Abstract
Gut bacteria that produce urease, the enzyme hydrolysing urea, contribute to nitrogen balance in diverse vertebrates, although the presence of this system of urea-nitrogen recycling in Amphibia is as yet unknown. Our studies of the wood frog (Rana sylvatica), a terrestrial species that accrues urea in winter, documented robust urease activity by enteric symbionts and hence potential to recoup nitrogen from the urea it produces. Ureolytic capacity in hibernating (non-feeding) frogs, whose guts hosted an approximately 33% smaller bacterial population, exceeded that of active (feeding) frogs, possibly due to an inductive effect of high urea on urease expression and/or remodelling of the microbial community. Furthermore, experimentally augmenting the host's plasma urea increased bacterial urease activity. Bacterial inventories constructed using 16S rRNA sequencing revealed that the assemblages hosted by hibernating and active frogs were equally diverse but markedly differed in community membership and structure. Hibernating frogs hosted a greater relative abundance and richer diversity of genera that possess urease-encoding genes and/or have member taxa that reportedly hydrolyse urea. Bacterial hydrolysis of host-synthesized urea probably permits conservation and repurposing of valuable nitrogen not only in hibernating R. sylvatica but, given urea's universal role in amphibian osmoregulation, also in virtually all Amphibia.
Keywords: host-bacterial symbiosis, gut microbiome, urease, urea hydrolysis, nitrogen conservation, hibernation
1. Introduction
Osmoregulators adapt to stresses of dehydration or saline exposure by accumulating one or more small organic osmolytes, or ‘compatible solutes'. Urea is an important balancing osmolyte in some ectotherms despite its potentially destabilizing effects on macromolecular structure and function [1]. Amphibians respond to osmotic challenge by accruing urea (up to 0.3 mol l−1 in some species) by ceasing urination, reabsorbing urea from the renal tubules and bladder fluid, reducing renal filtration rate, and in some cases upregulating hepatic ureagenesis [2]. Hyperuraemia, which they readily tolerate, preserves the water potential gradient conducive to retaining body water while also limiting the injurious rise in ionic concentration. Given its crucial role in the water economy of amphibians—and ultimately in their survival of environmental extremes and exploitation of severe habitats—it is surprising that the ultimate fate of accrued urea is as yet unknown.
Amphibians exhibit a low-energy lifestyle, and thereby tolerate environmental circumstances that impose nutrient limitation [3]. They would benefit from recouping the nitrogen in surplus urea, although, as with other vertebrates, they lack endogenous urease, the enzyme needed to hydrolyse this metabolite. Nevertheless, various mammals, birds, reptiles, fishes and even some invertebrates recycle urea's nitrogen through symbiotic relationships with certain gut bacteria that produce urease. Although this system is best known for its role in maintaining nitrogen balance in ruminants, it also contributes to other functions, such as regulating blood pH, conserving water in desert herbivores and maintaining lean mass in hibernating mammals [4–7]. Regulation of this system to accommodate changes in the host's diet, nutritional and metabolic states, or gut morphology, presumably is achieved by remodelling the microbial community and/or altering the microbial expression of urease in response to changes in urea's availability [8,9].
Postulating that urea-nitrogen recycling would be especially beneficial in amphibians that accumulate urea during periods of activity and dormancy, we focused our research on the wood frog (Rana sylvatica), a terrestrial hibernator in which urea is not only an osmoprotectant but also a cryoprotectant [10] and metabolic depressant [11]. Catabolism of muscle proteins in the weeks preceding hibernation leads to accrual of large amounts of urea (reaching 0.25 mol l−1 in the blood of some individuals) that persist until late winter [12]. We hypothesize that the ultimate recovery of urea's constitutive nitrogen and its incorporation into biosynthetic compounds helps restore body condition well before frogs can resume feeding.
We here provide evidence for the presence of urea-nitrogen recycling in Amphibia by documenting a robust capacity for urea hydrolysis by bacteria in the gut of hyperuraemic, hibernating R. sylvatica. We compared bacterial load and gross morphology of guts from hibernating and active frogs because the anticipated downregulation and remodelling of the gastrointestinal tract in response to aphagia and seasonal dormancy (e.g. [13,14]) potentially influences the numbers and ureolytic capacity of enteric symbionts (e.g. [15–17]). We tested the hypothesis that the host's urea level influences ureolytic capacity in the gut, and also inventoried and compared the gut bacterial communities in hibernating and active frogs, predicting that hibernators would host a greater abundance of ureolytic taxa.
2. Material and methods
(a). Experimental subjects
Male wood frogs (R. sylvatica) were collected in late winter from breeding ponds in southern Ohio, housed individually in plastic tubs containing damp moss, and kept at 4°C in darkness. ‘Late-winter' frogs were used in experiments approximately four weeks later, whereas others were kept until April and then released in an outdoor enclosure. Situated in a mature, deciduous woodlot, this 48 m2 outdoor pen provided herbaceous cover, cool, moist conditions and a small pool of water [10]. Vitamin-fortified crickets (Gryllodes sigillatus; Ghann's Cricket Farm, Augusta, GA, USA) were stocked thrice weekly, although the frogs' diet was enriched with various arthropods drawn to a UV-A light. In June, some individuals (hereafter, ‘active' frogs) were collected, returned to the laboratory, and immediately sampled. Additional frogs were gathered after feeding ceased in early November and placed in simulated hibernation (4°C, darkness) as described above; these ‘hibernating' frogs were sampled two months later. Frogs were collected under permit issued by the Ohio Division of Wildlife; protocols for their husbandry, experimentation and euthanasia, which was carried out by double-pithing, were approved by the Institutional Animal Care and Use Committee of Miami University.
(b). Characterizing ureolytic capacity in guts of hibernating frogs
We determined the size, bacterial load and ureolytic capacity of gut segments sampled from hibernating frogs (n = 10). Procedures were conducted in a refrigerated room (4°C) using aseptic technique and filter-sterilized reagents. Frogs were purged of any bladder fluid, weighed, euthanized, measured to determine snout–ischium length and dissected. Blood was drawn from an incision in the aortic trunk into heparinized capillary tubes and centrifuged (2000g, approx. 5 min); resultant plasma was frozen in liquid nitrogen, stored at –80°C, and ultimately assayed for urea using a urea-nitrogen assay kit (B7551-120, Pointe Scientific, Canton, MI, USA). The foregut, midgut and hindgut were ligated at each end with suture silk, removed from the coelom, rinsed externally with phosphate-buffered saline (PBS) and opened longitudinally. We separately isolated the bacteria within each gut segment by collecting any luminal matter and gently scraping the mucosa with a spatula into 700 µl sodium phosphate buffer (10 mmol l−1; pH 7.0), centrifuging the thoroughly mixed suspension (400g, 5 min) to pellet coarse debris and reserving the supernatant. The pellet from this step was resuspended in 700 µl fresh buffer, thoroughly mixed and again centrifuged (400g, 5 min). Bacteria in the combined supernatants were coalesced by centrifugation (14 000g, 20 min) and resuspended in 800 µl fresh buffer. Cells in a 20 µl aliquot of this suspension were fixed with 1% glutaraldehyde [18] for enumeration (see below), whereas the remainder was centrifuged (14 000g, 20 min) and the resultant bacterial pellet was stored at –80°C for four to six weeks before we assayed urease activity (see below). Finally, we measured the resting length and, after drying at 65°C, the mass of each gut segment.
