Abstract
Tumor microenvironment plays a pivotal role in cancer progression; however, little is known regarding how differences in the microenvironment affect characteristics of cancer cells. Here, we investigated the effects of tumor microenvironment on cancer cells by using mouse tumor models. After three cycles of inoculation and extraction of human pancreatic cancer cells, including SUIT-2 and Panc-1 cells, from tumors, distinct cancer cell lines were established: 3P cells from the pancreas obtained using the orthotopic tumor model and 3sc cells from subcutaneous tissue obtained using the subcutaneous tumor model. On re-inoculation of these cells, the 3sc cells and, more prominently, the 3P cells, exhibited higher tumorigenic activity than the parental cells. The 3P cells specifically exhibited low E-cadherin expression and high invasiveness, suggesting that they were endowed with the highest malignant characteristics. RNA-sequence analysis demonstrated that distinct signaling pathways were activated in each cell line and that the 3P cells acquired a cancer stem cell-like phenotype. Among cancer stem cell-related genes, those specifically expressed in the 3P cells, including NES, may be potential new targets for cancer therapy. The mechanisms underlying the development of highly malignant cancer cell lines were investigated. Individual cell clones within the parental cells varied in tumor-forming ability, indicating the presence of cellular heterogeneity. Moreover, the tumor-forming ability and the gene expression profile of each cell clone were altered after serial orthotopic inoculations. The present study thus suggests that both selection and education processes by tumor microenvironment are involved in the development of highly malignant cancer cells.
Introduction
Although considerable advances have been made in technologies for diagnosis and treatment of cancer, the 5-year survival rate of patients with pancreatic cancer is still less than 10% [1, 2]. Pancreatic cancer cells exhibit high metastatic ability and resistance to antitumor drugs [2]. KRAS mutations, which are found in over 90% of pancreatic cancer cases, are considered to be a driver of the tumorigenesis in pancreatic cancer [3, 4]. In addition, deletions or inactivating mutations in several genes, including CDKN2A, TP53, and SMAD4, are frequently observed in advanced pancreatic cancer [3, 4]. Although the accumulation of these gene alterations contributes to development of pancreatic cancer, some other mechanisms, including the effects of tumor microenvironment on cancer cells, are thought to be involved in the acquisition of high malignant potential of pancreatic cancer cells.
Tumor microenvironment consists of cancer cells as well as various other types of cells in the tumor stroma, including fibroblasts, immune cells, and cells that comprise the blood vessels [5]. In addition, tumor microenvironment includes the proteins that are produced by these cells and support the growth of cancer cells, such as extracellular matrix (ECM) proteins [6]. The interactions between cancer cells and stromal cells have been considered important for cancer progression [5–7]. Abundant stroma, which is a histological hallmark of pancreatic cancer, contributes to the production of growth factors, secretion of ECM proteins, and the activation of fibroblasts [2, 8]. Tumor-promoting roles of the pancreatic tumor microenvironment have previously been reported. Fibroblasts stimulate the invasion and proliferation of cancer cells through the action of growth factors [9]. Co-injection of pancreatic cancer cells with pancreatic stellate cells has been reported to enhance their tumor-forming ability in vivo [10, 11]. In addition, the immune response to cancer cells is suppressed in the pancreatic tumor microenvironment by the infiltration of regulatory T cells [2]. Since ECM inhibits the delivery of antitumor drugs, the tumor stroma is thought to be a therapeutic target for cancer [2, 12]. However, recent studies using genetically engineered mouse tumor models have also demonstrated a tumor-suppressive role of the pancreatic microenvironment [13, 14]. Thus, the roles of tumor microenvironment in pancreatic cancer are complicated and still remain controversial.
To investigate the interactions between cancer cells and stromal cells, co-culture assays have been frequently used [9]. Recently, the usefulness of orthotopic tumor models have been recognized, because tumor microenvironments affect the properties of cancer cells, and orthotopic tumor models mimic the effects of tumor microenvironments more effectively than ectopic transplantation models. However, only a few studies have shown how interactions among cancer cells and different tissue microenvironments in vivo affect the properties of cancer cells. In this study, we compared the characteristics of cancer cells growing in different tumor microenvironments. In addition, the mechanisms by which pancreatic cancer cells gain highly malignant characteristics in the pancreatic tumor microenvironment were investigated.
Results
Monitoring pancreatic cancer progression in different tumor microenvironments
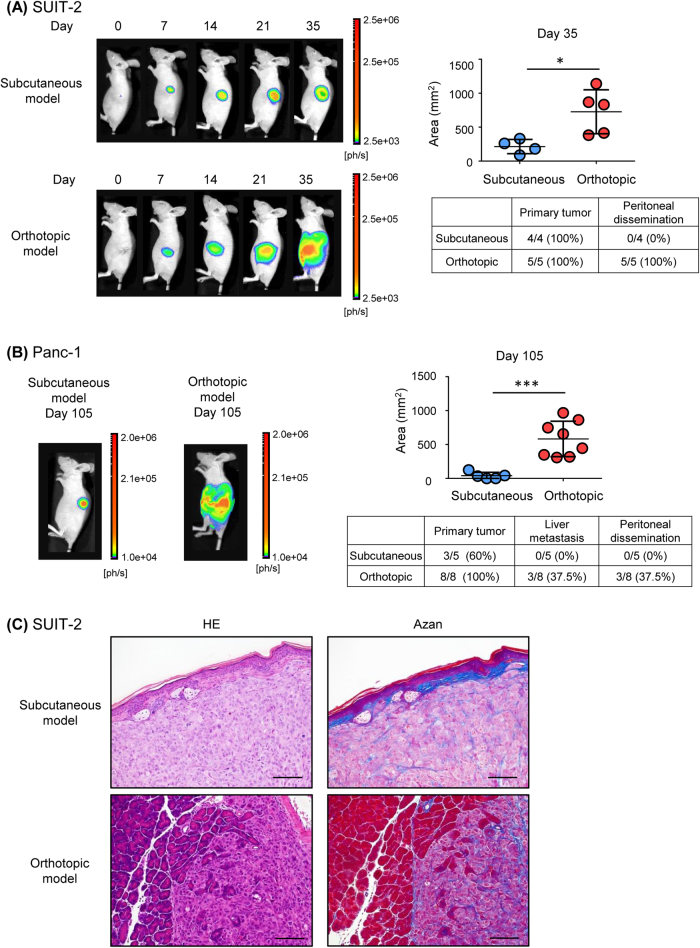
We first compared the pancreatic cancer progression in two different mouse tumor models. Human pancreatic cancer SUIT-2 cells, constitutively expressing firefly luciferase and green fluorescent protein (GFP), were ectopically inoculated into subcutaneous tissues (subcutaneous tumor model) or orthotopically into pancreatic tissues (orthotopic tumor model) in BALB/c-nu/nu mice. Although in vivo bioluminescence imaging revealed formation of primary tumors in both models, peritoneal dissemination was observed only in the orthotopic tumor model (Fig. 1a). Similar results were obtained in mouse tumor models with human pancreatic cancer Panc-1 cells (Fig. 1b). Primary tumors were observed in all mice in the orthotopic tumor model of Panc-1 cells, whereas not all mice developed primary tumors in the subcutaneous model of Panc-1 cells. In addition, liver metastasis and peritoneal dissemination were observed in some mice in the orthotopic tumor model with Panc-1 cells (Fig. 1b). Histological examination revealed that dermal tissue was located next to the inoculated cancer cells in the subcutaneous tumor model with SUIT-2 cells, while cancer cells in pancreatic tissue were close to normal pancreatic acinar cells in the orthotopic tumor model with SUIT-2 cells (Fig. 1c). Although the histological features were distinct between the two models, the proportion of Azan-positive areas did not apparently differ between the two tumor models (Fig. 1c). These observations suggested that interactions between cancer cells and surrounding stromal cells were activated in both tumor models.
