Abstract
To protect the biosecurity of research rodent colonies, research institutions frequently require a quarantine period for live animals transferred into their facilities. Quarantine practices often include antibiotic and antiparasitic treatment with drugs such as fenbendazole and macrolide lactones. The influence of these compounds on the resident gut microbiota of mice is unknown, and any effects might subsequently affect model reproducibility. To test the influence of standard quarantine procedures on the composition of the microbiota, C57BL/6 mice, purchased from 2 different commercial suppliers, were randomly assigned to treatment groups (n = 12) by vendor and treated with fenbendazole-supplemented feed, topical moxidectin, both treatments, or no treatment (control), according to our institution's standard treatment regimen and duration. Feces were collected on arrival, immediately after completing the 8-wk treatment, and at 2 and 4 wk after treatment. Fecal DNA was extracted, sequenced, and analyzed to compare the changes in the microbiota of treated and control groups. Although significant main effects of time and treatment and interactions between those variables were detected in comparisons of richness, α-diversity, and β-diversity, the effect sizes associated with any particular treatment were consistently much smaller than that associated with acclimation to a new facility in the absence of any quarantine treatments. This outcome, along with the visual evaluation of principal coordinate analysis based on multiple similarity indices, suggests that time or institution plays a larger role in alterations of the murine gut microbiota than do quarantine treatments on its composition.
Abbreviations: FBZ, fenbendazole; GM, gut microbiota; MOX, moxidectin; OTU, operational taxonomic unit; UC, uncultured
Despite the availability of alternative methods, such as transport of gametes or embryos, shipping live, unique, transgenic, potentially infected mice between institutions is commonplace and presents a potential biosecurity risk to receiving institutions.18,29,31 The biosecurity risk is frequently mitigated by using rederivation by embryo transfer, cross-fostering, or quarantine procedures.3,4,34,42 The transport of mice between institutions has previously been associated with changes in research models or, in some cases, complete loss of the model phenotype, both leading to concerns regarding research reproducibility.45 Although both interfacility movement and rederivation have been shown to alter the gut microbiota (GM), little has been done to evaluate the effect of quarantine procedures on the GM.13,15,26
Quarantine procedures are imperative to the protection of health status of rodent research colonies and subsequent research reproducibility.36 Procedures vary across institutions but frequently include physical isolation of the quarantined animals, treatment with fenbendazole-impregnated feed to prevent or treat pinworm infestation, and administration of a topical or oral treatment for fur mites.32,34 Both fenbendazole and the macrolide lactones (avermectin, ivermectin, moxidectin) have been shown to affect research outcomes through their pharmacologic mechanism of action, but no data are available regarding the potentially subtler effects on research models due to possible alterations in the GM.11,40,44 Quarantine at our facility (the University of Missouri) is typically at least 8 wk long, using fenbendazole-impregnated feed throughout the quarantine period and 2 treatments of topical moxidectin.
Given the continued need to use quarantine procedures to protect the biosecurity of our rodent colonies, balanced with the concern surrounding model continuity and reproducibility in the context of quarantine-induced changes in the GM, we evaluated the effects of common quarantine treatments on the GM of 6-wk-old C57BL mice from 2 different, popular sources. Previous work from our lab has demonstrated that mice of the same strain—but procured from different vendors—have significantly different gut microbiota.8 We chose these 2 particular vendors because their mice represent opposite ends of the spectrum of GM with regard to microbial richness and diversity. Regardless of genetic background, mice from vendor A consistently harbor fecal microbiota of significantly greater richness and diversity than that of the comparable substrain from vendor B.8 C57BL/6 substrains were chosen for this study because they are common background strains for genetically modified mice. To this end, we treated separate cohorts of mice with fenbendazole-impregnated feed, topical moxidectin, or a combination of fenbendazole-impregnated feed and topical moxidectin. Fecal samples were collected on arrival, immediately after completion of treatment, and at 2 and 4 wk after cessation of treatment. Once we better understand the effects of routine quarantine procedures on murine GM, we can take steps to mitigate any potential alterations, with the ultimate goal of enhanced research reproducibility and appropriate biosecurity.
Materials and Methods
Animals and husbandry.
Female C57BL/6NHsd mice (n = 48; age, 6 wk) were obtained from vendor A (Envigo, Indianapolis, IN) and female C57BL/6J mice (n = 48; age, 6 wk) from vendor B (The Jackson Laboratory, Bar Harbor, ME). All mice were housed at 4 per cage in IVC (Thoren, Hazelton, PA) with compressed paper bedding (Paperchips, Shepherd Specialty Papers, Watertown, TN); acidified, autoclaved water; and a 14:10-h light:dark cycle. Cages were assembled with bedding, wire bar lids, and filter tops and autoclaved as a unit prior to use. All animal and cage manipulations were performed in a class II A2 biosafety cabinet (LabGard ES NU-540, Nuaire, Plymouth, MN) disinfected with 10% bleach solution prior to use. All mice were housed in an AAALAC-accredited facility, and all animal use was performed according to the standards put forth in the Guide for the Care and Use of Laboratory Animals (8th ed.)17 and approved by the University of Missouri ACUC.
