Abstract
As an opportunistic pathogen, Aspergillus flavus is one of the major causes of food contamination around the world. In this study, pbsB gene knockout mutant (ΔpbsB) and pbsB overexpression strain (OE) of A. flavus were constructed by homologous recombination. The results showed that the mycelia growth, conidiation, and the formation of sclerotia in ΔpbsB mutant were significantly suppressed, and up-regulated in OE strian compared to wild-type strain (WT). Q-PCR analysis showed that PbsB regulated the sclerotia formation through sclerotia related gene nsdC. With TLC and qRT-PCR analysis, it was found that PbsB up-regulated the bio-synthesis of aflatoxin B1 (AFB1) through regulatory gene aflR and structural gene aflC, aflD, aflK, and aflQ in the aflatoxin gene cluster. In osmotic stress response analysis, ΔpbsB mutant was significantly more sensitive to osmotic pressure with 1.2 mol/L sorbitol, compared to WT and OE strains. In virulence analysis, the infection capacity of ΔpbsB strain to peanut and maize kernels decreased dramatically, and significantly fewer spores and lesser mycelia were produced in ΔpbsB strain on the surface of peanut and maize kernels, and the infection capacity of OE strain to kernels increased significantly compared with WT strain. The AFB1 bio-synthesis ability of A. flavus in crop invasion models was also found to be coincide with the expression level of pbsB. All the results of the study shows that, as a MAPKK, PbsB is critical for growth and virulence in A. flavus, and lay a theoretical foundation for the prevention and control of A. flavus contamination.
Keywords: pbsB, aflatoxin B1, MAPKK, crop invasion, Aspergillus flavus
Introduction
The soil-borne pathogen Aspergillus flavus is an opportunistic pathogen, which is one of the major causes of the mycotoxins contamination to crops (such as peanuts, maize kernel, and cotton) around the world (Satterlee et al., 2016). The most agriculturally important mycotoxins known are aflatoxins, in which aflatoxin B1 (AFB1) is the most toxic, carcinogenic, mutagenic, and teratogenic secondary metabolite (Xing et al., 2016). AFB1 has been classified by the International Agency for Research on Cancer (IARC) as a Group 1 carcinogen (Wu et al., 2014). AFB1 is extensively linked to liver cancer, reports indicated that chronic exposure to AFB1 through daily diets lead to immunosuppression, fatty liver, hepatic lesions, and even hepatomas (Yu, 2012). And high dose aflatoxins could cause death from aflatoxicosis (Misihairabgwi et al., 2017).
The development and secondary metabolite production of filamentous fungi are found to be regulated by a number of orthodox regulatory factors (Cary et al., 2012). For example, the global regulator, VeA, mediates aflatoxin production via AflR, and regulates the development of cleistothecia in Aspergillus nidulans and sclerotia in A. flavus (Cary et al., 2006). Environmental factors (such as nutrition and stresses) are also found to be involved in the morphogenesis and secondary metabolites bio-synthesis in Aspergillus spp. (Han et al., 2003; Fountain et al., 2016). MAP (mitogen-activated protein) kinase cascade is one of the mechanisms for eukaryotic cell to transfer the extracellular environment information through plasma membrane-associated receptors to the expression of target genes in the nucleus. Exposure of budding yeast to increased extracellular osmolarity activates one of the MAP kinase cascades—high osmolarity glycerol (HOG) response pathway, in which the MAP kinase (MAPK) HOG1 is activated with the phosphorylation of Thr174 and Tyr176 by MAP kinase kinase (MAPKK) Pbs2 (Ferrigno et al., 1998). The activation of Pbs2 requires to be phosphorylated by a MAP kinase kinase kinase (MAPKKK) STE11, or one of two partially redundant MAPKKKs, SSK2, and SSK22 (Posas et al., 1998). A. nidulans pbsB or hogA (pbs2p or hog1 in yeast) deletion mutant showed growth inhibition under the stress of high osmolarity, and HOGA MAPK in these mutants could not be phosphorylated under the stress of osmolality or oxidation (Furukawa et al., 2005). Similar to HOG1, Pbs2 was reported to wildly involve in stress responses in yeast and various pathogenic fungus (Gustin et al., 1998; Bahn et al., 2006; Cheetham et al., 2007). Bahn et al. (2005) reported that the human pathogenic fungus Cryptococcus neoformans Serotype A hog1 and pbs2 mutants are attenuated in virulence. The study from Cheetham et al. (2011) indicated that both Candida albicans with nonphosphorylatable Pbs2 and with mutation of the consensus sites of Pbs2 displayed the impaired stress resistance and attenuated virulence in a mouse model.
The development and mycotoxin bio-synthesis of fungi were also regulated by signaling pathways which interacting with MAP kinase cascade. By systematic disruption the MAP kinase genes in yeast, Kawasaki et al. (2008) found that kinase Ste20p and Ste50p interacted with G protein subunits. FadA is the alpha subunit of heterotrimeric G protein in A. nidulans, Hicks et al. (1997) reported that both asexual sporulation and ST (sterigmatocystin) production of A. nidulans require the inhibition of FadA-dependent signaling, which is also a conserved mechanism to regulate aflatoxins biosynthesis in aflatoxins producing fungi. Yu (2016) reported that the conidiation, vegetative growth, stress response, and toxigenesis of A. nidulans were governed by the components of G protein (including FadA, GanB, SfaD, and GpgA). And, (Shimizu and Keller, 2001) found sporulation was decreased in the pkaA (the cAMP-dependent protein kinase catalytic subunit, a downstream target of FadA) overexpression strain, as occurs in fadA-dominant active strains.
A. flavus distributes widely in the world as a pathogenic fungus. The pathogenicity, infection, and toxicity of A. flavus seriously impacts the safety of human society globally. It is very important to reveal the regulation mechanism of A. flavus pathogenicity. But the role of PbsB (Gene bank No. in NCBI: AFLA_083380) in the aflatoxin bio-synthesis and virulence of A. flavus keeps unknown. Therefore, this study was designed to explore the PbsB functions in mycelia growth, conidiation, sclerotia formation, aflatoxin production, and pathogenicity of A. flavus.
