Abstract
Neural tube defects (NTDs) are considered to be a complex genetic disorder, although the identity of the genetic factors remains largely unknown. Mouse model studies suggest a multifactorial oligogenic pattern of inheritance for NTDs, yet evidence from published human studies is surprisingly absent. In the present study, targeted next-generation sequencing was performed to screen for DNA variants in the entire coding regions and intron-exon boundaries of targeted genes using DNA samples from 510 NTD cases. These candidate genes were PCP genes, including VANGL1, VANGL2, CELSR1, SCRIB, DVL2, DVL3 and PTK7. Candidate variants were validated using Sanger sequencing. A total of 397 single nucleotide variants(SNVs) were identified with a mean depth of approximately 570×. Of these identified SNVs, 74 were predicted to affect protein function and had a minor allele frequency of < 0.01 or unknown. Among these 74 missense SNVs, 10 were identified from six NTD cases that carried two mutated genes. Of the six NTD cases, three spina bifida cases and one anencephaly case carried digenic variants in the CELSR1 and SCRIB gene; one anencephaly case carried variants in the CELSR1 and DVL3 gene; and one spina bifida case carried variants in the PTK7 and SCRIB genes. Three cases that parental samples were available were confirmed to be compound heterozygous. None of the digenic variants were found in the 1000 genome database. The findings imply that genetic variation might interact in a digenic fashion to generate the visible NTD phenotypes and emphasize the importance of these genetic interactions in the development of NTDs in humans.
Keywords: Digenic variants, Neural tube defects, PCP pathway
1. Introduction
Neural tube defects (NTDs), such as anencephaly and spina bifida, are congenital malformations of the central nervous system caused by a partial or complete failure of the neural tube to close during embryogenesis [1]. Although the etiology of human NTDs has been intensively studied for over 40 years [1–5], the causative genetic mechanism of NTD development is largely unknown. It is estimated that up to 70% of the risk of NTDs is attributed to genetic factors [6]; yet the main predisposing genetic factors for human NTDs remain unknown. Given that NTDs present a mainly sporadic pattern and exhibit a relatively high prevalence across the world, it is thought that the etiology of these disorders represents a multifactorial oligogenic or polygenic pattern of inheritance, together with an important role for non-genetic factors such as the environment [2].
It is now well established that over 300 genes were causally linked to the expression of a NTD phenotype in mice [7–10], indicating the complex genetic requirements for neurulation during the period of neural tube closure. Interestingly, a disproportionately large number of these genes are localized in the planar cell polarity (PCP) pathway. The PCP pathway, also called the non-canonical Frizzled/Dishevelled pathway, controls the process by which cells become polarized within the plane of an epithelium in numerous tissues in both Drosophila and vertebrates [11,12]. Genetic studies in Drosophila have initially identified a group of “core” PCP genes including: frizzled (fz), dishevelled (dvl), vang gogh/strabismus (vang/stbm), flamingo (fmi), prickle (pk), and diego (dg) [12]. These genes encode proteins that are highly conserved in vertebrates, where they mediate a complex morphogenetic process called convergent extension during gastrulation and neurulation. Convergent extension is a polarized cellular rearrangement that leads to the narrowing of the mediolateral axis and lengthening of the anteroposterior axis for gastrulation and neural tube formation. Mouse model studies show that mutants that disrupt core components of the PCP pathway, including Vangl1, Vangl2, Celsr1, Fzd3, Fzd6, Dvl1, Dvl2 and Dvl3, can cause NTDs [7,13–16].
As suggested from the mouse NTD literature, PCP genes were subsequently determined to have potentially a causal role in humans with NTDs [7]. Single-nucleotide variants (SNVs) in the core PCP genes CELSR1, FZD6, PRICKLE1, DVL2, VANGL1 and VANGL2, and the PCP associated genes SEC24B, DACT1, FUZ and SCRIB [7,17–23] have been proposed as human NTD risk factors. In contrast to the association of mouse PCP gene mutations with craniorachischisis in homozygotes, most of the variants identified in human NTDs are heterozygous variants, and the PCP-mutation-associated NTD phenotypes vary widely ranging from the “open NTD” of craniorachischisis, myelomeningocele, and anencephaly, to the “closed NTD” of lipomyelomeningocele, lipomyelocele, and lipoma [2,7]. Unlike mouse models, variants in single genes may not explain the genetic mechanism underlying the development of human NTDs.
Recently, a digenic or polygenic pattern of inheritance was suggested to contribute to the cause of NTDs. Actually, mouse studies showed that several digenic combinations involving the core PCP gene Vangl2 and other genes (Sec24b, Sfrp1/Sfrp2/Sfrp5, Dvl3, Scrib, Celsr1, Ptk7, Vangl1), most of which were double heterozygotes, could cause either open spina bifida or exencephaly or craniorachischisis, in contrast to only the craniorachischisis phenotype when homozygous mutants were detected in some of these PCP genes [2,24–28]. Additionally, double mutants of ScribCrc/+and Celsr1Crsh/+, Dvl2−/− and Dvl3+/−, Dvl2+/− and Dvl3−/− induced a phenotype of craniorachischisis in mice [2,26]. Although these findings raise the possibility of similar genetic combinatorial mechanisms in human NTDs, little evidence was available in support of this hypothesis in human studies except a small chohort study including 90 patients with cranial NTDs in England [29]. Therefore, the present study aimed to find double or multiple heterozygous variant combinations of critical PCP genes in a large number of human NTD cases, which may provide novel insight into the comprehensive genetic mechanisms in humans.
