Abstract
Big data are a major driver in the development of precision medicine. Efficient analysis methods are needed to transform big data into clinically-actionable knowledge. To accomplish this, many researchers are turning toward machine learning (ML), an approach of artificial intelligence (AI) that utilizes modern algorithms to give computers the ability to learn. Much of the effort to advance ML for precision medicine has been focused on the development and implementation of algorithms and the generation of ever larger quantities of genomic sequence data and electronic health records. However, relevance and accuracy of the data are as important as quantity of data in the advancement of ML for precision medicine. For common diseases, physiological genomic readouts in disease-applicable tissues may be an effective surrogate to measure the effect of genetic and environmental factors and their interactions that underlie disease development and progression. Disease-applicable tissue may be difficult to obtain, but there are important exceptions such as kidney needle biopsy specimens. As AI continues to advance, new analytical approaches, including those that go beyond data correlation, need to be developed and ethical issues of AI need to be addressed. Physiological genomic readouts in disease-relevant tissues, combined with advanced AI, can be a powerful approach for precision medicine for common diseases.
Keywords: artificial intelligence, functional genomics, machine learning, physiological genomics, precision medicine
INTRODUCTION
Big data are a major driver of precision medicine.1 In 2011, the US National Research Council developed a comprehensive plan for how to advance precision medicine, specifying the development of a “New Taxonomy” for human diseases, to integrate molecular, environmental, and phenotypic data (41a). Big data alone are limited without analysis methods to transform the data into clinically actionable knowledge. Thanks to the advances in parallel computing, many researchers are turning to artificial intelligence (AI) to develop robust data analysis approaches (5, 42). In this article, we will first give a broad introduction to machine learning (ML), which is a major approach of AI, and then discuss how physiological genomics, combined with ML, can be a critical and powerful approach to advancing precision medicine for common diseases.
MACHINE LEARNING
ML is an approach to achieving AI that utilizes modern computer and mathematic algorithms to build models based on available data sets, in which the model itself can improve with experience. ML can recognize complex combinations of variables that reliably predict an outcome (42). ML is useful for analyzing large, heterogeneous data sets, such as genomic or epigenomic data (34). Based on how the machine “learns” and depending on the question the “learning” is trying to answer, ML can be categorized into supervised, unsupervised, and reinforcement learning. The major application of ML in precision medicine comes from supervised learning where the patient outcome is known (labeled). Unsupervised learning utilizes uncategorized data sets to find hidden patterns or grouping in data. Reinforcement learning utilizes simulated data and is not generally used in medicine.
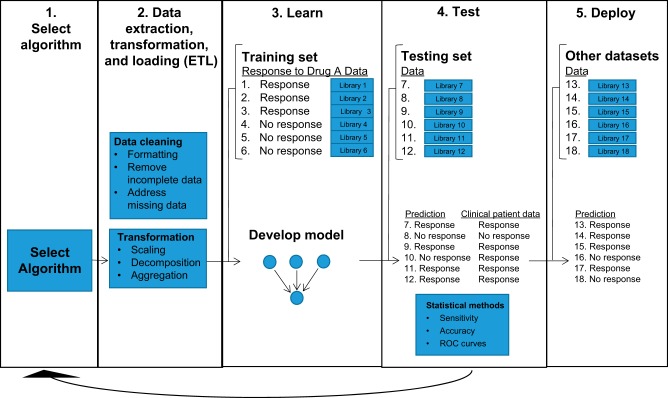
Traditionally, supervised learning is conducted in five phases (Fig. 1). First, an algorithm is selected with the goal of maximum learning of the training set. Several examples of ML software libraries used with Python and R are shown in Table 1. Second, preprocess treatment of the data, or data extraction, transformation, and loading (ETL), is performed. The data are formatted and cleaned to remove incomplete data and address missing information. Then the data may be transformed; common transformations include scaling (taking attributes with a mixture of scales and putting them on the same scale such as between 0 and 1), decomposition (splitting attributes into constituent parts), or aggregation (combining related attributes into a single feature). Third, the data are split into training and testing sets, with a majority of data assigned to the training set. The training set is then used to develop a model or “train” the AI. The parameters in the model are tuned to optimize the final model built. Fourth, the model is used to make predictions on the testing set, and the predictability of the model is assessed. K-fold cross-validation can be used to assess the stability of the model. In the cross-validation, the data are divided into k subsets. Now the built model is repeated k times, such that each time, one of the k subsets is used as the test set and the other k − 1 subsets are put together to form a training set. The prediction error rate is averaged over all k trials to obtain total effectiveness of the built model. Other statistical methods may be utilized to evaluate the predictability of the model, including sensitivity, accuracy, and receiver operating characteristic (ROC) curves. Area under the curve (AUC) may be calculated from the ROC curve, though major limitations of AUC utilization have been identified (21, 54). AUC is a measure of how accurate the model is in predicting the actual patient outcome, with an AUC of 1 representing perfect accuracy. Steps 1 through 4 may be conducted for many different algorithms to select an algorithm with optimum predictability. Lastly, the model is deployed to make predictions on other clinical data sets (1, 34).
