Abstract
Personalized medicine (PM) aims to harness a wave of ‘omics’ discoveries to facilitate research and discovery of targeted diagnostics and therapies and increase the efficiency of healthcare systems by predicting and treating individual predispositions to diseases or conditions. Despite significant investment, limited progress has been made bringing PM to market. We describe the major perceived regulatory, intellectual property, and reimbursement challenges to the development, translation, adoption, and implementation of PM products into clinical care. We conducted a scoping review to identify (i) primary challenges for the development and implementation of PM identified in the academic literature; (ii) solutions proposed in the academic literature to address these challenges; and (iii) gaps that exist in that literature. We identified regulatory barriers to PM development and recommendations in 344 academic papers. Regulatory uncertainty was a cross-cutting theme that appeared in conjunction with other themes including: reimbursement; clinical trial regulation; regulation of co-development; unclear evidentiary requirements; insufficient incentives for research and development; incompatible information systems; and different regulation of different diagnostics. To fully realize the benefits of PM for healthcare systems and patients, regulatory, intellectual property, and reimbursement challenges need to be addressed in lock step with scientific advances.
Keywords: personalized medicine, precision medicine, device regulation, drug regulation, ‘omics’, diagnostics, biomarkers, intellectual property, reimbursement
INTRODUCTION
Personalized medicine (PM)1 aims to harness a wave of ‘omics’2 discoveries to tailor drug choices, dosages, and interventions to the biology of individual patients. Its goals are to target better healthcare, facilitate research and discovery of diagnostics and therapies, and predict individual predispositions to diseases or conditions.3 The Precision Medicine Initiative (PMI),4 announced by President Obama in January 2015 to ‘enable a new era of medicine through research, technology and policies that empower patients researchers and providers to work together toward development of individualized care’, fits within this broad definition of PM. The term precision medicine, however, is more mechanistic: its goal is to integrate individual-level molecular and clinical data to develop a more accurate taxonomy of diseases to enhance diagnosis, treatment, and disease management.5
The promise of PM is both therapeutic and economic. Many countries have invested financial, human, and infrastructure resources into the development and delivery of PM, hoping to provide better, more individualized healthcare and to use healthcare dollars more efficiently. High-throughput sequencing and bioinformatics infrastructure are facilitating PM research and development (R&D). However, while infrastructure costs are rapidly decreasing, human capacity to interpret and clinically apply ‘omics’ results remains a costly barrier to the widespread implementation of PM.6
A further goal of PM is to reinvigorate stagnating therapeutic R&D.7 Despite substantial increases in R&D investments to US$50 billion per year, the number of new drugs approved annually in the USA has remained constant over the past 60 years.8 The high cost of many new therapies is in part driven by the cost of failures in clinical development: only 15.3% of drugs traverse the pipeline from phase I to market authorization for lead indications, a percentage that drops to 10.4% for all indications.9 The greatest rate of failure occurs due to a lack of evidence of efficacy in phase II. Specifically, for cancer R&D, a key target for PM, billions of dollars of investment have produced median gains in progression-free survival of only 2.1 months and median gains in overall survival of only 2.5 months for new cancer drugs developed between 2002 and 2014.10 PM promises to improve the current R&D environment with a more targeted approach. It will refine our understanding of disease taxonomy and thereby enable us to design clinical trials that enroll participants more likely to benefit from experimental interventions. This promises to improve efficacy and thus decrease clinical trial failure rates.
Despite significant investment, limited progress has been made to date in bringing PM products to the market.11 While there have been some successes in precision medicine, notably in the field of oncology, its products are not currently in use for most diseases.12 Challenges to progress in PM are both scientific and non-scientific. This paper focuses on the latter: we detail the major perceived non-science barriers to the creation, translation, and introduction of PM products into clinical care. We focus especially on those challenges that exist in the regulatory approvals arena, such as clinical trial design, the legal environment that impacts incentives for PM R&D, and the regulations that impact reimbursement of medical therapies, for example, a requirement for evidence of clinical utility. Despite harmonized rules for regular pharmaceutical clinical trials in the International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH),13 a significant challenge to the international adoption of PM products is a lack of internationally accepted best practice standards, coupled with a lack of clear evidentiary standards for regulatory approval or reimbursement (in health insurance schemes). Existing standards are not harmonized across jurisdictions to facilitate cross-border movement or adoption of PM products. In large part, this reflects the confusion or inadequacy of regulatory and practice standards for PM within national borders.
Even though our research was international in scope, in this paper we focus on regulation of PM in the USA. This focus reflects the dominance in medical research with respect to funding and volume in that country. For example, President Obama's ambitious PMI includes the PMI Cohort Program, renamed under the current administration to All of Us, which will collect health and biospecimen data from over a million volunteers to facilitate research on individual variations in genetics, lifestyle, and environment. This research should facilitate delivery on the promise of PM. Furthermore, developments in the USA influence global norms for human subjects research through the ICH; and most PM products will seek approvals from the US Federal Drug Administration (FDA).14 Finally, there is considerable political will15 to create regulatory and policy incentives, including regulatory pathways to facilitate the introduction of PM products into American healthcare markets.
RESEARCH QUESTION AND METHODS
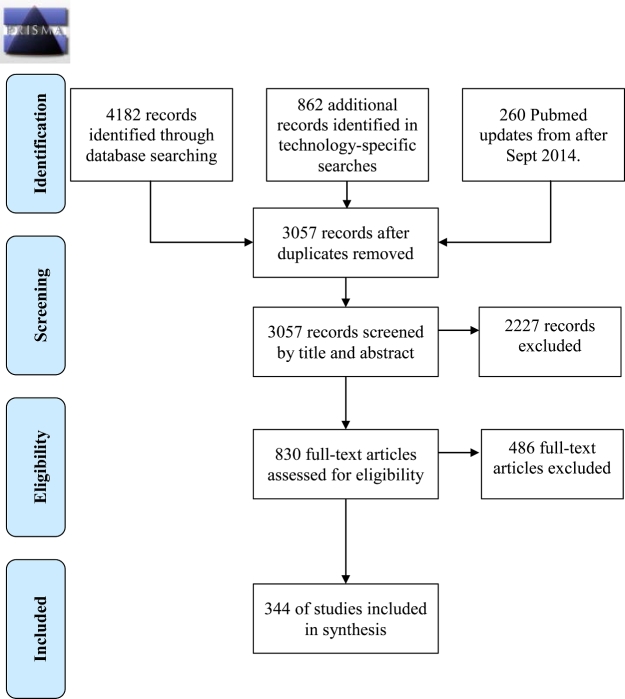
We used a scoping review to answer the following research questions: (i) What are the primary legal and regulatory challenges for the development and implementation of PM identified in the academic literature? and (ii) What solutions have been proposed in the academic literature to address these challenges? A scoping review provides a systematic overview of the type, extent, and quantity (but not the quality) of literature in a given research field.16 It also provides a mechanism to summarize and disseminate research findings to policy makers, practitioners, and consumers. We adapted the Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement17 to report the flow of information through the different phases of our scoping review (Figure 1).
Figure 1.
PRISMA diagram of personalized medicine scoping review method, illustrating inclusion/exclusion criteria. Adapted from Moher et al.17
We defined PM as seeking ‘to improve stratification and timing of healthcare by utilizing biological information and biomarkers on the level of molecular disease pathways, genetics, proteomics as well as metabolomics’.18 We defined barrier as a regulatory structure identified by the study author as an impediment to PM development. We identified recommendations by looking for ‘ought’ statements about how regulatory systems could facilitate PM development. Based on this vocabulary and a list of synonyms for PM, developed with the input of a research librarian, we searched PubMed, Scopus, and LexisNexis for papers published between January 1, 2010 and April 1, 2014 (see Appendix). On September 30, 2014, the FDA published draft guidelines on a laboratory developed test (LDT) regulatory oversight framework. By then, LDT regulation had emerged as a topic of interest in our analysis. To capture responses to the proposed guidelines in the literature, we therefore extended our search period until February 6, 2015. For logistical reasons, we only conducted the extended search in PubMed. We imported all search results into Endnote 7 and removed duplicates. Of 5304 articles identified by our initial searches, 3057 remained. In consultation with economic and legal experts, we developed inclusion and exclusion standards to apply to our search results. We included English language papers that addressed both our definition of PM and regulatory challenges/barriers to PM development or proposed solutions to regulatory challenges/barriers to PM development. We included papers written on the regulatory systems affecting PM development in the European Union (EU), the USA, Canada, Japan, Singapore, and China.
Two researchers, WL and a research assistant, evaluated the titles and abstracts of search results for inclusion in the analysis. To ensure validity, the two researchers evaluated the same subset of titles and articles, and then compared their results. We calculated a kappa score, a standard measure for intercoder reliability, of >0.9, indicating consistent application of the inclusion and exclusion criteria.19 Eight hundred and thirty articles met our inclusion criteria. We retrieved full texts versions of these 830 articles, and two researchers (WL and LK) re-applied the inclusion and exclusion criteria to the full papers. They independently reviewed the same papers, representing 10% of the total dataset, and met to compare findings. Discrepancies were resolved through discussion to consensus. The authors then independently reviewed the remaining 746 papers for inclusion in the study, resulting in 344 articles that met our inclusion criteria.
We used NVivo10, a qualitative analysis software package, to assign relevant text from these 344 articles to subthemes and to organize subthemes into main themes. We constructed separate categories for barriers and recommendations. Using an inductive approach, WL and LK based their initial subthemes on the article text. We then organized these subthemes into main themes that minimized redundancy and overlap without losing important nuance. WL and LK assigned text to subthemes and main themes based on consensus discussion. When new subthemes emerged, we re-checked already analysed articles to apply the new subthemes. This iterative process is known in qualitative research as the constant comparison method.20 We then summarized the features of the subthemes and main themes and provided these to experts in law and economics. We asked these experts to contextualize the barriers and recommended solutions we identified. This paper summarizes the themes for challenges and associated recommendations. Note that due to the time-consuming nature of a scoping review, we have added updated discussion of articles from targeted searches conducted after February 6, 2015.
OVERCOMING CHALLENGES FOR THE DEVELOPMENT AND IMPLEMENTATION OF PM
We identified eight main themes in our set of 344 articles related to challenges for the development and implementation of PM.21 These themes were unevenly represented across the literature set. Of the 344 articles, 40.1% discussed regulatory uncertainty; 25.3% discussed reimbursement; 20.1% discussed clinical trial regulations and design; 17.7% discussed regulations for co-development of pharmaceuticals and companion diagnostics (CDx); 16.0% discussed conflicting and unclear evidentiary standards in regulations; 14.0% discussed lack of or ineffective legal incentives for PM R&D; 12.5% discussed problematic or incompatible information systems and privacy concerns; and 11.3% discussed regulation of LDTs and direct-to-consumer (DTC) genetic tests. Table 1 lists the acronyms used in our discussion.
Table 1.
List of acronyms.
| Acronym | Term |
|---|---|
| CDx | Companion diagnostics |
| CER | Comparative effectiveness research |
| CLIA | Clinical Laboratories Improvement Amendments |
| CMS | Center for Medicare and Medicaid Services |
| DTC | Direct to consumer |
| EHR | Electronic health records |
| EU | European Union |
| FDA | Food and Drug Administration |
| FTC | Federal Trade Commission |
| GINA | Genetic Information Non-Discrimination Act |
| HIPAA | Health Insurance Portability and Accountability Act |
| HTA | Health technology assessment |
| ICH | International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use |
| IND | Investigational New Drug Application |
| IRB | Institutional review board |
| LDT | Laboratory developed test |
| NIH | National Institutes of Health |
| ODA | Orphan Drug Act |
| PHI | Protected health information |
| PM | Personalized medicine |
| PMA | Premarket approval |
| PMI | Personalized Medicine Initiative |
| PMR | Postmarket research studies |
| R&D | Research and development |
| RCT | Randomized controlled trial |
| SAE | Serious adverse events |
| SSRD | Single subject research design |
| USA | United States of America |
Regulatory uncertainty was the most discussed challenge for PM R&D and implementation. This was not surprising given that governments and healthcare systems were developing new regulations, guidance documents, and regulatory initiatives for PM. Because regulatory uncertainty was a cross-cutting theme, we discuss it in the context of each of the other themes, rather than in a stand-alone section. However, the key issue was that existing regulations appeared to be inapplicable to PM and suffered from a lack of harmonization, and thus interfered with PM development. Regulatory reform, whether to facilitate PM or to ensure greater safety and harmonization, created a climate of uncertainty, which created difficulties for research and approvals. There was a shared view that regulations were in flux, both in terms of frequent changes during the life cycle of drug or diagnostic development, and in terms of the flexibility with which those regulations might be applied for different PM products and services. For some stakeholders, changes to regulations were occurring too quickly,22 while for others a lack of clarity and transparency in applicable regulations created uncertainty.23
An exemplar of regulatory reform that generated uncertainty in PM R&D and implementation is the 21st Century Cures Act24 (the Act), passed by US Congress on December 7, 2016, and which President Obama signed into effect on December 13, 2016. The sweeping bipartisan legislation was widely supported by pharmaceutical industry and medical device manufacturers but was opposed by many consumer protection and patient safety groups.25 The act provides increased funding of nearly 5 billion dollars for biomedical research through the NIH, the PMI, the BRAIN initiative (research into the human brain), and cancer research (referred to as the Cancer Moonshot).26 Further, the act provides states with funding for opioid abuse prevention and funding at the federal level for increased research on and prevention of mental illness.27
Central to the act's aim of facilitating the adoption of new therapies are controversial provisions that provide incentives for experimental therapies and faster and easier approvals for innovative drugs and medical devices.28 In particular, the use of patient experience data and ‘real-world evidence’ (including clinical experience data) in the approvals process, in addition to the standard data from randomized clinical trials, has raised concerns.29 Primary among these concerns is that the loosening of evidentiary requirements for FDA approval will come at the cost of patient safety.30
The act also facilitates a review pathway for biomarkers and other tools (such as animal model research) that help facilitate the development of new drugs.31 It provides regulatory clarity on the best path for approvals of combination products.32 The FDA has been directed to publish guidance on novel or adaptive clinical trial designs to ease burdens on companies submitting applications for approvals.33
The ripple effects of broader regulatory changes also generate uncertainty for PM R&D and implementation by compounding the effects of other regulatory changes. Under the current administration of President Trump, the implementation of the 21st Century Cures Act faces uncertainty given both a hiring freeze for federal employees that will make it difficult for the FDA to hire the technicians and staff necessary to undertake the work of the act.34 Additionally, in an effort to eliminate much of the federal regulatory bureaucracy, President Trump signed an executive order requiring the elimination of two regulations for each new regulation passed.35 The order also applies to federal guidances of the kind used by the FDA to explain how companies can meet the requirements of the agency. To implement the act, the FDA would have to pass a number of regulations and publish several guidances. It remains unclear, therefore, whether President Trump will exempt the FDA from the executive order so that it can implement the act.
Further, the Trump administration has proposed cutting the budget of the NIH by approximately 18%, which puts in jeopardy research programs such as the Cancer Research Moonshot, and substance abuse (opioid abuse) programs.36 In addition, the majority of the spending in the act was to have come from the Prevention and Public Health Fund, created under the Affordable Care Act (ACA).37 With legislation proposed to repeal the ACA before the Senate at the time of writing, 38 it is unclear what impact such a repeal might have on the implementation of the act. What is clear is that both funding and implementation of the FDA initiatives and other biomedical research programs of the 21st Century Cures Act face increased uncertainty and delays in implementation.
In an environment of significant regulatory change, regulatory uncertainty is a barrier to realizing the potential of PM. PM developers weigh the risks posed by regulatory uncertainty when investing in PM R&D. And regulatory uncertainty hinders the implementation of PM, delaying potential benefits of PM to patients and healthcare systems. Specific recommendations for creating certainty and clarity in specific regulatory areas are embedded in the sections below.
Challenge: Clinical Trials
Randomized controlled trials (RCTs) provide the evidentiary gold standard of safety and efficacy required for regulatory approvals in most jurisdictions, with some exceptions for rare diseases.39 While differences exist, jurisdictions such as the USA, EU,40 and Canada41 require similar evidence of safety and efficacy to satisfy regulators. While clinical trials are necessary to ensure patient safety, numerous commentators posited that they created a significant barrier to the timely and efficient translation of research to therapy,42 especially in the area of PM. There is a profound tension between the goals of PM to provide stratified, smaller populations of patients with tailored therapies and standard clinical trial designs, which examine efficacy in a large segment of the generalizable patient population.43
Some concerns about clinical trials in the context of PM echoed general concerns about the regulation of clinical trials expressed by R&D firms in pharmaceutical and biotechnology sectors, researchers, and patient organizations. These concerns related to the long timelines (10–15 years) and high costs of clinical trials ($2.87 billion dollars (in 2013 dollars)).44 These debates highlighted tensions between the regulatory process, which legislatively mandated regulators to approve only safe and efficacious products, and the interests of R&D firms and patients, which wished to bring potential therapies to market quickly and at lower cost. In the following sections, we expand on the PM subthemes in the 69 articles that discussed clinical trials, namely expense and delay (21 articles); uncertainties in the approvals process (13 articles); the use of biomarkers in clinical trials (17 articles); and clinical trial design (26 articles).
Expense and Delay
Twenty-one articles discussed the time and expense required to generate both the preclinical and clinical data for FDA approval in the USA.45 Regulatory requirements drive up R&D costs, which are then reflected in the cost of approved therapeutics.46 Where market size is reduced due to stratification of patients into small populations likely to benefit from a given therapy, the additional cost of running a clinical trial may create a financial disincentive. The high cost of clinical trials can also create barriers to physician and patient uptake in the form of higher prices, and difficulties with downstream reimbursement, for the same aforementioned reason.47
Biomarkers
A major impediment for PM developers is the high cost and complexity of establishing and validating predictive biological markers (biomarkers). Biomarkers are objectively measurable biological characteristics, including genetic variants, or medical signs that indicate a pathogenic process or pharmacological response to a particular therapy.48 Biomarkers may answer the following questions: ‘Does the drug reach the target? Does it have the desired biological effect? Does it have an influence on other expected or unexpected targets? Does the drug affect characteristics that predict desired or undesired effects?’49 The presence or absence of a biomarker may predict whether and how a particular patient or subset of patients will respond to an experimental drug. Genetic biomarkers are particularly useful because genetic factors account for 15% to 30% of differences in drug metabolism and response between individuals.50 Pharmacogenetic tests use genetic variations between individuals to determine drug absorption and disposition, or drug activity.51 The ability to predetermine, with the use of biomarkers, the targeted patient population and therapeutic dosage should shorten drug development timelines, reduce expenses and provide a greater likelihood of bringing a drug successfully through the regulatory process. The development of appropriate biomarkers is therefore a major strategy for reinvigorating a lagging pharmaceutical business model focused on the development of blockbuster drugs that can be used in large portions of the population.52
There are, however, problems with the identification and the validation of biomarkers that have undermined their utility thus far. First, scientifically identifying and establishing biomarkers is difficult.53 Second, the regulatory criteria necessary to validate biomarkers are neither agreed upon nor clear. Third, there are difficulties in collecting high-quality data to show that the biomarker is clinically valid and useful, as biomarkers are often identified late in the drug development process when it becomes apparent that only a small subset of the patient population is reacting to the drug under development.54 Indeed, PM has the potential to ‘rescue’ a drug that has failed to perform as expected in clinical trials by using a biomarker to identify the subset population in which the drug is effective. This ‘rebrands’ the drug as a PM for patients with that biomarker;55 however, in this situation, where further investigation determines what biological or genetic factor is at work in the responding subpopulation, the drug is normally well along the drug validation and clinical development pipeline. Consequently, due to issues of timing, and difficulty generating adequate evidence, regulatory demands for biomarker validation data can often be greater than the evidence that emerges from the trials involving the biomarkers.
In the context of genetic biomarkers, differences exist between the evidentiary standards that are required to use a genetic test for a biomarker in a laboratory setting and those required by regulators before that test can be used in a clinical setting. Internal laboratory biomarker tests do not require independent validation and vary considerably from laboratory to laboratory and from jurisdiction to jurisdiction.56 Regulatory standards for the use of biomarkers in a clinical setting are generally higher, requiring analytical validation that can be difficult to provide to the FDA without prior planning and early identification.57
Finally, while PM developers see the opportunity to further reduce clinical trial sizes by identifying biomarker-positive participants, regulators are concerned with the risks posed to biomarker-negative patients. Regulators may therefore require both biomarker-positive and negative participants in clinical trials, thus reducing or nullifying any potential savings for the trial sponsor.58 While regulatory approval is still possible without this evidence, the approval will limit clinical application to biomarker-positive patients,59 unless physicians use it off-label.60 The manufacturer may further have to undertake postmarketing studies or phase IV clinical trials in biomarker-negative populations, which raises costs and uncertainty from a manufacturer's perspective.61
Clinical Trial Design
Regulations require that clinical trials are designed with specific hypotheses and therapeutic targets in mind and proceed with prospective analysis to demonstrate safety and efficacy in a generalizable population. There are, therefore, significant tensions between standard clinical trial designs and designs adapted to the smaller patient subsets that are the hallmark of PM. First, smaller clinical trial designs provide inferior evidence of safety and efficacy due to the small numbers of patients involved.62 Small trials do not have the statistical power to detect efficacy, especially if the magnitude of the effect is predicted to be small. Second, without a large generalizable population study, it is difficult to fully understand the benefit/risk analysis of the drug.63 This is concerning because any off-label64 use by clinicians once the drug is approved can lead to unexpected and serious adverse events (SAE) not discovered in a phase III study. In addition, analysis of the subgroups enrolled in a clinical trial often comprises a retrospective analysis that takes place as the trial is ongoing or after a suboptimal or failed clinical trial.65 Such post hoc analysis requires a mid-stream course correction with new patient targets and a different hypothesis during or after the clinical trial to ‘rescue’ the drug candidate, and ‘rebrand’ it as a stratified therapeutic for a specific, smaller population.66 This type of post hoc analysis is not permitted under the current regulations, and it is not clear whether in all cases an entirely new clinical trial would be required.67
Uncertainty in the Approval Process
In light of the preceding discussion, uncertainty about the clinical trial process for PM leads to uncertainty about the likelihood of regulatory approval. In addition to the high cost associated with the regulatory approval process, failure to gain approval means those costs cannot be recouped. Regulatory uncertainty in the clinical trial process exists with respect to the precise information companies must present to regulators for a successful application, and how that information will be used;68 the use of biomarkers;69 and the design of clinical trials that incorporate CDx.70 PM developers desire better guidance on how best to design a successful clinical trial for a personalized therapy, because absent guidance, they risk presenting suboptimal evidence regarding stratification options. Designing clinical trials for differently responding subgroups (for example, biomarker-positive and biomarker-negative groups) requires additional time and resources.71 Companies are reluctant to make this investment without a commensurate increase in the certainty of regulatory approval.
Meanwhile, regulators struggle to make evidence-based risk and benefit decisions from clinical trials with very small sample sizes, which do not capture rare SAE.72 While existing standards for clinical trial design may be suboptimal for PM,73 using novel clinical trial designs may not improve a therapeutic's chance for approval.74 In the absence of functional regulatory guidelines on clinical trial designs, PM developers and regulators will remain frustrated with the quality of evidence generated by clinical trials and the uncertain regulatory outcomes that follow.