Quarantine treatment.
To best replicate quarantine conditions, mice were randomly placed into treatment groups according to vendor immediately on arrival. Animals were divided into 4 groups (n = 12 mice per group). The FBZ group received irradiated fenbendazole-impregnated feed (150 ppm fenbendazole, Lab Diet, St Louis, MO) for 8 consecutive weeks. The MOX group received 3 µL topical moxidectin (Cydectin 5% moxidectin topical, Boehringer Ingelheim Vetmedica, St Joseph, MO) between the shoulder blades on arrival and at 2 wk after arrival and was fed 5LOD chow (the base diet for fenbendazole-impregnated feed; Lab Diet). The FBZ+MOX group received both fenbendazole-impregnated feed and moxidectin as described earlier. The control group did not receive any treatment and was on irradiated 5LOD chow for the duration of the study. At 8 wk, all mice were switched to 5LOD for the remainder of the study.
Sample collection.
Feces were collected from each mouse on arrival (baseline), immediately after 8 wk of treatment (week 0), and at 2 and 4 wk afterward cessation of treatment (weeks 2 and 4, respectively). To collect feces, mice were placed individually in sterile, empty cages and allowed to defecate voluntarily. After defecation, mice were returned to their home cage, and 1 or 2 fecal pellets were removed from the collection cage floor by using an autoclaved toothpick. Samples were placed in a sterile 2.0-mL round-bottom tubes with stainless steel beads (diameter, 0.5 cm) and stored at –80 °C until further analysis. After collection, gross debris was removed from the fecal collection cages, and the cages were cleaned with 10% bleach prior to use with the next animal. All feces were collected between 1500 and 1800.
Fecal DNA extraction.
All fecal DNA extraction was performed as previously described.8 Lysis buffer was added to the sample, and the sample was homogenized for 3 min by using a TissueLyser II (Qiagen, Venlo, Netherlands). After homogenization, samples were incubated at 70 °C for 20 min, with periodic vortexing. The samples were centrifuged at 5000 × g for 5 min at room temperature, and the supernatant was transferred into a sterile 1.5-mL microfuge tube. Ammonium acetate (10 mM, 200 μL) was added to the supernatant, vortexed, and incubated on ice for 5 min. Samples were centrifuged as described earlier. After centrifugation, samples were combined with an equal volume of chilled isopropanol and incubated on ice for 30 min. The samples were then centrifuged at 16,000 × g for 15 min at 4 °C. The supernatant was collected and discarded. The remaining DNA pellet was washed multiple times with 70% ethanol and resuspended in 150 µL of Tris–EDTA. DNA was isolated from the sample by using a DNeasy kit (Qiagen) according to the manufacturer's recommendations. DNA was quantified by fluorometry (Qubit 2.0, Life Technologies, Carlsbad CA) by using quant-iT BR dsDNA reagent kits (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
16S rRNA library preparation and sequencing.
All processing of extracted DNA was performed at the University of Missouri DNA Core as previously described.8 A bacterial 16S rDNA amplicon library was generated through amplification of the V4 hypervariable region of the 16s rDNA gene with universal primers (U515F/806R), flanked by Illumina standard adapter sequences.5,41 PCR amplification of the variable regions used the following parameters: 98 °C for 3 min; followed by 25 cycles of 98° for 15 s, 50 °C for 30 s, and 72 °C for 30 s; with a final incubation at 72 °C for 7 min. Amplified products were pooled and mixed. The pooled samples were purified by using Axygen AxyPrep MagPCR Clean-up beads (Corning Life Sciences, Tewksbury, MA) and incubated at room temperature for 15 min. The products were washed multiple times with 80% ethanol and resuspended in EB buffer (Qiagen) at room temperature for 2 min. The solution was then placed on a magnetic stand for 5 min. The amplicon pool was evaluated by using the Fragment Analyzer automated electrophoresis system (Advanced Analytical, Ankeny, IA), quantified with a Qubit fluorometer using the quant-iT HS dsDNA reagent kit (Invitrogen), diluted according to Illumina's standard protocol, and sequenced on an automated sequencer (MiSeq, Illumina, San Diego, CA).
Informatics.
The University of Missouri Informatics Research Core Facility performed all trimming, assembly, binning, and annotation of all sequences as previously described.8 Contiguous sequences were assembled by using FLASH software,27 with culling of sequences with a base quality of less than 31. Qiime v1.921 was used to identify and remove de novo and reference-based chimeras. All remaining contigs were placed in operational taxonomic units (OTU) according to a criterion of 97% identity. OTU were assigned to taxonomic groups by using BLAST2 against the SILVA database of 16s rRNA gene sequences.35 Principal coordinate analyses were performed by using 1/4 root-transformed OTU relative abundance data, and indices of richness and α-diversity were determined by using Past 3.1614 software downloaded on 10 August 2017.
