Abstract
The ability to respond to changes in pH is critical for all organisms. In Salmonella enterica, the sensor PhoQ responds to a mildly acidic pH by phosphorylating, and thereby activating, the virulence regulator PhoP. This PhoP/PhoQ two-component system is conserved in a subset of Gram-negative bacteria. PhoQ has been thought to be sufficient to activate PhoP in mildly acidic pH. However, we found that the Salmonella-specific protein UgtL, which was horizontally acquired by Salmonella before the divergence of S. enterica and S. bongori, was also necessary for PhoQ to activate PhoP under mildly acidic pH conditions, but not for PhoQ to activate PhoP in response to low Mg2+ or the antimicrobial peptide C18G. UgtL increased the abundance of phosphorylated PhoP by stimulating autophosphorylation of PhoQ, thereby increasing the amount of the phosphodonor for PhoP. Deletion of ugtL attenuated Salmonella virulence and further reduced PhoP activation in a strain bearing a form of PhoQ that is not responsive to acidic pH. Thus, when Salmonella experiences mildly acidic pH, PhoP activation requires PhoQ to detect pH and UgtL to amplify the PhoQ response. Our findings reveal how acquisition of a foreign gene can strengthen signal responsiveness in an ancestral regulatory system.
INTRODUCTION
pH governs critical biological processes including gene expression, energy generation, and enzymatic functions (1). The failure to control or respond to changes in pH results in organelle malfunction, leading to disease (2–4) and cell death (5). Thus, organisms monitor changes in extracytoplasmic or cytoplasmic H+, or both, or chemical or physical modifications triggered by acidification (6, 7). For example, the Escherichia coli CadC, Helicobacter pylori ArsS, and Salmonella enterica PmrB proteins detect acidic pH in the periplasm (8–10), whereas the E. coli Tsr and Tar proteins and the S. enterica EnvZ and PhoQ proteins respond to a decrease in cytoplasmic pH (11–13). Here, we report that full activation of the Salmonella sensor PhoQ by a mildly acidic pH requires a protein to amplify the response to this signal.
PhoP/PhoQ is a two-component system that controls virulence, Mg2+ homeostasis, and resistance to antimicrobial peptides in several species of Gram-negative bacteria (14, 15). In the intracellular pathogen S. enterica serovar Typhimurium (16), mildly acidic pH (13, 17), low Mg2+ (18), or certain antimicrobial peptides (19) activate the sensor PhoQ, which promotes the phosphorylated state of its cognate response regulator PhoP (PhoP-P). PhoP-P is the active form of the PhoP protein: PhoP-P binding to promoter regions (20) stimulates or represses gene transcription (20, 21), depending on the particular target gene, in vivo. In the absence of inducing conditions, PhoQ operates as a phosphatase that dephosphorylates PhoP-P. PhoQ harbors two transmembrane regions that define a periplasmic domain (18) that is required for sensing Mg2+ (18, 22) and the antimicrobial peptide C18G (19), but not for sensing acidic pH (13, 23). The cytosolic domain of PhoQ mediates the response to a mildly acidic pH and is sufficient to do so, albeit not as efficiently as the full-length protein (13). The curtailed response to acidic pH by a mutant bearing only the cytoplasmic domain of PhoQ implies that another region of PhoQ or an additional factor(s), or both, is required for full PhoQ activation by acidic pH.
The PhoP protein controls the expression of ~9% of Salmonella genes (24). PhoP-activated genes include several that encode proteins that modify the lipid A portion of lipopolysaccharide (25, 26) or confer resistance to antimicrobial peptides (26–28), or both. XXXXXXX. Curiously, a strain lacking the horizontally acquired PhoP-activated gene ugtL has a lipid A profile resembling that of a phoP null mutant (29) and exhibits hypersusceptibility to the antimicrobial peptide magainin 2, although neither phenotype is as strong as in the phoP null (29). Transcription of ugtL from a heterologous promoter restores magainin 2 resistance to a mutant defective in slyA (30), a PhoP-activated gene required for transcription of many PhoP-activated horizontally acquired genes (31–33). The deduced amino acid sequence of the Salmonella-specific ugtL gene bears no similarity to predicted proteins in sequence databases.
We report that activation of the ancestral PhoP/PhoQ system by acidic pH requires the horizontally acquired UgtL protein and that this activation is necessary for Salmonella virulence. UgtL promoted PhoQ autophosphorylation in vitro and enhanced PhoP phosphorylation in vivo, resulting in transcription of PhoP-activated genes. UgtL was specifically required for PhoP activation by a mildly acidic pH and was dispensable for activating PhoP under other PhoQ-stimulating conditions. Our findings indicate that PhoP activation under acidic pH requires a protein for signal sensing (PhoQ) and a separate protein (UgtL) for amplification of that response. Moreover, they argue that activation of the PhoP-dependent virulence program is a derived state resulting from the acquisition of ugtL and potentially additional genes.
RESULTS
UgtL is required for PhoP/PhoQ activation by mildly acidic pH
Because the transcriptional regulatory targets of a two-component system may be subject to additional levels of control, the abundance of transcripts controlled by the response regulator is not necessarily a reliable indicator of activation of the system. Instead, the direct output of a two-component system is the amount of phosphorylated response regulator (34). Therefore, to investigate activation of the PhoP/PhoQ system, we examined the amount of PhoP-P by using Phos-tag gels (35) and Western blotting with antibodies recognizing PhoP. Phos-tag gels allow the separation of the phosphorylated and unphosphorylated forms of the PhoP protein in crude extracts from bacteria grown under different conditions (36).
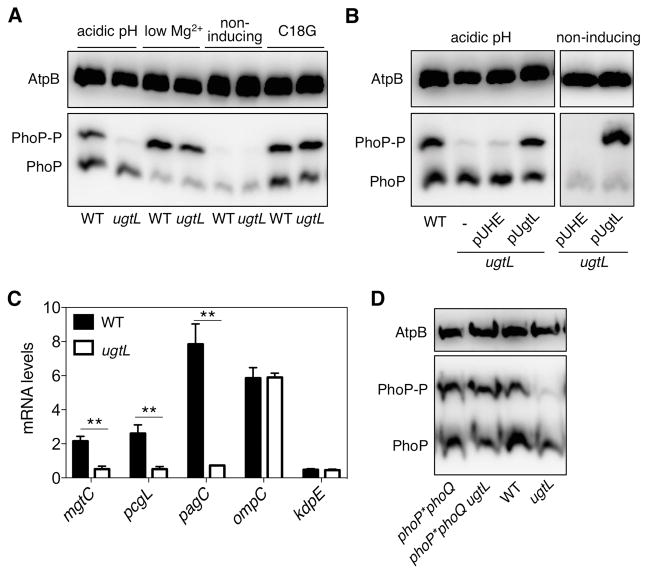
We examined the amounts of PhoP-P in isogenic wild-type and ugtL Salmonella strains grown under different conditions that induce the sensor PhoQ. These experiments were performed in a hns-FLAG derivative background in which the PhoP/PhoQ system was activated in mildly acidic pH similarly to wild-type Salmonella (fig. S1). When wild-type Salmonella was exposed to a mildly acidic pH, PhoP-P represented ~40% of the total PhoP protein and unphosphorylated PhoP the remaining ~60% (Fig. 1A). By contrast, the fraction of PhoP-P was very much reduced in the ugtL mutant (Fig. 1A) under mildly acidic conditions. The defect of the ugtL mutant is due to lack of UgtL protein, as opposed to the ugtL mutation compromising expression of a gene(s) located downstream, because a plasmid expressing the ugtL gene from a heterologous promoter restored the abundance of PhoP-P but the empty vector control did not (Fig. 1B). UgtL was required for normal PhoP-P abundance during growth specifically in mildly acidic pH because the ugtL mutant displayed normal PhoP activation when PhoQ was induced by the antimicrobial peptide C18G or low Mg2+ conditions (Fig. 1A). As expected, PhoP-P was not detected in wild-type or ugtL Salmonella grown under non-inducing conditions (neutral pH, high Mg2+ and no C18G) (Fig. 1A), and both strains contained similar amounts of unphosphorylated PhoP (Fig. 1A).
Fig. 1. The ugtL gene is required for promoting the phosphorylated state of PhoP in acidic conditions in a PhoQ-dependent manner.