Recommendations to Overcome Clinical Trial Challenges
Given the numerous challenges posed by clinical trial regulations, many proponents of PM recommend changing or improving clinical trial design to aid PM development and clinical translation. In general, PM developers would like greater clarity and guidance about how information on biomarkers should be presented and how it will be used in the approvals process.75 Strengthening postmarket study and surveillance (PMR) requirements,76 using postmarketing authorization, encouraging phase IV clinical trials,77 gathering ongoing evidence78 and using limited or conditional approvals79,80 are all frequently mentioned recommendations. Recommendations involving PMR include the creation of an ‘approval with conditions’—a regulatory option that would fast track some approvals in combination with mandatory phase IV PMR and the creation of a centrally managed database for SAE reporting by physicians.81 It is thought that strengthening PMR could alleviate some of the difficulty companies have meeting evidentiary burdens, particularly with respect to bringing prospective population-segmented data to regulators.82 PMR might evaluate a drug's effect in a specific patient population or identify new uses for the drug.
Recommendations favoring mandatory PMR also reflect a general desire to capture SAE, and provide more rigorous feedback of clinical results into the regulatory process.83 PMR might be used to identify the variability that underlies serious event rates in different patient groups.84 To that end, recommendations range from using postmarking data collection pilot projects created by the Center for Medicare and Medicaid Services (CMS) and Centers for Disease Controls and Prevention as models for the FDA’s Sentinel program,85 to standardizing reporting of SAE independent of pharmaceutical companies.86 This would foster greater transparency, learning, and quicker turnover of innovation, which, while desirable, is foreign to a strong trade secret and confidentiality ethos in medical product development. There is little consensus, however, on how mandatory PMR should be instituted, as an FDA power through legislation,87 or under a public/private funding partnership.88 The latter option leaves all information publicly owned and appears to be at odds with existing data protection laws. In addition, recommendations aim to mitigate the difficulties, both scientific and evidentiary, with the validation of biomarkers.89 Some propose the use of unvalidated biomarkers, so long as the sponsor commits to a phase IV clinical trial,90 or the use of biomarkers that emerge during the early phases of the trial.91
Innovative Clinical Trial Design
Other recommendations relate to clinical trial design and several authors suggest a number of innovative or alternative trial designs to aid PM products through the regulatory approval stage. Several articles recommend that regulators work with PM manufacturers to create new models and provide greater guidance on permissible clinical trial design.92 Recommendations include innovations or alternatively designed studies that reduce costs,93 do not rely on animal models,94 or involve modified or conditional approval paired with a postapproval requirement to develop additional evidence.95 Adaptive trial design could reduce drug approval time and cost by recruiting only ‘likely responders’ based on biomarkers into clinical trials.96 Alternatively, adopting an ‘adaptive group sequential design’ involving rolling admissions of sensitive subgroups could overcome some of the current clinical trial design barriers.97
Authors also recommend regulatory acceptance of smaller clinical trial populations in lieu of prospective RCTs.98 Reliance on prospective-only trial design invalidates the use of retrospective analysis or identification of biomarkers during the course of clinical trials. Adaptive trial design that involves contemporaneous biomarker identification would overcome this regulatory barrier.99 Along with trial design that overcomes the difficulties with biomarker validation, several articles recommend moving from the traditional clinical trial group experimental design to Single Subject Research Design (SSRD) or ‘N of 1’ studies.100 This move would be difficult and controversial as SSRD studies are not sufficient to satisfy current clinical trial information and design for regulatory approval.101 Despite the difficulty, SSRD would help identify highly reactive patients and speed up evidence collection to meet regulatory requirements.102
Challenge: CDx and Co-Development
A second challenge facing PM development is the significant uncertainty that exists around the optimal, possible, and acceptable clinical trial design for therapeutics intended to be used with CDx. This uncertainty negatively impacts the advantages predicted for PM in the combination of tailored therapies and diagnostics that identifies the optimal patient population. Such CDx, commonly regulated as medical devices, are most often in vitro assays103 or genetic tests. These assays and tests help identify the presence or absence of a biomarker, indicating the patient's suitability as the recipient of a particular therapy, such as a small-molecule drug, biologic, or gene/cell therapy. To fully exploit the therapeutic and economic advantages of the therapeutic–diagnostic partnership, ideally the CDx is marketed together and cross-labeled with its therapeutic to indicate paired use. Here, we expand on the subthemes related to co-development from 61 articles, namely issues with mandated co-development by regulators (32 articles); labeling issues (19 articles); lack of clarity on regulatory pathways (15 articles); the co-dependence of the therapeutic and the diagnostic threatens the success of both (8 articles); and the lack of appropriate business models for co-development (4 articles).
Regulatory preference to facilitate the pairing of a therapeutic and its CDx means that both should navigate the clinical trial and regulatory process at the same time. This will establish evidence that their use together is safe and effective. In a 2014 Guidance, the FDA indicated that co-development was preferred for CDx, including pharmacogenetic tests, 104 thereby making co-development all but mandated except for rare cases.105 The regulatory preference for co-development, as well as ongoing uncertainty about how to achieve this, is a significant barrier in PM development. Relatively few concurrently developed therapeutics and CDx exist to date.106
According to the FDA, a CDx should be identified and developed in parallel with a therapy. This is very difficult for developers, especially those in the private sector, to achieve, as R&D timelines for therapeutics and diagnostics do not support concurrent development.107 Since biomarkers that form the basis of most in vitro assays and tests are normally identified retrospectively and late in the therapy's validation process, 108 developers that identify useful CDx for a therapy after phase II clinical trials face regulatory challenges that increase time and cost of development.109 Additionally, achieving cooperation between different developers whose business interests may not be aligned is complicated and requires an unusual degree of transparency. Unlike large pharmaceutical developers, diagnostic developers are commonly small biotechnology companies that tend to have leaner business models and marketing budgets.110 These differences make full cooperation difficult.
Co-development may threaten the success of each company's product. Where the diagnostic is tied to the use of a particular drug, the success or failure of that drug in clinical trials determines the success or failure of the diagnostic.111 In situations in which the diagnostic might have multiple uses, co-development with a single drug may mean that the diagnostic will not clear regulatory hurdles for other uses. The converse occurs when rejection of the diagnostic leads to the rejection of the drug, which might otherwise be effective.112 Co-development can also delay the introduction of the drug or diagnostic while the other remains in the approval process. This can have profound economic consequences for both companies.113 Furthermore, pairing the use of a drug with a diagnostic may reduce market size for drug development companies.114 Finally, it is less complicated for a company to develop a stand-alone diagnostic.115 Thus, a myriad of disincentives contribute to the lack of co-developed drugs and diagnostics.
In the USA, diagnostics and therapeutics have traditionally gone through different regulatory streams with different evidentiary standards. Moreover, regulatory streams at the FDA were designed well before the advent of PM. Two separate and distinct regulatory streams, therefore, exist for therapeutics and medical devices, respectively. Through the FDA, the Center for Drug Evaluation and Research116 and the Center for Biologics Evaluation and Research117 evaluate drugs, biologics, and combination products, 118 whereas the Center for Devices and Radiological Health oversees medical devices.119 (see Figure 2). This separation creates uncertainty about how best to satisfy regulatory requirements for co-development, what data should be submitted and how best to time the submission of relevant regulatory requirements.120 The existence of multiple paths for regulatory approval makes it confusing to determine which regulators approve CDx,121 especially if the test may have multiple intended uses.122 However, the appeal of having a paired therapeutic and diagnostic has led some companies engaged in the development of PM to attempt co-development, despite the myriad of difficulties they face.123
Figure 2.
National Human Genome Research Institute (NHGR) Regulation of Genetic Tests. Adapted from http://www.genome.gov/10002335 accessed July, 23, 2015.
Recommendations to Overcome Barriers for Co-development
Recommendations to overcome the barriers to co-development of therapeutics and diagnostics focus on implementing regulatory reforms that incentivize the co-development process, something the FDA attempted in its 2014 guidance.124 In general, stakeholders seek the alignment of regulations between therapeutics and diagnostics,125 greater regulatory clarity,126 and recognition of the complications of the co-development process.127 The creation of a single regulatory stream would ease the burden on companies seeking regulatory approvals in the PM space.128 While co-development is strongly encouraged by the FDA, greater guidance on how to proceed with non-parallel development would aid the sizable majority of companies for which co-development is not possible.129
Recommendations also focus on improving clarity on cross-labeling requirements.130 Commentators note the lack of clear and common standards indicating what evidence is required to label drugs for a particular subgroup or for a particular genetic mutation.131 Greater clarity in labeling requirements would enhance patient care and the uptake of CDx (for example, pharmacogenetic tests).132 Clarity is needed on when cross-labeling is required and when it is simply recommended for informational purposes.133 Greater flexibility in labeling or in making labeling changes could create a more responsive system in which additional tests or diagnostics could be added to labels as improvements and innovations in diagnostics appear.134 Successful pairing of therapeutics and diagnostics could be enhanced through PMR to provide evidentiary support for labeling claims and by greater specificity in labeling requirements.135 For example, labels could provide consumers and prescribers with further information and assessment on predictive claims,136 the requirement for testing prior to taking a prescription,137 and information on the actions required depending on the results of the CDx test.138
Challenge: Regulation of Genetic Testing
As a subset of in vitro diagnostic tests, genetic tests may determine therapeutic choices or other personalized interventions.139 Genetic tests fall into two categories: LDTs and genetic test kits. LDTs are the most common and are generally developed in a single laboratory to which patient samples must be sent for analysis.140 LDTs can be thought of as ‘in-house’ genetic tests. Genetic test kits, on the other hand, comprise a set of reagents and analytical information sold together to multiple testing laboratories.141 Some genetic tests are marketed directly to consumers; such DTC genetic tests have been the subject of considerable controversy.142 In the following sections, we expand on the subthemes relevant to PM in 39 articles on genetic testing, namely: lack of regulations on LDTs (17 articles); lack of guidance on LDTs in the approvals process (9 articles); inadequate regulation of DTC genetic tests (6 articles); an underappreciation for the potential harms DTC genetic tests pose (6 articles); the problem of misleading advertising associated with DTC genetic tests (5 articles); the unsuitability of current regulations for DTC genetic tests (4 articles) and LDTs (2 articles); perceived difficulties with regulating LDTs (3 articles); a lack of clarity on how to regulate DTC genetic tests (3 articles); and the lack of clinical utility measures that hinder the uptake of LDTs (2 articles).
Three measures determine the validity and utility of genetic tests. Analytic validity determines whether and how well the test measures or determines the presence of the biomarker.143 Clinical validity determines whether there is a correlation between the targeted clinical condition and the biomarker.144 The third and more difficult measurement to determine is clinical utility. It asks: Does the test lead to increased human health, or a beneficial medical outcome?145 Quality tests should therefore be analytically and clinically valid, as well as clinically useful.
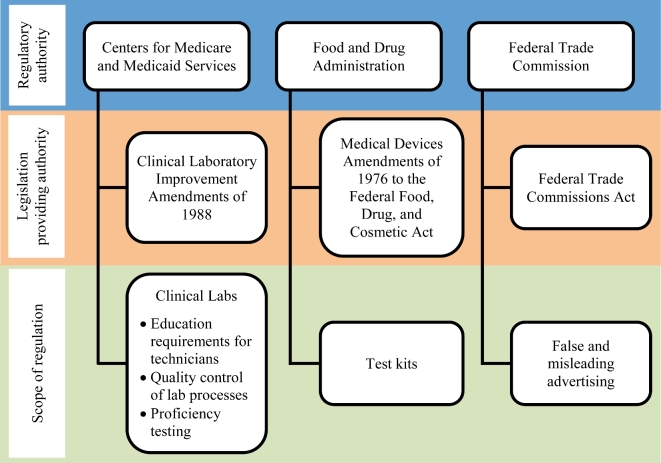
These three measurements are applied differently and unequally to LDTs and to genetic test kits, due in part to the different regulatory regimes that govern the different types of tests.146 This creates significant regulatory challenges.147 In the USA, regulatory oversight of genetic testing is split between three federal government agencies: the FDA, the CMS, and the Federal Trade Commission (FTC), which is the federal consumer protection agency. Each of these agencies has a different mandate and applies different standards. This has led to numerous complaints of a lack of adequate, rigorous, and consistent regulation.148 Despite the novel issues that genetic tests raise, no new regulatory system has been created for their approval. The existing regulatory structure creates false separations based on where and how genetic tests are used, rather than focusing on their validity and utility regardless of how they are applied.
The FDA has the authority to regulate genetic test kits as medical devices based on safety and efficacy under the Food, Drug and Cosmetic Act.149 The regulations in that act create a stratified or tiered system of approval requirements based on the level of risk posed by the device to the patient or the user based on intended use and the indications for use. Class I devices are low risk and generally exempt from the requirement for a 510(k) premarket notification (a process establishing that the device is substantially equivalent to a device already marketed). As such, class I devices generally require only broad controls such as registration, labeling, and good manufacturing practices. Class II devices are classified as moderate risk and generally require a 510(k) premarket notification prior to marketing. Devices considered class III (highest risk classification) require a premarket approval (PMA) for clearance by the FDA to market the device. The PMA process is more complicated and requires the submission of clinical evidence to support the claims being made about the device.150
With respect to genetic tests, FDA regulations require evidence that genetic test kits are analytically and clinically valid, although evidence of their clinical utility is not required.151 This is a point of concern, given that ensuring genetic testing leads to positive health outcomes (clinical utility) is critical for both patient welfare and reimbursement decisions. Until recently, the FDA has exercised its regulatory authority sparingly and only as it applies to genetic test kits—choosing to leave LDTs regulated only by the less stringent Clinical Laboratories Improvement Amendments (CLIA) regulations.152 The imposition of different standards on genetic test kits and LDTs is the most pressing issue for those who want to have equal, reliable testing for all genetic tests. Ongoing criticism of the differences between FDA and CLIA regulation has led to significant policy action at the federal level.
The CMS oversees clinical laboratories and regulates LDTs under the authority of CLIA.153 The CMS ensures that LDTs are analytically valid but does not require evidence that LDTs meet the standards of either clinical validity or utility.154 CLIA regulations categorize laboratory tests as either ‘waived’ or ‘non-waived’ according to whether they are low, moderate, or high complexity tests. Tests that are either moderate or high complexity are subject to additional requirements to ensure their safety. Higher complexity tests are generally grouped into specialty areas that are subject to more rigorous external proficiency testing to verify their ongoing analytic validity. While molecular and genetic tests are considered high complexity, they have not been designated as a subspecialty and, therefore, are not subjected to the most stringent testing available under CLIA.155
In addition, significant concern exists about a lack of rigorous and consistent regulation of DTC genetic testing, which enables consumers or patients to access their genetic information directly from a testing company.156 For many years the FDA did not exercise its jurisdiction over DTC genetic tests, creating yet another group of genetic tests without rigorous regulatory oversight. This situation is changing as the technology advances and as the FDA asserts its authority over genetic testing. Whether the FDA regulatory approach is appropriate for DTC testing is an open question. Proponents claim that many of the risks associated with DTC genetic testing are speculative and that it has intangible benefits, including patient autonomy and privacy.157 DTC genetic tests continue to create controversy, however, because they are advertised directly to consumers and the advertising may be fraudulent and/or may misrepresent the benefits for the consumer.158 Misleading claims are under the regulatory purview of the FTC, which protects consumers from unfair and deceptive business practices and from false and misleading advertising claims.159 Concerns about the claims made by DTC genetic testing providers have prompted calls for the FTC to exercise its authority to protect consumers, some of whom may make medical decisions to either forgo treatment, based on a false-negative test results, or undertake drastic medical procedures, such as prophylactic surgery, based on false-positive test results.160 The FTC has largely ignored evaluating the truth of health marketing claims made by DTC genetic testing companies, which leaves the field open to fraudulent claims.161
Current perceptions persist that advertising of DTC genetic tests is false and/or misleading to such an extent as to undermine consumer and provider confidence in the veracity and utility of all genetic tests.162 The perception that the FTC is not taking action against DTC companies challenges the uptake of legitimate genetic tests.163 Exacerbating this situation, there is a plethora of reasons for an individual to purchase a DTC test outside of health, including recreational, educational, and ancestry. This can be exploited by DTC companies in marketing and promotional materials.
Recommendations for Regulation of Genetic Testing
Regulatory recommendations for improving genetic testing in PM include greater involvement of healthcare professionals, 164 greater regulation of both DTC165 tests and LDT166 at the federal level to ensure quality, and the creation of a comprehensive genetic test registry.167
LDT Recommendations
The most common recommendations concerning LDT testing are to increase regulatory oversight168 and evidentiary standards.169 Recommendations include expanding FDA oversight to cover LDTs170 (likely to occur in 2016); expanding evidentiary standards to include evidence of clinical validity and utility;171 and ensuring that LDT laboratories undergo proficiency testing, certification, or increased accreditation requirements.
Some critics who have complained that CLIA regulations are inadequate to ensure the quality of genetic tests advocate for the creation of a genetic subspecialty. This would involve an external review and a more rigorous examination of tests resulting in higher quality review.172 Furthermore, commentators over the last 15 years have criticized the FDA’s unwillingness to hold DTC genetic tests173 and LDTs174 to the same standards it uses for test kits. After numerous studies and reports calling for increased oversight action,175 the FDA issued draft guidance on October 3, 2014, thus asserting its statutory authority and signaling its intention to exercise regulatory oversight over LDT. The draft guidance outlines the phasing in of a plan that extends the stratified risk-based framework applied to medical devices for premarket notifications and approvals to all genetic tests, regardless of where they are developed. Tests for rare diseases that meet the definition of a Humanitarian Use Device would be subject to discretionary authority regarding the need for PMA.176 The new guidance requires notification to the FDA of LDTs as a no-fee alternative to registration and listing of LDTs. It further requires SAE reporting and a demonstration of clinical validity of LDTs.177
The guidance has not been met with universal approval. Some articles express concern that increasing FDA oversight of LDTs will increase cost and delay for manufacturers. Others are concerned with the chilling effect on swift and ongoing improvement and innovation due to oversight bureaucracy, and some assert that the FDA lacks statutory authority to expand its oversight to LDT.178 Statutory oversight aside, concerns about the FDA’s workload and its ability to oversee the large number of LDTs will need to be weighed against the greater evidence of validity and utility derived from FDA oversight. Following a series of FDA workshops, several hearings before the House Energy and Commerce Committee Subcommittee on Health, and a lengthy public comment period, the FDA has indicated it will issue its final guidance in 2016.179
On November 18, 2016, the FDA announced that it would not issue a final guidance for the oversight of LDTs.180 This leaves the current uneven patchwork of oversight on DTC, LDT, and genetic test kits in place, and the concomitant problems discussed above. In January 2017, the FDA released a discussion paper laying out issues that could guide a legislative solution and signaling that the issue has not yet been put to rest.181 In particular, the FDA points to the need for ‘well-curated databases’ of scientific evidence on clinical validity and other measurements that could be used to establish the accuracy and validity of LDTs.182
While backing away from guidance on LDTs, in July 2016, the FDA issued draft guidances on next generation sequencing (NGS) genetic tests as part of the PMI183. While these guidances signal the FDA’s intention to regulate IVD based on NGS as part of the PMI, the FDA has taken an approach aimed at facilitating the use of these tests and easing the scientific regulatory requirements.184 The first guidance called for more public databases of genetic variants and their use in establishing clinical validity of NGS tests.185 The second guidance discussed the need for standards in the designing, developing, and validating of NGS-based tests for germline diseases as part of the PMI.186
In addition to greater quality assurances of LDTs, a number of authors advocate for the creation of a mandatory genetic test data registry.187 Currently, there is a voluntary test registry hosted by the National Institutes of Health's (NIH) National Center for Biotechnology Information.188 Despite recommendations by the Secretary's Advisory Committee on Genetics, Health, and Society to make registration mandatory for all genetic tests, it does not include all genetic tests. Additionally, the test registry may be more useful to consumers if standardized fields, such as the molecular basis of the tests and the methods used for testing, are included in the registry and uniformly entered to be easily searchable.189
DTC Recommendations
Although there are a number of US states that currently either prohibit DTC testing or mandate professional involvement in ordering or interpreting genetic test results, many states do not regulate provision of DTC tests.190 Recommendations include federal regulation of DTC tests through mandatory inclusion of professionals, including genetic counselors, in either the test ordering or the provision of test results.191 DTC companies should be responsible for providing greater quality assurances and more accurate information about their tests.192 Failing these changes, the numerous calls for increased comprehensive federal regulation by the FDA using its stratified risk-based approach193 and FTC194 oversight of DTC testing are likely to continue. In November 2013, the FDA signaled a new readiness to flex its authority over DTC tests in a way it had not done previously. It barred 23andMe, a DTC genetic testing company, from using its broad-based genetic screen without regulatory approval.195 In October 2015, 23andMe became the first DTC company to be granted FDA approval for its DTC test kits, a fact it heralded on its website.196 Continuing the trend to greater scrutiny of DTC tests by the FDA, in November 2015 the FDA sent letters to three DTC genetic testing companies, DNA4Life, DNA-CardioCheck, and Interleukin Genetics, for marketing unapproved genetic tests.197 However, on April 6, 2017, the FDA softened its initial decision, permitting 23andme to offer DTC genetic testing for 10 diseases or conditions, including Parkinson's disease, late-onset Alzheimer's disease, Celiac disease, and several hereditary conditions.198 While paving the way for future genetic predisposition tests from 23andme and similar DTC companies, the FDA clarified its expectations with regard to the accuracy, reliability, and clinical relevance of such tests through special controls.199 Further, the FDA has created a regulatory pathway for DTC genetic predisposition tests that will expedite approvals from premarket review after an initial de novo application.200 Despite relaxing its stance on some genetic tests, DTC diagnostic tests for conditions such as BRCA, for example, are not included in the authorization granted to 23andme.201
In addition to FDA regulation of DTC testing, expansion of other federal protections such as privacy protections under The Genetic Information Non-Discrimination Act (GINA),202 or the FDA Sentinel (postmarketing surveillance)203 program,204 could further bring DTC testing into the regulatory fold. Calls for additional oversight of DTC communications and advertising by the FTC are joined by calls to ‘regulate’ misleading or harmful claims on DTC tests by using a Tort law standard or reasonableness in negligence law,205 and by requiring DTC testing information and risk predictions to be harmonized with industry standards.206 The creation of uniform standards for risk predictions and other DTC communications could bring DTC genetic tests in line with other medical device regulation and hold riskier tests to a higher standard of examination.207
Finally, concerns about lax federal regulation and misleading advertising of DTC genetic testing has led several states to mandate that genetic test results be returned to a physician or learned intermediary with the skill to meaningfully interpret test results. The hope is that such meaningfully interpretation of genetic test results will lead to better healthcare decision making.208 While physicians have a duty, in many cases a legal duty, to impart incidental findings from genetic tests to a patient, the regulations are unclear regarding the duties of generators, providers, and interpreters of genetic test results.209 This confusion with respect to communication leads to quality control issues and inconsistent standards.
Challenge: Uncertain Regulations on Evidentiary Standards
In the following sections, we expand on the subthemes relevant to PM in 55 articles that discuss issues with evidence standards, namely existing standards that are unreasonable (11 articles) or unclear (11 articles) in a PM context; the failure of evidence produced by companies to meet regulatory requirements (12 articles), including the inability of many companies to carry out phase IV trials (8 articles); the increased cost associated with meeting regulatory data standards (7 articles); the inadequacy of data to expand applications to broader indications (4 articles); the delay that meeting evidentiary standards can cause (4 articles); and the unclear standards for clinical utility (2 articles).
