Abstract
B cells are responsible for protective antibody production after differentiation into antibody-secreting cells during humoral immune responses. From early B cell development in the bone marrow, to their maturation in the periphery, activation in the germinal center, and differentiation into plasma cells or memory B cells, B cells display ever-changing functions and properties. Autophagy and mitochondria play important roles in B cell development, activation, and differentation to accommodate the phenotypic and environmental changes encountered over the lifetime of the cell. Among their many functions, mitochondria and autophagy generate energy, mediate cell survival, and produce/eliminate reactive oxygen species that can serve as signal molecules to regulate differentiation. As B cells mature and differentiate into plasma or memory cells, both autophagic and mitochondrial functions undergo significant changes. In this review, we aim to provide an overview of the role of the autophagosome and mitochondria in regulating B cell fate, survival, and function. Moreover, we will discuss the interplay between these two highly metabolic organelles during B cell development, maturation, and differentiation.
Keywords: Mitochondria, germinal center, B cells, plasma cells, memory B cells, ROS, metabolism, autophagy, mitophagy
INTRODUCTION
Vertebrates have developed various strategies to counter the constant and myriad barrage of pathogens they face throughout their lifespans. The adaptive arm of the immune system is capable of discriminating between different pathogens by recognizing the unique molecular markers, or antigens, that distinguish the various pathogens (Litman et al., 2010; Parra et al., 2013). This allows immune responses to tailor their response to specific pathogens, to remember previous pathogen encounters, and to mount a rapid and potent response upon subsequent exposure to the same pathogen (Litman et al., 2010; Parra et al., 2013). Lymphocyte activation, proliferation, and effector functions, as well as the formation of memory cells, are linked to dynamic changes in cellular metabolism (Buck et al., 2017). The metabolic functions of lymphocytes rely heavily on the mitochondria, the cellular metabolic hub that regulates energy production through coordination of the electron transport chain (ETC) and the tricarboxylic acid (TCA) cycle. Moreover, mitochondria catabolize nutrients, including glucose, amino acids, and fatty acids, to produce building blocks for cell activation and expansion (Ahn and Metallo, 2015). In order for the cell to meet its metabolic demands, they have to change their mitochondrial volume, membrane potential (ΔΨm), and location in response to nutrient availability and growth stimuli. The engagement of various metabolic pathways is controlled by growth factors and nutrient availability dictated by competition between other interacting cells, as well as the balance of internal metabolites, reactive oxygen species (ROS), and reducing and oxidizing substrates (Buck et al., 2017).
Mitochondria couple metabolite oxidation to aerobic respiration, making them a major energy producer within a cell. Glucose and fatty acids, after being catabolized through glycolysis and β-oxidation, form acetyl-CoA, which fuels the TCA cycle. Acetyl-CoA is further oxidized into carbon dioxide to generate NADH and FADH2, the main sources of electrons for the electron transport chain (ETC). The ETC transfers electrons provided by NADH and FADH2 to oxygen, while generating ΔΨm with proton gradient across the mitochondrial inner membrane. This proton gradient is further utilized to produce ATP (Weinberg et al., 2015). Mitochondria also contribute to lipid and amino acid synthesis to build macromolecules. Glutamine uptake provides another source of carbons that can be used either for oxidative metabolism or anabolism after conversion into nucleic acid precursors and other amino acids (Boothby and Rickert, 2017). At present, the balances among these varied processes have not yet been fully elucidated in specific B lineages. Mitochondria bridge nutrient metabolism to fulfill the bioenergetic demands of the cell through the coordination of the TCA cycle and ETC (Boothby and Rickert, 2017; Chao et al., 2017). It will be important to determine the functions of mitochondria in the regulation of B cells at different stages of differentiation.
During differentiation into effector and memory cells, deleterious products, such as oxidized proteins and lipids, can accumulate in the cells (Bhattacharya and Eissa, 2015; Bullon et al., 2016; Puleston and Simon, 2013; Rathmell, 2012). Autophagy is an intracellular homeostatic mechanism important for the degradation of waste components from the cytoplasm in acidic lysosomal compartments (Yang and Klionsky, 2010). Originally, surplus parts of the cytoplasm that acted as targets for autophagy were thought to comprise only cellular organelles and proteins, but this has now been extended to include a range of pathogens (Kuballa et al., 2012). Autophagy is involved in the regulation of cell survival and homeostasis in B and T cells (Bhattacharya and Eissa, 2015; Puleston and Simon, 2013; Rathmell, 2012). In the thymus, autophagy can modulate the selection of certain CD4+ T cell clones, while in the bone marrow autophagy is needed for B cell differentiation and survival at specific stages (Arnold et al., 2016; Bhattacharya and Eissa, 2015; Puleston and Simon, 2013).
T and B cells are the major lymphocyte lineages for adaptive immunity (Litman et al., 2010; Parra et al., 2013). B cells have a semi-nomadic life cycle due to their role in patrolling the body for foreign antigens. However, cell activation and differentiation can lead to the retention of B cells in distinct microenvironments that differ in nutrient availability, oxygen, and redox species (Boothby and Rickert, 2017). B cells are specifically responsible for mediating the humoral arm of the adaptive immune system (Hoffman et al., 2016). Humoral immunity involves the production of antibodies that recognize antigens via a unique antigen-binding pocket (Schroeder and Cavacini, 2010; Sela-Culang et al., 2013). Antibodies, also known as immunoglobulins, exist as in a membrane-bound form before secretion, where they serve as the B cell antigen receptor (BCR) (Hoehn et al., 2016; Hoffman et al., 2016). The immunoglobulins expressed by a given B cell have the same antigenic specificity to recognize a particular antigen (Hoehn et al., 2016). In this section, we will summarize how mitochondria and autophagy control B cell fate and long-term maintenance.
Mitochondria in early B cell development and maturation
In mammals, conventional B cells are generated from the common lymphoid progenitor and undergo early development in the bone marrow (Hoffman et al., 2016). One of the essential tasks of a developing B cell is to assemble a complete immunoglobulin gene from a diverse pool of gene segments in order to express a functional pre-B cell receptor (BCR) (Melchers, 2015). Early B cell development is characterized by alternating states of quiescence and activation in response to BCR and IL-7 receptor (IL-7R) signaling (Heizmann et al., 2013; Herzog et al., 2009; Stein et al., 2017). Pro-B cells represent the earliest stage of B cell development, and transition to the pre-B cell stage following the successful rearrangement of their immunoglobulin heavy chain gene loci to form the pre-BCR (Figure 1), one of the first major checkpoints in B cell development (Herzog et al., 2009). Pre-B cells can be further subdivided into the immature, actively dividing large pre-B cell stage and the more mature, quiescent small pre-B cell stage (Herzog et al., 2009; Stein et al., 2017). Pro-B cells develop in IL-7-rich regions in the bone marrow, with IL-7 serving to induce cell division and survival (Clark et al., 2014; Hamel et al., 2014; Heizmann et al., 2013). As pro-B cells transition to large pre-B cells, they undergo a burst of proliferation (Clark et al., 2014; Hamel et al., 2014). However, pre-BCR signaling antagonizes IL-7R signaling, constraining the clonal expansion of large pre-B cells to several divisions (Clark et al., 2014; Hamel et al., 2014; Smart and Venkitaraman, 2000). Large pre-B cells start to lose IL-7 responsiveness and migrate towards IL-7-poor regions, causing them to exit the cell cycle and develop into small pre-B cells (Clark et al., 2014; Hamel et al., 2014; Smart and Venkitaraman, 2000).