Statistics.
Only samples returning greater than 10,000 high-quality reads were included in the analysis. Two-way repeated-measures ANOVA with posthoc comparisons with control treatment group or pretreatment time point according to the Holm–Sidak method was used to test for main effects of treatment and time (and interactions) on the mean number of OTU (richness) and mean Simpson index (α-diversity) within each vendor by using SigmaPlot 12.3 (Systat Software, San Jose, CA). Past 3.16 software was used to evaluate differences in β-diversity between groups by using 2-way permutational ANOVA of Jaccard and ranked Bray–Curtis distances. After principal coordinate analysis, one-way ANOVA was used to compare the intrasubject Bray–Curtis similarity indices between the baseline and week 0 time points for all treatment groups. The data failed a test for normality; therefore ANOVA on ranks was used to compare the intrasubject Jaccard similarity indices between the baseline and week 0 time points for all treatment groups. Differences with a P value less than 0.05 were considered significant.
Results
Of the 384 samples (96 mice at 4 time points) processed and sequenced, only 2 failed to achieve a threshold of 10,000 reads. Overall, a total of 27,391,847 reads were included in the final dataset, resulting in a coverage (mean ± 1 SD) for the remaining 382 samples of 71706 ± 11890 reads.
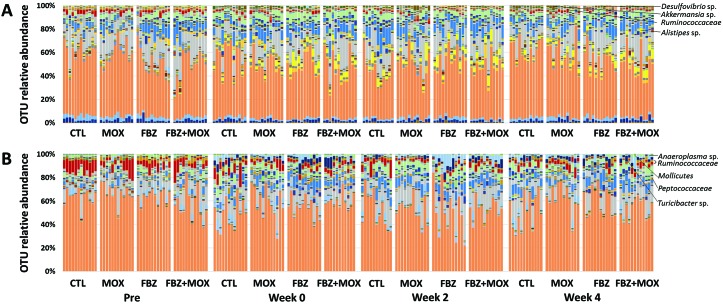
To facilitate visualization of the differences in the GM before and after treatment, we first generated stacked bar charts showing the relative abundance of bacteria (and archaea) at the OTU level for both vendors’ groups of mice (Figure 1). At the level of the OTU (that is, groups of sequences sharing a minimum of 97% nucleotide identity), there is an apparent decrease in the relative abundance of several specific taxa, such as OTU197 (uncultured [UC] member within the family Ruminococcaceae), and a subjective increase in relative abundance of other taxa, such as OTU32 (Alistipes sp.), OTU245 (Desulfovibrio sp.), and OTU267 (Akkermansia sp.), in mice across all treatment groups. Other than a subtle increase in the relative abundance of microbes in the family Tenericutes, minimal change was present in the taxonomic composition of samples from vendor B mice at the phylum level. At the level of the OTU, there were however apparent differences between the pretreatment samples and samples from all of the posttreatment time-points for all treatment groups, including controls, suggesting that these changes over time are due to unidentified institutional factors or simply effects of aging. Some of the most striking changes noted include decreases in the relative abundance of OTU197 and OTU201 (UC family Ruminococcaceae) and OTU263 (UC order Mollicutes, group RF9) and concurrent increases in the relative abundance of OTU147 (UC family Peptococcaceae), OTU179 (Ruminoclostridium sp.), OTU214 (Turicibacter sp.), and OTU258 (Anaeroplasma sp.).
Figure 1.
Stacked bar charts showing the fecal microbiota of C57BL/6 mice from (A) vendor A and (B) vendor B prior to treatment (Pre) with moxidectin (MOX), fenbendazole (FBZ), both compounds (FBZ+MOX), or neither compound (CTL); immediately after 8 wk of treatment (week 0); and 2 wk (week 2) and 4 wk (week 4) after completion of treatment, annotated to the level of operational taxonomic unit (OTU).
Influence of quarantine procedures on richness and α-diversity of GM.
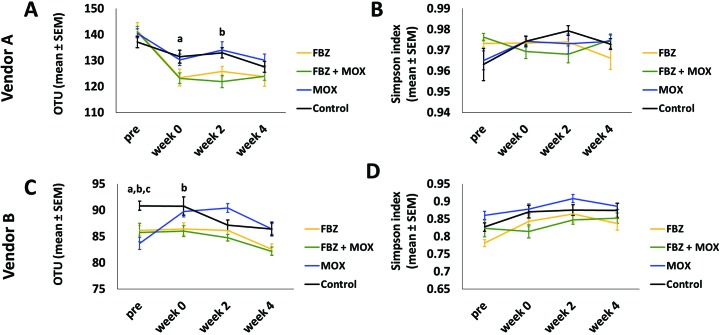
To test for treatment-dependent changes in the fecal microbiota, we first determined the overall richness and α-diversity of each sample and, with stratification according to mouse supplier, tested for main effects and compared treated and control groups at each time point. Richness was measured as the number of OTU detected, whereas α-diversity (a metric comprising both richness and evenness of distribution) was assessed by using the Simpson index.