PM developers find the data and evidentiary requirements for regulatory approval unclear.210 Vague evidentiary standards and requirements act as a barrier to the translation of PM products as companies struggle to develop appropriate evidence for approval and reimbursement.211 Whereas the FDA requires evidence of safety and efficacy, it does not require measures of clinical utility—that is, evidence that overall health outcomes are improved in a clinical setting. Payers, making choices about which therapies to cover, require this additional layer of evidence as a basis of payment decisions. In other words, what matters to a payer is whether a therapy is ultimately proven to be clinically useful and that it is a cost-effective alternative to existing therapies. These considerations are currently outside the evidence required for regulatory approval.
Lack of uniformity and clarity in data requirements between statutes, regulators, and payers poses challenges for developers. Not only is there lingering uncertainty about what data are required, but also when that data should be incorporated into the FDA investigational new drug application (IND) process.212 This is especially evident when data about a potential effect in a subpopulation or a relevant biomarker for that population emerge during a clinical trial. Further, it is not clear how the FDA will use the data that are submitted, and whether the data will be sufficient to validate a biomarker, or to permit the subpopulation effects to be generalized to the wider population. Manufacturers are concerned that evidence of subpopulation effects may be used to limit the application of a drug or device, rather than expand its application.
In addition to these concerns, the FDA can require companies to conduct PMR, otherwise known as phase IV studies, to collect evidence on how the therapeutic or device is working in clinical practice, and what SAE might have been reported with its use.213 PMR are undertakings by companies to continue to collect evidence following regulatory approval; however, research shows that there is very low compliance with these commitments.214 Such low compliance masks previously unknown SAE, but also does not reveal new uses for a drug or how the drug functions in different patient subgroups.215 While regulators are reluctant to extrapolate from subpopulation data to other populations, groups, and ethnicities,216 the expense involved in providing evidence on additional populations (and potentially undertaking an additional clinical trial on the subgroup) may be prohibitive to developing PM products.217 In the case of pharmacogenetic tests, developers remain uncertain whether they will be required to undertake phase IV studies, and whether that evidence might lead to restrictions on approvals in genetic subpopulations.218
There are a number of reasons that companies do not fulfill their PMC and do not provide ongoing data to the regulators. First, PMR are expensive and time consuming.219 Second, they are notoriously difficult to conduct, requiring data gathering of physician and patient reporting of SAE, which are widely underreported.220 Such detailed assessments of clinical efficacy are very difficult to provide.
Recommendations to Improve Evidentiary Requirements
Recommendations for bringing better quality evidence to regulators generally fall on the manufacturers. There is a call for submissions to regulators to provide higher quality evidence that is not only useful to payers and providers, but also that anticipates the need to show cost-effectiveness for reimbursement.221 Additional recommendations for regulators include the use of less restrictive, more flexible, and innovative approaches to PM clinical trials and approvals.222 This might be achieved by allowing alternative forms of evidence,223 initially using exploratory or modeling analysis combined with corroborative evidence,224 or permitting the use of comparison or equivalency data which relies on prior, similar submissions225 (as in follow on biologics). In particular, clear technical guidance on generation and submission of genomic data, clinical evidence standards, and the use of biomarkers in both drug development and pharmacogenetic studies is needed as an incentive to stimulate manufacturers to design their studies to capture these data.226 Pathways to make the generation of these data less expensive and time consuming might include leveraging public–private partnerships and large cross-institutional studies involving non-industry actors as a means of validating biomarkers. Developing industry guidelines to create more accurate methods for predicting drug safety and pooling resources227 to address the costs associated with bringing PM products to market would help reduce the time and costs of evidence generation.
Non-compliance with PMR commitments has become such a problem that Congress mandated the FDA to work through the backlog of PMR commitments and provide annual reports on progress.228 To date, the backlog has been reduced, and the FDA is working to ensure greater compliance.229 In furtherance to fulfilling these obligations, the FDA has continued its commitment to the Sentinel program which tracks performance of medical products by accessing a number of data sources, including electronic health records (EHR), patient registries, and insurance datasets to better identify SAE and anticipates much better PMR of medical products.230
Challenge: Information Systems and Privacy
The literature on regulation of information systems reveals three main subthemes highlighting a tension between the interests of patients and developers of PM therapies and devices. In this section, we expand on privacy concerns (26 articles); sample storage and collection concerns (14 articles); and the heterogeneous nature of sample storage and collection regulations (9 articles). Issues of patient and research subject consent are mentioned in all three subthemes.
Human tissue samples, housed in hospital pathology laboratories and biobanks, support PM R&D. The samples are often used to determine genetic contributions to diseases when linked with phenotypic information and to test for biomarkers. The value of tissue samples is greatly enhanced when linked to medical health records or other data on disease histories and phenotypes. An ability to aggregate data across multiple national and international research facilities is crucial for scientific research, especially for rare disease phenotypes where information may be scarce. Consequently, pathological samples and EHR containing personal health information are simultaneously private patient information and valuable resources for drug companies and PM developers. Information systems that manage PM research data and samples are in tension, trying to balance the access and use requirements of researchers and developers with the autonomy and privacy interests of research participants.
Informed Consent and Privacy
Information and data management systems and the associated legal and ethical issues raised with respect to consent and privacy of personal or protected health information (PHI) are central to PM R&D. This is because much PM development involves use of biospecimens and knowledge of patient genetics or biomarkers. Since genetic information may reveal both personal and familial health or ancestry information, issues of consent and privacy are paramount in genetic studies and tests used in PM R&D. The legal requirement for researchers to obtain informed consent from research subjects is imbedded in US federal regulations that deal with human subjects research supported by the federal government.231 The Common Rule governs human subjects research on products regulated by the FDA. These regulations also cover research on human tissues and associated information that can be linked to an identifiable individual.232 In addition to the Common Rule, federal regulations governing the use of PHI were implemented in 2003 under the Health Insurance Portability and Accountability Act (HIPAA) of 1996.233 HIPAA applies to three categories of healthcare institutions, known as HIPAA covered entities: most healthcare providers, most health plans (insurers), and healthcare clearinghouses. HIPAA imposes limitations on the research uses and disclosures of identifiable patient information, requiring patient authorization for specific uses of certain PHI. HIPAA privacy rules do not apply to health information held by data brokers, websites, credit bureaus, disease registries, health researchers, disease advocacy organizations, law enforcement agencies, or others as defined under the act.234
The scope of informed consent as it relates to PM, in particular, is complex and important. Generally, consent to conduct research on a person or that person's tissues relates to a specific research activity that can be well described and for which consent can be meaningfully given or withheld based on knowledge of risks and alternatives. A problem for the PM research enterprise relates to whether a patient can give a non-specific consent to future research, without knowing what the nature and attendant risks of that research might be. Many argue that consent to research that is not yet defined cannot be informed and does not respect patient autonomy as required.235 While the acceptability of consent to future research is hotly debated, the ability to obtain consent to future undefined research is central to PM development. Much PM research comes from a retrospective analysis of clinical trials to determine what subpopulations might derive a therapeutic effect. Consequently, the need to reexamine tissue samples for a different research outcome than originally described often arises. Re-consent of tissue donors for a different research objective may be neither feasible nor possible, and it is unclear whether it is required in all circumstances.
PM developers complain that the balance of regulations often emphasize individual autonomy and control over personal health information rather than the research enterprise and creation of a research platform of genetic information and resources, creating a barrier to PM development.236 Indeed, the regulations reflect a tension between balancing privacy and patient autonomy against a need for transparency, greater information gathering and sharing of research resources to advance PM.237
Complex layers of oversight from local institutional review boards (IRBs) to federal regulations protect individual autonomy and privacy, necessitating re-consent of research subjects, allowing subjects to withdraw from studies, and preferring anonymity and coded samples that make it more difficult to link PHI to tissue samples.238 Finding the right balance between researcher needs and subject protections is an ongoing regulatory exercise. In 2015, the Department of Health and Human Services proposed changes to the Common Rule239 that would require informed consent for research on biospecimens even if not linked to identifiable information. In addition, the changes would permit a broad consent to unspecified future research, heretofore not permitted. While the stated purpose for the changes includes increased simplicity and transparency for researchers, and additional protections for individual research subjects, the proposed rules are very controversial and some implicate individual privacy concerns. Some argue that they unduly favor researchers over patients, and ‘demand that patients accept a one-time grab at all data, for any purpose, in order to provide broad access to others with no promised informational or other return and no mechanism to reciprocate patients’ altruism’.240 On January 19, 2017, the final Common Rule was published241. Changes that required consent for non-identifiable biospecimens were eliminated, but changes that provided the ability to use a broad consent for future research were retained242.
A further regulatory barrier to PM is the heterogeneous and inconsistent regulations relating to human subjects research and biobanking, particularly the collection, sharing, and storage of data.243 There is a lack of harmonization both nationally and internationally between regulations covering the storage and use of biospecimens or use of PHI. PM developers and advocates argue that inconsistent regulations and conflicting standards create a climate of uncertainty that impedes the flow of both research materials and information between researchers and laboratories.244 Further barriers are created by inadequate and conflicting regulations to protect and use PHI, leading to improper use of that information and potential discrimination.245 Conflicting regulations exist not just between countries but within national systems.246 In the USA, federal regulations have differing definitions of and rules about what constitutes human subjects research, and whether and how informed consent for the use of tissue samples may be obtained. Adding to the complexity, both the federal Common Rule and HIPAA specifically leave room for individual states to create stricter standards on disclosure and use of biospecimens and PHI. State laws affecting the use of tissue and associated data in scientific research are found in a variety of sources, including medical records laws, privacy and health privacy laws, genetic testing/genetic information laws, and human subject protection laws. These laws are often different and inconsistent both among and within states. Regulations may impose different limits on uses of biospecimens and associated data and may offer different scopes of protection.247
The lack of regulatory clarity on how biospecimens and PHI can be shared and with whom has led to real concerns about individual privacy. This is particularly acute in genetic research since sensitive personal health information can now be revealed through genetic analysis of tissue samples using increasingly powerful sequencing tools that often reveal incidental health information. In the USA, despite the protections of the GINA, which forbids health insurers and employers from misuse of genetic information, patients continue to be fearful of discrimination after genetic testing. Where health insurers control access to care and testing in the USA, concerns about improper use of genetic information have a chilling effect on research into genetics of diseases so critical to PM.248
Of continued concern is the lack of HIPAA protection for the medical and personal health information collected by the PMI. It is, as yet, unclear exactly how the biospecimens and associated data will be collected and stored, whether in a federal registry or in different states (each with different and sometimes conflicting regulations). Federal agencies including the NIH are not ‘covered entities’ under HIPAA and therefore the privacy rules do not apply, even if the PHI originated from a HIPAA covered entity. Once an HIPAA-covered entity shares PHI with a non-covered entity, the information generally passes outside the scope of the HIPAA privacy rule and beyond the jurisdiction of HIPAA oversight. The question of how the PHI of millions of volunteers will be adequately protected under the PMI is a pressing question.249
Recommendations on Management of Information
Recommendations to remove information management barriers include creating, improving, and harmonizing practices, guidelines, and policies for information sharing (4 articles). Most recommendations focus on improving biospecimen collection and storage (14 articles), and on creating stronger consent and privacy protections (7 articles). While some recommendations focus on systemic solutions, such as encrypting PHI between researchers and physicians (1 article), others focus on getting consent from patients to archive genetic data for future use (1 article), and moving toward the use of research advance directives (1 article). These recommendations support broader consents aimed at facilitating future research uses of biospecimens and PHI.
Collection procedures could be improved by ensuring samples are collected and sequenced in advance of clinical trial submissions. This should help with prospective biomarker identification, and ensure that enough genetic variants are represented and that samples from the intended patient population are available for submission in clinical trials. Strong protocols that include sample collection and storage procedures would also enable retrospective associations with safety and efficacy outcomes and retrospective identification of biomarkers.
In response to the PMI, the FDA created precisionFDA, an ‘online, cloud-based portal that will allow scientists from industry, academia, government and other partners to come together to foster innovation and develop the science behind a method of “reading” DNA known as next-generation sequencing’.250 Using NGS, precisionFDA aims to create a web platform to aid researchers in sharing and learning about individual genetic variations, with the hope that these actions will lead to PM knowledge and innovations. PrecisionFDA hopes to leverage tools for greater information sharing and to help with the validation of genetic sequences and ultimately biomarker identification. Tools include the creation of reference genomes that will be posted online.
To address privacy concerns, authors recommend more transparent privacy laws with legal supports that protect individual interests in health information.251 These might take the form of stronger federal regulations that cover more than just federally funded research (as in the Common Rule) and policies to handle genetic discrimination.252 Recommendations to address the misuse of genetic information in contexts currently not covered by GINA, such as life, disability, and long-term care insurance or coverage of existing conditions with a genetic component, could strengthen PHI protections.253 Extending coverage to include full medical histories in the statutory definition of ‘genetic information’ and to misuse of that information to other contexts, like schools and biobanking, would provide patients with greater protections.254
In the context of the PMI, in November 2015 the White House published Privacy and Trust Principles, which recommended, inter alia, that ‘[m]ultiple tiers of data access—from open to controlled—based on data type, data use and user qualifications should be employed to ensure that a broad range of interested communities can utilize data while ensuring that privacy is safeguarded and public trust is maintained’.255 On May 25, 2016, the White House published its PMI Data Security Policy Principles and Framework.256 The recommendations require each organization involved in the PMI to create its own data security system, but call for ‘processes and controls to address both internal and external threats, and assure the confidentiality, integrity, and availability of data generated and contributed during precision medicine activities’.257 While requiring organizations embrace standard best practices, the Data Security Policy Principles and Framework does not create a harmonized system across the diverse set of organizations involved in the PMI. Instead, organizations must ensure that their security framework adequately addresses ‘the security risks they face and is consistent with the PMI Data Security Policy Principles and Framework’. The heterogeneous and myriad of organizations involved in the PMI are a particular challenge for data security in the PMI. With regard to a framework for data security, the report outlines a framework developed by the National Institute for Standards and Technology for Improving Critical Infrastructure Cybersecurity, Version 1.0, that enables five simultaneous and continuous functions—Identify, Protect, Detect, Respond and Recover—to assess cybersecurity and data security performance.258
In addition, articles recommend increasing federal funding of biobanks as a means to achieving higher standards and more harmonization of rules and practices relating to data management and security. With regard to the PMI, on May 26, 2016 the NIH awarded $142 million over five years to the Mayo Clinic in Minnesota to establish the world's largest research-cohort biobank. The PMI cohort biobank will collect, store, and distribute biospecimens for research.259 Finally, numerous articles recommend greater adoption and use of EHR and of systems to facilitate protected transfer and storage of those EHR.260 Use of EHR is a cornerstone of the PMI as well.
Challenge: Incentives to Enter PM space and IP barriers
According to the literature, intellectual property rights, particularly patents, create barriers to PM innovation, investment, and development in two distinct ways. First, PM developers argue that the patent system does not provide certain, predictable, strong rights that protect the substantial investment in PM drug and device development.261 PM developers call for stronger incentives where the market may not provide adequate financial reward. In contrast, patients, healthcare providers, and those unable to innovate due to blocking patents argue for a more open system, because existing patents on drugs and diagnostics stifle innovation, investment, and development of PM drugs and devices. These stakeholders argue that strong patent rights block further innovation,262 create patent thickets,263 and function as barriers to provider and patient access to drugs and devices.264 Limited empirical evidence supports either side of the patent debate.265 Here, we expand on the subthemes related to intellectual property (32 articles) and a lack of incentives for developers (20 articles).
Numerous articles assert that the high cost of drug development generally coupled with smaller financial returns for niche or personalized therapies necessitates strong and certain incentives to develop PM products. PM developers, in particular, decry a lack of certainty in scope and strength of patent protection.266 Industry stakeholders assert that consistent moves to amend and reform patent law have led to unpredictability in patent application and protections, 267 which in turn undermine investment and innovation in PM.268
However, not all commentators favor stronger patent rights for PM therapeutics. Several authors argue that patents hinder innovation in the PM space and can create barriers to patient access of drugs and genetic testing. Patent holders may block development of better tests, can prevent second opinion testing (especially through licensing practices, granting overly broad or exclusive license to underlying technology), may cover gene sequences such that development of genetic tests is hindered, and may create lack of innovation space through overly broad patent claims. In other words, broadly drafted or multiple overlapping patents can create an anticommon that stifles innovation because follow-on developers fear liability for patent infringement.269 What is clear from the literature is a lack of consensus on whether (i) patents act as necessary incentives to PM investment, innovation, and development such that they should be strengthened, or (ii) patents stifle innovation and investment, particularly in the device space, such that novel incentives are needed and patent rights should be curtailed. This tension runs through the literature and through the recommendations.
Some see the use of patents on subpopulations as a means of ‘evergreening’ or extending patent protection over a drug that has been repackaged as a PM for a smaller patient population. Even novel regulatory initiatives to entice innovation in the event of market failures may have an ‘evergreening’ effect such that they encourage unnecessary patient stratification (known as ‘salami-slicing’) in drug development without scientific justification270 and, thereby, stifle innovation in the PM space.
Unlike in pharmaceuticals, stronger and clearer patent law is not seen as the most significant incentive needed to support innovation and investment in PM tests and devices.271 One of the greatest barriers to PM device development with respect to intellectual property is general uncertainty about the applicability, strength, scope, and enforceability of patents on medical devices due to shifting patent law. The validity of many diagnostic patents has recently been called into question; therefore, it is unclear whether novel genetic tests will encounter patent thickets or infringe existing gene patents.272 Recent judicial rulings by the Supreme Court of the United States have created uncertainty as to the status of patents on both diagnostic and medical methods.273 Patents on gene-based molecular diagnostics may be problematic and fall afoul of recent rulings invalidating patent claims on naturally occurring molecules, including DNA sequences, and biomarker associations with drug dosing.274 Given the uncertainty in patent incentives, other forms of incentives may be needed to support the business model for device and diagnostic makers, which is leaner than that of pharmaceutical companies.275 Nevertheless, the current curtailing of the scope of PM-relevant patents serves as a natural experiment on the role of patent incentives in the diagnostics industry.
Recommendations on Incentive Systems and Patent Regulation
Where the market fails to provide adequate financial incentives for investment and development of PM drugs and devices, authors recommend stimulating entry into the PM device market via additional incentives. Articles recommend economic and regulatory incentives, ranging from using existing statutes and regulations in innovative ways276 to creating entirely new incentive structures.277
Incentivizing PM development using existing patent systems is a recommendation many favor.278 Increased market exclusivity or extended patent periods for drugs marketed with a CDx could incentivize PM co-development.279 Other authors advocate for additional market exclusivity for diagnostics and novel manufacturing innovations.
However, recommendations for using patent incentives reflect the tension between proprietary rights and freedom to operate and access. Recommendations therefore range from calls for stronger, clearer patent laws280 to calls for limits on patent protection.281 Recommended mechanisms to enhance freedom to operate include the creation of gene-patent clearinghouses282 and patent pools.283 For genetic testing in particular, advocates for accessibility recommend using compulsory licensing,284 requiring reasonable royalties,285 or instituting march-in rights to overcome patent barriers on genetic test development.286 Several authors recommend the creation of a research exemption in the Patent Act287 that would insulate physicians and researchers from patent infringement liability, which could stimulate R&D in academic centers.288 Other exemptions such as protections from patent infringement suits for all diagnostic use289 or genetic testing for patient care purposes290 would likewise ease fears of infringement liability. Recent developments in case law, specifically Alice Corp v. CLS Bank on the application of patent protections on abstract ideas,291 continue to contribute to a lack of clarity about the applicability of patent protection for diagnostic tests. This continued uncertainty undermines patent protections as incentives on genetic diagnostics.
Several authors point to incentives for the development of pharmaceuticals for rare diseases under the US Orphan Drug Act292 (ODA)293 as a model. Incentives under the ODA have led to the development of drugs for smaller or less-lucrative markets; more than 400 drugs for rare diseases have been developed since 1983.294 Recommendations include repurposing the ODA for PM to incentivize market entry and provide market exclusivity while avoiding the uncertainties of the patent system.295 Others caution regulators to revisit provisions of the ODA to ensure that incentives are not misappropriated as science advances and disease categories shift.296 Additional recommendations include creating either an ODA-like regulatory pathway for PM-based drugs297 or a sui generis framework for PM products.298
Initiatives like the Humanitarian Device Exemption299 also provide a mechanism for device manufacturers to short-cut some evidentiary requirements normally associated with PMA from the FDA. However, some authors note that no period of market exclusivity is provided under the Exemption and suggest that market exclusivity is a necessary incentive for PM device development. Others recommend implementing incentive programs like the Priority Review Voucher Program,300 which provides incentives for R&D in drugs and biologics for rare pediatric diseases. The Voucher Program does not currently apply to medical devices.301
However, the 21st Century Cures Act provides medical device manufacturers in the PM space with additional incentives in the form of a quicker pathway through FDA approvals. Building on the priority review device pathway, the FDA will have a fast track for breakthrough medical technologies aimed at patients with life-threatening or irreversibly debilitating diseases or conditions, and limited alternatives.302 In addition, the Humanitarian Device Exemption has been expanded to include devices that treat conditions that affect up to 8000 people, up from the previous level of 4000 people.303
There was also support for more predictable funding for PM, for example, from a centralized fund or government funding of studies for smaller or underserved populations.304 President Obama's PMI is intended to create this kind of incentive. Other authors call on the FDA to create incentives for PM developers, including reduced FDA user fees for pharmaceutical companies that work with device manufacturers to incentivize co-development.305 Tax breaks and other incentives for investment in diagnostic companies to ‘rescue’ drugs that fail in phase II trials might stimulate creation of PGx data for smaller groups of patients306 and boost overall investment in PM. Finally, one author suggests forgoing incentive schemes in favor of increasing drug prices to reflect the costs involved in drug development and allowing the market to create incentives.307 Noting the current political climate in the USA regarding high drug prices308 and the need to keep drugs affordable in the CMS and in other national healthcare systems,309 this seems an unworkable policy recommendation.
Challenge: Reimbursement
Uncertain or inadequate reimbursement by public and private payers of diagnostics and therapeutics creates one of the greatest barriers to the development and adoption of PM.310 PM developers and their investors already face a high chance of failure either in proof of concept or regulatory approval.311 However, following regulatory approval, additional uncertainty exists as to whether a PM product will be reimbursed. Lack of a positive reimbursement decision disincentivizes investment and creates barriers to use by prescribing and authorizing clinicians and patients.312 Clinicians may account for the financial state of a patient in recommending an expensive therapy, and, if no billing mechanism for a service (eg to order a specific PM diagnostic test) exists, clinicians have no financial incentive to perform that service.313 In public health systems, like those in Canada and the UK, the government is the payer and simply does not purchase or provide access to all existing therapies.314 The same is true in the USA, where health insurance plans are determinative of which treatments will be reimbursed. At a federal level, the CMS makes similar decisions with regard to Medicare and Medicaid. Here, we expand on the subthemes related to reimbursement from 87 articles, including diagnostics and drugs are reimbursed separately (22 articles); insufficient reimbursement (17 articles); the ineffectiveness of current health technology assessment (HTA) for evaluating PM (14 articles); clinical utility is not required for reimbursement decisions (13 articles); the mismatch between evidence required by payers and regulators (13 articles); a general lack of data (10 articles), more specifically, a lack of evidence on clinical utility (6 articles); a sense of uncertainty about the reimbursement of PM (6 articles); fixed reimbursement for technologies treating some diseases (5 articles); an unwillingness on developers’ parts to invest without assured reimbursement (3 articles); a mismatch between evidence produced by companies and the payers’ expectations (2 articles); and uncertainty over reimbursement for predictive genotyping (2 articles).
