Abstract
Inflammatory bowel diseases (IBD) are chronic inflammatory conditions of the gastrointestinal tract with multifactorial etiology. Both dietary factors and the microbe Campylobacter concisus have been found to be associated with the condition. The current study examined the effects of sodium fumarate, a neutralized product of the food additives fumaric acid and monosodium fumarate when in the intestinal environment, on the growth of C. concisus to determine the effects of these food additives on IBD-associated bacterial species. Through culture methods and quantification, it was found that neutralized fumaric acid, neutralized monosodium fumarate, and sodium fumarate increased the growth of C. concisus, with the greatest increase in growth at a concentration of 0.4%. Further examination of 50 C. concisus strains on media with added sodium fumarate showed that greatest growth was also achieved at a concentration of 0.4%. At a concentration of 2% sodium fumarate, all strains examined displayed less growth in comparison with those cultured on media without sodium fumarate. Using mass spectrometry, multiple C. concisus proteins showed significant differential expression when cultured on media with and without 0.4% sodium fumarate. The findings presented suggest that patients with IBD should consider avoiding excessive consumption of foods with fumaric acid or its sodium salts, and that the addition of 0.4% sodium fumarate alone to media may assist in the isolation of C. concisus from clinical samples.
Keywords: Campylobacter concisus, sodium fumarate, enteric bacteria, food additives, inflammatory bowel disease
Introduction
Inflammatory bowel disease (IBD) refers to the inflammatory disorders affecting the gastrointestinal tract, including Crohn’s disease (CD) and ulcerative colitis (UC) (Cosnes et al., 2011). The symptoms of CD and UC are very similar, though the pathology of each differs, allowing for diagnosis. Inflammation in CD may occur in any part of the gastrointestinal tract with a discontinuous pattern, while the inflammation in UC is continuous with diffuse and superficial lesions (Cosnes et al., 2011). Currently, the etiology of IBD is unknown, and several factors are suggested to contribute to its pathogenesis, such as host microbiota and diet (Sartor, 2006). Campylobacter concisus is an oral bacterium that has been linked to IBD. There was a significantly higher prevalence of C. concisus detected in intestinal biopsies of patients with IBD as opposed to healthy controls (Zhang et al., 2010; Mahendran et al., 2011; Mukhopadhya et al., 2011; Kirk et al., 2016). Studies have also shown that some oral C. concisus strains possess the ability to damage the intestinal epithelial barrier, suggesting that translocation of these strains to the lower gastrointestinal tract may increase the risk of the individual developing IBD (Nielsen et al., 2011; Ismail et al., 2012; Zhang, 2015; Mahendran et al., 2016). C. concisus strains are comprised of two genomospecies (Aabenhus et al., 2005; Engberg et al., 2005; Kalischuk and Inglis, 2011; Miller et al., 2012; On et al., 2013; Mahendran et al., 2015; Chung et al., 2016; Nielsen et al., 2016) and recently we have found that C. concisus genomospecies 2 strains were better adapted to the gastrointestinal environment (Wang et al., 2017).
Dietary factors have long been associated with a range of chronic illnesses. In IBD, there was a positive correlation of high intake of carbohydrates and fats to IBD incidence (Sakamoto et al., 2005). Non-Western countries historically have a low incidence and prevalence of IBD, but are showing an increasing incidence and prevalence of IBD coinciding with industrialization and westernization (Drewnowski and Popkin, 1997; Cosnes et al., 2011). The societal changes in these non-Western countries shift the diet toward high fat, high sugar, and processed foods, which are characteristic of Western diets, suggesting that dietary factors are likely to contribute to IBD incidence (Pingali, 2007). In murine models, it was shown that a high fat and high sugar diet changes the gut microbiota composition and promoted the colonization of IBD-associated adherent-invasive Escherichia coli (Martinez-Medina et al., 2014). With processed foods becoming increasingly consumed due to low cost and convenience achieved by food additives and industrial ingredients (Monteiro et al., 2013), their effects on IBD-related bacterial species require investigation.
Fumaric acid (E297) is a widely used acidity regulator (Smith and Hong-Shum, 2011), with its sodium salts, monosodium fumarate and sodium fumarate (E365) used less commonly for the same purposes. While the use of sodium fumarate as a food additive has been discontinued in Europe, it is allowed in Australia and New Zealand. There is no limit on the amount of fumaric acid used in both United States and Australia (CFR 172.350 in the United States, Standard 1.3.1 in Australia), while in the European Union a maximum of 4000 mg/L has been suggested with voluntary enforcement [Regulation (EC) no 1333/2008]. The neutralization of fumaric acid (C4H4O4) via contact with the naturally occurring sodium bicarbonate in the intestinal tract can produce both monosodium fumarate and sodium fumarate. At less acidic pH, fumaric acid dissociates to form monosodium fumarate (NaC4H3O4) and then sodium fumarate (Na2C4H2O4) at almost neutral pH (Roa Engel et al., 2013).
Previous studies utilized the combined supplementation of sodium formate and sodium fumarate into culture media to isolate C. concisus (Tanner et al., 1981; Macuch and Tanner, 2000). This suggests that sodium fumarate can increase C. concisus yield. Therefore, we hypothesize that sodium fumarate, the neutralized product of the food additive fumaric acid, affects the growth of C. concisus and protein expression. This hypothesis was examined in this study.
We found that sodium fumarate supplementation into culture media alone affected the growth of C. concisus and some other enteric bacterial species. In addition, sodium fumarate altered the protein expression in C. concisus.
Materials and Methods
Bacterial Strains Used in This Study
A total of 50 C. concisus strains were examined in this study. Of these, 49 strains were isolated from previous studies with ethics approval granted by the Ethics Committees of the University of New South Wales and the South East Sydney Area Health Service, Australia [HREC09237/SESIAHS 09/078, HREC08335/SESIAHS(CHN)07/48, HREC06233/SESAHS(ES)06/164, HREC09237/SESIAHS09/078, and HREC08335/SESIAHS(CHN)07/48] (Zhang et al., 2010; Mahendran et al., 2011). C. concisus strain 13826 was purchased from the American Type Culture Collection. The details of the C. concisus strains used in this study and genomospecies, if known (Mahendran et al., 2015; Chung et al., 2016; Wang et al., 2017), are shown in Table 1. Additionally, four other enteric species obtained from the American Type Culture collection were included in this study as representatives of other species in the enteric environment (Bachmann, 1972; Gregory et al., 1978; Moschetti et al., 1998): Bacteroides fragilis ATCC 25285, Bacteroides vulgatus ATCC 8482, Enterococcus faecalis ATCC 19433, and E. coli.
Table 1.
Bacterial strains used in this study.
| Strain ID | Health status | Source | Genomospecies |
|---|---|---|---|
| P10CDO-S1 | CD | Saliva | 1 |
| P10CDO-S2 | CD | Saliva | 1 |
| P11CDO-S1 | CD | Saliva | 1 |
| P19CDO-S1 | CD | Saliva | 1 |
| P20CDO-S4 | CD | Saliva | 1 |
| P25CDO-S3 | CD | Saliva | 1 |
| P27CDO-S2 | CD | Saliva | 1 |
| P26UCO-S2 | UC | Saliva | 1 |
| P3UCB1 | UC | Intestinal biopsy | 1 |
| P3UCLW1 | UC | Fecal sample | 1 |
| P3UCO1 | UC | Saliva | 1 |
| H1O1 | Healthy | Saliva | 1 |
| H10O-S1 | Healthy | Saliva | 1 |
| H12O-S1 | Healthy | Saliva | 1 |
| H15O-S1 | Healthy | Saliva | 1 |
| H17O-S1 | Healthy | Saliva | 1 |
| H21O-S3 | Healthy | Saliva | 1 |
| H24O-S1 | Healthy | Saliva | 1 |
| H25O-S1 | Healthy | Saliva | 1 |
| H26O-S1 | Healthy | Saliva | 1 |
| H26O-S1 | Healthy | Saliva | 1 |
| H27O-S1 | Healthy | Saliva | 1 |
| H28O-S1 | Healthy | Saliva | 1 |
| P1CDO2 | CD | Saliva | 2 |
| P1CDO3 | CD | Saliva | 2 |
| P2CDO4 | CD | Saliva | 2 |
| P6CDO1 | CD | Saliva | 2 |
| P12CDO-S1 | CD | Saliva | 2 |
| P18CDO-S1 | CD | Saliva | 2 |
| P20CDO-S2 | CD | Saliva | 2 |
| P20CDO-S3 | CD | Saliva | 2 |
| P21CDO-S1 | CD | Saliva | 2 |
| P21CDO-S2 | CD | Saliva | 2 |
| P24CDO-S2 | CD | Saliva | 2 |
| P24CDO-S3 | CD | Saliva | 2 |
| P27CDO-S1 | CD | Saliva | 2 |
| P26UCO-S1 | UC | Saliva | 2 |
| P7UCO-S2 | UC | Saliva | 2 |
| P8UCO1 | UC | Saliva | 2 |
| P13UCO-S3 | UC | Saliva | 2 |
| P16UCO-S1 | UC | Saliva | 2 |
| P16UCO-S2 | UC | Saliva | 2 |
| H3O1 | Healthy | Saliva | 2 |
| H7O-S1 | Healthy | Saliva | 2 |
| H9O-S2 | Healthy | Saliva | 2 |
| H11O-S1 | Healthy | Saliva | 2 |
| H14O-S1 | Healthy | Saliva | 2 |
| H22O-S1 | Healthy | Saliva | 2 |
| H29O-S1 | Healthy | Saliva | 2 |
| 13826 | Gastroenteritis | Fecal sample | 2 |
The 50 strains of C. concisus were previously isolated from our studies. The letters P and H under Strain ID indicate a patient with IBD or a healthy control, respectively, and the following number is unique for separate individuals. UC, CD, and H under Health status indicate whether the strain was isolated from a patient with ulcerative colitis, Crohn’s disease, or a healthy control, respectively. Genomospecies status known from previous studies is listed (Mahendran et al., 2015; Chung et al., 2016; Liu et al., 2018).
