Abstract
Brain arteriovenous malformations (AVMs) are abnormal connections between arteries and veins that can result in hemorrhagic stroke. A genetic basis for AVMs is suspected, and we investigated potential mutations in a 14-year-old girl who developed a recurrent brain AVM. Whole-exome sequencing (WES) of AVM lesion tissue and blood was performed accompanied by in silico modeling, protein expression observation in lesion tissue and zebrafish modeling. A stop-gain mutation (c.C739T:p.R247X) in the gene SMAD family member 9 (SMAD9) was discovered. In the human brain tissue, immunofluorescent staining demonstrated a vascular predominance of SMAD9 at the protein level. Vascular SMAD9 was markedly reduced in AVM peri-nidal blood vessels, which was accompanied by a decrease in phosphorylated SMAD4, a downstream effector protein of the bone morphogenic protein signaling pathway. Zebrafish modeling (Tg kdrl:eGFP) of the morpholino splice site and translation-blocking knockdown of SMAD9 resulted in abnormal cerebral artery-to-vein connections with morphologic similarities to human AVMs. Orthogonal trajectories of evidence established a relationship between the candidate mutation discovered in SMAD9 via WES and the clinical phenotype. Replication in similar rare cases of recurrent AVM, or even more broadly sporadic AVM, may be informative in building a more comprehensive understanding of AVM pathogenesis.
Introduction
Brain arteriovenous malformations (AVMs) are a significant cause of hemorrhagic stroke in children and adults.1 They are defined as an abnormal tangle of blood vessels, where arteries feed blood into a nest of disorganized and dysmorphic vascular tissue known as a nidus, which ultimately drains into abnormal veins. While their natural history is debated, AVMs are known to rupture unpredictably, and these hemorrhages can result in severe permanent neurological disability and death.2 They can occur as part of hereditary syndromes, such as capillary malformation-AVM syndrome (OMIM 608354) and hereditary hemorrhagic telangiectasia (OMIM 187300, 600376 and 175050), where they result from mutations in genes that have known or plausible roles in angiogenesis and vascular remodeling, such as RASA1, ENG, SMAD4 and ACVRL1, among others.3–5 Similarly, familial inheritance of isolated brain AVMs has been ascribed to mutations in ACVRL1.6 Less is known about the cause of sporadically occurring AVMs, which account for the vast majority (>95%) of disease burden in the general population. Genotyping from cohorts of patients with sporadic brain AVMs has failed to reliably identify any common single-nucleotide polymorphisms or chromosomal structural variation conferring susceptibility,7–9 and the genesis of these potentially devastating malformations remains enigmatic.10,11
Brain AVMs have historically been referred to as congenital lesions; however, observations of de novo formation and progressive AVM growth have shifted views regarding their pathogenesis toward a cryptic genetic mechanism.12,13 Furthermore, a subset of AVMs recur following complete surgical removal, suggesting that germline genetic alterations may be present.14,15 Next-generation sequencing of paired blood and lesion tissue represents a powerful and unbiased method to identify potential germline and somatic mutations in brain AVMs. In this study, we investigated the potential genetic basis of a sporadic brain AVM that recurred in a child following complete surgical resection by performing whole-exome sequencing (WES) on paired lesion-blood samples. The highlight of our study is the discovery of a rare stop-gain mutation in SMAD9, which resulted in downstream signaling defects in the BMP signaling pathway. In addition, we showed that zebrafish (Danio rerio) knockdown modeling of this mutation resulted in abnormal artery-to-vein connections in the cerebral vasculature, thereby recapitulating the human disease and attributing further in vivo functional significance to this mutation.
Materials and methods
Sequencing and mutation analysis
To identify the molecular etiology of sporadic brain AVM, we performed WES on purified DNA from the proband’s lesion tissue and peripheral blood. Written informed consent to participate in this study was obtained, and the study was approved by the institutional review board at the REDACTED. The clinical course and features of the patient are highlighted in Figure 1. In brief, she suffered a hemorrhagic stroke from a brain AVM at age 12 and underwent surgical AVM resection. Two years later, she was observed to have local regrowth of the AVM that was previously completely removed, and a second surgery was performed. The patient had no other known health problems. DNA from tissue obtained at the time of the second surgery was used for WES. Sequencing was performed on the Illumina HiSeq2000 platform using an Illumina Nextera Rapid Capture Exome Kit (Illumina, San Diego, CA, USA).
Figure 1.
