Abstract
Background/Objective
Obesity is a complex and multifactorial disease resulting from the interactions among genetics, metabolic, behavioral, sociocultural and environmental factors. In this sense, the aim of the present study was to identify phenotype and genotype variables that could be relevant determinants of body mass index (BMI) variability.
Subjects/Methods
In the present study, a total of 1050 subjects (798 females; 76%) were included. Least angle regression (LARS) analysis was used as regression model selection technique, where the dependent variable was BMI and the independent variables were age, sex, energy intake, physical activity level, and 16 polymorphisms previously related to obesity and lipid metabolism.
Results
The LARS analysis obtained the following formula for BMI explanation: (64.7 + 0.10 × age [years] + 0.42 × gender [0, men; 1, women] + −40.6 × physical activity [physical activity level] + 0.004 × energy intake [kcal] + 0.74 × rs9939609 [0 or 1–2 risk alleles] + −0.72 × rs1800206 [0 or 1–2 risk alleles] + −0.86 × rs1801282 [0 or 1–2 risk alleles] + 0.87 × rs429358 [0 or 1–2 risk alleles]. The multivariable regression model accounted for 21% of the phenotypic variance in BMI. The regression model was internally validated by the bootstrap method (r2 original data set = 0.208, mean r2 bootstrap data sets = 0.210).
Conclusion
In conclusion, age, physical activity, energy intake and polymorphisms in FTO, APOE, PPARG and PPARA genes are significant predictors of the BMI trait.
Introduction
In the past 50 years, the prevalence of obesity has steadily raised becoming a global public health problem contributing for a huge increase of health-care costs1. It has been estimated that 2.16 billion adults will be overweight and 1.12 billion adults will be obese by 2030, if the present trends continue2. An increase in the global burden of overweight and obesity will translate into an increase of the risk of several other health conditions, including type 2 diabetes, cardiovascular disease or certain types of cancer1. Although obesity is generally attributed to an imbalance between the energy consumed and the energy expenditure, it is also accepted that it is a complex and a multifactorial disease resulting from genetic, physiological, behavioral, sociocultural and environmental factors3–7.
Heritability studies indicate that genetic factors could account for 31–90% of the body inter-individual weight variability8. However, the large number of single-nucleotide polymorphisms (SNPs) identified by genome-wide association studies (GWAS) and candidate gene studies, appeared to explain only 2–4% of the obesity status9. Even taken together such polymorphisms, they seemed to provide very little risk prediction of the disease10. In one of the last GWAS related to adiposity, the 97 genome-wide significant loci identified associated with obesity accounted for 2.7% of the body mass index (BMI) variance11.
In addition, a limited predictive value of genetic markers have been described, specifically when they are compared to classical non-genetic risk factors12,13. In this context, the design and development of a multivariable regression model based on phenotype and genotype variables could lead us toward the development of more effective precision preventive and treatment dietary interventions14. Therefore, the aim of the present study was to identify, in an adult population, phenotype and genotype variables, that combined in a multivariable model, could be associated with BMI variability.
Subjects and methods
Study population
The data set included men and women of Caucasian ancestry, who voluntarily attended community pharmacies in Spain. Genotype information of 1065 individuals was available. Of these, 7 subjects were excluded due to missing values for dietary intake, physical activity and/or anthropometric measurements, and 8 subjects were removed because they were <18 years old. Therefore, a total of 1050 subjects were included in the present study.
Individuals were specifically asked if they would be willing to take part anonymously in the research study. After ensuring that participants had understood the information, only those who provided written informed consent for participation were enrolled. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation and with the Helsinki Declaration of 1975, as revised in 2000. The Research Ethics Committee of the University of Navarra gave confirmation of fulfillment of the ethical standards and deontological criteria affecting the present survey (Ref. 2410/2014).
Data collection
Anthropometrics, habitual dietary intake and physical activity measurements were collected by trained nutritionists using a standardized protocol previously described15. Briefly, weight and height were measured with a digital scale (Tanita BF-522W, Tanita Corporation, Tokyo, Japan) and a portable stadiometer (Leicester Tanita), respectively. BMI was calculated as weight (kg)/height2 (m2).
Habitual dietary intake was determined using a validated food groups frequency questionnaire, where basic foods were categorized into 19 food groups. Subjects were asked to report how often (daily, weekly, monthly or never) they had consumed a choice of each food group16. Physical activity was estimated by a short 24 h physical activity questionnaire in which subjects were asked about the number of hours resting and practicing activities during a weekday and a weekend day17.