Fig. 1.
Schematic of machine learning (ML) steps for supervised learning. ML is conducted in five phases. 1) An algorithm is selected. 2) Data extraction, transformation, and loading (ETL) are performed. The data are formatted and then cleaned to remove incomplete data and address missing data. The data may also be transformed by transformations such as scaling, decomposition, or aggregation. 3) The data are split into training and testing sets. The training set is then used to develop a model or “train” the artificial intelligence (AI). The parameters in the model may be tuned to optimize the final model built. 4) The model is used to make predictions on the testing set, and the predictability of the model is assessed. Cross-validation can be performed to compare the predicted outcome to the actual patient outcome. Sensitivity, accuracy, and receiver operating characteristic (ROC) curves may also be used to evaluate the predictability of the model. 5) The model is deployed to make prediction for other data sets.
Table 1.
Examples of software libraries used with Python and R to perform various ML tasks
| Task | Example Software Libraries Used with Python | Example Software Libraries Used with R |
|---|---|---|
| Data preprocessing | NumPy, SciPy, pandas, Matplotlib, seaborn | R(data.frame), dplyr, ggplot2 |
| Training testing data split | scikit-learn | Caret, mtcars, caTools, dplyr |
| Linear regression | scikit-learn | Stats, H2O |
| Classification_K-NN | scikit-learn | Class |
| Classification_tree | scikit-learn | C50 |
| Classification_SVM | scikit-learn | e1017 |
| Classification_naive bayes | scikit-learn | e1017 |
| Classfication_random forest | scikit-learn | randomForest, H2O |
| Clustering PCA/K-Means | scikit-learn | Stats, H2O |
| Association analysis | scikit-learn | arules |
| Dimensional reduction | scikit-learn | Stats, H2O |
| Deep learning_ANN | Keras, Tensorflow, Theano | H2O, neuralnet |
| Deep learning_CNN | Keras, Tensorflow, Theano | KerasR, MXNetR |
| Deep learning_RNN | Keras, Tensorflow, Theano | rnn |
ML, machine learning.
Games have frequently served as milestones for large advancements in ML (41). Early accomplishments were in backgammon (50), checkers (44), chess (6), Jeopardy! (14), and Atari (40). These games are perfect-information games, meaning that every player has identical, complete information about the current game status and can see all possible scenarios for every move. Once simple perfect-information games were mastered, programmers turned their focus toward the game Go, which is considered the most complex perfect-information game.
Go is an ancient Chinese game with simple rules, but it requires highly complex strategy because of the 19×19 grid board, which contains 10170 possible configurations (18). Google researchers developed an algorithm named AlphaGo. AlphaGo was first trained with supervised learning using 30 million iterations from professional games played by humans. Then AlphaGo played against itself using reinforcement learning, simulating thousands of random games using Monte Carlo tree search programs. AlphaGo’s success was due to its ability to overcome the complexity of the game by developing prioritization methods that reduced the number of possible moves the algorithm had to consider for any one given move. In 2015 and 2016, AlphaGo successfully defeated European champion Fan Hui and 18-time world champion Lee Sedol (46). More recently, Google developed AlphaGo Zero, an AI that utilizes a novel reinforcement learning algorithm that initiates training from completely random play against itself rather than through observation of human games. AlphaGo Zero was clearly stronger than previous versions of AlphaGo (47).
Imperfect-information games like poker, which involve chance or incomplete information, are more challenging to master with ML because they require more complex reasoning. All possible cards are known in poker, but players have uncertainty in the cards their opponents have and asymmetric knowledge about the game status due to each player’s private hand. DeepStack is an AI trained in heads-up no-limit Texas hold’em, a two-person poker game. DeepStack used reinforcement learning to play thousands of simulated poker hands against itself. The key to DeepStack’s success was incorporating recursive reasoning methods that effectively addressed issues with incomplete and asymmetric information. Although DeepStack has beaten several professional poker players, the technology is still ineffective in the standard 10-person no-limit Texas hold ’em game because of increased incomplete information, since the number of cards in opponent hands are 2/52 in two-person games and 18/52 in 10-person games (41).
