126 moieties, respectively. We compared the specificity and efficiency of CpG-miR-126 inhibitor uptake with a nanoparticle (NP) delivery method previously reported by our group6. We incubated K562 cells with fluorescently-labeled CpG-miR-126 inhibitor-Cy3 (CpG), human CD45 antibody (Ab)- or transferrin (TF)-conjugated NPs containing miR-126 inhibitor-Cy3 (Ab-NP or TF-NP), or naked miR-126 inhibitor-Cy3 (control), in the absence of any reagents routinely used for nucleic acid transfection. Flow cytometric analysis at 4 h and 24 h after treatment (Supplementary Fig. 6a–b) showed that CpG-miR-126 inhibitor-Cy3 was taken up by 99% of the K562 cells at both 4 h and 24 h, compared with 24% and 30% of the cells incubated with Ab-NP and 74% and 88% of the cells incubated with TF-NP at 4h and 24 h, respectively. K562 cells did not take up naked miR-126 inhibitor (control) (Supplementary Fig. 6b). Efficient miR-126 down-regulation by CpG-miR-126 inhibitor-Cy3 in K562 cells was shown at 24 h (Supplementary Fig. 6c). We further showed by flow cytometry that, even without routinely used transfection reagents, the CpG-miR-126 inhibitor-Cy3 was internalized by HUVECs as well as by human normal and CML Lin−CD34+CD38− cells at 4 h (Supplementary Fig. 6d–f); >95% cells were positive for CpG-miR-126 inhibitor-Cy3 uptake in all three cell types. CpG-miR-126 inhibitor-Cy3 uptake led to efficient miR-126 down-regulation in HUVECs and HSCs (Supplementary Fig. 6g–i). We also observed increased cell cycling in both CpG-miR-126 inhibitor-treated normal and CML HSCs, as compared to CpG-scrambled RNA (scrRNA)-treated controls (Supplementary Fig. 6j–k).
Next, we evaluated CpG-miR-126 inhibitor-Cy3 uptake in mouse LT-HSCs and ECs in vitro and in vivo. Following in vitro exposure to CpG-miR-126 inhibitor-Cy3, efficient uptake at 4 h and miR-126 down-regulation at 24 h were shown by flow cytometry and QPCR, respectively (Supplementary Fig. 6l–m). We also observed increased cell cycling in normal and CML BM LT-HSCs treated with the CpG-miR-126 inhibitor (Supplementary Fig. 6n). We treated normal and CML mice with one dose (5 mg/kg, iv injection) of CpG-miR-126 inhibitor-Cy3. At 16h post-treatment, efficient in vivo uptake was demonstrated by flow cytometry in both LT-HSCs (56±5%) and ECs (62±3%) isolated from femurs (Supplementary Fig. 6o). After CpG-miR-126 inhibitor treatment (5 mg/kg/day, iv, daily) for 3 days, we sorted LT-HSCs and ECs and found significant miR-126 down-regulation (Supplementary Fig. 6p–q).
CpG-miR-126 inhibitor enhances in vivo targeting of CML LSCs in combination with TKI treatment
We next tested the effects of the CpG-miR-126 inhibitor in normal mice to ensure that the compound does not have hematologic toxicity. We treated WT B6 mice with CpG-scrRNA (scrRNA) or CpG-miR-126 inhibitor (inhibitor, 5 mg/kg/day i.v.) for 3 weeks, after which we collected their BM cells and transplanted them into recipient mice (3×105 BM cells/mouse). Compared with scrRNA-treated control mice, inhibitor-treated mice showed increased numbers of red blood cells (RBC), but no significant differences in the numbers of WBCs or platelets (PLT) in PB (Supplementary Fig. 7a–c), or in the numbers of mononuclear cells, LT-HSCs, or ECs in the BM (Supplementary Fig. 7d–f). These findings are in line with the observation that miR-126 down-regulation in normal HSCs increases hematological output4. In the recipient mice receiving BM cells from donor mice treated with scrRNA or inhibitor, we observed no significant differences in donor cell engraftment in PB, BM or spleen (Supplementary Fig. 7g–h) or in donor LT-HSC numbers in BM (Supplementary Fig. 7i) at 16 weeks after transplantation. These data demonstrate that the inhibitor lacks pre-clinical toxicity for normal hematopoiesis.
We then tested the effects of the inhibitor alone and in combination with NIL on human and mouse CML LT-HSCs in vivo. First, we transplanted human CD34+ cells from CP CML patients into NSG-SGM3 mice. At 6 weeks after transplantation, the mice were divided into 4 groups and treated with scrRNA (5 mg/kg i.v. 4 times a week), inhibitor (5 mg/kg, i.v. 4 times a week), scrRNA + NIL (50 mg/kg, daily by oral gavage), or inhibitor + NIL for 3 weeks, followed by assessment of human cell engraftment in PB, BM and spleen. We observed significantly reduced human CD45+, CD45+CD34+CD38− HSC and CD45+CD34+CD38−CD90+ LT-HSC engraftment in the BM of inhibitor+NIL treated mice as compared with scrRNA alone, inhibitor alone or scrRNA+NIL treated mice (Fig. 6f–h, Supplementary Fig. 7j). QPCR analyses confirmed that the engrafted human CD45+ cells were BCR-ABL positive (Supplementary Fig. 7k).
Next, we transplanted BM cells from SCLtTA/BCR-ABL mice (CD45.2) into congenic B6 mice (CD45.1). Following confirmation of CML development at 4 weeks after transplantation, mice were treated as above with scrRNA, inhibitor, scrRNA + NIL, or inhibitor + NIL for 3 weeks. As EC-derived miR-126 plays a key role in LSC maintenance, we sorted BM ECs from treated mice and confirmed significantly lower miR-126 levels in total ECs, Sca-1+ ECs and Sca-1− ECs from inhibitor-treated mice, as compared with scrRNA-treated mice (Supplementary Fig. 7l–m). Mice receiving the combination of inhibitor + NIL had a significant reduction in the percentage of CD45.2+ CML cells in PB, spleen and BM (Fig. 6i–k), and a significant reduction in the numbers of CML LSK cells and LT-HSCs in spleen and BM, as compared with all other groups (Fig. 6l–o). We followed a cohort of mice for survival studies after 3 weeks of treatment, and found that all of the mice treated with scrRNA alone died of leukemia within 60 days after treatment discontinuation, whereas 50% of the mice treated with inhibitor alone or scrRNA+NIL and 90% of the mice treated with the combination of inhibitor + NIL survived (p=0.0012; Fig. 6p).
