Abstract
The CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) adaptive immune systems use small guide RNAs, the CRISPR RNAs (crRNAs), to mark foreign genetic material, e.g. viral nucleic acids, for degradation. Archaea and bacteria encode a large variety of Cas proteins that bind crRNA molecules and build active ribonucleoprotein surveillance complexes. The evolution of CRISPR-Cas systems has resulted in a diversification of cas genes and a classification of the systems into three types and additional subtypes characterized by distinct surveillance and interfering complexes. Recent crystallographic and biochemical advances have revealed detailed insights into the assembly and DNA/RNA targeting mechanisms of the various complexes. Here, we review our knowledge on the molecular mechanism involved in the DNA and RNA interference stages of type I (Cascade: CRISPR-associated complex for antiviral defense), type II (Cas9) and type III (Csm, Cmr) CRISPR-Cas systems. We further highlight recently reported structural and mechanistic themes shared among these systems.
Keywords: viruses, CRISPR, DNA interference, guide crRNAs, Cascade, Cas9, tracrRNA, ribonucleoprotein complexes
This review details and compares the assembly and the DNA/RNA targeting mechanisms of the various surveillance complexes of prokaryotic CRISPR-Cas immune systems.
INTRODUCTION
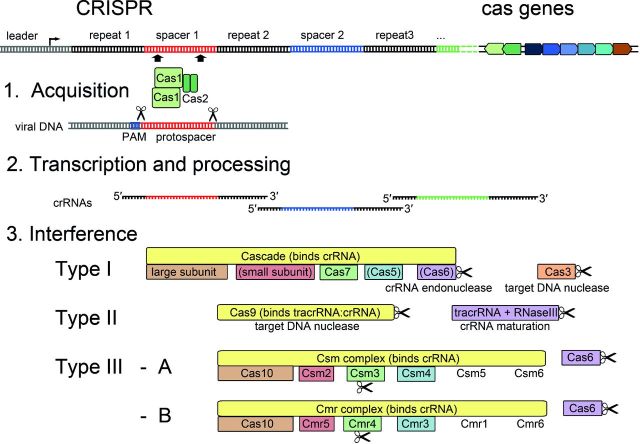
Viruses are the most abundant biological entities on Earth and have been found to quickly adapt to their respective hosts in any environmental niche (Wasik and Turner 2013). As a consequence, all organisms have evolved a wide range of specific antiviral measures that are classified as either innate or adaptive immunity systems. Prokaryotes protect themselves from viruses by using distinct innate immune systems, e.g. restriction-modification systems, the modification of receptors or abortive infection (Samson et al.2013). In addition, RNA-mediated adaptive immune systems, termed CRISPR-Cas are prevalently distributed in bacterial and archaeal genomes (Barrangou et al.2007). CRISPR-Cas has largely evolved into three types (type I, II, III) of systems that are characterized by several conserved features and by a common functionality. The hallmarks of these systems are CRISPR loci that consist of a series of short identical repeat sequences separated by spacer sequences that are mostly originated from mobile genetic elements including viruses (Mojica et al.2005; Pourcel, Salvignol and Vergnaud 2005; Grissa, Vergnaud and Pourcel 2009) (Fig. 1). In most cases, CRISPR arrays are genome encoded, but they can also be located on plasmids or megaplasmids (Mojica et al.2005; Godde and Bickerton 2006). Additionally, cas genes are often located in close proximity to the CRISPR loci and the encoded Cas proteins fulfill essential roles in three defined stages of CRISPR-Cas-mediated immunization and the protection of the prokaryotic cell (Jansen et al.2002; Haft et al.2005; Makarova et al.2011b). In the first stage of immunity, termed acquisition, a protospacer sequence of DNA from a defective viral infection is recognized by a complex of the universal Cas proteins Cas1–Cas2 and inserted into the host CRISPR array, generating a new spacer as well as a duplication of the repeat in the extended locus (Barrangou et al.2007; Swarts et al.2012; Yosef, Goren and Qimron 2012; Savitskaya et al.2013; Nunez et al.2014, 2015). Protospacer selection in type I and type II relies on short conserved sequences (2–5 bp) that are defined as the protospacer adjacent motif (PAM). PAM sequences are not required for type III systems. Thus, any sequence that flanks correct PAMs has the potential to be integrated into the CRISPR array on the host genome (Mojica et al.2009; Shah et al.2013). In most cases, CRISPR immunity is activated by the transcription of the repeat-spacer array into a long precursor-crRNA (pre-crRNA) that is further processed into short crRNA molecules (Carte et al.2008; Hale et al.2008; Haurwitz et al.2010). The mature crRNAs that contain the acquired viral sequence are then incorporated into CRISPR ribonucleoprotein complexes (crRNP) and guide the sequence-specific degradation of viral DNA or RNA upon a second infection (Brouns et al.2008; Sapranauskas et al.2011; Rouillon et al.2013). In this review, we focus mostly on recent structural and mechanistic insights into the crRNP complexes that promote DNA or RNA interference. The diversification of core Cas components and the recruitment of specific Cas proteins to the complexes have resulted in significant differences in structure and composition of these crRNP assemblies in bacteria and archaea. Yet, several mechanistic principles are uniformly shared.
Figure 1.
CRISPR-Cas systems and conserved stages of CRISPR-Cas activity. The general organization of a CRISPR-Cas locus is indicated. In the first stage of CRISPR-Cas activity—acquisition—the universal proteins Cas1 and Cas2 recognize viral DNA that is flanked by a PAM. The protospacer is excised and integrated as a spacer sequence into the extending CRISPR array. The CRISPR array is transcribed from the leader sequence and processed into mature crRNAs that are incorporated into crRNP surveillance complexes. The Cas protein composition of the complexes is schematically depicted for the three different CRISPR-Cas types. Nucleases are indicated by scissors and proteins proposed to fulfill similar roles are colored accordingly.
CRISPR-CAS COMPLEXITY
In the last decade, the identification and classification of cas genes illustrates the evolution of diversified CRISPR-Cas systems. A first link between CRISPR loci and the accompanying cas genes, cas1-cas4, was uncovered in 2002 (Jansen et al.2002). Comparative analyses of available microbial genomes detected numerous associated cas genes and revealed major differences in their sequence and organization (Makarova et al.2002, 2006; Haft et al.2005). Comprehensive analyses of cas sequences in bacteria and archaea demonstrated their highly dynamic evolution. Recombined cas gene sequences were identified, indicating a frequent horizontal transfer of single genes within a CRISPR-Cas module or even entire cas gene cassettes (Millen et al.2012; Takeuchi et al.2012; Jiang et al.2013). Such directed evolution has resulted in a rather challenging classification as well as functional comparison, and was recently highlighted by the branching of Cas proteins into more than 100 families (Koonin and Makarova 2013). Computational classification studies combined with sequence information and prediction of structural similarities have led to the identification of major crRNP building blocks and the detection of many shared domains among Cas protein families in the different CRISPR-Cas types (Makarova et al.2011a; Koonin and Makarova 2013). These defined building blocks compose the three functional stages of the CRISPR-Cas immunity: (i) spacer insertion, (ii) crRNA processing, (iii) crRNP assembly and target cleavage (Makarova, Wolf and Koonin 2013) (Fig. 1).
DIVERSIFICATION OF CRISPR-CAS INTERFERENCE MECHANISMS
The current nomenclature classifies the CRISPR-Cas systems of bacteria and archaea into three main types (I, II and III) and 11 subtypes (I-A to F, II-A to C, III-A to B) based on phylogenetic and often limited functional studies (Makarova et al.2011b; Chylinski, Le Rhun and Charpentier 2013; Koonin and Makarova 2013). The types are defined by a conserved signature protein (Cas3 in type I, Cas9 in type II and Cas10 in type III) and mainly differ in crRNP assembly and target cleavage mechanisms. All type I crRNP complexes are termed Cascade (CRISPR-associated complex for antiviral defense), while in type II the stand-alone Cas9 nuclease is responsible for target cleavage. Type III systems encode the Csm (III-A) or Cmr (III-B) crRNP complex (Makarova et al.2011b) (Fig. 1). Only Cas1 and Cas2, which are involved in spacer acquisition, are conserved in the majority of CRISPR-Cas systems (Yosef, Goren and Qimron 2012; Makarova, Wolf and Koonin 2013). In the following sections, we will focus on the distinctions of crRNA processing, crRNP assembly and target cleavage within the different CRISPR-Cas types.
Composition of type I Cascade effector complexes
The type I-E DNA interference complex
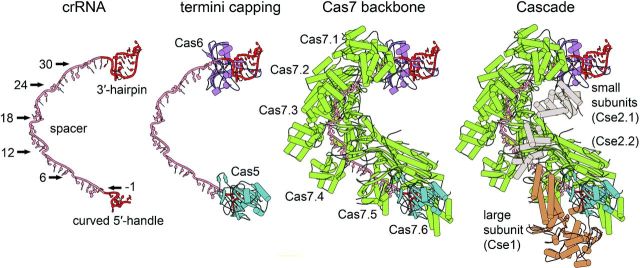
The type I-E system is widespread among bacterial phyla, common for Gammaproteobacteria, and rarely found in individual euryarchaeal organisms (Brouns et al.2008; Makarova, Wolf and Koonin 2013). This is the best studied type I CRISPR-Cas system and was originally found in Escherichia coli. This type has been termed I-E (for Escherichia) and encodes the first DNA interference complex to be named Cascade (Brouns et al.2008). The I-E Cascade is built up by a single 61-nt long crRNA and five different Cas proteins in an uneven subunit stoichiometry: (Cse1)1-(Cse2)2-(Cas5)1-(Cas7)6-(Cas6e)1 resulting in a total crRNP mass of 405 kDa (Brouns et al.2008; Jore et al.2011). The mature 61-nt crRNA is generated via specific cleavage by the Cas6e endoribonuclease within the repeat sequence of a pre-crRNA transcript (Gesner et al.2011; Sashital, Jinek and Doudna 2011). A former nomenclature of type I-E refers to Cas6e as Cse3 or CasE. For the sake of clarity, we will use Cas6e and related terminology throughout this review. Cas6e-mediated cleavage of repeat sequences yields a 8-nt 5′ handle with a hydroxyl group, a 32-nt spacer sequence and a 21-nt 3′ hairpin structure with a cyclic 2′-3′ phosphate end (Jore et al.2011). After cleavage, Cas6e stays bound to the 3′ hairpin of the mature crRNA (Niewoehner, Jinek and Doudna 2014). Cascade then assembles with Cas5 (known as Cas5e or CasD) binding to the 5′ handle of the crRNA and six copies of Cas7 (known as Cse4 or CasC) binding to the spacer sequence (Brouns et al.2008; Jore et al.2011) (Fig. 2). Two additional proteins are Cse1 (known as CasA) and the Cse2 dimer (known as CasB), which are defined as the large and small Cascade subunits (Brouns et al.2008). Both subunits are involved in DNA binding, while the large subunits also functions in the target selection (Jore et al.2011; Wiedenheft et al.2011a; Sashital, Wiedenheft and Doudna 2012). In the last step of the interference mechanism, the Cas3 helicase-endonuclease is recruited to degrade the target DNA (Brouns et al.2008; Westra et al.2012b). In Streptococcus thermophilus, a rarer orthologous type I-E CRISPR-Cas system was identified. Its structural study revealed a similar crRNP assembly with an observed overrepresentation of the backbone forming Cas7 subunit (Horvath and Barrangou 2010; Sinkunas et al.2011, 2013). The isolated mature crRNA of 61-nt length of this system included a 7-nt 5′ handle with a hydroxyl group, a 33-nt spacer sequence and a 21-nt 3′ handle with a Pi terminal group (Sinkunas et al.2013).
Figure 2.
Assembly of the type I-E Cascade structure. The I-E Cascade complex has a seahorse-shaped structure and consists of 11 protein subunits ((Cse1)1-(Cse2)2-(Cas5)1-(Cas7)6-(Cas6e)1) and a single 61-nt crRNA (pdb: 4TVX). Cas6e is tightly bound to the 3′ stem-loop structure of the mature crRNA and positioned at the head of the complex. Cas5e directly caps the 5′ handle of the crRNA, which leads to the hook-like structure of the crRNA. The structure of Cas5 and Cas7 displays a conserved palm-thumb domain arrangement, highlighting the intertwined assembly of the Cascade backbone. The thumb of either Cas5 or each of the six Cas7 subunits (Cas7.1-Cas7.6) kinks the crRNA at position −1 in the 5′ handle and every sixth position in the spacer sequence, and buries the base between the thumb and the palm of the adjacent Cas7 subunit. The two small Cse2 subunits (Cse2.1–Cse2.2) are connected to the crRNA backbone via protein:protein interactions to the Cas7 subunits. The large subunit Cse1 is positioned at the Cascade tail and interacts with Cas5, Cas7 and Cse2.
The type I-A DNA interference complex
The type I-A systems are predominantly found in thermophilic archaeal species and are rarely seen in bacteria (Makarova et al.2011b). The mechanism of crRNA generation shows clear differences to Cas6e-mediated cleavage, which results in a tight association of crRNA product and Cas6e (Niewoehner, Jinek and Doudna 2014). In contrast, a Cas6 enzyme of type I-A was shown to catalyze a multiple turnover reaction with the release of free crRNA (Sokolowski, Graham and White 2014). Structural studies revealed a dimeric composition of Cas6 with the unstructured repeat RNA substrate bound in a sequence-dependent manner that is essential for catalytic activity (Wang et al.2012; Reeks et al.2013b). The crRNAs have a broad distribution of ∼60–70 nt lengths and contain the characteristic 8 nt 5′ handle and a 16–17 nt 3′ handle. However, spacer length is more variable ranging from 38 to 44 nts in Sulfolobus solfataricus or 37 to 57 nts in Thermoproteus tenax (Lintner et al.2011; Plagens et al.2012). The 3′ handles are often nearly absent in the cellular crRNA pool (Plagens et al.2014). One explanation for this phenomenon is the observed weak association of Cas6 with the Cascade crRNP, which suggests that Cas6 is not an integral part of the complex. This weak association renders the 3′ handle accessible for chemical and/or enzymatic trimming in I-A Cascade that is assembled around the mature crRNA (Plagens et al.2014; Sokolowski, Graham and White 2014). The core backbone of I-A Cascade is presumably assembled via a multimerization of Cas7 units that form a helical structure along the crRNA and interact with Cas5 (Lintner et al.2011; Plagens et al.2014). It has been proposed that the roles of large and small Cascade subunits of the I-A complex are fulfilled by the proteins Cas8a and Csa5, respectively (Lintner et al.2011; Plagens et al.2014). The small subunit Csa5 of The. tenax was shown to preferably bind to ssDNA, suggesting its involvement in target DNA interaction (Daume, Plagens and Randau 2014). Another characteristic feature of type I-A systems is the split of the signature protein Cas3 into two proteins containing either the helicase (Cas3′) or the nuclease domain (Cas3′′). In contrast to I-E Cas3, that is recruited to cleave target DNA, both Cas3′ and Cas3′′ subunits are an integral part of the I-A Cascade crRNP (Plagens et al.2014).