Fig. 1.
Effects of the tumor microenvironment on tumor progression in pancreatic cancer cells. a Time-course analysis of mouse tumor models of SUIT-2 cells. An equal number of SUIT-2 cells was inoculated into subcutaneous tissue (subcutaneous tumor model; top left) or the pancreas (orthotopic tumor model; bottom left). Tumor progression was monitored using in vivo bioluminescence imaging. After the mice were killed, incidence of primary tumor formation and metastasis was confirmed by autopsy. The signal area (top right) and incidence (bottom right) of primary tumor formation and peritoneal dissemination at 35 d after inoculation are shown. b Analysis of the mouse tumor models of Panc-1 cells. An equal number of Panc-1 cells was inoculated into subcutaneous tissue (subcutaneous tumor model) or the pancreas (orthotopic tumor model; left). Tumor progression was monitored using in vivo bioluminescence imaging 105 d after inoculation. After the mice were killed, incidence of primary tumor formation and metastasis was confirmed by autopsy. The signal area in the primary tumor (top right) and the incidence of the primary tumor, liver metastasis, and peritoneal dissemination (bottom right) are shown. c Primary tumors were subjected to hematoxylin–eosin (HE) staining and Azan staining. Representative images are shown. Scale bars are 100 µm. Data are presented as mean ± SD (a, b). *P < 0.05, ***P < 0.001
Establishment of distinct pancreatic cancer cell lines from each mouse tumor model
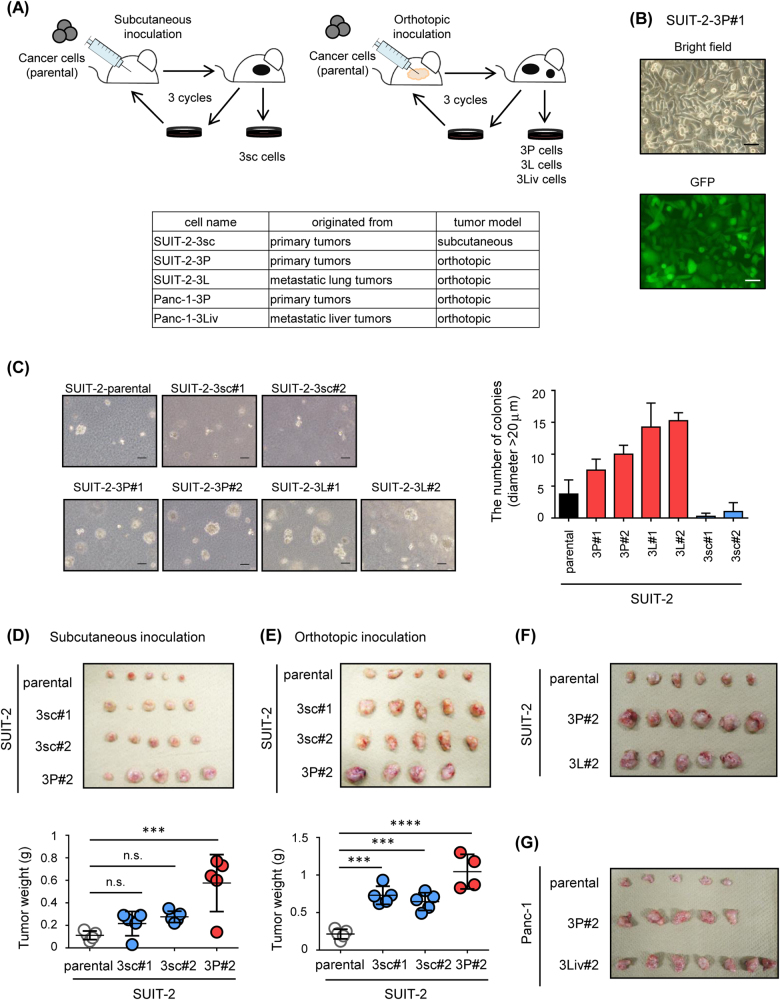
To investigate the characteristics of pancreatic cancer cells that interacted with the tumor microenvironments, distinct cancer cell lines were established with these mouse tumor models using serial transplantations (Fig. 2a). Cancer cells were obtained from primary tumors in the subcutaneous or orthotopic tumor model mice injected with parental SUIT-2 and Panc-1 cells, and then subjected to secondary transplantation. By performing three serial transplantations, the cancer cell lines were exposed to the pancreatic or subcutaneous microenvironment for a certain period. Elimination of host-derived fibroblasts was confirmed by the presence of GFP expression in the cancer cells (Fig. 2b and Supplementary Fig. S1).
Fig. 2.
Establishment of distinct pancreatic cancer cell lines using mouse tumor models. a Scheme of establishment of cancer cell lines using subcutaneous (left) and orthotopic tumor models (right). SUIT-2 cells were inoculated into subcutaneous tissue or the pancreas, and the primary tumor was excised. These processes were repeated three times. After three serial transplantations, distinct cell lines were established. The models of inoculation, the names of established cells, and the derived tissues are shown. b Expression of GFP in the SUIT-2-3P#1 cells was determined by fluorescence microscopy. Scale bars are 50 µm. c Colony formation assay of parental SUIT-2 cells and established cell lines. Cells were cultured in soft agar for 8 d. Representative image (left) and the number of colonies (diameter > 20 μm; right) are shown. Scale bars are 100 µm. d, e Tumor-forming ability of parental SUIT-2 cells and established cell lines. An equal number of cells was inoculated into subcutaneous tissue (d) or the pancreas (e). One month after inoculation, the primary tumor was excised (top) and tumor weight was measured (bottom). f, g Tumor-forming ability of established cell lines. Parental SUIT-2 cells and established cell lines (f) or parental Panc-1 cells and established cell lines (g) were inoculated into the pancreas. One month after inoculation, the primary tumor was excised. Image of primary tumors is shown. Data are presented as mean + SEM (c) and mean ± SD (d, e), respectively. ***P < 0.001, ****P < 0.0001, n.s., not significant
To maintain heterogeneity during serial transplantation procedures, cells obtained from two or three independent primary tumors were mixed, and inoculated into mice. We established more than two cell lines from each tumor model (#1, #2, etc.), and termed them as follows: SUIT-2-3P and Panc-1-3P cells from primary tumors in the orthotopic tumor model, SUIT-2-3L cells from metastatic lung tumors in the orthotopic tumor model, Panc-1-3Liv cells from metastatic liver tumors in the orthotopic tumor model, and SUIT-2-3sc cells from primary tumors in the subcutaneous tumor model (Fig. 2a). To determine whether the cancer cell lines established exhibited high malignant potential, their tumor-forming ability was examined. Colony formation assay revealed that SUIT-2-3P cells and SUIT-2-3L cells formed larger colonies than the parental SUIT-2 cells and SUIT-2-3sc cells (Fig. 2c). Moreover, when these cells were inoculated into mice, SUIT-2-3P cells exhibited the highest tumorigenic ability in the subcutaneous and orthotopic tumor models (Figs. 2d,e). The tumorigenic ability of SUIT-2-3L cells was similar to that of SUIT-2-3P cells in vivo (Fig. 2f). Similarly, Panc-1-3P and Panc-1-3Liv cells had higher tumorigenic ability in vivo than the parental Panc-1 cells (Fig. 2g). These results suggested that the pancreatic cancer cells, which interacted with the pancreatic microenvironment, but not with the ectopic microenvironment, acquired high malignant potential.