To quantify the frequency of leukemia-initiating cells (LICs) after treatment, BM cells from leukemic mice treated with scrRNA, inhibitor, scrRNA + NIL, or inhibitor + NIL for 3 weeks were transplanted in limiting dilution assays into secondary congenic CD45.1 recipient mice. Treatment with the combination of inhibitor + NIL resulted in a significantly higher levels of depletion of LICs, as assessed by leukemia development in secondary recipient mice after 16 weeks of follow-up, as compared with treatment with scrRNA alone, inhibitor alone or scrRNA + NIL (Fig. 6q). None of the secondary recipients that received BM cells from the mice treated with the combination of inhibitor + NIL developed leukemia. These results indicate that, compared with NIL treatment, treatment with a combination of NIL and the CpG-miR-126 inhibitor enhances the eradication of CML LSCs capable of engraftment in secondary recipients.
Discussion
We report here that miR-126 expression levels in both human and mouse CML cells follow the hierarchy of hematopoietic differentiation, with more primitive hematopietic stem cells or progenitors expressing higher levels of miR-126 than mature cells. Moreover, quiescent CML LT-HSCs have higher levels of miR-126 and a higher leukemia engraftment capacity than proliferating CML LT-HSCs, in line with findings reported for normal hematopoiesis4. Unexpectedly, we found that miR-126 levels are significantly lower in CML LT-HSCs than their normal counterparts, consistent with previous findings that a lower frequency of long-term repopulating cells are observed within CML LT-HSCs as compared to normal LT-HSCs2. Indeed, we demonstrated that BCR-ABL expression lowers mature miR-126 levels and increases pri- and pre-miR-126 levels, whereas pharmacologic BCR-ABL inhibition by a TKI increases mature miR-126 levels and decreases pri- and pre-miR-126 levels. Altogether, these data support a role for BCR-ABL in altering the biogenesis of endogenous miR-126.
To our knowledge, BCR-ABL-dependent down-regulation of miR-126 in CML cells and its mechanistic basis has not been previously reported. SPRED1, a validated miR-126 target4,7,9,14,15, is a tyrosine kinase substrate known to inhibit GF-mediated activation of RAS protein family members and, in turn, the RAS/MAPK/ERK pathway16. Tyrosine residue phosphorylation is required for SPRED1 inhibition of RAS/MAPK/ERK activation17. We show here that SPRED1 is a substrate for BCR-ABL and that BCR-ABL-induced SPRED1 phosphorylation is critical for miR-126 biogenesis in CML. We found that BCR-ABL-phosphorylated SPRED1 binds with RAN, a RAS family member, disrupts the RAN/Exp-5/RCC1 complex, which is involved in pre-miRNA nucleus-to-cytoplasm shuttling, increases nuclear levels of pri- and pre-miR-126 and decreases cytoplasmic levels of mature miR-126. Conversely, BCR-ABL inhibition disrupts the binding of SPRED1 with RAN, enhances formation of the RAN/Exp-5/RCC1 complex, increases mature miR-126 levels, and decreases pri- and pre-miR-126 levels (Supplementary Fig. 1j).
Because miR-126 is necessary for normal and clonal HSC quiescence and continuous down-regulation of miR-126 can cause clonal exhaustion4,8, we reasoned that this autoregulatory loop must be circumvented in order to maintain a reservoir of quiescent CML LSCs. Previous reports suggested that BM ECs participate in the regulation of normal hematopoiesis10,26. Here, we showed that among cell populations in the leukemic BM niche, ECs express the highest miR-126 levels and supply miR-126 to CML cells, likely through EV trafficking. Furthermore, consistent with previous reports showing that Sca-1+ ECs are associated with quiescent normal HSCs and that Sca-1− ECs are associated with proliferating normal HSC in the marrow10, we found that endosteal Sca-1+ ECs express higher levels of miR-126 than central marrow Sca-1− ECs and are associated with a larger fraction of quiescent BCR-ABL+ LT-HSCs, which also express higher miR-126 levels than proliferating BCR-ABL+ LT-HSCs. Our data support a functional interplay between ECs and HSCs in CML, resulting in a non-random BM distribution of the quiescent CML LSC fraction that is more likely to be found proximal to the high-level miR-126-expressing ECs from the endosteal marrow than to the low-level miR-126-expressing ECs from the central marrow. The functional relevance of the exchange of miR-126 between ECs and LT-HSCs to leukemia growth was demonstrated by showing a decreased engraftment of CML LSCs and improved survival observed in recipient mice with miR-126 KO in the endothelial compartment transplanted with BCR/ABL+ LT-HSCs.
Our results may be clinically relevant to CML patients treated with TKI. Persistence of CML LSCs during TKI treatment is an active area of investigation, as these agents are remarkably potent against cycling cells, but fail to eliminate quiescent CML LSCs3,27. Using primary human CML cells and CML mouse models, we showed that the resistance of CML LSCs to TKI treatment is likely mediated by decreased levels of phosphorylated SPRED1 due to BCR-ABL inhibition, leading to increased endogenous miR-126 levels, which pushes LSCs into a relatively treatment-refractory quiescent state. Furthermore, miR-126 up-regulation in TKI-treated CML CD34+ cells resulted in decreased SPRED1 levels, activation of the MAPK/ERK pathway and increased cell survival25,28. Accordingly, miR-126 KD in CML cells and/or ECs enhances the anti-leukemic activity of TKI treatment by counteracting the undesired TKI-induced miR-126 up-regulation. In vivo, all NIL-treated CML mice with genetic miR-126 KD in ECs survived, demonstrating the therapeutic potential of targeting miR-126 in CML.
For clinical translation of this concept, we designed a novel CpG-miR-126 ODN inhibitor that could be efficiently taken-up by both hematopoietic and non-hematopoietic cells in the BM niche. We have previously shown that the uptake of CpG–ODN conjugated molecules depends on endocytosis by scavenger family dextran sulfate-sensitive receptors (SRs)29,30, which are expressed on the surface of normal and malignant myeloid cells31,32. Following SR-mediated internalization, CpG-conjugates bind to endosomal TLR9, triggering their cytoplasmic release29. SRs and TLR9 are both expressed on hematopoietic cells and ECs33–36, and likely facilitate the efficient intracellular delivery of CpG-miR-126 inhibitor and its subsequent endosomal release and pharmacologic activity. We found that the CpG-miR-126 inhibitor was efficiently taken up by both LT-HSCs and ECs in vitro and in vivo, down-regulated miR-126 expression and reduced LT-HSC quiescence and frequency. Combination treatment with the CpG-miR-126 inhibitor and a TKI in CML mice resulted in increased survival, compared to treatment with either agent alone; moreover, no leukemia development in secondary recipients transplanted with BM cells from combination-treated mice was observed, suggesting that combination treatment results in the elimination of CML LSCs. In support of the possibility of clinical translation of this treatment strategy, we observed no hematologic toxicity in normal mice treated with CpG-miR-126 inhibitor.