Ideally, health system payers, in making coverage decisions, require evidence that a therapy or test is clinically useful—that evidence demonstrates measurable increases in health or well-being in clinical application.315 This evidentiary issue is at the crux of the reimbursement barrier. Evidence of clinical utility is not required for regulatory approvals and yet it is necessary to meet the threshold for reimbursement.316 As mentioned above, CLIA regulations require that genetic tests meet a threshold of analytic validity317 and FDA regulations require evidence of safety and effectiveness in comparison with existing standard therapies, which includes data of analytical and clinical validity.318 Neither set of approval regulations requires evidence of clinical utility.
The same problems of inadequate data and insufficient evidence for successful navigation of the regulatory approvals system create difficulties for the reimbursement process. Evidence of clinical utility is often forthcoming only after PMR and longitudinal studies and is generally lacking in the field of PM.319 Data on clinical utility of genetic tests are particularly hard to develop, in part due to the difficulties correlating genetic tests and therapies with positive medical outcomes resulting from the underlying limitations of genetic predictors.320 Since all genetic variants are not directly associated with a disease or clinical condition, it is difficult to make clear medical predictions based on genetic information.321 Absent a clearer relationship between biomarkers, CDx, and improved health outcomes, payers remain skeptical and reimbursement remains uncertain.322
Questions remain about the level of evidence that is required to demonstrate clinical utility. Data required for regulatory approval differ not only in kind (analytical and clinical validity compared to clinical utility), but also in application of standards required for reimbursement decisions.323 There is no standardized method of preparing evidence of clinical utility, and uncertainty about what technical standards will be used creates confusion for test and drug manufacturers. This adds to the general uncertainty about the financial viability of PM therapies324 that already results from the expense of R&D and clinical trials, uncertain success of pharmaceuticals, and the smaller markets to which PM may be targeted.
Once a medical intervention is proven to be clinically useful, determining its reimbursement value is another barrier, one that is particularly troublesome with respect to CDx.325 First, the value of genetic information to overall health outcomes is indeterminate, let alone the value of tests that uncover that information.326 There is no agreed-upon set of parameters used to evaluate the reimbursement rate of CDx and little transparency in the reimbursement process.327 As a nascent field, it is unclear how to economically evaluate the worth of predictive testing for drug response.328
HTA and economic evaluation are traditionally used to evaluate the value of new pharmaceuticals and medical technologies relative to existing standards of care. HTA bodies have little experience with CDx and uses cost-effectiveness (a measure of the improvement in health outcomes relative to cost) and clinical utility measures, which may not be directly applicable.329 The interrelationship between PM therapy and CDx means the tests often follow the therapies, resulting in different timelines, different evidence, and interdependence, all of which complicate HTA and reimbursement decisions.330 Complicating the valuation exercises are multiple potential CDx, whose value may be linked to, or differ from, the value of the therapy.331 Traditionally reimbursement rates for diagnostics are much lower than pharmaceuticals, and are based on cost, as low as $50 to $100 per test, rather than value.332 Reimbursement policies based on cost do not recognize the clinical value or development costs of CDx333 thus, the CDx reimbursement rate is magnitudes lower than for its therapy. This is a major barrier to independent investment in the development of new and innovative CDx.334 In addition, the development of value-based analysis for reimbursement for PM is being developed on an ad hoc, case by case basis, leading to inconsistencies.335
Comparative Effectiveness Research in Healthcare
Comparative effectiveness research (CER) compares the relative effectiveness of different therapies at improving health outcomes. It is particularly useful in identifying ineffective therapies. By contrast, cost-effectiveness measures the improvement in health outcomes relative to cost.336 CER has become an important part of the US healthcare industry. This follows the authorization of $1.1 billion in The American Recovery and Reinvestment Act of 2009337 to conduct research comparing ‘clinical outcomes, effectiveness, and appropriateness of items, services, and procedures that are used to prevent, diagnose, or treat diseases, disorders, and other health conditions’.338 Competing needs for scarce healthcare dollars has increased government and healthcare purchaser initiatives to determine the most effective use of those resources. CER compares the effectiveness as well as benefits and harms of existing healthcare interventions, including tests, surgeries, and drugs. As such, its objective is to find the most effective interventions between medical alternatives. Purchasers, governments, and consumers can then determine which treatments are most effective and least expensive between competing options. Cost-effectiveness measures remain controversial in the USA, because they are the basis for decisions about rationing of healthcare expenses.339
Since CER uses aggregate outcomes data to determine whether one treatment option is less effective than another, it can be very difficult for advocates of PM interventions with smaller groups of patients to contribute data for CER comparisons.340 Outcomes data for a particular condition may overwhelm a smaller subgroup of patients, unless the subgroup becomes the basis for comparative analysis.341 While CER can use different subgroups for analysis, doing so would require validating the subgroup as appropriate and looking at different data research tools.342 Exacerbating this research difficulty is the extremely high price of some PM drugs and tests, which, coupled with a small patient population, disadvantages them in CER. Those making reimbursement decisions are faced with questions about whether a test, drug, or treatment is worth purchasing. To justify higher prices, developers of PM will be required to provide more evidence than is required to support regulatory approval, which excludes considerations of cost.343
Companion Diagnostic Reimbursement
Historical structures for reimbursement of diagnostics pose a barrier to investment in and adoption of CDx. In part, this barrier is the result of the different reimbursement structures, payers, and codes that exist for drugs and diagnostics.344 Twenty-two articles raise the disparate treatment of drugs and diagnostics at the payer level; however, these articles do not discuss the role of hospitals in decision making about diagnostics. Traditionally, diagnostic tests have been developed in hospital laboratories and the addition of tests has been under the purview of the director of the laboratory. As such, tests were part of the hospital budget with no explicit or formal process for their development or reimbursement. Thus, a non-transparent adoption process for diagnostics evolved. As more diagnostics are developed outside the hospital laboratory and sold as kits to hospitals, they become part of the formal budgeting process and must compete for those dollars. The prices for commercialized genetic tests and CDx are very expensive, compared to in-house developed diagnostics, even though diagnostic tests cost less than drugs. This makes it very difficult for a hospital diagnostic budget to absorb a CDx and puts pressures on laboratory directors to expand their budgets to accommodate new CDx.
Where insurance plans are concerned, pharmaceuticals often have a separate plan from medical services and are generally reimbursed at a higher rate than diagnostics.345 Drugs are reimbursed according to their value, whereas diagnostics are reimbursed according to their cost, depending on the materials and type of test.346 Pricing of drugs presents a balance between volume and price; in other words, the lower the volume, the higher the price that may be charged to make a profit. However, if the patient population is small, different incentives must be found or the cost to patients will be high and hospital formulary restrictions may reduce access.347 Medical diagnostics on the other hand are generally reimbursed according to Current Procedural Terminology codes managed by the American Medical Association, and the reimbursement rates established by the CMS.348 The existing reimbursement system has established codes with fixed rates for certain disease states that would need to be changed for all types of PM diagnostics to be reimbursed according to their value based on evidence.349 In addition, Medicare, the American government's largest health insurance plan, has declared that it will not pay for most preventative genetic testing.350 Under statute, CMS cannot reimburse for preventative services without Congressional approval. It is therefore unclear what evidence or reimbursement changes will spur additional coverage of CDx at the federal level.351
Recommendations for Reimbursement of PM
Realizing the potential of PM by overcoming reimbursement barriers requires the creation of closer working relationships and clearer communications between drug and device manufacturers, regulators, and reimbursement authorities earlier in the drug development process.352 This would ensure that manufacturers understand what kind of evidence is necessary for reimbursement decisions.353 While earlier involvement of reimbursement agencies requires a shift in traditional timelines, it would enable HTA and cost-effectiveness considerations to be incorporated earlier in the design of clinical programs.354 Increasing the predictability of reimbursement for PM products should facilitate the development of PM.355 Clarifying current reimbursement guidelines and the basis on which risk-benefit and effectiveness determinations are made would illuminate decision criteria and valuation interpretations,356 thereby facilitating business decisions and preparation for regulatory and reimbursement submissions.
Some articles recommend greater alignment between safety regulators and reimbursement authorities.357 In the USA, such alignment would broaden the remit of the FDA beyond safety and efficacy concerns to include considerations of comparative effectiveness and clinical utility. Numerous commentators consider this reform not as a wholesale change but as a novel approach to regulatory approvals specifically aimed at assessing PM drugs and their CDx. Many stakeholders believe that regulators and researchers should consider clinical utility and cost-effectiveness data in the development of drugs, biomarkers, and diagnostics.358
The creation of shared models of valuation between regulatory and reimbursement authorities, coupled with adoption of complementary evidentiary standards that include data on clinical utility,359 would facilitate a more predictable return on investment for PM developers. If regulators require effectiveness data to be generated in clinical trials, these data could create an evidence base for PM to satisfy reimbursement authorities. Pathways for these data to be passed on to hospitals, clinicians, and payers could further facilitate adoption of PM products. In addition, where complete effectiveness evidence is not available, conditional reimbursement could be granted contingent on generation of additional effectiveness evidence and ongoing CER.360 This would require alignment between conditional regulatory approvals and conditional reimbursement, both of which would be reliant on postmarket data collection. Reimbursement authorities might require submission of a strategy for how postmarket evidence will be generated prior to unconditional approval.361 Consequently, drug and device manufacturers need to develop standardized methods to assess validity and utility. This will provide data to reimbursement authorities in a standardized fashion.362 Developing and harmonizing clinical best practices and practices for modeling economic outcomes of PM will be an important part of this exercise.
Other recommendations advocate greater change to reimbursement policies, including a move away from cost-based reimbursement of diagnostics to value-based reimbursement.363 Focusing on value rather than on technology or process will create incentives for investment in CDx and move diagnostics onto a more level playing field with drug reimbursement.364 While this would certainly create incentives to invest in diagnostics, it is unclear what the overall impact of such a change would be on healthcare costs and resource allocation. Keeping costs under control in this scenario might involve federal price controls on patented tests and pharmaceuticals.365 Other systemic changes include changing the reimbursement codes that are used by clinicians, hospitals, and insurance companies to keep up with new advances in diagnostics.366 This would enable reimbursement of precision therapeutics and incentivize next-generation diagnostics that may be more cost-effective than standard treatments that do not segment the population. However, as whole genome sequencing increases in use for diagnosis, greater evidence of its analytic and clinical validity as well as clinical utility should be required.367 Finally, a number of authors underline the need for the HTA system to be changed to respond to PM.368 HTA needs to be adapted for the evaluation of combination products and those diagnostics aimed at smaller segments of the patient population.
CONCLUSION
PM promises to be at the forefront of government and scientific agendas for the foreseeable future. Since beginning our research, the PMI has motivated significant US health research and policy action; there will be continued spending, development of infrastructure, and data-sharing systems. The FDA final guidance on LDTs is expected at any time. Revisions to the federal Common Rule will continue to be debated. The open reference platform of precision FDA will generate information that will help inform future regulatory directions and decision making. Pharmaceutical and biotechnology companies will continue to work toward developing personalized treatments and diagnostics, not just in the USA but around the globe.369 While the reviewed articles express concern about barriers to PM, they all assume that PM would bring tangible benefits to patients and/or healthcare systems (a limitation of our study).
Our research revealed the significant regulatory and non-regulatory barriers to the realization of PM, which will need to be addressed in tandem. In the first instance, while PM R&D has focused almost exclusively on genetics, it has become clear that genes are not as predictive as once believed. Indeed, highly prevalent and predictive genetic mutations are uncommon. Thus, the science of PM requires broadening to include fields such as proteomics, metabolomics, and cell therapies. In addition, there is a lack of clinical uptake of PM by healthcare providers resulting from both structural and human factors. The structural barriers include the traditional focus on treatment of disease rather than its prevention in the current US healthcare system.370 PM tests will require greater resources and expertise for interpretation and explanation of the meaning of test results for patients.371 Human factors include inadequate clinician education and/or training in PM, especially genetics, and, therefore, lack of expertise and comfort with interpretation of test results372 and how best to counsel patients. Lack of training inhibits clinical uptake, which, in turn, will impede the generation of data on clinical utility of PM products and services.
Addressing the structural, financial, and regulatory barriers to PM needs to proceed in lock-step with scientific advances if PM is to reach its promised potential. Indicators are that reforms will be propelled by the scientific community, industry, political will, and, most importantly, the patient advocacy community—the ultimate beneficiary of PM technologies and services.
Personalized medicine search terms: theranostic* OR personalized medicine* OR personalised medicine* OR individualized medicine* OR individualised medicine* OR pharmacogenetic* OR targeted therap* OR pharmacogenomic* OR companion diagnostic* OR genomic test* OR stratified medicine* OR precision medicine* OR biotherap* OR co-dependent technolog* OR co-dependent technolog* OR codependent technolog* OR hybrid technolog* OR genomic medicine* OR customized medicine* OR customised medicine* OR integrated diagnostic* OR integrated therapeutic* OR molecular targeted therap* OR P4 medicine* OR patient specific therap* OR personalized molecular cancer therap* OR personalised molecular cancer therap* OR personalised molecular cancer therap* OR personalized cancer therap* OR personalised cancer therap* OR individualized cell therap* OR individualised cell therap* OR PGX OR pharmacoproteomic* OR pharmacometabolomic* OR stratified therap* OR tailored therap* OR targeted drug therap* OR (diagnosti* OR prognosti* AND personalized OR personalised OR companion OR codependent OR stratified OR genomic OR individual* OR pharmacogenomic*).
ACKNOWLEDGEMENTS
The authors are grateful for support from PACEOMICS (PIs: Drs Christopher McCabe and Tania Bubela) funded by Genome Canada, Genome Alberta, Genome Quebec, the Canadian Institutes for Health Research (CIHR) and Alberta Innovates-Health Solutions (AI-HS) and the Alberta Ocular Gene Therapy Team (Co-leads: Drs Ian MacDonald and Tania Bubela) funded by the CRIO program of AI-HS. We would like to thank Emma Camicioli and Mark Bieber for research assistance and data management, respectively. We would like to thank Drs Shelagh Genuis and Christopher McCabe for expert comments on the manuscript. Finally, Yael Mansour, John Minkley and Kiah Van der Loos took on the herculean task of footnoting and editing the manuscript.
Footnotes
Bubela (BSc (Hons), PhD, JD) is a Professor and Dean of the Faculty of Health Sciences at Simon Fraser University, Canada. Prior to 2017, she was a Professor in the School of Public Health and Adjunct Professor in the Alberta School of Business at the University of Alberta, Canada. Her research program focuses on large collaborative science networks in genomics, gene therapy, and stem cell biology, addressing barriers to the effective translation of new technologies and the introduction of precision medicine. These are varied and include ethical issues, commercialization, and regulation. She provides advice for Government Health and Science agencies, life sciences research communities, and patient organizations. Her research is funded by the Canadian Institutes of Health Research, the Canadian Stem Cell Network, BioCanRx, and Genome Canada, among others. She co-leads the PACEOMICS program on the development of cost-effective personalized medicine.
Other terms associated and often used interchangeably with personalized medicine include individualized medicine, molecular medicine, personalized genomic/genetic medicine, stratified medicine, tailored therapeutics, targeted therapeutics; What is Personalized Medicine, Paceomics, http://paceomics.org/index.php/issues/ (accessed Aug. 2, 2016). Each of these terms has a slightly different meaning. A useful taxonomy of the terms, along with their definitions and first usages, is provided in Anna Pokorska-Bocci et al., ‘Personalized Medicine’: What's in a name?, 11 Pers. Med. 197, 199–200 (2014). Despite the announcement of President Obama's Precision Medicine Initiative, which likely increased the global usage of the term ‘Precision Medicine’, we used personalized medicine at the time we conducted the searches for our scoping review.
‘“Omics” is a term encompassing multiple molecular disciplines that involve the characterization of global sets of biological molecules such as DNAs, RNAs, proteins, and metabolites’. Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials, Institute of Medicine (U.S.), Evolution of Translational Omics: Lessons Learned and the Path Forward 1, 1 (Christine M. Micheel, Sharyl J. Nass & Gilbert S. Omenn eds., Nat’l Academies Press 2012).
Edward Abrahams et al., The Personalized Medicine Coalition: Goals and Strategies, 5 Am. J. Pharmacogenomics 345, 345 (2005); Robert Langreth & Michael Waldholz, New Era of Personalized Medicine: Targeting Drugs for Each Unique Genetic Profile, 4 Oncologist 426, 426 (1999).
The Precision Medicine Initiative includes multiple components with efforts from the federal government and was budgeted at $215 million in the fiscal year of 2016 by the President. NIH will lead efforts in cancer genomics, as well as the development of the large research participant cohort consisting of over one million participants; White House, FACT SHEET: President Obama's Precision Medicine Initiative, https://www.whitehouse.gov/the-press-office/2015/01/30/fact-sheet-president-obama-s-precision-medicine-initiative.
Committee on a Framework for Developing a New Taxonomy of Disease, National Research Council (U.S.), Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease, 12 (Steve Olson ed., Nat’l Academies Press 2011).
William G. Feero, Clinical Application of Whole-Genome Sequencing: Proceed with Care, 311 J. Am. Med. Ass’n 1017, 1018 (2014).
Matthew Avery, Personalized Medicine and Rescuing Unsafe Drugs with Pharmacogenomics: A Regulatory Perspective, 65 Food & Drug L. J. 37, 41 (2010).
Bernard Munos, Lessons from 60 Years of Pharmaceutical Innovation, 8 Nat. Rev. Drug Discov. 959, 961 (2009).
Michael Hay et al., Clinical Development Success Rates for Investigational Drugs, 32 Nat. Biotechnol. 40, 41 (2014).
Tito Fojo et al., Unintended Consequences of Expensive Cancer Therapeutics—The Pursuit of Marginal Indications and a Me-Too Mentality That Stifles Innovation and Creativity: The John Conley Lecture, 140 JAMA Otolaryngol. Head & Neck Surg. 1225, 1225 (2014).
The FDA approved a total of 22 therapeutics with companion diagnostic tests that are ‘essential for the therapeutic product's safe and effective use’. Thirty-six products defined by their parent companies as companion diagnostics are marketed in the EU with approval.
National Health Institute, About the Precision Medicine Initiative Cohort Program, https://www.nih.gov/precision-medicine-initiative-cohort-program (accessed July 20, 2016).
ICH Harmonised Tripartite Guideline: Guideline For Good Clinical Practice E6(R1) (June 10, 1996), http://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Efficacy/E6/E6_R1_Guideline.pdf.
Rumiko Shimazawa & Masayuki Ikeda, Are There Any Differences in the Regulations of Personalized Medicine Among the USA, EU and Japan?, 75 Brit. J. Clin. Pharmacol. 1365, 1365 (2013).
President Barack Obama, Remarks at the East Room of the White House (Jan. 30, 2015) (transcript https://www.whitehouse.gov/the-press-office/2015/01/30/remarks-president-precision-medicine); Julie Steenhuysen, NIH Takes Next Steps in Obama's Precision Medicine Plan, Reuters (Sept. 17, 2015), http://www.reuters.com/article/2015/09/17/us-health-precisionmedicine-idUSKCN0RH2K720150917
Hilary Arksey & Lisa O’Malley, Scoping Studies: Towards a Methodological Framework, 8 Int’l J. Soc. Res. Methodol. 19, 20 (2005).
David Moher et al., Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement, 6 Plos Med. (July 21, 2009), http://journals.plos.org/plosmedicine/article?id=10.1371/journal.pmed.1000097#s9
Sebastian Schleidgen et al., What is Personalized Medicine: Sharpening a Vague Term Based on a Systematic Literature Review, 14 BMC Med. Ethics 55, 64 (2013).
Mary L. McHugh, Interrater Reliability: The Kappa Statistic, 22 Biochem. Med. 276, 279 (2012).
Kathy Charmaz, Constructing Grounded Theory 54 (2d ed. 2014).
Note that articles could discuss more than one main theme.
Stuart Hogarth, Regulatory Experiments and Transnational Networks: The Governance of Pharmacogenomics in Europe and the United States, 25 Innovation: Eur. J. Soc. Sci. 441, 452 (2012); James Mittra & Joyce Tait, Analysing Stratified Medicine Business Models and Value Systems: Innovation-Regulation Interactions, 29 New Biotechnol. 709, 717 (2012); Kate Traynor, Targeted Drug Therapy Remains a Challenge, 68 Am. J. Health-Sys. Pharmacy 2320, 2324 (2011).
James P. Evans & Michael S. Watson, Genetic Testing and FDA Regulation Overregulation Threatens the Emergence of Genomic Medicine, 313 J. Am. Med. Ass’n 669, 669 (2015); John D. Johnston & Peter Feldschreiber, Challenges Posed to the European Pharmaceutical Regulatory System by Highly Personalized Medicines, 77 Brit. J. Clin. Pharmacol. 421, 425 (2014); Kelly A. McClellan et al., Personalized Medicine and Access to Health Care: Potential for Inequitable Access?, 21 Eur. J. Hum. Genet. 143, 145 (2013); Jessica E. Palmer, Genetic Gatekeepers: Regulating Direct-to-Consumer Genomic Services in an Era of Participatory Medicine, 67 Food & Drug L.J. 475, 494 (2012); M. Pirmohamed, Acceptance of Biomarker-Based Tests for Application in Clinical Practice: Criteria and Obstacles, 88 Clin. Pharmacol. & Ther. 862, 865 (2010); Randy J. Prebula, The Promise and Personalized Medicine: Regulatory Controls and Tort Influences in the Context of Personalized Risks and Benefits, 26 J. Contemp. Health L. & Pol’y 343, 367 (2010); Agnese Querci, Advanced Therapy Medicinal Products: The Regulation 1394/2007/CE, its Inurement in Italy and Connected Liability Regimes, 12 Pharmaceuticals Pol’y & L. 259, 265 (2010); Rashmi R. Shah & Devron R. Shah, Personalized Medicine: Is it a Pharmacogenetic Mirage?, 74 Brit. J. Clin. Pharmacol. 698, 710 (2012); L. Shahmirzadi et al., Patient Decisions for Disclosure of Secondary Findings Among the First 200 Individuals Undergoing Clinical Diagnostic Exome Sequencing, 16 Genet. Med. 395, 396 (2014); Kayte Spector-Bagdady & Elizabeth Pike, Consuming Genomics: Regulating Direct-to-Consumer Genetic and Genomic Information, 92 Neb. L. Rev. 677, 731 (2014); Thomas Tursz & Rene Bernards, Hurdles on the Road to Personalized Medicine, 9 Mol. Oncol. 935, 937 (2015); A. W. Warner et al., Challenges in Obtaining Adequate Genetic Sample Sets in Clinical Trials: The Perspective of the Industry Pharmacogenomics Working Group, 89 Clin. pharmacol. & Ther. 529, 532 (2010).
21st Century Cures Act, H.R. 34, 114th Cong. (2015).
Mike DeBonis, Congress Passes 21st Century Cures Act, Boosting Research and Easing Drug Approvals, The Washington Post, Dec. 7, 2016, https://www.washingtonpost.com/news/powerpost/wp/2016/12/07/congress-passes-21st-century-cures-act-boosting-research-and-easing-drug-approvals/?utm_term=.453cd4204838.