APPLICATION OF AI IN MEDICINE
Many of the techniques and algorithms developed for games have been extrapolated to develop AI to assist in medicine. Watson, an AI developed by IBM originally intended to play Jeopardy!, is a cognitive natural-language processing system, meaning it has the ability to read and interpret words. In a recent study, the degree of treatment recommendation agreement was compared between Watson for Oncology and a 15-member multidisciplinary tumor board of physicians for 638 breast cancer patients. Treatment recommendations made by Watson for Oncology were highly concordant (93%) with the multidisciplinary tumor board, except for in cases of stage IV triple negative breast cancer (P < 0.05) (48).
Some other notable advances have been made using image recognition strategies for disciplines that utilize image diagnostic tools such as ophthalmology, pathology, and radiology. In a few recent studies that utilize image recognition, ML was compared with human doctors performing the standard of care to assess our progress in ML. Deep neural networks were used to train AI to make diagnoses on images of skin cancer (15) and diabetic retinopathy (19). When compared with professional dermatologists and ophthalmologists, the AI performed just as well and in some cases slightly better than their human counterparts.
In another study, multiple types of ML algorithms were compared with the most widely accepted guidelines in clinical practice, the American College of Cardiology/American Heart Association (ACC/AHA) guidelines, to predict the first cardiovascular event over a 10 yr period (52). Compared with ACC/AHA guidelines (AUC 0.728), the four ML algorithms improved predictions slightly: random forest (AUC 0.745), logistic regression (AUC 0.760), gradient boosting (AUC 0.761), and neural networks (AUC 0.764), with neural networks preforming the best. In-depth reviews of advances made in genomics (34), epigenomics (23), and proteomics (31) using ML can be found elsewhere.
KEY TO THE SUCCESS OF AI FOR PRECISION MEDICINE: DATA QUALITY AND RELEVANCE
While the performance of AI was impressive in the medical studies discussed above, one must ask why AI is not performing even better. For instance, in the study of cardiovascular risk prediction, the AUC was only around 0.75 for each of the AI algorithms even though the AI was provided with practically all available data. It is certainly possible that AI needs even more data or better algorithms to improve its performance. The quantity of data used for training sets is essential for maximizing the performance of ML algorithms. ML algorithms are extremely data-hungry, frequently requiring millions of data points to perform optimally. Additionally, programmers have found that using more data with simple algorithms produces more effective models than using less data with complex algorithms (20). Indeed, much of the current effort in precision medicine is focused on obtaining ever larger sample sizes and ever more DNA sequence data and electronic health records including mobile health records.
However, it is just as important to ensure that the data obtained are accurate and relevant. Data accuracy is important because it significantly affects the predictability of the model. The common “garbage in, garbage out” principle applies to ML, meaning that the quality of the predictions made by the model will never exceed the quality of the training set used to build the model. While conventional statistical methods and ML methods can be employed during ETL to check data quality and remove outliers, these methods have limited effectiveness due to the size of the data sets. Most of the time, ML itself does not judge the data quality and instead assesses all of the data equally to build the model. Thus, inaccuracy of data points will greatly impact the integrity of the final model built. Incomplete or even wrong data points will introduce biases. For example, one study that assessed the impact of data accuracy on model predictability found that using a training set with 10% inaccurate data increased the incorrect edges in a Bayesian network by 325% (27).
It is also important to collect the types of data that are relevant to the disease of interest from disease-applicable tissues. Many recent attempts to use ML to answer precision medicine questions have relied heavily on DNA sequence data (10), even though the vast majority of common diseases are widely believed to result from both (epi)genetic and environmental factors as well as their interactions. Unfortunately, causative environmental and lifestyle factors for many of these complex diseases are difficult to quantify, and the specific environmental exposures driving disease progression for a particular patient and their effect size are unknown. For these reasons, functional genomics are an appropriate surrogate, since they measure cumulative effects of environmental factors on the genome and provide biological links between DNA sequences, environmental factors, and the disease. Functional genomic readouts can take the forms of epigenetic modifications of the DNA, profiles of RNAs (mRNA, microRNA, lncRNA, etc.), proteins, and small metabolites, and physiological function (32, 37).
The logistic bottleneck for utilizing functional genomic readouts for precision medicine in common diseases is the difficulty in obtaining patient tissues directly relevant to these diseases. Because of this limitation, researchers have devoted significant effort toward identifying surrogate genomic readouts in body fluids or other readily obtainable materials. While this method may be useful, disease-applicable tissues are more likely to possess clinically actionable molecular information (51). There are important exceptions to the difficulty of obtaining disease-applicable tissues. One example is the kidney needle biopsy. Although it is an invasive procedure, it is done at a high enough frequency that it provides nephrology a significant advantage in driving forward functional genomics-based precision medicine. Importantly, it is possible to isolate and analyze specific tissue types or substructures in kidney biopsy samples, such as glomeruli, which would substantially mitigate concerns about tissue complexity. Another example is resistance arterioles, which are important determinants of cardiovascular physiology and can be readily obtained through biopsy (53). Physicians and scientists need to work closely together to collect tissues relevant to common diseases for testing the functional genomic approach to precision medicine. Importantly, the tissues need to be collected, processed, and stored in a manner appropriate for the intended analysis. While it is challenging to do so, the payoff is potentially very high.
