Graphical abstract
Keywords: Kazal-type proteinase inhibitors, Frog skin secretion, Peptidomic, Molecular cloning, Tandem mass spectrometry, Cruziohyla calcarifer
Highlights
-
•
18 novel Kazal proteins were identified in skin secretions of Cruziohyla calcarifer.
-
•
CCKPs share the C-X(7)-C-X(6,7)-C-X(6,7)-Y-X(3)-C-X(2)-C-X(15-21)-C pattern.
-
•
Trypsin and chymotrypsin inhibitory activity was proposed for 5 types of CCKPs.
-
•
CCKP-1 has trypsin inhibitory activity and molecular mass of [M+H]+ = 5926.43 Da.
Abstract
Peptidase inhibitors have an important role controlling a variety of biological processes. Here, we employed a peptidomic approach including molecular cloning, tandem mass spectrometry and enzymatic assays to reveal 7 Kazal-type proteinase inhibitors (CCKPs) (18 variants) in the skin secretion of the unexplored frog, Cruziohyla calcarifer. All 18 proteins shared the Kazal pattern C-X(7)-C-X(6,7)-C-X(6,7)-Y-X(3)-C-X(2)-C-X(15-21)-C and 3 disulphide bridges. Based on structural comparative analysis, we deemed trypsin and chymotrypsin inhibitory activity in CCKP-1, 4 and CCKP 2, 5, 7, respectively. These peptidase inhibitors presumably play a role to control the balance between other functional peptides produced in the amphibian skin secretions.
1. Introduction
Amphibian skin contains granular glands that are responsible for synthesizing a complex mixture of different biologically active peptides. Among these peptides are tachykinins, bradykinins, sauvagines, caeruleins, bombesins, dermophins, dermaseptins, and other peptides [1]. These peptides are synthetized as inactive precursors often containing a signal peptide, an acidic spacer, and the mature sequence. In addition, they usually have one or more predicted processing sites within the acidic spacer, such as lysine-arginine (KR) or two arginines (RR), which are a target for peptidases with trypsin-like specificity [2], [3], [4].
Peptidases are hydrolases that are crucial in controlling a variety of biological process, such as digestion, blood clotting, wound healing, and host defence; but they are also involved in pathogenic infection, viral replication, and disease progression [5], [6], [7]. Besides, proteolytic enzymes play and important role in releasing active mature peptides upon the surface of the frog, and for this reason these enzymes must be carefully controlled. Among the processes that could be involved in their control are spatial and temporal regulation, zymogen activation, peptidase degradation, and macromolecular peptidase inhibitors. However, these mechanism have not been proven in amphibians up to now [5], [6], [7], [8], [9], [10], [11], [12].
Peptidase inhibitors as well as peptidases are ubiquitous in nature. Due to the fact that peptidase inhibitors show a high level of homology in their active sites, they can inhibit multiple peptidases at a rate of 1:5. The mechanism of inhibition in the majority of peptidase inhibitors is competitive, interacting with the active site of the peptidase in a substrate-like manner [6].
Proteinase inhibitors of different families and specificities have been found in some amphibian species. The first proteinase inhibitors were found in the skin secretions of five species of bombinid toads, including the following trypsin/thrombin inhibitors: BSTI of Bombina bombina; BMSI1 and BMSI2 of B. microdeladigitora; BOTI of B. orientalis; BVTI of B. variegata; and BMTI of B. maxima. They have 60 amino acids and masses between 6345 and 6446.56 Da. Noticeably, they all contain 10 cysteine residues similar to the distribution pattern of the peptidase inhibitor of the nematode worm, Ascaris suum. In addition, they share the reactive centre motif -CDKKC-, and their Ki is in the range of 0.1–1 μM [8], [9], [13], [14]. Another species, Rana areolata contains three peptidase inhibitors: one trypsin inhibitor of 61 amino acids that also contains the 10 cysteine residue motif (6751 Da) and is similar to the protein inhibitor from A. suum; a 48 residue elafin-related peptide (of mass 5164 Da) with 8 cysteine residues distinctive for whey acidic protein (WAP) motif; and a secretory leukocyte peptidase inhibitor [15].
Other proteinase inhibitors found in amphibian skin secretions can be classified based on the presence of their structural motifs such as Kunitz, Bowman-Birk and Kazal types. Two Kunitz type proteinase inhibitors have been isolated from Dyscophus guineti and Kassina senegalensis. The first is a trypsin inhibitor of 57 residues and 6301 Da, which belongs to the Kunitz/bovine pancreatic trypsin inhibitor family; and the second a chymotrypsin inhibitor (KSCI) of 62 residues and 6776.24 Da, which is structurally similar to other chymotrypsin inhibitors from silkworms to snakes. KSCI has its P1 site occupied by a phenylalanine residue and contains 6 cysteines that form 3 disulphide bridges [16], [17].
The next group of proteinase inhibitors contain the Bowman-Birk motif. They include the following trypsin inhibitors: HV-BBI isolated from Odorrrana versabilis; HJTI isolated from O. hejiangensis; and pLR-HL isolated from Hylarana latouchii. These inhibitors have 17–18 residues, their masses range between 1804.83 and 2013.95 Da, and their Ki varies between 19 and 388 nM. Besides, they share a disulphide loop between 2 cysteines spaced by 9 residues, and have a lysine in the third position inside the loop, which corresponds to the P1 site of the inhibitor [18], [19]. In addition, OGTI isolated from O. grahami, is another trypsin inhibitor similar to those described above. OGTI contains 17 amino acids and its molecular mass is 1949.4 Da. However, OGTI contains a smaller disulphide loop formed by 2 cysteines that are 4 residues apart, and it has a lysine in the P1 site as the immediate residue following the first cysteine [20].
Finally, Kazal-type protein inhibitors seem to be more taxa-specific. Until now, they have only been reported in the Phyllomedusinae clade. Two prolyl endopeptidase inhibitors, PSKP-1 and PSKP-2 of 58 residues and 6695.87 and 6548.65 Da, respectively, have been isolated from Phyllomedusa sauvagii. PSKP-1 and PSKP-2 share two prolines in the P1 and P2 positions of the putative active sites, making them unable to inhibit trypsin, chymotrypsin, v8 peptidase, and proteinase K [21]. In contrast, ACKTI of 5892.82 Da – a trypsin inhibitor isolated from Agalychnis callidryas – has proline in P2 but arginine in the P1 position consistent with other trypsin inhibitors [22]. In addition, another two Kazal-type peptidase inhibitors – PI01 and PI02–were identified in P. nordestina by ETS analysis; however, their specificity has not been elucidated [23].
Although the biological roles of the amphibian proteinase inhibitors have not been established with certainty, they may include defence against extrinsic peptidases produced by pathogenic microorganisms to prevent damage of host tissue and evasion of host defences. Proteinase inhibitors could also prevent degradation of bioactive peptides, so they can target cell receptors. In addition, proteinase inhibitors might act indirectly as regulators of the processing reactions of bioactive peptides, including cationic α-helical antimicrobial peptides, allowing them to be released onto the skin, so they can display their activity and protect the skin from invading microorganisms [8], [9], [16].
