Abstract
Wolbachia is a common heritable bacterial symbiont in insects. Its evolutionary success lies in the diverse phenotypic effects it has on its hosts coupled to its propensity to move between host species over evolutionary timescales. In a survey of natural host–symbiont associations in a range of Drosophila species, we found that 10 of 16 Wolbachia strains protected their hosts against viral infection. By moving Wolbachia strains between host species, we found that the symbiont genome had a much greater influence on the level of antiviral protection than the host genome. The reason for this was that the level of protection depended on the density of the symbiont in host tissues, and Wolbachia rather than the host‐controlled density. The finding that virus resistance and symbiont density are largely under the control of symbiont genes in this system has important implications both for the evolution of these traits and for public health programmes using Wolbachia to prevent mosquitoes from transmitting disease.
Keywords: Drosophila, symbiont‐mediated protection, viruses, Wolbachia
1. INTRODUCTION
Wolbachia is a maternally transmitted bacterial symbiont that produces a remarkably diverse array of phenotypes in arthropods. In many cases, it manipulates its host's reproduction to increase its transmission to future generations, for example, by distorting sex ratios or inducing cytoplasmic incompatibility (CI) (Werren, Baldo, & Clark, 2008). More recently, it was discovered that many Wolbachia strains can protect their hosts against viral pathogens (Hedges, Brownlie, O'Neill, & Johnson, 2008; Teixeira, Ferreira, & Ashburner, 2008). Other Wolbachia infections have been associated with an array of other phenotypes, ranging from being mutualists that synthesize essential nutrients (Hosokawa, Koga, Kikuchi, Meng, & Fukatsu, 2010) to causing reductions in survival and fecundity (Martinez et al., 2015).
This phenotypic variation across host–Wolbachia associations could be caused by genetic differences in the hosts, the symbionts or both partners. Understanding the determinants of this variation is important because, over evolutionary timescales, Wolbachia jumps between host species (Vavre, Fleury, Lepetit, Fouillet, & Boulétreau, 1999; Werren, Zhang, & Guo, 1995; Zhang, Han, & Hong, 2013). Whether a phenotype is controlled by the host or the symbiont genome will determine if Wolbachia‐induced phenotypes are transferred along with the infection to the new host and therefore affect the success of the host shift. From an applied perspective, artificially moving the bacterium between host species allows Wolbachia to be used as a biocontrol agent. Strains of Wolbachia have been transferred from Drosophila to the mosquito Aedes aegypti with the aim of preventing the transmission of dengue virus (Frentiu et al., 2014; Joubert et al., 2016; Moreira et al., 2009; Walker et al., 2011; Yeap et al., 2011). Understanding what governs changes in phenotype following a host shift can thus help predict the success of such symbiont‐based applications, and will determine whether model species like Drosophila melanogaster can be used to identify the best symbiont strains to transfer to mosquitoes.
The role of the host genome in determining the phenotype of Wolbachia infections has been investigated by experimentally moving Wolbachia between host species. Many of these studies have investigated reproductive manipulations such as cytoplasmic incompatibility and sex ratio distortion (Fujii, Kageyama, Hoshizaki, Ishikawa, & Sasaki, 2001; Jaenike, 2007; Poinsot, Bourtzis, Markakis, & Savakis, 1998; Sakamoto et al., 2005; Veneti et al., 2012). Here, host shifts have been shown to be associated with changes in the intensity of the phenotype (Poinsot et al., 1998), a complete loss of the phenotype (Veneti et al., 2012) or even a switch in the type of reproductive alteration (Jaenike, 2007).
The roles of host and symbiont genomes in determining whether Wolbachia blocks viral replication are especially important as considerable effort is being put into transferring symbiont strains to mosquitoes to prevent the transmission of viral pathogens (Hoffmann, Ross, & Rašić, 2015). The antiviral phenotype of Wolbachia was first observed in D. melanogaster (Hedges et al., 2008; Teixeira et al., 2008), and later in other Drosophila species (Cattel, Martinez, Jiggins, Mouton, & Gibert, 2016; Osborne, Leong, O'Neill, & Johnson, 2009; Unckless & Jaenike, 2011) and mosquitoes (Bian, Zhou, Lu, & Xi, 2013; Blagrove, Arias‐Goeta, Failloux, & Sinkins, 2012; Glaser & Meola, 2010; Moreira et al., 2009). The ability of Wolbachia to spread by manipulating host reproduction in combination with its antiviral properties makes it a promising tool for the control of mosquito‐borne viruses like dengue and Zika (Aliota, Peinado, Velez, & Osorio, 2016; Dutra et al., 2016; Moreira et al., 2009). Currently, large‐scale field trials are evaluating whether releasing Ae. aegypti mosquitoes infected with a Wolbachia strain from D. melanogaster prevents dengue transmission (Frentiu et al., 2014; Hoffmann et al., 2011). There is extensive genetic variation among symbiont strains in the level of antiviral protection (Bian et al., 2013; Blagrove et al., 2012; Chrostek, Marialva, Yamada, O'Neill, & Teixeira, 2014; Chrostek et al., 2013; Martinez et al., 2014; Osborne et al., 2009). However, little is known about the role of the host genotype in affecting this trait. Two strains of Wolbachia have been transferred from D. melanogaster to Ae. aegypti, and in both host species, an over‐replicating laboratory mutant called wMelPop had the strongest antiviral effects (Chrostek et al., 2013; van den Hurk et al., 2012; Hussain et al., 2013). In a different system, moving a Spiroplasma symbiont between Drosophila species determined whether it protected its host against parasitic nematodes (Haselkorn, Cockburn, Hamilton, Perlman, & Jaenike, 2013).
Here, we compared Wolbachia strains in their native host and a new host to test whether the host and/or symbiont genome determines the level of antiviral protection. We first assessed the frequency of antiviral protection in 16 natural host–symbiont associations. We then compared the level of protection induced by eight of these Wolbachia strains in both their original host and a line of D. simulans to which they have been artificially transferred. We find that the level of antiviral protection is largely determined by the Wolbachia strain rather than the host species.
2. METHODS
2.1. Drosophila stocks and Wolbachia strains
All Drosophila species were maintained on a cornmeal diet (see recipe in Longdon et al., 2015) at 25°C, under a 12‐hr light/dark cycle and 70% relative humidity. Ten Drosophila species infected with their native Wolbachia strains were used in this study (Tables 1 and S1). Of these, more than one line of D. melanogaster and D. simulans was used, each infected with a different Wolbachia strain. For each Wolbachia‐infected fly line, we had a matching Wolbachia‐free control. Wolbachia‐infected D. melanogaster and the uninfected control were created using balancer chromosomes to homogenize their nuclear background as described in Chrostek et al. (2013). For all the other fly lines, a Wolbachia‐free line was created from Wolbachia‐infected flies by raising them on Ready Mix Dried Food (Philip Harris) supplemented with 0.03% w/v tetracycline for two generations. In order to homogenize the gut microbiota between Wolbachia‐infected lines and their tetracycline‐treated counterparts, the tetracycline‐treated lines were then raised for one generation on standard cornmeal food on which ten males of the respective Wolbachia‐infected line had been kept for 1 day and removed (as in Martinez et al., 2016). Experiments were all performed more than twenty generations after tetracycline treatment. The Wolbachia infection status of all fly lines was checked by PCR and Sanger sequencing as described below.
Table 1.
