Abstract
Dopamine is critical for processing of reward and etiology of drug addiction. Astrocytes throughout the brain express dopamine receptors, but consequences of astrocytic dopamine receptor signaling are not well established. We found that extracellular dopamine triggered rapid concentration-dependent stellation of astrocytic processes that was not a result of dopamine oxidation but instead relied on both cAMP-dependent and cAMP-independent dopamine receptor signaling. This was accompanied by reduced duration and increased frequency of astrocytic Ca2+ transients, but little effect on astrocytic voltage-gated potassium channel currents. To isolate possible mechanisms underlying these structural and functional changes, we used whole-genome RNA sequencing and found prominent dopamine-induced enrichment of genes containing the CCCTC-binding factor (CTCF) motif, suggesting involvement of chromatin restructuring in the nucleus. CTCF binding to promoter sites bidirectionally regulates gene transcription and depends on activation of poly-ADP-ribose polymerase 1 (PARP1). Accordingly, antagonism of PARP1 occluded dopamine-induced changes, whereas a PARP1 agonist facilitated dopamine-induced changes on its own. These results indicate that astrocyte response to elevated dopamine involves PARP1-mediated CTCF genomic restructuring and concerted expression of gene networks. Our findings propose epigenetic regulation of chromatin landscape as a critical factor in the rapid astrocyte response to dopamine.
SIGNIFICANCE STATEMENT Although dopamine is widely recognized for its role in modulating neuronal responses both in healthy and disease states, little is known about dopamine effects at non-neuronal cells in the brain. To address this gap, we performed whole-genome sequencing of astrocytes exposed to elevated extracellular dopamine and combined it with evaluation of effects on astrocyte morphology and function. We demonstrate a temporally dynamic pattern of genomic plasticity that triggers pronounced changes in astrocyte morphology and function. We further show that this plasticity depends on activation of genes sensitive to DNA-binding protein CTCF. Our results propose that a broad pattern of astrocyte responses to dopamine specifically relies on CTCF-dependent gene networks.
Keywords: astrocyte, chromatin, CTCF, dopamine, PARP1, RNA sequencing
Introduction
Astrocytes are a major subpopulation of the brain glial cells that mediate multiple functions, including metabolic, ionic and pH homeostasis, trophic support of neurons, antioxidant defense, the establishment and maintenance of the blood–brain barrier, and regulation of neuronal activity (Bushong et al., 2002; Fattore et al., 2002; Perea et al., 2009; Freeman, 2010; Kandel et al., 2012; Oberheim et al., 2012; Zamanian et al., 2012; Clark et al., 2013; Clarke and Barres, 2013). Such functional diversity is accompanied by morphological heterogeneity that varies with development and across brain regions (Raff et al., 1983; Miller and Raff, 1984; Khakh and Sofroniew, 2015; Chai et al., 2017). For example, astrocytic domain boundaries are observed later, but not early in development (Bushong et al., 2004), and astrocytes in the striatum have significantly more processes than hippocampal astrocytes (Khakh and Sofroniew, 2015; Chai et al., 2017)
In cultured conditions, two astrocytic phenotypes have been distinguished based on morphological characteristics: Type I cells have a flat polygonal appearance with a “mesh” of intertwined fine processes and resemble astrocytes populating gray matter; and Type II cells have clearly defined thicker “stellated” processes and resemble the astrocytes arranged along white matter tracts (Raff et al., 1983; Oberheim et al., 2012). Standard protocols that use serum-supplemented culture media result in predominantly polygonal, Type 1 astrocytes. However, these astrocytes can be transformed into a stellated morphology by exposure to cAMP analogs (Moonen et al., 1975; Won and Oh, 2000; Paco et al., 2016). The cAMP-induced morphological transformation occurs within 2 h and is associated with decreased cell body area and extension of distinct thick processes.
Dopamine (DA) may trigger cAMP production via activation of D1-like DA receptors, expressed by astrocytes along with other DA receptor subtypes (Khan et al., 2001; Miyazaki et al., 2004; Mladinov et al., 2010; Shao et al., 2013; Nagatomo et al., 2017). D1-induced production of cAMP has been linked to release of astrocytic Ca2+ from internal stores, an important hallmark of astrocyte-to-astrocyte, astrocyte-to-neuron, and astrocyte-to-vasculature signaling (Chai et al., 2017; Jennings et al., 2017). Indeed, both structural and functional astrocyte adaptations have been reported in models of neurologic disease linked to dysregulated DA signaling. For example, Parkinson's disease is associated with hypertrophy of astrocytic processes, a feature of reactive astrocytosis (Booth et al., 2017), whereas cocaine use has been linked to a reduction of astrocyte surface area and reduced interaction with synapses (Scofield et al., 2016). Furthermore, a number of studies have demonstrated that DA-induced increase in astrocytic Ca2+ impacts cellular respiration mechanisms coupled to PARP1 and sirtuin activation (Requardt et al., 2010, 2012; Verdin, 2015; Gupte et al., 2017). PARP1 is a critical cofactor for NAD+-dependent poly-ADP-ribosylation of CCCTC binding factor (CTCF), which promotes CTCF binding to target genes (Yu et al., 2004; Ong et al., 2013; Han et al., 2017). Recent studies have shown that CTCF regulates both chromatin remodeling (Wright et al., 2016) and chromatin insulation (Yu et al., 2004; Ong et al., 2013), thereby organizing transcriptional regulation of distinct gene suites in response to cellular signaling. However, it is unknown whether DA- or DA receptor-induced structural or genomic changes are linked to PARP1/CTCF interaction.
In this study, we demonstrate that DA receptor signaling generates pronounced morphological changes in cultured astrocytes, which rely on PARP1 activation and are accompanied by a dynamic regulation of the astrocyte transcriptome, including, most prominently, specific enrichment of genes containing CTCF motifs in promoter regions. Our results highlight a novel PARP1/CTCF-dependent mechanism that drives DA-induced morphological changes and associated transcriptional profile in astrocytes.
Materials and Methods
Astrocyte cultures.
Primary astrocytes were dissociated from hippocampi of neonatal (postnatal day 1–2) Sprague Dawley rat pups. The heads were removed and hippocampi dissected in Petri dishes filled with ice-cold dissection medium composed of the following: Hanks Balanced Salt Solution (without Ca2+, Mg2+, NaHCO3, or phenol red) supplemented with 10 mm HEPES (both from Sigma-Aldrich). The dissected hippocampi were finely chopped and dissociated by trypsin treatment (10 μl/ml; Invitrogen), followed by trituration with sterile glass pipettes. Cells were plated on 12 mm glass coverslips, 6-well, or 24-well plates coated with poly-L-lysine (50 μg/ml; Sigma-Aldrich). Cultures were maintained in DMEM/F12 (Invitrogen) media supplemented with 10% FBS and stored in an incubator at 37°C (5% O2/95% CO2) for 10–14 d (DIV 10–14). This protocol produces astrocyte-enriched cultures with 86 ± 0.02% of all cells staining positive for the astrocyte marker GFAP.
Immunocytochemistry.
The cells were washed twice in PBS, pH 7.4, fixed in 4% PFA (10–20 min; Affymetrix), and then washed with 1× PBS three times. The cells were next treated with 0.3% Triton-X (Sigma-Aldrich) for 15 min and washed once with 1× PBS. Nonspecific staining was blocked with PBS, containing 0.1% Tween 20 (Thermo Fisher Scientific) and 10% donkey or goat serum (Sigma-Aldrich) for 1–2 h at room temperature. Coverslips were incubated with primary (mouse, polyclonal) antibodies against GFAP (1:1000; 2HR RT, Millipore; Sigma-Aldrich, MAB360), and then washed with 1× PBS three times. Fluorescent staining was then performed by incubating with secondary (donkey anti-mouse) antibodies (1:1000; Invitrogen) conjugated to AlexaFluor-488 (A11008) or AlexaFluor-594 (A21203) at room temperature in the dark for 1 h. Cells were then washed three times with 1× PBS and counterstained with DAPI (5 μg/ml; Invitrogen) for 5 min. Finally, coverslips were rinsed once with distilled H2O and mounted on glass slides with Prolong Antifade mounting medium (Invitrogen). Negative controls omitting primary or secondary antibodies were included in all experiments. Cells were examined under a widefield fluorescent microscope (Eclipse E200, Nikon). Morphological analyses were conducted with National Institutes of Health ImageJ software. For cell area measurements, GFAP-positive cells were manually outlined and isolated from background by applying a color threshold. For analysis of stellation, all distinct primary processes extending directly from the cell body, thinner than 40 pixels wide, and at least 30 pixels in length were counted.
RNA extraction/sequencing.
Total RNA was extracted using TRIzol (Invitrogen) and an RNeasy Mini Kit (QIAGEN) according to the manufacturers' instructions. For slice experiments, acute hippocampal and striatal brain slices were prepared from 6- to 8-week-old Sprague Dawley rats as described by Ortinski et al. (2015). Hippocampal and striatal tissue was microdissected from slices before flash-freezing for further processing. Samples were homogenized and incubated in TRIzol for 5 min before addition of chloroform and brief vigorous shaking for 30 s. Following a 3 min incubation, samples were centrifuged at 4°C for 10 min at maximum speed (≥10,000 rpm). The aqueous phase was aspirated and transferred to a microcentrifuge tube before addition of 70% EtOH, centrifugation at ≥10,000 rpm for 15 s and collection of the precipitate from the RNeasy mini column. This step was repeated after adding 700 μl of Buffer RW1 and next 500 μl of Buffer RPE to the mini column. Another 500 μl of Buffer RPE was centrifuged for 2 min before the sample/mini column underwent a 2 min “dry” spin and transferred to the final collection tube. Last, 30 μl of DEPC water was used to elute the sample. RNA samples were quantified using Spectrophotometer ND-2000 (Nanodrop Technologies) and checked for quality and degradation by the 2100 Bioanalyzer (Agilent Technologies). All samples were of high quality (RNA integrity number between 9.9 and 10). Strand-specific mRNA libraries were prepared using the TruSeq Stranded mRNA Library Prep Kit (Set B, Illumina) and sequenced on the Illumina NextSeq500 in a paired-end mode with read length of 2 × 75 bp.
Sequencing data preprocessing and analyses.