Characterization of the highly malignant pancreatic cancer cell lines
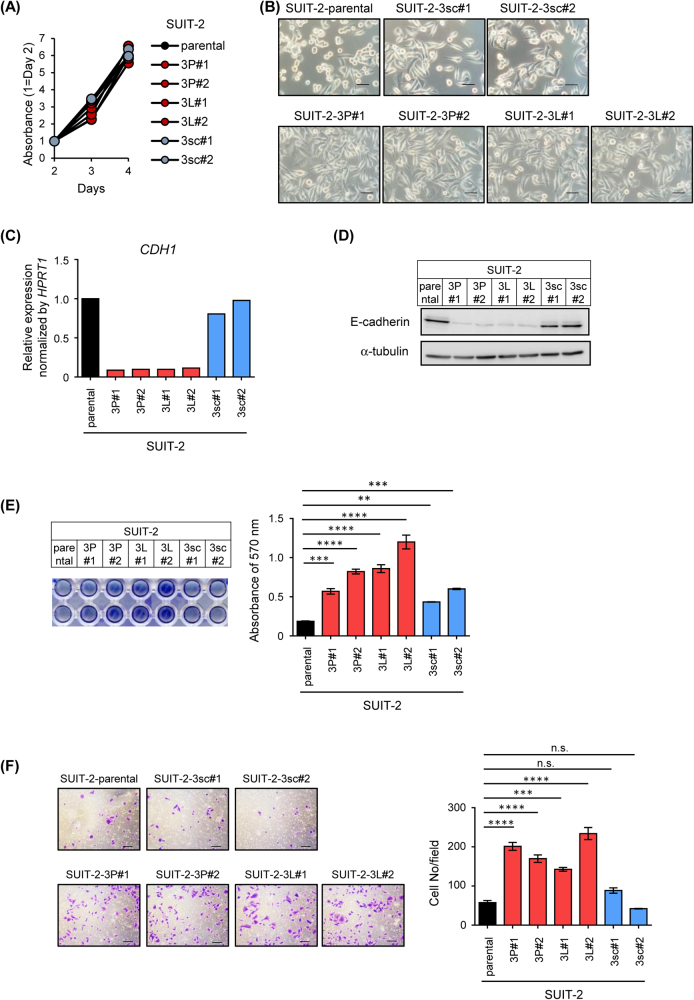
Characteristics of the 3sc and 3P cell lines from each pancreatic cancer cell were further investigated. The proliferative ability in cell culture did not significantly differ between the two cell lines and the parental SUIT-2 cells (Fig. 3a). The proliferative ability of Panc-1-3P cells and Panc-1-3Liv cells also did not differ from that of parental Panc-1 cells in cell culture (Supplementary Fig. S2A). However, microscopic examination revealed that the half of parental SUIT-2 cells were round-shaped cells, whereas the population of spindle-shaped cells increased in SUIT-2-3P and SUIT-2-3L cells (Fig. 3b). In addition, decrease in E-cadherin expression, a representative marker of the epithelial phenotype, was observed at mRNA and protein levels in SUIT-2-3P and SUIT-2-3L cells, but not in SUIT-2-3sc cells (Figs. 3c,d). Increased adhesive ability and cell motility were observed in SUIT-2-3P and SUIT-2-3L cells (Figs. 3e,f). Highly malignant cancer cell lines derived from Panc-1 were also characterized. In accordance with the results in SUIT-2 cells, the morphological features of Panc-1-3P cells differed from those of parental Panc-1 cells, and low E-cadherin expression was also observed in Panc-1-3P and Panc-1-3Liv cells (Supplementary Fig. S2B–D). These findings suggest that pancreatic cancer cells underwent epithelial–mesenchymal transition (EMT) and obtained invasive ability through interactions with the pancreatic microenvironment.
Fig. 3.
Characterization of highly malignant cancer cell lines derived from SUIT-2 cells. a Cell proliferation assay of the cell lines derived from SUIT-2 cells. Cells were seeded into 96-well plates and cultured for 2–4 d. Relative absorbance (450–595 nm) at the indicated days is shown. b Morphological features of the cell lines derived from SUIT-2 cells. Scale bars are 50 µm. c, d Expression of E-cadherin in the cell lines derived from SUIT-2 cells. Expression levels of CDH1 mRNA and amounts of E-cadherin protein were determined by qRT-PCR analysis (c) and immunoblotting (d), respectively. e Adhesion assay of the cell lines derived from SUIT-2 cells. Cells were seeded into fibronectin-coated 96-well plates under the FBS-free conditions and cultured for 30 min. The images of adhered cells (left) and the absorbance at 570 nm (right) are shown. f Chamber migration assay of the cell lines derived from SUIT-2 cells. Cells were seeded into the chamber and incubated for 24 h. The representative images (left) and the number of migrated cells (right) are shown. Scale bars are 100 µm. Data are presented as mean (duplicate; c) and mean ± SD (e, f), respectively. **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s., not significant
Gene signature of pancreatic cancer cell lines derived from mouse tumor models
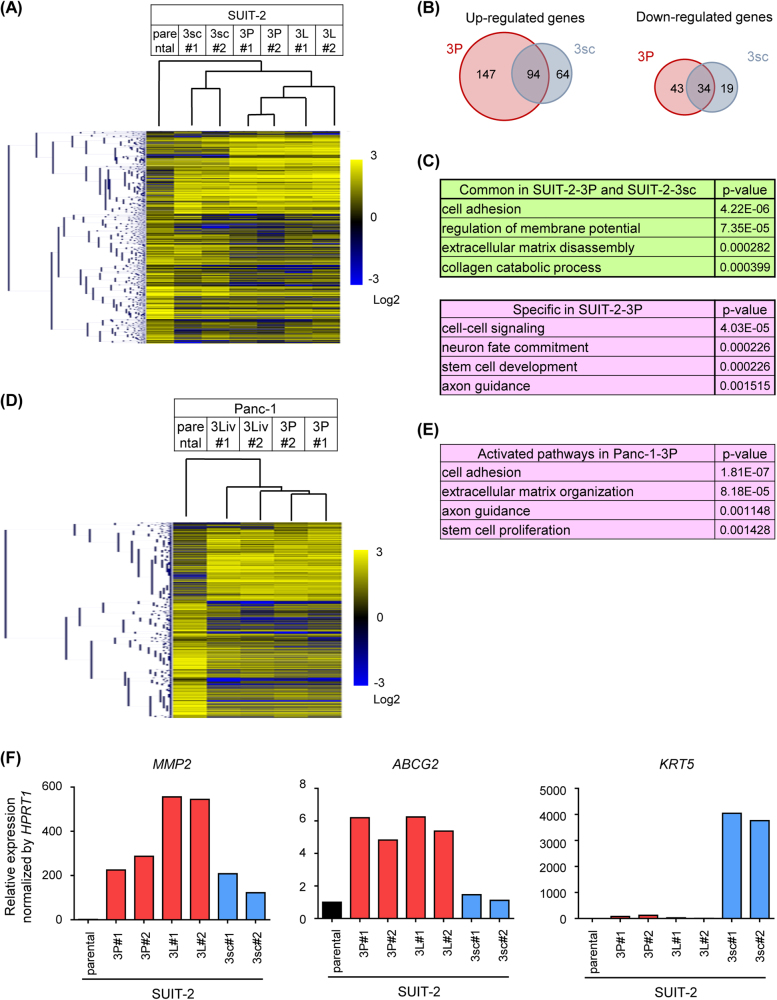
Gene expression in cell lines established from SUIT-2 cells and Panc-1 cells was profiled using RNA-sequence (RNA-seq) analysis. The gene expression signatures highly differed between SUIT-2-3sc cells and SUIT-2-3P cells (Fig. 4a). Clustering analysis showed that the profiles of parental SUIT-2 cells were similar to those of SUIT-2-3sc, SUIT-2-3P, and SUIT-2-3L cells in that order (Fig. 4a). The number of upregulated or downregulated genes specific to SUIT-2-3P cells was higher than that for SUIT-2-3sc cells (Fig. 4b). Among them, 94 genes were upregulated and 34 genes were downregulated in both SUIT-2-3P and SUIT-2-3sc cells. Biological processes extracted from this analysis revealed that distinct pathways were activated in each cell line. Pathways associated with ECM disassembly or collagen catabolic processes were commonly activated in SUIT-2-3P and SUIT-2-3sc cells, whereas pathways associated with stem cell development or axon guidance were activated specifically in SUIT-2-3P cells (Fig. 4c, see Supplementary Table S1). Clustering analysis of the cell lines derived from Panc-1 cells also showed that expression patterns in Panc-1-3Liv and Panc-1-3P cells highly differed from those in the parental Panc-1 cells (Fig. 4d). Similar results with SUIT-2-3P cells were obtained for ontology analysis of the activated pathways of Panc-1-3P cells (Fig. 4e, see Supplementary Table S2). These data suggested that different biological pathways were activated in each microenvironment, which may result in the acquisition of malignant traits of each cell line. The genes that were highly expressed in established cell lines can be categorized into three groups as shown in Fig. 4f. The genes in the first group, including matrix metalloproteinase-2 (MMP2), were expressed in SUIT-2-3P, SUIT-2-3L, and SUIT-2-3sc cells. The genes in the second group, including ATP-binding cassette subfamily G member 2 (ABCG2), were highly expressed only in SUIT-2-3P or SUIT-2-3L cells. The genes in the third group, including keratin-5 (KRT5), were highly expressed specifically in SUIT-2-3sc cells. Considering the previous data that SUIT-2-3P and SUIT-2-3L cells showed the highest malignant phenotype, the genes in the second group may be important for pancreatic cancer progression.