Materials and methods
Fungal strains and growth conditions
A. flavus strains used in the study were listed in Table 1. The primers used in the study were showed in Table 2. YPD (1% yeast extract, 2% peptone, 2% glucose, and 1.5% agar) and YES (2% yeast extract, 150 g/L sucrose, 1 g/L MgSO4·7H2O) were prepared for the cultivation of A. flavus strains in the study. Supplements (Uracil, and uridine) for auxotrophic marker (pyrG-) were added as required (Yang et al., 2016; Li et al., 2017). Strains were stored in 30% glycerol at −80°C.
Table 1.
A. flavus strains used in this study.
| Strain name | Related genotype | Source |
|---|---|---|
| CA14 Δku70ΔpyrG | Wild type (pyrG-) | Purchased from ATCC |
| CA14 Δku70 | Wild type (pyrG+) | Prepared in our lab |
| ΔpbsB | Δku70, ΔpbsB::pyrG | This study |
| OE | Δku70, pyrG::gpdA(p)::pbsB | This study |
Table 2.
Primers used in this study.
| Primer name | Sequence (5′ → 3′) |
|---|---|
| pbsB-AF | TAGTGCGTGCGTCCGTTTA |
| pbsB-AR | GGGTGAAGAGCATTGTTTGAGGCAGCGATGTCGCAAATCCAG |
| pbsB-BF | GCATCAGTGCCTCCTCTCAGACGCCTCCAAGGATGATGATGA |
| pbsB-BR | GAATGGTGTATCCGTAGTGC |
| pbsB-OrfF | ATCAGTGCAGCTCGAAGA |
| pbsB-OrfR | AGGCACCGTAGAAATCAA |
| pyrG-PF | GCCTCAAACAATGCTCTTCACCC |
| pyrG-PR | GTCTGAGAGGAGGCACTGATGC |
| pyrG-R | CAGGAGTTCTCGGGTTGTCG |
| pyrG-F | ATCGGCAATACCGTCCAGAAGC |
| OverlapF | CTGACATAGTCCATTGGCTG |
| OverlapR | GTTCCAGGTCATCTTCTTCG |
| gpdA-F | GCATCAGTGCCTCCTCTCAGACGTACAGTGACCGGTGACTCTTTCTGG |
| gpdA-R | GTGATGTCTGCTCAAGCGGGGTAG |
| OE-F | CTACCCCGCTTGAGCAGACATCACATGGCATCCGAAATCGATCCTGTAGC |
| OE-R | CCACCATCCATATACTCAACGCAGATGTAGAC |
| OE-overlap-F | AAGTCATAGAAGGATTCTGTTCGCCTACG |
| OE-overlap-R | TTATCCGTGACCCCATCGGAGCCA |
Bioinformatics analysis
The homologs of PbsB from (A. flavus, A. oryzae, A. clavatus, A. fumigatus, A. niger, A. terreus, A. nidulans, N. crassa, and S. cerevisiae) were downloaded from NCBI (http://www.ncbi.nlm.nih.gov), and further aligned with Clustal X. Protein domains of above 9 species were analyzed with InterPro (http://www.ebi.ac.uk/interpro/scan.html) and edited with IBS 1.0. Phylogenetic tree of PbsB homologs from these 9 species was constructed with MEGA5.1 by an algorithm of 1,000 times Neighbering comparison.
Preparation of pbsB deletion and over expression strains of A. flavus
The pbsB deletion mutants were constructed according to Han et al. (2016) with minor modification. The deletion cassette was constructed by fusion PCR. In details, 5′- untranslated regions (5′-UTR) and 3′-UTR were amplified from A. flavus genomic DNA with two pairs of primers (pbsB-AF and pbsB-AR, pbsB-BF and pbsB-BR in Table 2). The intermediate A. fumigatus pyrG was amplified from A. fumigatus genomic DNA by primers pyrG-PF and pyrG-PR (Table 2) as transformation selecting marker. 5′-UTR and 3′-UTR of pbsB and pyrG were fused together by fusion PCR with nesting primers OverlapF and OverlapR (Table 2). Polyethylene glycol-mediated transformation of CA14Δku70ΔpyrG protoplasts was carried out as described by Cary et al. (2005). The pyrG prototroph strain (ΔpbsB: Δku70, ΔpbsB::pyrG showed in Table 1), in which the entire pbsB coding region was replaced by pyrG, was further tested with PCR (primers listed in Table 2), and the flanking regions of confirmed ΔpbsB strain was further sequenced by BioSune (Shanghai, China). The pbsB overexpression strains were prepared by homologous recombination (Nie et al., 2016). After four DNA fragments were amplified with four pairs of primers: pbsB-AF and pbsB-AR for 5′HR, pyrG-PF and pyrG-PR for pyrG, gpdA-F and gpdA-R for gpdA(p), and OE-F and OE-R for 3′HR, they were overlapped with nesting primers: OE-overlap-F and OE-overlap-R (Table 2). And pbsB overexpression strains (OE) were prepared by Polyethylene glycol-mediated transformation of CA14Δku70ΔpyrG protoplasts with the overlap PCR production as mentioned above. The OE strains were further confirmed with primer gpdA-F and OE-R for 2,000 bp OE-ORF fragment, and with primer OE-overlap-F and OE-R for 4,730 bp OR-ORF fragment as shown in Table 2. Finally, the expression level of pbsB in A. flavus strains mentioned above was further assayed with qRT-PCR analysis.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis
The qRT-PCR was conducted according to the protocol provided by Zhang et al. (2016). Three micrograms of total RNA were treated with DNase I (Thermo Fisher Scientific, Waltham, MA, USA) to remove possible G-DNA contamination. One microgram G-DNA free RNA was reverse transcribed by using the Revert Aid First-strand cDNA Synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA). qRT-PCR was carried out by using the SYBR Green Premix kit (Takara, Dalian, China) with Mx3000p thermocycler (Agilent Technologies). The expression levels of target gene were evaluated with the 2′ ΔΔCt method.