2. Materials and methods
2.1. Study subjects
The subjects were recruited from five rural counties (Xiyang, Shouyang, Taigu, Pingding, and Zezhou) of Shanxi Province in northern China, utilizing a population-based birth defect surveillance program, which has been previously described elsewhere [30]. Birthing hospitals report to the system newborns with major external structural birth defects or fetuses that are terminated due to prenatal diagnosis of such defects. Maternal venous blood samples, cord blood samples and umbilical cord tissues were collected at delivery or at the time of termination of NTD-affected pregnancies. Venous blood samples were collected from NTD case fathers. Dried blood spots were made using cord blood and paternal blood samples. The tissue samples were stored at −80 °C and the dried blood spots were stored at −20 °C until analysis. The study protocol was approved by the institutional review board of Peking University, and written informed consent was obtained from the mothers prior to the investigation.
2.2. DNA extraction
Fetal DNA from umbilical cord tissues or cord blood samples was extracted with QIAamp DNA Mini Kit Tissue kit (Qiagen, Germany). The concentration of DNA was measured by NanoDrop2000 Ultra-micro spectrophotometer (Thermo Fisher Scientific, USA). All the DNA samples were stored at −80 °C until utilized for analysis.
2.3. Target gene selection
We selected candidate PCP genes for sequencing according to the following criteria: heterozygous mutants that have been reported as NTD-causing factors when they interact with other variants in a digenic or polygenic fashion. The NTD-causing digenic mutant combinations of non-homologous PCP genes in mice are shown in Table 1. These candidate genes sequenced are: VANGL1, VANGL2, CELSR1, SCRIB, DVL2, DVL3, and PTK7.
Table 1.
Mouse mutants of key PCP Genes in a digenic fashion and NTD phenotypes.
| Gene and mutant | NTD Type | Penetrance | References |
|---|---|---|---|
| Vangl2Lp/+, Dvl2−/− | CRN | 100% | Wang et al. (2006) [25] |
| Vangl2Lp/+, Dvl3+/− | CRN, EX | 30% | Etheridge et al. (2008) [26] |
| Vangl2Lp/+,ScribCrc/+ | CRN or SB | 55% CRN, 5% SB | Murdoch et al. (2001) [27] |
| Vangl2Lp/+,ScribCrc/+ | CRN | 54% | Murdoch et al. (2014) [2] |
| Vangl2Lp/+,Celsr1Crsh/+ | CRN | not stated | Copp et al. (2003) [41] |
| Vangl2Lp/+,Celsr1Crsh/+ | CRN | 54% | Murdoch et al. (2014) [2] |
| Vangl2Lp/+, Ptk7+/− | SB | 95% | Lu et al. (2004) [47] |
| Vangl2Lp/+,Vangl1+/− | CRN | 60% | Torban et al. (2008) [28] |
| Dvl2−/−,Dvl3+/− | CRN | ~100% | Etheridge et al. (2008) [26] |
| Dvl2+/−,Dvl3−/− | CRN | some | Etheridge et al. (2008) [26] |
| ScribCrc/+,Celsr1Crsh/+ | CRN | 8% | Murdoch et al. (2014) [2] |
PCP, planar cell polarity; NTD, neural tube defect; CRN, craniorachischisis; SB, spina bifida; EX, exencephaly.
2.4. Multiplex PCR amplification and next-generation sequencing
Multiplex PCR amplification and next-generation sequencing was used to screen for DNA variants along the entire coding regions and intron-exon boundaries of targeted genes. Primers were designed using primer3. One hundred oligonucleotide pairs were constructed to cover all of the coding sequences and intron-exon boundaries of the targeted genes. After the first round of primer design, under the most stringent conditions (no SNPs in primer annealing region, amplicon length between 200 and 270 bp, GC content between 30 and 80%), the 100 oligonucleotide pairs were put into 6 multiplex PCR panels that amplified all of the target regions. The amplification reactions were carried out on an AB 2720 Thermal Cycler (Life Technologies Corporation, USA). The cycling program was 95 °C for 2 min; 11 cycles of 94 °C for 20 s, 63 °C–0.5 °C per cycle for 40 s, 72 °C for 1 min; 24 cycles of 94 °C for 20 s, 65 °C for 30 s, 72 °C for 1 min; 72 °C for 2 min.
The PCR product of each sample was labeled with 8 bp barcode; all the libraries of each sample were pooled. After cluster generation and hybridization of sequencing primer, base incorporation was carried out on a MiSeq Benchtop Sequencer (Illumina, Inc., San Diego, CA) in one single lane following the manufacturer’s standard cluster generation and sequencing protocols. The sequencing reactions ran for 300 cycles per read to generate paired-end reads including 300 bp at each end and 8 bp of the index tag.
2.5. Bioinformatics analysis
Sequencing reads were aligned to hg19 using the Burrows–Wheeler Aligner (BWA) [31]. SNV calling was performed using both GATK [32] and Varscan programs [33], and the called SNV data were then combined. The Annovar program was used for SNV annotation [34]. The potential effect of non-synonymous SNVs was assessed by the PolyPhen-2, SIFT, and MutationTaster [35–37]. Non-synonymous SNVs with SIFT score of < 0.05, Polyphen-2 score of > 0.85, or MutationTaster score of > 0.85 were considered as significant evidence that the variants were not likely to be benign. To sort potentially deleterious variants from benign polymorphisms, perl scripts were used to filter the SNVs against those of dbSNP135. Any SNV with a minor allele frequency of ≥0.01 in 1000 genome database (http://www.1000genomes.org/, Han Chinese in Bejing) or ExAC (http://exac.broadinstitute.org/) databases or ESP6500 (http://evs.gs.washington.edu/EVS) databases was considered as a benign polymorphism and therefore removed from subsequent analysis. Clustal-Omega 1.2.1 software was used to estimate the conservation of amino acids that were changed by a variant. Residues were considered to be highly conserved if there was no variation in amino acid properties observed across the compared six orthologous proteins, including five mammalian orthologous proteins plus zebra-fish. Localization of a variant in protein domains was assessed by Uniprot (http://www.uniprot.org/).