Natural Drosophila–Wolbachia associations used in this study
| Drosophila group | Drosophila subgroup | Drosophila species | Wolbachia strain |
|---|---|---|---|
| melanogaster | ananassae | D. ananassae | wAnaa |
| melanogaster | D. melanogaster | wMelCSa | |
| D. melanogaster | wMelPop | ||
| D. melanogaster | wMela | ||
| D. sechellia | wSha | ||
| D. simulans | wHa | ||
| D. simulans | wMa | ||
| D. simulans | wNo | ||
| D. simulans | wAua | ||
| D. simulans | wRi | ||
| D. teissieri | wTeia | ||
| montium | D. triauraria | wTri | |
| suzukii | D. suzukii | wSuz | |
| saltans | saltans | D. prosaltans | wProa |
| sturtevanti | D. sturtevanti | wStv | |
| willistoni | willistoni | D. tropicalis | wTroa |
Wolbachia strains that were also used in the D. simulans line STCP (see Methods).
In order to compare the Wolbachia strains in their original host and in a new host, we also used the D. simulans line STCP into which some of the Wolbachia strains were previously transferred through backcrossing or microinjection (Martinez et al., 2014; Poinsot et al., 1998; Zabalou et al., 2008; Table S1). In order to minimize inbreeding depression, before each experiment STCP females were crossed to males of a different Wolbachia‐free isofemale line (14021–0251.175, Dsim\wild‐type, San Diego Drosophila Species Stock Center). All measurements were carried out on the emerging F1 adults from this cross, as in Martinez et al. (2015).
2.2. Virus production
To test antiviral protection, we used Flock House virus, which has a positive‐sense single‐stranded RNA genome. FHV belongs to the family Nodaviridae and was initially isolated from a beetle (Scotti, Dearing, & Mossop, 1983). We chose to use FHV instead of a native virus such as Drosophila C virus (Comendador et al., 1986; Plus, Croizier, Jousset, & David, 1975) as we have found that there is less genetic variation among hosts in susceptibility to FHV (Magwire et al., 2012). FHV was produced in Schneider Drosophila line 2 (DL2) cells. Cells were cultured at 26.5°C in Schneider's Drosophila medium with 10% foetal bovine serum, 100 U/ml penicillin and 100 mg/ml streptomycin (all Invitrogen, UK). Cells were then freeze‐thawed twice to lyse cells and centrifuged at 4,000 g for 10 min at 4°C to remove cellular debris. Virus was then aliquoted and frozen at −80°C. Virus infectivity was calculated using serial dilutions of virus in Schneider's medium added to wells of a plate of DL2 cells as described in Longdon, Cao, Martinez, and Jiggins (2013). After 7 days, the wells were visually examined under the microscope and classed as “infected” when cell death (presence of cell debris) and cytopathic effects were visible (lysing, shrinking or losing of compartmentation of cells). The Tissue Culture Infective Dose 50 (TCID50) was calculated by the Reed–Muench endpoint method (Reed & Muench, 1938).
2.3. Wolbachia screening
The Wolbachia infection status of fly lines was checked by PCR using the diagnostic primers wsp81F and wsp691R (Zhou, Rousset, & O'Neill, 1998). DNA from ten female flies per fly line was first extracted by crushing the flies in 150 μl of a 5% w/v suspension of Chelex 100 resin (Sigma‐Aldrich) and 1 μl of proteinase K (20 mg/ml, Fermentas). Extracts were incubated for 5 hr at 56°C. After 10 min at 95°C, samples were centrifuged and stored at −20°C. PCR conditions were as described in Ref. (Zhou et al., 1998). For the Wolbachia‐infected lines, the PCR products of the genes wsp and 16S (16Swol F: 5′‐TTGTAGCCTGCTATGGTATAACT‐3′; 16SWol R: 5′‐GAATAGGTATGATTTTCATGT‐3′, O'Neill, Giordano, Colbert, Karr, & Robertson, 1992) were Sanger‐sequenced to identify the Wolbachia infections at the strain level.
2.4. Survival assay
To infect flies with FHV, 3‐ to 6‐day‐old females were anaesthetized with CO2 and then stabbed into the left pleural suture of the thorax with a 0.15‐mm‐diameter anodized steel needle (Austerlitz Insect Pins) bent 0.25 mm from the end (half of the dorsal width of the thorax). The needle was either dipped into viral suspension or with a control solution produced from a virus‐free cell culture medium. The FHV stock was defrosted on the day of infection and then disposed of. Following infection, replicates of fifteen to twenty flies were placed in vials with standard cornmeal food. Survival was recorded every day. Flies were transferred into a fresh vial of food every 3 days.
Our first survival experiment was performed using all the Wolbachia strains in their original host species or background (i.e., not including the Wolbachia strains transferred into the D. simulans STCP line) and a virus dose of 3.6 × 1010 TCID50/ml. In this experiment, flies were placed at 22°C following virus infection in order to minimize the mortality that occurs in mock‐infected controls for some of the species (based on a pilot experiment). In a second experiment, eight of the Wolbachia strains were compared in parallel in their original host line and in the D. simulans line STCP (outcrossed as explained above). In this second experiment, flies were kept at 25°C following virus infection and the virus dose used was 3.6 × 108 TCID50/ml. In both experiments, infections were carried out over 5–9 days. On each day, one biological replicate (vial of flies) per treatment (virus/mock infection, Wolbachia‐infected/uninfected, host line) was infected. The order of treatments was randomized between days. In total, five vials of flies were prepared for each treatment.
2.5. Wolbachia density
To measure the Wolbachia density within fly tissues, DNA was extracted using the Gentra Puregene kit (Qiagen) from a pool of ten 2‐ to 5‐day‐old females reared at 25°C. Flies from each Wolbachia‐infected fly line were collected every day from the same cohorts used in the second survival experiment. Five biological replicates (independent pools of females) were extracted for each Wolbachia‐infected line and the DNA was then diluted 1:10 with nuclease‐free water. For each Drosophila species, we sequenced the fly gene RpL32 as in Longdon, Hadfield, Webster, Obbard, and Jiggins (2011) and designed species‐specific primers in two conserved regions for quantitative PCR (qPCR) (Table S2). The copy number of the Wolbachia gene atpD (atpDQALL_F: 5′‐CCTTATCTTAAAGGAGGAAA‐3′; atpDQALL_R: 5′‐AATCCTTTATGAGCTTTTGC‐3′) relative to the endogenous Drosophila control gene RpL32 (species‐specific primers; Table S2) was quantified with the SensiFAST SYBR and Fluorescein kit (Bioline). For each gene, all samples were run on the same qPCR plate and a second technical replicate was performed on a different plate. The efficiency with which each set of primers amplified the product was checked using a dilution series. In all cases, the efficiency was >95% (with 100% efficiency equating to a doubling of the PCR product concentration every cycle). The Wolbachia density was estimated as 2ΔCt, where Ct is the mean cycle threshold of the two technical replicates and ΔCt = Ct RpL32 − Ct atpD. The qPCR cycle was 95°C for 2 min, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s.