TLC analysis of AFB1 production
The effect of PbsB in AFB1 production was performed after 5 d cultivation according to the method by Dhingra et al. (2013). 25 mL of liquid YES medium was inoculated with 104 conidia/mL, and the cultures were shaking at 180 r/min at 28°C. The supernatant samples were analyzed by thin-layer chromatography (TLC). A volume of 3 μL supernatant was spotted onto a TLC plate (Si250F, J.T. Baker), and the plate was developed in acetone-chloroform (2:8, vol/vol), and dried at 80°C for 10 min, then the aflatoxin B1 was examined under UV light.
Kernel assays
Peanut and maize kernels were prepared to discover the role of PbsB in plant infection following the method described by Yang et al. (2016) with minor modification. Each peanut and maize kernel was sterilized with 0.05% sodium hypochlorite. Viable cotyledon was dried and placed on sterile Petri dish plate. The cultures were incubated at 28°C in dark for 7 d. For AFB1 analysis, a Petri dish plate of kernels was soaked in double distill water for 10 h. After chloroform (half of the water volume) was added, the mixture was shacked at 180 r/min for 1 h, and kept still for another 2 h. The chloroform layer was collected and dried by evaporation. The sediment was re-suspended with 1 mL chloroform, and re-dissolved in 30 μL chloroform after dried by evaporation. Finally, 5 μL sample was analysis by TLC.
Statistical analysis
The data in the study was presented as the means ± standard deviation (SD). The presence of statistical differences was determined by one-way ANOVA, and statistical significance was recognized when P < 0.05.
Results
Bioinformatics analysis of PbsB
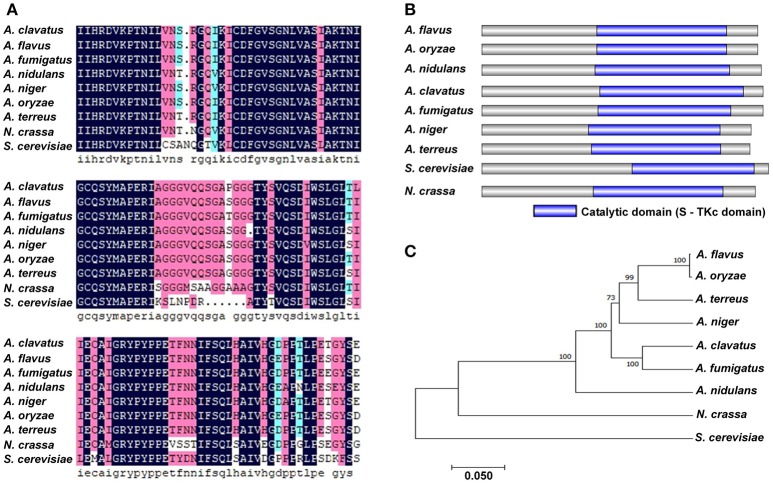
PbsB protein in A. flavus and its orthologs in other 8 species were aligned by Clustal X, and the result showed that PbsB in A. flavus and A. oryzae had the highest similarity (83.27%), and the lowest homology was found between A. flavus and S. cerevisiae (29.70%) in Figure 1A. The protein domain in PbsB was further analyzed with IBS 1.0, and a catalytic domain (Serine/Threonine protein kinases) was found in all 9 species (Figure 1B), which meant that the catalytic domain was very conservative. Phylogenetic tree among these 9 species was further constructed with MEGA5.1 as shown in Figure 1C. The genetic relationship of PbsB from A. flavus and A. oryzae was found to be the closest, compared with the relationship of the protein from A. flavus and any other species, and the protein sequences of PbsB from Aspergillus spp. were classified into one cluster compared with the species from other genera (N. crassa and S. cerevisiae).
Figure 1.
Bioinformatics analysis of PbsB. (A) Amino acid alignment of A. flavus PbsB and other 8 putative orthologs. Clustal X was used in this analysis. (B) Diagram shows the domains in PbsB among above 9 species. InterPro (http://www.ebi.ac.uk/interpro/scan.html) and IBS 1.0 were used in the analysis. (C) The diagram of the phylogenetic tree among 9 species.