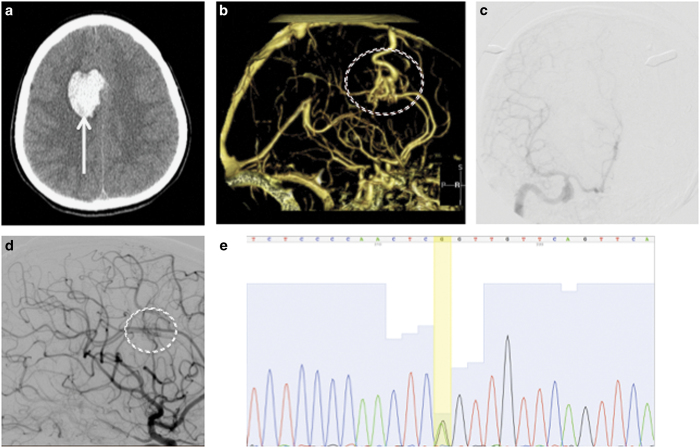
Clinical features. Initial axial head computed tomography (a) demonstrated a brain hemorrhage (arrow) in the right frontal lobe. Further investigation with computed tomography angiography (b) identified an arteriovenous malformation (circled) as the causative etiology for the hemorrhage. The lesion was resected surgically, and digital subtraction angiography (c) demonstrated no residual arteriovenous shunting. Two years later, a surveillance angiogram was performed (d) that demonstrated regrowth of the arteriovenous malformation (circled). The patient underwent another craniotomy, and the recurrent lesion was removed. Whole-exome sequencing of the tissue was performed, and Sanger sequencing (e) confirmed a mutation at chr13:37439827,G,A (yellow bar) corresponding to a stop codon in SMAD9.
Sequences were aligned to NCBI human reference GRCh37 using the Burrows-Wheeler Aligner.16 The aligned reads were converted to SAM format (Sequence Alignment/Map) and sorted. PCR duplicates were then removed and alignments were recalibrated. Single-nucleotide variant and insertion/deletion calling was performed using HaplotypeCaller from the Genome Analysis Toolkit, and the output was in the gVCF file format.17 Variant annotation was performed using the ANNOVAR program with references to GRCh37/hg19, dbSNP version 132, 1000 Genomes project (1KGP; 2012 data release), 69 Complete Genomics (2012 update) and exome variant server with ~6,500 exomes (NHLBI-ESP project, 2013 update).
Somatic variants were filtered out from changes observed in matched normal DNA to rule out artifacts and generate a list of true somatic and germline variants. The sequencing quality was determined by Genome Analysis Toolkit’s DepthOfCoverage Walker. Quality control steps excluded variants with a sequencing depth <10 or with genotype quality <90, those that were found in a selection of frequently mutated genes, and pseudo-genes, as well as those in repetitive regions of genes.
The rare nonsense mutation in SMAD9 (c.C739T:p.R247X, rs553369182) was then confirmed with Sanger sequencing with the following primer sequences: 5ʹ- GTGGTCACGTGCACTTCTACACA-3ʹ; and 5ʹ- ACATCTCAGGTGTGCTAGTGAAAT-3ʹ (Elim Biopharmaceuticals, Hayward, CA, USA).
Within the Exome Aggregation Consortium database, the minor allele frequency was reported at 0.00002 (n=3). To estimate the damaging character of the stop mutation, we used MutationTaster18,19 and Combined Annotation Dependent Depletion.20 Evolutionary conservation was estimated using genome conservation scores measured by PhyloP.21
Immunofluorescence staining
On the basis of the disease-causing prediction, we investigated the role of the SMAD9 mutation in altering protein expression within the BMP signaling pathway. To determine the effect of the mutation on translation, immunofluorescence staining of AVM samples removed from the patient was used to qualitatively assess both the localization and expression of protein products. Control samples were obtained from samples of a normal human temporal lobe neocortex that were removed as part of a routine protocol for temporal lobe epilepsy. Once isolated, the frozen brain and AVM samples were embedded in an optimal cutting temperature compound (Tissue-Tek, Torrance, CA, USA). Embedded tissues were cryosectioned at a thickness of 14 μm with a microtome and subsequently fixed with immersion in ice-cold acetone on positively charged glass slides in preparation for staining. All tissue sections were stored at −20 °C and rehydrated with phosphate-buffered saline on the day of staining. Sections were blocked in 10% swine serum (Vector Laboratories, Burlingame, CA, USA) and incubated overnight in primary antibody at 4 °C with the following antibodies: sheep anti-human SMAD9 antibody (R&D systems, Minneapolis, MN, USA); and rabbit anti-human phospho SMAD4 Thr 277 (Thermo Scientific, Waltham, MA, USA). To detect brain endothelial cells, sections were also incubated overnight at 4 °C with biotinylated Ulex Europaeus Agglutinin I lectin (Vector Laboratories). Sections were subsequently washed in phosphate-buffered saline with 0.05% Triton X-100 and incubated for 1 h at room temperature with the appropriate secondary antibody. To visualize SMAD9, phospho-SMAD4 and brain endothelial cells, sections were incubated with a Cy3-conjugated donkey anti-sheep antibody (Jackson ImmunoResearch Laboratories, West Grove, PA, USA), Alexa Fluor 488-conjugated donkey anti-rabbit antibody (Jackson ImmunoResearch) and DyLight 649-conjugated streptavidin (Vector Laboratories). Following immunodetection, sections were incubated in 1% Sudan Black B (Sigma Aldrich, St. Louis, MO, USA) to quench autofluorescence and counterstained with Hoechst 33342 solution (Thermo Scientific) to visualize cell nuclei. Sections were mounted with fluorescent mounting media (Dako, Carpinteria, CA, USA) and coverslipped. All imaging was performed with a Leica TCS SP5X confocal microscope, and representative images were prepared with NIH U.S. National Institutes of Health, Bethesda, MD, USA, ImageJ software.