2.6. Viral titre
In order to estimate FHV titre, flies were raised and females were infected with virus under the same conditions as the survival experiments. As the first experiment was performed at 22°C and the second one at 25°C, flies were collected 5 and 3 days post‐infection, respectively, in order to allow sufficient time for the viral replication before any significant mortality occurs. Flies were snap‐frozen in liquid nitrogen in pools of ten females (five to ten biological replicates from separate pools of flies per fly line). Flies were then homogenized in TRIzol. Total RNA was extracted using TRIzol (Invitrogen) and reverse‐transcribed with Promega GoScript reverse transcriptase (Promega) and random hexamer primers, and then diluted 1:10 with nuclease‐free water. The FHV RNA copy number (forward: 5′‐ACCTCGATGGCAGGGTTT‐3′; reverse: 5′‐CTTGAACCATGGCCTTTTG‐3′) relative to the endogenous control gene RpL32 (species‐specific primers; Table S2) was quantified as for the Wolbachia density with two technical replicates per sample. As for the Wolbachia density, the species‐specific primers for RpL32 were designed in two conserved regions, except that the forward primer was designed on an exon–exon junction in order to amplify only mRNA. This exon–exon junction was previously confirmed in several Drosophila species (Longdon et al., 2011). For a given sample, the Ct values were averaged between the two technical replicates and the relative FHV titre was calculated as ΔCt = Ct RpL32 − Ct FHV.
2.7. Statistical analysis of survival data
Statistical analyses were performed in the r software (R Core Team 2013). Survival rates were analysed using Cox's proportional hazard mixed‐effect models (package coxme). This allowed estimating the antiviral protection conferred by each Wolbachia strain as a hazard ratio. The hazard ratio for a given Wolbachia‐infected line is the probability of death occurring at a given time point divided by the probability of death in the respective Wolbachia‐free line. Flies that were alive at the end of the experiment were treated as censored data.
To estimate the level of antiviral protection provided by each Wolbachia strain in the original hosts, we fitted the model:
| (1) |
where λ0 is a baseline hazard, H i is a fixed effect of host species i, W j is a fixed effect of Wolbachia infection status j (infected or Wolbachia‐free), and H i:W j is an interaction between host species and infection status. The vial in which each fly was found was treated as a random effect (v k) and εijkl was the residuals.
The survival data in D. simulans STCP line were analysed with the simpler model:
| (2) |
where S i is the Wolbachia strain. The effect of Wolbachia in each host–Wolbachia association was tested using multiple pairwise comparisons (glht function, package multcomp, Hothorn, Bretz, & Westfall, 2008).
2.8. Statistical analysis of viral titres and Wolbachia density
Wolbachia density and viral titre data were analysed using a series of linear models. The Wolbachia density data were log‐transformed to reach the assumptions of normality and homoscedasticity. The effect of Wolbachia on viral titres in each host–Wolbachia association was further tested using multiple pairwise comparisons to compare the Wolbachia‐infected flies to the appropriate Wolbachia‐free control (glht function, package multcomp, Hothorn et al., 2008).
Viral titres (T) in the original host species were analysed as:
| (3) |
where the parameters are defined in Model (1). Viral titres (T) in D. simulans STCP were analysed as:
| (4) |
where the parameters are defined in Model (2). Wolbachia density (D) in D. simulans STCP and the original hosts was analysed as:
| (5) |
2.9. The relative importance of host and symbiont genomes
To quantify the relative importance of the host and symbiont genomes, we used an ANOVA to analyse trait data from both the original hosts and D. simulans STCP. The response variable was either relative survival (see below), relative viral titre (see below) or Wolbachia density of Wolbachia‐infected flies (R). This allowed us to fit the linear model:
| (6) |
In this data set, there is no cross‐factoring of the different hosts and symbiont strains. Therefore, we cannot distinguish a main effect of the host (an effect of the host on all Wolbachia strains) from a host‐by‐Wolbachia interaction (an effect of the host on specific Wolbachia strains). For this reason, the effect of the host j (H j) was nested within the effect of Wolbachia strain i (S i). As these were both treated as fixed effects, this is equivalent to fitting one main effect (S i) and one interaction (S i: H j).
For the survival data, the response variable R ijk was the hazard of a vial of Wolbachia‐infected flies relative to the mean hazard of the Wolbachia‐free flies (R). This hazard ratio of each vial was estimated as a Best Linear Unbiased Predictor (BLUP) from a coxme model, with a separate model fitted to each host species. This model was identical to Model 2 except that the fixed effect was Wolbachia infection status (W) rather than strain (S).
For the viral titre data, the response variable R ijk was the viral titre from each qPCR sample of Wolbachia‐infected flies relative to their respective Wolbachia‐free counterparts. This was calculated by normalizing each sample i to the average titre in the Wolbachia‐free controls as ΔΔCt i = mean(ΔCt control) − ΔCt Wolbachia i. Here, mean(ΔCt control) is the mean of all the Wolbachia‐free vials, and ΔCt Wolbachia i is the titre of Wolbachia‐infected sample i.
3. RESULTS
3.1. Symbiont‐mediated protection against viruses is common across natural Drosophila–Wolbachia associations
To assess how common Wolbachia‐mediated antiviral protection is, we tested whether a panel of 16 Wolbachia strains protected their natural host species against viral infection (Table 1). The 16 symbiont strains were in ten Drosophila species, and we created matched Wolbachia‐free lines. Following infection with the highly pathogenic RNA virus FHV, high rates of mortality were observed in all fly hosts (Figure 1a). The Wolbachia strains conferred varying levels of protection (Figure 1a; Model 1, host main effect: χ2 = 654.26, df = 30, p < .0001; Wolbachia main effect: χ2 = 396.06, df = 16, p < .0001; host‐by‐Wolbachia interaction: χ2 = 242.35, df = 15, p < .0001) with 10 of 16 Wolbachia strains significantly increasing the survival of their respective fly host after virus infection. While two Wolbachia strains prevented any virus‐induced mortality, many of the other protective strains only modestly increased survival (Figure 1a). The protective Wolbachia strains are found in five Drosophila species: D. simulans, D. melanogaster, D. prosaltans, D. teissieri and D. tropicalis. As found in previous studies (Chrostek et al., 2013; Martinez et al., 2014; Osborne et al., 2009), within D. simulans and D. melanogaster, different Wolbachia strains were associated with varying levels of protection against viruses.
Figure 1.