The present study was focused on the Splendid leaf frog, Cruziohyla calcarifer, which belongs to the Phyllomedusinae clade—a known source of pharmacological and antimicrobial peptides. Recently, one insulin-releasing peptide RK-13, and 18 cruzioseptins with antimicrobial activity have been described from C. calcarifer [24], [25]. Here, we describe a group of 7 peptidase inhibitors (with 18 variants) in C. calcarifer skin secretion which belong to the Kazal–type family. One of these, CCKP-1, showed trypsin inhibitory activity and possessed a lysine in its P1 site and unusually, an asparagine in its P2 site. Based on their structural homology, it is predicted that CCKP-2, CCKP-5 and CCKP-7 have chymotrypsin inhibitory activity, while CCKP-4 has trypsin inhibitory activity. Therefore, the proteinase inhibitors of Kazal-type from C. calcarifer are the most diverse group of proteinase inhibitors found to date in a single amphibian species.
2. Material and methods
2.1. Sourcing of samples
The skin secretions of Cruziohyla calcarifer employed in this study came from two different geographical locations, Costa Rica and Ecuador.
The first Costa Rican sample consisted of a pool of two adults secretions collected in 1999. While the Ecuadorian sample consisted of a pool of four juvenile captive breed and one wild adult secretion collected in 2013. The four juvenile captive bred frogs (n = 4) (from Esmeraldas Province, Reserve Otokiki) were provided by Centro Jambatu for Research and Conservation of Amphibians in Ecuador and the wild specimen of Cruziohyla calcarifer (n = 1) was collected in northwestern Ecuador (Esmeraldas Province, Durango). Skin secretions were extracted from each frog by lightly stressing the animal – massaging the dorsal area of the frog – then washing off the secretion with distilled water. Secretions from the same geographical location were pooled, equally split into two 50 mL conical tubes, and then freeze dried. Samples were transported to Queen’s University Belfast at room temperature and dried samples were stored at −20 °C prior to their analysis.
Moreover, twelve additional samples were extracted from a group of 13-month-old captive bred frogs in 2015. The parental line of these frogs came from a Costa Rican population and the animals were housed in terraria as pets in Belgium and Austria. Skin secretions were extracted in the same way as described above but kept individually. In contrast to the previous samples, the twelve samples were acidified with TFA and transported at room temperature to the laboratory facilities in Queen's University Belfast where they were freeze-dried.
Collection and rearing of frogs in Ecuador and transportation of samples were done under permits of the Ecuadorian Ministerio de Ambiente (MAE) (described in acknowledgments).
2.2. Molecular cloning
One aliquot containing half of the dried secretion material of the Ecuadorian sample was dissolved in 1 mL of cell lysis/binding buffer and polyadenylated mRNA was isolated using magnetic Dynabeads Oligo (dTs), as described by the manufacturer (Dynal Biotec, UK). Isolated mRNA was subjected to 3′-rapid amplification of cDNA by using the SMART-RACE kit (Clontech, UK). Briefly, the 3′-RACE reaction used a nested universal primer (NUP), provided with the kit, and two senses primers: S1: 5′-AGCAGCAAAAGAAGAAGAAGCCATG-3′ and S2: 5′- GAGAAGAAGCCATGAAGACTCTGA-3′, that were complementary to the signal sequence of the ACKTI gene precursor of Agalychnis callidryas. The 3′-RACE product was purified and cloned using a pGEM-T vector system (Promega Corporation), and later sequenced using an ABI 3100 automated sequencer. Nucleotide sequences were analysed with MEGA 6.0 and Vector NTI software (Life technologies) [26]. Sequences were compared with databases in the NCBI using the BLAST tool [27]. Theoretical peptide masses were calculated with the peptide mass calculator 3.2 and secondary structure was predicted with GOR IV method [28], [29]. Nucleotide sequences were submitted to the GenBank of the NCBI, accession numbers are included in Table 1.
Table 1.
Primary structures of 18 variants of Kazal type proteins from Cruziohyla calcarifer as confirmed by tandem mass spectrometry. Data represent the best scores of 13 repetitions.
| Peptide | Sequence | Theoretical Average MW (Da) | LCQ MW (Da) | # Peptide fragments | #AAs | Coverage% | Score | Accession number |
|---|---|---|---|---|---|---|---|---|
| CCKP-1 | AVSAECARYGLACNKMLAPVCGTDGTTYSNQCMLCYYNRKNKKNIEIRSRGRC | 5920.82 | 5922.79 | 1631 | 53 | 100 | 17.80 | KX065060 |
| CCKP-2 | ATEPDCKKYPGKCPLAQNPVCGTDGRMYYNECALCVFMRDSKNKVKIQIKKMGKC | 6198.33 | 6199.99 | 902 | 55 | 100 | 8.68 | KX065061 |
| CCKP-3 | ATKPKCPSLFSSGCPSTQDFVCGTDGNSYMNECVMCKMNKNNGGKVKVVKKGYC | 5780.69 | 5782.63 | 2335 | 54 | 100 | 42.37 | KX065062 |
| CCKP-4a | GGVVLLDCRPYGPVCSKIFDPVCGTNFITYDNTCELCKAQRENPRISMRTKGKC | 5992.94 | 5994.90 | 699 | 54 | 100 | 7.52 | KX065063 |
| CCKP-4b | VVRLDCRPYGPVCSKIFDPVCGTNFITYDNTCELCKAQRENPRISMRTKGKC | 5921.87 | 5923.87 | 647 | 52 | 100 | 8.90 | KX065064 |
| CCKP-4c | VVRLDCRPYGPVCSKVLDPVCGTNFKTYDNTCELCKAQRENPRISMRTKGDCRKPYLIPENFRR | 7446.60 | 7447.70 | 864 | 64 | 100 | 20.52 | KX065065 |
| CCKP-5a | VIEPNCKKYEGKKCDLNPNPVCGTNGREYFNECALCVFIRDSKKKADKMCKIKKWGKC | 6667.84 | 6669.29 | 649 | 58 | 100 | 12.57 | KX065066 |
| CCKP-5b | VIEPNCKKYEGKKCDLNPCPVCGTNGREYYNECALCVFIRDSKKKADKMVKIKKWGKC | 6668.87 | 6670.31 | 480 | 58 | 100 | 14.77 | KX065067 |
| CCKP-5c | VIEPNCKKYEGKKCDLNPNPVCGTNGREYFNECALCVFIKDSKKKADKMVKIKKWGKC | 6635.82 | 6637.34 | 684 | 58 | 100 | 11.40 | KX065068 |
| CCKP-5d | VIEPNCKKYEGKKCDLNPNPVCGTNGREYFNECALCVFIRDSTKKADKMVKIKKWGKC | 6636.76 | 6638.30 | 681 | 58 | 100 | 10.53 | KX065069 |
| CCKP-6a | EEDVACPWYYVFGCHDKYTVCGTDGCTYPNKCTLCKINGEDNIKIRKWGNC | 5849.58 | 5851.58 | 426 | 51 | 100 | 8.46 | KX065070 |
| CCKP-6b | EEDVTCPWYYVFGCHDKYTVCGTDGVTYPNKCTLCKINGEDNIKIRKWGNC | 5875.59 | 5877.65 | 617 | 51 | 100 | 7.86 | KX065071 |
| CCKP-7a | PLPSQPQFFKKVLKTLAEPNCKKYEGKKCDLNLNPVCGTNGRTYYNECALCVFIRDSTKKSDKMVKIHKWGKC | 8377.81 | 8378.27 | 865 | 73 | 100 | 13.16 | KX065072 |
| CCKP-7b | PPPSQPQFSNKVLKTLAEPNCKKYEGKKCDLNLNPVCGTNGRTYYNECALCVFIRDSTKKADKMVKIHKWGEC | 8272.54 | 8273.11 | 1015 | 73 | 100 | 18.61 | KX065073 |
| CCKP-7c | PLPSQPQFFKKVLKTLAEPNCKKYEGKKCDLNLNPVCGTNGRTYYNECALCVFIRNSTKKSDKMVKIHKWGKC | 8376.83 | 8377.29 | 1001 | 73 | 100 | 13.46 | KX065074 |
| CCKP-7d | PLPTQPQFFKKVLKTLAEPNCKKYEGKKCDLNLNPVCGTNGRTYYNECALCVFIRDSTKKSDKMVKIHKWGKC | 8391.84 | 8392.29 | 868 | 73 | 100 | 17.68 | KX065075 |
| CCKP-7e | PLPSQPQFFKKVLKTLAEPNCKKYEGKKCDLNLNPVCGTNGGTYYNECALCVFIRDSTKKSDKMVKIHKWGKC | 8278.68 | 8279.19 | 1113 | 73 | 100 | 14.51 | KX065076 |
| CCKP-7f | PLPSQPQFSDKVLKTLAEPNCKKYEGKKCDLNLNPVCGTNGRTYYNECALCVFIRDSTRKADKMVKIHKWGKC | 8316.64 | 8317.18 | 961 | 73 | 100 | 21.25 | KX065077 |
2.3. Reverse phase HPLC fractionation of skin secretion
Different experimental conditions were necessary for the separations as follow:
The Ecuadorian sample (remaining half of the dried secretion) was dissolved in 1.2 mL of solution A (99.95% H2O, 0.05% trifluoroacetic acid) and clarified by centrifugation. 1 mL supernatant was subjected to reverse phase HPLC employing a Waters binary pump HPLC system fitted with an analytical column: Phenomenex Jupiter C-18 (4.6 × 250 mm). Peptides were eluted with a gradient formed from 100% solution A (99.95% H2O, 0.05% trifluoroacetic acid) to 100% solution B (80.00% acetonitrile, 19.95% H2O, 0.05% trifluoroacetic acid) for 240 min at a flow rate of 1 mL/min. Fractions of 1 mL were collected every minute. Detection at 214 and 280 nm was performed continuously.