The type I-B DNA interference complex
The type I-B systems are present in diverse archaeal and bacterial lineages and show some characteristics of both type I-A and type I-C systems (Makarova et al.2011b). Mature crRNAs of bacterial and archaeal species are generated by the endoribonuclease Cas6b and contain a clearly defined 8-nt 5′ handle, a 36–40 nt spacer and a 3′ handle that is gradually shortened to a minimal 2-nt tag (Richter et al.2012b; Elmore et al.2013; Li et al.2013; Richter et al.2013). Type I-B systems further contain Cas7 and Cas5 for building the Cascade backbone which interacts with Cas6b (Brendel et al.2014). The large subunit is represented by the subtype-specific protein Cas8b and is predicted to have the small subunit fused to its C-terminal end (Makarova et al.2011a). In addition, type I-B systems contain Cas3 versions that either have the typical nuclease-helicase fusion arrangement or are occasionally split into two discrete subunits (Makarova et al.2011b).
The type I-C DNA interference complex
The type I-C system can be found in various bacterial species, predominantly within the Firmicutes group (Haft et al.2005). This system was characterized in Bacillus halodurans and, in contrast to other type I and III systems, does not encode a Cas6 protein for crRNA maturation. Instead, the pre-crRNA cleavage activity is performed by a Cas5 (known also as Cas5d) homolog (Makarova et al.2011b). Cas5d was shown to cleave pre-crRNA resulting in a mature crRNA with an 11-nt 5′ handle that has a hydroxyl group, a 33-nt spacer sequence and a 21-nt 3′ handle containing a cyclic 2′-3′ phosphate end (Garside et al.2012; Nam et al.2012a). Following the processing event, one Cas5d subunit remains associated with the 3′ hairpin handle of the mature crRNA, a mechanistic feature that was also observed for Cas6e (Nam et al.2012a). Furthermore, the type I-C Cas5 is proposed to act as a bifunctional protein and a second subunit that presumably binds to the 5′ handle of the crRNA, similar to Cas5 from E. coli type I-E (Nam et al.2012a). The I-C Cascade crRNP also contains the backbone-forming subunit Cas7 (known as Csd2) and the large subunit Cas8 (known as Cas8c or Csd1) (Nam et al.2012a). Computational analyses predicted a fusion of large and small subunits in the type I-C Cas8 protein with its C-terminal region showing homology to the small subunit Cse2 of type I-E (Makarova et al.2011a; Punetha, Sivathanu and Anand 2014). The overall architecture of the I-C Cascade resembles I-E Cascade with a mass of ∼400 kDa and a proposed stoichiometry (Cas8)1-(Cas7)6-(Cas5)2 (Nam et al.2012a). The type I-C system additionally encodes Cas3 with a conserved nuclease-helicase domain involved in the final target DNA degradation step (Makarova et al.2011b).
The type I-D DNA interference complex
Type I-D systems are mainly found in cyanobacteria and euryarchaeal species (Makarova, Wolf and Koonin 2013). They feature the type I signature protein Cas3, but also a Cas10 protein, the signature protein for type III systems, suggesting an evolutionary link between I-C and III-B systems (Makarova et al.2011b). The maturation of pre-crRNA in the cyanobacterium Synechocystis sp. PCC6803 is mediated by a conserved Cas6 enzyme generating the common 8-nt 5′ handle with varying spacer length of 31–47 nt (Scholz et al.2013). Similar to type I-A and I-B, crRNAs of type I-D show a stepwise trimming of the 3′ end that might indicate a release of Cas6 from the crRNA after pre-crRNA cleavage (Hein et al.2013). The Thermofilum pendens I-D Cas7 protein was crystallized, revealing structural similarities with other Cas7 proteins and ssRNA binding activity. These features suggest that it builds up the I-D crRNP backbone (Makarova et al.2011a; Hrle et al.2014). A second protein involved in backbone formation might be the subtype-specific protein Csc1 that was previously grouped into the Cas5 family (Makarova et al.2011a). The large subunit of I-D Cascade is predicted to be the protein Cas10d, which shows a similar structural organization as the large subunits of the Cas8 family and Cse1 of type I-E (Makarova et al.2011a). Additionally, the HD nuclease domain of Cas3 (Cas3′′), essential for target DNA degradation, is fused to Cas10d and the helicase domain (Cas3′) is encoded by a stand-alone gene (Makarova et al.2011b).
The type I-F DNA interference complex
The occurrence of type I-F CRISPR-Cas systems is restricted to bacterial organisms. They are often found in Gammaproteobacteria and show remarkable similarities with the type I-E system (Makarova, Wolf and Koonin 2013). The maturation of crRNAs is mediated by the repeat-specific endoribonuclease Cas6f (known as Csy4) (Haurwitz et al.2010; Przybilski et al.2011). The 60-nt mature crRNA is characterized by the typical 8-nt 5′ handle, a repeat stem containing a 20-nt 3′ handle and 5′ OH as well as cyclic 2′-3′ phosphate termini (Haurwitz et al.2010; Sternberg, Haurwitz and Doudna 2012). It was observed that Cas6f binds to the 3′ handle of the crRNA, which is essential for RNA protection and Cascade I-F crRNP assembly (Haurwitz, Sternberg and Doudna 2012; Sternberg, Haurwitz and Doudna 2012). The I-F Cascade consists of four Cas proteins with a subunit stoichiometry that is similar to other Cascade complexes ((Csy1)1-(Cas5)1-(Cas7)6-(Cas6f)1) and a mass of 350 kDa (Wiedenheft et al.2011b; Richter et al.2012a). The crescent-shaped crRNA backbone is formed by six copies of a Cas7 family protein (known as Csy3), the terminal Cas5 (known as Csy2) at the 5′ end and Cas6f at the 3′ end of the crRNA (Wiedenheft et al.2011b). Sequence analysis and secondary structure predictions could not identify a clear homolog to other large Cascade subunits of the Cas8 family or Cse1, but distinct interactions of Cas5 and Csy1 suggest that Csy1 is fulfilling the role of the large and small subunits for target recognition and DNA binding in the I-F Cascade (Makarova et al.2011a; Richter et al.2012a). The conserved protein Cas3 shows the typical helicase-nuclease arrangement and is N-terminally fused to a Cas2-like domain (Richter et al.2012a). This Cas2-Cas3 fusion forms a complex with Cas1 and also interacts with the I-F Cascade subunits, suggesting a dual function in spacer acquisition and target DNA degradation (Richter et al.2012a; Richter and Fineran 2013).
Composition of type II DNA interference complexes
While the interference process in types I and III CRISPR-Cas systems involves the formation of a multi-Cas protein complex, Cas9 (formerly Csn1) is the only protein that is required in the DNA targeting event of the type II systems (Barrangou et al.2007; Garneau et al.2010; Deltcheva et al.2011; Sapranauskas et al.2011). In the first CRISPR-Cas classification, every system possessing a Cas9 protein (formerly COG3515) was grouped into the Neisseria (Nmeni) subtype (Haft et al.2005). The classification currently followed by the community is based on the identification of significant differences among Nmeni-type systems from various organisms, yielding a further sub-classification into subtypes II-A, II-B and II-C (Chylinski, Le Rhun and Charpentier 2013; Koonin and Makarova 2013; Zhang et al.2013; Chylinski et al.2014; Fonfara et al.2014). The discrimination relies mainly on the presence or absence of the adaptation module proteins, Cas4 and Csn2. While Csn2 is found in type II-A, type II-B contains Cas4 and type II-C possesses neither of the two. The type II Cas9 signature proteins show significant diversity in sequence; however, three common features are shared: conserved split HNH and RuvC nuclease domains, an arginine-rich motif and a similar globular architecture (Makarova et al.2006, 2011b; Chylinski, Le Rhun and Charpentier 2013; Koonin and Makarova 2013; Sampson et al.2013; Chylinski et al.2014; Jinek et al.2014; Nishimasu et al.2014) (Fig. 3). Based on the predicted nuclease domains, it was proposed that Cas9 would act as a nuclease during the invading nucleic acid interference reaction (Haft et al.2005; Makarova et al.2006).
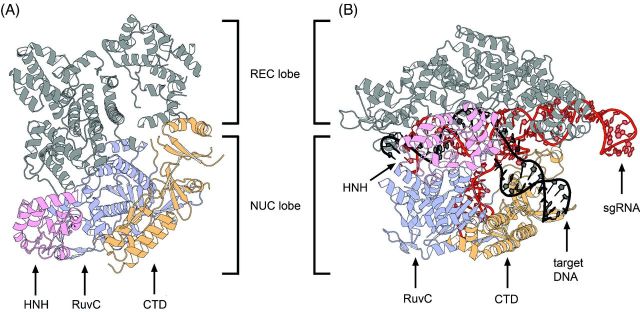
Figure 3.
Structures of S. pyogenes (Spy) type II-A Cas9. (A) Crystal structure of the apoenzyme SpyCas9 resolved at 2.6 Å (pdb: 4CMP) (Jinek et al.2014). (B) Structure of SpyCas9 bound to sgRNA and target DNA resolved at 2.5 Å (pdb: 4UN3) (Anders et al.2014). While the CTD and the RuvC domains remain in their positions, the REC lobe of Cas9 accommodates its position to facilitate sgRNA and target DNA binding. At the same time, the disordered HNH domain of inactive Cas9 (A) undergoes a conformational change for cleavage of the targeted strand.
The first interference activity of a type II system was demonstrated in phage challenge experiments. Streptococcus thermophilus, containing a type II-A system, acquired immunity against phages following the acquisition of one or more phage sequences that were integrated as spacers in the CRISPR array (Barrangou et al.2007). It was observed that this acquired bacterial immunity against phage attack was lost upon disruption of the cas9 gene. Disruption of the csn2 gene additionally indicated that Cas9 is the only protein required for type II interference (Barrangou et al.2007; Deveau et al.2008; Garneau et al.2010). This was further confirmed when deletions of the conserved adaptation cas1 and cas2 genes were shown to also retain the interference function (Sapranauskas et al.2011; Zhang et al.2013). In vivo experiments later identified dsDNA as the sole target of type II systems (Garneau et al.2010; Deltcheva et al.2011; Sapranauskas et al.2011), in which Cas9 uses HNH and RuvC nuclease domains (Sapranauskas et al.2011) to cleave the DNA sequence yielding blunt-ended double-strand breaks (Garneau et al.2010; Magadan et al.2012).
A second hallmark of the type II systems is the trans-activating CRISPR RNA (tracrRNA). While the surveillance machinery of types I and III consists of crRNPs composed of one single crRNA guiding a multi-Cas protein complex to the target nucleic acids, type II systems use a duplex of RNAs (dual-tracrRNA-crRNA) to guide Cas9 to the invading target DNA. The tracrRNA was identified as an abundant small RNA containing an anti-CRISPR repeat and located in the vicinity of type II cas genes and CRISPR repeat-spacer arrays (Deltcheva et al.2011). Initial experiments performed in S. pyogenes showed that tracrRNA acts as a trans-activator of crRNA maturation (Deltcheva et al.2011). However, subsequent work demonstrated that tracrRNA also forms a critical component of the Cas9 cleavage complex (Jinek et al.2012). With respect to maturation, type II systems lack genes encoding Cas6 endoribonucleases that are used by types I and III systems to process crRNAs. Instead, maturation of type II crRNAs involves tracrRNA, Cas9 and the effector endoribonuclease III from the bacterial host. tracrRNA binds via its anti-repeat sequence to each of the repeats of the pre-crRNA forming heteroduplexes that are stabilized by Cas9 (Deltcheva et al.2011). Each duplex RNA is then recognized and cleaved by the endoribonuclease III, yielding an intermediate form of crRNA and a mature form of tracrRNA (77 nt length) (Deltcheva et al.2011; Chylinski, Le Rhun and Charpentier 2013; Karvelis et al.2013). An additional processing of the crRNA, by a so far unknown endo- and/or exonuclease, yields the mature crRNA (44 nt length) composed of spacer sequence in 5′ and repeat sequence in 3′ (Deltcheva et al.2011).
With respect to cleavage, it was found that mature crRNA and Cas9 alone were incapable of cleaving target DNA, but the addition of tracrRNA resulted in DNA targeting and cleavage (Jinek et al.2012). Biochemical analysis of the DNA targeting mechanism of the S. pyogenes type II CRISPR-Cas system (Deltcheva et al.2011) thus demonstrated that Cas9 is a DNA endonuclease that uses a tracrRNA: crRNA duplex to direct DNA cleavage site specifically (Jinek et al.2012). The HNH domain of Cas9 cleaves the DNA strand that is complementary to the spacer region of crRNA while the RuvC-like domain cleaves the DNA strand opposite the complementary strand (Gasiunas et al.2012; Jinek et al.2012).
In a type II-C system (e.g. Neisseria meningitidis), an alternative crRNA maturation pathway has been described (Zhang et al.2013). In this particular system, the repeat units of the CRISPR array contain promoter sequences that can initiate the production of short crRNAs in an endoribonuclease III-independent manner. However, duplex formation of tracrRNA and the short crRNAs were shown to still be required for interference with DNA (Zhang et al.2013).
A variant Cas9 protein with single-stranded DNA cleavage (nickase) activity can be generated by mutating either the HNH or the RuvC-like domain (Gasiunas et al.2012; Jinek et al.2012). Mutating both domains creates an RNA-guided DNA-binding protein with deficient cleavage activity (dead-Cas9) (Gasiunas et al.2012; Jinek et al.2012). DNA target recognition requires both base pairing of the crRNA sequence to the protospacer region and the presence of the PAM adjacent to the targeted sequence in the DNA (Gasiunas et al.2012; Jinek et al.2012). While it was found that some nucleotide changes in the protospacer sequences can be tolerated, the PAM and the region proximal to the PAM in the protospacer are requirements critical for site-specific Cas9 targeting. Single mutations in the PAM led to escape phages that were not targeted any longer. Changes close to the PAM yielded also non-cleavable targets, indicating the presence of a seed sequence similar to type I systems (Deveau et al.2008; Garneau et al.2010; Sapranauskas et al.2011; Semenova et al.2011; Jinek et al.2012; Martel and Moineau 2014). Further experiments revealed that the PAM is only recognized in a dsDNA context and that cleavage by the HNH and RuvC domains of Cas9 occurs within the protospacer, exactly 3 nt away from the PAM (Garneau et al.2010; Gasiunas et al.2012; Jinek et al.2012; Magadan et al.2012). It was also shown, that the HNH domain appears to have a fixed cleavage site, while the targeting position for the RuvC domain is defined by a ruler mechanism, in which a linker between the PAM and the protospacer influences the cleavage site, yielding non-blunt-ended cleavage in very rare cases (Chen, Choi and Bailey 2014).