To ensure there were no sequencing errors, raw sequences were checked for quality using FastQC, and then aligned to the rat genome (downloaded from iGenomes, Illumina) using the STAR aligner program (Dobin et al., 2013). Aligned SAM files from STAR were converted to BAM files using SAMtools (Li et al., 2009). BAM files were processed for read summarization using featureCounts (Liao et al., 2014), and the resulting read counts were preprocessed by filtering out low read counts (read counts < 5) in R software. Processed data were then analyzed for differential expression using DESeq2 (Love et al., 2014) in R software. False discovery rate (FDR < 0.05) was used to determine the threshold of p value for the analysis.
Functional annotation/gene ontology analyses for biological function were conducted using Panther classification system (www.pantherdb.org) accessed in September-October, 2017. CTCF binding motif analysis was performed using CTCFBSDB 2.0 (Ziebarth et al., 2013), a database for CTCF binding sites and genome organization. Top 30 differentially expressed genes (15 downregulated and 15 upregulated) from each of the treatment groups were extracted and then matched with the downloaded CTCF binding motif database. This analysis was completed in R software.
qRT-PCR.
Oligonucleotide primers were designed using the NCBI Primer-Designing tool, checked for specificity via the BLAST database, and supplied by Eurofins Genomics. The following primers were used: FosB (forward: ctcccctaccctcgatatttg; reverse: gcccatctccccaatattaaa); Pde10A (forward: tgtgggacatcctgttttga; reverse: cgacagacaggcaattagca); Ap1s3 (forward: ctctaagggtccgctgtacg; reverse: agcacccagaacaaaacacc), Gpr83 (forward: acaggggttgactgaaccag; reverse: aagtggtgggagagggttct), Nr4a3 (forward: tgctctggtcctcatcactg; reverse: tccagcttgaggcaaaagat), and Klf4 (forward: gggagggaagaccagagttc; reverse: accaagcaccatcgtttagg). For each primer tested, 1000 μl of primer mix contained the following: 200 μl of 100 μm forward primer, 200 μl of 100 μm reverse primer, and 600 μl of DEPC H2O. Samples were diluted to 500 ng/μl with DEPC water before undergoing cDNA synthesis. For each sample, 10 μl of 2× SYBR Green Mix (Applied Biosystems, Thermo Fisher Scientific), 4 μl of DEPC H2O, and 1 μl of cDNA sample were prepared and subsequently mixed with 5 μl of primer mix for a total volume of 20 μl before pipetting into a 385-well plate. Real-time PCR was performed using Bio-Rad CFX384 Real-Time PCR Detection System. For all genes, the cycling parameters were as follows: initial denaturation at 95°C for 3 min, 40 cycles of 95°C for 20 s, and final cycle of 60°C for 20 s. Transcript measurements in each sample were done in triplicates. The mRNA levels were determined using the 2−ΔΔCT method (Livak and Schmittgen, 2001) and target genes were normalized to the housekeeping genes GAPDH or hypoxanthine phosphoribosyltransferase. Expression values were normalized to GAPDH and reported as a fold change.
Ca2+ imaging.
Astrocyte cultures were incubated with 5 μl of GFAP-driven GCamp6f virus (AAV5.GfaABC1D.cytoGCamp6f.SV50, Addgene #52925, packaged by the University of Pennsylvania Vector Core) for 3–5 d before imaging at DIV 10–14. Coverslips were transferred to a recording chamber continuously perfused (2–3 ml/min) with recording solution composed of the following (in mm): 145 NaCl, 5 KCl, 1 MgCl2, 1 CaCl2, 5 HEPES, 5 glucose, and 15 sucrose (all from Sigma-Aldrich) adjusted to pH 7.4 with NaOH. Two-minute videos were acquired with ORCA-Flash 4.0 (V2) digital camera mounted on an Olympus BX51WI upright microscope equipped with an LED light source (X-Cite XLED1, Excelitas Technologies). Videos were binned at 1024 × 1024 pixels and collected at 25 frames/s. Background subtraction and fluorescent signal measurements were done using ImageJ based on manual isolation of individual cells as ROIs. Relative fluorescence intensity within each ROI was calculated as dF/F0, where F0 is average fluorescence intensity in a 1 s window of baseline fluorescence for each ROI identified in a background-subtracted image. Average amplitude and duration of Ca2+ transients exceeding three SDs of baseline noise were calculated following manually curated identification of individual events in Clampfit 10.4 (Molecular Devices). Ca2+ transient duration was expressed for each cell as half-width, the time between two frames on either side of the peak that are first to reach 50% of peak amplitude relative to baseline. Event frequency for each cell was calculated as total number of identified events per 2 min video.
Electrophysiology.
Coverslip-plated astrocytes were viewed in a perfusion chamber under an upright microscope (Olympus BX51WI) with infrared differential interference contrast optics and a 40× water-immersion objective. Recording solution was the same as described for Ca2+ imaging. Recording pipettes were pulled from borosilicate glass capillaries (World Precision Instruments) to a resistance of 4–7 mΩ when filled with the intracellular solution. The intracellular solution contained the following (in mm): 145 potassium gluconate, 2 MgCl2, 2.5 KCl, 2.5 NaCl, 0.1 BAPTA, 10 HEPES, 2 Mg-ATP, and 0.5 GTP-Tris, adjusted to pH 7.2–7.3 with KOH, osmolarity 280–290 mOsm. Currents were low-pass filtered at 2 kHz and digitized at 20 kHz using Digidata 1440A acquisition board (Molecular Devices) and pClamp10 software (Molecular Devices). Potassium (K+) currents were recorded in voltage-clamp mode (Vholding = −80 mV). Access resistance (10–30 mΩ) was monitored throughout each recording by injection of 10 mV hyperpolarizing pulses; data were discarded if access resistance changed >25% over the course of data collection. Following seal rupture, series resistance was compensated (65%–75%). Inward rectifying K+ currents were evoked by a series of 10 mV, 40-ms-long steps from −180 mV to −80 mV following a 200 ms prestep to 0 mV. Outward K+ currents were evoked by a series of 10 mV, 500-ms-long voltage steps from −80 to 80 mV following a 100 ms prestep to −110 mV or −50 mV. Outward delayed rectifier K+ currents were measured following presteps to −50 mV. Outward A-type K+ currents were isolated by subtracting traces recorded following presteps to −50 mV from those recorded following presteps to −110 mV. Inward rectifying K+ currents were measured at the end of the 40 ms step. Delayed rectifier and A-type K+ currents were measured at current peak. All measurements were acquired using Clampfit 10 (Molecular Devices).
Data analyses and statistics.
Statistical analyses were performed with Excel 2016 (Microsoft) or Prism 6 (GraphPad Software). For immunocytochemical, Ca2+ imaging, and electrophysiological experiments, cells from a minimum of 3 different coverslips were analyzed for each reported condition. For RNA seq, cells from 3 or 4 experimental replicates were processed for analysis as described above. For qPCR validation of RNA-seq data, one-way ANOVAs were used for each gene. In the hippocampal and striatal slice experiments, planned comparisons predicted effects of DA and/or DA antagonists compared with control condition; thus, Student's t tests were performed to determine significance. For electrophysiological studies, mean peak K+ current amplitude was analyzed with two-way repeated-measures ANOVAs with current amplitude as a within-subject factor and drug treatment as a between-subject factor followed by Bonferroni post hoc tests. Data were reported as mean ± SEM, and significance threshold was set at p < 0.05.
Results
Elevated DA leads to stellation of astrocyte processes
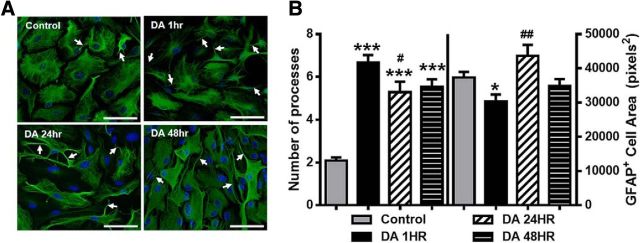
To examine effects of DA on astrocyte morphology, we evaluated GFAP immunostaining following treatment with DA (75 μm) for 1, 24, and 48 h. This revealed a rapid and persistent DA-induced stellation (Fig. 1A). Following 1 h of exposure, DA induced a 3.2-fold increase in the number of primary processes and also significantly reduced mean GFAP-positive (GFAP+) cell area to 81 ± 5.6% of control values (Fig. 1A,B). The increase in process number was maintained following 24 and 48 h of treatment (253 ± 23.8% and 266 ± 16.8%, respectively). However, reduction in GFAP+ area recovered to control levels by the 48 h time point. Astrocytes exposed to control media for 1, 24, or 48 h had similar stellation and GFAP+ area and were pooled together for analysis (Fig. 1A,B).
Figure 1.
Elevated extracellular DA induces morphological changes in astrocytes. A, Widefield images of astrocytes double-stained for GFAP (green) and DAPI (blue) in control conditions and after exposure to DA (75 μm) for 1, 24, or 48 h (DA 1 h, DA 24 h, DA 48 h). DA induces a prominent increase in the number of primary processes (arrows). Scale bars, 75 μm. B, Quantification of the number of primary processes and average GFAP+ cell area. One-way ANOVA for number of processes (F(3,469) = 56.64, p < 0.0001). One-way ANOVA for GFAP+ area (F(3,469) = 5.52, p = 0.001). Bonferroni post hoc: *p < 0.05 versus control. ***p < 0.001 versus control. #p < 0.05 versus DA 1 h. ##p < 0.01 versus DA 1 h. Control, n = 179 cells; DA 1 h, n = 152 cells; DA 24 h, n = 58 cells; DA 48 h, n = 84 cells.