We identify digenic or multiple variants in the targeted genes in individual NTD cases. Any variant combination that was present in an NTD case but not in the 1000 genome project database was considered pathogenic.
2.6. Sanger sequencing validation
Sanger sequencing was used to validate all protein-altering variants in the target genes. We used NCBI/Primer-BLAST online tool to design the PCR primers (Supplemental Material, Table S1). PCR reaction volume is 50 μl, each of which contains: 2 × Taq PCR Green Mix 25ul, double distilled H2O 15 μl, the forward and reverse primers 2 μl, and template DNA 6 μl (20 ng/μl). PCR reaction program details were: 37 cycles of 94 °C for 12 min (94 °C for 30s; 58 °C for 30s; 72 °C for 30s); 72 °C for 10 min; Sequencing was performed using the BigDye Terminator v3.1 Cycle Sequencing Kit and a 3130XL Genetic Analyzer (Applied Biosystems). The results were processed by Mutation Surveyor 4.0.8 software. Parental genotypes of the identified variants were also determined by using Sanger sequencing. Paternity testing was carried out by genotyping a panel of polymorphic short tandem repeats to exclude false paternity and to determine if a parent is an individual’s biological parent.
3. Results
A total of 510 DNA samples from NTD fetuses/newborns were used for targeted next-generation sequencing. The phenotypic diagnosis of the NTD cases were as follows: 125 cases with anencephaly, 232 with spina bifida, 46 with encephalocele, 99 with multiple phenotypes, and 8 with other NTD subtypes. The phenotype information of the NTD cases is shown in Table 2.
Table 2.
Characteristics of neural tube defects.
| Neural tube defect phenotypes | No. of Cases |
|---|---|
| Anencephaly | 125 |
| Spina bifida | 232 |
| Encephalocele | 46 |
| Anencephaly & Spina bifida | 79 |
| Anencephaly & Encephalocele | 1 |
| Spina bifida & Encephalocele | 15 |
| Anencephaly & Spina bifida & Encephalocele | 4 |
| Other | 8 |
| Total | 510 |
AN, Anencephaly; SB, Spina bifida; EN, Encephalocele; Other, NTDs with missing subtype information.
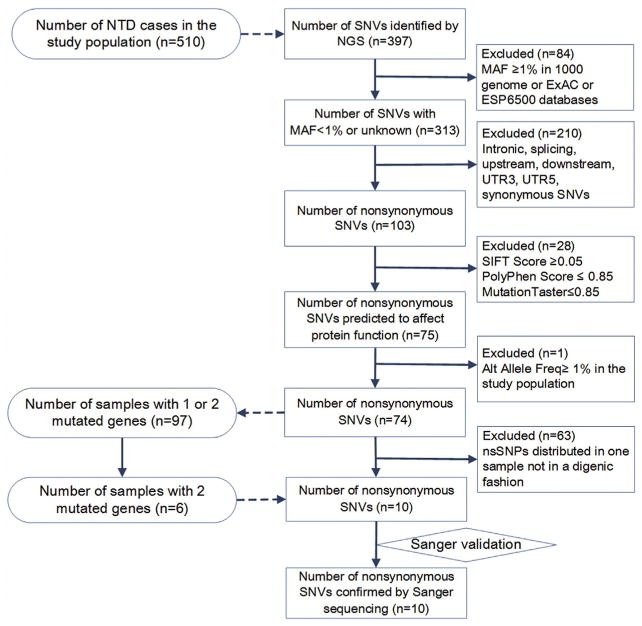
The mean sequencing depth was 570×. The proportion of target bases with read depths of 2×, 10×, 20× and 30× was 98.2%, 96.5%, 94.6% and 92.5%, respectively. Overall, 98.5% of the reads had a Phred-like quality score (Q score) > 20, and 88.8% of the reads had a Q score > 30. We identified 397 potential SNVs in 510 NTD cases. After we excluded the SNVs with a minor allele frequency of ≥0.01 in 1000 genome database (Han Chinese in Bejing) or ExAC databases or ESP6500 databases, we obtained a total of 313 SNVs. We focused on those changes that were predicted to affect the protein-CDS, including 103 non-synonymous substitutions. Of these non-synonymous SNVs, 75 were predicted to affect protein function. One SNV with alternative allele frequency > 0.01(in the database of the present study) was excluded and 74 SNVs were left (Supplemental Material Table S2). These 74 SNVs were identified from 97 cases. Six NTD cases were doubly heterozygous, i.e., they carried 2 variants each in a different target gene, involving a total of 10 SNVs. The identification flow is described in Fig. 1. For verification, we amplified the corresponding genomic region directly from the original samples using PCR and Sanger sequencing. All of these 10 variants were confirmed.
Fig. 1.
Flowchart for selecting samples and SNVs.