Examination of the Effects of Sodium Fumarate and Sodium Formate on C. concisus Growth
We first examined whether individual supplementation of fumarate is comparable to supplementation with both sodium fumarate (Sigma-Aldrich, St. Louis, MO, United States) and sodium formate (Sigma-Aldrich), five strains of C. concisus were randomly chosen. These strains were first cultured on horse blood agar (HBA, Oxoid, Hampshire, United Kingdom) plates consisting of 6% defibrinated horse blood (Oxoid) in anaerobic conditions with 5% hydrogen as previously described (Lee et al., 2014). Cultures with an optical density of 0.1 at a wavelength of 595 nm (OD595 0.1) were prepared (Ma et al., 2015), and 5 μL was inoculated onto HBA, HBA with 0.2% sodium formate (HBAform), HBAfum0.4, and HBA plates with both 0.2% sodium formate and 0.4% sodium fumarate (HBAfum+form) and streaked for single colonies. After incubation, all plates were examined for colony size differences by visual inspection and plate photographs were obtained through a Samsung HZ30W digital camera attached to the stereomicroscope at a magnification of 20×.
The effects of sodium fumarate and sodium formate on the growth of C. concisus were also quantified through CFU determination of the randomly chosen C. concisus strain, P26UCO-S2. P26UCO-S2 was cultured first cultured on HBA as described above before inoculation onto HBA, HBAfum0.4, HBAform, and HBAfum+form in triplicates and incubated for 72 h. After incubation, cultures were collected and serially diluted for bacterial enumeration using the drop plate method as previously described (Ma et al., 2015). Experiments were repeated three times.
Quantitative Assessment of the Effect of Neutralized Fumaric Acid and Neutralized Monosodium Salt on C. concisus
The pH of H2O solutions containing 0.05, 0.2, 0.4, 1, and 2% fumaric acid, monosodium fumarate, or disodium fumarate was measured with a pH meter (Eutech pH700, Eutech Instrument, Singapore). The solutions of fumaric acid and monosodium fumarate were acidic and it was previously shown that low pH abolishes the viability of C. concisus (Ma et al., 2015). However, the ingested fumaric acid and monosodium fumarate would be neutralized in the lower gastrointestinal tract. Therefore, to determine the possible effects of the neutralized ingested fumaric acid and sodium monofumarate, neutralized solutions of fumaric acid and monosodium fumarate were examined.
Solutions of 20% (w/v) fumaric acid (Sigma-Aldrich) and monosodium fumarate (Nippon Shokubai, Osaka, Japan) were neutralized with sodium bicarbonate (Sigma-Aldrich) and used as the stock solution. C. concisus strains P26UCO-S1 and P2CDO4 were chosen randomly to be examined. Cultures of OD595 0.1 were prepared as described in the Section “Examination of the Effects of Sodium Fumarate and Sodium Formate on C. concisus Growth” and 5 μL was inoculated onto HBA and HBA supplemented with 0, 0.05, 0.2, and 0.4% fumaric acid adjusted to pH 7 (HBAFA0.05, HBAFA0.2, and HBAFA0.4) or monosodium fumarate adjusted to pH 7 (HBAMF0.05, HBAMF0.2, and HBAMF0.4) in triplicate and spread in a radial pattern with a sterile hockey stick. Plates were incubated for 72 h and colony-forming units (CFU) were quantified as described in the Section “Examination of the Effects of Sodium Fumarate and Sodium Formate on C. concisus Growth.” Experiments were conducted three times.
C. concisus Cultivation for the Observation of Colony Size Differences in the Presence of Sodium Fumarate
As both neutralized monosodium fumarate and neutralized fumaric acid increased growth of C. concisus, further experiments were performed with the neutralized final product, sodium fumarate. All 50 C. concisus strains were cultured and suspensions of OD595 0.1 were prepared as described in the Section “Examination of the Effects of Sodium Fumarate and Sodium Formate on C. concisus Growth.” From these bacterial suspensions, 5 μL was streaked for single colonies onto a HBA plate and HBA plates supplemented with 0.05% (HBAfum0.05), 0.4% (HBAfum0.4), 1% (HBAfum1), and 2% (HBAfum2) sodium fumarate in triplicates and the plates were incubated for 72 h. After incubation, plates were examined and photographed as described in the Section “Examination of the Effects of Sodium Fumarate and Sodium Formate on C. concisus Growth.”
Quantitative Assessment of the Effects of Different Concentrations of Sodium Fumarate on the Growth of C. concisus and Other Enteric Bacterial
From the 50 strains of C. concisus examined for morphological colony changes above, 7 strains were randomly selected for quantitative assessment of growth in the presence of different concentrations of sodium fumarate, including P2CDO4, P11CDO-S1, and P20CDO-S3 from patients with CD, P26UCO-S1 from a patient with UC, H12O-S1, and H17O-S1 from healthy controls and C. concisus strain 13826. Furthermore, the growth of B. fragilis, B. vulgatus, E. faecalis, and E. coli were also quantified.
Four different concentrations of sodium fumarate were supplemented into HBA plates, including the HBAfum0.05, HBAfum0.4, HBAfum1, and HBAfum2 used previously in this Section. C. concisus strains were first cultured on HBA plates as described in the Section “Examination of the Effects of Sodium Fumarate and Sodium Formate on C. concisus Growth.” Bacterial cells were harvested, diluted to an OD595 of 0.1 and 5 μL of the suspension was spread plated onto HBA plates and HBAfum0.05, HBAfum0.4, HBAfum1, and HBAfum2 plates in triplicate and incubated for 72 h. After incubation, cultures were collected and serially diluted for bacterial enumeration using the drop plate method as previously described (Ma et al., 2015). Experiments were repeated three times.
Bacteroides fragilis, B. vulgatus, E. faecalis, and E. coli were cultured as previously described (Liu et al., 2017). Bacterial pellets were then collected, diluted and cultured as described for all C. concisus strains above. Due to their faster growth rates, these species were incubated for 24 h instead of 48 h (Liu et al., 2017).
Quantitative Assessment of the Growth of C. concisus When Cultured With and Without Sodium Fumarate Over 96 h
As the previous experiment found that a concentration of 0.4% sodium fumarate was preferable for the growth of most strains, this concentration was then used to examine the growth of C. concisus over a period of 94 h to examine the effect of sodium fumarate on growth rate. Three strains were randomly chosen to observe their growth when cultured on media with and without sodium fumarate.
All three strains were cultured on HBA plates as previously described and bacteria were collected. To prevent bacterial oversaturation of the agar plates, a lower OD595 of bacteria were used. Each strain was diluted to an OD595 of 0.025 and 5 μL of each were spread plated onto HBA and HBAfum0.4 in triplicates for enumeration at 0, 12, 24, 48, 72, and 96 h. After incubation for the respective amount of time, plates were removed from incubation and the bacteria were collected and quantified as previously described (Ma et al., 2015). Additionally, wet mounts of each strain were also prepared at each time point to observe the morphology of C. concisus.