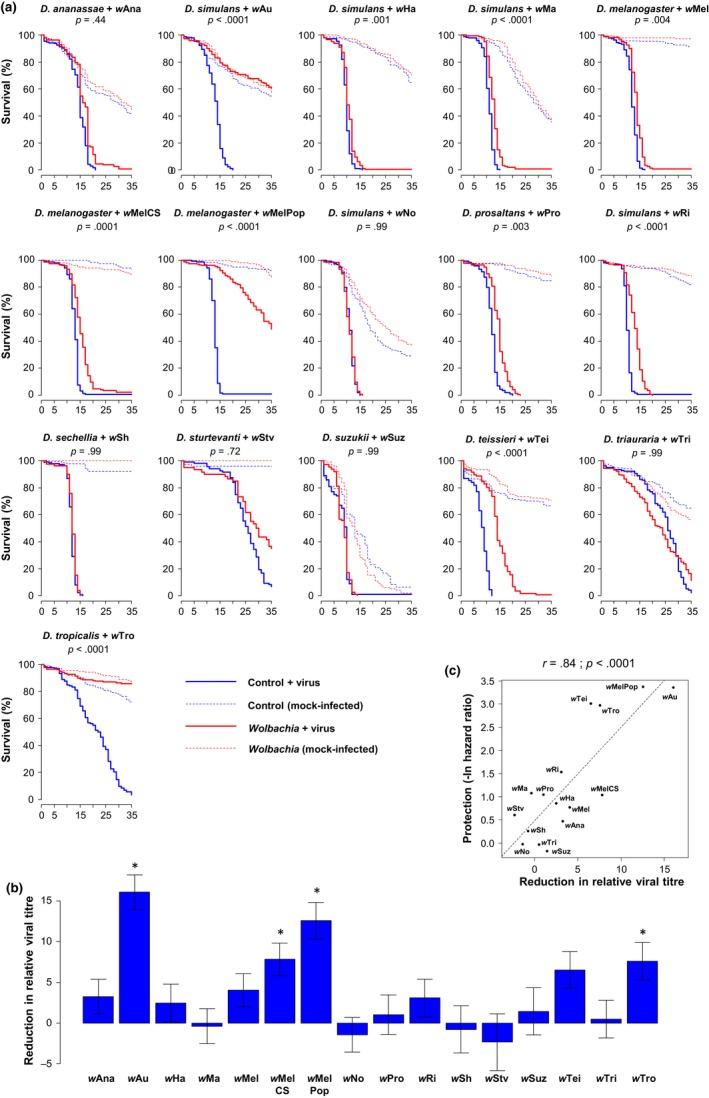
Antiviral protection in natural Drosophila–Wolbachia associations. (a) Survival curves following infection with FHV. p‐values indicate the significance of the difference between Wolbachia‐infected flies and their respective Wolbachia‐free counterparts (Model 1, see Methods). When this analysis was repeated on the mock‐infected flies, none of the Wolbachia strains significantly affected survival (Model 1; p > .05 in all cases). (b) Wolbachia‐induced reduction in viral titre calculated as the difference between Wolbachia‐free and Wolbachia‐infected flies. Positive values correspond to lower viral titres in Wolbachia‐infected flies on a log2 scale (ΔΔCt). Stars indicate significant differences between Wolbachia‐infected flies and their respective uninfected controls based on a multiple comparison test (Model 3, p < .05). Means, standard errors and p‐values were estimated from the Model 3 using the glht function to perform multiple comparisons. (c) Correlation between the increase in the survival of FHV‐infected flies caused by Wolbachia and the reduction in viral titre. The dashed line shows predicted values from a linear regression. r is Pearson's correlation coefficient between traits [Colour figure can be viewed at http://wileyonlinelibrary.com]
The Wolbachia strains also varied in their effects on viral titre, measured as relative viral RNA copy number (Model 3, host main effect: F 15,95 = 12.18, p < .0001; Wolbachia main effect: F 1,95 = 43.07, p < .0001; host‐by‐Wolbachia interaction: F 15,95 = 2.51, p = .004). Four of the symbiont strains significantly reduced titres (Figure 1b). Furthermore, the reduction in viral titre caused by Wolbachia was positively correlated with increases in survival after infection (Figure 1c). Overall, the change in titre explained 70% (r 2) of the variance in protection (Figure 1c).
3.2. Most variation in antiviral protection is explained by the symbiont strain rather than the host species
We next investigated the relative importance of the host genetic background and symbiont strain in determining whether Wolbachia protects its host against FHV. To this end, in a single experiment, we compared eight Wolbachia strains in both their original host species and a common genotype of D. simulans (STCP line, see Methods). As in the previous experiment, we followed fly survival upon infection with FHV and observed varying levels of protection among the original hosts as well as in the common genotype of D. simulans (Figure 2a). Both the Wolbachia strain and host background significantly affected the level of antiviral protection (Table 2a). However, while the Wolbachia strain explained more than 90% of the variance in protection, less than 5% was explained by symbionts behaving differently in the different hosts (Model 6, Table 2a). This suggests that antiviral protection mostly depends on the symbiont strain. In the case of the strain wAu, the original and new hosts are different genotypes of D. simulans. Because this could affect the correlation between the original and the new host, we ran the same analysis without wAu and found similar results (Wolbachia strain explains 91% of variation in protection). Accordingly, levels of protection were strongly correlated between the original hosts and the common genotype of D. simulans (Figure 2b), even when discarding wAu from the analysis (without wAu: r = .77; p = .04).
Figure 2.
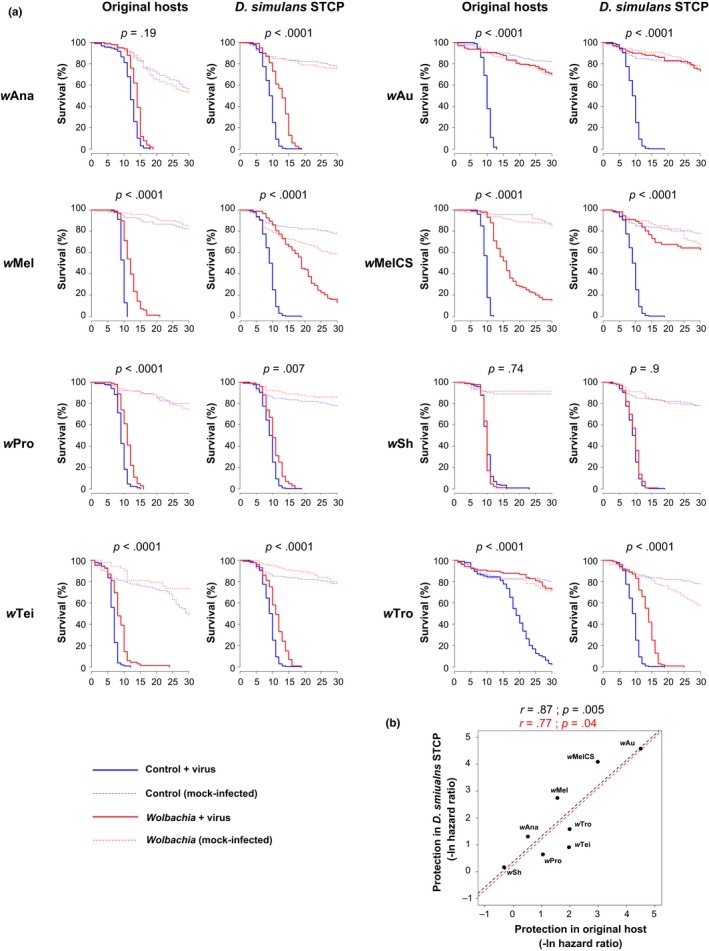
Wolbachia‐mediated protection in original hosts and a common genotype of D. simulans. (a) Survival curves following infection with FHV. p‐values for the comparisons of Wolbachia‐infected and Wolbachia‐free flies after infection with FHV are shown (Model 1 and 2, see Methods). When this analysis was repeated on the mock‐infected flies, none of the Wolbachia strains significantly affected survival (models 1 and 2; p > .05 in all cases). (b) Correlation in Wolbachia‐mediated increases in survival after FHV infection in the original hosts and the common genotype of D. simulans. The dashed lines show predicted values from linear regressions with (black) and without (red) wAu. r is Pearson's correlation coefficient between traits [Colour figure can be viewed at http://wileyonlinelibrary.com]
Table 2.
