Abstract
Nitro-conjugated linoleic acid (NO2-CLA) is formed by metabolic and inflammatory reactions of nitric oxide and nitrite, and represents the most abundant nitro-fatty acid species in humans. These electrophilic fatty acid nitroalkene derivatives mediate pleiotropic cell signaling responses. Here, we report a systematic approach to investigate the effect of NO2-CLA on human coronary artery smooth muscle cells (hCASMC), based on the RNA-Seq and bioinformatics analysis. There were extensive differentially expressed genes in NO2-CLA vs. control (510) and NO2-CLA vs. CLA (272) treatment groups, respectively. Notably, only minimal alterations were observed in CLA vs. control conditions, indicating that the electrophilic character of NO2-CLA is requited to induce differential gene expression responses independently from native CLA. Functional enrichment analysis of differentially expressed genes reveals multiple cellular processes to be affected under NO2-CLA treatment, including cell proliferation, lipid metabolism, antioxidant and inflammatory-related gene expression responses. These findings reveal that nitro-fatty acid derivatives such as NO2-CLA regulate a broad array of adaptive gene expression responses by hCASMC.
Keywords: conjugated linoleic acid, nitro-fatty acid, nitroalkene, RNA-Seq, vascular smooth muscle cell
INTRODUCTION
Unsaturated fatty acids are nitrated by nitrogen dioxide (·NO2), a species abundantly formed by nitric oxide (·NO) and nitrite ()-dependent metabolic and inflammatory reactions (18, 22, 23). These reactions, resulting in ·NO2 addition to fatty acid double bonds, yield fatty acid derivatives having electrophilic nitroalkene substituents. This functionality rapidly and reversibly reacts with biological nucleophiles, such as cysteine and histidine, via Michael addition. Nitro-fatty acids (NO2-FA) are derived from the major unsaturated fatty acids and include nitro-oleic (OA-NO2), nitro-linoleic (L-NO2), nitro-linolenic and nitro-arachidonic acids, products that are detectable in a broad array of living organisms including plant and mammalian tissues (2).
Along with other reactive species, electrophilic NO2-FA derivatives modulate diverse gene expression and enzymatic and cell signaling processes via the posttranslational modification of proteins (6, 10, 16). For example, NO2-FA will alkylate functionally significant thiols of the transcription-regulatory proteins Kelch-like ECH-associated protein-1 (Keap1)-nuclear factor-erythroid 2-related factor 2 (Nrf2) (17, 19, 31), which regulate the expression of ~100 antioxidant proteins (6, 14, 34), nuclear factor-κB (NFkB), peroxisome proliferator activating receptor- γ (PPARγ), and heat shock factor-1 (HSF-1) (24). Notably, NO2-FA are endogenous mediators of anti-inflammatory and adaptive tissue signaling responses, thus playing protective roles in cardiovascular diseases and diabetes (6, 29, 30).
Conjugated linoleic acid (CLA) has been identified as the primary endogenous substrate for fatty acid nitration, both in vitro and in vivo (4). Fatty acids such as CLA that have a conjugated diene (every other carbon), rather than a bis-allylic (every third carbon) double bond configuration, are highly susceptible to the addition of ·NO2 to the hyperreactive external flanking carbons of the conjugated double bond system (4). In healthy humans, “free” NO2-CLA levels in plasma and urine range from 2 to 20 nM (7), Additionally, there is extensive deposition of NO2-FA in both complex lipids, such as phospholipids and triglycerides, and as Michael adducts with low-molecular-weight thiols and proteins containing nucleophilic amino acids such as cysteine and histidine. Diet can affect NO2-CLA levels, with increased NO2-CLA produced by dietary CLA and supplementation in rodents (4). Also, humans, supplemented with nitrate (), , and CLA promotes the generation of NO2-CLA at concentrations sufficient to exert systemic effects before being catabolized or excreted (7). Because the biochemical reactivities of NO2-CLA differs from the more simple monounsaturated NO2-OA (28) and because of its endogenous abundance, we viewed it important to better define the biochemical and physiological responses to NO2-CLA.
In the present study, we performed RNA sequencing (RNA-Seq) analysis of human coronary artery smooth muscle cells (hCASMCs) treated with and without NO2-CLA. We discovered an extensive number of differentially expressed genes (DEGs) in response to NO2-CLA in hCASMCs and identified novel potential targets that NO2-FA would be expected to modulate. By using bioinformatics tools, we examined the functional distribution of DEGs and report a diverse impact of NO2-CLA on key metabolic and inflammatory-related cellular functions and signaling pathways. Our study provides a useful resource for investigating and understanding the role of NO2-FA in modulating the biological functions of hCASMCs and other cells.
MATERIALS AND METHODS
hCASMC culture.
hCASMCs were purchased from PromoCell (Germany) and cultured in Smooth Muscle Cell Growth Medium 2 from same company at 37°C, 5% CO2 in a standard cell culture incubator. The hCASMCs were used at passage 5–6 in this study. The cells at 80% confluence were maintained in 0.5% FBS medium for overnight and subsequently treated with 1 μM NO2-CLA, 1 μM CLA solvated in DMSO, or vehicle for 6 h. NO2-CLA was synthesized and purified as previously (33).
