Abstract
Objective:
To determine whether indeterminate (Likert-score 3/5) peripheral zone (PZ) multiparametric MRI (mpMRI) studies are classifiable by prostate-specific antigen (PSA), PSA density (PSAD), Prostate Imaging Reporting And Data System version 2 (PI-RADS_v2) rescoring and morphological MRI features.
Methods:
Men with maximum Likert-score 3/5 within their PZ were retrospectively selected from 330 patients who prospectively underwent prostate mpMRI (3 T) without an endorectal coil, followed by 20-zone transperineal template prostate mapping biopsies +/- focal lesion-targeted biopsy. PSAD was calculated using pre-biopsy PSA and MRI-derived volume. Two readers A and B independently assessed included men with both Likert-assessment and PI-RADS_v2. Both readers then classified mpMRI morphological features in consensus. Men were divided into two groups: significant cancer (≥ Gleason 3 + 4) or insignificant cancer (≤ Gleason 3 + 3)/no cancer. Comparisons between groups were made separately for PSA & PSAD using Mann–Whitney test and morphological descriptors with Fisher’s exact test. PI-RADS_v2 and Likert-assessment were descriptively compared and percentage inter-reader agreement calculated.
Results:
76 males were eligible for PSA & PSAD analyses, 71 for PI-RADS scoring, and 67 for morphological assessment (excluding significant image artefacts). Unlike PSA (p = 0.915), PSAD was statistically different (p = 0.004) between the significant [median: 0.19 ng ml–2 (interquartile range: 0.13–0.29)] and non-significant/no cancer [median: 0.13 ng ml–2 (interquartile range: 0.10–0.17)] groups. Presence of mpMRI morphological features was not significantly different between groups. Subjective Likert-assessment discriminated patients with significant cancer better than PI-RADS_v2. Inter-reader percentage agreement was 83% for subjective Likert-assessment and 56% for PI-RADS_v2.
Conclusion:
PSAD may categorize presence of significant cancer in patients with Likert-scored 3/5 PZ mpMRI findings.
Advances in knowledge:
PSAD may be used in indeterminate PZ mpMRI to guide decisions between biopsy vs monitoring.
INTRODUCTION
Multiparametric MRI (mpMRI) has the potential to be the modality of choice for ruling out clinically significant prostate cancer with reported negative predictive values (NPV) as high as 89% (83–94%).1, 2 However, prostate mpMRI is assessed as indeterminate (Likert-score 3/5) in almost one-third of cases (163 of 576 patients enrolled in the recent multicentre prospective study assessing the 'diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer' - the PROMIS study) where the radiologist is unable to confirm or refute the presence of clinically significant cancer.
Two scoring systems are commonly used to evaluate the likelihood of prostate cancer on mpMRI.3–6 First, the Prostate Imaging Reporting And Data System version 2 (PI-RADS_v2) which uses explicit criteria based on a zonal mpMRI dominant sequence (e.g. diffusion-weighted imaging in the peripheral zone (PZ) and T2 weighted imaging in the transition zone) to rate the suspicion of prostate cancer on a 5-point scale7 and second, a subjective 5-point Likert assessment,8 based on each mpMRI sequence equally (unlike the use of dominant sequence in PIRADS_v2) and adapted to the radiologist’s experience for overall impression. Both 5-point scales define the likelihood for the presence of prostate cancer, as follows (1) highly unlikely, (2) unlikely, (3) equivocal, (4) likely, (5) highly likely. The Likert-assessment has recently been prospectively validated in the multicentre, multireader PROMIS trial.1 PI-RADS_v2 remains more widely accepted where prospectively scored cohorts have also been reported9 but studies with head-to-head comparisons of PI-RADS_v2 and Likert-assessment as scoring systems are lacking.
Indeterminate mpMRI poses both a management dilemma and potential unnecessary increase in healthcare cost. If all patients within this group were biopsied, a majority of men without significant disease would be exposed to potentially unnecessary risks of haemorrhage and urinary tract infections, which can lead to hospitalization10 further increasing healthcare costs; conversely, if all males were not biopsied then, a significant proportion of men would have significant cancer missed. This cohort of patients has been scarcely studied as an independent group11, 12 since most attention so far, has been focused on both extremes of the scale.13–16 Decreasing the number of equivocal mpMRI scans remains an unmet clinical challenge and represents a determinant factor for widespread global adoption of mpMRI.
The prevalence of prostate cancer is higher in the PZ than the transition zone and zone-specific molecular and imaging phenotypes exist, prompting separate zonal assessment.17–19
The aim of this work was to determine whether indeterminate (Likert-score 3/5) PZ multiparametric MRI (mpMRI) studies can be categorized into significant/insignificant cancer by prostate-specific antigen (PSA), PSA density (PSAD), PI-RADS_v2 rescoring and morphological MRI features.
METHODS and materials
Local institutional review board waived the requirement for individual consent for this single-centre retrospective analysis of prospectively enrolled patients from a previous study cohort20—Research Ethics Committee reference 11/LO/1657.