EXAMPLES OF ML APPLICATION IN FUNCTIONAL GENOMICS STUDIES USING DISEASE-APPLICABLE TISSUES
Functional genomic data have been used to advance the precision medicine of cancer (25, 26). Recent progress has been made in utilizing functional genomics to inform precision medicine of common noncancer diseases including kidney disease. The following examples illustrate how functional genomics data obtained from disease-applicable tissues can be analyzed with ML in addition to other statistical methods to work toward the development of precision medicine for common noncancer diseases. Reeve et al. (43) collected 1,208 kidney transplant biopsies to assess rejection-related disease by using archetypal analysis of mRNA phenotypes combined with principal component analysis (PCA), a dimensionality reduction method. Archetypal analysis is a type of unsupervised learning that identifies extreme or pure phenotypes within a training set. They calculated bootstrap corrected C-statistics, which are analogous to AUCs, and found that their archetypal analysis scores (C-statistics 0.73) were more predictive of allograft loss than histological diagnosis using the Banff classification (C-statistics 0.60) (P = 3 × 10−6).
In another study, Liu et al. (35) examined cell type-specific differential mRNA expression in glomeruli isolated from IgA nephropathy patients. They used “in silico nanodissection,” an ML algorithm that predicts cell type-specific transcripts based on large data sets, to identify mesangial and podocyte cell-specific genes that were differentially expressed between IgA nephropathy and healthy control patients. They concluded that the mesangial cell genes were highly correlated to serum creatinine (P = 0.024) and estimated glomerular filtration rate (eGFR) (P = 0.025) progression for up to 4 yr following biopsy. Mass spectrometry was used to examine differentially expressed pathways in these samples as well as human mesangial cells that were incubated with serum from IgA nephropathy patients. They found several inflammatory pathways that were differentially expressed both in vivo and in vitro, along with pathways related to inducible nitric oxide synthase and endothelial nitric oxide synthase.
In a recent study from our laboratory, we used human formalin-fixed paraffin-embedded renal cortex samples to examine differentially expressed microRNAs among four different types of chronic kidney disease (2). MicroRNA are small noncoding RNA molecules that regulate gene expression through targeting specific mRNA for translational repression or degradation, thus leading to change in protein expression that can greatly affect the structure and function of a cell (3, 16, 55). Laser capture microdissection was used to collect glomeruli and proximal tubules, allowing tissue-specific whole genome microRNA libraries to be generated. This approach could elucidate diagnostic biomarkers within a particular nephron segment affected by disease pathogenesis that would not have otherwise been detected had the RNA been extracted from whole kidney cortex tissue.
We identified a number of microRNAs that were differentially expressed in glomeruli and proximal tubule samples between the control group vs. each disease, and in glomeruli between any two disease cohorts. Interestingly, we didn’t detect any differentially expressed microRNAs between any two disease cohorts in the proximal tubule samples. This may suggest that since the diseases studied were all glomerular diseases, the disease-specific effects on microRNA occur in the glomeruli and the pathological changes in the proximal tubules represent changes that are common across all proteinuric renal diseases.
ML also comes with its own limitations. Overfitting occurs when the model makes predictions on spurious correlations rather than causal correlations. Classification and regression trees are particularly at risk for overfitting, since these methods can continue to split data sets until a perfect correlation is observed (17). Another limitation with ML is the “black box” problem. With many types of ML, the data and model specifications are entered into the ML software, then the algorithm makes predictions without elucidating the algorithm’s decision-making process. Without this information, researchers can miss warning signs of defective models.
Effective validation strategies are needed to mitigate risks associated with overfitting and black box analysis. One method is to evaluate the predictability of models by using multiple independent patient cohorts to assess the generalizability of study findings (13). Another approach that we have utilized in our laboratory is to verify study results in in vitro or in vivo model systems. In a recent study of human salt sensitivity, we identified five urinary metabolites significantly associated with systolic blood pressure and one urinary metabolite significantly associated with diastolic blood pressure by using random forest and generalized linear mixed model analysis. To validate our results, we performed a proof-of-principle experiment to examine the effects of β-aminoisobutyric acid, a metabolite that was found to be significantly correlated to systolic blood pressure, on high-salt-induced hypertension in Dahl salt-sensitive (SS) rats. Treatment with β-aminoisobutyric acid significantly attenuated high-salt-induced hypertension in SS rats, supporting the relevance of the inverse relationship between β-aminoisobutyric acid and blood pressure we observed in humans (9).
