Abstract
CRISPR-Cas is an RNA-mediated adaptive immune system that defends bacteria and archaea against mobile genetic elements. Short mature CRISPR RNAs (crRNAs) are key elements in the interference step of the immune pathway. A CRISPR array composed of a series of repeats interspaced by spacer sequences acquired from invading mobile genomes is transcribed as a precursor crRNA (pre-crRNA) molecule. This pre-crRNA undergoes one or two maturation steps to generate the mature crRNAs that guide CRISPR-associated (Cas) protein(s) to cognate invading genomes for their destruction. Different types of CRISPR-Cas systems have evolved distinct crRNA biogenesis pathways that implicate highly sophisticated processing mechanisms. In Types I and III CRISPR-Cas systems, a specific endoribonuclease of the Cas6 family, either standalone or in a complex with other Cas proteins, cleaves the pre-crRNA within the repeat regions. In Type II systems, the trans-acting small RNA (tracrRNA) base pairs with each repeat of the pre-crRNA to form a dual-RNA that is cleaved by the housekeeping RNase III in the presence of the protein Cas9. In this review, we present a detailed comparative analysis of pre-crRNA recognition and cleavage mechanisms involved in the biogenesis of guide crRNAs in the three CRISPR-Cas types.
Keywords: crRNA biogenesis, Cas5d, Cas6, Cas9, tracrRNA, RNase III
This review presents a detailed comparative analysis of pre-crRNA recognition and cleavage mechanisms involved in the biogenesis of guide crRNAs in the different bacterial and archaeal CRISPR-Cas immune systems.
INTRODUCTION
CRISPR-Cas are RNA-mediated adaptive immune systems that protect bacteria and archaea from invading mobile genetic elements (Reeks, Naismith and White 2013; Charpentier and Marraffini 2014; van der Oost et al.2014). The systems are composed of an operon of CRISPR-associated (cas) genes and a CRISPR array consisting of a leader sequence followed by a series of short identical repeats interspaced by short unique spacer sequences. The spacers originate from mobile genetic elements memorized upon a first infection, and enable recognition of the invading elements upon a second infection (Barrangou et al.2007). The CRISPR-Cas systems are highly variable in their cas gene composition, and a classification has resulted into three main CRISPR-Cas types that are further divided into subtypes (Makarova et al.2011a,b) (Fig. 1). Despite the cas gene diversification, all systems share a common molecular principle for genome silencing in which the mature CRISPR RNAs (crRNAs) contain a (partially) unique spacer (invader-derived) sequence that guides one or more Cas protein(s) to cognate invading nucleic acids for their eventual destruction after sequence-specific recognition.
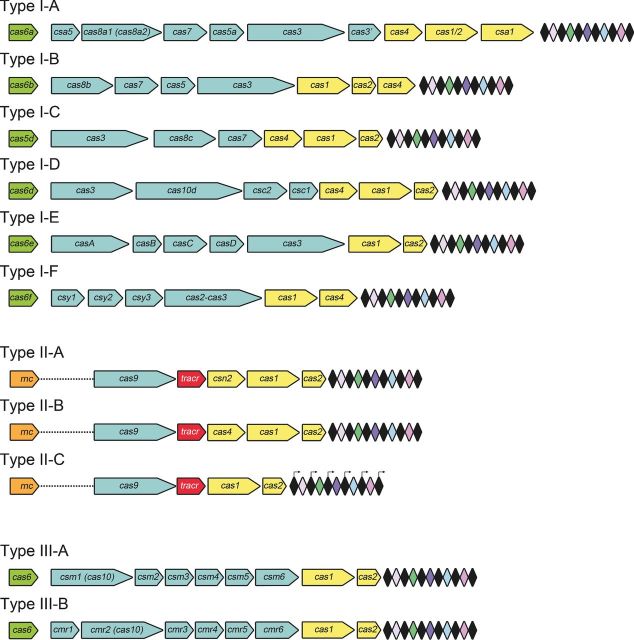
Figure 1.
cas gene composition of the CRISPR-Cas systems. Loci from Types I-A to I-F, Types II-A to II-C and Types III-A and II-B CRISPR-Cas systems are represented. The CRISPR arrays are composed of a series of repeats (black diamonds) interspaced by invading genome-targeting spacers (colored diamonds). An operon of cas genes is located in the close vicinity of the CRISPR array. The Cas proteins involved in the crRNA biogenesis in Types I-A, I-B, I-D, I-E and I-F and Types III-A and III-B belong to the Cas6 family. An exception is the gene product Cas5d responsible for the processing of pre-crRNA in Type I-C. In Type II systems, tracrRNA, and the proteins Cas9 and RNase III are the three components responsible for pre-crRNA maturation.
The maturation of the crRNAs is critical for the activity of the system and the biogenesis of mature crRNAs can be divided into three steps. First, a long primary transcript or precursor crRNA (pre-crRNA) is generated from a promoter located within the leader sequence that precedes the CRISPR repeat-spacer array. Next, primary cleavage of the pre-crRNA occurs at a specific site within the repeats to yield crRNAs that consist of the entire spacer sequence flanked by partial repeat sequences. In some cases, an additional secondary cleavage step is required to generate the active mature crRNAs.
Distinct mechanisms of crRNA biogenesis have evolved, reflected by the diversification of CRISPR-Cas into various subtypes and the large panel of distinct Cas proteins. A common theme among the CRISPR-Cas types is the transcription of the pre-crRNA and the first processing event within the repeats. In Types I and III, a protein of the Cas6 family or alternatively Cas5d catalyzes this step (Figs 2 and 4). In Type II, a trans-acting small RNA directs pre-crRNA dicing by housekeeping endoribonuclease III-mediated cleavage within the repeats in the presence of Cas9 (Fig. 3). The processed crRNAs from Types I-C, I-E and I-F do not undergo further maturation, whereas in at least Types I-A, I-B and I-D, as well as in Types II and III, a second maturation step produces the active crRNAs, the components and mechanisms of which are yet to be determined (Figs 2–4). In this review, we describe and provide a comparative analysis of the remarkable crRNA maturation processes that have evolved in the three CRISPR-Cas types.
Figure 2.
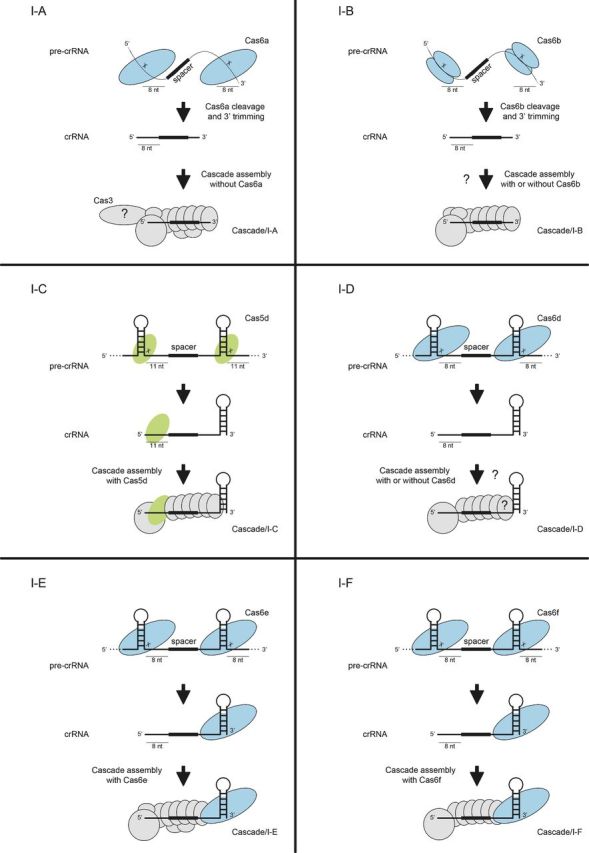
crRNA processing pathways in Type I CRISPR-Cas systems. In Type I systems, the palindromic repeats in the pre-crRNA are either unstructured (Cascade/I-A, Cascade/I-B) or form hairpin structures (Cascade/I-C, Cascade/I-D, Cascade/I-E, Cascade/I-F) that are recognized by the nuclease Cas6 (Cas6a, Cascade/I-A; Cas6b, Cascade/I-B; Cas6d, Cascade/I-D; Cas6e, Cascade/I-E; Cas6f, Cascade/I-F) or Cas5 (Cas5d, Cascade/I-C). After cleavage, the crRNA hairpin remains associated with Cas6 or Cas5 whilst other subunits bind the 5′ handle and spacer, which is used for the recognition of cognate genetic element sequences by the respective Cascade complexes.