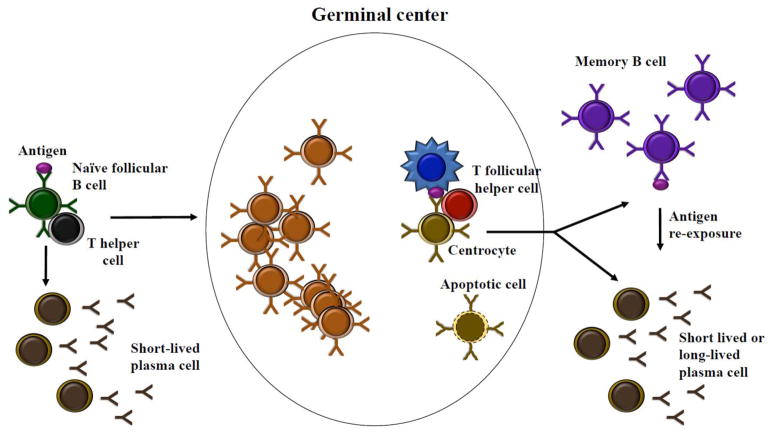
Figure 1. Development and maintenance of memory B cells.
Naïve B cells activated by cognate antigen and T cell help migrate to secondary lymphoid organs and seed germinal centers (GCs). GC B cells rapidly divide and mutate their immunoglobulin genes to improve their affinity to the immunizing antigen. GC B cells with favorable mutations are selected to survive, while those with unfavorable mutations undergo apoptosis. Multiple rounds of proliferation, mutation, and affinity-based selection generate a B cell population with increased average affinity for the antigen. The majority of GC B cells die, but a small fraction survives to differentiate into antibody-secreting plasma cells or memory B cells. In response to re-challenge with the same antigen, memory B cells can rapidly differentiate into plasma cells or re-seed germinal centers. Memory B cells are long-lived, and maintenance of mitochondrial homeostasis by autophagy is thought to contribute to their persistence.
The development of pro-B cells into small pre-B cells is driven by pre-BCR and IL-7R-induced metabolic changes involving the mitochondria (Heizmann et al., 2013; Stein et al., 2017). Withdrawal of IL-7 in pre-B cell line cultures results in down-regulation of genes related to mitochondrial function (Heizmann et al., 2013). Compared to small pre-B cells, large pre-B cells have increased mitochondrial membrane potential, glucose uptake, and reactive oxygen species (ROS) levels, consistent with their proliferative state (Stein et al., 2017). Swiprosin-2, a calcium-binding inner mitochondrial membrane protein, regulates a metabolic program triggered as pro-B cells develop into small pre-B cells (Stein et al., 2017). Swiprosin-2 is expressed in proB cells but is repressed in pre-B cells due to the expression of the pre-BCR (Stein et al., 2017). Knockout of swiprosin-2 in a pro-B cell line results in decreased oxidative phosphorylation, but increased glycolysis, relative to wild-type controls (Stein et al., 2017). A metabolic switch regulated by swiprosin-2 is triggered as dividing pro-B cells develop into quiescent small pre-B cells.
After the initial development stages in the bone marrow, B cells migrate to the spleen and undergo maturation (Hoffman et al., 2016). Maturing B cells can develop into follicular B cells or marginal zone B cells (Hoffman et al., 2016; Pillai and Cariappa, 2009). Follicular B cells are circulating cells that represent the majority of mature B cells, whereas marginal zone B cells are non-circulating cells that reside in the marginal zone of the spleen (Hoffman et al., 2016; Pillai and Cariappa, 2009). The choice of becoming a follicular B cell or a marginal zone B cell depends upon the strength of BCR signaling, with strong signaling favoring a follicular B cell fate, while weak signaling favoring a marginal zone B cell fate (Hoffman et al., 2016; Pillai and Cariappa, 2009). Additionally, signaling through the Notch2 receptor is crucial to drive marginal zone B cell development (Hoffman et al., 2016; Pillai and Cariappa, 2009). Compared to follicular B cells, marginal zone B cells have increased size and a lower threshold for activation (Jellusova et al., 2017; Pillai and Cariappa, 2009). Moreover, marginal zone B cells are also long-lived and can undergo self-renew (Pillai and Cariappa, 2009). Activated B cells increase glucose uptake and mitochondrial mass, resulting in increased glycolysis and oxidative phosphorylation (Table 1) (Caro-Maldonado et al., 2014). Marginal zone B cells seem to exhibit increased glucose uptake compared to follicular B cells (Jellusova et al., 2017). The metabolic demands are high in the proliferative early B cell stages and decrease in the pre-B, immature, and transitional stages. Glucose uptake, which increases substantially after B cell activation (Caro-Maldonado et al., 2014; Cho et al., 2011; Dufort et al., 2007), provides the substrate for glycolysis (Table 1). Whether marginal zone B cells also increase their mitochondrial mass and oxidative phosphorylation remains to be investigated.
Table 1.
Comparison of the metabolic characteristics of selected B cell lineages
| Lineage | Glucose uptake | OXPHOS | ROS | Type of autophagy | Fatty acid oxidation |
|---|---|---|---|---|---|
| Naïve | -- | -- | -- | Canonical | ?? |
| Germinal center |
|
|
|
Non-Canonical | ?? |
| Short-lived plasma |
|
|
|
Canonical | ?? |
| Long-lived plasma |
|
|
|
Canonical | ?? |
| Memory | ?? | ?? |
|
Canonical | ?? |
Key: -- Baseline,
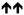 Increased,
Increased,
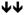 Decreased and ?? Unknown
Decreased and ?? Unknown
Mitochondria in activated B cells
Upon encountering cognate antigen and receiving costimulation from T cells, antigen-specific naïve B cells become activated and migrate to secondary lymphoid tissues, where they undergo rapid growth and proliferation in regions called germinal centers (GC) (Litman et al., 2010; Parra et al., 2013; Wykes, 2003). In the GC, B cells undergo class switch recombination (CSR) to different classes of immunoglobulins, as well as editing their immunoglobulin genes through somatic hypermutation (SHM) to generate high affinity BCRs, a process known as affinity maturation (Litman et al., 2010; Parra et al., 2013). GC B cells that have successfully completed affinity maturation undergo differentiation into either antibody-secreting plasma cells or long-lived memory B cells (Bhattacharya et al., 2007). Additionally, in response to certain types of antigens, naïve B cells can become activated without T cell help and differentiate directly into short-lived antibody-secreting plasma cells (Tangye et al., 2003).