(A) Phos-tag Western blot analysis of crude extracts prepared from wild-type (WT) Salmonella enterica (JC805) and the isogenic ugtL mutant (JC925) strains grown in N-minimal media with 1 mM of Mg2+ at pH 4.9 (acidic pH), 10 μM Mg2+ at pH 7.6 (low Mg2+), 1mM Mg2+ at pH 7.6 (non-inducing), or the antimicrobial peptide C18G to mid-log phase using antibodies recognizing PhoP or the loading control AtpB. (B) Phos-tag Western blot analysis of crude extracts prepared from wild-type Salmonella (JC805) and ugtL mutants (JC925) harboring either the empty vector (pUHE) or a plasmid expressing UgtL (pUgtL) grown in inducing conditions (acidic pH) or non-inducing conditions to mid-log phase using antibodies recognizing PhoP or the loading control AtpB. (C) Abundance of mgtC, pcgL, pagC, ompC, and kdpE transcripts in wild-type (JC805) and ugtL (JC925) Salmonella grown in acidic pH to mid-log phase. The mean and SD from three independent experiments are shown. Unpaired students T-test were performed between wild-type and isogenic ugtL mutant strains; ** p < 0.01. (D) Phos-tag Western blot analysis of crude extracts prepared from wild-type (JC805), ugtL (JC925), phoP*phoQ (JC1014), and phoP*phoQ ugtL (JC1056) Salmonella grown in acidic pH to mid-log phase using antibodies directed against PhoP or the loading control AtpB. Data are representative of three independent experiments.
The ugtL mutant displayed reduced mRNA abundance of the PhoP-activated horizontally acquired genes mgtC, pcgL and pagC when cells were grown in mildly acidic pH (Fig. 1C), in agreement with the notion that PhoP-P is the form of PhoP that promotes gene transcription (20). By contrast, the utgL mutant retained wild-type mRNA abundance of the ompC and kdpE genes, which are under transcriptional control of the response regulators OmpR (37) and KdpD (38), respectively (Fig. 1C). Altogether, these results establish that Salmonella requires UgtL to achieve wild-type PhoP-P amounts when the PhoQ inducing signal is a mildly acidic pH.
UgtL enhances PhoP-P abundance even in neutral pH
If UgtL activates PhoP by helping PhoQ sense a mildly acidic pH, then ugtL expression under non-inducing conditions would not be expected to increase the abundance of PhoP-P. However, transcription of the ugtL gene from a heterologous promoter increased PhoP-P amounts even when Salmonella was grown under non-inducing conditions for PhoQ (Fig. 1B). These results indicate that stimulation of PhoP-P by UgtL is not dependent on a mildly acidic pH.
UgtL increases PhoP-P abundance in a PhoQ-dependent manner
UgtL may increase PhoP-P amounts by stimulating PhoP phosphorylation or by decreasing PhoP-P dephosphorylation, or both. Thus, PhoQ is a likely UgtL target because PhoQ-P is both the only known phosphodonor for wild-type PhoP (39) and the only known PhoP-P phosphatase (40). Alternatively or in addition, UgtL may prevent the PhoP-activated protein MgrB from inhibiting PhoQ’s autokinase activity (41), increase the basal amount of the PhoP protein, which could facilitate PhoP phosphorylation from a non-cognate phosphodonor, or both.
If UgtL targets PhoQ, then inactivation of the ugtL gene should not alter PhoP-P amounts in the absence of PhoQ. Because PhoQ-P is the only known phosphodonor for wild-type PhoP (39), it is not possible to evaluate the effects of ugtL mutation PhoP phorphorylation in a phoQ mutant background. Therefore, we investigated the role of ugtL in a phoP*phoQ strain, which lacks the phoQ gene but produces a PhoP variant that autophosphorylates from the small molecular weight phosphoryl donor acetyl phosphate (39). Isogenic ugtL+ and ugtL− strains in the phoP*phoQ background displayed similar PhoP-P amounts (Fig. 1D), which is in contrast to the lower PhoP-P amounts displayed by the ugtL mutant in a phoP+phoQ+ (wild-type) genetic background (Fig. 1D). Thus, UgtL acts in a PhoQ-dependent manner.
The UgtL-dependent increase in PhoP-P is independent of MgrB because the ugtL mgrB double mutant displayed lower PhoP-P amounts than the mgrB single mutant (fig. S2). In agreement with previous reports that examined transcription of PhoP-activated genes in E. coli during growth in low Mg2+ (42), PhoP-P amounts were higher in the mgrB mutant than in wild-type Salmonella grown in mildly acidic pH (fig. S2). Moreover, amounts of PhoP-P were higher in the mgrB ugtL double mutant than the ugtL single mutant (fig. S2). These results indicate that MgrB activity is not dependent on the presence of UgtL or specific to a particular PhoQ-inducing condition, and that the UgtL-dependent enhancement of PhoP-P does not require the presence of MgrB.
The positive feedback loop that PhoP exerts on its own promoter (43) was also dispensable for UgtL activity because PhoP-P amounts were still lower in the ugtL mutant than in the isogenic ugtL+ strain when the chromosomal phoPphoQ operon was transcribed from a PhoP-independent promoter (21) (fig. S3). Cumulatively, these data indicate that UgtL enhances PhoP-P abundance by altering one or more PhoQ activities.
UgtL binds to the sensor PhoQ
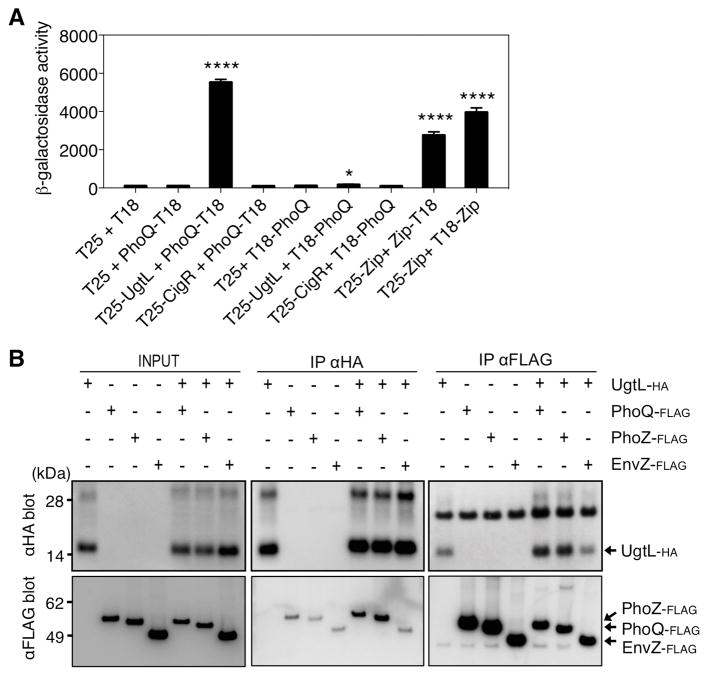
Independent approaches demonstrated that UgtL interacted directly with PhoQ. First, we used the bacterial two-hybrid system wherein β-galactosidase activity was examined in an E. coli strain lacking its own adenylate cyclase but expressing two fragments (T25 and T18) of the Bordetella pertussis adenylate cyclase that, when brought in close proximity by interactions of chimeric proteins, restore activity (44). The β-galactosidase activity of a strain co-expressing the T25 adenylate fragment fused to the N terminus of UgtL (T25-UgtL) and the T18 adenylate fragment fused to the C terminus of PhoQ (PhoQ-T18) was ~50 times higher than that produced by bacteria carrying the negative control plasmids (Fig. 2A). This activity was even higher than that produced by the strain with the positive control plasmids (Fig. 2A). By contrast, there was little β-galactosidase activity when the UgtL fusion protein (T25-UgtL) was co-expressed with T18 fused to the N terminus of PhoQ (T18-PhoQ) (Fig. 2A). Likewise, no β-galactosidase activity was produced by strains expressing T18 fusions to either the N or C terminus of PhoQ (T18-PhoQ or PhoQ-T18) along with a T25 fusion to CigR (T25-CigR) (Fig. 2A), an inner membrane protein used as a negative control.
Fig. 2. The UgtL and PhoQ proteins interact.