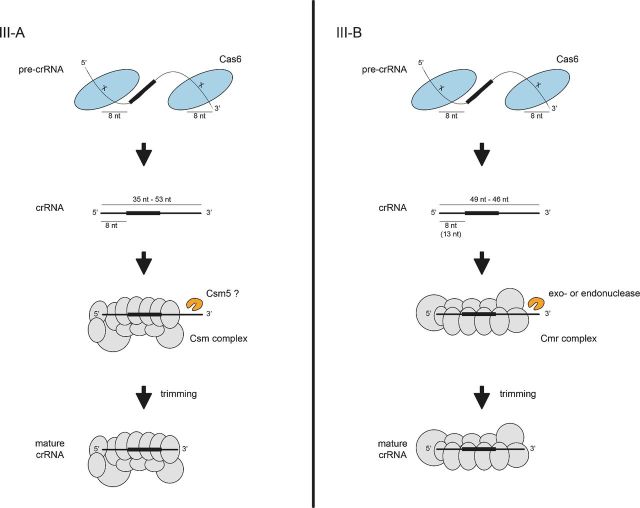
Figure 4.
crRNA processing pathways in Type III CRISPR-Cas systems. In Type III-A and III-B systems, the standalone Cas6 endonuclease binds unstructured pre-crRNA and cleaves within each repeat to generate intermediate crRNAs with 5′ and 3′ repeat-derived termini. The crRNAs are loaded into the Csm (Type III-A) or Cmr (Type III-B) complex and undergo further maturation through trimming of the 3′ repeat-derived sequence by nucleases that are yet to be identified.
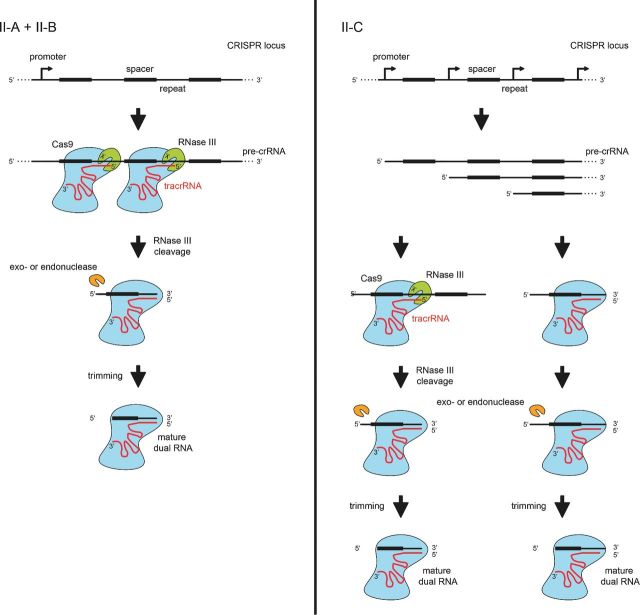
Figure 3.
crRNA processing pathways in Type II CRISPR-Cas systems. In Type II systems, the precursor transcript of the CRISPR repeat-spacer array forms duplexes with the trans-activating tracrRNA through pre-crRNA repeat:tracrRNA anti-repeat interactions. The duplex RNAs stabilized by the protein Cas9 are recognized and cleaved by the bacterial endoribonuclease III (RNase III). A second processing by unknown nucleases (trimming by an exonuclease and/or cleavage by an endoribonuclease) generates the mature crRNAs. An alternative pathway for the production of mature crRNAs was described in a Type II-C of N. meningitidis. Here, the transcription of short crRNAs occurs directly from promoters contained within the repeats of the array, and thus independently of cleavage by RNase III. The mature dual tracrRNA:crRNAs complexed with the protein Cas9 form the interference complex that target and cleave site specifically double-stranded DNA.
crRNA BIOGENESIS IN TYPE I SYSTEMS
Type I systems are present in both bacteria and archaea (Makarova et al.2011a,b). Like all CRISPR-Cas systems, Types I have been shown to target mobile genetic sequences. First, experimental evidence for spacer acquisition by Type I systems has been provided in Escherichia coli (Type I-E), with the correlating resistance against plasmids (Swarts et al.2012; Yosef et al.2012) and phages (Datsenko et al.2012). The Type I-F system of Pseudomonas aeruginosa has been linked to inhibition of biofilm formation, the effect being most probably indirect and depending on an integrated bacteriophage (Cady and O'Toole 2011), whereas its role in the maintenance of phage resistance is yet to be demonstrated (Cady et al.2012). Type I systems are characterized by the CRISPR-associated ribonucleoprotein (crRNP) complex for antiviral defense (Cascade) and a nuclease/helicase (Cas3) that are both required for interference (Brouns et al.2008). Processing of the pre-crRNA transcript is catalyzed by the family of Cas6 metal-independent endoribonucleases that cleave the repeat sequence at a conserved position typically 8 nt upstream of the repeat-spacer boundary (Brouns et al.2008; Carte et al.2008). Once maturated, the crRNAs bound to Cascade play the crucial role of guiding the complex to a complementary target DNA. In Type I-E and I-F systems, the Cas6 enzymes are a subunit of a Cascade-like complex (Jore et al.2011; Wiedenheft et al.2011a,b). This is different from the apparent standalone version of Cas6 that most likely supplies the intermediate or mature crRNAs to different complexes in Type I-A and Type III systems (see below, ‘crRNA biogenesis in Type III’). The crRNAs of Types I-C, I-D, I-E and I-F have stable hairpin structures, which function to initially expose the cleavage site to the Cas6 (or Cas5d in Type I-C) catalytic domain, and to subsequently assist in the stable interaction between guide crRNA and Cascade. Following Cas6-mediated cleavage within the repeats, crRNAs of Types I-C, I-E and I-F are not processed any further (Jore et al.2011; Wiedenheft et al.2011a,b; Nam et al.2012).
Type I crRNAs are expressed and processed in vivo
Expression of Type I crRNAs has been demonstrated amongst others in Sulfolobus solfataricus and Thermoproteus tenax (I-A), Clostridium thermocellum and Methanococcus maripaludis (I-B), E. coli and Thermus thermophilus (I-E), P. aeruginosa (I-F) and Nanoarchaeum equitans (Brouns et al.2008; Haurwitz et al.2010; Jore et al.2011; Lintner et al.2011; Juranek et al.2012; Randau 2012; Richter et al.2012; Zoephel and Randau 2013; Plagens et al.2014). Type I-A loci are characterized by the presence of cas6a, located in proximity to an operon typically composed of cas1, cas2, cas4, csa1, csa5, cas8a1 or cas8a2, cas7 (csa2), cas5, cas3′ and cas3″. The archaeon S. solfataricus was shown to express Type I-A crRNAs of 60–70 nt bound to a Cascade-like protein complex (Lintner et al.2011). Expression of Type I-A crRNAs processed from larger transcripts with subsequent trimming events was also detected in the hyperthermophilic crenarchaeon T. tenax (Plagens et al.2012, 2014). A Type I-B locus contains the gene cas6b followed by the genes cas8b, cas7, cas5, cas3, cas1, cas2 and cas4. Expression and processing of Type I-B pre-crRNAs were detected in the bacterial species C. thermocellum and the archaeal species M. maripaludis (Richter et al.2012; Zoephel and Randau 2013), Haloferax volcanii (Fischer et al.2012), H. mediterranei (Li et al.2013) and M. mazeii (Nickel et al.2013). Interestingly, RNAs antisense to crRNAs, transcribed from spacer elements, were detected in C. thermocellum, as previously described for the Type III-B system of S. acidocaldarius (Lillestol et al.2009) and Pyrococcus furiosus (Hale et al.2012) (see below). In Type I-D, expression of crRNAs of varying length was detected in the cyanobacterium Synechocystis sp. PCC6803 (Scholz et al.2013) and was shown to be dependent on environmental conditions (Hein et al.2013). Type I-E found in E. coli, for example, is specified by the presence of the Cascade genes cse1 (casA), cse2 (casB), cas7 (casC), cas5 (casD), cas6e (casE), the adaptation genes cas1 and cas2 and the nuclease/helicase gene cas3. In 2008 and 2011, Brouns and Jore identified crRNAs of 61 nt as mature species produced from the Type I-E array (Brouns et al.2008; Jore et al.2011). The expression (i) of the Cascade (see below)-encoding cse1-cse2-cas7-cas5-cas6e operon, (ii) of an antisense transcript to cas3 mRNA and to a certain extent (iii) of the CRISPR array is controlled by an interplay of the global transcriptional regulators H-NS (heat-stable nucleoid-structuring) and LeuO (Hommais et al.2001; Oshima et al.2006; Pougach et al.2010; Pul et al.2010; Westra et al.2010). In addition, the response regulator BaeR of the two-component system BaeSR positively regulates expression of the E. coli Cascade operon (Baranova and Nikaido 2002; Perez-Rodriguez et al.2011). The Type I-F cas operon consists of the genes cas1, a cas2-cas3 fusion, csy1, csy2, csy3 and cas6f (csy4). In P. aeruginosa, mature crRNAs of this type were visualized as 60-nt fragments by Northern blot analysis of RNAs co-purified with Cas6f (Haurwitz et al.2010).