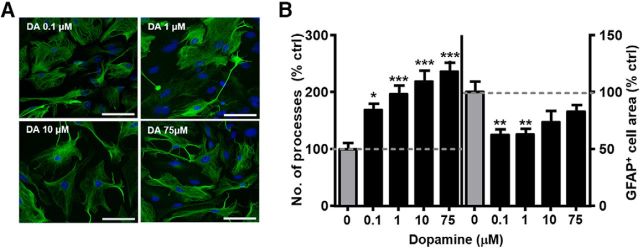
We next examined astrocyte response to DA concentrations in the low nanomolar to low micromolar range, similar to the reported brain DA levels (Smith et al., 1992; Zhang et al., 2009). Process number was significantly increased to 169 ± 12.6% of control values following a 1 h treatment with the lowest DA concentration (0.1 μm) and continued to increase at a more modest rate following treatments with 1, 10, and 75 μm DA (196 ± 15.9%, 219 ± 19.9%, and 236 ± 17.6% of control values, respectively; Fig. 2). Interestingly, the decrease in GFAP+ area was strongest after a 1 h treatment with 0.1 and 1 μm DA (62.4 ± 4.4% and 62.9 ± 5.1% of control values, respectively) but failed to reach statistical significance after treatment with 10 or 75 μm DA in this experiment (73.5 ± 10.6% and 82.9 ± 6.2% of control values, respectively; Fig. 2). These results indicated that exposure to elevated DA triggered a rapid, pronounced, and sustained concentration-dependent stellation of astrocyte processes. DA-induced reduction in GFAP+ area was more variable between cultures and had a distinctly different concentration dependence profile.
Figure 2.
DA-induced stellation of astrocyte processes is concentration-dependent. A, Widefield images of GFAP+ astrocytes following a 1 h exposure to 0.1, 1, 10, and 75 μm DA. Scale bars, 75 μm. B, Quantification of average number of primary processes and average GFAP+ cell area. One-way ANOVA for number of processes (F(4,325) = 11.54, p < 0.001). One-way ANOVA for GFAP cell area (F(4,325) = 5.266, p < 0.001). Bonferroni post hoc: *p < 0.05 versus control. **p < 0.01 versus control. ***p < 0.001 versus control. Control, n = 67 cells; DA 0.1 μm, n = 63 cells; DA 1 μm, n = 67 cells; DA 10 μm, n = 54 cells; DA 75 μm, n = 79 cells.
Astrocyte stellation is mediated by DA receptors
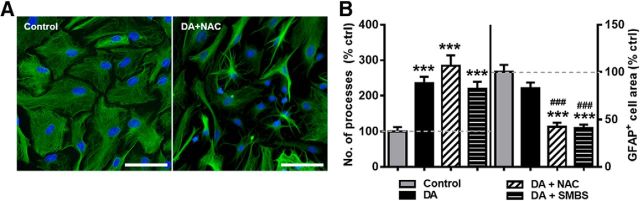
DA can undergo oxidation in cell culture media, and oxidative processes have been linked to changes in astrocyte morphology, including stellation (Clement et al., 2002; Pekny and Nilsson, 2005; Sofroniew, 2009; Vaarmann et al., 2010). Sodium metabisulfite (SMBS) and N-acetyl-L-cysteine (NAC) can be used to prevent oxidation of DA (Ortinski et al., 2015; Goldstein et al., 2017). To examine whether oxidative processes play a role in DA-induced astrocyte stellation, we pretreated the astrocyte cultures with NAC (5 mm) in culture media for 15 min before addition of DA (75 μm) for 1 h. This treatment did not prevent stellation of astrocyte processes. We observed an average of 236 ± 17.6% increase in number of processes for cells treated with DA alone versus an average increase of 285 ± 29.6% in number of processes for cells treated with DA and NAC (Fig. 3A,B). GFAP+ cell area was reduced following NAC pretreatment (42 ± 4.5% of control levels) in excess of what was observed with DA alone (82.9 ± 6.2% of control levels). This suggests that, in the absence of NAC, DA oxidation or background oxidative processes may oppose DA-induced effects on GFAP+ area. Pretreatment with SMBS (50 μm), a different antioxidant, had similar effects: SMBS failed to prevent DA-induced stellation but did facilitate reduction of GFAP+ area (Fig. 3B).
Figure 3.
Oxidation of DA does not promote rapid stellation of astrocyte processes. A, Representative images of GFAP+ astrocytes in the control conditions and following exposure to DA (75 μm) and N-acetylcysteine (5 mm; DA + NAC). Scale bars, 75 μm. B, Quantification of number of primary processes and average GFAP+ cell area following a 1 h exposure to control media, DA, DA with NAC, or DA with SMBS (50 μm; DA + SMBS). One-way ANOVA for number of processes (F(3,222) = 17.56, p < 0.001). One-way ANOVA for GFAP+ area (F(3,222) = 17.99, p < 0.001). Bonferroni post hoc: ***p < 0.001 versus control. ###p < 0.001 versus DA. Control, n = 67 cells; DA, n = 79 cells; DA + NAC, n = 41 cells; DA + SMBS, n = 39 cells.
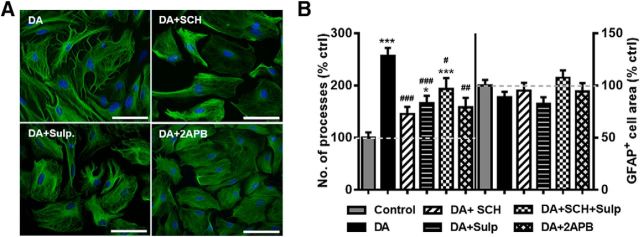
To test whether DA-induced stellation required activation of DA receptors, we examined the effects of pretreatment with the D1-like DA receptor antagonist, SCH23390 (50 μm), or the D2-like DA receptor antagonist, sulpiride (20 μm). Cells were pretreated with one or both antagonists for 15 min before addition of DA (75 μm) for 1 h. The results indicated that DA-induced stellation could be blunted by either D1-like or D2-like DA receptor antagonists. Pretreatment with SCH23390 rescued a 257 ± 15.7% DA-induced increase in process number to 145 ± 13.6% of control levels, whereas pretreatment with sulpiride rescued DA-induced increase in process number to 166 ± 14% of control levels (Fig. 4A,B). Pretreatment with both SCH23390 and sulpiride, surprisingly, was less effective in blunting DA-induced stellation (193 ± 21.2% of control values) than treatment with either antagonist plus DA. We investigated the possibility that DA could drive stellation through the Gq-PLC pathway-coupled D1/D2 heterooligomers (Lee et al., 2004; Rashid et al., 2007; Hasbi et al., 2010; Beaulieu and Gainetdinov, 2011). A 15 min preincubation with the membrane-permeable IP3 inhibitor, 2-aminoethox6ydiphenylborane (2-APB; 50 μm), before the 1 h treatment with DA blunted DA-induced stellation to 158 ± 18.2% of control values (Fig. 4B). Application of 2-APB alone had no effect on astrocyte stellation (119 ± 21.3% of control values) or GFAP area (77 ± 7% of control values). GFAP+ area was also not significantly affected by any of the other treatments in this experiment (Fig. 4B). Together, these findings indicate that DA effects on astrocyte stellation are attributable to DA receptor activation with both D1- and D2-like receptors playing a role in this process. Additionally, we demonstrate a role for the IP3 receptor activation downstream of DA receptor signaling. Finally, while oxidative processes may play a role in DA-induced regulation of GFAP+ area, these effects are likely dissociable from DA-receptor mediated stellation.
Figure 4.
DA-induced morphological changes depend on activation of D1-like and D2-like DA receptors. A, Representative images of GFAP+ astrocytes after a 1 h exposure to DA (75 μm) or after exposure to DA in the presence of D1-like DA receptor antagonist SCH23390 (50 μm; DA + SCH), D2-like DA receptor antagonist, sulpiride (20 μm; DA + Sulp), or DA with the IP3 receptor antagonist, 2-APB (50 μm; DA + 2-APB). Scale bars, 75 μm. B, Quantification of number of primary processes and average GFAP+ cell area. One-way ANOVA for number of processes (F(5,502) = 14.15, p < 0.0001). One-way ANOVA for GFAP area (F(5,503) = 1.777, p = 0.1160). Bonferroni post hoc: *p < 0.05 versus control. ***p < 0.001 versus control. #p < 0.05 versus DA. ##p < 0.01 versus DA. ###p < 0.001 versus DA. Control, n = 105 cells; DA, n = 116 cells; DA + SCH, n = 84 cells; DA + Sulp, n = 70 cells; DA + SCH + Sulp, n = 84 cells; DA + 2-APB, n = 50 cells.
DA treatment elicits broad changes in astrocyte transcriptome
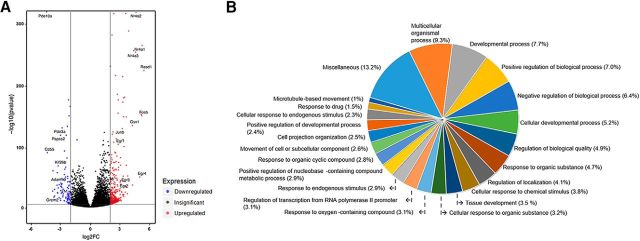
Considering that DA treatment can trigger activation of multiple pathways to induce rapid structural morphogenesis, we profiled genome-wide transcriptional changes using RNA seq. We examined astrocyte transcriptomic changes following brief (20 min) and extended (60 min) exposure to exogenous DA (75 μm). Brief treatment with DA resulted in enrichment (log2fold values > 2) of 569 (116 downregulated, 453 upregulated) of 17,327 identified genes. Extended treatment with DA resulted in enrichment of 159 (37 downregulated, 122 upregulated) of the 17,327 genes relative to control levels. Figure 5A (volcano plot) shows RNA-seq results from all treatment groups (control, 20 min, 60 min). Significantly enriched genes (log2 fold change >2) are color-coded in red (upregulated genes) and blue (downregulated genes) with FDR < 0.0001 (horizontal line). A number of genes with especially large magnitude changes and p values are labeled. Gene ontology analysis indicated that biological and developmental process regulation accounted for 21.1% of differentially enriched transcripts in the 20 min treatment condition and for 18.8% of differentially enriched transcripts in the 60 min treatment condition. Relevant to our morphological findings, 6.2% of differentially enriched genes (DEGs) in the 20 min condition were accounted for by “microtubule-based,” “cell or subcellular component movement,” and “cell projection organization” categories (Fig. 5B). None of these categories was represented in the 60 min condition, although 3.5% of all DEGs were classified as responsible for “structure morphogenesis.” Relevant to astrocyte function, 40 genes were classified as responsible for ion transport in the 20 min condition and 14 genes as responsible for calcium ion homeostasis in the 60 min condition. Prominently represented within the ion transport category were potassium channel genes: 8 of 40 total. Of those eight, six genes coded for voltage-gated potassium channels.