PbsB in A. flavus enhances the growth of mycelia
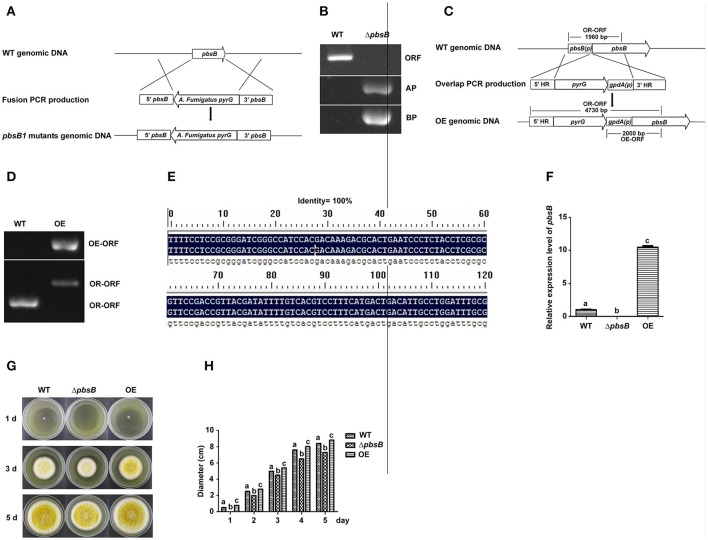
The A. flavus pbsB deletion strains (ΔpbsB) and over expression strains (OE) were constructed by transforming the protoplasts of WT (pyrG-) with the fusion PCR productions, respectively (Figures 2A,C), and the resulting transformants were confirmed by PCR analysis (Figures 2B,D). The result of PCR showed that the ORF of pbsB has been deleted from pbsB deletion mutants, and the amplification of AP and BP from ΔpbsB meant that ORF of pbsB has been replaced by pyrG from A. fumigatus (Figure 2B). The franking regions between pyrG and 5′-UTR and 3′-UTR of pbsB of the PCR confirmed ΔpbsB mutant were further sequenced and aligned with DNAMAN (Version: 6.0.40), the results (100% identity, Figure 2E) showed pbsB had been deleted as the scheme showed in Figure 2A. The results in Figure 2D showed that 2,000 bp DNA fragment (OE-ORF) only could be amplified from OE strain, and the OR-ORF fragment from OE strain was 4,730 bp, but it was only 1,960 bp for WT strain (Figures 2C,D). By qRT-PCR analysis, it was found that not pbsB activity could be detected in ΔpbsB, and the expression level of pbsB in OE was dramatically improved compared with WT (Figure 2F). These results showed that both ΔpbsB and OE strains of A. flavus were successfully constructed. In order to evaluate the role of PbsB played in the growth of mycelium, the conidia (104 spores/mL) from A. flavus strains (WT, ΔpbsB and OE) were point inoculated onto YES agar in dark at 37°C for 5 d. The results showed that the colony diameter of ΔpbsB mutant was obvious smaller than that of WT from 1 to 5th d, and opposite situation was found in OE strain compared to WT strain (Figure 2G). The diameter of each mycelium colony was measured from 1 to 5 day, and the results of the histogram in Figure 2H revealed that the PbsB significantly improved the growth of A. flavus.
Figure 2.
PbsB involved in the growth of A. flavus. (A) The scheme for ΔpbsB strain construction by homologous recombination. 5′-UTR (5′pbsB) and 3′-UTR (3′pbsB) of pbsB and 1.89 kb pyrG from A. fumigatus were amplified, respectively, and they were fused together with nesting primers. The pbsB deletion strain was prepared with pyrG to replace pbsB in WT (CA14Δku70ΔpyrG) through transformation the protoplast with the fusion production of 5′-UTR-pyrG-3′-UTR by homologous recombination. (B) The results of PCR analysis, in which ORF was amplified with primer pbsB-OrfF and pbsB-OrfR, AP was with pbsB-AF and pyrG-R, and BP was with pyrG-F and pbsB-AR. (C) The scheme for OE strain construction by homologous recombination. (D) OE strain was detected with PCR analysis. OE-ORF was amplified with primer gpdA-F and OE-R, and OR-ORF was with primer OE-overlap-F and OE-R. (E). The alignment of flanking regions of pyrG and 5′ and 3′UTR of pbsB in ΔpbsB strain between sequencing results and constructed sequence map. (F) The expression level of pbsB in WT, ΔpbsB, and OE strain were analyzed with qRT-PCR. (G) Colony of WT A. flavus, ΔpbsB mutant and OE strains were cultured in YES medium for 1 d to 5 d. (H). Comparative analysis of colony diameters, the letters “a,” “b,” and “c” used in the histogram of this study represented the significant difference among WT, ΔpbsB mutant, and OE strain (P < 0.05).
PbsB up-regulates A. flavus conidation
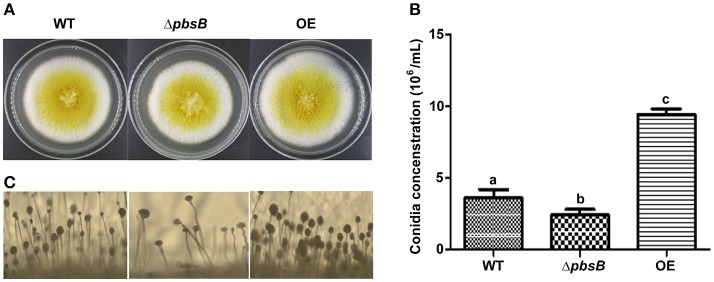
Aspergillus spp. always produces a massive number of conidia which are easily dispersed in the air in breeze. Conidiation is an important means to spread contamination for A. flavus, so the capacity of asexual reproduction—conidiation is a critical index to amplify the detriment of A. flavus. To explore the role of PbsB in conidiation, A. flavus strains (WT, ΔpbsB, and OE) were point inoculated onto YES at 37°C in dark, and phenotype was observed after 4 d cultivation. ΔpbsB mutant showed the decreased conidial production compared to WT strain (Figures 3A,B). Meanwhile, more conidiophores were observed in WT strain when compared to ΔpbsB mutant, and the conidiophores from ΔpbsB mutant were obvious dysplasia (Figure 3C). When pbsB overexpressed, significantly (P < 0.005) more conidia and conidiophores was observed in OE strain compared with WT strain (Figure 3). The result showed that PbsB improved the production of conidia, and the absence of PbsB significantly (P < 0.05) repressed the asexual reproduction of A. flavus.
Figure 3.
PbsB up-regulates A. flavus asexual reproduction. (A) Colony of WT, ΔpbsB mutant and OE strains were cultured in YES for 4 d. (B) The conidia numbers of WT and ΔpbsB, and OE strains were calculated with hemocytometer. (C) The conidiophores of WT, ΔpbsB, and OE strains were observed under microscope.