Type-I-associated Cas6 endoribonucleases cleave the pre-crRNA within the repeats
Cas6a
Cas6 of the Type I-A system of the archaeon S. solfataricus has a metal-independent ribonuclease activity, that is specifically used for generating crRNAs by cleavage of template pre-crRNAs at a single position within the repeat, consistent with the cleavage site used by other Cas6 enzymes (Lintner et al.2011). This is also consistent with the sequencing analysis of crRNAs associated with Type I-A Cascade that revealed a composition of an 8-nt 5′ repeat fragment followed by a complete spacer sequence and a varying repeat fragment at the 3′ end (Lintner et al.2011). The apparent differences between the Cascade subcomplex of S. solfataricus (Lintner et al.2011) and the complete complex of T. tenax (Plagens et al.2014) may suggest that Cas6 is only transiently associated to Type I-A Cascade and only delivers the mature crRNA to a pre-preformed subcomplex. Type I-A Cascade complexes from the archaea S. solfataricus and T. tenax have been analyzed in detail (Lintner et al.2011; Plagens et al.2014). In S. solfataricus, Cas7 was shown to co-purifiy with the proteins Cas5a, Cas6, Csa5 and processed forms of crRNAs, with the dominant protein Cas7 forming a stable complex with Cas5a (Lintner et al.2011). For T. tenax, however, in vitro reconstitution of a functional Cascade did not require Cas6. The latter was also not co-purified with Csa5 (Plagens et al.2014). Transmission electron microscopy revealed helical structures of variable length (Lintner et al.2011; Plagens et al.2014), perhaps because of substoichiometric amounts of other Cascade components, similar to that observed with E. coli Cascade samples (Brouns, Jore and Van der Oost unpublished). Cas7 (Csa2) was structurally analyzed and shown to have a crescent-shape structure composed of a modified RNA-recognition motif (RRM; Lintner et al.2011), in perfect agreement with the role of Cas7 in binding crRNAs (Wiedenheft et al.2011a,b; Jackson et al.2014; Mulepati et al.2014).
Cas6b
Cas6 proteins from Type I-B of the bacterium C. thermocellum and the archaeon M. maripaludis were recently demonstrated to act as endoribonucleases cleaving pre-crRNA yielding the canonical 8-nt 5′ handle (Richter et al.2012). In these species, RNA-seq data indicate a further trimming of the 3′ end. Biochemical analysis showed that Cas6b requires two histidine residues for catalysis, which is in contrast to other Cas6 family proteins that utilize only one histidine residue (see below), suggesting more flexibility in the catalytic core of Cas6b endoribonucleases (Richter et al.2012). Additionally, it was shown that Cas6b forms dimers upon substrate binding although the native form of the protein is monomeric (Richter et al.2013). Oligomerization of Cas6 proteins was also shown for Type III enzymes of P. horikoshii and S. solfataricus (see below) (Wang et al.2012; Reeks et al.2013). The formation of dimers is not unusual as other endoribonucleases were shown to be active as multimers (Li et al.1998; Calvin et al.2005; Randau et al.2005).
Cas6d
In the cyanobacterium Synechocystis sp. PCC6803, crRNAs contain a typical 8-nt tag generated from cleavage of the pre-crRNA by Cas6d through recognition of the repeat structure (Scholz et al.2013). The crRNAs in this Type I-D are of 39–45 nt in size. The 6-nt gap between the two species may indicate that, as observed in Type III systems, the 3′ handle of the guide is dissociated from the Cas6-like ribonuclease, after which secondary trimming occurs depending on the size of the Cas7 backbone of the complex.
Cas6e
In E. coli Type I-E, Brouns et al. (2008) were first to identify a Cas protein complex formed by Cse1, Cse2, Cas7, Cas5 and Cas6e, which was named CRISPR associated complex for antiviral defense (Cascade). A subsequent combined genetic and biochemical approach was used to demonstrate that mature crRNAs were only produced when all proteins forming the Cascade complex were present (Brouns et al.2008; Jore et al.2011). It was shown that the conserved nucleotide sequence of the repeats within pre-crRNA is essential for recognition and processing by Cas6e (Brouns et al.2008). RNA cleavage was demonstrated to be independent of divalent metal ions or adenosine triphosphate. In 2006, Ebihara et al. (2006) provided the crystal structure of Cas6e from the bacterium T. thermophilus that revealed two independently folded domains exhibiting a ferredoxin-like fold and adopting an RRM-like domain. Based on this, the protein was predicted to function as a nucleic acid-binding protein (Ebihara et al.2006). In 2011, the structure of Cas6e from T. thermophilus bound to repeat RNAs (3′ handle) was determined (Gesner et al.2011; Sashital et al.2011). Recently, the structures of two Cas6e enzymes of T. thermophilus were solved and showed dimerization with two RNA substrates bound in the resulting crRNP, further displaying the differences in RNA recognition and processing by various Cas6-like enzymes (Niewoehner et al.2014).
Based on the first Cas6e structure, an invariant histidine residue (H20) in Cas6e was demonstrated to be essential for the catalytic process (Brouns et al.2008). Initially some heterogeneity at the 3′ end of the isolated crRNAs was reported (Brouns et al.2008), but a later study demonstrated that mature crRNAs of Type I-E are the result of a single processing step, typically resulting in 61-nt fragments (see below; Jore et al.2011). Sequence analysis of crRNA species associated to Cascade demonstrated that the mature crRNAs are composed of (i) an 8-nt repeat fragment (5′ handle), (ii) a complete spacer sequence (32-nt) and (iii) a 21-nt repeat fragment consisting of a stable stem loop of seven base pairs and a four nucleotide loop (3′ handle) (Brouns et al.2008). Subsequent ESI-MS/MS analysis of the Cascade-bound crRNAs revealed 5′-hydroxyl and 2′-3′ cyclic phosphate termini (Jore et al.2011); likewise, crRNAs associated to T. thermophilus Cas6e have the same 5′ and 3′ termini (Gesner et al.2011; Sashital et al.2011). It was demonstrated that crRNA-mediated guiding of Cascade to the target DNA relies on the specific base pairing between crRNA and its complementary DNA strand with displacement of the non-complementary strand, resulting in an R-loop (Jore et al.2011). Cryoelectron microscopy analysis and crystal structures of the crRNA-Cascade complex revealed the display of crRNA along a backbone of six Cas7 subunits (Wiedenheft et al.2011a,b; Jackson et al.2014; Mulepati et al.2014; Zhao et al.2014). This arrangement protects crRNA from degradation and positions the crRNA to allow high-affinity base pairing of invading DNA, initially with the seed sequence at the 5′ end of cognate crRNA (Semenova et al.2011; Wiedenheft et al.2011b).
Cas6f
In P. aeruginosa Type I-F, the Csy proteins Csy1, Csy2, Csy3 and Cas6f assemble into a ribonucleoprotein complex, the function of which is to facilitate recognition of target DNA by enhancing crRNA-DNA sequence-specific hybridization (Haurwitz et al.2010; Rollins et al.2015). Similar to E. coli Cascade, the complex has a crescent shape (Haurwitz et al.2010; Rollins et al.2015). The structure of Cas6f bound to crRNA revealed that Cas6f makes sequence-specific interactions in the major groove of the crRNA repeat stem loop (Haurwitz et al.2010). Cas6f binds tightly to pre-crRNA sequences by exclusive interactions with the hairpin upstream of the scissile phosphate, allowing Cas6f to generate crRNA guides for subsequent targeting of DNA (Haurwitz et al.2010). As observed for the Cas6e (Brouns et al.2008), binding of Cas6f to RNA is substrate specific and requires RNA major groove contacts that are highly sensitive to helical geometry. A strict preference for guanosine adjacent to the scissile phosphate in the active site was reported to contribute to the selectivity mechanism (Haurwitz et al.2010). Cas6f employs a serine and an histidine residue to facilitate cleavage of the pre-crRNA within the repeat at the 3′ side of a stable RNA stem-loop structure (Haurwitz et al.2010). Interestingly, unlike the crRNA processing by E. coli or T. thermophilus Cas6e, crRNAs produced by P. aeruginosa Cas6f have a non-cyclic phosphate at the 3′ end (Wiedenheft et al.2011b).