In summary, we report that BCR-ABL-mediated SPRED1 phosphorylation down regulates miR-126 biogenesis in CML LSCs, such that CML LSC quiescence and leukemogenic capacity relies on trafficking of miR-126 from ECs to LSCs in the BM niche (Supplementary Fig. 7n). Consistent with this model, TKI treatment inhibits BCR-ABL-induced SPRED1 phosphorylation, leading to the undesired increase in miR-126 levels. Based on the proof-of-concept findings reported here, which showed that in vivo treatment of CML mice with a newly-developed CpG-miR-126 inhibitor enhances TKI activity in vivo and results in LSC elimination; this CpG-miR-126 inhibitor is now being translated to the clinic for the treatment of CML patients.
Online Methods (also see Life Sciences Reporting Summary for details)
Human samples
Normal PB and BM samples were obtained from donors at the City of Hope National Medical Center (COHNMC). CP CML samples were obtained from patients who had not received prior TKI treatment from the COHNMC, the Glasgow Experimental Cancer Medicine Centre and UK SPIRIT2 clinical trial patient samples (http://spirit-cml.org). All CML samples used in this study are P210 BCR-ABL positive, as confirmed by FISH analysis and QPCR. Mononuclear cells were isolated using Ficoll separation. CD34+ cells were then isolated using a positive magnetic bead selection protocol (Miltenyi Biotech, Germany). All CML patients and healthy donors signed an informed consent form. Sample acquisition was approved by the Institutional Review Boards at the COHNMC and the Greater Glasgow and Clyde NHS Trust, in accordance with an assurance filed with and approved by the Department of Health and Human Services, and met all requirements of the Declaration of Helsinki.
Animal studies
Inducible transgenic SCLtTA/BCR-ABL mice in the FVB/N background37, 38 were backcrossed to the B6 background (CD45.2) for 10 generations. Transgenic BCR-ABL mice were maintained on tetracycline water at 0.5 g/L. Withdrawal of tetracycline results in expression of BCR-ABL and generation of a CML-like disease in these mice37, 38. Unless otherwise indicated, BCR-ABL expression was induced for 2–3 weeks by tetracycline withdrawal in 6–8 weeks old male and female mice and then BM cells (from both tibias and femurs) were collected for experiments. SCLtTA/BCR-ABL mice (CD45.2, B6) were also bred with CD45.1 B6 mice to produce CD45.1/CD45.2 SCLtTA/BCR-ABL mice as donors. miR-126flox/flox mice (B6, from Dr. Kuo, Stanford) were crossed with Mx1-cre, Tie2-cre (both from The Jackson Laboratory) and SCLtTA/BCR-ABL mice (all B6) to obtain the following strains: miR-126flox/flox/Mx1-cre, SCLtTA/BCR-ABL/miR-126flox/flox/Mx1-cre, miR-126flox/wt/Tie2-cre and miR-126flox/flox/Tie2-cre. Recipient mice in the CD45.1 B6 background (from Charles River) were used to allow tracking of donor CD45.2 cells after transplantation. Recipients were 6 to 8 weeks old male and female mice and were irradiated at 900 cGy within 24 h before transplantation. The number of mice for each study group was chosen based on the expected endpoint variation (i.e., engraftment rate and latency period of leukemia) and on the availability of mice of different strains. Mice of the same gender and age were randomly divided into groups. Investigators were blinded to mouse genotype while performing treatment or monitoring engraftment or survival. Mouse care and experimental procedures were performed in accordance with federal guidelines and protocols and were approved by the Institutional Animal Care and Use Committee at the City of Hope.
Engraftment of human cells in immunodeficient mice
GFP+ cells (2×105 cells/mouse) selected from CML Lin−CD34+CD38− cells transduced with miRZip anti-miR-126 (126 KD) or control (Ctrl) lentiviruses were cultured with or without NIL (5 μM) for 96 h. Cells were then harvested, washed and transplanted via tail vein injection into sublethally irradiated (300cGy) 6–8 week old NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ mice ( NSG-SGM3, The Jackson Laboratory). Engraftment of human CD45+ cells in PB was monitored at 6 weeks. Mice were euthanized after 16 weeks and femur marrow contents, spleen cells and blood cells were obtained at necropsy. To assess human cell engraftment, cells were labeled with anti-mouse CD45 and anti-human CD45 and CD33 antibody and analyzed by flow cytometry. To assess engraftment of malignant BCR-ABL expressing cells, BM cells obtained were evaluated for BCR-ABL mRNA levels by QPCR39.
In vivo treatment of normal and CML mice
BM cells (CD45.2) were obtained from SCLtTA/BCR-ABL mice at 4 weeks after induction of BCR-ABL expression by tetracycline withdrawal and were then transplanted by tail vein injection (106 cells/mouse) into irradiated (900 cGy) recipient mice (CD45.1). Blood samples were obtained 4 weeks after transplantation to confirm development of neutrophilic leukocytosis. Mice were treated with scrRNA (5 mg/kg, 4 times a week by vein injection), CpG-miR-126 inhibitor (inhibitor, 5 mg/kg, 4 times a week by vein injection), scrRNA + NIL (50 mg/kg, daily by oral gavage), or inhibitor + NIL for 21 days. After 3 weeks of treatment, mice were euthanized and BM cells from the right femur and spleen cells were analyzed for CML cell output. BM cells from the left femur of the treated mice were pooled and 4×106, 2×106, 1×106 and 5×105 cells/mouse (6 mice/dose × 4 doses × 4 conditions=96 mice) were transplanted into irradiated (900 cGy) recipient mice (CD45.1). WBC counts and CML cell engraftment were monitored every 4 weeks. The mice were euthanized at 16 weeks, followed by assessment of donor CML cell engraftment in PB, BM and spleen. The fraction of mice showing evidence of CML development at 16 weeks after secondary transplantation was determined and the frequency of LICs was calculated using L-Calc software. Another cohort of mice was followed for survival up to 60 days after discontinuation of treatment. To determine the in vivo effect of the CpG-miR-126 inhibitor on normal hematopoiesis, normal mice were treated with scrRNA or inhibitor for 3 weeks, followed by assessment of WBC, RBC, PLT counts in PB and BM subpopulations in BM.
Isolation of cells from different marrow regions
Tibias and femurs were excised from 8–12 week old mice. After removing the muscle and connective tissue, the bones were flushed 5 times using a 23-gauge needle and 3 ml of cold IMDM medium and collected as central marrow. The marrow-depleted bones were crushed gently with a mortar and pestle in cold IMDM medium and the bone fragments were incubated at 37 °C with 3ml of 3mg/ml collagenase I (Sigma) and gently agitated for 45 min. The digested bones were then filtered through a 40 μm strainer (BD Bioscience) and collected as endosteal marrow.