Early observations of the conserved ability to form tracrRNA:crRNA duplexes despite the large diversity in tracrRNA anti-repeat and crRNA repeat sequences indicated coevolution of tracrRNA and crRNA (Deltcheva et al.2011). Further analysis of phylogenetic trees and sequence variability of tracrRNA anti-repeat, CRISPR repeat and Cas9 orthologs led to the proposal that the dual-tracrRNA:crRNAs have functionally coevolved with the Cas9 proteins (Deltcheva et al.2011; Jinek et al.2012; Chylinski, Le Rhun and Charpentier 2013; Chylinski et al.2014; Fonfara et al.2014). DNA cleavage by dual-tracrRNA:crRNA-guided Cas9 was reported for Cas9 orthologs from various bacterial species, closely or distantly related to the S. pyogenes Cas9 (Jinek et al.2012; Karvelis et al.2013; Fonfara et al.2014). Orthologous Cas9 proteins can utilize non-cognate tracrRNA:crRNAs as guide sequences only when these RNAs originate from loci with highly similar Cas9 sequences (Jinek et al.2012; Fonfara et al.2014), demonstrating orthologonality in CRISPR-Cas9 activities. The involvement of a secondary structure of the tracrRNA anti-repeat:crRNA repeat duplex in the specific recognition by Cas9 orthologs was proposed based on similar structure features shared among the exchangeable RNAs (Jinek et al.2012; Briner et al.2014; Fonfara et al.2014).
Although the cleavage complex in type II systems was shown to essentially include three components: tracrRNA, crRNA and Cas9 endonuclease (Jinek et al.2012), a custom-made single-guide RNA (sgRNA) that combined both tracrRNA and crRNA into one molecule was shown to be capable of complexing with Cas9 to promote effective cleavage of target DNA, paving the way for RNA programmable genome editing using a single guide CRISPR-Cas9 system (Jinek et al.2012). The sgRNA retains two main characteristics of the natural dual-RNA: a nucleotide sequence at the 5′ end that forms specific base pairing with the target DNA and the double-stranded anti-repeat–repeat structure at the 3′ end that binds to Cas9. sgRNA-Cas9 forms thus a two-component system in which changes in the guide sequence can program the system to target any DNA sequence of interest owing to the presence of a PAM adjacent to the sequence to be targeted (Jinek et al.2012). Programmable CRISPR-Cas9 using the S. pyogenes system has rapidly and widely been recognized as an effective technology to target, edit or modify the genomes of a large variety of cells and organisms (Doudna and Charpentier 2014; Hsu, Lander and Zhang 2014). The technology was also recently harnessed for programmable RNA recognition and cleavage (O'Connell et al.2014). In addition to its role in crRNA maturation and interference with DNA, recent studies show Cas9 is also required for the selection of spacers by recognizing the PAM of the protopacers during the phase of adaptation (Heler et al.2015; Wei, Terns and Terns 2015).
Following the identification of CRISPR-Cas, suggestions indicated that the system could be involved in cellular pathways other than interference with mobile genetic elements (Westra, Buckling and Fineran 2014). The type II-B system of Francisella novicida has provided the first evidence for another targeting function of CRISPR-Cas. In this specific case, scaRNA (small CRISPR-Cas associated RNA) is a small RNA that pairs with tracrRNA to form a heteroduplex, similar to the dual-tracrRNA:crRNA (Sampson et al.2013; Sampson and Weiss 2013). A model was proposed whereby tracrRNA:scaRNA guides Cas9 to target the mRNA of a bacterial lipoprotein. The formation of the targeting complex results in the reduction of lipoprotein production, which in turn enables F. novicida to evade the host immune response (Sampson et al.2013; Sampson and Weiss 2013). Thus, the mRNA-targeting function of the type II-B system via the tracrRNA:scaRNA-Cas9 complex confers to CRISPR-Cas an alternative function in endogenous gene regulation and virulence.
Composition of type III nucleic acid interference complexes
The type III-A DNA/RNA interference complex
Similar to type I CRISPR-Cas, the type III systems are present in a wide range of phylogenetically diverse bacterial and archaeal species (Makarova et al.2011a). Both types share significant similarities in the mechanism of crRNA maturation and encode a crRNP interference complex that contains multiple Cas protein subunits with a conserved RNA recognition motif (RRM) fold. The mature crRNAs of type III are primarily processed by the endoribonuclease Cas6 generating the common 8-nt 5′ handle. Extensive nucleolytic trimming of the 3′ end is observed, producing a pool of two major cellular crRNA species (39–45 nt) with a specific 6-nt length difference (Carte et al.2008; Hale et al.2009; Zhang et al.2012). In contrast to type I and II systems, type III systems do not rely on the presence of a PAM sequence during interference (Staals et al.2014; Tamulaitis et al.2014).
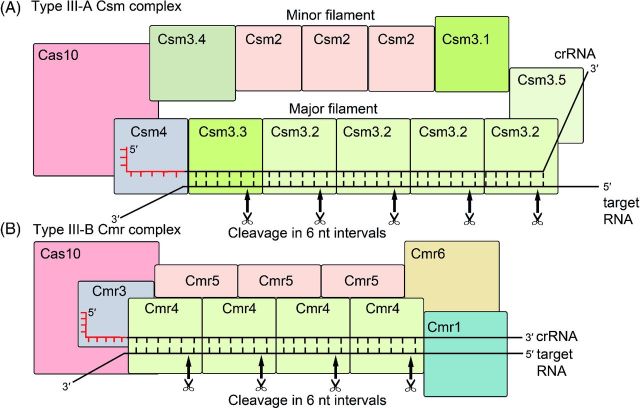
The Csm complex (type III-A) from Su. solfataricus with a molecular weight of ∼428 kDa is composed of eight different proteins with the stoichiometry of (Csm2)3-(Csm3.1)1-(Csm3.2)4-(Csm3.3)1-(Cas10)1-(Csm3.4)1-(Csm4)1-(Csm3.5)1. Several Csm3 subunits and one Csm4 subunit form the crRNA-binding backbone (Rouillon et al.2013). It was suggested that the function of large and small subunits is fulfilled by Cas10 and a trimer of Csm2 proteins based on their location in a Csm complex structure (Rouillon et al.2013). The purified Csm complex of Thermus thermophilus has a nearly identical molecular weight of ∼427 kDa with a slightly differing composition. Here, six Csm3 and two Csm4 subunits form the crRNA-interacting backbone and additionally the protein Csm5 was identified as an integral part of the complex (Staals et al.2014). This crRNP complex, as well as the Csm complex of S. thermophilus, was shown to target complementary ssRNA and cleave it at multiple sites in vitro and in vivo (Staals et al.2014; Tamulaitis et al.2014). In Staphylococcus epidermidis, the type III-A system was shown to target plasmid DNA, as well as temperate phages, in a transcription-dependent manner (Marraffini and Sontheimer 2008; Goldberg et al.2014).
The type III-B RNA interference complex
The Cmr complex (type III-B) was analyzed in the bacterium T. thermophilus and was shown to have an estimated molecular weight of ∼365 kDa and a complex stoichiometry of (Cmr1)1-(Cas10)1-(Cmr3)1-(Cmr4)4-(Cmr5)3-(Cmr6)1 (Staals et al.2013). A similar subunit composition was found in Pyrococcus furiosus with either three (Spilman et al.2013) or four Cmr4 subunits (Hale et al.2014). The crRNP backbone is formed by Cmr3 and multiple copies of Cmr4. The large and small subunits are proposed to be represented by Cas10 and three subunits of Cmr5 (Spilman et al.2013; Staals et al.2013). The type III-B systems exclusively target ssRNA and not DNA sequences that are complementary to the crRNA (Hale et al.2009, 2012, 2014; Zhang et al.2012).
STRUCTURE AND ASSEMBLY OF THE BACTERIAL TYPE I-E DNA INTERFERENCE COMPLEX
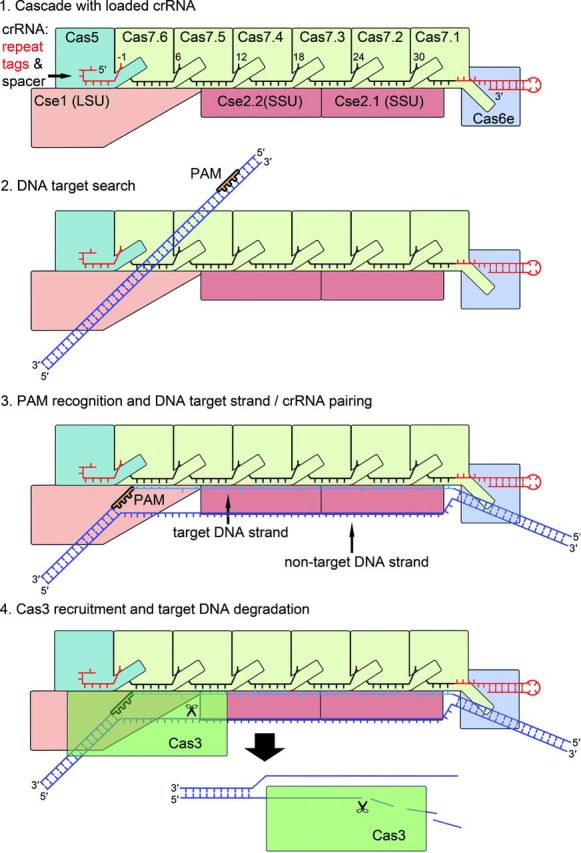
A first overview of the general morphology of the I-E Cascade structure at a resolution of 8 Å was obtained by single-particle cryoelectron microscopy in combination with further structural and biochemical studies (Brouns et al.2008; Jore et al.2011; Wiedenheft et al.2011a). Recently, the crystal structure of different I-E Cascade complexes with resolution between 3.03 and 3.24 Å was solved by several groups, providing important insights into Cascade assembly and the mechanism of target DNA recognition (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014; Zhao et al.2014). Overall, the complex consists of 11 protein subunits ((Cse1)1-(Cse2)2-(Cas5)1-(Cas7)6-(Cas6e)1) and a single 61-nt crRNA (Fig. 2). The shape of the structure was described to resemble a seahorse (Jore et al.2011). Accordingly, Cas6e is tightly bound to the 3′ stem-loop structure of the mature crRNA and positioned at the head of the complex (Wiedenheft et al.2011a). The 5′ handle of the crRNA is placed between Cas5 and the large subunit Cse1 at the tail of the seahorse. The head and tail of the crRNA are bridged by six Cas7 copies (Cas7.1–Cas7.6) that form a helical backbone, while the belly is represented by two Cse2 subunits (Jackson et al.2014a).
Interactions between crRNA and Cas6e at the Cascade head
The structure of Cas6e consists of two RRMs that are connected by an eight-residue linker. The typical RRM-fold consists of four anti-parallel beta-strands and two alpha-helices that are arranged in a β1-α1-β2-β3-α2-β4 pattern. The β-strands are ordered in a four-stranded antiparallel β-sheet with two α-helices packed on one side (Ebihara et al.2006). The two β-sheets face one another and create a funnel-shaped cleft. Cas6e comprises a positively charged basic patch opposite of this cleft that interacts with the crRNA 3′ end. The major groove of the crRNA stem loop is wrapped around a positively charged Cas6e groove-loop (i.e. the β6–β7 hairpin, residues 90–119) (Jackson et al.2014a; Zhao et al.2014). This positioning of the crRNA stem loop directs the scissile phosphate group into the active site of Cas6e. Thus, Cas6e recognizes bases on both sides of the stem loop and interacts tightly with the 3′ handle of the crRNA after cleavage (Sashital, Jinek and Doudna 2011).
Structure of the Cas7:crRNA backbone
The prominent backbone of Cascade is assembled via the oligomerization of six Cas7 subunits around the mature crRNA (Fig. 2). The intertwined structure is arranged in six discrete segments, in which one nucleotide is buried, followed by five accessible bases that are coordinated in a pseudo A-form configuration. The typical structure of Cas7 resembles a right hand and consists of distinct regions termed fingers (residues 59–180), a palm (residues 1–58, 181–189 and 224–263) and a thumb (residues 190–223) domain (Mulepati, Heroux and Bailey 2014). The palm contains a modified RRM and two smaller loops inserted in this RRM are forming a web between the thumb and the fingers (Jackson et al.2014a). The five crRNA base segments that are accessible for target DNA hybridization are in contact with the palm domain via several conserved polar and positively charged residues (K27, S40, Q42 and K45). Additionally, a conserved M166 residue intercalates with the third and fourth base in each segment, which keeps the nucleotides apart and helps to distort the A-form configuration (Jackson et al.2014a; Zhao et al.2014). In contrast to the classical RRM arrangement, the α1-helix is not located on the back of the β-sheets, but is positioned perpendicular on the central ß-sheet. This helix contains several conserved residues (W199, F200, T201 and V203) that interact with three consecutive phosphates of the crRNA (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014). These interactions introduce two successive ∼90° turns in the crRNA backbone, which causes every sixth base in the segment (crRNA positions: 6, 12, 18, 24 and 30) to flip outwards (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014; Zhao et al.2014). This flipped base is further buried between the α1-helix of the palm and the thumb of an adjacent Cas7 subunit. Therefore, the conformations of the Cas7.2–Cas7.6 subunits are identical and the neighboring subunits display two pronounced protein:protein interactions sites next to the crRNA contact (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014). A first area (∼1500 Å2) of interaction is formed between the thumb-base and the palm back of one Cas7 with the palm front of the adjacent Cas7 subunit. The second area (∼400 Å2) of interaction is generated between the thumb-tip and the fingers of neighboring subunits (Mulepati, Heroux and Bailey 2014). The Cas7:crRNA backbone interacts with the Cascade head via protein:protein contacts between Cas6e and Cas7.1 at the 3′ end of the crRNA (Fig. 2). In contrast to the other five Cas7 subunits, a short helix located on the thumb-tip of Cas7 (contact residues: W199, F200 and V203) is inserted into the hydrophobic funnel-shaped cleft of Cas6e, which lies opposite of the crRNA interaction site. As a result, the flexible Cas7 thumb domain is rotated outwards by 73° in comparison to the thumb domains of the other five Cas7 subunits (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014; Zhao et al.2014).