In Type I-C, Cas5d acts as the pre-crRNA endoribonuclease
The Type I-C locus is characterized by the presence of cas3, cas5d, cas8c, cas7, cas4, cas1 and cas2 genes, and by the absence of a cas6-like gene. The molecular basis of pre-crRNA processing in Type I-C was investigated in Bacillus halodurans and Mannheimia succiniciproducens (Garside et al.2012; Nam et al.2012). Cas5d of the locus was identified as the endoribonuclase that cleaves pre-crRNA within the repeats. Cas5d recognizes both the base of the pre-crRNA stem loop and the 3′ single-stranded overhang in the pre-crRNA repeat. Following recognition, Cas5d then cleaves the substrate into unit length in a metal-independent manner (Nam et al.2012). Thus, recognition of the 3′ overhang, which corresponds to the 5′ handle in the mature crRNA, distinguishes Cas5d from the Cas6-like enzymes. The cleavage by Cas5d yields an 11-nt 5′ tag instead of the canonical 8 nt generated by Cas6 enzymes (Garside et al.2012; Nam et al.2012; Koo et al.2013). Cleavage was reported to generate crRNA products with a 5′ OH and a 2′,3′-cyclic phosphate. The crystal structure of Cas5d revealed a ferredoxin-based architecture and a catalytic triad consisting of residues Y46, K116 and H117, indicative of a general acid-base mechanism (Garside et al.2012; Nam et al.2012). Additional biochemical and structural analysis showed that following pre-crRNA cleavage, Cas5d assembles into a 400-kDa complex together with the mature crRNA and Cas8c (Csd1) and Cas7 (Csd2), the other two Cas proteins specific to Type I-C. Similar to Cascade, the Type I-C crRNA-Cas complex would subsequently act in interference with DNA. Nam et al. also suggested that pre-crRNA processing by Cas5d and formation of the Type I-C Cascade-like complex may be spatially and temporally coupled. Taken together the structural features of Cas5d and the cleavage site on pre-crRNA show that Cas5d is distinct from the Cas6-like endoribonuclases, although the canonical general acid-base mechanism is applied for processing.
crRNA BIOGENESIS IN TYPE II SYSTEMS
In addition to the adaptation modules Cas1 and Cas2, Type I and III CRISPR-Cas systems encode CRISPR-specific ribonucleases (Cas6, Cas5d) responsible for crRNA biogenesis and interference. In contrast, Type II CRISPR-Cas systems are characterized by a minimal locus: the CRISPR repeat-spacer array, a unique cas9 gene as the first gene in an operon containing two or three cas adaptation modules (cas1, cas2, csn2 or cas4) and a small RNA, tracrRNA (Deltcheva et al.2011; Makarova et al.2011a,b; Chylinski et al.2013, 2014). Types II are present in bacteria but absent in archaea (Makarova et al.2011a,b), and phylogenetic studies have resulted in a classification into Types II-A, II-B and II-C (Koonin and Makarova 2013; Chylinski et al.2014; Fonfara et al.2014). The first biological evidence for CRISPR-Cas immunity was demonstrated in a Type II-A system of Streptococcus thermophilus against lytic phages (Barrangou et al.2007). Subsequently, studies have shown (i) a role of a Type II-A in the limitation of horizontal gene transfer (immunity against temperate phages encoding virulence factors) in the human pathogen S. pyogenes (Deltcheva et al.2011), (ii) a role of a Type II-C in preventing mobile genetic element acquisition via natural transformation in Neisseria meningitidis (Zhang et al.2013) and (iii) an immunity-independent unexpected role of a Type II-B system in the downregulation of endogenous expression of a virulence factor encoding mRNA in Francisella novicida (Sampson et al.2013). In 2011, it was demonstrated that Type II CRISPR-Cas systems use a unique crRNA biogenesis pathway distinct from Type I and III CRISPR-Cas systems that involve the coordinated action of three factors: the trans-acting tracrRNA, the host-encoded RNase III and the Cas9 protein (Deltcheva et al.2011). Later in 2013, a study in a Type II-C in N. meningitidis identified an alternative pathway for guide RNA biogenesis. In absence of RNase III, the production of crRNA 5′ termini occurs through promoter sequences located within the repeats of the CRISPR array (Zhang et al.2013)
tracrRNA trans-activates pre-crRNA cleavage by the housekeeping endoribonuclease III in the presence of Cas9
A genome-wide computational analysis aiming to reveal new small RNAs in a clinical isolate of S. pyogenes revealed tracrRNA located upstream of the cas genes of a Type II-A system on the opposite strand. Northern blot followed by differential RNA sequencing (dRNA-seq) analysis demonstrated in vivo expression of precursor and mature forms of the Type II-A tracrRNA and pre-crRNA (Deltcheva et al.2011). Low abundance of unique intermediate crRNA forms of 66 nt composed of 5′-partial repeat-spacer-partial repeat-3′ and high abundance mature forms of 39–42 nt consisting of spacer-derived guide sequence in 5′ and repeat-derived sequence in 3′ were detected. It was proposed that crRNA biogenesis in Type II-A occurs as a two-step process with a first cleavage within the repeats and a second maturation of spacer sequences by either cleavage within the spacers at a specific distance from the first cleavage site and/or by trimming (Deltcheva et al.2011). In the same clinical isolate of S. pyogenes, tracrRNA is expressed in three main forms with two primary species (181–89 nt) transcribed from two distinct promoters and a processed form (75 nt), the three species sharing the same transcriptional terminator. Both primary tracrRNAs share a 25-nt stretch of almost perfect (one mismatch) complementarity with each of the pre-crRNA repeats. Genetic and dRNA-seq analysis concluded that tracrRNA and pre-cRNA undergo co-processing through base pairing of tracrRNA anti-repeat and pre-crRNA repeats (Deltcheva et al.2011). Moreover, the study showed that the 89-nt tracrRNA was the least stable of the two primary forms of tracrRNA, an indication that it may be the primary species preferentially processed in vivo. Both co-processed 75-nt tracrRNA and 66-nt intermediate crRNA species carried short overhangs at the 3′ end, typical for cleavage by the endoribonuclease RNase III (Deltcheva et al.2011). Further genetic and biochemical analysis confirmed that the endogenous RNase III—a general RNA processing factor in bacteria—was recruited to cleave tracrRNA and pre-crRNA upon base pairing and that stabilization of the duplex RNA by the protein Cas9 was required in the process (Deltcheva et al.2011). These findings represented the first description of RNase III-mediated co-processing of two small non-coding RNAs and consisted of the first example of a non-Cas protein being recruited to CRISPR activity.
Subsequent work demonstrated that tracrRNA not only plays a key role in the processing of crRNA in Type II systems but also forms an essential component of the Cas9 cleavage complex (Jinek et al.2012). In particular, following a second maturation event of still uncharacterized nature, a mature duplex comprising both crRNA and tracrRNA bound to Cas9 guide the protein to the invading DNA in a recognition process involving base-pairing complementarity between the guide crRNA sequence of the dual-RNA and the cognate target DNA sequence (Jinek et al.2012). Cas9 was also shown recently to be required during the phase of adaptation for the selection of spacers by recognizing the PAM of the protopacers (Heler et al.2015; Wei et al.2015). Cas9 is the signature protein of the Type II systems and does not share any obvious similarity with the Type I and III Cas proteins (Makarova et al.2006, 2011a,b). It is a large protein containing two nuclease domains, an HNH domain and a split RuvC-like (RNase H-fold) domain responsible for DNA target cleavage, a domain for the recognition of the target DNA and an arginine-rich motif initially suggested to be involved in RNA recognition (Makarova et al.2006, 2011a,b; Sapranauskas et al.2011; Gasiunas et al.2012; Sampson et al.2013; Anders et al.2014; Chylinski et al.2014; Jinek et al.2014). tracrRNA is the second signature of the Type II systems. Analysis of bacterial genomes demonstrated already in 2011 an association of tracrRNA to Type II CRISPR-Cas loci in a number of commensal and pathogenic bacteria (Deltcheva et al.2011; Chylinski et al.2013, 2014). Expression and RNase III-mediated co-processing of tracrRNA and pre-crRNAs were demonstrated in selected bacterial species of Types II-A, II-B and II-C (Deltcheva et al.2011; Chylinski et al.2013, 2014). Anti-repeat and repeat sequences differ significantly in the analyzed genomes, and the repeat sequences analyzed share a certain degree of similarity, especially in the terminal regions and around the putative cleavage site (Deltcheva et al.2011; Chylinski et al.2013, 2014). Notably, despite sequence differences, the sequence complementarity in anti-repeat:repeat base pairing is conserved and co-evolution of tracrRNA, crRNA and the Cas9 protein was further proposed (Deltcheva et al.2011; Chylinski et al.2013, 2014).
