Abstract
Tumor viruses have provided relatively simple genetic systems, that can be manipulated for understanding the molecular mechanisms of the cellular transformation process. A growing body of information in the tumor virology field provides several prospects for rationally targeted therapies. However, further research is needed to better understand the multiple mechanisms utilized by these viruses in cancer progression in order to develop therapeutic strategies. Initially viruses were believed to be associated with cancers as causative agents only in animals. It was almost half a century before the first human tumor virus, Epstein-Barr virus (EBV), was identified in 1964. Subsequently, several human tumor viruses have been identified including Kaposi sarcoma associated herpesvirus (KSHV), human papillomaviruses (HPV), hepatitis B virus (HBV), Hepatitis C virus (HCV), human T lymphotropic virus (HTLV-1) and recently identified Merkel cell polyomavirus (MCPyV). Tumor viruses are sub-categorized as either DNA viruses, which include EBV, KSHV, HPV, HBV and MCPyV or RNA viruses such as HCV and HTLV-1. Tumor-viruses induce oncogenesis through manipulating an array of different cellular pathways. These viruses initiate a series of cellular events, which lead to immortalization and proliferation of the infected cells by disrupting the mitotic checkpoint upon infection of the host cell. This is often accomplished by functional inhibition or proteasomal degradation of many tumor suppressor proteins by virally encoded gene products. The virally infected cells can either be eliminated via cell-mediated apoptosis or persist in a state of chronic infection. Importantly, the chronic persistence of infection by tumor viruses can lead to oncogenesis. This review discusses the major human tumor associated viruses and their ability to modulate numerous cell signaling pathways, which can be targeted for potential therapeutic approaches.
Keywords: EBV, HTLV-III, hepatitis B, hepatitis C, JAK/STAT, PI-3K, IRF signaling
Tumor Viruses
Epstein-Barr virus (EBV)
EBV belongs to the genus lymphocryptovirus of the human γ-herpesvirus family and infects more than 90% of the world wide adult population.1 Once a person is infected with EBV, the virus remains latent, mainly in B lymphocytes, for the lifetime of the infected host.1 The EBV genome is 184 Kb of linear, double stranded DNA, which is maintained in the nucleus as an episome via tethering to the host cell chromosome after infection.1 Primary exposure of EBV, which occurs during infancy, is usually asymptomatic. However, when a person is first infected with EBV during adolescence, it usually results in a clinical condition known as infectious mononucleosis (IM).1 EBV has powerful transforming potential for B lymphocytes in vitro and thus contributes to the development of numerous acute diseases and cancers, such as Burkitt lymphoma, nasopharyngeal carcinoma, natural killer cell lymphoma, Hodgkin disease and X-linked lymphoproliferative disease.1 Moreover, immunocompromised patients including AIDS or post-organ transplant patients, also have a high probability of obtaining EBV associated lymphomas.1
One of the biological hallmarks of EBV-cell communication is the establishment of latency.1 Three major types of latency have been described, each having its own distinct viral gene expression pattern.1 EBV latency proteins include EBV nuclear antigens (EBNA) 1, 2, 3A/3, 3B/4, 3C/6 and LP/5, and latent membrane proteins (LMP) 1, 2A and 2B.1 These proteins are expressed in type III latency, also referred to as the growth program, which is seen in AIDS associated lymphoma, post-transplant lymphoma patients and lymphoblastoid cell lines (LCLs) generated from in vitro EBV infection in primary B-cells.1 In short, four of the EBV encoded latent proteins including LMP1, EBNA2, EBNA3A and EBNA3C have been shown to be essential for B cell immortalization in vitro.1 EBNA-LP functions like a co-stimulator of EBNA-2 mediated transactivation of many cellular and viral genes shown to be critical for B cell immortalization.1 EBNA1 is essential for the maintenance and segregation of the EBV genome.1,2 LMP2A has been shown to block normal B cell receptor signaling.1 EBNA3A and 3C are also critical for B cell immortalization while EBNA3B enhances the survival of cells.1 All three EBNA3 proteins are shown to bind with RBP-Jκ/CBF1 and regulate cellular gene transcription important for transforming B-cells into immortalized LCLs.1
Kaposi sarcoma associate herpesvirus (KSHV)
KSHV also belongs to the human γ-herpesvirus family and can establish lifelong persistence in the host after primary infection similar to EBV.3,4 KSHV is etiologically linked to Kaposi sarcoma, and persists as the most common cancer in AIDS patients, in primary effusion lymphomas (PEL), and some forms of multicentric Castleman disease.3,4 Despite marked differences between KSHV and EBV, both viruses have been shown to target many similar cellular pathways, although they utilize diverse strategies to achieve the same goals. KSHV encoded lytic and latent antigens have been shown to block cell cycle regulatory checkpoints, apoptosis control machinery and importantly, the immune response regulatory mechanisms.3,4 Thus, inhibition of these cellular regulatory networks appears to be a defensive means that allows the virus to escape from innate antiviral immune responses. However, because of the overlapping nature of the innate immune system and tumor-suppressor pathways, inhibition of these regulatory networks can lead to unregulated cell proliferation and may contribute to virus-induced tumorigenesis.
There are over 90 open reading frames (ORFs) identified in KSHV genome, but only a small number of these genomes are expressed during latency,3,4 including LANA (latency-associated nuclear antigen), vCyclin, vFLIP/K13, K12/Kaposin and an miRNA cluster.3,4 One of the major latent proteins, LANA, encoded by ORF73, is a multifunctional nuclear antigen4 and functional homolog to the EBV EBNA1 protein. LANA has been shown to play a central role in deregulating various cellular functions including maintenance of the viral episome,3 degradation of the p53 and pRb tumor suppressors,3,4 transactivation of the telomerase reverse-transcriptase promoter, promotion of chromosome instability in KSHV-infected B cells,3,5 and accumulation of the intracellular domain of Notch3,6 in KSHV mediated tumorigenesis. LANA also inhibits the expression of RTA, another critical viral encoded transcriptional activator required for regulating the switch from the latent to the lytic cycle and thus, the maintenance of latency.4 Importantly, LANA tethers the viral episomal DNA to the host chromosomes, which helps in the efficient partitioning of the viral DNA in the daughter cells. Therefore, disruptions in LANA expression lead to reduction in the episomal copies, suggesting the importance of LANA in KSHV-mediated pathogenesis.3,4
Human papillomaviruses (HPV)
HPVs belong to the papovaviridae family and are the most common sexually transmitted disease in the United States.7 So far, more than 130 HPV types have been identified and subsequently classified into low- or high-risk groups according to their potential for causing cervical cancer.7 HPV and other papillomaviruses have an exceptional mechanism of infection that has evolved to limit infection to the basal cells of the stratified epithelium, the only tissue in which they replicate.7 Infection with some types of HPVs causes genital warts and some other HPV types cause cervical cancer. HPV-16 and HPV-18 are tightly associated with 70% of cervical cancers and HPV-6 and HPV-11 are linked to 90% of genital warts cases.7,8
The HPV genome is divided into long control regions (LCR), which play a role in regulating gene expression and DNA replication; open reading frames (ORF), which are involved in the expression of early gene proteins known as E1–E8; and late genes, which express the structural proteins known as L1–L2. Viral genes encode proteins responsible for replication, cellular transformation, control of viral transcription, and those necessary for the generation of viral progeny. Encoding two oncoproteins, E6 and E7, HPV establishes cancer through the ubiquitin-proteasome mediated degradation of two major tumor suppressor proteins, p53 and pRb.7 Additional studies have shown that E6 and E7 can directly interact with several host proteins and therefore further contribute to genomic instability. However, the expression of the E6 and E7 oncoproteins alone is insufficient for cellular transformation, indicating the requirement for additional genetic alterations. Greater understanding of the role of HPV antigens in cancer propagation will eventually aid in the development of antiviral treatment, as well as to unveil general mechanisms of HPV mediated oncogenesis.
Hepatitis B virus (HBV) and hepatitis C virus (HCV)
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third leading cause of cancer death worldwide.9 In spite of recent developments in the treatment for HCC, its prognosis still remains grim. Infection with hepatitis B virus (HBV) and hepatitis C virus (HCV) is the major contributor to HCC development, accounting for over 80% of all HCC globally; however, the oncogenic mechanisms of these two viruses are significantly different at the molecular level.9 Most HCC develop from liver cirrhosis, but the linkage between cirrhosis and HCC is likely to be multi-factorial. HBV belongs to the Hepadnaviridae family, a DNA virus which integrates into the host DNA and directly transforms hepatocytes.9 The role for integration in transformation is supported by the fact that integrated HBV sequences can be found in approximately 80% of HBV-related HCC although HBV integration can also be found in non-HCC tissue.10 HBV integration can have several mutagenic consequences, including large inverted duplications, deletions, amplifications and translocations resulting in chromosomal instability.11 The HBx gene is the most commonly integrated and over 95% of patients with HBV-related cirrhosis and dysplasia are positive for HBx.12 HBx is a transcription activator through its interactions with epidermal growth factor receptor, c-myc, c-jun, c-fos, p53, AP-1, NFκB and SP1.9
In contrast to HBV, HCV belongs to the Flaviviridae family, a positive-stranded RNA virus that lacks reverse transcriptase activity.9,12 Both HBV and HCV encoded proteins have been implicated in the disruption of cellular signal transduction pathways leading to unrestricted cell proliferation.9 The HCV structural proteins consist of the core protein and envelope proteins 1 and 2. The core protein has been shown to affect the modulation of cellular gene products and several cellular regulatory pathways by binding to p53 and pRb tumor suppressor proteins; modulating the expression of p21WAF1/CIP1, which is involved in cell cycle control; and interacting with cytoplasmic signal transduction molecules to regulate transcription.9 Importantly, the advancement of an effective vaccine against HCV-mediated hepatocarcinomas has been challenging due to the weak immune response elicited by HCV infection.9 Nevertheless, using the HCV replicon cell culture system as a model for HCV RNA replication in the high-throughput screening of pharmacological inhibitors, recent studies have revealed that the obstruction of NS5A functions is a promising therapeutic strategy for the treatment of HCV.13
Human T lymphotropic virus (HTLV-1)
HTLV-I was the first human retrovirus to be associated with malignant adult T-cell leukemia/lymphoma (ATL) and is also associated with a variety of lymphocyte-mediated diseases.14 HTLV-1 has elaborate strategies that allow it to persist and replicate in the presence of a strong immune response and most HTLV-1 carriers have a lifelong infection without developing any major clinical manifestation.14 HTLV-1 expresses multiple gene products by using both strands of its proviral genome and complex mRNA splicing patterns.15 Among all of the regulatory proteins encoded by HTLV-1, the Tax and HBZ proteins appear to have particularly important roles in viral persistence and pathogenesis, presumably through stimulating the continuous cell growth of infected cells in the presence of strong immune surveillance.15 Tax has been shown to be the major oncogenic determinant of HTLV-1.15 It augments cell survival via positive modulation of the NFκB and AKT signaling cascades and negatively regulates the tumor suppressor proteins, p53 and pRb.15
Merkel cell polyomavirus (MCPyV)
MCPyV has been found to be associated with approximately 80% of Merkel cell carcinoma (MCC) by digital transcriptome subtraction and high throughput sequencing of cDNA libraries constructed from MCC tumors, which are rare, aggressive carcinomas of cutaneous neuroendocrine cells.16,17 On the other hand, this virus is not present in many cutaneous neoplasms that are histologically parallel to MCC, including small cell carcinoma of the lung and other high-grade neuroendocrine tumors.18 Only discovered in 2008, little is known about the virus’ distribution, transmission dynamics or natural history. MCPyV, which has characteristics that could contribute to neoplastic transformation, is found in most MCC tumors. The transcripts expressed by MCPyV in MCCs are similar to the large T (LT), small T (ST) and the 17 Kb transcripts of SV40.16 Moreover, the viral genome in MCC tumors is reported to have mutations that truncate the product of the large T antigen thereby preventing autoactivation of integrated virus replication, which would be detrimental to cell survival.19 A mutation in the VP1 gene, possibly related to incomplete integration of the virus in MCC has also been reported. Interestingly there is a striking correlation between MCC development and hypermethylation of the p14ARF promoter and p63 expression.16 The near future should bring an improvement in our fundamental understanding of MCC pathogenesis, which will fuel therapeutic advances.
Deregulation of Cellular Pathways
To explore viral-mediated oncogenesis, a complex series of genomic studies illustrated the marked complexity of cellular deregulation induced by the expression of viral-oncoproteins and also identified numerous signaling pathways involved in cancer development as shown in Figure 1. Similarly, proteomic and various biochemical approaches have identified many novel cellular targets associated with viral oncoproteins (Table 1) and thus allowed correlations with the findings from genomic analyses of viral infected cells.
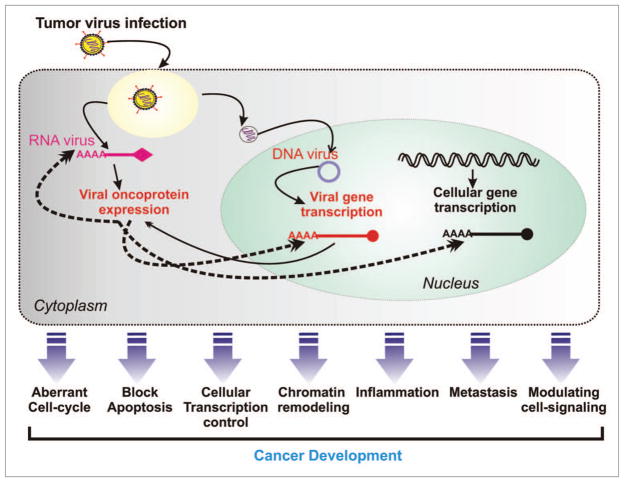
Figure 1.
Tumor virus infection leads to immortalization of the infected cell through deregulation of cellular homeostasis. Via expression of many potent oncoproteins tumor virus promote an aberrant cell-proliferation via modulating cellular cell-signaling pathways and escape from cellular defense system such as blocking apoptosis. Subsequently primary cancerous cells become metastatic through inhibiting cellular metastasis suppressor proteins.
Table 1.