DNA isolation and genotyping
Genomic DNA was obtained from oral epithelial cells collected by ORAcollect DNA® (DNAGenotek, Kanata, Ont., Canada). It was isolated by QIAcube using QiAmp DNA Mini QIAcube Kit (Qiagen, Hilden, Germany), following the manufacturer’s procedures. Sixteen polymorphisms previously associated in the scientific database with body weight regulation and lipid metabolism (rs9939609 (FTO), rs17782313 (MC4R), rs1801133 (MTHFR), rs1800206 (PPARA), rs1801282 (PPARG), rs662799 (APOA5), rs429358 (AAPOE), rs7412 (APOE), rs1800588 (LIPC), rs894160 (PLIN1), rs1799983 (NOS3), rs1260326 (GCKR), rs328 (LPL), rs12740374 (CELSR2), rs1800777 (CETP) and rs4939883 (LIPG)) were genotyped using Luminex® 100/200TM System (Luminex Corporation, Austin, Texas), which is based on the principles of xMAP® Technology18–32. This method uncompressed polystyrene microspheres internally dyed with various ratios of spectrally distinct fluorophores, which are detected by a flow cytometry-based instrument33.
Statistical analyses
Deviation from Hardy–Weinberg equilibrium (HWE) was tested using χ2 test and allele frequencies were estimated. Least angle regression (LARS) analysis was used as regression model selection technique due to its advantages in speed, interpretability and predictive accuracy34. In the current study, the dependent variable was BMI. The independent variables were age, sex, energy intake, physical activity level and the 16 selected polymorphisms. Because LARS algorithm is designed for linear regression with continuous or binary covariates, polymorphisms were recoded in binary variables according to the association between each polymorphism and BMI tested by using dummy linear regression models. In those cases, where there was no significant association and due to the limited frequency of the variant allele, homozygotes of the minor allele (aa) and heterozygotes (Aa) were grouped and compared with major allele homozygotes (AA). Stagewise regression and Lasso were also performed to confirm the selection of the independent variables established by LARS34. The independent variables selected by LARS method were combined to generate the regression function. The formula was constructed by adding each genotype or phenotype variable multiplied by its beta coefficient, and the constant of the regression model. To test potential gene–gene and gene–phenotype interactions among the factors selected by LARS, genotype-by-genotype and genotype-by-phenotype product terms were included in the model. Bootstrapping was performed to internally validate the regression model. It was implemented by constructing a number of resamples (K = 1000) of the data set that was obtained by random sampling with replacement from the original data set. For multiple comparisons, Benjamini–Hochberg correction was applied. Statistical analyses were performed using Stata SE, version 12.1 (StataCorp, College Station, TX, USA) and R, version 3.3.2 (R Foundation for Statistical Computing, Vienna, Austria). A p value of p < 0.05 was considered as statistically significant.
Results
Baseline phenotypic characteristics of the individuals according to gender have been described (Table 1). The genotypes distribution, minor allele frequencies (MAF) and HWE for each polymorphism are listed (Table 2). MAF ranged from 0.02 to 0.45. The distributions of all the polymorphisms alleles were in HWE except the rs1800588 polymorphism located in LIPC gene even after Benjamini–Hochberg correction for multiple comparisons.
Table 1.
Anthropometrical and nutritional characteristics
| Total population (n = 1050) | Males (n = 252) | Females (n = 798) | |
|---|---|---|---|
| Age (years) | 50.0 (12.8) | 47.4 (12.9) | 50.8 (12.7) |
| Height (cm) | 164 (9.0) | 174 (7.2) | 160 (6.6) |
| Weight (kg) | 79.5 (17.1) | 91.4 (16.4) | 75.7 (15.5) |
| BMI (kg/m2) | 29.6 (5.9) | 29.9 (5.0) | 29.4 (6.1) |
| Nutritional status | |||
| Normal weight (BMI: 18.5–24.9 kg/m2) | 232 (22.1) | 32 (12.7) | 200 (25.1) |
| Overweight (BMI: 25–29.9 kg/m2) | 377 (35.9) | 106 (42.1) | 271 (34.0) |
| Obesity (BMI: ≥30 kg/m2) (%) | 441 (42.0) | 114 (45.2) | 327 (41.0) |
| Energy intake (kcal) | 2138 (453) | 2438 (492) | 2043 (395) |
| Physical activity level | 1.23 (0.03) | 1.24 (0.03) | 1.23 (0.03) |
The t test for continuous variables are expressed as medium (s.d.); and Χ2 test for qualitative variables are expressed as n (%)
BMI Body mass index
Table 2.