An RNase III-independent alternative pathway for crRNA biogenesis in a Type II-C CRISPR-Cas system
A Type II-C CRISPR-Cas system in N. meningitidis is characterized by the presence of an operon of only three cas genes (cas9, cas1 and cas2) displaying a unique pathway for crRNA biogenesis (Deltcheva et al.2011; Zhang et al.2013). In this system, promoter sequences were predicted embedded within each CRISPR repeat. It was shown that some of these promoters initiate transcription in the spacer regions of the CRISPR array yielding intermediate forms of crRNAs containing 5′PPP termini (Zhang et al.2013). Further genetic and dRNA-seq analysis demonstrated that following annealing to tracrRNA through antirepeat:repeat interaction, RNase III cleaves both strands of the tracrRNA:pre-crRNA duplex (Chylinski et al.2013; Zhang et al.2013). However, the authors of this study show that pre-crRNA processing is dispensable. When RNase III is not available or fails to cleave, Cas9 can still form functional complexes with tracrRNA and crRNA. Similar promoters present within the repeats of a Type II-C CRISPR array were also observed and described in Campylobacter jejuni (Dugar et al.2013; Zhang et al.2013).
crRNA BIOGENESIS IN TYPE III SYSTEMS
Type III CRISPR-Cas systems are present in both bacteria and archaea (Makarova et al.2011a,b). This variant has initially been studied in the archaeon P. furiosus (Type III-B) by the Terns laboratory (Carte et al.2008,2010; Hale et al.2008). Later, the biogenesis of crRNAs has also been investigated in the Gram-positive bacterial pathogen Staphylococcus epidermidis (Type III-A) (Hatoum-Aslan et al.2011). Interestingly, it was shown that Type III-B systems do not target DNA sequences but exclusively target ssRNA (Hale et al.2012,2014; Zhang et al.2012). In one of the first demonstrations of CRISPR-Cas activity, the Type III-A system from S. epidermidis was shown to target conjugative plasmid DNA in vivo (Marraffini and Sontheimer 2008). Recently, it was demonstrated by several groups that Type III-A systems also target ssRNA in vitro (Staals et al.2014; Tamulaitis et al.2014) and in vivo (Tamulaitis et al.2014).
Like the Type I systems, crRNA production in Type III systems is dependent on the activity of proteins of the Cas6 family. Cas6 enzymes are normally an integral subunit of some Type I (Cascade) systems (for example Cas6e and Cas6f in E. coli and P. aeruginosa, respectively) (Brouns et al.2008; Haurwitz et al.2010). In contrast, Cas6 enzymes of Types III appear to function independently of the Cas protein complexes and have not been observed to co-purify with them. crRNA maturation in Types III occurs in two steps. In these systems, processing involves cleavage of pre-crRNA by Cas6 within the repeats, generating 1X intermediate units that undergo further processing at the 3′ end of the crRNA to produce the active mature crRNAs (Carte et al.2008,2010), similarly to the trimming of crRNAs in Type I-A (Plagens et al.2014) and I-B (Richter et al.2012). Type III systems have a backbone of Cas7-like proteins in both Type III-A (Rouillon et al.2013) and III-B systems (Staals et al.2013). In both types, the proteins were shown to assemble around the crRNAs to form interference complexes (Csm and Cmr), similar to Cascade of Type I. After complex formation, the crRNA is facilitated to guide the crRNP to target ssRNA/dsDNA for Csm (Staals et al.2014; Tamulaitis et al.2014) and ssRNA for Cmr (Hale et al.2012,2014; Zhang et al.2012), respectively.
Type III crRNAs are expressed and processed in vivo
The bacterial Type III-A system
In 2008, Marraffini and Sontheimer showed that initial crRNA processing generated products of 71 nt in S. epidermidis, suggestive of pre-crRNA cleavage at the base of a potential stem-loop structure within each repeat. These products were in turn further trimmed to mature crRNA of 49-nt species by 3′-end processing (Marraffini and Sontheimer 2008, 2010). Differential RNA-seq and Northern blot analysis confirmed crRNA production and maturation in the T. thermophilus Type III-A and III-B systems (Juranek et al.2012).
The archaeal Type III-B system
In 2002, Tang et al. (2002) showed that small RNAs derived from CRISPR repeats, although then known as SRSRs (short regularly spaced repeats), were transcribed in the archaeon Archaeoglobus fulgidus. Ladders of RNA corresponding in length to 1, 2, 3 or more repeat-spacer units were detected by Northern blot analysis. Similar ladders were subsequently observed in the crenarchaeon S. solfataricus (Tang et al.2005) and in S. acidocaldarius (Chen et al.2005; Lillestol et al.2006, 2009). The authors proposed that SRSRs were transcribed as a precursor RNA that was further processed to generate the unit length small RNAs. These studies represented the first experimental evidence for crRNA processing, although the endonuclease, Cas6, was not yet discovered. Interestingly, Northern blotting and RNA mapping experiments in S. acidocaldarius and S. solfataricus revealed expression and processing of RNA molecules from complementary strands of repeat-spacer arrays into discrete short RNAs of length distinct from that of the mature crRNAs (Lillestol et al.2009). The authors of the study suggested that the antisense RNAs could either serve as neutralizers of crRNAs in the absence of invading elements or alternatively be required for the slicing activity of the invaders (Lillestol et al.2009). The presence of anti-sense RNAs was also shown for the bacterial I-B system of C. thermocellum (Richter et al.2012) and led to the speculation of regulatory functions by the anti-sense crRNAs (Zoephel and Randau 2013).
In 2008, pre-crRNA expression and processing was investigated in P. furiosus by the Terns lab (Hale et al.2008). Small RNA species primarily of lengths 39 nt and 45 nt were the predominant, mature crRNA forms identified. An intermediate of about 65 nt corresponded to pre-crRNA cleaved within the repeat sequences, prior to 3′-end processing (Hale et al.2008). The same mature species were subsequently identified in the purified Type III-B complex from P. furiosus (Hale et al.2012). Analysis of crRNA co-purifying with the Type III-B complex from S. solfataricus showed the presence of RNA molecules with variable sizes centered on 46 nt consistent with a first cleavage within each repeat followed by exonucleolytic digestion at the 3′ end (Zhang et al.2012). Small amounts of RNA corresponding to the reverse complement of pre-crRNA were also identified in this experiment; however, they constituted just 0.01% of the RNA sequenced (Zhang et al.2012). In addition, pre-crRNA antisense transcription, probably driven by the presence of functional promoter sequences within spacers, was detected at a significant level compared to crRNA products in P. furiosus (Hale et al.2012). These are thought to function as endogenous target RNA of the system (Hale et al.2012).
The endoribonuclease Cas6 cleaves pre-crRNA within the repeats
The bacterial Type III-A system
Using primer extension and conjugation experiments with a series of pre-crRNA mutants, the Marraffini group showed that both the RNA hairpin formation within the repeats and the sequence 5′-GGGACG-3′ at the base of the stem-loop structure were needed for efficient primary processing of pre-crRNA (Hatoum-Aslan et al.2011). Furthermore, it was shown that not only Cas6 but also Cas10 (the large subunit of Type III systems) and Csm4 (the Cas5 subunit of Type III-A systems) were required for the production of crRNAs in stable form in vivo, suggesting that the latter maintain the stability of crRNAs (Hatoum-Aslan et al.2011). The recent advances in structural analysis of the Type III-A showed a flexible composition of the Csm complex based on the length of the crRNA. Flexibility is achieved by varying amounts of the subunits Csm3 and Csm4 that display the backbone of the crRNP. In these studies it is speculated that Csm5, potentially an integral part of the Csm complex is involved in the 3′ processing of the crRNA (Rouillon et al.2013; Staals et al.2014).