Zebrafish modeling
To further investigate the functional significance of the SMAD9 mutation, we utilized a transient knockdown strategy using translation-blocking and splice-blocking morpholino oligonucleotides in zebrafish (D. rerio) Tg(kdrl:eGFP) that were linked to a green florescent endothelial cell marker. Zebrafish were raised and maintained under routine husbandry conditions, and experiments were approved and conducted in accordance with the Institutional Animal Care and Use Committee of University of California San Francisco standard procedures and guidelines. Morpholino oligonucleotides were obtained commercially (Gene Tools, Philomath, OR, USA) and were targeted to the SMAD9 splice site (5ʹ- AGTCTGGACTGTCACCTCTTTGTGG-3ʹ) and translation blocking site (5ʹ- TCGTGAGACGGGTTGATTTTAAATC-3ʹ) based on previously validated sequences.22,23 Morpholino oligonucleotides were injected at the single-cell stage into hundreds of embryos using varying concentrations from 5 to 15 ng before optimizing the injections at 7.2 ng. This concentration allowed for survival of over 95% of embryos that had the observed phenotype. Morphometric assessment was then performed on the embryos via microscopy at 68–72 h post fertilization (h.p.f.). Image acquisition from zebrafish embryos in the dorsal and lateral positions was achieved using a Nikon confocal microscope, and merged Z-stack images were analyzed with ImageJ.24 Comparison to normal developmental vascular anatomy was performed on corresponding time points from reference atlas images and uninjected embryos.25
Results
Identification of a rare SMAD9 mutation
WES was performed on DNA extracted from the patient’s brain AVM tissue and circulating leukocytes from peripheral blood (Table 1). Nine potential loss-of-function variants from nine genes were found in both germline and somatic DNA of the patient, which included four single-nucleotide variants and five insertions/deletions (Table 2). Six of the genes were involved in the immune-response or tumor development, two of which had unknown function. Among these, the most interesting was a rare single-nucleotide variant at chr13:37439827, introducing a stop codon in SMAD9 (NM_005905:exon4:c.C739T:p.R247X; NM_001127217:exon5:c.C850T:p.R284X) (Figure 1). The total coverage of the germline and somatic mutations at this location were 129- and 112-fold with mutation frequencies 0.57 and 0.5, respectively. Both genotype quality scores were 99. This mutation was then confirmed by Sanger sequencing. A battery of in silico testing with MutationTaster predicted it to be a disease-causing mutation with a probability of 1, indicating a high “security” of the prediction. Evolutionary conservation of the amino-acid sequence was high, as estimated by the PhyloP score, which ranged from 2.128 to 4.188 between different transcripts (Table 3). The score generated by the Combined Annotation Dependent Depletion tool was 40, which suggested a strong deleterious effect. Analysis of somatic variations in this patient (insertion-deletion mutation, missense mutation, nonsense mutation and loss of heterozygosity) did not demonstrate any significant changes in genes associated with vascular development (Supplementary Information 1, 2, 3).
Table 1. Capture methods and coverage summary of exome data.
| Blood | AVM | |
|---|---|---|
| Capture method | Illumina Nextera Rapid Capture Exome Kit (transposase-based, bait set containing 38 Mb target territory) |
|
| Mean coverage | 91.36 | 89.03 |
| % Covered>10× | 96.2 | 95.5 |
| % Covered>20× | 93.3 | 92 |
| % Covered>30× | 88.5 | 86.6 |
| % Zero coverage targets | 1.9 | 2 |
Abbreviation: AVM, arteriovenous malformation.
DNA obtained from formalin-fixed, paraffin-embedded tissue and whole blood.
Table 2. Summary of germline mutations.
| Chr | Position | Reference allele | Mutant allele | Variant class | Gene symbol | Coding variant type |
|---|---|---|---|---|---|---|
| 13 | 37439827 | G | A | SNP | SMAD9 | Stopgain |
| 16 | 31374391 | A | AG | Insertion | ITGAX | Frameshift_insertion |
| 22 | 32875242 | GT | G | Deletion | FBXO7 | Frameshift_deletion |
| 22 | 37882124 | GA | G | Deletion | MFNG | Frameshift_deletion |
| 22 | 42473987 | G | A | SNP | FAM109B | Stopgain |
| 4 | 175443135 | CAG | C | Deletion | HPGD | Frameshift_deletion |
| 5 | 637664 | C | T | SNP | CEP72 | Stopgain |
| 6 | 154428918 | CG | C | Deletion | OPRM1 | Frameshift_deletion |
| 9 | 125288771 | T | A | SNP | OR1N1 | Stopgain |
Abbreviation: SNP, single-nucleotide polymorphism.