B cells can be activated by a variety of signals, including ligation of the BCR, Toll-like receptors (TLRs), CD40, and cytokine receptors. The type and extent of activation depends upon the specific signal(s) involved, with some of these signals capable of synergizing to produce a more potent or different activation than they would individually. Different signals may exert their effect in B cells by differentially modulating mitochondrial status and metabolic activity. Activated B cells likely have increased energy requirements and switch to a more anabolic metabolism compared to their quiescent naïve B cell counterparts. Metabolism in B cell has long been considered to be similar to that in T cells. Activated T cells are known to preferentially increase glycolysis over oxidative phosphorylation (van der Windt and Pearce, 2012). However, B cells activated through toll-like receptor 4 (TLR4) or the BCR increase both oxidative phosphorylation and glycolysis in a balanced manner (Table 1) (Caro-Maldonado et al., 2014). TLR4 and BCR-induced upregulation of glycolysis relies upon activation of phosphoinositide 3 kinase (PI3K), a key regulator of glucose metabolism (Donahue and Fruman, 2003). IL-4, a B cell survival factor, also increases glucose uptake and glycolytic activity in B cells, though in a PI3K-independent manner (Dufort et al., 2007). PI3K signaling antagonizes expression of the transcription factor Foxo1 (Sander et al., 2015). The interplay between PI3K and Foxo1 regulates B cell development at different stages (Dominguez-Sola et al., 2015; Inoue et al., 2017; Ochiai et al., 2012; Sander et al., 2015). During early B cell development, in the bone marrow, PI3K promotes proliferation, while Foxo1 promotes cell cycle arrest and differentiation (Ochiai et al., 2012). IL-7 signaling in pre-B cells downregulates Foxo1 through the PI3K pathway; failure to attenuate IL-7 signaling thus inhibits the differentiation of pre-B cells (Ochiai et al., 2012). The PI3K/Foxo1 signaling axis also regulates the GC B cell compartment (Dominguez-Sola et al., 2015; Inoue et al., 2017; Sander et al., 2015). The germinal center can be divided into a dark zone, containing proliferating, somatically hypermutating cells known as centroblasts, and a light zone, containing quiescent cells known as centrocytes (Figure 1) (Klein and Dalla-Favera, 2008; Zhang et al., 2016). Centroblasts differentiate into centrocytes and migrate to the light zone; centrocytes undergo positive selection for re-entry into the dark zone as centroblasts or germinal center exit and differentiation into plasma cells or memory B cells (Figure 1) (Klein and Dalla-Favera, 2008; Zhang et al., 2016). Interestingly, Foxo1 expression is highest in the proliferating centroblasts, whereas PI3K signaling is highest in the quiescent centrocytes (Dominguez-Sola et al., 2015; Sander et al., 2015). Deletion of Foxo1 or forced expression PI3K signaling in GC B cells both result in impaired CSR and loss of the germinal center dark zone (Dominguez-Sola et al., 2015; Inoue et al., 2017; Sander et al., 2015).
Consistent with increased oxidative phosphorylation activity, activated B cells have also been shown to have increased mitochondrial mass and mitochondrial ROS production (Table 1) (Jang et al., 2015; Wheeler and Defranco, 2012). Although mitochondrial ROS is best known as an inducer of oxidative stress, it has also been identified as a signaling molecule for T cell activation. ROS has been reported to regulate self-renewal and differentiation of hematopoietic stem cells (Ho et al., 2017; Sena et al., 2013). Similarly, B cell activation and fate decisions have been linked to changes in mitochondrial activity and ROS levels (Jang et al., 2015). Neutralizing ROS with the antioxidant N-acetylcysteine (NAC) attenuates activation and proliferation of BCR-stimulated B cells (Wheeler and Defranco, 2012). The transcription factor Blimp1 reduces mitochondrial mass in B cells, which results in lower levels of mitochondrial ROS, antagonizing CSR and promoting commitment to the plasma cell lineage (Jang et al., 2015). B cells stimulated in vitro with LPS and IL-4 undergo CSR and plasma cell differentiation (Jang et al., 2015). Inhibition of ROS with the antioxidant ascorbic acid enhances plasma cell differentiation while inhibiting CSR in B cells stimulated with LPS and IL4 (Jang et al., 2015). In contrast, treatment with the glycolysis inhibitor 2-deoxyglucose or the Krebs cycle substrate methyl pyruvate, increases the percentage of cells that display high mitochondrial content and activity, as well as mitochondrial ROS levels in LPS- and IL-4-stimulated B cells. As a result, these treatments result in reduced formation of plasma cells (Table 1) (Jang et al., 2015).
Glycogen synthase kinase 3 (Gsk3) has been shown to regulate GC B cell survival and differentiation by acting as a metabolic sensor (Jellusova et al., 2017). The light zones of germinal centers are regions where GC B cells reduce division and compete to undergo positive selection to survive and differentiate (Figure 1) (Klein and Dalla-Favera, 2008; Zhang et al., 2016). Light zones are hypoxic regions where nutrient and cytokine availability is limited (Cho et al., 2016). This harsh environment places a constraint upon GC B cell survival and proliferation (Jellusova et al., 2017). In response to these conditions, GC B cells undergo metabolic reprogramming, increasing mitochondrial mass, glucose uptake, and HIF1-α-dependent glycolysis (Table 1) (Jellusova et al., 2017). In the absence of Gsk3, GC B cells exhibit increased metabolic activity and proliferation, suggesting that Gsk3 serves to constrain GC B cell growth and proliferation when nutrients are limited (Jellusova et al., 2017). Gsk3 has been reported to suppress mammalian target of rapamycin complex 1 (mTORC1), a nutrient sensor that inhibits autophagy but promotes cell growth and proliferation (Jellusova et al., 2017). However, Gsk3-deficient GC B cells do not exhibit increased mTORC1 signaling, suggesting that this pathway is not responsible for the elevated growth and proliferation of Gsk3-deficient GC B cells. The phenotype of Gsk3-deficient GC B cells may be in part due to increased activity of c-Myc, a transcription factor that is well characterized for its role in mitochondrial biogenesis and cell growth, proliferation and differentiation (Jellusova et al., 2017). Therefore, Gsk3 plays an important role in linking mitochondrial metabolic functions to the regulation of GC B cells.
Recently, non-canonical autophagy has been shown to play an important role during the GC B cell response (Martinez-Martin et al., 2017). Autophagy (which translates to “self-eating”) is a process in which cells engulf a portion of their own cytoplasm via a double membrane-bound organelle called the autophagosome, and deliver it to the lysosome for degradation (Codogno et al., 2011). Canonical autophagy requires the coordinated action of a core set of autophagy proteins to assemble the autophagosome (Codogno et al., 2011). In non-canonical autophagy, autophagosome formation is able to proceed using only a subset of the autophagy proteins (Codogno et al., 2011). Naïve follicular B cells activated in vitro by BCR or CD40 stimulation transition from canonical to non-canonical autophagy (Table 1) (Martinez-Martin et al., 2017). Similarly, naïve follicular B cells in vivo adopt a non-canonical autophagy program as they develop into GC B cells (Martinez-Martin et al., 2017). The subsequent differentiation of GC B cells into plasma cells or memory B cells involves a return to canonical autophagy (Martinez-Martin et al., 2017). The switch between canonical and non-canonical autophagy in B cells is regulated by WD repeat domain phosphoinositide interacting 2 (WIPI2), a protein involved in early autophagosome biogenesis (Martinez-Martin et al., 2017). In the absence of WIPI2, B cells exhibit a loss of canonical autophagy and a concomitant increase in non-canonical autophagy (Martinez-Martin et al., 2017). WIPI2 is highly upregulated in GC B cells versus other B cell subsets, and WIPI2 knockout mice have significantly reduced numbers of GC B cells and plasma cells post-immunization (Martinez-Martin et al., 2017). Interestingly however, WIPI2 knockout B cells more readily undergo differentiation to plasma cells in vitro, compared to their wild-type counterparts (Martinez-Martin et al., 2017). This indicates a metabolic control of B cell fate decisions, similar to what has been reported in T cells. Mitochondrial status and metabolic activity have been implicated as signals regulating B cell differentiation (Jang et al., 2015). In the absence of WIPI2, naïve B cells have increased mitochondrial content, mitochondrial ROS, and mitochondrial membrane potential, and also show elevated basal oxidative phosphorylation and glycolytic activity (Martinez-Martin et al., 2017). Thus, WIPI2, in its role as a regulator of autophagy and mitochondria, serves to modulate B cell activation and differentiation.