The archaeal Type III-B system
It was demonstrated by the Terns lab that the endoribonuclease responsible for crRNA processing in the Type III-B of P. furiosus was Cas6, one of the core Cas proteins (Carte et al.2008). The Cas6 cleavage site was mapped to a defined position 8 nt from the 3′ end of the repeat sequence, generating unit length crRNAs (1X intermediates) with a central spacer typically flanked by 8 nt of repeat-derived sequence at the 5′ end (13-nt 5′ tag in the case of the cyanobacterium Synechocystis (Scholz et al.2013) and a longer repeat sequence (∼ 22 nt) at the 3′ end (Carte et al.2008). Mature crRNAs isolated from the Type III-B (Cmr) complex from S. solfataricus also began with the 8-nt 5′ handle derived from the CRISPR repeat with spacer-derived sequence at the 3′ end (Zhang et al.2012). The 3′ termini of the sequenced crRNAs showed some variability, with some spacer-derived sequences displaying short 3′ handle and others containing little repeat-derived sequences (Zhang et al.2012). A similar pattern was observed for the crRNA isolated from the Type III-A (Csm) complex (Rouillon et al.2013). This was in contrast to mature crRNAs isolated from S. solfataricus Cascade complexes (Type I-A), which include longer 3′ repeat-derived handles (Lintner et al.2011). The reasons for these differences are not yet understood, but may relate to differing extents of protection of the crRNA intermediates following binding by Type I and Type III effector complex subunits.
Insights into the structure of the endoribonuclease Cas6
The crystal structure of P. furiosus Cas6 revealed a duplicated RRM (ferredoxin-like) fold, with the two halves of the protein separated by a cleft (Carte et al.2010). Cas6 is distinguishable from the other members of the RAMP family of proteins by the presence of a predicted G-rich loop motif (consensus GhGxxxxxGhG, where h is hydrophobic and xxxxx has at least one lysine or arginine) at the C-terminus (Makarova et al.2002; Haft et al.2005). Within the cleft of Cas6, a catalytic triad, consisting of Y31, H46 and K52, which is conserved in some other Cas6 proteins, was detected and its importance in the catalytic mechanism was confirmed by mutagenesis (Carte et al.2008, 2010). Overall, the fold is related to the Cas6e subunit of the Type I-E Cascade complex (van der Oost et al.2009), which performs the same function and produces unit length crRNAs with the canonical 8-nt repeat-derived 5′ tag (Brouns et al.2008). Like Cas6, Cas6e also cleaves RNA in a metal-independent manner. In contrast to Cas6 having a duplicated ferredoxin fold, the RNA-bound Cas6f of the Type I-F contains a single ferredoxin fold (Haurwitz et al.2010). An active site histidine has also been implicated in the Cas6b, Cas6e and Cas6f nucleases (Brouns et al.2008; Haurwitz et al.2010; Richter et al.2012). Curiously however, there is no conserved histidine in the crenarchaeal Cas6 orthologs from S. solfataricus (Lintner et al.2011), suggesting a different catalytic mechanism may operate in these enzymes. Site directed mutagenesis coupled with kinetic analyses have shown that a constellation of basic residues positioned near the base of the small hairpin formed by bound crRNA contribute to efficient catalysis (Reeks et al.2013). Interestingly, Cas6 enzymes are not always monomers. One form of Cas6 from S. solfataricus is a dimer (Reeks et al.2013; Shao and Li 2013), and this is also the case for Cas6b of M. maripaludis (Richter et al.2013). The functional significance of these dimeric structures is still unclear.
The structure of P. furiosus Cas6 bound to crRNA revealed that the first 10 nt of crRNA, which was the only part observed in the crystal structure, makes sequence-specific interactions with a conserved binding interface in Cas6 on the face opposite the catalytic site (Wang et al.2011). The RNA was predicted to loop around the protein, before re-engaging at the active site, resulting in cleavage of the crRNA between nucleotides A22 and A23. In the middle, a linker region of the crRNA between residues 10 and 20 can accommodate point mutations, insertions and deletions without abrogating Cas6 activity, suggesting that it may not be recognized by the protein (Wang et al.2011). In contrast, the structure of S. solfataricus Cas6 bound to a crRNA revealed specific recognition and stabilization of a short hairpin structure in the repeat, with cleavage at the base of the hairpin (Shao and Li 2013) similar to the bacterial Cas6 enzymes. The mode of crRNA recognition by the P. furiosus Cas6 enzyme thus appears to be an outlier. Several families of Cas6 exist in S. solfataricus, which differ in their specificity for the two types of CRISPR repeat encoded in the genome. This may provide a mechanism for specific loading of crRNAs from particular CRISPR loci into specific effector complexes (Sokolowski et al.2014). A similar situation may exist in the cyanobacterium Synechocystis sp. PCC6803, which has three CRISPR loci, each associated with genes encoding an effector complex (one Type I-D and two Type III) and two Cas6 paralogs, each specific for a particular CRISPR repeat sequence (Scholz et al.2013).
CONCLUSIONS
The core components of the CRISPR-Cas defense machinery are the short mature crRNAs that contain signature sequences of mobile genetic elements and associate with one or more Cas proteins to target and destroy invading nucleid acids through crRNA:target sequence specific recognition. The CRISPR repeat-spacer array is transcribed as a long pre-crRNA that undergoes a first cleavage within the repeats sometimes followed by an additional maturation step. Although this principle is commonly shared, CRISPR-Cas types have evolved distinct mechanisms for the biogenesis of mature crRNAs.
Different Cas proteins characteristic for the subtype play distinct catalytic or assisting functions in the first step of pre-crRNA processing. Types I and III both use endoribonucleases of the Cas6 family to cleave the pre-crRNA within the repeats. Both types encode also a module of several additional Cas proteins, which in the case of some Type I subsystems form complexes with the respective Cas6 enzyme. For example, Type I-E encodes Cse1, Cse2, Cas7 and Cas5, which together with Cas6e and crRNA form Cascade (Ebihara et al.2006; Brouns et al.2008; Gesner et al.2011; Jore et al.2011; Sashital et al.2011; Wang et al.2011; Wiedenheft et al.2011a). The trans-acting nuclease Cas3 is then recruited to the complex to cleave invading DNA (Beloglazova et al.2011; Howard et al.2011; Mulepati and Bailey 2011; Sinkunas et al.2011; Wiedenheft et al.2011a; Westra et al.2012). Type I-F (Ypest or CASS3) encodes Csy1, Csy2 and Csy3, which together with Cas6f and crRNA form a crRNP complex, which is likely to recruit the DNA-cleaving enzyme Cas3 as for Type I-E (Haurwitz et al.2010; Wiedenheft et al.2011b; Rollins et al.2015). The Type III systems encode a set of Cas proteins that include the signature protein, Cas10 (formerly Csm1, Cmr2 and Csx11). In Type III-B, Cas6 functions as a standalone endoribonuclease, and the associated proteins Cmr1, Cas10, Cmr3, Cmr4, Cmr5 and Cmr6 interfere downstream of the Cas6-mediated processing event in target RNA interference (Carte et al.2008, 2010; Hale et al.2008, 2009, 2012, 2014;Wang et al.2011). In Type III-A, it was shown that Cas10, Csm2, Csm3 and Csm4 form a complex and that the action of Csm5 may be required for further processing of the Cas6-generated intermediate crRNAs to produce the mature crRNAs (Hatoum-Aslan et al.2011; Rouillon et al.2013; Staals et al.2014). Interestingly, no Cas6 endoribonuclease is found in Type I-C. Instead, the protein Cas5d is the endoribonuclease that processes the pre-crRNA within the repeats, using a mechanism distinct from that of Cas6 (Garside et al.2012; Nam et al.2012; Koo et al.2013). Similar to Cas6 proteins of other Types I, Cas5d assembles with crRNA and two other Cas proteins, Cas8c and Cas7, to form a Cascade-like interference complex (Nam et al.2012). In contrast, the minimal Type II system uses Cas9 as the only Cas protein for the steps of crRNA biogenesis and interference with invading DNA. The system has evolved a trans-acting small RNA, tracrRNA, which takes advantage of the housekeeping endoribonuclease III to catalyze tracrRNA-directed cleavage within the pre-crRNA repeats, involving the stabilization of the RNA duplex by Cas9 (Deltcheva et al.2011). The tracrRNA also forms an essential component of the Cas9 target recognition and cleavage complex (Jinek et al.2012). Type II systems are found exclusively in bacteria and the absence of these systems in archaea may be explained by the absence of genes encoding endoribonuclease III-like activities. The description of a Type II-C in N. meningitidis that does not require the activity of RNase III for the maturation of crRNAs is an interesting alternative strategy evolved by bacteria. In this particular case, crRNA forms are expressed from promoter sequences located within the repeats of the CRISPR arrays.