H.R. 34, § 1001.
H.R. 34, § 1003.
H.R. 34.
H.R. 34, § 3001.
Debonis, supra note 25.
H.R. 34, § 3011.
H.R. 34, § 3038.
H.R. 34, § 3021.
US Office of Management and Budget. M-17-22, Comprehensive Plan for Reforming the Federal Government and Reducing the Federal Civilian Workforce (Apr. 12, 2017).
Exec. Order No. 13771, 82 Fed. Reg. 9339 (Jan. 30, 2017).
Id.
US Office of Management and Budget, Budget of the U.S. Government: A New Foundation for American Greatness. Fiscal Year 2018 (2017).
Better Care Reconciliation Act of 2017. H.R. 1628. 115th Cong. (2017).
U.S. Food & Drug Administration, FDA and Clinical Drug Trials: A Short History, http://www.fda.gov/AboutFDA/WhatWeDo/History/Overviews/ucm304485.htm (accessed Apr. 11, 2016).
In the EU, medicinal products may only be marketed after receiving marketing authorization from the Union through the centralized market authorization process or from a competent authority in a member state. Certain medicines (medicines for the treatment of HIV/AIDS, cancer, diabetes, neurodegenerative diseases, autoimmune and other immune dysfunctions, and viral diseases; medicines derived from biotechnology processes; advanced-therapy medicines, including gene-therapy, somatic cell-therapy or tissue-engineered medicines; orphan medicines) must be approved through a centralized marketing authorization application. Other medicines (new active substances not authorized in the European Community before May 20, 2004; medicinal products that contribute significant therapeutic, scientific, or technical innovation or are in the interests of patient health; generic copies of centrally authorized products) can be approved through the centralized approval procedure, or can be approved through national marketing authorizations and mutual recognition procedures. Through the centralized marketing procedure, companies submit their application to the EMA. The EMA strongly encourages prospective applicants to schedule a presubmission meeting to obtain regulatory, procedural, and legal advice. Once the application is submitted, it is subject to a scientific evaluation and after consultation with the Standing Committee for Medicinal Products for Human Use, the European Commission may grant a market authorization for the product. Applications submitted to a reference member state must include a draft assessment report, summary of product characteristics (SmPC), labeling and package leaflet. Identical applications must also be sent to concerned member state(s). If the national application is successful, a harmonized marketing approval will be issued in the reference member state and concerned member state(s). The regulatory agency involved may seek additional information or clarity from the sponsor during the application process, Eudralex - Volume 2 - Pharmaceutical Legislation Notice to Applicants and Regulatory Guidelines Medicinal Products for Human Use, Europa, http://ec.europa.eu/health/documents/eudralex/vol-2/index_en.htm (accessed July 29, 2016).
Prior to filling out an application for a New Drug Submission, sponsors can submit a presubmission package and meet with the Health Canada review staff to discuss the presentation of the data in support of the submission. After submission, Health Canada may issue a Clarification Request to expand on, add precision, or re-analyze existing data, or a Notice of Noncompliance if the submission is deficient. The sponsor has these opportunities to submit the necessary additional information in order to complete their application. Health Canada issues a notice of compliance for successful applications, indicating that the product has met Health Canada's requirements for safety, efficacy and quality; Guidance for Industry Management of Drug Submissions, Health Canada, http://www.hc-sc.gc.ca/dhp-mps/prodpharma/applic-demande/guide-ld/mgmt-gest/mands_gespd-eng.php (accessed Dec. 19, 2013).
Alan Devlin, Systemic Bias in Patent Law, 61 DePaul L. Rev. 57, 70 (2011); Anna B. Laakmann, Collapsing the Distinction Between Experimentation and Treatment in the Regulation of New Drugs, 62 Ala. L. Rev. 305, 314 (2011); George A. Neyarapally, A Review of Recent Federal Legislative and Policy Initiatives to Enhance the Development and Evaluation of High Value Drugs in the United States, 14 DePaul J. Health Care L. 503, 510 (2013); Matthew Piehl, Regulating Hype and Hope: A Business Ethics Model Approach to Potential Oversight of Direct-to-Consumer Genetic Tests, 16 Mich. St. U. J. Med. & L. 59, 70 (2011); Andrew S. Robertson, The Role of DNA Patents in Genetic Test Innovation and Access, 9 Nw. J. Tech. & Intell. Prop. 377, 392 (2011).
There are typically four or five phases to clinical testing of a new therapeutic. Phase 0 trials are exploratory studies with very limited human exposure to the therapeutic. Phase I is a safety trial to identify frequent and serious adverse events, as well as how the therapeutic is metabolized and excreted. The focus of phase II is preliminary effectiveness in the specific indication, with continuing collection of safety data and data collection on short-term adverse events. In phase III, the therapeutic is tested in different populations and at different dosages, as well as in conjunction with other therapeutics. Phase IV studies, or postmarket surveillance, occur after the regulator approves the therapeutic for marketing and generate data on safety, efficacy, and/or optimal use; Glossary Definition: Phase, Clinical Trials, https://clinicaltrials.gov/ct2/help/glossary/phase (accessed July 20, 2016).
Joseph A. DiMasi et al., The Price of Innovation: New Estimates of Drug Development Costs, 22, J. Health Econ. 151, 164–5 (2003); Joseph A. DiMasi, Innovation in the Pharmaceutical Industry: New Estimates of R&D Costs, 47 J. Health Econ. 20, 20 (2016).
Jonathan J. Darrow, Crowdsourcing Clinical Trials, 98 Minn. L. Rev. 805 (2014); Devlin, supra note 42, at 854; Avery, supra note 7, at 38; Rena Conti et al., Personalized Medicine and Genomics: Challenges and Opportunities in Assessing Effectiveness, Cost-Effectiveness, and Future Research Priorities, 30 Med. Dec. Making 328, 336 (2010); Evans & Watson, supra note 23, at 669; Jennifer S. Geetter, Another Man's Treasure: The Promise and Pitfalls of Leveraging Existing Biomedical Assets for Future Use, 4 J. Health & Life Sci. 1, 39 (2011); John Hudson & Marta Orviska, European Attitudes to Gene Therapy and Pharmacogenetics, 16 Drug Discov. Today 843, 844 (2011); K Ichimaru et al., PMDA’s Challenge to Accelerate Clinical Development and Review of New Drugs in Japan, 88 Clin. Pharmacol. & Ther. 454, 454 (2010); Robert Jones & Maria DeSantis, Access to Targeted Therapies in Renal Cell Cancer, 40 Semin. Oncol. 521, 522 (2013); Gottfried E. Konecny, The Path to Personalized Medicine in Women's Cancers: Challenges and Recent Advances, 27 Curr. Opin. Obstet. & Gynecol. 45, 46 (2015); Laakmann, supra note 42, at 314; Janet L. MacPherson & John E.J. Rasko, Cellular Therapy in the Asia-Pacific Region. A Guide for the Future Pathologist, 43 Pathology 616, 623 (2011); Michael McCarthy, Obama Promises to Defend Health Law and Promote ‘Precision Medicine’, 350 BMJ 385, 385 (2015); Mittra & Tait, supra note 22, at 717; Richard A. Montagna, Meeting the Technical Challenges of Personalized Medicine and Companion Diagnostics, 44 Med. Lab. Obs. 16, 18 (2012); Bryn Nelson, Ensuring Quality in Genomic Medicine: Amid the Rise in Complex Laboratory-Developed Tests, Regulatory Officials are Seeking the Right Balance on Quality Assurance, 122 Cancer Cytopathol. 855, 856 (2014); Neyarapally, supra note 42, at 510; Piehl, supra note 42, at 70; Scott D. Ramsey et al., How Comparative Effectiveness Research Can Help Advance ‘Personalized Medicine’ in Cancer Treatment, 30 Health Aff. 2259, 2264 (2011); Robertson, supra note 42, at 392; Richard L. Schilsky et al., Commentary: Tackling the Challenges of Developing Targeted Therapies for Cancer, 15 Oncologist 484, 485 (2010).
Avery, supra note 7, at 38; Evans & Watson, supra note 23, at 669; Hudson & Orviska, supra note 45, at 844; Ichimaru et al., supra note 45, at 454; Jones & DeSantis, supra note 45, at 522; Konecny, supra note 45, at 46; MacPherson & Rasko, supra note 45, at 623; Jeanette J. McCarthy et al., Genomic Medicine: A Decade of Successes, Challenges, and Opportunities, 5 Sci. Transl. Med. 189, 207 (2013); Mittra & Tait, supra note 22, at 710; Montagna, supra note 45 at 18; Nelson, supra note 45, at 856; Ramsey et al., supra note 45, at 2264; Schilsky et al., supra note 45, at 485.
Joshua P. Cohen & Abigail E. Felix, Personalized Medicine's Bottleneck: Diagnostic Test Evidence and Reimbursement, 4 J. Pers. Med. 163, 171 (2014); Leonard M. Fleck, Just Caring: Assessing the Ethical and Economic Costs of Personalized Medicine, 32 Urol. Oncol. 202, 206 (2014); Shannon G. Gibson & Trudo Lemmens, Niche Markets and Evidence Assessment in Transition: A Critical Review of Proposed Drug Reforms, 22 Med. L. Rev. 200, 211 (2014); Christof Koelsch et al., Towards a Balanced Value Business Model for Personalized Medicine: An Outlook, 14 Pharmacogenomics 89, 92 (2013); Christophe Le Tourneau et al., Designs and Challenges for Personalized Medicine Studies in Oncology: Focus on the SHIVA Trial, 7 Target. Oncol. 253, 264 (2012); Pierre Miossec et al., Biomarkers and Personalised Medicine in Rheumatoid Arthritis: A Proposal for Interactions Between Academia, Industry and Regulatory Bodies, 70 Ann. Rheum. Dis. 1713, 1716 (2011); Katelin E. Petersen et al., Personalized Medicine, Availability, and Group Disparity: An Inquiry into How Physicians Perceive and Rate the Elements and Barriers of Personalized Medicine, 17 Pub. Health Genom. 209, 216 (2014); Mark R. Trusheim et al., Quantifying Factors for the Success of Stratified Medicine, 10 Nat. Rev. Drug Discov. 817, 823 (2011); Tursz & Bernards, supra note 23, at 938.
‘A biomarker is a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or biological responses to a therapeutic intervention’; U.S. Food & Drug Administration, From our Perspective: Clinical Biomarker Qualification, http://www.fda.gov/Drugs/NewsEvents/ucm424545.htm (accessed Jan. 5, 2015).
Lasse Tengbjerg Hansen & Adam Heathfield, Personalized Medicine: Translation of Concepts, Genengnews (Mar. 20, 2013), http://www.genengnews.com/bioperspectives/personalized-medicine-translation-of-concepts/4783.
Giuseppe Novelli et al., Genetic Tests and Genomic Biomarkers: Regulation, Qualification and Validation, 5 Clin. Cases Miner. & Bone Metab. 149, 152 (2008).
Lawrence J. Lesko & Janet Woodcock, Translation of Pharmacogenomics and Pharmacogenetics: A Regulatory Perspective, 3 Nat. Rev. Drug Discov. 763, 763 (2004).
The forerunner of this new generation of drugs is Herceptin, which the FDA approved, with a companion diagnostic to identify women with HER2 overexpression, as a treatment for metastatic HER2-overexpressing breast cancer. Genentech, the developer of Herceptin, has led the way in seeking approval for ever-broadening indications for its targeted therapeutics. This new generation of drugs begins its lifecycle in a small population with unmet medical needs, in which the developer can charge premium rates while also seeking evidence to expand into broader indications. While the growth rates of biologics have grown 20%, the growth rate of US pharmaceutical market grew only 6%–8%. Saurabh Aggarwal, What's fueling the biotech engine?, 25 Nat. Biotech. 1097, 1098 (2007).
Christopher-Paul Milne & Rachel Zuckerman, Biopharmaceutical Industry Perspectives on the Business Prospects for Personalized Medicine, 8 Pers. Med. 541, 544 (2011); Eric E. Walk, Improving the Power of Diagnostics in the Era of Targeted Therapy and Personalized Healthcare, 13 Curr. Opin. Drug Discov. & Dev. 226, 230 (2010).
Hansen & Heathfield, supra note 49.
Ignacio I. Wistuba et al., Methodological and Practical Challenges for Personalized Cancer Therapies, 8 Nat. Rev. Clin. Oncol. 135, 140 (2011).
Jean-Claude Libeer & Sharon Ehrmeyer, ISO 15189: A Worldwide Standard for Medical Laboratories, 3 Point Care 5, 6 (2004).
Michael Nohaile, The Biomarker is Not the End, 16 Drug Discov. Today 878, 881 (2011).
Koelsch et al., supra note 47, at 91; Yann Joly et al., Regulatory Approval for New Pharmacogenomic Tests: A Comparative Overview, 66 Food & Drug L.J. 1, 4 (2011); Marie Loh & Richie Soong, Challenges and Pitfalls in the Introduction of Pharmacogenetics for Cancer, 40 Ann. Acad. Med. 369, 369–70 (2011); Yasuto Otsubo et al., Regulatory Perspective on Remaining Challenges for Utilization of Pharmacogenomics-Guided Drug Developments, 14 Pharmacogenomics 195, 198 (2013); M.A. Pacanowski, Next-Generation Medicines: Past Regulatory Experience and Considerations for the Future, 95 Clin. Pharmacol. & Ther. 247, 248 (2013); Trusheim et al., supra note 47, at 824.
Elizabeth A. Mansfield, FDA Perspective on Companion Diagnostics: An Evolving Paradigm, 20 Clin. Cancer Res. 1453, 1455 (2014).
Off-label use is defined as ‘unapproved use of an approved drug’. The FDA approves drugs for specific indications; its approval indicates that the benefits of using the drug for that particular use outweigh the risks. Once a drug is approved, a healthcare provider may prescribe the drug for an unapproved use that they judge as medically appropriate for their patient; U.S Food & Drug Administration, Understanding Unapproved Use of Approved Drugs ‘Off Label’, http://www.fda.gov/ForPatients/Other/OffLabel/default.htm (accessed June 2, 2016).
Francesco Pignatti et al., Cancer Drug Development and the Evolving Regulatory Framework for Companion Diagnostics in the European Union, 20 Clin. Cancer Res. 1458, 1462 (2014).
Robert L. Cohen & Jeff Settleman, From Cancer Genomics to Precision Oncology—Tissue's Still an Issue, 157 Cell 1509, 1512 (2014); Sean A. McGhee, How the Practice of Allergy Shows the Promise and Challenge of Personalized Medicine, 104 Mol. Genet. & Metab. 3, 4 (2011); Henrik Winther & Jan Trøst Jørgensen, Drug-Diagnostic Co-Development in Cancer, 24 Pharmceutical Med. 363, 363 (2010).
Efthimios Parasidis, Patients over Politics: Addressing Legislative Failure in the Regulation of Medical Products, 5 Wis. L. Rev. 929, 931 (2011).
See footnote 60.
Anup Malani et al., Accounting for Heterogeneous Treatment Effects in the FDA Approval Process, 67 Food & Drug L.J. 23, 28 (2012).
Id. at 29.
Id.
Gibson & Lemmens, supra note 47, at 208; Chiara Piana et al., Integration of Pharmacogenetics and Pharmacogenomics in Drug Development: Implications for Regulatory and Medical Decision Making in Pediatric Diseases, 52 J. Clin. Pharmacol. 704, 704 (2012); Francesco Pignatti et al., Assessment of Benefits and Risks in Development of Targeted Therapies for Cancer – The View of Regulatory Authorities, 9 Mol. Oncol. 1034, 1037 (2015).
Jane Fridlyand et al., An Industry Statistician's Perspective on PHC Drug Development, 36 Contemp. Clin. Trials 624, 744 (2013); Malani, supra note 65, at 29.
Sarah Blankstein, Pharmacogenomics- History, Barriers, and Regulatory Solutions, 69 Food & Drug L.J. 273, 283–4 (2014).
Avery, supra note 7, at 39; Gibson & Lemmens, supra note 47, at 204; Malani, supra note 65, at 28.
Blankstein, supra note 70, at 281; Cohen & Settleman, supra note 62, at 1512; McGhee, supra note 62, at 4; Parasidis, supra note 63, at 931; Winther & Jørgensen, supra note 62, at 363.
Jaap Verweij et al., Moving Molecular Targeted Drug Therapy Towards Personalized Medicine: Issues Related to Clinical Trial Design, 6 Mol. Oncol. 196, 201 (2012); J. S. de Bono & Alan Ashworth, Translating Cancer Research into Targeted Therapeutics, 467 Nature 543, 543 (2010).
Cohen & Settleman, supra note 62, at 1512; Jin Huang et al., Emerging Trends in US Oncological Approvals: A 13-Year Review (1999-2011), 46 Drug Info. J. 344, 353 (2012); Le Tourneau et al., supra note 47, at 264; Nohaile, supra note 57, at 881; Manish R. Sharma & Richard L. Schilsky, Role of Randomized Phase III Trials in an Era of Effective Targeted Therapies, 9 Nat. Rev. Clin. Oncol. 208, 211 (2011); Nigel Stallard et al., Adaptive Designs for Confirmatory Clinical Trials with Subgroup Selection, 24 J. Biopharm. Stat. 167, 183 (2014).
Blankstein, supra note 70, at 277; Malcolm Rowland et al., Impact of the Pharmaceutical Sciences on Health Care: A Reflection over the Past 50 Years, 101 J. Pharm. Sci. 4075, 4089 (2012); Sharma & Schilsky, supra note 74, at 212 (2011); Takeki Uehara et al., The Japanese Toxicogenomics Project: Application of Toxicogenomics, 54 Mol. Nutr. & Food Res. 218, 226 (2010); F. Randy Vogenberg et al., Personalized Medicine Part 2: Ethical, Legal, and Regulatory Issues, 35 Pharm. & Ther. 624, 631 (2010); Walk, supra note 53, at 230.
Laakmann, supra note 42, at 334; Parasidis, supra note 63, at 935.
Avery, supra note 7, at 39; Cohen & Settleman, supra note 62, 1512; Joly et al., supra note 58, at 4-5.
Zeinab Awada & Nathalie Khoueiry Zgheib, Pharmacogenovigilance: A Pharmacogenomics Pharmacovigilance Program, 15 Pharmacogenomics 845, 847 (2014); Gibson & Lemmens, supra note 47, at 208; J.W. Lee et al., The Emerging Era of Pharmacogenomics: Current Successes, Future Potential, and Challenges, 86 Clin. Genet. 21, 25 (2014); N. A. Meadows et al., An Evaluation of Regulatory and Commercial Barriers to Stratified Medicine Development and Adoption, 15 Pharmacogenomics J. 6, 8 (2015); Ramsey et al., supra note 45, at 2267–68; Renee Ahrens Thomas et al., APhA 2010 House of Delegates: Paving the Way for the Profession's Best Practices, 50 J. Am. Pharm. Ass’n. 450, 452 (2010).
Under provisions for limited, accelerated, or conditional approval, regulators may approve pharmaceutical products based on limited datasets while requiring the company to perform confirmatory tests post approval. Conditional approvals are a compromise between earlier patient access who suffer diseases with unmet medical needs and patient safety, Arna H. Arnardottir et al., Additional Safety Risk to Exceptionally Approved Drugs in Europe?, 72 Brit. J. Clin.Pharmacol. 490, 491 (2011).
Blankstein, supra note 70, at 313; Cohen & Felix, supra note 47, at 172; Cohen & Settleman, supra note 62, at 1513; William S. Dalton et al., The 2010 Health Care Reform Act: A Potential Opportunity to Advance Cancer Research by Taking Cancer Personally, 16 Clin. Cancer Res. 5987, 5994 (2010); de Bono & Ashworth, supra note 73, at 545; Hans-Georg Eichler et al., Bridging the Efficacy-Effectiveness Gap: A Regulator's Perspective on Addressing Variability of Drug Response, 10 Nat. Rev. Drug Discov. 495, 502 (2011); Gibson & Lemmens, supra note 47, at 208; Joly et al., supra note 58, at 4; Jan Trøst Jørgensen, Companion Diagnostics in Oncology – Current Status and Future Aspects, 85 Oncology 59, 65 (2013); Laakmann, supra note 42, at 334; D. O’Kane, An Outsider's Viewpoint: The FDA Should Regulate Clinical Pharmacogenetic/ Genomic Tests, But..., 88 Clin. Pharmacol. & Ther. 746, 747 (2010); George Poste et al., Leveling the Playing Field: Bringing Development of Biomarkers and Molecular Diagnostics up to the Standards for Drug Development, 18 Clin. Cancer Res. 1515, 1520 (2012); Ramsey et al., supra note 45, at 2267-68; Schilsky et al., supra note 45, at 486; Carla G. van El & Martina C. Cornel, Genetic Testing and Common Disorders in a Public Health Framework, 19 Eur. J. Hum. Genet. 377, 380 (2011); S. J.H. Vijverberg et al., Ethical and Social Issues in Pharmacogenomics Testing, 16 Curr. Pharm. Des. 245, 250 (2010); Vogenberg et al., supra note 75, at 631; Wistuba et al., supra note 55, at 140.
Laakmann, supra note 42, at 340; Gibson & Lemmens, supra note 47, at 215; Schilsky et al., supra note 45, at 486; Poste et al., supra note 80, at 1521.
Laakmann, supra note 42, at 340.
Awada & Zgheib, supra note 78, at 847; Gibson & Lemmens, supra note 47, at 215; Laakmann, supra note 42, at 343.
Awada & Zgheib, supra note 78, at 847.
Laakmann, supra note 42, at 340.
Awada & Zgheib, supra note 78, at 847; Gibson & Lemmens, supra note 47, at 215.
Parasidis, supra note 63, at 934-935.
Cohen & Settleman, supra note 62, at 1512; Laakmann, supra note 42, at 344.
Avery, supra note 7, at 51; Eichler et al., supra note 80, at 501; Federico M. Goodsaid, Voluntary Exploratory Data Submissions to the US FDA and the EMA: Experience and Impact, 9 Nat. Rev. Drug Discov. 435, 436 (2010); Otsubo et al., supra note 58, at 195; Mittra & Tait, supra note 22, at 716.
Avery, supra note 7, at 54.
Mittra & Tait, supra note 22, at 716.
Lidia Becla et al., Health Technology Assessment in the Era of Personalized Health Care, 27 Int’l J. Tech. Assess. Health Care 118, 125 (2011); Cinnamon S. Bloss et al., Genomics for Disease Treatment and Prevention, 34 Psychiatr. Clin. N. Am. 147, 154–55 (2011); Craig P. Carden et al., Can Molecular Biomarker-Based Patient Selection in Phase I Trials Accelerate Anticancer Drug Development?, 15 Drug Discov. Today 88, 93-4 (2010); Zubin J. Eapen et al., The Imperative of Overcoming Barriers to the Conduct of Large, Simple Trials, 311 JAMA 1397, 1397 (2014); Eric Faulkner et al., Challenges in the Development and Reimbursement of Personalized Medicine—Payer and Manufacturer Perspectives and Implications for Health Economics and Outcomes Research: A Report of the ISPOR Personalized Medicine Special Interest Group, 15 Value Health 1162, 1167 (2012); Judith M. Fontana et al., Translational Research in Infectious Disease: Current Paradigms and Challenges Ahead, 159 Transl. Res. J. Lab. & Clin. Med. 430, 446 (2012).