Mitochondria and autophagy in plasma cell survival and function
GC B cells that survive the germinal center reaction have the choice of differentiating into plasma cells or memory B cells (Bhattacharya et al., 2007). Plasma cells are the effector cells of the humoral adaptive immune system. They are terminally differentiated B cells that downregulate most B cell markers and secrete large quantities of antibodies to neutralize pathogens (Holmes et al., 2008; Klein and Dalla-Favera, 2008). Plasma cells can be broadly divided into short-lived plasma cells that die soon after the contraction of the primary antibody response, and long-lived plasma cells, which migrate to the bone marrow and continue to secrete antibodies long after clearance of the immunizing antigen (Connor et al., 2002). For most of their lifespans, B cells express membrane-bound immunoglobulin, and differentiation into plasma cells dedicated to immunoglobulin secretion demands significant phenotypic changes (Pengo et al., 2013). Plasma cells significantly increase their endoplasmic reticulum (ER) network, and concomitantly reduce their mitochondrial content and activity (Jang et al., 2015; Pengo et al., 2013). Plasma cells face heightened ER stress compared to other B cell subsets due to their elevated synthesis and secretion of immunoglobulins, and the inevitable accumulation of misfolded immunoglobulins during this process (Pengo et al., 2013). Autophagy has been reported to degrade superfluous ER, as well as to eliminate misfolded proteins (Hoyer-Hansen and Jaattela, 2007). Through a mouse model with B cell-conditional deletion of an essential autophagy gene Atg5, both short-lived and long-lived plasma cells have been demonstrated to require autophagy in order to manage and survive the ER stress induced by their role as dedicated immunoglobulin-secreting cells (Pengo et al., 2013). Atg5−/−plasma cells exhibit an expanded ER network and increased immunoglobulin synthesis compared to their wild-type counterparts (Pengo et al., 2013). However, Atg5−/−plasma cells also display increased ER stress signaling, reduced ATP levels and decreased survival, indicating that plasma cells use autophagy both to generate metabolic substrates and to constrain immunoglobulin production in order to maintain viability (Oliva and Cenci, 2014; Pengo et al., 2013). These findings are also applicable to various disease settings where plasma cells play an important role. In infectious and non-infectious intestinal inflammation models, protective antibody responses by plasma cells are defective in the absence of autophagy (Conway et al., 2013). Loss of autophagy impairs plasma cell differentiation and autoantibody production in lupus models, indicating that autophagy can be targeted to ameliorate certain autoimmune disease phenotypes (Arnold et al., 2016; Clarke et al., 2015).
Zbtb32, a transcription factor critical for the proliferative burst of NK cells responding to viral infection, is highly expressed in memory B cells compared to naïve B cells (Jash et al., 2016). Zbtb32−/− mice exhibit normal primary antibody responses, but undergo more rapid and prolonged production and maintenance of secondary plasma cells, indicating that Zbtb32 serves to keep secondary antibody responses under control (Jash et al., 2016). Compared to wild-type controls, Zbtb32−/− secondary plasma cells in the bone marrow were found to express elevated levels of genes encoding mitochondrial ribosomal proteins, which are required for translation of electron transport chain proteins (Jash et al., 2016). B cells activated under conditions of plasma cell differentiation increase glucose uptake through up-regulation of the glucose transporter Glut1, which stimulates glycolysis and increases electron transport chain activity (Table 1) (Caro-Maldonado et al., 2014). Long-lived plasma cells undergo further metabolic adaptation distinct from short-lived plasma cells, which allows them to prolong their survival (Lam et al., 2016). Relative to short-lived plasma cells, long-lived plasma cells have similar basal oxidative phosphorylation activity, but substantially greater maximal capacity for oxidative phosphorylation (Lam et al., 2016). Long-lived plasma cells and short-lived plasma cells have similar levels of mitochondria, so increased spare respiratory capacity of long-lived plasma cells is due to a difference in substrate uptake (Lam et al., 2016). Indeed, long-lived plasma cells can be distinguished from short-lived plasma cells by their increased expression of Glut1 and the mitochondrial pyruvate carrier (Lam et al., 2016). As a result, long-lived plasma cells exhibit constitutively increased glucose uptake and mitochondrial pyruvate import compared to short-lived plasma cells (Table 1) (Lam et al., 2016). Increasing the diversity and quantity of substrates for mitochondrial activity allows long-lived plasma cells to switch between different metabolic states, and therefore survive when there are fluctuations in the local environment.
Gut mucosal humoral immunity is dominated by IgA-secreting plasma cells, which are generated in the intestinal mucosa in response to intestinal antigens (Kunisawa et al., 2015). The differentiation of naive B cells in Peyer’s patches to IgA+ plasma cells in the lamina propria of the intestine involves a metabolic shift; intestinal naïve B cells and plasma cells both use the Krebs cycle, but only intestinal plasma cells exhibit high glycolytic activity (Kunisawa et al., 2015). Interestingly, intestinal IgA+ plasma cells also have much higher levels of glycolysis compared to splenic plasma cells, indicating that their metabolic program is a product of their local environment, rather than being intrinsic to plasma cells (Kunisawa et al., 2015). Dietary depletion of vitamin B1 in mice inhibits the Krebs cycle without suppressing glycolysis (Kunisawa et al., 2015). Thus diet-induced vitamin B1 deficiency in mice results in a loss of naive B cells in Peyer’s patches, while IgA+ plasma cell maintenance is normal (Kunisawa et al., 2015).
Mitochondria and autophagy in memory B cell survival
The majority of GC B cells undergo apoptosis when antigens are cleared (Klein et al., 2003). However, a small fraction of the GC B cells are selected to survive and exit the germinal center to become long-lived, quiescent, antigen-experienced cells known as memory B cells (Chen et al., 2014; Klein et al., 2003). Memory B cells form the basis of humoral immunological memory, acting as sentinels primed to recognize and respond to previously encountered antigens (Chen et al., 2014; Tangye et al., 2003). Upon antigen re-exposure, memory B cells rapidly expand and differentiate into antibody-secreting plasma cells (Chen et al., 2014; Tangye et al., 2003). Naïve B cells, like memory B cells, are quiescent cells that can be stimulated by cognate antigen to divide and differentiate into plasma cells (Tangye et al., 2003). However, memory B cells differ from naïve B cells in a number of important respects. Memory B cells have a lower threshold for activation, requiring less antigen and costimulation. Additionally, memory B cells have a greater proliferative capacity, entering the cell cycle more rapidly upon activation and undergoing more rounds of division. Moreover, memory B cells are able to persist up to decades after formation, even in the absence of antigen stimulation (Arpin et al., 1997; Maruyama et al., 2000; Tangye et al., 2003). Therefore, the formation of memory B cells can lead to long-lasting protection against new infections after previous infections are cleared.