Tumor virus and associated cell-signaling molecules
| Tumor virus | Viral genome | Viral oncoproteins* | Important cellular binding partners | Deregulated signaling-pathways | Ref. |
|---|---|---|---|---|---|
| HPVs (16, 18) | DNA | E6 | p53, p73, E6AP, CBP/p300, c-Myc | p53, cell-cycle | 188 |
|
| |||||
| E7 | pRb, pRb pocket proteins, p21CIP1, p27KIP1, IRF-1, Cyclin A and E | pRb, cell-cycle, ub-proteasome | 188 | ||
|
|
|||||
| EBV | DNA | EBNA2 | RBP-Jκ, PU.1, AUF1, DDX20, SMN | Notch, cellular transcription, metastasis | 189, 190 |
|
| |||||
| EBNA3C | p53, Mdm2, pRb, p300, RBP-Jκ, Chk2, Nm23-H1, c-Myc, HDAC1, SUMO-1, SUMO-3, SCFSkp2-complex, DDX20, SMN, CtBP, Cyclin A, E and D1 | Cell-cycle, Notch, ub-proteasome, metastasis, chromatin remodeling, cellular transcription, apoptosis, inflammation | 31, 52, 60, 82, 99, 100, 128, 190, 191 | ||
|
| |||||
| LMP1 | TRAFs 1, 2, 3 and 5, TRADD, RAS, JAK | NFκB, cell-cycle, cellular transcription, apoptosis, inflammation, autophagy, metastasis, MAPK, PI3K/Akt, JAK/STAT, TNF | 51, 75, 81, 98, 142, 147, 151, 152, 190, 192, 193 | ||
|
| |||||
| LMP2 | TNFR associated factors, RAS, JAK | Apoptosis, metastasis, MAPK, PI3K/Akt, JAK/STAT, TNF, BCR signaling | 83, 135, 142, 194 | ||
|
|
|||||
| KSHV | DNA | LANA | p53, pRb, c-Myc, GSK3β, MAPK2, FADD, core histones, Transcriptional activators-Brd2, Brd4, Sp1, AP-1, and CBP and transcriptional inhibitors HP1, Dnmt3 and mSin3 | cell-cycle, cellular transcription, apoptosis, Notch, Wnt/β-catenin, ub-proteasome, chromatin remodeling | 3–6, 50, 57, 67, 153 |
|
| |||||
| vFLIP | TRAF2 | Apoptosis, NFκB, JNK/AP1 | 85, 86, 148, 153 | ||
|
|
|||||
| HBV | DNA | HBx | NFκB, p53, c-jun, c-fos, PKC, c-myc, SP1, HIF-1α | Cell-cycle, apoptosis, cellular transcription, NFκB Wnt/β-catenin, TGFβ, JAK/STAT, metastasis | 40, 63, 68, 90, 195 |
|
| |||||
| MCPyV | DNA | LT | p53, pRb | Cell-cycle | 17 |
|
| |||||
| HCV | RNA | NS3 | p53, Arginine methyltransferase 1, PKA, H2B, H4 | PKC, inflammation | 158, 184, 196, 197 |
|
| |||||
| NS5A | p53, Bax, IFN-induced dsRNA activated protein kinase (PKR), growth factor receptor-binding protein 2 (Grb2), PI3K p85 subunit, TRADD, CDK1, TRAF2, TBP | Cell-cycle, apoptosis, Ras-Erk MAPK pathway, PI3K, NFκB | 13, 41, 198, 199 | ||
|
|
|||||
| HTLV-1 | RNA | Tax | Cyclic AMP, p300/CBP, MAD-1, MAD-2, cyclin D1, Chk1 and 2 | Cell-cycle, apoptosis, cellular transcription, NFκB, PI3K/AKT, chromatin remodeling | 15, 186, 200, 201 |
Most potent and studied viral oncoproteins.
Targeting tumor suppressors
In cancer, there are fundamental alterations in the genetic control of cell division, resulting in uncontrolled cellular proliferation. Genetic mutations primarily occur in two major classes of genes: proto-oncogenes and tumor suppressor genes. In the case of normal cells, the proto-oncogene products (such as cyclin D1 and c-myc) act at various levels along the pathways responsible for stimulating cell propagation. Mutated versions of proto-oncogenes or oncogenes can promote tumor expansion while the inactivation of tumor suppressor genes, like pRb and p53, resulting in the dysfunction of proteins that, in general, block cell cycle progression (Fig. 2). The cell cycle dysregulation associated with cancerous growth occurs typically through the mutation of proteins that function at various stages of the cell cycle (Fig. 2). In human cancers, mutations have been observed in genes encoding cyclins, cyclin dependent kinases (CDKs), CDK-activating enzymes, CDK inhibitors (CKI), CDK substrates and checkpoint proteins.20,21
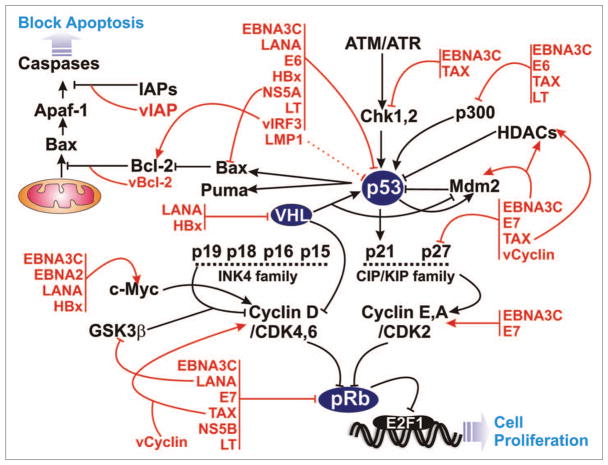
Figure 2.
Tumor virus encoded oncoproteins deregulate components of the cell cycle and apoptotic pathways. Viral-oncoproteins (red) interact with and manipulate normal functions of numerous cellular important proteins (black) to hijack entire cell cycle and apoptotic machineries.
p53—the master regulator
The tumor suppressor protein, p53, is a sequence-specific DNA-binding protein, which is able to induce either cell cycle arrest or apoptosis at cell cycle checkpoints in response to genotoxic stress (Fig. 2).22–24 Identified as an interacting partner of the SV40 encoded large T antigen in virally transformed cells, p53 is the most frequently mutated gene in human cancer.23 Point and missense mutations also lead to conformational changes and functional inactivation of the protein.25 p53, in response to DNA damage or viral infection, can arrest cellular growth and promote apoptosis through transcriptional activation of its downstream target genes, such as p21WAF1/CIP1cip1, GADD45, cyclin G, bax, IGF-BP3 and Mdm2.23 The p53 gene has been demonstrated to be mutated or deleted in half of all malignant tumors. The other half of human cancers express wild-type p53 protein, which is competent to induce apoptosis in malignant cells after genotoxic stress, thus offering a potential therapeutic opportunity applicable to a wide range of human tumors expressing wild-type p53.26 In general, tumors that retain wild-type p53 have a better prognosis and in addition, have a better response to chemotherapy.27 Tumor cells are prone to p53-mediated apoptosis as a result of oncogene activation. Therefore, it is likely that p53-based anticancer strategies may not require selective targeting of tumor cells.28
The importance of Mdm2 in the regulation of p53
Mdm2 (Hdm2 for the human homolog) is an E3 ubiquitin ligase that binds to p53 and facilitates its ubiquitin-proteasome dependent degradation.26 It has been reported that mechanisms of enhancing Mdm2 activity, such as gene amplification of the mdm2 gene or enhancing its E3 ligase activity towards p53, may represent an alternative to p53 mutation as a means to escape p53-mediated growth arrest in malignant cells. Elevated expression of Mdm2 has also been shown to be associated with approximately 10% of all human cancers29 and the interaction of Mdm2 with p53 regulates the transactivation activity and stability of p53.30 Moreover, Mdm2 facilitates the nuclear export of the Mdm2-p53 complex and also regulates its sumoylation, neddylation and acetylation status, which negatively modify the stability and biological function of p53.31 Surprisingly, the mdm2 gene is amongst those that are upregulated by p53; thus not only is Mdm2 required for keeping p53 in check under normal conditions, it is also part of an auto-regulatory feedback loop.30 Overexpression of the Mdm2 protein abolishes p53-mediated cell cycle arrest and apoptosis.30 These studies are consistent with clinical observations that implicate a dysfunctional p53-Mdm2 system in nearly 60% of all malignant samples.30 For approximately 7% of all cancers, wild-type p53 is present and the problem lies in a surplus of Mdm2.30 For example, a single nucleotide polymorphism (SNP309) in the Mdm2 promoter leads to an increase of both mRNA and protein levels, which accelerates tumor formation in both hereditary and spontaneous cancers via blocking p53-dependent pathways.32 Thus, in tumors with wild-type p53, an attractive target for therapeutic development would be to reduce the inhibitory effects of Mdm2 by blocking its interaction with p53.33 Interestingly, recent studies have identified potent and selective small-molecule inhibitors of the p53-Mdm2 interaction, such as Nutlin-3a, which specifically triggers p53-mediated pathways in cells with wild-type p53 and causes cell cycle arrest and induces apoptosis via induction of p21.34 Although the pathways by which Nutlin-3a acts are not fully elucidated, the fact that Nutlin-3a has shown potent antitumor activity in mouse xenograft models, suggests that it is a potential treatment option for tumor cells expressing wild-type p53.
Tumor viruses and p53
In general, oncogenic viruses are able to disrupt cell cycle checkpoints induced by genotoxic stress.35 Among the human DNA tumor virus oncoproteins, the SV40 encoded large T antigen, adenovirus encoded E1A and HPV encoded E6 have all been shown to physically interact with p53 and disrupt p53-mediated transcriptional activity and apoptosis through distinct mechanisms.36,37 Large T antigen and E1B form complexes with p53 and enhance the protein stability but inhibit its functional activity,38 while the HPV encoded E6 antigen facilitates p53 degradation through the ubiquitin-proteasome mediated pathway.39 In addition, the HBV encoded HBx has been shown to interact with p53 and inhibit its functional activity in multiple ways in order to contribute to the molecular pathogenesis of human hepatocellular carcinoma (HCC).40 Whereas, in the case of RNA tumor viruses, HCV encoded NS5A has been shown to abrogate the p53-mediated transactivation function by interfering with the DNA binding activity of p53.41 Thus, using different approaches, these viruses can contribute to oncogenesis by blocking p53-mediated function.
In response to viral infection, p53 protects normal cells from malignant transformation by inducing either cell cycle arrest or apoptosis.42 Therefore, it is not surprising that either p53 itself or the connecting cellular proteins that are involved in downstream activities are inactivated by viral-antigens via either releasing cells from cell cycle checkpoints or protecting cells from the p53-dependent apoptotic pathway.43,44 Cell cycle arrest depends on the ability of p53 to initiate the transcription of target genes such as the CDK inhibitor, p21,45 while apoptosis depends on the transcriptional activation of cellular genes including bax, puma, perp, among others.46
Similar to smaller DNA tumor viruses, large DNA herpesvirus family members have also been shown to manipulate p53 function either by direct physical interaction or by some other activities. For example, the CMV encoded IE2 protein,47 the KSHV encoded ORF K8 protein,48 and the HHV6 encoded ORF 1 protein49 block p53-mediated host cell death through their interaction with p53. Additionally, the KSHV-encoded LANA and the EBV-encoded proteins, LMP1 and EBNA3C, have been shown to interfere with p53 functional activity to block apoptosis.50–52 LANA and EBNA3C modulate p53 function by repressing its transcriptional activity50,51 and LMP1 blocks p53-mediated apoptosis through the induction of the A20 gene.52 In addition, the EBV encoded EBNA1 protein interacts directly with the cellular deubiquitination enzyme, USP7, to regulate the normal p53-Mdm2 pathway, resulting in a reduction of p53 levels, increased cell survival and proliferation as seen in most forms of EBV-latent infection.53,54 Interestingly, the T-cell leukemia-associated virus HTLV-1 encodes the transforming transcriptional activator Tax, which does not bind directly to p53 but inactivates it through an as yet unexplained mechanism.15
Nutlin-3a targets the p53-Mdm2 interaction: a therapeutic strategy
DNA double-stranded break initiators, such as gamma irradiation or treatment with doxorubicin, lead to a normal p53 response in EBV-transformed cells and KSHV-infected primary effusion lymphoma (PEL) cell lines respectively.55 It is therefore conceivable that p53 is wild-type and fully functional in either EBV or KSHV infected cells. The degradation as well as inactivation of p53 functions by its negative regulator, Mdm2, represents a critical circuit in the regulation of p53 both in response to acute DNA damage and in its tumor suppressor functions.56 Recently, we have shown that the EBV-encoded latent antigen EBNA3C stabilizes Mdm2 function and so enhances the degradation of p53 through the ubiquitin-proteasome degradation pathways.31 In agreement with these results, the growth of PEL cell-lines latently infected with KSHV as well as in vitro EBV-transformed lymphoblastoid cell-lines were shown to be sensitive to Nutlin-3a-mediated growth suppression.55,57 The antitumoral function of Nutlin-3a in combination with various chemotherapy or radiotherapy has been verified by both human cells and animal models. Interestingly, since p53 is rarely mutated in hematologic malignancies, numerous studies have been attempted to check p53-dependent functions on various types of tumor cells including chronic lymphocytic leukemia B cells (B-CLL), myeloma, KHSV-induced lymphoma and EBV transformed tumor cells.55,57 In some cases, Nutlin-3a can induce apoptosis in p53-inactivated cells through a p73-dependent pathway as shown recently in a mantle cell lymphoma cell line with mutated p53.57 These studies suggest that Nutlin-3a may indeed be considered, either alone or in combination with various genotoxic drugs, as a novel therapeutic approach for curing viral associated human cancers.
pRb negatively regulates the G1-S phase transition
Tumor viruses have evolved diverse mechanisms to abrogate the function of the retinoblastoma tumor suppressor protein (pRb).58,59 Studies of these viruses have been invaluable in uncovering the central role of the pRb family of pocket proteins in mitotic cell cycle control.58,59 While the molecular mechanisms by which the viral oncoproteins inactivate the pRb family are still being elucidated, it is clear that targeting of this family is required both for viral replication and for virus-induced transformation of mammalian cells.58,59 The Rb family of so-called pocket proteins, which includes pRb, p107 and p130, negatively regulates cell cycle progression from G1 to S-phase through binding with the E2F family of transcription factors to abrogate abnormal cell proliferation (Fig. 2).58,59 Viral oncoproteins as well as E2Fs have been shown to interact through the pocket region of the Rb family of proteins. The pocket proteins are regulated in part via phosphorylation by CDKs including cyclin D/CDK4/6, cyclin E/CDK2 and cyclin A/CDK2. Hyperphosphorylation of pRb results in loss of binding to both E2F and chromatin remodeling factors and reverses pRb-mediated cell cycle arrest.58,59 As a result, cyclins and pRb family proteins represent key factors for the development of cancer.
Tumor viruses and pRb
A large body of evidence has shown that several tumor virus encoded antigens efficiently inactivate Rb family members, which ultimately leads to an increase in E2F activity.3,59,60 Adenovirus 12 was the first human virus demonstrated to induce tumors in an animal model; however, to date there has been no association with any human tumors.59 A number of studies have shown that the adenovirus encoded E1A oncoprotein directly antagonizes pRb function by disrupting the interaction between pRb and E2F.59 Interestingly, only adenovirus gene expression was found to be dependent on E2F;59 therefore, inactivation of the Rb family proteins by the E1A oncoprotein primarily serves to induce transcription of E2F-controlled cell cycle and DNA synthesis genes to establish an environment permissive for viral replication. Like E1A, the HPV 16 encoded E7 oncoprotein possesses an pRb binding LxCxE motif and bypasses cell cycle arrest at G0/G1 through binding to the hypophosphorylated form of pRb, thereby inducing proteasome-mediated degradation of pRb, p107 and p130.59 Similarly, EBNA3C, an essential EBV-encoded latent antigen has been shown to induce pRb degradation.60 To date, the studies probing EBNA3C functions provide the best link between latent EBV infection and the pRb regulated checkpoint, which controls the G1-S transition.61 EBNA3C was previously shown to indirectly target pRb regulated pathways by activating E2F-dependent promoters and bypassing the G1-S restriction point.61 EBNA3C also recruits the SCFSkp2 E3 ligase to facilitate pRb’s degradation in an ubiquitin-proteasome mediated fashion.60 Furthermore, unlike E7, EBNA3C does not have any activity on the other Rb family proteins.60 Interestingly, the same regions of E1A, E7 and EBNA3C antigens required for targeting pRb are also required for the transforming activity of these oncoproteins. The KSHV encoded LANA can also directly interact with pRb and enhance E2F-dependent transactivation activity, indicating that LANA contributes to KSHV-induced oncogenesis by targeting the pRb-E2F transcriptional regulatory pathway.3 Unlike other DNA tumor virus oncoproteins possessing cell immortalizing activity, the EBV-encoded essential oncoprotein LMP1 does not bind to pRb, but instead blocks expression of the p16INK4a tumor suppressor gene, perturbing the pRb-regulated pathways in development of cancer.62 Moreover, the HBV encoded HBx oncoprotein upregulates E2F1 promoter activity63 and the HCV core protein destabilizes pRb and upregulates E2F1 protein levels.64
pVHL-multifunctional tumor suppressor protein
The von Hippel-Lindau (VHL) tumor-suppressor gene is frequently inactivated in VHL disease and in sporadic cases of renal cell carcinoma.65 VHL protein (pVHL) acts as an E3 ubiquitin ligase complex that targets many cellular proteins for proteasomal degradation.65 The best-characterized cellular target is HIF-α, a hypoxia inducible transcription factor, whose activity is critically controlled by pVHL.65 Reduction of pVHL and subsequent upregulation of HIF-α targeted genes has been attributed to the highly vascular nature in these cancers.65 It is clear that loss of pVHL can result in the activation of different cellular pathways that are strongly associated with both tumor initiation and progression. pVHL forms a stable complex with p53 and prevents Mdm2-mediated ubiquitination/degradation.66 Stabilization of p53 thus resulted in induction of p53 transactivation function.66 Interestingly, KSHV encoded LANA has been shown to facilitate pVHL degradation along with p53 via recruitment of the EC5S ubiquitin complex.67 In addition, the HBV-encoded oncoprotein HBx was shown to directly interact with HIFα and increase its transcriptional activity.68 HBx also inhibits the interaction between pVHL and HIF-α and prevents its degradation as well as stimulates angiogenesis.68 Recent evidence suggests that multiple deregulating events are likely to co-operate with pVHL in the development of renal cancers as well as a potential role in KSHV and HBV mediated epithelial cancers. Further research is necessary in order to comprehend fully the tumor-suppressive capabilities of pVHL and its critical targets in other tumor virus mediated cancers.