Flow cytometry analyses
Human Lin−CD34+CD38+ committed progenitors, Lin−CD34+CD38− and Lin−CD34+CD38−CD90− primitive progenitors, and Lin−CD34+CD38−CD90+ stem cells were obtained by flow cytometry sorting. The following human antibodies were used: human biotinylated lineage antibodies against CD2 (clone RPA-2.10, cat 555325, BD), CD7(124-1D1, 13-0079-80, ebioscience), CD10(CB-CALLA, 15259439, Thermo Fisher), CD11b (C67F154, 13019682, Thermo Fisher), CD19 (eBio1D3 (1D3), 13-0193-82, Thermo Fisher), CD33 (HIM3-4, MA1-19522, Thermo Fisher), CD235a (HIR2 (GA-R2), 13-9987-82, Thermo Fisher); human antibodies against CD34 (PE-Cy7, 581, 560710; FITC, 581, 555821; APC, 581, 555824; all from BD), CD38 (PE, HIT2, 560981; APC, HIT2, 555462, both from BD), CD90 (PerCP-Cy5.5, eBio5E10 (5E10), 45-0909-42, Thermo Fisher), CD33 (PE, P67.6, 347787, BD), CD45 (FITC, 2D1,11-9459-42; PerCP-Cy5.5, 2D1,45-9459-42, Thermo Fisher), CD31 (PE, 390, 50-103-20, Thermo Fisher), Ki-67 (PE, B56, 556027; FITC, B56, 556026, BD), CD63 (PE-Cy7, H5C6, 561982, BD), CD9 (FITC, H19a, 312104, Biolegend), and CD81 (APC, JS81, 561958, BD). Mouse cells were obtained from PB, BM (from both tibias and femurs), or spleen. For analysis of stem and progenitor cells, c-kit+ cells were selected using anti-mouse CD117 microbeads or Lin− cells were selected using Lineage depletion microbeads (both from Miltenyi Biotec, San Diego, CA). The following mouse antibodies were used: mouse biotinylated lineage antibodies (all from ebioscience) against: CD3 (clone 17A2, cat 13-0031-85), CD4 (GK1.5, 13-0041-85), CD8 (53-6.7, 13-0083-85), B220 (RA3-6B2, 13-0452-85), CD19 (eBio1D3 (1D3), 13-0193-85), IgM (eB121-15F9, 13-5790-85), Gr-1 (RB6-8C5, 13-5931-85), CD11b (M1/70, 13-0112-85), NK1.1 (PK136, 13-5941-85), Ter119 (clone TER-119, cat 13-5921-85), Flt3 (A2F10, 13-1351-85); mouse antibodies against Flt3 (PE, A2F10, 12-1351-82, ebioscience), Sca-1 (PE-Cy7, D7, 25-5981-82, ebioscience), CD117 (APC-eflu780, ACK2, 47-1172-82, ebioscience), CD16/32 (PE-Cy7, 2.4G2, 560829, BD), CD34 (Alexa Fluor 647, RAM34, 560230, BD), CD150 (PerCP-Cy5.5, TC15-12F12.2, 115922, Biolegend), CD48 (APC, HM48-1,17-0481-82, ebioscience; Pacific blue, HM48-1, 103418, Biolegend), CD45.1(PE-Cy7, A20, 25-0453-82; PerCP-Cy5.5, A20, 45-0453-80, both from ebioscience), CD45.2 (FITC, 104, 11-0454-85; eFluor450, 104,48-0454-82, both from ebioscience), CD45 (PE-Cy7, 30-F11, 25-0451-82, ebioscience), Ter119 (APC-eflu780, TER-119, 50-162-15, ebioscience), CD31 (APC, 390, 17-0311-82; PE, 390, 12-0311-82, both from ebioscience), CD166 (PE, FAB1172P, R&D Systems). Other antibodies include anti-streptavidin (PE, 12-4317-87; FITC, 11-4317-87; PerCP-Cy5.5, 45-4317-80, ebioscience) and Annexin V (PE, 559763, BD). Myeloid progenitors were identified as Lin−Sca-1−c-Kit+CD34+Fc≥RII/IIIlo(CMP), Lin−Sca-1−c-Kit+CD34+Fc≥RII/IIIhi (GMP), or Lin−Sca-1−c-Kit+CD34−Fc≥RII/IIIlo(MEP)38,40. Stem and progenitor populations were identified as LSK cells (Lin−Sca-1hic-Kithi) and long-term hematopoietic stem cells (LT-HSCs; LSK Flt3−CD150+CD48−)38, 41. Endothelial cells were identified as CD45−Ter119−CD31+. All analyses were performed on a LSRII flow cytometer (BD Biosciences) and sorting was performed on ARIAIII or ARIA SORP instruments (BD Biosciences).
Cell Culture
Human HPCs (Lin−CD34+ ) and HSCs (Lin−CD34+CD38−) were cultured in Stemspan serum-free medium II (SFEM II, StemCell Technologies), supplemented with low concentrations of growth factors (GFs) similar to those present in long-term BM culture stroma-conditioned medium [granulocyte-macrophage colony-stimulating factor (GM-CSF) 200pg/mL, leukemia inhibitory factor (LIF) 50pg/mL, granulocyte colony-stimulating factor (G-CSF) 1ng/mL, stem cell factor (SCF) 200pg/mL, macrophage-inflammatory protein-1α (MIP-1α) 200pg/mL, and interleukin-6 (IL-6) 1ng/mL]42. Mouse BM LT-HSCs were cultured in SFEM II supplemented with 10ng/ml SCF and 10ng/ml TPO. Human Umbilical Vein Endothelial Cells (HUVEC) and K562 cells were recently purchased from Lonza and ATCC respectively. We confirmed that HUVEC cells are human CD31 positive by flow cytometry and that K562 cells are BCR-ABL positive by QPCR. These two cell lines were tested for mycoplasma contamination and both were negative. HUVEC cells were cultured in complete EGM-2 medium (Lonza) and mouse BM ECs were cultured in complete mouse endothelial cell medium (Cell biologics). K562 cells were cultured in RPMI-1640 medium supplemented with 10% FBS (Life Technologies). All cells were cultured at 37°C with 5% CO2 and high humidity.
Differential Ultracentrifugation
Conditioned medium was made by centrifuging the culture medium containing FBS for 8–10 h at 100,000 × g, 4°C to remove the FBS-derived EV contamination and the supernatant was collected (leaving the FBS-derived EVs/protein pellet). The cells were cultured with the conditioned medium for 2 days. EVs were isolated by differential ultracentrifugation43–45. Briefly, the supernatant was collected and centrifuged for 5 min at 500 × g, 4°C ; the supernatant was then collected and centrifuged for 10 min at 2,000 × g, 4°C ; and the supernatant was then collected and centrifuged for 15 min at 10,000 × g, 4°C. It was important that none of the pellets consisting of cells and cell debris was collected. The supernatant was then transferred to a new tube and centrifuged for 60 min at 100,000 × g, 4°C. The EV pellet was washed once with PBS and centrifuged again for 60 min at 100,000 × g, 4°C to obtain the final EV pellet. The EV pellet was dissolved in 50ul of PBS and stored at 4°C for up to one week.
