Abstract
Nm23-H1 is a well-known tumor metastasis suppressor, which functions as a nucleoside-diphosphate kinase converting nucleoside diphosphates to nucleoside triphosphates with an expense of ATP. It regulates a variety of cellular activities, including proliferation, development, migration and differentiation known to be modulated by a series of complex signaling pathway. Few studies have addressed the mechanistic action of Nm23-H1 in the context of these cellular processes. To determine the downstream pathways modulated by Nm23-H1, we expressed Nm23-H1 in a Burkitt lymphoma derived B-cell line BJAB and performed pathway specific microarray analysis. The genes with significant changes in expression patterns were clustered in groups which are responsible for regulating cell cycle, p53 activities and apoptosis. We found a general reduction of cell cycle regulatory proteins including cyclins and cyclin dependent kinase inhibitors (anti proliferation), and upregulation of apoptotic genes which included caspase 3, 9 and Bcl-x. Nm23-H1 was also found to upregulate p53 and downregulate p21 expression. A number of these genes were validated by real time PCR and results from promoter assays indicated that Nm23-H1 expression downregulated cyclin D1 in a dose responsive manner. Further, we show that Nm23-H1 forms a complex with the cellular transcription factor AP1 to modulate cyclin D1 expression levels. BJAB cells expressing Nm23-H1 showed reduced proliferation rate and were susceptible to increased apoptosis which may in part be due to a direct interaction between Nm23-H1 and p53. These results suggest that Nm23-H1 may have a role in the regulation of cell cycle and apoptosis in human B-cells.
Keywords: Nm23-H1, B cells, cyclin D1, p53, oligo GE array
Introduction
The nm23 gene family is a closely related group of nucleoside diphosphate kinases for which eight distinct genes are known in humans (Nm23-H1 to H8).1 Traditionally, Nm23/NDP kinases were ascribed a single physiological function, namely the synthesis of nucleoside triphosphates, other than ATP.2 However mounting evidence now suggests that these proteins are multifunctional with diverse biological roles that include differentiation,3 proliferation4 and development.5 The nm23-H1 gene is the first known gene shown to regulate tumor metastasis and was identified using differential hybridization analysis between murine K-1735 melanoma cell lines that varied in metastatic potential in vivo.6 The expression of Nm23-H1 is divergent in malignant tumors; however there is sufficient data indicating that increased levels of Nm23-H1 correlated with decreased metastasis in most cancers including breast, gastric and cervical cancers.6 This data is supported by in vivo studies carried out in mice lacking the nm23-M1 gene, the murine homologue of nm23-H1.7 Nm23-M1 knockout mice had a significantly higher incidence of lung metastases compared to nm23-M1+/+ mice, when these mice were induced to develop hepatocellular carcinoma.8 However, it appears that the anti-metastatic effects of Nm23-H1 are independent of the tumor cell growth rate as the primary tumor size of the knockout mice did not change significantly.7
Although the nm23 family of genes has been studied extensively for more than a decade, the exact mechanism whereby Nm23-H1 suppresses metastasis has yet to be fully defined. Most studies have focused on its ability to inhibit motility using in vitro assays, as the ability to migrate is the hallmark of metastasis. Overexpression of Nm23-H1 almost abolished the cell motility of human MDA-MB-435 cell line in response to multiple chemo-attractants and reduced their anchorage-independent growth and invasion.9 This was also found to be true for other cell lines including cell lines of colon and prostate carcinoma origin.10 When injected into nude mice, Nm23-H1 expressing MDA-MB-435 cell line produced metastasis in significantly fewer mice than the control cell lines.11
Biochemically, the metastasis suppressing function of Nm23-H1 is independent of its NDP kinase enzymatic activity.12 However, its histidine protein kinase activity may contribute towards suppression of motility as revealed by the mutational analysis of Nm23-H1. Transfection of substitution mutants P96S and S120G which were deficient only in histidine-dependent serine auto-phosphorylation did not inhibit the motility and invasion of MDA-MB-435.13 Recent studies indicate that stimulation of phosphodiesterase activity may also correlate with the ability of Nm23-H1 to suppress MDA-MB-435 cell motility.14
Nm23 has been shown to interact with several proteins including the integrin cytoplasmic domain-associated protein 1α (ICAP-1α),15 the prune protein (Pn),16 the centrosomal kinase Aurora-A/STK15,17 the Lbc proto-oncogene,18 intermediate filaments,19 map kinases20 and telomeres.21 Although the biological significance of these interactions is not very well characterized, these studies clearly suggest that Nm23-H1 may target multiple downstream pathways.
Nm23-H1 can modulate the gene expression in several ways. It was found to repress the transcription of platelet-derived growth factor (PDGF-A) promoter through DNA binding to nuclease-hypersensitive transcriptional elements.22 The C-terminal half of Nm23-H1 exhibits strong transactivation activity in Gal4 transcriptional assays.23 Previous studies in our laboratory also suggested that Nm23-H1 can function through interaction with cellular factors like Dbl.24 The objective of this study was to explore the role of Nm23-H1 during malignant transformation of the human B cells by identifying genes, groups of genes or signaling pathways targeted by Nm23-H1. We compared the gene expression profiles of BJAB cells stably transfected with a myc tagged Nm23-H1 expression construct, with the vector alone transfected cells as a control. The results obtained from the DNA microarrays analysis confirmed the significance of some pathways previously reported, but also opened new questions about the functional role of Nm23-H1 in regulating different cellular events in normal and malignant transformed cells as well as the potential for development of therapeutic strategies in tumor regression.
Results
Characterization of nm23-H1 transfected cell lines and genes associated with Nm23-H1 expression in BJAB cell line
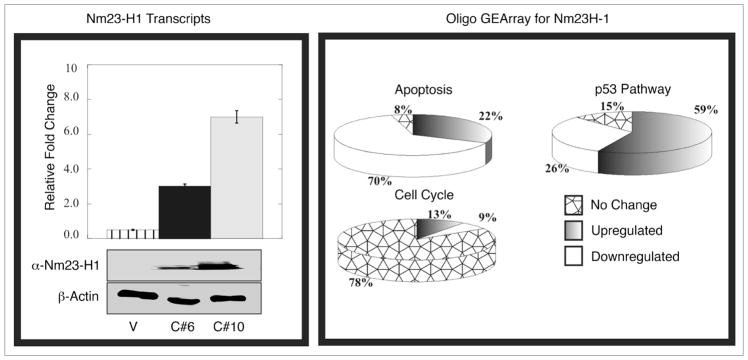
BJAB (vector control) and BJAB (pA3M-Nm23-H1) cell phenotypes are not distinguishable by morphologic variations. To determine the expression of Nm23-H1 in specific cell lines, before initiating the microarray experiments, we verified the expression of RNA and corresponding protein levels of the nm23-H1 gene in two clones C#6 and C#10 and showed that both clone 6 and 10 were expressing nm23-H1 although clone 10 was more robust in its expression (Fig. 1, Left). Gene expression of vector control profile BJAB cell line was compared with that of BJAB clones expressing nm23-H1 from an exogenous promoter by Oligo GE array. The expression profile of different genes identified within specific cellular pathways revealed the possible involvement of some previously well characterized genes. We used three different pathway specific arrays named p53, cell cycle and apoptosis oligo GE array (Fig. 1, Right). The data clearly shows downregulation of the majority of apoptotic genes up to about 70%, and close to 80% of the genes upregulated shown to be involved in cell cycle proliferation. Some genes responsible for p53 dependent cell cycle arrest were also found to be upregulated (Fig. 1, Right). These results were validated for a number of these genes by semi quantitative real time PCR. A number of these genes that were found to be up or downregulated by significant levels after normalization are shown in Table 1. In fact some of these genes significantly upregulated included the caspases 3 and 9, tumor necrosis factor receptor superfamily member 25 and the p53 tumor suppressor as well as the regulator Bcl2 associated X protein. Additionally, we saw a dramatic decrease in levels of cyclins and cyclin associated proteins (Table 1). These proteins were all associated with regulation of cell proliferation and survival in the nm23-H1 expressing BJAB cell clones investigated suggesting an important role of nm23-H1 in contributing to these activities. We decided to focus our efforts on CyclinD1 and p53 as we were able to easily obtain reagents to validate and study the regulation of their expression in this setting.
Figure 1.
Expression of Nm23-H1 in different BJAB cell lines and the Oligo GE array analysis. Western blot showed about 20-fold overexpression of Nm23-H1 protein in clone 10 cells (used for experiments) compared to clone 6 cells (highly metastatic). The same blot was stripped and probed with actin to demonstrate equal loading. As shown in same Figure, RT-PCR analysis showed 2.2-fold overexpression of nm23 transcript in clone 10 cell line compared to clone 6. The right panel showing the trend of the individual array of the specific pathway (Apoptosis, cell cycle and p53 pathway).
Table 1.