Quantification of the Growth of 50 Strains of C. Concisus on Media With and Without Sodium Fumarate
Based on the results of the Section “Quantitative Assessment of the Growth of C. concisus When Cultured With and Without Sodium Fumarate Over 96 h,” the biggest effect of 0.4% sodium fumarate on C. concisus could be observed at 24 h; consequently, this condition was utilized to quantify the growth of all C. concisus strains in this study. All strains of C. concisus were cultured as per the Section “Examination of the Effects of Sodium Fumarate and Sodium Formate on C. concisus Growth” and diluted to an OD595 of 0.025. From the bacterial suspensions, 5 μL were spread plated onto HBA and HBAFum0.4. Plates were incubated for 24 h before bacteria were harvest and serially diluted for enumeration as described previously in the Section “Examination of the Effects of Sodium Fumarate and Sodium Formate on C. concisus Growth.”
Analysis of the Proteins of C. concisus 13826 When Cultured With and Without Sodium Fumarate
From the C. concisus strains examined, strain 13826 (accession number: NC_009802) was chosen as the representative strain for protein analysis to examine changes in protein profile when cultured with and without sodium fumarate as its genome is readily accessible from the National Center for Biotechnology Information. First, C. concisus strain 13826 was cultured on HBA for 48 h. The bacterial cells were then collected, prepared and cultured on HBA and HBAfum0.4 as described in the Section “Examination of the Effects of Sodium Fumarate and Sodium Formate on C. concisus Growth.”
After incubation, bacteria were harvested and washed according to the Section “Examination of the Effects of Sodium Fumarate and Sodium Formate on C. concisus Growth.” Whole cell lysate proteins were prepared by three cycles of rapid freezing and thawing. Protein concentration was determined via the bicinchoninic acid method according to manufacturer’s instructions (Pierce, Rockford, IL, United States) and 20 μg of proteins were subjected to SDS–PAGE. Mass spectrometry was performed on proteins as previously described (Ismail et al., 2012; Luu et al., 2017). Experiments were repeated three times.
Peak lists were generated using mascot daemon/extract_msn (Matrix Science, London, United Kingdom), with default parameters, and all MS/MS spectra were searched against the NCBIprot_3_11_15 database (68653660 entries, specificity selected for bacteria) using MASCOT (version 2.5.1, Matrix Science, London, United Kingdom). Data search parameters were as follows: precursor tolerance was 4 ppm, and product ion tolerances were ± 0.4 Da; oxidation of methionine, carbamidomethyl alkylation of cysteine, and propionamide on cysteine was specified as a variable modification, enzyme specificity was trypsin and one missed cleavage was possible.
Scaffold Q+ (v.4.7.3, Proteome software, Portland, OR, United States) was used to validate peptide and protein identities (Searle, 2010). Peptide identifications were accepted if they could be established at greater than 95% probability by the Protein Prophetalgorithm (Nesvizhskii et al., 2003). Proteins were accepted if they could be established at greater than 99% probability and contained at least 2 identified peptides. Only proteins present in two or more biological replicates were included. Normalized spectral counts in Scaffold provided an estimation of relative protein quantification. Protein function was derived from the eggNOG database (Muller et al., 2010).
Statistical Analysis
The comparisons of CFU and total protein variation of C. concisus strains cultured on the varying media with the CFU on the control HBA plates were obtained through t-tests. To compare relative abundances of proteins, t-tests were also performed with multiple test correction using the Benjamini–Hochberg method (Benjamini and Hochberg, 1995). A p-value of less than 0.05 was considered significant.
Results
Comparison of C. concisus Growth on HBA Plates Supplemented With Sodium Fumarate Alone With the Combined Supplementation of Sodium Fumarate and Sodium Formate
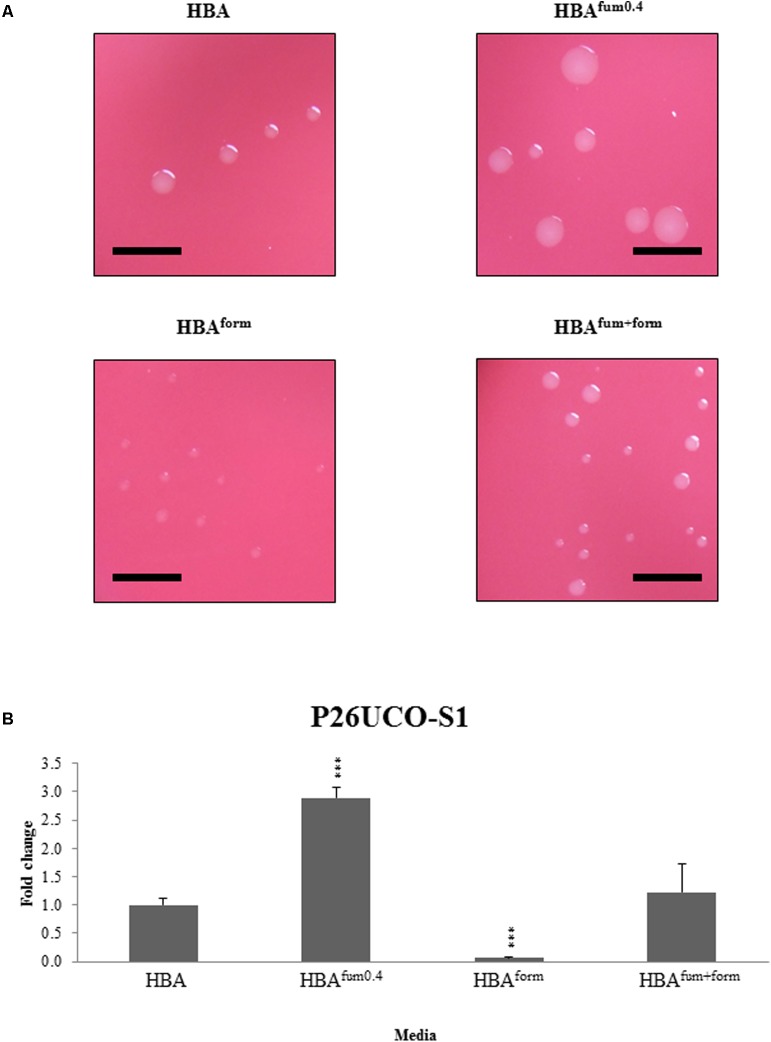
To investigate the effects of sodium fumarate and sodium formate on colony size, the growth of five C. concisus strains were examined. In all five strains, there was a visible decrease in colony size on HBAform in comparison with HBA, while all C. concisus on HBAfum0.4 showed an increased colony size. On HBAfum+form, there was no visible difference in colony sizes compared to HBA plates.
To further investigate the effects of the supplements, the CFU of strain P26UCO-S2 was quantified. The CFU of P26UCO-S2 when supplemented with 0.4% sodium fumarate and 0.2% sodium formate was not significantly different to HBA. When cultured on HBAfum0.4, the CFU increased significantly (p < 0.05) in comparison with HBA. On HBAform, the CFU showed a significant decrease (p < 0.05 in comparison with HBA) (Figure 1).
FIGURE 1.
Quantitative analysis of sodium fumarate and sodium formate on the growth of C. concisus. (A) Colonies of representative C. concisus strain P26UCO-S2. A total of five C. concisus strains were streaked onto HBA plates, HBA plates with 0.4% sodium fumarate (HBAfum0.4), HBA with 0.2% sodium formate (HBAform), and HBA with both sodium fumarate and sodium formate (HBAfum+form). After incubation the colonies were observed under a stereomicroscope and photographs were captured at 20× magnification. Scale bars represent 5 mm. Of the five strains examined, all showed increased colony size when cultured on HBAfum0.4 and decreased colony size on HBAform. All strains showed no visible difference in colony size when cultured on HBAfum+form. (B) The fold changes in CFU of C. concisus strain P26UCO-S2 cultured on HBA, HBAfum0.4, HBAform, and HBAfum+form is shown. Fold changes were calculated relative to the CFU of the same strain cultured on plates without fumarate or formate from the mean of quadruplicate counts on each media. ∗∗∗ indicates p < 0.001.