RNA extraction and RNA-Seq sample preparation.
After 6 h treatment with NO2-CLA, CLA, or vehicle, the total RNA from hCASMCs were isolated and purified using RNeasy mini kit (Qiagen) and subjected to on-column RNase-free DNase treatment following the manufacturer’s recommendation (Qiagen). Total RNA samples were assessed for quality with the BioAnalyzer (Agilent). An RNA-Seq library was prepared with the EnCore Complete RNA Seq Library System (NuGen, San Carlos, CA). Twelve samples were sequenced on a HiSeq2000 (Illumina, Madison, WI) to generate 50 bp single-end reads, with TruSeq SBS v3 reagents (12) according to manufacturer's protocols.
Read mapping and differential expression analysis.
For each sample, the fastq files from multiple runs were combined and subjected to quality check by FastQC. The high-quality reads were mapped to the human genome with TopHat (26). Genome reference and index files (Ensembl GRCh37) were downloaded from the Tophat website. After read-mapping, Cufflinks (27) was used for transcriptome assembly and quantification of gene expression. Next, differential expression analyses were performed between NO2-CLA vs. control, NO2-CLA vs. CLA, and CLA vs. control with DESeq (1). DEGs were defined at a minimum fold change of 1.3, and false discovery rate (FDR) at 0.05. The principal component analysis (PCA), volcano plots, and heat maps were generated with R scripts.
Gene ontology and KEGG pathway enrichment analysis.
The gene ontology (GO) enrichment analysis was performed using ClueGO (3). The “GO biological process,” “GO molecular function,” “GO cellular component,” and “KEGG” were selected as background databases for testing. The GO terms were further integrated by using the “GO term fusion” option, based on similarly associated genes. P values were corrected by Benjamini-Hochberg’s approach.
Gene network analysis.
The gene network of proliferation-associated genes was constructed with GeneMinia (32) and visualized by Cytoscape (25). Edges were constructed based on evidence of direct physical interactions and colocalizations between two genes.
Quantitative real-time PCR.
Total RNA was reverse-transcribed into cDNA pools with SuperScript III RT-PCR kit (ThermoFisher). Target RNA expression was determined by quantitative real-time PCR (qPCR) using iQ SYBR Green Supermix (Bio-Rad). The gene expression level was normalized by the internal control, 18S rRNA. The primer sequences are shown in Supplemental Table S1. (The online version of this article contains supplemental material.)
RESULTS
Transcriptomic profiling of hCASMCs.
To comprehensively investigate the impact of NO2-FA on smooth muscle cell gene expression, we compared the gene expression profiles of hCASMCs in three conditions: NO2-CLA, native CLA treatment and vehicle control. We performed RNA-Seq of 12 hCASMC samples, including four replicates of each condition. A total of ~400 million sequence reads were generated for these samples. After quality checking, the reads were mapped to the human genome assembly GRCh37 (also known as hg19) and gene expressions were calculated based on alignment per sample (see materials and methods). In total, we detected 14,472 genes that were abundantly expressed in hCASMCs. The raw data were deposited to the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE100676.
Identification of DEGs regulated by NO2-CLA.
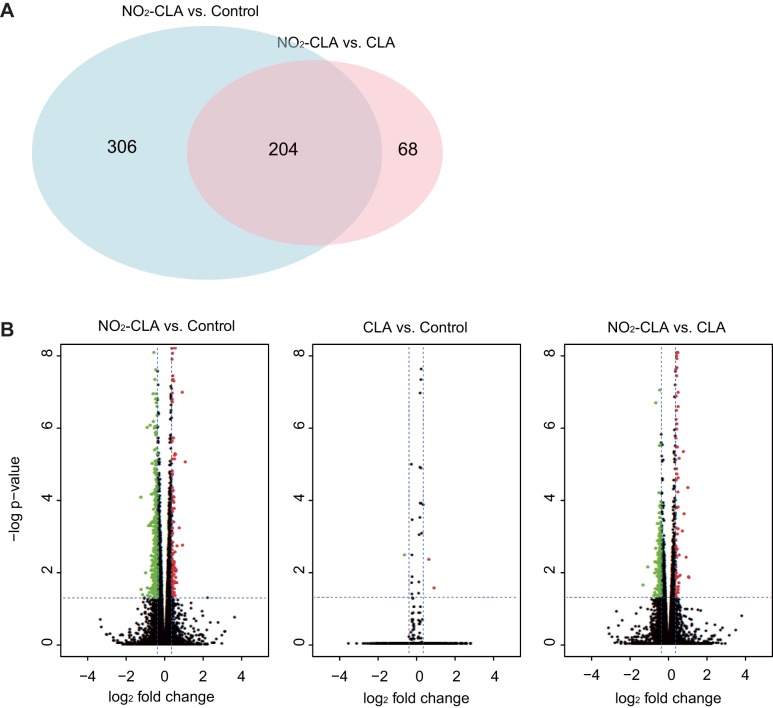