Dysregulation of the cell cycle machinery
Aberrant cell proliferation and disruption of cell cycle checkpoints are hallmarks of viral mediated oncogenesis (Fig. 2).21,35,69,70 Tumor viruses use the host cell machinery to replicate and drive quiescent cells to become active and proliferate.35,70 Tumor virus encoded oncoproteins have evolved a range of strategies to deregulate cell cycle progression and bypass cell cycle checkpoints (Fig. 2).35,70 As indicated earlier, tumor virus-encoded oncoproteins contribute to the cellular immortalization process through targeting and inactivating two prime tumor suppressors, pRb and p53. By using different tactics to prevent cell cycle arrest at the G1-S and/ or G2-M transition points, they induce inappropriate entry to S phase and apoptosis (see ‘targeting tumor suppressors—p53 and pRb’ section for more details) (Fig. 2).69
Common pathways in cell cycle control
The cyclin-CDK complexes positively regulate DNA replication and cell cycle progression.35,69,70 Cell cycle progression from G1 to S and G2 to M is accomplished through a complex interplay of regulatory signals that ensures accurate duplication of DNA during the synthesis (S) phase and proper segregation of chromosomes during mitosis (M).69 This regulation is mediated by sequential activation of the kinase activity of CDKs, which are regulated by interaction with specific cyclins at specific stages of the cell cycle.69 While CDK levels remain constant throughout the cell cycle, cyclin levels oscillate as a result of post-transcriptional as well as post-translational modifications.69 Different cyclins are required at different phases of the cell cycle: cyclin D in G1, cyclin E and A in S phase and cyclin B and A in M phase.69 As mentioned earlier, active cyclin/CDK phosphorylates the Rb family of proteins, resulting in the release of the E2F family of transcriptional factors, which ultimately activates the transcription of genes responsible for cell cycle control, initiation of replication and DNA synthesis.69 In the absence of mitogenic signals, CDKs remain inactive and so prevent aberrant proliferation. This negative control is achieved by a family of proteins termed CDK inhibitors (CKIs), which include p15INK4B, p16 INK4A, p21WAF1/CIP1 and p27KIP1.69
Viral oncoproteins and deregulation of cell cycle
Tumor viruses have developed numerous sophisticated strategies to ensure the continuous cell cycle proliferation of the infected cell.35,71,72 Importantly, aberrant alterations in the regulation of several cell cycle components, including cyclins and associated CDKs as well as CDK inhibitors, are characteristic of tumor virus mediated oncogenesis.35 Usually, viral oncoproteins act directly on either the CDKs or their activating cyclins, to enhance the kinase activity of the complex.35 Functional inactivation of the CDK inhibitors, in particular p21WAF1/CIP1 and p27KIP1, represents yet another critical mechanism exhibited by several tumor viruses, including HPV, HTLV1 and EBV.35 The HPV-encoded E6 oncoprotein has been shown to decrease p21CIP1 expression at the mRNA level, via regulating p53, which normally transactivates the p21 promoter.73 Similarly, the adenovirus-encoded E1A oncoprotein also blocks transcriptional activation of p21WAF1/CIP1 by sequestering the activity of transcriptional coactivator p300/CBP.27,74 Another oncoprotein encoded by HPV, E7, can bypass the p21WAF1/CIP1 mediated growth arrest either by blocking its interaction with the cyclin/CDK complex or altering the sub-cellular localization of p21WAF1/CIP1.35,59 Similarly, E1A and E7 also directly interact with p27KIP1, which appears to be cell-type dependent.35 Interestingly, the HTLV-1 encoded Tax protein represses p27KIP1 transcription but also induces transcriptional activation of p21WAF1/CIP1 In cells expressing Tax, p21WAF1/CIP1 does not accumulate following DNA damage, which suggests that its regulation may be at the post-translational level.15
The INK4 family of CDK inhibitors are also targeted by viral oncoproteins.35 For instance, E1A modulates the expression of both p15INK4B and p16INK4A in different contexts.35 Tax blocks the interaction of p16INK4A with CDK4 or CDK6.35 In addition, Tax also suppresses the expression of p15INK4B, p18INK4C and p19INK4D.15 The level of p21WAF1/CIP1 protein is maintained in EBV-infected cells, possibly due to its rapid proteasome-mediated degradation.35 Interestingly, EBV encoded EBNA3C targets another CDK inhibitor, p27KIP1, for ubiquitin-proteasome dependent degradation through the recruitment of the SCFSkp2 E3 ligase activity similar to pRb degradation.61 By disrupting p27KIP1 from cyclin A/CDK2 complexes, EBNA3C enhances CDK activity.61 EBNA3C can also override p16INK4A-mediated suppression during EBV-mediated in vitro transformation, consistent with EBNA3C targeting the checkpoint at the G1-S transition regulated by pRb.61 As a result, similar to the E1A and E7 oncoproteins, EBNA3C also cooperates with oncogenic mutant H-ras for immortalization and transformation of rat embryonic fibroblasts (REFs).61 LMP1, another EBV oncoprotein, represses transcription from the p16INK4A promoter but did not have any significant effect on the p21WAF1/CIP1 promoter.35,75 It is well known that constitutive expression of the c-Myc oncoprotein in B lymphocytes induces overall protein synthesis and cell cycle division.35 In addition, c-Myc can also stimulate expression of D-type cyclins and cyclin E and downregulate p21WAF1/CIP1 and p27KIP1.35 The EBV-encoded latent antigen, EBNA2, directly activates c-Myc, which further transactivates cyclin D2, whose enhanced expression is generally found in EBV associated lymphomas.35
Association between viral oncoproteins and cyclin/CDK complexes is well documented. For example, E1A, E7 and EBNA3C all form complexes with cyclin A/CDK2 and stimulate its kinase activity.35 The HTLV-1 encoded Tax adopts a more direct approach; it transcriptionally activates expression of the cyclin D gene and also increases the association between cyclin D and CDK4.15 LANA stabilizes β-catenin by binding to the negative regulator GSK-3β, resulting in cyclin D1 accumulation.3 As a result, both β-catenin and cyclin D1 have increased expression in both PEL cells and KS tissue.3
A novel mechanism for controlling the cell cycle machinery is illustrated in several herpesviruses including herpesvirus saimiri (HVS), murine herpesvirus (MHV-68) and KSHV, in which they all encode a protein with cyclin homology.35 The KSHV encoded v-cyclin, which is a homolog of cyclin D1, is the most extensively characterized of the viral encoded cyclins.76 Like its cellular counterparts, v-cyclin can bind and activate the kinase partner, CDK6, increasing phophorylation of pRb and facilitating the G1-S transition.76 In addition, the association of v-cyclin/CDK6 induces phosphorylation of a wide range of important cellular substrates including histone H1, p27KIP1 and cdc25A as well as components of the DNA replication apparatus.76 The v-cyclin/CDK6 phosphorylation of p27KIP1 induces its proteasome mediated degradation and increases activation of endogenous cyclin-CDK complexes.76
Finally, at least one virally-encoded oncoprotein disrupts the control of mitosis. Tax interacts with two proteins that regulate mitosis, MAD-1 and MAD-2, which leads to aneuploidy and genomic instability.15
Deregulation of apoptotic pathways
Virally encoded oncoproteins have been shown to have a profound effect on apoptosis by interacting directly with the components involved in the highly conserved biochemical pathway that regulates cell death (Fig. 2).77,78 It appears that tumor viruses can efficiently block apoptosis to prevent premature host cell death and so facilitate persistent infection and the development of oncogenesis (Figs. 1 and 2).77,78 Thus, impairment of apoptotic pathways represents a major causative factor in cancer progression (Fig. 2).77,78 Cellular Bcl-2-related proteins function as critical regulators of apoptosis by modulating the release of pro-apoptotic signaling molecules from the mitochondria. Interestingly, DNA tumor viruses encode homologs of cellular anti-apoptotic Bcl-2 proteins (vBcl-2s) and the role of vBcl-2s in viral infection and the mechanisms by which they function are beginning to emerge.79 It is now apparent that inhibition of mitochondrial apoptosis by vBcl-2s can prevent premature death of the host cell and subsequently, the development of cancer.79
A recent report has shown that EBV positive Burkitt lymphoma (BL) cells express significant amounts of a viral latent protein, BHRF1, which is a homolog of the cellular anti-apoptotic protein Bcl-2.80 The hallmark of BL tumors appears to be the deregulated expression of c-myc, which induces cellular proliferation as well as the apoptotic process.80 Studies have clearly suggested that BHRF1 blocks apoptosis and acts as a survival factor for EBV positive BL tumors.80 One of the major oncoproteins encoded by EBV, LMP1, forms a complex with the pro-apoptotic tumor necrosis factor (TNF)-receptor 1-associated death domain protein (TRADD), which dictates TRADD-mediated NFκB signaling and subverts TRADD’s potential to induce apoptosis.81 LMP1 has also been shown to upregulate several cellular anti-apoptotic proteins, including mcl-1 and bcl-2, to promote EBV-mediated cell survival.81 Two EBV encoded oncoproteins, EBNA3A and EBNA3C, functionally cooperate to transcriptionally downregulate the expression of Bim (Bcl-2-interacting mediator), a pro-apoptotic tumor-suppressor protein.82 More recently, EBNA3C has been shown to directly interact with p53 and block its apoptotic activities.52 One study showed that another EBV encoded antigen, LMP2A, protects B cells from apoptosis by blocking B-cell receptor (BCR) signaling. However, studies in animal models suggest that LMP2A makes resting B cells sensitive to NFκB inhibition and apoptosis.83
Similar to EBV, the KSHV-encoded vBcl-2 also efficiently blocks apoptosis.84 KSHV encodes an anti-apoptotic, viral Fas-associated death domain-like interleukin-1β-converting enzyme-inhibitory protein (vFLIP/K13) whose anti-apoptotic activity is attributed to an inhibition of caspase 8 activation84 and to its capability to induce expression of anti-apoptotic proteins via activation of NFκB.85 vFLIP also inhibits superoxide-induced apoptosis in endothelial cells via upregulation of MnSOD expression.86 The KSHV protein, K1, also suppresses apoptosis induced by the anti-Fas antibody CH-11 or Fas ligand.87 Increased expression of the HPV E2 protein in HPV-transformed cells represses transcription of E6 and E7, thus inducing apoptosis.88 The HPV-16 E5 protein impairs CD95L- and TRAIL-mediated apoptosis in HaCaT cells by downregulating the total amount of CD95 receptor and reducing the amount of CD95 on the cell surface; and altering the formation of the DISC triggered by TRAIL.89 The HPV E6 oncoprotein plays a role in the proteolytic inactivation of pro-apoptotic proteins including p53, Bak, FADD, procaspase-8 or c-Myc.73 The HBV-encoded HBx protein has been shown to prevent the apoptosis of HCC cells by upregulating SATB1 and HURP expression.90 Additionally, both HBV and HCV replication upregulate the expression of the apoptosis inhibitor, SPIK.91
Autophagy
Autophagy is one of two major mechanisms for protein degradation. By controlling both the quality and quantity of proteins, autophagy plays a central role in many cellular processes, including the immune response, development, survival and aging.92 Recently, an essential role for autophagy in tumor suppression has also been clearly demonstrated.92 For example, radiation and many antitumor molecules induce autophagy in tumors, resulting in cell death. Many oncoproteins and proto-oncoproteins, such as AKT, mTOR, PI3K and IKK, are potent inhibitors of autophagy while many tumor suppressors, such as PTEN, are activators of autophagy.92 Most significantly, studies have shown that depletion of the autophagy regulatory genes Beclin 1, UVRAG and Bif-1 leads to tumor formation in both animals and humans whereas ectopic expression of these genes reverses this effect to increase the efficacy of antitumor drugs in vivo.92 Although the mechanisms that regulate autophagy and those by which autophagy blocks oncogenesis are largely unknown, it is clear that autophagy is imperative in the regulation of cancer development and progression and in determining the response of tumor cells to anti-cancer therapy.93 A number of studies have shown that viruses can evade or modulate the host cell autophagic pathway to enhance their own replication. The EBV-encoded LMP1 oncoprotein regulates autophagy in EBV-infected B cells.81 Inhibition of autophagy reduces the ability of KSHV to reactivate the lytic cycle and an enhancement of autophagy can be detected during KSHV lytic replication. Importantly, KSHV RTA increases the activation of autophagy to facilitate KSHV lytic replication.94 The HBV-encoded HBx protein sensitizes cells to starvation-induced autophagy via upregulation of beclin 1 expression.40 Autophagy has also been shown to enhance the replication of HBV DNA, suggesting the possibility of targeting the autophagic pathway for the treatment of infected HBV patients.95 Autophagy proteins Beclin-1, Atg4B, Atg5 and Atg12 are among the proviral factors required for initiation of HCV replication.96 Thus, inhibition of autophagy is an attractive strategy for the treatment of virally-mediated cancers.
Chromatin remodeling and regulation of cellular transcription
In eukaryotic cells, DNA methylation at CpG dinucleotides within or near various promoters efficiently represses gene transcription and thus has a central role in the regulation of gene expression during tumor virus-mediated cancer development (Fig. 3).97 CpG dinucleotides in the human genome are mostly methylated, whereas the unmethylated dinucleotides are a signature of human pathogens. Eukaryotic genomic material is packaged as chromatin, containing double stranded DNA wrapped with core histone proteins.97 Covalent modifications of these core histones play a key role in regulating gene transcription by altering chromatin structure and condensation (Fig. 3). For example, acetylation of core histones, by histone acetyltransferases (HATs), leads to the unraveling of chromatin and transcriptional activation, whereas, deacetylation by histone deacetylases (HDACs), leads to chromatin condensation and transcriptional silencing (Fig. 3).97 In addition, modified histones also recruit several transcription factors and chromatin remodeling complexes to control the transcriptional activity (Fig. 3).97 A large body of evidence has demonstrated that tumor virus-mediated malignant transformation includes both genetic and epigenetic alterations. Methylation and histone modification patterns of many targeted cellular genes are markedly altered after viral infection, which in turn, manipulates chromatin structure and subsequent gene expression leading to tumorigenesis.97
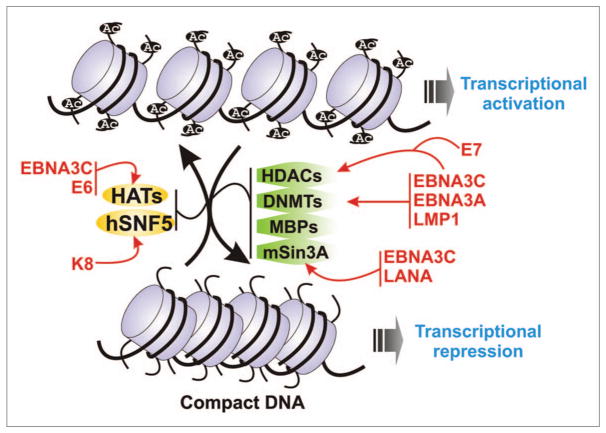
Figure 3.