Considering first the C57BL/6 mice from vendor A, which typically harbor a richer GM relative to those from vendor B, significant time- and treatment-dependent (time: P < 0.001, F = 43.13); treatment: P < 0.047, F = 2.87) differences and a significant time×treatment interaction (P = 0.004, F = 2.87) were present (Figure 2 A). Note, however, that the F values ascribed to each main effect suggest a much larger effect size due to time, and main effects of treatment will also reflect minor preexisting differences between treatment groups and individual mice due to random chance. Post hoc tests revealed significant decreases in richness between the pretreatment sample and the week 4 sample in all 4 groups (FBZ and FBZ+MOX, P < 0.001; MOX, P = 0.001; control, P = 0.003). No significant differences in microbial richness were detected between treatment groups compared with control at the baseline or week 4 time points; significant, albeit subtle, differences were detected between the FBZ and control groups at week 0 (P = 0.041) and between the FBZ+MOX and control groups at week 2 (P = 0.010).
Figure 2.
Number of OTU (mean ± SEM; indicative of diversity of microbiota) across all time points for each treatment group from (A) vendor A and (B) vendor B. Simpson index (mean ± SEM); α-diversity) across all time points for each treatment group from (C) vendor A and (D) vendor B. Significant (P < 0.05) differences of FBZ (a), FBZ+MOX (b), and MOX (c) groups from the control group are noted.
Conversely, no significant effects of time (P = 0.316, F = 1.19) or treatment (P = 0.994, F = 0.026) on α-diversity were detected in samples from mice from vendor A (Figure 2 B).
With regard to the C57BL/6 mice from vendor B, 2-way ANOVA detected significant effects of time (P < 0.001, F = 10.19) and treatment (P < 0.001, F = 7.18) and their interaction (P = 0.010, F = 2.55) on richness (Figure 2 C). That said, the significant main effect of treatment again was based on all samples (including baseline) and thus, at least partially, reflects inherent differences between groups. Supporting this interpretation, post hoc tests detected significant differences in the number of OTU between controls and all other groups at the baseline time-point (P < 0.05) and between the control and FBZ+MOX groups at week 0 (P = 0.033), with no other group-dependent differences at week 2 or 4. Within the MOX group, richness was significantly greater at weeks 0 and 2 relative to baseline (P < 0.001), whereas there were no differences between week 4 and any of the preceding time points. In the control group, richness was significantly decreased at week 4 relative to pretreatment (P = 0.011).
Similarly, significant effects of both time (P < 0.001, F = 7.68) and treatment (P < 0.001, F = 8.04), with no significant interaction (P = 0.665, F = 0.75), were detected in α-diversity in samples from the mice from vendor B (Figure 2 D). Post hoc testing indicated that, within the FBZ group, values at all posttreatment time points were significantly greater than the pretreatment values (P < 0.05) and, within the control group, the week 0 and 4 time points were increased relative to pretreatment (P = 0.046 and 0.045, respectively). These findings, in combination with the relatively modest F score associated with the main effect of treatment, suggest that the effect of time was much greater than any specific quarantine treatment under study. Collectively, we interpreted the analyses of richness and α-diversity as evidence that any effect of FBZ, MOX, or FBZ and MOX in combination on the GM are far outweighed by the changes that occur in untreated mice arriving at, acclimating to, and aging in a new facility.
Influence of quarantine procedures on GM composition.
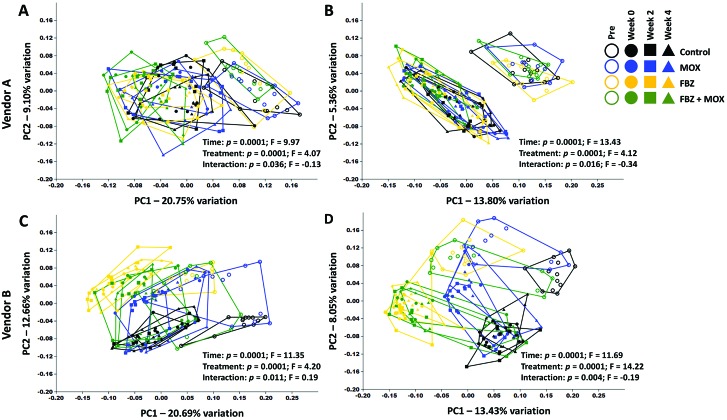
To determine whether significant treatment-dependent effects on the composition of the GM were present, data were visualized by using principal coordinate analysis, based on both weighted (Bray–Curtis) and unweighted (Jaccard) similarity indices and tested by using 2-way permutational multivariate ANOVA. Weighted indices determine the similarity between sample pairings based on their agreement with regard to the relative abundance of shared taxa, whereas unweighted indices are based on agreement between samples regarding the presence or absence of taxa.