The Costa Rican sample (0.5 mL of 3 mL) from 1999 was injected onto an HPLC system fitted with a semi-preparative C-18 Phenomenex Jupiter Column employing a gradient of 0–80% solution B for 80 min at a flow rate of 2 mL/min. Fractions were collected every minute. This analysis was performed on 08/03/1999.
2.4. MALDI-TOF MS
The molecular masses of peptides and proteins in each chromatographic fraction were analysed by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) using a linear time of flight Voyager DE mass spectrometer (Perseptive Biosystems, MA, USA). First, the instrument was set in positive detection mode and all fractions were analysed with α-cyano-4-hydroxycinnamic acid (CHCA) matrix (10 mg/mL) for low mass peptides. The settings of the instrument were as follows: mode of operation: linear, accelerating voltage: 2000 V, acquisition mass range: 500–5000 Da, number of laser shots: 50/spectrum, laser intensity: 1813. Later, fractions were analysed for high mass proteins employing sinapinic acid (10 mg/mL) as matrix. The settings of the instrument were as follow: mode of operation: linear, accelerating voltage: 25000 V, acquisition mass 500–10,000 Da, number of laser shots: 50/spectrum, laser intensity: 3119. In both cases, two microliters of sample plus 1 μL of matrix (10 mg/mL) were allowed to dry and later analysed for mass/charge.
2.5. MS/MS sequencing
20 μL of the remaining skin secretion of the Ecuadorian sample dissolved in solution A were applied directly to an analytical HPLC column (Phenomenex C-18; 4.6 × 150 mm) connected to a linear ion-trap mass spectrometer (LCQ-Fleet, Thermo Fisher). The linear elution gradient was formed from 100% solution A (99.90% H2O, 0.1% formic acid) to 100% solution B (80% acetonitrile, 19.0% H2O, 0.1% formic acid) for 135 min at a flow rate 20 μL/min. The mass spectrometer was controlled by Xcalibur software (Thermo). The mass analysis was performed in positive ion mode, and the acquired spectra were in the range of m/z 500–2000 Da. The parameters for electrospray ionization ion-trap mass spectrometry (ESI/MS) were: spray voltage +4.5 kV, drying gas temperature 320 °C, drying gas flow 200 μL/min, and maximum accumulation time – for the ion trap – 350 ms. After the first mass analysis in full scan mode, peptide ions with >50% relative intensity were fragmented by collision induced dissociation (CID), in order to generate b and y ions that were detected in a second mass analysis. Fragment ion profiles were analysed by Proteome Discoverer 1.0 software (Thermo). In order to confirm the amino acid sequences of individual peptides we used the Sequest™ algorithm which compares the acquired fragment ion profiles with the theoretical fragment ions generated from a FASTA file containing the specific sequences obtained by molecular cloning of this species.
In addition, twelve individual samples from the Costa Rican population were analysed in the same way as described above but instead of 135 min these samples were run for only 120 min.
2.6. In solution Trypsin digestion
The HPLC fraction 91 was freeze-dried and later reconstituted in 5 μL of Solution A (99.95% H2O; 0.05% TFA). The sample was denatured with 8 M Urea, 25 mM Tris and 20 mM DTT (final concentrations) and incubated at 60 °C for 1 h. The alkylation of the sample was performed with Iodoacetemide (IAA) at 40 mM and incubated in dark at room temperature for 30 min. The alkylation reaction was quenched with DTT solution at 10 mM (final concentration). Later, the sample was diluted 10 times with 200 mM ammonium blcarbonate and digested with 0.5 μL of trypsin solution (1 μg/μL) at 37 °C for 8 h. The digestion reaction was stopped by adding 10 μL of 2.5% TFA solution and then cleaned up with C18 ZipTip. In brief, the trypsin-digested sample was drawed to a previously wet C18 ZipTip at least 10 times. Then, the ZipTip was washed with 20 μL of 0.1% TFA for three times. Tryptic fragments were eluted with 20 μL of 80% ACN/0.1% TFA by pipetting up and down for 10 times. Ten microliter of eluted fragments were subjected for LC–MS/MS analysis as described above.
2.7. Trypsin inhibitor assay
HPLC fractions (400 μL), containing proteins with molecular masses higher that 5 kDa, were dried in a vacuum concentrator. Fractions were reconstituted in 22 μL of PBS and screened for trypsin inhibition. In brief, 180 μL of substrate working concentration (50 μM Z-Gly-Gly-Arg-AMC) were placed in 4 rows of a black 96-well plate. Then, 10 μL of each fraction were placed in the plate in duplicates and the first 5 cycles of fluorescence were measured to establish a baseline. Next, 10 μL of trypsin solution (0.001 mg/mL) were added to each well, excepting the first two that constituted negative controls and contained only substrate. The next two columns contained substrate and trypsin as positive controls. Finally, hydrolysis of the substrate by trypsin was monitored by the fluorescence generated for 60 min employing a Fluostar Optima plate reader (BMG LABTECH spectrofluorimeter) set to 460 nm emission and 395 nm excitation.
2.8. Chymotrypsin inhibition assay
The chymotrypsin inhibition assay was performed in the same way as the trypsin inhibition assay described previously with the following differences: the substrate Suc-Ala-Ala-Pro-Phe-AMC (50 μM), and chymotrypsin solution (0.001 mg/mL) were employed for this assay.