Nanoparticle tracking analysis
NanoSight measurements were carried out in 0.2 μm filtered PBS. The concentration and size distribution profile of the particles isolated by differential ultracentrifugation were evaluated using a NanoSight NS300 instrument (Malvern, Worcestershire, UK) and NTA 3.2 software. Videos were recorded at camera level 15. Samples were diluted 1:100 in PBS to achieve a measured particle concentration of 5–15×108/mL in accordance with the manufacturer’s recommendations. For each sample, three 60 sec videos were recorded and analyzed in the batch processing mode.
Electron Microscopy (EM)
Specimens at an optimal concentration were placed onto a 300-mesh carbon/formvar coated grids and allowed to absorb to the formvar for a minimum of 1 min. Grids were rinsed with ddH2O and stained for contrast using 1% uranyl acetate. The samples were viewed with an FEI Tecnai T12 transmission electron microscope at 120keV and images were taken with a Gatan Ultrascan 2K CCD camera.
Cytofluorimetric analysis
A fraction was isolated from HUVEC-derived EVs using magnetic beads coated with anti-CD63 antibody (10606D, ThermoFisher). Briefly, EVs isolated from HUVEC by differential ultracentrifugation were incubated with magnetic beads coated with anti-CD63 antibody overnight. The bead-bound CD63+ EVs were selected using a DynaMag-5 magnetic separator (12303D, ThermoFisher), stained with PE-Cy7-anti-human CD63, FITC-anti-human CD9 and APC-anti-human CD81 (BD) antibodies and then analyzed by flow cytometry.
Lentiviral or retroviral transduction of human and mouse cells
GFP-expressing miRZip anti-miR-126-3p (126 KD, CS940MZ-1, a custom order from System Biosciences, with EF1a promoter for anti-miR-126-3p), miR-126 precursor (126 OE, CS940MR-1, a custom order from System Biosciences, with EF1a promoter for miRNA and PGK promoter for GFP-T2A-Puro expression) and control [MZIP000-PA-1, miRZip negative control; CD813A-1, pCDH-EF1-MCS-(PGK-GFP-T2A-Puro); both from System Biosciences] lentiviruses were produced and used for transduction of human and/or mouse HSCs, HUVECs and mouse ECs. Briefly, human HPCs or HSCs were cultured overnight in SFEM II supplemented with IL-3 (25ng/ml), IL-6 (10ng/ml), SCF (50ng/ml), TPO (100ng/ml) and Flt-3 ligand (100ng/ml). Mouse BM LT-HSCs were cultured overnight in SFEM II supplemented with mouse SCF (10ng/ml) and mouse TPO (10ng/ml). The next day, cells were resuspended in SFEM II and lentiviral supernatant [multiplicity of infection (MOI) = 10–20], supplemented with the above GFs and 1xTransDux virus transduction reagent (System Biosciences), and centrifuged at 1500 g for 90 minutes for transduction by spinoculation. We observed 30–60% of GFP+ cells in human HSCs and 90–100% in mouse LT-HSCs transduced with 126 KD lentivirus (MOI = 20), and 10–30% of GFP+ cells in human HSCs and 30–50% in mouse LT-HSCs transduced with 126 OE lentivirus (MOI = 10) at 48 h. HUVECs and mouse BM ECs were exposed to 126 KD or control lentiviral supernatant (MOI = 10) with 1xTransDux virus transduction reagent, and 100% of GFP+ cells were detected at 48 h. Normal BM LSK cells were transduced with BCR-ABL or control retroviral supernatant (MOI=5) with polybrene (5 μg/ml, American Bioanalytical) by spinoculation, and 20–30% of GFP+ cells were detected at 48 h. GFP+ cells from the samples with low transduction efficiency (<80%) were selected by flow cytometry at 48h for further studies.
Apoptosis, cell cycle, cell growth and colony-forming cell assays
After transduction performed as above, GFP+ cells selected at 48 h were exposed to NIL (2 μM and 5 μM, Novartis) for another 72h, and analyzed using assays for cell growth, apoptosis, cell cycle, CFC and CFC replating. Human HSCs and mouse LT-HSCs were also treated with CpG-miR-126 inhibitor or CpG-scrRNA (500nM), with or without NIL (5 μM), for 72h and analyzed for cell growth, apoptosis, cell cycle and CFC. Human CML HSCs co-cultured with control or miR-126 KD HUVECs and mouse CML LT-HSCs co-cultured with control or miR-126 KD ECs for 96h, with or without NIL (5 μM), were also analyzed for cell growth, apoptosis, cell cycle and CFC. Cell growth was measured by Lumino Glo (Promega). Apoptosis was measured by labeling cells with Annexin V-PE or FITC or APC and 4, 6-diamidino-2-phenylindole (DAPI) (all from BD-PharMingen, San Diego, CA) and analyzed by flow cytometry. Cell cycle was analyzed by Ki-67-Alexa Fluor 647 (B56, BD) and DAPI labeling based on the manufacturer’s protocol. Cells were also exposed to EdU (C10640, Invitrogen) for 2h and EdU staining was analyzed according to the manufacturer’s protocol. Cell proliferation was also measured by CFSE staining (Molecular probes) based on the manufacturer’s protocol. For CFC, Lin−CD34+CD38− cells cultured with or without NIL (5 μM) were plated in methylcellulose progenitor culture, and burst-forming unit-erythroid and colony-forming unit-granulocyte and macrophage cells were counted after 14 days. Colony replating assays were performed by collecting and pooling colonies from primary CFC assays and plating 10,000 cells in secondary CFC assays.
QPCR analysis
To measure the miRNA and mRNA expression, total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Valencia, CA). For miRNA expression, reverse transcription using MultiScribe™ Reverse Transcriptase and Q-PCR analysis using Taqman assays (Applied Biosystems) were performed according to the manufacturer’s protocol. RNU44 and snoRNA234 was used as internal controls for human and mouse miRNA respectively. For mRNA expression, first-strand cDNA was synthesized using the SuperScript III First-Strand Kit and then QPCR was performed using TaqMan Gene Expression Assays (Thermo Fisher). BCR-ABL expression in human and mouse samples were measured with primer and probe sequences for BCR-ABL (B3A2 or B2A2), as previously described46. Results are presented as log2-transformed ratio according to the 2−ΔCt method (ΔCt=Ct of target −Ct of reference).
| Gene name | Assay ID |
|---|---|
| miR-126 | 2228 |
| RNU44 | 111094 |
| RNU48 | 1006 |
| snoRNA234 | 1234 |
| Pri-miR-126 | Hs03303230_pri |
| Pre-miR-126 | Hs04273250_s1 |
| PIK3R2 | Hs00178181_m1 |
| SPRED1 | Hs01084559_m1 |
| BCL-2 | Hs00608023_m1 |
| β2M | Hs00187842_m1 |
| Pik3r2 | Mm00435694_m1 |
| Spred1 | Mm01277511_m1 |
| β2m | Mm00437762_m1 |
miRNA labeling and analysis
K562, HUVEC, normal and CML CD34+CD38− cells were cultured and incubated with miR126 SmartFlare RNA probe (EMD Millipore) for 16h. To ensure that the cell types, including K562, HUVEC and primary CML cells, were able to effectively endocytose the SmartFlare probes, we examined the uptake of probes in these cells using SmartFlare uptake control, scramble control and housekeeping 18S control (according to the manufacturer’s guidelines). Cells were then washed in 1× phosphate buffered saline (PBS) and fixed in 4% paraformaldehyde for 3 min. Nuclei were counterstained with DAPI and the images were analyzed using a confocal microscope (Carl Zeiss).