PbsB is important in the formation of sclerotia in A. flavus
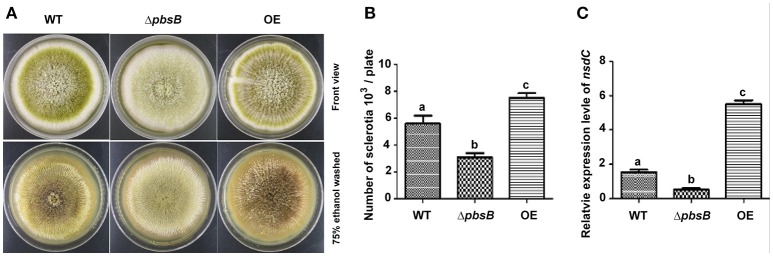
Sclerotia are readily produced by single strain of A. flavus, and sclerotia formation is commonly considered to be survival structure of A. flavus against unfavorable conditions. To reveal the bio-function of PbsB in the formation of sclerotia, A. flavus strains (WT, ΔpbsB, and OE) were point inoculated onto YPD at 37°C in dark for 7 d. After spraying with 75% ethanol, and the number of sclerotia on each plate was counted, respectively. The result showed that lack of PbsB significantly (P < 0.01) decreased the production of sclerotia, and the overexpression of pbsB obviously (P < 0.01) increased sclerotia production compared with WT (Figures 4A,B). The expression level of sclerotia regulator (nsdC) was also analyzed by qRT-PCR, and the results showed that nsdC was significantly (P < 0.01) down-regulated at 72 h when PbsB was absent, and was dramatically (P < 0.005) up-regulated when PbsB was overexpressed (Figure 4C). All these results indicate that PbsB is important in sclerotia formation in A. flavus.
Figure 4.
PbsB played an important role in sclerotia formation. (A) Point inoculated cultures of A. flavus WT, ΔpbsB mutant, and OE strain in YPD. Strains were cultivated for 7 d at 37°C. (B) The numbers of sclerotia in each plate were calculated. (C) The qRT-PCR analysis of gene expression levels of nsdC in WT strain, ΔpbsB mutant, and OE strain.
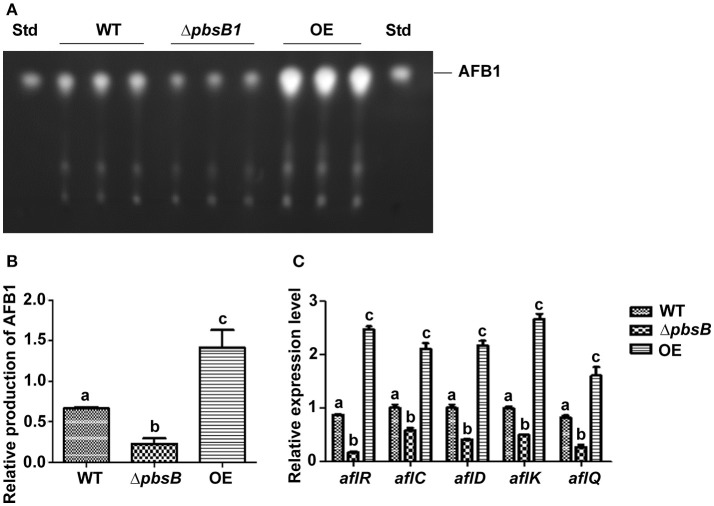
PbsB plays a vital role in AFB1 bio-synthesis of A. flavus
The role of PbsB in AFB1 production was analyzed by cultivating the WT, ΔpbsB and OE strains in the YES liquid medium, and the samples were collected at 5th day. The result of TLC analysis showed that AFB1 production was significantly (P < 0.005) decreased when pbsB was deleted, and the mycotoxin was obviously (P < 0.005) elevated when pbsB overexpressed in OE strain compared with WT strain (Figures 5A,B). The expression levels of aflatoxin bio-synthesis gene aflC, aflD, aflK, and aflQ, and regulatory gene aflR at 48 h were further analyzed by qRT-PCR, and the results showed that the expression levels of the aflatoxin bio-synthesis and regulatory genes were all significantly down-regulated in ΔpbsB mutant and up-regulated in OE strain compared to WT strain (Figure 5C). These results reflected that PbsB positively regulated the aflatoxin bio-synthesis through aflatoxin bio-synthesis and regulatory genes (aflR, aflC, aflD, aflK, and aflQ).
Figure 5.
The role of PbsB in the AFB1 bio-synthesis of A. flavus. (A) TLC analysis of AFB1 production from WT, ΔpbsB mutant, and OE strain after 5 d of incubation. (B) Relative quantification of AFB1 production of flavus strain as mentioned in (A). (C) Relative expression levels of aflatoxin bio-synthesis regulatory and structural genes aflR, aflC, aflD, aflK, and aflQ monitored by qRT-PCR at 48 h.
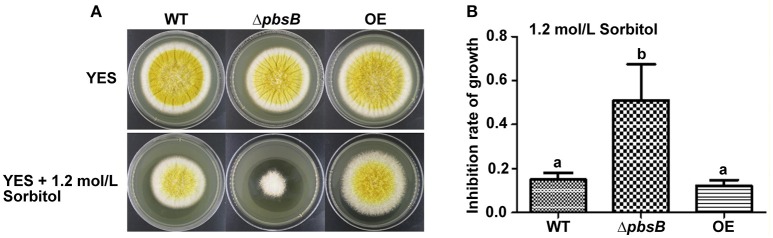
PbsB involves in the resistance of A. flavus to hyperosmotic stress
PbsB was reported to play an important role in the resistance of C. albicans to hyperosmotic conditions (Arana et al., 2005). To assess the role of PbsB under hyperosmotic stress in A. flavus, WT, ΔpbsB and OE strains were inoculated onto YES agar with 1.2 mol/L sorbitol for 4 d. It was found that the inhibition rate of ΔpbsB mutants dramatically (P < 0.01) greater than that of WT and OE strains (Figure 6). The results revealed that pbsB deletion mutant was more susceptible to hyperosmotic stress, and PbsB was one of the critical factors in A. flavus to fight against hyperosmotic conditions.
Figure 6.