CRISPR-Cas systems have evolved mature crRNAs with distinct subtype-dependent composition and length. In Types I-A (Cas6a), I-B (Cas6b), I-D (Cas6d), I-E (Cas6e), I-F (Cas6f), and Types III-A (Cas6) and III-B (Cas6), mature crRNAs are composed of 8 nt of repeat sequence in 5′ directly followed by invader-targeting spacer-derived sequence (Brouns et al.2008; Carte et al.2008; Marraffini and Sontheimer 2008; Haurwitz et al.2010; Plagens et al.2014). Accordingly, C. thermocellum and M. maripaludis Cas6b, E. coli, S. solfataricus and T. thermophilus Cas6e, P. aeruginosa Cas6f and P. furiosus Cas6 all cleave exactly 8 nt upstream of the repeat-spacer junction within the pre-crRNA repeats (Ebihara et al.2006; Brouns et al.2008; Haurwitz et al.2010; Gesner et al.2011; Sashital et al.2011). In contrast to Types II and III, Cas6-like-generated crRNAs of Types I-E and I-F do not undergo additional maturation and are composed of the 8-nt repeat tag at the 5′ end, complete sequence of the spacer in the middle and the remainder of the repeat fragment, generally forming a hairpin structure, at the 5′ end (Brouns et al.2008; Haurwitz et al.2010). This does not seem to be a feature of all Type I systems since processing of the 3′ end of the crRNAs was observed for I-A (Plagens et al.2014) and I-B (Richter et al.2012) systems. Furthermore, Cas6 is not an integral part of the I-A Cascade of T. tenax (Plagens et al.2014), leading to the speculation that crRNAs produced by standalone Cas6 enzymes are generally 3′ trimmed before being loaded to their respective interference complex. Type III (S. epidermidis, P. furiosus) mature crRNAs have repeat-derived sequences at the 5′ end and spacer-derived sequence at the 3′ end (Carte et al.2008; Marraffini and Sontheimer 2008). A reverse configuration characterizes Type II mature crRNAs that are composed of a spacer-derived sequence in 5′ and a repeat-derived sequence in 3′ (Deltcheva et al.2011). Furthermore, Type I, Type II and Type III systems produce mature crRNAs of distinct sizes (Carte et al.2008; Marraffini and Sontheimer 2008). Intriguingly, maturation in both Types III-A and III-B generates two distinct crRNA species. Finally, the crRNAs have different terminal configurations, Type I-C crRNAs in B. halodurans and Type I-E crRNAs in E. coli have 5′-hydroxyl group and 2′-3′ cyclic phosphate (Jore et al.2011) while in P. aeruginosa Type I-F crRNAs terminate with 5′-hydroxyl group and 3′ phosphate (not cyclic) (Haurwitz et al.2010; Richter et al.2012; Plagens et al.2014). Type III-A crRNAs (S. epidermidis) contain 3′-hydroxyl groups (Hatoum-Aslan et al.2011) whereas Type III-B crRNAs end with either 3′-hydroxyl or 2′-3′-cyclic phosphate ends (Carte et al.2008). Several reports also describe differential expression levels of the individual mature crRNAs produced from a same CRISPR array. Deep dRNA-seq studies in Types I and III indicate that the most recently acquired sequences at the leader end of the CRISPR loci appear to correspond to the most abundant crRNA species (Wurtzel et al.2010; Hale et al.2012; Juranek et al.2012; Randau 2012; Richter et al.2012; Nickel et al.2013; Soutourina et al.2013; Su et al.2013; Plagens et al.2014). It has been suggested that differences in pre-crRNA transcription rates, processing and/or stability could provide plausible explanations for this observation.
An interesting additional characteristic is the property of pre-crRNA repeats to fold or not to fold. In 2007, a systematic analysis of the sequences and RNA folding stabilities of CRISPR repeats was reported (Kunin et al.2007). The CRISPR repeats were classified into 12 major clusters on the basis of conserved sequence features. The authors noted that the repeats in some clusters had a pronounced ability to fold into a stable hairpin structure whilst others lacked this property, and divided CRISPRs into ‘folded’ and ‘unfolded’ categories. The authors further suggested that the hairpin structures of the repeats might serve as a motif for Cas protein recognition. With some exceptions, most of the Type I CRISPR repeats fall into the ‘folded’ category whereas Type II and Type III repeats are considered ‘unfolded’. Type I repeats mostly contain palindromic sequences predicted to form stable hairpin structures ending upstream of the cleavage site. Structural analysis demonstrated that P. aeruginosa Cas6f interacts specifically with the hairpin to place the cleavage site at the base of the stem loop within the enzyme active site (Haurwitz et al.2010). In 2010, Carte et al. (2010) suggested that the CRISPR repeats of Type III-B in P. furiosus belong to a group of repeat sequences considered unstructured with the potential to form weak stem loops. Along these lines, the same authors showed that in absence of proteins, the pre-crRNA is predominantly unstructured in solution (Carte et al.2010). Analysis of the crRNA-bound Cas6 structure also indicate that pre-crRNA wraps around the surface of the endoribonuclease, consistent with the lack of folded structure (Wang et al.2011). Even though Cas6 orthologs share extremely low sequence identity, the ‘wrap around’ mechanism involved in Cas6 recognition and cleavage of unstructured crRNA could also apply to Type III-A and potentially to Type I systems with unstructured repeats. However, it was suggested that Type III-A repeats of S. epidermidis form internal hairpins that would enhance crRNA processing at the binding and/or nucleolytic level (Hatoum-Aslan et al.2011). In the case of Type II, base pairing of unstructured pre-crRNA to tracrRNA may compensate this deficiency by providing an intermolecular structure that directs the processing within pre-crRNA repeats (Deltcheva et al.2011; Chylinski et al.2013; Briner et al.2014).
To conclude, there are numerous variations of crRNA biogenesis, mediated by distinct components and mechanisms, which we have begun to understand only recently. Unique RNA recognition mechanisms enable to discriminate pre-crRNAs from other cytosolic RNAs. Distinct RNA cleavage mechanisms specifically produce the mature guide crRNAs that associate to respective interference complexes. Future studies will certainly provide additional details on the crRNA maturation complexes of the multiple rapidly evolving CRISPR-Cas subtypes and should shed some light on the molecular mechanisms involved in the second maturation events.
FUNDING
EC is supported by the Alexander von Humboldt Foundation, the German Federal Ministry for Education and Research, the Helmholtz Association, the Göran Gustafsson Foundation, the Swedish Research Council, the Kempe Foundation and Umeå University. HR is supported by an Helmholtz Post-doctoral Fellowship. JO is supported by the Netherlands Organization for Scientific Research (NWO).
Conflict of interest. None declared.