Figure 5.
Transcriptomic effects of DA. A, Volcano plot of all DEGs in control, 20 min, and 1 h DA treatment conditions. Significantly enriched genes (log2 fold change >2) are color-coded in red (upregulated genes) and blue (downregulated genes) with FDR < 0.0001 (horizontal line). There is an abundance of immediate early gene family members among the highly upregulated genes. B, A gene ontology analysis of over-represented “biological process” categories in the 20 min condition. The fraction of the total number of all over-represented (upregulated and downregulated) genes is indicated for each category in parentheses.
Functional effects of chronic DA
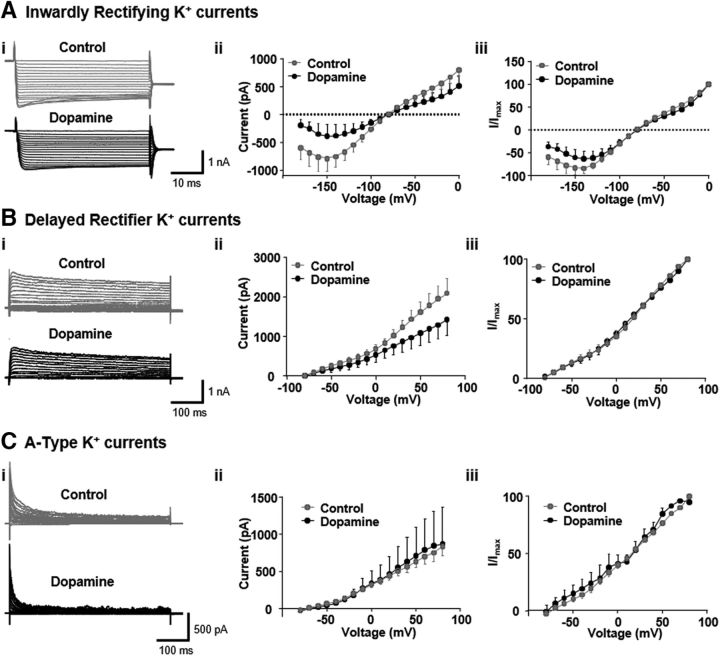
We investigated whether astrocytic voltage-gated potassium channels, involved in regulation of K+ ion homeostasis (Sontheimer, 1994), are affected by DA treatment. In whole-cell patch-clamp recordings of cells exposed to control or DA-containing media for 60 min, we found no main effect of DA treatment on the amplitude of the following: inwardly rectifying (Kir) currents (F(1,13) = 1.5, p = 0.24), delayed rectifier (Kdr) currents (F(1,12) = 1.44, p = 0.25), or the A-type (IA) currents (F(1,11) = 0.01, p = 0.9) (Fig. 6Ai,Aii,Bi,Bii,Ci,Cii). The only significant effect was an interaction between DA treatment and the injected voltage step for Kir currents (F(18,234) = 1.79, p = 0.03) (Fig. 6Ai–Aii). When Kir, Kdr, and IA data were normalized to the maximal current amplitude in each cell, we observed no significant main effects of DA treatment or treatment by voltage interactions (Fig. 6Aiii,Biii,Ciii). The resting membrane potential (RMP) was not affected by DA treatment (control RMP = −67.58 ± 1.72 mV; DA RMP = −66.8 ± 0.93 mV, t(15) = 0.3, p = 0.75, Student's t test). A decrease in membrane capacitance after DA failed to reach statistical significance (control = 17.1 ± 1.8 pF; DA = 11.72 ± 1.9 pF, t(16) = 1.99, p = 0.06). However, DA treatment significantly increased membrane resistance, although this was highly variable between cells (control Rm = 77.6 ± 36.35 mΩ; DA Rm = 312.87 ± 103.77 mΩ, t(13) = 2.43, p = 0.03, Student's t test). The increase in Rm and the trend toward decreased Cm are consistent with a reduction in GFAP+ cell area seen in immunocytochemical experiments.
Figure 6.
DA treatment does not affect astrocytic voltage-gated potassium channels. Ai, Bi, Ci, Representative traces from astrocytes incubated in control- or DA (75 μm)-containing media showing Kir (inwardly-rectifying), Kdr (delayed-rectifier), and IA (A-type) K+ currents. Aii, Bii, Cii, Corresponding I–V plots illustrate mean amplitude ± SEM of currents for each voltage-gated conductance. Aiii, Biii, Ciii, Same as in Aii, Bii, Cii, but with currents normalized to maximal recorded current in each cell: Kir control (n = 9 cells), DA (n = 6 cells); Kdr control (n = 8 cells), DA (n = 6 cells); IA control (n = 7 cells), DA (n = 6 cells). The only significant difference is an interaction in Aii (see Results).
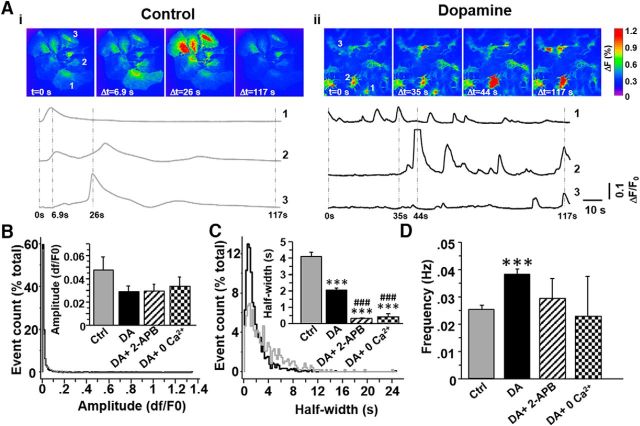
Given the contribution of genes that regulate Ca2+ ion homeostasis in the RNA seq data and functional relevance of intracellular Ca2+, we also evaluated astrocytic Ca2+ transients. To do so, we monitored fluorescence of the GFAP-driven Ca2+ sensor, GCamp6f, after exposing the cells to control or DA-containing media for 60–90 min. DA exposure had only a mild effect on the amplitude of spontaneous Ca2+ events reducing them from 4.7 ± 1.1% of baseline fluorescence in control conditions to 2.9 ± 0.5% of baseline fluorescence after incubation with DA (p = 0.08, Student's t test; Fig. 7A,B). However, we observed pronounced effects on duration and frequency of astrocytic Ca2+ transients. In control conditions, spontaneous Ca2+ events were long, lasting an average of 4.1 ± 0.25 s and occurred at a frequency of 0.025 ± 0.001 Hz. Incubation with DA (75 μm) significantly reduced the duration of spontaneous Ca2+ transients to 2.05 ± 0.11 s (p = 1 × 10−16, Student's t test), but increased their occurrence to 0.038 ± 0.002 Hz (p = 4.6 × 10−7, Student's t test; Fig. 7A,C,D). A 10 min preincubation with the IP3 receptor blocker 2-APB (50 μm) before the 60 min treatment with DA + 2-APB reduced the number of cells displaying spontaneous Ca2+ transients (from 68% of cells after DA alone to 19% of cells after DA + 2-APB), had no effect on event amplitude or frequency, and led to a marked decrease in event duration (Fig. 7B–D). Broad reduction in number of spontaneously active astrocytes is consistent with the IP3-mediated Ca2+ release, although additive effects of DA and 2-APB on event duration may suggest also an IP3 receptor-independent component (Agarwal et al., 2017). In Ca2+-free media, a very small number of cells (5%, 4 cells) continued to display spontaneous GCamp6f fluorescence. We observed a total of 11 fluorescence transients in Ca2+-free media with amplitude, frequency, and event duration similar to those measured in the presence of 2-APB (Fig. 7B–D). Together, these results indicate that a 60 min exposure to DA modulates duration and frequency of astrocyte Ca2+ signals. However, DA treatment has little to no effect on activity of astrocyte voltage-gated K+ channels.
Figure 7.
Effects of DA treatment on astrocytic Ca2+ signals. A, Representative “heat map” frames of GCamp6f fluorescence after a 1 h exposure to control (Ai) or DA-containing (Aii) media. Ca2+ fluorescence traces for the three numbered astrocytes are presented below the image frames. Vertical dotted lines are drawn at intervals corresponding to the image frames. B, C, Cumulative distribution histograms summarizing the effects of DA on amplitude and duration (half-width) of all recorded spontaneous Ca2+ transients: control (n = 473 evens); DA (n = 1429 events); DA + 2-APB (n = 53 events); DA + 0 Ca2+ (n = 11 events). Insets, Mean amplitude and duration of Ca2+ events averaged across all identified astrocytes: control (n = 172 cells); DA (n = 325 cells); DA + 2-APB (n = 15 cells); DA + 0 Ca2+ (n = 3 cells). ***p < 0.001 versus control (Student's t test). ###p < 0.001 versus DA (Student's t test). D, Bar histogram illustrating DA-induced increase in frequency of spontaneous Ca2+ transients. ***p < 0.001 (Student's t test, t(504) = 5.1).
DA signaling in astrocytes triggers transcription of CTCF target genes
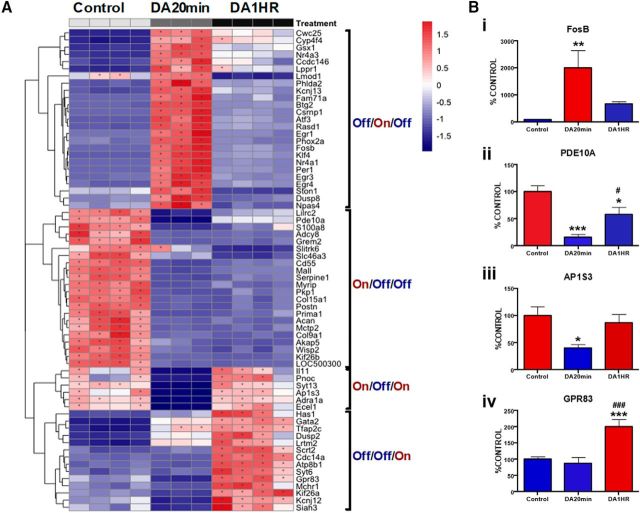
Having confirmed gene ontology results that both morphological and ionic regulatory processes were selectively induced by DA, we further analyzed our RNA-Seq data for mechanistic indicators of potential regulatory processes. To do this, we first plotted the top differentially enriched transcripts from the control, DA 20 min, and DA 60 min conditions in relation to each other (30 genes from each category resulting in 67 distinct genes). This revealed a clear effect of DA treatment and four temporally discrete patterns of transcriptomic regulation (“off/on/off,” “on/off/off,” “on/off/on,” and “off/off/on”; Fig. 8A). To support these results, we used RT-PCR to evaluate levels of select differentially enriched transcripts in a biological replicate. In line with RNA seq data, we observed the expected regulation of FosB (off/on/off), Pde10a (on/off/off), Ap1s3 (on/off/on), and Gpr83 (off/off/on) following treatment with DA (Fig. 8Bi–Biv). It is notable that, in addition to FosB, the “off/on/off” category prominently featured transient activation of other immediate early gene family members (e.g., Nr4a3, Atf3, Egr1, Nr4a1, Egr4) among the top differentially enriched transcripts.