The ΔpbsB mutants were more sensitive to hyperosmotic stress. (A) The colonies of WT, ΔpbsB mutant and OE strain in YES medium with or without 1.2 mol/L Sorbitol. (B) Inhibition rate of growth with 1.2 mol/L Sorbitol. The inhibition rate = (diameter of colony without inhibitor—diameter of colony with inhibitor)/diameter of colony without inhibitor.
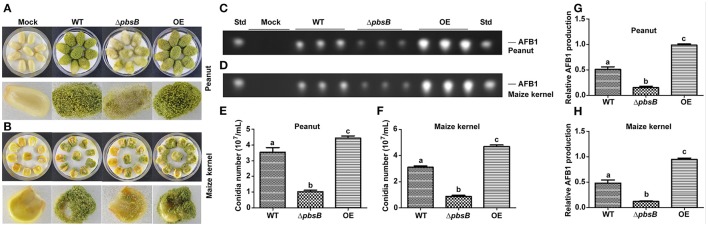
PbsB is essential for A. flavus pathogenicity
Peanut and maize kernels were inoculated with 104 spores/mL of WT, ΔpbsB and OE strains in Petri dish plate for 7 d, respectively. It was shown in Figures 7A,B,E,F that dramatically (P < 0.005) reduced conidiation was observed in ΔpbsB mutant, and significant (P < 0.05) increased sporulation yield was found in OE strain compared to that in WT strain. TLC analysis showed that AFB1 bio-synthesis capacity of A. flavus on peanuts was significantly (P < 0.005) repressed when PbsB was absent from ΔpbsB mutant, and was obviously (P < 0.005) improved when PbsB was over-produced in OE strain (Figures 7C,G). To maize kernel, similar results were observed (Figures 7D,H). All these results of the study indicated that PbsB played an essential role in crops infection.
Figure 7.
Crop grains colonization of WT, ΔpbsB mutant, and OE strain. (A,B) A. flavus strains cultured on peanut and maize kernels. (C,D) TLC analysis of AFB1 levels in infected peanut and maize kernels. (E) The number of conidia on peanut seeds counted after 7 d inoculation. (F) The number of conidia on maize kernel. (G) AFB1 level produced from A. flavus invaded peanut seeds from the results of (C,H). AFB1 level produced from A. flavus invaded maize kernel from the results of (D).
Discussion
MAP kinase cascades are important pathway for yeast and filamentous fungus to respond to the stimuli from outside environment (Gustin et al., 1998; Furukawa et al., 2005; Bahn et al., 2006).
As an MAPKK, PbsB catalyzes the phosphorylation of MAPK under the stress of osmolality or oxidation (Furukawa et al., 2005). In A. flavus, pbsB (AFLA_083380) was made up by 2002 nucleotide residues, composing of 2 extrons and 1 introns, which were translated into protein PbsB (XP_002373253) with 643 amino acid residues. The bio-information analysis on PbsB showed that it harbored a conservative Serine/Threonine protein kinases domain, and the PbsB protein sequences in all selected Aspergillus spp. were grouped into one cluster, which suggested that the sequence of PbsB was very conservative, and it play a very important role in MAP kinase cascades.
PbsB involves in the growth of mycelium, conidiation, and sclerotia formation. Arana et al. reported 2005 that Pbs2 repressed hyphal formation in C. albicans, and deletion of pbs2 from C. albicans increased hyphal growth under different conditions. On the contrary, our study found that PbsB in A. flavus positively participated the regulation of mycelium growth (Figures 2G,H). Liu et al. (2017) reported that vegetative growth of Beauveria bassiana Δpbs2 strains on minimal media with different carbon/nitrogen sources was suppressed, and suffered severe conidiation defects. In the study, sever asexual reproduction defects were also observed in A. flavus pbsB mutant (Figure 3,P < 0.05) on YES medium and on the surface of peanut and maize kernels (Figures 7A,B,E,F,P < 0.005). And it was also found that the overexpression of pbsB in OE strain significantly improved its conidiation capacity on both conditions (Figures 3,7). Cary et al. (2012) and Kim et al. (2009) reported that nsdC gene (encoding a putative transcription factor) was found up-regulating vegetative growth in A. nidulans. Our Q-PCR results for nsdC gene (Figure 4C) showed that when pbsB was deleted, the expression level of nsdC was down-regulated significantly (P < 0.01), and when pbsB was overexpressed, the activity of nsdC was up-regulated dramatically (P < 0.005), which meant that pbsB regulated the asexual reproduction (conidiation) of A. flavus at the upstream of nsdC. No reports on the role of PbsB in sclerotia development was found, yet. The formation of sclerotia is considered to survive the unfavorable conditions for A. flavus (Wicklow, 1987). In the study, we found that the absence of PbsB in A. flavus obviously (P < 0.01) down-regulated the formation of sclerotia, and the increase of PbsB in OE strain significantly (P < 0.01) improved sclerotia formation. It is reported that nsdC gene is also required for production of sclerotia (Cary et al., 2012). From the result of Figure 4, it was concluded that PbsB positively regulated the formation of sclerotia in A. flavus through nsdC. The results of our study disclosed that PbsB plays an important role in the mycelium growth, development, and reproduction of A. flavus.
PbsB up-regulates the bio-synthesis of AFB1 in A. flavus. No reports which directly linked the role of PbsB with production of mycotoxin is found till now. Our study on A. flavus revealed that the absence of PbsB dramatically &(P < 0.005) reduced the level of AFB1 production in liquid YES medium and on the surface of both peanut and maize kernels. And when the expression level of pbsB was up-regulated in OE, the production of AFB1 increased dramatically (Figures 5,7,P < 0.005). The expression of regulatory gene aflR is required for transcription of most structural genes in the aflatoxin gene cluster, and structural gene aflC, aflD, aflK, and aflQ are involved in the conversion of acetate to AFB1 in AFB1 bio-synthesis pathway (Yu, 2012). The results in Figure 5 showed that the expression level of aflR, aflC, aflD, aflK, and aflQ were significantly down-regulated in pbsB deletion mutant and up-regulated in OE strain, which reflected that PbsB increased AFB1 bio-synthesis level through up-regulating the expression levels of regulatory and structural genes in the aflatoxin gene cluster (Figure 5C). Our research revealed that PbsB took part in the regulation of AFB1 bio-synthesis at the up-stream of the orthodox pathway of aflatoxin bio-synthesis (such as aflR and aflQ). Further exploration of the bio-function of PbsB in mycotoxin synthesis in pathogenic fungus would reveal the role of MAP kinase cascades in secondary metabolism in filamentous fungus.