(A). β-galactosidase activity in bacterial two-hybrid system assays in E. coli BTH101 expressing the indicated fusion proteins. In each experimental condition, the bacteria carried the two Bordetella pertussis adenylate cyclase fragments T25 and T18 either alone or fused to UgtL, CigR, or PhoQ in the indicated combinations. The adenylate cyclase fragment was fused to the N terminus in fusion proteins T25-UgtL, T25-CigR, and T18-PhoQ and to the C terminus in fusion protein PhoQ-T18. T25-Zip, Zip-T18, and T18-Zip were used as positive controls. The mean and SD from three independent experiments are shown. Unpaired students T-test were performed between strains harboring empty vectors with the other combinations; * p < 0.05, **** p <0.0001. (B) Pulldown assays showing interactions between in vitro–synthesized UgtL-HA, PhoQ-FLAG, PhoZ-FLAG, and EnvZ-FLAG proteins. Samples were analyzed by Western blotting using antibodies recognizing the HA epitope and the FLAG epitope. Densitometry of each blot with ImageJ software is shown below the blots in the same order using arbitrary units (AU). Dashed lines in the densitometry graphs indicate signals from nonspecific binding of the UgtL-HA and FLAG-tagged proteins to antibodies recognizing the FLAG and HA epitopes, respectively. The data are representative of two independent experiments, which produced similar results.
As an independent test for the interaction between UgtL and PhoQ, we used immunoprecipitation of the UgtL-HA and PhoQ-FLAG proteins and corresponding controls synthesized with the PURExpress in vitro transcription-translation system (45). These tagged proteins were reconstituted into liposomes (46) because PhoQ and UgtL are integral membrane proteins (18, 29). Antibodies recognizing the FLAG epitope pulled down UgtL-HA, and antibodies recognizing the HA epitope pulled down PhoQ-FLAG (Fig. 2B, S4). In these experiments, there was a background signal that indicated a small amount of nonspecific binding of the UgtL-HA and PhoQ-FLAG proteins to antibodies recognizing the FLAG and HA epitopes, respectively. The UgtL-PhoQ interaction appears to be specific because the antibodies recognizing the HA epitope did not pull down EnvZ-FLAG, a sensor kinase used as a negative control, and the antibodies recognizing the FLAG epitope did not pull down UgtL-HA, from liposomes harboring UgtL-HA and EnvZ-FLAG (Fig. 2B, S4).
UgtL interacts with a region of PhoQ not involved in acidic pH sensing
The PhoQ protein harbors two transmembrane domains that define a periplasmic region necessary for sensing Mg2+ (22) and C18G (19). The large C-terminal region of PhoQ is required for the response to acidic pH and harbors the catalytic and dimerization domains of PhoQ. To determine whether UgtL interacts with regions of PhoQ implicated in signal sensing, we investigated the behavior of a strain expressing PhoZ in place of PhoQ. PhoZ is a chimeric protein consisting of the N-terminal portion of PhoQ including its periplasmic, transmembrane and HAMP (i.e., Histidine kinase, adeylate cyclases, Methyl-accepting proteins and Phosphatases, mediating signaling output) domains, fused to the catalytic and dimerization domains from EnvZ (Fig. 2B, S4). The N-terminal 266 amino acid region from PhoQ present in PhoZ was sufficient for interaction with UgtL-HA (Fig. 2B, S4). Therefore, UgtL binds to a region of PhoQ that is distinct from the cytosolic region previously implicated in sensing a mildly acidic pH (13).
UgtL promotes PhoQ autophosphorylation from ATP
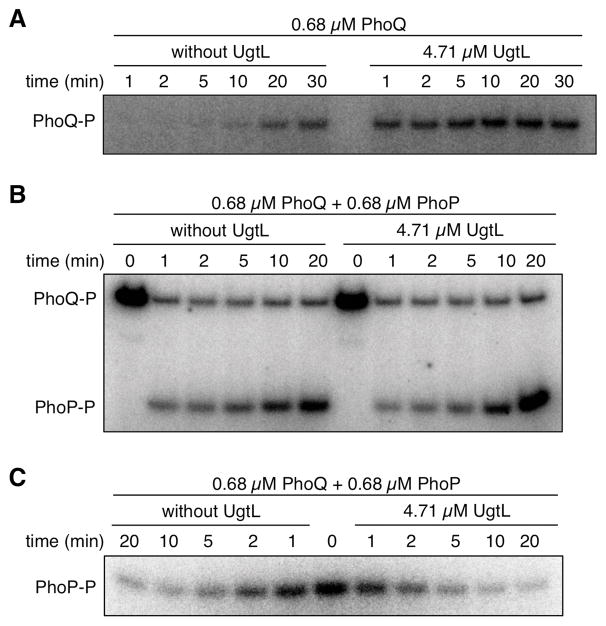
We wondered how UgtL binding to PhoQ (Fig. 2) increases PhoP-P abundance in vivo (Fig. 1A). UgtL could promote PhoP phosphorylation by stimulating PhoQ autophosphorylation, by enhancing phosphotransfer from PhoQ-P to PhoP, by inhibiting dephosphorylation of PhoP-P by PhoQ, or by modifying several of these activities. In phosphorylation assays using in vitro–synthesized proteins, UgtL accelerated autophosphorylation of the full length PhoQ protein (Fig. 3A). In contrast, neither phosphotransfer from PhoQ to PhoP (Fig. 3B) nor PhoP-P dephosphorylation by PhoQ (Fig. 3C) were affected by the presence of UgtL in the reaction. These results assign a biochemical function to UgtL, a protein with no homologs in sequence databases.
Fig. 3. UgtL promotes autophosphorylation of PhoQ in vitro.
(A) Amounts of PhoQ-P at the indicated time points in the presence or absence of UgtL and 32P-labeled ATP. (B) Amounts of PhoQ-P and PhoP-P at the indicated times after addition of PhoP to reaction mixtures containing PhoQ-P or PhoQ-P + UgtL. (C) Amounts of PhoP-P at the indicated times after addition of PhoP-P to reaction mixtures containing PhoQ or PhoQ + UgtL. The data are representative of two independent experiments, which produced similar results.
The transmembrane and periplasmic regions of UgtL are required to enhance PhoP-P abundance
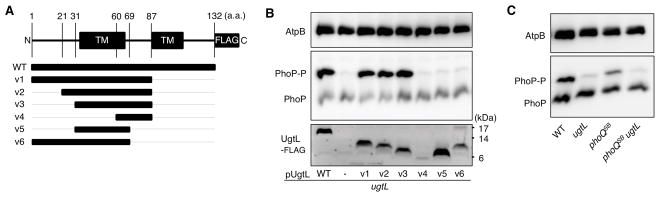
UgtL is an inner membrane protein (29). The deduced amino acid sequence of the ugtL gene predicts two transmembrane domains that define a 24 amino acid periplasmic region (47) (Fig. 4A). To define the portion of UgtL required for PhoQ activation, we examined PhoP phosphorylation in a ugtL deletion strain expressing various truncated variants of a C-terminally FLAG-tagged UgtL protein (Fig. 4A). The use of FLAG-tagged variants enabled us to probe the amounts of the investigated proteins. A strain expressing the UgtL-FLAG protein exhibited the same proportion of PhoP-P:PhoP proteins as a strain expressing the wild-type, untagged UgtL protein (fig. S5).)
Fig. 4. UgtL and the cytoplasmic domain of PhoQ respond independently to mildly acidic pH.
(A) Schematic of the full-length (FL) UgtL-FLAG protein and the truncated variants v1–v6. (B) Phos-tag Western blot (top and middle) or Western blot (bottom) analysis of crude extracts prepared from Salmonella ugtL mutant (EG13682) harboring plasmids expressing FL FLAG-tagged UgtL or the truncated variants as indicated. Phos-tag Western blots were probed with antibodies recognizing PhoP and AtpB (loading control), and the Western blot was probled with antibodies recognizing the FLAG epitope. Data are representative of three independent experiments, which produced similar results. (C) Phos-tag Western blot analysis of crude extracts prepared from wild-type (WT) Salmonella (JC805), ugtL mutant Salmonella (JC925), Salmonella expressing a PhoQ variant (PhoQSB) that is insensitive to acidic pH (JC1102; phoQSB), and phoQSB ugtL Salmonella (JC1123) grown in acidic pH to mid-log phase. Blots were probed with antibodies recognizing PhoP and AtpB. Data are representative of three independent experiments, which produced similar results.