List of cellular genes which showed modulation of expression in presence of Nm23-H1 on micro array analysis and were validated by real time PCR
| Genes | Fold Change |
|---|---|
| Selected Genes Upregulated by Nm23-H1 | |
| Caspase 9, apoptosis-related cysteine protease | 16.20 |
| Tumor necrosis factor receptor superfamily, member 25 | 8.36 |
| p53 Tumor protein (Li-Fraumeni syndrome) | 2.16 |
| BCL2-associated X protein | 2.14 |
| Caspase 3, apoptosis-related cysteine protease | 2.67 |
| Selected Genes Downregulated by Nm23-H1 | |
| Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 16 |
| Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 11 |
| Cyclin D | 6 |
| Cyclin A2 | 5.88 |
| Cyclin B | 5 |
| Cyclin E2 | 2.7 |
Nm23-H1 reduces transcriptional activity of the cyclin D1 promoter in human cells
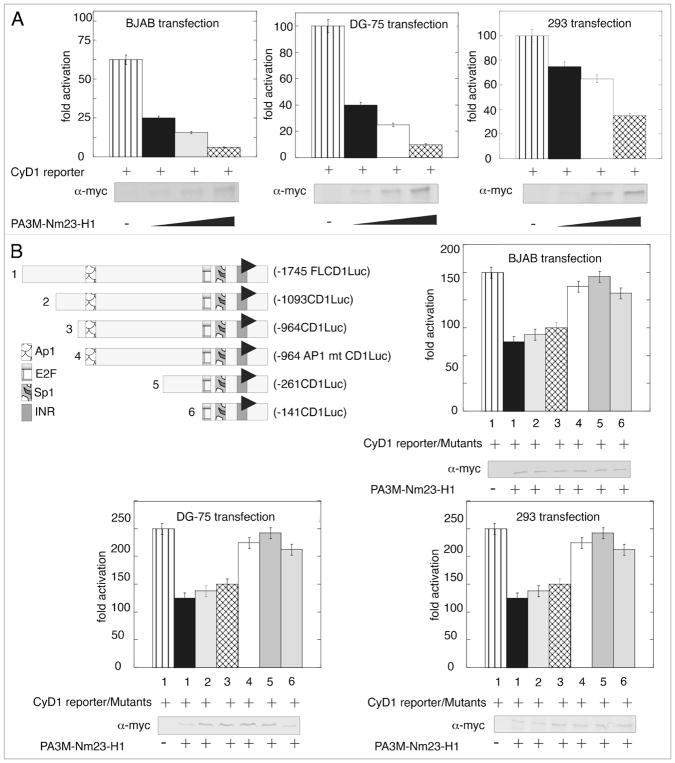
Previous studies have shown that Nm23-H1 can function as a transcription factor in regulating gene expression.25 The microarray data and the subsequent Real Time PCR validation showed that Cyclin D1 similar to other cyclins was clearly downregulated. The effect of Nm23-H1 expression on the promoter activity of full length Cyclin D1 promoter cloned into pGL2 basic vector was assessed by luciferase reporter assay. The expression vector pA3M-Nm23-H1 along with either the empty pGL2 vector, or pGL2 with Cyclin D1 promoter were transfected in BJAB, DG-75 and 293 cell lines. The total amount of DNA in each transfection was normalized by adding the appropriate amount of empty vector. The results from this assay demonstrated that the Nm23-H1 effectively downregulated the Cyclin D1 promoter in a dose-dependent manner in the three cell lines which were tested (Fig. 2A). Nm23-H1 expression was confirmed by western blot using specific antibodies against the myc epitope tag which was fused in frame at the carboxy terminus to the nm23-H1 open reading frame. Interestingly, the level of repression was more dramatic in the B cell lines compared to the epithelial cell line 293 to about 2-fold greater based on the data obtained.
Figure 2.
Nm23-H1 reduces the transcriptional activity of the Cyclin D1 promoter in B cells and binding to the specific site in the promoter. (A) pGL2-Cyclin D1 promoter was co-transfected with increasing amounts of pA3M-Nm23-H1, the pCMV-Rlu promoter and 24 h post-transfection BJAB or DG75 or 293 cells were harvested for dual luciferase assay. Increasing amounts of Nm23-H1 showed a dose-dependent decrease of promoter activity. (B) Different truncation along with the full length Cyclin D1 was shown in the left top of the figure. Either the full length or the truncated one pGL2-Cyclin D1 promoter was co-transfected with pA3M-Nm23-H1 and the CMV-Rlu promoter and 24 h post-transfection BJAB or DG 75 or 293 cells were harvested for dual luciferase assay. The promoters lacking AP1 sites (mutant number 4, 5 and 6) showed a lack of downregulation activity in presence of Nm23-H1.
The cellular transcription factor AP1 is important for modulation of the cyclin D1 promoter
To date, Nm23-H1 has not been shown to regulate transcription by directly binding DNA. Therefore we focused our attention on transcription factors which were likely to be targeted by Nm23-H1 as has previously been shown for regulation of the specific promoter like integrins, MMP 9 and Cox2.26–28 Transcription factor AP1 and Sp1 have been previously shown to activate Cyclin D1 expression.29,30 The pGL2-Cyclin D1 promoter reporter plasmid with targeted deletions which removed the Sp1, AP1 and E2F-1 binding sites were used in luciferase reporter assays (Fig. 2B). The results of these assays showed that the reporter constructs deleted for AP1 had little or no apparent change in activity when compared with the vector alone control. However, when assayed in the context of Nm23-H1 expression, an approximately 50% reduction in the transcription activity levels was seen (Fig. 2B and compare lines 1 with 4, 5 and 6). To further address the role of AP1, we generated truncated deletions of the cyclin D1 promoter reporter element here the AP1 transcription binding site was clearly deleted and when the activities were compared to the AP1 specific mutated reporter promoter plasmid the results showed that there was negligible change in the activity in the presence of Nm23-H1 in the three cell lines (Fig. 2B and compare lanes 4 with 5 and 6).
Nm23-H1 forms a complex with the cellular transcription factor AP1
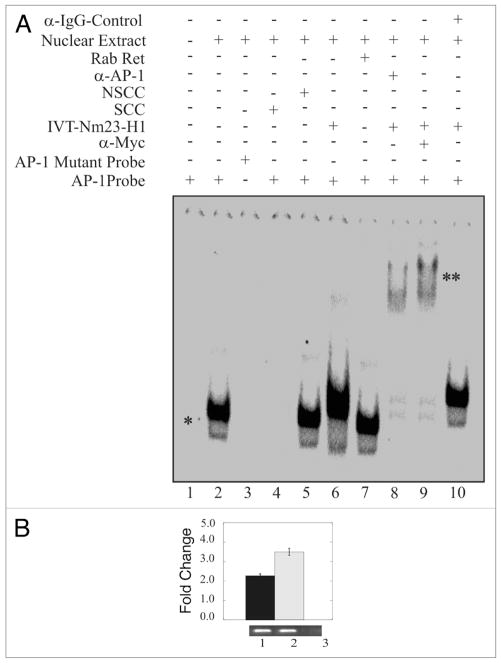
The results of the luciferase reporter promoter assays above indicated that the AP1 binding sites within the cyclin D1 promoter were critical for Nm23-H1-mediated downregulation. Therefore, we performed an electrophoretic mobility shift assay (EMSA) to determine if Nm23-H1 and AP1 formed a complex with each other bound to the cis-acting DNA binding site for AP1. In addition specific antibodies against AP1 and myc-Nm23-H1 were also used to supershift the specific complex containing AP1 and myc-Nm23-H1. The results of the EMSA showed that a shift specific for AP1 was observed when nuclear extract was added with labeled AP1 specific probe indicating that the AP1 transcription factor can bind to its specific cis-acting element at the Cyclin D1 promoter (Fig. 3A, lane 2). The AP1 specificity of the shift was confirmed because we did not see a specific shift when we used the mutant AP1 (Fig. 3A and compare lanes 1, 2 and 3). Furthermore, the specificity of this shift was also verified through the disappearance of the shift in the presence of specific cold competitor probe (Fig. 3A and compare lanes 1, 2 and 4). Importantly, the shift was not disrupted when we used a non-specific cold competitor probe (Fig. 3A, lane 5). In addition, the mobility of the AP1 probe was also reduced in the presence of in vitro translated myc-Nm23-H1 protein (Fig. 3A lane 6). A similar shift as the AP1 shift was seen when we used unprogrammed rabbit reticulocytes which was used as a control (Fig. 3A, lane 7). The presence of Nm23-H1 in the complex was verified by addition of anti-myc antibody to complex with the myc tagged Nm23-H1 protein resulting in super-shifting of the AP1 probe (Fig. 3A, lane 8). The addition of AP1-specific antibody also showed a specific supershift indicating the presence of AP1 in the complex (Fig. 3A, lane 9). The antibody specificity was shown when we use anti IgG control antibody and no supershift was observed (Fig. 3A and compare lanes 8, 9 and 10). These results suggest that Nm23-H1 can target AP1 bound to the distinct AP1 element within the Cyclin D1 promoter and therefore may regulate expression through interaction with one or more factors which is bound to the promoter element.
Figure 3.
Nm23-H1 forms a complex with the AP1 transcription factor bound to its binding site. EMSA showed a specific AP1 shift observed when nuclear extract was added to reaction with labeled AP1 specific probe. The specificity of this shift was verified through the disappearance of the shift in the presence of mutant AP1 probe or specific cold competitor (A and compare lanes 1, 2 with 3 or 4). The shift was not disrupted when we used a non-specific cold competitor (lane 5). The mobility of the AP1 probe was reduced by the presence of in vitro translated Nm23-H1 (lane 6). There was no apparent change on effect when we used un-programmed the rabbit reticulocytes (lane 7). The presence of Nm23-H1 in the complex was verified by additional supershifting in the presence of anti-myc antibody (1 μg) used to detect Nm23-H1 (lane 8). The presence of AP1 was shown by AP1-specific supershift (lane 9). Additionally, no specific supershift was observed with when the IgG control antibody was used (lane 10). (B) Chromatin from Nm23-H1 overexpressing cells was cross-linked with 1% formaldehyde followed by washing with PBS. Cell nuclei were released from these cells followed by sonication of chromatin to approximately 700 bp. Sonicated chromatin were diluted with chromatin immunoprecipitation dilution buffer. Chromatin was immunoprecipitated using antibodies specific for Nm23-H1. The primer sets mentioned in material and methods were used to amplify the regions 100 bp sequence on CyD1 promoter containing the AP1 cognate sequence. Amplified bands were quantitated and the relative amounts were calculated. Lane 1, DNA purified from 10% of the total chromatin; lane 2, DNA from chromatin immunoprecipitated with rabbit polyclonal anti-Nm23-H1; lane 3, DNA from chromatin immunoprecipitated with control immunoglobulin G antibody (Sigma, Inc.).