We estimated the bacterial population within each gut segment by staining the glutaraldehyde-fixed samples for 40 min with 10 µg ml−1 DAPI [19] and counting bacteria in a Bright-line Petroff-Hausser chamber viewed at 1000× [20]. The average of counts obtained from three separate chamber loadings was taken to represent each sample.
Urease activity was assayed in bacterial pellets that were thawed on ice, mixed with cold sodium phosphate buffer and homogenized for 4 min at 4°C using 0.1 mm glass beads and a bead mill (BBY24M; Next Advance, Averill Park, NY, USA). We centrifuged the homogenate (14 000g, 5 min) and assayed the clear lysate solution (approx. 250 µl) using a urease activity assay kit (MAK120, Sigma Aldrich, MO, USA) that quantifies ammonia produced from urea hydrolysis after subtracting background absorbance, per the manufacturer's instructions. The incubation time necessary to obtain an observable signal was determined in preliminary experiments; except as otherwise noted, lysate was incubated with substrate (urea) at 20°C for 1 h (hindgut), 5–12 h (midgut) or 30 h (foregut). Urease is reported in mU, where one unit (U) is the amount of enzyme hydrolysing 1.0 µmol urea min−1. Urease measured in lysate samples was extrapolated to the entire gut segment from which the bacteria were collected; we also normalized urease activity to protein concentration of the lysate, which is a sensitive and reliable proxy for bacterial density [21]. Protein was measured using the NanoDrop 2000 protocol for the Coomassie Plus (Bradford) protein assay (23236, Pierce, Rockford, IL, USA) with bovine serum albumin as a standard.
We used separate aliquots of lysates of hindgut bacteria from half of these frogs in order to validate that the ammonia produced in urease assay results from enzymatic hydrolysis of urea. These samples were removed from frozen storage (–80°C), thawed, divided into three portions and assayed as described above after preincubation with a urease inhibitor (acetohydroxamic acid; 15 mmol l−1, 30 min); after heating (95°C, 10 min); or without treatment. Additional hibernating frogs (n = 9) were used to investigate the thermal sensitivity of urease activity in hindgut bacterial lysates. We assayed activity at 20°C, 5°C or 0°C, extending incubation time (up to 5 h) to accommodate the reduced catalytic activity occurring at low temperature. Combining lysates from three individuals was necessary to provide sufficient material to assay at each temperature; results are reported for three separate pools. For context, we performed identical tests on lysates prepared from bacteria harvested from the caeca of euthanized laboratory mice (n = 3 lysate pools, each prepared from three individuals).
(c). Ureolytic capacity in frog gut influenced by host's activity status
We compared morphometrics and bacterial load of individual gut segments, as well as the ureolytic capacity of hindgut bacteria, of the hyperuraemic, hibernating frogs mentioned above with results for normouraemic, active frogs (n = 10), which were treated in the same manner. Hibernating and active frogs were indistinguishable with respect to standard body mass (15.7 ± 0.5 versus 14.8 ± 0.5 g, respectively; p = 0.79) and snout–ischium length (53.6 ± 0.9 versus 52.7 ± 0.6 mm, respectively; p = 0.43).
(d). Ureolytic capacity in frog gut influenced by host's urea level
We tested the hypothesis that ureolytic capacity of hindgut bacteria is enhanced under hyperuraemia by experimentally manipulating urea levels in late-winter frogs, which are aphagic and maintain low levels of urea [10]. Following Muir et al. [11], we injected a volume (approx. 3% of standard body mass) of PBS, or PBS containing 1.5 mol l−1 urea, into the dorsal lymph pad. Frogs were held in darkness at 4°C for 10 d before being sampled. Lysates prepared from hindgut bacteria were assayed for urease activity and protein, and plasma urea level was determined, as described above. Sham-treated (n = 7) and urea-loaded (n = 6) frogs were well matched for standard body mass (14.6 ± 0.6 versus 13.6 ± 0.7 g; p = 0.35) and snout–ischium length (52.6 ± 1.2 versus 53.4 ± 0.7 mm; p = 0.56).
(e). Bacterial inventory
We investigated whether seasonal variation in ureolytic capacity in the frog hindgut is associated with differences in the bacterial community by comparing inventories obtained for hibernating (n = 5) and active (n = 8) frogs not used in aforementioned experiments. Hindgut contents were collected as described above, suspended in 200 µl PBS and frozen at –20°C. Total DNA was extracted using the QIAamp PowerFecal DNA Kit (12830, MO BIO Laboratories, Carlsbad, CA, USA) following the kit's instructions, except that we repeated the elution to increase the DNA yield. Quantity and quality of the isolated DNA were determined using a NanoDrop 2000 spectrophotometer. Samples were shipped under dry ice to LC Sciences, LLC (Houston, TX, USA) for amplification of the V3–V4 region of the 16S rRNA gene and sequencing using the Illumina MiSeq platform [22].
We used QIIME 1.9.1 [23] to analyse the sequences. After implementing standard quality control measures, sequences were grouped into operational taxonomic units (OTUs) using the open reference method against the Greengenes core set [24]. Sequences were grouped with UCLUST [25] using a minimum sequence identity of 99%. We aligned the most abundant sequences within each OTU against the Greengenes core set [24], removed the hypervariable regions and classified the OTUs using UCLUST [25]. Phylogenetic trees of representative sequences were constructed with FastTree [26]. All microbial sequences have been uploaded to the NCBI Short Read Archive (SRA) under accession PRJNA432152.
We attempted to determine which among the observed genera had at least one member that potentially can catabolize urea by querying the Kyoto Encyclopedia of Genes and Genomes (KEGG), an online resource that contains gene catalogues from sequenced organisms, for entries having urease-encoding genes using the KEGG orthology (KO) identifiers K01427, K01428, K01429, K01430 and K14048. For genera for which these genes were not reported, we combed the scholarly literature using Internet search engines for evidence of ureolytic activity or the presence of urease in any member taxon, ultimately qualifying 17 genera on this basis (electronic supplementary material, table S1).