On visual inspection, samples from vendor A mice demonstrate similar patterns of clustering regardless of the similarity index used. Specifically, samples collected prior to any treatment clustered together, partially (Figure 3 A) or completely (Figure 3 B) separate from samples collected at all 3 posttreatment time points, depending on the similarity index used. Samples collected at week 0 and beyond demonstrated a comparable directional shift along PC1 in all groups, indicating that the compositional changes observed in Figure 1 were similar in nature and not dependent on treatment. Whereas permutational ANOVA detected significant main effects of both time and treatment, the F values associated with those tests suggests that the observed changes predominantly reflect differences across time in all groups (Table 1). Similarly, samples collected from vendor B mice show a relatively uniform shift from baseline to week 0, regardless of treatment group or similarity index used (Figure 3 C and D). Although preexisting differences between treatment groups are more apparent in the vendor B C57BL/6 mice (due to random chance), all groups again evinced similar directional shifts at week 0, followed by minimal change between weeks 0, 2, and 4. Testing by using permutational ANOVA based on the Bray–Curtis distances indicated a much greater effect size of time than treatment, which, in conjunction with the complete separation of treatment groups at the pre time-point, suggests that any compositional changes in the GM induced by FBZ or MOX were negligible (Table 1).
Figure 3.
Principal coordinate analysis plots showing β-diversity in C57BL/6 mice purchased from (A, B) vendor A or (C,D) vendor B, by using (A, C) Bray–Curtis distances and (B, D) Jaccard distances. Main effects of time and treatment (and interactions between those variables) were determined by using 2-way permutational ANOVA are shown.
Table 1.
P and F values from tests of Bray–Curtis and Jaccard similarity indices evaluating the significance of the effects of time and treatment on the gut microbiota of mice according to vendor
| Bray–Curtis |
Jaccard |
||||
| P | F | P | F | ||
| Vendor A | Time | 0.0001 | 9.97 | 0.0001 | 11.35 |
| Treatment | 0.0001 | 4.07 | 0.0001 | 4.20 | |
| Interaction | 0.036 | −0.13 | 0.011 | −0.34 | |
| Vendor B | Time | 0.0001 | 9.97 | 0.0001 | 11.69 |
| Treatment | 0.0001 | 4.07 | 0.0001 | 14.22 | |
| Interaction | 0.036 | 0.19 | 0.004 | −0.19 | |
P values were determined from one-way ANOVA (Bray–Curtis) and one-way permutational ANOVA (Jaccard) to ascertain differences in the effects of treatment, time, and their interaction. Differences with P < 0.05 are considered significant.
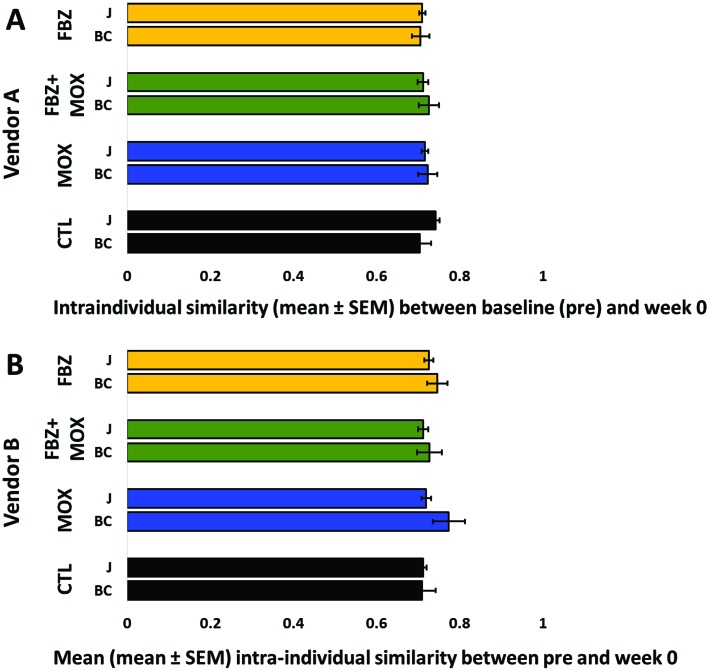
To incorporate individual mouse variation in the comparisons, we determined the intraindividual similarity between the pre and week 0 samples from each mouse, again using both weighted and unweighted indices. Regardless of the similarity metric, no significant treatment-dependent differences were detected in the intraindividual sample similarity between the baseline time point and the end of treatment 8 wk later in mice from vendor A (Figure 4 A) or vendor B (Figure 4 B; vendor A: Bray–Curtis P = 0.799, Jaccard P = 0.095; vendor B: Bray–Curtis P = 0.453, Jaccard P = 0.778).