How are memory B cells able to survive over a prolonged period of time, in contrast to other B cell subsets? At least part of the answer can be found in the mitochondria. The Bcl-2 family of proteins regulates mitochondrial apoptosis, and includes both anti-apoptotic and pro-apoptotic members (Gross et al., 1999). Bcl-2 family proteins control permeabilization of the outer mitochondrial membrane, which culminates in the release of apoptosis-inducing molecules, such as cytochrome c, apoptosis-inducing factor (AIF), and endonuclease G (EndoG) (Arnoult et al., 2003; Chipuk and Green, 2008; Cregan et al., 2004). Bcl-2 inhibits outer mitochondrial membrane permeabilization, and thus acts as an anti-apoptotic member of the Bcl-2 family (Dlugosz et al., 2006). It is expressed at low levels in GC B cells, but becomes highly up-regulated in memory B cells (Bhattacharya et al., 2007; Smith et al., 1994). In mice, transgenic overexpression of Bcl-2 boosts recruitment of GC B cells into the memory B cell compartment, increases the size of the memory B cell compartment, and enhances the survival of adoptively transferred memory B cells in recipient mice (Nunez et al., 1991; Smith et al., 1994).
Memory B cells must balance long-term survival in a quiescent state with the need to be poised for rapid reactivation. This requires active and tightly regulated suppression of the cell cycle and differentiation programs, an energy-consuming process. Thus, in addition to blocking mitochondrial apoptosis, memory B cells are likely to alter their metabolic programs to prolong cell survival and to maintain their quiescence. Recently, the roles of AIF in B cell development, functions and cell death have been examined (Milasta et al., 2016). AIF is a mitochondrial intermembrane space protein that supports normal oxidative phosphorylation by promoting the assembly of complex I (NADH-ubiquinone oxidoreductase) of the electron transport chain (Milasta et al., 2016). However, AIF also mediates caspase-independent apoptosis when released from the mitochondria. After translocating to the nucleus, AIF induces widespread chromatin condensation and DNA fragmentation (Milasta et al., 2016). In a mouse model with B cell-specific deletion of AIF, early B cell development in the bone marrow and subsequent maturation in the spleen were found to be normal, despite a partial deficiency in complex I (Milasta et al., 2016). B cell functions, including proliferation and primary antibody production, were also found to be normal in the absence of AIF (Milasta et al., 2016). Moreover, despite the role of AIF as a cell death molecule, AIF deficiency did not rescue B cells from normal caspase-dependent and caspase-independent cell death (Milasta et al., 2016). These findings are in contrast with T cells which are regulated by AIF in their survival and function (Milasta et al., 2016). However, memory B cell survival and function in the absence of AIF has not been studied. Compared to naïve follicular B cells, memory B cells appear to have increased energy demands. As a result, memory B cells may be more susceptible to AIF deficiency and the consequent electron transport chain defects compared to naïve follicular B cells.
Unlike GC B cells, memory B cells cannot rely upon antigen, T cells and cytokine-rich niches in the germinal center for their maintenance. Instead, memory B cells have to survive in comparatively barren environments in the periphery (Maruyama et al., 2000). Autophagy, an evolutionarily conserved process that involves the sequestration of a portion of cytoplasm for lysosomal degradation, has been demonstrated to play a critical role in the long-term survival of memory B cells (Chen et al., 2014). Memory B cells exhibit increased basal levels of autophagy compared to both naïve and GC B cells (Chen et al., 2014). TLR ligands and cytokines may preferentially stimulate memory B cells compared to other B cell subsets; since TLR signaling has been reported to promote autophagy in B cells, it promote the long-term maintenance of the memory B cell compartment, though it does not appear to be required (Bernasconi et al., 2002; Meyer-Bahlburg et al., 2007; Pengo et al., 2013). In a mouse model with B cell-specific deletion of the essential autophagy gene Atg7, early B cell development and naïve and GC B cell numbers are normal. However, memory B cell survival is severely impaired in these mice, although memory B cell formation is not affected (Chen et al., 2014; Chen et al., 2015). The decreased survival of Atg7−/− memory B cells is not due to an increase in caspase activity. Instead, mitochondrial function appears to be perturbed in these cells, which exhibit reduced mitochondrial membrane potential and increased mitochondrial ROS production (Chen et al., 2014). In vivo treatment with the ROS scavenger NAC partially rescues the survival of Atg7−/− memory B cells, indicating that elevated mitochondrial ROS levels contribute to the impaired maintenance of Atg7−/ −memory B cells (Chen et al., 2014). The role of autophagy in memory B cells can be compared and contrasted to its role in another important population of memory lymphocytes, memory CD8+ T cells. Impaired autophagy, in both genetic and aging models, results in decreased numbers of memory CD8+ T cells, similar to what is observed with memory B cells (Puleston et al., 2014; Schlie et al., 2015; Xu et al., 2014). However, whereas autophagy is required for the survival of memory B cells, but not their formation, loss of autophagy does impair the formation of memory CD8+ T cells, due to compromised survival at the effector T cell stage (Chen et al., 2015; Puleston et al., 2014; Schlie et al., 2015; Xu et al., 2014). The differentiation of effector CD8+ T cells into memory CD8+ T cells involves a metabolic switch from glycolysis to mitochondrial fatty acid oxidation (Cui et al., 2015; O'Sullivan et al., 2014). In contrast, the metabolic transition of GC B cells to memory B cells is unclear. However, if memory B cells also rely upon fatty acid oxidation, autophagy may, in addition to an antioxidant role, also generate the lipid substrates required to drive this pathway.
While the molecular mechanisms by which autophagy protects memory B cells from cell death remain to be fully elucidated, the evidence suggests that autophagy regulates mitochondrial quality and quantity to maintain memory B cell survival (Figure 2). For many years, autophagy was thought to be a non-selective, bulk degradation process to recycle cellular components and generate nutrients under stress conditions (Zaffagnini and Martens, 2016). However, many specific forms of autophagy have been discovered over the past couple of decades, including mitochondrial autophagy, the selective clearance of mitochondria by autophagy (Sandoval et al., 2008; Schweers et al., 2007; Zaffagnini and Martens, 2016). Mitochondrial autophagy is critical for the development and survival of various cell types, including reticulocytes and memory natural killer (NK) cells (O'Sullivan et al., 2015; Sandoval et al., 2008; Schweers et al., 2007). Deficiencies in mitochondrial autophagy may lead to accumulation of dysfunctions mitochondria to induce accelerated cell death in memory B cells (Figure 2). Recently, hematopoietic stem cells have been found to prune healthy mitochondria through autophagy, maintaining mitochondrial homeostasis in order to prevent excessive metabolic activity that could trigger unwanted differentiation (Ho et al., 2017). Memory B cells may be comparable to hematopoietic stem cells due to their common properties of prolonged lifespan, self-renewal, and quiescence (Lucky et al., 2006). Given their molecular and phenotypic similarities to hematopoietic stem cells, it is probable that memory B cells also depend upon mitochondrial homeostasis for their survival and self-renewal. Mitochondrial homeostasis has been implicated in T and B cell differentiation (Adams et al., 2016). LPS-stimulated B cells can differentiate into plasma cells, which repress the B cell lineage commitment transcription factor Pax5 (Adams et al., 2016). However, LPS-stimulated B cell populations are not homogenous, consisting of both Pax5hi and Pax5lo populations (Adams et al., 2016). The Pax5lo subset is irreversibly committed to plasma cell differentiation, whereas the Pax5hi subset can undergo self-renew and generate Pax5lo progeny. Pharmacological inhibition of mitochondrial clearance in LPS-activated B cells increases the proportion of Pax5lo cells (Adams et al., 2016). This phenotype can be countered by treatment with the ROS scavenger NAC (Adams et al., 2016). Therefore, mitochondrial ROS may be a powerful signal to regulate B cell differentiation (Table 1).