(f). Statistical analyses
Summary statistics for morphological and physiological variables are presented as mean ± s.e. Data from different groups were compared using a Student's t-test or analysis of variance (ANOVA), followed by Student–Newman–Keuls procedure. Two-factor ANOVA was used to compare morphometric variables of gut segments between hibernating and active frogs, with pairs of means distinguished using Bonferroni's tests. Datasets failing to meet assumptions of normality and homoscedasticity were transformed or, if necessary, analysed using a non-parametric equivalent (Mann–Whitney U-test or Kruskal–Wallis/Dunn's test). Analyses were performed with R v. 3.2.2 (https://www.R-project.org; α = 0.05).
We compared the number of 16S rRNA gene sequences in each group using Student's t-tests. Metrics of α diversity (Shannon Index, evenness, observed OTUs and Faith's Phylogenetic Diversity) were also compared between groups using Student's t-tests; here, we calculated the mean of 20 iterations of a random sub-sampling of 17 800 sequences for each sample. Beta-diversity metrics of community membership and community structure were calculated from unweighted and weighted Unifrac distances, respectively, using 17 800 sequences per sample, and compared using adonis, a permutational MANOVA [27]. We used principal coordinate analysis to visually compare these results. Relative abundances of bacterial phyla and genera underwent a variance-stabilizing transformation of arcsin(abundance0.5) [28,29] and were compared using the Response Screening function in JMP 12.0, which performs multiple t-tests. p-values were adjusted using the false discovery rate correction for multiple comparisons [30]; α = 0.05.
3. Results
(a). Ureolytic capacity in guts of hibernating frogs
The gastrointestinal tracts of hibernating frogs (n = 10) contained small amounts of mucus and presumably autochthonous matter, but were largely devoid of recognizable ingesta. Mass and length of the foregut, midgut and hindgut varied markedly, the latter being the tract's smallest segment (table 1). Despite its diminutive size, the hindgut harboured 2.4-fold more bacteria than the midgut and 40-fold more bacteria than the foregut. Accordingly, abundant urease (approx. 5 mU) occurred only in this segment. Indeed, despite the exquisite sensitivity of our assay (lower limit of detection, 0.0005 mU ml−1), we did not detect urease activity in the foreguts of two frogs and the midguts of three frogs.
Table 1.
Morphometrics, number of bacteria and urease in gut segments of hibernating frogs. Within each row, means (±s.e.) denoted by different letters were statistically distinguishable (p < 0.05).
| foregut | midgut | hindgut | p | |
|---|---|---|---|---|
| mass (mg) | 24.3 ± 1.3a | 15.4 ± 0.8b | 7.4 ± 0.4c | <0.0001 |
| length (mm) | 21.3 ± 1.3a | 46.2 ± 3.6b | 12.5 ± 1.5c | <0.0001 |
| bacteria (×108) | 0.03 ± 0.01a | 0.5 ± 0.3b | 1.2 ± 0.2c | 0.0003 |
| urease (mU) | 0.007 ± 0.004a | 0.01 ± 0.01a | 4.87 ± 0.95b | <0.0001 |
| n | 9–10 | 9–10 | 9–10 |
Validation tests, using residual lysates prepared from hindgut bacteria sampled from five of the hibernating frogs, suggested that the ammonia accrued in urease assays was produced by an enzymatic process. Relative to results for freshly prepared lysates, urease activity in frozen/thawed samples was reduced by 24% (range: 10–37%; p = 0.027). Pre-treating the lysate with the urease inhibitor acetohydroxamic acid before assay reduced its activity by 97% (range: 95–98%; p = 0.003); heating it before assay reduced its activity by 99% (range: 98–100%; p = 0.003).
Urease activity in lysates prepared from the hindgut bacteria of additional hibernating frogs was strongly dependent on assay incubation temperature (repeated-measures ANOVA; p = 0.0002), with the activity measured at 0°C (37.2 ± 6.9 mU mg−1 lysate protein) being only one-third of that measured at 20°C (106.9 ± 11.2 mU mg−1 lysate protein). The overall temperature coefficient (Q10) was 1.69 (figure 1). The Q10 of urease activity in lysates prepared from mouse caecal bacteria was similar; however, the urease activity measured at 20°C in these samples, 71.7 ± 6.1 mU mg−1 lysate protein, was approximately 33% lower than that determined for frogs (p = 0.050; figure 1).
Figure 1.
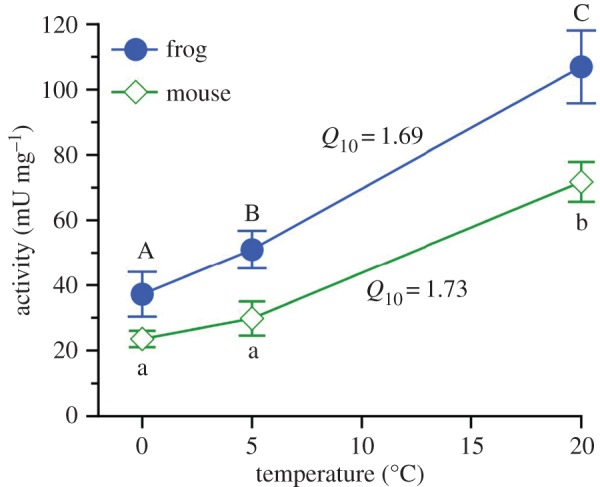
Thermal sensitivity of urease activity in a lysate prepared from bacteria collected from frog hindgut or mouse caecum. Mean ± s.e.; n = 3 samples (each a composite of three individuals; see Material and methods section for details) were tested at each temperature. Means identified by dissimilar letters were distinguishable (repeated-measures ANOVA; p < 0.05). (Online version in colour.)
(b). Ureolytic capacity in frog gut influenced by host's activity status
The austere gut of the aforementioned hibernating frogs contrasted with that of active frogs (n = 10), in which the foregut usually contained insect parts, epithelium of the midgut supported well-formed villi, and the hindgut contained faeces. Emptied gastrointestinal tracts of hibernators weighed 61% less (p < 0.0001) and were 25% shorter (p = 0.005) and half as dense (i.e. mass per unit length; p < 0.0001) as those of active frogs (figure 2a). However, activity state × gut segment interaction for both mass (p = 0.004) and density (p = 0.0001) attested that such disparities were non-uniform among the tract's components. Indeed, intergroup variation in mass and density of hindgut was relatively small relative to that of foregut and midgut, and, furthermore, hindgut length was indistinguishable between hibernating and active frogs (figure 2b).
Figure 2.