Table 3. Conservation of the Arg284 amino acid throughout evolution.
| 284 | |||||||||
| Homo sapiens | E | L | N | N | R | V | G | E | T |
| Pan troglodytes | E | L | N | N | R | V | G | E | T |
| Macaca mulatta | E | L | N | N | R | V | G | E | T |
| Mus musculus | E | L | N | N | R | V | G | E | T |
| Gallus gallus | E | L | N | N | R | V | G | E | T |
| Takifugu rubripes | E | L | N | S | R | V | G | E | T |
| Danio rerio | E | L | N | N | R | V | G | E | T |
| Drosophila melanogaster | E | L | N | C | R | V | G | E | V |
| Xenopus tropicalis | E | L | N | N | R | V | G | E | T |
Bold values indicates amino acid position (#284).
Cellular localization and reductions of SMAD9 in the peri-nidal brain AVM vasculature
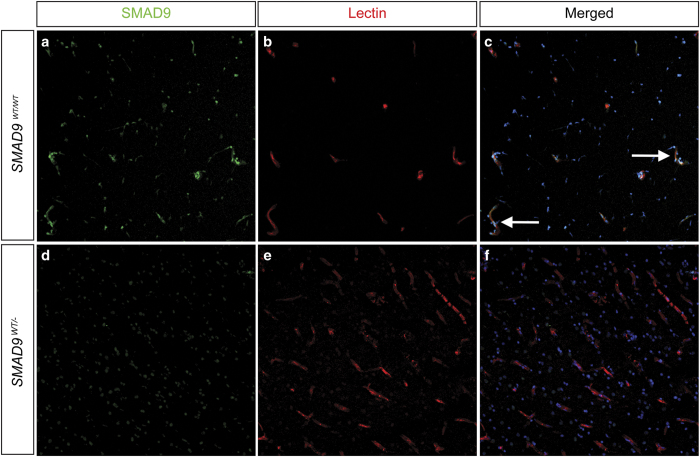
Immunofluorescent staining in control brain tissue revealed prominent SMAD9 expression in the vasculature, including both endothelial and peri-vascular vascular mural cells (Figure 2).
Figure 2.
Effect of the SMAD9 mutation on target protein expression. Immunohistochemical staining was performed to determine the localization and expression of SMAD9 in both control (a, b, c) and lesion (d, e, f) tissue. There was prominent expression of SMAD9 in control tissue (a) that co-localized (arrows) to vascular structures identified by lectin staining of the endothelium (b) and Hoechst nuclear counterstaining (c). Compared to control tissue, SMAD9 expression was greatly reduced in arteriovenous malformation tissue, with the most noticeable loss appearing in perivascular tissue.
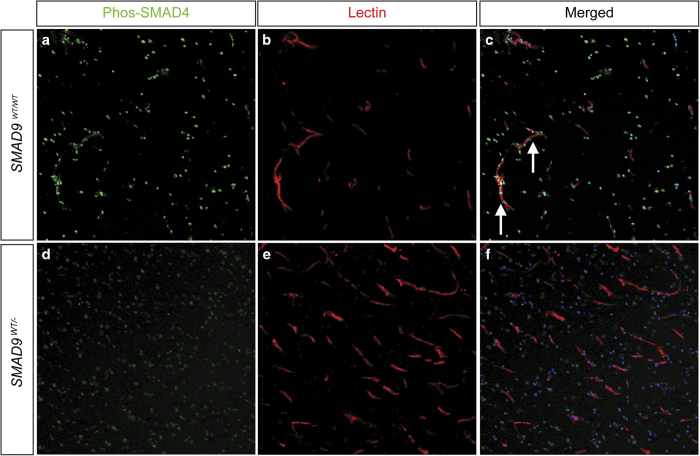
SMAD9 expression was found far less in surrounding neuronal and glial parenchymal cells (data not shown). In the peri-nidal AVM vessels, SMAD9 protein expression was markedly reduced. To demonstrate whether this may impact downstream signal transduction, we next performed immunofluorescent analysis of phosphorylated-SMAD4. In control tissues, there was a ubiquitous and robust phosphorylated-SMAD4 signal in multiple cell types, including vascular cell types (endothelial cells and perivascular mural cells), glia and neurons (Figure 3). In the brain AVM specimen, however, the vascular phosphorylated SMAD4 was nearly absent, with gross reductions also observed in the surrounding parenchyma.
Figure 3.
Effect of the SMAD9 mutation on downstream pathway protein phosphorylated SMAD4. Immunohistochemical staining was performed to determine the localization and expression of p-SMAD4 in both control (a, b, c) and lesion (d, e, f) tissue. Similar to SMAD9 staining, there was prominent expression of p-SMAD4 in control tissue (a) that co-localized (arrows) to vascular structures identified by lectin staining of the endothelium (b) and Hoechst nuclear counterstaining (c). Decreased expression of p-SMAD4 was observed in arteriovenous malformation tissue, and its expression appeared to be dissociated from vascular structures.