Interactions between crRNA and Cas5 at the Cascade tail
Type I-E mature crRNAs are characterized by a conserved 8-nt 5′ handle, generated by Cas6e cleavage within the repeat sequence. The respective nucleotides (numbered from position −8 to −1) are protected by the Cas5 subunit within Cascade (Jore et al.2011). The Cas5 structure reveals a palm domain that includes a modified RRM (residues 1–78 and 115–224) and a thumb (residues 79–114) domain (Fig. 2). Thus, structural similarities between Cas5 and Cas7 are apparent (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014). Additionally, the positioning of the Cas5 thumb supports a function analogous to the Cas7 thumb. The Cas5 thumb folds over the kinked base at position −1, employing similar residues in the binding pocket that Cas7 uses for binding (L89 and T87) and thus ensures the crRNA A-form configuration of the first base of the spacer (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014; Zhao et al.2014). This assembly guarantees the segmentation of the Cas5-protected 5′ handle from the accessible crRNA guide region that is clamped by the subunit Cas7.6. The Cas5 thumb contacts the adjacent Cas7.6 subunit at the fingers domain, which leads to a ∼180° rotation of the fingers in comparison to the fingers of the other Cas7 subunits (Jackson et al.2014a). Consequently, a broader 28 Å gap appears between the proximate Cas7.5 and Cas7.6 fingers (Jackson et al.2014a). The buried crRNA bases of positions −8 to −2 form a hook-like structure and are sandwiched between Cas5 and the web of Cas7.6 via extensive contacts of charged and polar residues. The terminal three bases (A-8, U-7 and A-6) interact with binding pockets on top of the glycine-rich α1-helix of Cas5 with sequence-specific contacts between Y145 and U-7. The three central bases (A-5, A-4, C-3) form a triplet stack orthogonal to A-6 positioned between the palm domains of Cas5 and Cas7.6. The position C-2 forms sequence-specific hydrogen bonds with R108 of the Cas5 thumb (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014). Mutation of residue R108 showed that this position is essential for the interference mechanism (Zhao et al.2014).
Positioning of small and large subunits in Cascade
The E. coli large subunit protein Cse1 has a unique globular fold that contains a zinc-ion coordinated by four cysteine residues (C140, C143, C250 and C253) and a C-terminal four-helix bundle (Mulepati, Orr and Bailey 2012; Jackson et al.2014a). The interaction of the large subunit Cse1 with Cas5 and the crRNA 5′ handle at the tail is mediated via a short α-helix within a loop (termed L1, residues 130–143) that inserts into a Cas5 helix-binding pore (Fig. 2) (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014). This cylindric pore is formed when the Cas5 thumb reaches the Cas7.6 subunit and thereby allows base-specific contacts of Cse1 (residues F129, V130, N131 and Q132) to the accessible A-A-C triplet stack of the crRNA 5′ handle. Additional contact sites are observed between the globular domain of Cse1 and the RRM of Cas5 (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014). Furthermore, the external α-helix of the four-helix bundle on top of the globular domain interacts with the C-terminal domain of the second small subunit Cse2.2 via salt bridge formation (Cse1 R483:Cse2.2 E150). The two Cse2 subunits that form the Cascade belly are assembled as a head-to-tail dimer, and the Cse2.1 protein interacts with Cas6e (Fig. 2) (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014). Several crystal structures of Cse2 were solved that revealed its α-helical bundle scaffold and identified multiple basic patches on the protein surface (Agari et al.2008; Nam et al.2012a). Direct contacts of Cse2 to the crRNA are not observed, but the attachment of both Cse1 and Cse2 to the Cas7 backbone is mediated by five unique binding spots that are all formed by a triad of (i) the negatively charged residue D22 of Cas7, (ii) a positively charged residue of Cse1 and Cse2 (R27 and R101 of Cse2.1 or Cse2.2, as well as K474 of Cse1) and (iii) a stabilizing aromatic side chain of W199 of the following Cas7 subunit (Mulepati, Heroux and Bailey 2014; Zhao et al.2014). Thus, each Cse2 subunit is connected to three Cas7 subunits (Cse2.1:Cas7.1–3 and Cse2.2:Cas7.3–5) and Cse1 is connected to two Cas7 subunits (Cse1:Cas7.5–6) (Zhao et al.2014).
Interaction of Cascade with a target DNA
The crystal structure of the Cascade-crRNA in complex with a 32-nt ssDNA protospacer has facilitated further insights into the DNA targeting mechanism and the localization of the DNA strands within the assembly (Mulepati, Heroux and Bailey 2014). The two strands of the crRNA:target hybrid shape as a ribbon-like structure that does not form a helix, but is underwound due to the crRNA kinks at every Cas7 segment (Mulepati, Heroux and Bailey 2014). Cas7 inhibits the base pairing of complementary nucleotides at the kink, as the thumb of Cas7 extends towards the finger of the adjacent Cas7 subunit passing directly between the strands of the hybrid and shielding the crRNA base. This kink is observed at five crRNA positions in the guide region (position: 6, 12, 18, 24 and 30) due to the linkage of the Cas7 subunits and in one position of the 5′ handle due to Cas5 thumb–Cas7 interactions (position: −1) (Mulepati, Heroux and Bailey 2014). Consequently, each 5-bp segment is distorted from a canonical A-form ensuring continuous accessibility of the guide region. The Cas7 subunits are not only interacting with the crRNA backbone via the palm, but are also contacting the target DNA across the minor groove via the thumb-tip (H213 and L214) and the fingers of the adjacent Cas7 (residue 109–111, 163–169) (Mulepati, Heroux and Bailey 2014). Interestingly, the same binding residues that are observed to attach the large (K474) and small subunit (R27 and R101) to the Cas7 backbone are involved in contacting the displaced DNA bases of the target strand. Their function is likely to hold the target strand in position for crRNA hybridization between the Cas7 backbone and Cse1, Cse2.2 and Cse2.1 (Mulepati, Heroux and Bailey 2014).
Insights into the localization of the non-target DNA strand were obtained by cryo-EM studies of Cascade-dsDNA complexes. The PAM-proximal end of the DNA was found to bind between Cas7.5, Cas7.6 and Cse1 (Westra et al.2012b; Hochstrasser et al.2014). The combination of crystal structure and cryo-EM density data revealed potential interactions between the dsDNA and several basic residues (Cas7.5: K137, K138, K141 and Cas7.6: H67, K105) that are exposed in the broader gap of the Cas7.6 and Cas7.5 fingers (Mulepati, Heroux and Bailey 2014). A conserved structural motif accessibly located on the L1 loop of Cse1 was identified that influenced PAM recognition, resulting in destabilization of the target DNA duplex and crRNA-directed strand invasion (Sashital, Wiedenheft and Doudna 2012; Tay, Liu and Yuan 2015). This region is disordered in the Cascade:ssDNA structure, suggesting its flexibility in the absence of dsDNA (Mulepati, Heroux and Bailey 2014). The non-target strand is displaced during crRNA:ssDNA pairing (Jore et al.2011). A distinctive basic groove spanning from Cse1 via the Cse2 dimer to Cas6e provides a binding site for the displaced strand (Mulepati, Heroux and Bailey 2014; Tay, Liu and Yuan 2015). Mutations in the conserved positive patch of Cse1 from Thermobifida fusca abolished the DNA binding (Tay, Liu and Yuan 2015). Mechanistically, after PAM recognition and target pairing, the non-target strand has to loop around the four-helix bundle of Cse1 and is then directed via basic residues of Cse2 (R53, R110, R142, R143) as well as of Cas7 (K34, K299, K301) (Mulepati, Heroux and Bailey 2014). The cryo-EM studies of Cascade bound to dsDNA identified structural rearrangements of the large and small subunit upon DNA targeting (Wiedenheft et al.2011a). During DNA binding, the Cse2 dimer moves ∼16 Å relative to Cse1, which leads to a ∼30° rotation of the Cse1 four-helix bundle and a ∼15° rotation of the Cse1 base. These rearrangements make the binding sites of Cse2 within the basic groove accessible for non-target DNA binding and coordinate a platform for Cas3 recruitment (Hochstrasser et al.2014; Mulepati, Heroux and Bailey 2014).
STRUCTURES AND FUNCTIONAL DOMAINS OF THE TYPE I SIGNATURE PROTEIN CAS3
Cas3 is the signature protein of type I CRISPR-Cas systems. This protein plays a crucial role in the viral defense reaction, as it is responsible for target degradation (Brouns et al.2008). Phylogenetic analyses of Cas3 proteins from all type I systems revealed a common helicase domain core and diverse N-terminal and C-terminal accessory domains (CTD; Jackson et al.2014b) (Fig. 4). The core helicase domain shows highly conserved residues of superfamily 2 (SF2) helicases including the NTP-binding Walker A and Walker B motifs (Jansen et al.2002; Makarova et al.2002). SF2 helicases contain a tandem RecA-like fold, which forms a cleft coordinating the amino acids responsible for the binding of NTP, Mg2+ ions and nucleic acid substrates (Cordin et al.2006; Fairman-Williams, Guenther and Jankowsky 2010). Biochemical studies show that Cas3 enzymes of type I-E are ATP dependent and unwind duplex DNA in a 3′-5′ direction via an inchworm-like mechanism (Sinkunas et al.2011; Mulepati and Bailey 2013). Additionally, all type I systems encode an HD nuclease that is either fused as an N-terminal accessory domain to the helicase core (type I-B, I-C, I-E and I-F) or encoded by a separate gene (type I-A, type I-B and I-D). The HD nuclease is characterized as a metal-dependent exo- and endonuclease in the presence of divalent metals that are coordinated by the active site HD motif (Beloglazova et al.2011; Mulepati and Bailey 2011; Sinkunas et al.2011). Typically, a CTD is fused to the helicase core. This domain is suggested to connect Cas3 and Cascade (Gong et al.2014; Huo et al.2014). Noteworthy, Cas3 of type I-F is additionally fused to a Cas2-like domain at its N-terminus and interacts with Cas1 (Makarova et al.2011a; Richter et al.2012a).
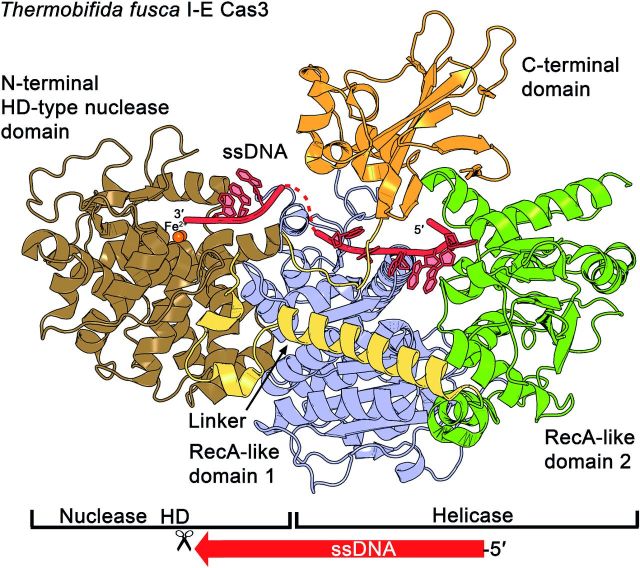
Figure 4.
Structure of the DNA nuclease Cas3. The type I-E Cas3 crystal structure of Th. fusca (pdb: 4QQW) reveals two tandem RecA-like domains, one HD-type nuclease domain and a CTD located at the top of the ensemble. The core helicase, containing two RecA-like domains, forms a cleft that locates the residues for the binding of NTP, Mg2+ ions and the ssDNA substrate. Two Fe(II) ions are located at the catalytic center's HD motif. The 5′ end of the ssDNA enters Cas3 from the RecA2 side and is further threaded to RecA1 and the HD-type nuclease domain (indicated by a scissor). The CTD is proposed to close the ssDNA channel and to contact the Cascade complex.
In 2011, the crystal structures of the Cas3 nuclease domains from T. thermophilus (type I-E) and Methanocaldococcus jannaschii (type I-A) were published (Beloglazova et al.2011; Mulepati and Bailey 2011). More recently, two crystal structures of the entire type I-E Cas3 protein were solved, one from Thermobaculum terrenum in complex with ATP, and one from Th. fusca in complex with an ssDNA substrate and ATP (Gong et al.2014; Huo et al.2014). The Cas3 structure shows the typical arrangement of tandem RecA-like domains and an HD domain docked at RecA1 with the CTD domain located at the top of the helicase core (Fig. 4). The contact sites for the essential cofactors ATP and Mg2+ are present within the RecA1 domain, in the cleft towards the RecA2 domain. In Th. fusca, the binding of Cas3 to ATP involves the residues Q284 (Q motif) and in addition G308, E309 and G310 (Walker A motif). The Mg2+ ion is coordinated via D451 and E452 (DEAH, Walker B motif) (Huo et al.2014).
All available HD domain structures reveal a globular shape with a concave surface (Mulepati and Bailey 2011). The HD domain:RecA1 contact area in Th. fusca is provided via a hydrophobic interface of ∼4200 Å2, which includes several conserved residues within the inner concave site (HD: W216, L217, L260 and RecA1: W406, R412, L415, F441, W470) (Huo et al.2014). Different metal ions were found to be coordinated by the invariant HD residues in the three available HD domain structures. In Th. fusca, two Fe(II) ions are positioned by the residues of this HD motif (H83 and D84) and several conserved histidines (H37, H115, H149, H150) (Huo et al.2014). In contrast, the second type I-E HD domain structure from T. thermophilus contained one Ni2+ ion in the HD motif (residues: H69, D70 and H24, D205) (Mulepati and Bailey 2011). In the type I-A HD domain structure from M. jannaschii, two Ca2+ ions were identified to interact with residues of the HD motif (residues: H66, D67 and H91, H123, H124) (Beloglazova et al.2011). Furthermore, ssDNase activity was observed with several transition-metal ions including Mg2+, Mn2+, Co2+, Cu2+ and Zn2+ indicating a broad range of functional metal cofactors (Mulepati and Bailey 2011; Sinkunas et al.2011; Gong et al.2014; Huo et al.2014). The positioning of these transition-metal ions in the catalytic center suggests that their role in the cleavage mechanism is to coordinate a deprotonated water molecule for nucleophilic attack of the ssDNA (Huo et al.2014).
The cocrystallized ssDNA molecule in the Th. fusca Cas3 provides further insights into the coordinated path of the target DNA during its degradation (Huo et al.2014) (Fig. 4). The 5′ end of the ssDNA gets incorporated into Cas3 from the RecA2 side via a postulated separation hairpin (position: 715–727) that is conserved in many SF2 helicases and intercalates into the dsDNA. The CTD contacts surface loops of both RecA domains on top and a closed ssDNA channel is formed. The ssDNA is then further threaded from RecA2 to RecA1 by contacts of several salt-bridge and hydrogen-bond interactions (Gong et al.2014; Huo et al.2014). Finally, the positioning of the ssDNA in the catalytic center of the HD nuclease is supported by K411 and W216, resulting in a sharp bend of the DNA backbone (Huo et al.2014).