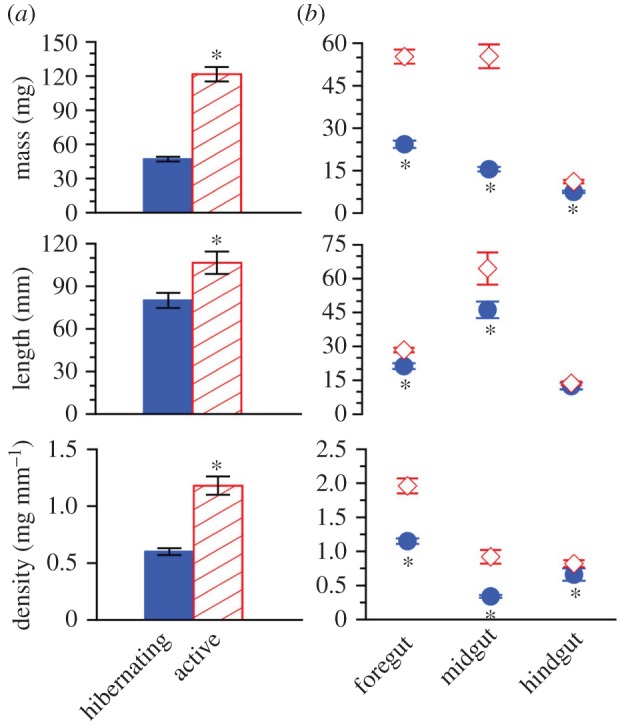
Variation in morphometrics of (a) the entire gastrointestinal tract and (b) individual gut segments sampled from hibernating (closed circle) and active (open diamond) frogs. Mean ± s.e.; n = 10. Asterisk signifies that the means within a pair differed (Bonferroni; p < 0.05). (Online version in colour.)
Expectedly, plasma urea concentration was markedly higher (p < 0.0001) in hibernating frogs as compared to active frogs (23.5 ± 2.9 versus 3.7 ± 0.5 mmol l−1). The hindgut of hibernators harboured 33% fewer bacteria (p = 0.020) but, nevertheless, held twice the urease (p = 0.024); moreover, urease activity was 2.8-fold higher (p < 0.0001) in hibernators than in active frogs (figure 3).
Figure 3.
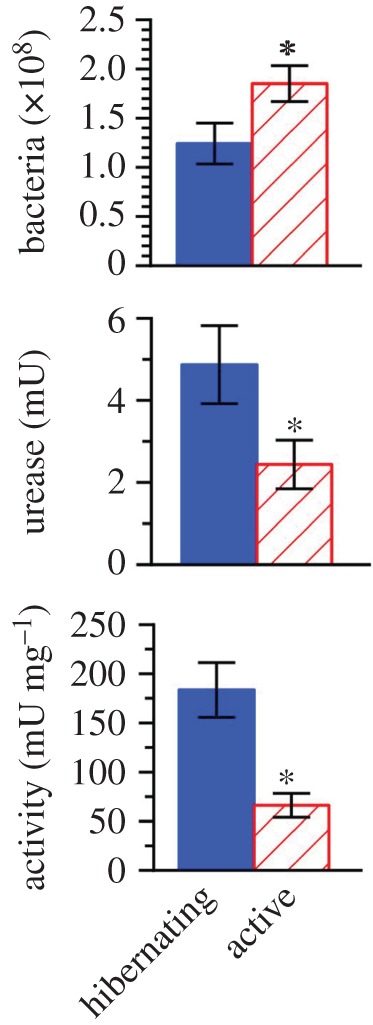
Number of bacteria and urease in hindgut, and urease activity in lysates prepared from hindgut bacteria, in hibernating and active frogs. Mean ± s.e.; n = 9–10. Asterisk signifies that the means differed (p < 0.05). (Online version in colour.)
(c). Ureolytic capacity in frog gut influenced by host's urea level
Compared with their sham-treated counterparts (n = 7), late-winter frogs injected with urea solution (n = 6) had plasma urea levels that were fivefold higher (42.2 ± 4.5 versus 8.4 ± 1.1 mmol l−1; p < 0.0001). Urea augmentation did not raise the number of hindgut bacteria, as the complements in urea-loaded frogs (1.0 ± 0.2 × 108) and controls (1.0 ± 0.1 × 108) were indistinguishable (p = 0.357). However, results suggested that hyperuraemia enhanced ureolytic capacity, as the urease in hindgut was nominally (albeit not significantly; p = 0.126) greater in urea-loaded frogs (1.7 ± 2.0 versus 0.6 ± 0.2 mU), and urease activity in bacterial lysates was 2.7-fold higher in urea-loaded frogs as compared with controls (215.7 ± 68.3 versus 79.9 ± 29.2 mU mg−1 lysate protein; p = 0.037).
(d). Bacterial inventories from hibernating and active frogs
We obtained 26 679 ± 1692 sequences per sample, finding no difference (p = 0.929) in the number of sequences between hibernating (n = 5) and active (n = 8) frogs. These groups did not differ (t-tests: p > 0.261, all cases) in metrics of alpha diversity: Shannon Index (hibernating: 6.42 ± 0.26 versus active: 6.56 ± 0.29), observed OTUs (1,370 ± 101 versus 1,180 ± 124), evenness (0.62 ± 0.02 versus 0.64 ± 0.02) and Faith's Phylogenetic Diversity (43.89 ± 3.98 versus 43.35 ± 3.22). However, they differed markedly in bacterial community membership (adonis: R2 = 0.166; p = 0.001; figure 4a) and structure (adonis: R2 = 0.168; p = 0.028; figure 4b). Clustering suggested that inter-individual variation among hibernating frogs was relatively high with respect to community membership, but (except for one individual) extremely low with respect to community structure. Of the 9056 observed bacterial OTUs, 2042 (22.5%) occurred in both groups, whereas 3024 (33.4%) were exclusive to hibernating frogs and 3990 (44.1%) were exclusive to active frogs.
Figure 4.
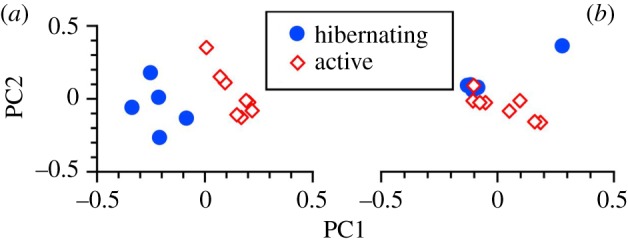
(a) Principal coordinate analysis of bacterial community membership using unweighted Unifrac distances to identify the presence/absence of bacterial lineages. Principal coordinates 1 and 2 explain 17.9% and 12.1%, respectively, of the variation. (b) Principal coordinate analysis of bacterial community structure using weighted Unifrac distances and taking relative abundances into account. Principal coordinates 1 and 2 explain 33.0% and 29.2%, respectively, of the variation. Closed circles represent individual hibernating frogs (n = 5); open diamonds represent individual active frogs (n = 8). (Online version in colour.)