Figure 8.
DA regulation of astrocyte transcriptome is heterogeneous and temporally dynamic. A, Clustergram characterizing top DEGs based on expression changes between treatments. Bi–Biv, qPCR validation of representative genes from each class of the DEGs indicated in A. Data are mean ± SEM. One-way ANOVAs: FosB (F(2,21) = 7.2, p < 0.01); Pde10A (F(2,22) = 14.6, p < 0.001); Ap1s3 (F(2,19) = 4.4, p < 0.05); Gpr83 (F(2,20) = 19.9, p < 0.001). Bonferroni post hoc: ***p < 0.001 versus control. **p < 0.01 versus control. *p < 0.05 versus control. ###p < 0.001 versus DA 20 min. #p < 0.05 versus DA 20 min.
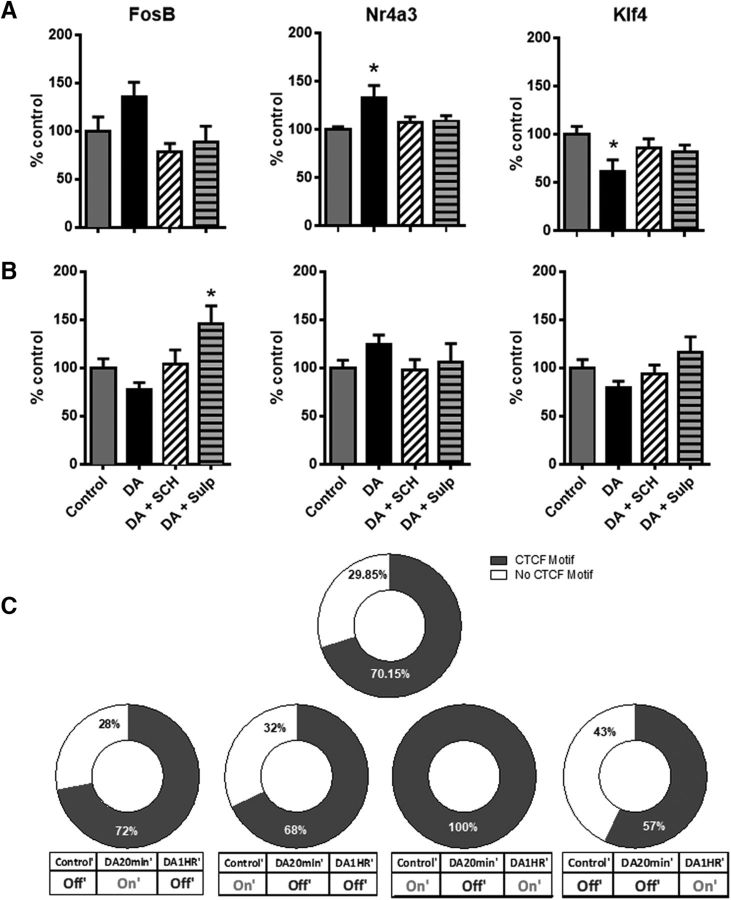
We next examined whether DA regulates astrocytic gene expression ex vivo in hippocampal and striatal brain slices. To do this, we examined genes present in our in vitro RNA sequencing dataset but selectively enriched in astrocytes ex vivo as reported by Zhang et al. (2014). Acute hippocampal and striatal slices were exposed to DA (75 μm + 50 μm SMBS) for 20 min with or without 10 min pretreatment with D1-like DA receptor antagonist, SCH23390 (50 μm), or the D2-like DA receptor antagonist, sulpiride (20 μm). The results support our in vitro observations that DA induces distinct patterns of transcriptomic regulation of astrocyte-enriched genes. Exposure to DA led to a significant increase in expression of Nr4a3 and significantly downregulated expression of Klf4 in hippocampal slices (Fig. 9A). However, in striatal slices, treatment with DA did not result in significant differential regulation of any of the genes screened (Fig. 9B), suggesting that there may be inherent differences among astrocytes from discrete brain regions. Furthermore, all DA-induced alterations in gene expression were absent in those slices preincubated with either D1-like or D2-like DA receptor antagonists, confirming the role of both D1- and D2-like receptors in mediating DA effects on the astrocyte transcriptome (Fig. 9A,B).
Figure 9.
DA exposure regulates astrocytically enriched CTCF response genes in rat hippocampal and striatal slices. A, qPCR analysis of astrocyte-enriched genes from hippocampal slices (n = 9–12 slice experiments from 7 rats). Data are mean ± SEM. *p < 0.05. B, qPCR screening of astrocyte-enriched genes from striatal slice (n = 9–12 slice experiments from 7 rats). Data are mean ± SEM. *p < 0.05. C, CTCF binding motif analysis. Top, Donut chart represents CTCF binding motif analysis of all top DEGs. Bottom, Four charts represent similar analysis, but for each of the DEG classes indicated in Figure 8A, with the validated genes in Figure 8B, Figure 9A, and Figure 9B included.
In neurons, discrete gene activation patterns have been attributed to modifications in the chromatin accessibility landscape (Su et al., 2017). One protein shown to dynamically coordinate chromatin accessibility and genomic organization in the brain is CTCF, which both regulates the 3D structure of chromatin as well as defines the boundaries between active and heterochromatic DNA (Phillips and Corces, 2009; Sams et al., 2016; Su et al., 2017). To evaluate whether CTCF could mediate DA-elicited transcriptomic changes in astrocytes, we examined promoters of the top DEGs (Fig. 8A) for CTCF motifs. As shown in Figure 9C, the results indicated that 70.15% of the top DEGs possessed a binding motif for CTCF, a substantial increase from the expected genome-wide CTCF binding site prevalence of ∼33% (Holwerda and de Laat, 2013). Notably, genes with the CTCF motif included those validated by qPCR in vitro (Figs. 8Bi–Biv) and ex vivo (Fig. 9A,B). Furthermore, all of the genes with the “on/off/on” pattern of DA-induced transcriptional regulation possessed the CTCF binding motif, whereas genes with the “off/off/on” pattern had the lowest CTCF motif prevalence.
PARP1-mediated CTCF regulation recapitulates morphogenic effects of DA
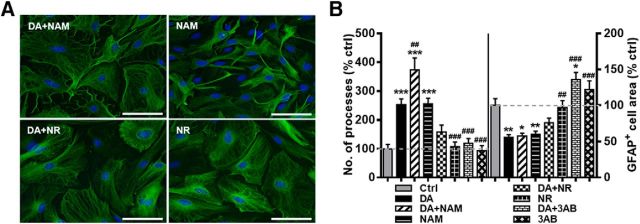
CTCF triggers transcriptional changes by interacting with PARP1 (Yu et al., 2004; Ong et al., 2013; Zhao et al., 2015; Han et al., 2017), which has been implicated in behavioral and molecular effects of cocaine (Scobie et al., 2014). PARP1 both facilitates NAD+-dependent generation of ATP for chromatin remodeling (Wright et al., 2016) and regulates CTCF binding to target genes via NAD+-dependent poly-ADP-ribosylation of CTCF (Yu et al., 2004; Ong et al., 2013; Han et al., 2017). Although there are no direct pharmacological tools for CTCF, commonly used NAD+ precursors, nicotinamide and nicotinamide riboside, exhibit opposite effects on PARP1 activity due to their disparate effects on SIRT1, the negative regulator of PARP1 (Bitterman et al., 2002; Rajamohan et al., 2009; Sajish and Schimmel, 2015; Trammell et al., 2016). This results in nicotinamide riboside acting as an “antagonist” of PARP1 and, conversely, nicotinamide acting as an “agonist” of PARP1. We took advantage of this distinction to determine whether DA-induced morphological changes could be linked to PARP1 activation. To accomplish this, astrocytes were preincubated with nicotinamide (5 mm) or nicotinamide riboside (1 mm) for 15 min before a 60 min incubation with DA (75 μm) or control media. Nicotinamide increased astrocyte stellation and decreased GFAP+ area in the absence of DA (Fig. 10). In the presence of DA, nicotinamide enhanced stellation beyond that seen with DA alone but maintained reduction in GFAP+ area at levels similar to that seen with DA alone. Nicotinamide riboside had no effect on astrocyte stellation or GFAP+ area when applied on its own, but occluded changes in both stellation and GFAP+ area when applied in combination with DA (Fig. 10). Similarly, preincubation with PARP inhibitor, 3-aminobenzamide (3-AB; 5 mm), completely blocked DA-induced stellation, but 3-AB had no effect when applied on its own. However, 3-AB increased GFAP+ cell area when applied on its own or in combination with DA (Fig. 10B). Another, more potent PARP1 inhibitor, AG-14361 (10 μm), reduced DA-induced stellation (160 ± 21.4% of control values) but had no effect on GFAP+ cell area (101 ± 9.5% of control values). When applied alone, AG-14361 had similar effects on astrocyte stellation and GFAP+ area as it did in combination with DA (stellation: 142 ± 18.7% of control; GFAP+ area: 127 ± 38.1% of control). Together, these results indicate that DA-induced morphological changes in cultured astrocytes rely on PARP1 activation, resulting in recruitment of CTCF to target genes and remodeling of astrocyte transcriptome.
Figure 10.