Genotype, minor allele frequency and Hardy–Weinberg equilibrium calculations of the 16 SNPs included in LARS analysis
| Gene | SNP | Major/minor allele | Major allele homozygote (%) | Heterozygote (%) | Minor allele homozygote (%) | MAF | HWE p Value |
|---|---|---|---|---|---|---|---|
| FTO | rs9939609 | T/A | 329 (31.3) | 519 (49.4) | 202 (19.2) | 0.44 | 0.92 |
| MC4R | rs17782313 | T/C | 646 (61.5) | 355 (33.8) | 49 (4.7) | 0.22 | 0.98 |
| MTHFR | rs1801133 | C/T | 380 (36.2) | 511 (48.7) | 159 (15.1) | 0.39 | 0.55 |
| PPARA | rs1800206 | C/G | 881 (83.9) | 162 (15.4) | 7 (0.7) | 0.08 | 0.88 |
| PPARG | rs1801282 | C/G | 889 (84.7) | 152 (14.5) | 9 (0.9) | 0.08 | 0.38 |
| APOA5 | rs662799 | T/C | 919 (87.5) | 126 (12.0) | 5 (0.5) | 0.06 | 0.46 |
| APOE | rs429358 | T/C | 842 (80.2) | 199 (18.9) | 9 (0.9) | 0.10 | 0.76 |
| APOE | rs7412 | C/T | 920 (87.6) | 127 (12.1) | 3 (0.3) | 0.06 | 0.53 |
| LIPC | rs1800588 | C/T | 617 (58.8) | 360 (34.3) | 73 (6.9) | 0.24 | 0.04* |
| PLIN1 | rs894160 | G/A | 560 (53.3) | 404 (38.5) | 86 (8.2) | 0.27 | 0.28 |
| NOS3 | rs1799983 | G/T | 418 (39.8) | 488 (46.5) | 144 (13.7) | 0.37 | 0.93 |
| GCKR | rs1260326 | C/T | 315 (30.0) | 527 (50.2) | 208 (19.8) | 0.45 | 0.64 |
| LPL | rs328 | C/G | 765 (72.9) | 264 (25.1) | 21 (2.0) | 0.15 | 0.75 |
| CELSR2 | rs12740374 | G/T | 658 (62.3) | 250 (33.3) | 42 (4.0) | 0.21 | 0.59 |
| CETP | rs1800777 | G/A | 1002 (95.4) | 46 (4.4) | 2 (0.2) | 0.02 | 0.06 |
| LIPG | rs4939883 | C/T | 756 (72.0) | 260 (24.8) | 34 (3.2) | 0.16 | 0.05 |
MAF minor allele frequency, HWE Hardy–Weinberg equilibrium
*p value < 0.05 after Benjamini–Hochberg correction for multiple comparisons
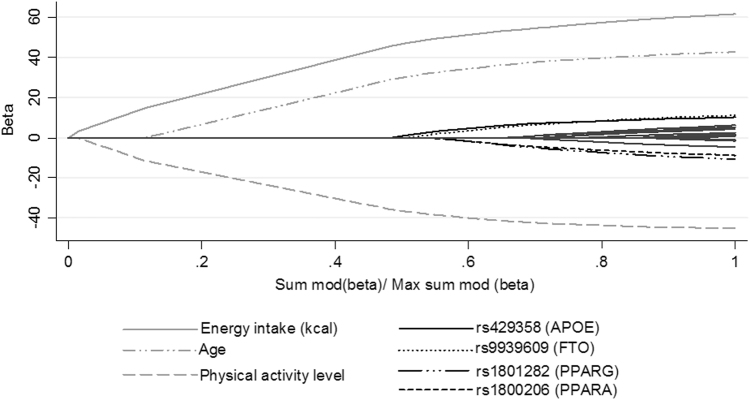
According to the LARS analysis age, physical activity, energy intake and 4 polymorphisms were associated with BMI variability (Table 3 and Fig. 1). Although gender was not selected by LARS it was included in the model as a common cofounding factor. The LARS analysis obtained the following formula for explain BMI: (64.7 + 0.10 × age [years] + 0.42 × gender [0, men; 1, women] + −40.6 × physical activity [physical activity level] + 0.004 × energy intake [kcal] + 0.74 × rs9939609 [0 or 1–2 risk alleles] + −0.72 × rs1800206 [0 or 1–2 risk alleles] + −0.86 × rs1801282 [0 or 1–2 risk alleles] + 0.87 × rs429358 [0 or 1–2 risk alleles]. The multivariable regression model accounted for 21% of the phenotypic variance in BMI. Energy intake, physical activity level, age, and gender, explained 8.3, 7.3, 4.2 and 0.04%, of the BMI variance, respectively. Among the genotypic variables, FTO polymorphism explained 0.1% of the BMI variance, APOE polymorphism 0.3%, PPARG polymorphism 0.1%, and PPARA polymorphism 0.2%. The selection of the independent variables established by LARS was confirmed by stagewise regression and Lasso (data not shown).