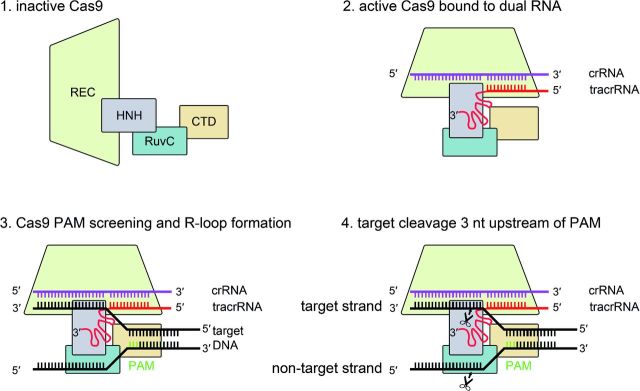
Several biochemical studies revealed that Cascade recruits Cas3 after it formed the crRNA:target DNA hybrid structure, the so-called R-loop (Westra et al.2012b; Mulepati and Bailey 2013; Sinkunas et al.2013). Negative-stain EM visualization of I-E Cascade:dsDNA complexes that were incubated with Cas3 revealed that Cas3 binds between the four-helix bundle and the base of Cse1, a region where conformational changes during DNA binding were observed (Hochstrasser et al.2014). Mapping this information onto the Cascade crystal structure identified several residues (E192, E280, N376 and T383) and two loops at the Cse1 base (residues 288–294, 318–323) that might mediate the Cas3 interaction (Mulepati, Heroux and Bailey 2014). Additionally, the Cas3 protein of Th. fusca, lacking the CTD domain, showed a decreased affinity for Cascade (Huo et al.2014). These structural insights into the target degradation pathway support a model of concerted events: (i) Cascade assembly, (ii) target search and R-loop formation, (iii) Cas3 recruitment and finally (iv) target DNA cleavage (Fig. 5). In the following section, we review these individual stages as the combination of available biochemical data and the Cascade structures considerably expanded our knowledge of the Cascade-mediated DNA interference mechanism.
Figure 5.
Mechanism of type I Cascade-mediated DNA interference. After the assembly of the crRNA-loaded Cascade, the surveillance complex (SSU: small subunits, LSU large subunit) scans DNA sequences. Potential DNA targets are identified via PAM recognition. This triggers the destabilization of the DNA duplex and allows the crRNA to pair with the target strand, while the non-target strand is displaced and spanned via the large and small subunit. Following R-loop formation, interaction sites at the base of the large subunit enable a stable interaction with Cas3. The HD domain of Cas3 nicks the DNA strand downstream of the PAM and the duplex is further unwound in 3′–5′ direction and degraded. The remaining single-stranded target DNA is proposed to be cleaved by the stand-alone Cas3 enzyme.
MECHANISM OF CASCADE-MEDIATED DNA INTERFERENCE
Cascade assembly
Cas6e cleaves pre-crRNA within the repeats at the level of the RNA stem-loop structure and remains tightly bound to the 3′ handle, protecting the RNA stem loop from unspecific nucleolytic trimming (Brouns et al.2008; Sashital, Jinek and Doudna 2011). This initial reaction is proposed to serve as a platform for the coordinated Cascade complex formation (Jore et al.2011). However, the order of the following Cascade backbone and tail assembly steps is not known. It is possible that Cas5e directly caps the crRNA's 5′ handle via base-specific interactions, which leads to the hook-like structure of the crRNA 5′ tag (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014; Zhao et al.2014). The structure of Cas5e and Cas7 revealed a conserved palm-thumb domain arrangement that explains the intertwined assembly of the Cascade backbone. The thumb of either Cas5 or each of the six Cas7 subunits kinks the crRNA at position −1 in the 5′ handle and every sixth position in the spacer sequence, and buries the base between the thumb and the palm in a positively charged pocket of the adjacent Cas7 subunit (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014). Accordingly, the 5-nt segments of the crRNA guide are stretched along the Cas7 palm and are therefore accessible for complementary base pairing (Fineran et al.2014). The thumb of the subunit Cas7.1 positioned at the end of the crRNA guide then folds into a cleft of Cas6e and links the whole crRNA backbone (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014). It remains to be seen if Cas7 oligomerization at the crRNA is either started or stopped at one of the two terminal caps. These steps ensure that the crRNA is fully protected in the cell. The final assembly is suggested to involve the subunits that interact with the DNA. The two small Cse2 subunits are connected to the crRNA backbone via protein:protein interactions between one Cse2 and three Cas7 subunits (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014). The large subunit Cse1 is positioned at the Cascade tail and interacts with Cas5, Cas7 and Cse2 (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014). This step completes Cascade assembly and ensures the presence of a surveillance complex that constantly screens the cell for potential target DNAs.
Cascade interactions with PAM sequences
Target DNAs are identified via different quality checkpoints to ensure that only harmful DNA is degraded and that e.g. the host genome is not targeted (van der Oost et al.2014). First, Cascade scans the dsDNA for a potential PAM sequence (Westra et al.2012b; Hochstrasser et al.2014; Rollins et al.2015). The PAM is a conserved type-specific short sequence (2–5 nt) that directly flanks the protospacer sequence on the mobile genetic element. PAMs are not inserted in the CRIPSR array during the stage of spacer acquisition. This specific signature of the invading DNA allows the interference complex to differentiate between the spacer of the CRISPR locus on the host chromosome (self) and the invading DNA (non-self) (Deveau et al.2008; Mojica et al.2009; Shah et al.2013). Thus, PAM sequences of the invading DNA do not base pair with the respective positions of the CRISPR repeat flanking the corresponding spacer sequence in the array (Westra et al.2013). Initially, the PAM is used for the selection of new spacers during acquisition to guarantee the integration of functional spacers in the host CRISPR locus (Datsenko et al.2012; Swarts et al.2012; Yosef, Goren and Qimron 2012). Since spacer acquisition and DNA interference are performed by two different molecular machineries, analyses showed that the respective motifs are not necessarily identical for these two processes. The PAM recognition is an exact process during the DNA interference reaction, as typically only a single 2–3 bp motif is tolerated (Westra et al.2013; Rollins et al.2015). One exception is the type I-B system of Haloferax volcanii, in which different PAM sequences were found to be functional in DNA targeting (Fischer et al.2012). Usually, several different PAM sequences of 2–5 bp length are tolerated during the spacer acquisition process (Yosef, Goren and Qimron 2012; Fineran et al.2014). Therefore, the additional terms spacer acquisition motif and target interference motif for the respective recognition sites were proposed (Shah et al.2013). In type I systems, the PAM is located on the target crRNA strand at the 3′ end of the protospacer (Mojica et al.2009; Westra et al.2013). For type I-E, the dsDNA enters Cascade in the gap between Cas7.5 and Cas7.6 and is then transferred to the large subunit Cse1, which has several, mostly non-specific, interactions with the target dsDNA. It is shown that a conserved structural motif of the L1 loop in Cse1 is mediating the PAM identification (Sashital, Wiedenheft and Doudna 2012; Hochstrasser et al.2014; Mulepati, Heroux and Bailey 2014; Tay, Liu and Yuan 2015).
Target DNA binding and R-loop formation
The PAM recognition by Cse1 triggers the destabilization of the adjacent DNA duplex and allows the crRNA to access the target DNA strand (Szczelkun et al.2014). Effective R-loop formation requires the full complementarity of a crRNA seed region and the protospacer, while a mismatch inhibits Cascade-mediated targeting. This seed region covers the positions 1–5 and 7–8 at the 5′ end of the crRNA guide adjacent to the PAM (Semenova et al.2011; Wiedenheft et al.2011b; Fineran et al.2014). The intertwined architecture of the Cascade backbone explains this phenomenon, as the Cas7 and Cas5 folding shields the crRNA base at positions −1 and 6. This makes the adjacent 5-bp segment accessible for target hybridization (Jackson et al.2014a; Mulepati, Heroux and Bailey 2014; Zhao et al.2014). The crRNA-guided strand invasion proceeds further throughout the entire guide sequence, in which the later positions affect primarily the R-loop stability and a limited number of single mismatches between crRNA and ssDNA target are tolerated (Semenova et al.2011; Fineran et al.2014; Szczelkun et al.2014). The non-target strand is displaced during R-loop formation and is spanned from Cse1 and the Cse2 dimer to Cas6e via a basic groove (Tay, Liu and Yuan 2015). At the same time, the large and small subunits rotate upon targeting, which creates binding pockets for R-loop stabilization accessible on the surfaces of Cse2 (Westra et al.2012a; Mulepati, Heroux and Bailey 2014). Furthermore, a locking mechanism of the established R-loop structure after the recruitment of Cas3 to Cascade was shown (Rutkauskas et al.2015). The base pairing of the later crRNA guide nucleotides (positions 24–30), that are accessible upon Cse2.1 movement, might prevent this subunit from retraction (Mulepati, Heroux and Bailey 2014; Szczelkun et al.2014).
Cascade-mediated Cas3 recruitment and target DNA degradation
Following R-loop formation and the conformational changes of the large and small subunit, interaction sites at the base of Cse1 are accessible for creating a stable interaction to the CTD of Cas3 (Westra et al.2012b; Hochstrasser et al.2014; Mulepati, Heroux and Bailey 2014). This recruitment at the dsDNA fork site of the R-loop might trigger the CTD to transiently dissociate from the Cas3 core to open the ssDNA channel localized within the helicase domains RecA2 and RecA. After CTD repositioning and closing of the DNA channel, the non-target strand is then positioned in the catalytic center of the HD nuclease domain (Gong et al.2014; Huo et al.2014). The HD nuclease nicks the non-target DNA strand at position ∼11–15 downstream of the PAM with the use of two catalytic transition-state metal ions (Mulepati and Bailey 2013; Sinkunas et al.2013). This nick triggers a change of conformation of the helicase into an active stage, which allows ATP binding and hydrolysis (Sinkunas et al.2011). The dsDNA is unwound in 3′-5′ direction at a separation hairpin of RecA2. The movement of the helicase domain translocates the HD domain to a new substrate position for exonucleolytical degradation of the non-target strand (Gong et al.2014; Huo et al.2014). It is not known if Cascade is still involved in the dsDNA unwinding or if it is displaced by the progression of the Cas3 helicase (Hochstrasser et al.2014). The remaining single-stranded target DNA is then exonucleolytically degraded by a stand-alone Cas3 enzyme (Mulepati and Bailey 2013; Sinkunas et al.2013). Thus, the target DNA is effectively removed from the host cell and the Cascade crRNP can be recycled for another round of target DNA recognition.
MECHANISM OF TYPE II DNA INTERFERENCE
Three crystal structures (2.2–2.6 Å) of S. pyogenes Cas9, one as apoenzyme (Jinek et al.2014) or two as Cas9 bound to an sgRNA and target DNA (Anders et al.2014; Nishimasu et al.2014) were recently resolved. A fourth structure of Cas9 from Actinomyces naeslundii was also reported (Jinek et al.2014). The three studies independently show a bilobal structure of Cas9 proteins in a crescent-shaped conformation (100 × 100 × 50 Å) (Fig. 3).
In S. pyogenes, a Cas9 recognition lobe (REC, residues 60–718) consists mainly of α-helices and is involved in both target recognition and binding. The REC lobe is composed of three regions: (i) a long α-helix (bridge-helix, residues 60–94), (ii) a Rec1 domain (residues 94–180, 308–718) and (iii) a Rec2 domain (residues 180–308). REC was reported to be a specific feature of Cas9 proteins since no structural similarities with other proteins could be identified. This lobe is also the least conserved portion of the protein. While the Rec2 domain is dispensable for target cleavage, the Rec1 domain contains a region that is crucial for the recognition of the tracrRNA anti-repeat:crRNA repeat duplex and is therefore necessary for Cas9 activity. Additionally, the Cas9 crystal structure shows that the Rec1 domain and the arginine-rich motif (RRM) on the bridge-helix bind the guide sequence of the sgRNA (Nishimasu et al.2014). Both motifs interact with the backbone phosphates of the sequence and not with the nucleobases, indicating that recognition of the guide portion of the sgRNA is sequence independent. The recognition of the tracrRNA anti-repeat:crRNA repeat duplex by the REC lobe on the other hand is sequence specific since Cas9 directly interacts with the respective nucleobases. The Rec1 and RRM interaction with the phosphate backbone of the guide RNA leads to an exposure of the eight PAM proximal nucleotides to the solvent. This observation led to the visualization of a seed region, which serves as a nucleation start site enabling target binding and finally cleavage by the nuclease lobe (Jinek et al.2012). Furthermore, the RRM motif, involved in guide RNA recognition and RNA strand invasion, is a feature that is conserved amongst Cas9 proteins of all three subtypes of type II systems, and is one of the two linkers that connect the two lobes of Cas9 (Nishimasu et al.2014).
The Cas9 nuclease lobe (NUC) is composed of the HNH (residues 775–909) and the split RuvC (residues 1–60, 718–775, 909–1099) nuclease domains and additionally contains a C-terminal topoisomerase homology domain (CTD, residues 1099–1368). The NUC lobe is involved in PAM recognition by the CTD and target cleavage by the domains RuvC and HNH (Anders et al.2014; Jinek et al.2014; Nishimasu et al.2014). Interestingly, it was observed that the HNH domain is disordered in the apoenzyme structure leading to the assumption that this domain is flexible with respect to its position during target DNA recognition and cleavage (Jinek et al.2014). The target bound structures confirm this hypothesis and show that the HNH domain undergoes a major conformational change yielding an active state of the Cas9 protein (Fig. 6). During this conformational reorientation, the REC lobe also alters its position leading to the formation of a positively charged central channel that harbors the substrate DNA (Anders et al.2014; Nishimasu et al.2014). A second but significantly smaller positively charged cleft is situated directly within the NUC lobe. This surface is formed between the CTD and the RuvC domain and harbors the 3′ tail of the sgRNA (Jinek et al.2014; Nishimasu et al.2014). The elongated CTD (or PAM interacting) is composed of seven α-helices, a three-stranded, a five-stranded and a two-stranded β-sheet. It displays another Cas9-specific fold that lacks structural similarities to other proteins. This part of Cas9 alone recognizes the PAM and it was shown to be sufficient to change this domain to alter PAM specificities (Anders et al.2014; Nishimasu et al.2014).
Figure 6.
Mechanism of type II Cas9-mediated DNA interference. In the absence of type II specific dual-RNA, Cas9 is in an inactive state. Upon binding to tracrRNA and crRNA, Cas9 undergoes conformational changes, subsequently enabling dual-RNA guided binding to the target DNA. After successful DNA interrogation for a PAM and subsequent nucleation, the guide RNA pairs with the seed sequence on the DNA. This is followed by activation of the Cas9 nuclease domains HNH and RuvC for cleavage of the target and non-target DNA strand, respectively.