Statistical analysis of Wolbachia‐mediated protection in original hosts and the common genotype of D. simulans
| Traita | Fixed effectsb | df | Sum Sq | F‐values | p‐values | % variance explained |
|---|---|---|---|---|---|---|
| (A) Survival | Wolbachia strain | 7 | 136.3 | 484.2 | <.0001 | 93.6 |
| Host within Wolbachia strain | 8 | 6.9 | 21.5 | <.0001 | 4.7 | |
| Residuals | 61 | 2.5 | 1.7 | |||
| (B) Viral titre | Wolbachia strain | 7 | 920.1 | 49.6 | <.0001 | 70.9 |
| Host within Wolbachia strain | 8 | 172.9 | 8.2 | <.0001 | 13.3 | |
| Residuals | 77 | 204.2 | 15.7 | |||
| (C) Wolbachia density | Wolbachia strain | 7 | 68.7 | 60.0 | <.0001 | 81.2 |
| Host within Wolbachia strain | 8 | 7.6 | 5.8 | <.0001 | 8.9 | |
| Residuals | 51 | 8.3 | 9.9 |
We next examined the roles of host and symbiont genomes in determining the effect of Wolbachia on viral titres, and found similar patterns to our analysis of survival rates. Wolbachia had varying effects on viral titres, with 71% of the variation in our viral titre measurements being explained by the Wolbachia strain compared to the 13% explained by strains having different effects in different hosts (Model 6; Table 2b; Figure 3a, b). This was reflected in a strong correlation between the effect of Wolbachia on viral titre in the original hosts and the common genotype of D. simulans (Figure 3b), even if wAu is excluded (correlation without wAu: r = .89; p = .02).
Figure 3.
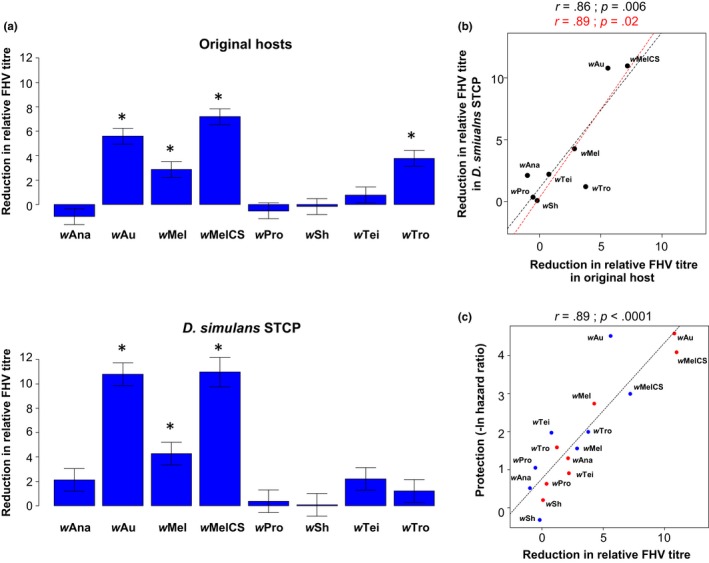
Wolbachia‐mediated reductions in viral titre in their original hosts and the common genotype of D. simulans. (a) Differences in the reduction in viral titre between Wolbachia‐free and Wolbachia‐infected flies. Positive values correspond to lower viral titres in Wolbachia‐infected flies on a log2 scale (ΔΔCt). Stars indicate significant differences between Wolbachia‐infected flies and their respective Wolbachia‐free controls based on a multiple comparison test (p < .05). Means, standard errors and p‐values were estimated using models 3 and 4 (see Methods). (b) Correlation between the effects of Wolbachia on FHV titre in original hosts and the common genotype of D. simulans. (c) Correlation between the effects of Wolbachia on FHV titre and survival after FHV infection. The blue and red points are the mean trait values per Wolbachia strain in the original host and the common genotype of D. simulans, respectively. In panels b and c, the dashed lines show predicted values from linear regressions with (black) and without (red) wAu and r is Pearson's correlation coefficient between traits [Colour figure can be viewed at http://wileyonlinelibrary.com]
The extent to which Wolbachia reduced viral titre was strongly correlated to increases in survival after FHV infection (Figure 3c). If the data from the common genotype of D. simulans and the original hosts are combined, changes in titre explain 80% of the variance in survival (Figure 3c; r 2 = .80). Furthermore, the strength of this correlation is similar if the data from the common genotype of D. simulans and the original hosts are analysed separately (original hosts: r = .83 and p = .01; common genotype of D. simulans: r = .96 and p = .0001).
3.3. Symbiont density is conserved when strains are transferred between host species and explains most of the variation in antiviral protection
Within a single host species, Wolbachia‐mediated protection is known to be tightly linked to the density of the symbiont in host tissues (Chrostek et al., 2013; Martinez et al., 2014; Osborne, Iturbe‐Ormaetxe, Brownlie, O'Neill, & Johnson, 2012; Osborne et al., 2009). To explain why the host genetic background has little effect on the level of antiviral protection that a given Wolbachia strain provides, we tested whether symbiont densities were conserved when a Wolbachia strain was moved between different hosts. We found significant variation in density between Wolbachia strains in both the original hosts and the common genotype of D. simulans (Figure 4a). As for protection, the Wolbachia strain explained far more of the variance in symbiont density than the host genetic background (Model 6; Table 2c), and there was a positive correlation between the density in the original hosts and the common genotype of D. simulans (Figure 4b; correlation excluding wAu: r = .86; p = .01). Wolbachia density was also correlated to the extent to which Wolbachia increased survival after viral infection (Figure 4c) as well as to the reduction in viral titre (Figure 4d). The strength of these correlations with symbiont density was similar in the original hosts and in the common genotype of D. simulans for both survival (original hosts: r = .81 and p = .01; D. simulans: r = .85 and p = .007) and reduction in viral titre (original hosts: r = .70 and p = .05; D. simulans: r = .86 and p = .006).
Figure 4.
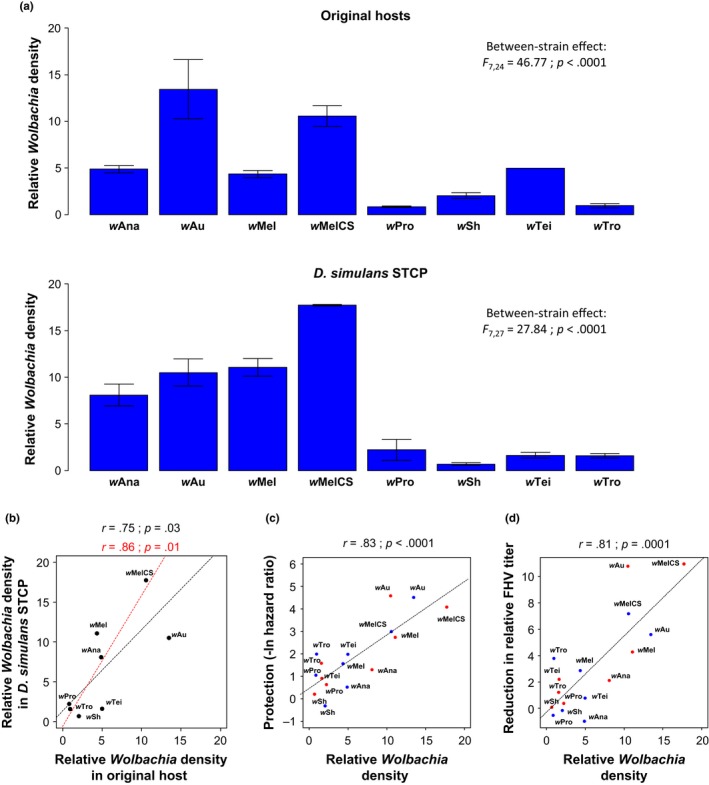
Wolbachia density and antiviral protection. (a) Mean Wolbachia density on a linear scale expressed as the copy number of the Wolbachia gene atpD relative to the fly gene RpL32. Between‐Wolbachia strain differences were tested using Model 5 on ln‐transformed data (see Methods). Error bars are standard errors. (b) Correlation in Wolbachia density between original hosts and the common genotype of D. simulans. (c) Correlation between the Wolbachia‐mediated increase in survival after FHV infection and Wolbachia density. (d) Correlation between the Wolbachia‐mediated reduction in FHV titre and Wolbachia density. The blue and red points are the mean trait values per Wolbachia strain in the original host and the common genotype of D. simulans, respectively. The dashed lines show predicted values from linear regressions with (black) and without (red) wAu and r is Pearson's correlation coefficient between traits [Colour figure can be viewed at http://wileyonlinelibrary.com]
4. DISCUSSION
The extent to which Wolbachia protects insects against viruses varies greatly among host–symbiont associations. While symbiont strain is known to be a key determinant of protection (Chrostek et al., 2013; Martinez et al., 2014; Osborne et al., 2009), the role of the host genome has been poorly investigated. By comparing several Wolbachia strains in different host species, we found that the symbiont genome was far more important than the host genome in determining the level of protection. This was due to the density that a given Wolbachia strain reaches being conserved when it is moved to a new host.