Measurement of C. concisus Growth on HBA Plates Supplemented With Neutralized Fumaric Acid or Neutralized Monosodium Fumarate
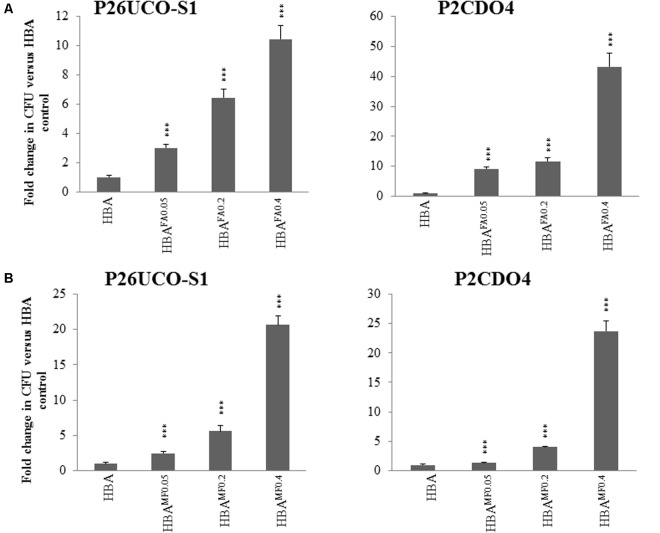
The pH of different concentrations of fumaric acid, monosodium fumarate, and sodium fumarate were measured and are shown in Table 2. C. concisus strains P26UCO-S1 and P2CDO4 representing C. concisus genomospecies 1 and 2, respectively, were examined for growth on HBA plates supplemented with neutralized monosodium fumarate or neutralized fumaric acid. Both strains had significantly increased growth on HBAMF0.05 compared with HBA (p < 0.05), with P26UCO-S1 showing a fold change of 2.5 and P2CDO4 showing a fold change of 1.3. On HBAMF0.2, both P26UCO-S1 and P2CDO4 showed a further increase in fold change, with a fold change of 5.6 and 4, respectively. The greatest increase in fold change could be seen on HBAMF0.4 for both strains, with P26UCO-S1 increasing to 20.1 times the CFU on HBA and P2CDO4 increasing 23.7 times (Figure 2).
Table 2.
The pH of fumaric acid and its sodium salts at different concentrations.
| Concentration of solute | Fumaric acid | Monosodium fumarate | Sodium fumarate |
|---|---|---|---|
| 0.05% | 2.96 | 3.81 | 7.54 |
| 0.2% | 2.76 | 3.67 | 7.22 |
| 0.4% | 2.55 | 3.68 | 7.75 |
| 1% | 2.5 | 3.65 | 7.76 |
| 2% | 2.48 | 3.6 | 7.92 |
Fumaric acid, monosodium fumarate, and sodium fumarate solutions were made by suspension in distilled water at concentrations of 0.05, 0.2, 0.4, 1, and 2%.
FIGURE 2.
Quantitative analysis of neutralized fumaric acid and neutralized monosodium fumarate on the growth of C. concisus. P26UCO-S1 and P2CDO4 are both C. concisus strains, the former isolated from a patient with UC and the latter from a patient with CD. Both strains were quantified after growth on HBA plates supplemented with varying concentrations of (A) neutralized fumaric acid and (B) neutralized monosodium fumarate. Fold changes were calculated relative to the CFU of the same strain cultured on plates without supplementation from the mean of quadruplicate counts on each media. ∗∗∗ indicates p < 0.001.
When cultured on HBAFA0.05, both C. concisus strains P26UCO-S1 and P2CDO4 showed a significant increase in CFU compared to on HBA plates, with fold changes of 3 and 9.1, respectively. As the concentration of neutralized fumaric acid increased, the CFU of P26UCO-S1 also increased with fold changes of 6.4 on HBAFA0.2 and 10.4 on HBAFA0.4. Similarly, for P2CDO4, the fold changes on HBAFA0.2 and HBAFA0.4 were significantly increased P26UCO-S1 and P2CDO4, at 11.7 and 43.3, respectively.
Changes in Colony Size on HBA Plates With and Without Sodium Fumarate
When cultured on HBAfum0.05, 27 of the 50 C. concisus strains examined showed an increase in colony size compared to the same strain on HBA. The remaining 23 stains showed no visible change in colony size compared to the same strain on HBA. On HBAfum0.4, all strains of C. concisus tested showed visibly larger colonies as compared to that of the same strain on HBA, although the colony size increments varied in different strains. When cultured on HBAfum1, 45 of 50 strains examined noticeably decreased in colony size as compared to that of the same strain cultured on HBA. From the five remaining strains, four showed no obvious difference in colony size compared to the controls and the final strain was P11CDO-S1, which showed an increase in colony size compared to the control. When the concentration of sodium fumarate was increased to 2%, 6 of the examined strains showed complete inhibition of growth. All of the remaining strains had a clearly smaller colony size in comparison with HBA. Photos of the results of a randomly chosen representative strain, H14O-S1, are shown in Figure 3.
FIGURE 3.
The effects of sodium fumarate on the colony size of C. concisus. Colonies of representative C. concisus strain H14O-S1 on HBA plates (A) without supplementation and (B–E) with supplementation of sodium fumarate at 0.05, 0.4, 1, and 2%, respectively. Scale bars represent 5 mm. A total of 50 C. concisus strains were streaked onto HBA, HBAfum0.05, HBAfum0.4, HBAfum1, and HBAfum2. After incubation, colonies were observed under a stereomicroscope and photographs were captured at 20× magnification.
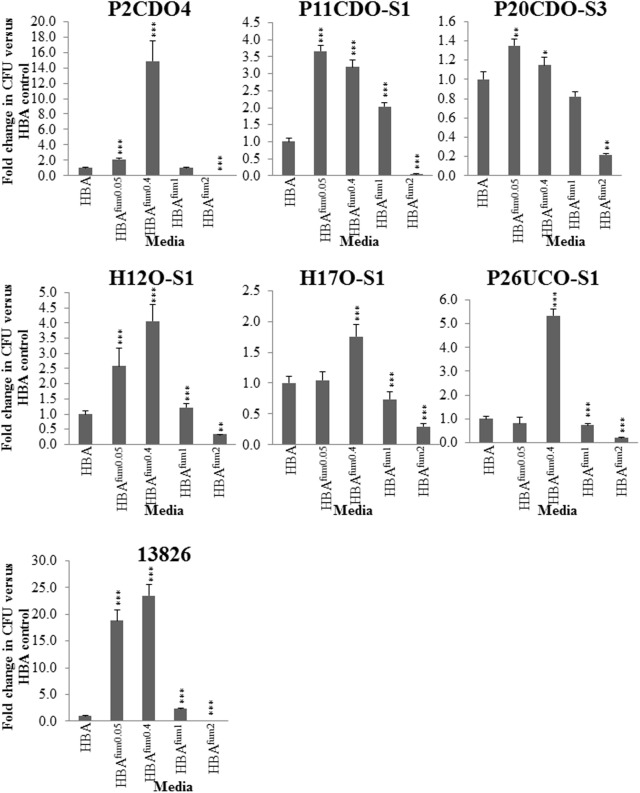
Quantitative Comparison of the Effects of Sodium Fumarate on C. concisus Growth
To further characterize the impact of sodium fumarate on the growth of C. concisus, the growth of seven strains of C. concisus were quantified after culturing on HBA supplemented with varying concentrations of sodium fumarate. When cultured on HBAfum0.05, the CFU of five C. concisus strains (P2CDO4, P11CDO-S1, P20CDO-S3, H12O-S1, and 13826) were significantly increased compared to that on HBA (Figure 4). The CFU of C. concisus strain 13826 was increased significantly (p < 0.05), with an 18.8-fold change compared to its CFU on HBA plates. The remaining 4 strains had fold changes of between 1.3 and 2.7 on HBAfum0.05 compared to the same strain on HBA. All strains of C. concisus examined showed a significant increase in CFU (p < 0.05) when cultured on HBAfum0.4 as compared to HBA, with fold changes of 1.2- to 24.5-fold in CFU when cultured on HBAfum0.4 as compared to HBA. When cultured on HBAfum1, strains showed a differential response with strains 13826, H12O-S1 and P11CDO-S1 showing an increase of 1.2- and 3.2-fold in CFU, respectively, P26UCO-S1 and H17O-S1 both showing 0.7-fold decrease in CFU, and P2CDO4 and P20CDO-S3 showing no change (p > 0.05) in CFU as compared to that on HBA (Figure 4). However, when cultured on HBAfum2, all examined strains of C. concisus consistently showed a decrease in CFU as compared to the same strain cultured without sodium fumarate, with fold changes between 0.05 and 0.3.