Collectively, these data suggest that AVM-specific reductions in SMAD9 may influence downstream signal transduction, as seen by reductions in phosphorylated SMAD4.
In vivo SMAD9 knockdown results in aberrant vasculogenesis and brain AVMs
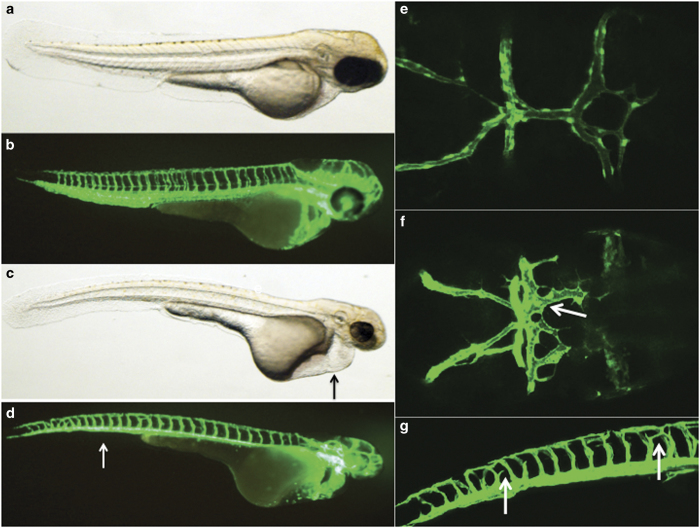
To further investigate whether reductions in SMAD9 contribute to brain AVM formation in vivo, we next utilized morpholino knockdown in transgenic zebrafish expressing green fluorescent protein in endothelial cells (Tg(kdrl:eGFP)). Following morpholino injection, embryos at 68–72 h.p.f. were systematically analyzed for both gross developmental abnormalities, as well as for vascular abnormalities. Overall, the SMAD9 morphants demonstrated slightly smaller heads, smaller eyes and thin, elongated trunks. There was a prominent amount of cardiac edema. Trunk circulation was markedly reduced or absent. Although the cardinal vein was often present (albeit faintly visible with florescence), the caudal vein plexus was markedly reduced (Figure 4).
Figure 4.
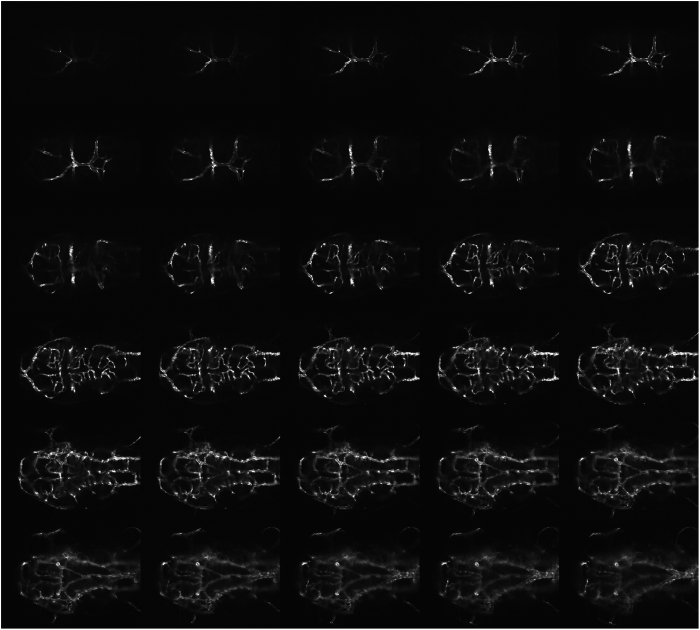
Zebrafish modeling of the SMAD9 mutation. Zebrafish morphants resulting from an injection of a translation blocking morpholino were created and imaged with a confocal microscope at 68–72 h post fertilization. Uninjected (a) light and (b) confocal lateral imaging of Tg(kdrl:eGFP) embryos was performed; the embryos were oriented laterally, cranial-right. Comparative images of SMAD9 knockdown (c, d) were typified by smaller heads, smaller eyes and thin trunks. The embryos also demonstrated cardiac edema (c, arrow) and had a near-absence of the caudal vein plexus (d, arrow). Cranial circulation was also imaged in uninjected (e) and morphants (f) with embryos oriented dorsal, cranial-right. Numerous abnormal arteriovenous connections between the dorsal longitudinal vein, mesencephalic vein and metencephalic artery were present (arrow). Further evidence of abnormal artery–vein specification during development was observed in the intersegmental vessels, where ectopic branches (g, arrow) were observed above the myoseptum.
The vascular phenotype of the SMAD9 morphant was typified by both developmental abnormalities in the trunk vessels and cranial circulation. There was abnormal intersegmental vessel formation, with ectopic branches above the myoseptum near the dorsal longitudinal anastomotic vessel. Within cranial circulation, there were abnormal arteriovenous connections between the dorsal longitudinal vein, mesencephalic vein and metencephalic artery (Figure 4). There was also a paucity of central arteries that penetrate the hindbrain and usually connect the basilar artery and posterior hindbrain channel. The lateral dorsal aorta was also significantly dilated and irregular (Figures 5 and 6).