Patients
330 males [median age: 63 years, interquartile range, IQR (42-83); median PSA: 7.4 ng ml−1, IQR (0.7–58.05)], with prior negative/non-significant prostate disease on TRUS biopsies, but in whom a clinical suspicion of prostate cancer remained (elevated PSA or PSA kinetics, abnormal digital rectal examination etc.) were consecutively enrolled from January 2012 to 2014.20 They all underwent prostate mpMRI without an endorectal coil at 3 T (unless contraindicated). A radiologist (Reader A) with 10 years of experience in prostate imaging, blinded to histopathological results, prospectively reported mpMRI findings and scored the likelihood of significant cancer on a subjective 5-point Likert scale. All patients underwent 20-zone transperineal template prostate mapping (TPM) biopsies.21 In addition, when a focal lesion was identified on mpMRI, its location was mapped on a prostate gland representative diagram and its biopsy template grid co-ordinates noted.21 MR-guided targeted biopsies were performed by experienced urologists (≥10 years of experience), aware of the radiologist’s report, as described within the previous study protocol.20
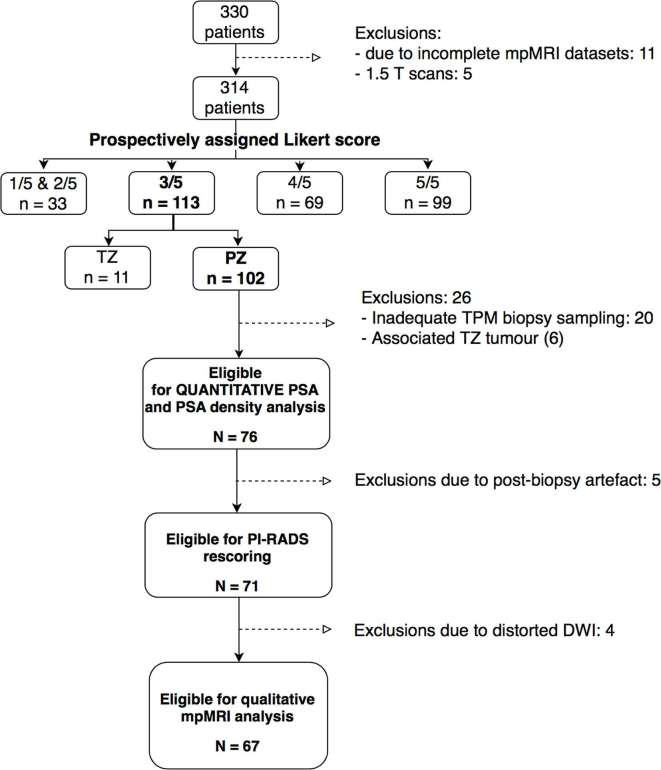
Men with complete 3 T mpMRI data sets, full template biopsy ± targeted biopsy and maximum Likert-score 3/5 PZ were eligible for inclusion. 107/330 patients fulfilled these criteria. Patients with a concurrent Likert-assessment 3/5 in the transition zone (TZ) subsequently identified as clinically significant tumour were excluded (n = 6), males with a lack of complete gland sampling/inadequate sampling density were excluded (n = 20), and five men who underwent 1.5 T scans were excluded. The final cohort comprised of 76/107 PZ Likert-score 3/5 mpMRI studies for analysis. Figure 1 summarizes the patient selection.
Figure 1.
Flowchart diagram illustrating the patient selection process for this study. DWI, diffusion-weighted imaging; mpMRI, multiparametric MRI; PI-RADS, Prostate Imaging Reporting And Data System version 2; PSA, prostate-specific antigen; PZ, peripheral zone; TPM, template prostate mapping; TZ, transition zone.
MpMRI protocol
The 3 T mpMRI protocol for included males (Table 1) has been previously described.22 All studies were performed with the same protocol on a single 3 T scanner (Achieva®, Philips Healthcare, Netherlands) using a 32-channel pelvic-phased array coil. Briefly, sequences included axial turbo spin echo and coronal T2W; axial DWI using a high b value at 2000 s mm–2; axial ADC map generated by diffusion gradients b0, b150, b500, and b1000 (s mm–2) and axial T1W dynamic–contrast enhanced sequences before and after intravenous administration of at least 0.1 mmol kg–1 gadolinium meglumine contrast agent (Dotarem®, Guerbet, France) at a rate of 3 ml s−1 via power injector, followed by 20 ml saline bolus at the same rate with a temporal resolution of 15 s.
Table 1.
Parameters of mpMRI at 3 T
| Parameter | Repetition time (ms) | Echo time (ms) | Flip angle (degree) | Orientation | Slice thickness (mm) | Matrix size | Field of view (mm) | Fat suppression | Time for scan (min) |
| T2 coronal | 6128 | 100 | 90 | Coronal | 3 | 300 × 290 | 180 | No | 05:55 |
| T2 axial | 5407 | 100 | 90 | Axial | 3 | 300 × 290 | 180 | No | 05:13 |
| DWI b 0, 150, 500, 1000 s mm–2 | 2753 | 80 | 90 | Axial | 5 | 168 × 169 | 220 | SPAIR | 05:16 |
| DWI b 2000 s mm–2 | 2000 | 78 | 90 | Axial | 5 | 168 × 169 | 220 | SPIR | 03:40 |
| DCE 20 dynamic mode sense | 5.8 | 28 | 10 | Axial | 3 | 140 × 162 | 180 | SPAIR | 04:14 |
DCE, dynamic contrast-enhanced; DWI, diffusion-weighted imaging; mpMRI, multiparametric MRI; SPAIR, spectral attenuated inversion recovery; SPIR, spectral pre-saturation with inversion recovery.
Cancer significance
Histology results were reported by a uropathologist with 12 years of experience. Recognizing that there still is an ongoing debate on what constitutes clinically significant prostate cancer, for the purposes of this study, the presence of any Gleason 7 pattern or higher (≥3 + 4), anywhere within the PZ was considered as clinically significant.23 The maximum cancer core length (MCCL) was measured and categorized as < 6 or ≥ 6 mm.