To determine if Nm23-H1 in fact can form a complex with AP-1 bound to its cis-acting DNA element within the cyclin D1 promotor. We performed chromatin immunoprecipitation (ChIP) analysis using anti-myc specific antibody as Nm23-H1 was tagged with the myc epitope. The results from the ChIP analysis indicated that myc tagged Nm23-H1 was in a complex with AP-1 as amplification of the AP-1 binding sequences showed positive signals (Fig. 3B). As expected, specific signals were obtained from total chromatin used as input as well as chromatin immunoprecipitated with anti myc-Nm23-H1 antibody. However, no signal was seen when control antibody was used (Fig. 3B compare lane 1 and 2 with lane 3).
Nm23-H1 enhances the transcriptional activity of a multimerized p53 responsive cis-acting DNA element in human cell lines
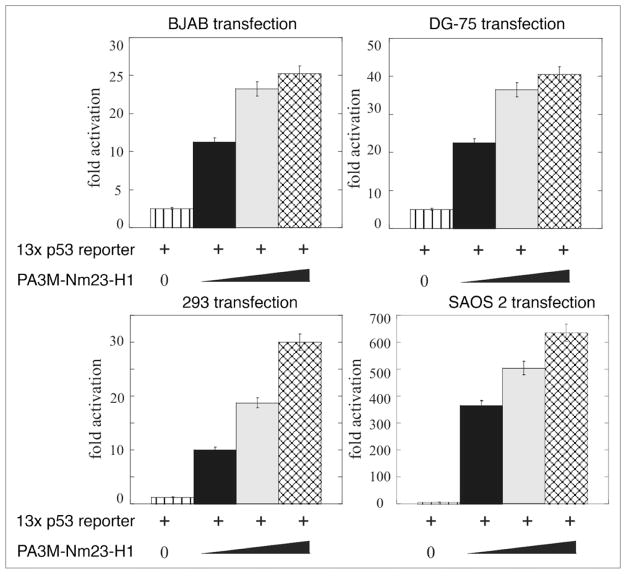
Approximately 60% of the genes from the p53 related pathway gene array were upregulated. Can Nm23-H1 modulate the transcriptional activity at p53 responsive genes? Therefore BJAB, DG-75 and 293 cells were co-transfected with the pG13 promoter construct containing the p53 cis-acting elements with increasing amounts of the Nm23-H1 expression construct. Vector control was used to normalize the total DNA being transfected. Results from this assay showed a dose dependent increase in luciferase activity in all three cell types (Fig. 4). There was a consistent pattern of increase between 25–40 fold activity in terms of RLU in these cells. The EBV negative B cell line DG75 showed the most characteristic increase with an approximately 40 fold level of activation (Fig. 4). Therefore, Nm23-H1 expression can lead to an increase in p53 transcription as seen by in vitro luciferase assay (Fig. 4). The results from in vitro luciferase assays clearly show that Nm23-H1 expression can lead to a dramatic increase in activation of the p53 responsive element suggesting that Nm23-H1 can activate the promoter possible through interaction with p53 and possibly additional factors which may form a complex with p53 bound to its cis-acting element.
Figure 4.
Nm23-H1 enhances the transcriptional activity of p53 in B cells. (A) pG13 promoter was co-transfected with increasing amounts of pA3M-Nm23-H1 and the CMV-Rlu promoter and 24 h post-transfection BJAB or DG 75 or 293 and SAOS2 cells were harvested for dual luciferase assay. Increasing amounts of Nm23-H1 showed a dose-dependent upregulation of promoter activity.
Nm23-H1 directly binds to the carboxy-terminal domain of the p53 tumor suppressor
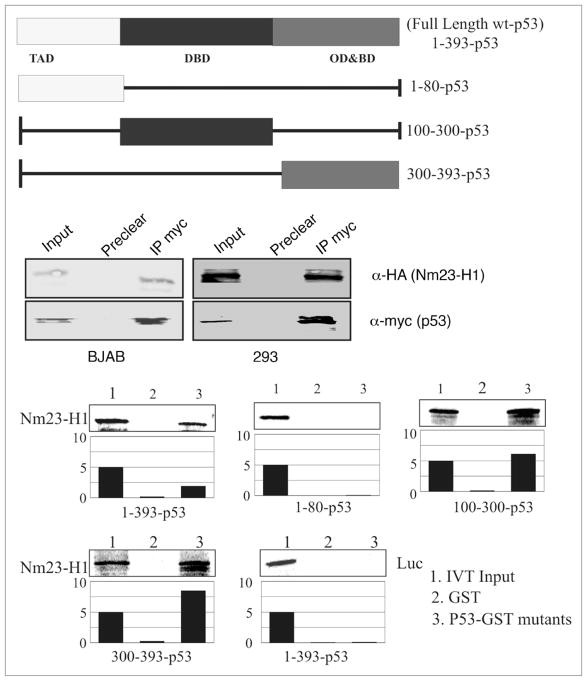
Our microarray analysis of the Nm23-H1 expressing BJAB cells demonstrated upregulation of p53 levels in BJAB cells (Table 1), which was validated by semi quantitative real time PCR. In addition the above transcription analyses suggested that Nm23-H1 may associate with p53 in complex with its binding element. To determine if there was an association between Nm23-H1 and p53, co-immunoprecipitation was performed in BJAB and 293 cells expressing Nm23-H1 and p53 from a heterologous system. Nm23-H1 tagged with the HA epitope was co-immunoprecipitated with myc tagged p53 in both cell types using anti-myc antibodies for immunoprecipitation (Fig. 5). This finding was corroborated by GST pull down assays using in vitro-translated Nm23-H1 protein labeled with 35S-Met/Cys mixture (Fig. 5). The labeled protein was incubated with a number of truncated mutants of GST-p53, as well as the full-length GST-p53 to map the binding region. The results of the binding assays indicate that the region of p53 that directly interacts with Nm23-H1 lies between amino acids 100 and 300, which primarily includes the DNA binding domain of p53 (Fig. 5). Importantly, the region of p53 between residues 300–393 also bind to Nm23-H1, which includes its basic domain and oligomerization domain. Additionally, the data indicated that the amino terminal region comprising amino acid 1–80 of p53 is not involved in binding, as there was little or no detectable interaction seen with Nm23-H1 (Fig. 5). Importantly, the luciferase control showed negligible binding to p53 fusion protein. Therefore, these results clearly show that Nm23-H1 directly interacts with p53 through its DNA binding domain and may also interact albeit with lesser affinity with the oligomerization domain. This is distinct from previous studies which show association with p53 in complex in the cells.31
Figure 5.
Nm23-H1 specifically interacts with the DNA-binding domain of p53. (A) A schematic representation of Full length domains along with the different truncations. (B) In vitro-translated Nm23-H1 protein was 35S labelled and tested for binding with a number of truncated mutants of GST-p53 constructs mapping the binding region (Full length, 1–80, 100–300 and 300–393). Relative binding levels are shown in the bar diagrams. (C) Lysates from Nm23-H1 expressing BJAB and 293 cells were incubated with the GST-p53 FL and the results of the binding are shown.
Cell expressing Nm23-H1 has reduced proliferation and are prone to apoptosis
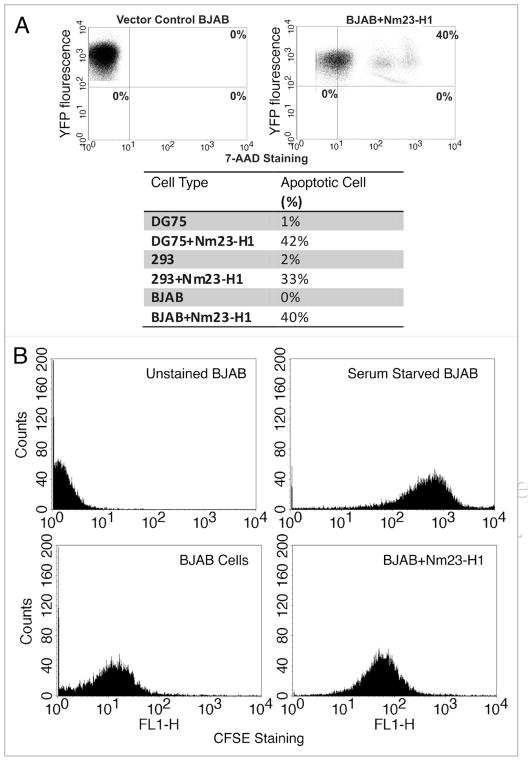
Since, we noticed decreased expression of cyclin D1 and p21, and an increased expression of p53 in Nm23-H1 expressing cell lines from our screen, we further wanted to correlate these findings with relevant biological and functional significance linked to cyclin D1 and p21 as well as p53 expression. The obvious corollary of these findings based on the functional relevance of these proteins, will be the decreased proliferation rate and increased susceptibility to apoptosis in cells expressing Nm23-H1. To determine whether or not, Nm23-H1 expressing cells would be more prone to apoptosis, we plated an equal number of BJAB cells expressing either pA3M-Nm23-H1 or pA3M vector control for 48 h and stained them with 7ADD. The data from the analysis showed that approximately 40% of cells expressing the YFP-Nm23-H1 protein took up the 7AAD stain compared to no detectable uptake in the vector control cell lines (Fig. 6A). The results of these data imply that cells expressing Nm23-H1 were significantly more prone to induction of apoptosis when compared to vector control. Moreover, we saw the same pattern when we performed the experiment in the background of other cell lines like DG75 and 293 cells (Fig. 6A, Lower). Therefore Nm23-H1 expressing cells were found to have a greater propensity for apoptosis when tested in three different cell lines independently.
Figure 6.