Immunofluorescence
K562 cells were collected and washed in PBS followed by spinning down onto slides using the CytoSpin4 Cytocentrifuge (600 rpm, 10 min). The cells were then fixed in 4% paraformaldehyde for 15 min and permeabilized in 0.5% Triton X-100 for 15 min. Non-specific epitopes were blocked with 5% bovine serum albumin (BSA) in PBS for 30 min. SPRED1 and RAN were visualized using anti-SPRED1 (ab64740, Abcam) and anti-RAN (ab4781, Abcam) antibodies and secondary anti-mouse/rabbit-Alexa 594/488 goat antibodies (Molecular Probes). 3D cell images were acquired using a Zeiss confocal Laser Scanning Microscope (Carl Zeiss). Nuclei were counterstained with DAPI.
Western Blotting and Immunoprecipitation analysis
Normal and CML CD34+ cells with or without miR-126 KD were lysed in buffer containing 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 1mM PMSF, 50mM NaF, 1mM Na3VO4, and a protease inhibitor cocktail (all from Sigma Diagnostics). Proteins were resolved on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nitrocellulose membrane. Membranes were sequentially reprobed with primary and secondary antibodies. Primary antibodies included anti-SPRED1 antibody (M23-P2G3, #ab64740, Abcam), anti-RAN antibody (C-20, #SC-1156, Santa Cruz), anti-Exportin5 antibody (D7W6W, #12565, Cell Signaling), anti-PARP antibody (#9542, Cell Signaling), anti-RCC1 antibody (F-2, #SC-376049, Santa Cruz), anti-Actin antibody (C-4, #SC-47778, Santa Cruz), anti-phospho-Tyrosine antibody (4G10, #05-321, Millipore), anti-BCL-2 antibody (124, #15071, Cell Signaling), anti-phospho-ERK (#9101, Cell Signaling), anti-normal mouse IgG (#SC-2025, Santa Cruz), anti-normal rabbit IgG (#SC-2027, Santa Cruz), CD63 (10628D, ThermoFisher), TSG101(SAB2702167, Sigma), HSP90 (2D12, Enzo Life Sciences), Cytochrome C (sc-13156, Santa Cruz). Horseradish peroxidase-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories (Westgrove, PA). Antibody detection was performed using the Superfemto kit (Pierce Biotechnology, Rockford, IL). Protein levels were determined by densitometry using Image-Quant software (Amersham Pharmacia Biotech, Piscataway, NJ).
For fractionation, the cells were collected and washed in PBS following fractionation into nuclear and cytoplasmic fractions using a subcellular fractionation kit (Thermo Fisher). Briefly, the cells were vigorously vortex in cytoplasmic extraction reagents and subsequently centrifuged to isolate the soluble cytoplasmic fraction. The remaining insoluble fraction, which contains nuclei, was suspended in nuclear extraction reagent and centrifuged to collect the nuclear fraction.
For IP, the cells were washed and harvested in ice-cold PBS and subsequently lysed in buffer containing 1mM PMSF and 10mM protease inhibitor cocktail. 500μg of cell lysate was incubated with the indicated antibody overnight at 4°C. 30μl of Protein A/G agarose beads (Calbiochem) were added and the mixture was inverted for 2h at 4°C. For immunoblotting, immunoprecipitated complex or 30μg of each cell lysate were separated on NuPAGE 4–12% gradient gels (Invitrogen) and immunocomplexes were visualized with enhanced chemiluminescence reagent (Thermo Scientific).
Kinase assay
One microgram of purified SPRED1 protein (Novus) was incubated with recombinant active c-Abl (Sigma) and 10μCi of [≥-32P]ATP (PerkinElmer Life Sciences) in 50μL of kinase buffer. Reactions were incubated at 30°C for 1 h. Protein was separated on NuPAGE 4–12% gradient gels (Invitrogen). The gel was then dried and phosphorylated protein was visualized by autoradiography. SPRED1 phosphorylated by c-Abl kinase was visualized on the autoradiogram.
Northern blot
Northern blot was performed using a Northern blot-based protocol (LED) for micro-RNA detection using digoxigenin (DIG)-labeled miR-126 probes containing locked nucleic acids (LNA) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide for cross-linking the RNA to the membrane. Briefly, total RNA was prepared using Trizol reagent (Life Technologies). 20 μg of total RNA was separated on a 15% TBE-Urea gel (Life Technologies) and transferred onto positively charged nylon membrane (Roche). The blots were hybridized with LNA miR-126 probes for 12h. After washing twice with 2×SSC and 0.1×SSC (containing 0.1% SDS), the blots were immunobloted with anti-DIG antibody (Roche) and exposed.
Small RNA deep sequencing using Illumina HiSeq2500
LSK cells from non-induced and induced CML mice were sorted and total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Valencia, CA). Small RNA sequencing was performed using Illumina HiSeq2500 at the COH Integrative Genomics Core following the manufacturer’s sample preparation protocol (TruSeq Small RNA Sample Prep kit, Illumina, Inc.) with some modifications. Briefly, 250ng of total RNA was used for smRNA sequencing library construction. Total RNA was ligated to the modified 3′ Adapter (5′ TCTGGAATTCTCGGGTGCCAAGGAACTCC) with T4 RNA Ligase 2, truncated (NEB, M0242L) for 1 h at 22°C. The unligated free 3′ adaptors were blocked by annealing with RT primer (5′ GGAGTTCCTTGGCACCCGAGAATTCCA) at 75°C for 5min, 37°C for 30 min and 25°C for 15 min. The product subsequently was ligated to the modified 5′ adaptor (5′GUUCAGAGUUCUACAGUCCGACGAUCNNN) with T4 RNA ligase1 (NEB, M0204L) for 1 h at 20°C. The constructed smRNA library was reverse-transcribed, then subjected to a PCR amplification for 12 cycles, using barcoded index primers GX1(CAAGCAGAA GACGGCATACGAGATNNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC) and GX2 (AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCG A); followed by 6% TBE PAGE gel purification with size selection (for targeted smRNAs of 17–35 nt). The final libraries were sequenced using the Illumina HiSeq2500 platform in the single read mode of 51 cycle of read1 and 7 cycles of index read. Real-time analysis (RTA) 2.2.38 software was used to process the image analysis and base calling.