Correlation of transperineal template mapping biopsies and mpMRI
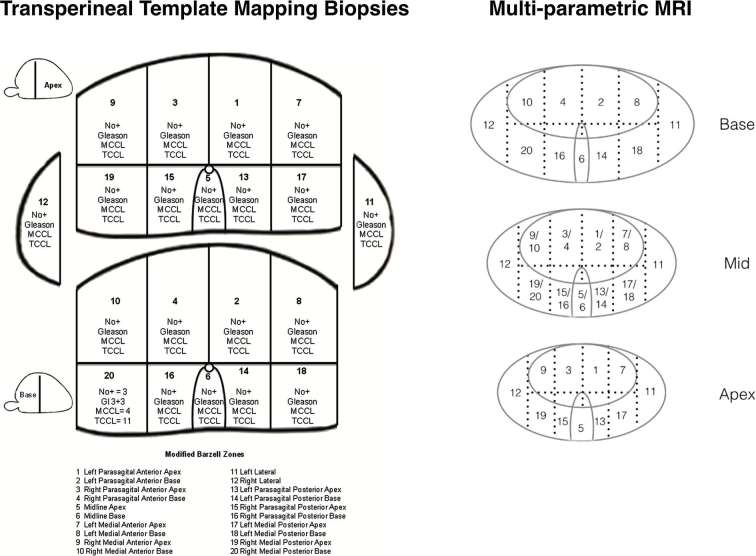
For mpMRI to histopathology matching, Likert-scores 3/5 at the apex and base were considered positive for significant cancer if the corresponding Barzell zone was positive on biopsy. Likert-scores 3/5 at the midgland level were considered positive if the corresponding apical or basal Barzell zone was positive on biopsy. A schema representing the correspondence of Barzell zones on template prostate mapping biopsies21 and prostate mpMRI regions is illustrated in Figure 2.
Figure 2.
Schema showing the correlation of the 20 Barzell zones from template mapping biopsy to the corresponding regions on mpMRI. MCCL, maximum cancer core length; mpMRI, multiparametric MRI.
Serum PSA and PSA density
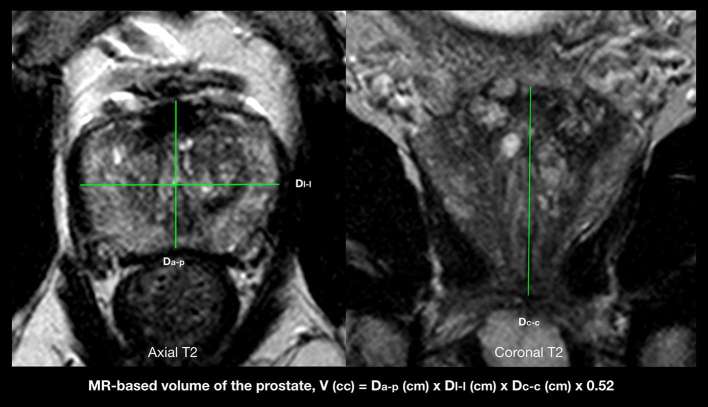
PSA prior to entry of patients into the previous study14 was recorded. To calculate PSA density, the prostate volume was measured on the mpMRI study. The maximum anteroposterior and laterolateral diameters of the prostate were manually measured by Reader A on mid axial T2W slice while its craniocaudal diameter was measured on mid coronal T2W slice as shown in Figure 3. Assuming an ellipsoid shape of the prostate, its volume was calculated by Reader A as the product of the three diameters and π/6 (approximately 0.52).24 PSAD was calculated by the quotient of serum PSA over the gland volume.
Figure 3.
Figure 3 illustrates T2 weighted MRI images where the anteroposterior (Dap) and laterolateral diameters (Dll) of the prostate gland are measured on the axial plane and the craniocaudal diameter or height (Dcc) is measured on the coronal plane to calculate the volume of the prostate gland by using the prolate ellipse formula (Dap × Dll × Dcc × 0.52).
Likert-assessment to PI-RADS scoring
Reader A, who reported the mpMRI of the prostate as Likert-score 3/5 in the previous study,22 rescored them with PI-RADS_v2 criteria.8 A second radiologist, Reader B (4 years of experience in reading prostate mpMRI) independently rescored the scans using first Likert-assessment, then at different time points with PI-RADS_v2. Both readers were blinded to histopathological reports, unaware of each other’s scores.
Peripheral zone mpMRI morphology
As PZ Likert-scored 3/5 exhibit varied appearances on mpMRI, the presence/absence of the morphological descriptors (described below i-iii and in Figure 4) of PZ signal changes on all combined sequences were considered by Readers A and B in consensus (as no pre-defined morphological validated classification scheme has yet been reported for Likert-score 3/5 cohorts). This was assessed only after the Likert-assessment and PI-RADS_v2 scores of both readers had been locked.
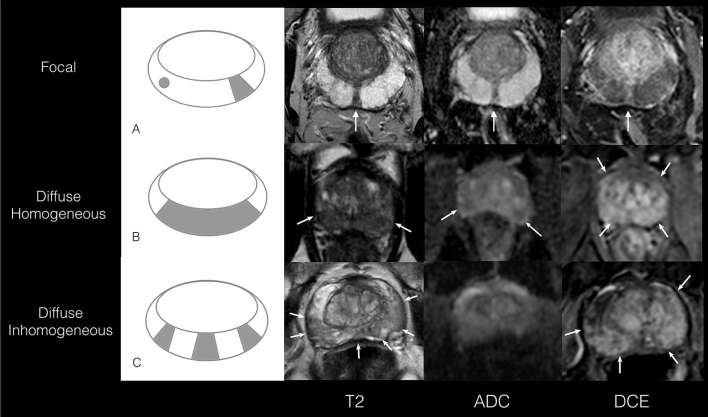
Figure 4.
(A–C) Illustrations of morphological mpMRI descriptors for PZ scored 3/5. (A) A schematic representation and example of a focal lesion (occupying <50% of the slice) in the PZ (axial plane) in a 70-year-old male with PSA of 13 ng ml−1 and PSAD of 0.14 ng ml–2. The lesion is seen at 6 o’clock, hypointense on T2, low signal on ADC and enhances on DCE images. Histology results were benign. (B) A schematic representation and example of diffuse homogeneous signal changes in a 51-year-old male with PSA of 6 ng ml−1 and PSAD of 0.25 ng ml–2. The signal changes seen are hypointense on axial T2 and the whole PZ enhances uninterruptedly on axial DCE images. Histology results revealed Gleason 3 + 4 with MCCL <6 mm on the right lateral side whereas the left PZ was benign. (C) A schematic illustration and example of diffuse inhomogeneous changes in the axial plane in a 66-year-old male with PSA of 5 ng ml−1 and PSAD of 0.09 ng ml–2. Areas of low T2 signal interspersed by normal high T2 signal intensities are observed. The low T2 signal intensities of the PZ are seen to enhance (on >50% of the gland) from 1 to 5 o’ clock and 7 to 8 o’ clock on DCE images. Histology results were benign. ADC, apparent diffusion coefficient; DCE, dynamic contrast-enhanced; MCCL, maximum cancer core length; mpMRI, multiparametric MRI; PSA, prostate-specific antigen; PSAD, PSA density; PZ, peripheral zone.
