ABSTRACT
Efficient degradation of abnormal or aggregated proteins is crucial to protect the cell against proteotoxic stress. Selective targeting and disposal of such proteins usually occurs in a ubiquitin-dependent manner by proteasomes and macroautophagy/autophagy. Whereas proteasomes are efficient in degrading abnormal soluble proteins, protein aggregates are typically targeted for degradation by autophagic vesicles. Both processes require ubiquitin-binding receptors, which are targeted to proteasomes via ubiquitin-like domains or to phagophores (the precursors to autophagosomes) via Atg8/LC3 binding motifs, respectively. The use of substrate modification by ubiquitin in both pathways raised the question of how degradative pathway choice is achieved. In contrast to previous models, proposing different types of ubiquitin linkages for substrate targeting, we find that pathway choice is a late event largely determined by the oligomeric state of the receptors. Monomeric proteasome receptors bind soluble substrates more efficiently due to their higher affinity for ubiquitin. Upon substrate aggregation, autophagy receptors with lower ubiquitin binding affinity gain the upper hand due to higher avidity achieved by receptor bundling. Thus, our work suggests that ubiquitination is a shared signal of an adaptive protein quality control system, which targets substrates for the optimal proteolytic pathway.
KEYWORDS: autophagy, Cue5, Dsk2, proteasome, ubiquitin
To maintain cellular homeostasis, various protein quality control pathways are used to either repair misfolded proteins or target them for degradation if misfolding or aggregation persists. The best pathway for degradation of proteins depends on their physical state within the cell. Soluble substrates are mainly degraded by proteasomes, whereas large insoluble aggregated proteins are engulfed by phagophores targeting substrates to the vacuole/lysosome for their destruction. A shared feature of both proteolytic pathways is substrate modification by ubiquitin ultimately raising the question of how pathway choice is achieved.
Key to the degradation of ubiquitinated proteins are proteasomal and autophagy-specific receptors that recognize their substrates via ubiquitin-binding domains. To separate both pathways, it was suggested that soluble and aggregated proteins are modified with different ubiquitin chain types. Lys48-linked chains are well established in targeting proteins for proteasomal degradation whereas it was initially suggested that Lys63-linked poly-ubiquitin chains would facilitate degradation by autophagy. However, autophagy receptors show no obvious preference binding Lys63-linked poly-ubiquitin chains over Lys48-linked chains. Thus, how substrates are sorted to their designated degradation pathway remained unclear.
To elucidate how pathway choice is made, we took advantage of the budding yeast Saccharomyces cerevisiae as a model organism in which both pathways are present and are mainly dependent on a single ubiquitin binding receptor, Dsk2 for proteasomal and Cue5 for autophagic degradation, respectively. To compare the 2 pathways, we used ubiquitin fused to β-galactosidase (Ub-β-gal), which is soluble, and a truncated version of this protein (Ub-β-gal-X90), which is aggregation prone, as substrates.
We observed that indeed Dsk2 mediates the proteasomal degradation of soluble substrates, whereas Cue5 is required for clearance of aggregated proteins. We therefore tested previous views on how pathway choice is achieved, focusing first on early events including the enzymes involved in ubiquitination and different types of ubiquitin chains. We found that degradation of both soluble Ub-β-gal-WT and aggregation-prone Ub-β-gal-X90 depend on the same set of enzymes, the E2 conjugating enzymes Ubc4/Ubc5 and the E3 ligase Rsp5. Moreover, we could exclude the previously suggested requirement of Lys63-linked for autophagic degradation. Both receptors did not show any ubiquitin linkage specificity, however, we observed a 10-fold higher affinity of Dsk2 compared with Cue5 toward ubiquitin.
Having excluded different types of ubiquitin-linkages as a signal for pathway choice, we continued analyzing how proteasome and autophagy receptors differ. The most remarkable difference between both types of receptors is the strong capacity of the autophagy receptor Cue5 to form higher order oligomers, as opposed to Dsk2, which did not show self-interaction. To evaluate the features required for correct targeting of substrates to their designated proteolytic pathway, we constructed a set of artificial receptors. All receptors harbor a ubiquitin-binding UBA domain for substrate recognition but differ in their oligomeric state due to the presence of 1 or 2 oligomerization domains, creating monomeric, dimeric and oligomeric molecules. These receptors were additionally modified either with a proteasome-targeting UBL domain, an Atg8-interacting motif (AIM), or both. We then analyzed the ability of the artificial receptors in mediating degradation of soluble and aggregated proteins. As expected, monomeric receptors were able to support degradation of soluble proteins by the proteasome and failed to facilitate degradation of aggregated proteins. On the contrary, artificial oligomeric receptors could support autophagy-mediated degradation of insoluble protein aggregates. Importantly, even for receptors harboring both UBL (proteasome) and AIM (autophagy) targeting domains, the supported pathway was strictly dependent on the oligomeric state of the receptor, demonstrating that receptor oligomerization is the main determinant for proteolytic pathway choice.
Soluble ubiquitinated substrates are efficiently targeted for proteasomal degradation by monomeric Dsk2 due to its higher affinity to ubiquitin compared with Cue5. However, when ubiquitinated substrates aggregate, the oligomerization of Cue5 confers higher avidity toward the substrates due to bundling of several ubiquitin-binding domains, which then allows for autophagic degradation (Fig. 1). These findings show that pathway choice is a late event dictated by the physical properties of the receptors rather than by substrate recognition or modification. Importantly, the pathway chosen depends on the solubility of the substrates themselves and ensures that substrates are targeted to the appropriate machinery best suited for their degradation. The shared upstream enzymes guarantee that ubiquitinated substrates, which fail to be degraded by the proteasome and thus aggregate, can efficiently be degraded by autophagy without further modification. Herein, ubiquitination of substrates serves as a common signal for 2 branches of cellular protein quality control. This ensures that potentially harmful misfolded, soluble or aggregated proteins are eventually targeted for degradation. Therefore, the mechanism we have uncovered provides an explanation as to how pathway choice is achieved revealing that misfolded proteins are partitioned to their appropriate degradative pathway based on solubility and, thus, selective receptor binding.
Figure 1.
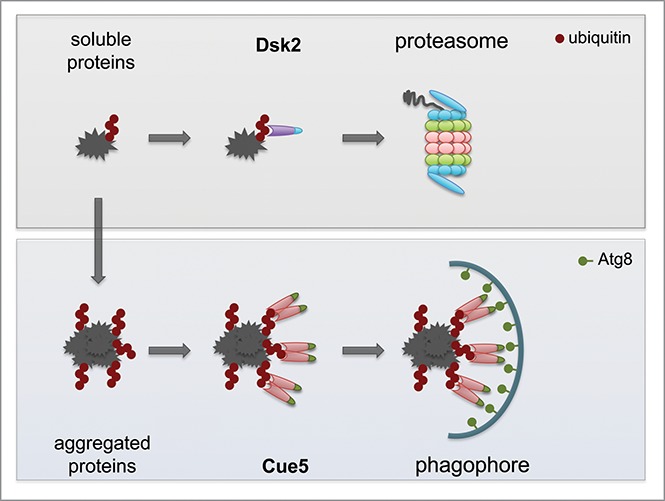
Proteasome receptor Dsk2 and autophagy receptor Cue5 function in the clearance of soluble and aggregated substrates. Monomeric Dsk2 with high affinity for ubiquitin binds soluble ubiquitinated substrates and targets them for proteasomal degradation. When ubiquitinated proteins accumulate in aggregates they are bound by Cue5 receptors with low affinity for ubiquitin but high avidity due to receptor bundling and subsequently targeted for degradation by autophagy.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.