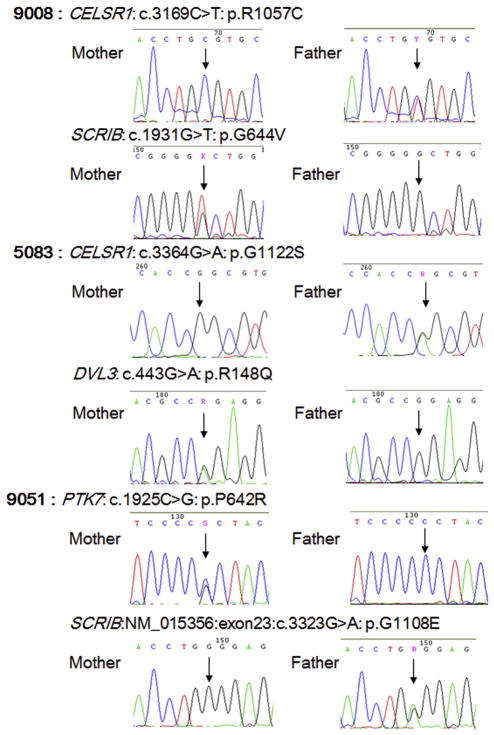
The identified digenic variants in 6 NTD cases are shown in Table 3 and Supplemental Material Fig. S1. Three spina bifida cases and one anencephaly case carried digenic variants in CELSR1and SCRIB genes. One anencephaly case had digenic variants in the CELSR1and DVL3 genes; and one spina bifida case had digenic variants in the PTK7 and SCRIB genes. Because the parental samples were available for three NTD cases, parental genotypes of the identified variants were also determined and the results are presented in Fig. 2 and Supplemental Material Table S3. No mother or father carried two variants which were identified in one NTD case.
Table 3.
Digenic variants of planar cell polarity genes in human neural tube defects.
| Sample | Phenotype | Variant 1 | Variant 2 | Variant Gene |
|---|---|---|---|---|
| B4004 | SB | CELSR1:NM_014246:exon1:c.2305C > T:p.R769W | SCRIB:NM_015356:exon23:c.3323G > A:p.G1108E | CELSR1, SCRIB |
| B9004 | SB | CELSR1:NM_014246:exon33:c.8772G > C:p.Q2924H | SCRIB:NM_015356:exon15:c.1853A > G:p.K618R | CELSR1, SCRIB |
| B9008 | SB | CELSR1:NM_014246:exon1:c.3169C > T:p.R1057C | SCRIB:NM_015356:exon15:c.1931G > T:p.G644 V | CELSR1, SCRIB |
| B0958 | AN | CELSR1:NM_014246:exon1:c.3364G > A:p.G1122S | SCRIB:NM_015356:exon23:c.3131G > A:p.R1044Q | CELSR1, SCRIB |
| B5083 | AN | CELSR1:NM_014246:exon1:c.3364G > A:p.G1122S | DVL3:NM_004423:exon4:c.443G > A:p.R148Q | CELSR1, DVL3 |
| B9051 | SB | PTK7:NM_152882:exon13:c.1925C > G:p.P642R | SCRIB:NM_015356:exon23:c.3323G > A: p.G1108E | PTK7, SCRIB |
AN, Anencephaly; SB, Spina bifida.
Fig. 2.
DNA sequence electropherograms of parental genotypes for the identified variants.
Table 4 shows detailed information on the 10 rare missense variants, including: CELSR1 c.8772G > C (p.Q2924H); CELSR1 c.3364G > A (p.G1122S); CELSR1 c.3169C > T (p.R1057C); CELSR1 c.2305C > T (p.R769W); DVL3 c.443G > A (p.R148Q); PTK7 c.1925C > G (p.P642R); SCRIB c.3323G > A (p.G1108E); SCRIB c.3131G > A (p.R1044Q); SCRIB c.1931G > T (p.G644 V); and SCRIB c.1853A > G (p.K618R). A novel variant SCRIB c.1853A > G (p.K618R) was not found in the dbSNP database, 1000 genome data, or ExAC database, while the other 10 variants were found with a minor allele frequency lower than 0.0005 in the ExAC population database, or a minor allele frequency of lower than 0.01 in the 1000 genome data of Han Chinese population in Beijing. The alternative allele frequencies of these variants in 510 NTD cases were all lower than 0.01.
Table 4.
Annotation of identified SNVs of planar cell polarity genes in human neural tube defects cases.