Morphological descriptors (i) “Discrete focal—”single or multiple focal changes occupying <50% of PZ, (ii) “diffuse homogeneous—”uninterrupted signal changes occupying >50% of PZ, and without focal intensity variation, (iii) “diffuse inhomogeneous”—signal changes occupying >50% of PZ interrupted by focal intensity variation or stranded changes.
Statistical analyses
The Mann–Whitney tests were used to compare each PSA and PSAD between clinically significant and non-significant/no cancer groups. From a receiver operating characteristic curve, sensitivity and specificity of various PSAD thresholds were obtained and the highest Youden’s J index was determined to identify PSAD threshold for significant cancer in our cohort.25 Proportions of upscored, downscored and unchanged Likert and PI-RADS scores per reader were descriptively compared. Inter-reader percentage agreement for Likert and PI-RADS were calculated. The Kappa agreement coefficient, κ, between both readers was also computed for PI-RADS (κ < 0.4: fair, 0.4 < κ < 0.8: moderate, κ > 0.8: strong agreement). PZ morphological descriptors were compared between significant and insignificant cancer groups with Fisher’s exact test. Statistical significance was set at p < 0.05. GraphPad Prism statististical software (v. 6, GraphPad Software, San Diego, CA) was used.
RESULTS
Of 76 men [median age 61 years (IQR 57–66); median PSA 7.2 ng ml−1 (IQR 4.9–10.3); median gland volume 52 cc (IQR 33–63), 21 (27%) had a clinically significant cancer at biopsy, 31 (41%) harboured low grade (Gleason 3 + 3) disease, and 24 (32%) had no cancer (including high-grade prostate intra-epithelial neoplasm, atypical acini, inflammation, atrophy, and/or benign cores).
Serum PSA and PSA density
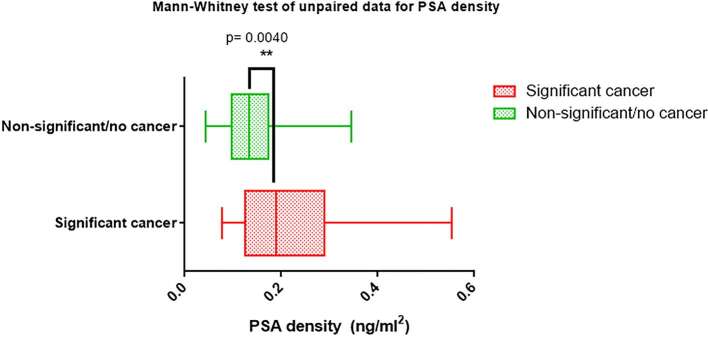
Median PSA and PSAD were 7.17 ng ml−1 (IQR: 5.55–8.69) and 0.19 ng ml–2 (IQR: 0.13–0.29) in the significant cancer group while in the non-significant/no cancer group, these were 7.20 ng ml−1 (IQR: 4.31–10.7) and 0.13 ng ml–2 (IQR: 0.10–0.17) respectively. PSAD was significantly higher in the significant cancer group (p = 0.004) as represented in Figure 5; PSA was not significantly different between the two groups (p = 0.915). A PSAD threshold of >0.17 ng ml–2 {sensitivity: 67% [95% CI (43–85)], specificity: 75% [95% CI (61–85)], NPV: 85% [95% CI (72–94)]} for significant cancer was identified in our cohort with the Youden’s index. To maximize sensitivity, the use of >0.10 ng ml–2 as PSAD threshold would yield 90% sensitivity [95% CI (70–99)], a reduced specificity of 36% [95% CI (23–50)] but NPV would increase to 89% [95% CI (67–99)].
Figure 5.
Bar charts showing the median PSA density comparison between the significant cancer and non-significant/no cancer groups with a statistical difference of p < 0.01. PSA, prostate-specific antigen.
Likert-assessment to PI-RADS scoring
Of 76 patients, 5 had extensive post-biopsy artefact, leaving 71 patients eligible for PZ PI-RADS scoring. Among them, four had non-diagnostic quality DWI, due to air in the rectum, and PI-RADS “assessment without adequate DWI” was applied.8
The set of 71 patients all assessed as Likert-score 3/5 by Reader A comprised of 18 (18/71, 25%) clinically significant cancer at biopsy. Reader B assessed 59/71 patients as Likert-score 3/5, of whom 15 (15/59, 25%) had clinically significant cancer; 4 were assessed Likert-score 2/5 and none had clinically significant cancer; 8 were assessed Likert-score 4/5, 3 of which had clinically significant cancer (3/8, 38%) where 2 had Gleason 3 + 4 pattern and MCCL <6 mm, 1 male had Gleason 4 + 3, MCCL ≥6 mm.
On PI-RADS_v2 rescoring, Readers A and B downscored to PI-RADS ≤2, 34/71 (48%) and 34/59 (58%) patients respectively; with 27/34 (79%) and 26/34 (76%) demonstrating non-significant/no cancer at biopsy. For Reader A, 6/34 (18%) PI-RADS ≤2 scored males had Gleason 3 + 4 disease, MCCL <6 mm and 1/34 male Gleason 3 + 4, MCCL ≥6 mm. For Reader B, 7/30 (23%) PI-RADS ≤2 scored males had Gleason 3 + 4 pattern with MCCL <6 mm, 1/34 male Gleason 3 + 4, MCCL ≥6 mm, and 1/34 Gleason 4 + 3 disease.