FURTHER DEVELOPMENT OF ANALYTICAL APPROACHES INCLUDING THOSE BEYOND DATA CORRELATION
As we improve the data that we use in ML, there is also a great need to develop new analytical approaches that take into account the challenges and limitations of the study design and the data used for the training set, similar to how AlphaGo and DeepStack were built to address the challenges in data complexity and imperfect information, respectively. Current ML algorithms are based essentially on analysis of correlation. Correlation-based approaches require high data density relative to dimensionality, which is difficult to satisfy in functional genomic studies.
This challenge is compounded by the n of 1 problem in precision medicine where data relevant to a given patient may be limited to the data available from that patient or a small group of highly similar patients. For this reason, an integrative approach is needed to build more complete training sets that incorporate genomic data, functional genomic data, and relevant information from electronic health records such as demographics and laboratory reports. While it may never be possible to solve the n of 1 problem in precision medicine for most diseases, an integrative approach may allow us to do “deep grouping” for diseases, where we are able to identify a set of markers that can stratify patients to predict outcomes and determine optimal treatment plans.
While correlation-based analysis can be valuable for deep grouping of patients, biological processes underlying disease operate with kinetic properties that may not be easily captured by correlation-based analysis. Construction of kinetic regulatory networks of molecular and physiological events would be complex, but critical for understanding disease and ultimately improving precision medicine, including repurposing of existing therapeutics. However, similar to ML, most of the data analysis methods that have been applied in attempts to construct molecular and physiological regulatory networks are based on correlation or some variant thereof.
There is an urgent need to develop new analytical approaches that can effectively construct regulatory networks utilizing functional genomic readouts. Any analytical approaches developed for this purpose would need to perform well in the context of the disease and disease-applicable tissues, not just in highly reductionist systems. Integration of correlation-based methods and mechanistic modeling could be one such fruitful approach (28, 33). Another approach may be through the study of neuroscience, similar to how convoluted neural networks were developed. The algorithms, architectures, and functions utilized by the human brain can be a source of inspiration for the development of new algorithms that incorporate methods for complex reasoning and decision making (22).
ETHICS OF AI
While AI has many potential benefits in medicine, researchers must also consider the potential social, ethical, and economic impacts of AI (39). The safety of AI is an important concern, especially as AI gains increasing levels of autonomy. A more fundamental challenge that AI poses is the legal definition of personhood (29) and the psychological perception of human identity as AI becomes increasingly human-like. The issue is further compounded by the inevitable development of increasingly complex human-machine hybrids. A full discussion of these issues is beyond the scope of this article.
However, there are more immediate concerns about the use of AI in precision medicine. For example, AI can learn bias and prejudice values when they are present within the data set, leading to unfair or inaccurate predictions (4, 12, 24). Bias can be introduced when a specific patient population is not included in the training set. For instance, a study that predicted the probability of death for pneumonia patients to determine whether the patient were to be admitted into the hospital or treated as an outpatient mistakenly predicted treating asthma patients as outpatients. This error occurred because the training set used included patients who required further care, but a majority of asthma patients were immediately transferred to intensive care, so they were not adequately represented in the data set (7). While the bias involved in this example was related to disease representation, incorporation of prejudice values can occur in a similar manner. AI can learn preexisting inequalities from society represented in the training set, resulting in bias toward historically disadvantaged populations (4). Additionally, data can be manipulated and misinterpreted. Issues like these highlight the importance of integration of data quality and high ethical standards to mitigate bias when AI is used for precision medicine.
Preliminary efforts are underway to address the potential social, ethical, and economic impacts of AI. In 2016, the White House, the European Parliament, and the UK House of Commons each released reports summarizing how government and society should prepare for the development of AI and its widespread use. While these reports identify the potential negative impacts of AI, they fail to outline long-term implementation strategies to assess risk and harm (8). Much work is left to develop strategies to identify and address the negative impacts of AI on society, so that we can work toward the development of a “good AI society” (41b) and unleash the tremendous benefits of AI including advancement in big data-based precision medicine.
GRANTS
The authors acknowledge grant support from the National Institutes of Health (HL-121233, HL-125409, GM-066730, HL-116264, HL-082798), the Advancing a Healthier Wisconsin Endowment, and the American Heart Association (15SFRN23910002).
DISCLAIMERS
The funding sources had no role in the preparation, review, approval, or decision to submit the manuscript for publication.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
A.M.W. and Y.L. prepared figures; A.M.W. and M.L. drafted manuscript; A.M.W., Y.L., K.R.R., F.J., P.L., and M.L. edited and revised manuscript; A.M.W., Y.L., K.R.R., F.J., P.L., and M.L. approved final version.