Like full-length UgtL, one specifying the UgtL variant UgtL(1–87), which includes only the first 87 amino acids of UgtL, retained normal PhoP activation when cells were grown in mildly acidic pH conditions (Fig. 4B). Expression of UgtL(21–87) or UgtL(31–87) also increased PhoP-P amounts in mildly acidic pH (Fig. 4B), although these latter two UgtL variants displayed slightly lower activity than did full-length UgtL or UgtL(1–87) (Fig. 4B). By contrast, plasmids expressing UgtL(60–87), UgtL(31–69), or UgtL(1–69) failed to increase PhoP-P amounts in acidic pH (Fig. 4B). The FLAG-tagged UgtL(31–69) and UgtL(1–69) proteins were produced in similar abundance as the wild-type UgtL-FLAG protein (Fig. 4B). Therefore, the failure of these variants to enhance PhoP-P abundance is likely due to loss of activity as opposed to decreased expression or stability. Taken together, these results indicate that the first predicted transmembrane domain and periplasmic region of UgtL are necessary to increase PhoP-P abundance, but the 30 N-terminal and 45 C-terminal amino acids of UgtL are not. These results leave open the possibility that the periplasmic domain of UgtL alone, if artificially targeted to the periplasm, could be able to bind to PhoQ and enhance PhoP phosphorylation.
UgtL functions independently of the cytoplasmic pH-sensing domain of PhoQ
The UgtL protein appears to stimulate PhoQ’s autokinase activity independently of PhoQ’s ability to sense a mildy acidic pH, because expression of the ugtL gene from a heterologous promoter increased PhoP-P amounts even when Salmonella was grown under non-inducing conditions (neutral pH) for PhoQ (Fig. 1B). If this is the case, PhoP-P amounts should be lower in a double mutant lacking the ugtL gene and carrying a PhoQ mutation (phoQSB) that renders the protein insensitive to mildly acidic pH (13) compared to either single mutant. As hypothesized, the ratio of PhoP-P to PhoP was the highest in wild-type Salmonella, somewhat reduced in the phoQSB mutant, and greatly reduced in the ugtL single mutant and the acidic pH–blind phoQSB ugtL double mutant (Fig. 4C). Thus, PhoQ-promoted phosphorylation of PhoP triggered by an acidic pH entails PhoQ detecting a change in pH (or a downstream effect of a change in pH) and the UgtL protein enhancing PhoQ autophosphorylation.
PhoP promotes ugtL transcription in mildly acidic pH
Given that UgtL enhances PhoQ activation in mildly acidic pH (Fig. 1), we wondered how the ugtL gene is expressed under different PhoQ-inducing conditions. The abundance of ugtL transcritps was much higher in wild-type Salmonella experiencing low Mg2+ or a mildly acidic pH than when the organism was grown under non-inducing conditions (fig. S6). The transcriptional induction of the ugtL gene is PhoP-dependent because little ugtL mRNA accumulation was observed in the isogenic phoP mutant (fig. S6). The transcriptional induction of the ugtL gene in mildly acidic pH implies that the UgtL protein is present and therefore available to activate the sensor PhoQ under acidic conditions.
UgtL-dependent resistance to magainin 2 requires PhoP
Our laboratory reported growth in low Mg2+ renders both ugtL and phoP null mutants hypersensitive to the antimicrobial peptide magainin 2 (29). A plasmid in which the ugtL gene is transcribed from a heterologous promoter complemented the ugtL mutant but not a phoP mutant in this context (fig. S7). These results suggest that UgtL-mediated magainin 2 resistance requires PhoP itself, a PhoP-dependent product(s), or both.
ugtL is required for full virulence
It has been proposed that activation of the PhoP/PhoQ system by mildly acidic pH is critical for Salmonella virulence because inhibition of acidification of the Salmonella-containing vacuole prevents the expression of PhoP-activated genes in phagocytic (48, 49) and non-phagocytic cells (50), limits replication inside macrophages (51, 52), and attenuates virulence in mice (53). Given that the ugtL gene is necessary for full PhoP activation under a mildly acidic pH, we reasoned that inactivation of the ugtL gene would attenuate Salmonella virulence.
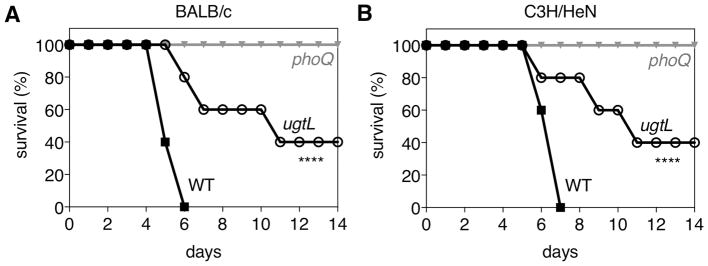
Intraperitoneal inoculation of BALB/c and C3H/HeN mice with ugtL mutant Salmonella demonstrated that virulence of the ugtL strain was indeed attenuated compared to wild-type Salmonella (Fig. 5A–B). Natural resistance-associated macrophage protein 1 (NRAMP1) is a divalent metal ion transporter that is present in macrophage phagosomal membranes and has been implicated in mammalian resistance to several intracellular pathogens that remain within acidic phagosomes, including Salmonella (54). Whereas C3H/HeN mice are wild-type with respect to NRAMP1, BALB/c mice harbor mutant alleles of SCL11A1 (54, ref). Both BALB/c and C3H/HeN mice were susceptible to infection by wild-type Salmonella and resistant to infection by phoQ mutants. Whereas no mice survived more than seven days after inoculation with wild-type Salmonella, mice infected with ugtL Salmonella survived longer, with some surviving more than 14 days post inoculation (Fig. 5A–B). These results reinforce the notion that the ability of the PhoP/PhoQ system to respond to a mildly acidic pH is necessary for Salmonella pathogenesis.
Fig. 5. UgtL is required for maximal Salmonella virulence in mice.
Survival of BALB/c (A) or C3H/HeN (B) mice inoculated intraperitoneally with wild-type (14028s), ugtL (EG13682) or phoQ (MS5996s) Salmonella. Data are representative of N = 2 independent experiments, which produced similar results, n=5 mice per each experimental group. Mantel-Cox test was performed between wild-type and ugtL Salmonella infected mice; **** p < 0.0001.
DISCUSSION
We have established that activation of the response regulator PhoP during growth under mildly acidic pH requires two proteins providing two different functions: the sensor PhoQ is responsible for detecting changes in pH (or a downstream effector of such changes), and the protein UgtL boosts the response of PhoQ to mildly acidic pH (Fig. 1 and 6). In addition, UgtL confers resistance to the antimicrobial peptide magainin 2 (29). This activity of UgtL is independent of the promotion of PhoP-P abundance (Fig. 1) because wild-type and ugtL Salmonella display similar PhoP-P amounts during growth in low Mg2+ (Fig. 1A), a condition in which the ugtL mutant exhibits hypersusceptibility to magainin 2 (fig. S7). That a given protein may have more than one target is also illustrated by another PhoP-activated protein, MgtC, which binds to and inhibits the F1Fo ATP synthase (55) and binds to and stabilizes PhoP (56).
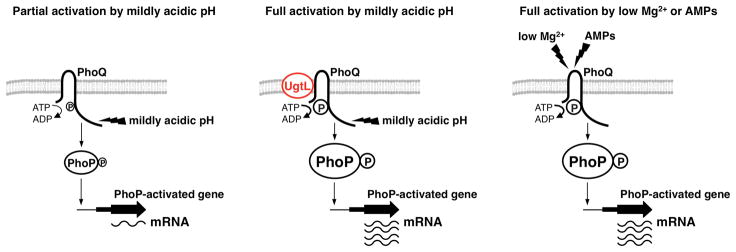
Fig. 6. Activation of the sensor PhoQ in mildly acidic pH requires the Salmonella-specific UgtL protein to intensify the response of PhoQ to a mildly acidic pH.