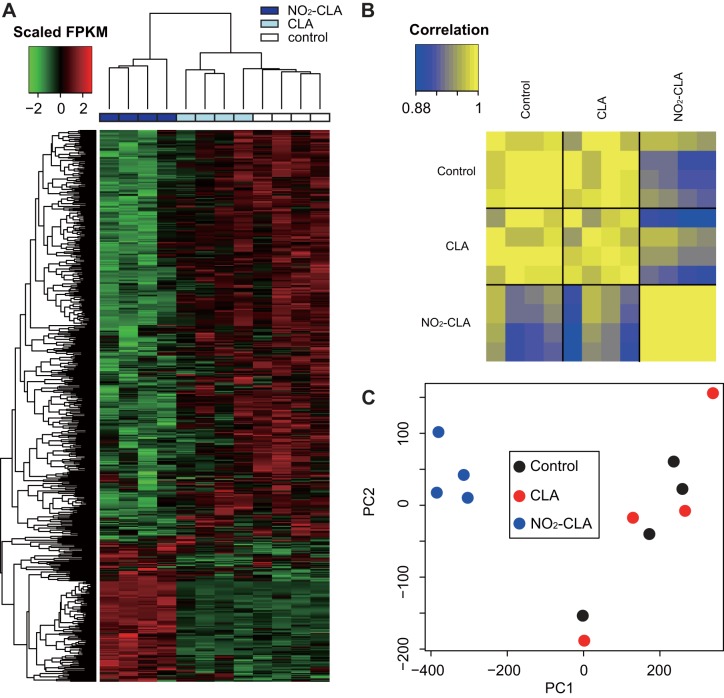
With a cut-off at a 30% expression level change and an FDR < 0.05, we obtained 510 DEGs by comparing NO2-CLA with vehicle control, and 272 DEGs by comparing NO2-CLA with CLA (Fig. 1A) (Supplemental Tables S2, S3). There are 204 genes that overlapped in both conditions. Notably, CLA treatment showed no significant impact on gene expression (Fig. 1B) compared with vehicle control. PCA and Pearson’s correlation coefficient analysis also confirmed that NO2-CLA-treated cells exhibited unique gene expression profiles (Fig. 2, A and B). Taken together, these results reveal that DEGs induced by NO2-CLA treatment are triggered in response to the electrophilic nitroalkene-containing fatty acid but not the nonnitrated native fatty acid.
Fig. 1.
Transcriptomic profiling of human coronary artery smooth muscle cells (hCASMCs) treated by nitro-conjugated linoleic acid (NO2-CLA). A: Venn diagram of differentially expressed genes (DEGs) identified in NO2-CLA vs. conjugated linoleic acid (CLA) and NO2-CLA vs. control. B: pairwise volcano plot of gene expression differences between conditions. The threshold of DEGs is defined at false discovery rate (FDR) > 0.05, and 1.3 fold up- (red) or down- (green) regulated upon treatment.
Fig. 2.
Differentially expressed genes (DEGs) in NO2-CLA-treated hCASMCs. A: heat map of 578 DEGs expression levels. Color bar denotes the row-scaled fragments per kilobase of transcript per million mapped reads (FPKM) value, representing the Z score of gene expression across samples. B: correlation matrix of 12 samples constructed based on DEGs. C: plot of the 1st and 2nd principal component by principal component analysis (PCA) of sample variations.
Among the 578 DEGs, ~25% (143 genes) were upregulated in NO2-CLA-treated cells (Fig. 2C). These genes include key factors induced by oxidative stress, such as HMOX1, GCLM, GCLC, TXNRD1, and SLC7A11. In addition, genes downstream of transcriptional regulation by the PPAR nuclear lipid receptor family signaling mediators such as ABCA1 and ANGPTL4 were also upregulated. These findings are consistent with previous discoveries from other nitro-fatty acid species, such as the bis-allylic nitro-linoleic acid (LNO2), which induces oxidative stress response through the Nrf2 pathway and also acts as a ligand of PPARγ (15). On a different note, the majority of the DEGs (~75%, 435 genes) were downregulated by NO2-CLA, when compared with either CLA or control. The top responding genes included the nuclear receptor NR4A1, the transcription factor JUNB, and the proinflammatory cytokine MIF (Supplemental Tables S2, S3). Expression of genes related to lipid storage after NO2-CLA treatment was also found to be significantly altered, which has not been observed previously: PLIN2, member of the perilipin family that coats intracellular lipid storage droplets, was upregulated.
Although the 578 DEGs comprised only 25% of the upregulated genes induced by NO2-CLA, they were strongly enriched in the top significant changes (NO2-CLA vs. control, 18 out of 20 top genes were upregulated; NO2-CLA vs. CLA, 20 out of 20 top genes were upregulated). This indicates a strong and immediate gene expression response of hCASMC upon stimulation by NO2-CLA and included the expression of essential genes involved in oxidative stress response and PPAR signaling pathways.
qPCR validation of DEGs.