Glossary
- Area under the curve (AUC)
A measure of the predictability of a statistical model by calculating the probability that the prediction for a random data point will match the actual outcome across all possible thresholds on an ROC curve. An AUC of 1 represents perfect accuracy (21).
- Big data
Data sets that are high volume (quantity, variety of sources), high velocity (speed of data generation and processing), and/or high variety (type, variety of formats), making traditional data processing techniques inadequate (38).
- Machine learning (ML)
An application of AI used to build models that can learn from data sets to make predictions. The model itself can improve with experience without being explicitly programmed to do so (45).
- Precision medicine
A customized medical approach used to prevent, diagnose, or treat disease based on a patient’s unique genetic, environmental, and lifestyle factors (36).
- Reinforcement learning
A type of ML that specifies how the AI should take actions for a given environment to work toward maximum benefit of a cumulative reward. This type of machine learning is often used in simulation-based optimization of games (1).
- Supervised learning
A type of ML in which the training set is labeled, with the goal of predicting a known outcome (1, 30, 34).
- Unsupervised learning
A type of ML in which the training set is unlabeled, with the goal of finding naturally occurring patterns or groupings within the data (1, 34).
Footnotes
This article is based, in part, on the Distinguished Lecture in Physiological Genomics Research given by M. Liang at Experimental Biology 2017.
REFERENCES
- 1.Alanazi HO, Abdullah AH, Qureshi KN. A critical review for developing accurate and dynamic predictive models using machine learning methods in medicine and health care. J Med Syst 41: 69, 2017. doi: 10.1007/s10916-017-0715-6. [DOI] [PubMed] [Google Scholar]
- 2.Baker MA, Davis SJ, Liu P, Pan X, Williams AM, Iczkowski KA, Gallagher ST, Bishop K, Regner KR, Liu Y, Liang M. Tissue-specific microRNA expression patterns in four types of kidney disease. J Am Soc Nephrol 28: 2985–2992, 2017. doi: 10.1681/ASN.2016121280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balaram P, Bothner-By AA, Breslow E. Nuclear magnetic resonance studies of the interaction of peptides and hormones with bovine neurophysin. Biochemistry 12: 4695–4704, 1973. doi: 10.1021/bi00747a024. [DOI] [PubMed] [Google Scholar]
- 4.Barocas S, Selbst AD. Big data’s disparate impact. Calif Law Rev 104: 671–732, 2016. [Google Scholar]
- 5.Beckmann JS, Lew D. Reconciling evidence-based medicine and precision medicine in the era of big data: challenges and opportunities. Genome Med 8: 134, 2016. doi: 10.1186/s13073-016-0388-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell M, Hoane AJ Jr, Hsu F-H. Deep Blue. Artif Intell 134: 57–83, 2002. doi: 10.1016/S0004-3702(01)00129-1. [DOI] [Google Scholar]
- 7.Caruana R, Lou Y, Gehrke J, Koch P, Sturm M, Elhadad N. Intelligible models for healthcare: predicting pneumonia risk and hospital 30-day readmission. In: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Sydney, NSW, Australia: ACM, 2015, p. 1721–1730. [Google Scholar]
- 8.Cath C, Wachter S, Mittelstadt B, Taddeo M, Floridi L. Artificial intelligence and the ‘good society’: the US, EU, and UK approach. Sci Eng Ethics: 1–24, 2017. doi: 10.1007/s11948-017-9901-7. [DOI] [PubMed] [Google Scholar]
- 9.Cheng Y, Song H, Pan X, Xue H, Wan Y, Wang T, Tian Z, Hou E, Lanza I, Liu P, Liu Y, Laud P, Usa K, He Y, Liang M. Urinary metabolites associated with blood pressure on a low- or high-sodium diet. Theranostics 8: 1468–1480, 2018. doi: 10.7150/thno.22018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 372: 793–795, 2015. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford K, Calo R. There is a blind spot in AI research. Nature 538: 311–313, 2016. doi: 10.1038/538311a. [DOI] [PubMed] [Google Scholar]
- 13.Doyle OM, Mehta MA, Brammer MJ. The role of machine learning in neuroimaging for drug discovery and development. Psychopharmacology (Berl) 232: 4179–4189, 2015. doi: 10.1007/s00213-015-3968-0. [DOI] [PubMed] [Google Scholar]
- 14.Epstein EA, Schor MI, Iyer BS, Lally A, Brown EW, Cwiklik J. Making Watson fast. IBM J Res Develop 56: 15:1–15:12, 2012. doi: 10.1147/JRD.2012.2188761. [DOI] [Google Scholar]