Blankstein, supra note 70, at 277; Fontana et al., supra note 92, at 446.
Fontana et al., supra note 92, at 446.
Gibson & Lemmens, supra note 47, at 208.
Blankstein, supra note 70, at 277.
Cohen & Settleman, supra note 62, at 1512; Malani, supra note 65, at 25.
Gibson & Lemmens, supra note 47, at 210; Tursz & Bernards, supra note 23, at 938.
Eichler et al., supra note 80, at 502; Denis Horgan et al., An Index of Barriers for the Implementation of Personalised Medicine and Pharmacogenomics in Europe, 17 Pub. Health Genom. 287, 295 (2014).
In general, ‘N of 1’ studies consist of a single patient as the whole trial who serves as his/her own control. In oncology, an ‘N of 1’ trial refers to the use of pathological and molecular characteristics to select an individualized therapeutic regimen. Michael J. Malinowski & Grant G. Gautreaux, All that is Gold Does Not Glitter in Human Clinical Research: A Law– Policy Proposal to Brighten the Global ‘Gold Standard’ for Drug Research and Development, 45 Cornell Int’l L.J. 185, 197 (2012); Funda Meric-Bernstam & Gordon B. Mills, Overcoming Implementation Challenges of Personalized Cancer Therapy, 9 Nat. Rev. Clin. Oncol. 542, 545 (2012).
Meric-Bernstam & Mills, supra note 100, at 545.
An in-depth discussion of SSRD is beyond the scope of this paper. See Malinowski & Gautreaux, supra note 100.
In vitro diagnostic tests are those reagents, instruments, and systems intended for use in the diagnosis of disease or other conditions, including a determination of the state of health, in order to cure, mitigate, treat, or prevent disease or its sequelae, 21 C.F.R. § 809.3 (1980). They are defined as any medical device which is a reagent, reagent product, calibrator, control material, kit, instrument, apparatus, equipment, or system, whether used alone or in combination, intended by the manufacturer to be used in vitro for the examination of specimens, including blood and tissue donations, derived from the human body. The label of in vitro diagnostics must state that the product is ‘For In Vitro Diagnostic Use’, Council Directive 98/79, 1998 O.J. (L 331) (EC). Labels for in vitro diagnostics must also include the intended use or uses of the diagnostic and a statement of warnings or precautions and any other warnings appropriate to user hazards. Note that labels of in vitro diagnostics are not required to identify specific therapeutic products, unlike companion diagnostics (Food and Drug Administration (2014)), In Vitro Diagnostic Device Labeling Requirements, Fda, http://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/Overview/DeviceLabeling/InVitroDiagnosticDeviceLabelingRequirements/ (accessed July 16, 2014).
Pharmacogentics is the study of interindividual variations in whole-genome or candidate gene single-nucleotide polymorphism maps, haplotype markers, and alterations in gene expression or inactivation that might be correlated with pharmacological function and therapeutic response. A pharmacogenetic test is, therefore, a genetic test that predicts pharmacological function or therapeutic response in a patient, Lesko & Woodcock, supra note 51, at 763.
U.S. Food & Drug Administration, In Vitro Companion Diagnostic Devices: Guidance for Industry and Food and Drug Administration Staff (Aug. 6, 2014), http://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm262327.pdf.
Amit Agarwal et al., The Current and Future State of Companion Diagnostics, 8 Pharmacogenomics & Pers. Med. 99, 100 (2015).
Blankstein, supra note 70, at 283; Abdel-Baset Halim, The Biggest Challenges Currently Facing Companion Diagnostic Advancement, 14 Expert Rev. Mol. Diagn. 27, 33 (2014); Horgan et al., supra note 99, at 289; Koelsch et al., supra note 47, at 92.
Avery, supra note 7, at 56; Carden et al., supra note 92, at 90; Joshua P. Cohen, Overcoming Regulatory and Economic Challenges Facing Pharmacogenomics, 29 New Biotechnol. 751, 753 (2012); de Bono & Ashworth, supra note 73, at 546; Patricia A. Deverka, Economic Opportunities and Challenges for Pharmacogenomics, 50 Ann. Rev. Pharmacol. & Toxicol. 423, 427 (2010); Elizabeth Drucker & Kurt Krapfenbauer, Pitfalls and Limitations in Translation from Biomarker Discovery to Clinical Utility in Predictive and Personalised Medicine, 4 EMPA J. 7, 10 (2013); Eichler et al., supra note 80 at 502 ; Faulkner et al., supra note 92, at 1167; Joly et al., supra note 58, at 17.
Mittra & Tait, supra note 22, at 714.
Koelsch et al., supra note 47, at 94.
Joly et al., supra note 58, at 17; Jan Trøst Jørgensen, Companion Diagnostics and the Drug-Diagnostic Codevelopment Model, 73 Drug Dev. Res. 390 (2012); Jørgensen, supra note 80, at 65; Christine Leopold et al., Personalised Medicine as a Challenge for Public Pricing and Reimbursement Authorities – A Survey Among 27 European Countries on the Example of Trastuzumab, 113 Health Pol’y 313, 314 (2013); Meadows et al., supra note 78, at 10; Tracy Merlin et al., Assessing Personalized Medicines in Australia: A National Framework for Reviewing Codependent Technologies, 33 Med. Dec. Making 333, 340 (2013); Krishna Prasad & Alasdair Breckenridge, Pharmacogenomics: A New Clinical or Regulatory Paradigm? European Experiences of Pharmacogenomics in Drug Regulation and Regulatory Initiatives, 16 Drug Discov. Today 867, 870–71 (2011); Richard Simon, Clinical Trial Designs for Evaluating the Medical Utility of Prognostic and Predictive Biomarkers in Oncology, 7 Pers. Med. 33, 47 (2010).
Jørgensen, supra note 80, at 65; Jørgensen, supra note 111, at 392.
Milne & Zuckerman, supra note 53, at 542; Adrian M. Senderowicz & Otmar Pfaff, Similarities and Differences in the Oncology Drug Approval Process between FDA and European Union with Emphasis on In Vitro Companion Diagnostics, 20 Clin. Cancer Res. 1445, 1448 (2014).
Blankstein, supra note 70, at 283; Special Report: Companion Diagnostics–Example of BRAF Gene Mutation Testing to Select Patients with Melanoma for Treatment with BRAF Kinase Inhibitors, 26 Tech. Eval. Center Assess. Program Exec. Summ. 1, 12 (2011).
Milne & Zuckerman, supra note 53, at 542; Mittra & Tait, supra note 22, at 716.
U.S. Food & Drug Administration, About the Center for Drug Evaluation and Researchhttp://www.fda.gov/AboutFDA/CentersOffices/OfficeofMedicalProductsandTobacco/CDER/ (accessed Dec. 9, 2014).
U.S. Food & Drug Administration, About the Center for Biologics Evaluation and Research, http://www.fda.gov/AboutFDA/CentersOffices/OfficeofMedicalProductsandTobacco/CBER/ (accessed Dec. 9, 2014).
U.S. Food & Drug Administration, Transfer of Therapeutic Products to the Center for Drug Evaluation and Research (CDER), http://www.fda.gov/AboutFDA/CentersOffices/OfficeofMedicalProductsandTobacco/CBER/ucm133463.htm (accessed Apr. 15, 2015).
U.S. Food & Drug Administration, Overview of Device Regulation, http://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/Overview/ (accessed Aug. 14, 2015).
Blankstein, supra note 70, at 283.
Eric Bender, Catch-22 for Cancer Tests, 3 Cancer Discov. 1090, 1090 (2013); Id.; F. Ciardiello et al., Delivering Precision Medicine in Oncology Today and in Future—The Promise and Challenges of Personalised Cancer Medicine: A Position Paper by the European Society For Medical Oncology (ESMO), 25 Ann. Oncol. 1673, 1676 (2014); Hudson & Orviska, supra note 45, at 844; Joly et al., supra note 58, at 20; Y.W. Francis Lam, Scientific Challenges and Implementation Barriers to Translation of Pharmacogenomics in Clinical Practice, 2013 ISRN Pharmacol. 1, 7 (2013); Meric-Bernstam & Mills, supra note 100, at 547; Milne & Zuckerman, supra note 53 at 544; David R. Parkinson et al., Making Personalized Cancer Medicine a Reality: Challenges and Opportunities in the Development of Biomarkers and Companion Diagnostics, 18 Clin. Cancer Res. 619, 620 (2012); Pignatti et al., supra note 61, at 1466; Elizabeth A. Punnoose & Mark R. Lackner, Challenges and Opportunities in the Use of CTCs for Companion Diagnostic Development, 195 Recent Results Cancer Res. 241, 248 (2012); Donald R.J. Singer & John Watkins, Using Companion and Coupled Diagnostics Within Strategy to Personalize Targeted Medicines, 9 Pers. Med. 751, 753 (2012); Janette Thomas et al., Companion Diagnostics: Emerging Strategies and Issues in Pharmaceutical Development, 12 Expert Rev. Mol. Diagn. 561, 561 (2014); Mark R. Trusheim et al., Uncertain Prognosis for High-Quality Diagnostics: Clinical Challenges, Economic Barriers and Needed Reforms, 14 Pharmacogenomics 325, 327 (2013).
Pignatti et al., supra note 61, at 1466.
For example: Talazoparib (BioMarin) and myChoice HRD (Myriad Genetics); PRAME ASCI (astuprotimut-r) (GlaxoSmithKline), and a qPCR-based diagnostic assay (Abbott Molecular); PC-MAb (Athera Biotechnologies/Boehringer Ingelheim) and CVDefine Kit (Electra-Box Diagnostica).
In Vitro Companion Diagnostic Devices: Guidance for Industry and Food and Drug Administration Staff, supra note 105.
Koelsch et al., supra note 47, at 96; Eunice Y. Lee & Hsin-Chieh Jennifer Shen, Regulatory Considerations for Companion Diagnostic Devices, 9 Biomark. Med. 67, 69 (2015); Yann Joly et al., Diagnostic Testing for Vaccinomics: Is the Regulatory Approval Framework Adequate? A Comparison of Canada, the United States, and Europe, 15 OMICS 597, 604 (2011); Mittra & Tait, supra note 22, at 711; Adrian Thomas et al., Comparative Effectiveness, Personalized Medicine and Innovation: The Path Forward, 28 PharmacoEconomics 923, 925 (2010).
Blankstein, supra note 70, at 310; Cohen, supra note 108, at 755; Singer & Watkins, supra note 121, at 755.
Avery, supra note 7, at 57; Cohen, supra note 108, at 753; Fridlyand et al., supra note 69, at 631; Mittra & Tait, supra note 22, at 716.
Joly et al., supra note 125, at 604.
Avery, supra note 7, at 57; Cohen, supra note 108, at 753.
Blankstein, supra note 70, at 282; Rianne M.F. van Schie et al., Implementation of Pharmacogenetics in Clinical Practice Is Challenging, 12 Pharmacogenomics 1231, 1232 (2011).
Blankstein, supra note 70, at 282; Prasad & Brekenridge, supra note 111, at, 871; Samuya Pant et al., Navigating the Rapids: The Development of Regulated Next-Generation Sequencing-Based Clinical Trial Assays and Companion Diagnostics, 4 Front. Oncol. 1, 13 (2014); Apostolia M. Tsimberidou et al., Strategies to Overcome Clinical, Regulatory, and Financial Challenges in the Implementation of Personalized Medicine, 2013 Am. Soc’y Clin. Oncol. 118, 124 (2013); Charlie Schmidt, Challenges Ahead for Companion Diagnostics, 104 J. Nat’l Cancer Inst. 14, 14 (2012).
Kathy L. Hudson, Genomics, Health Care, and Society, 365 New Eng. J. Med. 1033, 1036 (2011).
Blankstein, supra note 70, at 310; Pant et al., supra note 131, at 13.
Blankstein, supra note 70, at 307; Mansfield, supra note 59, at 1455.
Mansfield, supra note 59, at 1455.
Id.
Singer & Watkins, supra note 121, at 751.
Id.
U.S. Food & Drug Administration, Paving the Way for Personalized Medicine: FDA’s Role in a New Era of Medical Product Development (Oct. 2013), http://www.fda.gov/downloads/ScienceResearch/SpecialTopics/PersonalizedMedicine/UCM372421.pdf.
Id.
Id.
Palmer, supra note 23, at 476; Piehl, supra note 42, at 63-64; Serra J. Schlanger, Putting Together the Pieces: Recent Proposals to Fill in the Genetic Testing Regulatory Puzzle, 21 Ann. Health L. 384, 388 (2012); Spector-Bagdady & Pike, supra note 23, at 691; Trevor Woodage, Gatekeepers and Goalposts: The Need for a New Regulatory Paradigm for Whole Genome Sequence Results, 11 Nw. J. Tech & Intell. Prop. 1, 3 (2012).
National Human Genome Research Institute, Regulation of Genetic Tests, http://www.genome.gov/10002335 (accessed June 21, 2016).
Id.
Id.
Id.
Blankstein, supra note 70, at 284; Joly et al., supra note 58, at 13-14; Meadows et al., supra note 78, at 9; McCarthy et al., supra note 46, at 199; O’Kane, supra note 80, at 746; Parkinson et al., supra note 121, at 621; Prebula, supra note 23, at 370; Charles Schmidt, Larger Companies Dominate Cancer Companion Diagnostic Approvals, 29 Nat. Biotechnol. 955, 956 (2011); Special Report: Companion Diagnostics–Example of BRAF Gene Mutation Testing to Select Patients with Melanoma for Treatment with BRAF Kinase Inhibitors, 26 Tech Eval. Center Assess. Program Exec. Summ. 1, 8 (2011).
Joly et al., supra note 58, at 19; Jaimy Lee, Missing the Target? Personalized Medicine Advances, but Questions Remain on Outcomes, Cost, 43 Mod. Healthcare 38, 40 (2013); Meadows et al., supra note 78, at 19; Conti et al., supra note 45, at 330; Faulkner et al., supra note 92, at 1164; Nelson, supra note 45, at 855; Pant et al., supra note 131, at 16; Piehl, supra note 42, at 71-73; Ramsey et al., supra note 45, at 2263; Charles L. Sawyers & Laura J. van ‘t Veer, Reliable and Effective Diagnostics Are Keys to Accelerating Personalized Cancer Medicine and Transforming Cancer Care: A Policy Statement from the American Association for Cancer Research, 20 Clin. Cancer Res. 4978, 4979 (2014); Schlanger, supra note 142, at 386-388; Schmidt, supra note 131, at 15; Yoshiaki Tazawa, Perspective for the Development of Companion Diagnostics and Regulatory Landscape to Encourage Personalized Medicine in Japan, 23 Breast Cancer 19, 23 (2016); Winther & Jørgensen, supra note 62, at 373; Wistuba et al., supra note 55, at 140; Woodage, supra note 142, at 4–5.
21 U.S.C. § 301–92 (1976).
Overview of Device Regulation, supra note 119.
U.S. Food & Drug Administration, Framework for Regulatory Oversight of Laboratory Developed Tests (LDTs) (Oct. 3, 2014), http://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm416685.pdf.
Regulation of Genetic Tests, supra note 143.
Clinical Laboratory Improvement Amendments of 1988, Pub. L. No. 100-578, 102 Stat. 2903 (1988).
Regulation of Genetic Tests, supra note 143.
Bin Chen et al., Good Laboratory Practices for Molecular Genetic Testing for Heritable Diseases and Conditions, 58 MMWR Recomm. & Rep. 1, 2 (2009).
Stephanie Bair, Direct-to-Consumer Credit Genetic Testing: Learning from the Past and Looking Toward the Future, 67 Food & Drug L.J. 413, 413 (2012); Michelle D. Irick, Age of an Information Revolution: The Direct-to-Consumer Genetic Testing Industry and the Need for a Holistic Regulatory Approach, 49 San Diego L. Rev. 279, 284 (2012); McCarthy et al., supra note 46, at 199; Piehl, supra note 42, at 64; van El & Cornel, supra note 80, at 378; Vijverberg et al., supra note 80, at 251.
Palmer, supra note 23, at 476; Kathryn Schleckser, Physician Participation in Direct-to-Consumer Genetic Testing: Pragmatism or Paternalism, 26 Harv. J.L. & Tech. 695, 719–20 (2013); Woodage, supra note 142, at 9.
Palmer, supra note 23, at 489; Piehl, supra note 42, at 77; Schlanger, supra note 142, at 388; Spector-Bagdady & Pike, supra note 23, at 695; Woodage, supra note 142, at 6; United States Government Accountability Office, Nutrigenetic Testing Tests Purchased from Four Web Sites Mislead Consumers (July 27, 2006), http://www.gao.gov/assets/120/114612.pdf.
Federal Trade Commission, Direct-to-Consumer Genetic Tests (Jan. 2014), http://www.consumer.ftc.gov/articles/0166-direct-consumer-genetic-tests ; 15 U.S.C. § 45a.
Palmer, supra note 23, at 490; Piehl, supra note 42, at 84-85; Schleckser et al., supra note 157, at 720-22; Spector-Bagdady & Pike, supra note 23, at 705; Woodage, supra note 142, at 5.
In 2013, the FTC took its first actions to protect consumers of genetic tests. The FTC filed charges against two companies in January (Genelink, Inc. and its former subsidiary foru International Corporation) and against a third company in June (L’Oreal USA, Inc.) for ‘purported personalized genomics products’. The charges were related to the marketing of nutrigenetic and dermagenetic products. The matters against the first two companies were resolved by settlement approved on May 12, 2014. The FTC announced a proposed settlement of the charges against L’Oreal on June 30, 2014.
Palmer, supra note 23, at 489; Piehl, supra note 42, at 77; Schlanger, supra note 142, at 388; Spector-Bagdady & Pike, supra note 23, at 691; Woodage, supra note 142, at 6.
Irick, supra note 156, at, 286; Schleckser et al., supra note 157, at 705–6; Palmer, supra note 23, at 476.
Helen C. Dick, Risk and Responsibility: State Regulation and Enforcement of the Direct-to-Consumer Genetic Testing Industry, 6 St. Louis U. J. Health L. Pol’y 167, 177 (2012); Piehl, supra note 42, at 86; Spector-Bagdady & Pike, supra note 23, at 718.
Bair, supra note 156, at 431–32; Dick, supra note 164, at 194; Irick, supra note 156, at 298; Palmer, supra note 23, at 523; Piehl, supra note 42, at 93; Schleckser et al., supra note 157, at 713; Spector-Bagdady & Pike, supra note 23, at 715; Woodage, supra note 142, at 5.
Euan A. Ashley et al., Genetics and Cardiovascular Disease: A Policy Statement from the American Heart Association, 126 Circulation 142, 145 (2012); Bair, supra note 156, at 413-14; Pascale Bourret et al., Regulating Diagnosis in Post-Genomic Medicine: Re-Aligning Clinical Judgment?, 73 Soc. Sci. & Med. 816, 822 (1982); Evans & Watson, supra note 23, at 669; Joseph D. Khoury & Daniel V. T. Catenacci, Next-Generation Companion Diagnostics: Promises, Challenges, and Solutions, 139 Arch. Pathol. & Lab. Med. 11, 12 (2015); Pant et al., supra note 131, at 15; Schlanger, supra note 142, at 398; Spector-Bagdady & Pike, supra note 23, at 703; Mark E. Robson et al., American Society of Clinical Oncology Policy Statement Update: Genetic and Genomic Testing for Cancer Susceptibility, 28 J. Clin. Oncol. 893, 898 (2010); Mark A. Rothstein, Currents in Contemporary Bioethics: The Case Against Precipitous, Population-Wide, Whole-Genome Sequencing, 40 J. L. Med. & Ethics 682, 684 (2012).
Dick, supra note 164, at 198; Piehl, supra note 42, at 91; Spector-Bagdady & Pike, supra note 23, at 694.
Bair, supra note 156, at 416; Lawrence J. Lesko & Issam Zineh, DNA, Drugs and Chariots: On a Decade of Pharmacogenomics at the US FDA, 11 Pharmacogenomics 507, 510 (2010); Schlanger, supra note 142, at 398; Spector-Bagdady & Pike, supra note 23, at 703.
Bair, supra note 156, at 429–30.
Lesko & Zineh, supra note 168, at 510; Schlanger, supra note 142, at 398–99.
Bair, supra note 156, at 428; Spector-Bagdady & Pike, supra note 23, at 703.
Piehl, supra note 42, at 93; Schlanger, supra note 142, at 399; Spector-Bagdady & Pike, supra note 23, at 743; Woodage, supra note 142, at 7.
Bair, supra note 156, at 427; Irick, supra note 156, at 286; McCarthy et al., supra note 46, at 199; Piehl, supra note 42, at 63; van El & Cornel, supra note 80, at 378; Vijverberg et al., supra note 80, at 251.
Conti et al., supra note 45, at 330; Faulkner et al., supra note 92, at 1164; Joly et al., supra note 58, at 14; Lee, supra note 148, at 38; Meadows et al., supra note 78, at 9; Nelson, supra note 45, at 855; Pant et al., supra note 131, at 16; Piehl, supra note 42, at 72; Ramsey et al., supra note 45, at 2263; Sawyers & van ’t Veer, supra note 148, at 4979; Schlanger, supra note 142, at 387-389; Schmidt, supra note 131, at 15; Tazawa, supra note 148, at 24; Winther & Jørgensen, supra note 62, at 373; Wistuba et al., supra note 55, at 140; Woodage, supra note 142, at 19.
National Institute of Health, U.S. System of Oversight of Genetic Testing: A Response to the Charge of the Secretary of Health and Human Services (Apr. 2008), http://osp.od.nih.gov/sites/default/files/SACGHS_oversight_report.pdf.
21 C.F.R § 814.102a(5).
Framework for Regulatory Oversight of Laboratory Developed Tests (LDTs), supra note 151.
Alan Mertz, Re: Framework for Regulatory Oversight of Laboratory Developed Tests; Draft Guidance for Industry, Food and Drug Administration Staff, and Clinical Laboratories, ACLA (Feb. 2, 2015), http://www.acla.com/wp-content/uploads/2015/02/ACLA-Comments-to-FDA-on-LDT-Draft-Guidance-2015.02.021.pdf.
Jamie K. Wolszon & Jeffrey N. Gibbs, FDA Plans to Issue Final LDT Framework in 2016; Subcommittee Members, CMS and FDA Officials Critique Proposed Legislative Approach that Would Give CMS LDT Premarket Review Authority, FDA Law Blog (Dec. 20, 2015), http://www.fdalawblog.net/fda_law_blog_hyman_phelps/2015/12/fda-plans-to-issue-final-ldt-framework-in-2016-subcommittee-members-cms-and-fda-officials-critique-p.html.
FDA, Discussion Paper on Laboratory Developed Tests (LDTs) (2017), https://www.fda.gov/downloads/medicaldevices/productsandmedicalprocedures/invitrodiagnostics/laboratorydevelopedtests/ucm536965.pdf.
Id.
Id.
FDA, Draft guidance: Use of Standards in FDA Regulatory Oversight Of Next Generation Sequencing (NGS)-Based In Vitro Diagnostics (IVD) Used For Diagnosing Germline Diseases (2016), http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/fGuidanceDocuments/UCM509838.pdf (Accessed Feb. 5, 2017); FDA, Draft guidance: Use Of Public Human Genetic Variant Databases To Support Clinical Validity For Next Generation Sequencing (NGS)-based In Vitro Diagnostics (2016), https://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM509837.pdf.