Oligonucleotide design and synthesis
The partially phosphothioated oligodeoxyribonucleotide(ODN) and miR-126 inhibitor or scrRNA was linked using 5 units of C3 carbon chain linker, (CH2)3 (indicated by x). The constructs were also conjugated with Cy3 to track the internalization in cells by flow cytometry. The sequences were as follows:
-
CpG-miR-126 inhibitor
5′ G*G*T GCA TCG ATG CAGG*G*G* G*G xxxxx mCmGmC mAmUmU mAmUmU mAmCmU mCmAmC mGmGmU mAmCmG mA -3′
-
CpG-scrRNA
5′ G*G*T GCA TCG ATG CAGG*G*G* G*G xxxxx mGmUmA mGmAmA mCmCmG mUmAmC mUmCmG mUmCmA mCmUmU mA 3′
‘*’ – phosphorothioation. One none bridging oxygen on phosphate replaced with sulfur.
‘m’ – 2′-O-methyl analogue of the nucleotide
Transferrin or anti-CD45.2 antibody conjugated nanoparticle preparation
Previously we developed a transferrin(TF)-targeted neutral NP delivery system47,48. Briefly, positively charged polyethylenimine and negatively charged miR-126 inhibitor-Cy3 or scrRNA-Cy3 form a polyplex core. This core was then loaded into pre-made anionic liposomal NPs to form lipopolyplex NPs. The formulation consisted of 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dimyristoyl-sn-glycerol, methoxypolyethylene glycol (DMG-PEG) and linoleic acid. TF or anti-human CD45 antibody conjugated with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (DSPEPEG2000 maleimide) was then post-inserted to the surface of lipopolyplex nanoparticles (TF-NP and Ab-NP). The molar ratio of lipids to TF was 2000 as previously described47,48 and the molar ratio of lipids to anti-CD45 antibody was optimized to 10,000.
Statistics
Comparison between groups was performed by two-tailed, paired or unpaired Student’s t-test. The log-rank test was used to assess significant differences between survival curves. All statistical analyses were performed using Prism version 6.0 software (GraphPad Software). Sample sizes chosen are indicated in the individual figure legends and were not based on formal power calculations to detect prespecified effect sizes. All of the in vitro experiments were performed 3–6 times using biologically independent samples; the in vivo experiments were performed using 6–16 mice in each group. P values ≤0.05 were considered significant. Results shown represent mean ± SEM. *p ≤ 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Data availability
miRNA deep sequencing data produced in our laboratory and analysed in this study are available at the Gene Expression Omnibus (GEO) repository of the National Center for Biotechnology Information, under the accession number GSE107431. Supplementary Figs 1–7 and Source Data for Fig 3 and Supplementary Fig 3c and 5h, k, m, p, shown in Supplementary Figs 8–10, are provided with the online version of this paper. All other datasets generated during this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
This work was supported in part by National Cancer Institute grants: CA205247 (YHK), CA102031 (GM), CA201184 (GM), CA213131 (MK), CA180861 (GM), CA158350 (GM), CA184411 (LL); the Gehr Family Foundation and George Hoag Family Foundation; Cancer Research UK programme grant 11074/A11008; and The Howat Foundation. We acknowledge the support of the Animal Resources Center, Analytical Cytometry, Pathology (Liquid Tumor), Bioinformatics, Electron Microscopy, Light Microscopy, Integrative Genomics and DNA/RNA Cores at City of Hope Comprehensive Cancer Center supported by the National Cancer Institute of the National Institutes of Health under award number P30CA33572. We are grateful to COH Comprehensive Cancer Center, Glasgow Experimental Cancer Medicine Centre and SPIRIT trials, together with the patients and their physicians for providing primary patient material for this study.
Footnotes
Conflict of interest: none
Author contributions
BZ and L.X.T.N designed and conducted experiments, analyzed data and wrote the manuscript; LL, D.Z, BK, HW, FP, Y.-L.S, CB, HW, TM and ET conducted experiments; AL, D.S.S, HA and A.S.S provided samples and reviewed the patients’ data; PS and MK designed the CpG-miR-126 inhibitor, reviewed data and the manuscript; LH, C.-C.C, AD, VP, Y.-C.Y, DP, and NC analyzed data; C.J.K provided the miR-126(c-KO) mouse model; RB provided the B6 SCLtTA/BCR-ABL CML mouse model; MC, TLH and SJF provided patient samples, designed experiments and reviewed the manuscript; MK and Y.-H.K designed experiments, analyzed data and reviewed manuscript; GM designed experiments, analyzed data, wrote manuscript and provided administrative support.