Table 3.
Regression coefficients of the variables selected by LARS
| Variables selected | β | 95% CI | Adjusted r2 | p value |
|---|---|---|---|---|
| Energy intake (kcal) | 0.004 | 0.003;0.005 | 0.083 | <0.001 |
| Physical activity level | −40.6 | −49.9;−31.3 | 0.073 | <0.001 |
| Age (years) | 0.10 | 0.08;0.13 | 0.042 | <0.001 |
| rs429358 (APOE)a | 0.86 | 0.06;1.66 | 0.003 | 0.035 |
| rs9939609 (FTO)a | 0.74 | 0.05;1.42 | 0.001 | 0.035 |
| rs1800206 (PPARA)a | −0.72 | −1.58;0.15 | 0.002 | 0.104 |
| rs1801282 (PPARG)a | −0.86 | −1.74;0.043 | 0.001 | 0.058 |
| Gender | 0.42 | −0.39;1.24 | 0.000 | 0.304 |
| Constant | 64.7 | 43.3;74.9 | — | <0.001 |
aDominant model
Fig. 1.
LARS analysis
Additionally, gene–phenotypic factors and gene–gene interactions were tested. Trend toward significance interactions were found for FTO polymorphism and energy intake and for PPARA genetic variant and energy intake. When both product terms of the interactions were included in the regression model the adjusted r2 did not improve significantly (adjusted r2 for regression model 0.208; adjusted r2 for the regression model, including interactions 0.212).
In order to evaluate the accuracy of the model, the relationship between the observed and the predicted BMI was plotted (Fig. 2). The predicted BMI agrees with the observed or “real” BMI by checking the parameters of the linear regression. The intercept of the model is very close to zero and the slope is almost 1, meaning that the change in both variables can be considered proportional.
Fig. 2.

Correlation coefficient between observed BMI and predicted BMI based on the multivariable regression model obtained by LARS
The internal validation was performed by the bootstrap method, whose estimates agreed closely with the parameters obtained by LARS (r2 original data set = 0.208, mean r2 bootstrap data sets = 0.210).
Discussion
Because common obesity is a multifactorial disease, where genetic, metabolic, physiological, behavioral, sociocultural, and environmental factors are involved, in the current study, a regression model based on phenotype and genotype determinants of BMI has been defined. The regression model includes a total of 4 phenotypic characteristics (age, gender, energy intake, and physical activity) and 4 polymorphisms located next to or in FTO, APOE, PPARG, and PPARA genes.
The LARS analysis reported 4 polymorphisms significantly or marginally associated with BMI located in FTO, APOE, PPARG, and PPARA genes. FTO is a nuclear protein, which is a member of the AlkB related non-haem iron and 2-oxoglutarate-dependent oxygenase superfamily35. Although the relationship between FTO genetic variant and obesity-related traits (BMI, obesity risk, waist circumference, body fat mass) has been confirmed in several populations, the physiological function of this gene in body weight regulation seems unclear36,37.
As far as we know our group reported for the first time, a significant association between rs429358 APOE genetic variant and BMI15. In the present study, such relationship has been verified in a large sample. The APOE gene plays a major role in maintaining plasma lipids homeostasis and it is implicated in adipogenesis38–40. APOE genetic variants have been associated with several metabolic disorders including high obesity risk41–44.