Fig. 4.
Gene expression profiles of pancreatic cancer cell lines. a Gene expression profiles of the cells lines derived from SUIT-2 cells using RNA-seq analysis. The result of the cluster analysis is shown by a dendrogram. b Number of upregulated and downregulated genes (left and right, respectively). The genes were purified as follows: upregulated genes: the fragments per kilobase of exon per million mapped sequence reads (FPKM) in SUIT-2-3P cells or SUIT-2-3sc cells were >3 and increased more than threefold, compared to the FPKM in parental SUIT-2; downregulated genes: the FPKM in parental SUIT-2 were >3 and decreased more than twofold, compared to the FPKM in SUIT-2-3P cells or SUIT-2-3sc cells. c Gene ontology analysis of upregulated genes in (b). Biological processes activated in both SUIT-2-3P and SUIT-2-3sc cells (top) and specifically in SUIT-2-3P cells (bottom) are shown. P-values for comparison between parental SUIT-2 and SUIT-2-3P cells are shown. d Gene expression profiles of the cell lines derived from Panc-1 cells obtained by RNA-seq analyses. The result of the cluster analysis is shown by a dendrogram. e Gene ontology analysis of upregulated genes in Panc-1 cells. Biological processes activated in Panc-1-3P cells are shown. P-values for comparison between parental Panc-1 and Panc-1-3P cells are shown. f Three patterns for upregulation of gene expression in each cancer cell line. Expression levels of MMP2, ABCG2, and KRT5 mRNA were determined by qRT-PCR analysis. Data are presented as mean (duplicate; f)
Roles of Nestin in pancreatic cancer progression
Gene ontology analyses showed that biological processes related to stem cell development or proliferation were activated specifically in the cell lines derived from the orthotopic models of SUIT-2 and Panc-1 (Figs. 4c,e). Specifically, RNA-seq analysis demonstrated that the expression of some stem cell markers increased, including CD24 in the cell lines from SUIT-2 and sex-determining region Y (SRY)-box 2 (SOX2) in the cell lines from Panc-1 (Supplementary Fig. S3A and B).
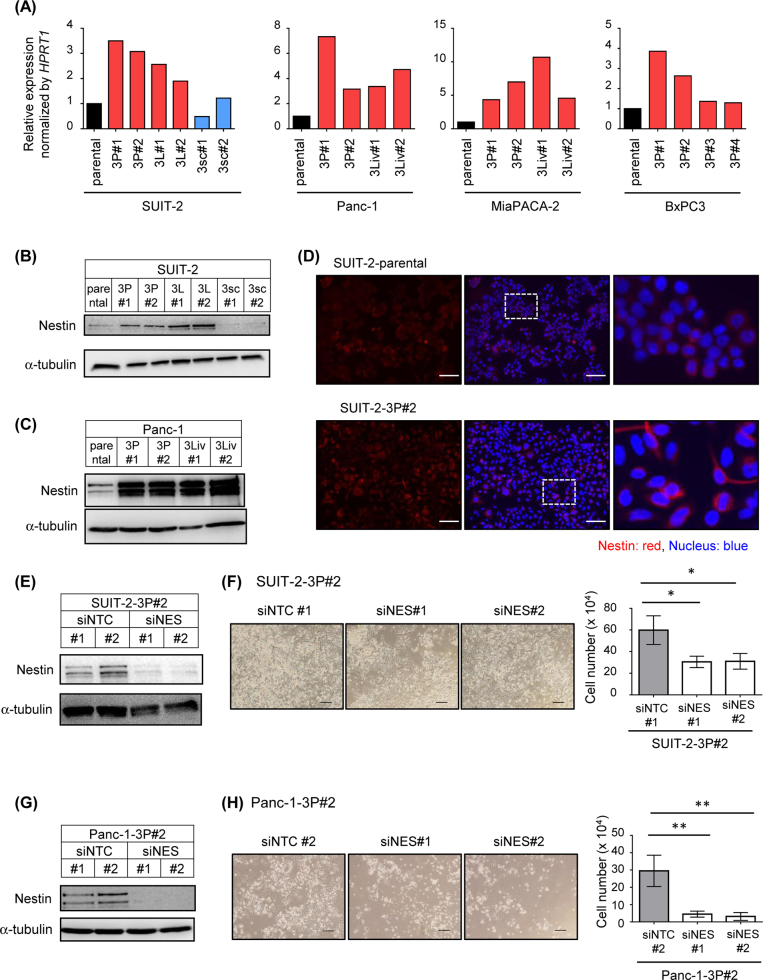
Among them, we focused on Nestin, a type VI intermediate filament that is expressed in central neurons [15]. Amplification of the NES gene has been reported in ~17% of clinical specimens of pancreatic cancer [16]. Highly malignant cancer cell lines were also established from other pancreatic cancer cells (MiaPACA-2 and BxPC3) through serial transplantations with the orthotopic model, as described in Fig. 2a. Elevated expression of NES mRNA was also observed in these cell lines in quantitative real-time reverse transcription-PCR (qRT-PCR) analysis (Fig. 5a). Immunoblot and immunocytochemical analyses demonstrated that the amounts of Nestin protein increased in cells obtained from the orthotopic tumor models (Figs. 5b–d and Supplementary Fig. S3C and D). In addition, the number of living cells in 3P cells decreased when the expression of Nestin was silenced with small interfering RNAs (siRNAs; Figs. 5e–h). These observations suggest that the genes that were highly expressed specifically in cell lines from the orthotopic tumor model may be new targets for treatment or serve as biomarkers for pancreatic cancer.
Fig. 5.