FIGURE 4.
Quantitative analysis of C. concisus cultured in the presence of varying concentrations of sodium fumarate. Seven randomly chosen strains of C. concisus were grown on HBA plates with varying concentrations of sodium fumarate; the number following Fum indicates the percentage of sodium fumarate in the HBA plate. Each strain was then collected and the CFU were enumerated. Fold changes were calculated relative to the CFU of the same strain cultured on HBA from quadruplicate counts. ∗ indicates p < 0.05, ∗∗ indicates p < 0.01, and ∗∗∗ indicates p < 0.001.
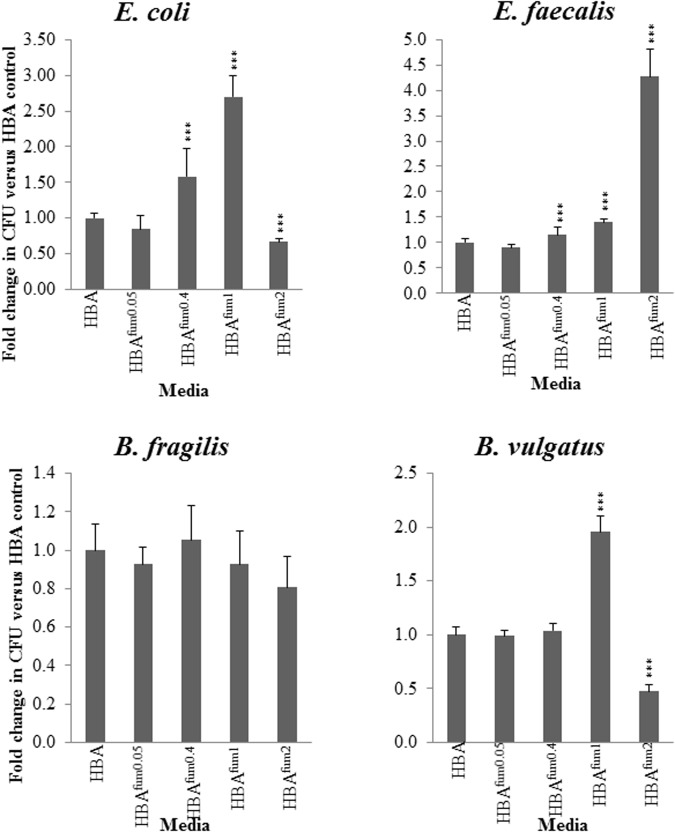
Effects of Sodium Fumarate on the Growth of Other Enteric Species
For all the examined enteric species, there was no significant change between the CFU when cultured on HBAfum0.05 as compared to HBA. E. coli and E. faecalis showed similar results on HBAfum0.4 with growth significantly increased (p < 0.05) compared to HBA. B. vulgatus showed no change in CFU (Figure 5). When the concentration of sodium fumarate is increased to 1%, E. coli, E. faecalis, and B. vulgatus showed a significant increase (p < 0.05) in CFU. On HBAfum2, E. coli and B. vulgatus displayed a significant decrease in CFU. E. faecalis showed a significantly increased (p < 0.05) CFU compared to HBA plates.
FIGURE 5.
Quantitative effects of varying concentrations of sodium fumarate on enteric bacterial species. B. fragilis, B. vulgatus, E. faecalis, and E. coli were chosen as representative bacteria of the enteric environment. Each species was cultured on HBA plates with varying concentrations of sodium fumarate; the number following Fum indicates the percentage of sodium fumarate in the HBA plate. Bacteria were then collected and the CFU were quantified. Fold changes were calculated relative to the CFU of the same species cultured on HBA from quadruplicate counts. ∗∗∗ indicates p < 0.001.
At all concentrations of sodium fumarate examined, B. fragilis showed no significant difference in CFU compared to the CFU on HBA plates (p > 0.05).
Growth Curve of C. concisus Cultured on HBA and HBAFum0.4
In the growth curves of all three strains of C. concisus examined, marked increase in CFU could be observed when cultured on HBAfum0.4 as compared to HBA as early as after 12 h of incubation (Figure 6). All three strains maintained a higher overall CFU when cultured on HBAfum0.4 as compared to HBA at the same incubation time. While there was an increase in CFU on HBAfum0.4, the trend of CFU increase and decrease followed the same pattern as that on HBA in all three strains. From the wet mount preparations, no difference in bacterial morphology could be seen in the same strains on HBA as compared to HBAfum0.4 at the same time point.
FIGURE 6.
Growth of C. concisus strains P2CDO4, H14O-S1, and 13826 cultured on HBAfum0.4 and HBA over 96 h. A. Each strain was cultured on HBA and HBA with 0.4% sodium fumarate (HBAfum0.4) and the total bacteria were enumerated at 0, 12, 24, 48, 72, and 96 h after incubation. Results are represented as a CFU fold change relative to the CFU at 0 h of incubation from quadruplicate counts.
Fold Change in Growth of 50 Strains of C. concisus When Cultured on Media With and Without Sodium Fumarate
All 50 strains of C. concisus examined displayed an increase in growth when cultured on HBAfum0.4 as compared to HBA. The increase in CFU varied depending on the strain with the lowest fold change in comparison with HBA on HBAfum0.4 being 1.1 ± 0.1 for P20CDO-S3 and the highest fold change being 28.8 ± 1.2 for P10CDO-S2 (Table 3). There was no significant difference in the average CFU fold changes from C. concisus strains isolated from patient groups (CD nor UC) versus those isolated from healthy controls (p > 0.05). No difference in average CFU fold changes were also observed between genomospecies 1 or 2 C. concisus (p > 0.05).
Table 3.
CFU fold changes of all 50 strains of C. concisus cultured on HBAfum0.4 compared to HBA plates.
| Strain ID | Genomospecies | Fold change (mean ± SD) |
|---|---|---|
| P10CDO-S1 | 1 | 12.7 ± 1.3 |
| P10CDO-S2 | 1 | 28.8 ± 1.2 |
| P11CDO-S1 | 1 | 3.2 ± 0.3 |
| P19CDO-S1 | 1 | 1.4 ± 0.0 |
| P20CDO-S4 | 1 | 2.3 ± 0.3 |
| P25CDO-S3 | 1 | 4.0 ± 0.6 |
| P27CDO-S2 | 1 | 16.0 ± 1.9 |
| P26UCO-S2 | 1 | 1.6 ± 0.2 |
| P3UCB1 | 1 | 17.4 ± 1.9 |
| P3UCLW1 | 1 | 3.4 ± 0.1 |
| P3UCO1 | 1 | 17.1 ± 2.2 |
| H1O1 | 1 | 2.2 ± 0.2 |
| H10O-S1 | 1 | 6.5 ± 1.3 |
| H12O-S1 | 1 | 1.8 ± 0.4 |
| H15O-S1 | 1 | 6.5 ± 1.0 |
| H17O-S1 | 1 | 1.3 ± 0.2 |
| H21O-S3 | 1 | 20.8 ± 2.0 |
| H24O-S1 | 1 | 5.5 ± 0.3 |
| H25O-S1 | 1 | 2.1 ± 0.1 |
| H26O-S1 | 1 | 9.4 ± 1.5 |
| H26O-S1 | 1 | 14.5 ± 0.6 |
| H27O-S1 | 1 | 13.6 ± 0.7 |
| H28O-S1 | 1 | 22.7 ± 1.4 |
| P1CDO2 | 2 | 18.2 ± 11.6 |
| P1CDO3 | 2 | 8.5 ± 1.6 |
| P2CDO4 | 2 | 5.9 ± 0.6 |
| P6CDO1 | 2 | 11.9 ± 2.1 |
| P12CDO-S1 | 2 | 10.2 ± 0.8 |
| P18CDO-S1 | 2 | 2.0 ± 0.4 |
| P20CDO-S2 | 2 | 5.2 ± 0.6 |
| P20CDO-S3 | 2 | 1.1 ± 0.1 |
| P21CDO-S1 | 2 | 2.8 ± 0.3 |
| P21CDO-S2 | 2 | 4.3 ± 0.8 |
| P24CDO-S2 | 2 | 7.1 ± 1.4 |
| P24CDO-S3 | 2 | 1.4 ± 0.3 |
| P27CDO-S1 | 2 | 3.1 ± 0.2 |
| P26UCO-S1 | 2 | 14.3 ± 1.6 |
| P7UCO-S2 | 2 | 5.8 ± 0.5 |
| P8UCO1 | 2 | 1.2 ± 0.1 |
| P13UCO-S3 | 2 | 15.8 ± 2.8 |
| P16UCO-S1 | 2 | 11.4 ± 1.8 |
| P16UCO-S2 | 2 | 10.1 ± 0.8 |
| H3O1 | 2 | 6.9 ± 1.2 |
| H7O-S1 | 2 | 2.4 ± 0.3 |
| H9O-S2 | 2 | 3.6 ± 0.5 |
| H11O-S1 | 2 | 18.7 ± 3.4 |
| H14O-S1 | 2 | 2.0 ± 0.3 |
| H22O-S1 | 2 | 2.2 ± 0.5 |
| H29O-S1 | 2 | 10.5 ± 2.3 |
| 13826 | 2 | 15.5 ± 0.5 |
Fold changes were calculated from the CFU of the same strain on HBAfum0.4 plates divided by the CFU on HBA plates. Results displayed are the mean fold changes ± the standard deviation (SD). The genomospecies which each strain belongs to is listed beside the Strain ID.