Nicotinamide recapitulates morphological effects of DA. A, Representative images of GFAP+ astrocytes following exposure to DA and nicotinamide (DA+NAM), DA and nicotinamide riboside (DA+NR), nicotinamide alone (NAM), or nicotinamide riboside alone (NR). Scale bars, 75 μm. B, Quantification of average number of primary processes and average GFAP+ cell area. One-way ANOVA for number of processes: F(7,324) = 19.89, p < 0.001; one-way ANOVA for GFAP area: F(7,324) = 15.79, p < 0.001; Bonferroni post hoc: *p < 0.05 versus control. **p < 0.01 versus control. ***p < 0.001 versus control. ##p < 0.01 versus DA. ###p < 0.001 versus DA.
Discussion
In this study, we used primary cultures and brain slices to characterize morphological, molecular, genetic, and functional effects of DA exposure on astrocytes. We found that elevated extracellular DA induced pronounced stellation of astrocytic processes and a reduction in astrocyte area positive for the intermediate filament marker, GFAP. These DA effects were blunted by exposure to DA receptor antagonists or by inhibition of PARP1, suggesting a DA receptor-mediated, PARP1-dependent signaling as the source of morphological changes. DA exposure altered astrocytic Ca2+ signaling but did not disrupt astrocyte regulation of K+ ion homeostasis at the level of voltage-gated K+ channel function. Critically, we demonstrated, for the first time, that CTCF target genes are a prominent marker of astrocytic transcriptional response to DA.
DA-mediated morphological changes
This study focused on two morphological features of cultured astrocytes, primary process stellation and GFAP+ area, as notable responses to DA treatment (Fig. 1). Subsequent experimentation revealed these two morphological measures to be dissociable. For example, process stellation was strongest after treatment with high DA concentrations, whereas increase in GFAP+ area was strongest after treatment with low DA concentrations (Fig. 2). Similarly, profound suppression of GFAP+ cell area following combined treatment with DA and antioxidants contrasted with the lack of changes in process number following a similar treatment (Fig. 3).
Treatments with cAMP analogs have been observed to trigger morphological changes in cultured astrocytes (Moonen et al., 1975; Won and Oh, 2000; Paco et al., 2016). However, sensitivity of astrocyte morphology to elevated DA has, to our knowledge, only been reported in one other study (Koppel et al., 2018). In that report, DA-induced stellation of cortical astrocytes was blocked by propranolol, a β adrenoreceptor antagonist. Our results support a direct role of DA receptors in regulation of astrocyte morphology. Thus, we found that both D1-like receptor antagonist, SCH23390, or D2-like receptor antagonist, sulpiride, were effective at blunting DA-induced stellation of astrocyte processes (Fig. 4). The results with sulpiride were surprising given the classical notion that D2-like receptor activation decreases cytoplasmic cAMP levels. A possible explanation could involve a novel phosphatidylinositol-linked D1-like receptor found in both astrocytes and neurons, which couples to PLC by Gαq and increases intracellular Ca2+ levels (Ming et al., 2006; Liu et al., 2009). Another explanation could involve an interaction of the antagonists with Gq-PLC pathway-coupled D1/D2 receptor heterooligomers (Lee et al., 2004; Rashid et al., 2007; Hasbi et al., 2010; Beaulieu and Gainetdinov, 2011; Jennings and Rusakov, 2016). Indeed, we show that blockade of IP3 receptors activated downstream of PIP2 hydrolysis by PLC reduced DA-induced astrocyte stellation. Therefore, our results suggest that cAMP increase may not be necessary for DA-induced stellation.
Does DA induce astrocyte reactivity?
DA can oxidize in culture media (Clement et al., 2002), and reactive oxygen species can trigger morphological changes in astrocytes, including cell hypertrophy, swelling/thickening of processes, and increased GFAP expression, collectively referred to as reactive astrocytosis (Vaarmann et al., 2010). We also found individual processes to be thicker following DA treatment; however, morphological changes associated with DA exposure persisted in the presence of antioxidants, NAC or SMBS, arguing against oxidative process-mediated reactivity. Moreover, we observed variable, but consistent, downregulation of GFAP+ area (Figs. 1–4, 10), arguing against swelling of existing processes as mediators of DA-induced morphological changes. Furthermore, RNA seq indicated only small changes in GFAP transcript (log2fold value of 1.2 at 60 min of treatment with DA), a result that was confirmed with qRT-PCR (data not shown). Finally, cross-referencing with published reactive astrocyte transcriptome, we found little to no overlap with our RNA seq data. Of the top 100 reactive astrocyte genes in Zamanian et al. (2012), three were enriched at above log2fold level in our dataset (Gdf15, Srxn1, and Asns). Lcn2 and Serpina3n identified as the prominent novel markers of astrocytic reactivity (Zamanian et al., 2012) displayed minimal changes in our dataset after 1 h of treatment with DA (log2fold values of 0.1 and −0.9, for Lcn2 and Serpina3n, respectively). Therefore, in conjunction with prior studies that support anti-inflammatory effects of DA (Beck et al., 2004; Shao et al., 2013; Yan et al., 2015), we conclude that DA treatment did not promote a reactive state that could account for the observed morphological or transcriptome changes in our cultured astrocytes. This, however, could be different for astrocytes that experience withdrawal from elevated DA as reported in animals withdrawn from experimenter or self-administered cocaine (Fattore et al., 2002; Bowers and Kalivas, 2003).
Astrocyte sensitivity to duration of DA exposure
One line of evidence that supports duration of exposure as a factor relevant to DA-mediated response comes from our functional analysis of astrocyte Ca2+ signaling. Substantial evidence indicates that DA plays a role in modulation of Ca2+ signaling in astrocytes. Specifically, acute application of DA or DA receptor agonists, such as SKF83959, to astrocytes in culture has generally been found to induce elevations in cytosolic Ca2+ via an IP3-mediated release from intracellular stores (Parpura and Haydon, 2000; Lee et al., 2004; Liu et al., 2009; Vaarmann et al., 2010; Jennings and Rusakov, 2016; Jennings et al., 2017). Further, astrocytes from cortical and hippocampal cultures express a distinct phosphatidylinositol-linked D1-like receptor coupled to PLC by Gαq, which upon activation leads to an increase in intracellular calcium levels (Liu et al., 2009; Ming et al., 2006). Our results indicate that a 1 h treatment with DA decreased the duration and amplitude of Ca2+ transients, the majority of which were dependent on IP3 receptor activation (Fig. 7). However, a 1 h DA treatment increased Ca2+ event frequency (Fig. 7). The distinction between acute and chronic effects of DA receptor stimulation has been reported in studies of NMDA-receptor mediated neuronal signaling (Schilström et al., 2006; Ortinski et al., 2013, 2014). For example, Schilström et al. (2006) indicated that a brief exposure to a D1 agonist, SKF81297, produces a delayed increase in NMDA receptor-mediated currents with an even longer delay observed after blockade of DA transport. Given the well-documented behavioral and molecular differences driven by temporal pattern of exposure to elevated DA (Knackstedt et al., 2010; Ortinski et al., 2012), understanding the extent of astrocytic contribution to these differences is an important future direction. Our sequencing data indicate that, even at relatively short timescales (20 vs 60 min), several distinct patterns of gene regulation can be distinguished at the genomic level (Fig. 8). This is characterized most prominently by a sustained upregulation of genes involved in structure morphogenesis, kinase signaling, and signal transduction following a long exposure to DA and a transient upregulation of immediate early genes after a brief exposure to DA.
DA regulation of astrocyte transcriptome
DA had distinct effects on the transcriptome of cultured astrocytes at 20 and 60 min after treatment. Additionally, we observed that DA regulates expression of astrocyte-enriched transcripts in acutely isolated brain slices and that these effects may be attenuated by antagonists at D1- and D2-DA receptors. The differences in DA effects on RNA levels between striatal and hippocampal slices reported here extend previous work from Chai et al. (2017), which detailed DEG profiles in these two structures. Approximately three-fourths of the top 40 genes enriched in both hippocampus and striatum in Chai et al. (2017) experiments are represented in our RNA-Seq dataset, indicating substantial overlap between ex vivo and in vitro observations at the transcriptome level. Furthermore, the genes that we validated by qPCR (FosB, Klf4, Nr4a3, Pde10a, Gpr83, Ap1s3) all possess a CTCF binding motif. Indeed, our RNA-Seq data analysis reported over-representation of CTCF binding motifs, indicating a coordinated gene network response to DA signaling.
DA and NAD+/PARP1-dependent regulation of CTCF target genes
We find that the dramatic alterations in astrocyte morphology triggered by DA treatment may be due to significant modifications in chromatin accessibility. Across the genome, the expected genome-wide binding site prevalence of CTCF is ∼33% (Holwerda and deLaat, 2013). However, prevalence of CTCF binding motifs in the top genes significantly altered by DA treatment was found to be between 70% and 100%. Complementing the evidence showing that CTCF orchestrates genomic reorganization underlying learning and memory (Sams et al., 2016), we highlight a possible NAD+/PARP1-role linking DA signaling in astrocytes to epigenetic regulation of the chromatin landscape.
Our data suggest that DA activates PARP1, a NAD+-dependent poly-ADP-ribose polymerase that promotes CTCF binding to transcription initiation sites (Yu et al., 2004; Ong et al., 2013; Zhao et al., 2015; Han et al., 2017), leading to changes in astrocyte morphology through alterations in 3D chromatin architecture. We speculate that protein kinase C is a likely mediator of coupling between DA receptors and PARP1/CTCF activation. Both D1- and D2-like DA receptors can stimulate protein kinase C (Neve et al., 2004; Ortinski et al., 2015) and protein kinase C is, in turn, known to promote PARP1 production and formation of PARP1/CTCF complexes (Henderson et al., 2017). We find that PARP1 agonist, nicotinamide, recapitulates, whereas PARP1 antagonists, nicotinamide riboside, 3-AB, or AG-14361, occlude DA-induced stellation and DA-induced decrease in GFAP+ area (Fig. 10). Our observation, that coadministration of DA and nicotinamide results in more pronounced effects than treatment with either DA or nicotinamide alone, suggests that DA may not recruit all of the available PARP1 pool. Furthermore, the nicotinamide/DA synergism supports PARP1-dependent generation of nicotinamide (Sajish and Schimmel, 2015) rather than regulation of PARP1 protein levels. This is indirectly supported by the sequencing data indicating a lack of PARP1 transcript enrichment following either 20 or 60 min of DA treatment (log2fold values of −0.130 and −0.134, respectively). Our observation that nicotinamide riboside or AG-14361 did not evoke any morphological changes by themselves and only partially suppressed morphological effects of DA treatment suggests that DA effects on astrocyte morphology involve both PARP1/NAD+-dependent as well as PARP1/NAD+-independent processes. Indeed, a recent finding demonstrated that a 3 h DA treatment of cortical astrocytes induced expression of BDNF (Koppel et al., 2018), a SIRT1-dependent process (Zocchi and Sassone-Corsi, 2012). SIRT1 and PARP1 are mutually inhibitory (Sajish and Schimmel, 2015), but both SIRT1 and PARP1 rely on NAD+ as a substrate, potentially leading to a cyclical pattern of SIRT1/PARP1 activation, which could serve as the basis of astrocyte sensitivity to duration of DA exposure. DA-induced inhibition of specific sets of genes that lack CTCF binding sites in their promoter regions, highlighted by our sequencing data, may be an indicator of such a relationship.