Epigenetic regulation by tumor virus encoded oncoproteins. Schematic showing epigenetic control of gene transcription by alterations in DNA methylation status and chromatin modifications by tumor virus encoded oncoproteins (red). Transcriptionally active DNA represents an open chromatin structure with widely spaced nucleosomes, associated with unmethylated CpG residues and specific post translational histone modifications, including increased levels of acetylation as well as other enzymes and co-factors. On the other hand, transcriptionally repressive conformation of DNA is characterized by methylated CpG islands, compacted nucleosomes, with deacetylated histones, and further post translational modifications, including methylation of specific histone H3 lysine residues. This repressive conformation renders the DNA inaccessible to the transcriptional machinery. MBPs, methyl-CpG binding proteins; HDACs, histone deacetylases; DNMTs, DNA methyltransferases; HATs, histone acetyl transferase.
In EBV-infected cells, epigenetic modifications represent one of the mechanisms regulating expression of cell cycle and apoptosis-related genes.98 For example, in nasopharyngeal carcinoma, the hypermethylation of certain cellular promoters is attributed to the upregulation of DNA methyltransferases by LMP1 via the JNK/AP1-signaling pathway. EBNA3A and EBNA3C are shown to block the transcriptional expression of the pro-apoptotic protein, Bim, by increasing methylation of the CpG island in the Bim promoter.82 Similarly, these viral oncoproteins are shown to repress transcriptional activation of the CDK inhibitor, p16INK4A, via epigenetic chromatin modifications.99 Epigenetic repression of p16INK4A by EBNA3A and EBNA3C requires the cooperative interaction with the C-terminal binding protein-1 (CtBP-1), a transcriptional corepressor.99 EBNA3C functions as a prime regulator of both viral and cellular gene transcription by recruiting HAT and HDAC activities.61 EBNA3C not only binds to transcriptional repressor complexes that include HDAC-1 and HDAC-2 but also interacts with the transcriptional coactivator complex p300/prothymosin α, modulating its HAT activity.100 The EBV encoded BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II to provide more extrachromosomal space for viral DNA replication and successful egress of the nucleocapsid from the nucleus.101 Another EBV protein, EBNA2, activates cellular and viral transcription through interactions with the cellular DNA-binding proteins RBP-Jκ and PU.1 that bind consensus sites at responsive promoters. Similar to EBNA3C, EBNA2 also contains an acidic activation domain and can interact with a number of general transcription factors and co-activators.102
Chromatin modifications have also been found to be connected with KSHV mediated pathogenesis.103 For instance, several HDACs are associated with the KSHV RTA promoter during latency for efficient repression. However, enforced KSHV reactivation leads to the dissociation of these repressor complexes and subsequent association of hSNF5 a component of the Swi/Snf family of chromatin remodeling factors.103 In addition, the KSHV encoded K8 protein functions as a transcriptional activator by interacting with hSNF5.104 LANA, a potent, multifunctional oncoprotein encoded by KSHV, appears to be involved in modulating activation and repression of both cellular and viral gene transcription, in addition to its role in tethering the KSHV genome to the host chromosome.3 LANA associates with the transcription factor 4/cyclic AMP response element-binding protein 2 and mSin3A, as well as CREB-binding protein and RING3, to repress transcription.3 LANA also modulates transcription by altering the subcellular distribution of GSK3β, a negative regulator of β-catenin.4 In addition, LANA represses the transcriptional activity of p53 and stimulates transcription from the cyclin E promoter.3 Interestingly, both RTA and LANA have been shown to interact with HAT protein CBP indicating their roles in chromatin modification.4
Studies on the HPV E6 and E7 have shown that these oncoproteins may also modulate the host chromatin structure driving the establishment of cancer.7 In vitro studies showed that E6 inhibits p300-mediated HAT activity on both p53 and core histones, suggesting a possible mechanism for the repression of p53-dependent transcription.7 In human foreskin keratinocytes, HPV16 E7 has been shown to increase histone acetylation in a manner dependent upon its binding to both pRb and HDAC.7 This functional interaction may therefore indirectly create a transcriptionally active chromatin structure to promote expression of genes vital for cell cycle progression. During mitosis, high-risk HPV16 E7 interacts with microtubule-associated nuclear mitotic apparatus protein 1 (NuMA).7 The interaction between HPV16 E7 and NuMA correlates with the induction of chromosome alignment defects during prometaphase.7 It has been proposed that this abrogation of mitotic events by HPV E7 may contribute to viral maintenance and propagation by disrupting the differentiation program of infected epithelia cells.7 The high-risk HPV E7 protein also interacts with BRG-1, a component of the human SWI/SNF complex that either activates or represses cellular promoters by modulating the chromatin structure. This protein-protein interaction deregulates BRG-1 mediated transcriptional silencing and abolishes cell cycle control.7
Although the initial studies using DNA demethylating agents in hematologic neoplasia and hemoglobinopathies were begun more than twenty years ago, advancement of this type of chemotherapy has only been encouraged in the last five years by the discovery of many genes that are specifically hypermethylated in several human cancers. The exciting breakthrough that compounds that suppress HDAC activity also have antitumor activities has focused attention on their use as anti-cancer drugs.105 As a result, there is ongoing evaluation of several HDAC inhibitors in different clinical trials with promising early results. It is possible that many of the enzymes involved in the control of histone modification would provide future therapeutic opportunities for viral associated cancers.
Modulation of the inflammatory response and role of COX-2 in regulating this response
Inflammation has been associated with the development of cancer; tumor viruses have been shown to modulate cellular pathways related to inflammation. Cyclooxygenases (COX) play a major role in inflammatory responses.106 Prostaglandin (PG) G/H endoperoxidase synthase, also known as COX, is a key enzyme in synthesis of prostanoids (PG and thromboxanes).106 Of the two isoforms of COX, COX-1 is constitutively expressed in many cells and tissues; COX-2 is the inducible form with a central role in the inflammatory response.106 COX-2 is highly induced in a variety of inflammatory diseases and in response to proinflammatory cytokines, growth factors and other tumor inducers.107–109 Elevated expression of COX-2 has been reported in a wide range of human cancers110 and pharmacological inhibitors of COX-2 have been shown to reduce tumorigenesis or tumor cell growth,111 suggesting that COX-2 plays a critical role in development of these cancers. We and others have shown that COX-2 is frequently expressed in EBV-positive nasopharyngeal tumors, as well as detected at higher levels in EBV-positive LCLs upon lipopolysaccharide (LPS) induction when compared to EBV negative nasopharyngeal tumors or LPS-induced, EBV-negative BL lines, suggesting a role for COX-2 in EBV pathogenesis.112,113 The EBV encoded oncoproteins, LMP1 and EBNA3C, have been shown to upregulate COX-2 expression utilizing different mechanistic pathways. LMP1 induces COX-2 expression in an NFκB-dependent manner,113 whereas EBNA3C, in conjunction with Nm23-H1, a potent metastasis suppressor, enhances the expression of COX-2.112 Importantly, COX-2 also plays a role in de novo infection of various DNA and RNA viruses including herpesviruses, such as herpes simplex virus (HSV), human cytomegalovirus (HCMV), EBV and murine gamma-herpesvirus 68 (MHV-68).114 Rhesus cytomegalovirus encodes a COX-2 homolog from its genome, emphasizing the importance of this enzyme.115 De novo infection by herpesviruses MHV-68 and KSHV induce COX-2 expression.116,117 Interestingly, the presence of COX-2 specific inhibitors (indomethacin) reduces viral gene expression, which can be rescued by addition of exogenous PGE2, a downstream product of COX-2 suggesting that the elevated levels of COX-2 in response to de novo infection play an important role in viral gene expression.116,117 Although no therapeutic strategies to prevent or treat cancers based on insights into inflammatory pathways are currently approved for common epithelial malignancies, there remains substantial interest in the development of anti-cancer drugs targeting COX-2.118 Colville-Nash and colleagues have shown that COX-2 is associated with the early phase of an inflammatory response, however, later in the inflammatory process COX-2 has been shown to have anti-inflammatory effects.119 While the new COX-2 inhibitors and older NSAIDs (non-steroidal anti-inflammatory drugs) serve to interrupt inflammation early on, it appears they may aggravate the inflammatory process later. This provides an explanation as to why older NSAIDs, although having anti-inflammatory properties, fall short of preventing the disease progression. The new findings contribute to better application of COX-2 inhibitors and NSAIDs.120 However, these conclusions may not directly apply to humans, which can hardly be extrapolated from lung inflammation in rats. Hence, a more thorough study utilizing these inhibitors into human clinical trials is warranted for virally associated cancers.
Tumor viruses can promote cell invasion and metastasis
Tumor viruses are known to induce invasiveness and metastasis factors in transformed cells.121,122 This includes the ability to secrete enzymes for the digestion of cell to cell adhesion molecules so that the transformed cells may travel to new locations.122 Several metastasis suppressor proteins are normally expressed in cells, however, these proteins lose their activity, or their expression is inhibited in malignant cells.122 Metastasis suppressor proteins are generally not associated with tumorgenicity and have almost no effect on development of primary tumors. Twenty metastasis suppressor proteins have been identified so far and most act by altering aspects of signal transduction. Nm23-H1 is the most extensively characterized metastasis suppressor protein, shown to be inactivated in melanomas, breast and colon cancers and can also inhibits the kinase activity important for promoting cell division.123 The expression level of Nm23-H1 protein or mRNA has an inverse association with lymph node status and patient survival in human breast carcinoma as well as the metastatic potential of several human tumors. Intriguingly, a positive association between Nm23-H1 levels and metastatic potential has also been shown in neuroblastoma, osteosarcoma and pancreatic carcinoma. Furthermore, a unique point mutation, S120G, was also reported in 21% of advanced neuroblastomas.124 The nm23 gene family is highly conserved and eight genes have been identified in humans. Both Nm23-H1 and Nm23-H2 are hetero-hexameric enzymes while the other members are less studied.123
A few studies have investigated the role of Nm23-H1 in the biology of tumor viruses. Our group has shown that the EBV encoded oncoproteins, EBNA3C and EBNA1, interact with Nm23-H1 in vitro as well as in vivo.125 Nm23-H1 primarily localizes to the cytoplasm, but when co-expressed with EBNA3C, most of Nm23-H1 is translocated into nucleus.125 This interaction and translocation of Nm23-H1 plays a major role in modulation of cell signaling because of the critical role of Nm23-H1 NDP kinase and histidine kinase activities.125 EBV-infected cells have also been shown to synthesize Matrix Metalloproteinase (MMP-9), which plays a role in degradation of the basement membrane component, a prerequisite of metastasis.126 EBV encoded LMP1 increases MMP9 level and invasiveness and MMP9 expression is also regulated by EBNA3C and Nm23-H1.125 Necdin, which interacts with Nm23-H1, plays an important role in the regulation of angiogenesis critical for metastasis. Both EBNA3C and Nm23-H1 not only rescue Necdin-mediated transcriptional repression of the downstream vascular endothelial growth factor promoter but also Necdin-mediated growth suppression and anti-angiogenic effects on cancer cells.127 Additionally, αv integrins, which are primarily expressed on migratory cells including metastatic melanomas and breast cancer cells, play a major role in promoting epithelial cell migration and cell growth. EBV encoded antigens LMP1, LMP2 and EBNA2 have also been shown to transcriptionally activate the αv integrin promoter, and their expression in LCLs each correlate with increased levels of αv integrin.122 Knock-down of αv integrin has been shown to reduce cell-growth, invasiveness and, importantly, significantly reduce MMP-9 expression, suggesting a critical role of αv integrins in EBV-induced B-lymphocyte proliferation and invasion.122 Furthermore, interaction between EBNA3C and Nm23-H1 leads to increased expression of the αv integrin subunit.128
Interestingly, in a similar manner to the EBV nuclear antigens, the HPV encoded E7 oncoprotein also interacts with and nullifies the metastatic activity of Nm23-H1 via both transcriptional downregulation and protein degradation.122 The HBV encoded HBx oncoprotein also plays a key role in progression of HCC by promoting cell detachment from the primary tumor and migration towards a new site.122 HBx induces morphological alterations and cytoskeletal remodeling in hepatoma cells and alters integrin-mediated ECM attachments.122 Furthermore, HBx-expressing cells migrate more rapidly in collagen-invasion assays by inducing MT1-MMP and MMP-9 expression levels. HBx also promotes metastasis by downregulating E-cadherin expression, which is frequently deregulated in HBV infected HCC.122
Metastatic disease causes the majority of cancer-related deaths. The regulation of cancer metastasis by viral proteins through the modulation of metastasis suppressors provides an attractive target for therapeutic interventions especially in viral cancers that have yet to metastasize. Therapeutic approaches to restore the anti-metastatic function of Nm23-H1 have been attempted using a range of different strategies including Nm23-H1 promoter activation by medroxyprogesterone acetate (MPA) treatment.129 A recent study involving nanoparticle delivery of the Nm23-H1 gene was shown to improve chemotherapy in a mouse tumor model.130 Overall, it is clear that tumor viruses have evolved multiple strategies to interfere with the normal cellular processes that regulate cell-adhesion and motility and so enhance pathogenesis. Future therapies aimed at targeting the signaling pathways engaged by tumor virus encoded oncoproteins may provide therapies against these virus-associated cancers.
Hijacking Major Cell-Signaling Pathways
Despite many differences among human oncogenic viruses, there is at least one common feature to efficiently transform the infected cells: targeting important cellular signaling pathways through virus encoded oncoproteins. There are many cellular pathways that regulate cell-fate, with numerous opportunities for tumor viruses to manipulate them to promote virus-mediated cancers (Fig. 4 and Table 1). It would not be surprising if oncogenic viruses can hijack all possible cell-signaling pathways that are linked to development of oncogenesis.
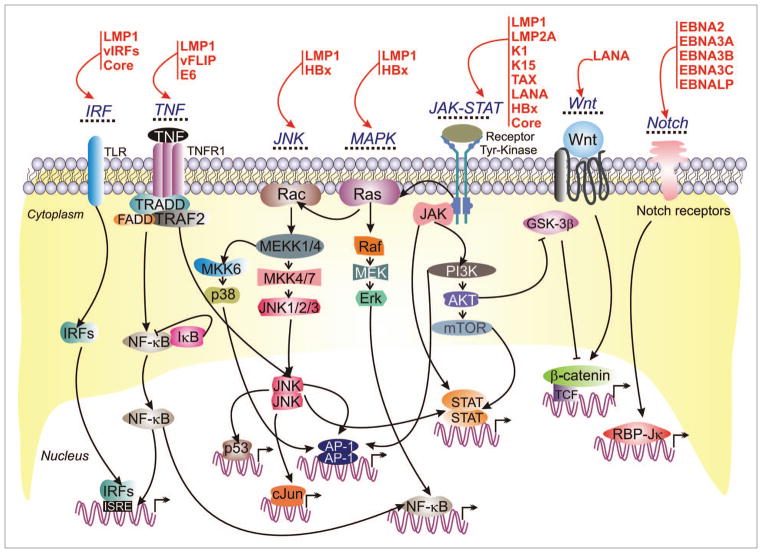
Figure 4.