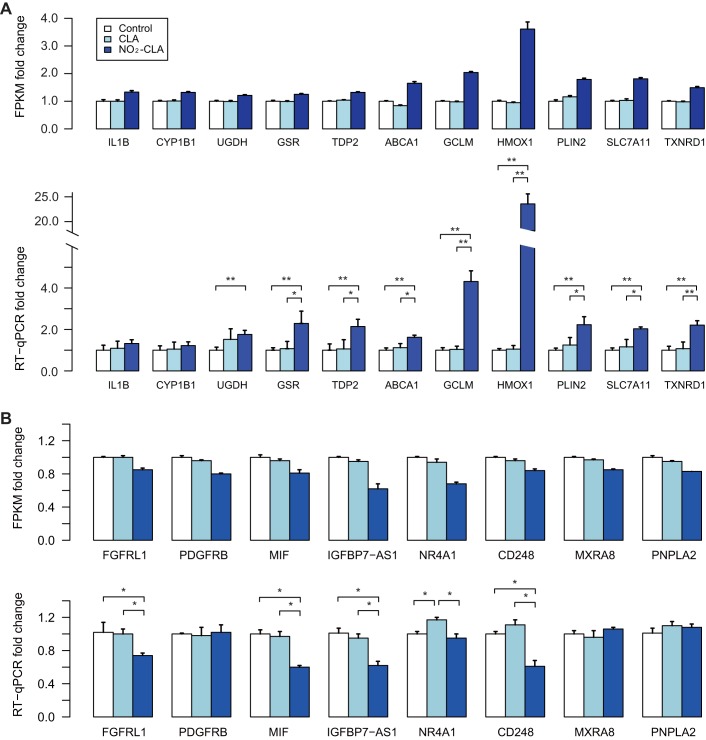
We selected 19 DEGs (11 up- and 8 downregulated genes) with abundant expression in hCASMC and performed qPCR under the three different conditions for validation of the RNA-Seq data. The new RNA samples used for qPCR were isolated from cells processed independently from those in the RNA-Seq experiments. Among the 19 genes, 13 (9 up- and 4 downregulated genes) showed significant change in expression under NO2-CLA treatment, consistent with the fragments per kilobase of transcript per million mapped reads measured by RNA-Seq analysis (Fig. 3, A and B), while IL1B and CYP1B1 exhibited the same pattern as expected from the RNA-Seq findings but failed to reach statistical significance (Fig. 3A).
Fig. 3.
Quantitative real-time PCR (qPCR) validation of 19 differentially expressed genes. Expression levels of 11 upregulated genes (A) and 8 downregulated genes (B) measured by RNA-Seq and qPCR in NO2-CLA, CLA, and control groups. Data are expressed as means ± SE. For qPCR groups, Student’s t-tests were performed to search for comparisons with statistical significance: *P < 0.05, **P < 0.001; n = 4 for upregulated and n = 6 for downregulated genes.
Although most of the 19 genes showed similar expression profiles between the two experimental approaches, it is worth noting that HMOX1 and GCLM have exhibited inconsistent response to NO2-CLA between different batches of cells. Remarkably, HMOX1 was ~23-fold induced in the cells used for the ensuing validation, about six times higher than the levels detected by RNA-Seq (Fig. 3A). Moreover, we failed to confirm four out of eight downregulated genes (Fig. 3B). These inconsistencies might be caused by multiple factors, including batch effects of cells and reagents, the gene expression normalizing method, and genetic background variations (cells were derived from different individuals).
Functional distribution of DEGs.
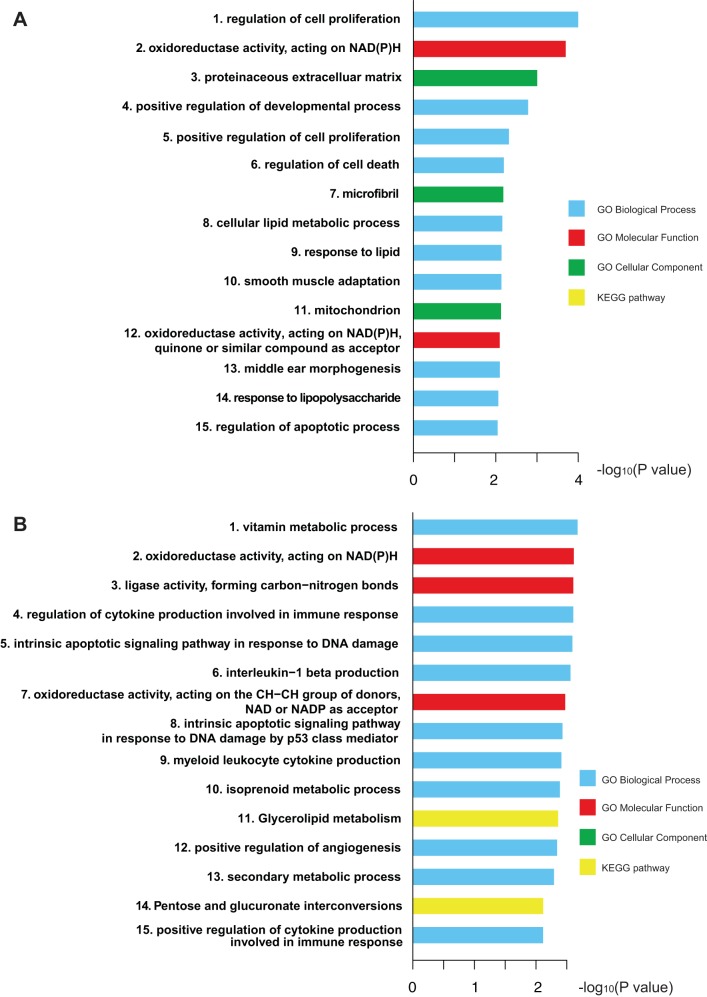
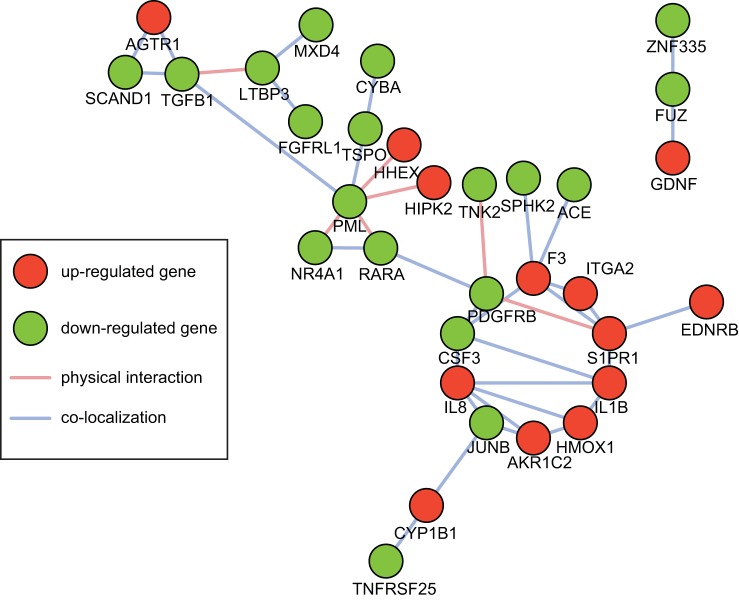
To further understand the pathways and biological processes being regulated by NO2-CLA treatment, we performed the GO and KEGG pathway enrichment analysis of the DEGs. Among all the GO terms, “regulation of cell proliferation” was most significantly enriched in DEGs (P = 9.77 × 10−5), associated with 77 out of the 578 genes (Fig. 4A, Supplemental Table S4). We also displayed the knowledge-based network of these genes to see whether they are involved in compact functional modules. Of the 77 genes, 32 genes were closely connected by either direct physical interactions or colocalization, and 29 genes formed a primary cluster (Fig. 5). NO2-CLA potentially regulates the proliferation of hCASMCs through one or multiple transcriptional regulatory mechanisms that affect these genes.