Nm23-H1 expressing cells have reduced proliferation and prone to apoptosis. (A) BJAB cells transfected with either with pEYFP or with pEYFP-Nm23-H1 and stained with 7AAD to detect the apoptotic cell. About 40% of the cells were both Nm23-H1 and 7AAD positive. Similarly, the results from 293 and DG75 assays shown in the table had increased 7AAD suggesting increased apoptosis. (B) BJAB cells transfected either with vector alone or with pA3M-Nm23-H1 and stained with CFSE for determination of the level of proliferation. The Nm23-H1 expressing cells showed a reduced level of proliferation.
Next we investigated the proliferation potential of BJAB cells stably expressing Nm23-H1 and compared it to cells containing vector alone using carboxyfluorescein diacetate succinimidyl ester (CFSE) assay. As a control for CFSE staining, BJAB cells were serum starved. The results showed that these cells had increased uptake of CFSE compared to BJAB vector control, which was shifted to the left. This indicated a greater level of proliferation in BJAB vector control cells when compared to the isogenic background cell line BJAB Nm23-H1 (Fig. 6B). The BJAB cells expressing Nm23-H1 had an increased level of CFSE uptake suggesting a definite decrease in the proliferation. Similar results were observed for multiple clones of Nm23-H1-BJAB and when using DG75 repeated and 293 cells (data not shown).
Nm23-H1 can promote apoptosis in a p53 dependent manner
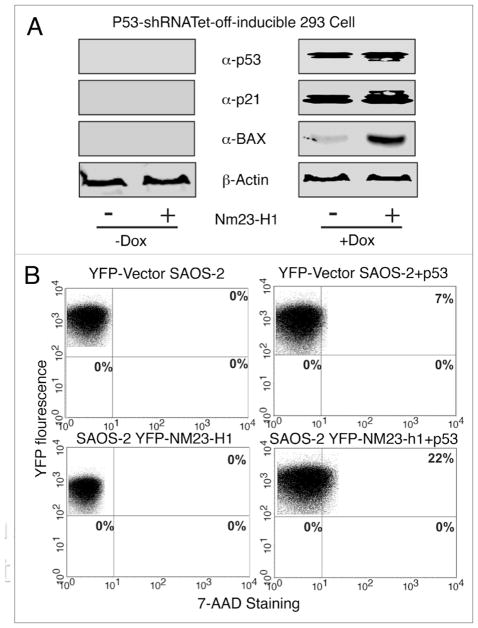
To investigate whether increased Nm23-H1 expression can induce apoptosis through utilization of the p53 dependent pathway, we used doxycycline-regulated knockdown of wild-type p53 expressing 293 cell lines. In the absence of doxycycline, the shRNA specific for p53 was expressed and the cells showed knockdown of p53 protein levels. Whereas in the presence of doxycycline, the shRNA specific for p53 was not produced resulting in expression of wild type p53 at endogenous levels (Fig. 7). We checked the status of two downstream targets of p53 i.e., Bax and p21.32–34 Both of these molecules were undetectable when the p53 specific shRNA was induced to knockdown endogenous p53. Additionally, when we transfected these cells with Nm23-H1, the p21 and Bax expression remained undetectable (Fig. 7A). However, upon doxycycline induction, the expression of the p53 specific shRNA was shut off and resulted in expression of wild-type p53 with a simultaneous increase in Bax and p21 (Fig. 7A, right). Bax along with p21 expression in the p53-expressing cells showed a dramatic increase in protein levels with increased expression of Nm23-H1 (Fig. 7A). To demonstrate that p53 is specifically involved in Nm23-H1-induced apoptosis, we used the p53-null cell line SAOS-2.35 In these p53-null cells, an increase in Nm23-H1 expression failed to induce apoptosis (Fig. 7B). However, when we introduced wild type p53 in the SAOS2 cell line from an exogenous system, apoptosis was observed in about 7% of the cells. Furthermore co-transfecting the SAOS2 cells with both p53 and YFP-Nm23-H1 resulted in a relatively high amount of approximately 22% of the cells undergoing apoptosis. These results strongly suggest that enhanced Nm23-H1 expression induces apoptosis through a p53-dependent mechanism.
Figure 7.
Nm23-H1 can induce apoptosis in a p53 dependent manner (A) Tet-off inducible p53-expressing 293 cells were transfected with Nm23-H1 and p53. Bax and p21 levels were detected by western blot in the absence and presence of tetracycline. (B) YFP tagged Nm23-H1 and pGEYFP were transfected and stained with 7AAD. The result showed there was reduced apoptosis in cells were knocked down for p53 using the shRNA strategy. Again cotransfection with p53 and Nm23-H1 showed a marked increase of apoptotic cell population.
Discussion
Nm23-H1 mediated suppression of metastasis is a well documented phenomenon. However, recent studies implicate Nm23-H1 in other aspects of carcinogenesis including anchorage independent growth and promoting tumor cell differentiation.36 It seems likely that Nm23-H1 mediates signaling that involves the coordinated regulation of multiple cellular genes and pathways. As mentioned earlier in this text the complete picture of functions, targets, and partners of Nm23-H1 is still incomplete. Different groups of investigators have reported its involvement in cell proliferation, differentiation, signaling and other cellular events.24,26–28,37 Moreover, earlier studies have also reported Nm23-H1 modulation of cellular gene expression.26,38 Zhao et al. had reported gene expression profiles of a high metastatic cell line MDA-MB-435 transfected with Nm23-H1 while Bosnar et al. overexpressed Nm23 in oral squamous cell carcinoma (OSSC) derived CAL27 cells.39,40 More recently an expression microarray analysis identified EDG2 as a candidate gene that is targeted by Nm23-H1 to suppress metastasis in MDA-MB-435 cancer cell line.14,41 The present study was done to elucidate downstream pathways and identify potential new genes involved in Nm23-H1 mediated signaling in B cells. Gene expression comparisons were made by cDNA microarray analysis to determine genes that are upregulated or downregulated by Nm23-H1 over-expression in a Burkitt lymphoma derived B-cell line, BJAB. The focus of this study was to investigate signaling involved in the regulation of cell cycle and apoptosis, two key processes during cancer development.
Gene expression analysis revealed that there was a general reduction of cell cycle regulatory proteins including cyclins and cyclin dependent kinase inhibitors and upregulation of apoptotic genes which included p53, caspase 3, 9 and Bcl-x. Promoter assays revealed that Nm23-H1 indeed reduced the transcriptional activity of the Cyclin D1 promoter in BJAB cells suggesting that the functional consequence may be a lower growth rate in cells expressing Nm23-H1. Proliferation assays showed that BJAB cells expressing Nm23-H1 had a lower growth rate than vector transfected cells. Why would a gene that suppresses metastasis, inhibit growth? Perhaps the ability to suppress metastasis and the ability to inhibit growth are regulated through two distinct pathways. The answer may lie in the fact that cell cycle withdrawal is essential for terminal cell differentiation and that Nm23-H1 induces cell differentiation.42,43 Transfection of Nm23-H1 in MDA-MB-435 cells resulted in reduced growth rate and influenced the differentiation state with the formation of organized acinus like structures and synthesis and secretion of basement membrane components in a three-dimensional matrix.44 Similarly, PC12 cells overexpressing Nm23-M1 underwent growth arrest accumulating in the G0/G1 phase of cell cycle in response to nerve growth factor (NGF) treatment.45 However, when Nm23-M1 expression was inhibited by anti-sense cDNA transfection, the cells displayed a remarkably higher proliferation rate and did not differentiate despite NGF treatment.45
A recent finding that MDA-MB-435 cells that express Nm23 showed a reduced ERK activation levels compared to control cells, may provide some insight on the molecular level as to why these cells have lower proliferation rate.46 Nm23-H1 was shown to interact and phosphorylate an ERK1/2 MAP kinase (extracellular signal-regulated kinase mitogen-activated protein kinase) scaffold protein, kinase suppressor of RAS (KSR).46 It was hypothesized that the phosphorylation of KSR by Nm23-H1 alters its scaffold function which could lead to reduced ERK activation in response to signaling.46 Since, ERK signaling regulates cell cycle progression from G0/G1 to S phase by inducting cyclin D1 followed by phosphorylation of the retinoblastoma (Rb) protein, it is plausible that Nm23-H1 targets both upstream event (e.g., ERK activation), and the downstream event (e.g., cyclin D induction) to lower the proliferation rate of cells.