Tumor virus encoded oncoproteins hijack major cell-signaling pathways. Viral-oncoproteins (red) interact with and control major cellular signaling pathways which ultimately lead to immortalization and continuous proliferation of the virus infected cells.
Notch signaling
The Notch gene family encodes evolutionarily conserved trans-membrane receptors with a key role in the normal development of many tissue and cell types through diverse effects on differentiation, survival, and/or proliferation.131 Due to its role in regulation of these important cellular activities, deregulated Notch signaling has been suggested to contribute to cancer development.131 Numerous viral oncoproteins target the Notch signaling pathway (Fig. 4), emphasizing its significance in regulating normal cell growth and differentiation.131 Through modulation of RBP-Jκ activity, EBNA2 stimulates the expression of cellular genes including c-myc, CD21, Hes-1, EBI 1/2 and Runx3.1 This complex activates various targets including the LMP1 promoter.1,35 The EBNA2/RBP-Jκ complex can similarly activate gene transcription by recruiting several components of the gene transcription machinery. Thus, through interaction with RBP-Jκ, EBNA2 can constitutively activate Notch signaling activity.35 The EBV antigens EBNA3A, 3B and 3C can also bind to RBP-Jκ.132 These interactions are competitive with that of EBNA2 and lead to downregulation of EBNA2-mediated promoter activation.132 Binding of the EBNA3 proteins to RBP-Jκ also inhibits the ability of RBP-Jκ to bind to DNA, thus modulating EBNA2 function both through access to RBP-Jκ and through CSL promoter targeting.132 Interestingly, in primary keratinocytes, Notch1 can function as a tumor suppressor. Similarly, in HPV-positive cervical cancer cells, constitutively active Notch signaling was found to cause growth suppression by repressing viral E6 and E7 oncoprotein expression through AP-1 down-modulation, resulting in increased p53 expression and a block of pRb hyperphosphorylation, suggesting that Notch1-signaling pathways can counteract the transforming potential of HPV.133 The activated intracellular domain of Notch (ICN) accumulates aberrantly in PEL cells with a latent KSHV infection and results in increased proliferation.134 ICN also plays an essential role in KSHV’s ability to prolong the life-span of KSHV-infected human primary B cells, which is likely to be due to the upregulation of cyclin D1 by ICN.134 However, LMP2A of EBV constitutively activates the Notch1 pathway to auto-regulate the LMP2A promoter. In addition, LMP2A requires the Notch pathway to alter levels of the B cell specific transcription factors, E2A and EBF.135 Cervical keratinocytes undergo neoplastic transformation induced by HPV-encoded oncoproteins either by cooperating with Ras or Notch.136 Moderate levels of Notch can upregulate c-Myc, activate PKB/Akt and induce transformation although elevated levels of activated Notch can also induce apoptosis.136 A number of studies have shown that chronic HBV infection is dependent on the imbalance of T-helper Th1/Th2 cells and Notch signaling is directly involved in the proliferation and differentiation of T lymphocytes.137 Interestingly, a recent study has shown that blockage of the Notch signaling pathway inhibits the production of Th2-type cytokines and GATA-3 expression, suggesting a possible mechanism for proliferation in chronic HBV infected patients.137 The ability of Notch signaling to influence both proliferation and differentiation responses makes this pathway an attractive target for manipulation by tumor viruses.
Wnt signaling
For several years, the Wnt signaling pathway has been the object of intense attention in diverse biological areas.72,132 β-catenin is the central modulator of the Wnt pathway (Fig. 4). Elevated levels of β-catenin are observed in EBV-infected B-cells with type III latency as well as in the nasophryngeal carcinomas (NPC).132 The EBV-encoded membrane protein, LMP2A, induces β-catenin accumulation in NPC epithelial cells via activation of the phosphatidylinositol 3-kinase (PI3K)-Akt signaling pathway.132 Interestingly, another EBV encoded oncoprotein, LMP1, has been suggested to affect β-catenin stabilization in EBV infected B cells through transcriptional repression of Siah-1, an E3 ubiquitin ligase that binds to APC and promotes degradation of cytoplasmic β-catenin in a GSK-3β-independent manner.132 Also, KSHV encoded LANA activates the β-catenin pathway via regulation of the intracellular redistribution of GSK-3β. LANA increases nuclear accumulation of GSK-3β and paradoxically also diminishes nuclear GSK-3β activity through a stable interaction.132
JAK/STAT signaling
JAK/STAT pathway is one of the major cytokine-signaling pathways regulating T-lymphocyte function. In response to IL-2, STAT proteins, a family of transcription factors essential for cytokine-regulated activation of downstream genes, are activated by JAKs, a group of RTK (receptor tyrosine kinase)-associated proteins (Fig. 4).138 STAT5 is hyper-activated in both HTLV-1-transformed human T-cell lines and lymphocytes from HTLV-1 patients.138,139 Recent results have shown that STAT5-inducible genes (including IL-5, IL-9, IL-13) were uniquely upregulated by IL-2 in the presence of Tax.138,139 These results are consistent with the previous findings that Tax can directly activate STAT5 transcription.138 In contrast to HTLV-1, transformation of T cell lines by HTLV-2 do not appear to involve the activation of JAK/STAT signaling.138 Similarly, the JAK/STAT signal transduction pathway is also constitutively active in EBV positive B cell lymphomas (PTLD) or NPC cell lines.140,141 In order to contribute to cellular transformation, EBV encoded LMP1 and LMP2A induce survival signals in EBV infected cells by interacting with a number of cell-signaling molecules, such as non-receptor protein tyrosine kinases, TNFR-associated factors, Ras and Janus kinase (JAK), thereby initiating the downstream signaling cascades: MAPK, PI3K/Akt, NFκB and JAK/STAT signaling pathways.142 In NPC cell lines, LMP1 stimulates STAT3 activation by increasing its phosphorylation status.140 Moreover, a recent report has shown that LMP1-induced phosphorylation and subsequent activation of STAT-1 in EBV positive LCLs is almost exclusively due to the NFκB-dependent secretion of α- and γ-interferons.142 The activation of similar pathways by KSHV-encoded membrane proteins (K1 and K15), expressed during lytic replication, may extend the lifespan of virus-producing cells and modulate antiviral immune responses.142 Furthermore, the KSHV encoded latent protein LANA has been shown to directly associate with STAT3 and also enhance its transcriptional activity.143 Recently, we have shown that the IL-4/STAT6 signaling network is precisely controlled by KSHV for survival and maintenance of latency where LANA reduces STAT6 activity by decreasing its phosphorylation.144 Similarly, HBV encoded HBx was also found to sensitize protein kinases such as Ras/Raf/MAPK, ERK, JNK, Akt and JAK/STAT signaling pathways.145
TNF signaling
Tumor necrosis factor (TNF) is a proinflammatory cytokine whose role has been established in the pathogenesis of both acute and chronic inflammatory diseases. When administered in large amounts, it can induce hemorrhagic necrosis of various tumors in mice model. The innate and adaptive immune responses against foreign pathogens are mediated by the TNF super-family of ligands and receptors by regulating cell death and survival.146 Tumor viruses have also evolved to target various aspects of TNF signaling (Fig. 4). The role of EBV encoded LMP1 on TNF signaling is well documented. EBV expresses LMP during viral latency and it displays the properties of a constitutively activated member of the TNF receptor (TNFR) family.147 Both TNFR1 and LMP1 share a similar set of proximal adapters and signaling pathways although they display different biological responses.147 TNFR-associated factors (TRAFs) are direct mediators of NFκB signaling by TNF family receptors. The KSHV oncoprotein vFLIP directly binds to TRAF2 and uses TRAF2 and TRAF3 for signaling to NFκB, which is crucial for KSHV-associated lymphomagenesis.148 HPV-mediated TNF resistance is a key event in the multi-step process leading to cervical cancer, even though the role of TNF seems to be more critical in HPV18 immortalized keratinocytes and HPV transformed cell lines.149 Activation of the TNFα system also has a pivotal role in the inflammatory process linked to chronic HCV-mediated hepatocellular carcinomas.150
JNK signaling
The c-Jun NH2-terminal kinase (JNK) is a member of an evolutionarily conserved sub-family of mitogen-activated protein (MAP) kinases (Fig. 4). EBV-encoded LMP1 can potentiate cisplatin-triggered apoptosis, which suggests that both JNK and NFκB signaling pathways are involved.151 LMP1 also auto-activates its own promoter through the JNK signaling pathway, which is a key mechanism for maintaining LMP1 expression during the development EBV mediated cancers.152 KSHV vFLIP modulates the JNK/AP1 pathway and induces gene expression from the cIL-6 promoter in a JNK/AP1-dependent fashion.153 The activation of JNK and p38 mitogen-activated protein kinase (MAPK) pathways is important for KSHV-mediated primary cell infection as well as KSHV reactivation from latency.154,155 Also, the HBV encoded HBx differentially activates the Ras-Raf-MAPK and JNK Pathways.156 Similarly, the HCV encoded core protein is also known to activate ERK, JNK and p38 MAP kinases157 and activation of JNK signaling pathway is crucial for HCV non-structural protein NS3-mediated cell growth.158 The diverse functions of JNKs on cell proliferation and on the induction of cell death in the context of virus mediated cancers provide an opportunity to explore the use of small molecules for targeted therapies.159 JAK is one of the cellular targets of the HCV core protein. Studies have shown that HCV core-mediated modulation of the JAK-STAT signaling pathway under IL-6 and IFNγ stimuli may also have a substantial role in the pathogenesis of HCV-related liver diseases.160 The activation of p38, which works independently of JAK-STAT signaling pathway, can also play a substantial role in the generation of the anti-HCV effects of IFNα.161 The MK2 protein, which works as a downstream kinase of p38 under IFNα stimulation, is responsible for the IFNα-mediated antiviral activity in HCV positive cells.161 These observations suggest that the activation of the p38-MK2 pathway is a major contributor to the generation of anti-HCV activity by IFNα.
IRF signaling
Members of the interferon regulatory factor (IRF) family are transcription factors that have been implicated in the regulation of a variety of biological processes (Fig. 4). The expression of IRF1 is regulated throughout the cell cycle and decreased expression of IRF1 mRNA is seen in several types of cancers.162 The EBV immediate-early protein, BZLF1, inhibits the IFNγ signaling pathway by decreasing the ability of IFNγ to activate a variety of important downstream target genes, such as IRF-1.163 EBV LMP-1 stimulates IRF-4 expression in B lymphocytes via NFκB signaling pathways.164 Interestingly, KSHV encodes four viral IRF homologues: vIRF-1 to vIRF-4. The KSHV gene ORF K9 encodes for vIRF, which is a protein with low but significant homology to members of the IRF family responsible for regulating intracellular interferon signal transduction.165 KSHV vIRF3 specifically interacts with either the DNA binding domain or the central IRF association domain of IRF7, and this interaction leads to the inhibition of IRF7 DNA binding activity and suppression of IFNα production and IFN-mediated immunity.166 Also, the vIRF-2 inhibits both IFNα and IFNγ-driven transactivation of a reporter promoter containing the interferon stimulated response element (ISRE).167 The HPV E6 oncoprotein has been shown to inhibit IRF-3 activity while the E7 gene product has been shown to inhibit both the IRF-1 and IRF-9.168 IRF-2 serves as a baseline transactivator of the HPV-16 major early promoter, P97.169 Interestingly, the HCV core protein represses IRF-1 and may be a mechanism to boost a protective innate and adaptive immune response against HCV.170 HCV and disrupted interferon signaling are believed to promote lymphoproliferation via type II CD95 and interleukins.171 The requirement of IRFs in multiple signaling pathways in which they control a cascade of events makes them attractive candidates and targets for future medical therapies for virus mediated cancers.172
Cell-signaling pathways and therapeutic intervention
Understanding the different signal transduction pathways altered in tumor virus associated malignant cells is important for detecting novel targets for cancer therapy. The EBV oncoprotein LMP1 has been shown to modulate telomerase activity by inducing a direct binding of the NFκB p65 subunit to hTERT, which facilitates the translocation of both proteins from the cytoplasm to the nucleus in NPC cells.173 An NFκB inhibitor or a dominant-negative mutant of NFκB can significantly block these effects.173 A number of tumor virus antigens can manipulate the ubiquitin-proteasome machinery to facilitate cell-proliferation by regulating the pRb, p53, pVHL, Wnt and NFκB signaling pathways. The ubiquitin system provides a platform that many viruses hijack in order to facilitate their survival and evade the immune response.174 As described earlier, small molecule drugs such as nutlin-3a targeting the p53-Mdm2 interaction shows promising results against EBV as well as KSHV mediated cancer cell lines expressing wild-type p53.55,57 A study has shown that bortezomib (Velcade), a proteasome inhibitor, can be used as a therapeutic agent against relapsed and refractory multiple myeloma, and it has been shown that Bortezomib can inhibit proliferation and induce apoptosis in KSHV-infected PEL cells.175 Several companies have efforts underway to develop inhibitors that target distinct activities of the proteasome. The most promising therapeutic targets in KS are the downstream signaling pathways upregulated by a viral G-protein coupled receptor (vGPCR).176 Cells expressing constitutively active vGPCR have a high level of activated Akt, inactivated TSC2 (a tumor suppressor which is inactivated by Akt) and activated mTOR.176 This has been reversed with either a PI3K inhibitor (LY 294002) or an mTOR inhibitor (rapamycin) in vitro and in a murine animal model.176 The use of VEGF inhibitors is also an area of active investigation for patients with KS.176 Another molecule which has been investigated as a potential treatment for KS is IL-12, which upregulates interferon-induced-protein-10 (IP-10), a known negative regulator of vGPCR.176 Furthermore, a promising therapeutic strategy for KS is the inhibition of matrix metalloproteinases (MMPs).176 Clinical trials using COL3, a known MMP inhibitor, has shown promising results in treating KS.176 Recombinant interferons (IFN), especially IFNα, has been found to be partially effective against HBV associated diseases.177 The conditional expression of human and murine IFNα and IFNγ under the control of the HIV-LTR promoter has been used to test the responsiveness of IFN to HBV and the ability of these IFNs to inhibit HBV transcripts and protein production and to activate IFN signaling in neighboring untransduced cells.177 The delivery of whole HBV genome to cells by transfection to simulate HBV infection activated IFN expression.177 IFNs were produced and secreted and protected the cells from HBV.177 The secretion of IFNs was able to activate IFN-induced signaling pathways in neighboring untransduced cells, which could provide protection to these cells. Additionally, the role of COX-2 in HBV-associated HCC has also been investigated.178 Upregulation of COX-2 correlates with VEGF expression and tumor angiogenesis in HBV-associated hepatocellular carcinoma.178 Based on these observations, the selective inhibition of COX-2 was suggested to block HCC-induced angiogenesis for treatment of the associated malignancy. Purified human soluble death receptor 5 (sDR5) could also alleviate liver damage in a mouse model by blocking tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis of HBV-transfected hepatocytes.179
Recently, a pilot study involved short-term (four week long) multiple daily intravenous doses of IFNβ for patients with chronic HBV infection.180 This is in contrast to conventional therapy, which normally lasts from twelve weeks to a year or more. The short therapy produced therapeutic effects similar to those achieved by IFNα or pegylated-IFNα (peg-IFN).180 This study showed fewer adverse effects, greater efficacy and a shorter treatment period, which led to an improvement in patients’ quality of life.180
Resveratrol is a natural phenolic compound produced by certain plants such as grapes. It has been shown to be a potent antiviral compound against numerous DNA and RNA viruses including EBV, HSV, HIV and influenza A.181 Interestingly, p53 and NFκB are the key cellular pathways that are affected by resveratrol.181
In addition to viral factors, host factors, such as obesity, have also been found to interfere with the normal activities of signaling pathways resulting in reduced biological response to therapeutic agents in chronic HCV infected patients.182 For example, increased expression of factors such as TNFα and SOCS3, which inhibit interferon signaling, may be one mechanism by which obesity can reduce the biological response to IFNα.182 More recently, SOCS3 was found to be consistently expressed in chronic HCV patients with a negative response to antiviral combination therapy, which suggests that SOCS3 and the pathways it regulates are important targets for future investigations toward the development of new therapies and more effective ways of predicting treatment responses.183
The non-structural protein NS3 of HCV enhances COX-2 gene promoter activity, its mRNA expression, COX-2 protein production and prostaglandin E2 (PGE2) release in HepG2 cells in a concentration-dependent fashion.184 Moreover, the non-structural protein NS4B of HCV has been identified as a factor that inhibits the antiviral activity by IFNα.185 HIF-1α is highly expressed in HTLV-1-infected T-cell lines and is associated with HIF-1 DNA-binding and transcriptional activity.186 Increased HIF-1α expression in these cells is dependent on Tax and the PI3K/Akt signaling pathway.186 The PI3K inhibitor, LY294002, suppressed HIF-1α protein expression, HIF-1 DNA-binding and HIF-1 transcriptional activity in HTLV-1-infected T-cell lines and represents yet another molecular target for the development of novel ATL therapeutics.