Role of Nestin in highly malignant pancreatic cancer cell lines. a Expression of NES mRNA in the cell lines derived from human pancreatic cancer cell lines was determined by qRT-PCR analysis. b, c Expression of Nestin protein in the cell lines derived from SUIT-2 cells (b) and Panc-1 cells (c) was determined by immunoblotting. d Expression of Nestin protein in parental SUIT-2 and SUIT-2-3P#2 cells was determined by immunocytochemistry. Enlarged pictures of the middle panels are also shown in the right panels. Scale bars are 100 µm. e Expression of Nestin in SUIT-2-3P#2 cells upon treatment with control or Nestin siRNAs. Cells were transfected with control siRNA or siRNAs targeting Nestin. Forty-eight hours after transfection, the expression of Nestin protein was determined by immunoblotting. f Cell proliferation assay of the SUIT-2-3P#2 cells. Cell number was counted 6 d after the transfection of siRNAs. Representative images (left) and the number of living cells (right) are shown. Scale bars are 200 µm. g Expression of Nestin in Panc-1-3P#2 cells. Cells were transfected with control siRNA or siRNAs targeting Nestin. Forty-eight hours after transfection, expression of Nestin protein was determined by immunoblotting. h Survival of the Panc-1-3P#2 cells. Cell number was counted 6 d after the transfection of siRNAs. Representative images (left) and the number of living cells (right) are shown. Scale bars are 200 µm. Data are presented as mean (duplicate; a) and mean ± SD (f, h), respectively. *P < 0.05, **P < 0.01
Mechanisms underlying the development of highly malignant cancer cell lines in the pancreatic microenvironment
We investigated the mechanisms underlying the development of highly malignant cancer cell lines in the pancreatic microenvironment. Two hypotheses may be involved: small fractions of highly malignant cancer cells within parental cancer cells adapted to the pancreatic microenvironment (“selection” process), and the characteristics of parental cancer cells were altered through interaction with the pancreatic microenvironment (“education” process). As shown in Figs. 1a,b, the incidence of tumor formation in primary and metastatic sites differed between the orthotopic and subcutaneous models.
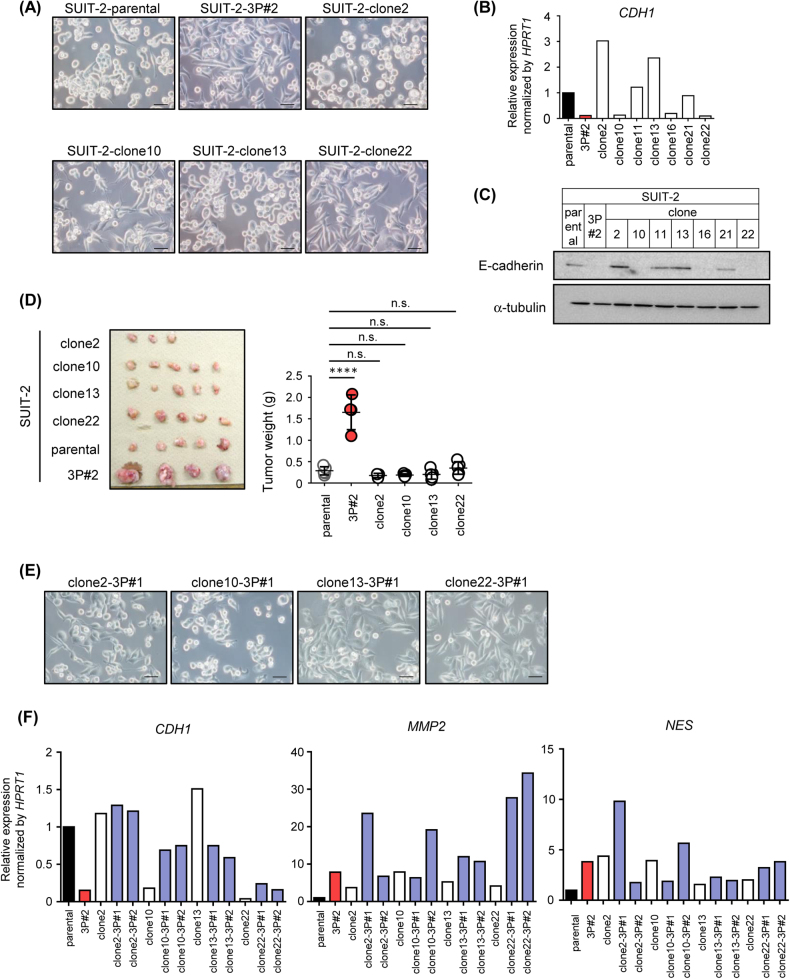
To investigate whether cellular heterogeneity exists in primary tumors, several cell clones were isolated from parental SUIT-2 cells. Although some clones, such as clones 2 and 13, had round-shaped cells, similar to parental SUIT-2 cells, clones 10 and 22 had spindle-shaped cells, similar to SUIT-2-3P cells (Fig. 6a). Moreover, qRT-PCR and immunoblot analyses revealed that expression levels of E-cadherin varied in each clone from parental SUIT-2 cells; clones 2, 11, 13, and 21 expressed similar or elevated levels of E-cadherin compared to parental SUIT-2 cells, while clones 10, 16, and 22 exhibited decreased levels of E-cadherin, like SUIT-2-3P cells (Figs. 6b,c). These results suggested that cellular heterogeneity was present in parental SUIT-2 cells, and that certain clones, such as clones 10 and 22, might have become a dominant population through the selection process in the pancreatic environment. However, when each clone was applied to the orthotopic tumor model, all the four clones tested exhibited tumor-forming ability, similar to parental SUIT-2 cells, in vivo (Fig. 6d). These observations suggested that the education process was associated with the formation of primary tumors and acquisition of highly malignant traits.
Fig. 6.
Mechanisms of acquisition of the malignant phenotype in vivo. a Morphological features of clones isolated from parental SUIT-2 cells. Scale bars are 50 µm. b, c Expression of E-cadherin in the clones from SUIT-2 cells. Expression of CDH1 mRNA and that of E-cadherin protein were determined by qRT-PCR analysis (b) and immunoblotting (c), respectively. d Tumor-forming ability of the clones isolated from SUIT-2 cells. An equal number of each clone from parental SUIT-2 cells was inoculated into the pancreas. One month after inoculation, the primary tumor was excised (left) and tumor weight was measured (right). e Morphological features of the 3P cells from each clone. The clone-3P cells were established from each clone with three cycles of orthotopic inoculation. Representative images are shown. Scale bars are 50 µm. f Expression levels of E-cadherin, MMP2, and Nestin in the SUIT-2 clones and 3P clones were determined by qRT-PCR analysis. Data are presented as mean (duplicate; b,f) and mean ± SD (d), respectively. ****P < 0.0001, n.s., not significant
To determine whether the characteristics of the clonal cells growing in vivo were altered, each clone (clones 2, 10, 13, and 22) was subjected to serial orthotopic transplantations by the same strategy as shown in Fig. 2a. Tumorigenicity of each cell clone was enhanced through serial transplantations (Supplementary Fig. S4), suggesting that clone cells could acquire the malignant trait through the interaction with orthotopic environment. Morphological features of the clone-derived 3P cells were distinct from those of the corresponding parental clone (Figs. 6a,e). In particular, the population of spindle-shaped cells in clones 2-3P and 13-3P cells was higher than that in the parental clonal cells. Although the expression levels of E-cadherin in clones 2, 10, and 22 were not downregulated, or even upregulated, clone 13 showed decreased E-cadherin expression after the serial orthotopic transplantations (Fig. 6f). In addition, the expression of MMP2 and NES in each clone often increased after serial transplantations (Fig. 6f). The gene expression profiles in the clone-derived 3P cells were distinct from those in parental clones, suggesting that these alterations were attributed to the education process, but not to the selection process.