The results of our screen also identified the tumor suppressor p53 as a target which was transcriptionally upregulated in Nm23-H1 transfected cells. We found that BJAB cells stably transfected with Nm23-H1 had an increased susceptibility to apoptosis and that the apoptosis seen was p53 dependent as evidenced by the expression of Bax, as well as the downstream effector of p53, p21. One of the most interesting questions in the p53 field is how a cell makes the decision to undergo growth arrest or apoptosis. It has been proposed that p53 may induce two sets of genes upon stress signals.47,48 One set mainly functions in cell growth control, such as p21/Waf-1 and GADD45, and the other set acts on apoptosis, such as Bax.49 In this study, we observed that Nm23-H1 is capable of inducing apoptosis in a B-cell line where expression of wild-type p53 is enhanced. The resulting apoptosis was accompanied by an increase in p53 levels as well as its DNA-binding activity followed by Bax expression at the protein level. Further experiments using p53-null cells as well as increased expression induced apoptosis in tumor cells via a p53-dependent pathway and expression of Bax the downstream effector of p53. Interestingly, upregulation of p21, an inhibitor of cyclin/cdk supports the anti-proliferative effect of the Nm23-H1. These findings are similar to a recent work where expression of Nm23-H1 in HeLa cells resulted in increased levels of p53 triggering the downstream antioxidative pathway.50 Interestingly p53 has been shown to be a positive regulator of Nm23-H1 in MCF-7 and J7B cells.48,51 This raises the possibility whether there is a negative feedback loop between these two proteins as our results indicate that Nm23-H1 is able to modulate p53 expression. Here in support of the previous finding by Jung et al. we now show that Nm23-H1 can directly interact with p53 and its DNA binding domain and oligomerization domain and may do so in the absence of the intermediate any molecule.31
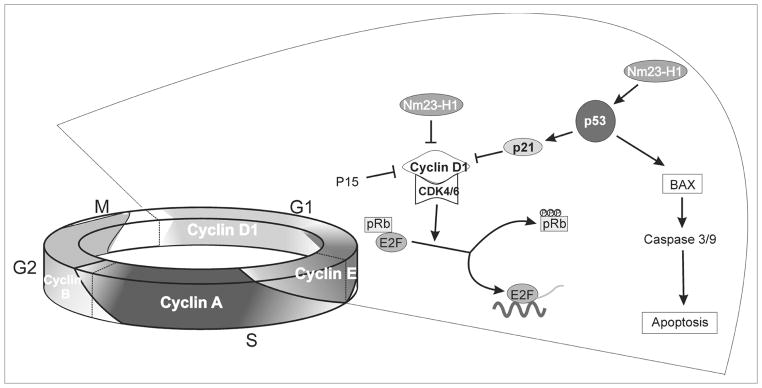
In conclusion, our microarray data analysis have indentified two important proteins targeted by Nm23-H1 that merit investigation in Nm23-H1 mediated pathways in B cells (Fig. 8). Overall, our results indicate that Nm23-H1 may have a role in the development of B cell cancers. We also show utilization of the p53 dependent pathway resulting in growth arrest and apoptosis. By using the pathway specific microarray technology we have now elucidated a number of cellular processes which can be modulated by Nm23-H1. Though our proposed model (Fig. 8) is not totally complete still we able to propose a specific pathway in our current data. Importantly, a thorough investigation of these cellular processes and their interaction with Nm23-H1 may be a step forward in enlightening its functional role in the context of cell cycle regulation, growth, proliferation and apoptosis.
Figure 8.
Schematic showing the role of Nm23-H1 in cell cycle regulation and apoptosis. A Hypothetical model shows the putative mechanisms for Nm23-H1 induced cell cycle arrest and subsequent apoptosis in p53 dependent pathway.
Materials and Methods
Cell lines and antibodies and constructs
EBV negative Burkitt’s lymphoma BJAB, DG75 cells were provided by Elliott Kieff (Brigham and Women’s Hospital, Boston, MA). BJAB-pA3M stable (pZp) and BJAB-pA3M-Nm23-H1 (clone #6 and clone #10) stable cell lines were grown in RPMI 1640 medium (Hyclone, Logan, UT) supplemented with 10% bovine growth serum, 2 mM Glutamine and 25 U/ml Penicillin/Streptomycin and 300 μg/ml G418 (active concentration).
The p53 reporter plasmid pGL-3 contains 13 copies of p53-binding sites upstream of the luciferase gene and was constructed by insertion of p53-binding sequences at EcoRV site of the pGL-3 luciferase reporter plasmid.52 The p53 expression vector pC53-C1N3 carries a wild-type human p53 gene with a proline polymorphism at residue 73 and is controlled by the cytomegalovirus promoter (gift from Gary J. Nabel, National Institutes of Health, Bethesda, MD). The pGEX-p53 construct expresses an N-terminal Glutathione S-transferase (GST)-p53 fusion protein and was derived from pGEX-2T (Amersham Pharmacia, Inc., Piscataway, NJ) by insertion of human p53 cDNA (gift from Gary J. Nabel, National Institutes of Health, Bethesda, MD) at the BamHI and EcoRI sites. The pA3M-p53 expression construct was generated by cloning PCR-amplified p53 cDNA using pGEX-p53 as a template into the previously described vector pA3M53 at EcoRI and NotI sites. GST-p53 deletion constructs were constructed by insertion of PCR fragments into the pGEX-5x-1 backbone (gift from Shelley L. Berger, The Wistar Institute, Philadelphia, PA).
Two hundred ninety-three cell line was obtained from Jon Aster (Brigham and Woman’s Hospital, Boston, MA).54 The p53-null cell line SAOS-2 was derived from a human osteosarcoma and was obtained from Jon Aster (Brigham and Women’s Hospital, Boston, MA, USA). 293 and SAOS2 cells were grown in DMEM medium (Hyclone, Logan, UT) supplemented with 10% bovine growth serum, 2 mM glutamine and 25 U/ml Penicillin/Streptomycin. The doxycycline regulated p53 inducible 293 cells were grown in DMEM medium (Hyclone, Logan, UT) supplemented with 10% bovine growth serum, 2 mM Glutamine and 25 U/ml Penicillin/Streptomycin and Puromycin. 9E10 and 12CA5 hybridoma supernatant was used as an antibody for detection myc-tag and HA-tagged proteins, respectively. Anti-p53 antibody was purchased from Santa Cruz Inc., and anti-rabbit Nm23-H1 antibody was purchased from Seikagaku Corp (Tokyo, Japan). Anti-p21 and anti-Bax antibodies were purchased from Santa Cruz, CA.
Stable transfection
BJAB cells were transfected by electroporation using a BioRad Gene Pulser II electroporator. Ten million cells were collected and washed once in phosphate buffered saline. The cells were then resuspended in 400 μl of RPMI 1640 containing DNA (pA3M or pA3M-Nm23-H1) normalized to balance total DNA and transfection efficiency was determined by GFP expression as an internal control. Once resuspended the cells were transferred to a 0.4 cm electroporation cuvettes and electroporated at 975 μF and 220 V. Following electroporation the cells were plated in 10 ml of supplemented media and grown at 37°C with 5% CO2. Cells were harvested twenty four hours post-transfection, resuspended in 1:40 ratio in DMEM supplemented with 1 mg/ml G418, and seeded on 12-well plates. The antibiotic supplemented medium was changed every 2 d until the development of stable, resistant colonies. After 3 w of selection, five resistant, pA3M-Nm23-H1, and two vector control clones were isolated. The presence of pA3M-Nm23-H1 was confirmed by western blotting using anti Nm23-H1 and anti-myc specific antibodies as well as by florescent microscopy. One of the pA3M-Nm23-H1 clone and one of the vector control clone were subjected to further analysis. The pEYFP-Nm23-H1 fusion and pEYFP control vector were also used in some experiments. We also use HA-tagged pCDNA-Nm23-H1 in some of the experiments.
Total cellular RNA isolation, microarray hybridization and analysis
Total cellular RNA was extracted from the clone expressing pA3M-Nm23-H1 and the vector control clone. The selected cell clones were thawed incubated in DMEM supplemented with 10% fetal bovine serum and 300 μg/ml G418 and seeded on Petri dishes, split and collected upon 90% confluency. The RNA was isolated using TRIzol Reagent (Invitrogen, Inc., Carlsbad, CA) following the manufacturer’s instructions. Isolated total RNA was electrophoresed through 1% agarose gel to verify the presence and integrity of rRNA. The concentration and purity of RNA were determined from absorbance measurements at 260 and 280 nm. Additional PCR using primers for intron sequences was preformed to exclude possible DNA contamination. For microarray analyses, the concentration of total RNA was adjusted to 2 μg/μl and the standard procedure for preparing the total RNA (15 μg) to be hybridized (first- and second-strand cDNA synthesis, synthesis of biotin-labeled cRNA—in vitro transcription, fragmentation), was followed as recommended by OligoGEarray standard protocol (www.superarray.com/OligoGEArray.php). Labeled and fragmented cRNA was further hybridized to probes on oligoGEArray membrane (SuperArray Bioscience Corporation, Frederick, MD) according to manufacturer’s instructions. The mRNA expression levels were evaluated using the GE Array Expression analysis Suite 5.0 Software. A comparison analysis was carried out which evaluated the relative change in abundance for each transcript.
Generation of doxycycline inducible cell lines for p53 silencing
To generate pSuper-puro-p53 for inducible expression of small interfering RNA targeting p53, the annealed oligonucleotides GAT CCC CGA CTC CAG TGG TAA TCT ACT TCA AGA GAG TAG ATT ACC ACT GGA GTC TTT TTG GAA A and AGC TTT TCC AAA AAG ACT CCA GTG GTA ATC TAC TCT CTT GAA GTA AGA TTA CCA CTG GAG TCG GG were ligated into the pSIREN-RetroQ-TetP vector (BD Bioscience, San Jose, CA). The constructs, pSuper-puro-p53 along with the control vector pSuper-puro-Luciferase, were transfected in 293 cells and then selected with 0.5 μg/ml puromycin (Sigma, St. Louis, MO) to get the stable clone.
Real-time PCR
To verify the results of the microarray analysis real-time PCR was preformed (Applied Biosystems, Foster City, CA) according the manufacturer’s instructions. Total RNA was isolated as described earlier. The cDNA was prepared with 2 μg of total RNA in a total volume of 100 μl, using a Superscript II RT kit (Invitrogen, Inc., Carlsbad, CA) according to the manufacturer’s instructions. Real-time PCR reactions were performed in 25 μl volume with 2.5 μl cDNA, 0.2 μM each primer and SYBR Green PCR Master Mix (Applied Biosystems). All PCR reactions were run at 95°C for 10 min, followed by 40 cycles 95°C for 15 s, 62°C for 30 s and 60°C for 30 s. Each sample was done in triplicate. The melting curve analysis was performed after each run. Validation experiments were done by making serial dilution of the cDNA, and calculated using the 2−ΔΔCt method (ΔΔCt = ΔCt (sequence of interest) − ΔCt (GAPDH DNA), and ΔCt = Ct (immunoprecipitated DNA) − Ct (input DNA), Ct = threshold cycle).55 Target genes Ct values were normalized against endogenous control GAPDH. The real-time PCR was performed in triplicates in two independent experiments.