REFERENCES
- Anders C, Niewoehner O, Duerst A, et al. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–73. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranova N, Nikaido H. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J Bacteriol. 2002;184:4168–76. doi: 10.1128/JB.184.15.4168-4176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Beloglazova N, Petit P, Flick R, et al. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30:4616–27. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briner AE, Donohoue PD, Gomaa AA, et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56:333–9. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady KC, Bondy-Denomy J, Heussler GE, et al. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol. 2012;194:5728–38. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady KC, O'Toole GA. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol. 2011;193:3433–45. doi: 10.1128/JB.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvin K, Hall MD, Xu F, et al. Structural characterization of the catalytic subunit of a novel RNA splicing endonuclease. J Mol Biol. 2005;353:952–60. doi: 10.1016/j.jmb.2005.09.035. [DOI] [PubMed] [Google Scholar]
- Carte J, Pfister NT, Compton MM, et al. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–8. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, et al. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Gene Dev. 2008;22:3489–96. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier E, Marraffini LA. Harnessing CRISPR-Cas9 immunity for genetic engineering. Curr Opin Microbiol. 2014;19C:114–9. doi: 10.1016/j.mib.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Brugger K, Skovgaard M, et al. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J Bacteriol. 2005;187:4992–9. doi: 10.1128/JB.187.14.4992-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylinski K, LeRhun A, Charpentier E. The tracrRNA and Cas9 families of Type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–37. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylinski K, Makarova KS, Charpentier E, et al. Classification and evolution of Type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:6091–105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–7. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugar G, Herbig A, Forstner KU, et al. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 2013;9:e1003495. doi: 10.1371/journal.pgen.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara A, Yao M, Masui R, et al. Crystal structure of hypothetical protein TTHB192 from Thermus thermophilus HB8 reveals a new protein family with an RNA recognition motif-like domain. Protein Sci. 2006;15:1494–9. doi: 10.1110/ps.062131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S, Maier LK, Stoll B, et al. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem. 2012;287:33351–63. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, LeRhun A, Chylinski K, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous Type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–90. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garside EL, Schellenberg MJ, Gesner EM, et al. Cas5d processes pre-crRNA and is a member of a larger family of CRISPR RNA endonucleases. RNA. 2012;18:2020–8. doi: 10.1261/rna.033100.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. P Natl Acad Sci USA. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesner EM, Schellenberg MJ, Garside EL, et al. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–92. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, et al. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, et al. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–9. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Cocozaki A, Li H, et al. Target RNA capture and cleavage by the Cmr Type III-B CRISPR-Cas effector complex. Gene Dev. 2014;28:2432–43. doi: 10.1101/gad.250712.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. P Natl Acad Sci USA. 2011;108:21218–22. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, et al. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–8. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein S, Scholz I, Voss B, et al. Adaptation and modification of three CRISPR loci in two closely related cyanobacteria. RNA Biol. 2013;10:852–64. doi: 10.4161/rna.24160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JW, et al. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommais F, Krin E, Laurent-Winter C, et al. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- Howard JA, Delmas S, Ivancic-Bace I, et al. Helicase dissociation and annealing of RNA-DNA hybrids by Escherichia coli Cas3 protein. Biochem J. 2011;439:85–95. doi: 10.1042/BJ20110901. [DOI] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PB, et al. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–9. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–36. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Juranek S, Eban T, Altuvia Y, et al. A genome-wide view of the expression and processing patterns of Thermus thermophilus HB8 CRISPR RNAs. RNA. 2012;18:783–94. doi: 10.1261/rna.031468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo Y, Ka D, Kim EJ, et al. Conservation and variability in the structure and function of the Cas5d endoribonuclease in the CRISPR-mediated microbial immune system. J Mol Biol. 2013;425:3799–810. doi: 10.1016/j.jmb.2013.02.032. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS. CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013;10:679–86. doi: 10.4161/rna.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Trotta CR, Abelson J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science. 1998;280:279–84. doi: 10.1126/science.280.5361.279. [DOI] [PubMed] [Google Scholar]
- Li M, Liu H, Han J, et al. Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei. J Bacteriol. 2013;195:867–75. doi: 10.1128/JB.01688-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillestol RK, Redder P, Garrett RA, et al. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillestol RK, Shah SA, Brugger K, et al. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009;72:259–72. doi: 10.1111/j.1365-2958.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- Lintner NG, Kerou M, Brumfield SK, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–56. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Grishin NV, et al. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–96. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, et al. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011a;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, et al. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011b;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) J Biol Chem. 2011;286:31896–903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Heroux A, Bailey S. Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–84. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Haitjema C, Liu X, et al. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subType I-C/Dvulg CRISPR-Cas system. Structure. 2012;20:1574–84. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel L, Weidenbach K, Jager D, et al. Two CRISPR-Cas systems in Methanosarcina mazei strain Go1 display common processing features despite belonging to different Types I and III. RNA Biol. 2013;10:779–91. doi: 10.4161/rna.23928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, Jinek M, Doudna JA. Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases. Nucleic Acids Res. 2014;42:1341–53. doi: 10.1093/nar/gkt922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T, Ishikawa S, Kurokawa K, et al. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–53. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- Perez-Rodriguez R, Haitjema C, Huang Q, et al. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol Microbiol. 2011;79:584–99. doi: 10.1111/j.1365-2958.2010.07482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A, Tjaden B, Hagemann A, et al. Characterization of the CRISPR/Cas subType I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J Bacteriol. 2012;194:2491–500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A, Tripp V, Daume M, et al. In vitro assembly and activity of an archaeal CRISPR-Cas Type I-A Cascade interference complex. Nucleic Acids Res. 2014;42:5125–38. doi: 10.1093/nar/gku120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pougach K, Semenova E, Bogdanova E, et al. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol. 2010;77:1367–79. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pul U, Wurm R, Arslan Z, et al. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol. 2010;75:1495–512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- Randau L. RNA processing in the minimal organism Nanoarchaeum equitans. Genome Biol. 2012;13:R63. doi: 10.1186/gb-2012-13-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randau L, Calvin K, Hall M, et al. The heteromeric Nanoarchaeum equitans splicing endonuclease cleaves noncanonical bulge-helix-bulge motifs of joined tRNA halves. P Natl Acad Sci USA. 2005;102:17934–9. doi: 10.1073/pnas.0509197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeks J, Naismith JH, White MF. CRISPR interference: a structural perspective. Biochem J. 2013;453:155–66. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeks J, Sokolowski RD, Graham S, et al. Structure of a dimeric crenarchaeal Cas6 enzyme with an atypical active site for CRISPR RNA processing. Biochem J. 2013;452:223–30. doi: 10.1042/BJ20130269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter H, Lange SJ, Backofen R, et al. Comparative analysis ofCas6b processing and CRISPR RNA stability. RNA Biol. 2013;10:700–7. doi: 10.4161/rna.23715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter H, Zoephel J, Schermuly J, et al. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012;40:9887–96. doi: 10.1093/nar/gks737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins MF, Schuman JT, Paulus K, et al. Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res. 2015;43:2216–22. doi: 10.1093/nar/gkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C, Zhou M, Zhang J, et al. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol Cell. 2013;52:124–34. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Saroj SD, Llewellyn AC, et al. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–7. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–82. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–7. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- Scholz I, Lange SJ, Hein S, et al. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PLoS One. 2013;8:e56470. doi: 10.1371/journal.pone.0056470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. P Natl Acad Sci USA. 2011;108:10098–103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Li H. Recognition and cleavage of a nonstructured CRISPR RNA by its processing endoribonuclease Cas6. Structure. 2013;21:385–93. doi: 10.1016/j.str.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, et al. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–42. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski RD, Graham S, White MF. Cas6 specificity and CRISPR RNA loading in a complex CRISPR-Cas system. Nucleic Acids Res. 2014;42:6532–41. doi: 10.1093/nar/gku308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina OA, Monot M, Boudry P, et al. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 2013;9:e1003493. doi: 10.1371/journal.pgen.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Agari Y, Maki-Yonekura S, et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell. 2013;52:135–45. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Zhu Y, Taylor DW, et al. RNA targeting by the Type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell. 2014;56:518–30. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AA, Tripp V, Randau L. RNA-Seq analyses reveal the order of tRNA processing events and the maturation of C/D box and CRISPR RNAs in the hyperthermophile Methanopyrus kandleri. Nucleic Acids Res. 2013;41:6250–8. doi: 10.1093/nar/gkt317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Mosterd C, van Passel MW, et al. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Kazlauskiene M, Manakova E, et al. Programmable RNA shredding by the Type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol Cell. 2014;56:506–17. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, et al. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. P Natl Acad Sci USA. 2002;99:7536–41. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Polacek N, Zywicki M, et al. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–81. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, et al. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–7. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, et al. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–92. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Preamplume G, Terns MP, et al. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–64. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zheng H, Preamplume G, et al. The impact of CRISPR repeat sequence on structures of a Cas6 protein-RNA complex. Protein Sci. 2012;21:405–17. doi: 10.1002/pro.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Terns RM, Terns MP. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Gene Dev. 2015;29:356–61. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Pul U, Heidrich N, et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol. 2010;77:1380–93. doi: 10.1111/j.1365-2958.2010.07315.x. [DOI] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Kunne T, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011a;477:486–9. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. P Natl Acad Sci USA. 2011b;108:10092–7. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Sapra R, Chen F, et al. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–41. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–76. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–13. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Heidrich N, Ampattu BJ, et al. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, et al. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014;515:147–50. doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
- Zoephel J, Randau L. RNA-Seq analyses reveal CRISPR RNA processing and regulation patterns. Biochem Soc Trans. 2013;41:1459–63. doi: 10.1042/BST20130129. [DOI] [PubMed] [Google Scholar]