PPARG modulates the expression of target genes involved in adipocyte differentiation, insulin sensitivity and inflammatory processes45,46, whereas PPARA regulates fatty acid oxidation systems46. Although in most of candidate gene studies, Pro12Ala has been associated with higher BMI, other authors reported the opposite association or have not found any association at all23,47–51. These controversial results suggest that, if this variant does influence obesity predisposition, it may do so through environment-dependent mechanisms. In fact, several studies have reported interactions between PPARG and environmental factors such as gender, dietary fat intake, or breast feeding on obesity traits52–55. Although the association between genetic variants of the PPARG gene and obesity traits has been widely studied, as far as we know there is limited evidence regarding the relationship between PPARA variants and obesity phenotype. Meanwhile, Costa-Urrutia et al. (2017) reported a positive association between the rs1800206 PPARA polymorphism and obesity risk, Sirbelnagel et al. (2009) did not find a relationship between such genetic variant and BMI or body fat composition56,57. We hypothesized that our opposite results regarding PPARG and PPARA could be due partly to the fact that we have carried out the analysis in the presence of other genetic variants.
Interestingly, 21% of the phenotypic variance in BMI was accounted using the regression model obtained by LARS, including gender, age, energy intake, physical activity and four genetic variants located near or in FTO, APOE, PPARG, and PPARA genes. When the polymorphisms were included in the regression model as a genetic risk score, summing the number of risk alleles, the model accounted for 20% of the phenotypic variance. The four polymorphisms accounted for 0.5% of the BMI variability. This finding is in accordance with the studies by Martínez-García et al. (2013), Belsky et al. (2013) and Li et al. (2010), in which a small number of SNPs explained <1% of the BMI heritability10,12,58. In this sense, it should be highlighted that when Locke et al. (2015) included a total of 97 SNPs in a prediction model of BMI the authors found a BMI explanation of 2.7%11. As far as we know, prediction models that added energy intake and physical activity have not been reported up to date, so we cannot be able to compare our results. However, some authors have observed that, when phenotypical factors are included in the genetic model (such as socioeconomic or depression status), the percentage of the explanation of the BMI significantly increases10,59.
Several potential explanations can be offered for the low predictive value of the regression model, but are mainly related to the fact that obesity is characterized for being a multifactorial disease. Although we have included in the model the two main factors that characterized obesity, energy intake and physical activity, there are other features that have not been taken into account such as social determinants (education level, economic status), endocrine disorders (hypothyroidism) or use of certain medications4,7,60,61. Another explanation for the low predictive value of the regression model could be related with the marginal effect sizes of the tested variants and the skewed distribution of the effect sizes. In addition, predictive models could include other sources of variation known or hypothesized to influence BMI such as rare variants, gene–gene and gene-environment interactions, copy number variation, and epigenetic and metagenomic effects14. Finally, it should be mentioned that in the present study BMI instead of body fat mass was selected as dependent variable. Although BMI is the adiposity measurement most widely used in epidemiological studies, its interpretation does not differ between gender and race, and neither distinguishes between degree of fatness, muscle mass, and skeletal mass62. Therefore, it can lead to errors in the estimation of adiposity, over or underestimating adiposity depending on subject complexion; such as athletes or metabolic obese normal weight individuals.
To the best of our knowledge, this is the first study that applies LARS analysis to select phenotype and genotype variables for explain BMI status. However, the study bears some limitations that need to be mentioned. First, the regression model may need to be replicated in an external population. However, the regression model was internally validated by bootstrapping. Second, the present study included only subjects of Caucasian ancestry, so the findings may not be generalizable to other ethnic groups. Third, the model developed in this study used BMI as the response variable instead of body fat mass. Although BMI has some limitations in its interpretability, it is the adiposity measurement more used in epidemiological studies.
In conclusion, significant predictors of BMI included age, energy intake, physical activity, and polymorphisms located near or in FTO (rs9939609), APOE (rs429358), PPPARG (rs1801282), and PPARA (rs1800206). Although 4 polymorphisms were selected by LARS, it should be mentioned that they explain a small percentage of BMI variation as has found other authors. Moreover, the proposed statistical method, LARS analysis, could help to implement new criteria for the identification of BMI predictors since obesity is a multifactorial disease in which a large number of phenotypic and genotypic features are involved.
Acknowledgements
We thank all the users of the Nutrigenetic Service who voluntarily offer her data to the study. We are grateful to the nutritionists for the data collection and to Amaia Ibañez for excellent technical assistance. Moreover, we wish to thank Enrique Goñi for excellent statistical assistance in the University of Navarra. The predoctoral research grant to Leticia Goni from the Spanish Ministry of Education, Culture and Sport is gratefully acknowledged. The authors also wish to thank the support from Linea Especial (University of Navarra; LE/97). Spanish Ministry of Economy and Competitiveness (AGL2013-45554-R project). CIBERobn/RETICS schedules (Institute of Health Carlos III), and CINFA concerning the genetic tools and general logistic.