3. Results
3.1. Molecular cloning of Kazal protein precursor encoding cDNA
A group of 7 novel Kazal-type proteins (18 variants) with proteinase inhibitory activity was identified by molecular cloning of the skin secretion of Cruziohyla calcarifer. Their nucleotide and translated amino acid sequences are described in Fig. 1. The translated open reading frames of these precursors contain 76 to 93 amino acids. The first 24–26 residues constitute a putative signal peptide, predicted by the SignalP 4.0 server, while the remaining 51–73 amino acids correspond to the mature Kazal-type inhibitor protein (Fig. 2). Table 1 shows the sequences of the 18 different variants found within these Kazal proteinase inhibitors and their theoretical average masses fall in the range of 5782–8392 Da. In addition, all seven Kazal-types proteins and their variants share the Kazal pattern C-X(7)-C-X(6,7)-C-X(6,7)-Y-X(3)-C-X(2)-C-X(15-21)-C, as highlighted in Table 2. Proteins were labelled Cruziohyla calcarifer Kazal Protein 1-7 (CCKP-1-7) adding letters for their variants. Sequences were submitted to GenBank (NBCI) with accession numbers: KX065060–KX065077.
Fig. 1.
Nucleotide and translated open-reading frame amino acid sequences of cloned cDNAs that encode the biosynthetic precursors of the Kazal-type proteins from Cruziohyla calcarifer. A–G) Representative seven types of Kazal-type proteins with a range of 51–73 amino acids. The putative signal peptides are double-underlined, the mature sequences are single-underlined and the stop codons are indicated by asterisks.
Fig. 2.
Precursor structures of Kazal-type proteins from Cruziohyla calcarifer. All amino acid sequences contain a signal peptide (19–26 residues)-predicted by the SignalP4.1 server, followed by the mature sequence (51–73 residues).
Table 2.
Alignment of the mature sequences of Kazal proteins indicating the canonical Kazal motif.
 |
*Conserved sites, (x) number of clones with the same sequence. The Kazal motif is highlighted: 6 Cysteines are in yellow, one Tyrosine in pink, and P1 and P2 sites in red. Conectors indicate predicted disulfide bond formation.
3.2. Identification, isolation, and structural characterization of the trypsin inhibitory Kazal-type protein
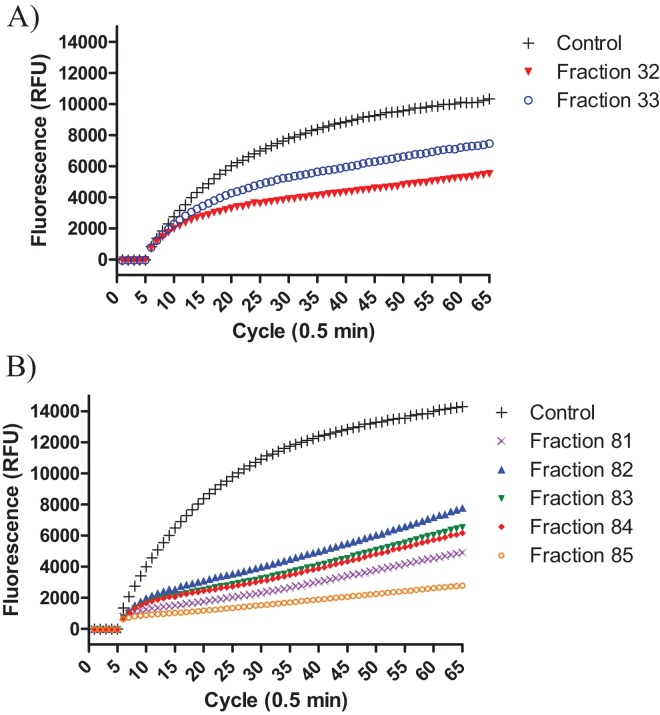
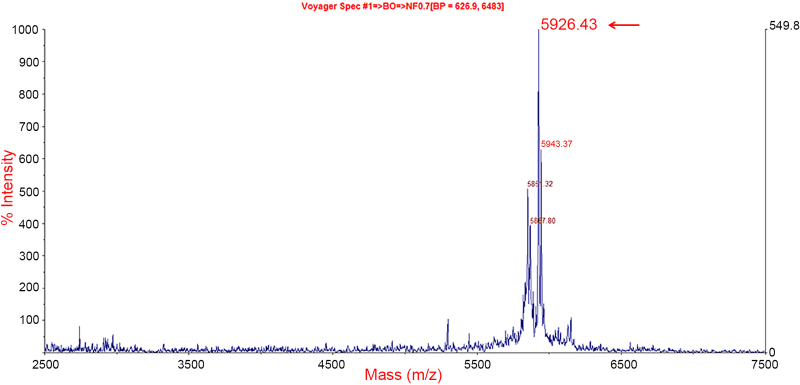
The HPLC fractions of the Costa Rican population obtained in 1999 were analysed by MALDI-TOF mass spectrometry in 2013, and all fractions containing m/z peaks between 5 and 8 kDa were selected. The selected fractions were: 29–33. Each fraction was tested for trypsin and chymotrypsin inhibitory activity, however only fractions # 32 and 33 showed trypsin inhibitory activity (Fig. 3). MALDI-TOF mass spectrometric analysis of trypsin inhibitor fractions: #32 and 33 revealed a singly-charged ion of m/z 5926.43 that was confirmed by LCQ analysis (data not shown). This mass was consistent with the sequence of CCKP-1 (Table 1 and Fig. 4).
Fig. 3.
Trypsin inhibitory activity of HPLC fractions. A) Fractions 32–33 from the Costa Rican sample. B) Fractions 81–85 from the Ecuadorian sample.
Fig. 4.
MALDI-TOF MS spectrum of HPLC fraction 32 with trypsin inhibitory activity. The arrow denotes CCKP-1 with a molecular mass of 5926.43 Da.
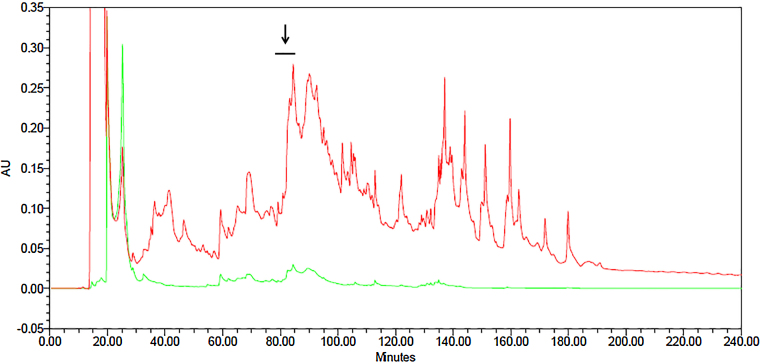
Recently, the Ecuadorian sample of Cruziohyla calcarifer skin secretions was fractionated by reverse phase HPLC for 240 min, and they were found to be a complex mixture of different peptides due to the numerous peaks observed in the chromatogram at 214/280 nm (Fig. 5). Besides, it was also noticeable that these peaks were very low compared to other chromatograms obtained in our research group that were 8 times higher. Usually 5–10 mg of dried secretion is employed for HPLC fractionation; however, the secretions of C. calcarifer were very scarce and could not be weighed. MALDI-TOF analysis was performed on each fraction and those fractions containing peptides with high molecular mass (5–8 kDa) were analysed for proteinase inhibitory activity (trypsin and chymotrypsin inhibition). The fractions tested were #81–85, 109, 110, 121–127, and 129; however, only fractions # 81–85 showed trypsin inhibition (Fig. 3). MALDI TOF MS analysis and LCQ MS confirmed a mass of 5953 Da that did not coincide with any of the sequences found by molecular cloning to date. As a consequence of the initial low amount starting material, it was not possible to determine the Ki value of these peptidase inhibitors.