Comparison of Proteins Identified in the Whole Cell Lysates of C. concisus 13826 Cultured on HBA With and Without Sodium Fumarate
C. concisus strain 13826 was used to examine protein expression when cultured on HBA and HBAfum0.4. In total, 391 proteins were identified, with 355 being commonly expressed by C. concisus cultured on HBA and HBAfum0.4. On HBA, there were 9 unique proteins that were not detected when cultured on HBAfum0.4 (Table 4). When C. concisus was cultured on HBAfum0.4, there were 27 unique proteins identified (Table 5).
Table 4.
Proteins only identified in C. concisus 13826 when cultured on HBA plates without fumarate.
| Protein | Accession number | Functional category |
|---|---|---|
| Heavy metal translocating P-type ATPase | EAT98303.2 | Inorganic ion transport and metabolism |
| Peptidylprolyl isomerase | WP_002940164.1 | Post-translational modification, protein turnover, and chaperones |
| Peptidase | WP_012000970.1 | Amino acid transport and metabolism |
| Flavocytochrome c | WP_012001109.1 | Energy production and conversion |
| C4-dicarboxylate ABC transporter | WP_012140574.1 | Carbohydrate transport and metabolism |
| Hypothetical protein | WP_012140660.1 | Cell wall/membrane/envelope biogenesis |
| LemA protein | WP_021084760.1 | Function unknown |
| C4-dicarboxylate ABC transporter | WP_048809748.1 | Energy production and conversion |
| UDP-N-acetylmuramate- alanine ligase | WP_048809780.1 | Signal transduction mechanisms |
Table 5.
Proteins only identified in C. concisus 13826 when cultured on HBAfum0.4.
| Protein | Accession number | Functional category |
|---|---|---|
| Cell division protein FtsA | WP_002940409.1 | Cell cycle control, cell division, chromosome partitioning |
| ATP-dependent protease ATP-binding subunit HslU | WP_002940444.1 | Post-translational modification, protein turnover, and chaperones |
| Twitching motility protein PilT | WP_002941386.1 | Cell motility, intracellular trafficking, secretion, and vesicular transport |
| 3-isopropylmalate dehydratase small subunit | WP_002941843.1 | Amino acid transport and metabolism |
| 3-isopropylmalate dehydrogenase | WP_002941890.1 | Amino acid transport and metabolism |
| Elongation factor P | WP_004317246.1 | Translation, ribosomal structure and biogenesis |
| Cation ABC transporter substrate-binding protein | WP_004317362.1 | Inorganic ion transport and metabolism |
| DNA-binding response regulator | WP_009294394.1 | Signal transduction mechanisms |
| Amino acid ABC transporter substrate-binding protein | WP_012001169.1 | Amino acid transport and metabolism |
| Competence/damage-inducible domain-containing protein | WP_012001188.1 | Function unknown |
| Thioredoxin | WP_012001226.1 | Post-translational modification, protein turnover, and chaperones |
| Hypothetical protein | WP_012001228.1 | Function unknown |
| 6,7-dimethyl-8-ribityllumazine synthase | WP_012001235.1 | Coenzyme transport and metabolism |
| Iron ABC transporter ATP-binding protein | WP_012001386.1 | Inorganic ion transport and metabolism |
| Hypothetical protein | WP_012001503.1 | Function unknown |
| 3,4-dihydroxy-2-butanone-4-phosphate synthase | WP_012139903.1 | Coenzyme transport and metabolism |
| UDP-glucose 4-epimerase | WP_012140196.1 | Cell wall/membrane/envelope biogenesis |
| Hypothetical protein | WP_012140308.1 | Function unknown |
| Hypothetical protein | WP_012140384.1 | Function unknown |
| Membrane protein | WP_012140437.1 | Post-translational modification, protein turnover, and chaperones |
| ABC transporter substrate-binding protein | WP_012140548.1 | Inorganic ion transport and metabolism |
| Superoxide dismutase | WP_012140576.1 | Inorganic ion transport and metabolism |
| Transcription elongation factor GreA | WP_035142507.1 | Transcription |
| Phosphoribosylformylglycinamidine synthase | WP_048809794.1 | Nucleotide transport and metabolism |
| Hypothetical protein | WP_048809867.1 | Function unknown |
| Endoribonuclease | WP_048809890.1 | Translation, ribosomal structure and biogenesis |
| Protein-export membrane protein SecD | WP_054196430.1 | Intracellular trafficking, secretion, and vesicular transport |
The normalized total spectral count was used to quantitatively compare protein expression between C. concisus 13826 cultured on HBA and HBAfum0.4. Of the 355 proteins detected in both conditions, 31 proteins displayed a significant difference in fold change (p < 0.05, Table 6). Of these, 14 proteins were detected to be significantly less abundant in C. concisus when cultured on HBAfum0.4 as compared to the same C. concisus strain cultured on HBA, and the remaining 17 proteins were significantly more abundant (Table 6).
Table 6.
Proteins with a significant changed of expression profile when cultured on HBA plates and on HBAfum0.4.
| Protein | Accession number | t-Test (p < 0.05) | Fold change | Functional category |
|---|---|---|---|---|
| Flavocytochrome c | WP_012140366.1 | 0.0002 | 0.06 | Energy production and conversion |
| Thiosulfate reductase | WP_012140134.1 | 0.0022 | 0.07 | Energy production and conversion |
| Dehydrogenase | WP_012140508.1 | 0.0097 | 0.2 | Energy production and conversion |
| Trimethylamine-N-oxide reductase | WP_012001034.1 | 0.0001 | 0.3 | Energy production and conversion |
| Aryl-sulfate sulfotransferase | WP_012140335.1 | 0.0059 | 0.3 | Function unknown |
| L-asparaginase | WP_012140623.1 | 0.0002 | 0.4 | Amino acid transport and metabolism |
| Methyl-accepting chemotaxis protein | WP_012001460.1 | 0.0058 | 0.4 | Cell motility, signal transduction mechanisms |
| Flavocytochrome c | WP_004317416.1 | 0.0025 | 0.5 | Energy production and conversion |
| Peptidase M20 | WP_012001703.1 | 0.0078 | 0.5 | Amino acid transport and metabolism |
| Biotin attachment protein | WP_012001365.1 | 0.0380 | 0.5 | Energy production and conversion |
| Flagellin | WP_012140492.1 | 0.0250 | 0.6 | Cell motility |
| Hypothetical protein | WP_012001270.1 | 0.0160 | 0.7 | Function unknown |
| DNA-binding protein | WP_012001719.1 | 0.0250 | 0.7 | Replication, recombination and repair |
| Long-chain-fatty-acid-CoA ligase | WP_012001496.1 | 0.0260 | 0.8 | Cell wall/membrane/envelope biogenesis |
| 50S ribosomal protein L2 | WP_002941522.1 | 0.0400 | 1.4 | Translation, ribosomal structure, and biogenesis |
| 30S ribosomal protein S2 | WP_021091647.1 | 0.0015 | 1.5 | Translation, ribosomal structure, and biogenesis |
| Hydrogenase accessory protein HypB | WP_002941014.1 | 0.0160 | 1.5 | Transcription, post-translational modification, protein turnover, and chaperones |
| Hypothetical protein | WP_002941798.1 | 0.0260 | 1.5 | Function unknown |
| 50S ribosomal protein L15 | WP_002941506.1 | 0.0250 | 1.6 | Translation, ribosomal structure and biogenesis |
| Phenylalanine-tRNA ligase subunit beta | WP_048809769.1 | 0.0130 | 1.7 | Translation, ribosomal structure and biogenesis |
| ABC transporter substrate-binding protein | WP_012001287.1 | 0.0150 | 1.7 | Carbohydrate transport and metabolism |
| Glutamyl-tRNA amidotransferase | WP_048809879.1 | 0.0160 | 1.7 | Translation, ribosomal structure and biogenesis |
| Peptide ABC transporter substrate-binding protein | WP_012001388.1 | 0.0072 | 1.9 | Inorganic ion transport and metabolism |
| 3-oxoacyl-[acyl-carrier-protein] reductase | WP_012140278.1 | 0.0160 | 1.9 | Lipid transport and metabolism |
| Iron transporter | WP_009294901.1 | 0.0080 | 2.3 | Inorganic ion transport and metabolism |
| Succinate dehydrogenase | WP_012001794.1 | 0.0120 | 2.5 | Energy production and conversion |
| Heterodisulfide reductase subunit B | WP_012001792.1 | 0.0430 | 2.6 | Energy production and conversion |
| Hypothetical protein | WP_012001093.1 | 0.0130 | 3.4 | Function unknown |
| Molybdopterin molybdenumtransferase MoeA | WP_002942864.1 | 0.0160 | 5.5 | Coenzyme transport and metabolism |
| 2-Cys peroxiredoxin | WP_012140581.1 | 0.0058 | 7 | Post-translational modification, protein turnover, and chaperones |
| (2Fe-2S)-binding protein | WP_012001793.1 | 0.0120 | 14 | Energy production and conversion |
The fold change reflects the spectral counts of a protein in C. concisus 13826 cultured on HBA plates divided by the spectral counts of the same protein in C. concisus 13826 cultured on HBAfum0.4.