Western blotting
Western blotting assays were performed as previously reported.26 Western blots were performed using antibodies specific to Nm23-H1, p53, p21 and Bax along with antibodies specific to the HA and myc tag and infra red labeled secondary antibodies (Rockland, Inc., Gilbertsville, PA). Blots were scanned and signals were detected with an Odyssey Imager (LiCor, Inc., Lincoln, NE).56–59
Electrophoresis mobility shift assays (EMSA)
The probes for the AP-1 binding site within the Cyclin D1 promoter have been previously described.26 The probes were end-labeled by Klenow fill-in reaction with [α-32P] dCTP and purified with NucTrap Probe Purification Columns (Stratagene, Inc., La Jolla, CA). Radioactive probes were diluted in STE (100 mM NaCl, 10 mM Tris pH 7.5, 1 mM EDTA) to a final concentration of 100,000 cpm/μl. DNA binding reactions and the preparation of nuclear extracts were performed as described previously.26 BJAB cells were used for preparing nuclear extract. Invitro translated myc-Nm23-H1 was used. Fifteen micrograms of protein from nuclear extracts was mixed with 1 μg poly (dI-dC) (Sigma) in DNA binding buffer (20 mM HEPES pH 7.5, 0.01% NP-40, 5.0% glycerol, 10 mM MgCl2, 100 μg of bovine serum albumin, 1 mM DTT, 1 mM PMSF, 40 mM KCl) to a total volume of 50 μl and incubated at room temperature for 5 min. One microliter of labeled probe was added to each reaction followed by an additional 15 min at room temperature. Cold competitors (200x) were added prior to the initial incubation at room temperature. Rabbit polyclonal antibodies against AP1 and anti-myc ascites were used for supershifting the specific bands. DNA-protein complexes were resolved on a non-denaturing 6% PAGE run in 0.5x TBE buffer at a constant voltage of 150 V. Following electrophoresis, the gels were dried and exposed to a Phosphor Imager Screen (Amersham Biosciences Inc., Piscataway, NJ) for 48–72 h.
Chromatin immunoprecipitation assay
Nm23-H1 (overexpressed with myc-Nm23-H1) expressing cells were cross-linking with 1% formaldehyde for 10 min at room temperature. Cross-linking was stopped by adding 125 mM glycine to the culture medium. Cells were washed twice with unlabeled phosphate-buffered saline (PBS) and resuspended in cell lysis buffer containing 5 mM piperazine-N, N-bis(2-ethanesulfonic acid) (PIPES), KOH, pH 8.0, 85 mM KCl, 0.5% NP-40 and protease inhibitors and incubated on ice for 10 min. Cells were dounced for efficient lysis followed by centrifugation at 5,000 rpm for 5 min at 4°C. Nuclei were resuspended in nuclei lysis buffer, 50 mM Tris, pH 8.0/10 mM EDTA/1% SDS containing protease inhibitors and incubated for 10 min. Chromatin were sonicated to an average length of 700 bp and cell debris were removed by centrifugation at high speed for 15 min at 4°C. Supernatant containing the sonicated chromatin was diluted fivefold with chromatin immunoprecipitation dilution buffer containing 0.01% SDS/1.0% Triton X-100/1.2 mM EDTA, 16.7 mM Tris, pH 8.1/167 mM NaCl including protease inhibitors. Samples were precleared with salmon sperm DNA/protein A/Sepharose slurry for 30 min at 4°C with rotation. Supernatants were collected after brief centrifugation; 10% of the total supernatant was saved for input control and the remaining 90% was divided into fractions: (i) control antibody (Sigma, Inc.,), (ii) Rabbit-polyclonal anti Nm23-H1 from Seikagaku Corp (Tokyo, Japan). Immune complex was precipitated using salmon sperm DNA/Protein A/Protein G slurry. Beads were then washed consecutively with low-salt buffer containing 0.1% SDS/1.0% Triton X-100/2 mM EDTA-20 mM Tris, pH 8.1/150 mM NaCl; high-salt buffer containing 0.1% SDS/1.0% Triton X-100/2 mM EDTA-20 mM Tris, pH 8.1/500 mM NaCl; LiCl wash buffer containing 0.25 M LiCl/1.0%NP-40/1% deoxycholate-1 mM EDTA-10 mM Tris, pH 8.0; and twice in Tris-EDTA. The complex was eluted using elution buffer containing 1% SDS/0.1 M NaHCO3 and reverse cross-linked by adding 0.3 M NaCl at 65°C for 4–5 h. Eluted DNA was precipitated and treated with proteinase K at 45°C for 2 h and was subjected to purification. The primers [Forward primer 5′ TCT GAA TGG AAA GCT GAG AAA CAG 3′ and Reverse Primer 5′ CCC AGG CAG AGG GGA CTA ATA 3′] and it targets 100 bp sequence on CyD1 promoter and has AP1 site in the centre Amplified bands were quantified using KODAK 1D 3.6 imaging software (Kodak Gel Logic, Rochester, NY).
Glutathione S-transferase (GST) fusion protein preparation, in vitro binding assays, and cellular lysate binding
Escherichia coli strain BL21 cells were transformed with pGEX2T-p53 plasmids expressing three different truncations of p53 and selected on an ampicillin plate. A culture grown overnight from a single colony was inoculated into 500 ml of Luria-Bertani medium and grown to mid-exponential phase with shaking. The cells were induced with 1 mM isopropyl-D-thiogalactopyranoside (IPTG) overnight at 30°C with shaking. The cells were subsequently harvested and sonicated, and the protein was solubilized. The lysate was then incubated with glutathione-Sepharose beads overnight at 4°C with rotation. The beads were collected by centrifugation and washed four times with NETN (20 mM Tris HCl [pH 8.0]-100 mM NaCl-1 mM EDTA-0.5% Nonidet P-40) containing protease inhibitors. The protein-bound beads were stored at 4°C in NETN containing protease inhibitors. The full-length pA3M clone of the Nm23-H1 gene was transcribed and translated in vitro with [35S]-methionine-cysteine in the T7 TNT system (Promega, Inc., Madison, WI). The in vitro translated proteins were first precleared with glutathione-Sepharose beads in binding buffer (1X PBS, 0.1% NP-40, 0.5 mM dithiothreitol, 10% glycerol, supplemented with protease inhibitors, 1 mM phenyl-methylsulfonyl fluoride, 2 μg of aprotinin per ml, 1 μg of pepstatin A per ml, 2 μg of leupeptin per ml) for 30 min at 4°C with rotation, and the beads were removed by centrifugation. A second preclearing was done with GST-bound Glutathione-Sepharose beads followed for 1 h at 4°C with rotation, with the beads removed by centrifugation. The precleared protein was incubated with truncation mutant constructs of GST-p53 for 16 h at 4°C with rotation. The beads were then pelleted by centrifugation and washed four times with the binding buffer. The beads and bound protein were then denatured with sodium dodecyl sulfate (SDS)-β-mercaptoethanol lysis buffer with boiling, followed by SDS-polyacrylamide gel electrophoresis (PAGE). The gel was then dried and exposed to a storage phosphor screen (Amersham Biosciences Inc., Piscataway, NJ). Western blotting using the specific anti-myc antibody was performed to detect Nm23-H1.
Coimmunoprecipitation
Nm23-H1 and p53 were sub-cloned in pCDNA3.1-HA and pA3M-myc respectively by PCR amplification of the respective template. The clones were analyzed for integrity by restriction digestion and sequence analysis. The pA3M-p53 (Myc-tagged) and pCDNA3.1-HA-Nm23-H1 constructs were transfected separately into 293 and BJAB cells. Cells were harvested 36 h post transfection and lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40, 1 mM EDTA, pH 8.0) with protease inhibitors (1 mM phenylmethyl sulfonyl fluoride (PMSF), 10 μg/μl pepstatin, 10 μg/ml leupeptin and 10 μg/ml aprotinin). Lysates were centrifuged to remove cell debris and precleared using control antibody. Precleared lysates were then incubated with anti-Myc antibody (Myc ascites) overnight at 4°C with rotation followed by incubation with protein A/G Sepharose beads at 4°C for 1 h. The resulting immunoprecipitates were collected by centrifugation at 2,000 g for 3 min at 4°C and the pellets were washed four times with 1 ml of ice-cold RIPA buffer and resuspended in 30 μl of 2 μl SDS protein sample buffer (62.5 mM Tris, pH 6.8, 40 mM dithiothreitol, 2% SDS, 0.025% bromophenol blue and 10% glycerol). The proteins were resolved on 10% SDS-PAGE, transferred to nitrocellulose membranes, and subjected to immunodetection of Nm23-H1 using specific HA antibody. The same membrane was stripped for the detection of p53 in immunoprecipitation.
Reporter assays
Ten million cells (BJAB or DG75 or 293 cells) were co-transfected with 5 μg Cyclin D1 or pG13 promoter construct containing 13 copies of the p53 responsive element kindly provided by Dr. Wafik El-Deiry, along with pCMV-Rulc (10:1 ratio) constructs and the indicated amounts of pA3M-Nm23-H1. The differences in amount of the Nm23-H1 construct were adjusted with pA3M vector to keep the total amount of transfected DNA constant. Twenty-four h post-transfection, the cells were harvested, washed with PBS, and lysed in 300 μl of reporter lysis buffer. A 40 μl aliquot of the lysate was transferred to a 96-well plate. Luciferase activity was measured using an LMaxII384 luminometer (Molecular Devices, Sunnyvale, CA) by injecting 25 μl of luciferase substrate into each well and integrating the luminescence for 10 s post-injection. The total protein was measured after using stop and glow reagent (Promega Pvt. Ltd., Singapore).