Fig. 4.
Function and pathway enrichment analysis of DEGs. Top GO terms and KEGG pathways enriched in the 578 DEGs (A) and overlapping 204 DEGs that were consistently altered under NO2-CLA treatment compared with CLA or control. (B). Terms from 4 major categories are shown in different color, ranked by adjusted P values.
Fig. 5.
Network of cell proliferation-associated DEGs. Network of closely related DEGs was constructed based on evidences of direct physical interaction and colocalization, which reflects either coexpression across tissue types or coexistence in similar cellular compartment. Only clusters with >3 nodes are displayed.
Terms related to programming cell death and lipid metabolism were also found in the top 15 ontologies, including “regulation of cell death” (ranked as 6th, P = 6.21 × 10−3), “regulation of apoptotic process” (ranked as 15th, P = 8.77 × 10−3), “cellular lipid metabolic process” (ranked as 8th, P = 6.76 × 10−3), and “response to lipid” (ranked as 9th, P = 7.07 × 10−3). Moreover, “oxidoreductase activity acting on NAD(P)H” was identified as the top term in the “GO molecular function” category (P = 1.96 × 10−4). This term is associated with several oxidoreductases including GSR and TXNRD1, which catalyze the reduction of glutathione and thioredoxin respectively, and protect cells from oxidative and inflammatory stress.
To exclude genes that were merely responding to the intake of fatty acids, we also did enrichment analysis of 204 genes that were consistently altered under NO2-CLA vs. CLA and NO2-CLA vs. control. Interestingly, four out of the 15 top terms were related to cytokine production and immune responses (Fig. 4B, Supplemental Table S5). “Regulation of cytokine production involved in immune response” was the third most significant term (P = 2.48 × 10−3). Its related genes showed higher levels of enrichment in the overlapping genes (before adjustment, Punadjust = 3.23 × 10−4) than the total set (before adjustment, Punadjust = 1.30 × 10−3), suggesting that cytokine production was regulated in hCASMCs by the treatment of NO2-CLA, independently from its fatty acid substrate.