Sequence analysis identified 15 bacterial phyla, of which three (Acidobacteria, Deferribacteres, WPS-2) were exclusive to hibernating frogs and one (Fusobacteria) exclusive to active frogs. We identified 96 genera, 20 of which were exclusive to hibernating frogs and 16 of which were exclusive to active frogs; thus, approximately 38% of all genera occurred only in one or the other group.
Relative abundance data for all observed taxa are presented in electronic supplementary material, table S2. Group differences were found for two phyla, Actinobacteria (FDR-corrected p = 0.004) and Acidobacteria (FDR-corrected p = 0.004), which were more abundant in hibernating frogs. Five phyla accounted for approximately 95% of all bacteria identified from the frog hindgut (table 2). Bacteroidetes comprised greater than 56% of the bacteria from both hibernating and active frogs, and Proteobacteria and Firmicutes jointly accounted for much of the remainder (24%, hibernating; 40%, active). Actinobacteria comprised approximately 5% of bacteria from hibernating frogs, but was poorly represented in active frogs.
Table 2.
Five most predominant bacterial phyla and genera comprising at least 1% of all bacteria in the hindgut microbial community of hibernating (n = 5) and active (n = 8) frogs. n.d., not detected. Relative abundance data for all observed taxa are presented in electronic supplementary material, table S2. Bold typeface signifies that relative abundance (%; mean ± s.e.) of the indicated taxon differed (*p < 0.05; **p < 0.01) between groups.
| hibernating | active | |
|---|---|---|
| phyla | ||
| Bacteroidetes | 63.16 ± 12.61 | 56.16 ± 7.44 |
| Firmicutes | 10.29 ± 1.90 | 22.77 ± 3.93 |
| Proteobacteria | 14.05 ± 10.42 | 16.89 ± 5.88 |
| Actinobacteria** | 4.50 ± 1.93 | 0.22 ± 0.08 |
| Verrucomicrobia | 1.27 ± 0.75 | 0.08 ± 0.07 |
| genera | ||
| Bacteroides | 51.56 ± 11.58 | 40.48 ± 7.57 |
| Parabacteroides | 4.49 ± 1.71 | 8.84 ± 1.83 |
| Desulfovibrio | 1.36 ± 0.45 | 9.16 ± 5.35 |
| Arthrobacter* | 3.31 ± 1.64 | n.d. |
| Pseudomonas* | 4.63 ± 4.10 | 0.005 ± 0.004 |
| Oscillospira | 1.35 ± 0.36 | 2.04 ± 0.31 |
| Bilophila** | 0.28 ± 0.16 | 2.13 ± 0.71 |
| Akkermansia | 1.26 ± 0.74 | 0.07 ± 0.07 |
| Anaerovorax** | 1.15 ± 0.24 | 0.16 ± 0.06 |
| Clostridium | 0.008 ± 0.004 | 1.06 ± 0.58 |
| Citrobacter | 0.03 ± 0.03 | 1.02 ± 0.73 |
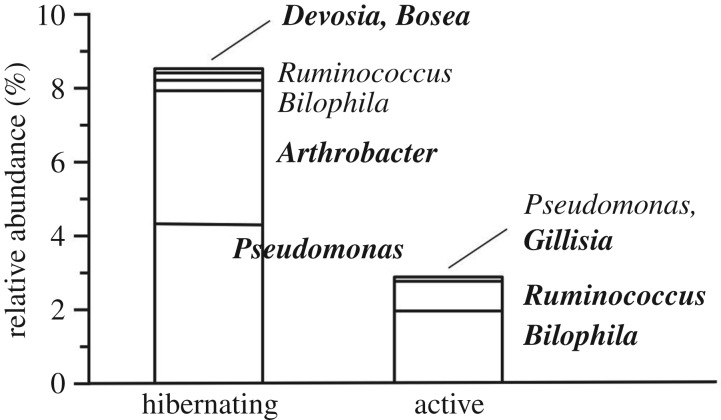
Fourteen of the 96 genera observed differed in relative abundance (electronic supplementary material, table S2), six being more abundant (FDR-corrected p < 0.029, all cases) in hibernating frogs and eight being more abundant (FDR-corrected p < 0.049, all cases) in active frogs. Eleven genera were particularly well represented (i.e. relative abundance ≥ 1%) overall, including Bacteroides, which comprised 40–50% of all bacteria (table 2). Desulfovibrio, Parabacteroides and Oscillospira collectively accounted for approximately 20% of bacteria from active frogs, but only 7% of bacteria from hibernating frogs. Approximately, 9% of bacteria from hibernating frogs belonged to three genera (Pseudomonas, Anaerovorax and Arthrobacter) that were poorly represented in active frogs. Ureolytic potential was recognized for 56 (58.3%) of the observed genera (electronic supplementary material, table S2), more of which were hosted by hibernating frogs (27.0 ± 3.1 versus 19.5 ± 2.3; p = 0.036). Relative abundance differed between the groups for seven of the ureolytic genera, which overall were better represented in hibernating frogs (figure 5).
Figure 5.
Genera comprising one or more taxa having potential for urea hydrolysis that differed in mean relative abundance between hibernating (n = 5) and active (n = 8) frogs. Bold typeface signifies that the depicted mean was significantly higher (p < 0.05) as compared with the other group.
4. Discussion
Ubiquitous in nature, urease-producing bacteria benefit ecosystems by making nitrogen from urea available to life [31], and many are gut symbionts that contribute to nitrogen balance in their host [5]. Our demonstration of robust ureolytic activity in bacteria residing within the frog gut provides seminal evidence that amphibians can potentially benefit by retaining nitrogen reclaimed from the urea they produce.
Urea's catabolism to carbon dioxide and ammonia mainly occurs within the rumen of foregut fermenters, the midgut of some non-ruminants, or the hindgut (caecum and/or proximal colon) of monogastric herbivores, omnivores and carnivores [5,6], but potentially also occurs in other alimentary organs, which host distinct bacterial communities [32–34]. In the domestic goat, for example, urease activity occurs not only in the rumen but also in the small intestine, caecum and colon [35]. Limited urea hydrolysis occurs in the stomach and/or small intestine of rats [36,37], rabbits [38] and grouse [39]. In our hibernating frogs, bacterial urease was scanty or absent from the foregut and midgut but highly abundant in the hindgut, suggesting the latter is the primary site of urea hydrolysis in amphibians, as is the case with non-ruminant mammals, birds, fish and probably reptiles [5]. Nevertheless, the possibility that this function also occurs parenterally cannot be excluded, given that autochthonous ureolytic bacteria flourish in multiple organs of some vertebrates [40].