Conclusion
Cancer involves the dysregulation of multiple cell-signaling pathways governing fundamental cellular processes such as death, proliferation, differentiation and migration.187 The biologic pathways that lead to cancer are intertwined. The characteristics of viral associated cancers and their metastasized cells can also change over time. Tumor associated viruses provide a unique opportunity to understand the role played by viral proteins in transformation and to identify pathways critical for tumorigenesis and metastasis. A clear understanding of the pathways most critically involved in tumor formation and progression and the consequences of altered cell behavior in the tissue micro-environments will provide nuggets of information which will help us in formulating better therapeutic approaches. It is likely that a combination of therapeutic agents targeting multiple signal transduction pathways will be needed for maximum therapeutic benefits.
References
- 1.Bajaj BG, Murakami M, Robertson ES. biology of EBV in relationship to AIDS-associated oncogenesis. Cancer Treat Res. 2007;133:141–62. doi: 10.1007/978-0-387-46816-7_5. [DOI] [PubMed] [Google Scholar]
- 2.Nayyar VK, Shire K, Frappier L. Mitotic chromosome interactions of Epstein-Barr nuclear antigen 1 (EBNA1) and human EBNA1-binding protein 2 (EBP2) J Cell Sci. 2009;122:4341–50. doi: 10.1242/jcs.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verma SC, Lan K, Robertson E. Structure and function of latency-associated nuclear antigen. Curr Top Microbiol Immunol. 2007;312:101–36. doi: 10.1007/978-3-540-34344-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verma SC, Robertson ES. Molecular biology and pathogenesis of Kaposi sarcoma-associated herpesvirus. FEMS Microbiol Lett. 2003;222:155–63. doi: 10.1016/S0378-1097(03)00261-1. [DOI] [PubMed] [Google Scholar]
- 5.Si H, Robertson ES. Kaposi’s sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen induces chromosomal instability through inhibition of p53 function. J Virol. 2006;80:697–709. doi: 10.1128/JVI.80.2.697-709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lan K, Kuppers DA, Verma SC, Sharma N, Murakami M, Robertson ES. Induction of Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen by the lytic transactivator RTA: a novel mechanism for establishment of latency. J Virol. 2005;79:7453–65. doi: 10.1128/JVI.79.12.7453-7465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 8.Munoz N, Bosch FX, Castellsague X, Diaz M, de Sanjose S, Hammouda D, et al. Against which human papillomavirus types shall we vaccinate and screen?. The international perspective. Int J Cancer. 2004;111:278–85. doi: 10.1002/ijc.20244. [DOI] [PubMed] [Google Scholar]
- 9.Tsai WL, Chung RT. Viral hepatocarcinogenesis. Oncogene. 29:2309–24. doi: 10.1038/onc.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonilla Guerrero R, Roberts LR. The role of hepatitis B virus integrations in the pathogenesis of human hepatocellular carcinoma. J Hepatol. 2005;42:760–77. doi: 10.1016/j.jhep.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene. 2006;25:3823–33. doi: 10.1038/sj.onc.1209559. [DOI] [PubMed] [Google Scholar]
- 12.Fung J, Lai CL, Yuen MF. Hepatitis B and C virus-related carcinogenesis. Clin Microbiol Infect. 2009;15:964–70. doi: 10.1111/j.1469-0691.2009.03035.x. [DOI] [PubMed] [Google Scholar]
- 13.Schmitz U, Tan SL. NS5A—from obscurity to new target for HCV therapy. Recent Pat Antiinfect Drug Discov. 2008;3:77–92. doi: 10.2174/157489108784746597. [DOI] [PubMed] [Google Scholar]
- 14.Gallo RC. History of the discoveries of the first human retroviruses: HTLV-1 and HTLV-2. Oncogene. 2005;24:5926–30. doi: 10.1038/sj.onc.1208980. [DOI] [PubMed] [Google Scholar]
- 15.Boxus M, Willems L. Mechanisms of HTLV-1 persistence and transformation. Br J Cancer. 2009;101:1497–501. doi: 10.1038/sj.bjc.6605345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker JC, Schrama D, Houben R. Merkel cell carcinoma. Cell Mol Life Sci. 2009;66:1–8. doi: 10.1007/s00018-008-8483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busam KJ, Jungbluth AA, Rekthman N, Coit D, Pulitzer M, Bini J, et al. Merkel cell polyomavirus expression in merkel cell carcinomas and its absence in combined tumors and pulmonary neuroendocrine carcinomas. Am J Surg Pathol. 2009;33:1378–85. doi: 10.1097/PAS.0b013e3181aa30a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci USA. 2008;105:16272–7. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald ER, 3rd, El-Deiry WS. Cell cycle control as a basis for cancer drug development)\ Int J Oncol. 2000;16:871–86. [PubMed] [Google Scholar]
- 21.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–7. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 22.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–15. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farnebo M, Bykov VJ, Wiman KG. The p53 tumor suppressor: a master regulator of diverse cellular processes and therapeutic target in cancer. Biochem Biophys Res Commun. 396:85–9. doi: 10.1016/j.bbrc.2010.02.152. [DOI] [PubMed] [Google Scholar]
- 24.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–72. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 25.Nataraj AJ, Trent JC, 2nd, Ananthaswamy HN. p53 gene mutations and photocarcinogenesis. Photochem Photobiol. 1995;62:218–30. doi: 10.1111/j.1751-1097.1995.tb05262.x. [DOI] [PubMed] [Google Scholar]
- 26.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–67. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–98. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 29.Juven-Gershon T, Oren M. Mdm2: the ups and downs. Mol Med. 1999;5:71–83. [PMC free article] [PubMed] [Google Scholar]
- 30.Momand J, Wu HH, Dasgupta G. MDM2—master regulator of the p53 tumor suppressor protein. Gene. 2000;242:15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 31.Saha A, Murakami M, Kumar P, Bajaj B, Sims K, Robertson ES. Epstein-Barr virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and degradation by deubiquitinating Mdm2. J Virol. 2009;83:4652–69. doi: 10.1128/JVI.02408-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 33.Zheleva DI, Lane DP, Fischer PM. The p53-Mdm2 pathway: targets for the development of new anticancer therapeutics. Mini Rev Med Chem. 2003;3:257–70. doi: 10.2174/1389557033488178. [DOI] [PubMed] [Google Scholar]
- 34.Vassilev LT. MDM2 inhibitors for cancer therapy. Trends Mol Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 35.O’Nions J, Allday MJ. Deregulation of the cell cycle by the Epstein-Barr virus. Adv Cancer Res. 2004;92:119–86. doi: 10.1016/S0065-230X(04)92006-4. [DOI] [PubMed] [Google Scholar]
- 36.Steegenga WT, van Laar T, Riteco N, Mandarino A, Shvarts A, van der Eb AJ, et al. Adenovirus E1A proteins inhibit activation of transcription by p53. Mol Cell Biol. 1996;16:2101–9. doi: 10.1128/mcb.16.5.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massimi P, Banks L. Repression of p53 transcriptional activity by the HPV E7 proteins. Virology. 1997;227:255–9. doi: 10.1006/viro.1996.8315. [DOI] [PubMed] [Google Scholar]
- 38.Martin ME, Berk AJ. Adenovirus E1B 55K represses p53 activation in vitro. J Virol. 1998;72:3146–54. doi: 10.1128/jvi.72.4.3146-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mietz JA, Unger T, Huibregtse JM, Howley PM. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 1992;11:5013–20. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via upregulation of beclin1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- 41.Gong GZ, Jiang YF, He Y, Lai LY, Zhu YH, Su XS. HCV NS5A abrogates p53 protein function by interfering with p53-DNA binding. World J Gastroenterol. 2004;10:2223–7. doi: 10.3748/wjg.v10.i15.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 43.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary arrest in human cancer cells. for the p53-mediated G1. Cancer Res. 1995;55:5187–90. [PubMed] [Google Scholar]
- 44.Yin C, Knudson CM, Korsmeyer SJ, Van Dyke T. Bax suppresses tumorigenesis and stimulates apoptosis in vivo. Nature. 1997;385:637–40. doi: 10.1038/385637a0. [DOI] [PubMed] [Google Scholar]
- 45.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 46.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 47.Castillo JP, Yurochko AD, Kowalik TF. Role of human cytomegalovirus immediate-early proteins in cell growth control. J Virol. 2000;74:8028–37. doi: 10.1128/jvi.74.17.8028-8037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park J, Seo T, Hwang S, Lee D, Gwack Y, Choe J. The K-bZIP protein from Kaposi’s sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J Virol. 2000;74:11977–82. doi: 10.1128/jvi.74.24.11977-11982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doniger J, Muralidhar S, Rosenthal LJ. Human cytomegalovirus and human herpesvirus 6 genes that transform and transactivate. Clin Microbiol Rev. 1999;12:367–82. doi: 10.1128/cmr.12.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–94. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 51.Fries KL, Miller WE, Raab-Traub N. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J Virol. 1996;70:8653–9. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi F, Saha A, Murakami M, Kumar P, Knight JS, Cai Q, et al. Epstein-Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities. Virology. 2009;388:236–47. doi: 10.1016/j.virol.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, Frappier L. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J Biol Chem. 2003;278:47753–61. doi: 10.1074/jbc.M307200200. [DOI] [PubMed] [Google Scholar]
- 54.Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol Cell. 2005;18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 55.Forte E, Luftig MA. MDM2-dependent inhibition of p53 is required for Epstein-Barr virus B-cell growth transformation and infected-cell survival. J Virol. 2009;83:2491–9. doi: 10.1128/JVI.01681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meek DW. The p53 response to DNA damage. DNA Repair (Amst) 2004;3:1049–56. doi: 10.1016/j.dnarep.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 57.Sarek G, Kurki S, Enback J, Iotzova G, Haas J, Laakkonen P, et al. Reactivation of the p53 pathway as a treatment modality for KSHV-induced lymphomas. J Clin Invest. 2007;117:1019–28. doi: 10.1172/JCI30945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Felsani A, Mileo AM, Paggi MG. Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene. 2006;25:5277–85. doi: 10.1038/sj.onc.1209621. [DOI] [PubMed] [Google Scholar]
- 59.Helt AM, Galloway DA. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis. 2003;24:159–69. doi: 10.1093/carcin/24.2.159. [DOI] [PubMed] [Google Scholar]
- 60.Knight JS, Sharma N, Robertson ES. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci USA. 2005;102:18562–6. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.West MJ. Structure and function of the Epstein-Barr virus transcription factor, EBNA 3C. Curr Protein Pept Sci. 2006;7:123–36. doi: 10.2174/138920306776359777. [DOI] [PubMed] [Google Scholar]
- 62.Ohtani N, Brennan P, Gaubatz S, Sanij E, Hertzog P, Wolvetang E, et al. Epstein-Barr virus LMP1 blocks p16INK4a-RB pathway by promoting nuclear export of E2F4/5. J Cell Biol. 2003;162:173–83. doi: 10.1083/jcb.200302085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jung JK, Arora P, Pagano JS, Jang KL. Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res. 2007;67:5771–8. doi: 10.1158/0008-5472.CAN-07-0529. [DOI] [PubMed] [Google Scholar]
- 64.Cho J, Baek W, Yang S, Chang J, Sung YC, Suh M. HCV core protein modulates Rb pathway through pRb downregulation and E2F-1 upregulation. Biochim Biophys Acta. 2001;1538:59–66. doi: 10.1016/s0167-4889(00)00137-3. [DOI] [PubMed] [Google Scholar]
- 65.Nyhan MJ, O’Sullivan GC, McKenna SL. Role of the VHL (von Hippel-Lindau) gene in renal cancer: a multifunctional tumour suppressor. Biochem Soc Trans. 2008;36:472–8. doi: 10.1042/BST0360472. [DOI] [PubMed] [Google Scholar]
- 66.Semenza GL. VHL and p53: tumor suppressors team up to prevent cancer. Mol Cell. 2006;22:437–9. doi: 10.1016/j.molcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 67.Cai QL, Knight JS, Verma SC, Zald P, Robertson ES. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2006;2:116. doi: 10.1371/journal.ppat.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moon EJ, Jeong CH, Jeong JW, Kim KR, Yu DY, Murakami S, et al. Hepatitis B virus X protein induces angiogenesis by stabilizing hypoxia-inducible factor-1alpha. FASEB J. 2004;18:382–4. doi: 10.1096/fj.03-0153fje. [DOI] [PubMed] [Google Scholar]
- 69.Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36:131–49. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar P, Saha A, Robertson ES. Epstein-Barr virus hijacks cell cycle machinery. Microbe. 2010:5. [Google Scholar]
- 71.Javier RT, Butel JS. The history of tumor virology. Cancer Res. 2008;68:7693–706. doi: 10.1158/0008-5472.CAN-08-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shackelford J, Pagano JS. Tumor viruses and cell signaling pathways: deubiquitination versus ubiquitination. Mol Cell Biol. 2004;24:5089–93. doi: 10.1128/MCB.24.12.5089-5093.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lagunas-Martinez A, Madrid-Marina V, Gariglio P. Modulation of apoptosis by early human papillomavirus proteins in cervical cancer. Biochim Biophys Acta. 1805:6–16. doi: 10.1016/j.bbcan.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 74.Attardi LD, Lowe SW, Brugarolas J, Jacks T. Transcriptional activation by p53, but not induction of the p21 gene, is essential for oncogene-mediated apoptosis. EMBO J. 1996;15:3693–701. [PMC free article] [PubMed] [Google Scholar]
- 75.Middeldorp JM, Pegtel DM. Multiple roles of LMP1 in Epstein-Barr virus induced immune escape. Semin Cancer Biol. 2008;18:388–96. doi: 10.1016/j.semcancer.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 76.Dittmer DP, Krown SE. Targeted therapy for Kaposi’s sarcoma and Kaposi’s sarcoma-associated herpesvirus. Curr Opin Oncol. 2007;19:452–7. doi: 10.1097/CCO.0b013e3281eb8ea7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kasibhatla S, Tseng B. Why target apoptosis in cancer treatment? Mol Cancer Ther. 2003;2:573–80. [PubMed] [Google Scholar]