DISCUSSION
Posttranslational modifications induced by metabolic and inflammatory-derived reactive species provide a mechanism for organisms to respond to dietary and environmental factors. This process is facilitated by the fact that many transcriptional regulatory proteins possess functionally significant, redox-sensitive moieties such as cysteine. The present results reveal that there are a broad array of genomic responses to chemically reactive electrophilic lipid oxidation and nitration products. We report that >500 genes, comprising multiple important gene families, are regulated by NO2-CLA in hCASMCs. These genes include some targets previously appreciated to be responsive to other NO2-FA species, such as HMOX1, which is rapidly and highly induced in various cell types by NO2-FA (4, 6, 31). HMOX1 was also identified as one of the most significantly upregulated genes by NO2-CLA in hCASMCs. Notably, L-NO2 promotes the nuclear translocation of Nrf2 without impacting Nrf2 mRNA levels to mediate activation of the Nrf2-regulated antioxidant response element (5). Consistent with this, Nrf2 expression was not modulated by NO2-CLA in hCASMC. Also, a cluster of genes involved in maintaining glutathione (GCLM, GCLC, GSR, SLC7A11) and thioredoxin homeostasis (TXNRD1), also downstream of transcriptional regulation by Keap1/Nrf2 (17, 21), were significantly induced by NO2-CLA. Increased expression of these enzymes, along with the cysteine-glutamate transport channel SLC7A11, promotes concerted reductive responses that will limit oxidative stress (8, 13, 20).
Moreover, novel gene targets regulated by NO2-CLA were also identified for the first time. For example, PLIN2 is associated with lipid storage and were regulated by NO2-CLA but not CLA. This indicates a more complex pattern of responses than those merely induced by elevated concentrations of a fatty acid substrate. ANGPTL4, which modulates wound healing and angiogenesis (9, 11, 35), was also highly induced. The expression of this gene is regulated by the PPAR receptor class (11). In these latter instances, it is not fully understood as to how the expression of these genes was modulated by NO2-CLA, with the mechanisms accounting for the regulation of these novel targets remaining to be determined. GO enrichment analysis of DEGs indicated four major functional modules whose expression was under NO2-CLA regulation (Fig. 6). The upregulation of antioxidant defenses was the primary cellular response to the treatment (or stimulation) with NO2-CLA. DEGs related with this process are typically involved in reductive and oxidant scavenging reactions. Lipid metabolism, especially genes related with lipid storage, were also affected by NO2-CLA treatment. Moreover, NO2-CLA regulates a group of genes that are critical for the control of cell proliferative (e.g., AGTR1, EDNRB, FGFRL1), and inflammatory responses (e.g., MIF, IL1B). These gene expression responses are consistent with prior observations that NO2-FA inhibit proliferation of vascular smooth muscle cells (VSMCs) (31). Both L-NO2 and OA-NO2 have also been revealed to display pleiotropic anti-inflammatory signaling properties (6). In aggregate, these gene expression responses reveal that NO2-CLA, the predominant endogenous NO2-FA, regulates VSMC functions via an array of both integrated and pleiotropic mechanisms.
Fig. 6.
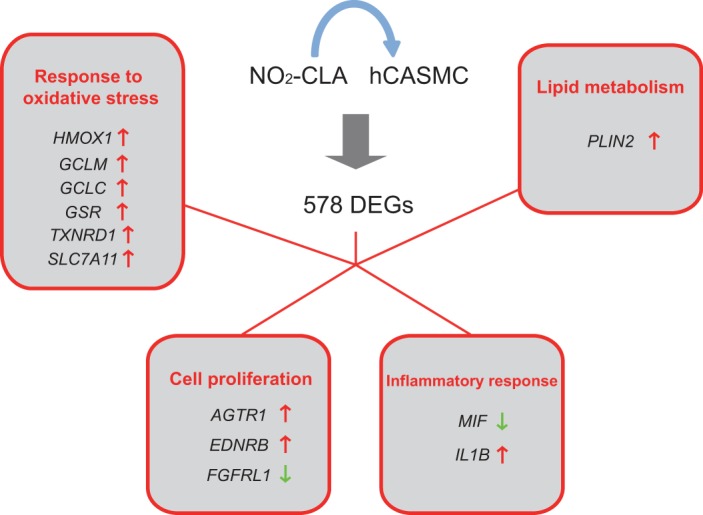
Functional distribution of NO2-CLA effects in vascular smooth muscle cell biology. A brief summary of the functional categories of DEGs after NO2-CLA treatment in the present analysis. Representative genes of each category are listed.
In summary, the present study presents a systematic analysis of the hCASMC transcriptome that responds uniquely to electrophilic nitroalkene derivatives of CLA. In turn, NO2-CLA regulates critical cellular functions, including cell proliferation, anti-oxidant responses, lipid metabolism and inflammatory responses. In addition to previously identified nitrated fatty acid-responsive genes, this study reveals additional novel target genes and pathways modulated by NO2-CLA, providing new insight into our understanding of the complex role electrophilic products of metabolism and inflammation in cell signaling responses.