Fig. 5.
Reverse phase HPLC chromatogram of skin secretion from Cruziohyla calcarifer fractionated over 240 min. The arrow denotes fractions 81–85 showing trypsin inhibitory activity. Detection at 214 nm (red line) and detection at 280 nm (green line).
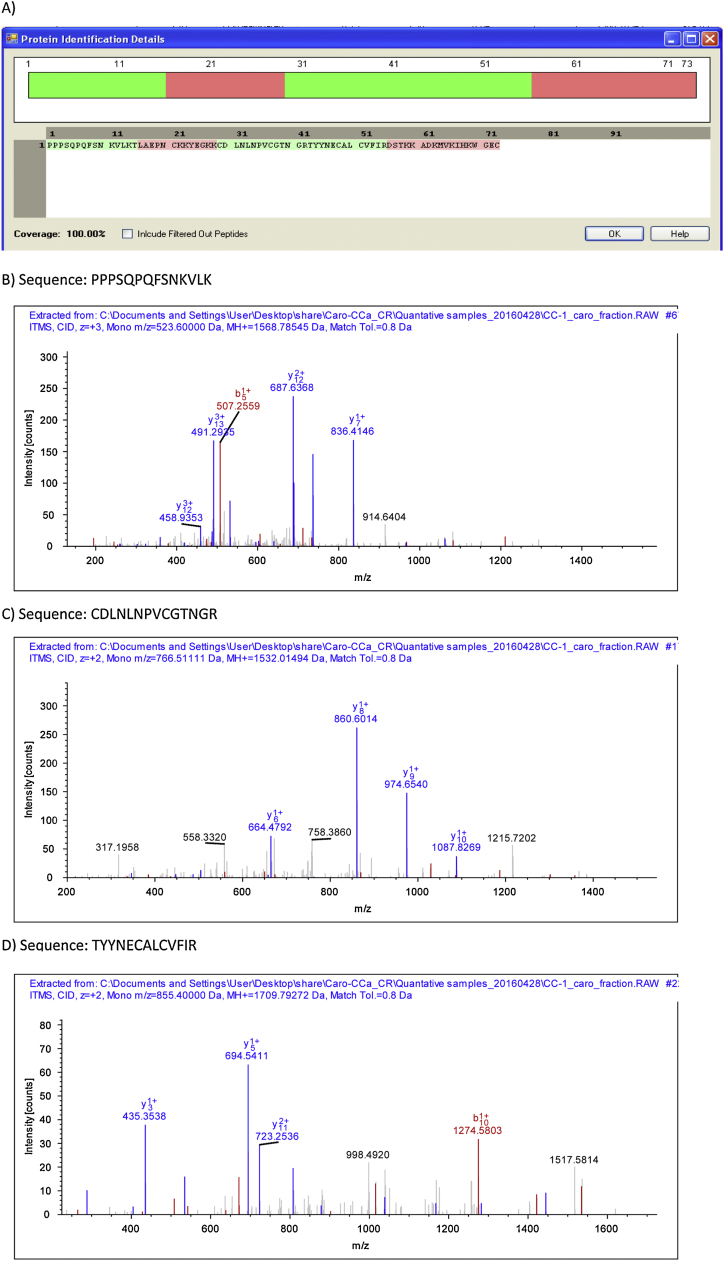
In addition, we analysed 13 skin secretion samples (1 Ecuadorian and 12 Costa Rican samples) directly by tandem mass spectrometry. By using Sequest algorithm, we confirmed the 100% coverage of amino acid sequences of the 18 variants of Kazal type proteins initially found by molecular cloning (Table 1 include the best scores of these analysis). Moreover, the fraction 91 was trypsin digested and later analysed by LC–MS/MS which confirmed the sequence of CCKP-7b (Fig. 6).
Fig. 6.
Tryptic fragments sequenced by tandem mass spectrometry corresponding to the Cruziohyla calcarifer Kazal protein 7b (CCKP-7b). A) Sequence coverage of tryptic fragments, B–D) MS/MS profiles of the tryptic peptides sequenced by tandem mass spectrometry. Fragmented peptide ions b+ and y+ are highlighted in red and blue respectively.
3.3. Bioinformatic analyses on Kazal-type proteinase inhibitors
The Kazal motif was identified in the 7 novel proteins and 18 variants of Cruziohyla calcarifer by BLAST analysis using databases from the NCBI.
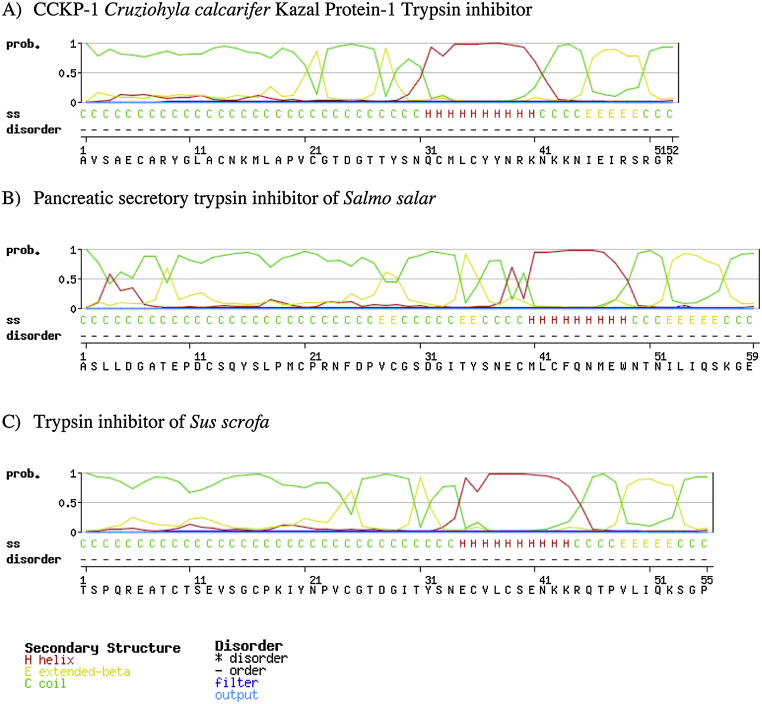
The precursor sequence of Cruziohyla calcarifer Kazal protein-1 (CCKP-1) was compared with NCBI databases (BLAST/n tool), and it showed 76% similarity with the trypsin inhibitor, ACKTI, of Agalychnis callidryas (accession number HE653907.1). However, the translated mature protein only shared 52–55% similarity with trypsin inhibitor proteins from the fulmar Fulmarus glacialis, the falcon Falco cherrug, the egret Egretta garzetta, and A. callidryas (accession numbers: XP_009582735.1, XP_005436761.1, XP_009633154.1, and CCF72386.1 respectively) (Table 3). Subsequent bioinformatic analysis of the CCKP-1 predicted secondary structure showed additional similarities with the pancreatic secretory trypsin inhibitor of the salmon Salmo salar and the trypsin inhibitor of the boar Sus scrofa, especially at their C-terminal sites, as shown in Fig. 7.
Table 3.
Comparison of CCKP-1 with other Kazal trypsin inhibitors according to BLAST/p (protein–protein blast).
 |
*Conserved sites, The Kazal motif is highlighted: 6 Cysteines are in yellow, one Tyrosine in pink, and P1 and P2 sites in red.
Fig. 7.