Readers A and B upscored to PI-RADS 4 in 31/71 (44 %) and 20/59 (34%) patients respectively, and none to PI-RADS 5; with 23/31 (74%) and 13/20 (65%) demonstrating non-significant/no cancer at biopsy. For Reader A, 6/31 (19%) PI-RADS 4 scored males had Gleason 3 + 4 and MCCL <6 mm, 1/31 had a Gleason 3 + 4 with MCCL ≥6 mm, and 1/31 had Gleason 4 + 3 pattern. For Reader B, 5/20 (25%) PI-RADS 4 scored males had Gleason 3 + 4 and MCCL <6 mm, and 2/20 males had Gleason 3 + 4, MCCL ≥6 mm. Reader B scored five patients PI-RADS 3 (none with significant cancer) while Reader A scored six patients PI-RADS 3 where three had clinically significant cancer (3/6, 50%)—two of which were Gleason 3 + 4, MCCL <6 mm and one Gleason 3 + 4, MCCL ≥6 mm. These results are summarized in Figure 6.
Figure 6.
Bar charts illustrating the results of Likert and PI-RADS scoring of the PZ by Readers A and B. PI-RADS segregate Likert-score 3/5 lesions mostly into PI-RADS ≤2 and PI-RADS 4 by both Readers A and B. The number of Likert-indeterminate lesions decreased from 71 to 6 (8%) by Reader A and 5 (7%) by Reader B with PI-RADS scoring. Readers A and B respectively upscored 31/71 (44%) and 20/59 (34%) to PI-RADS 4; 8 of 31 (26%) and 7 of 25 (28%) had significant cancer. They downscored 34/71 (48%) and 34/59 (58%) to PI-RADS ≤2; 7 of 34 (24%) and 8 of 31 (26%) had significant cancer. PI-RADS, Prostate Imaging Reporting And Data System version 2.
The percentage agreement between Readers A and B for Likert-score 3/5 was (59/71) 83% and (40/71) 56% for PI-RADS_v2. The latter had an inter-reader agreement coefficient κKappa of 0.27 [95% CI (0.10–0.44)].
Peripheral zone mpMRI morphology
67 males were included in this analysis after excluding non-diagnostic DWI studies. Results of qualitative mpMRI assessment are summarized in Table 2. 13 of 47 (28%) patients with discrete focal change [median volume 0.17 cc (IQR: 0.05–0.16)] demonstrated significant cancer; and 3/10 patients with diffuse homogeneous changes also demonstrated significant cancer. No patient with diffuse inhomogeneous signal changes had significant cancer. However, differences between groups did not reach statistical significance (p = 0.21 to 1.00).
Table 2.
Qualitative mpMRI assessment
| Qualitative mpMRI descriptor | Significant cancer | Non-significant cancer/no cancer | Total |
| Focal lesion | 13 (28%) | 34 (72%) | 47 (70%) |
| Diffuse homogeneous changes | 3 (30%) | 7 (70%) | 10 (15%) |
| Diffuse inhomogeneous changes | 0 (0%) | 10 (100%) | 10 (15%) |
Table 2 shows the number of significant cancer and non-significant cancer/no cancer, in each of the qualitative mpMRI descriptor groups.
mp MRI, multiparametric MRI.
DISCUSSION
This paper assessed whether serum PSA & PSAD, PI-RADS_v2 rescoring and morphological features of Likert-score 3/5 PZ signal changes could help identify patients harbouring significant cancer.
Firstly, we found serum PSA level by itself, was not able to identify patients with significant cancer. Yet, when combined with gland volume assessment, PSAD was the best predictor of patients with significant cancer. Rais-Bahrami et al reported that PSAD coupled to the number of MR suspicious lesions on biparametric MRI (T2W and DWI) improve categorization of Gleason score ≥7 upon TRUS or MR/ultrasound fusion biopsies.26 Recently, the use of PSA density with mpMRI has gained further interest in improving mpMRI accuracy.27–29 Our study complements this work, and provides evidence that PSAD can specifically address the problem of indeterminate mpMRI studies.
Furthermore, we reported two thresholds of PSAD for classification of patients with Likert-score 3/5. Various PSAD thresholds have been previously proposed to select patients with significant disease.27,28,30–33 Epstein et al found >0.15 ng ml–2 to be associated with significant disease upon prostatectomy with a 66% NPV.21 The National Comprehensive Cancer Network has adopted this value34 while the Prostate Cancer Research International Active Surveillance programme has adopted >0.2 ng ml–2 33 as predictors of significant disease. In our study, Youden’s cut-off of PSAD >0.17 ng ml–2 in determining the presence of significant cancer offers a 75% specificity [95% CI (61–85)] and 85% NPV [95% CI (72–94)]. This would provide a prudent approach to management by accepting a higher number of patients to be biopsied to minimize the chances of missing patients with significant tumour. Were this threshold applied to our cohort of 76 patients, it would have correctly avoided biopsy in 40/76 (53%) and led to 7/76 (9%) patients with cancer not being immediately diagnosed.
Secondly, we found a relatively poor performance of PI-RADS_v2 as a classifier of Likert 3/5 patients. PI-RADS_v2 rescoring from both readers, up or downscored almost all patients into PI-RADS ≤2 and PI-RADS4 score groups. However, as approximately three-quarter of men upscored to PI-RADS 4 had no significant disease (and hence would undergo unnecessary biopsy) and almost one-quarter of males downscored to PI-RADS ≤2 had significant cancer (and hence significant cancer would be missed if no biopsy was performed) we conclude that within our patient cohort PI-RADS scoring was not a good classifier of patients. Our results are not unique in highlighting some current deficiencies within PI-RADS_v2 reporting schema;35 Besides, Vargas et al have also reported that it offered limited assessment of Gleason 4 + 3 pattern of volume ≤0.5 ml.36
Interestingly, whilst Likert-assessment demonstrated 83% inter-reader concordance, PI-RADS_v2 showed only 56% concordance—in line with Greer et al who reported an overall inter-observer concordance of 58% for PI-RADS_v2,37 while Renard-Penna et al reported higher concordance values (92%) for Likert-assessment.5 While no other study has compared Likert-assessment and PI-RADS_v2 yet, some studies have compared Likert-assessment to PI-RADS_v14, 5 with Vaché et al showing a more accurate performance of Likert-assessment.4 For PI-RADS_v2, Rosenkrantz et al found expert inter-reader agreement, κ in the PZ score Groups of ≥4 and ≥3 to be 0.59 and 0.53 respectively.38 Muller et al39 showed an overall κ = 0.47 for PI-RADS_v2 scoring with mixed reader experience. Within our study, we found a smaller κ of 0.27 for PI-RADS_v2. We do not believe this to be surprising as we specifically assessed a subgroup (Likert-assessment 3/5) of patients where radiological assessment is inherently more challenging.