PbsB is an essential toxic factor to encounter severe environmental stresses in A. flavus. In the crop models of the study, attenuated toxicity of A. flavus was found when PbsB was absent. ΔpbsB mutant was found to produce significantly fewer conidia and AFB1, but more conidia and AFB1 were produced in OE strain when pbsB was overexpressed. When pathogenic fungi invade host plant, they face a serials of challenges from outside environment, including hyperosmotic conditions and peroxide stress. Arana et al. (2005) revealed that the human pathogen C. albicans lacking Pbs2 was defective in growth under hyperosmotic conditions mediated by sorbitol. Mkc1 is involved in the response to oxidative stress and cell wall integrity (Correia et al., 2017). And the Western blot signal of phosphorylated Mkc1 in response to hydrogen peroxide is reduced significantly in pbs2 mutants (Arana et al., 2005). Esquivel-Naranjo et al. (2016)reported that Trichoderma atroviride pbs2 mutants were highly sensitive to cellular insults, such as osmotic and oxidative stress, cell wall damage, high temperature, cadmium, and UV irradiation. Our results from the study showed PbsB played an important role in the resistance of A. flavus to severe outside environmental conditions, as well as to the environment inside its hosts.
The results of our study revealed the bio-function of pbsB in mycelia growth, conidiation, sclerotia formation, aflatoxin production, and pathogenicity of filamentous pathogenic fungus A. flavus, and might provide potential targets in the control of the contamination from pathogenic fungus.
Author contributions
ZZ designs and writes the manuscript, takes part in all experiments of the whole project, and provided funds support. JY takes part in all experiments designs, and constructed Aspergillus flavus deletion strains in the study, took part in the experiments on stress resistance of A. flavus, and provided funds support. ZC took part in the experiments on aflatoxin production analysis, and morphogenesis analysis. FZ took part in the experiments of peanut seeds and corn grains model construction. JS took part in the experiments of corn grains model construction. SW took part in the designation of the project, manuscript correction, and provided funds support. ZG repeated all the experiments (from Figures 2–7), and analysis of the data for the work. DL prepared the over-expression strain and was responsible for the DNA sequencing of the flanking regions of the deletion strain. YZ took part in the preparation of the over-expression strain.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research was supported by the grants of the National Natural Science Foundation of China (No.31600118), the Nature Science Foundation of Fujian Province (2017J01426), the fund of Cultivation of Outstanding Youth Science and Technology Talents in Fujian Agriculture and Forestry University (xjq201410, xjq201412).
References
- Arana D. M., Nombela C., Alonso-Monge R., Pla J. (2005). The Pbs2 MAP kinase kinase is essential for the oxidative-stress response in the fungal pathogen Candida albicans. Microbiology 151, 1033–1049. 10.1099/mic.0.27723-0 [DOI] [PubMed] [Google Scholar]
- Bahn Y. S., Kojima K., Cox G. M., Heitman J. (2005). Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16, 2285–2300. 10.1091/mbc.E04-11-0987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn Y. S., Kojima K., Cox G. M., Heitman J. (2006). A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell 17, 3122–3135. 10.1091/mbc.E06-02-0113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary J. W., Ehrlich K. C., Bland J. M., Montalbano B. G. (2005). The aflatoxin biosynthesis cluster gene, aflX, encodes an oxidoreductase involved in conversion of vesicolorin A to demethylsterigmatocystin. Appl. Environ. Microbiol. 72, 1096–1101. 10.1128/AEM.72.2.1096-1101.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary J. W., Ehrlich K. C., Kale S. P., Calvo A. M., Bhatnagar D., Cleveland T. E. (2006). Regulatory elements in aflatoxin biosynthesis. Mycotoxin Res. 22, 105–109. 10.1007/BF02956773 [DOI] [PubMed] [Google Scholar]
- Cary J. W., Harris-Coward P. Y., Ehrlich K. C., Mack B. M., Kale S. P., Larey C., et al. (2012). NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot. Cell 11, 1104–1111. 10.1128/EC.00069-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham J., MacCallum D. M., Doris K. S., da Silva Dantas A., Scorfield S., Odds F., et al. (2011). MAPKKK-independent regulation of the Hog1 stress-activated protein kinase in Candida albicans. J. Biol. Chem. 286, 42002–42016. 10.1074/jbc.M111.265231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham J., Smith D. A., da Silva Dantas A., Doris K. S., Patterson M. J., Bruce C. R., et al. (2007). A single MAPKKK regulates the Hog1 MAPK pathway in the pathogenic fungus Candida albicans. Mol. Biol. Cell 18, 4603–4614. 10.1091/mbc.E07-06-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia I., Alonso-Monge R., Pla J. (2017). The Hog1 MAP Kinase Promotes the Recovery from Cell Cycle Arrest Induced by Hydrogen Peroxide in Candida albicans. Front. Microbiol. 7:2133. 10.3389/fmicb.2016.02133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra S., Lind A. L., Lin H. C., Tang Y., Rokas A., Calvo A. M. (2013). The fumagillin gene cluster, an example of hundreds of genes under veA control in Aspergillus fumigatus. PLoS ONE 8:e77147. 10.1371/journal.pone.0077147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquivel-Naranjo E. U., García-Esquivel M., Medina-Castellanos E., Correa-Pérez V. A., Parra-Arriaga J. L., Landeros-Jaime F., et al. (2016). A Trichoderma atroviride stress-activated MAPK pathway integrates stress and light signals. Mol. Microbiol. 100, 860–876. 10.1111/mmi.13355 [DOI] [PubMed] [Google Scholar]