Discussion
To identify the key molecules responsible for cancer progression, highly malignant cancer cell lines were established from tumor tissues using mouse tumor models, and their characteristics were analyzed [17, 18]. In the present study, highly malignant pancreatic cancer cell lines, that is, 3P cells, were obtained from primary tumors in the pancreatic orthotopic tumor model. We then applied the high-throughput RNA-seq approach for characterization of the 3P cells and focused on the function of Nestin in these cells. Previous reports have demonstrated that expression of Nestin was elevated by epigenetic modifications or miRNA-mediated regulatory mechanisms and that it was correlated with poor prognosis in patients with various types of cancers [19–24]. Other studies showed that the depletion of Nestin induced spontaneous DNA damage accumulation, delayed DNA damage repair, and enhanced sensitivity to ionizing radiation in nasopharyngeal carcinoma [24]. Since Nestin acts as a signaling scaffold for the cyclin-dependent kinase 5 (Cdk5)/p35 complex and protects from oxidative stress-induced apoptosis by negative regulation of Cdk5 and p35, a cytoprotective function of Nestin has also been found in neural progenitor cells [25]. In addition, Nestin has been regarded as a cancer stem cell (CSC) marker [15]. Nestin promotes survival of CSCs through the Wnt/β-catenin pathway, and silencing of NES expression induces cell cycle arrest and promotes apoptosis of breast CSCs [26]. Previous studies on pancreatic cancer have shown that knockdown of Nestin with siRNA suppressed tumor progression in Panc-1-xenografted mice [23]. These observations are consistent with our results, suggesting that analysis of highly expressed genes in cancer cells obtained from mouse orthotopic tumor models may provide an attractive screening system for identification of potential targets of cancer therapy.
We also partially elucidated the mechanism underlying the generation of highly malignant cancer cell lines in vivo. We hypothesized that two processes are important for the development of highly malignant cancer cells, that is, the selection and education processes. In the selection process, certain populations within cancer cells become dominant under selective pressures, including the host immune response, proliferative or survival ability of cancer cells, physiological restraints, and cancer therapy [27], which may account for the “seed and soil” theory originally documented by Stephen Paget [28]. In the present study, we showed that parental SUIT-2 cells were heterogeneous in morphological features and E-cadherin expression (Figs. 6a–c). Yachida et al. reported that primary pancreatic tumors contained various subclones, parts of which had metastatic ability [29]. In addition, a previous report demonstrated that CD133+CXCR4+ subpopulation determined tumor growth and metastatic ability [30]. These data support the idea that the selection process might be responsible for the emergence of highly malignant cancer cells in vivo.
Although a limited number of clones were investigated in this study, none of the cell clones from parental SUIT-2 cells exhibited high tumor-forming ability compared to that of the SUIT-2-3P cells (Fig. 6d), suggesting that the education process was involved in tumorigenesis in vivo. Functional diversity within cancer cells is present during the proliferation of clone-originated cancer cells [31]. In the present study, we showed that the gene expression profiles in parental cells changed after serial transplantations in vivo, and these results were reproduced by the transplantations of the cloned cells (Figs. 4 and 6f). These observations suggested that the functional diversity of cancer cells was induced by their interactions with the tumor microenvironment in our experimental model. It is considered that this functional diversity of cancer cells is not attributable to gene mutations but is due to epigenetic regulation of DNA and histones, or to modifications in protein production or degradation [32]. In addition, Takeda et al. showed that the immune response induced genetic instability in cancer cells, which might contribute to cancer immunoediting [33]. These reports indicate that the education process is another important process enabling cancer cells to acquire malignant properties in vivo. Since the morphological features and gene expression profiles of the 3P cells appeared to be conserved during in vitro cell culture, some underlying mechanisms, including epigenetic modification, may be involved in fixation of the malignant traits of 3P cells during the education process.
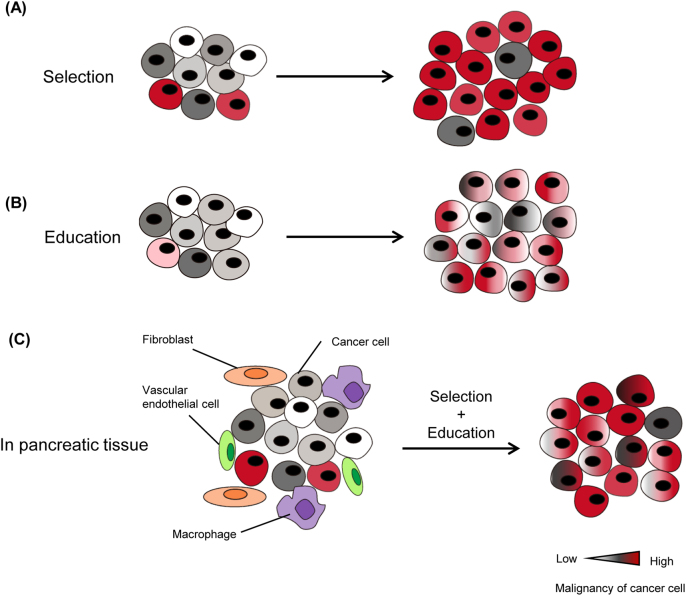
In the present study, RNA-seq analysis revealed that the 3P cells possessed a stem cell-like phenotype (Figs. 4c,e). Since the CSC theory is essentially based on the heterogeneity within cancer cells, the selection process might account for the formation of 3P cells [34]. However, recent studies indicated that more malignant CSC cells emerged during cancer progression rather than initially existed in tumor tissues [34]. In addition, several studies showed that cancer cells that have undergone EMT obtained a CSC-like phenotype [35]. Our data also showed that parental cells obtained the mesenchymal phenotype as well as the CSC-like phenotype in the pancreatic microenvironment. We thus conclude that the establishment of 3P cells is attributed to not only the selection but also the education process, and that the pancreatic microenvironment is favorable for the maintenance of a CSC-like phenotype (Fig. 7).
Fig. 7.
Tumor microenvironment determines the characteristics of pancreatic cancer cells through the selection and education processes. Two hypotheses for the acquisition of highly malignant characteristics in the tumor microenvironment (“selection” and “education”) and a working model for the current pancreatic tumor model are shown. a In the selection model, only highly malignant cancer clones can survive and adapt to the surrounding microenvironment. They become dominant during progression. b In the education model, many clones might have the potential to acquire the malignant phenotype by interactions with the tumor microenvironment. The phenotype of each clone is altered during progression and shows highly malignant characteristics. c In the current pancreatic tumor model, both selection and education processes appear to be involved in the acquisition of malignant characteristics
Materials and methods
Cell culture
The human pancreatic adenocarcinoma cell lines SUIT-2 (Japanese Cancer Research Resource Bank) and Panc-1 (American Type Culture Collection) were maintained in Dulbecco’s Modified Eagle Medium containing 10% fetal bovine serum (FBS), 50 U/ml penicillin, and 50 μg/ml streptomycin, as previously described [36, 37].
Establishment of stable transfectants expressing firefly luciferase and GFP with the lentiviral expression system
Firefly luciferase and GFP were introduced into pancreatic cancer cells by infection of lentiviral vectors. The lentiviral expression system was kindly provided by Dr. Hiroyuki Miyoshi (Keio University) as previously described [38]. To prepare an expression vector construct, the firefly luciferase gene (originated from pGL4.10; Promega) was inserted into the entry vector pENTR201. Recombination between pENTR201 and the destination vector CSII-CMV-RfA was performed with Gateway LR Clonase II enzyme (Invitrogen). For the production of lentiviral vectors, 293FT cells (Invitrogen) were transfected with an expression vector construct (CS-CDF-CG-PRE and CSII-CMV-Luc2), a VSV-G- and Rev-expressing construct (pCMV-VSV-G-RSV-Rev), and a packaging vector construct (pCAG-HIVgp). The culture supernatants containing lentiviral particles were collected and concentrated with Lenti-X Concentrator (Clontech).