Finally, we investigated whether any particular pattern of PZ signal change could help classify patients. We found the group of patients with “diffuse inhomogeneous” pattern did not include any patient with significant cancer. However, the number of patients within our cohort was too small to confirm the statistical significance of this observation.
The results of our study are relevant to directing clinical practice following the recent PROMIS study publication with Likert-scored mpMRI.1 When 3/5-scores are classified as positive, the specificity of the mpMRI study is reduced.5, 40 If these scores are classified as negative, the sensitivity of the mpMRI test reduces. We would advocate that the Likert 3/5 score should be treated as a separate indeterminate group which needs further classification with secondary features. Using PSAD provides a simple method to manage patients with indeterminate scores: males with high PSAD would undergo biopsy; those with low PSAD may benefit from further observation (perhaps PSA surveillance) as our results suggest that some of them (albeit a small percentage) will have significant tumour. Our results support the necessity for continued iteration of PI-RADS reporting schema based on ongoing research to improve classification, minimize subjectivity and promote inter-reader agreement.
Our study has several limitations. Firstly, it was necessary to use a template biopsy-based reference in our cohort as it comprised many patients without significant cancer/any cancer and, therefore, could not have a prostatectomy. We acknowledge the limitations of template biopsy—(which has a 95% detection rate for significant tumours against 100% at prostatectomy41)—nevertheless, unavoidable within our cohort. Secondly, although we propose PSAD thresholds to aid management of Likert-score 3/5 patients, the clinical impact of these thresholds should be prospectively validated. Thirdly, it would be prudent to replicate our study in other cohorts to confirm generalizability of our findings, both in terms patient cohorts being imaged with mpMRI (e.g. pre-biopsy vs delayed post-biopsy) and also different MRI scanning platforms (e.g. 1.5 vs 3 T).
CONCLUSIONS
MR-adjusted PSAD may help classify patients with PZ Likert-scored 3/5 on mpMRI who have clinically significant cancer and could be used to select patients for biopsy over observation. Prospective studies are further required to validate the use of PSA density in indeterminate mpMRI cohorts.
Contributor Information
Mrishta Brizmohun Appayya, Email: mrishta@gmail.com.
Harbir S Sidhu, Email: harbir.sidhu@nhs.net.
Nikolaos Dikaios, Email: n.dikaios@ucl.ac.uk.
Edward W Johnston, Email: edward.johnston@ucl.ac.uk.
Lucy AM Simmons, Email: l.simmons1@nhs.net.
Alex Freeman, Email: alex.freeman@uclh.nhs.uk.
Alexander PS Kirkham, Email: alexkirkham@yahoo.com.
Shonit Punwani, Email: shonit.punwani@gmail.com.
REFERENCES
- 1.Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. The Lancet 2017; 389: 815–22. doi: https://doi.org/10.1016/S0140-6736(16)32401-1 [DOI] [PubMed] [Google Scholar]
- 2.Fütterer JJ, Briganti A, De Visschere P, Emberton M, Giannarini G, Kirkham A, et al. Can clinically significant prostate cancer be detected with multiparametric magnetic resonance imaging? A systematic review of the literature. Eur Urol 2015; 68: 1045–53. doi: https://doi.org/10.1016/j.eururo.2015.01.013 [DOI] [PubMed] [Google Scholar]
- 3.Rastinehad AR, Waingankar N, Turkbey B, Yaskiv O, Sonstegard AM, Fakhoury M, et al. Comparison of multiparametric MRI scoring systems and the impact on cancer detection in patients undergoing MR US fusion guided prostate biopsies. PLoS One 2015; 10: e0143404. doi: https://doi.org/10.1371/journal.pone.0143404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaché T, Bratan F, Mège-Lechevallier F, Roche S, Rabilloud M, Rouvière O. Characterization of prostate lesions as benign or malignant at multiparametric MR imaging: comparison of three scoring systems in patients treated with radical prostatectomy. Radiology 2014; 272: 446–55. doi: https://doi.org/10.1148/radiol.14131584 [DOI] [PubMed] [Google Scholar]
- 5.Renard-Penna R, Mozer P, Cornud F, Barry-Delongchamps N, Bruguière E, Portalez D, et al. Prostate imaging reporting and data system and Likert scoring system: multiparametric MR imaging validation study to screen patients for initial biopsy. Radiology 2015; 275: 458–68. doi: https://doi.org/10.1148/radiol.14140184 [DOI] [PubMed] [Google Scholar]
- 6.Rosenkrantz AB, Lim RP, Haghighi M, Somberg MB, Babb JS, Taneja SS. Comparison of interreader reproducibility of the prostate imaging reporting and data system and likert scales for evaluation of multiparametric prostate MRI. AJR Am J Roentgenol 2013; 201: W612–W618. doi: https://doi.org/10.2214/AJR.12.10173 [DOI] [PubMed] [Google Scholar]
- 7.Rensis L. A technique for the measurement of attitudes. Archives of Psychology 1932; 22: 55. [Google Scholar]