(A) The sensor PhoQ responds to acidification of the cytoplasm through its cytoplasmic domain. Mildly acidic pH causes PhoQ to autophosphorylate, after which it functions as a phosphodonor to the response regulator PhoP. Phosphorylated PhoP (PhoP-P) binds to promoters and promotes transcription of PhoP-activated genes. (B) UgtL enhances PhoQ autophosphorylation in response to acidic conditions, which results in increased abundance of PhoP-P and of PhoP-activated mRNAs. UgtL is required for Salmonella to achieve full PhoP-dependent gene transcription when the PhoQ-inducing signal is mildly acidic pH. (C) The periplasmic domain of PhoQ mediates PhoQ activation in response to conditions of low Mg2+ or the antimicrobial peptide C18G. In response to either of these stimuli, PhoQ undergoes autophosphorylation and then functions as a phosphodonor to PhoP. In this context, PhoQ-mediated activation of PhoP is sufficient for full transcription of PhoP-activated genes and is not dependent on UgtL.
Horizontal acquisition of foreign gene broadens the responsiveness of an ancestral regulatory system
The UgtL protein is highly conserved in the pathogenic species S. enterica (≥98% amino acid sequence identity), which includes the human pathogen serovar Typhi. Because the phoP gene is required for virulence of S. enterica serovar Typhi (57) and the PhoQ proteins from the Typhi and Typhimurium serovars are 99% identical, ugtL may also contribute to typhoid pathogenesis.
The UgtL protein from the non-pathogenic species S. bongori exhibits 55% amino acid identity to S. enterica UgtL. This unusually low amino acid identity is intriguing given that S. bongori PhoQ is insensitive to mildly acidic pH but can be activated by other PhoQ-inducing signals (13). The presence of ugtL in both S. enterica and S. bongori suggests that this gene was acquired before the two Salmonella species diverged from a common ancestor. This also suggests that the ability of Salmonella to respond to acid through PhoQ activation and UgtL-mediated enhancement of acid-induced PhoQ activation may be an important determinant of a species’ virulence.
Mildly acidic pH also stimulates the transcription of PhoP-activated genes in E. coli (42), yet E. coli lacks ugtL. Instead, the E. coli–specific protein SafA mediates this activation (58). Like UgtL, the SafA protein enhances the abundance of PhoP-P under non-inducing conditions for PhoQ when expressed from a heterologous promoter (58). The absence of sequence similarity between SafA and UgtL makes these proteins functional paralogues rather than homologues.
The safA gene is under transcriptional control of the E. coli–specific EvgA/EvgS two-component system, which is also activated by acidic pH (58). In contrast, the Salmonella ugtL gene is under transcriptional control of the PhoP/PhoQ system (fig. S6) (30, 59). Therefore, SafA enables E. coli to activate PhoP-dependent genes in response to the signals acting on the sensor EvgS, whereas UgtL enables Salmonella to heighten the response to a particular PhoQ-inducing signal (acidic pH). Our findings identify an instance of convergent evolution whereby a gene horizontally acquired through foreign DNA enhances the responsiveness of an ancestral regulatory system.
UgtL promotes sensor phosphorylation without participating in signal sensing
The process of signal transduction typically involves sensors that modify the enzymatic activity of proteins directly responsible for transmission of a signal. For example, upon binding to (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuryl borate, the sensor LuxP stimulates the phosphatase activity of the LuxQ protein in Vibrio harveyi (60, 61). Likewise, G protein–coupled receptors respond to receptor-specific stimuli by activating a downstream G protein that, in turn, initiates an intracellular signaling cascade (62). In contrast, other proteins limit signal transduction processes. For instance, binding of the E. coli LysP protein to the pH sensor CadC renders it unresponsive to acidic pH (63). The mode of action of UgtL differs from those described above. That is, the output of the sensor PhoQ reflects both its direct response to a midly acidic pH and tuning of that response by UgtL.
Activation of PhoQ by acidic pH is required for Salmonella virulence
The PhoQ protein responds to a mildly acidic pH (13, 17), low Mg2+ (18), certain antimicrobial peptides (19), unsaturated long chain fatty acids (64), acetate concentrations (65), and the redox state in the periplasmic space (66). Which of these signals activates PhoQ during Salmonella infection of its mammalian hosts? It has been proposed that acidic pH and low Mg2+ are not relevant signals because a Salmonella strain with amino acid substitutions in PhoQ that render PhoQ less responsive to these two signals retained wild-type virulence in mice (67).
The data presented here strongly suggest that PhoQ activation by mildly acidic pH is critical for Salmonella virulence because a ugtL null mutant is defective for both PhoP/PhoQ activation by mildly acidic pH (Fig. 1) and virulence in mice (Fig. 5). This notion is supported by the virulence attenuation exhibited by a Salmonella strain with a PhoQ variant impaired in its ability to respond to a mildly acidic pH (13), which, like the ugtL mutant (Fig. 1), responds normally to both low Mg2+ and the antimicrobial peptide C18G (13). Furthermore, Salmonella replication within macrophages and virulence in mice are compromised upon inhibition of phagosome acidification (51–53). An additional signal(s), protein(s), or both must activate PhoP/PhoQ during infection because the Salmonella strains lacking ugtL or bearing amino acid substitutions in PhoQ that hinder the response to mildly acidic pH are not as attenuated for virulence as the phoQ null mutant (13, 28) (Fig. 5).
MATERIALS AND METHODS
Bacterial strains, plasmids, oligodeoxynucleotides, and growth conditions
Bacterial strains and plasmids used in this study are listed in Table S1. All S. enterica serovar Typhimurium strains were derived from the wild-type strain 14028s (28) and constructed by phage P22-mediated transductions as described (68). Bacteria were grown at 37°C in Luria-Bertani (LB) broth or N-minimal media (69) supplemented with 0.1% casamino acids, 38 mM glycerol, and the indicated pH (pH 7.6 or pH 4.9) and concentrations of MgCl2. E. coli DH5α was used as the host for preparation of plasmid DNA (70). E. coli BTH101 cya was used for the bacterial two-hybrid assay (44). To induce plasmid expression, isopropyl β-D-1-thiogalactopyranoside (IPTG) was added at 20 μM for experiments with the UgtL truncation variants and at 200 μM for the magainin 2 assays. When necessary to select for plasmid maintenance, appropriate antibiotics were added at the following final concentrations: ampicillin at 50 μg/ml, chloramphenicol at 20 μg/ml, kanamycin at 50 μg/ml, and tetracycline at 10 μg/ml. The antimicrobial peptide C18G was used at a final concentrations of 5 μg/ml, and bacteria were treated for 90 min. DNA oligonucleotides used in this study are listed in Table S2.
In vivo detection of phosphorylated PhoP
PhoP and PhoP-P were separated on 12.5% polyacrylamide gels containing acrylamide-Phos-tag™ ligand (Phos-tag™ Consortium) as described by the manufacturer. Gels were copolymerized with 50 μM Phos-tag™ acrylamide and 100 μM MnCl2. Whole-cell extracts were prepared as described and normalized by OD600 (35). The samples were electrophoresed on Phos-tag™ gels with standard running buffer [0.4% (w/v) SDS, 25 mM Tris, 192 mM glycine] at 4°C under 20 mAmp for 5 h, transferred to nitrocellulose membranes, and analyzed by immunoblotting using polyclonal rabbit antibodies recognizing PhoP (1:2,000) and polyclonal mouse antibodies recognizing AtpB (1:5,000). Secondary horseradish peroxidase-conjugated antisera recognizing rabbit and mouse antibodies (GE healthcare) were used at 1:5,000 dilution. The blots were developed with the Amersham ECL Western Blotting Detection Reagents (GE Healthcare) or SuperSignal West Femto Chemiluminescent system (Pierce). The data are representative of three independent experiments, which produced similar results.
Construction of mutant strain
To generate an hns-FLAG strain, a cat cassette was introduced in the 3′end of the hns gene as follows: the cat fragment was amplified from plasmid pKD3 using primers 15748/15749, then introduced into wild-type Salmonella (14028s) harboring plasmid pKD46 as described (71). The cat cassette was removed using plasmid pCP20 (71).