Ureolysis in lysates of bacteria sampled from the hindgut of hibernating frogs was robust. Measured at 20°C, urease activity in these samples was approximately 1.5-fold higher than that determined for bacteria sampled from mouse caecum, and sixfold to 11-fold higher than activities measured at 37°C for bacteria sampled from bovine rumen [41]. Amphibians can accumulate substantial quantities of urea for osmoprotection [2] and, in some species, also cryoprotection [10] and metabolic inhibition [11], and therefore could possess a greater ureolytic capacity than mammals, which do not normally accrue this ‘waste' metabolite. The relatively low temperature coefficient, 1.69, for urease activity in our samples is comparable with, if not slightly lower than, that previously reported [42,43], and probably facilitates urea hydrolysis in frogs even at winter temperatures.
Digestive organs of amphibians undergo profound transformations in structure and function during adaptation to altered physiological and nutritional states in dormancy [13,14]. Accordingly, the gastrointestinal tract of our hibernating frogs was reduced in mass (25%) and length (61%), and also in density (approx. 50%), perhaps owing to degeneration of the mucosa and musculature, particularly of the midgut. Remodelling responses vary among alimentary organs, as, for example, the hindgut is only slightly altered, whereas size of the foregut and/or midgut is reduced by 50–85% in estivating frogs [44] and salamanders [45], and in hibernating toads [46]. Results for our hibernating frogs were comparable. Selectively maintaining the hindgut's morphology in dormancy despite the added energetic cost presumably benefits the host by providing symbiotic bacteria with diverse colonization niches [34].
Populations of gut bacteria commonly are reduced in dormancy and, accordingly, in R. pipiens and R. catesbeiana they fall by 80–90% during underwater hibernation [47–50]. By contrast, the reduction in our R. sylvatica, a terrestrial hibernator, was only approximately 33%. The smaller population of hindgut bacteria in these frogs nevertheless had twice the urease and achieved nearly three times the hydrolytic activity as the bacteria in active frogs. Greater abundance of enzyme (as inferred from activity assays) in these hyperuraemic frogs potentially derived from the inductive effect of high urea on urease expression and/or remodelling of the microbial community [8,9,51,52]; future research should elucidate the specific causes. Indeed, relative to active frogs, hibernators hosted a higher relative abundance and richer diversity of genera that contain ureolytic members, including, notably, Pseudomonas and Arthrobacter. The putative positive effect of increasing substrate availability on ureolytic capacity is underscored by our finding that experimentally augmenting the host's urea levels markedly increased urease activity in hindgut bacteria. The considerable inter-individual variation in hindgut urease observed in our frogs conceivably derived from natural variability in underlying determinants, such as size of the bacterial load and abundance of ureolytic microbes and factors (such as urea availability) that influence urease expression in gut symbionts.
Comprehensive analyses of the gut bacterial community in amphibians are scarce. The principal phyla hosted by R. sylvatica—Bacteroidetes, Firmicutes, Proteobacteria—were the same as those found in other anurans [32,53–55], and the predominant genus, Bacteroides, which comprised more than 40% of hindgut bacteria, is also well represented in other ranids [47–49,55]. Complexity of the bacterial community may be reduced in hibernating frogs [48,49,54,56], much as it is in hibernating mammals (e.g. [57]). We observed no change in α diversity metrics, albeit marked alterations in community membership and structure of the bacteria hosted by hibernating R. sylvatica. Indeed, the bacterial assemblage of hibernators uniquely included several phyla (Acidobacteria, Deferribacteres and WPS-2) and 21% of the observed genera. Pseudomonas, Anaerovorax and Arthrobacter predominated in hibernators but were rare in active frogs. Pseudomonas (Proteobacteria), a well-known psychrophile, is highly abundant in cold-acclimated R. sylvatica and other ranids, but otherwise is uncommon [47,48,50,58]. Extensive remodelling of the microflora in hibernating animals, including frogs, seems essential to maintain the host–bacteria symbiosis under altered biotic and abiotic conditions within the gut.
5. Concluding remarks
Urea-nitrogen recycling is presently unknown in amphibians despite the universal role of urea accrual in their osmoregulation [2]. The hindgut of R. sylvatica, a species that remains hyperuraemic throughout the winter, harbours a rich diversity of urease-containing bacteria capable of hydrolysing the host's urea. Nitrogen liberated by ureolytic bacteria presumably is incorporated into the biosynthetic compounds needed to restore body condition at or before hibernal emergence; however, this system potentially is of general importance to nitrogen balance in virtually all Amphibia.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank C. do Amaral, A. Rosendale and M. Wright for assistance collecting frogs; M. Brake for extracting bacterial DNA; K. Wilkins for counting bacteria; Y. Tomoyasu for sharing equipment; M. Lee for technical advice; and K. Ash for researching the ureolytic potential of gut bacteria. C. do Amaral and K. Killian advised on the experimental design and or statistical analysis. We thank three anonymous reviewers for providing comments and suggestions that strengthened this paper.
Ethics
All experimental and euthanasia procedures complied with protocols approved by Miami University Institutional Animal Care and Use Committee (Research Protocol no. 931). Animals were collected from the wild under permit (20-015) issued to J.P.C. by the Ohio Division of Wildlife.
Data accessibility
All microbial sequences have been uploaded to the NCBI SRA under accession PRJNA432152. Supporting data can be found in the electronic supplementary material.
Authors' contributions
J.P.C. and J.M.W. conceived, designed and performed the experiments. J.M.W, K.D.K. and J.P.C. analysed and interpreted the data. K.D.K. and R.E.L. contributed reagents, material and/or analytical tools. J.M.W. and J.P.C. wrote the paper with contributions from K.D.K. and R.E.L. All authors edited drafts of the manuscript and gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the National Science Foundation (grant IOS1022788 to J.P.C.) and Sigma Xi grants-in-aid of research to J.M.W.