Funding
Linea Especial (University of Navarra; LE/97). Spanish Ministry of Economy and Competitiveness (AGL2013-45554-R project). CIBERobn/RETICS schedules (Institute of Health Carlos III).
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Wang YC, McPherson K, Marsh T, Gortmaker SL, Brown M. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet. 2011;378:815–825. doi: 10.1016/S0140-6736(11)60814-3. [DOI] [PubMed] [Google Scholar]
- 2.Kelly T, Yang W, Chen CS, Reynolds K, He J. Global burden of obesity in 2005 and projections to 2030. Int. J. Obes. 2008;32:1431–1437. doi: 10.1038/ijo.2008.102. [DOI] [PubMed] [Google Scholar]
- 3.Singh RK, Kumar P, Mahalingam K. Molecular genetics of human obesity: a comprehensive review. C. R. Biol. 2017;340:87–108. doi: 10.1016/j.crvi.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Weaver JU. Classical endocrine diseases causing obesity. Front. Horm. Res. 2008;36:212–228. doi: 10.1159/000115367. [DOI] [PubMed] [Google Scholar]
- 5.Forouzanfar MH, et al. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386:2287–2323. doi: 10.1016/S0140-6736(15)00128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.St-Onge MP. Sleep-obesity relation: underlying mechanisms and consequences for treatment. Obes. Rev. 2017;18:34–39. doi: 10.1111/obr.12499. [DOI] [PubMed] [Google Scholar]
- 7.Kim TJ, Roesler NM, von dem Knesebeck O. Causation or selection - examining the relation between education and overweight/obesity in prospective observational studies: A meta-analysis. Obes. Rev. 2017;18:660–672. doi: 10.1111/obr.12537. [DOI] [PubMed] [Google Scholar]
- 8.Min J, Chiu DT, Wang Y. Variation in the heritability of body mass index based on diverse twin studies: A systematic review. Obes. Rev. 2013;14:871–882. doi: 10.1111/obr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Sayed Moustafa JS, Froguel P. From obesity genetics to the future of personalized obesity therapy. Nat. Rev. 2013;9:402–413. doi: 10.1038/nrendo.2013.57. [DOI] [PubMed] [Google Scholar]
- 10.Belsky DW, et al. Development and evaluation of a genetic risk score for obesity. Biodemography Soc. Biol. 2013;59:85–100. doi: 10.1080/19485565.2013.774628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, et al. Cumulative effects and predictive value of common obesity-susceptibility variants identified by genome-wide association studies. Am. J. Clin. Nutr. 2010;91:184–190. doi: 10.3945/ajcn.2009.28403. [DOI] [PubMed] [Google Scholar]
- 13.Loos RJF, Janssens ACJW. Predicting polygenic obesity using genetic information. Cell. Metab. 2017;25:535–543. doi: 10.1016/j.cmet.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Goni L, Cuervo M, Milagro FI, Mart JA. Future perspectives of personalized weight loss interventions based on nutrigenetic, epigenetic, and metagenomic data. J. Nutr. 2016;146:905–912. doi: 10.3945/jn.115.218354. [DOI] [PubMed] [Google Scholar]
- 15.Goni L, Cuervo M, Milagro FI, Martinez JA. A genetic risk tool for obesity predisposition assessment and personalized nutrition implementation based on macronutrient intake. Genes Nutr. 2015;10:445. doi: 10.1007/s12263-014-0445-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goni L, Aray-Miranda M, Martínez JA, Cuervo M. Validation of a food groups frequency questionnaire based in an exchange system. Nutr. Hosp. 2016;33:1391–1399. doi: 10.20960/nh.800. [DOI] [PubMed] [Google Scholar]
- 17.Institute of Medicine (US) Panel on Macronutrients, Institute of Medicine (US) Standing Committee on the Scientific Evaluation of Dietary Reference Intakes. The Standing Committee on the Scientific Evaluation of DRI for Energy, Carbohydrate, Fiber, Fat, Fatty Acids, Cholesterol, Protein, and Amino Acids (Macronutrients) (National Academies Press, Washington, DC, USA, 2005).