Construction of plasmids
Plasmid pUgtL(1–87) was constructed as follows: the ugtL gene was amplified from wild-type Salmonella (14028s) using primers 2240/16060, then introduced between the BamHI and HindIII sites of pUHE21–2lacIq (43). Plasmid pUgtL-FLAG was constructed as follows: the ugtL coding region was amplified from wild-type Salmonella (14028s) using primers 2240/3143, then introduced between the BamHI and HindIII sites of pUHE21–2lacIq (43). Plasmids for expressing truncated UgtL-FLAG were constructed as follows: various truncated FLAG-tagged ugtL genes were amplified using primers 2240/16096 (for 1–87), 16096/16098 (for 21–87), 16096/16099 (for 31–87), 16096/16104 (for 61–87), 16098/16105 (for 21–69), 16099/16105 (for 31–69), or 2240/16105 (for 1–69) from strain 14028s, then introduced between the BamHI and HindIII sites of pUHE21–2lacIq (43). Plasmid pPhoZ, constructed by a former lab member, harbors the DNA sequence specifying the N-terminal 266 amino acids of PhoQ fused in frame to the C-terminal 218 amino acids of EnvZ between the BamHI and HindIII sites of pUHE21–2lacIq (43). Plasmid pUT18-PhoQ and pUT18C-PhoQ were constructed as follows: the phoQ gene was amplified from wild-type Salmonella (14028s) using primers 11824/11825, then introduced between BamHI and KpnI sites of pUT18 or pUT18C (72). Plasmid pKT25-UgtL was constructed as follows: the ugtL gene was amplified from wild-type Salmonella (14028s) using primers 16041/16042, then introduced between XbaI and KpnI sites of pKT25 (72). Plasmid pKT25-CigR was constructed as follows: the cigR gene was amplified from wild-type Salmonella (14028s) using primers 16108/16109, then introduced between XbaI and KpnI sites of pKT25 (72).
Quantitative RT–PCR
Total RNA was isolated using the RNeasy Kit (Qiagen) according to the manufacturer’s instructions. The purified RNA was quantified using a Nanodrop machine (NanoDrop Technologies). Complementary DNA (cDNA) was synthesized using High Capacity RNA-to cDNA Master Mix (Applied Biosystems). The mRNA abundance of the mgtC, phoP, ugtL, pmrD and rrs genes were measured by quantification of cDNA using Fast SYBR Green PCR Master Mix (Applied Biosystems) and following primers (mgtC: 6962/6963; pagC: 6964/6965; pcgL: 6627/6628; ompC: W2033/W2034; kdpE: 7923/7924) and monitored using a QuantStudio6 machine (Applied Biosystems). Data were normalized to the abundance of 16S ribosomal RNA amplified with primers 3023 and 3024.
Bacterial two-hybrid assay
E. coli BTH101 harboring derivatives of plasmids pUT18 and pKT25 were grown overnight in LB broth containing ampicillin (100 μg/ml) and kanamycin (50 μg/ml), added at 1:100 dilution to 1 ml of the same fresh media containing isopropyl β-D-1-thiogalactopyranoside (IPTG; 0.5 mM) and grown with shaking at 30°C overnight as described (44, 72). β-galactosidase activities were determined using a kinetic assay as described (73).
Pull-down assay with in vitro transcription-translation system
Pull-down assay was performed as described with some modifications (55). All proteins for in vitro synthesis were produced using the cell-free PURExpress in vitro protein synthesis system (New England Biolabs) in the presence of 0.12 mg/ml proteoliposomes at 37°C for 3 h. Proteoliposomes were prepared using soybean L-α-phosphatidylcholine (Sigma) in buffer (20 mM Tricine, 20 mM succinic acid, 80 mM NaCl and 0.6 mM KOH, adjusted to pH 8.0) to a concentration of 32 mg/ml as described (46). DNA templates were prepared according to the manufacturer’s instructions. To synthesize UgtL-HA, PhoQ-FLAG, PhoZ-FLAG, and EnvZ-FLAG, we used primers 16061/16062 for UgtL-HA, 16063/16064 for PhoQ-FLAG, 16063/16065 for PhoZ-FLAG, 16065/16066 for EnvZ-FLAG. At the end of the reaction, samples were diluted in TBS buffer (Tris-buffered saline) with 20 times the reaction volumes. Diluted reactions were mixed in 500 μL TBS containing 0.12 mg/ml proteoliposomes and incubated at room temperature for 2 h. Then samples were pulled down with magnetic beads conjugated to antibodies recognizing the HA epitope at 4°C for 1 h or with magnetic beads conjugated to antibodies recognizing the FLAG epitope at room temperature for 2 h. Pulled down samples were analyzed by Western blotting using antibodies recognizing the HA or FLAG epitopes.
Autophosphorylation, phosphotransfer and dephosphorylation assays
For autophosphorylation assays, 0.68 μM PhoQ-Strep was incubated with 4.71 μM of UgtL-HA in TKM buffer (50 mM Tris-HCl [pH 8.0], 50 mM KCl, 1 mM MgCl2) at 37°C for 15 min. The reaction was started by addition of 30 μM of ATP containing 30 μCi of [γ-32P] ATP in a 50 μl reaction mixture at room temperature. An 8 μl aliquot was mixed with 5 × SDS loading buffer at different times to stop the reaction. Samples were kept on ice until they were loaded onto a 10% Bis-Tris SDS-PAGE gel to separate the protein from free nucleotides.
For phosphotransfer assays, phosphorylated PhoQ-Strep was generated by incubating 0.68 μM of PhoQ-Strep with or without 4.71 μM of UgtL-HA in TKM buffer with 30 μM of ATP containing 30 μCi of [γ-32P] ATP at room temperature for 1 h. The PhoQ-P was recovered using a Micro Bio-Spin 6 chromatography column (Bio-Rad) by removing excess ATP and ADP generated from ATP. The phosphotransfer reaction was started by adding 0.68 μM of PhoP proteins in a 50 μl reaction mixture. An 8 μl aliquot was mixed with 5 × SDS loading buffer at different time points to stop the reaction. The samples were kept on ice until they were loaded onto 10% Bis-Tris SDS-PAGE gel.
For dephosphorylation assays, phosphorylated PhoP-His6 was prepared as described (74). Briefly, 300 pmol of GST-YgiYc beads were incubated with 20 μCi [γ-32P] ATP in 50 μl of TBS-MD buffer at room temperature for 12 h, and then the beads were washed with TBS buffer five times to remove free ATP. 3 nmol of purified PhoP-His6 was incubated with phosphorylated GST-YgiYc beads in 60 μl of TBS-MD buffer at room temperature for 4 h. After incubation, the PhoP-P was recovered using a Micro Bio- Spin 6 chromatography column that had been pre-equilibrated with TBS. The dephosphorylation reaction was started by adding 0.68 μM of PhoP-P to 0.68 μM of PhoQ-Strep with or without 4.71 μM of UgtL-HA pre-incubated with 100 μM of ADP in 50 μl reaction mixture. An 8 μl aliquot was mixed with 5 × SDS loading buffer at different times to stop the reaction. Samples were kept on ice until they were loaded onto a 10% Bis-Tris SDS-PAGE gel.
Amounts of PhoP-P and PhoQ-P were quantified using a BAS-5000 imaging system (FujiFilm) and a phosphorimaging plate (FujiFilm).
Mouse virulence assay
Six-week-old female BALB/c or C3H/HeN mice were purchased from Charles River Laboratories. Five mice in each group were infected intraperitoneally with 0.1 ml of PBS containing ~102 (for BALB/c) or ~2 × 104 (for C3H/HeN) Salmonella that had been grown overnight in LB broth and resuspended and diluted in PBS. Mouse survival was monitored every 12 hours for three weeks. Virulence assays were conducted twice with similar outcomes, and data for each experimental group correspond to groups of five mice. All animals were housed in temperature- and humidity-controlled rooms and maintained on a 12 h light/12 h dark cycle. All procedures complied with regulations of the Institutional Animal Care and Use Committee of the Yale School of Medicine.
Antimicrobial peptide killing assay
Antimicrobial peptide susceptibility assays were conducted as described (29). Bacteria were grown in N-minimal media with 10 μM Mg2+ and pH 7.6 with IPTG (200 μM) to mid-log phase. Bacterial cells were then diluted to 1~2 × 105 colony forming units (CFU)/ml in the same media. Magainin 2 was dissolved and diluted with autoclaved distilled water. Samples of 5 μl of peptide solution were placed in wells of 96-well plate and 45 μl of the bacteria samples were added. After 1 h incubation at 37°C with aeration, samples were serially diluted in LB and plated on LB agar plates for enumeration. The percentage survival was calculated as following: survival (%) = CFU of peptide-treated culture/CFU of no peptide culture × 100.