References
- 1.Yancey PH. 2005. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208, 2819–2830. ( 10.1242/jeb.01730) [DOI] [PubMed] [Google Scholar]
- 2.Shoemaker VH, Hillman SS, Hillyard SD, Jackson DC, McClanahan LL, Withers PC, Wygoda ML. 1992. Exchange of water, ions, and respiratory gases in terrestrial amphibians. In Environmental physiology of the amphibians (eds ME Feder, WW Burggren), pp. 125–150. Chicago, IL: University of Chicago Press. [Google Scholar]
- 3.Pough FH. 1980. The advantages of ectothermy for tetrapods. Am. Nat. 115, 92–112. ( 10.1086/283547) [DOI] [Google Scholar]
- 4.Atkinson DE, Camien MN. 1982. The role of urea synthesis in the removal of metabolic bicarbonate and the regulation of blood pH. In Current topics in cellular regulation (eds Horecker BL, Stadtman ER), pp. 261–302. Amsterdam, The Netherlands: Elsevier. [DOI] [PubMed] [Google Scholar]
- 5.Singer MA. 2003. Do mammals, birds, reptiles and fish have similar nitrogen conserving systems? Comp. Biochem. Physiol. B Biochem. Mol. Biol. 134, 543–558. ( 10.1016/S1096-4959(03)00027-7) [DOI] [PubMed] [Google Scholar]
- 6.Stewart GS, Smith CP. 2005. Urea nitrogen salvage mechanisms and their relevance to ruminants, non-ruminants and man. Nutr. Res. Rev. 18, 49–62. ( 10.1079/NRR200498) [DOI] [PubMed] [Google Scholar]
- 7.Riedesel ML, Steffen JM. 1980. Protein metabolism and urea recycling in rodent hibernators. Fed. Proc. 39, 2959–2963. [PubMed] [Google Scholar]
- 8.Collins CM, D'Orazio SE. 1993. Bacterial ureases: structure, regulation of expression and role in pathogenesis. Mol. Microbiol. 9, 907–913. ( 10.1111/j.1365-2958.1993.tb01220.x) [DOI] [PubMed] [Google Scholar]
- 9.Mobley HL, Island MD, Hausinger RP. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59, 451–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costanzo JP, Lee RE. 2005. Cryoprotection by urea in a terrestrially hibernating frog. J. Exp. Biol. 208, 4079–4089. ( 10.1242/jeb.01859) [DOI] [PubMed] [Google Scholar]
- 11.Muir TJ, Costanzo JP, Lee RE. 2007. Osmotic and metabolic responses to dehydration and urea-loading in a terrestrially-hibernating frog. J. Comp. Physiol. B. 177, 917–926. ( 10.1007/s00360-007-0190-3) [DOI] [PubMed] [Google Scholar]
- 12.Costanzo JP, do Amaral MCF, Reynolds AM, Rosendale AJ, Lee RE. 2015. Cryoprotectants and extreme freeze tolerance in a subarctic population of the wood frog. PLoS ONE 10, e0117234 ( 10.1371/journal.pone.0117234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Secor SM. 2005. Physiological responses to feeding, fasting and estivation for anurans. J. Exp. Biol. 208, 2595–2609. ( 10.1242/jeb.01659) [DOI] [PubMed] [Google Scholar]
- 14.Secor SM, Lignot J-H. 2010. Morphological plasticity of vertebrate aestivation. In Aestivation: molecular and physiological aspects, vol. 49 (eds Navas CA, Carvalho JE), pp. 183–208. Berlin, Germany: Springer. [DOI] [PubMed] [Google Scholar]
- 15.Steffen J, Rigler G, Moore A, Riedesel M. 1980. Urea recycling in active golden-mantled ground squirrels (Spermophilus lateralis). Am. J. Physiol. Regul. Integr. Comp. Physiol. 239, R168–RR73. ( 10.1152/ajpregu.1980.239.1.R168) [DOI] [PubMed] [Google Scholar]
- 16.Bintz GL, Torgerson GE. 1981. The metabolism of [14 C] urea by control and starved Richardson's ground squirrels. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 69, 551–555. ( 10.1016/0300-9629(81)93018-8) [DOI] [Google Scholar]
- 17.Harlow HJ. 1987. Urea-hydrolysis in euthermic hibernators and non-hibernators during periods of food availability and deprivation. J. Therm. Biol. 12, 149–154. ( 10.1016/0306-4565(87)90055-6) [DOI] [Google Scholar]
- 18.Kepner RL, Pratt JR. 1994. Use of fluorochromes for direct enumeration of total bacteria in environmental samples: past and present. Microbiol. Rev. 58, 603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu W, Dodds WK, Banks MK, Skalsky J, Strauss EA. 1995. Optimal staining and sample storage time for direct microscopic enumeration of total and active bacteria in soil with two fluorescent dyes. Appl. Environ. Microbiol. 61, 3367–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pryor GS. 2008. Anaerobic bacteria isolated from the gastrointestinal tracts of bullfrog tadpoles (Rana catesbeiana). Herpetol. Conserv. Biol. 3, 176–181. [Google Scholar]
- 21.Nittayajarn A, Baker DD. 1989. Methods for the quantification of Frankia cell biomass. Plant Soil. 118, 199–204. ( 10.1007/BF02232807) [DOI] [Google Scholar]
- 22.Caporaso JG, et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. ( 10.1038/ismej.2012.8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. ( 10.1038/nmeth.f.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. ( 10.1128/AEM.03006-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. ( 10.1093/bioinformatics/btq461) [DOI] [PubMed] [Google Scholar]
- 26.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. ( 10.1093/molbev/msp077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. ( 10.1111/j.1442-9993.2001.01070.pp.x) [DOI] [Google Scholar]
- 28.Shchipkova AY, Nagaraja HN, Kumar PS. 2010. Subgingival microbial profiles of smokers with periodontitis. J. Dent. Res. 89, 1247–1253. ( 10.1177/0022034510377203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar PS, Mason MR, Brooker MR, O'brien K. 2012. Pyrosequencing reveals unique microbial signatures associated with healthy and failing dental implants. J. Clin. Periodontol. 39, 425–433. ( 10.1111/j.1600-051X.2012.01856.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57, 289–300. [Google Scholar]