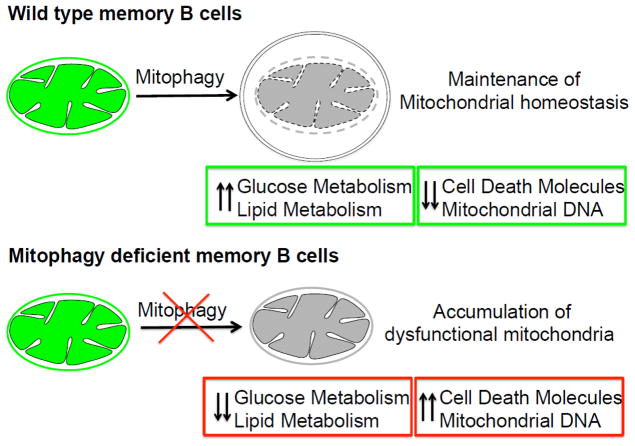
Figure 2. Model of maintenance of memory B cells by mitochondrial autophagy.
Maintenance of mitochondrial homeostasis by autophagy contributes to the long-term persistence of memory B cells. Mitochondrial autophagy possibly serves as a regulator of mitochondrial homeostasis by removing superfluous or dysfunctional mitochondria to prevent the release of cell death molecules, mitochondrial DNA, and to maintain a quiescent metabolic state of glucose and lipid metabolism. In autophagy-deficient memory B cells, accumulation of mitochondria could lead to increased release of cell death molecules and the imbalance of quiescent metabolic program.
Future Directions
B cells can transit between rapidly proliferating and quiescent states to meet the demands of the adaptive immune response. Memory B cells and long-lived plasma cells, the mediators of long-term protective immunity, can persist for many years in humans. Memory B cells can recirculate and also undergo self-renewal, similar to long-lived hematopoietic stem cells, to prolong their survival (Boothby and Rickert, 2017; Luckey et al., 2005). Regulation of mitochondrial homeostasis and functions is likely to be critical for the development of B cell lineages, as well as maintenance of the quiescent status of memory B cells and B cells at other differentiation stages. Mitochondria respond to a variety of nutritional cues and metabolic demands to generate sufficient energy to either drive differentiation or maintenance. Mitochondria and autophagy work in concert to respond to both catabolic and anabolic demands of the cells. Together they provide the metabolic milieu that can promote, inhibit, or possibly kill the cells.
We still have a number of questions to address in terms of the metabolic needs for B cell lineages development and the maintenance of B cell quiescence. Changing the metabolic programming from catabolism to anabolism and vice versa shifts the B cell lineages to generate either more plasma cells or memory cells. Interestingly, Gsk3 inhibits the mTOR pathway in a manner dependent on the serine/threonine kinase AMP-activated protein kinase (AMPK) phosphorylation (Inoki et al., 2006). Although in Gsk3-deficient GC B cells mTOR was not affected, it raises the question whether AMPK pathway is altered (Jellusova et al., 2017). AMPK is a great candidate for the regulation of metabolic programming in B cell lineages due to its ability to regulate lipid and glucose metabolism, autophagy, and mitochondrial homeostasis/mitophagy (Herzig and Shaw, 2017).
Also, the interplay of both the mitochondria and autophagosomes in different B cell fate decisions need to be further elucidated. For example, is there a regulatory role for mitochondrial autophagy in B cell effector/memory maintenance (Figure 2)? Again, of both these organelles are in the metabolic trenches allowing B cells to shift and survive under different developmental and environmental stresses. Interestingly, both organelles contact other organelles such as the ER, lysosomes, and lipid droplets to allow for an effective and precise metabolic response. These contacts allow for signaling platforms to accelerate the metabolic needs of the cells. Further studies will need to be performed to understand the sophisticated nature of such contact-based signaling.
Understanding the metabolic requirements for individual B cell lineages will open new therapeutic avenues to induce and maintain individual B cell lineages for treatment of different disorders and vaccine development.
Highlights.
Differentiating B cells undergo lineage-dependent metabolic reprogramming regulated by mitochondria and autophagy
Mitochondrial ROS serves as a signaling molecule to regulate fate commitment
The regulation of mitochondrial activity and autophagy by metabolic regulators
Local environment modulates the metabolic program of B cells
Autophagy is essential for survival of long-lived plasma and memory B cells
B cell lineages rely on the interplay of mitochondria and autophagy for development, activation, and survival
Acknowledgments
Our work was supported by grants from the NIH (R01AI116644 and R01AI123221) and the Cancer Research Institute of Texas (RP160384).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams WC, Chen YH, Kratchmarov R, Yen B, Nish SA, Lin WW, Rothman NJ, Luchsinger LL, Klein U, Busslinger M, et al. Anabolism-Associated Mitochondrial Stasis Driving Lymphocyte Differentiation over Self-Renewal. Cell Rep. 2016;17:3142–3152. doi: 10.1016/j.celrep.2016.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn CS, Metallo CM. Mitochondria as biosynthetic factories for cancer proliferation. Cancer Metab. 2015;3:1. doi: 10.1186/s40170-015-0128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J, Murera D, Arbogast F, Fauny JD, Muller S, Gros F. Autophagy is dispensable for B-cell development but essential for humoral autoimmune responses. Cell Death Differ. 2016;23:853–864. doi: 10.1038/cdd.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult D, Gaume B, Karbowski M, Sharpe J, Cecconi F, Youle R. Mitochondrial release of AIF and EndoG requires caspase activation downstream of Bax/Bak-mediated permeabilization. The EMBO Journal. 2003;22:4385–4399. doi: 10.1093/emboj/cdg423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpin C, Banchereau J, Liu Y. Memory B cells are biased towards terminal differentiation: a strategy that may prevent repertoire freezing. Journal of Experimental Medicine. 1997;186:931–940. doi: 10.1084/jem.186.6.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi NL, Traggiai E, Lanzavecchi A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298:2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Eissa NT. Autophagy as a Stress Response Pathway in the Immune System. Int Rev Immunol. 2015;34:382–402. doi: 10.3109/08830185.2014.999156. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Cheah MT, Franco CB, Hosen N, Pin CL, Sha WC, Weissman IL. Transcriptional Profiling of Antigen-Dependent Murine B Cell Differentiation and Memory Formation. The Journal of Immunology. 2007;179:6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothby M, Rickert RC. Metabolic Regulation of the Immune Humoral Response. Immunity. 2017;46:743–755. doi: 10.1016/j.immuni.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, Sowell RT, Kaech SM, Pearce EL. Metabolic Instruction of Immunity. Cell. 2017;169:570–586. doi: 10.1016/j.cell.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullon P, Marin-Aguilar F, Roman-Malo L. AMPK/Mitochondria in Metabolic Diseases. EXS. 2016;107:129–152. doi: 10.1007/978-3-319-43589-3_6. [DOI] [PubMed] [Google Scholar]