- 8.Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, et al. PI-RADS prostate imaging - reporting and data system: 2015, version 2. Eur Urol 2016; 69: 16–40. doi: https://doi.org/10.1016/j.eururo.2015.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mertan FV, Greer MD, Shih JH, George AK, Kongnyuy M, Muthigi A, et al. Prospective evaluation of the prostate imaging reporting and data system version 2 for prostate cancer detection. J Urol 2016; 196: 690–6. doi: https://doi.org/10.1016/j.juro.2016.04.057 [DOI] [PubMed] [Google Scholar]
- 10.Loeb S, Vellekoop A, Ahmed HU, Catto J, Emberton M, Nam R, et al. Systematic review of complications of prostate biopsy. Eur Urol 2013; 64: 876–92. doi: https://doi.org/10.1016/j.eururo.2013.05.049 [DOI] [PubMed] [Google Scholar]
- 11.Rosenkrantz AB, Meng X, Ream JM, Babb JS, Deng FM, Rusinek H, et al. Likert score 3 prostate lesions: association between whole-lesion ADC metrics and pathologic findings at MRI/ultrasound fusion targeted biopsy. J Magn Reson Imaging 2016; 43: 325–32. doi: https://doi.org/10.1002/jmri.24983 [DOI] [PubMed] [Google Scholar]
- 12.Liddell H, Jyoti R, Haxhimolla HZ. mp-MRI prostate characterised PIRADS 3 lesions are associated with a low risk of clinically significant prostate cancer - a retrospective review of 92 biopsied PIRADS 3 lesions. Curr Urol 2015; 8: 96–100. doi: https://doi.org/10.1159/000365697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grey AD, Chana MS, Popert R, Wolfe K, Liyanage SH, Acher PL. Diagnostic accuracy of magnetic resonance imaging (MRI) prostate imaging reporting and data system (PI-RADS) scoring in a transperineal prostate biopsy setting. BJU Int 2015; 115: 728–35. doi: https://doi.org/10.1111/bju.12862 [DOI] [PubMed] [Google Scholar]
- 14.Thompson JE, van Leeuwen PJ, Moses D, Shnier R, Brenner P, Delprado W, et al. The diagnostic performance of multiparametric magnetic resonance imaging to detect significant prostate cancer. J Urol 2016; 195: 1428–35. doi: https://doi.org/10.1016/j.juro.2015.10.140 [DOI] [PubMed] [Google Scholar]
- 15.Reisüter LA, Fütterer JJ, Halvorsen OJ, Nygærd Y, Biermann M, Andersen E, et al. 1.5-T multiparametric MRI using PI-RADS: a region by region analysis to localize the index-tumor of prostate cancer in patients undergoing prostatectomy. Acta Radiol 2015; 56: 500–11. doi: https://doi.org/10.1177/0284185114531754 [DOI] [PubMed] [Google Scholar]
- 16.Renard-Penna R, Roupret M, Compérat E, Rozet F, Granger B, Barkatz J, et al. Relationship between non-suspicious MRI and insignificant prostate cancer: results from a monocentric study. World J Urol 2016; 34: 673–8. doi: https://doi.org/10.1007/s00345-015-1685-2 [DOI] [PubMed] [Google Scholar]
- 17.Sinnott JA, Rider JR, Carlsson J, Gerke T, Tyekucheva S, Penney KL, et al. Molecular differences in transition zone and peripheral zone prostate tumors. Carcinogenesis 2015; 36: 632–8. doi: https://doi.org/10.1093/carcin/bgv051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dikaios N, Alkalbani J, Abd-Alazeez M, Sidhu HS, Kirkham A, Ahmed HU, et al. Zone-specific logistic regression models improve classification of prostate cancer on multi-parametric MRI. Eur Radiol 2015; 25: 2727–37. doi: https://doi.org/10.1007/s00330-015-3636-0 [DOI] [PubMed] [Google Scholar]
- 19.McNeal JE, Redwine EA, Freiha FS, Stamey TA. Zonal distribution of prostatic adenocarcinoma. Correlation with histologic pattern and direction of spread. Am J Surg Pathol 1988; 12: 897–906. [DOI] [PubMed] [Google Scholar]
- 20.Simmons LA, Ahmed HU, Moore CM, Punwani S, Freeman A, Hu Y, et al. The PICTURE study - prostate imaging (multi-parametric MRI and Prostate HistoScanning™) compared to transperineal ultrasound guided biopsy for significant prostate cancer risk evaluation. Contemp Clin Trials 2014; 37: 69–83. doi: https://doi.org/10.1016/j.cct.2013.11.009 [DOI] [PubMed] [Google Scholar]
- 21.Barzell WE, Melamed MR. Appropriate patient selection in the focal treatment of prostate cancer: the role of transperineal 3-dimensional pathologic mapping of the prostate—a 4-year experience. Urology 2007; 70(6 Suppl): S27–S35. doi: https://doi.org/10.1016/j.urology.2007.06.1126 [DOI] [PubMed] [Google Scholar]
- 22.Simmons LA, Ahmed HU, Moore CM, Punwani S, Freeman A, Hu Y, et al. The PICTURE study - prostate imaging (multi-parametric MRI and Prostate HistoScanning™) compared to transperineal ultrasound guided biopsy for significant prostate cancer risk evaluation. Contemp Clin Trials 2014; 37: 69–83. doi: https://doi.org/10.1016/j.cct.2013.11.009 [DOI] [PubMed] [Google Scholar]
- 23.Eggener SE, Badani K, Barocas DA, Barrisford GW, Cheng JS, Chin AI, et al. Gleason 6 prostate cancer: translating biology into population health. J Urol 2015; 194: 626–34. doi: https://doi.org/10.1016/j.juro.2015.01.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss BE, Wein AJ, Malkowicz SB, Guzzo TJ. Comparison of prostate volume measured by transrectal ultrasound and magnetic resonance imaging: is transrectal ultrasound suitable to determine which patients should undergo active surveillance? Urol Oncol 2013; 31: 1436–40. doi: https://doi.org/10.1016/j.urolonc.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 25.youden wj. index for rating diagnostic tests. CANCER 1950; 3: 32–5. doi: 10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2-3https://doi.org/10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2-3 [DOI] [PubMed]