- 18.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loos RJF, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis SJ, et al. The methylenetetrahydrofolate reductase C677T genotype and the risk of obesity in three large population-based cohorts. Eur. J. Endocrinol. 2008;159:35–40. doi: 10.1530/EJE-08-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tai ES, et al. Association between the PPARA L162V polymorphism and plasma lipid levels: the Framingham Offspring Study. Arterioscler. Thromb. Vasc. Biol. 2002;22:805–810. doi: 10.1161/01.ATV.0000012302.11991.42. [DOI] [PubMed] [Google Scholar]
- 23.Galbete C, et al. Pro12Ala variant of the PPARG2 gene increases body mass index: an updated meta-analysis encompassing 49,092 subjects. Obesity. 2013;21:1486–1495. doi: 10.1002/oby.20150. [DOI] [PubMed] [Google Scholar]
- 24.Soenen S, et al. Relationship between perilipin gene polymorphisms and body weight and body composition during weight loss and weight maintenance. Physiol. Behav. 2009;96:723–728. doi: 10.1016/j.physbeh.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson JF, et al. NOS3 gene polymorphisms are associated with risk markers of cardiovascular disease, and interact with omega-3 polyunsaturated fatty acids. Atherosclerosis. 2010;211:539–544. doi: 10.1016/j.atherosclerosis.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 26.Chrysohoou C, et al. Evidence for association between endothelial nitric oxide synthase gene polymorphism (G894T) and inflammatory markers: the ATTICA study. Am. Heart J. 2004;148:733–738. doi: 10.1016/j.ahj.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 27.Lu Y, et al. Multiple genetic variants along candidate pathways influence plasma high-density lipoprotein cholesterol concentrations. J. Lipid Res. 2008;49:2582–2589. doi: 10.1194/jlr.M800232-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Bennet AM, et al. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA. 2007;298:1300–1311. doi: 10.1001/jama.298.11.1300. [DOI] [PubMed] [Google Scholar]
- 29.Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kathiresan S, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandhu MS, et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kettunen J, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 2012;44:269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunbar SA. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Zamar RH. Least angle regression for model selection. WIREs Comput. Stat. 2014;6:116–123. doi: 10.1002/wics.1288. [DOI] [Google Scholar]
- 35.Gerken T, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao X, Yang Y, Sun BF, Zhao YL, Yang YG. FTO and obesity: Mechanisms of association. Curr. Diab. Rep. 2014;14:486–495. doi: 10.1007/s11892-014-0486-0. [DOI] [PubMed] [Google Scholar]
- 37.Speakman JR. The “Fat Mass and Obesity Related” (FTO) gene: mechanisms of impact on obesity and energy balance. Curr. Obes. Rep. 2015;4:73–91. doi: 10.1007/s13679-015-0143-1. [DOI] [PubMed] [Google Scholar]
- 38.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem. Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Kypreos KE, et al. Mechanisms of obesity and related pathologies: role of apolipoprotein E in the development of obesity. FEBS J. 2009;276:5720–5728. doi: 10.1111/j.1742-4658.2009.07301.x. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Liu L. Apolipoprotein E synthesized by adipocyte and apolipoprotein E carried on lipoproteins modulate adipocyte triglyceride content. Lipids Health Dis. 2014;13:136. doi: 10.1186/1476-511X-13-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volcik KA, et al. Apolipoprotein E polymorphisms predict low density lipoprotein cholesterol levels and carotid artery wall thickness but not incident coronary heart disease in 12,491 ARIC study participants. Am. J. Epidemiol. 2006;164:342–348. doi: 10.1093/aje/kwj202. [DOI] [PubMed] [Google Scholar]