- Caro-Maldonado A, Wang R, Nichols AG, Kuraoka M, Milasta S, Sun LD, Gavin AL, Abel ED, Kelsoe G, Green DR, et al. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J Immunol. 2014;192:3626–3636. doi: 10.4049/jimmunol.1302062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao T, Wang H, Ho PC. Mitochondrial Control and Guidance of Cellular Activities of T Cells. Front Immunol. 2017;8:473. doi: 10.3389/fimmu.2017.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Hong MJ, Sun H, Wang L, Shi X, Gilbert BE, Corry DB, Kheradmand F, Wang J. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nat Med. 2014;20:503–510. doi: 10.1038/nm.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Kodali S, Jang A, Kuai L, Wang J. Requirement for autophagy in the long-term persistence but not initial formation of memory B cells. J Immunol. 2015;194:2607–2615. doi: 10.4049/jimmunol.1403001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18:157–164. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Ahn AK, Bhargava P, Lee CH, Eischen CM, McGuinness O, Boothby M. Glycolytic rate and lymphomagenesis depend on PARP14, an ADP ribosyltransferase of the B aggressive lymphoma (BAL) family. Proc Natl Acad Sci U S A. 2011;108:15972–15977. doi: 10.1073/pnas.1017082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Raybuck AL, Stengel K, Wei M, Beck TC, Volanakis E, Thomas JW, Hiebert S, Haase VH, Boothby MR. Germinal centre hypoxia and regulation of antibody qualities by a hypoxia response system. Nature. 2016;537:234–238. doi: 10.1038/nature19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MR, Mandal M, Ochiai K, Singh H. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat Rev Immunol. 2014;14:69–80. doi: 10.1038/nri3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AJ, Ellinghaus U, Cortini A, Stranks A, Simon AK, Botto M, Vyse TJ. Autophagy is activated in systemic lupus erythematosus and required for plasmablast development. Ann Rheum Dis. 2015;74:912–920. doi: 10.1136/annrheumdis-2013-204343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2011;13:7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- Connor B, Cascalho M, Noelle R. Short-lived and long-lived bone marrow plasma cells are derived from a novel precursor population. Journal of Experimental Medicine. 2002;195:737–745. doi: 10.1084/jem.20011626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway KL, Kuballa P, Khor B, Zhang M, Shi HN, Virgin HW, Xavier RJ. ATG5 regulates plasma cell differentiation. Autophagy. 2013;9:528–537. doi: 10.4161/auto.23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregan SP, Dawson VL, Slack RS. Role of AIF in caspase-dependent and caspase-independent cell death. Oncogene. 2004;23:2785–2796. doi: 10.1038/sj.onc.1207517. [DOI] [PubMed] [Google Scholar]
- Cui G, Staron MM, Gray SM, Ho PC, Amezquita RA, Wu J, Kaech SM. IL-7-Induced Glycerol Transport and TAG Synthesis Promotes Memory CD8+ T Cell Longevity. Cell. 2015;161:750–761. doi: 10.1016/j.cell.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugosz P, Billen L, Annis M, Zhu W, Zhang Z, Lin J, Leber B, Andrews D. Bcl-2 changes conformation to inhibit Bax oligomerization. The EMBO Journal. 2006;25:2287–2296. doi: 10.1038/sj.emboj.7601126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D, Kung J, Holmes AB, Wells VA, Mo T, Basso K, Dalla-Favera R. The FOXO1 Transcription Factor Instructs the Germinal Center Dark Zone Program. Immunity. 2015;43:1064–1074. doi: 10.1016/j.immuni.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Donahue AC, Fruman DA. Proliferation and Survival of Activated B Cells Requires Sustained Antigen Receptor Engagement and Phosphoinositide 3-Kinase Activation. The Journal of Immunology. 2003;170:5851–5860. doi: 10.4049/jimmunol.170.12.5851. [DOI] [PubMed] [Google Scholar]
- Dufort FJ, Bleiman BF, Gumina MR, Blair D, Wagner DJ, Roberts MF, Abu-Amer Y, Chiles TC. Cutting edge: IL-4-mediated protection of primary B lymphocytes from apoptosis via Stat6-dependent regulation of glycolytic metabolism. J Immunol. 2007;179:4953–4957. doi: 10.4049/jimmunol.179.8.4953. [DOI] [PubMed] [Google Scholar]
- Gross A, McDonnell J, Korsmeyer S. BCL-2 family members and the mitochondria in apoptosis. Genes and Development. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- Hamel KM, Mandal M, Karki S, Clark MR. Balancing Proliferation with Igkappa Recombination during B-lymphopoiesis. Front Immunol. 2014;5:139. doi: 10.3389/fimmu.2014.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heizmann B, Kastner P, Chan S. Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J Exp Med. 2013;210:2823–2832. doi: 10.1084/jem.20131735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2017 doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- Ho TT, Warr MR, Adelman ER, Lansinger OM, Flach J, Verovskaya EV, Figueroa ME, Passegue E. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543:205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehn KB, Fowler A, Lunter G, Pybus OG. The Diversity and Molecular Evolution of B-Cell Receptors during Infection. Mol Biol Evol. 2016;33:1147–1157. doi: 10.1093/molbev/msw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman W, Lakkis FG, Chalasani G. B Cells, Antibodies, and More. Clin J Am Soc Nephrol. 2016;11:137–154. doi: 10.2215/CJN.09430915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes ML, Pridans C, Nutt SL. The regulation of the B–cell gene expression programme by Pax5. Immunol Cell Biol. 2008;86:47–53. doi: 10.1038/sj.icb.7100134. [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Inoue T, Shinnakasu R, Ise W, Kawai C, Egawa T, Kurosaki T. The transcription factor Foxo1 controls germinal center B cell proliferation in response to T cell help. J Exp Med. 2017;214:1181–1198. doi: 10.1084/jem.20161263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KJ, Mano H, Aoki K, Hayashi T, Muto A, Nambu Y, Takahashi K, Itoh K, Taketani S, Nutt SL, et al. Mitochondrial function provides instructive signals for activation-induced B-cell fates. Nat Commun. 2015;6:6750. doi: 10.1038/ncomms7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jash A, Wang Y, Weisel FJ, Scharer CD, Boss JM, Shlomchik MJ, Bhattacharya D. ZBTB32 Restricts the Duration of Memory B Cell Recall Responses. J Immunol. 2016;197:1159–1168. doi: 10.4049/jimmunol.1600882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellusova J, Cato MH, Apgar JR, Ramezani-Rad P, Leung CR, Chen C, Richardson AD, Conner EM, Benschop RJ, Woodgett JR, et al. Gsk3 is a metabolic checkpoint regulator in B cells. Nat Immunol. 2017;18:303–312. doi: 10.1038/ni.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Klein U, Tu Y, Stolovitzky G, Keller J, Haddad J, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. Transcriptional analysis of the B cell germinal center reaction. Proceedings of the National Academy of Sciences. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012;30:611–646. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- Kunisawa J, Sugiura Y, Wake T, Nagatake T, Suzuki H, Nagasawa R, Shikata S, Honda K, Hashimoto E, Suzuki Y, et al. Mode of Bioenergetic Metabolism during B Cell Differentiation in the Intestine Determines the Distinct Requirement for Vitamin B1. Cell Rep. 2015;13:122–131. doi: 10.1016/j.celrep.2015.08.063. [DOI] [PubMed] [Google Scholar]