DNA interrogation and PAM recognition by Cas9:sgRNA
The binding of tracrRNA to crRNA, the subsequent duplex formation and co-processing of the dual-RNA converts an inactive single Cas9 into a DNA surveillance active complex (Deltcheva et al.2011; Jinek et al.2012). Once activated, Cas9 is ready to screen for targeted sites on an invading DNA. In type II systems, as well as in type I systems, the discrimination of target and non-target DNA is mediated via the recognition of the PAM (Deveau et al.2008; Garneau et al.2010; Sapranauskas et al.2011). It was shown that binding to target DNA, as well as the subsequent cleavage, requires a PAM (Gasiunas et al.2012; Jinek et al.2012; Sternberg et al.2014). The PAM screening process by Cas9 was investigated in single molecule experiments using DNA curtains. In these experiments, Cas9 displayed unspecific binding to the target, which strongly correlated with the presence of a PAM. Substrates without a PAM but containing the sgRNA-targeted sequences are ignored by Cas9, confirming the importance of the motif (Sternberg et al.2014). Once Cas9 has identified the dsDNA PAM, it binds to the target. This binding results in a local melting of the target DNA and enables the formation of a RNA-DNA heteroduplex that begins at the PAM (Sternberg et al.2014). PAM recognition is facilitated by the CTD of Cas9 proteins. In the case of S. pyogenes Cas9 (SpyCas9), the 5′-NGG-3′ PAM is recognized by two arginine residues (R1333 and R1335) that reside in a DRKRY motif that is conserved amongst the 5′-NGG-3′ recognizing Cas9 proteins (Anders et al.2014). Interestingly, arginine residues typically pair with guanine nucleotides, while for example glutamine residues usually pair with adenines. Cas9 proteins that recognize the 5′-NAAAA-3′ PAM contain a QLQ motif instead of the RKR of SpyCas9 (Anders et al.2014). The recognition of the dG2 and dG3 on the non-complementary (non-target) strand of the target DNA correlates with strand separation since the following interaction of the lock loop (Lys1107-Ser1109) results in a rotation of the first phosphate. This rotation enables pairing with the guide RNA and displays the start of guide RNA and target DNA binding (Anders et al.2014).
The following R-loop formation is a unidirectional kinetic event in which the PAM is only required for licensing of the DNA distortion but not for R-loop stability. Following the melting event, the guide RNA searches for homology within the 8 – 12 nt seed sequence and H-bonding is formed between the guide RNA and the target DNA sequence (Sternberg et al.2014; Szczelkun et al.2014). Only when the seed sequence can pair, the R-loop will continue to propagate from here on (Szczelkun et al.2014). While Cascade of type I locks the R-loop for Cas3 recruitment, no such stabilization is needed for type II as Cas9 itself is the nuclease cleaving the target. Despite the locking step that is required for type I R-loop formation, the overall kinetics are faster compared to the Cas9 driven event (Szczelkun et al.2014). In addition to the discrimination between self and non-self, it is likely that binding to the PAM activates the nuclease activity of Cas9 (Sternberg et al.2014). The analysis of the Cas9 structure bound to its target DNA could show that van der Waals interactions with the C2′ atoms of the target help to discriminate between DNA and RNA (Nishimasu et al.2014).
Cas9-mediated DNA cleavage
DNA targeting of type II CRISPR-Cas systems requires tracrRNA and crRNA, which can be combined into a single molecule (Jinek et al.2012). The tracrRNA of S. pyogenes contains three regions resulting in stem-loop hairpins located 3′ to the tracrRNA:crRNA duplex-forming segment (Deltcheva et al.2011). Truncations of the wild-type tracrRNA directed at identifying minimal requirements for target DNA binding and cleavage showed that a substantially shortened tracrRNA retaining only 25 nucleotides but comprising the first hairpin could still support cleavage (Jinek et al.2012). As would be expected, longer sequences more closely resembling the wild type were effective (Jinek et al.2012), and the second and third loops have been confirmed to enhance stability, while the first loop is indispensable (Nishimasu et al.2014). The formation of a ternary complex occurs upon target DNA binding. In this event, the sgRNA and the dsDNA target form a T-shaped structure, which yields a four-way junction in which the heteroduplex of guide and target strand is placed into the major groove formed by the REC lobe and the NUC lobe. The double-stranded PAM is located within the C-terminal domain enabling target recognition and R-loop initiation (Anders et al.2014).
The following target cleavage by Cas9 proteins is performed by the NUC lobe. The two nuclease domains HNH and RuvC cleave the complementary and the non-complementary strand, respectively (Sapranauskas et al.2011; Gasiunas et al.2012; Jinek et al.2012). During this reaction, a double-strand break is introduced within the protospacer, exactly 3 nt upstream from the PAM (Gasiunas et al.2012; Jinek et al.2012). With a ββα-fold (or ββα-metal motif) that resembles the active site of T4 endonuclease, the HNH domain employs a single metal mechanism with one Mg2+ coordinated by an aspartate and an asparagine residue. An histidine residue serves as general base in the cleavage reaction of the complementary strand and completes the active site of the HNH domain. The RuvC domain is characterized by an RNase H-fold in which two aspartates, a glutamate and an histidine residue coordinate Mg2+ ions to facilitate a two-metal reaction to cleave the non-complementary strand of the target DNA (Jinek et al.2014; Nishimasu et al.2014). The cleavage activity of both domains yields a blunt-ended double-strand break (Sapranauskas et al.2011; Gasiunas et al.2012; Jinek et al.2012). Following cleavage, Cas9 remains tightly associated to the ends of the targeted DNA, characterizing the protein as a single turnover enzyme (Sternberg et al.2014).
STRUCTURES AND TARGETING MECHANISMS OF TYPE III DNA AND RNA INTERFERENCE COMPLEXES
The Csm complex
The structures of several type III crRNP complexes were recently visualized by electron microscopy, enabling the identification of striking similarities to the Cascade I-E structure (Zhang et al.2012; Rouillon et al.2013; Spilman et al.2013; Staals et al.2013, 2014).
The type III-A Csm complex of Su. solfataricus is composed of 13 subunits that are arranged in a basal body and two intertwined filaments (Fig. 7). Several Csm3 paralogs were identified and numbered Csm3.1–Csm3.5 based on their position in the csm operon. Located at the base of the Csm complex, the large subunit Cas10 serves as an anchor for the major and the minor filaments (Rouillon et al.2013). The major filament consists of four Csm3.2 subunits, two different Csm3 subunits (Csm3.5 and Csm3.3) and Csm4. The crRNA is located within this helical assembly (Rouillon et al.2013). The minor filament consists of three Csm2 subunits and two additional Csm3 subunits (Csm3.1 and Csm3.4). Directly adjacent to the large subunit Cas10 at the base are Csm4 and Csm3.4, while the head of the complex is formed by three Csm3 subunits (Csm3.1, Csm3.2 and Csm3.5). The EM map comparison of the Csm complex and Cascade revealed that the assembly of the Cascade backbone with its six Cas7 subunits correlates with the Csm3 subunit composition of the major filament (Csm3.3, 4 monomers of Csm3.2 and Csm3.5) with an identical pitch of both backbones (Rouillon et al.2013). In agreement, the crystal structure of the Methanopyrus kandleri Csm3 provided evidence that its domain architecture resembles the one of Cas7 (Hrle et al.2013). As mentioned earlier, Csm4 is located at the base of the Csm complex and its position and overall structure correlates with Cas5 within the Cascade structure (Rouillon et al.2013). The type III-A crRNPs show some variability in the composition of subunits, as one Csm3 subunit in the minor filament can be replaced by the similar RRM-fold protein Csm5 in T. thermophilus or S. thermophilus. Overall, five to ten Csm3 and one or two Csm4 subunits, depending on the crRNA length, form the crRNA-interacting backbone (Staals et al.2014; Tamulaitis et al.2014).
Figure 7.
Comparison of type III crRNP-mediated RNA interference. (A) The type III-A Csm complex of Su. solfataricus is composed of 13 subunits that are arranged as a basal body and two intertwined filaments. Located at the base of the Csm complex is the large subunit Cas10, serving as an anchor for the major and the minor filament. The major filament consists of Csm4 and six Csm3 subunits and binds the crRNA. The minor filament consists of three Csm2 subunits and two additional Csm3 subunits. The Csm3 units that form the backbone of the Csm complex were shown to act as target RNA nucleases (indicated by a scissor) in S. thermophilus. (B) The type III-B Cmr complex of P. furiosus contains an extended helical backbone composed of Cmr4 and Cmr5 subunits. The tail is composed of the stable heterodimer Cas10 and Cmr3, while the curled head contains Cmr1 and Cmr6. The crRNA-binding backbone is formed by several Cmr4 subunits, while the Cas10-Cmr3 heterodimer plays a role in the recognition of the crRNA 5′ handle. A second helical structure is formed by three Cmr5 subunits and is inserted alongside the helical Cmr4 filament. Cleavage of target RNA by Cmr4 was observed in 6-nt intervals (indicated by a scissor).
Similar to type I systems, Cas6 endonuclease cleavage in type III-A generates crRNAs with an 8-nt 5′ handle derived from the repeat sequence (Hatoum-Aslan, Maniv and Marraffini 2011; Reeks et al.2013b; Shao and Li 2013). This handle might directly be protected by Csm4 and the large subunit Cas10 (Staals et al.2014; Tamulaitis et al.2014; Numata et al.2015). Remarkably, a distinctively shortened repeat-derived crRNA 3′ handle was observed that might correspond to a defined protection of the crRNA based on the fixed number of Csm3 subunits in the backbone, while the unprotected RNA overhangs are accessible for exonucleolytical trimming (Hatoum-Aslan et al.2013; Rouillon et al.2013; Staals et al.2014). Thus, variations in the number of Csm3 subunits and the trimming of crRNA 3′ handles might be consequences of different spacer lengths or a heterogeneous pool of assembled Csm complexes (Staals et al.2014; Tamulaitis et al.2014). Consequently, a stabilizing hairpin structure of the crRNA 3′ handle or a stable interaction of Cas6 with the Csm complex was not observed (Rouillon et al.2013). A requirement for PAM recognition during the targeting of nucleic acid in III-A systems appears to be absent (Staals et al.2014; Tamulaitis et al.2014). Instead, the base pairing of the crRNA 5′ handle with the host chromosome seems to block the propagation of the interference reaction leading to ‘self-inactivation’ (Marraffini and Sontheimer 2010). The large subunit of the Csm complex is proposed to be involved in targeting, followed by a complementary binding of the crRNA and the matching nucleic acid (Hatoum-Aslan et al.2014). The minor filament includes the putative small subunit Csm2 and is proposed to be involved in target binding based on the morphological architecture of the two intertwined filaments and the structural similarities between Csm2 and the small subunits of the type I system (Reeks et al.2013a; Rouillon et al.2013). Recently, the binding and cleavage of complementary target ssRNAs was observed in vivo and in vitro. The Csm complexes generated multiple cuts at 6-nt intervals within the protospacer RNA (Staals et al.2014; Tamulaitis et al.2014). The multicopy subunit Csm3, which forms the backbone of the Csm complex, acts as endoribonuclease and thus a mutation of D33 in S. thermophilus abolished the RNA cleavage (Tamulaitis et al.2014). In contrast, the previous reported activity on target DNAs could not be shown in vitro yet. Two possible scenarios for DNA degradation are discussed. One candidate for DNA cleavage is Cas10 as it has a permuted HD domain and showed 3′-5′ exonuclease activity for ssDNA and RNA in vitro (Cocozaki et al.2012; Ramia et al.2014a; Jung et al.2015). Another possibility is that the Csm6 protein needs to be recruited to the Csm complex, as a homologous protein was shown to be involved in the interference mechanism of the III-B system (Deng et al.2013). A transcription-dependent DNA targeting mechanism was also proposed by Goldberg et al., which may provide a possible explanation for RNA cleavage observed in in vitro studies (Goldberg et al.2014; Staals et al.2014; Tamulaitis et al.2014).
The Cmr complex
Detailed structural information of the RNA-targeting type III-B Cmr complex is available for T. thermophilus and P. furiosus (Spilman et al.2013; Staals et al.2013; Benda et al.2014). The T. thermophilus Cmr complex is composed of 11 subunits with four copies of Cmr4 and three Cmr5 subunits, while in P. furiosus either three or four Cmr4 subunits were detected (Spilman et al.2013; Staals et al.2013; Benda et al.2014; Ramia et al.2014a). In all cases, two major crRNA species (T. thermophilus: 40 and 46 nt or P. furiosus: 39 and 45 nt) were identified that were associated with the complex and contained the characteristic 8-nt 5′ handle and a trimmed 3′ end (Juranek et al.2012; Staals et al.2013). In reference to the Cascade sea-horse structure, the Cmr complex was described as a ‘sea-worm’ structure with an extended helical backbone consisting of Cmr4 and Cmr5 subunits (Fig. 7). The tail is composed of the stable heterodimer Cas10 and Cmr3, while the curled head contains Cmr1 and Cmr6 (Spilman et al.2013; Staals et al.2013; Benda et al.2014). Structural analysis of Cas10 revealed a triangular four-domain protein core that has two adenylyl cyclase-like domains arranged as a homodimer and two smaller α-helical domains (Cocozaki et al.2012). A characteristic treble-clef zinc-finger motif and a degenerated GGDD motif for binding of a Mn2+ ion were identified. Additionally, an N-terminal, permuted HD domain, containing two Mn2+ ions, is fused to Cas10 but showed no nuclease activity (Cocozaki et al.2012; Benda et al.2014). The N-terminal triangular side of Cas10 binds Cmr3, a subunit with two RRM domains (Osawa, Inanaga and Numata 2013; Shao et al.2013; Benda et al.2014). At this interface of Cas10-Cmr3, a cleft is formed that serves as a binding pocket for nucleotides embedded within the conserved adenylyl cyclase domain of Cas10. These embedded nucleotides play a role in the recognition of the crRNA 5′ handle (Osawa, Inanaga and Numata 2013; Shao et al.2013; Hale et al.2014). A helical filament of four Cmr4 subunits closely interacts with the Cas10-Cmr3 base. The Cmr4 protein contains one central RRM core with several inserted elements and a flexible thumb domain, which results in a more elongated structure in comparison to several other members of the Cas7 family (Benda et al.2014; Ramia et al.2014a). The Cmr4 helical structure is capped by Cmr6 whose single RRM domain shows a remarkable similarity to the N-terminal RRM of Cmr1, while the Cmr6 thumb domain superimposes with the thumb domain of Cmr4 (Benda et al.2014). The last protein in this sequential RRM arrangement of the Cmr backbone is Cmr1. Cmr1 is composed of two close-fitting RRM domains that form a groove of conserved basic and hydrophobic residues (including W38, W39, R154 in P. furiosus) for RNA binding of the crRNA 3′ region (Benda et al.2014; Hale et al.2014; Sun et al.2014). A second helical structure is formed by three Cmr5 subunits and inserted alongside the helical Cmr4 filament (Benda et al.2014). Cmr5 is a globular α-helical protein, in which the adjacent subunits interact in a head-to-tail arrangement forming the double-helical Cmr complex body (Park et al.2013; Benda et al.2014).