Figure 5.
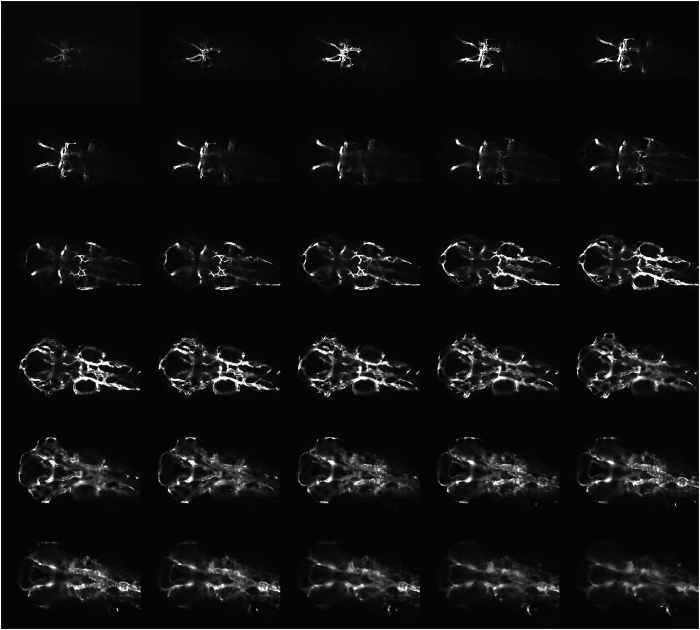
Wild-type control imaging of zebrafish cranial circulation. Reference confocal source dataset images of cranial circulation in a representative uninjected Tg(kdrl:eGFP) zebrafish embryo at 72 h post fertilization.
Figure 6.
SMAD9 zebrafish morphant cranial circulation. Confocal source data set images of the cranial circulation in a representative SMAD9 translation-blocking morpholino-injected Tg(kdrl:eGFP) zebrafish embryo at 72 h post fertilization. In addition to the numerous abnormal arteriovenous connections previously observed in Figure 4f, other cerebrovascular changes were also present. There was a paucity of central arteries that penetrate the hindbrain and usually connect the basilar artery and posterior hindbrain channel. The lateral dorsal aorta was also significantly dilated and irregular.
Discussion
Sporadic brain AVMs are a rare disease,2 and lesions that recur following complete removal are exceedingly rare.15,26,27 These unique cases provide an opportunity to evaluate the suspected genetic basis of their pathogenesis, which shares a vascular phenotype with syndromic forms of AVM, such as hereditary hemorrhagic telangiectasia. In this report, WES was used to survey the genetic landscape of sporadic brain AVM in an unbiased manner, and a candidate causative mutation in a sporadic AVM was discovered in SMAD9. The nature of the discovery was further investigated by tissue-level protein expression studies, in silico modeling and in vivo vertebrate modeling.
By drawing on the vast experiences of genetic variation in humans represented in the 1000 Genomes Project and others, we were able to establish the rarity of this mutation, which was only observed in 3 alleles out of 121,314 (minor allele frequency=2.473e-05) in the Exome Aggregation Consortium database v0.3.1.28 This establishes that the discovered variant is not generally present in healthy humans. While no genotype–phenotype relationship has been previously established with this variant, the majority of brain AVMs are clinically silent unless they produce symptoms from hemorrhage. It is plausible that the three individuals with this rare variant unknowingly harbored occult brain AVMs. Without neuroimaging, it is not possible to exclude the presence of clinically silent AVM. In addition, it is possible that other low-frequency mutations, other than in SMAD9, existed in the studied patient, but were not detected because of limitations of the sequencing depth and cellular heterogeneity of the samples sequenced (resulting in potential false negatives).
As other alterations in the BMP signaling pathway have been implicated in genetic AVM syndromes,29 we hypothesized that altered SMAD9 expression could alter downstream signal transduction, resulting in abnormal brain vasculogenesis and/or vascular remodeling. As SMAD9 is routinely phosphorylated and subsequently associates with SMAD4 to translocate to the nucleus,30 we sought to identify changes in phosphorylated SMAD4 as an indicator of BMP signaling pathway inhibition, as demonstrated by a potential reduction in downstream target activation. Surgical specimens allowed for a unique, direct qualitative observation of the decrease in downstream protein expression. The profound decrease in protein expression despite only a heterozygous loss is not clear, but may be related to a dominant negative effect of nonsense-mediated mRNA decay escape or a truncated protein.