In conclusion, we provide evidence that DA signaling drives pronounced structural and functional adaptations in cultured astrocytes. These adaptations are associated with distinct short- and long-term changes in the astrocyte genome that are consistent with transcriptional effects of the PARP1/CTCF complex and PARP1-mediated NAD+ signaling. The impact of dynamic astrocyte response to DA on neuronal energy homeostasis, neuronal signaling, and behavioral output in the context of DA-related disorders remains to be examined.
Footnotes
This work was supported by the National Institutes of Health Grants DA031747 and DA041513 to P.I.O., and Grant DA032681 to J.R.T. We thank Dr. Rosemarie Booze and Dr. Marina Aksenova (University of South Carolina) for help with generation of hippocampal astrocyte cultures.
The authors declare no competing financial interests.
References
- Agarwal A, Wu PH, Hughes EG, Fukaya M, Tischfield MA, Langseth AJ, Wirtz D, Bergles DE (2017) Transient opening of the mitochondrial permeability transition pore induces microdomain calcium transient in astrocyte processes. Neuron 93:587–605.e7. 10.1016/j.neuron.2016.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Gainetdinov RR (2011) The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev 63:182–217. 10.1124/pr.110.002642 [DOI] [PubMed] [Google Scholar]
- Beck GC, Brinkkoetter P, Hanusch C, Schulte J, van Ackern K, van der Woude FJ, Yard BA (2004) Clinical review: immunomodulatory effects of dopamine in general inflammation. Crit Care 8:485–491. 10.1186/cc2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA (2002) Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem 277:45099–45107. 10.1074/jbc.M205670200 [DOI] [PubMed] [Google Scholar]
- Booth HD, Hirst WD, Wade-Martins R (2017) The role of astrocyte dysfunction in Parkinson's disease pathogenesis. Trends Neurosci 40:358–370. 10.1016/j.tins.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers MS, Kalivas PW (2003) Forebrain astroglial plasticity is induced following withdrawal from repeated cocaine administration. Eur J Neurosci 17:1273–1278. 10.1046/j.1460-9568.2003.02537.x [DOI] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Jones YZ, Ellisman MH (2002) Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J Neurosci 22:183–192. 10.1523/JNEUROSCI.22-01-00183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Ellisman MH (2004) Maturation of astrocyte morphology and the establishment of astrocyte domains during postnatal hippocampal development. Int J Dev Neurosci 22:73–86. 10.1016/j.ijdevneu.2003.12.008 [DOI] [PubMed] [Google Scholar]
- Chai H, Diaz-Castro B, Shigetomi E, Monte E, Octeau JC, Yu X, Cohn W, Rajendran PS, Vondriska TM, Whitelegge JP, Coppola G,Khakh BS (2017) Neural circuit-specialized astrocytes: transcriptomic, proteomic, morphological, and functional evidence. Neuron 95:531–549.e9. 10.1016/j.neuron.2017.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KH, Wiley CA, Bradberry CW (2013) Psychostimulant abuse and neuroinflammation: emerging evidence of their interconnection. Neurotox Res 23:174–188. 10.1007/s12640-012-9334-7 [DOI] [PubMed] [Google Scholar]
- Clarke LE, Barres BA (2013) Emerging roles of astrocytes in neural circuit development. Nat Rev Neurosci 14:311–321. 10.1038/nrn3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement MV, Long LH, Ramalingam J, Halliwell B (2002) The cytotoxicity of dopamine may be an artefact of cell culture. J Neurochem 81:414–421. 10.1046/j.1471-4159.2002.00802.x [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattore L, Puddu MC, Picciau S, Cappai A, Fratta W, Serra GP, Spiga S (2002) Astroglial in vivo response to cocaine in mouse dentate gyrus: a quantitative and qualitative analysis by confocal microscopy. Neuroscience 110:1–6. 10.1016/S0306-4522(01)00598-X [DOI] [PubMed] [Google Scholar]
- Freeman MR. (2010) Specification and morphogenesis of astrocytes. Science 330:774–778. 10.1126/science.1190928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DS, Jinsmaa Y, Sullivan P, Sharabi Y (2017) N-acetylcysteine prevents the increase in spontaneous oxidation of dopamine during monoamine oxidase inhibition in PC12 cells. Neurochem Res 42:3289–3295. 10.1007/s11064-017-2371-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte R, Liu Z, Kraus WL (2017) PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev 31:101–126. 10.1101/gad.291518.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Chen Q, Shi J, Zhang F, Yu X (2017) CTCF participates in DNA damage response via poly(ADP-ribosyl)ation. Sci Rep 7:43530. 10.1038/srep43530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasbi A, O'Dowd BF, George SR (2010) Heteromerization of dopamine D2 receptors with dopamine D1 and D5 receptors generates intracellular calcium signaling by different mechanism. Curr Opin Phamacol 10:93–99. 10.1016/j.coph.2009.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson DJ, Miranda JL, Emerson BM (2017) The β-NAD+ salvage pathway and PKC-mediated signaling influence localized PARP-1 activity and CTCF poly(ADP)ribosylation. Oncotarget 8:64698–64713. 10.18632/oncotarget.19841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holwerda SJ, de Laat W (2013) CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos Trans R Soc Lond B Biol Sci 368:20120369. 10.1098/rstb.2012.0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings A, Rusakov DA (2016) Do astrocytes respond to dopamine? Med Physiol 2:10. [Google Scholar]
- Jennings A, Tyurikova O, Bard L, Zheng K, Semyanov A, Henneberger C, Rusakov DA (2017) Dopamine elevates and lowers astroglial Ca2+ through distinct pathways depending on local synaptic circuitry. Glia 65:447–459. 10.1002/glia.23103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel E, Jessell T, Siegelbaum S (eds) (2012) Principles of neural science, Ed 5 McGraw Hill, New York, USA. [Google Scholar]
- Khakh BS, Sofroniew MV (2015) Diversity of astrocyte functions and phenotypes in neural circuits. Nat Neurosci 18:942–952. 10.1038/nn.4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan ZU, Koulen P, Rubinstein M, Grandy DK, Goldman-Rakic PS (2001) An astroglia-linked dopamine D2-receptor action in prefrontal cortex. Proc Natl Acad Sci U S A 98:1964–1969. 10.1073/pnas.98.4.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knackstedt LA, Moussawi K, Lalumiere R, Schwendt M, Klugmann M, Kalivas PW (2010) Extinction training after cocaine self-administration induces glutamatergic plasticity to inhibit cocaine seeking. J Neurosci 30:7984–7992. 10.1523/JNEUROSCI.1244-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel I, Jaanson K, Klasche A, Tuvikene J, Tiirik T, Pärn A, Timmusk T (2018) Dopamine cross-reacts with adrenoreceptors in cortical astrocytes to induce BDNF expression, CREB signaling and morphological transformation. Glia 66:206–216. 10.1002/glia.23238 [DOI] [PubMed] [Google Scholar]
- Lee SP, So CH, Rashid AJ, Varghese G, Cheng R, Lança AJ, O'Dowd BF, George SR (2004) Dopamine D1 and D2 receptor co-activation generates novel phospholipase C-mediated calcium signal. J Biol Chem 279:35671–35678. 10.1074/jbc.M401923200 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G,Durbin R (2009) The sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu J, Wang F, Huang C, Long LH, Wu WN, Cai F, Wang JH, Ma LQ, Chen JG (2009) Activation of phosphatidylinositol-linked novel D1 dopamine receptor contributes to the calcium mobilization in cultured rat prefrontal cortical astrocytes. Cell Mol Neurobiol 29:317–328. 10.1007/s10571-008-9323-9 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-δ C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RH, Raff MC (1984) Fibrous and protoplasmic astrocytes are biochemically and developmentally distinct. J Neurosci 4:585–592. 10.1523/JNEUROSCI.04-02-00585.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming Y, Zhang H, Long L, Wang F, Chen J, Zhen X (2006) Modulation of Ca2+ signals by phosphatidylinositol-linked novel D1 dopamine receptor in hippocampal neurons. J Neurochem 98:1316–1323. 10.1111/j.1471-4159.2006.03961.x [DOI] [PubMed] [Google Scholar]
- Miyazaki I, Asanuma M, Diaz-Corrales FJ, Miyoshi K, Ogawa N (2004) Direct evidence for expression of dopamine receptors in astrocytes from basal ganglia. Brain Res 1029:120–123. 10.1016/j.brainres.2004.09.014 [DOI] [PubMed] [Google Scholar]
- Mladinov M, Mayer D, Brcic L, Wolstencroft E, Man NT, Holt I, Simic G (2010) Astrocyte expression of D2-like dopamine receptors in the prefrontal cortex. Transl Neurosci 1:238–243. [Google Scholar]
- Moonen G, Cam Y, Sensenbrenner M, Mandel P (1975) Variability of the effects of serum-free medium, dibutyryl-cyclic AMP or theophylline on the morphology of cultured new-born rat astroblasts. Cell Tissue Res 163:365–372. [DOI] [PubMed] [Google Scholar]
- Nagatomo K, Suga S, Saitoh M, Kogawa M, Kobayashi K, Yamamoto Y, Yamada K (2017) Dopamine D1 receptor immunoreactivity on fine processes of GFAP-positive astrocytes in the substantia nigra pars reticulata of adult mouse. Front Neuroanat 11:3. 10.3389/fnana.2017.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve KA, Seamans JK, Trantham-Davidson H (2004) Dopamine receptor signaling. J Recept Signal Transduct Res 24:165–205. 10.1081/RRS-200029981 [DOI] [PubMed] [Google Scholar]