In natural host–symbiont associations, we found that Wolbachia commonly protects Drosophila against viral infection. Wolbachia significantly decreased virus‐induced mortality in more than half (10/16) of the Drosophila–Wolbachia associations tested, although in most cases the increase in survival was only modest. This is similar to the patterns we have reported from a panel of Wolbachia strains that we transferred into D. simulans, where about half of the strains provided protection (Martinez et al., 2014). Other studies of single species found that Wolbachia protects against FHV in Drosophila innubila (Unckless & Jaenike, 2011), D. suzukii (Cattel et al., 2016) and D. melanogaster (Hedges et al., 2008; Teixeira et al., 2008), but not D. bifasciata (Longdon, Fabian, Hurst, & Jiggins, 2012). Unlike our results, interpreting these different studies can be difficult due to publication biases towards positive results and differences in experimental conditions and statistical power. For example, antiviral protection and symbiont density are affected by temperature and diet (Caragata et al., 2013; Mouton, Henri, Charif, Boulétreau, & Vavre, 2007; Serbus et al., 2015; Ulrich, Beier, Devine, & Hugo, 2016). This may explain why the strain wHa was previously found to be nonprotective (Osborne et al., 2009) but conferred low levels of protection in our study using the same fly stock. Similarly, Cattel et al. (2016) found weak protection against FHV in D. suzukii but we did not.
By comparing the same symbionts in different hosts, we found that the symbiont strain was far more important than the host species in determining whether Wolbachia protects Drosophila against FHV. This was true in terms of both survival and viral titre. The large differences between Wolbachia strains have been reported before (Chrostek et al., 2013; Martinez et al., 2014; Osborne et al., 2009), but the small effect of the host was unexpected given that the host genetic background is critical to the expression of other Wolbachia phenotypes (Jaenike, 2007; Poinsot et al., 1998; Veneti et al., 2012).
As found in previous studies (Chrostek et al., 2013; Martinez et al., 2014; Osborne et al., 2009), the symbiont strains varied greatly in their densities and this correlated with antiviral protection. Critically, when the symbionts were transferred between host species, the strain‐specific densities were mostly conserved. Therefore, symbionts appear to regulate their density independently of the host, and this in turn determines the level of antiviral protection. Previous work found that the host genotype affects Wolbachia density (Kondo, Shimada, & Fukatsu, 2005; Mouton et al., 2007; Veneti et al., 2012). Our study does not contradict these results as we also found host effects on symbiont density, but these were small compared with the Wolbachia strain effect. Our results differ from studies of another Drosophila symbiont. As is the case for Wolbachia, the density of Spiroplasma symbionts that protect some Drosophila species against parasitic nematodes is similar between the native and the novel hosts (Haselkorn et al., 2013). However, the protective effect of different symbiont strains is decoupled from their density and strongly depends on the host species (Haselkorn et al., 2013). Therefore, the host genetic background is a strong determinant of the protective phenotype of Spiroplasma but not Wolbachia.
During its evolution, Wolbachia has frequently jumped between host species (Vavre et al., 1999; Werren et al., 1995; Zhang et al., 2013). Our results suggest the protective phenotype will often be transferred to the newly infected host. This could drive up the frequency of Wolbachia in the new host, potentially making protective strains more likely to move between species. This may be especially important for CI‐inducing Wolbachia strains, as these need to reach a minimum frequency in the population to be able to spread (Turelli, 1994). The benefit conferred by antiviral protection to the new host may promote the spread of the newly acquired Wolbachia infection allowing it to reach this threshold. The importance of this effect will depend on RNA viruses being a strong selective pressure, as highly protective Wolbachia strains are costly for the insect owing to their high density within host's tissues (Chrostek et al., 2013; Martinez et al., 2015). RNA viruses are extremely prevalent in Drosophila populations (Webster et al., 2015), but their effects on fitness in nature are unknown.
The observation that the host genome has comparatively little effect on antiviral protection or Wolbachia density is interesting. It seems likely that there is selection on hosts to control Wolbachia density to some optimal level, as RNA viruses are common Drosophila pathogens (Webster et al., 2015) and high Wolbachia densities substantially reduce host fitness (Martinez et al., 2015). Our finding that hosts have not evolved to modulate symbiont densities suggests there may be constraints that prevent flies from altering Wolbachia density. For example, Wolbachia may occupy an intracellular niche that protects it from insect immune defences (Bourtzis, Pettigrew, & O'Neill, 2000; Siozios, Sapountzis, Ioannidis, & Bourtzis, 2008). This could mean that hosts might be more likely to evolve tolerance to Wolbachia infections rather than mechanisms controlling the symbiont density.
Being able to predict the antiviral effects of a Wolbachia strain in a new host is useful for public health programmes that are releasing Wolbachia‐infected mosquitoes to prevent disease transmission. The Zika and dengue vector Ae. aegypti does not harbour Wolbachia in nature and therefore needs to be artificially infected with Wolbachia strains found in other host species (Hoffmann et al., 2015). These transfers are laborious and time‐consuming. Finding the optimal Wolbachia strains for disease control would be greatly facilitated by screening symbiont strains in Drosophila where the artificial transfer of Wolbachia between species has become routine (Chrostek et al., 2014; Martinez et al., 2014; Poinsot et al., 1998; Veneti et al., 2012). Our results suggest that such studies are likely to be a powerful way to select symbiont strains to be used as biocontrol agents.
We conclude that the extent to which Wolbachia protects different Drosophila species against viral infection depends primarily on the symbiont strain and not the host genome. This is due to Wolbachia regulating its density to similar levels in different host species. Wolbachia density in turn determines whether Wolbachia protects the host against viruses.
AUTHOR CONTRIBUTIONS
J.M. and F.M.J. conceived the experiments. I.T., S.O., S.S., K.S., J.P.D. and J.M. conducted the experiments. J.M. and F.M.J. analysed the data and wrote the manuscript.
DATA ACCESSIBILITY
The raw survival and qPCR data (FHV titre and Wolbachia density) are available at Dryad: https://doi.org/10.5061/dryad.869j5.
Supporting information
ACKNOWLEDGEMENTS
We thank Kostas Bourtzis, Luis Teixeira, Wolfgang Miller and the East Malling Research Station for providing some of the fly lines used in our study. This study was funded by the Wellcome Trust grant WT094664MA (http://www.wellcome.ac.uk/) and the European Research Council (ERC) grant 281668 DrosophilaInfection.