In addition, we provided evidence of the pathogenic effect of this mutation by creating a vertebrate model (zebrafish). While morpholino knockdown of SMAD9 has been conducted using previously validated oligomer sequences,22,23 this report is the first to describe the effects of SMAD9 knockdown on vascular development. Zebrafish have traditionally been used as model organisms because of the ease with which their gene expression can be manipulated combined with their high fecundity and the short time-course in which phenotypes can be established.31,32 For vascular disease, in particular, the translucent nature of embryos, along with transgenic endothelial cell reporter lines (such as kdrl:eGFP), allows for real-time visualization of vascular development from the single-cell stage onward. As vascular development in zebrafish embryos follows a defined, stereotypic anatomical pattern, aberrations in these steps secondary to gene product knockdown can be readily visualized in vivo.25,33,34 Indeed, genetic manipulation in zebrafish that targets an ortholog of a known genetic cause of human AVM (ACVRL1) creates a model of cranial arteriovenous shunting.35,36
Not surprisingly, the SMAD9 morphant in the presented experiments has pleotropic manifestations on overall organism morphology and extracranial vasculature, which reflects the essential role of the BMP pathway in development.37 However, these morphants develop abnormal cranial artery-to-vein connections that are strikingly reminiscent of human AVMs, more so than some other models. It is possible that targeting SMAD9, which is downstream of ACVRL1, may reduce the ability of other components of the BMP signaling pathway to compensate for deficiencies, thereby displaying a more robust AVM phenotype.
From a wider perspective, other mutations in SMAD9 have primarily been associated with a rare heritable form of pulmonary hypertension resulting from a heterozygous deletion (OMIM 615342),38,39 although the apparent lack of complete penetrance in some of these cases further complicates the interpretation of the genotype–phenotype relationship. Patients with these mutations have also demonstrated a reduction in expression of downstream target genes in the canonical BMP signaling pathway.40 The index patient presented here demonstrated no signs or symptoms of pulmonary dysfunction. The heterogeneity of phenotypes resulting from various truncating mutations in SMAD9 in humans, either associated with brain AVMs or with hyperplasia of pulmonary artery smooth muscle cells, may relate to variable microRNA processing or an inability to form various protein products to dimerize with SMAD4.40–43
Conclusion
In silico modeling, protein expression in lesion tissue and zebrafish modeling provide orthogonal trajectories of evidence that establish a relationship between the candidate gene mutation discovered in SMAD9 via WES and the clinical phenotype. Replication in similar rare cases of recurrent AVM, or even more broadly sporadic AVM, may be informative in building a more comprehensive understanding of AVM pathogenesis.
Additional infomation
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
Financial support for the research in this report was provided by the Congress of Neurological Surgeons (Christopher Getch, MD Research Fellowship) to BPW. MTL is the principal investigator of multiple projects funded by NIH grant U54 NS065705-07.
Footnotes
Supplemental Information for this article can be found on the Human Genome Variation website (http://www.nature.com/hgv).
The authors declare no conflict of interest.
References
- Lawton MT, Rutledge WC, Kim H, Stapf C, Whitehead KJ et al. Brain arteriovenous malformations. Nat Rev Dis Primers 2015; 1: 15008. [DOI] [PubMed] [Google Scholar]
- Al-Shahi R, Warlow C. A systematic review of the frequency and prognosis of arteriovenous malformations of the brain in adults. Brain 124: 1900–19262001. [DOI] [PubMed] [Google Scholar]
- Walcott BP, Smith ER, Scott RM, Orbach DB. Pial arteriovenous fistulae in pediatric patients: associated syndromes and treatment outcome. J Neurointerv Surg 2013; 5: 10–14. [DOI] [PubMed] [Google Scholar]
- Guttmacher AE, Marchuk DA, White RI Jr. Hereditary hemorrhagic telangiectasia. N Engl J Med 1995; 333: 918–924. [DOI] [PubMed] [Google Scholar]
- Eerola I, Boon LM, Mulliken JB, Burrows PE, Dompmartin A et al. Capillary malformation-arteriovenous malformation, a new clinical and genetic disorder caused by RASA1 mutations. Am J Hum Genet 2003; 73: 1240–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz B, Toktaş ZO, Akakın A, Işık S, Bilguvar K et al. Familial occurrence of brain arteriovenous malformation: a novel ACVRL1 mutation detected by whole exome sequencing. J Neurosurg 2016; 126: 1879–1883, 1–5. [DOI] [PubMed] [Google Scholar]