GRANTS
This work was supported in part by National Institutes of Health Grants HL-138139 (to J. Zhang), HL-123333 (to L. Villacorta), and HL-134569 to Y. E. Chen) and by the National Key R&D Program of China (2016YFC0901704 and 2017YFA0505500 to Y. Li).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
S.L., Z.C., Y.E.C., and J.Z. conceived and designed research; S.L., Z.C., T.Z., and J.Z. performed experiments; S.L., Z.C., and J.Z. analyzed data; S.L., Z.C., Y.L., B.A.F., Y.E.C., and J.Z. interpreted results of experiments; S.L., Z.C., and J.Z. prepared figures; S.L., Z.C., and J.Z. drafted manuscript; S.L., Z.C., Y.L., B.A.F., Y.E.C., and J.Z. edited and revised manuscript; S.L., Z.C., T.Z., L.V., Y.L., B.A.F., Y.E.C., and J.Z. approved final version of manuscript.
Supplemental Data
Supplemental tables (1-5) - .xlsx (148 KB)
REFERENCES
- 1.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 11: R106, 2010. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker PR, Lin Y, Schopfer FJ, Woodcock SR, Groeger AL, Batthyany C, Sweeney S, Long MH, Iles KE, Baker LM, Branchaud BP, Chen YE, Freeman BA. Fatty acid transduction of nitric oxide signaling: multiple nitrated unsaturated fatty acid derivatives exist in human blood and urine and serve as endogenous peroxisome proliferator-activated receptor ligands. J Biol Chem 280: 42464–42475, 2005. doi: 10.1074/jbc.M504212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093, 2009. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonacci G, Baker PR, Salvatore SR, Shores D, Khoo NK, Koenitzer JR, Vitturi DA, Woodcock SR, Golin-Bisello F, Cole MP, Watkins S, St Croix C, Batthyany CI, Freeman BA, Schopfer FJ. Conjugated linoleic acid is a preferential substrate for fatty acid nitration. J Biol Chem 287: 44071–44082, 2012. doi: 10.1074/jbc.M112.401356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckley BJ, Marshall ZM, Whorton AR. Nitric oxide stimulates Nrf2 nuclear translocation in vascular endothelium. Biochem Biophys Res Commun 307: 973–979, 2003. doi: 10.1016/S0006-291X(03)01308-1. [DOI] [PubMed] [Google Scholar]
- 6.Cui T, Schopfer FJ, Zhang J, Chen K, Ichikawa T, Baker PR, Batthyany C, Chacko BK, Feng X, Patel RP, Agarwal A, Freeman BA, Chen YE. Nitrated fatty acids: Endogenous anti-inflammatory signaling mediators. J Biol Chem 281: 35686–35698, 2006. doi: 10.1074/jbc.M603357200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delmastro-Greenwood M, Hughan KS, Vitturi DA, Salvatore SR, Grimes G, Potti G, Shiva S, Schopfer FJ, Gladwin MT, Freeman BA, Gelhaus Wendell S. Nitrite and nitrate-dependent generation of anti-inflammatory fatty acid nitroalkenes. Free Radic Biol Med 89: 333–341, 2015. doi: 10.1016/j.freeradbiomed.2015.07.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espinosa-Diez C, Miguel V, Mennerich D, Kietzmann T, Sánchez-Pérez P, Cadenas S, Lamas S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol 6: 183–197, 2015. doi: 10.1016/j.redox.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goh YY, Pal M, Chong HC, Zhu P, Tan MJ, Punugu L, Tan CK, Huang RL, Sze SK, Tang MB, Ding JL, Kersten S, Tan NS. Angiopoietin-like 4 interacts with matrix proteins to modulate wound healing. J Biol Chem 285: 32999–33009, 2010. doi: 10.1074/jbc.M110.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groeger AL, Cipollina C, Cole MP, Woodcock SR, Bonacci G, Rudolph TK, Rudolph V, Freeman BA, Schopfer FJ. Cyclooxygenase-2 generates anti-inflammatory mediators from omega-3 fatty acids. Nat Chem Biol 6: 433–441, 2010. doi: 10.1038/nchembio.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo L, Li SY, Ji FY, Zhao YF, Zhong Y, Lv XJ, Wu XL, Qian GS. Role of Angptl4 in vascular permeability and inflammation. Inflamm Res 63: 13–22, 2014. doi: 10.1007/s00011-013-0678-0. [DOI] [PubMed] [Google Scholar]
- 12.Illumina. https://hsls.nextbio.com. Ontology Editor.