The postulated mechanisms of Cmr complex formation and RNA targeting highlight remarkable similarities to the previously described type III-A system. These encompass Cas6-mediated crRNA maturation and the direct protection of the 5′ handle by the Cas10-Cmr3 heterodimer (Spilman et al.2013; Hale et al.2014). The crRNA-binding backbone is then assembled by several Cmr4 subunits and the crRNA 3′ end is sandwiched between the Cmr1 and the Cmr6 subunit (Benda et al.2014; Hale et al.2014). The variable number of three or four Cmr4 subunits observed for different organisms correlates with the two incorporated crRNA species that differ in length by 6 nt (Benda et al.2014). The assembled Cmr4 backbone supports target interaction via direct contacts of the Cmr4 palm domain to the ssRNA target (Ramia et al.2014a). Cas10 is proposed to fulfill the role of the large subunit and, together with the three copies of the small subunit Cmr5, might be involved in complex stability and RNA targeting (Spilman et al.2013; Staals et al.2013). The observed target ssRNA cleavage mechanism occurs in three to four 6-nt intervals. The first cut was shown 5 nt away from the 5 ′ end and the last cut 14 nt away from the 3′ end of the target ssRNA/crRNA duplex (Hale et al.2009, 2014; Staals et al.2013; Zhu and Ye 2015). Modeling a target ssRNA into the pseudoatomic Cmr complex model identified a potential catalytic center (H15, D26 and E227) within the Cmr4 subunits, whose distances would be able to explain the observed cleavage pattern (Benda et al.2014). Accordingly, the mutation of the D26 residue abolished endonucleolytical activity without a loss of crRNA interaction or Cmr complex formation (Benda et al.2014; Hale et al.2014; Ramia et al.2014a). In addition, the Cmr4 residue K46 was identified to be important for crRNA binding (Ramia et al.2014a).
COMMON THEMES AMONG CRISPR-CAS SURVEILLANCE COMPLEXES
The recent elucidation of the structures of several Cas proteins and crRNP complexes, together with biochemical studies, revealed a common architectural core of type I and type III CRISPR-Cas surveillance complexes. Individual subunits often fulfill similar functions, despite their remarkable sequence diversity. Basically, each of these crRNP complexes is built up by a crRNA-binding helical backbone that is composed of at least seven RRM domain proteins, one large subunit and two or three small subunits. The protein backbone responsible for crRNA recognition and interaction is structurally more conserved than the small and large subunits that display more divergent architectures. In the following section, we focus on the comparison of common themes among crRNP surveillance complexes.
Structural conservation of crRNA binding in type I and type III crRNP complexes
All of the identified proteins that interact with the crRNAs in type I and type III CRISPR-Cas systems contain the RRM domain, an ssRNA-binding domain that is widely distributed in all domains of life and involved in many aspects of RNA binding, regulation and maintenance (Maris, Dominguez and Allain 2005). This group of Cas proteins that contain a conserved RRM domain was previously classified as the RAMP (repeat associated mysterious proteins) superfamily of Cas proteins and further divided into three main groups: Cas7, Cas5 and Cas6 (Makarova et al.2011a; Koonin and Makarova 2013). The observed structural similarities support a shared evolutionary origin of all crRNA-interacting Cas proteins. Subsequently, Cas proteins evolved with specialized functions of their individual RRM domains (Koonin and Makarova 2013).
Apart from the type I-E Cas7 structure described above, two other structures of Cas7 enzymes from type I-A (Csa2) and I-D (Csc2) were solved (Lintner et al.2011; Hrle et al.2014). Additionally, the Csm3 protein of type III-A as well as the Cmr4 protein of type III-B could be functionally and structurally linked to members of the Cas7 family (Hrle et al.2013; Ramia et al.2014a; Numata et al.2015). All these structures reveal a similar domain arrangement, consisting of the modified RRM (palm domain), the helical finger domain and the thumb domain. The exact structure of the I-A Cas7 and III-A Csm3 thumb domain is not known as the disordered region suggests the presence of a flexible loop. Mutations within the thumb domain decreased the crRNA affinity in both proteins (Lintner et al.2011; Hrle et al.2013). In contrast, the thumb of I-D Cas7 showed no direct influence on crRNA interaction, while mutations in the conserved palm domain interfered with RNA binding (Hrle et al.2014). All Cas7 members are suggested to provide the crRNP platform by oligomerizing on the crRNA, which protects the crRNA from degradation and enables hybridization with the target nucleic acids (van der Oost et al.2014). Furthermore, the Cas7 members of type III systems are endoribonucleases that catalyze the target cleavage (Ramia et al.2014a; Staals et al.2014; Tamulaitis et al.2014). Other members of the Cas7 family are found in type I systems (I-B, I-C and I-F) as well as type III systems (Csm5, Cmr6 and Cmr1) that share structural and sequence similarities with Cas7 (Makarova et al.2011a; Benda et al.2014).
The superimposition of available Cas5 structures of I-E (Cas5e), I-C (Cas5d), III-A (Csm4) and III-B (Cmr3) highlights a conserved core consisting of a modified RRM and a thumb domain (Nam et al.2012a; Koo et al.2013; Shao et al.2013; Jackson et al.2014a; Numata et al.2015). In all cases, a conserved binding pocket was identified that is most likely involved in the recognition of the specific crRNA 5′ handle (Jackson et al.2014a). Remarkably, some of the Cas proteins in the CRISPR-Cas systems can functionally be replaced. One example is the substitution of Cas5d for Cas6 in type I-C crRNP complexes. The evolution of this subunit is reflected in the modulation of the Cas5d structure as the Cas5 core is extended at its C-terminus with a repeat-specific endonuclease active site (Nam et al.2012a). A similar phenomenon is observed for the structure of Cmr3, which contains a second RRM domain at the C-terminus that is likely involved in the crRNP assembly process (Shao et al.2013). Additional candidates for Cas5 family members are found in the other type I systems (I-A, I-B, I-D and I-F). These proteins share a similar structural core, are proposed to recognize the crRNA 5′ handle and closely interact with the large subunit (Cas8, Cse1, Csy1 or Cas10) in the respective systems (Makarova et al.2011a; Shao et al.2013; Jackson et al.2014a).
Members of the Cas6 family show a two RRM domain arrangement and are experimentally characterized as crRNA-processing endonucleases in all type I and III systems, even though they show structural variations and a high-sequence divergence (Reeks, Naismith and White 2013; Niewoehner, Jinek and Doudna 2014). In the Cascade context, the I-E Cas6 is delivering the mature crRNA and tightly caps the 3′ handle (Sashital, Jinek and Doudna 2011; Jackson et al.2014a). This protection results in crRNAs with a complete 3′ handle, which was also shown for type I-F (Haurwitz et al.2010). In contrast, crRNAs from other type I (I-A, I-B, I-D) systems and all type III systems were shown to harbor trimmed 3′ ends. Consequently, the corresponding Cas6 enzymes are not likely to be permanent subunits of their crRNP complexes, but are rather stand-alone ribonucleases (Hatoum-Aslan, Maniv and Marraffini 2011; Richter et al.2012b; Plagens et al.2014). The varying affinities of Cas6 to mature crRNA products might be a consequence of the presence or absence of stable stem-loop structures in the crRNA 3′ handle or structural variations of Cas6 (Kunin, Sorek and Hugenholtz 2007; Niewoehner, Jinek and Doudna 2014). The diversification of Cas6 enzymes correlates with repeat sequence variation. However, the reasons for the evolution of different mechanisms of Cas6-mediated repeat cleavage and crRNA delivery to the crRNP complexes are not yet understood.
Diversification of Cas proteins responsible for interference in type I and III CRISPR-Cas systems
The large and small subunits of different type I and type III crRNP complexes show limited sequence conservation and little structural similarities (Makarova et al.2011a; Makarova, Wolf and Koonin 2013). The large subunits comprise members of the Cas8 (I-A, I-B, I-C) and Cas10 (I-D, III-A, III-B) family as well as Cse1 and Csy1. In general, the classification of the large subunits is mainly based on their size, their function in PAM recognition and R-loop formation and/or their position in the crRNP complex (van der Oost et al.2014). Comparison of the two available structures of Cse1 of the I-E system and Cas10 of subtype III-B did not reveal structural similarities. The overall Cse1 structure has a unique globular fold with a coordinated zinc-ion and a C-terminal four-helix bundle (Mulepati, Orr and Bailey 2012; Jackson et al.2014a). In contrast, Cas10 has a four-domain protein core with two adenylyl cyclase-like domains and a permuted HD domain (Cocozaki et al.2012). Computational analyses predicted a common arrangement of core cyclase/polymerase-like domains including finger, palm and thumb domains for all large subunits (Makarova et al.2011a). Additionally, numerous members of the Cas8, Cas10d, Cas10 and Cse1 groups were suggested to contain a little conserved RRM-fold inside the palm domain (Makarova et al.2011b). However, Cse1 lacks a palm-like RRM and shared no common architectural layout with Cas10. One reason for the missing structural similarities of the diverse large subunits might be extensive structural rearrangements of the proteins (Koonin and Makarova 2013).
Similarly, only very limited structural conservation was observed for the small subunits of different crRNP complexes. Structures of the small subunits have been solved for type I-E (Cse2), I-A (Csa5) and subtype III-B systems (Cmr5) (Agari et al.2008; Sakamoto et al.2009; Park et al.2013; Reeks et al.2013a). In general, all these proteins are characterized by the presence of several α-helices, but show only very limited conservation between the N-terminal domains of Cse1 and Cmr5 and the C-terminal domains of Cse1 and Csa5 (Reeks et al.2013a). To further complicate this issue, it has been proposed that the small subunit is fused to the large subunits in several subtypes (I-B, I-C, I-D, I-F). Even within the same subtype, small subunits can show only very limited sequence similarity as observed for Csa5 of type I-A (Makarova et al.2011a; Daume, Plagens and Randau 2014).
Several reasons can be discussed for this phenomenon of Cas protein diversification. The proteins that interact with the crRNA, namely members of the Cas5, Cas6 and Cas7 families, likely show structural similarities, as the crRNA component is similar in all different subtypes (Makarova et al.2011a). In all type I and III systems, the 8-nt 5′ handle and a similar crRNA length is observed, which indicates an evolutionary conservation of the primary repeats within the host CRISPR loci (Kunin, Sorek and Hugenholtz 2007). Minor structural differences might be a consequence of the adaptation to variable prokaryotic growth parameters in the environment. In contrast, the large and small subunits interact with the respective targets in the surveillance complex. One obvious reason for different Cas protein target recognition mechanisms is the presence of either dsDNA or ssRNA as target molecules (van der Oost et al.2014). To effectively scan a compatible target, the respective proteins have to evolve specialized binding patches for either R-loop formation or the hybridization of RNA:RNA duplexes (Benda et al.2014; Mulepati, Heroux and Bailey 2014). Another aspect to consider is the high selective pressure for the continuous effectiveness of the crRNP surveillance. Different studies report evidence for the arms race of viruses and their prokaryotic host (Levin et al.2013; Seed et al.2013; Fineran et al.2014). This means that diverse crRNP complexes have likely evolved in reaction to viral measures to avoid crRNA-mediated targeting. One way to escape targeting is the introduction of mutation in the region targeted by the crRNA guide, which is most efficient for the seed sequence region. Alternatively, the introduction of variations in the PAM can also inhibit base pairing with the target (Deveau et al.2008; Westra et al.2013). Interestingly, recent experiments show that Cascade does not only bind targets tightly, but also establishes ‘low fidelity’ interactions with mutated targets independent of PAM or seed sequences. This process is proposed to allow the acquisition of new spacers (Blosser et al.2015). Another striking example of this arms race are phage-encoded anti-CRISPR genes that enable viruses to evade the CRISPR-Cas systems (Bondy-Denomy et al.2013; Pawluk et al.2014). Thus, the large and small subunits, as a central point in the surveillance machinery, have to adapt to these target variations and viral evasion strategies, possibly providing an explanation for their high degree of diversification. Finally, different mechanisms of target degradation are identified in the type I and III systems. In type I-E systems, Cas3 docks to the large subunit Cse1 of Cascade after R-loop formation, while the type I-A Cas3 is proposed to be an integral part of the complex (Hochstrasser et al.2014; Plagens et al.2014). The interaction of Cas3 to Cascade is mediated by its CTD. As a possible consequence of structural variations of the large subunits, Cas3 sequences show remarkable diversification of the CTD, despite harboring a highly conserved common core (Huo et al.2014; Jackson et al.2014b). In type III systems, the cleavage of the target is executed by the Cas7-like protein Cmr4 in the Cmr complex or Csm3 in the Csm complex (Rouillon et al.2013; Benda et al.2014; Staals et al.2014; Tamulaitis et al.2014). These observations highlight examples for striking differences among the crRNP complexes. Future studies on both CRISPR-Cas systems and their natural virus and plasmid targets should provide us with a better understanding of the coevolution and diversification of these antagonists.
Acknowledgments
We thank Ines Fonfara for helpful comments.
FUNDING
This work was supported by funding from the DFG (FOR1680). EC is supported by the Alexander von Humboldt Foundation, the German Federal Ministry for Education and Research, the Helmholtz Association, the Göran Gustafsson Foundation, the Swedish Research Council, the Kempe Foundation and Umeå University. HR is supported by the Helmholtz Post-doctoral Programme.
Conflict of interest. None declared.