Supplementary Material
Fig. S1. UgtL-dependent activation of the PhoP protein under mildly acidic pH is similar in wild-type and hns-FLAG Salmonella.
Fig. S2. UgtL enhances PhoP-P abundance in an MgrB-independent manner.
Fig. S3. UgtL enhances PhoP-P abundance posttranscriptionally.
Fig. S4. The UgtL and PhoQ proteins interact.
Fig. S5. FLAG-tagged UgtL functions as wild-type UgtL in vivo.
Fig. S6. PhoP activates transcription of the ugtL gene in mildly acidic pH and low Mg2+ conditions.
Fig. S7. UgtL promotes Salmonella resistance to magainin 2 in a PhoP-dependent manner.
Table S1. Bacterial strains and plasmids used in this study
Table S2. Primers used in this study
Acknowledgments
We thank Wonsik Yeo for purifying the PhoQ-strep and PhoP-His6 proteins, and Jinki Yeom for providing plasmid pKT25-CigR.
Funding: This research was supported by NIH grant R01 AI120558 to EAG.
Footnotes
Author contributions: JC conducted the experiments reported in this paper. JC and EAG analyzed the data and wrote the manuscript.
Competing interests: The authors do not have any competing interests.
Data and materials availability: Materials and Data are available from the authors.
REFERENCES AND NOTES
- 1.Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 2.Piwon N, Gunther W, Schwake M, Bosl MR, Jentsch TJ. ClC-5 Cl--channel disruption impairs endocytosis in a mouse model for Dent’s disease. Nature. 2000;408:369–373. doi: 10.1038/35042597. [DOI] [PubMed] [Google Scholar]
- 3.Obara M, Szeliga M, Albrecht J. Regulation of pH in the mammalian central nervous system under normal and pathological conditions: facts and hypotheses. Neurochem Int. 2008;52:905–919. doi: 10.1016/j.neuint.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Vaughan-Jones RD, Spitzer KW, Swietach P. Intracellular pH regulation in heart. J Mol Cell Cardiol. 2009;46:318–331. doi: 10.1016/j.yjmcc.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 5.Schneider D, Gerhardt E, Bock J, Muller MM, Wolburg H, Lang F, Schulz JB. Intracellular acidification by inhibition of the Na+/H+-exchanger leads to caspase-independent death of cerebellar granule neurons resembling paraptosis. Cell Death Differ. 2004;11:760–770. doi: 10.1038/sj.cdd.4401377. [DOI] [PubMed] [Google Scholar]
- 6.Damaghi M, Wojtkowiak JW, Gillies RJ. pH sensing and regulation in cancer. Front Physiol. 2013;4:370. doi: 10.3389/fphys.2013.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lund P, Tramonti A, De Biase D. Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol Rev. 2014;38:1091–1125. doi: 10.1111/1574-6976.12076. [DOI] [PubMed] [Google Scholar]
- 8.Haneburger I, Eichinger A, Skerra A, Jung K. New insights into the signaling mechanism of the pH-responsive, membrane-integrated transcriptional activator CadC of Escherichia coli. J Biol Chem. 2011;286:10681–10689. doi: 10.1074/jbc.M110.196923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller S, Gotz M, Beier D. Histidine residue 94 is involved in pH sensing by histidine kinase ArsS of Helicobacter pylori. PLoS One. 2009;4:e6930. doi: 10.1371/journal.pone.0006930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez JC, Groisman EA. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol Microbiol. 2007;63:283–293. doi: 10.1111/j.1365-2958.2006.05512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakraborty S, Mizusaki H, Kenney LJ. A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLoS Biol. 2015;13:e1002116. doi: 10.1371/journal.pbio.1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umemura T, Matsumoto Y, Ohnishi K, Homma M, Kawagishi I. Sensing of cytoplasmic pH by bacterial chemoreceptors involves the linker region that connects the membrane-spanning and the signal-modulating helices. J Biol Chem. 2002;277:1593–1598. doi: 10.1074/jbc.M109930200. [DOI] [PubMed] [Google Scholar]
- 13.Choi J, Groisman EA. Acidic pH sensing in the bacterial cytoplasm is required for Salmonella virulence. Mol Microbiol. 2016;101:1024–1038. doi: 10.1111/mmi.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst RK, Guina T, Miller SI. Salmonella Typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 2001;3:1327–1334. doi: 10.1016/s1286-4579(01)01494-0. [DOI] [PubMed] [Google Scholar]
- 15.Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. 2001;183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunol Cell Biol. 2007;85:112–118. doi: 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- 17.Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit RE, Miller SI. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell. 2007;26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Garcia Vescovi E, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 19.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 20.Shin D, Groisman EA. Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J Biol Chem. 2005;280:4089–4094. doi: 10.1074/jbc.M412741200. [DOI] [PubMed] [Google Scholar]
- 21.Shin D, Lee EJ, Huang H, Groisman EA. A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science. 2006;314:1607–1609. doi: 10.1126/science.1134930. [DOI] [PubMed] [Google Scholar]
- 22.Chamnongpol S, Cromie M, Groisman EA. Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica. J Mol Biol. 2003;325:795–807. doi: 10.1016/s0022-2836(02)01268-8. [DOI] [PubMed] [Google Scholar]
- 23.Prost LR, Daley ME, Bader MW, Klevit RE, Miller SI. The PhoQ histidine kinases of Salmonella and Pseudomonas spp. are structurally and functionally different: evidence that pH and antimicrobial peptide sensing contribute to mammalian pathogenesis. Mol Microbiol. 2008;69:503–519. doi: 10.1111/j.1365-2958.2008.06303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colgan AM, Kroger C, Diard M, Hardt WD, Puente JL, Sivasankaran SK, Hokamp K, Hinton JC. The Impact of 18 Ancestral and Horizontally-Acquired Regulatory Proteins upon the Transcriptome and sRNA Landscape of Salmonella enterica serovar Typhimurium. PLoS Genet. 2016;12:e1006258. doi: 10.1371/journal.pgen.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. Regulation of lipid A modifications by Salmonella Typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 26.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 27.Groisman EA, Kayser J, Soncini FC. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J Bacteriol. 1997;179:7040–7045. doi: 10.1128/jb.179.22.7040-7045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fields PI, Groisman EA, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 29.Shi Y, Cromie MJ, Hsu FF, Turk J, Groisman EA. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol Microbiol. 2004;53:229–241. doi: 10.1111/j.1365-2958.2004.04107.x. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y, Latifi T, Cromie MJ, Groisman EA. Transcriptional control of the antimicrobial peptide resistance ugtL gene by the Salmonella PhoP and SlyA regulatory proteins. J Biol Chem. 2004;279:38618–38625. doi: 10.1074/jbc.M406149200. [DOI] [PubMed] [Google Scholar]
- 31.Perez JC, Latifi T, Groisman EA. Overcoming H-NS-mediated transcriptional silencing of horizontally acquired genes by the PhoP and SlyA proteins in Salmonella enterica. J Biol Chem. 2008;283:10773–10783. doi: 10.1074/jbc.M709843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarre WW, Halsey TA, Walthers D, Frye J, McClelland M, Potter JL, Kenney LJ, Gunn JS, Fang FC, Libby SJ. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol Microbiol. 2005;56:492–508. doi: 10.1111/j.1365-2958.2005.04553.x. [DOI] [PubMed] [Google Scholar]