Figure 4.
Intraindividual Jaccard (J) and Bray–Curtis (BC) similarity indices from pretreatment to immediately after 8 wk of treatment with fenbendazole (FBZ), fenbendazole and moxidectin (FBZ+MOX), moxidectin (MOX), or neither compound (CTL) in C57BL/6 mice from (A) vendor A or (B) vendor B. Higher values indicate greater similarity between baseline (pre) and week 0 time point. No significant differences between treatment groups in the gut microbiota of mice from either vendor A or B according to either similarity index (Jaccard or Bray–Curtis) were detected.
Collectively, the data we have presented complement earlier reports of changes in the GM after transport to a new facility or institution and suggest that such changes in institution or mere aging have a greater influence on the GM of laboratory mice than do the routine quarantine treatments we evaluated in the current study.8
Discussion
The GM of research mice has been shown to undergo compositional changes after shipping. Individually, the treatment of mice with moxidectin and fenbendazole have been demonstrated to alter the behavior of multiple rodent models of disease, primarily through direct action on specific receptors. The effects of moxidectin on internal and external parasites through its mechanism of action on GABA- and glutamate-mediated ion channels, causing a flaccid paralysis, is well established.7,28 When delivered topically, moxidectin distributes to most tissues, including the gastrointestinal tract.23,38 Moxidectin is effective against some bacteria either individually or synergistically.25,43 Because this agent is not typically considered antibacterial, we anticipated that the effects of moxidectin on the GM would be minimal. Fenbendazole is considered a parasiticide, and it inhibits the binding of tubulin subunits thus disrupting organelle movement, cell division, and motility and decreases glucose uptake.28 Although not typically considered antibacterial, fenbendazole has been shown to influence bacterial antimicrobial resistance.39 The effects of fenbendazole on the GM are most like to be secondary and become apparent if parasites, such as pinworms, are removed.10,19,24 Although the current study was not designed to evaluate the individual effects of time, transport, or change in institution on the murine GM, any change associated with the quarantine procedures evaluated was minor when compared with those effects. This outcome is demonstrated by the changes in the variation of the CTL group mirroring the changes of all 3 treatment groups across all time points. This effect is especially evident when evaluating the relative abundance of the OTU in Figures 1 and 2. All baseline groups were visually very similar by vendor, as expected due to the random assignment of animals to treatment groups. Changes in OTU relative abundance in the control group are visually similar to the changes noted in all 3 groups at the 0-, 2-, and 4-wk posttreatment time points, suggesting that time, shipping, or change in institution plays a much more important role in the variation in richness and α-diversity than the evaluated quarantine treatments.
Similar results are apparent when comparing richness and by using the average number of OTUs and α-diversity according to the Jaccard similarity indices. Some variation was present between the controls and treatment groups for both measures on arrival (vendor B, Figure 2 C) and throughout treatment (Figure 2 A–D). However, at the 4-wk time point, none of the treatment groups are statistically significantly different from the controls. This finding is in line with results from other groups showing that the GM stabilizes over time after a change in institution, further supporting time, shipping, and institutional changes as larger sources of variation than the evaluated treatment effects.26
Alterations in GM can affect research models.30 The proliferation of the genus Alistipes has been noted as mice age and has been associated with increased frailty indices, depression, and leaness in rodent models.19,22,37 Whereas Akkermansia demonstrates the opposite trend in relation to age, an increase is associated with colitis and exacerbated inflammatory responses in gnotobiotic mice and is seen for several generations following facility change.6,9,12,22 An increase in Turicibacter has also been noted following facility change and is seen in sedentary mice.1,6 An increase in Tenericutes has been noted when mice are placed on high-fat diets or there is a change in diet.16,33 Although many of these changes can be associated with the change in facility or diet, their effects on individual models may need to be discussed with investigators prior to the movement and quarantine of animals.
Significant main effects of treatment and time and significant interactions were noted at all time points (Figure 2 and Table 1). Nevertheless, the relative P values and associated F or q values for the effect of time were larger than those associated with effects of the treatment at most time points, further supporting that time or institution change account for more of the variation noted than the treatment. This conclusion is supported by lack of significant change noted between the baseline time point and immediately after treatment at week 0 for all treatment groups including controls (Figure 4). Intuitively any changes in the GM due to treatment would be most visible at week 0, with institutional stabilization occurring at the following time points.