REFERENCES
- Agari Y, Yokoyama S, Kuramitsu S, et al. X-ray crystal structure of a CRISPR-associated protein, Cse2, from Thermus thermophilus HB8. Proteins. 2008;73:1063–7. doi: 10.1002/prot.22224. [DOI] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, et al. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–73. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Beloglazova N, Petit P, Flick R, et al. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30:4616–27. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benda C, Ebert J, Scheltema RA, et al. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell. 2014;56:43–54. doi: 10.1016/j.molcel.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Blosser TR, Loeff L, Westra ER, et al. Two distinct DNA binding modes guide dual roles of a CRISPR-Cas protein complex. Mol Cell. 2015;58:60–70. doi: 10.1016/j.molcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL, et al. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–32. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel J, Stoll B, Lange SJ, et al. A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii. J Biol Chem. 2014;289:7164–77. doi: 10.1074/jbc.M113.508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briner AE, Donohoue PD, Gomaa AA, et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56:333–9. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, et al. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Gene Dev. 2008;22:3489–96. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Choi J, Bailey S. Cut site selection by the two nuclease domains of the Cas9 RNA-guided endonuclease. J Biol Chem. 2014;289:13284–94. doi: 10.1074/jbc.M113.539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–37. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylinski K, Makarova KS, Charpentier E, et al. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:6091–105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocozaki AI, Ramia NF, Shao Y, et al. Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex. Structure. 2012;20:545–53. doi: 10.1016/j.str.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, et al. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- Daume M, Plagens A, Randau L. DNA binding properties of the small cascade subunit csa5. PLoS One. 2014;9:e105716. doi: 10.1371/journal.pone.0105716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–7. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Garrett RA, Shah SA, et al. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol. 2013;87:1088–99. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Ebihara A, Yao M, Masui R, et al. Crystal structure of hypothetical protein TTHB192 from Thermus thermophilus HB8 reveals a new protein family with an RNA recognition motif-like domain. Protein Sci. 2006;15:1494–9. doi: 10.1110/ps.062131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JR, Yokooji Y, Sato T, et al. Programmable plasmid interference by the CRISPR-Cas system in Thermococcus kodakarensis. RNA Biol. 2013;10:828–40. doi: 10.4161/rna.24084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–24. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Gerritzen MJ, Suarez-Diez M, et al. Degenerate target sites mediate rapid primed CRISPR adaptation. P Natl Acad Sci USA. 2014;111:E1629–38. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S, Maier LK, Stoll B, et al. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem. 2012;287:33351–63. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, LeRhun A, Chylinski K, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–90. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Garside EL, Schellenberg MJ, Gesner EM, et al. Cas5d processes pre-crRNA and is a member of a larger family of CRISPR RNA endonucleases. RNA. 2012;18:2020–8. doi: 10.1261/rna.033100.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. P Natl Acad Sci USA. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesner EM, Schellenberg MJ, Garside EL, et al. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–92. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–29. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- Goldberg GW, Jiang W, Bikard D, et al. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–7. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Shin M, Sun J, et al. Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3. P Natl Acad Sci USA. 2014;111:16359–64. doi: 10.1073/pnas.1410806111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. Clustered regularly interspaced short palindromic repeats (CRISPRs) for the genotyping of bacterial pathogens. Methods Mol Biol. 2009;551:105–16. doi: 10.1007/978-1-60327-999-4_9. [DOI] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, et al. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, et al. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–9. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Cocozaki A, Li H, et al. Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex. Gene Dev. 2014;28:2432–43. doi: 10.1101/gad.250712.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. P Natl Acad Sci USA. 2011;108:21218–22. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Samai P, et al. Genetic characterization of antiplasmid immunity through a type III-A CRISPR-Cas system. J Bacteriol. 2014;196:310–7. doi: 10.1128/JB.01130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Samai P, Maniv I, et al. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J Biol Chem. 2013;288:27888–97. doi: 10.1074/jbc.M113.499244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, et al. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–8. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Sternberg SH, Doudna JA. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 2012;31:2824–32. doi: 10.1038/emboj.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein S, Scholz I, Voss B, et al. Adaptation and modification of three CRISPR loci in two closely related cyanobacteria. RNA Biol. 2013;10:852–64. doi: 10.4161/rna.24160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JW, et al. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Taylor DW, Bhat P, et al. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. P Natl Acad Sci USA. 2014;111:6618–23. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Hrle A, Maier LK, Sharma K, et al. Structural analyses of the CRISPR protein Csc2 reveal the RNA-binding interface of the type I-D Cas7 family. RNA Biol. 2014;11:1072–82. doi: 10.4161/rna.29893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrle A, Su AA, Ebert J, et al. Structure and RNA-binding properties of the type III-A CRISPR-associated protein Csm3. RNA Biol. 2013;10:1670–8. doi: 10.4161/rna.26500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Nam KH, Ding F, et al. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol. 2014;21:771–7. doi: 10.1038/nsmb.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PB, et al. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014a;345:1473–9. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Lavin M, Carter J, et al. Fitting CRISPR-associated Cas3 into the Helicase Family Tree. Curr Opin Struct Biol. 2014b;24:106–14. doi: 10.1016/j.sbi.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, van Embden JDA, Gaastra W, et al. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–75. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Jiang W, Maniv I, Arain F, et al. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet. 2013;9:e1003844. doi: 10.1371/journal.pgen.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–36. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Jung TY, An Y, Park KH, et al. Crystal structure of the Csm1 subunit of the Csm complex and its single-stranded DNA-specific nuclease activity. Structure. 2015 doi: 10.1016/j.str.2015.1001.1021. [DOI] [PubMed] [Google Scholar]
- Juranek S, Eban T, Altuvia Y, et al. A genome-wide view of the expression and processing patterns of Thermus thermophilus HB8 CRISPR RNAs. RNA. 2012;18:783–94. doi: 10.1261/rna.031468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis T, Gasiunas G, Miksys A, et al. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 2013;10:841–51. doi: 10.4161/rna.24203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo Y, Ka D, Kim EJ, et al. Conservation and variability in the structure and function of the Cas5d endoribonuclease in the CRISPR-mediated microbial immune system. J Mol Biol. 2013;425:3799–810. doi: 10.1016/j.jmb.2013.02.032. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS. CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013;10:679–86. doi: 10.4161/rna.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Moineau S, Bushman M, et al. The population and evolutionary dynamics of phage and bacteria with CRISPR-mediated immunity. PLoS Genet. 2013;9:e1003312. doi: 10.1371/journal.pgen.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Liu H, Han J, et al. Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei. J Bacteriol. 2013;195:867–75. doi: 10.1128/JB.01688-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintner NG, Kerou M, Brumfield SK, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–56. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadan AH, Dupuis ME, Villion M, et al. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS One. 2012;7:e40913. doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Grishin NV, et al. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–96. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, et al. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011a;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, et al. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011b;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. The basic building blocks and evolution of CRISPR-cas systems. Biochem Soc T. 2013;41:1392–400. doi: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–31. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–71. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel B, Moineau S. CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 2014;42:9504–13. doi: 10.1093/nar/gku628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen AM, Horvath P, Boyaval P, et al. Mobile CRISPR/Cas-mediated bacteriophage resistance in Lactococcus lactis. PLoS One. 2012;7:e51663. doi: 10.1371/journal.pone.0051663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–82. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–40. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Mulepati S, Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) J Biol Chem. 2011;286:31896–903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem. 2013;288:22184–92. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Heroux A, Bailey S. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–84. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Orr A, Bailey S. Crystal structure of the largest subunit of a bacterial RNA-guided immune complex and its role in DNA target binding. J Biol Chem. 2012;287:22445–9. doi: 10.1074/jbc.C112.379503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Haitjema C, Liu X, et al. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure. 2012a;20:1574–84. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Huang Q, Ke A. Nucleic acid binding surface and dimer interface revealed by CRISPR-associated CasB protein structures. FEBS Lett. 2012b;586:3956–61. doi: 10.1016/j.febslet.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, Jinek M, Doudna JA. Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases. Nucleic Acids Res. 2014;42:1341–53. doi: 10.1093/nar/gkt922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–49. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata T, Inanaga H, Sato C, et al. Crystal structure of the Csm3-Csm4 subcomplex in the type III-A CRISPR-Cas interference complex. J Mol Biol. 2015;427:259–73. doi: 10.1016/j.jmb.2014.09.029. [DOI] [PubMed] [Google Scholar]
- Nunez JK, Kranzusch PJ, Noeske J, et al. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. 2014;21:528–34. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Lee AS, Engelman A, et al. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–8. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MR, Oakes BL, Sternberg SH, et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–6. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Inanaga H, Numata T. Crystal structure of the Cmr2-Cmr3 subcomplex in the CRISPR-Cas RNA silencing effector complex. J Mol Biol. 2013;425:3811–23. doi: 10.1016/j.jmb.2013.03.042. [DOI] [PubMed] [Google Scholar]
- Park JH, Sun J, Park SY, et al. Crystal structure of Cmr5 from Pyrococcus furiosus and its functional implications. FEBS Lett. 2013;587:562–8. doi: 10.1016/j.febslet.2013.01.029. [DOI] [PubMed] [Google Scholar]
- Pawluk A, Bondy-Denomy J, Cheung VH, et al. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. MBio. 2014;5:e00896. doi: 10.1128/mBio.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A, Tjaden B, Hagemann A, et al. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J Bacteriol. 2012;194:2491–500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A, Tripp V, Daume M, et al. In vitro assembly and activity of an archaeal CRISPR-Cas type I-A Cascade interference complex. Nucleic Acids Res. 2014;42:5125–38. doi: 10.1093/nar/gku120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–63. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Przybilski R, Richter C, Gristwood T, et al. Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol. 2011;8:517–28. doi: 10.4161/rna.8.3.15190. [DOI] [PubMed] [Google Scholar]
- Punetha A, Sivathanu R, Anand B. Active site plasticity enables metal-dependent tuning of Cas5d nuclease activity in CRISPR-Cas type I-C system. Nucleic Acids Res. 2014;42:3846–56. doi: 10.1093/nar/gkt1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramia NF, Spilman M, Tang L, et al. Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR-Cas complex. Cell Rep. 2014a;9:1610–7. doi: 10.1016/j.celrep.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramia NF, Tang L, Cocozaki AI, et al. Staphylococcus epidermidis Csm1 is a 3′-5′ exonuclease. Nucleic Acids Res. 2014b;42:1129–38. doi: 10.1093/nar/gkt914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeks J, Graham S, Anderson L, et al. Structure of the archaeal Cascade subunit Csa5: relating the small subunits of CRISPR effector complexes. RNA Biol. 2013a;10:762–9. doi: 10.4161/rna.23854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeks J, Naismith JH, White MF. CRISPR interference: a structural perspective. Biochem J. 2013;453:155–66. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeks J, Sokolowski RD, Graham S, et al. Structure of a dimeric crenarchaeal Cas6 enzyme with an atypical active site for CRISPR RNA processing. Biochem J. 2013b;452:223–30. doi: 10.1042/BJ20130269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Fineran PC. The subtype I-F CRISPR-Cas system influences pathogenicity island retention in Pectobacterium atrosepticum via crRNA generation and Csy complex formation. Biochem Soc T. 2013;41:1468–74. doi: 10.1042/BST20130151. [DOI] [PubMed] [Google Scholar]
- Richter C, Gristwood T, Clulow JS, et al. In vivo protein interactions and complex formation in the Pectobacterium atrosepticum subtype I-F CRISPR/Cas System. PLoS One. 2012a;7:e49549. doi: 10.1371/journal.pone.0049549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter H, Lange SJ, Backofen R, et al. Comparative analysis ofCas6b processing and CRISPR RNA stability. RNA Biol. 2013;10:700–7. doi: 10.4161/rna.23715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter H, Zoephel J, Schermuly J, et al. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012b;40:9887–96. doi: 10.1093/nar/gks737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins MF, Schuman JT, Paulus K, et al. Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res. 2015;43:2216–22. doi: 10.1093/nar/gkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C, Zhou M, Zhang J, et al. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol Cell. 2013;52:124–34. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkauskas M, Sinkunas T, Songailiene I, et al. Directional R-loop formation by the CRISPR-Cas surveillance complex cascade provides efficient off-target site rejection. Cell Rep. 2015;10:1534–43. doi: 10.1016/j.celrep.2015.01.067. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Agari Y, Agari K, et al. X-ray crystal structure of a CRISPR-associated RAMP module Cmr5 protein from Thermus thermophilus HB8. Proteins. 2009;75:528–32. doi: 10.1002/prot.22358. [DOI] [PubMed] [Google Scholar]
- Sampson TR, Saroj SD, Llewellyn AC, et al. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–7. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Weiss DS. Alternative roles for CRISPR/Cas systems in bacterial pathogenesis. PLoS Pathog. 2013;9:e1003621. doi: 10.1371/journal.ppat.1003621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson JE, Magadan AH, Sabri M, et al. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol. 2013;11:675–87. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–82. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–7. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–15. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitskaya E, Semenova E, Dedkov V, et al. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol. 2013;10:716–25. doi: 10.4161/rna.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz I, Lange SJ, Hein S, et al. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PLoS One. 2013;8:e56470. doi: 10.1371/journal.pone.0056470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed KD, Lazinski DW, Calderwood SB, et al. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature. 2013;494:489–91. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. P Natl Acad Sci USA. 2011;108:10098–103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SA, Erdmann S, Mojica FJ, et al. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–9. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Cocozaki AI, Ramia NF, et al. Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA silencing complex. Structure. 2013;21:376–84. doi: 10.1016/j.str.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Li H. Recognition and cleavage of a nonstructured CRISPR RNA by its processing endoribonuclease Cas6. Structure. 2013;21:385–93. doi: 10.1016/j.str.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, et al. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–42. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, et al. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–94. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski RD, Graham S, White MF. Cas6 specificity and CRISPR RNA loading in a complex CRISPR-Cas system. Nucleic Acids Res. 2014;42:6532–41. doi: 10.1093/nar/gku308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilman M, Cocozaki A, Hale C, et al. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell. 2013;52:146–52. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Agari Y, Maki-Yonekura S, et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell. 2013;52:135–45. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Zhu Y, Taylor DW, et al. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell. 2014;56:518–30. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–72. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, et al. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–7. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Jeon JH, Shin M, et al. Crystal structure and CRISPR RNA-binding site of the Cmr1 subunit of the Cmr interference complex. Acta Crystallogr D. 2014;70:535–43. doi: 10.1107/S1399004713030290. [DOI] [PubMed] [Google Scholar]
- Swarts DC, Mosterd C, van Passel MW, et al. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD, Tikhomirova MS, Sinkunas T, et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. P Natl Acad Sci USA. 2014;111:9798–803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi N, Wolf YI, Makarova KS, et al. Nature and intensity of selection pressure on CRISPR-associated genes. J Bacteriol. 2012;194:1216–25. doi: 10.1128/JB.06521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Kazlauskiene M, Manakova E, et al. Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol Cell. 2014;56:506–17. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- Tay M, Liu S, Yuan YA. Crystal structure of Thermobifida fusca Cse1 reveals target DNA binding site. Protein Sci. 2015;24:236–45. doi: 10.1002/pro.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, et al. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–92. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zheng H, Preamplume G, et al. The impact of CRISPR repeat sequence on structures of a Cas6 protein-RNA complex. Protein Sci. 2012;21:405–17. doi: 10.1002/pro.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasik BR, Turner PE. On the biological success of viruses. Annu Rev Microbiol. 2013;67:519–41. doi: 10.1146/annurev-micro-090110-102833. [DOI] [PubMed] [Google Scholar]
- Wei Y, Terns RM, Terns MP. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Gene Dev. 2015;29:356–61. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Buckling A, Fineran PC. CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Microbiol. 2014;12:317–26. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- Westra ER, Nilges B, van Erp PB, et al. Cascade-mediated binding and bending of negatively supercoiled DNA. RNA Biol. 2012a;9:1134–8. doi: 10.4161/rna.21410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Semenova E, Datsenko KA, et al. Type I-E CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013;9:e1003742. doi: 10.1371/journal.pgen.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Kunne T, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012b;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011a;477:486–9. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. P Natl Acad Sci USA. 2011b;108:10092–7. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–76. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–13. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Heidrich N, Ampattu BJ, et al. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, et al. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014;515:147–50. doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
- Zhu X, Ye K. Cmr4 is the slicer in the RNA-targeting Cmr CRISPR complex. Nucleic Acids Res. 2015;43:1257–67. doi: 10.1093/nar/gku1355. [DOI] [PMC free article] [PubMed] [Google Scholar]