- Ferrigno P., Posas F., Koepp D., Saito H., Silver P. A. (1998). Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 17, 5606–5614. 10.1093/emboj/17.19.5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain J. C., Bajaj P., Pandey M., Nayak S. N., Yang L., Kumar V., et al. (2016). Oxidative stress and carbon metabolism influence Aspergillus flavus transcriptome composition and secondary metabolite production. Sci. Rep. 6:38747. 10.1038/srep38747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Hoshi Y., Maeda T., Nakajima T., Abe K. (2005). Aspergillus nidulans HOG pathway is activated only by two-component signaling pathway in response to osmotic stress. Mol. Microbiol. 56, 1246–1261. 10.1111/j.1365-2958.2005.04605.x [DOI] [PubMed] [Google Scholar]
- Gustin M. C., Albertyn J., Alexander M., Davenport K. (1998). MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62, 1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K. H., Lee D. B., Kim J. H., Kim M. S., Han K. Y., Kim W. S., et al. (2003). Environmental factors affecting development of Aspergillus nidulans. J. Microbiol. 41, 34–40. [Google Scholar]
- Han X., Qiu M., Wang B., Yin W. B., Nie X., Qin Q., et al. (2016). Functional analysis of the nitrogen metabolite repression regulator gene nmrA in Aspergillus flavus. Front. Microbiol. 7:1794. 10.3389/fmicb.2016.01794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J. K., Yu J. H., Keller N. P., Adams T. H. (1997). Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J. 16, 4916–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki L., Castañeda-Bueno M., Sánchez-Paredes E., Velázquez-Zavala N., Torres-Quiroz F., Ongay-Larios L., et al. (2008). Protein kinases involved in mating and osmotic stress in the yeast Kluyveromyces lactis. Eukaryot. Cell 7, 78–85. 10.1128/EC.00362-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. R., Chae K. S., Han K. H., Han D. M. (2009). The nsdC gene encoding a putative C2H2-type transcription factor is a key activator of sexual development in Aspergillus nidulans. Genetics 182, 771–783. 10.1534/genetics.109.101667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., He Y., Li X., Fasoyin O. E., Hu Y., Liu Y., et al. (2017). Histone Methyltransferase aflrmtA Gene Is Involved in the Morphogenesis, Mycotoxin Biosynthesis, and Pathogenicity of Aspergillus flavus. Toxicon 127, 112–121. 10.1016/j.toxicon.2017.01.013 [DOI] [PubMed] [Google Scholar]
- Liu J., Wang Z. K., Sun H. H., Ying S. H., Feng M. G. (2017). Characterization of the Hog1 MAPK pathway in the entomopathogenic fungus Beauveria bassiana. Environ. Microbiol. 19, 1808–1821. 10.1111/1462-2920.13671 [DOI] [PubMed] [Google Scholar]
- Misihairabgwi J. M., Ezekiel C. N., Sulyok M., Shephard G. S., Krska R. (2017). Mycotoxin contamination of foods in Southern Africa: a 10-year review (2007-2016). Crit. Rev. Food Sci. Nutr. [Epub ahead of print]. 10.1080/10408398.2017.1357003 [DOI] [PubMed] [Google Scholar]
- Nie X., Yu S., Qiu M., Wang X., Wang Y., Bai Y., et al. (2016). Aspergillus flavus SUMO contributes to fungal virulence and toxin attributes. J. Agric. Food Chem. 64, 6772–6782. 10.1021/acs.jafc.6b02199 [DOI] [PubMed] [Google Scholar]
- Posas F., Takekawa M., Saito H. (1998). Signal transduction by MAP kinase cascades in budding yeast. Curr. Opin. Microbiol. 1, 175–182. 10.1016/S1369-5274(98)80008-8 [DOI] [PubMed] [Google Scholar]
- Satterlee T., Cary J. W., Calvo A. M. (2016). RmtA, a putative arginine methyltransferase, regulates secondary metabolism and development in Aspergillus flavus. PLoS ONE 11:e0155575. 10.1371/journal.pone.0155575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K., Keller N. P. (2001). Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicklow D. T. (1987). Survival of Aspergillus flavus sclerotia in soil. Trans. Br. Mycol. Soc. 89, 131–134. 10.1016/S0007-1536(87)80073-6 [DOI] [Google Scholar]
- Wu F., Groopman J. D., Pestka J. J. (2014). Public health impacts of foodborne mycotoxins. Annu. Rev. Food Sci. Technol. 5, 351–372. 10.1146/annurev-food-030713-092431 [DOI] [PubMed] [Google Scholar]
- Xing F., Ding N., Liu X., Selvaraj J. N., Wang L., Zhou L., et al. (2016). Variation in fungal microbiome (mycobiome) and aflatoxins during simulated storage of in-shell peanuts and peanut kernels. Sci. Rep. 6:25930. 10.1038/srep25930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Liang L., Ran F., Liu Y., Li Z., Lan H., et al. (2016). The DmtA methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Sci. Rep. 6:23259. 10.1038/srep23259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. (2012). Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins 4, 1024–1057. 10.3390/toxins4111024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. H. (2016). Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J. Microbiol. 44, 145–154. [PubMed] [Google Scholar]
- Zhang F., Xu G., Geng L., Lu X., Yang K., Yuan J., et al. (2016). The stress response regulator AfSkn7 infuences morphological development, stress response, and pathogenicity in the fungus Aspergillus favus. Toxins 8:E202 10.3390/toxins8070202 [DOI] [PMC free article] [PubMed] [Google Scholar]