- 78.Vermeulen K, Berneman ZN, Van Bockstaele DR. Cell cycle and apoptosis. Cell Prolif. 2003;36:165–75. doi: 10.1046/j.1365-2184.2003.00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Galluzzi L, Brenner C, Morselli E, Touat Z, Kroemer G. Viral control of mitochonedrial apoptosis. PLoS Pathog. 2008;4:e1000018. doi: 10.1371/journal.ppat.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Desbien AL, Kappler JW, Marrack P. The Epstein-Barr virus Bcl-2 homolog, BHRF1, blocks apoptosis by binding to a limited amount of Bim. Proc Natl Acad Sci USA. 2009;106:5663–8. doi: 10.1073/pnas.0901036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee DY, Sugden B. The latent membrane protein oncogene modifies B-cell physiology by regulating autophagy. Oncogene. 2008;27:2833–42. doi: 10.1038/sj.onc.1210946. [DOI] [PubMed] [Google Scholar]
- 82.Anderton E, Yee J, Smith P, Crook T, White RE, Allday MJ. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt’s lymphoma. Oncogene. 2008;27:421–33. doi: 10.1038/sj.onc.1210668. [DOI] [PubMed] [Google Scholar]
- 83.Swanson-Mungerson M, Bultema R, Longnecker R. Epstein-Barr virus LMP2A imposes sensitivity to apoptosis. J Gen Virol. doi: 10.1099/vir.0.021444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Belanger C, Gravel A, Tomoiu A, Janelle ME, Gosselin J, Tremblay MJ, et al. Human herpesvirus 8 viral FLICE-inhibitory protein inhibits Fas-mediated apoptosis through binding and prevention of procaspase-8 maturation. J Hum Virol. 2001;4:62–73. [PubMed] [Google Scholar]
- 85.Efklidou S, Bailey R, Field N, Noursadeghi M, Collins MK. vFLIP from KSHV inhibits anoikis of primary endothelial cells. J Cell Sci. 2008;121:450–7. doi: 10.1242/jcs.022343. [DOI] [PubMed] [Google Scholar]
- 86.Thurau M, Marquardt G, Gonin-Laurent N, Weinlander K, Naschberger E, Jochmann R, et al. Viral inhibitor of apoptosis vFLIP/K13 protects endothelial cells against superoxide-induced cell death. J Virol. 2009;83:598–611. doi: 10.1128/JVI.00629-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang S, Maeng H, Young DP, Prakash O, Fayad LE, Younes A, et al. K1 protein of human herpesvirus 8 suppresses lymphoma cell Fas-mediated apoptosis. Blood. 2007;109:2174–82. doi: 10.1182/blood-2006-02-003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Desaintes C, Demeret C, Goyat S, Yaniv M, Thierry F. Expression of the papillomavirus E2 protein in HeLa cells leads to apoptosis. EMBO J. 1997;16:504–14. doi: 10.1093/emboj/16.3.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kabsch K, Alonso A. The human papillomavirus type 16 E5 protein impairs TRAIL- and FasL-mediated apoptosis in HaCaT cells by different mechanisms. J Virol. 2002;76:12162–72. doi: 10.1128/JVI.76.23.12162-12172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuo TC, Chao CC. Hepatitis B virus X protein prevents apoptosis of hepatocellular carcinoma cells by upregulating SATB1 and HURP expression. Biochem Pharmacol. 2010;80:1093–102. doi: 10.1016/j.bcp.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 91.Lamontagne J, Pinkerton M, Block TM, Lu X. Hepatitis B and hepatitis C virus replication upregulates serine protease inhibitor Kazal, resulting in cellular resistance to serine protease-dependent apoptosis. J Virol. 84:907–17. doi: 10.1128/JVI.01249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brech A, Ahlquist T, Lothe RA, Stenmark H. Autophagy in tumour suppression and promotion. Mol Oncol. 2009;3:366–75. doi: 10.1016/j.molonc.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hippert MM, O’Toole PS, Thorburn A. Autophagy in cancer: good, bad or both? Cancer Res. 2006;66:9349–51. doi: 10.1158/0008-5472.CAN-06-1597. [DOI] [PubMed] [Google Scholar]
- 94.Wen HJ, Yang Z, Zhou Y, Wood C. Enhancement of autophagy during lytic replication by the Kaposi’s sarcoma-associated herpesvirus replication and transcription activator. J Virol. 84:7448–58. doi: 10.1128/JVI.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci USA. 107:4383–8. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:14046–51. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–59. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Niller HH, Wolf H, Minarovits J. Epigenetic dysregulation of the host cell genome in Epstein-Barr virus-associated neoplasia. Semin Cancer Biol. 2009;19:158–64. doi: 10.1016/j.semcancer.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 99.Skalska L, White RE, Franz M, Ruhmann M, Allday MJ. Epigenetic repression of p16(INK4A) by latent Epstein-Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog. 6:e1000951. doi: 10.1371/journal.ppat.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Subramanian C, Knight JS, Robertson ES. The Epstein-Barr nuclear antigen EBNA3C regulates transcription, cell transformation and cell migration. Front Biosci. 2002;7:704–16. doi: 10.2741/subraman. [DOI] [PubMed] [Google Scholar]
- 101.Lee CP, Chen JY, Wang JT, Kimura K, Takemoto A, Lu CC, et al. Epstein-Barr virus BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II. J Virol. 2007;81:5166–80. doi: 10.1128/JVI.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Palermo RD, Webb HM, Gunnell A, West MJ. Regulation of transcription by the Epstein-Barr virus nuclear antigen EBNA 2. Biochem Soc Trans. 2008;36:625–8. doi: 10.1042/BST0360625. [DOI] [PubMed] [Google Scholar]
- 103.Pantry SN, Medveczky PG. Epigenetic regulation of Kaposi’s sarcoma-associated herpesvirus replication. Semin Cancer Biol. 2009;19:153–7. doi: 10.1016/j.semcancer.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hwang S, Lee D, Gwack Y, Min H, Choe J. Kaposi’s sarcoma-associated herpesvirus K8 protein interacts with hSNF5. J Gen Virol. 2003;84:665–76. doi: 10.1099/vir.0.18699-0. [DOI] [PubMed] [Google Scholar]
- 105.Stimson L, Wood V, Khan O, Fotheringham S, La Thangue NB. HDAC inhibitor-based therapies and haematological malignancy. Ann Oncol. 2009;20:1293–302. doi: 10.1093/annonc/mdn792. [DOI] [PubMed] [Google Scholar]
- 106.Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular and molecular biology. Annu Rev Biochem. 2000;69:145–82. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 107.Herschman HR. Prostaglandin synthase 2. Biochim Biophys Acta. 1996;1299:125–40. doi: 10.1016/0005-2760(95)00194-8. [DOI] [PubMed] [Google Scholar]
- 108.Needleman P, Turk J, Jakschik BA, Morrison AR, Lefkowith JB. Arachidonic acid metabolism. Annu Rev Biochem. 1986;55:69–102. doi: 10.1146/annurev.bi.55.070186.000441. [DOI] [PubMed] [Google Scholar]
- 109.Levy GN. Prostaglandin H synthases, nonsteroidal anti-inflammatory drugs and colon cancer. Faseb J. 1997;11:234–47. [PubMed] [Google Scholar]
- 110.Marks F, Furstenberger G, Muller-Decker K. Tumor promotion as a target of cancer prevention. Recent Results Cancer Res. 2007;174:37–47. doi: 10.1007/978-3-540-37696-5_3. [DOI] [PubMed] [Google Scholar]
- 111.Reddy BS, Hirose Y, Lubet R, Steele V, Kelloff G, Paulson S, et al. Chemoprevention of colon cancer by specific cyclooxygenase-2 inhibitor, celecoxib, administered during different stages of carcinogenesis. Cancer Res. 2000;60:293–7. [PubMed] [Google Scholar]
- 112.Kaul R, Verma SC, Murakami M, Lan K, Choudhuri T, Robertson ES. Epstein-Barr virus protein can upregulate cyclo-oxygenase-2 expression through association with the suppressor of metastasis Nm23-H1. J Virol. 2006;80:1321–31. doi: 10.1128/JVI.80.3.1321-1331.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Murono S, Inoue H, Tanabe T, Joab I, Yoshizaki T, Furukawa M, et al. Induction of cyclooxygenase-2 by Epstein-Barr virus latent membrane protein 1 is involved in vascular endothelial growth factor production in nasopharyngeal carcinoma cells. Proc Natl Acad Sci USA. 2001;98:6905–10. doi: 10.1073/pnas.121016998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ray N, Bisher ME, Enquist LW. Cyclooxygenase-1 and −2 are required for production of infectious pseudorabies virus. J Virol. 2004;78:12964–74. doi: 10.1128/JVI.78.23.12964-12974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. Complete sequence and genomic analysis of rhesus cytomegalovirus. J Virol. 2003;77:6620–36. doi: 10.1128/JVI.77.12.6620-6636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Symensma TL, Martinez-Guzman D, Jia Q, Bortz E, Wu TT, Rudra-Ganguly N, et al. COX-2 induction during murine gammaherpesvirus 68 infection leads to enhancement of viral gene expression. J Virol. 2003;77:12753–63. doi: 10.1128/JVI.77.23.12753-12763.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sharma-Walia N, Raghu H, Sadagopan S, Sivakumar R, Veettil MV, Naranatt PP, et al. Cyclooxygenase 2 induced by Kaposi’s sarcoma-associated herpesvirus early during in vitro infection of target cells plays a role in the maintenance of latent viral gene expression. J Virol. 2006;80:6534–52. doi: 10.1128/JVI.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Demaria S, Pikarsky E, Karin M, Coussens LM, Chen YC, El-Omar EM, et al. Cancer and inflammation: promise for biologic therapy. J Immunother. 33:335–51. doi: 10.1097/CJI.0b013e3181d32e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Colville-Nash PR, Gilroy DW. COX-2 and the cyclopentenone prostaglandins—a new chapter in the book of inflammation? Prostaglandins Other Lipid Mediat. 2000;62:33–43. doi: 10.1016/s0090-6980(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 120.Khan MN, Lee YS. Cyclooxygenase inhibitors: Scope of their use and development in cancer chemotherapy. Med Res Rev. 2009 doi: 10.1002/med.20182. In press. [DOI] [PubMed] [Google Scholar]
- 121.Sakakibara S, Tosato G. Regulation of angiogenesis in malignancies associated with Epstein-Barr virus and Kaposi’s sarcoma-associated herpes virus. Future Microbiol. 2009;4:903–17. doi: 10.2217/fmb.09.49. [DOI] [PubMed] [Google Scholar]
- 122.Morris MA, Young LS, Dawson CW. DNA tumour viruses promote tumour cell invasion and metastasis by deregulating the normal processes of cell adhesion and motility. Eur J Cell Biol. 2008;87:677–97. doi: 10.1016/j.ejcb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 123.Gilles AM, Presecan E, Vonica A, Lascu I. Nucleoside diphosphate kinase from human erythrocytes. Structural characterization of the two polypeptide chains responsible for heterogeneity of the hexameric enzyme. J Biol Chem. 1991;266:8784–9. [PubMed] [Google Scholar]
- 124.Lascu I, Schaertl S, Wang C, Sarger C, Giartosio A, Briand G, et al. A point mutation of human nucleoside diphosphate kinase A found in aggressive neuroblastoma affects protein folding. J Biol Chem. 1997;272:15599–602. doi: 10.1074/jbc.272.25.15599. [DOI] [PubMed] [Google Scholar]
- 125.Murakami M, Kaul R, Kumar P, Robertson ES. Nucleoside diphosphate kinase/Nm23 and Epstein-Barr virus. Mol Cell Biochem. 2009;329:131–9. doi: 10.1007/s11010-009-0123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kuppers DA, Lan K, Knight JS, Robertson ES. Regulation of matrix metalloproteinase 9 expression by Epstein-Barr virus nuclear antigen 3C and the suppressor of metastasis Nm23-H1. J Virol. 2005;79:9714–24. doi: 10.1128/JVI.79.15.9714-9724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kaul R, Murakami M, Lan K, Choudhuri T, Robertson ES. EBNA3C can modulate the activities of the transcription factor Necdin in association with metastasis suppressor protein Nm23-H1. J Virol. 2009;83:4871–83. doi: 10.1128/JVI.02286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Choudhuri T, Verma SC, Lan K, Robertson ES. Expression of alphaV integrin is modulated by Epstein-Barr virus nuclear antigen 3C and the metastasis suppressor Nm23-H1 through interaction with the GATA-1 and Sp1 transcription factors. Virology. 2006;351:58–72. doi: 10.1016/j.virol.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 129.Palmieri D, Halverson DO, Ouatas T, Horak CE, Salerno M, Johnson J, et al. Medroxyprogesterone acetate elevation of Nm23-H1 metastasis suppressor expression in hormone receptor-negative breast cancer. J Natl Cancer Inst. 2005;97:632–42. doi: 10.1093/jnci/dji111. [DOI] [PubMed] [Google Scholar]
- 130.Li Z, Xiang J, Zhang W, Fan S, Wu M, Li X, Li G. Nanoparticle delivery of anti-metastatic NM23-H1 gene improves chemotherapy in a mouse tumor model. Cancer Gene Ther. 2009;16:423–9. doi: 10.1038/cgt.2008.97. [DOI] [PubMed] [Google Scholar]
- 131.Wang Z, Li Y, Sarkar FH. Notch signaling proteins: legitimate targets for cancer therapy. Curr Protein Pept Sci. 2010 Sep 1;11:398–408. doi: 10.2174/138920310791824039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hayward SD, Liu J, Fujimuro M. Notch and Wnt signaling: mimicry and manipulation by gamma herpesviruses. Sci STKE. 2006;2006:e4. doi: 10.1126/stke.3352006re4. [DOI] [PubMed] [Google Scholar]
- 133.Jayshree RS, Sreenivas A, Tessy M, Krishna S. Cell intrinsic & extrinsic factors in cervical carcinogenesis. Indian J Med Res. 2009;130:286–95. [PubMed] [Google Scholar]
- 134.Lan K, Choudhuri T, Murakami M, Kuppers DA, Robertson ES. Intracellular activated Notch1 is critical for proliferation of Kaposi’s sarcoma-associated herpesvirus-associated B-lymphoma cell lines in vitro. J Virol. 2006;80:6411–9. doi: 10.1128/JVI.00239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Anderson LJ, Longnecker R. Epstein-Barr virus latent membrane protein 2A exploits Notch1 to alter B-cell identity in vivo. Blood. 2009;113:108–16. doi: 10.1182/blood-2008-06-160937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Maliekal TT, Bajaj J, Giri V, Subramanyam D, Krishna S. The role of Notch signaling in human cervical cancer: implications for solid tumors. Oncogene. 2008;27:5110–4. doi: 10.1038/onc.2008.224. [DOI] [PubMed] [Google Scholar]
- 137.Pei J, Tang Z, Zang G, Yu Y. Blockage of Notch1 signaling modulates the T-helper (Th)1/Th2 cell balance in chronic hepatitis B patients. Hepatol Res. 40:799–805. doi: 10.1111/j.1872-034X.2010.00680.x. [DOI] [PubMed] [Google Scholar]
- 138.Hall WW, Fujii M. Deregulation of cell-signaling pathways in HTLV-1 infection. Oncogene. 2005;24:5965–75. doi: 10.1038/sj.onc.1208975. [DOI] [PubMed] [Google Scholar]
- 139.Grassmann R, Aboud M, Jeang KT. Molecular mechanisms of cellular transformation by HTLV-1 Tax. Oncogene. 2005;24:5976–85. doi: 10.1038/sj.onc.1208978. [DOI] [PubMed] [Google Scholar]
- 140.Vaysberg M, Lambert SL, Krams SM, Martinez OM. Activation of the JAK/STAT pathway in Epstein-Barr virus+-associated posttransplant lymphoproliferative disease: role of interferon-gamma. Am J Transplant. 2009;9:2292–302. doi: 10.1111/j.1600-6143.2009.02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu YP, Tan YN, Wang ZL, Zeng L, Lu ZX, Li LL, et al. Phosphorylation and nuclear translocation of STAT3 regulated by the Epstein-Barr virus latent membrane protein 1 in nasopharyngeal carcinoma. Int J Mol Med. 2008;21:153–62. [PubMed] [Google Scholar]