| Gene | RNA accession | Exon | Nucleotide change | Amino acid change | rs ID | Ref allele | Alt allele | Chrs | Position | SIFT | polyphen | Mutation Taster | 1000g_chbs | 1000g_all | ExAC03 | Alt Allele Freq |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CELSR1 | NM_014246 | exon33 | c.8772G > C | p.Q2924H | rs200116798 | C | G | 22 | 46,760,416 | 0.13 | 0.96 | 0.97 | 0.0025 | 0.0006 | 0.0003 | 0.00491 |
| CELSR1 | NM_014246 | exon1 | c.3364G > A | p.G1122S | rs200363699 | C | T | 22 | 46,929,704 | 0 | 0.99 | 0.66 | 0.0025 | 0.0002 | 0.00002 | 0.00196 |
| CELSR1 | NM_014246 | exon1 | c.3169C > T | p.R1057C | rs148349145 | G | A | 22 | 46,929,899 | 0.03 | 0.88 | 0.75 | 0 | 0.0006 | 0.0001 | 0.00196 |
| CELSR1 | NM_014246 | exon1 | c.2305C > T | p.R769W | rs201601181 | G | A | 22 | 46,930,763 | 0.03 | 1 | 0.77 | – | – | 0.0002 | 0.00098 |
| DVL3 | NM_004423 | exon4 | c.443G > A | p.R148Q | rs764021343 | G | A | 3 | 183,882,369 | 0.05 | 0.98 | 0.99 | – | – | 0.00001 | 0.00098 |
| PTK7 | NM_152882 | exon13 | c.1925C > G | p.P642R | rs148120569 | C | G | 6 | 43,111,200 | 0.01 | 0.91 | 1 | 0.0051 | 0.0012 | 0.0004 | 0.00594 |
| SCRIB | NM_015356 | exon23 | c.3323G > A | p.G1108E | rs529610993 | C | T | 8 | 144,885,908 | 0.02 | 1 | 0.99 | – | 0.0006 | 0.00006 | 0.00295 |
| SCRIB | NM_015356 | exon23 | c.3131G > A | p.R1044Q | rs782787420 | C | T | 8 | 144,886,100 | 0.26 | 1 | 0 | – | – | 0.0001 | 0.00197 |
| SCRIB | NM_015356 | exon15 | c.1931G > T | p.G644 V | rs201104891 | C | A | 8 | 144,890,963 | 0.02 | 1 | 0.38 | 0.0051 | 0.0008 | 0.0002 | 0.00492 |
| SCRIB | NM_015356 | exon15 | c.1853A > G | p.K618R | novel | T | C | 8 | 144,891,041 | 0.1 | 0.92 | 0.86 | – | – | – | 0.00098 |
SIFT, PolyPhen, and Mutation Taster are algorithms for predicting possible impact of an amino acid substitution on the structure and function of a human protein.
Non-synonymous SNVs with SIFT score of < 0.05, Polyphen-2 score of > 0.85 or MutationTaster score of > 0.85 were considered as significant of not being benign.
1000g_all, minor allele frequency of all population in the 1000 genomes.
1000g_chbs, minor allele frequency of the Chinese Han population in the 1000 genomes.
ExAC03, minor allele frequency in the Exome Aggregation Consortium database.
Alt Allele Freq, frequency of variant allele in the population of the present study.
Amino acid conservation analysis showed that seven of the 10 variants (CELSR1 p.G1122S, CELSR1 p.R769W, DVL3 p.R148Q, PTK7 p.P642R, SCRIB p.G1108E, SCRIB p.G644V and SCRIB p.K618R) were located at highly conserved nucleotides in human, dog, mouse, rat, and zebrafish. The four other variants (CELSR1 p.Q2924H, CELSR1 p.R1057C and SCRIB p.R1044Q) involved less conserved nucleotides (Supplemental material, Fig. S2). Among these variants, p.R769W and p.R1057C localized to the carbonic anhydrases subunits, named the CA domain of CELSR1, p.R1044Q was within the third PDZ domain of SCRIB, p.G1108E located very close to the fourth PDZ domain (1109–1192) of SCRIB, and p.P642R was within the fifth IGc2 domain of PTK7 (Supplemental Material, Fig. S3). Reviewing the 10 SNVs in the 1000 genome data of 2504 samples, we did not identify the same digenic variants in any of their samples.
4. Discussion
Several reports have shown that human NTD cases with missense PCP variants involved individuals carrying single heterozygous variants. According to the previous findings in mouse models, it was assumed that heterozygosity for one or more additional deleterious interacting variants might be present in human NTD cases. In this study, we analyzed doubly heterozygous variant combinations of critical PCP genes in a large group of 510 NTD cases to assess whether genetic mutations interacted in a digenic or polygenic fashion to generate the observed NTD phenotypes.
We identified a total of 10 missense SNVs from 6 samples that carried digenic variants. Of these samples, three spina bifida cases and two anencephaly cases carried digenic variants in CELSR1and other PCP genes (SCRIB and DVL3). CELSR1, homologs of the PCP core gene Fmi, is an evolutionarily conserved seven-pass transmembrane receptor that belong to the cadherin superfamily. In zebrafish, knockdown of the celsr1 homologue gene product with antisense morpholino oligonucleotides induced convergent extension defects manifested primarily by marked shortening of the anteroposterior axis [38]. In the mouse, homozygous mutants for Celsr1exhibit craniorachischisis, where the neural tube remains open from the midbrain/hindbrain boundary extending throughout the spinal region, and CELSR1 rare variants seem to have a significant role in the etiology of craniorachischisis [39]. Cases with myelomeningocele, lipomyelomeningocele, lipomyelocele, and lipoma were also found to have rare variants at CELSR1 [22]. There are three digenic combinations reported involving Celsr1 (with Vangl2, Ptk7 and Scrib), all of which are doubly heterozygous variants, and caused craniorachischisis and spina bifida in mice [2,40,41]. Two studies didn’t find any digenic cases when 20 human craniorachischisis cases were examined for rare variants in CELSR1, VANGL2 and PTK7 [39,42].