- 42.Zeljko HM, et al. E2 allele of the apolipoprotein E gene polymorphism is predictive for obesity status in Roma minority population of Croatia. Lipids Health Dis. 2011;10:9–14. doi: 10.1186/1476-511X-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazzotti DR, et al. Association of APOE, GCPII and MMP9 polymorphisms with common diseases and lipid levels in an older adult/elderly cohort. Gene. 2014;535:370–375. doi: 10.1016/j.gene.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 44.Petkeviciene J, et al. Associations between apolipoprotein E genotype, diet, body mass index, and serum lipids in Lithuanian adult population. PLoS ONE. 2012;7:2–7. doi: 10.1371/journal.pone.0041525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fajas L, et al. The organization, promoter analysis, and expression of the human PPARgamma gene. J. Biol. Chem. 1997;272:18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 46.Robinson E, Grieve DJ. Significance of peroxisome proliferator-activated receptors in the cardiovascular system in health and disease. Pharmacol. Ther. 2009;122:246–263. doi: 10.1016/j.pharmthera.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 47.Mansoori A, Amini M, Kolahdooz F, Seyedrezazadeh E. Obesity and Pro12Ala polymorphism of peroxisome proliferator-activated receptor-gamma gene in healthy adults: a systematic review and meta-analysis. Ann. Nutr. Metab. 2015;67:104–118. doi: 10.1159/000439285. [DOI] [PubMed] [Google Scholar]
- 48.Deeb SS, et al. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat. Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 49.Baldani DP, et al. Association of PPARG Pro12Ala polymorphism with insulin sensitivity and body mass index in patients with polycystic ovary syndrome. Biomed. Rep. 2014;2:199–206. doi: 10.3892/br.2013.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans D, et al. Variation in the gene for human peroxisome proliferator activated receptor gamma (PPARgamma) does not play a major role in the development of morbid obesity 2. IntJ Obes. Relat. Metab. Disord. 2000;24:647–651. doi: 10.1038/sj.ijo.0801214. [DOI] [PubMed] [Google Scholar]
- 51.Hsiao TJ, Lin E. The Pro12Ala polymorphism in the peroxisome proliferator-activated receptor gamma (PPARG) gene in relation to obesity and metabolic phenotypes in a Taiwanese population. Endocrine. 2015;48:786–793. doi: 10.1007/s12020-014-0407-7. [DOI] [PubMed] [Google Scholar]
- 52.Randall JC, et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9:e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamri A, et al. Dietary fat intake and polymorphisms at the PPARG locus modulate BMI and type 2 diabetes risk in the D.E.S.I.R. prospective study. Int. J. Obes. 2012;36:218–224. doi: 10.1038/ijo.2011.91. [DOI] [PubMed] [Google Scholar]
- 54.Dedoussis GV, et al. An age-dependent diet-modified effect of the PPARgamma Pro12Ala polymorphism in children. Metabolism. 2011;60:467–473. doi: 10.1016/j.metabol.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 55.Verier MDC, et al. Breast-feeding modulates the influence of the peroxisome proliferator-activated receptor-[gamma] (PPARG) Pro12Ala polymorphism on adiposity in adolescents. Diabetes Care. 2010;33:190–196. doi: 10.2337/dc09-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Costa-Urrutia P, et al. Genetic obesity risk and attenuation effect of physical fitness in Mexican-Mestizo population: a case-control study. Ann. Hum. Genet. 2017;81:106–116. doi: 10.1111/ahg.12190. [DOI] [PubMed] [Google Scholar]
- 57.Silbernagel G, et al. The L162V polymorphism of the peroxisome proliferator activated receptor alpha gene (PPARA) is not associated with type 2 diabetes, BMI or body fat composition. Exp. Clin. Endocrinol. Diabetes. 2009;117:113–118. doi: 10.1055/s-0028-1082069. [DOI] [PubMed] [Google Scholar]
- 58.Martínez-García F, et al. Impact of obesity-related genes in Spanish population. BMC Genet. 2013;14:111. doi: 10.1186/1471-2156-14-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hung CF, et al. A genetic risk score combining 32 SNPs is associated with body mass index and improves obesity prediction in people with major depressive disorder. BMC Med. 2015;13:86. doi: 10.1186/s12916-015-0334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.El-Sayed AM, Scarborough P, Galea S. Unevenly distributed: a systematic review of the health literature about socioeconomic inequalities in adult obesity in the United Kingdom. BMC Public. Health. 2012;12:18–30. doi: 10.1186/1471-2458-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martínez de Morentin-Aldabe BE, Hernández-Ruiz de Eguilaz M, Pérez-Díez S, Martínez JA. Influencia de la administración de psicofármacos en el aumento del peso corporal. Rev. Esp. Nutr. Hum. Y. Dietética. 2013;17:17–26. doi: 10.14306/renhyd.17.1.4. [DOI] [Google Scholar]
- 62.Gómez-Ambrosi J, et al. Body mass index classification misses subjects with increased cardiometabolic risk factors related to elevated adiposity. Int. J. Obes. 2012;36:286–294. doi: 10.1038/ijo.2011.100. [DOI] [PubMed] [Google Scholar]