Secondary structure prediction analysis of trypsin inhibitors using GOR IV method [22]. A) CCKP-1 Cruziohyla calcarifer Kazal Protein-1 Trypsin inhibitor. B) Pancreatic secretory trypsin inhibitor of Salmo salar. C) Trypsin inhibitor of Sus scrofa.
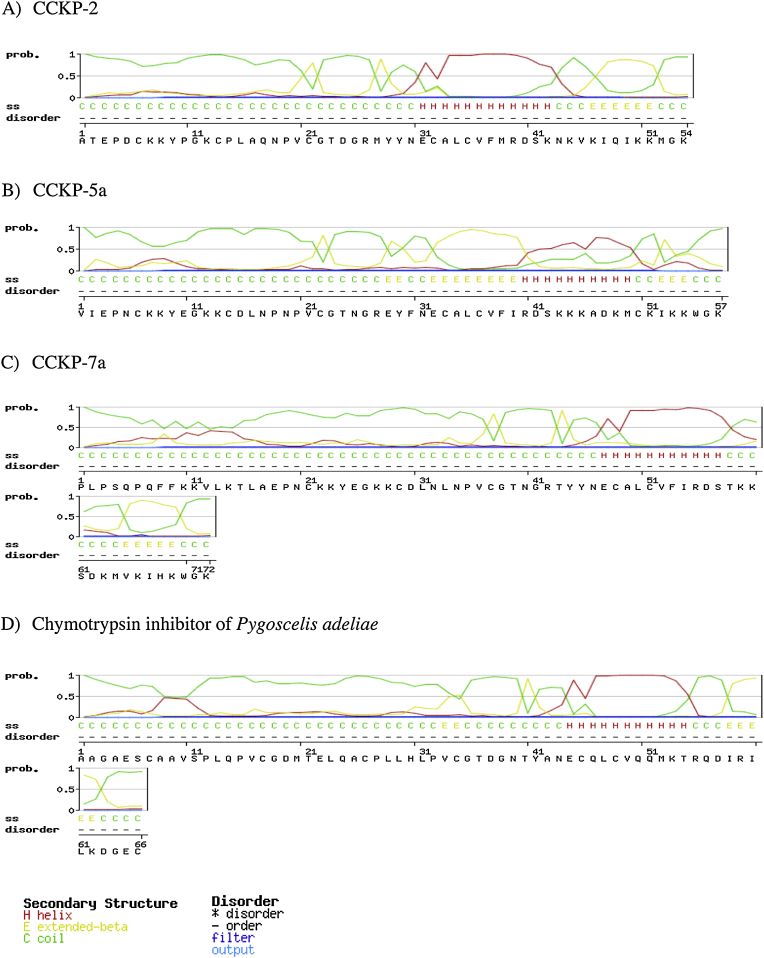
The mature sequence of Cruziohyla calcarifer Kazal protein-2 (CCKP-2) displayed 66–68% similarity with the proteinase inhibitors: PSKP-1 and PSKP-2 of Phyllomedusa sauvagii; and PI01 and PI02 of P. nordestina (accession numbers: P83579.2, P83578.1, AFY11406.1, and AFY11405.1 respectively). In addition, CCKP-2 was 47% similar to the chymotrypsin inhibitor from the tinamou Tinamus guttatus (accession number KGL85187.1). Moreover, its secondary structure was similar to the chymotrypsin inhibitor of the penguin Pygoscelis adeliae, especially in its N-terminal region (see Fig. 8).
Fig. 8.
Secondary structure prediction of chymotrypsin inhibitors using GOR IV method [22]. A) CCKP-2 Cruziohyla calcarifer Kazal Protein-2. B) Cruziohyla calcarifer Kazal Protein-5a) Cruziohyla calcarifer Kazal Protein-7a) Chymotrypsin inhibitor of Pygoscelis adeliae.
The mature sequence of Cruziohyla calcarifer Kazal protein-3 (CCKP-3) revealed 53–55% similarity with the pancreatic secretory trypsin inhibitor of the frog Xenopus (Silurana) tropicalis; the serine peptidase inhibitor Kazal-type 2 of the cormorant Phalacrocorax carbo; and the trypsin inhibitor CITI-1 of the ostrich Struthio camelus australis (accession numbers: XP_002939857.1, XP_009500666.1, and KFV85841.1 respectively).
The mature sequences of Cruziohyla calcarifer Kazal protein-4 (CCKP-4a, CCKP-4b, and CCKP-4c) showed 45–52% similarity with the sperm-activating protein of the herring Clupea pallasii, the pancreatic secretory trypsin inhibitor of Salmo salar, and the serine peptidase inhibitor Kazal-type 12-like of the shrew Tupaia chinensis (accession numbers: BAA14009.1, NP_001140094.1, and XP_006148122.1 respectively).
The mature sequences of Cruziohyla calcarifer Kazal Protein-5 (CCKP-5a, CCKP-5b, CCKP-5c, and CCKP-5d) and C. calcarifer Kazal protein-7 (CCKP-7a to CCKP-7f), similarly to CCKP-2, exhibited 64–78% similarity with proteinase inhibitor PSKP1 and PSKP-2 of Phyllomedusa sauvagii; and proteinase inhibitors PI01 and PI02 of P. nordestina (accession numbers: P83579.2, P83578.1, AFY11406.1, and AFY11405.1 respectively).
Finally, the mature sequences of Cruziohyla calcarifer Kazal protein-6 (CCKP-6a and CCKP-6b) revealed 48–52% similarity with the serine peptidase inhibitor Kazal-type 2 from the cuckoo Cuculus canorus; the pancreatic secretory trypsin inhibitor-like from the lizard Anolis carolinensis; and the trypsin inhibitor CITI-1-like of the bird Cariama cristata (accession numbers: XP_009565924.1, XP_008118717.1, and XP_009700547.1 respectively).
4. Discussion
Peptidase inhibitors with three different motifs – Kunitz, Bowman-Birk and Kazal – have been previously identified in some amphibians; however, Kazal-type inhibitors appeared to be specific to Phyllomedusinae. Among them are: the Kazal prolyl endopeptidase inhibitors PSKP-1 and PSKP-2 isolated from Phyllomedusa sauvagii; the proteinase inhibitor PI01 and PI02 from P. nordestina; and the trypsin inhibitor ACKTI isolated from Agalychnis callidryas [18], [19], [20]. Here, we report the finding of a group of seven distinguishable Kazal-type proteinase inhibitors in Cruziohyla calcarifer with 18 variants (Table 1). NCBI Database interrogation using the amino acid sequence of the mature proteins identified the Kazal motif C-X(7)-C-X(6,7)-C-X(6,7)-Y-X(3)-C-X(2)-C-X(15-21)-C in all 7 Kazal-type proteins and in their 18 different variants (Table 2).
It was found that the CCKP-1 nucleotide sequence was 75% similar to the precursor of the trypsin inhibitor Kazal-type of Agalychnis callidryas (ACKTI). In addition, the CCKP-1 mature sequence showed 50% similarity with a number of trypsin inhibitors from Fulmarus glacialis, Falco cherrug, Egretta garzetta, and A. callidryas. These trypsin inhibitors have a characteristic lysine (K) or arginine (R) in the P1 position and a proline (P) in the P2 position of the active site. A trypsin inhibitory activity was predicted for CCKP-1 because it also shared a lysine (K) in the P1 position; but, in contrast, it had an asparagine (N) instead of Proline (P) in the P2 position (Table 3). Moreover, CCKP-1 had a similar secondary structure to the trypsin inhibitors of Salmo salar and Sus scrofa (Fig. 7). In addition, CCKP-1 was identified in the archived fractions 32 and 33, from an HPLC fractionation performed 15 years ago, due to their monoisotopic molecular mass of 5926.43, as determined by MALDI-TOF MS analysis and confirmed by a LCQ ESI MS (Fig. 4). Additionally, trypsin inhibitory activity was detected in those fractions confirming the activity prediction as is shown in Fig. 3.