Id.
Draft guidance: Use of Standards in FDA Regulatory Oversight Of Next Generation Sequencing (NGS)-Based In Vitro Diagnostics (IVD) Used For Diagnosing Germline Diseases, supra note 183.
Draft guidance: Use Of Public Human Genetic Variant Databases To Support Clinical Validity For Next Generation Sequencing (NGS)-based In Vitro Diagnostics, supra note 183.
Dick, supra note 164, at 198; Schlanger, supra note 142, at 396; Spector-Bagdady & Pike, supra note 23, at 694.
GTR: Genetic Test Registry, NCBI, http://www.ncbi.nlm.nih.gov/gtr/ (accessed July 20, 2016).
Schlanger, supra note 142, at 402.
See, Dick, supra note 164.
Id. at 177; Piehl, supra note 42, at 83; Spector-Bagdady & Pike, supra note 23, at 718.
Piehl, supra note 42, at 81.
Dick, supra note 164, at 194; Irick, supra note 156, at 298; Spector-Bagdady & Pike, supra note 23, at 743; Woodage, supra note 142, at 5.
Dick, supra note 164, at 198; Schleckser et al., supra note 157, at 709.
Alberto Gutierrez, Warning Letter, FDA, http://www.fda.gov/ICECI/EnforcementActions/WarningLetters/2013/ucm376296.htm (accessed Sept. 24, 2015). On Feb. 19, 2015, the FDA announced that it had granted 23andMe permission to market its test for Bloom Syndrome (an autosomal recessive disease that can be passed on by asymptotic carriers), but not the broader genetic screen, Robert Hof, In Big Shift, FDA Plans To Let 23andMe Market Genetic Tests To Consumers, Forbes (Feb. 19, 2015), http://www.forbes.com/sites/roberthof/2015/02/19/in-big-shift-fda-plans-to-let-23andme-market-genetic-tests-to-consumers/.
23AndMe, http://mediacenter.23andme.com/en-ca/ (accessed Aug. 2, 2016).
James L. Woods, Document Number GEN1500296, FDA (Nov. 2, 2015), http://www.fda.gov/downloads/MedicalDevices/ResourcesforYou/Industry/UCM471784.pdf; James L. Woods, Document Number GEN 1500806, FDA (Nov 4, 2015), http://www.fda.gov/downloads/MedicalDevices/ResourcesforYou/Industry/UCM471788.pdf; James L. Woods, Document Number GEN1500800, FDA (Nov. 2, 2015), http://www.fda.gov/downloads/MedicalDevices/ResourcesforYou/Industry/UCM471785.pdf; Michael Mezher, FDA Warns Three Companies Over DTC Genetic Tests, RAPS (Nov. 9, 2015), http://www.raps.org/Regulatory-Focus/News/2015/11/09/23563/FDA-Warns-Three-Companies-Over-DTC-Genetic-Tests/.
U.S. Food & Drug Administration, FDA Allows Marketing of First Direct-to-Consumer Tests That Provide Genetic Risk Information for Certain Conditions (Apr. 6, 2017) https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm551185.htm.
Id.
Id.
Id.
Irick, supra note 156, at 292.
Palmer, supra note 23, at 523.
In the fall of 2007, Congress passed the FDA Amendments Act, mandating FDA to establish an active surveillance system for monitoring drugs by using electronic data from healthcare information holders. The FDA’s response to the mandate is the Sentinel Initiative. Its goal is to build and implement a new active surveillance system that will eventually be used to monitor all FDA-regulated products. The FDA has used administrative and insurance claims databases to investigate safety questions about agency-regulated products, but generally it has only worked with one particular healthcare system at a time to evaluate a given safety issue. Its current goal is to create a linked, sustainable system, which the FDA calls the Sentinel System, that will draw on existing automated healthcare data from multiple sources to actively monitor the safety of medical products continuously and in real-time. After a successful pilot program, Mini-Sentinel, the FDA activated the Sentinel System in February 2016; U.S. Food & Drug Administration, FDA’s Sentinel Initiative – Background, http://www.fda.gov/Safety/FDAsSentinelInitiative/ucm149340.htm (accessed June 27, 2016).
Piehl, supra note 42, at 90.
Id. at 81.
Id. at 93.
Judy G. Russell, NY and MD Limits on 23andMe, Legal Geneol. (Dec. 23, 2012), http://www.legalgenealogist.com/2012/12/23/ny-and-md-limits-on-23andme/#fn-4644-6; Karen L. Nickel, Notice to Cease and Desist Performing Genetic Testing Without Licensure or Physician Order, State of California Department of Public Health (June 9, 2008), http://www.wired.com/images_blogs/wiredscience/files/madrigal.PDF . California Business and Professions Code Section 1288 provides that ‘[a]ny person conducting or operating a clinical laboratory may accept assignments for tests only from and make reports only to persons licensed under the provisions of law relating to the healing arts or their representatives’ Cal. Bus. & Prof Code § 1288 (2012); Dick, supra note 164, at 177.
Palmer, supra note 23, at 483; Rothstein, supra note 166, at 684; Rebecca Suarez, Breaching Doctor-Patient Confidentiality: Confusion Among Physicians About Involuntary Disclosure of Genetic Information, 21 S. Cal. Interdisc. L.J. 491, 519 (2012); Susan M. Wolf, The Role of Law in the Debate over Return of Research Results and Incidental Findings: The Challenge of Developing Law for Translational Science, 13 Minn. J.L. Sci. & Tech. 435, 444 (2012).
Blankstein, supra note 70, at 308; Bloss et al., supra note 92, at 159; Cohen, supra note 108, at 631; Lam, supra note 121, at 11; Pant et al., supra note 131, at 13; Schilsky et al., supra note 45, at 485; Thomas et al., supra note 125, at 929; Debu Tripathy, FDA and Avastin: Crossroads in an Era of Targeted Therapies, 24 Oncology 989, 990 (2010); Tsimberidou et al., supra note 131, at 124; Vogenberg et al., supra note 75, at 630.
Varun Ahuja & Sharad Sharma, Drug Safety Testing Paradigm, Current Progress and Future Challenges: An Overview, 34 J. Appl. Toxicol. 576, 586 (2014); Awada & Zgheib, supra note 78, at 845; George P. Brownman, Special Series on Comparative Effectiveness Research Challenges to Real-World Solutions to Quality Improvement in Personalized Medicine, 30 J. Clin. Oncol. 4188, 4188 (2012); Wylie Burke et al., Essential Elements of Personalized Medicine, 32 Urol. Oncol. 193, 198 (2014); Carden et al., supra note 92, at 90; Jørgensen, supra note 80, at 63; Kenneth D. Levy et al., Prerequisites to Implementing a Pharmacogenomics Program in a Large Health-Care System, 96 Clin. Pharmacol. & Ther. 307, 307 (2014); Haridarshan N. Patel et al., Stakeholder Views on Pharmacogenomic Testing, 34 Pharmacotherapy 151, 163 (2014); Prasad & Brekenridge, supra note 111, at 869; Uehara et al., supra note 75, at 226.
Lam, supra note 121, at 11.
Glossary Definition: Phase, supra note 43.
Blankstein, supra note 70, at 284; Gibson & Lemmens, supra note 47, at 214; Laakmann, supra note 42, at 335; Malinowski et al., supra note 100, at 194; Milne & Zuckerman, supra note 53, at 542; W. Nicholson Price II, Making Do in Making Drugs: Innovation Policy & Pharmaceutical Manufacturing, 55 B.C.L. Rev. 491, 517 (2014); Sharma & Schilsky, supra note 74, at 211; Verweij et al., supra note 73, at 201.
Laakmann, supra note 42, at 337-38; Malinowski et al., supra note 100, at 194.
Otsubo et al., supra note 58, at 198; Piana et al., supra note 68, at 706; Meenu Wadhwa et al., WHO/KFDA Joint Workshop on Implementing WHO Guidelines on Evaluating Similar Biotherapeutic Products, Seoul, Republic of Korea 24–26 August, 2010, 39 Biologicals 349, 353 (2011); Warner et al., supra note 23, at 533.
Blankstein, supra note 70, at 284; Leopold et al., supra note 111, at 314; Milne & Zuckerman, supra note 53, at 543; O’Kane, supra note 80, at 747; Otsubo et al., supra note 58, at 195; Anup Patel, Tissue Banking for Research—Bench to Bedside and Back—Myth, Reality or Fast Fading Reality at the Dawn of a Personalised Healthcare Era, 12 Cell & Tissue Bank. 19, 20 (2011); Trusheim et al., supra note 47, at 827.
Blankstein, supra note 70, at 283.
Laakmann, supra note 42, at 336.
Gibson & Lemmens, supra note 47, at 214; Id. at 337-8; Milne & Zuckerman, supra note 53, at 542; Sharma & Schilsky, supra note 74, at 211; Verweij et al., supra note 73, at 201.
Meadows et al., supra note 78, at 11; Lisa M. Meckley & Peter J. Neumann, Personalized Medicine: Factors Influencing Reimbursement, 94 Health Pol’y 91, 96 (2010); Merlin et al., supra note 111, at 339; Pirmohamed, supra note 23, at 864; Thomas et al., supra note 121, at 561.
Pacanowski, supra note 58, at 294; Piana et al., supra note 68, 710; Pignatti et al., supra note 68, at 1037; Pirmohamed, supra note 23, at 864; Verweij et al., supra note 73, at 202.
Pacanowski, supra note 58, at 249.
Piana et al., supra note 68, at 710.
Chenggang Liang & Junzhi Wang, China's Perspective on Similar Biotherapeutic Products, 39 Biologicals 312, 316 (2011); Meadows et al., supra note 78, at 12; Reza Mirnezami et al., Preparing for Precision Medicine, 366 New Eng. J. Med. 489, 490 (2012); Wistuba et al., supra note 55, at 140.
Blankstein, supra note 70, at 284; Adam Falconi et al., Biomarkers and Receptor Targeted Therapies Reduce Clinical Trial Risk in Non-Small-Cell Lung Cancer, 9 J. Thorac. Oncol. 163, 168 (2014); Gibson & Lemmens, supra note 47, at 215; Joly et al., supra note 58, at 4; Myong-Jin Kim et al., Pharmacogenomics: The Regulatory Environment and Labeling Implications, in Pharmacogenomics and Personalized Medicine 55, 57 (Nadine Cohen ed., 2008); Mirnezami et al., supra note 225, at 490; Piana et al., supra note 68, at 709–10; Pignatti et al., supra note 68, at 1038 (2015); Tursz & Bernards, supra note 23, at 938; Zhiqun Zhang et al., Systems Biology and Theranostic Approach to Drug Discovery and Development to Treat Traumatic Brain Injury, 662 Methods Mol. Biol. 317, 325 (2010).
McCarthy et al., supra note 46, at 197; Uehara et al., supra note 75, at 226.
U.S. Food & Drug Administration, Postmarketing Requirements and Commitments: Reports, http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Post-marketingPhaseIVCommitments/ucm064436.htm (accessed Nov. 17, 2015).
FDA, Report to the Senate Committee on Health, Education, Labor, and Pensions and the House Committee on Energy and Commerce (2014), http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Post-marketingPhaseIVCommitments/UCM472973.pdf
Michael Mehzer, Woodcock: Drug Safety Surveillance System Ready for Full Operation, RAPS (Feb. 3, 2016), http://raps.org/Regulatory-Focus/News/2016/02/03/24248/Woodcock-Drug-Safety-Surveillance-System-Ready-for-Full-Operation/
These are set out in Code of Federal Regulations at 45 C.F.R. § 46 (a) (2009) (known as the ‘Common Rule’), and in 21 C.F.R. § 50, 56 (2015).
According to HHS regulations at 45 C.F.R. § 46.101(b)(4) (2009), the study or collection of ‘existing data’ (including stored samples, records, pathological specimens, or diagnostic specimens) is deemed exempt from the requirements of the Common Rule, if these sources are publicly available or if the information is recorded by the investigator in such a manner that subjects cannot be identified, either directly or through identifiers linked to subjects, R. Hakimian et al., 50-State Survey of Laws Regulating the Collection, Storage, and Use of Human Tissue Specimens and Associated Data for Research, National Cancer Institute (2004), http://m.acc.com/legalresources/resource.cfm?show=244011
Health Insurance Portability and Accountability Act of 1996, Pub.L. No. 104–191, 110 Stat. 1936 (1996).
Robert Gellman & Pam Dixon, The Precision Medicine Initiative and Privacy: Will Any Legal Protections Apply?, World Privacy Forum (May 18, 2016), http://www.worldprivacyforum.org/wp-content/uploads/2016/05/WPF_PrecisionMedicineInitiative_May2016_fs.pdf
Timothy Caulfield, Biobanks and Blanket Consent: The Proper Place of the Public Good and Public Perception Rationales, 18 King's L.J. 209, 213 (2007).
Bartha Maria Knoppers, Lecture, Genomics: From Persons to Populations and Back Again, 59 Genome 537, 537 (2013); Michelle L. McGowan et al., Integrating Genomics into Clinical Oncology: Ethical and Social Challenges from Proponents of Personalized Medicine, 32 Urol. Oncol. 187, 189 (2014); Charity Nofziger et al., Comment, Policies to Aid the Adoption of Personalized Medicine, 13 Nat. Rev. Drug Discov. 159, 160 (2014).
Gibson & Lemmens, supra note 47, at 216.
William K. Briggs, Freeing Archival Research from the Accidental and Overbearing IRB Regulation that Costs Human Lives, 14 J. Health Tech. L. 237, 238–39 (2014); Geetter, supra note 45, at 49 (2011); Knoppers, supra note 236, at 537; Valerie Gutmann Koch, PGTandME: Social Networking-Based Genetic Testing and the Evolving Research Model, 22 Health Matrix 33, 63 (2012); McGowan et al., supra note 236, at 189; Meric-Bernstam & Mills, supra note 100, at 545; Nofziger et al., supra 236 210, at 160; Matthew J. Piehl, The Brave New World of Genetic Biobanks: International Lessons for a Potential United States Biobank, 46 Val. U. L. Rev. 69, 77 (2011); Pablo Villoslada & Sergio Baranzini, Review, Data Integration and Systems Biology Approaches for Biomarker Discovery: Challenges and Opportunities for Multiple Sclerosis, 248 J. Neuroimmunol. 58, 59 (2012); Warner et al., supra note 23, at 533.
(NPRM) Notice of Proposed Rulemaking 2015 – Summary, National Archives and Records Administration (accessed Mar. 18, 2016), https://www.gpo.gov/fdsys/pkg/FR-2015-09-08/pdf/2015-21756.pdf
Patrick Taylor, Henrietta Lacks and the Great Health Data Giveaway, Harv. L.: Bill Health (May 19, 2016), http://blogs.harvard.edu/billofhealth/2016/05/19/henrietta-lacks-and-the-great-healthdata-giveaway/#more-18889 (accessed July 28, 2016).
Federal Policy for the Protection of Human Subjects; Final Rule, 82 Fed. Reg. 12 (Jan. 19, 2017).
Id.
Dalton et al., supra note 80, at 5992; Geetter, supra note 45, at 49; A. Harvey et al., Scientific Committee for the ESF Forward Look on Personalised Medicine for the European Citizen, France, 29 N. Biotechnol. 625, 630–31; Koch, supra note 238, at 63; William McGeveran et al., Deidentification and Reidentification in Returning Individual Findings from Biobank and Secondary Research: Regulatory Challenges and Models for Management, 13 Minn. J. L. Sci. & Tech. 485, 505–6 (2012); Sharon F. Terry, The Tension Between Policy and Practice in Returning Research Results and Incidental Findings in Genomic Biobank Research, 13 Minn. J. L. Sci. & Tech. 691, 699 (2012); Warner et al., supra note 23, 531; Wolf, supra note 205, at 442.
Mollie H. Ullman-Cullere & Jomol P. Mathew, Emerging Landscape of Genomics in the Electronic Health Record for Personalized Medicine, 32 Hum. Mutat. 512, 514 (2011); Tsimberidou et al., supra note 131, at 122. See, Simon Oxenham, Legal Confusion Threatens to Slow Data Science, 536 Nature 16, 16–17 (2017).
Kate Crawford & Jason Schultz, Big Data and Due Process: Toward a Framework to Redress Predictive Privacy Harms, 55 B.C. L. Rev. 93, 103 (2014); Geetter, supra note 45, at 43-4; Jonathan Kahn, Privatizing Biomedical Citizenship: Risk, Duty, and Potential in Circle of Pharmaceutical Life, 15 Minn. J. L. Sci. & Tech. 791, 814–5 (2014); McGeveran et al., supra note 243, at 505–6; Palmer, supra note 23, at 492; Piehl, supra note 238, at 90; Rothstein, supra note 166, at 686–7.
Dalton et al., supra note 80, at 5992; Geetter, supra note 45, at 43–4; Harvey et al., supra note 243, at 631; Koch, supra note 238, at 63; McGeveran et al., supra note 243, at 505–6; Terry, supra note 243, at 699; Tsimberidou et al., supra note 131, at 122; Warner et al., supra note 23, at 533; Wolf, supra note 205, at 442.
Hakimian et al., supra note 232.
Ashley et al., supra note 166, at 144; Conti et al., supra note 45, at 337; Kahn, supra note 245, at 813-4; Petersen et al., supra note 47, at 216; Thomas et al., supra note 125, at 926; Ullman-Cullere & Mathew, supra note 244, at 513; Vijverberg et al., supra note 80, at 247; Vogenberg et al., supra note 75, at 629.
Gellman & Dixon, supra note 234.
Taha A. Kass-Hout & Elaine Johanson, FDA Launches precision FDA to Harness the Power of Scientific Collaboration, FDA Voice (Dec. 15, 2015) http://blogs.fda.gov/fdavoice/index.php/2015/12/fda-launches-precisionfda-to-harness-the-power-of-scientific-collaboration/ (accessed July 28, 2016).
Ashley et al., supra note 166, at 144; Lynn G. Dressler, Commentary, Integrating Personalized Genomic Medicine into Routine Clinical Care: Addressing the Social and Policy Issues of Pharmacogenomic Testing, 74 N.C. Med. J. 509, 512 (2013); Andrew N. Freedman et al., Cancer Pharmacogenomics and Pharmacoepidemiology: Setting a Research Agenda to Accelerate Translation, 102 J. Nat’l Cancer Inst. 1698, 1703 (2010); Jason H. Karnes et al., Using Systems Approaches to Address Challenges for Clinical Implementation of Pharmacogenomics, 6 Wiley Interdisc. Rev. Sys. Biol. Med. 125, 126 (2014); Mirnezami et al., supra note 225, at 490.
Amal Alzu’bi, et al., Personal Genomic Information Management and Personalized Medicine Challenges, Current Solutions, and Roles of HIM Professionals, 11 Persp. Health Info. Mgmt. 1, 9 (2014); van El & Cornel, supra note 80, at 380; Vogenberg et al., supra note 75, at 629; Warner et al., supra note 23, at 535.
Anya E.R. Prince, Comprehensive Protection of Genetic Information: One Size Privacy or Property Models May Not Fit All, 79 Brook. L. Rev 175, 193 (2013).
Id.
White House, Precision Medicine Initiative: Privacy and Trust Principles (Nov. 9, 2015), https://www.whitehouse.gov/sites/default/files/microsites/finalpmiprivacyandtrustprinciples.pdf
White House, Precision Medicine Initiative: Data Security Policy Principles and Framework (May 25, 2016), https://www.whitehouse.gov/sites/whitehouse.gov/files/documents/PMI_Security_Principles_Framework_v2.pdf (accessed July 28, 2016).
Id.
National Institute of Standards and Technology, Framework for Improving Critical Infrastructure Cybersecurity (June 9, 2016), http://www.nist.gov/cyberframework/upload/cybersecurity-framework-021214.pdf
National Institutes for Health, NIH funds biobank to support Precision Medicine Initiative Cohort Program (May 26, 2016) https://www.nih.gov/news-events/news-releases/nih-funds-biobank-support-precision-medicine-initiative-cohort-program (accessed July 28, 3016).
Eichler et al., supra note 80, at 504; Sean X. Hu et al., Market Watch: Defining and Quantifying the Use of Personalized Medicines, 12 Nat. Rev. Drug Discov. 896, 897 (2013); Frank Pasquale, Grand Bargains for Big Data: The Emerging Law of Health Information, 72 Md. L. Rev. 382, 743 (2013).
Roger D. Klein, Opinion, Legal Developments and Practical Implications of Gene Patenting on Targeted Drug Discovery and Development, 87 Clin. Pharmacol. & Ther. 633, 633 (2010); Scott Parker & Ben Hall, Review, Patenting Personalized Medicines in the UK, Europe and USA, 3 Pharm. Pat. Anal. 163, 164 (2014); Price II, supra note 214, at 523.
Aura Bertoni, Open Source Models in Biomedicine: Workable Complementary Flexibilities Within the Patent System, 14 Wake For. J. Bus. & Intell. Prop. L. 126 (2013); Devlin, supra note 42, at 132; Kahn, supra note 245, at 877 (2014); Mini Kapoor, Comment, Proposal for Resolution to Challenges Posed by DNA Sequence Patents on the Development of Multiplex Genetic Tests, 49 Hous. L. Rev. 131, 133–4 (2012); Sapna Kumar, Life, Liberty and the Pursuit of Genetic Information, 65 Ala. L. Rev. 625, 629 (2013); William Lesser, Myriad & Prometheus, Laws & Products of Nature: Are the Courts Considering an Economic Non-Statutory Subject Matter Exclusion?, 53 IDEA: Intell. Prop. L. Rev. 173, 218 (2013); Yahong Li, Intellectual Property and Innovation: A Case Study of High-Tech Industries in China, 13 Or. L. Rev. 263, 275 (2011); Kali N. Murray & Esther van Zimmeren, Dynamic Patent Governance in Europe and the United States: The Myriad Example, 19 Cardozo J Int’l & Comp. L. 287, 291 (2011); W. Nicholson Price II, Unblocked Future: Why Gene Patents Won’t Hinder Whole-Genome Sequencing and Personalized Medicine, 33 Cardozo L. Rev. 1691, 1727 (2012); Robertson, supra note 42, at 388; Tiana Leia Russell, Unlocking the Genome: The Legal Case Against Genetic Diagnostic Patents, 16 Marq. Intell. Prop L. Rev. 81, 111 (2012); Spector-Bagdady & Pike, supra note 23, 723–4 (2014); Melissa F. Wasserman, The PTO’s Asymmetric Incentives: Pressure to Expand Substantive Patent Law, 72 Ohio St. L.J. 1, 11 (2011).
Murray & van Zimmeren, supra note 262, at 291.
Kapoor, supra note 262, at 133; Kumar, supra note 262, at 637; Robertson, supra note 42, at 386; Adriane Scola, Uncommon Genes, Unpatentable Subject Matter, 34 Seattle U. L. Rev. 909, 919 (2011).
Christopher M. Holman, Unpredictability in Patent Law and its Effect on Pharmaceutical Innovation, 76 Mo. L. Rev. 645, 651 (2011); Julia Carbone et al., Commentary, DNA patents and Diagnostics: Not a Pretty Picture, 28 Nat. Biotechnol. 784, 785 (2010).
Holman, supra note 265, at 645; Klein, supra note 261, at 635; Parker & Hall, supra note 261, at 168.
Holman, supra note 265, at 645.
Klein, supra note 261, at 633; Parker & Hall, supra note 261, at 164.