A wide variety of factors, such as transport and change in institution, affect the GM. The movement of animals between facilities is vital to collaboration and the forward movement of animal-based research. The potential changes in the BM can have a marked effect, including loss, on model phenotype. GM changes due to animal movement between facilities could be exacerbated by the treatments institutions require prior to including the animals in their vivaria, further hampering efforts to increase reproducibility in science. According to the data provided here, variation was observed in all factors examined. Although variation was seen among treatments of a standard quarantine regimen, the degree was much less than that seen with time or institutional change or their combination. Moreover, principal coordinate analysis revealed consistent overlap of treatment groups, suggesting that any treatment-associated variation is unlikely to affect studies where influences of differing GM on model phenotype or study reproducibility are of concern.
Acknowledgments
We thank the University of Missouri's Mutant Mouse Resource and Research Center (NIH U42OD010918) and Office of Animal Resources for funding for this project.
References
- 1.Allen JM, Berg Miller ME, Pence BD, Whitlock K, Nehra V, Gaskins HR, White BA, Fryer JD, Woods JA. 2015. Voluntary and forced exercise differentially alters the gut microbiome in C57BL/6J mice. J Appl Physiol (1985) 118:1059–1066. [DOI] [PubMed] [Google Scholar]
- 2.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Artwohl JE, Purcell JE, Fortman JD. 2008. The use of cross-foster rederivation to eliminate murine norovirus, Helicobacter spp., and murine hepatitis virus from a mouse colony. J Am Assoc Lab Anim Sci 47:19–24. [PMC free article] [PubMed] [Google Scholar]
- 4.Besselsen DG, Romero-Aleshire MJ, Munger SJ, Marcus EC, Henderson KS, Wagner AM. 2008. Embryo transfer rederivation of C.B-17/Icr-Prkdc(scid) mice experimentally infected with mouse parvovirus 1. Comp Med 58:353–359. [PMC free article] [PubMed] [Google Scholar]
- 5.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108 Suppl 1:4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choo JM, Trim PJ, Leong LEX, Abell GCJ, Brune C, Jeffries N, Wesselingh S, Dear TN, Snel MF, Rogers GB. 2017. Inbred mouse populations exhibit intergenerational changes in intestinal microbiota composition and function following introduction to a facility. Front Microbiol 8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobb R, Boeckh A. 2009. Moxidectin: a review of chemistry, pharmacokinetics and use in horses. Parasit Vectors 2 Suppl 2:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ericsson AC, Davis JW, Spollen W, Bivens N, Givan S, Hagan CE, McIntosh M, Franklin CL. 2015. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One 10:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA 110:9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ezenwa VO, Jolles AE. 2015. Opposite effects of anthelmintic treatment on microbial infection at individual versus population scales. Science 347:175–177. [DOI] [PubMed] [Google Scholar]
- 11.Gadad BS, Daher JP, Hutchinson EK, Brayton CF, Dawson TM, Pletnikov MV, Watson J. 2010. Effect of fenbendazole on 3 behavioral tests in male C57BL/6N mice. J Am Assoc Lab Anim Sci 49:821–825. [PMC free article] [PubMed] [Google Scholar]
- 12.Ganesh BP, Klopfleisch R, Loh G, Blaut M. 2013. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS One 8:e74963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. 2007. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131:33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammer O. [Internet]. 2017. Past 3.x—the Past of the future. [Cited 10 May 2016]. Available at: http://folk.uio.no/ohammer/past/.
- 15.Hart ML, Ericsson AC, Franklin CL. 2017. Differing complex microbiota alter disease severity of the IL10–/– mouse model of inflammatory bowel disease. Front Microbiol 8:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang EY, Leone VA, Devkota S, Wang Y, Brady MJ, Chang EB. 2013. Composition of dietary fat source shapes gut microbiota architecture and alters host inflammatory mediators in mouse adipose tissue. J Parenter Enteral Nutr 37:746–754. Erratum: J Parenter Enteral Nutr 2014. 38: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Institute for Laboratory Animal Research 2011. Guide for the care and use of laboratory animals, 8th ed. Washington (DC): National Academies Press. [Google Scholar]
- 18.Jacoby RO, Lindsey JR. 1998. Risks of infection among laboratory rats and mice at major biomedical research institutions. ILAR J 39:266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, Wang W, Tang W, Tan Z, Shi J, Li L, Ruan B. 2015. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun 48:186–194. [DOI] [PubMed] [Google Scholar]