References
- 1.Sawyers CL. Chronic myeloid leukemia. The New England journal of medicine. 1999;340:1330–1340. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- 2.Zhang B, et al. Altered microenvironmental regulation of leukemic and normal stem cells in chronic myelogenous leukemia. Cancer cell. 2012;21:577–592. doi: 10.1016/j.ccr.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu S, et al. Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood. 2011;118:5565–5572. doi: 10.1182/blood-2010-12-327437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lechman ER, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell stem cell. 2012;11:799–811. doi: 10.1016/j.stem.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Leeuw DC, et al. Attenuation of microRNA-126 expression that drives CD34+38− stem/progenitor cells in acute myeloid leukemia leads to tumor eradication. Cancer research. 2014;74:2094–2105. doi: 10.1158/0008-5472.CAN-13-1733. [DOI] [PubMed] [Google Scholar]
- 6.Dorrance AM, et al. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia. 2015;29:2143–2153. doi: 10.1038/leu.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, et al. Overexpression and knockout of miR-126 both promote leukemogenesis. Blood. 2015;126:2005–2015. doi: 10.1182/blood-2015-04-639062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lechman ER, et al. miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer cell. 2016;29:602–606. doi: 10.1016/j.ccell.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhnert F, et al. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 10.Itkin T, et al. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature. 2016;532:323–328. doi: 10.1038/nature17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lechman ER, et al. miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer cell. 2016;29:602–606. doi: 10.1016/j.ccell.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nucera S, et al. miRNA-126 Orchestrates an Oncogenic Program in B Cell Precursor Acute Lymphoblastic Leukemia. Cancer cell. 2016;29:905–921. doi: 10.1016/j.ccell.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Koschmieder S, et al. Inducible chronic phase of myeloid leukemia with expansion of hematopoietic stem cells in a transgenic model of BCR-ABL leukemogenesis. Blood. 2005;105:324–334. doi: 10.1182/blood-2003-12-4369. [DOI] [PubMed] [Google Scholar]
- 14.Fish JE, et al. miR-126 regulates angiogenic signaling and vascular integrity. Developmental cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Developmental cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintanar-Audelo M, Yusoff P, Sinniah S, Chandramouli S, Guy GR. Sprouty-related Ena/vasodilator-stimulated phosphoprotein homology 1-domain-containing protein (SPRED1), a tyrosine-protein phosphatase non-receptor type 11 (SHP2) substrate in the Ras/extracellular signal-regulated kinase (ERK) pathway. The Journal of biological chemistry. 2011;286:23102–23112. doi: 10.1074/jbc.M110.212662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circulation research. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 19.Welten SM, Goossens EA, Quax PH, Nossent AY. The multifactorial nature of microRNAs in vascular remodelling. Cardiovascular research. 2016;110:6–22. doi: 10.1093/cvr/cvw039. [DOI] [PubMed] [Google Scholar]
- 20.Chitteti BR, et al. CD166 regulates human and murine hematopoietic stem cells and the hematopoietic niche. Blood. 2014;124:519–529. doi: 10.1182/blood-2014-03-565721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houlihan DD, et al. Isolation of mouse mesenchymal stem cells on the basis of expression of Sca-1 and PDGFR-alpha. Nature protocols. 2012;7:2103–2111. doi: 10.1038/nprot.2012.125. [DOI] [PubMed] [Google Scholar]
- 22.Van Deun J, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. Journal of extracellular vesicles. 2014;3 doi: 10.3402/jev.v3.24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of extracellular vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu S, Holtz M, Gupta M, Bhatia R. BCR/ABL kinase inhibition by imatinib mesylate enhances MAP kinase activity in chronic myelogenous leukemia CD34+ cells. Blood. 2004;103:3167–3174. doi: 10.1182/blood-2003-04-1271. [DOI] [PubMed] [Google Scholar]
- 25.Galante JM, Mortenson MM, Bowles TL, Virudachalam S, Bold RJ. ERK/BCL-2 pathway in the resistance of pancreatic cancer to anoikis. The Journal of surgical research. 2009;152:18–25. doi: 10.1016/j.jss.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 26.Kunisaki Y, et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holyoake TL, Vetrie D. The chronic myeloid leukemia stem cell: stemming the tide of persistence. Blood. 2017;129:1595–1606. doi: 10.1182/blood-2016-09-696013. [DOI] [PubMed] [Google Scholar]
- 28.Boucher MJ, et al. MEK/ERK signaling pathway regulates the expression of Bcl-2, Bcl-X(L), and Mcl-1 and promotes survival of human pancreatic cancer cells. Journal of cellular biochemistry. 2000;79:355–369. [PubMed] [Google Scholar]
- 29.Nechaev S, et al. Intracellular processing of immunostimulatory CpG-siRNA: Toll-like receptor 9 facilitates siRNA dicing and endosomal escape. Journal of controlled release : official journal of the Controlled Release Society. 2013;170:307–315. doi: 10.1016/j.jconrel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q, et al. Serum-resistant CpG-STAT3 decoy for targeting survival and immune checkpoint signaling in acute myeloid leukemia. Blood. 2016;127:1687–1700. doi: 10.1182/blood-2015-08-665604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ewald SE, et al. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura N, et al. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature. 2014;509:240–244. doi: 10.1038/nature13133. [DOI] [PubMed] [Google Scholar]
- 33.Martin-Armas M, et al. Toll-like receptor 9 (TLR9) is present in murine liver sinusoidal endothelial cells (LSECs) and mediates the effect of CpG-oligonucleotides. Journal of hepatology. 2006;44:939–946. doi: 10.1016/j.jhep.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 34.Tamura Y, et al. Scavenger receptor expressed by endothelial cells I (SREC-I) mediates the uptake of acetylated low density lipoproteins by macrophages stimulated with lipopolysaccharide. The Journal of biological chemistry. 2004;279:30938–30944. doi: 10.1074/jbc.M313088200. [DOI] [PubMed] [Google Scholar]
- 35.Yeh YC, Hwang GY, Liu IP, Yang VC. Identification and expression of scavenger receptor SR-BI in endothelial cells and smooth muscle cells of rat aorta in vitro and in vivo. Atherosclerosis. 2002;161:95–103. doi: 10.1016/s0021-9150(01)00642-6. [DOI] [PubMed] [Google Scholar]
- 36.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nature immunology. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 37.Koschmieder S, et al. Inducible chronic phase of myeloid leukemia with expansion of hematopoietic stem cells in a transgenic model of BCR-ABL leukemogenesis. Blood. 2005;105:324–334. doi: 10.1182/blood-2003-12-4369. [DOI] [PubMed] [Google Scholar]
- 38.Zhang B, et al. Altered microenvironmental regulation of leukemic and normal stem cells in chronic myelogenous leukemia. Cancer cell. 2012;21:577–592. doi: 10.1016/j.ccr.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Branford S, Hughes TP, Rudzki Z. Monitoring chronic myeloid leukaemia therapy by real-time quantitative PCR in blood is a reliable alternative to bone marrow cytogenetics. British journal of haematology. 1999;107:587–599. doi: 10.1046/j.1365-2141.1999.01749.x. [DOI] [PubMed] [Google Scholar]
- 40.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 41.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 42.Bhatia R, McGlave PB, Dewald GW, Blazar BR, Verfaillie CM. Abnormal function of the bone marrow microenvironment in chronic myelogenous leukemia: role of malignant stromal macrophages. Blood. 1995;85:3636–3645. [PubMed] [Google Scholar]
- 43.Van Deun J, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. Journal of extracellular vesicles. 2014;3 doi: 10.3402/jev.v3.24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of extracellular vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lotvall J, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. Journal of extracellular vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Branford S, et al. Real-time quantitative PCR analysis can be used as a primary screen to identify patients with CML treated with imatinib who have BCR-ABL kinase domain mutations. Blood. 2004;104:2926–2932. doi: 10.1182/blood-2004-03-1134. [DOI] [PubMed] [Google Scholar]
- 47.Dorrance AM, et al. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia. 2015;29:2143–2153. doi: 10.1038/leu.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang X, et al. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: a novel therapeutic strategy in acute myeloid leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:2355–2367. doi: 10.1158/1078-0432.CCR-12-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
miRNA deep sequencing data produced in our laboratory and analysed in this study are available at the Gene Expression Omnibus (GEO) repository of the National Center for Biotechnology Information, under the accession number GSE107431. Supplementary Figs 1–7 and Source Data for Fig 3 and Supplementary Fig 3c and 5h, k, m, p, shown in Supplementary Figs 8–10, are provided with the online version of this paper. All other datasets generated during this study are available from the corresponding author on reasonable request.