Bertoni, supra note 262, at 132; Devlin, supra note 42, at 70; Kahn, supra note 245, at 877; Kapoor, supra note 262, at 147; Kumar, supra note 262, at 629; Lesser, supra note 262, at 217-8; Michael A. Heller & Rebecca S. Eisenberg, Can Patents Deter Innovation? The Anticommons in Biomedical Research, 280 Science 698, 699 (1998); Li, supra note 262, at 275; Murray & van Zimmeren, supra note 262, at 291; Price II, supra note 262, at 1727; Robertson, supra note 42, at 386; Russell, supra note 262, at 111; Spector-Bagdady & Pike, supra note 23, at 723; Wasserman, supra note 262, at 11.
Gibson & Lemmens, supra note 47, at 206; Milne & Zuckerman, supra note 53, at 543; Miossec et al., supra note 47, at 1717.
Munir Pirmohamed & Dyfrig A. Hughes, Comment, Pharmacogenetic Tests: The Need for a Level Playing Field, 12 Nat. Rev. Drug Discov. 3, 4 (2013).
Ashley et al., supra note 166, at 143; Wylie Burke, et al., Commentary, Extending the Reach of Public Health Genomics: What Should be the Agenda for Public Health in an Era of Genome-Based and ‘Personalized’ Medicine?, 12 Genet. Med. 785, 788 (2010); Ava Caffarini, Directed to or Encompassing a Human Organism: How Section 33 of the America Invents Act May Threaten the Future of Biotechnology, 12 J. Marshall Rev. Intell. Prop. L. 768, 784 (2013); Faulkner et al., supra note 92, at 1164; E. Richard Gold, Robert Cook-Deegan & Tania Bubela, Editorial, AMP v. Myriad: A Surgical Strike on Blockbuster Business Models, 5 Sci. Transl. Med. 192ed9, 2 (2013); Lesser, supra note 262, at 218; Price II, supra note 262, at 1727.
Caffarini, supra note 272, at 784; Lesser, supra note 262, at 175; Price II, supra note 262, at 1727. Note: in Canada, see Novartis Pharmaceuticals Canada Inc. v. Cobalt Pharmaceuticals Co., 236 A.C.W.S. (3d) 1001, 2014 FCA 17 (Can.), and Europe, The European Patent Convention art. 54 (Oct. 5, 1973), http://documents.epo.org/projects/babylon/eponet.nsf/0/00E0CD7FD461C0D5C1257C060050C376/$File/EPC_15th_edition_2013.pdf, medical methods are non-patentable.
Mayo Collaborative Servs. v. Prometheus Labs. Inc., 566 U.S. ___ (2012) (naturally occurring molecules); Ass’n for Molecular Pathology v. Myriad Genetics, Inc., 569 U.S. ___ (2013) (methods for determining biomarker-drug dose associations).
Koelsch et al., supra note 47, at 97.
Gibson & Lemmens, supra note 47, at 206; Avery, supra note 7, at 63; Holman, supra note 26, at 693.
Michael Abramowicz, Orphan Business Models: Towards a New Form of Intellectual Property, 124 Harv. L. Rev. 1362, 1387 (2011); David C. Babaian, Adopting Pharmacogenomics and Parenting Repurposed Molecules Under the Orphan Drug Act: A Cost Dilemma, 13 Marshall Rev. Intell. Prop. L. 668, 671 (2014); Price II, supra note 214, 496-7; Robertson, supra note 42, at 397.
Blankstein, supra note 70, at 283; Cynthia Hathaway, A Patent Extension Proposal to End Underrepresentation of Women in Clinical Trials and Secure Meaningful Drug Guidance for Women, 67 Food & Drug L.J.143, 172 (2012); Meadows et al., supra note 78, at 11; Mittra & Tait, supra note 22, at 717; Parker & Hall, supra note 261, at 164; Price II, supra note 262, at 698; Price II, supra note 214, at 527–8; Trusheim et al., supra note 121, at 328; van El & Cornel, supra note 80, at 380; Michael M. Ward, Personalized Therapeutics: A Potential Threat to Health Equity, 27 J. Gen. Intern. Med. 868, 870 (2010).
Blankstein, supra note 70, at 283; Meadows et al., supra note 78, at 11; Mittra & Tait, supra note 22, at 717; Parker & Hall, supra note 261, at 164; Trusheim et al., supra note 121, at 328; van El & Cornel, supra note 80, at 380; Ward, supra note 278, at 870.
Trusheim et al., supra note 121, at 328.
Kumar, supra note 262, at 669–70; Li, supra note 262, at 227; Price II, supra note 262, at 1727; John L. Ryan, Unlikely Splicing - the Myriad Decision, the Genomic Research and Accessibility Act, Orphan Diseases and the Future of Antisense Drugs, 28 J. Contemp. Health L. & Pol’y 144, 175 (2011).
A clearinghouse is ‘any agency that brings together seekers and providers of goods, services or information, thus matching demand and supply’, Lori Sheremeta & E. Richard Gold, Creating a Patent Clearinghouse in Canada: A Solution to Problems of Equity and Access, 11 Health L. Rev. 17, 18–9 (2003), http://www.hli.ualberta.ca/HealthLawJournals/∼/media/hli/Publications/HLR/11-3-04sheremetpstrfrm.pdf. A patent clearinghouse ‘would administer the rights of several patent owners; authority would be granted by the patent owner to the collective to set license terms to others who would be permitted to work the patent’, Id. at 17; Price II, supra note 262, at 1727.
A patent pool is ‘the aggregation of IP rights that are the subject of cross licensing arrangements, whether transferred directly by the patentee to a licensee or through some medium, such as a joint venture set up specifically to administer the patent pool’, Sheremeta & Gold, supra note 282, at 18, http://www.hli.ualberta.ca/HealthLawJournals/∼/media/hli/Publications/HLR/11-3-04sheremetpstrfrm.pdf . Kapoor, supra note 262, at 152.
Kumar, supra note 262, at 668.
Li, supra note 262, at 276; Simone A. Rose, Semiconductor Chips, Genes and Stem Cells: New Wine for New Bottles? 38 Am. J. L. & Med. 113, 152 (2012).
Ryan, supra note 281, at 175.
U.S. Patent Law, 35 U.S.C. §§ 1 et seq.
Kumar, supra note 262, at 637; Li, supra note 262, at 277; Price II, supra note 262, at 1727; Ryan, supra note 281, at 175.
Price II, supra note 262, at 1727.
Kumar, supra note 262, at 627.
Alice Corp. Pty. Ltd. v. CLS Bank International, 134 S. Ct. 2347 (2014).
Orphan Drug Regulations, 21 C.F.R. § 316 (2013).
U.S. Food and Drug Administration, Humanitarian Device Exemption, http://www.fda.gov/medicaldevices/deviceregulationandguidance/howtomarketyourdevice/premarketsubmissions/humanitariandeviceexemption/default.htm (accessed July 27, 2016).
U.S. Food and Drug Administration, Developing Products for Rare Diseases & Conditions, http://www.fda.gov/ForIndustry/DevelopingProductsforRareDiseasesConditions/default.htm (accessed July 27, 2016).
Avery, supra note 7, at 63.
Gibson & Lemmens, supra note 47, at 207.
Babaian, supra note 277, at 671.
Rose, supra note 285, at 121.
Developing Products for Rare Diseases & Conditions, supra note 294.
U.S. Food and Drug Administration, Rare Pediatric Disease Priority Review Voucher Program (Section 529), http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResources/ucm375479.htm (accessed July 27, 2016).
Food and Drug 126 Stat. 1094 21 U.S.C. 360ff. https://www.gpo.gov/fdsys/pkg/PLAW-112publ144/pdf/PLAW-112publ144.pdf
H.R. 34, § 3051.
H.R. 34, § 3052.
Blankstein, supra note 70, at 296; Hathaway, supra note 278, at 173; Lam, supra note 121, at 11; Trusheim et al., supra note 121, at 329.
Blankstein, supra note 70, at 296.
Trusheim et al., supra note 121, at 329.
Khrisna Chinthapalli, Higher Drug Prices Will Boost Development of Personalised Medicine, Says New Report, 347 BMJ, 4549, 4549 (2013); Rana Foroohar, Why Hillary Is Right To Take On Pharma's Price Gouging, TIME, Sept. 23, 2015, http://time.com/4046041/hillary-clinton-martin-shkreli/; Andrew Pollack & Sabrina Tavernise, Valeant's Drug Price Strategy Enriches It, but Infuriates Patients and Lawmakers, New York Times, Oct. 4, 2015, http://www.nytimes.com/2015/10/05/business/valeants-drug-price-strategy-enriches-it-but-infuriates-patients-and-lawmakers.html?_r=0 (accessed July 27, 2016).
Bianca DiJulio et al., Kaiser Health Tracking Poll: October 2015, Henry J. Kissinger Found. Oct. 28, 2015 http://kff.org/health-costs/poll-finding/kaiser-health-tracking-poll-october-2015/
Juliette Cubanski & Tricia Neuman, The Facts on Medicare Spending and Financing, Henry J. Kissinger Found. July 20, 2016 http://kff.org/medicare/issue-brief/the-facts-on-medicare-spending-and-financing/
Blankstein, supra note 70, at 282; Cohen & Felix, supra note 47, at 171; Louis D, Fiore & Leonard William D’Avolio, Commentary, Detours on the Road to Personalized Medicine: Barriers to Biomarker Validation and Implementation, 306 JAMA, 1914, 1914 (2011); Fleck, supra note 47, at 202; Hans-Joerg Fugel et al., Stratified Medicine and Reimbursement Issues, 3 Front. Pharmacol. 1, 2–3 (2012); Halim, supra note 107, at 34; Le Tourneau et al., supra note 47, at 264; J. Steven Leeder et al., Conference Scene: Pediatric Pharmacogenomics and Personalized Medicine, 11 Pharmacogenomics 1691, 1692 (2010); Miossec et al., supra note 47, at 1716; Petersen et al., supra note 47, at 217; Parkinson et al., supra note 121, at 622; Carrie Printz, Healthcare Reform Affects Cancer on Many Fronts: More Coverage for Treatment and Prevention Planned, 116 Cancer 3525, 3526 (2010); Rothstein, supra note 166, at 684; Trusheim et al., supra note 47, at 823; Tursz & Bernards, supra note 23, at 938; Walk, supra note 53, at 230; Wistuba et al., supra note 55, at 140.
Montagna, supra note 45, at 18.
Babaian, supra note 277, at 675; Evans & Watson, supra note 23, at, 670; Daniel Hayes et al., Personalized Medicine: Risk Prediction, Targeted Therapies and Mobile Health Technology, 12 BMC Med 1, 3 (2014); Holman, supra note 265, at 650.
Rothstein, supra note 166, at 684.
Blankstein, supra note 70, at 282; Le Tourneau et al., supra note 47, at 264.
Parkinson et al., supra note 121, at 620; Poste et al., supra note 80, at 1518–9.
Pignatti et al., supra note 68, at 1035; Parkinson et al., supra note 121, at 622; Poste et al., supra note 80, at 511.
Piehl, supra note 42, at 76; Schlanger, supra note 142, at 389.
Ramsey et al., supra note 45, at 2262.
Conti et al., supra note 45, at 333; Faulkner et al., supra note 92, at 1164.
Deverka, supra note 108, at 434.
Id. Merlin et al., supra note 111, at 338.
Cohen, supra note 108, at 754; Conti et al., supra note 45, at 333; Faulkner et al., supra note 92, at 1167; Lam, supra note 121, at 9.
Matthew Herder, Choice Patents, 52 IDEA: Intell. Prop. L. Rev. 309, 377–8 (2011); Jones & DeSantis, supra note 45, at 525; Meckley & Neumann, supra note 221, at 94; Parkinson et al., supra note 121, at 620; Pignatti et al., supra note 61, at 1466; Poste et al., supra note 80, at 1518-9; Rumiko Shimazawa & Masayuki Ikeda, Letter to the Editor, Approval gap of Pharmacogenomic Biomarkers and in Vitro Companion Diagnostics Between the United States and Japan, 39 Brit. J. Clin. Pharmacol. 210, 210 (2014); Cheryl A. Thompson, Regulations, Economics Hindering Adoption of Personalized Medicine, 68 Am. J Health-Sys. Pharmacy 372, 374 (2011); Issam Zineh & Shiew-Mei Huang, Biomarkers in Drug Development and Regulation: A Paradigm for Clinical Implementation of Personalized Medicine, 5 Biomark. Med. 705, 706 (2011).
Bender, supra note 121, at 1090; Meadows et al., supra note 78, at 10: Parker & Hall, supra note 261, at 164; Parkinson et al., supra note 121, at 622; Trusheim et al., supra note 47, at 823; Walk, supra note 53, at 230; Rachel Zuckerman & Christopher-Paul Milne, Market Watch: Industry Perspectives on Personalized Medicine, 11 Nat. Rev. Drug Discov. 178, 181 (2012).
Sarah K. Byron et al., The Health Technology Assessment of Companion Diagnostics: Experience of NICE, 20 Clin. Cancer Res. 1469, 1475 (2014); Ciardiello et al., supra note 121, at 1676; Merlin et al., supra note 111, at 334.
Ashley et al., supra note 166, at 150; Cohen, supra note 108, at 752; Conti et al., supra note 45, at 333; Dressler, supra note 251, at 511; Faulkner et al., supra note 92, at 1163; Merlin et al., supra note 111, at 338; Nelson, supra note 45, at 856.
Cohen & Felix, supra note 47, at 171.
Byron et al., supra note 325, at 1475; Bengt Jönsson, Technology Assessment for New Oncology Drugs, 19 Clin. Cancer Res. 6, 8 (2013).
Byron et al., supra note 325, at 1475; Conti et al., supra note 45, at 333; Faulkner et al., supra note 92, at 1164; Maggie H. Francis, Beyond Safe and Effective: The Role of the Federal Government in Supporting and Disseminating Comparative-Effectiveness Research, 21 Ann. Health L. 329, 335–6 (2012); Lam, supra note 121, at 9; Lesko & Zineh, supra note 168, at 510; Meadows et al., supra note 78, at 9; Merlin et al., supra note 111, at 334; Tibor van Rooij et al., Personalized Medicine Policy Challenges: Measuring Clinical Utility at Point of Care, 12 Expert Rev. Pharmacoecon. Outcomes Res. 289, 291 (2012); Richard S. Saver, Health Care Reform's Wild Card: The Uncertain Effectiveness of Comparative Effectiveness Research, 159 U. Pa. L. Rev. 2147, 2193 (2011); Thomas et al., supra note 121, at 561; Julia R. Trosman et al., Health Technology Assessment and Private Payers' Coverage of Personalized Medicine, 7 Am. J. Managed Care. 18s, 22s (2011); Ioannis S. Vizirianakis, Nanomedicine and Personalized Medicine Toward the Application of Pharmacotyping in Clinical Practice to Improve Drug-Delivery Outcomes, 7 Nanomed. Nanotechnol. Biol. & Med. 11, 14 (2011); Edie E. Zusman, Review, Comparative Effectiveness Research: Policy and Politics, 33 Neurosurg. Focus 6, 10 (2012).
Singer & Watkins, supra note 121, at 754.
Byron et al., supra note 325, at 1470.
Bender, supra note 121, at 1090; Byron et al., supra note 325, at 1475; Ciardiello et al., supra note 121, at 1676; Fiore & D’Avolio, supra note 310, at 1914; Meadows et al., supra note 78, at 10; Parker & Hall, supra note 261, at 164.
Bender, supra note 121, at 1090; Byron et al., supra note 325, at 1475; Ciardiello et al., supra note 121, at 1676; Fiore & D’Avolio, supra note 310, at 1914; Meadows et al., supra note 78, at 10; Parker & Hall, supra note 261, at 164.
Bender, supra note 121, at 1090; Pirmohamed & Hughes, supra note 271, at 4.
Cohen & Felix, supra note 47, at 171.
Jason Shafrin, Comparative Effectiveness vs. Cost Effectiveness Research, Healthcare Economist, Mar. 11, 2010, http://healthcare-economist.com/2010/03/11/comparative-effectiveness-vs-cost-effectiveness-research/
American Recovery and Reinvestment Act, H.R. 1, 111th Cong. (2009).
Milton C. Weinstein & Jonathan A. Skinner, Comparative Effectiveness and Health Care Spending — Implications for Reform, 362 New Eng. J. Med. 460, 460 (2010).
Id.
Francis, supra note 329, at 335-6; Pirmohamed & Hughes, supra note 271, at 4; Saver, supra note 329, at 2181-2; Zusman, supra note 329, at 10.
Saver, supra note 329, at 2180.
Id.
George Poste, Comment, Bring on the Biomarkers, 469 Nature 156, 157 (2011); Shimazawa & Ikeda, supra note 323, at 210.
Avery, supra note 7, at 38; Cohen, supra note 108, at 754; Dressler, supra note 251, at 511; Faulkner et al., supra note 92, at 1164; Fugel et al., supra note 310, at 2–3.
Bender, supra note 121, at 1090; Byron et al., supra note 325, at 1475; Ciardiello et al., supra note 121, at 1676; Fiore & D’Avolio, supra note 310, at 1914; Meadows et al., supra note 78, at 10; Parker & Hall, supra note 261, at 164.
Byron et al., supra note 325 at 1475; Ciardiello et al., supra note 121, at 1676; Fiore & D’Avolio, supra note 310, at 1914; Fugel et al., supra note 310, at 3; Leeder et al., supra note 310, at 1692; Meadows et al., supra note 78, at 10; Parkinson et al., supra note 121, at 622; Trusheim et al., supra note 121, 818.
Cohen & Felix, supra note 47, at 167.
Fugel et al., supra note 310, at 3.
Cohen, supra note 108, at 754; Dressler, supra note 251, at 511; Faulkner et al., supra note 92, at 1169; Fugel et al., supra note 310, at 3.
Blankstein, supra note 70, at 282.
Dressler, supra note 251, at 511.
Cohen & Felix, supra note 47, at 172; Deverka, supra note 108, at 434; Faulkner et al., supra note 92, at 1167; Thomas et al., supra note 121, at 561; Tsimberidou et al., supra note 131, at 123; Verweij et al., supra note 73, at 202.
Cohen & Felix, supra note 47, at 172.
Thomas et al., supra note 121, at 561.
Trusheim et al., supra note 121, at 329; Vogenberg et al., supra note 75, at 630; Walk, supra note 53, at 230.
Sinead Cuffe et al., Cancer Patients’ Acceptance, Understanding, and Willingness-to-Pay for Pharmacogenomic Testing, 24 Pharmacogenet. Genomics 348, 353; Fleck, supra note 47, at 205; Horgan et al., supra note 99, at 291; Joly et al., supra note 58, at 24; Pignatti et al., supra note 68, at 1040; Tazawa, supra note 148, at 22; Thomas et al., supra note 125, at 929.
Eichler et al., supra note 80, at 502; Nofziger et al., supra note 236, at 160; Pacanowski, supra note 58, at 249; Pirmohamed, supra note 23, at 864; Thomas et al., supra note 125, at 929; Walk, supra note 53, at 230.
Dick, supra note 164, at 195-6; Pignatti et al., supra note 61, at 1466; Thomas et al., supra note 125, at 929; Woodage, supra note 142, at 15-6.
Dick, supra note 164, at 195-196; Eichler et al., supra note 80, at 503; Faulkner et al., supra note 92, at 1164; Fugel et al., supra note 310, at 4; Nofziger et al., supra note 236, at 160; Pacanowski, supra note 58, at 249; Pirmohamed, supra note 23, at 864; Thomas et al., supra note 125, at 929; Walk, supra note 53, at 230; Woodage, supra note 142, at 13–4.
Conti et al., supra note 45, at 336; Horgan et al., supra note 99, at 291; Nelson, supra note 45, at 856; Ramsey et al., supra note 45, at 2268; Robertson, supra note 42, at 396-7; Trusheim et al., supra note 121, at 330; Tursz & Bernards, supra note 23, at 939.
Robertson, supra note 42, at 397.
Ramsey et al., supra note 45, at 2266.
Blankstein, supra note 70, at 283; Gibson & Lemmens, supra note 47, at 213; Lesko & Zineh, supra note 168, at 511; Meadows et al., supra note 78, at 11; Sawyers & van ’t Veer, supra note 148, at 4979.
Cohen & Felix, supra note 47, at 172; Khoury, supra note 166, at 1213; Meckley & Neumann, supra note 221, at 97.
Alzu’bi et al., supra note 252, at 9.
Cohen & Felix, supra note 47, at 172.
Woodage, supra note 142, at 15–16.
Byron et al., supra note 325, at 1475; Ciardiello et al., supra note 121, at 1676; Gibson & Lemmens, supra note 47, at 213; Jönsson, supra 328 297, at 8; van Rooij, supra note 329, at 292.
For example, in Europe the Innovative Medicines Initiative is a public–private partnership between the European Union and the European Federation of Pharmaceutical Industries and Associations, in order to encourage biopharmaceutical innovation in Europe. It supports projects like the Diabetes Research on Patient Stratification project and other personalized medicine projects. See DIRECT, DIRECT—Innovative Medicines Initiative: Diabetes Research on Patient Stratification, http://www.direct-diabetes.org/imi/index.php (accessed on July 25, 2016). In Canada, another public–private partnership, the Exactis Innovation initiative ‘Personalized my Treatment’, targets barriers in clinical trial design. This initiative will create a comprehensive database of tissue samples, genomic data, and clinical data from cancer patients to match patients with PM clinical trials. See Merck Canada, $4 Million Partnership to Reduce Barriers to the Development of Personalized Medicine in Oncology, McGill Fac. Med. Electronic Newsletter (Apr. 20, 2016) http://publications.mcgill.ca/medenews/2016/04/20/4-million-partnership-to-reduce-barriers-to-the-development-of-personalized-medicine-in-oncology/ (accessed July 25, 2016).
Geoffrey S. Ginsburg, Viewpoint, Realizing the Opportunities of Genomics in Health Care, 309 JAMA 1463, 1464 (2013).
Woodage, supra note 142, at 8.
Amy P. Abernethy et al., Rapid-Learning System for Cancer Care, 28 J. Clin. Oncol. 4268, 4273 (2010); Blankstein, supra note 70, at 274; Mafalda. M. Dias et al., Exploration of the Perceptions, Barriers and Drivers of Pharmacogenomics Practice Among Hospital Pharmacists in Adelaide, South Australia, 14 Pharmacogenomics J. 235, 238 (2014); Halim, supra note 107, at 31; Maxwell J. Mehlman, Professional Power and the Standard of Care in Medicine, 44 Ariz. St. L.J. 1109, 1218 (2012); Neyarapally, supra note 42, at 506; Palmer, supra note 23, at 482-3; Pasquale, supra note 260, at 742; Piehl, supra note 238, at 93; Piehl, supra note 42, at 79; Elizabeth R. Pike et al., Finding Fault? Exploring Legal Duties to Return Incidental Findings in Genomic Research, 102 Geo. L. J. 795, 811 (2014); Pirmohamed, supra note 23, at 864; Rothstein, supra note 166, at 684; Suarez, supra note 205, at 499; Eric J. Topol, Commentary, Pharmacy Benefit Managers, Pharmacies, and Pharmacogenomic Testing: Prescription for Progress? 2 Sci. Transl. Med. 44cm22, 2 (2010); Wolf, supra note 205, at 445; Woodage, supra note 142, at 11–2.