Martinez J, Tolosana I, Ok S, et al. Symbiont strain is the main determinant of variation in Wolbachia‐mediated protection against viruses across Drosophila species. Mol Ecol. 2017;26:4072–4084. https://doi.org/10.1111/mec.14164
REFERENCES
- Aliota, M. T. , Peinado, S. A. , Velez, I. D. , & Osorio, J. E. (2016). The wMel strain of wolbachia reduces transmission of Zika virus by Aedes aegypti . Scientific Reports, 6, 28792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, G. , Zhou, G. , Lu, P. , & Xi, Z. (2013). Replacing a native Wolbachia with a novel strain results in an increase in endosymbiont load and resistance to dengue virus in a mosquito vector. PLoS Neglected Tropical Diseases, 7, e2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagrove, M. S. C. , Arias‐Goeta, C. , Failloux, A.‐B. , & Sinkins, S. P. (2012). Wolbachia strain wMel induces cytoplasmic incompatibility and blocks dengue transmission in Aedes albopictus. Proceedings of the National Academy of Sciences of the United States of America, 109, 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis, K. , Pettigrew, M. M. , & O'Neill, S. L. (2000). Wolbachia neither induces nor suppresses transcripts encoding antimicrobial peptides. Insect Molecular Biology, 9, 635–639. [DOI] [PubMed] [Google Scholar]
- Caragata, E. P. , Rancès, E. , Hedges, L. M. , Gofton, A. W. , Johnson, K. N. , O'Neill, S. L. , & McGraw, E. A. (2013). Dietary cholesterol modulates pathogen blocking by Wolbachia. PLoS Pathogens, 9, e1003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattel, J. , Martinez, J. , Jiggins, F. , Mouton, L. , & Gibert, P. (2016). Wolbachia‐mediated protection against viruses in the invasive pest Drosophila suzukii . Insect Molecular Biology, 25, 595–603. [DOI] [PubMed] [Google Scholar]
- Chrostek, E. , Marialva, M. S. P. , Esteves, S. S. , Weinert, L. A. , Martinez, J. , Jiggins, F. M. , & Teixeira, L. (2013). Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: A phenotypic and phylogenomic analysis. PLoS Genetics, 9, e1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrostek, E. , Marialva, M. S. P. , Yamada, R. , O'Neill, S. L. , & Teixeira, L. (2014). High anti‐viral protection without immune upregulation after interspecies wolbachia transfer. PLoS ONE, 9, e99025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comendador, M. , Plus, N. , Louis, C. , Lopez‐Ferber, M. , Kuhl, A. , & Kuhl, G. (1986). Endemic microorganisms of a Drosophila simulans strain and their relationships with the non‐mendelian transmission of a character. Génétique, Sélection, Évolution, 18, 131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutra, H. L. C. , Rocha, M. N. , Dias, F. B. S. , Mansur, S. B. , Caragata, E. P. , & Moreira, L. A. (2016). Wolbachia blocks currently circulating Zika virus isolates in Brazilian Aedes aegypti mosquitoes. Cell Host & Microbe, 19, 771–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu, F. D. , Zakir, T. , Walker, T. , Popovici, J. , Pyke, A. T. , van der Hurk, A. , … O'Neill, S. L. (2014). Limited dengue virus replication in field‐collected Aedes aegypti mosquitoes infected with Wolbachia. PLoS Neglected Tropical Diseases, 8, e2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, Y. , Kageyama, D. , Hoshizaki, S. , Ishikawa, H. , & Sasaki, T. (2001). Transfection of Wolbachia in Lepidoptera: The feminizer of the adzuki bean borer Ostrinia scapulalis causes male killing in the Mediterranean flour moth Ephestia kuehniella . Proceedings of the Royal Society of London, Series B: Biological Sciences, 268, 855–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser, R. L. , & Meola, M. A. (2010). The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS ONE, 5, e11977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselkorn, T. S. , Cockburn, S. N. , Hamilton, P. T. , Perlman, S. J. , & Jaenike, J. (2013). Infectious adaptation: Potential host range of a defensive endosymbiont in Drosophila. Evolution, 67, 934–945. [DOI] [PubMed] [Google Scholar]
- Hedges, L. , Brownlie, J. , O'Neill, S. , & Johnson, K. (2008). Wolbachia and virus protection in insects. Science, 322, 702. [DOI] [PubMed] [Google Scholar]
- Hoffmann, A. A. , Montgomery, B. L. , Popovici, J. , Iturbe‐Ormaetxe, I. , Johnson, P. H. , Muzzi, F. , … O'Neill, S. L. (2011). Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature, 476, 454–457. [DOI] [PubMed] [Google Scholar]
- Hoffmann, A. A. , Ross, P. A. , & Rašić, G. (2015). Wolbachia strains for disease control: Ecological and evolutionary considerations. Evolutionary Applications, 8, 751–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, T. , Koga, R. , Kikuchi, Y. , Meng, X.‐Y. , & Fukatsu, T. (2010). Wolbachia as a bacteriocyte‐associated nutritional mutualist. Proceedings of the National Academy of Sciences, 107, 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn, T. , Bretz, F. , & Westfall, P. (2008). Simultaneous inference in general parametric models. Biometrical Journal, 50, 346–363. [DOI] [PubMed] [Google Scholar]
- van den Hurk, A. F. , Hall‐Mendelin, S. , Pyke, A. T. , Frentiu, F. D. , McElroy, K. , Day, A. , … O'Neill, S. L. (2012). Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti . PLoS Neglected Tropical Diseases, 6, e1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, M. , Lu, G. , Torres, S. , Edmonds, J. H. , Kay, B. H. , Khromykh, A. A. , & Asgari, S. (2013). Effect of Wolbachia on replication of West Nile virus in a mosquito cell line and adult mosquitoes. Journal of Virology, 87, 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike, J. (2007). Spontaneous emergence of a new Wolbachia phenotype. Evolution, 61, 2244–2252. [DOI] [PubMed] [Google Scholar]
- Joubert, D. A. , Walker, T. , Carrington, L. B. , De Bruyne, J. T. , Kien, D. H. T. , Hoang, N. L. T. , … O'Neill, S. L. (2016). Establishment of a Wolbachia superinfection in Aedes aegypti mosquitoes as a potential approach for future resistance management. PLOS Pathogens, 12, e1005434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, N. , Shimada, M. , & Fukatsu, T. (2005). Infection density of Wolbachia endosymbiont affected by co‐infection and host genotype. Biology Letters, 1, 488–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon, B. , Cao, C. , Martinez, J. , & Jiggins, F. M. (2013). Previous exposure to an RNA virus does not protect against subsequent infection in Drosophila melanogaster . PLoS ONE, 8, e73833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon, B. , Fabian, D. K. , Hurst, G. D. , & Jiggins, F. M. (2012). Male‐killing Wolbachia do not protect Drosophila bifasciata against viral infection. BMC Microbiology, 12(Suppl. 