Proliferation assays
For assessing cell proliferation we performed 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE) assay. BJAB, DG75, 293 and SAOS-2 cell lines transfected with pA3M-Nm23-H1 or pA3M as vector control in experiments. In carboxyfluorescein diacetate succinimidyl ester (CFSE) labeling experiments, briefly, 10 million cells were washed and incubated with 2.5 mM CFSE (Molecular Probes, Inc., Eugene, OR) in PBS for 10 min in the dark at room temperature. Unbound CFSE was quenched by the addition of BGS. The labeled cells were washed twice with 5% BGS in PBS and re-pelleted at a concentration of 300,000 cells per μl in RPMI 1640 medium containing 10% Bovine Growth Serum (BGS). The cells were incubated in culture media supplemented with 0.1% fetal bovine serum and assayed at 72 h post-staining using a flowcytometer. All flowcytometric analyses were conducted on a FACS-Caliber cytometer using cell quest software (Becton Dickinson Inc., San Jose, CA).
Apoptosis assays
BJAB, DG75, 293 and SAOS-2 cells were transfected with YFP-Nm23-H1 along with YFP vector clone. Transfected cells were incubated in growth medium for approximately 24 h to allow for sufficient levels of transgene expression. In cell viability analysis experiments, cells were collected, washed, and incubated with 10 μl 7-aminoactinomycin D (7AAD) (Pharmingen Inc., San Jose, CA) for 10 min in the dark at 4°C. Five hundred micro liters of PBS was then added, and cells were taken for FACS analysis immediately. 7AAD positive cells were counted as dead cells.
Acknowledgments
This work was supported by NIH grants NCI CA72150-07, NCI CA91792-01, NIDCR DE14136-01 and NCI CA108461 to E.S.R. E.S.R. is a scholar of the Leukemia and Lymphoma Society of America. Also partially supported by Institute of Life Sciences (Dept of Biotechnology, India) core grant.
References
- 1.Lacombe ML, Milon L, Munier A, Mehus JG, Lambeth DO. The human Nm23/nucleoside diphosphate kinases. J Bioenerg Biomembr. 2000;32:247–58. doi: 10.1023/a:1005584929050. [DOI] [PubMed] [Google Scholar]
- 2.Parks RE, Jr, Brown PR, Cheng YC, Agarwal KC, Kong CM, Agarwal RP, Parks CC. Purine metabolism in primitive erythrocytes. Comp Biochem Physiol B. 1973;45:355–64. doi: 10.1016/0305-0491(73)90070-9. [DOI] [PubMed] [Google Scholar]
- 3.Rosengard AM, Krutzsch HC, Shearn A, Biggs JR, Barker E, Margulies IM, et al. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature. 1989;342:177–80. doi: 10.1038/342177a0. [DOI] [PubMed] [Google Scholar]
- 4.Cipollini G, Berti A, Fiore L, Rainaldi G, Basolo F, Merlo G, et al. Downregulation of the nm23. h1 gene inhibits cell proliferation. Int J Cancer. 1997;73:297–302. doi: 10.1002/(sici)1097-0215(19971009)73:2<297::aid-ijc22>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 5.Lakso M, Steeg PS, Westphal H. Embryonic expression of nm23 during mouse organogenesis. Cell Growth Differ. 1992;3:873–9. [PubMed] [Google Scholar]
- 6.Steeg PS, Bevilacqua G, Pozzatti R, Liotta LA, Sobel ME. Altered expression of NM23, a gene associated with low tumor metastatic potential, during adenovirus 2 Ela inhibition of experimental metastasis. Cancer Res. 1988;48:6550–4. [PubMed] [Google Scholar]
- 7.Boissan M, Beurel E, Wendum D, Rey C, Lecluse Y, Housset C, et al. Overexpression of insulin receptor substrate-2 in human and murine hepatocellular carcinoma. Am J Pathol. 2005;167:869–77. doi: 10.1016/S0002-9440(10)62058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boissan M, Wendum D, Arnaud-Dabernat S, Munier A, Debray M, Lascu I, et al. Increased lung metastasis in transgenic NM23-Null/SV40 mice with hepatocellular carcinoma. J Natl Cancer Inst. 2005;97:836–45. doi: 10.1093/jnci/dji143. [DOI] [PubMed] [Google Scholar]
- 9.Leone A, Flatow U, VanHoutte K, Steeg PS. Transfection of human nm23-H1 into the human MDA-MB-435 breast carcinoma cell line: effects on tumor metastatic potential, colonization and enzymatic activity. Oncogene. 1993;8:2325–33. [PubMed] [Google Scholar]
- 10.Steeg PS, de la Rosa A, Flatow U, MacDonald NJ, Benedict M, Leone A. Nm23 and breast cancer metastasis. Breast Cancer Res Treat. 1993;25:175–87. doi: 10.1007/BF00662142. [DOI] [PubMed] [Google Scholar]
- 11.Kaul R, Murakami M, Choudhuri T, Robertson ES. Epstein-Barr virus latent nuclear antigens can induce metastasis in a nude mouse model. J Virol. 2007;81:10352–61. doi: 10.1128/JVI.00886-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freije JM, Blay P, MacDonald NJ, Manrow RE, Steeg PS. Site-directed mutation of Nm23-H1. Mutations lacking motility suppressive capacity upon transfection are deficient in histidine-dependent protein phosphotransferase pathways in vitro. J Biol Chem. 1997;272:5525–32. doi: 10.1074/jbc.272.9.5525. [DOI] [PubMed] [Google Scholar]
- 13.MacDonald NJ, Freije JM, Stracke ML, Manrow RE, Steeg PS. Site-directed mutagenesis of nm23-H1. Mutation of proline 96 or serine 120 abrogates its motility inhibitory activity upon transfection into human breast carcinoma cells. J Biol Chem. 1996;271:25107–16. doi: 10.1074/jbc.271.41.25107. [DOI] [PubMed] [Google Scholar]
- 14.Horak CE, Mendoza A, Vega-Valle E, Albaugh M, Graff-Cherry C, McDermott WG, et al. Nm23-H1 suppresses metastasis by inhibiting expression of the lysophosphatidic acid receptor EDG2. Cancer Res. 2007;67:11751–9. doi: 10.1158/0008-5472.CAN-07-3175. [DOI] [PubMed] [Google Scholar]
- 15.Fournier HN, Dupe-Manet S, Bouvard D, Lacombe ML, Marie C, Block MR, Albiges-Rizo C. Integrin cytoplasmic domain-associated protein 1alpha (ICAP-1alpha) interacts directly with the metastasis suppressor nm23-H2, and both proteins are targeted to newly formed cell adhesion sites upon integrin engagement. J Biol Chem. 2002;277:20895–902. doi: 10.1074/jbc.M200200200. [DOI] [PubMed] [Google Scholar]
- 16.D’Angelo A, Garzia L, André A, Carotenuto P, Aglio V, Guardiola O, et al. Prune cAMP phosphodiesterase binds nm23-H1 and promotes cancer metastasis. Cancer Cell. 2004;5:137–49. doi: 10.1016/s1535-6108(04)00021-2. [DOI] [PubMed] [Google Scholar]
- 17.Du J, Hannon GJ. The centrosomal kinase Aurora-A/STK15 interacts with a putative tumor suppressor NM23-H1. Nucleic Acids Res. 2002;30:5465–75. doi: 10.1093/nar/gkf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwashita S, Fujii M, Mukai H, Ono Y, Miyamoto M. Lbc proto-oncogene product binds to and could be negatively regulated by metastasis suppressor nm23-H2. Biochem Biophys Res Commun. 2004;320:1063–8. doi: 10.1016/j.bbrc.2004.06.067. [DOI] [PubMed] [Google Scholar]
- 19.Roymans D, Willems R, Vissenberg K, De Jonghe C, Grobben B, Claes P, et al. Nucleoside diphosphate kinase beta (Nm23-R1/NDPKbeta) is associated with intermediate filaments and becomes upregulated upon cAMP-induced differentiation of rat C6 glioma. Exp Cell Res. 2000;261:127–38. doi: 10.1006/excr.2000.5037. [DOI] [PubMed] [Google Scholar]
- 20.Kim MS, Lee EJ, Kim HR, Moon A. p38 kinase is a key signaling molecule for H-Ras-induced cell motility and invasive phenotype in human breast epithelial cells. Cancer Res. 2003;63:5454–61. [PubMed] [Google Scholar]
- 21.Nosaka K, Kawahara M, Masuda M, Satomi Y, Nishino H. Association of nucleoside diphosphate kinase nm23-H2 with human telomeres. Biochem Biophys Res Commun. 1998;243:342–8. doi: 10.1006/bbrc.1997.8097. [DOI] [PubMed] [Google Scholar]
- 22.Ma D, Xing Z, Liu B, Pedigo NG, Zimmer SG, Bai Z, et al. NM23-H1 and NM23-H2 repress transcriptional activities of nuclease-hypersensitive elements in the platelet-derived growth factor-A promoter. J Biol Chem. 2002;277:1560–7. doi: 10.1074/jbc.M108359200. [DOI] [PubMed] [Google Scholar]
- 23.Cho SJ, Lee NS, Jung YS, Lee H, Lee KJ, Kim E, Chae SK. Identification of structural domains affecting transactivation potential of Nm23. Biochem Biophys Res Commun. 2001;289:738–43. doi: 10.1006/bbrc.2001.6042. [DOI] [PubMed] [Google Scholar]
- 24.Murakami M, Meneses PI, Knight JS, Lan K, Kaul R, Verma SC, Robertson ES. Nm23-H1 modulates the activity of the guanine exchange factor Dbl-1. Int J Cancer. 2008;123:500–10. doi: 10.1002/ijc.23568. [DOI] [PubMed] [Google Scholar]