- 26.Rais-Bahrami S, Siddiqui MM, Vourganti S, Turkbey B, Rastinehad AR, Stamatakis L, et al. Diagnostic value of biparametric magnetic resonance imaging (MRI) as an adjunct to prostate-specific antigen (PSA)-based detection of prostate cancer in men without prior biopsies. BJU Int 2015; 115: 381–8. doi: https://doi.org/10.1111/bju.12639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Washino S, Okochi T, Saito K, Konishi T, Hirai M, Kobayashi Y, et al. Combination of prostate imaging reporting and data system (PI-RADS) score and prostate-specific antigen (PSA) density predicts biopsy outcome in prostate biopsy naïve patients. BJU Int 2017; 119: 225–33. doi: https://doi.org/10.1111/bju.13465 [DOI] [PubMed] [Google Scholar]
- 28.Hansen NL, Barrett T, Koo B, Doble A, Gnanapragasam V, Warren A, et al. The influence of prostate-specific antigen density on positive and negative predictive values of multiparametric magnetic resonance imaging to detect Gleason score 7-10 prostate cancer in a repeat biopsy setting. BJU Int 2017; 119: 724–30. doi: https://doi.org/10.1111/bju.13619 [DOI] [PubMed] [Google Scholar]
- 29.Distler FA, Radtke JP, Bonekamp D, Kesch C, Schlemmer HP, Wieczorek K, et al. The value of PSA density in combination with PI-RADS™ for the accuracy of prostate cancer prediction. J Urol 2017; 198: 575–82. doi: https://doi.org/10.1016/j.juro.2017.03.130 [DOI] [PubMed] [Google Scholar]
- 30.Epstein JI, Walsh PC, Carmichael M, Brendler CB. Pathologic and clinical findings to predict tumor extent of nonpalpable (stage T1c) prostate cancer. JAMA 1994; 271: 368–74. doi: https://doi.org/10.1001/jama.1994.03510290050036 [PubMed] [Google Scholar]
- 31.Tosoian JJ, JohnBull E, Trock BJ, Landis P, Epstein JI, Partin AW, et al. Pathological outcomes in men with low risk and very low risk prostate cancer: implications on the practice of active surveillance. J Urol 2013; 190: 1218–23. doi: https://doi.org/10.1016/j.juro.2013.04.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha YS, Yu J, Salmasi AH, Patel N, Parihar J, Singer EA, et al. Prostate-specific antigen density toward a better cutoff to identify better candidates for active surveillance. Urology 2014; 84: 365–71. doi: https://doi.org/10.1016/j.urology.2014.02.038 [DOI] [PubMed] [Google Scholar]
- 33.van den Bergh RC, Vasarainen H, van der Poel HG, Vis-Maters JJ, Rietbergen JB, Pickles T, et al. Short-term outcomes of the prospective multicentre 'prostate cancer research international: active surveillance' study. BJU Int 2010; 105: 956–62. doi: https://doi.org/10.1111/j.1464-410X.2009.08887.x [DOI] [PubMed] [Google Scholar]
- 34.Mohler J, Bahnson RR, Boston B, Busby JE, D'Amico A, Eastham JA, et al. NCCN clinical practice guidelines in oncology: prostate cancer. J Natl Compr Canc Netw 2010; 8: 162–200. [DOI] [PubMed] [Google Scholar]
- 35.Mehralivand S, Bednarova S, Shih JH, Mertan FV, Gaur S, Merino MJ, et al. Prospective evaluation of PI-RADS™ version 2 using the international society of urological pathology prostate cancer grade group system. J Urol 2017; 198: 583–90. doi: https://doi.org/10.1016/j.juro.2017.03.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vargas HA, Hötker AM, Goldman DA, Moskowitz CS, Gondo T, Matsumoto K, et al. Updated prostate imaging reporting and data system (PIRADS v2) recommendations for the detection of clinically significant prostate cancer using multiparametric MRI: critical evaluation using whole-mount pathology as standard of reference. Eur Radiol 2016; 26: 1606–12. doi: https://doi.org/10.1007/s00330-015-4015-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greer MD, Brown AM, Shih JH, Summers RM, Marko J, Law YM, et al. Accuracy and agreement of PIRADSv2 for prostate cancer mpMRI: A multireader study. J Magn Reson Imaging 2017; 45: 579–85. doi: https://doi.org/10.1002/jmri.25372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenkrantz AB, Ginocchio LA, Cornfeld D, Froemming AT, Gupta RT, Turkbey B, et al. Interobserver reproducibility of the PI-RADS version 2 lexicon: a multicenter study of six experienced prostate radiologists. Radiology 2016; 280: 793–804. doi: https://doi.org/10.1148/radiol.2016152542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller BG, Shih JH, Sankineni S, Marko J, Rais-Bahrami S, George AK, et al. Prostate cancer: interobserver agreement and accuracy with the revised prostate imaging reporting and data system at multiparametric MR imaging. Radiology 2015; 277: 741–50. doi: https://doi.org/10.1148/radiol.2015142818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao C, Gao G, Fang D, Li F, Yang X, Wang H, et al. The efficiency of multiparametric magnetic resonance imaging (mpMRI) using PI-RADS version 2 in the diagnosis of clinically significant prostate cancer. Clin Imaging 2016; 40: 885–8. doi: https://doi.org/10.1016/j.clinimag.2016.04.010 [DOI] [PubMed] [Google Scholar]
- 41.Crawford ED, Rove KO, Barqawi AB, Maroni PD, Werahera PN, Baer CA, et al. Clinical-pathologic correlation between transperineal mapping biopsies of the prostate and three-dimensional reconstruction of prostatectomy specimens. Prostate 2013; 73: 778–87. doi: https://doi.org/10.1002/pros.22622 [DOI] [PMC free article] [PubMed] [Google Scholar]