1), S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon, B. , Hadfield, J. D. , Day, J. P. , Smith, S. C. L. , McGonigle, J. E. , Cogni, R. , … Jiggins, F. M. (2015). The causes and consequences of changes in virulence following pathogen host shifts. PLOS Pathogens, 11, e1004728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon, B. , Hadfield, J. D. , Webster, C. L. , Obbard, D. J. , & Jiggins, F. M. (2011). Host phylogeny determines viral persistence and replication in novel hosts. PLoS Pathogens, 7, e1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwire, M. M. , Fabian, D. K. , Schweyen, H. , Cao, C. , Longdon, B. , Bayer, F. , & Jiggins, F. M. (2012). Genome‐wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster . PLoS Genetics, 8, e1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J. , Cogni, R. , Cao, C. , Smith, S. , Illingworth, C. , & Jiggins, F. M. (2016). Addicted? Reduced host resistance in populations with defensive symbionts. Proceedings of the Royal Society B‐Biological Sciences, 283, 20160778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J. , Longdon, B. , Bauer, S. , Chan, Y. S. , Miller, W. J. , Bourtzis, K. , … Jiggins, F. M. (2014). Symbionts commonly provide broad spectrum resistance to viruses in insects: A comparative analysis of Wolbachia strains. PLoS Pathogens, 10, e1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J. , Ok, S. , Smith, S. , Snoeck, K. , Day, J. P. , Jiggins, F. M. (2015). Should symbionts be nice or selfish? Antiviral effects of Wolbachia are costly but reproductive parasitism is not. PLOS Pathogens, 11, e1005021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira, L. A. , Iturbe‐Ormaetxe, I. , Jeffery, J. A. , Lu, G. , Pyke, A. T. , Hedges, L. M. , … O'Neill, S. L. (2009). A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell, 139, 1268–1278. [DOI] [PubMed] [Google Scholar]
- Mouton, L. , Henri, H. , Charif, D. , Boulétreau, M. , & Vavre, F. (2007). Interaction between host genotype and environmental conditions affects bacterial density in Wolbachia symbiosis. Biology Letters, 3, 210–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, S. L. , Giordano, R. , Colbert, A. M. , Karr, T. L. , & Robertson, H. M. (1992). 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proceedings of the National Academy of Sciences of the United States of America, 89, 2699–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, S. E. , Iturbe‐Ormaetxe, I. , Brownlie, J. C. , O'Neill, S. L. , & Johnson, K. N. (2012). Antiviral protection and the importance of Wolbachia density and tissue tropism in Drosophila simulans . Applied and Environmental Microbiology, 78, 6922–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, S. E. , Leong, Y. S. , O'Neill, S. L. , & Johnson, K. N. (2009). Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans . Plos Pathogens, 5, e1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plus, N. , Croizier, G. , Jousset, F. , & David, J. (1975). Picornaviruses of laboratory and wild Drosophila melanogaster: Geographical distribution and serotypic composition. Annales de Microbiologie, 126, 107–117. [PubMed] [Google Scholar]
- Poinsot, D. , Bourtzis, K. , Markakis, G. , & Savakis, C. (1998). Wolbachia transfer from Drosophila melanogaster into D. simulans: Host effect and cytoplasmic incompatibility relationships. Genetics, 150, 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2013). R: A language and environment for statistical computing (P. F. Pimenta, Ed.).
- Reed, L. , & Muench, H. (1938). A simple method of estimating fifty per cent endpoints. The American Journal of Hygiene, 27, 493–497. [Google Scholar]
- Sakamoto, H. , Ishikawa, H. , Sasaki, T. , Kikuyama, S. , Tatsuki, S. , & Hoshizaki, S. (2005). Transinfection reveals the crucial importance of Wolbachia genotypes in determining the type of reproductive alteration in the host. Genetical Research, 85, 205–210. [DOI] [PubMed] [Google Scholar]
- Scotti, P. , Dearing, S. , & Mossop, D. (1983). Flock house virus: A Nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeida). Archives of Virology, 75, 181–189. [DOI] [PubMed] [Google Scholar]
- Serbus, L. R. , White, P. M. , Silva, J. P. , Rabe, A. , Teixeira, L. , Albertson, R. , & Sullivan, W. (2015). The impact of host diet on Wolbachia titer in Drosophila. PLoS Pathogens, 11, e1004777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siozios, S. , Sapountzis, P. , Ioannidis, P. , & Bourtzis, K. (2008). Wolbachia symbiosis and insect immune response invited review Wolbachia symbiosis and insect immune response. Insect Science, 15, 89–100. [Google Scholar]
- Teixeira, L. , Ferreira, A. , & Ashburner, M. (2008). The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster . Plos Biology, 6, 2753–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M. (1994). Evolution of incompatibility‐inducing microbes and their hosts. Evolution, 48, 1500–1513. [DOI] [PubMed] [Google Scholar]
- Ulrich, J. N. , Beier, J. C. , Devine, G. J. , & Hugo, L. E. (2016). Heat Sensitivity of wMel Wolbachia during Aedes aegypti development. PLOS Neglected Tropical Diseases, 10, e0004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless, R. L. , & Jaenike, J. (2011). Maintenance of a male‐killing Wolbachia in Drosophila innubila by male‐killing dependent and male‐killing independent mechanisms. Evolution, 66, 678–689. [DOI] [PubMed] [Google Scholar]
- Vavre, F. , Fleury, F. , Lepetit, D. , Fouillet, P. , & Boulétreau, M. (1999). Phylogenetic evidence for horizontal transmission of Wolbachia in host‐parasitoid associations. Molecular Biology and Evolution, 16, 1711–1723. [DOI] [PubMed] [Google Scholar]
- Veneti, Z. , Zabalou, S. , Papafotiou, G. , Paraskevopoulos, C. , Pattas, S. , Livadaras, I. , … Bourtzis, K. (2012). Loss of reproductive parasitism following transfer of male‐killing Wolbachia to Drosophila melanogaster and Drosophila simulans . Heredity, 109, 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, T. , Johnson, P. H. , Moreira, L. A. , Iturbe‐Ormaetxe, I. , Frentiu, F. D. , McMeniman, C. J. , … Hoffmann, A. A. (2011). The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature, 476, 450–453. [DOI] [PubMed] [Google Scholar]
- Webster, C. L. , Waldron, F. M. , Robertson, S. , Crowson, D. , Ferrari, G. , Quintana, J. F. , … Obbard, D. J. (2015). The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster . PLoS Biology, 13, e1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren, J. H. , Baldo, L. , & Clark, M. E. (2008). Wolbachia: Master manipulators of invertebrate biology. Nature, 6, 741–751. [DOI] [PubMed] [Google Scholar]
- Werren, J. , Zhang, W. , & Guo, L. (1995). Evolution and phylogeny of Wolbachia: Reproductive parasites of arthropods. Proceedings of the Royal Society B‐Biological Sciences, 261, 55–71. [DOI] [PubMed] [Google Scholar]
- Yeap, H. L. , Mee, P. , Walker, T. , Weeks, A. R. , O'Neill, S. L. , Johnson, P. , … Hoffmann, A. A. (2011). Dynamics of the “popcorn” Wolbachia infection in outbred Aedes aegypti informs prospects for mosquito vector control. Genetics, 187, 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabalou, S. , Apostolaki, A. , Pattas, S. , Veneti, Z. , Paraskevopoulos, C. , Livadaras, I. , … Bourtzis, K. (2008). Multiple rescue factors within a Wolbachia strain. Genetics, 178, 2145–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K.‐J. , Han, X. , & Hong, X.‐Y. (2013). Various infection status and molecular evidence for horizontal transmission and recombination of Wolbachia and Cardinium among rice planthoppers and related species. Insect Science, 20, 329–344. [DOI] [PubMed] [Google Scholar]
- Zhou, W. , Rousset, F. , & O'Neill, S. (1998). Phylogeny and PCR‐based classification of Wolbachia strains using wsp gene sequences. Proceedings of the Royal Society B‐Biological Sciences, 265, 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw survival and qPCR data (FHV titre and Wolbachia density) are available at Dryad: https://doi.org/10.5061/dryad.869j5.