We report digenic variants in CELSR1and SCRIB in spina bifida and anencephaly cases, and CELSR1and DVL3 in anencephaly cases, involving 4 combinatorial variants that were uniquely present in cases but absent in the 1000G database. All of these variants had a minor allele frequency < 0.01 in the ExAC population database and the 1000 genome data of Han Chinese population in Beijing, save for the novel variant SCRIB c.1853A > G (p.K618R), which was not found in either the dbSNP database or 1000 genome data or ExAC database or among parental samples of the NTD cases. All of these rare variants were predicted to be detrimental in silico. During amino acid conservation analysis, over half of the variants were located at highly conserved nucleotides in human, dog, mouse, rat, and zebrafish, while the others involved less conserved nucleotides. Location analysis of the missense changes showed that the p.R769W and p.R1057C were mapped to the cadherin repeats (CA domain) of CELSR1. The two CELSR1 mouse mutants in the crash and spin cycle mice (referred to in Table 1) are also located in the cadherin repeat domain. These two SNVs may affect the function of cadherin repeat domain and lead to a failure of intercellular contact. Our findings are consistent with the disorders induced by the digenic variants of Celsr1 and Scrib in mice, and we provide novel evidence to support the notion that CELSR1 variants interacting with variants among other PCP genes may contribute to anencephaly and spina bifida phenotype.
SCRIB, as a PCP-associated gene in mammals, is a member of the LAP protein family containing 16 leucine-rich repeats and four PDZ domains [43]. In Drosophila, homozygous scrib mutations can cause loss of apicobasal cell polarity and neoplastic tissue overgrowth [44]. In the mouse, homozygous Scrib mutations can result in craniorachischisis [45,46]. The mouse heterozygote Scrib mutant in digenic combination with a heterozygote Vangl2 or Celsr1 mutation exhibits cranior-achischisis or sometimes spina bifida or severe craniorachischisis [40,45]. In humans, two variants in the SCRIB gene were found among 36 craniorachischisis cases, many of which had also been screened for VANGL1, VANGL2, CELSR1, PTK7, and PRICKLE1, but no digenic mutant combinations with SCRIB were found [39]. We report digenic variants in SCRIB and PTK7 associated with NTDs in addition to SCRIB and CELSR1 heterozygous variants in additional NTD cases. The combinatorial variation of PTK7 c.1925C > G (p.P642R) and SCRIB c.3323G > A (p.G1108E) only occurred in one spina bifida case, and was not found in the 1000G database or parental samples of NTD cases. Location analysis of missense changes showed that p.G1108E was located very close to the fourth PDZ domain (1109–1192) of SCRIB. The PDZ domains of human SCRIB are required for correct localization and physical interaction with other proteins, such as the core PCP protein VANGL2, which is required for transducing PCP signals. Herein we demonstrate that SCRIB variants combined with variants among other PCP genes might be associated with the observed NTD phenotypes in humans. However, pathogenic effect of these variants on protein function or on neural tube development need to be investigated in the future.
No reports are available in the literature on the contribution of digenic variants of CELSR1 and DVL3 gene in the pathogenesis of NTDs. Mouse studies with doubly heterozygous mutants for Ptk7 and Scrib showed no NTDs [40]. In fact, although all the involved missense variants were predicted to affect residues conserved across evolution, no functional studies have been done to look at the pathogenic effect of any of these variants on protein function or on neural tube development. Further studies are needed to examine whether these combinatorial variants affect expression of the PCP genes which may play a causal role in the development of NTDs.
In conclusion, the present study provides novel evidence of combinatorial PCP variants contributing to the etiology of NTDs. Our findings imply that genetic variation might interact in a digenic fashion to generate the observed NTD phenotypes, and emphasize the importance of these genetic interactions to the etiology of human NTDs. Furthermore, this study emphasizes the importance of performing comprehensive genetic screens in humans when searching for the genetic cause of birth defects, because disruption of multiple genes could combine in an individual to induce devastating consequences on embryonic development, although heterozygous variation in a single gene might not be sufficient to interfere with neural tube closure. Further studies including more reported PCP genes, such as FZD6, PRICKLE1, FUZZY, DACT1, SEC24B, DVL1 and LRP6, in other cohorts should be performed to validate our finding. Another possibility is that combinations between uncommon PCP variants and common PCP variants could contribute to NTDs genetic etiology which worth to study.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81472987 and 81773441); Beijing Natural Science Foundation (Grant No. 7162094); and the National Key Research and Development Program, Ministry of Science and Technology, P.R. China (Grant No. 2016YFC1000501). Drs. Lei and Finnell were supported by NIH grants HD081216, HD083809 and HD067244.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymgme.2018.03.005.
Footnotes
Conflicts of interest
All authors report no conflicts of interest.