- 20.Kreisinger J, Bastien G, Hauffe HC, Marchesi J, Perkins SE. 2015. Interactions between multiple helminths and the gut microbiota in wild rodents. Philos Trans R Soc Lond B Biol Sci 370:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, Knight R. 2011. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics 36:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langille MG, Meehan CJ, Koenig JE, Dhanani AS, Rose RA, Howlett SE, Beiko RG. 2014. Microbial shifts in the aging mouse gut. Microbiome 2:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leathwick DM, Miller CM. 2013. Efficacy of oral, injectable, and pour-on formulations of moxidectin against gastrointestinal nematodes in cattle in New Zealand. Vet Parasitol 191:293–300. [DOI] [PubMed] [Google Scholar]
- 24.Lee SC, Tang MS, Lim YA, Choy SH, Kurtz ZD, Cox LM, Gundra UM, Cho I, Bonneau R, Blaser MJ, Chua KH, Loke P. 2014. Helminth colonization is associated with increased diversity of the gut microbiota. PLoS Negl Trop Dis 8:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim LE, Vilcheze C, Ng C, Jacobs WR, Jr, Ramon-Garcia S, Thompson CJ. 2013. Anthelmintic avermectins kill Mycobacterium tuberculosis, including multidrug-resistant clinical strains. Antimicrob Agents Chemother 57:1040–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma BW, Bokulich NA, Castillo PA, Kananurak A, Underwood MA, Mills DA, Bevins CL. 2012. Routine habitat change: a source of unrecognized transient alteration of intestinal microbiota in laboratory mice. PLoS One 7:1–11. Erratum: PLoS One 2013. 8: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin RJ. 1997. Modes of action of anthelmintic drugs. Vet J 154:11–34. [DOI] [PubMed] [Google Scholar]
- 29.Marx JO, Gaertner DJ, Smith AL. 2017. Results of survey regarding prevalence of adventitial infections in mice and rats at biomedical research facilities. J Am Assoc Lab Anim Sci 56:527–533. [PMC free article] [PubMed] [Google Scholar]
- 30.Miller PG, Bonn MB, Franklin CL, Ericsson AC, McKarns SC. 2015. aTNFR2 deficiency acts in concert with gut microbiota to precipitate spontaneous sex-biased central nervous system demyelinating autoimmune disease. J Immunol 195:4668–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mook D, Taylor DK, Huerkamp MJ. 2009. The rodent quarantine quagmire. J Am Assoc Lab Anim Sci 48:472–474. [PMC free article] [PubMed] [Google Scholar]
- 32.Mook DM, Benjamin KA. 2008. Use of selamectin and moxidectin in the treatment of mouse fur mites. J Am Assoc Lab Anim Sci 47:20–24. [PMC free article] [PubMed] [Google Scholar]
- 33.Nagy-Szakal D, Mir SA, Ross MC, Tatevian N, Petrosino JF, Kellermayer R. 2013. Monotonous diets protect against acute colitis in mice: epidemiologic and therapeutic implications. J Pediatr Gastroenterol Nutr 56:544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otto G, Tolwani RJ. 2002. Use of microisolator caging in a risk-based mouse import and quarantine program: a retrospective study. Contemp Top Lab Anim Sci 41:20–27. [PubMed] [Google Scholar]
- 35.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41 D1:D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rehg JE, Toth LA. 1998. Rodent quarantine programs: purpose, principles, and practice. Lab Anim Sci 48:438–447. [PubMed] [Google Scholar]
- 37.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sallovitz JM, Lifschitz A, Imperiale F, Virkel G, Lanusse C. 2003. A detailed assessment of the pattern of moxidectin tissue distribution after pour-on treatment in calves. J Vet Pharmacol Ther 26:397–404. [DOI] [PubMed] [Google Scholar]
- 39.Tysnes KR, Luyckx K, Cantas L, Robertson LJ. 2015. Treatment of feline giardiasis during an outbreak of diarrhoea in a cattery: potential effects on faecal Escherichia coli resistance patterns. J Feline Med Surg 18:679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villar D, Cray C, Zaias J, Altman NH. 2007. Biologic effects of fenbendazole in rats and mice: a review. J Am Assoc Lab Anim Sci 46:8–15. [PubMed] [Google Scholar]
- 41.Walters WA, Caporaso JG, Lauber CL, Berg-Lyons D, Fierer N, Knight R. 2011. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics 27:1159–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson J, Thompson KN, Feldman SH. 2005. Successful rederivation of contaminated immunocompetent mice using neonatal transfer with iodine immersion. Comp Med 55:465–469. [PubMed] [Google Scholar]
- 43.Woerde DJ, Martin PA, Govendir M. 2015. Susceptibility of rapidly growing mycobacteria isolated from Australian cats to ivermectin, moxidectin, ceftiofur, and florfenicol. J Feline Med Surg 17:1065–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan S, Ci X, Chen N, Chen C, Li X, Chu X, Li J, Deng X. 2011. Antiinflammatory effects of ivermectin in mouse model of allergic asthma. Inflamm Res 60:589–596. [DOI] [PubMed] [Google Scholar]
- 45.Yang I, Eibach D, Kops F, Brenneke B, Woltemate S, Schulze J, Bleich A, Gruber AD, Muthupalani S, Fox JG, Josenhans C, Suerbaum S. 2013. Intestinal microbiota composition of interleukin-10-deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS One 8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]