The closest relatives of CCKP-2, based on their sequence similarities, are the proteinase inhibitors PI01 and PI02 from Phyllomedusa nordestina, and PSKP-1 and PSKP-2 from P. sauvagii. PI01 and PI02 have proline in P2 position and leucine or valine in position P1. However, PSKP-1 and PSKP-2 have two prolines in positions P1 and P2 displaying a prolyl endopeptidase activity and lacking any trypsin, chymotrypsin, V8-peptidase, and proteinase K inhibitory activity. Moreover, trypsin inhibitor activity can be acquired if the P1 Proline is changed for lysine. In contrast, CCKP-2 has a leucine in its P1 site and a proline in the P2 site of the active site. CCKP-2 also showed 47–53% similarity with chymotrypsin inhibitors from birds, such as Tinamus guttatus and Tauraco erythrolophus, which have a leucine in P1 and proline in P2 position. In addition, CCKP-2 showed a similar secondary structure to the chymotrypsin inhibitor of the penguin Pygoscelis adeliae (Fig. 8). Due to these sequence similarities and secondary structures, a chymotrypsin inhibitory activity is suspected for CCKP-2.
In a similar way, CCKP5 and CCKP-7 displayed a high similarity (64–81%) with the same peptidase inhibitors PI01, PI02 from Phyllomedusa nordestina and PSKP-1 and PSKP-2 from P. sauvagii. However, in contrast with CCKP-2, CCKP5 and CCKP-7 have an aspartic acid in the P2 position instead of proline, but share the leucine in P1 position. For this reason and their similarity to the secondary structure of the chymotrypsin inhibitor of Pygoscelis adeliae, especially in their N-terminal regions, it is presumed that CCKP-5 and CCKP-7 also have chymotrypsin inhibitory activity similar to CCKP-2 (Fig. 8).
In the same way, CCKP-4 has a lysine (K) in position P1 which leads to a suspicion of trypsin inhibitory activity; but instead of the P2 proline, present in most other trypsin inhibitors, CCKP-4 has a serine (S). In addition, CCKP-4 showed an unusual 47% similarity to the sperm-activation protein from the herring Clupea pallassi most likely by sharing the Kazal motif rather than for a functional relationship.
The other two proteins, CCKP-3 and CCKP-6, have a serine (S) and an aspartic acid (D) respectively in their P1 positions. Their sequences are similar to serine peptidase inhibitors in general, but we cannot infer any specificity based on their P1 residues.
To confirm the biological activity of CCKP-2 to CCKP-7, all peptides should be identified in the individual fractions and tested. Another option could be to overexpress the 5 proteins in Escherichia coli, so that the purified proteins could be tested over different peptidases to confirm our predictions. Unfortunately, the scarce material available did not allow the protein identification in the individual fractions. However, their primary structures were 100% confirmed by LCQ MS/MS sequencing employing the whole secretion of 13 individual samples including wild and captive frogs from different sources. It is noticeable, that differential expression was observed in the 12 Costa Rican samples in spite of being analysed at the same concentration (1 mg/mL). Would be interesting to test in the future how the developmental stages (juvenile vs. adults), habitats, and distributions affect the expression of these protease inhibitors.
In contrast with other species, Cruziohyla calcarifer has proven to be a rich source of Kazal-type proteins with proven trypsin inhibitor activity in at least one of them, and predicted trypsin and chymotrypsin inhibitory activity in some others. Moreover, their extraordinary diversity contains 2 Kazal-type proteins that show unusual P1 and P2 residues which preclude their activity prediction. The biological role of those inhibitors is uncertain; however, they probably regulate the balance of antimicrobial and other bioactive peptides on the skin surface of the frogs, to protect them against pathogenic microorganisms and predators. The extraordinary diversity of these Kazal-type protein inhibitors in the skin of C. calcarifer possibly mirrors the complexity of peptides and proteins still unknown in other species, and promotes research on this species in particular.
5. Conclusions
In conclusion, the combined strategy including tandem mass sequencing, molecular cloning and HPLC fragmentation allowed the identification of 18 variants of Kazal-type proteins of Cruziohyla calcarifer (CCKP). These proteins were classified in 7 types by sequence similarity. CCKP-1 and 4 were deemed of trypsin inhibitory activity while CCKP-2, 5, 7 were deemed of chymotrypsin inhibitory activity by sequence homology with other peptidase inhibitors. In addition, CCKP-1 was identified in the chromatographic fraction No.32 showing trypsin inhibitory activity and a molecular mass of [M+H]+ = 5926.43 Da. In this way, C. calcarifer has showed to be an important source of diverse peptidase inhibitors promoting further research of the skin secretions in this species.
Conflict of interest
The authors declare that there is no conflict of interest.
Authorship
This study was conceived and designed by CS, MZ, TC. Sample collections were performed by CPB, EET, and LAC. Data were acquired by CPB and RL. LC–MS/MS analysis was performed by LW and XX. The article was written by CPB and reviewed critically by CS and LAC.
Acknowledgments
Carolina Proaño-Bolaños is in receipt of a scholarship of the Ecuadorian Secretariat of Science and Technology (SENESCYT). This research was funded by the Natural drug discovery group, School of Pharmacy, Queen’s University Belfast and SENESCYT. The latter also supported field work to CPB. Collection and rearing of frogs, transportation and analysis of samples were done under permits of the Ecuadorian Ministerio de Ambiente (MAE): 001-13 IC-FAU-DNB/MA,003-11-IC-FAU-DNB/MA, 005-15 IC-FAU-DNB/MA.; Framework contract for access to genetic resources MAE-DNB-CM-2016-0051; and exportation permits: 003-13-EXP-CI-FAU-DNB/MA and 2015-003-FO-DPAP-MA. This research is part of the MAE project “Conservation of Ecuadorian amphibian diversity and sustainable use of its genetic resources”, which is promoted by The Global Environmental Facility (GEF) and Programa de las Naciones Unidas para el Desarrollo (PNUD), and that involves Ikiam-Universidad Regional Amazónica, Queen’s University Belfast, and Centro Jambatu.
Thanks to Mayra Rojas and David Narvaez of Universidad de las Américas in Quito, Ecuador, who helped to prepare freeze-dried samples. Christian Proy (Austria), and Stassen Raf (Belgium) generously provided access to their Costa Rican pet frogs to obtain samples for this research. Thanks to Dr. Catriona Arlow for reviewing the language of the first draft of this manuscript.
Contributor Information
Carolina Proaño-Bolaños, Email: cproanobolanos01@qub.ac.uk, carolina.proano@ikiam.edu.ec.
Chris Shaw, Email: chris.shaw@qub.ac.uk.