- Lam WY, Becker AM, Kennerly KM, Wong R, Curtis JD, Llufrio EM, McCommis KS, Fahrmann J, Pizzato HA, Nunley RM, et al. Mitochondrial Pyruvate Import Promotes Long-Term Survival of Antibody-Secreting Plasma Cells. Immunity. 2016;45:60–73. doi: 10.1016/j.immuni.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman GW, Rast JP, Fugmann SD. The origins of vertebrate adaptive immunity. Nat Rev Immunol. 2010;10:543–553. doi: 10.1038/nri2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckey CJ, Bhattacharya B, Goldrath AW, Weissman IL, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc Natl Acad Sci U S A. 2005;103:3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucky C, Bhattacharya D, Goldrath A, Weissman I, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proceedings of the National Academy of Sciences. 2006;103:3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Martin N, Maldonado P, Gasparrini F, Frederico B, Aggarwal S, Gaya M, Tsui C, Burbage M, Keppler SJ, Montaner B, et al. A switch from canonical to noncanonical autophagy shapes B cell responses. Science. 2017;355:641–647. doi: 10.1126/science.aal3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama M, Lam K, Rajewsky K. Memory B cell persistence is independent of persisting immunizing antigen. Nature. 2000;407:636–641. doi: 10.1038/35036600. [DOI] [PubMed] [Google Scholar]
- Melchers F. Checkpoints that control B cell development. J Clin Invest. 2015;125:2203–2210. doi: 10.1172/JCI78083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Bahlburg A, Khim S, Rawlings DJ. B cell intrinsic TLR signals amplify but are not required for humoral immunity. J Exp Med. 2007;204:3095–3101. doi: 10.1084/jem.20071250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milasta S, Dillon CP, Sturm OE, Verbist KC, Brewer TL, Quarato G, Brown SA, Frase S, Janke LJ, Perry SS, et al. Apoptosis-Inducing-Factor-Dependent Mitochondrial Function Is Required for T Cell but Not B Cell Function. Immunity. 2016;44:88–102. doi: 10.1016/j.immuni.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez G, Hockenberry D, McDonnell T, Sorensen C, Korsmeyer S. Bcl-2 maintains B cell memory. Nature. 1991;353:71–73. doi: 10.1038/353071a0. [DOI] [PubMed] [Google Scholar]
- O'Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, Qiu J, Smith AM, Lam WY, DiPlato LM, et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan TE, Johnson LR, Kang HH, Sun JC. BNIP3- and BNIP3L-Mediated Mitophagy Promotes the Generation of Natural Killer Cell Memory. Immunity. 2015;43:331–342. doi: 10.1016/j.immuni.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai K, Maienschein-Cline M, Mandal M, Triggs JR, Bertolino E, Sciammas R, Dinner AR, Clark MR, Singh H. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol. 2012;13:300–307. doi: 10.1038/ni.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva L, Cenci S. Autophagy in plasma cell pathophysiology. Front Immunol. 2014;5:103. doi: 10.3389/fimmu.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra D, Takizawa F, Sunyer JO. Evolution of B cell immunity. Annu Rev Anim Biosci. 2013;1:65–97. doi: 10.1146/annurev-animal-031412-103651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengo N, Scolari M, Oliva L, Milan E, Mainoldi F, Raimondi A, Fagioli C, Merlini A, Mariani E, Pasqualetto E, et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013;14:298–305. doi: 10.1038/ni.2524. [DOI] [PubMed] [Google Scholar]
- Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat Rev Immunol. 2009;9:767–777. doi: 10.1038/nri2656. [DOI] [PubMed] [Google Scholar]
- Puleston DJ, Simon AK. Autophagy in the immune system. Immunology. 2013;141:1–8. doi: 10.1111/imm.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, Watson AS, Cerundolo V, Townsend AR, Klenerman P, et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. Elife. 2014;3 doi: 10.7554/eLife.03706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell JC. Metabolism and autophagy in the immune system: immunometabolism comes of age. Immunol Rev. 2012;249:5–13. doi: 10.1111/j.1600-065X.2012.01158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander S, Chu VT, Yasuda T, Franklin A, Graf R, Calado DP, Li S, Imami K, Selbach M, Di Virgilio M, et al. PI3 Kinase and FOXO1 Transcription Factor Activity Differentially Control B Cells in the Germinal Center Light and Dark Zones. Immunity. 2015;43:1075–1086. doi: 10.1016/j.immuni.2015.10.021. [DOI] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlie K, Westerback A, DeVorkin L, Hughson LR, Brandon JM, MacPherson S, Gadawski I, Townsend KN, Poon VI, Elrick MA, et al. Survival of effector CD8+ T cells during influenza infection is dependent on autophagy. J Immunol. 2015;194:4277–4286. doi: 10.4049/jimmunol.1402571. [DOI] [PubMed] [Google Scholar]
- Schroeder HW, Jr, Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol. 2010;125:S41–52. doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela-Culang I, Kunik V, Ofran Y. The structural basis of antibody-antigen recognition. Front Immunol. 2013;4:302. doi: 10.3389/fimmu.2013.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, Schumacker PT, Licht JD, Perlman H, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart S, Venkitaraman A. Smart_Il-7 receptor BCR.pdf. Journal of Experimental Medicine. 2000;191:737–742. doi: 10.1084/jem.191.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Weiss U, Rajewsky K, Nossal G, Tarlinton D. Bcl-2 increases memory B cell recruitment but does not perturb selection in germinal centers. Immunity. 1994;1:803–813. doi: 10.1016/s1074-7613(94)80022-7. [DOI] [PubMed] [Google Scholar]
- Stein M, Dutting S, Mougiakakos D, Bosl M, Fritsch K, Reimer D, Urbanczyk S, Steinmetz T, Schuh W, Bozec A, et al. A defined metabolic state in pre B cells governs B-cell development and is counterbalanced by Swiprosin-2/EFhd1. Cell Death Differ. 2017 doi: 10.1038/cdd.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye SG, Avery DT, Deenick EK, Hodgkin PD. Intrinsic Differences in the Proliferation of Naive and Memory Human B Cells as a Mechanism for Enhanced Secondary Immune Responses. The Journal of Immunology. 2003;170:686–694. doi: 10.4049/jimmunol.170.2.686. [DOI] [PubMed] [Google Scholar]
- van der Windt GJ, Pearce EL. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol Rev. 2012;249:27–42. doi: 10.1111/j.1600-065X.2012.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42:406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler ML, Defranco AL. Prolonged production of reactive oxygen species in response to B cell receptor stimulation promotes B cell activation and proliferation. J Immunol. 2012;189:4405–4416. doi: 10.4049/jimmunol.1201433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykes M. Why do B cells produce CD40 ligand? Immunology and Cell Biology. 2003:328–331. doi: 10.1046/j.1440-1711.2003.01171.x. [DOI] [PubMed] [Google Scholar]
- Xu X, Araki K, Li S, Han JH, Ye L, Tan WG, Konieczny BT, Bruinsma MW, Martinez J, Pearce EL, et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat Immunol. 2014;15:1152–1161. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini G, Martens S. Mechanisms of Selective Autophagy. J Mol Biol. 2016;428:1714–1724. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Garcia-Ibanez L, Toellner KM. Regulation of germinal center B-cell differentiation. Immunological Reviews. 2016;270:8–19. doi: 10.1111/imr.12396. [DOI] [PMC free article] [PubMed] [Google Scholar]