Cell proliferation assay
Cells were seeded into 96-well plates and cultured for 2–4 d. At the indicated days after seeding, the number of living cells was determined with Cell Count Reagent SF (Nacalai Tesque). Absorbance at 450 nm was measured using a multiplate reader (Bio-Rad), followed by subtraction of the reference absorbance at 595 nm. For assays of the cells transfected with siRNAs, cells were stained with trypan blue, and non-stained cells were counted.
Colony formation assay
Colony formation assay was performed as previously described [36]. Cells were seeded in 0.33% agar in 12-well plates and cultured for 8–11 d (4000 cells/well). The colonies (diameter > 20 μm) were counted with the Cellsens Standard software (Olympus).
Adhesion assay
The surface of 96-well plates (0.5 μg/well) was coated with fibronectin (Wako Pure Chemicals) and blocked with 0.1% bovine serum albumin. Cancer cells in FBS-free medium were seeded and incubated in FBS-free medium for 30 min. Floating cells were washed with phosphate-buffered saline (PBS); then, the attached cells were fixed and stained with crystal violet. Stained cells were extracted with 33% acetic acid, and the absorbance at 570 nm was measured using a multiplate reader.
Chamber migration assay
Cells were seeded into the upper chamber (pore size, 8 μm; Corning) of 12-well plates (1 × 105 cells/chamber). After 24-h incubation, the cells were fixed and stained with crystal violet. The number of migrated cells was counted under a microscope.
Immunoblotting
Cells were lysed in cell lysis buffer (1% Nonidet P-40, 150 mM NaCl, and 20 mM Tris HCl). Proteins were applied to SDS-polyacrylamide gel electrophoresis and transferred to a membrane as previously described [38]. Anti-α-tubulin antibody (T9026; Sigma-Aldrich), anti-E-cadherin antibody (610181; BD), and anti-Nestin antibody (10C2 (MAB5326); Millipore) were used.
RNA isolation and qRT-PCR analysis
RNA isolation and qRT-PCR analysis were performed as previously described [36]. Briefly, total RNA was extracted using Isogen (Nippon Gene), and cDNA was synthesized using the PrimeScript II 1st strand cDNA synthesis kit (Takara). qRT-PCR analysis was performed with the Step One Plus Real time-PCR System and FastStart Universal SYBR Green Master with ROX (Roche Diagnostics). The expression levels of genes were normalized to that of hypoxanthine-guanine phosphoribosyltransferase 1 (HPRT1). Primer sequences are shown in Supplementary Table S3.
Mouse tumor models
BALB/c-nu/nu (nude) mice (age, 6 weeks, female) were purchased from CLEA Japan Inc. All experiments were approved by and carried out according to the guidelines of the Animal Care and the Use Committee of the Graduate School of Medicine, The University of Tokyo. In the orthotopic tumor model, the pancreas was surgically exposed through an abdominal excision under anesthesia with intraperitoneal injection (i.p.) of avertin (25 g of 2,2,2-tribromoethanol (Sigma-Aldrich) with 15.5 ml of 2-methyl-2-butanol (Sigma-Aldrich); 0.02 ml/g in Hank’s buffered salt solution (HBSS)). Human pancreatic cancer cells were inoculated directly into the pancreas in BALB/c-nu/nu mice (age, 6 weeks, female) using a 29 G needle (SUIT-2: 2.5 × 105 cells/50 μl HBSS/ mouse, Panc-1: 5 × 105 cells/50 μl HBSS/mouse) without any supplements. Bleeding was stopped by the pressure with cotton swab immediately after inoculation. After being confirmed that cancer cells did not leak out from the pancreas, excision was closed. In the subcutaneous tumor model, mice were anesthetized with inhalation of isoflurane, and subcutaneously inoculated with human pancreatic cancer cells using a 29 G needle (SUIT-2: 2.5 × 105 cells/50 μl HBSS/mouse, Panc-1: 5 × 105 cells/50 μl HBSS/mouse).
In vivo bioluminescence imaging
After the mice were anesthetized with avertin, D-luciferin potassium salt was injected (200 mg/kg in PBS, i.p.; Promega). Ten to fifteen minutes after luciferin injection, intensity of the bioluminescence signal was measured for 1–60 s once a week using the Night OWL II LB983 system (Berthold Technologies). Imaging analyses were performed with the IndiGO2 software (Berthold Technologies). All values are shown as photons per second.
RNA-seq analysis
RNA-seq analysis was performed with Ion Proton, Ion PI Template OT2 200 Kit v3, and Ion PI sequencing 200 Kit v3 (Thermo Fisher Scientific) as previously described [39]. Total RNA was extracted using the RNeasy Mini Kit (Qiagen), and genomic DNA was removed by the RNase-Free DNase Set (Qiagen). mRNA was extracted from 10 μg total RNA using the Dynabeads mRNA DIRECT Micro Kit (Life Technologies) according to the kit protocol. Fragmentation of mRNA, adaptor accession, reverse transcription, amplification, and establishment of cDNA libraries were conducted with the Ion Total RNA-Seq Kit v2 (Thermo Fisher Scientific). Sequenced reads were aligned to the human reference sequence with Tophat2. Gene expression was calculated using the Cuffdiff function of Cufflinks. Heat map of gene expression was drawn with TM4 Mev. Gene ontology analyses were performed with the CLC Genomics Workbench (Qiagen). Raw and processed data are available at GEO (GSE107960).
Histological examination
Histological examination was performed as previously described [40]. Briefly, samples were fixed with 10% formalin (Mildform, Wako Pure Chemicals), embedded in paraffin, and subjected to hematoxylin and eosin staining and Azan staining.
RNA interference
Stealth siRNAs against human Nestin (siNES#1 and siNES#2) and control siRNA (siNTC, Invitrogen) were used at a final concentration of 20 nM with Lipofectamine RNAiMax Reagent (Invitrogen) as previously described [41]. The siRNA sequences against human Nestin were 5’-CCACUCCAGUUUAGAGGCUAAGGAU-3’ (siNES#1) and 5’-CAGCCUUUCUUAAGAACCAAGAAUU-3’ (siNES#2).
Immunocytochemistry
Cells were fixed with 4% formaldehyde solution and permeabilized with 0.1% Triton X-100. The cells were stained with anti-Nestin antibody (10C2; 1:400–800, Merck Millipore). Stained cells were visualized using anti-mouse IgG H&L (Alexa Fluor 594; Invitrogen) and DAPI (Vector Laboratories Inc.). Images were captured with the All-in-One Fluorescence Microscope BZ-X710 (Keyence).
Statistical analysis
Student’s t-test and unpaired t-test with Welch’s correction were used to compare two groups. Tukey’s test was used for multiple comparisons of the data with GraphPad Prism (GraphPad Software). Significant differences were defined as P < 0.05.
Electronic supplementary material
Acknowledgements
We thank Dr. Yoko Katsuno, Dr. Yuichiro Yokoyama, and Dr. Yukari Hoshino (The University of Tokyo) for their technical assistance, and Hiroyuki Miyoshi (Keio University) for providing the lentiviral vectors. This work was supported by a Grand for Practical Research for Innovative Cancer Control (16ck0106193h0001) from the Japan Agency for Medical Research and Development (AMED; K.M.), a KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas, Integrative Research on Cancer Microenvironment Network (22112002) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT; K.M.), a KAKENHI Grant-in-Aid for Scientific Research (C; 15K08393; S.E.) and Research Fellowship for Young Scientists (25–10991; K.T.) from the Japan Society for the Promotion of Science (JSPS), Specific Research Grants from Yasuda Medical Foundation (K.M.), The Cell Science Research Foundation (S.E.), and The Tokyo Society of Medical Sciences (S.E.).