- 31.Mobley HL, Hausinger RP. 1989. Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 53, 85–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mashoof S, Goodroe A, Du CC, Eubanks JO, Jacobs N, Steiner JM, Tizard I, Suchodolski JS, Criscitiello MF. et al. 2013. Ancient T-independence of mucosal IgX/A: gut microbiota unaffected by larval thymectomy in Xenopus laevis. Mucosal Immunol. 6, 358–368. ( 10.1038/mi.2012.78) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Li D, Refaey MM, Xu W. 2017. High spatial and temporal variations of microbial community along the southern catfish gastrointestinal tract: insights into dynamic food digestion. Front. Microbiol. 8, 1531 ( 10.3389/fmicb.2017.01531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donaldson GP, Lee SM, Mazmanian SK. 2016. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32. ( 10.1038/nrmicro3552) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Šimůnek J, Skřivanová V, Hoza I, Březina P, Marounek M. 1995. Ontogenesis of enzymatic activities in the gastrointestinal tract of young goats. Small Rumin. Res. 17, 207–211. ( 10.1016/0921-4488(95)00678-E) [DOI] [Google Scholar]
- 36.Kim K-I, Lee W-S, Benevenga NJ. 1998. Feeding diets containing high levels of milk products or cellulose decrease urease activity and ammonia production in rat intestine. J. Nutr. 128, 1186–1191. ( 10.1093/jn/128.7.1186) [DOI] [PubMed] [Google Scholar]
- 37.Takebe S, Kobashi K. 1988. Acid urease from Lactobacillus of rat intesine. Chem. Pharm. Bull. 36, 693–699. ( 10.1248/cpb.36.693) [DOI] [PubMed] [Google Scholar]
- 38.Marounek M, Vovk S, Skřivanová V. 1995. Distribution of activity of hydrolytic enzymes in the digestive tract of rabbits. Br. J. Nutr. 73, 463–469. ( 10.1079/BJN19950048) [DOI] [PubMed] [Google Scholar]
- 39.Vecherskii MV, Kuznetsova TA, Stepan'kov AA. 2015. Activity of urealytic microorganisms in the gastrointestinal tract of the black grouse Lyrurus tetrix. Dokl. Biol. Sci. 462, 131–133. ( 10.1134/S0012496615030060) [DOI] [PubMed] [Google Scholar]
- 40.Knight IT, Crirnes DJ, Colwell QRR. 1988. Hydrolysis of urea in the tissues of carcharhinid sharks. Can. J. Fish Aquat. Sci. 45, 357–360. ( 10.1139/f88-043) [DOI] [Google Scholar]
- 41.Jin D, Zhao S, Zheng N, Bu D, Beckers Y, Denman SE, McSweeney CS, Wang J. 2017. Differences in ureolytic bacterial composition between the rumen digesta and rumen wall based on ureC gene classification. Front. Microbiol. 8, 385 ( 10.3389/fmicb.2017.00385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larson AD, Kallio RE. 1954. Purification and properties of bacterial urease. J. Bacteriol. 68, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magaña-Plaza I, Montes C, Ruiz-Herrera J. 1971. Purification and biochemical characteristics of urease from Proteus rettgeri. Biochim. Biophys. Acta 242, 230–237. ( 10.1016/0005-2744(71)90103-3) [DOI] [PubMed] [Google Scholar]
- 44.Cramp RL, Franklin CE. 2003. Is re-feeding efficiency compromised by prolonged starvation during aestivation in the green striped burrowing frog, Cyclorana alboguttata? J. Exp. Zool. A Ecol. Genet. Physiol. 300, 126–132. ( 10.1002/jez.a.10272) [DOI] [PubMed] [Google Scholar]
- 45.Smith ME, Secor SM. 2017. Physiological responses to fasting and estivation for the three-toed amphiuma (Amphiuma tridactylum). Physiol. Biochem. Zool. 90, 240–256. ( 10.1086/689216) [DOI] [PubMed] [Google Scholar]
- 46.Naya DE, Veloso C, Sabat P, Bozinovic F. 2009. The effect of short- and long-term fasting on digestive and metabolic flexibility in the Andean toad, Bufo spinulosus. J. Exp. Biol. 212, 2167–2175. ( 10.1242/jeb.030650) [DOI] [PubMed] [Google Scholar]
- 47.Banas JA, Loesche WJ, Nace GW. 1988. Classification and distribution of large intestinal bacteria in nonhibernating and hibernating leopard frogs (Rana pipiens). Appl. Environ. Microbiol. 54, 2305–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gossling J, Loesche WJ, Nace GW. 1982. Large intestine bacterial flora of nonhibernating and hibernating leopard frogs (Rana pipiens). Appl. Environ. Microbiol. 44, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gossling J, Loesche WJ, Nace GW. 1982. Response of intestinal flora of laboratory-reared leopard frogs (Rana pipiens) to cold and fasting. Appl. Environ. Microbiol. 44, 67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carr AH, Amborski RL, Culley DD, Amborski GF. 1976. Aerobic bacteria in the intestinal tracts of bullfrogs (Rana catesbeiana) maintained at low temperatures. Herpetologica 32, 239–244. [Google Scholar]
- 51.Wong J, Piceno YM, DeSantis TZ, Pahl M, Andersen GL, Vaziri ND. 2014. Expansion of urease- and uricase-containing, indole- and p-cresol-forming and contraction of short-chain fatty acid-producing intestinal microbiota in ESRD. Am. J. Nephrol. 39, 230–237. ( 10.1159/000360010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Z, Meng Q, Li S, Jiang L, Wu H. 2017. Effect of urea-supplemented diets on the ruminal bacterial and archaeal community composition of finishing bulls. Appl. Microbiol. Biotechnol. 101, 6205–6216. ( 10.1007/s00253-017-8323-4) [DOI] [PubMed] [Google Scholar]
- 53.Chang C-W, Huang B-H, Lin S-M, Huang C-L, Liao P-C. 2016. Changes of diet and dominant intestinal microbes in farmland frogs. BMC Microbiol. 16, 33 ( 10.1186/s12866-016-0660-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weng FC-H, Yang Y-J, Wang D. 2016. Functional analysis for gut microbes of the brown tree frog (Polypedates megacephalus) in artificial hibernation. BMC Genomics 17, 31–42. ( 10.1186/s12864-016-3318-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohl KD, Cary TL, Karasov WH, Dearing MD. 2013. Restructuring of the amphibian gut microbiota through metamorphosis. Environ. Microbiol. Rep. 5, 899–903. ( 10.1111/1758-2229.12092) [DOI] [PubMed] [Google Scholar]
- 56.Van der Waaij D, Cohen BJ, Nace GW. 1974. Colonization patterns of aerobic gram-negative bacteria in the cloaca of Rana pipiens. Lab. Anim. Sci. 24, 307–317. [PubMed] [Google Scholar]
- 57.Carey HV, Assadi-Porter FM. 2017. The hibernator microbiome: host-bacterial interactions in an extreme nutritional symbiosis. Annu. Rev. Nutr. 37, 447–500. ( 10.1146/annurev-nutr-071816-064740) [DOI] [PubMed] [Google Scholar]
- 58.Lee MR, Lee RE, Strong-Gunderson JM, Minges SR. 1995. Isolation of ice-nucleating active bacteria from the freeze-tolerant frog, Rana sylvatica. Cryobiology 32, 358–365. ( 10.1006/cryo.1995.1036) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All microbial sequences have been uploaded to the NCBI SRA under accession PRJNA432152. Supporting data can be found in the electronic supplementary material.