References
- 1.Wallingford JB, Niswander LA, Shaw GM, Finnell RH. The continuing challenge of understanding, preventing, and treating neural tube defects. Science. 2013;339:1222002. doi: 10.1126/science.1222002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murdoch JN, Damrau C, Paudyal A, Bogani D, Wells S, Greene ND, Stanier P, Copp AJ. Genetic interactions between planar cell polarity genes cause diverse neural tube defects in mice. Dis Model Mech. 2014;7:1153–1163. doi: 10.1242/dmm.016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lupo PJ, Mitchell LE, Canfield MA, Shaw GM, Olshan AF, Finnell RH, Zhu H National Birth S. Defects prevention, maternal-fetal metabolic gene-gene interactions and risk of neural tube defects. Mol Genet Metab. 2014;111:46–51. doi: 10.1016/j.ymgme.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loeken MR. Intersection of complex genetic traits affecting maternal metabolism, fetal metabolism, and neural tube defect risk: looking for needles in multiple haystacks. Mol Genet Metab. 2014;111:415–417. doi: 10.1016/j.ymgme.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heil SG, Van der Put NM, Waas ET, den Heijer M, Trijbels FJ, Blom HJ. Is mutated serine hydroxy methyl transferase (SHMT) involved in the etiology of neural tube defects? Mol Genet Metab. 2001;73:164–172. doi: 10.1006/mgme.2001.3175. [DOI] [PubMed] [Google Scholar]
- 6.Leck I. Causation of neural tube defects: clues from epidemiology. Br Med Bull. 1974;30:158–163. doi: 10.1093/oxfordjournals.bmb.a071187. [DOI] [PubMed] [Google Scholar]
- 7.Juriloff DM, Harris MJ. A consideration of the evidence that genetic defects in planar cell polarity contribute to the etiology of human neural tube defects. Birth Defects Res A Clin Mol Teratol. 2012;94:824–840. doi: 10.1002/bdra.23079. [DOI] [PubMed] [Google Scholar]
- 8.Harris MJ, Juriloff DM. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth Defects Res Part A Clin and Mol Teratol. 2010;88:653–669. doi: 10.1002/bdra.20676. [DOI] [PubMed] [Google Scholar]
- 9.Harris MJ, Juriloff DM. Mouse mutants with neural tube closure defects and their role in understanding human neural tube defects. Birth Defects Res Part A Clin and Mol Teratol. 2007;79:187–210. doi: 10.1002/bdra.20333. [DOI] [PubMed] [Google Scholar]
- 10.Wilde JJ, Petersen JR, Niswander L. Genetic, epigenetic, and environmental contributions to neural tube closure. Annu Rev Genet. 2014;48:583–611. doi: 10.1146/annurev-genet-120213-092208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray RS, Roszko I, Solnica-Krezel L. Planar cell polarity: coordinating morphogenetic cell behaviors with embryonic polarity. Dev Cell. 2011;21:120–133. doi: 10.1016/j.devcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simons M, Mlodzik M. Planar cell polarity signaling: from fly development to human disease. Annu Rev Genet. 2008;42:517–540. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kibar Z, Vogan KJ, Groulx N, Justice MJ, Underhill DA, Gros P. Ltap, a mammalian homolog of Drosophila strabismus/van Gogh, is altered in the mouse neural tube mutant loop-tail. Nat Genet. 2001;28:251–255. doi: 10.1038/90081. [DOI] [PubMed] [Google Scholar]
- 14.Hamblet NS, Lijam N, Ruiz-Lozano P, Wang J, Yang Y, Luo Z, Mei L, Chien KR, Sussman DJ, Wynshaw-Boris A. Dishevelled 2 is essential for cardiac outflow tract development, somite segmentation and neural tube closure. Development. 2002;129:5827–5838. doi: 10.1242/dev.00164. [DOI] [PubMed] [Google Scholar]
- 15.Curtin JA, Quint E, Tsipouri V, Arkell RM, Cattanach B, Copp AJ, Henderson DJ, Spurr N, Stanier P, Fisher EM, Nolan PM, Steel KP, Brown SD, Gray IC, Murdoch JN. Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr Biol. 2003;13:1129–1133. doi: 10.1016/s0960-9822(03)00374-9. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Guo N, Nathans J. The role of Frizzled3 and Frizzled6 in neural tube closure and in the planar polarity of inner-ear sensory hair cells. The J of Neurosci: the Off J of the Soc for Neurosci. 2006;26:2147–2156. doi: 10.1523/JNEUROSCI.4698-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei YP, Zhang T, Li H, Wu BL, Jin L, Wang HY. VANGL2 mutations in human cranial neural-tube defects. N Engl J Med. 2010;362:2232–2235. doi: 10.1056/NEJMc0910820. [DOI] [PubMed] [Google Scholar]
- 18.Kibar Z, Torban E, McDearmid JR, Reynolds A, Berghout J, Mathieu M, Kirillova I, De Marco P, Merello E, Hayes JM, Wallingford JB, Drapeau P, Capra V, Gros P. Mutations in VANGL1 associated with neural-tube defects. N Engl J Med. 2007;356:1432–1437. doi: 10.1056/NEJMoa060651. [DOI] [PubMed] [Google Scholar]
- 19.De Marco P, Merello E, Rossi A, Piatelli G, Cama A, Kibar Z, Capra V. FZD6 is a novel gene for human neural tube defects. Hum Mutat. 2012;33:384–390. doi: 10.1002/humu.21643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Marco P, Merello E, Consales A, Piatelli G, Cama A, Kibar Z, Capra V. Genetic analysis of disheveled 2 and disheveled 3 in human neural tube defects. J of Mol Neurosci: MN. 2013;49:582–588. doi: 10.1007/s12031-012-9871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosoi CM, Capra V, Allache R, Trinh VQ, De Marco P, Merello E, Drapeau P, Bassuk AG, Kibar Z. Identification and characterization of novel rare mutations in the planar cell polarity gene PRICKLE1 in human neural tube defects. Hum Mutat. 2011;32:1371–1375. doi: 10.1002/humu.21589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allache R, De Marco P, Merello E, Capra V, Kibar Z. Role of the planar cell polarity gene CELSR1 in neural tube defects and caudal agenesis. Birth Defects Res Part A Clin Mol Teratol. 2012;94:176–181. doi: 10.1002/bdra.23002. [DOI] [PubMed] [Google Scholar]