- 142.Brinkmann MM, Schulz TF. Regulation of intracellular signalling by the terminal membrane proteins of members of the Gammaherpesvirinae. J Gen Virol. 2006;87:1047–74. doi: 10.1099/vir.0.81598-0. [DOI] [PubMed] [Google Scholar]
- 143.Muromoto R, Okabe K, Fujimuro M, Sugiyama K, Yokosawa H, Seya T, et al. Physical and functional interactions between STAT3 and Kaposi’s sarcoma-associated herpesvirus-encoded LANA. FEBS Lett. 2006;580:93–8. doi: 10.1016/j.febslet.2005.11.057. [DOI] [PubMed] [Google Scholar]
- 144.Cai Q, Verma SC, Choi JY, Ma M, Robertson ES. Kaposi’ sarcoma Herpesvirus inhibits IL-4-mediated STAT6 phosphorylation to regulate apoptosis and maintain latency. J Virol. 2010;84:11134–44. doi: 10.1128/JVI.01293-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Matsuda Y, Ichida T. Impact of hepatitis B virus X protein on the DNA damage response during hepatocarcinogenesis. Med Mol Morphol. 2009;42:138–42. doi: 10.1007/s00795-009-0457-8. [DOI] [PubMed] [Google Scholar]
- 146.Benedict CA, Banks TA, Ware CF. Death and survival: viral regulation of TNF signaling pathways. Curr Opin Immunol. 2003;15:59–65. doi: 10.1016/s0952-7915(02)00018-3. [DOI] [PubMed] [Google Scholar]
- 147.Ndour PA, Ouk TS, Brocqueville G, Mougel A, Vanhecke E, Feuillard J, et al. Inhibition of tumor necrosis factor-induced phenotypes by short intracellular versions of latent membrane protein-1. Cell Signal. 22:303–13. doi: 10.1016/j.cellsig.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 148.Guasparri I, Wu H, Cesarman E. The KSHV oncoprotein vFLIP contains a TRAF-interacting motif and requires TRAF2 and TRAF3 for signalling. EMBO Rep. 2006;7:114–9. doi: 10.1038/sj.embor.7400580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Boccardo E, Manzini Baldi CV, Carvalho AF, Rabachini T, Torres C, Barreta LA, et al. Expression of human papillomavirus type 16 E7 oncoprotein alters keratinocytes expression profile in response to tumor necrosis factoralpha. Carcinogenesis. 31:521–31. doi: 10.1093/carcin/bgp333. [DOI] [PubMed] [Google Scholar]
- 150.Knobler H, Schattner A. TNF{alpha}, chronic hepatitis C and diabetes: a novel triad. QJM. 2005;98:1–6. doi: 10.1093/qjmed/hci001. [DOI] [PubMed] [Google Scholar]
- 151.Zhang X, Sanmun D, Hu L, Fadeel B, Ernberg I. Epstein-Barr virus-encoded LMP1 promotes cisplatin-induced caspase activation through JNK and NFkappaB signaling pathways. Biochem Biophys Res Commun. 2007;360:263–8. doi: 10.1016/j.bbrc.2007.06.043. [DOI] [PubMed] [Google Scholar]
- 152.Goormachtigh G, Ouk TS, Mougel A, Tranchand-Bunel D, Masy E, Le Clorennec C, et al. Autoactivation of the Epstein-Barr virus oncogenic protein LMP1 during type II latency through opposite roles of the NFkappaB and JNK signaling pathways. J Virol. 2006;80:7382–93. doi: 10.1128/JVI.02052-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.An J, Sun Y, Sun R, Rettig MB. Kaposi’s sarcoma-associated herpesvirus encoded vFLIP induces cellular IL-6 expression: the role of the NFkappaB and JNK/AP1 pathways. Oncogene. 2003;22:3371–85. doi: 10.1038/sj.onc.1206407. [DOI] [PubMed] [Google Scholar]
- 154.Pan H, Xie J, Ye F, Gao SJ. Modulation of Kaposi’s sarcoma-associated herpesvirus infection and replication by MEK/ERK JNK and p38 multiple mitogen-activated protein kinase pathways during primary infection. J Virol. 2006;80:5371–82. doi: 10.1128/JVI.02299-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Xie J, Ajibade AO, Ye F, Kuhne K, Gao SJ. Reactivation of Kaposi’s sarcoma-associated herpesvirus from latency requires MEK/ERK, JNK and p38 multiple mitogen-activated protein kinase pathways. Virology. 2008;371:139–54. doi: 10.1016/j.virol.2007.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tarn C, Lee S, Hu Y, Ashendel C, Andrisani OM. Hepatitis B virus X protein differentially activates RAS-RAF-MAPK and JNK pathways in X-transforming versus non-transforming AML12 hepatocytes. J Biol Chem. 2001;276:34671–80. doi: 10.1074/jbc.M104105200. [DOI] [PubMed] [Google Scholar]
- 157.Erhardt A, Hassan M, Heintges T, Haussinger D. Hepatitis C virus core protein induces cell proliferation and activates ERK JNK and p38 MAP kinases together with the MAP kinase phosphatase MKP-1 in a HepG2 Tet-Off cell line. Virology. 2002;292:272–84. doi: 10.1006/viro.2001.1227. [DOI] [PubMed] [Google Scholar]
- 158.Hassan M, Ghozlan H, Abdel-Kader O. Activation of c-Jun NH2-terminal kinase (JNK) signaling pathway is essential for the stimulation of hepatitis C virus (HCV) non-structural protein 3 (NS3)-mediated cell growth. Virology. 2005;333:324–36. doi: 10.1016/j.virol.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 159.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–49. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 160.Hosui A, Ohkawa K, Ishida H, Sato A, Nakanishi F, Ueda K, et al. Hepatitis C virus core protein differently regulates the JAK-STAT signaling pathway under inter-leukin-6 and interferon-gamma stimuli. J Biol Chem. 2003;278:28562–71. doi: 10.1074/jbc.M210485200. [DOI] [PubMed] [Google Scholar]
- 161.Ishida H, Ohkawa K, Hosui A, Hiramatsu N, Kanto T, Ueda K, et al. Involvement of p38 signaling pathway in interferon-alpha-mediated antiviral activity toward hepatitis C virus. Biochem Biophys Res Commun. 2004;321:722–7. doi: 10.1016/j.bbrc.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 162.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–84. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 163.Morrison TE, Mauser A, Wong A, Ting JP, Kenney SC. Inhibition of IFN-gamma signaling by an Epstein-Barr virus immediate-early protein. Immunity. 2001;15:787–99. doi: 10.1016/s1074-7613(01)00226-6. [DOI] [PubMed] [Google Scholar]
- 164.Xu D, Zhao L, Del Valle L, Miklossy J, Zhang L. Interferon regulatory factor 4 is involved in Epstein-Barr virus-mediated transformation of human B lymphocytes. J Virol. 2008;82:6251–8. doi: 10.1128/JVI.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gao SJ, Boshoff C, Jayachandra S, Weiss RA, Chang Y, Moore PS. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene. 1997;15:1979–85. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 166.Joo CH, Shin YC, Gack M, Wu L, Levy D, Jung JU. Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi’s sarcoma-associated herpesvirus viral IRF homolog vIRF3. J Virol. 2007;81:8282–92. doi: 10.1128/JVI.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fuld S, Cunningham C, Klucher K, Davison AJ, Blackbourn DJ. Inhibition of interferon signaling by the Kaposi’s sarcoma-associated herpesvirus full-length viral interferon regulatory factor 2 protein. J Virol. 2006;80:3092–7. doi: 10.1128/JVI.80.6.3092-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cordano P, Gillan V, Bratlie S, Bouvard V, Banks L, Tommasino M, et al. The E6E7 oncoproteins of cutaneous human papillomavirus type 38 interfere with the interferon pathway. Virology. 2008;377:408–18. doi: 10.1016/j.virol.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 169.Lace MJ, Anson JR, Haugen TH, Turek LP. Interferon regulatory factor (IRF)-2 activates the HPV-16 E6-E7 promoter in keratinocytes. Virology. 399:270–9. doi: 10.1016/j.virol.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 170.Ciccaglione AR, Stellacci E, Marcantonio C, Muto V, Equestre M, Marsili G, et al. Repression of interferon regulatory factor 1 by hepatitis C virus core protein results in inhibition of antiviral and immunomodulatory genes. J Virol. 2007;81:202–14. doi: 10.1128/JVI.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Machida K, Tsukiyama-Kohara K, Sekiguch S, Seike E, Tone S, Hayashi Y, et al. Hepatitis C virus and disrupted interferon signaling promote lymphoproliferation via type II CD95 and interleukins. Gastroenterology. 2009;137:285–96. doi: 10.1053/j.gastro.2009.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Battistini A. Interferon regulatory factors in hematopoietic cell differentiation and immune regulation. J Interferon Cytokine Res. 2009;29:765–80. doi: 10.1089/jir.2009.0030. [DOI] [PubMed] [Google Scholar]
- 173.Liu JP, Cassar L, Pinto A, Li H. Mechanisms of cell immortalization mediated by EB viral activation of telomerase in nasopharyngeal carcinoma. Cell Res. 2006;16:809–17. doi: 10.1038/sj.cr.7310098. [DOI] [PubMed] [Google Scholar]
- 174.Fujimuro M, Hayward SD, Yokosawa H. Molecular piracy: manipulation of the ubiquitin system by Kaposi’s sarcoma-associated herpesvirus. Rev Med Virol. 2007;17:405–22. doi: 10.1002/rmv.549. [DOI] [PubMed] [Google Scholar]
- 175.Sarosiek KA, Cavallin LE, Bhatt S, Toomey NL, Natkunam Y, Blasini W, et al. Efficacy of bortezomib in a direct xenograft model of primary effusion lymphoma. Proc Natl Acad Sci USA. 107:13069–74. doi: 10.1073/pnas.1002985107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sullivan RJ, Pantanowitz L, Dezube BJ. Targeted therapy for Kaposi sarcoma. BioDrugs. 2009;23:69–75. doi: 10.2165/00063030-200923020-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Matskevich AA, Cordelier P, Strayer DS. Conditional expression of IFNalpha and IFNgamma activated by HBV as genetic therapy for hepatitis B. J Interferon Cytokine Res. 2003;23:709–21. doi: 10.1089/107999003772084824. [DOI] [PubMed] [Google Scholar]
- 178.Cheng AS, Chan HL, To KF, Leung WK, Chan KK, Liew CT, et al. Cyclooxygenase-2 pathway correlates with vascular endothelial growth factor expression and tumor angiogenesis in hepatitis B virus-associated hepatocellular carcinoma. Int J Oncol. 2004;24:853–60. [PubMed] [Google Scholar]
- 179.Liu YG, Liu SX, Liang XH, Zhang Q, Gao LF, Han LH, et al. Blockade of TRAIL pathway ameliorates HBV-induced hepatocyte apoptosis in an acute hepatitis model. Biochem Biophys Res Commun. 2007;352:329–34. doi: 10.1016/j.bbrc.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 180.Okushin H, Ohnishi T, Morii K, Uesaka K, Yuasa S. Short-term intravenous interferon therapy for chronic hepatitis B. World J Gastroenterol. 2008;14:3038–43. doi: 10.3748/wjg.14.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Campagna M, Rivas C. Antiviral activity of resveratrol. Biochem Soc Trans. 38:50–3. doi: 10.1042/BST0380050. [DOI] [PubMed] [Google Scholar]
- 182.Walsh MJ, Jonsson JR, Richardson MM, Lipka GM, Purdie DM, Clouston AD, et al. Non-response to antiviral therapy is associated with obesity and increased hepatic expression of suppressor of cytokine signalling 3 (SOCS-3) in patients with chronic hepatitis C, viral genotype 1. Gut. 2006;55:529–35. doi: 10.1136/gut.2005.069674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Persico M, Capasso M, Russo R, Persico E, Croce L, Tiribelli C, et al. Elevated expression and polymorphisms of SOCS3 influence patient response to antiviral therapy in chronic hepatitis C. Gut. 2008;57:507–15. doi: 10.1136/gut.2007.129478. [DOI] [PubMed] [Google Scholar]
- 184.Lu L, Wei L, Peng G, Mu Y, Wu K, Kang L, et al. NS3 protein of hepatitis C virus regulates cyclooxygen-ase-2 expression through multiple signaling pathways. Virology. 2008;371:61–70. doi: 10.1016/j.virol.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Xu J, Liu S, Xu Y, Tien P, Gao G. Identification of the nonstructural protein 4B of hepatitis C virus as a factor that inhibits the antiviral activity of interferonalpha. Virus Res. 2009;141:55–62. doi: 10.1016/j.virusres.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 186.Tomita M, Semenza GL, Michiels C, Matsuda T, Uchihara JN, Okudaira T, et al. Activation of hypoxia-inducible factor 1 in human T-cell leukaemia virus type 1-infected cell lines and primary adult T-cell leukaemia cells. Biochem J. 2007;406:317–23. doi: 10.1042/BJ20070286. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 187.Kr eeger PK, Lauffenburger DA. Cancer systems biology: a network modeling perspective. Carcinogenesis. 31:2–8. doi: 10.1093/carcin/bgp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yim EK, Park JS. The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res Treat. 2005;37:319–24. doi: 10.4143/crt.2005.37.6.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Peng R, Gordadze AV, Fuentes Panana EM, Wang F, Zong J, Hayward GS, et al. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from non-human primate lymphocryptoviruses. J Virol. 2000;74:379–89. doi: 10.1128/jvi.74.1.379-389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Waltzer L, Perricaudet M, Sergeant A, Manet E. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-Jkappa-EBNA2-activated transcription by inhibiting the binding of RBP-Jkappa to DNA. J Virol. 1996;70:5909–15. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Subramanian C, Hasan S, Rowe M, Hottiger M, Orre R, Robertson ES. Epstein-Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J Virol. 2002;76:4699–708. doi: 10.1128/JVI.76.10.4699-4708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ikeda O, Sekine Y, Mizushima A, Oritani K, Yasui T, Fujimuro M, et al. BS69 negatively regulates the canonical NFkappaB activation induced by Epstein-Barr virus-derived LMP1. FEBS Lett. 2009;583:1567–74. doi: 10.1016/j.febslet.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 193.Kaye KM, Izumi KM, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–4. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Caldwell RG, Wilson JB, Anderson SJ, Longnecker R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9:405–11. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- 195.Wang XW, Forrester K, Yeh H, Feitelson MA, Gu JR, Harris CC. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–4. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tellinghuisen TL, Rice CM. Interaction between hepatitis C virus proteins and host cell factors. Curr Opin Microbiol. 2002;5:419–27. doi: 10.1016/s1369-5274(02)00341-7. [DOI] [PubMed] [Google Scholar]
- 197.Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci USA. 1999;96:14973–7. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Macdonald A, Harris M. Hepatitis C virus NS5A: tales of a promiscuous protein. J Gen Virol. 2004;85:2485–502. doi: 10.1099/vir.0.80204-0. [DOI] [PubMed] [Google Scholar]
- 199.Waris G, Siddiqui A. Regulatory mechanisms of viral hepatitis B and C. J Biosci. 2003;28:311–21. doi: 10.1007/BF02970150. [DOI] [PubMed] [Google Scholar]
- 200.Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–97. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 201.Nyborg JK, Egan D, Sharma N. The HTLV-1 Tax protein: revealing mechanisms of transcriptional activation through histone acetylation and nucleosome disassembly. Biochim Biophys Acta. 1799:266–74. doi: 10.1016/j.bbagrm.2009.09.002. [DOI] [PubMed] [Google Scholar]