Discussion
We investigated the effects of sodium fumarate on the growth of IBD associated bacterial species C. concisus and other enteric species. Additionally, proteprotein expression differences when cultured on HBA and HBAfum0.4 were also examined in C. concisus. We found that sodium fumarate supplementation into culture media alone affected the growth of C. concisus and some other enteric bacterial species. In addition, sodium fumarate altered the protein expression in C. concisus.
Fumaric acid is a widely used food additive, present in a variety of food products such as candy, jelly, juices, and flat breads. Both fumaric acid and monosodium fumarate are acidic (Table 2), which would inhibit the growth of C. concisus as shown by our previous study (Ma et al., 2015). However, the consumed fumaric acid will be neutralized in the intestinal tract. As the pH increases, the dissociation of fumaric acid causes a shift to monosodium fumarate and then ultimately to sodium fumarate at neutral pH. The human small intestine has a pH of 6.63 ± 0.53 (Evans et al., 1988); thus, the prevailing fumarate salt is likely to be sodium fumarate. The sodium fumarate in this study therefore represents the neutralized product of fumaric acid and monosodium fumarate in the intestinal tract.
Sodium fumarate at concentrations between 0.05 and 0.4% increased the growth of C. concisus. This finding suggests that fumaric acid from dietary sources may enhance the growth of C. concisus in the intestinal tract where it is neutralized. Sodium fumarate, although used less frequently as a food additive, may also enhance the growth of C. concisus in the gastrointestinal tract. As C. concisus was previously shown to be associated with IBD and that virulent strains could damage the intestinal epithelial barrier (Nielsen et al., 2011; Ismail et al., 2012), the data from our study suggests that patients with IBD should consider avoiding excessive consumption of foods containing fumaric acid or its sodium salts. C. concisus consists of two genomospecies and each genomospecies contains diverse strains (Chung et al., 2016). In our study, 0.4% sodium fumarate increased the growth of all C. concisus strains examined, with differences in the increments dependent on strain (Table 3).
At concentrations of 1% sodium fumarate, the growths of some C. concisus strains were inhibited, and at a concentration of 2%, the growth of all strains examined was inhibited. However, these concentrations of sodium fumarate are unlikely to be reach in the intestinal tract from dietary source.
Despite the association between C. concisus and IBD, the isolation rates of C. concisus from enteric samples are low (Mahendran et al., 2011; Mukhopadhya et al., 2011). Our finding that 0.4% sodium fumarate increases the growth of all C. concisus strains suggest that it can be used to enhance the isolation of C. concisus from enteric samples. We also found that 0.4% sodium fumarate affected C. concisus protein expression. Most of the proteins that decreased in expression when C. concisus was cultured on HBAfum0.4 were involved in metabolism and most of the proteins with increased expression were involved in nutrient acquisition (Tables 4, 5, 6). These findings indicate that sodium fumarate has increased the growth most likely by affecting energy generation.
We also examined the effects of sodium fumarate on the growth of additional enteric bacterial species including B. fragilis, B. vulgatus, E. faecalis, and E. coli. In general, sodium fumarate showed less effect on the growth of these bacterial species as compared to C. concisus. For example, sodium fumarate did not affect the growth of B. fragilis at all concentrations tested, and a concentration of 0.05% sodium fumarate did not affect the growth of all 4 enteric species. Although sodium fumarate increased the growth of E. coli and E. faecalis at a concentration of 0.4%, the fold change was only 1.58 and 1.2, respectively (Figure 5). These results suggest that sodium fumarate from dietary source is more like to affect the growth of C. concisus in the intestinal tract rather than these enteric species.
As this study was performed under laboratory conditions, future studies should be conducted using animal models or in human volunteers to more accurately reflect the effects of food additives on the bacterial species in the gastrointestinal tract. Clinical studies can also be conducted to examine whether the removal of fumaric acid and its salts from the diet of patients with IBD can help improve their condition.
In summary, the results from this study suggest that it may be prudent for patients with IBD to avoid the overconsumption of foods containing fumaric acid or its sodium salts. Additionally, HBAfum0.4 plates can be used for the increased probability of isolating C. concisus from clinical samples. This was the first study showing that sodium fumarate, the neutralized product of the food additives fumaric acid and monosodium fumarate, affects the growth of IBD associated C. concisus in a dose- and strain-dependent manner.
Ethics Statement
No patient samples were collected in this study and all C. concisus strains used in this study were isolated in previous studies. Ethics approval was not required as per institutional and national guidelines.
Author Contributions
RM, SY, HL, and FL performed the experiments and bioinformatics analyses. MG, SR, and RL provided an important feedback on clinical aspect. LZ and RM conceived the project. RM and LZ played a major role in writing the manuscript. All authors have read the manuscript and provided feedback. All authors have approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work is supported by Faculty Research Grant awarded to LZ from the University of New South Wales (Grant No: PS35329).