References
- 1.Erspamer V. Bioactive secretions of the amphibian integument. In: Heatwole H., Bartholameus G., editors. vol. 1. Surrey, Beatty and Sons; Chipping-Norton, N.S.W: 1994. pp. 178–350. (Amphibian Biology. The Integument). [Google Scholar]
- 2.Thompson A.H., Bjourson A.J., Orr D.F., Shaw C., McClean S. A combined mass spectrometric and cDNA sequencing approach to the isolation and characterization of novel antimicrobial peptides from the skin secretions of Phyllomedusa hypochondrialis azurea. Peptides. 2007;28:1331–1343. doi: 10.1016/j.peptides.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Chen X., Wang L., Wang H., Chen H., Zhou M., Chen T., Shaw C. A fish bradykinin (Arg0, Trp5, Leu8-bradykinin) from the defensive skin secretion of the European edible frog, Pelophylax kl. esculentus: structural characterization; molecular cloning of skin kininogen cDNA and pharmacological effects on mammalian smooth muscle. Peptides. 2011;32:26–30. doi: 10.1016/j.peptides.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 4.Apponyi M.A., Pukala T.L., Brinkworth C.S., Maselli V.M., Bowie J.H., Tyler M.J., Booker G.W., Wallace J.C., Carver J.A., Separovic F., Doyle J., Llewellyn L.E. Host-defence peptides of Australian anurans: structure, mechanism of action and evolutionary significance. Peptides. 2004;25:1035–1054. doi: 10.1016/j.peptides.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Laskowski M., Jr., Kato I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- 6.Farady C.J., Craik C.S. Mechanisms of macromolecular protease inhibitors. Chembiochem. 2010;11:2341–2346. doi: 10.1002/cbic.201000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bode W., Huber R. Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem. 1992;204:433–451. doi: 10.1111/j.1432-1033.1992.tb16654.x. [DOI] [PubMed] [Google Scholar]
- 8.Lai R., Liu H., Lee W.H., Zhang Y. Identification and cloning of a trypsin inhibitor from skin secretions of Chinese red-belly toad Bombina maxima. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 2002;131:47–53. doi: 10.1016/s1096-4959(01)00479-1. [DOI] [PubMed] [Google Scholar]
- 9.Chen T., Shaw C. Identification and molecular cloning of novel trypsin inhibitor analogs from the dermal venom of the Oriental fire-bellied toad (Bombina orientalis) and the European yellow-bellied toad (Bombina variegata) Peptides. 2003;24:873–880. doi: 10.1016/s0196-9781(03)00165-7. [DOI] [PubMed] [Google Scholar]
- 10.Liberio M., Bastos I., Pires J.O., Fontes W., Santana J.M., Castro M.S. The crude skin secretion of the pepper frog Leptodactylus labyrinthicus is rich in metallo and serine peptidases. PLoS One. 2014;9:e96893. doi: 10.1371/journal.pone.0096893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demesa-Balderrama G., Meneses E.P., Hernandez-Orihuela L., Pando-Robles V., Rodriguez M.C., Barrientos-Salcedo C., Aguilar M.B., Batista C.V. A comprehensive proteomic study of the skin secretions of the frog Lithobates spectabilis. Protein Pept. Lett. 2016;23:597–611. doi: 10.2174/0929866523666160505120853. [DOI] [PubMed] [Google Scholar]
- 12.Darby N.J., Lackey D.B., Smyth D.G. Purification of a cysteine endopeptidase which is secreted with bioactive peptides from the epidermal glands of Xenopus laevis. Eur. J. Biochem. 1991;195:65–70. doi: 10.1111/j.1432-1033.1991.tb15676.x. [DOI] [PubMed] [Google Scholar]
- 13.Mignogna G., Pascarella S., Wechselberger C., Hinterleitner C., Mollay C., Amiconi G., Barra D., Kreil G. BSTI, a trypsin inhibitor from skin secretions of Bombina bombina related to protease inhibitors of nematodes. Protein Sci. 1996;5:357–362. doi: 10.1002/pro.5560050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X., Ma Y., Wu J., Lai R. Two serine protease inhibitors from the skin secretions of the toad, Bombina microdeladigitora. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008;149:608–612. doi: 10.1016/j.cbpb.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Ali M.F., Lips K.R., Knoop F.C., Fritzsch B., Miller C., Conlon J.M. Antimicrobial peptides and protease inhibitors in the skin secretions of the crawfish frog, Rana areolata. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2002;1601:55–63. doi: 10.1016/s1570-9639(02)00432-6. [DOI] [PubMed] [Google Scholar]
- 16.Conlon J.M., Kim J.B. A protease inhibitor of the Kunitz family from skin secretions of the tomato frog, Dyscophus guineti (Microhylidae) Biochem. Biophys. Res. Commun. 2000;279:961–964. doi: 10.1006/bbrc.2000.4052. [DOI] [PubMed] [Google Scholar]
- 17.Wang H., Wang L., Zhou M., Yang M., Ma C., Chen T., Zhang Y., Zeller M., Hornshaw M., Shaw C. Functional peptidomics of amphibian skin secretion: a novel Kunitz-type chymotrypsin inhibitor from the African hyperoliid frog, Kassina senegalensis. Biochimie. 2012;94:891–899. doi: 10.1016/j.biochi.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Song G., Zhou M., Chen W., Chen T., Walker B., Shaw C. HV-BBI-a novel amphibian skin Bowman-Birk-like trypsin inhibitor. Biochem. Biophys. Res. Commun. 2008;372:191–196. doi: 10.1016/j.bbrc.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 19.Wang M., Wang L., Chen T., Walker B., Zhou M., Sui D., Conlon J.M., Shaw C. Identification and molecular cloning of a novel amphibian Bowman Birk-type trypsin inhibitor from the skin of the Hejiang Odorous Frog; Odorrana hejiangensis. Peptides. 2012;33:245–250. doi: 10.1016/j.peptides.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Li J., Wu J., Wang Y., Xu X., Liu T., Lai R., Zhu H. A small trypsin inhibitor from the frog of Odorrana grahami. Biochimie. 2008;90:1356–1361. doi: 10.1016/j.biochi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Gebhard L.G., Carrizo F.U., Stern A.L., Burgardt N.I., Faivovich J., Lavilla E., Ermacora M.R. A Kazal prolyl endopeptidase inhibitor isolated from the skin of Phyllomedusa sauvagii. Eur. J. Biochem. 2004;271:2117–2126. doi: 10.1111/j.1432-1033.2004.04127.x. [DOI] [PubMed] [Google Scholar]
- 22.Li R., Wang H., Jiang Y., Yu Y., Wang L., Zhou M., Zhang Y., Chen T., Shaw C. A novel Kazal-type trypsin inhibitor from the skin secretion of the Central American red-eyed leaf frog, Agalychnis callidryas. Biochimie. 2012;94:1376–1381. doi: 10.1016/j.biochi.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Neiva M., Vargas D.C., Conceicao K., Radis-Baptista G., Assakura M.T., Jared C., Hayashi M.A. Gene expression analysis by ESTs sequencing of the Brazilian frog Phyllomedusa nordestina skin glands. Toxicon. 2013;61:139–150. doi: 10.1016/j.toxicon.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 24.Abdel-Wahab Y.H., Marenah L., Orr D.F., Shaw C., Flatt P.R. Isolation and structural characterisation of a novel 13-amino acid insulin-releasing peptide from the skin secretion of Agalychnis calcarifer. Biol. Chem. 2005;386:581–587. doi: 10.1515/BC.2005.068. [DOI] [PubMed] [Google Scholar]
- 25.Proaño-Bolaños C., Zhou M., Wang L., Coloma L.A., Chen T., Shaw C. Peptidomic approach identifies cruzioseptins, a new family of potent antimicrobial peptides in the splendid leaf frog, Cruziohyla calcarifer. J. Proteom. 2016;146:1–13. doi: 10.1016/j.jprot.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.J. Rozenski, Peptide Mass Calculator v3.2 1999.
- 29.Combet C., Blanchet C., Geourjon C., Deleage G. NPS@ network protein sequence analysis. TIBB. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]