- 33.Kong W, Weatherspoon N, Shi Y. Molecular mechanism for establishment of signal-dependent regulation in the PhoP/PhoQ system. J Biol Chem. 2008;283:16612–16621. doi: 10.1074/jbc.M800547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Groisman EA. Feedback Control of Two-Component Regulatory Systems. Annu Rev Microbiol. 2016;70:103–124. doi: 10.1146/annurev-micro-102215-095331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbieri CM, Stock AM. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal Biochem. 2008;376:73–82. doi: 10.1016/j.ab.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SY, Groisman EA. Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system. Mol Microbiol. 2014;91:135–144. doi: 10.1111/mmi.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrett S, Taylor RK, Silhavy TJ. Isolation and characterization of chain-terminating nonsense mutations in a porin regulator gene, envZ. J Bacteriol. 1983;156:62–69. doi: 10.1128/jb.156.1.62-69.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polarek JW, Williams G, Epstein W. The products of the kdpDE operon are required for expression of the Kdp ATPase of Escherichia coli. J Bacteriol. 1992;174:2145–2151. doi: 10.1128/jb.174.7.2145-2151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chamnongpol S, Groisman EA. Acetyl phosphate-dependent activation of a mutant PhoP response regulator that functions independently of its cognate sensor kinase. J Mol Biol. 2000;300:291–305. doi: 10.1006/jmbi.2000.3848. [DOI] [PubMed] [Google Scholar]
- 40.Sanowar S, Martel A, Moual HL. Mutational analysis of the residue at position 48 in the Salmonella enterica Serovar Typhimurium PhoQ sensor kinase. J Bacteriol. 2003;185:1935–1941. doi: 10.1128/JB.185.6.1935-1941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salazar ME, Podgornaia AI, Laub MT. The small membrane protein MgrB regulates PhoQ bifunctionality to control PhoP target gene expression dynamics. Mol Microbiol. 2016;102:430–445. doi: 10.1111/mmi.13471. [DOI] [PubMed] [Google Scholar]
- 42.Lippa AM, Goulian M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 2009;5:e1000788. doi: 10.1371/journal.pgen.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soncini FC, Vescovi EG, Groisman EA. Transcriptional autoregulation of the Salmonella Typhimurium phoPQ operon. J Bacteriol. 1995;177:4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 46.Kuruma Y, Suzuki T, Ono S, Yoshida M, Ueda T. Functional analysis of membranous Fo-a subunit of F1Fo-ATP synthase by in vitro protein synthesis. Biochem J. 2012;442:631–638. doi: 10.1042/BJ20111284. [DOI] [PubMed] [Google Scholar]
- 47.Jones DT, Taylor WR, Thornton JM. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 48.Alpuche Aranda CM, Swanson JA, Loomis WP, Miller SI. Salmonella Typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci U S A. 1992;89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin-Orozco N, Touret N, Zaharik ML, Park E, Kopelman R, Miller S, Finlay BB, Gros P, Grinstein S. Visualization of vacuolar acidification-induced transcription of genes of pathogens inside macrophages. Mol Biol Cell. 2006;17:498–510. doi: 10.1091/mbc.E04-12-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nunez-Hernandez C, Tierrez A, Ortega AD, Pucciarelli MG, Godoy M, Eisman B, Casadesus J, Garcia-del Portillo F. Genome expression analysis of nonproliferating intracellular Salmonella enterica serovar Typhimurium unravels an acid pH-dependent PhoP-PhoQ response essential for dormancy. Infect Immun. 2013;81:154–165. doi: 10.1128/IAI.01080-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rathman M, Sjaastad MD, Falkow S. Acidification of phagosomes containing Salmonella Typhimurium in murine macrophages. Infect Immun. 1996;64:2765–2773. doi: 10.1128/iai.64.7.2765-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puiac S, Negrea A, Richter-Dahlfors A, Plant L, Rhen M. Omeprazole antagonizes virulence and inflammation in Salmonella enterica-infected RAW264.7 cells. Antimicrob Agents Chemother. 2009;53:2402–2409. doi: 10.1128/AAC.01483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arpaia N, Godec J, Lau L, Sivick KE, McLaughlin LM, Jones MB, Dracheva T, Peterson SN, Monack DM, Barton GM. TLR signaling is required for Salmonella Typhimurium virulence. Cell. 2011;144:675–688. doi: 10.1016/j.cell.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plant J, Glynn AA. Natural resistance to Salmonella infection, delayed hypersensitivity and Ir genes in different strains of mice. Nature. 1974;248:345–347. doi: 10.1038/248345a0. [DOI] [PubMed] [Google Scholar]
- 55.Lee EJ, Pontes MH, Groisman EA. A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium’s own F1Fo ATP synthase. Cell. 2013;154:146–156. doi: 10.1016/j.cell.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeom J, Wayne KJ, Groisman EA. Sequestration from Protease Adaptor Confers Differential Stability to Protease Substrate. Mol Cell. 2017;66:234–246 e235. doi: 10.1016/j.molcel.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hohmann EL, Oletta CA, Killeen KP, Miller SI. phoP/phoQ-deleted Salmonella Typhi (Ty800) is a safe and immunogenic single-dose typhoid fever vaccine in volunteers. J Infect Dis. 1996;173:1408–1414. doi: 10.1093/infdis/173.6.1408. [DOI] [PubMed] [Google Scholar]
- 58.Eguchi Y, Itou J, Yamane M, Demizu R, Yamato F, Okada A, Mori H, Kato A, Utsumi R. B1500, a small membrane protein, connects the two-component systems EvgS/EvgA and PhoQ/PhoP in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:18712–18717. doi: 10.1073/pnas.0705768104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hilbert F, Garcia-del Portillo F, Groisman EA. A periplasmic D-alanyl-D-alanine dipeptidase in the gram-negative bacterium Salmonella enterica. J Bacteriol. 1999;181:2158–2165. doi: 10.1128/jb.181.7.2158-2165.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 61.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 62.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 63.Tetsch L, Koller C, Haneburger I, Jung K. The membrane-integrated transcriptional activator CadC of Escherichia coli senses lysine indirectly via the interaction with the lysine permease LysP. Mol Microbiol. 2008;67:570–583. doi: 10.1111/j.1365-2958.2007.06070.x. [DOI] [PubMed] [Google Scholar]
- 64.Viarengo G, Sciara MI, Salazar MO, Kieffer PM, Furlan RL, Garcia Vescovi E. Unsaturated long chain free fatty acids are input signals of the Salmonella enterica PhoP/PhoQ regulatory system. J Biol Chem. 2013;288:22346–22358. doi: 10.1074/jbc.M113.472829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lesley JA, Waldburger CD. Repression of Escherichia coli PhoP-PhoQ signaling by acetate reveals a regulatory role for acetyl coenzyme A. J Bacteriol. 2003;185:2563–2570. doi: 10.1128/JB.185.8.2563-2570.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lippa AM, Goulian M. Perturbation of the oxidizing environment of the periplasm stimulates the PhoQ/PhoP system in Escherichia coli. J Bacteriol. 2012;194:1457–1463. doi: 10.1128/JB.06055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hicks KG, Delbecq SP, Sancho-Vaello E, Blanc MP, Dove KK, Prost LR, Daley ME, Zeth K, Klevit RE, Miller SI. Acidic pH and divalent cation sensing by PhoQ are dispensable for systemic salmonellae virulence. Elife. 2015;4:e06792. doi: 10.7554/eLife.06792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ronald DB, Davis W, Roth John R. A Manual for genetic engineering. Cold Spring Harbor Lab Press; Cold Spring Harbor, N.Y: 1980. Advanced Bacterial Genetics. [Google Scholar]
- 69.Snavely MD, Miller CG, Maguire ME. The mgtB Mg2+ transport locus of Salmonella Typhimurium encodes a P-type ATPase. J Biol Chem. 1991;266:815–823. [PubMed] [Google Scholar]
- 70.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 71.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karimova G, Ullmann A, Ladant D. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J Mol Microbiol Biotechnol. 2001;3:73–82. [PubMed] [Google Scholar]
- 73.Camp AH, Losick R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 2009;23:1014–1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kato A, Groisman EA. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 2004;18:2302–2313. doi: 10.1101/gad.1230804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. UgtL-dependent activation of the PhoP protein under mildly acidic pH is similar in wild-type and hns-FLAG Salmonella.
Fig. S2. UgtL enhances PhoP-P abundance in an MgrB-independent manner.
Fig. S3. UgtL enhances PhoP-P abundance posttranscriptionally.
Fig. S4. The UgtL and PhoQ proteins interact.
Fig. S5. FLAG-tagged UgtL functions as wild-type UgtL in vivo.
Fig. S6. PhoP activates transcription of the ugtL gene in mildly acidic pH and low Mg2+ conditions.
Fig. S7. UgtL promotes Salmonella resistance to magainin 2 in a PhoP-dependent manner.
Table S1. Bacterial strains and plasmids used in this study
Table S2. Primers used in this study