- 15.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542: 115–118, 2017. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol 48: 51–68, 2013. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster KR, Koprowski R, Skufca JD. Machine learning, medical diagnosis, and biomedical engineering research - commentary. Biomed Eng Online 13: 94, 2014. doi: 10.1186/1475-925X-13-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibney E. Google AI algorithm masters ancient game of Go. Nature 529: 445–446, 2016. doi: 10.1038/529445a. [DOI] [PubMed] [Google Scholar]
- 19.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316: 2402–2410, 2016. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 20.Halevy A, Norvig P, Pereira F. The unreasonable effectiveness of data. IEEE Intell Syst 24: 8–12, 2009. doi: 10.1109/MIS.2009.36. [DOI] [Google Scholar]
- 21.Halligan S, Altman DG, Mallett S. Disadvantages of using the area under the receiver operating characteristic curve to assess imaging tests: a discussion and proposal for an alternative approach. Eur Radiol 25: 932–939, 2015. doi: 10.1007/s00330-014-3487-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassabis D, Kumaran D, Summerfield C, Botvinick M. Neuroscience-inspired artificial intelligence. Neuron 95: 245–258, 2017. doi: 10.1016/j.neuron.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Holder LB, Haque MM, Skinner MK. Machine learning for epigenetics and future medical applications. Epigenetics 12: 505–514, 2017. doi: 10.1080/15592294.2017.1329068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvitz E. AI, people, and society. Science 357: 7, 2017. doi: 10.1126/science.aao2466. [DOI] [PubMed] [Google Scholar]
- 25.Huang C, Mezencev R, McDonald JF, Vannberg F. Open source machine-learning algorithms for the prediction of optimal cancer drug therapies. PLoS One 12: e0186906, 2017. doi: 10.1371/journal.pone.0186906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics 15: 41–51, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Hulse J. Data Quality in Data Mining and Machine Learning (PhD dissertation). Boca Raton, FL: Florida Atlantic University, 2007, p. 205. [Google Scholar]
- 28.Iyengar R, Altman RB, Troyanskya O, FitzGerald GA. MEDICINE. Personalization in practice. Science 350: 282–283, 2015. doi: 10.1126/science.aad5204. [DOI] [PubMed] [Google Scholar]
- 29.Jotterand F. The boundaries of legal personhood. In: Posthumanism: The Future of Homo sapiens, edited by Bess M and Pasulka DW Farmington Hills, MI: Macmillan Reference USA, 2018, p. 413–423. [Google Scholar]
- 30.Kan A. Machine learning applications in cell image analysis. Immunol Cell Biol 95: 525–530, 2017. doi: 10.1038/icb.2017.16. [DOI] [PubMed] [Google Scholar]
- 31.Kelchtermans P, Bittremieux W, De Grave K, Degroeve S, Ramon J, Laukens K, Valkenborg D, Barsnes H, Martens L. Machine learning applications in proteomics research: how the past can boost the future. Proteomics 14: 353–366, 2014. doi: 10.1002/pmic.201300289. [DOI] [PubMed] [Google Scholar]
- 32.Kotchen TA, Cowley AW Jr, Liang M. Ushering hypertension into a new era of precision medicine. JAMA 315: 343–344, 2016. doi: 10.1001/jama.2015.18359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang M. Integrative pathway knowledge bases as a tool for systems molecular medicine. Physiol Genomics 30: 209–212, 2007. doi: 10.1152/physiolgenomics.00002.2007. [DOI] [PubMed] [Google Scholar]
- 34.Libbrecht MW, Noble WS. Machine learning applications in genetics and genomics. Nat Rev Genet 16: 321–332, 2015. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu P, Lassén E, Nair V, Berthier CC, Suguro M, Sihlbom C, Kretzler M, Betsholtz C, Haraldsson B, Ju W, Ebefors K, Nyström J. Transcriptomic and proteomic profiling provides insight into mesangial cell function in IgA nephropathy. J Am Soc Nephrol 28: 2961–2972, 2017. doi: 10.1681/ASN.2016101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marson FAL, Bertuzzo CS, Ribeiro JD. Personalized or precision medicine? the example of cystic fibrosis. Front Pharmacol 8: 390, 2017. doi: 10.3389/fphar.2017.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattson DL, Liang M. Hypertension: From GWAS to functional genomics-based precision medicine. Nat Rev Nephrol 13: 195–196, 2017. doi: 10.1038/nrneph.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McAfee A, Brynjolfsson E. Big data: the management revolution. Harv Bus Rev 90: 60–66, 2012. [PubMed] [Google Scholar]