References
- Aabenhus R., On S. L., Siemer B. L., Permin H., Andersen L. P. (2005). Delineation of Campylobacter concisus genomospecies by amplified fragment length polymorphism analysis and correlation of results with clinical data. 43 5091–5096. 10.1128/JCM.43.10.5091-5096.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. (1972). Pedigrees of some mutant strains of Escherichia coli K-12. 36 525–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. 289–300. [Google Scholar]
- Chung H. K. L., Tay A., Octavia S., Chen J., Liu F., Ma R., et al. (2016). Genome analysis of Campylobacter concisus strains from patients with inflammatory bowel disease and gastroenteritis provides new insights into pathogenicity. 6:38442. 10.1038/srep38442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosnes J., Gower-Rousseau C., Seksik P., Cortot A. (2011). Epidemiology and natural history of inflammatory bowel diseases. 140 1785–1794. 10.1053/j.gastro.2011.01.055 [DOI] [PubMed] [Google Scholar]
- Drewnowski A., Popkin B. M. (1997). The nutrition transition: new trends in the global diet. 55 31–43. 10.1111/j.1753-4887.1997.tb01593.x [DOI] [PubMed] [Google Scholar]
- Engberg J., Bang D., Aabenhus R., Aarestrup F. M., Fussing V., Gerner-Smidt P. (2005). Campylobacter concisus: an evaluation of certain phenotypic and genotypic characteristics. 11 288–295. 10.1111/j.1469-0691.2005.01111.x [DOI] [PubMed] [Google Scholar]
- Evans D., Pye G., Bramley R., Clark A., Dyson T., Hardcastle J. (1988). Measurement of gastrointestinal pH profiles in normal ambulant human subjects. 29 1035–1041. 10.1136/gut.29.8.1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory E. M., Moore W. E., Holdeman L. V. (1978). Superoxide dismutase in anaerobes: survey. 35 988–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail Y., Mahendran V., Octavia S., Day A. S., Riordan S. M., Grimm M. C., et al. (2012). Investigation of the enteric pathogenic potential of oral Campylobacter concisus strains isolated from patients with inflammatory bowel disease. 7:e38217. 10.1371/journal.pone.0038217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalischuk L. D., Inglis G. D. (2011). Comparative genotypic and pathogenic examination of Campylobacter concisus isolates from diarrheic and non-diarrheic humans. 11:53. 10.1186/1471-2180-11-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk K. F., Nielsen H. L., Thorlacius-Ussing O., Nielsen H. (2016). Optimized cultivation of Campylobacter concisus from gut mucosal biopsies in inflammatory bowel disease. 8:27. 10.1186/s13099-016-0111-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Ma R., Grimm M. C., Riordan S. M., Lan R., Zhong L., et al. (2014). Examination of the anaerobic growth of Campylobacter concisus strains. 2014:476047. 10.1155/2014/476047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Ma R., Riordan S. M., Grimm M. C., Liu L., Wang Y., et al. (2017). Azathioprine, mercaptopurine, and 5-aminosalicylic acid affect the growth of IBD-associated campylobacter species and other enteric microbes. 8:527. 10.3389/fmicb.2017.00527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Ma R., Tay A., Octavia S., Lan R., Chung H. K. L., et al. (2018). Genomic analysis of oral Campylobacter concisus strains identified a potential bacterial molecular marker associated with active Crohn’s disease. 7:64. 10.1038/s41426-018-0065-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu L. D. W., Octavia S., Zhong L., Raftery M., Sintchenko V., Lan R. (2017). Characterisation of the Bordetella pertussis secretome under different media. 158 43–51. 10.1016/j.jprot.2017.02.010 [DOI] [PubMed] [Google Scholar]
- Ma R., Sapwell N., Chung H. K. L., Lee H., Mahendran V., Leong R. W., et al. (2015). Investigation of the effects of pH and bile on the growth of oral Campylobacter concisus strains isolated from patients with inflammatory bowel disease and controls. 64 438–445. 10.1099/jmm.0.000013 [DOI] [PubMed] [Google Scholar]
- Macuch P. J., Tanner A. C. R. (2000). Campylobacter species in health, gingivitis, and periodontitis. 79 785–792. 10.1177/00220345000790021301 [DOI] [PubMed] [Google Scholar]
- Mahendran V., Liu F., Riordan S. M., Grimm M. C., Tanaka M. M., Zhang L. (2016). Examination of the effects of Campylobacter concisus zonula occludens toxin on intestinal epithelial cells and macrophages. 8:18. 10.1186/s13099-016-0101-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahendran V., Octavia S., Demirbas O. F., Sabrina S., Ma R., Lan R., et al. (2015). Delineation of genetic relatedness and population structure of oral and enteric Campylobacter concisus strains by analysis of housekeeping genes. 161 1600–1612. 10.1099/mic.0.000112 [DOI] [PubMed] [Google Scholar]
- Mahendran V., Riordan S. M., Grimm M. C., Tran T. A. T., Major J., Kaakoush N. O., et al. (2011). Prevalence of campylobacter species in adult Crohn’s disease and the preferential colonization sites of campylobacter species in the human intestine. 6:e25417. 10.1371/journal.pone.0025417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Medina M., Denizot J., Dreux N., Robin F., Billard E., Bonnet R., et al. (2014). Western diet induces dysbiosis with increased E. coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. 63 116–124. 10.1136/gutjnl-2012-304119 [DOI] [PubMed] [Google Scholar]
- Miller W. G., Chapman M. H., Yee E., On S. L. W., McNulty D. K., Lastovica A. J., et al. (2012). Multilocus sequence typing methods for the emerging Campylobacter species C. hyointestinalis, C. lanienae, C. sputorum, C. concisus, and C. curvus. . 2:45. 10.3389/fcimb.2012.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro C. A., Moubarac J. C., Cannon G., Ng S. W., Popkin B. (2013). Ultra-processed products are becoming dominant in the global food system. 14 21–28. 10.1111/obr.12107 [DOI] [PubMed] [Google Scholar]
- Moschetti G., Blaiotta G., Aponte M., Catzeddu P., Villani F., Deiana P., et al. (1998). Random amplified polymorphic DNA and amplified ribosomal DNA spacer polymorphism : powerful methods to differentiate Streptococcus thermophilus strains. 85 25–36. 10.1046/j.1365-2672.1998.00461.x [DOI] [PubMed] [Google Scholar]
- Mukhopadhya I., Thomson J. M., Hansen R., Berry S. H., El-Omar E. M., Hold G. L. (2011). Detection of Campylobacter concisus and other campylobacter species in colonic biopsies from adults with ulcerative colitis. 6:e21490. 10.1371/journal.pone.0021490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J., Szklarczyk D., Julien P., Letunic I., Roth A., Kuhn M., et al. (2010). eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. 38(Suppl._1) D190–D195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. 75 4646–4658. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- Nielsen H. L., Nielsen H., Ejlertsen T., Engberg J., Günzel D., Zeitz M., et al. (2011). Oral and fecal Campylobacter concisus strains perturb barrier function by apoptosis induction in HT-29/B6 intestinal epithelial cells. 6:e23858. 10.1371/journal.pone.0023858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H. L., Nielsen H., Torpdahl M. (2016). Multilocus sequence typing of Campylobacter concisus from Danish diarrheic patients. 8:44. 10.1186/s13099-016-0126-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- On S., Siemer B., Chung P., Lastovica A. (2013). Characterisation of Campylobacter concisus strains from South Africa using amplified fragment length polymorphism (AFLP) profiling and a Genomospecies-specific Polymerase Chain Reaction (PCR) assay: Identification of novel genomospecies and correlation with clinical data. 7 1845–1851. 10.5897/AJMR12.2182 [DOI] [Google Scholar]
- Pingali P. (2007). Westernization of Asian diets and the transformation of food systems: implications for research and policy. 32 281–298. 10.1016/j.foodpol.2006.08.001 [DOI] [Google Scholar]
- Roa Engel C. A., ter Horst J. H., Pieterse M., van der Wielen L. A. M., Straathof A. J. J. (2013). Solubility of fumaric acid and its monosodium salt. 52 9454–9460. 10.1021/ie400794r [DOI] [Google Scholar]
- Sakamoto N., Kono S., Wakai K., Fukuda Y., Satomi M., Shimoyama T., et al. (2005). Dietary risk factors for inflammatory bowel disease a multicenter case-control study in Japan. 11 154–163. 10.1097/00054725-200502000-00009 [DOI] [PubMed] [Google Scholar]
- Sartor R. B. (2006). Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. 3 390–407. 10.1038/ncpgasthep0528 [DOI] [PubMed] [Google Scholar]
- Searle B. C. (2010). Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. 10 1265–1269. 10.1002/pmic.200900437 [DOI] [PubMed] [Google Scholar]
- Smith J., Hong-Shum L. (2011). Hoboken, NJ: John Wiley & Sons; 10.1002/9781444397741 [DOI] [Google Scholar]
- Tanner A. C. R., Badger S., Lai C. H., Lisgarten M. A., Visconti R. A., Socransky S. S. (1981). Wolinella gen. nov., Wolinella succinogenes (Vibrio succinogenes Wolin et al.) comb. nov., and description of Bacteroides gracilis sp. nov., Wolinella recta sp. nov., Campylobacter concisus sp. nov., and Eikenella corrodens from humans with periodontal disease. 31 432–445. 10.1099/00207713-31-4-432 [DOI] [Google Scholar]
- Wang Y., Liu F., Zhang X., Chung H. K. L., Riordan S. M., Grimm M. C., et al. (2017). Campylobacter concisus genomospecies 2 is better adapted to the human gastrointestinal tract as compared with Campylobacter concisus genomospecies 1. 8:543. 10.3389/fphys.2017.00543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. (2015). Oral Campylobacter species: Initiators of a subgroup of inflammatory bowel disease? 21 9239–9244. 10.3748/wjg.v21.i31.9239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Budiman V., Day A. S., Mitchell H., Lemberg D. A., Riordan S. M., et al. (2010). Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. 48 2965–2967. 10.1128/JCM.02391-09 [DOI] [PMC free article] [PubMed] [Google Scholar]