- 25.Lee E, Jeong J, Kim SE, Song EJ, Kang SW, Lee KJ. Multiple functions of Nm23-H1 are regulated by oxido-reduction system. PLoS One. 2009;4:7949. doi: 10.1371/journal.pone.0007949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choudhuri T, Verma SC, Lan K, Robertson ES. Expression of alphaV integrin is modulated by Epstein-Barr virus nuclear antigen 3C and the metastasis suppressor Nm23-H1 through interaction with the GATA-1 and Sp1 transcription factors. Virology. 2006;351:58–72. doi: 10.1016/j.virol.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 27.Kaul R, Verma SC, Murakami M, Lan K, Choudhuri T, Robertson ES. Epstein-Barr virus protein can upregulate cyclo-oxygenase-2 expression through association with the suppressor of metastasis Nm23-H1. J Virol. 2006;80:1321–31. doi: 10.1128/JVI.80.3.1321-1331.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuppers DA, Lan K, Knight JS, Robertson ES. Regulation of matrix metalloproteinase 9 expression by Epstein-Barr virus nuclear antigen 3C and the suppressor of metastasis Nm23-H1. J Virol. 2005;79:9714–24. doi: 10.1128/JVI.79.15.9714-9724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mechta F, Lallemand D, Pfarr CM, Yaniv M. Transformation by ras modifies AP1 composition and activity. Oncogene. 1997;14:837–47. doi: 10.1038/sj.onc.1200900. [DOI] [PubMed] [Google Scholar]
- 30.Tapias A, Ciudad CJ, Roninson IB, Noe V. Regulation of Sp1 by cell cycle related proteins. Cell Cycle. 2008;7:2856–67. doi: 10.4161/cc.7.18.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung H, Seong HA, Ha H. NM23-H1 tumor suppressor and its interacting partner STRAP activate p53 function. J Biol Chem. 2007;282:35293–307. doi: 10.1074/jbc.M705181200. [DOI] [PubMed] [Google Scholar]
- 32.Dushin NV, Radysh BB, Gonchar PA, Kutenev AV. Vision disorders as manifestations of transient ischemic attacks in the vertebrobasilar basin. Vestn Oftalmol. 2001;117:27–9. [PubMed] [Google Scholar]
- 33.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–30. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Z, Wang H, Zhang L, Tang A, Zhai Q, Wen J, et al. Both p53-PUMA/NOXA-Bax-mitochondrion and p53-p21cip1 pathways are involved in the CDglyTK-mediated tumor cell suppression. Biochem Biophys Res Commun. 2009;386:607–11. doi: 10.1016/j.bbrc.2009.06.083. [DOI] [PubMed] [Google Scholar]
- 35.Masuda H, Miller C, Koeffler HP, Battifora H, Cline MJ. Rearrangement of the p53 gene in human osteogenic sarcomas. Proc Natl Acad Sci USA. 1987;84:7716–9. doi: 10.1073/pnas.84.21.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tee YT, Chen GD, Lin LY, Ko JL, Wang PH. Nm23-H1: a metastasis-associated gene. Taiwan J Obstet Gynecol. 2006;45:107–13. doi: 10.1016/S1028-4559(09)60206-0. [DOI] [PubMed] [Google Scholar]
- 37.Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–72. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 38.Kaul R, Murakami M, Lan K, Choudhuri T, Robertson ES. EBNA3C can modulate the activities of the transcription factor Necdin in association with metastasis suppressor protein Nm23-H1. J Virol. 2009;83:4871–83. doi: 10.1128/JVI.02286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bosnar MH, Bago R, Gall-Troselj K, Streichert T, Pavelic J. Downstream targets of Nm23-H1: gene expression profiling of CAL 27 cells using DNA microarray. Mol Carcinog. 2006;45:627–33. doi: 10.1002/mc.20243. [DOI] [PubMed] [Google Scholar]
- 40.Zhao H, Jhanwar-Uniyal M, Datta PK, Yemul S, Ho L, Khitrov G, et al. Expression profile of genes associated with antimetastatic gene: nm23-mediated metastasis inhibition in breast carcinoma cells. Int J Cancer. 2004;109:65–70. doi: 10.1002/ijc.11676. [DOI] [PubMed] [Google Scholar]
- 41.Horak CE, Lee JH, Elkahloun AG, Boissan M, Dumont S, Maga TK, et al. Nm23-H1 suppresses tumor cell motility by downregulating the lysophosphatidic acid receptor EDG2. Cancer Res. 2007;67:7238–46. doi: 10.1158/0008-5472.CAN-07-0962. [DOI] [PubMed] [Google Scholar]
- 42.Mileo AM, Piombino E, Severino A, Tritarelli A, Paggi MG, Lombardi D. Multiple interference of the human papillomavirus-16 E7 oncoprotein with the functional role of the metastasis suppressor Nm23-H1 protein. J Bioenerg Biomembr. 2006;38:215–25. doi: 10.1007/s10863-006-9037-y. [DOI] [PubMed] [Google Scholar]
- 43.Lombardi D, Palescandolo E, Giordano A, Paggi MG. Interplay between the antimetastatic nm23 and the retinoblastoma-related Rb2/p130 genes in promoting neuronal differentiation of PC12 cells. Cell Death Differ. 2001;8:470–6. doi: 10.1038/sj.cdd.4400842. [DOI] [PubMed] [Google Scholar]
- 44.Howlett AR, Petersen OW, Steeg PS, Bissell MJ. A novel function for the nm23-H1 gene: overexpression in human breast carcinoma cells leads to the formation of basement membrane and growth arrest. J Natl Cancer Inst. 1994;86:1838–44. doi: 10.1093/jnci/86.24.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gervasi F, D’Agnano I, Vossio S, Zupi G, Sacchi A, Lombardi D. nm23 influences proliferation and differentiation of PC12 cells in response to nerve growth factor. Cell Growth Differ. 1996;7:1689–95. [PubMed] [Google Scholar]
- 46.Salerno M, Palmieri D, Bouadis A, Halverson D, Steeg PS. Nm23-H1 metastasis suppressor expression level influences the binding properties, stability and function of the kinase suppressor of Ras1 (KSR1) Erk scaffold in breast carcinoma cells. Mol Cell Biol. 2005;25:1379–88. doi: 10.1128/MCB.25.4.1379-1388.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choudhuri T, Pal S, Agwarwal ML, Das T, Sa G. Curcumin induces apoptosis in human breast cancer cells through p53-dependent Bax induction. FEBS Lett. 2002;512:334–40. doi: 10.1016/s0014-5793(02)02292-5. [DOI] [PubMed] [Google Scholar]
- 48.Choudhuri T, Pal S, Das T, Sa G. Curcumin selectively induces apoptosis in deregulated cyclin D1-expressed cells at G2 phase of cell cycle in a p53-dependent manner. J Biol Chem. 2005;280:20059–68. doi: 10.1074/jbc.M410670200. [DOI] [PubMed] [Google Scholar]
- 49.Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR. The p53 network. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 50.An R, Chu YL, Tian C, Dai XX, Chen JH, Shi Q, et al. Overexpression of nm23-H1 in HeLa cells provides cells with higher resistance to oxidative stress possibly due to raising intracellular p53 and GPX1. Acta Pharmacol Sin. 2008;29:1451–8. doi: 10.1111/j.1745-7254.2008.00902.x. [DOI] [PubMed] [Google Scholar]
- 51.Chen SL, Wu YS, Shieh HY, Yen CC, Shen JJ, Lin KH. p53 is a regulator of the metastasis suppressor gene Nm23-H1. Mol Carcinog. 2003;36:204–14. doi: 10.1002/mc.10110. [DOI] [PubMed] [Google Scholar]
- 52.Wang W, El-Deiry WS. Bioluminescent molecular imaging of endogenous and exogenous p53-mediated transcription in vitro and in vivo using an HCT116 human colon carcinoma xenograft model. Cancer Biol Ther. 2003;2:196–202. doi: 10.4161/cbt.2.2.347. [DOI] [PubMed] [Google Scholar]
- 53.Yi F, Saha A, Murakami M, Kumar P, Knight JS, Cai Q, et al. Epstein-Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities. Virology. 2009;388:236–47. doi: 10.1016/j.virol.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aiello L, Guilfoyle R, Huebner K, Weinmann R. Adenovirus 5 DNA sequences present and RNA sequences transcribed in transformed human embryo kidney cells (HEK-Ad-5 or 293) Virology. 1979;94:460–9. doi: 10.1016/0042-6822(79)90476-8. [DOI] [PubMed] [Google Scholar]
- 55.Cikos S, Bukovska A, Koppel J. Relative quantification of mRNA: comparison of methods currently used for real-time PCR data analysis. BMC Mol Biol. 2007;8:113. doi: 10.1186/1471-2199-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choudhuri T, Verma SC, Lan K, Murakami M, Robertson ES. The ATM/ATR signaling effector Chk2 is targeted by Epstein-Barr virus nuclear antigen 3C to release the G2/M cell cycle block. J Virol. 2007;81:6718–30. doi: 10.1128/JVI.00053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lan K, Choudhuri T, Murakami M, Kuppers DA, Robertson ES. Intracellular activated Notch1 is critical for proliferation of Kaposi’s sarcoma-associated herpesvirus-associated B-lymphoma cell lines in vitro. J Virol. 2006;80:6411–9. doi: 10.1128/JVI.00239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lan K, Murakami M, Choudhuri T, Kuppers DA, Robertson ES. Intracellular-activated Notch1 can reactivate Kaposi’s sarcoma-associated herpesvirus from latency. Virology. 2006;351:393–403. doi: 10.1016/j.virol.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 59.Verma SC, Lan K, Choudhuri T, Robertson ES. Kaposi’s sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen modulates K1 expression through its cis-acting elements within the terminal repeats. J Virol. 2006;80:3445–58. doi: 10.1128/JVI.80.7.3445-3458.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]