- 39.Mittelstadt BD, Allo P, Taddeo M, Wachter S, Floridi L. The ethics of algorithms: mapping the debate. Big Data Soc: 1–21, 2016. doi: 10.1177/2053951716679679. [DOI] [Google Scholar]
- 40.Mnih V, Kavukcuoglu K, Silver D, Rusu AA, Veness J, Bellemare MG, Graves A, Riedmiller M, Fidjeland AK, Ostrovski G, Petersen S, Beattie C, Sadik A, Antonoglou I, King H, Kumaran D, Wierstra D, Legg S, Hassabis D. Human-level control through deep reinforcement learning. Nature 518: 529–533, 2015. doi: 10.1038/nature14236. [DOI] [PubMed] [Google Scholar]
- 41.Moravčík M, Schmid M, Burch N, Lisý V, Morrill D, Bard N, Davis T, Waugh K, Johanson M, Bowling M. DeepStack: Expert-level artificial intelligence in heads-up no-limit poker. Science 356: 508–513, 2017. doi: 10.1126/science.aam6960. [DOI] [PubMed] [Google Scholar]
- 41a.National Research Council Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC: The National Academies Press, 2011, p. 142. [PubMed] [Google Scholar]
- 41b.National Science and Technology Council, Committee on Technology Preparing for the Future of Artificial Intelligence. Washington, DC: Executive Office of the President, 2016. [Google Scholar]
- 42.Obermeyer Z, Emanuel EJ. Predicting the future - big data, machine learning, and clinical medicine. N Engl J Med 375: 1216–1219, 2016. doi: 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reeve J, Böhmig GA, Eskandary F, Einecke G, Lefaucheur C, Loupy A, Halloran PF, MMDx-Kidney study group . Assessing rejection-related disease in kidney transplant biopsies based on archetypal analysis of molecular phenotypes. JCI Insight 2: 94197, 2017. doi: 10.1172/jci.insight.94197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaeffer J, Lake R, Lu P, Bryant M. Chinook: the world man-machine checkers champion. AI Mag 17: 21–29, 1996. doi: 10.1609/aimag.v17i1.1208. [DOI] [Google Scholar]
- 45.Senders JT, Zaki MM, Karhade AV, Chang B, Gormley WB, Broekman ML, Smith TR, Arnaout O. An introduction and overview of machine learning in neurosurgical care. Acta Neurochir (Wien) 160: 29–38, 2018. doi: 10.1007/s00701-017-3385-8. [DOI] [PubMed] [Google Scholar]
- 46.Silver D, Huang A, Maddison CJ, Guez A, Sifre L, van den Driessche G, Schrittwieser J, Antonoglou I, Panneershelvam V, Lanctot M, Dieleman S, Grewe D, Nham J, Kalchbrenner N, Sutskever I, Lillicrap T, Leach M, Kavukcuoglu K, Graepel T, Hassabis D. Mastering the game of Go with deep neural networks and tree search. Nature 529: 484–489, 2016. doi: 10.1038/nature16961. [DOI] [PubMed] [Google Scholar]
- 47.Silver D, Schrittwieser J, Simonyan K, Antonoglou I, Huang A, Guez A, Hubert T, Baker L, Lai M, Bolton A, Chen Y, Lillicrap T, Hui F, Sifre L, van den Driessche G, Graepel T, Hassabis D. Mastering the game of Go without human knowledge. Nature 550: 354–359, 2017. doi: 10.1038/nature24270. [DOI] [PubMed] [Google Scholar]
- 48.Somashekhar SP, Sepúlveda MJ, Puglielli S, Norden AD, Shortliffe EH, Rohit Kumar C, Rauthan A, Arun Kumar N, Patil P, Rhee K, Ramya Y. Watson for Oncology and breast cancer treatment recommendations: agreement with an expert multidisciplinary tumor board. Ann Oncol 29: 418–423, 2018. doi: 10.1093/annonc/mdx781. [DOI] [PubMed] [Google Scholar]
- 50.Tesauro G. Temporal difference learning and TD-Gammon. Commun ACM 38: 58–68, 1995. doi: 10.1145/203330.203343. [DOI] [Google Scholar]
- 51.Touyz RM, Montezano AC, Rios F, Widlansky ME, Liang M. Redox stress defines the small artery vasculopathy of hypertension: how do we bridge the bench-to-bedside gap? Circ Res 120: 1721–1723, 2017. doi: 10.1161/CIRCRESAHA.117.310672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weng SF, Reps J, Kai J, Garibaldi JM, Qureshi N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS One 12: e0174944, 2017. doi: 10.1371/journal.pone.0174944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Widlansky ME, Jensen DM, Wang J, Liu Y, Geurts AM, Kriegel AJ, Liu P, Ying R, Zhang G, Casati M, Chu C, Malik M, Branum A, Tanner MJ, Tyagi S, Usa K, Liang M. miR-29 contributes to normal endothelial function and can restore it in cardiometabolic disorders. EMBO Mol Med 10: e8046, 2018. doi: 10.15252/emmm.201708046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet 6: e1000864, 2010. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell 153: 516–519, 2013. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]