- Boshuisen K, Brundel M, de Kovel CGF, Letteboer TG, Rinkel GJE et al. Polymorphisms in ACVRL1 and endoglin genes are not associated with sporadic and HHT-related brain AVMs in Dutch patients. Transl Stroke Res 2013; 4: 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinsheimer S, Bendjilali N, Nelson J, Guo DE, Zaroff JG et al. Genome-wide association study of sporadic brain arteriovenous malformations. J Neurol Neurosurg Psychiatry 2016; 87: 916–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendjilali N, Kim H, Weinsheimer S, Guo DE, Kwok PY et al. A genome-wide investigation of copy number variation in patients with sporadic brain arteriovenous malformation. PLoS ONE 2013; 8: e71434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcott BP, Winkler EA, Rouleau GA, Lawton MT. Molecular, cellular, and genetic determinants of sporadic brain arteriovenous malformations. Neurosurgery 2016; 63(Suppl 1): 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturiale CL, Puca A, Sebastiani P, Gatto I, Albanese A et al. Single nucleotide polymorphisms associated with sporadic brain arteriovenous malformations: where do we stand? Brain 2012; 136(Pt 2), 665–681. [DOI] [PubMed] [Google Scholar]
- Akimoto H, Komatsu K, Kubota Y. Symptomatic de novo arteriovenous malformation appearing 17 years after the resection of two other arteriovenous malformations in childhood: case report. Neurosurgery 2003; 52: 228–231, discussion 231–232. [DOI] [PubMed] [Google Scholar]
- Minakawa T, Tanaka R, Koike T, Takeuchi S, Sasaki O. Angiographic follow-up study of cerebral arteriovenous malformations with reference to their enlargement and regression. Neurosurgery 1989; 24: 68–74. [DOI] [PubMed] [Google Scholar]
- Klimo P Jr, Rao G, Brockmeyer D. Pediatric arteriovenous malformations: a 15-year experience with an emphasis on residual and recurrent lesions. Childs Nervous System 2007; 23: 31–37. [DOI] [PubMed] [Google Scholar]
- Kader A, Goodrich JT, Sonstein WJ, Stein BM, Carmel PW et al. Recurrent cerebral arteriovenous malformations after negative postoperative angiograms. J Neurosurg 1996; 85: 14–18. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7: 575–576. [DOI] [PubMed] [Google Scholar]
- Schuelke M. Mutation Taster http://www.mutationtaster.org/cgi-bin/MutationTaster/MutationTaster69.cgi?start_insdel=55075&bases_inserted=T&end_insdel=55077&transcript_stable_id_text=ENST00000350148&sequence_type=gDNA. accessed 1 April 2016.
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 2010; 20: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JD, Kim J. Alk3/Alk3b and Smad5 mediate BMP signaling during lymphatic development in zebrafish. Mol Cells 2014; 37: 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CY, Wang HP, Zhu ZY, Sun YH. Transcriptional factors smad1 and smad9 act redundantly to mediate zebrafish ventral specification downstream of smad5. J Biol Chem 2014; 289: 6604–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai S, Horiguchi M, Weinstein BM. The vascular anatomy of the developing zebrafish: an atlas of embryonic and early larval development. Dev Biol 2001; 230: 278–301. [DOI] [PubMed] [Google Scholar]
- Codd PJ, Mitha AP, Ogilvy CS. A recurrent cerebral arteriovenous malformation in an adult. J Neurosurg 2008; 109: 486–491. [DOI] [PubMed] [Google Scholar]
- Gabriel EM, Sampson JH, Wilkins RH. Recurrence of a cerebral arteriovenous malformation after surgical excision. Case report. J Neurosurg 1996; 84: 879–882. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallione CJ, Repetto GM, Legius E, Rustgi AK, Schelley SL et al. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4). Lancet 2004; 363: 852–859. [DOI] [PubMed] [Google Scholar]
- Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J 2000; 19: 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov 2005; 4: 35–44. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’in zebrafish. Nat Genet 2000; 26: 216–220. [DOI] [PubMed] [Google Scholar]
- Walcott BP, Peterson RT. Zebrafish models of cerebrovascular disease. J Cereb Blood Flow Metab 2014; 34: 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 2002; 248: 307–318. [DOI] [PubMed] [Google Scholar]
- Walcott BP. BMP signaling modulation attenuates cerebral arteriovenous malformation formation in a vertebrate model. J Cereb Blood Flow Metab 2014; 34: 1688–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman BL, Pham VN, Lawson ND, Kulik M, Childs S et al. Disruption of acvrl1 increases endothelial cell number in zebrafish cranial vessels. Development 2002; 129: 3009–3019. [DOI] [PubMed] [Google Scholar]
- Wang RN, Green J, Wang Z, Deng Y, Qiao M et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis 2014; 1: 87–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani M, Yagi H, Nakayama T, Saji T, Matsuoka R. A new nonsense mutation of SMAD8 associated with pulmonary arterial hypertension. J Med Genet 2009; 46: 331–337. [DOI] [PubMed] [Google Scholar]
- Huang Z, Wang D, Ihida-Stansbury K, Jones PL, Martin JF. Defective pulmonary vascular remodeling in Smad8 mutant mice. Hum Mol Genet 2009; 18: 2791–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake KM, Zygmunt D, Mavrakis L, Harbor P, Wang L, Comhair SA et al. Altered MicroRNA processing in heritable pulmonary arterial hypertension: an important role for Smad-8. Am J Respir Crit Care Med 2011; 184: 1400–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin C-H, Miyazono K, Ten Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 1997; 390: 465–471. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003; 425: 577–584. [DOI] [PubMed] [Google Scholar]
- Yao G, Yin M, Lian J, Tian H, Liu L, Li X et al. MicroRNA-224 is involved in transforming growth factor-β-mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Mol Endocrinol 2010; 24: 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.