- 13.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47: 89–116, 2007. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 14.Khoo NK, Rudolph V, Cole MP, Golin-Bisello F, Schopfer FJ, Woodcock SR, Batthyany C, Freeman BA. Activation of vascular endothelial nitric oxide synthase and heme oxygenase-1 expression by electrophilic nitro-fatty acids. Free Radic Biol Med 48: 230–239, 2010. doi: 10.1016/j.freeradbiomed.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Zhang J, Schopfer FJ, Martynowski D, Garcia-Barrio MT, Kovach A, Suino-Powell K, Baker PR, Freeman BA, Chen YE, Xu HE. Molecular recognition of nitrated fatty acids by PPAR gamma. Nat Struct Mol Biol 15: 865–867, 2008. doi: 10.1038/nsmb.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim DG, Sweeney S, Bloodsworth A, White CR, Chumley PH, Krishna NR, Schopfer F, O’Donnell VB, Eiserich JP, Freeman BA. Nitrolinoleate, a nitric oxide-derived mediator of cell function: synthesis, characterization, and vasomotor activity. Proc Natl Acad Sci USA 99: 15941–15946, 2002. doi: 10.1073/pnas.232409599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 284: 13291–13295, 2009. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Donnell VB, Eiserich JP, Bloodsworth A, Chumley PH, Kirk M, Barnes S, Darley-Usmar VM, Freeman BA. Nitration of unsaturated fatty acids by nitric oxide-derived reactive species. Methods Enzymol 301: 454–470, 1999. doi: 10.1016/S0076-6879(99)01109-X. [DOI] [PubMed] [Google Scholar]
- 19.Paine A, Eiz-Vesper B, Blasczyk R, Immenschuh S. Signaling to heme oxygenase-1 and its anti-inflammatory therapeutic potential. Biochem Pharmacol 80: 1895–1903, 2010. doi: 10.1016/j.bcp.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Polewski MD, Reveron-Thornton RF, Cherryholmes GA, Marinov GK, Cassady K, Aboody KS. Increased expression of system xc- in glioblastoma confers an altered metabolic state and temozolomide resistance. Mol Cancer Res 14: 1229–1242, 2016. doi: 10.1158/1541-7786.MCR-16-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal 24: 981–990, 2012. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubbo H, Parthasarathy S, Barnes S, Kirk M, Kalyanaraman B, Freeman BA. Nitric oxide inhibition of lipoxygenase-dependent liposome and low-density lipoprotein oxidation: termination of radical chain propagation reactions and formation of nitrogen-containing oxidized lipid derivatives. Arch Biochem Biophys 324: 15–25, 1995. doi: 10.1006/abbi.1995.9935. [DOI] [PubMed] [Google Scholar]
- 23.Rubbo H, Radi R, Trujillo M, Telleri R, Kalyanaraman B, Barnes S, Kirk M, Freeman BA. Nitric oxide regulation of superoxide and peroxynitrite-dependent lipid peroxidation. Formation of novel nitrogen-containing oxidized lipid derivatives. J Biol Chem 269: 26066–26075, 1994. [PubMed] [Google Scholar]
- 24.Schopfer FJ, Cipollina C, Freeman BA. Formation and signaling actions of electrophilic lipids. Chem Rev 111: 5997–6021, 2011. doi: 10.1021/cr200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504, 2003. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111, 2009. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578, 2012. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turell L, Vitturi DA, Coitino EL, Lebrato L, Moller MN, Sagasti C, Salvatore SR, Woodcock SR, Alvarez B, Schopfer FJ. The chemical basis of thiol addition to nitro-conjugated linoleic acid, a protective cell-signaling lipid. J Biol Chem 292: 1145–1159, 2017. doi: 10.1074/jbc.M116.756288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villacorta L, Chang L, Salvatore SR, Ichikawa T, Zhang J, Petrovic-Djergovic D, Jia L, Carlsen H, Schopfer FJ, Freeman BA, Chen YE. Electrophilic nitro-fatty acids inhibit vascular inflammation by disrupting LPS-dependent TLR4 signalling in lipid rafts. Cardiovasc Res 98: 116–124, 2013. doi: 10.1093/cvr/cvt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villacorta L, Gao Z, Schopfer FJ, Freeman BA, Chen YE. Nitro-fatty acids in cardiovascular regulation and diseases: characteristics and molecular mechanisms. Front Biosci 21: 873–889, 2016. doi: 10.2741/4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villacorta L, Zhang J, Garcia-Barrio MT, Chen XL, Freeman BA, Chen YE, Cui T. Nitro-linoleic acid inhibits vascular smooth muscle cell proliferation via the Keap1/Nrf2 signaling pathway. Am J Physiol Heart Circ Physiol 293: H770–H776, 2007. doi: 10.1152/ajpheart.00261.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38, suppl_2: W214–W220, 2010. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodcock SR, Salvatore SR, Bonacci G, Schopfer FJ, Freeman BA. Biomimetic nitration of conjugated linoleic acid: formation and characterization of naturally occurring conjugated nitrodienes. J Org Chem 79: 25–33, 2014. doi: 10.1021/jo4021562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright MM, Schopfer FJ, Baker PR, Vidyasagar V, Powell P, Chumley P, Iles KE, Freeman BA, Agarwal A. Fatty acid transduction of nitric oxide signaling: nitrolinoleic acid potently activates endothelial heme oxygenase 1 expression. Proc Natl Acad Sci USA 103: 4299–4304, 2006. doi: 10.1073/pnas.0506541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu P, Goh YY, Chin HF, Kersten S, Tan NS. Angiopoietin-like 4: a decade of research. Biosci Rep 32: 211–219, 2012. doi: 10.1042/BSR20110102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental tables (1-5) - .xlsx (148 KB)