- Oberheim NA, Goldman SA, Nedergaard M (2012) Heterogeneity of astrocytic form and function. Methods Mol Biol 814:23–45. 10.1007/978-1-61779-452-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Van Bortle K, Ramos E, Corces VG (2013) Poly(ADP-ribosyl)ation regulates insulator function and intrachromosomal interactions in Drosophila. Cell 155:148–159. 10.1016/j.cell.2013.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortinski PI. (2014) Cocaine-induced changes in NMDA receptor signaling. Mol Neurobiol 50:494–506. 10.1007/s12035-014-8636-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortinski PI, Vassoler FM, Carlson GC, Pierce RC (2012) Temporally dependent changes in cocaine-induced synaptic plasticity in the nucleus accumbens shell are reversed by D1-like dopamine receptor stimulation. Neuropsychopharmacology 37:1671–1682. 10.1038/npp.2012.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortinski PI, Turner JR, Pierce RC (2013) Extrasynaptic targeting of NMDA receptors following D1 dopamine receptor activation and cocaine self-administration. J Neurosci 33:9451–9461. 10.1523/JNEUROSCI.5730-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortinski PI, Briand LA, Pierce RC, Schmidt HD (2015) Cocaine-seeking is associated with PKC-dependent reduction of excitatory signaling in accumbens shell D2 dopamine receptor-expressing neurons. Neuropharmacology 92:80–89. 10.1016/j.neuropharm.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paco S, Hummel M, Plá V, Sumoy L, Aguado F (2016) Cyclic AMP signaling restricts activation and promotes maturation and antioxidant defenses in astrocytes. BMC Genomics 17:304. 10.1186/s12864-016-2623-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parpura V, Haydon PG (2000) Physiological astrocytic calcium levels stimulate glutamate release to modulate adjacent neurons. Proc Natl Acad Sci U S A 97:8629–8634. 10.1073/pnas.97.15.8629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekny M, Nilsson M (2005) Astrocyte activation and reactive gliosis. Glia 50:427–434. 10.1002/glia.20207 [DOI] [PubMed] [Google Scholar]
- Perea G, Navarrete M, Araque A (2009) Tripartite synapses: astrocytes process and control synaptic information. Trends Neurosci 32:421–431. 10.1016/j.tins.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG (2009) CTCF: master weaver of the genome. Cell 137:1194–1211. 10.1016/j.cell.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff MC, Abney ER, Cohen J, Lindsay R, Noble M (1983) Two types of astrocytes in cultures of developing rat white matter: differences in morphology, surface gangliosides, and growth characteristics. J Neurosci 3:1289–1300. 10.1523/JNEUROSCI.03-06-01289.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Birukov KG, Samant S, Hottiger MO, Gupta MP (2009) SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol Cell Biol 29:4116–4129. 10.1128/MCB.00121-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid AJ, So CH, Kong MM, Furtak T, El-Ghundi M, Cheng R, O'Dowd BF, George SR (2007) D1–D2 dopamine receptor heterooligomers with unique pharmacology are coupled to rapid activation of Gq/11 in the striatum. Proc Natl Acad Sci U S A 104:654–659. 10.1073/pnas.0604049104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requardt RP, Wilhelm F, Rillich J, Winkler U, Hirrlinger J (2010) The biphasic NAD(P)H fluorescence response of astrocytes to dopamine reflects the metabolic actions of oxidative phosphorylation and glycolysis. J Neurochem 115:483–492. 10.1111/j.1471-4159.2010.06940.x [DOI] [PubMed] [Google Scholar]
- Requardt RP, Hirrlinger PG, Wilhelm F, Winkler U, Besser S, Hirrlinger J (2012) Ca2+ signals of astrocytes are modulated by the NAD(+)/NADH redox state. J Neurochem 120:1014–1025. 10.1111/j.1471-4159.2012.07645.x [DOI] [PubMed] [Google Scholar]
- Sajish M, Schimmel P (2015) A human tRNA synthetase is a potent PARP1-activating effector target for resveratrol. Nature 519:370–373. 10.1038/nature14028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sams DS, Nardone S, Getselter D, Raz D, Tal M, Rayi PR, Kaphzan H, Hakim O,Elliott E (2016) Neuronal CTCF is necessary for basal and experience-dependent gene regulation, memory formation, and genomic structure of BDNF and arc. Cell Rep 17:2418–2430. 10.1016/j.celrep.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Schilström B, Yaka R, Argilli E, Suvarna N, Schumann J, Chen BT, Carman M, Singh V, Mailliard WS, Ron D, Bonci A (2006) Cocaine enhances NMDA receptor-mediated currents in ventral tegmental area cells via dopamine D5 receptor-dependent redistribution of NMDA receptors. J Neurosci 26:8549–8558. 10.1523/JNEUROSCI.5179-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobie KN, Damez-Werno D, Sun H, Shao N, Gancarz A, Panganiban CH, Dias C, Koo J, Caiafa P, Kaufman L, Neve RL, Dietz DM, Shen L, Nestler EJ (2014) Essential role of poly(ADP-ribosyl)ation in cocaine action. Proc Natl Acad Sci U S A 111:2005–2010. 10.1073/pnas.1319703111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield MD, Li H, Siemsen BM, Healey KL, Tran PK, Woronoff N, Boger HA, Kalivas PW, Reissner KJ (2016) Cocaine self-administration and extinction leads to reduced glial fibrillary acidic protein expression and morphometric features of astrocytes in the nucleus accumbens core. Biol Psychiatry 80:207–215. 10.1016/j.biopsych.2015.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Zhang SZ, Tang M, Zhang XH, Zhou Z, Yin YQ, Zhou QB, Huang YY, Liu YJ, Wawrousek E, Chen T, Li SB, Xu M, Zhou JN, Hu G,Zhou JW (2013) Suppression of neuroinflammation by astrocytic dopamine D2 receptors via alphaB-crystallin. Nature 494:90–94. 10.1038/nature11748 [DOI] [PubMed] [Google Scholar]
- Smith AD, Olson RJ, Justice JB Jr (1992) Quantitative microdialysis of dopamine in the striatum: effect of circadian variation. J Neurosci Methods 44:33–41. 10.1016/0165-0270(92)90111-P [DOI] [PubMed] [Google Scholar]
- Sofroniew MV. (2009) Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci 32:638–647. 10.1016/j.tins.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer H. (1994) Voltage-dependent ion channels in glial cells. Glia 11:156–172. 10.1002/glia.440110210 [DOI] [PubMed] [Google Scholar]
- Su Y, Shin J, Zhong C, Wang S, Roychowdhury P, Lim J, Kim D, Ming GL,Song H (2017) Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat Neurosci 20:476–483. 10.1038/nn.4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trammell SA, Schmidt MS, Weidemann BJ, Redpath P, Jaksch F, Dellinger RW, Li Z, Abel ED, Migaud ME, Brenner C (2016) Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat Commun 7:12948. 10.1038/ncomms12948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaarmann A, Gandhi S, Abramov AY (2010) Dopamine induces Ca2+ signaling in astrocytes through reactive oxygen species generated by monoamine oxidase. J Biol Chem 285:25018–25023. 10.1074/jbc.M110.111450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E. (2015) NAD(+) in aging, metabolism, and neurodegeneration. Science 350:1208–1213. 10.1126/science.aac4854 [DOI] [PubMed] [Google Scholar]
- Won CL, Oh YS (2000) cAMP-induced stellation in primary astrocyte cultures with regional heterogeneity. Brain Res 887:250–258. 10.1016/S0006-8993(00)02922-X [DOI] [PubMed] [Google Scholar]
- Wright RH, Lioutas A, Le Dily F, Soronellas D, Pohl A, Bonet J, Nacht AS, Samino S, Font-Mateu J, Vicent GP, Wierer M, Trabado MA, Schelhorn C, Carolis C, Macias MJ, Yanes O, Oliva B, Beato M (2016) ADP-ribose-derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling. Science 352:1221–1225. 10.1126/science.aad9335 [DOI] [PubMed] [Google Scholar]
- Yan Y, Jiang W, Liu L, Wang X, Ding C, Tian Z, Zhou R (2015) Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell 160:62–73. 10.1016/j.cell.2014.11.047 [DOI] [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, Oshimura M, Feinberg AP, Lobanenkov V, Klenova E, Ohlsson R (2004) Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet 36:1105–1110. 10.1038/ng1426 [DOI] [PubMed] [Google Scholar]
- Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, Barres BA (2012) Genomic analysis of reactive astrogliosis. J Neurosci 32:6391–6410. 10.1523/JNEUROSCI.6221-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Doyon WM, Clark JJ, Phillips PE, Dani JA (2009) Controls of tonic and phasic dopamine transmission in the dorsal and ventral striatum. Mol Pharmacol 76:396–404. 10.1124/mol.109.056317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ (2014) An RNA-Seq transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sifakis EG, Sumida N, Millán-Ariño L, Scholz BA, Svensson JP, Chen X, Ronnegren AL, Mallet de Lima CD, Varnoosfaderani FS, Shi C, Loseva O, Yammine S, Israelsson M, Rathje LS, Németi B, Fredlund E, Helleday T, Imreh MP, Göndör A (2015) PARP1- and CTCF-mediated interactions between active and repressed chromatin at the lamina promote oscillating transcription. Mol Cell 59:984–997. 10.1016/j.molcel.2015.07.019 [DOI] [PubMed] [Google Scholar]
- Ziebarth JD, Bhattacharya A, Cui Y (2013) CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res 41(Database issue):D188–D194. 10.1093/nar/gks1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocchi L, Sassone-Corsi P (2012) SIRT1-mediated deacetylation of MeCP2 contributes to BDNF expression. Epigenetics 7:695–700. 10.4161/epi.20733 [DOI] [PMC free article] [PubMed] [Google Scholar]