- 23.Seo JH, Zilber Y, Babayeva S, Liu J, Kyriakopoulos P, De Marco P, Merello E, Capra V, Gros P, Torban E. Mutations in the planar cell polarity gene, Fuzzy, are associated with neural tube defects in humans. Hum Mol Genet. 2011;20:4324–4333. doi: 10.1093/hmg/ddr359. [DOI] [PubMed] [Google Scholar]
- 24.Greene ND, Stanier P, Copp AJ. Genetics of human neural tube defects. Hum Mol Genet. 2009;18:R113–129. doi: 10.1093/hmg/ddp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Wang X, Guan T, Xiang Q, Wang M, Zhang Z, Guan Z, Wang G, Zhu Z, Xie Q, Li G, Guo J, Wang F, Zhang Z, Niu B, Zhang T. Analyses of copy number variation reveal putative susceptibility loci in MTX-induced mouse neural tube defects. Dev Neurobiol. 2014;74:877–893. doi: 10.1002/dneu.22170. [DOI] [PubMed] [Google Scholar]
- 26.Etheridge SL, Ray S, Li S, Hamblet NS, Lijam N, Tsang M, Greer J, Kardos N, Wang J, Sussman DJ, Chen P, Wynshaw-Boris A. Murine dishevelled 3 functions in redundant pathways with dishevelled 1 and 2 in normal cardiac outflow tract, cochlea, and neural tube development. PLoS Genet. 2008;4:e1000259. doi: 10.1371/journal.pgen.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murdoch JN, Rachel RA, Shah S, Beermann F, Stanier P, Mason CA, Copp AJ. Circle tail, a new mouse mutant with severe neural tube defects: chromosomal localization and interaction with the loop-tail mutation. Genomics. 2001;78:55–63. doi: 10.1006/geno.2001.6638. [DOI] [PubMed] [Google Scholar]
- 28.Torban E, Patenaude AM, Leclerc S, Rakowiecki S, Gauthier S, Andelfinger G, Epstein DJ, Gros P. Genetic interaction between members of the Vangl family causes neural tube defects in mice. Proc Natl Acad Sci U S A. 2008;105:3449–3454. doi: 10.1073/pnas.0712126105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Ren A, Zhang L, Ye R, Li S, Zheng J, Hong S, Wang T. Extremely high prevalence of neural tube defects in a 4-county area in Shanxi Province China. Birth Defects Res A Clin Mol Teratol. 2006;76:237–240. doi: 10.1002/bdra.20248. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Zhang L, Li Z, Jin L, Zhang Y, Ye R, Liu J, Ren A. Prevalence and trend of neural tube defects in five counties in Shanxi province of northern China, 2000 to 2014. Birth Defects Res Part A Clin Mol Teratol. 2016;106:267–274. doi: 10.1002/bdra.23486. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: a Map Reduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 38.Formstone CJ, Mason I. Expression of the Celsr/flamingo homologue, c-fmi1, in the early avian embryo indicates a conserved role in neural tube closure and additional roles in asymmetry and somitogenesis. Dev Dyn. 2005;232:408–413. doi: 10.1002/dvdy.20228. [DOI] [PubMed] [Google Scholar]
- 39.Robinson A, Escuin S, Doudney K, Vekemans M, Stevenson RE, Greene ND, Copp AJ, Stanier P. Mutations in the planar cell polarity genes CELSR1 and SCRIB are associated with the severe neural tube defect craniorachischisis. Hum Mutat. 2012;33:440–447. doi: 10.1002/humu.21662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paudyal A, Damrau C, Patterson VL, Ermakov A, Formstone C, Lalanne Z, Wells S, Lu X, Norris DP, Dean CH, Henderson DJ, Murdoch JN. The novel mouse mutant, chuzhoi, has disruption of Ptk7 protein and exhibits defects in neural tube, heart and lung development and abnormal planar cell polarity in the ear. BMC Dev Biol. 2010;10:87. doi: 10.1186/1471-213X-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Copp AJ, Greene ND, Murdoch JN. The genetic basis of mammalian neurulation. Nat Rev Genet. 2003;4:784–793. doi: 10.1038/nrg1181. [DOI] [PubMed] [Google Scholar]
- 42.Doudney K, Ybot-Gonzalez P, Paternotte C, Stevenson RE, Greene ND, Moore GE, Copp AJ, Stanier P. Analysis of the planar cell polarity gene Vangl2 and its co-expressed paralogue Vangl1 in neural tube defect patients. Am J Med Genet A. 2005;136:90–92. doi: 10.1002/ajmg.a.30766. [DOI] [PubMed] [Google Scholar]
- 43.Montcouquiol M, Rachel RA, Lanford PJ, Copeland NG, Jenkins NA, Kelley MW. Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature. 2003;423:173–177. doi: 10.1038/nature01618. [DOI] [PubMed] [Google Scholar]
- 44.Bilder D, Perrimon N. Localization of apical epithelial determinants by the basolateral PDZ protein scribble. Nature. 2000;403:676–680. doi: 10.1038/35001108. [DOI] [PubMed] [Google Scholar]
- 45.Murdoch JN, Henderson DJ, Doudney K, Gaston-Massuet C, Phillips HM, Paternotte C, Arkell R, Stanier P, Copp AJ. Disruption of scribble (Scrb1) causes severe neural tube defects in the circle tail mouse. Hum Mol Genet. 2003;12:87–98. doi: 10.1093/hmg/ddg014. [DOI] [PubMed] [Google Scholar]
- 46.Zarbalis K, May SR, Shen Y, Ekker M, Rubenstein JL, Peterson AS. A focused and efficient genetic screening strategy in the mouse: identification of mutations that disrupt cortical development. PLoS Biol. 2004;2:E219. doi: 10.1371/journal.pbio.0020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu X, Borchers AG, Jolicoeur C, Rayburn H, Baker JC, Tessier-Lavigne M. PTK7/CCK-4 is a novel regulator of planar cell polarity in vertebrates. Nature. 2004;430:93–98. doi: 10.1038/nature02677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.