Author contributions:
K.T., S.E., and K.M. designed the study, analyzed the data, and wrote the manuscript. K.T., S.E., Y.M., and D.K. performed the experiments, and M.S. and H.M. analyzed the data.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
The online version of this article (10.1038/s41388-018-0144-0) contains supplementary material, which is available to authorized users.
References
- 1.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol. 2008;3:157–88. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:2140–1. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 3.Perera RM, Bardeesy N. Pancreatic cancer metabolism: breaking it down to build it back up. Cancer Discov. 2015;5:1247–61. doi: 10.1158/2159-8290.CD-15-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Cell. 2005;7:513–20. doi: 10.1016/j.ccr.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Nagai N, Ogura K, Tsuneyama K, Saiki I, Irimura T, et al. Mammary tissue microenvironment determines T cell-dependent breast cancer-associated inflammation. Cancer Sci. 2015;106:867–74. doi: 10.1111/cas.12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–17. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 9.Qian LW, Mizumoto K, Maehara N, Ohuchida K, Inadome N, Saimura M, et al. Co-cultivation of pancreatic cancer cells with orthotopic tumor-derived fibroblasts: fibroblasts stimulate tumor cell invasion via HGF secretion whereas cancer cells exert a minor regulative effect on fibroblasts HGF production. Cancer Lett. 2003;190:105–12. doi: 10.1016/S0304-3835(02)00517-7. [DOI] [PubMed] [Google Scholar]
- 10.Bachem MG, Schünemann M, Ramadani M, Siech M, Beger H, Buck A, et al. Pancreatic carcinoma cells induce fibrosis by stimulating proliferation and matrix synthesis of stellate cells. Gastroenterology. 2005;128:907–21. doi: 10.1053/j.gastro.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 11.Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008;68:918–26. doi: 10.1158/0008-5472.CAN-07-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–29. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–47. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Özdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–34. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neradil J, Veselska R. Nestin as a marker of cancer stem cells. Cancer Sci. 2015;106:803–11. doi: 10.1111/cas.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hippo Y, Yashiro M, Ishii M, Taniguchi H, Tsutsumi S, Hirakawa K, et al. Differential gene expression profiles of scirrhous gastric cancer cells with high metastatic potential to peritoneum or lymph nodes. Cancer Res. 2001;61:889–95. [PubMed] [Google Scholar]
- 18.Eguchi D, Ohuchida K, Kozono S, Ikenaga N, Shindo K, Cui L, et al. MAL2 expression predicts distant metastasis and short survival in pancreatic cancer. Surgery. 2013;154:573–82. doi: 10.1016/j.surg.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Piras F, Perra MT, Murtas D, Minerba L, Floris C, Maxia C, et al. The stem cell marker nestin predicts poor prognosis in human melanoma. Oncol Rep. 2010;23:17–24. [PubMed] [Google Scholar]
- 20.Strojnik T, Røsland GV, Sakariassen PO, Kavalar R, Lah T. Neural stem cell markers, nestin and musashi proteins, in the progression of human glioma: correlation of nestin with prognosis of patient survival. Surg Neurol. 2007;68:133–43. doi: 10.1016/j.surneu.2006.10.050. [DOI] [PubMed] [Google Scholar]
- 21.Ehrmann J, Kolár Z, Mokry J. Nestin as a diagnostic and prognostic marker: immunohistochemical analysis of its expression in different tumours. J Clin Pathol. 2005;58:222–3. doi: 10.1136/jcp.2004.021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, Chen B, Zhu J, Zhang R, Yao F, Jin F, et al. Clinical implications for nestin protein expression in breast cancer. Cancer Sci. 2010;101:815–9. doi: 10.1111/j.1349-7006.2009.01422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda Y, Ishiwata T, Yoshimura H, Yamashita S, Ushijima T, Arai T. Systemic administration of small interfering RNA targeting human nestin inhibits pancreatic cancer cell proliferation and metastasis. Pancreas. 2016;45:93–100. doi: 10.1097/MPA.0000000000000427. [DOI] [PubMed] [Google Scholar]
- 24.Ma J, Sun F, Li C, Zhang Y, Xiao W, Li Z, et al. Depletion of intermediate filament protein Nestin, a target of microRNA-940, suppresses tumorigenesis by inducing spontaneous DNA damage accumulation in human nasopharyngeal carcinoma. Cell Death Dis. 2014;5:e1377. doi: 10.1038/cddis.2014.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahlgren CM, Pallari HM, He T, Chou YH, Goldman RD, Eriksson JE. A nestin scaffold links Cdk5/p35 signaling to oxidant-induced cell death. EMBO J. 2006;25:4808–19. doi: 10.1038/sj.emboj.7601366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Z, Lu P, Zhang H, Xu H, Gao N, Li M, et al. Nestin positively regulates the Wnt/β-catenin pathway and the proliferation, survival and invasiveness of breast cancer stem cells. Breast Cancer Res. 2014;16:408. doi: 10.1186/s13058-014-0408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 29.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Kreso A, O’Brien CA, van Galen P, Gan OI, Notta F, Brown AM, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–8. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–91. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Takeda K, Nakayama M, Hayakawa Y, Kojima Y, Ikeda H, Imai N, et al. IFN-γ is required for cytotoxic T cell-dependent cancer genome immunoediting. Nat Commun. 2017;8:14607. doi: 10.1038/ncomms14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valent P, Bonnet D, De Maria R, Lapidot T, Copland M, Melo JV, et al. Cancer stem cell definitions and terminology: the devil is in the details. Nat Rev Cancer. 2012;12:767–75. doi: 10.1038/nrc3368. [DOI] [PubMed] [Google Scholar]
- 35.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 36.Hoshino Y, Nishida J, Katsuno Y, Koinuma D, Aoki T, Kokudo N, et al. Smad4 decreases the population of pancreatic cancer-initiating cells through transcriptional repression of ALDH1A1. Am J Pathol. 2015;185:1457–70. doi: 10.1016/j.ajpath.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 37.Horiguchi K, Shirakihara T, Nakano A, Imamura T, Miyazono K, Saitoh M. Role of Ras signaling in the induction of snail by transforming growth factor-β. J Biol Chem. 2009;284:245–53. doi: 10.1074/jbc.M804777200. [DOI] [PubMed] [Google Scholar]
- 38.Murai F, Koinuma D, Shinozaki-Ushiku A, Fukayama M, Miyaozono K, Ehata S. EZH2 promotes progression of small cell lung cancer by suppressing the TGF-β-Smad-ASCL1 pathway. Cell Discov. 2015;1:15026. doi: 10.1038/celldisc.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arase M, Horiguchi K, Ehata S, Morikawa M, Tsutsumi S, Aburatani H, et al. Transforming growth factor-β-induced lncRNA-Smad7 inhibits apoptosis of mouse breast cancer JygMC(A) cells. Cancer Sci. 2014;105:974–82. doi: 10.1111/cas.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katsuno Y, Hanyu A, Kanda H, Ishikawa Y, Akiyama F, Iwase T, et al. Bone morphogenetic protein signaling enhances invasion and bone metastasis of breast cancer cells through Smad pathway. Oncogene. 2008;27:6322–33. doi: 10.1038/onc.2008.232. [DOI] [PubMed] [Google Scholar]
- 41.Yokoyama Y, Watanabe T, Tamura Y, Hashizume Y, Miyazono K, Ehata S. Autocrine BMP-4 signaling is a therapeutic target in colorectal cancer. Cancer Res. 2017;77:4026–38. doi: 10.1158/0008-5472.CAN-17-0112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.