Abstract
Large scale consortia mapping the genomic risk architectures of schizophrenia provide vast amounts of molecular information, with largely unexplored therapeutic potential. We harnessed publically available information from the Psychiatric Genomics Consortium, and report MYOCYTE ENHANCER FACTOR 2C (MEF2C) motif enrichment in sequences surrounding the top scoring single nucleotide polymorphisms within risk loci contributing by individual small effect to disease heritability. Chromatin profiling at base pair resolution (ChIP-seq) in neuronal nucleosomes extracted from prefrontal cortex of 34 subjects, including 17 cases diagnosed with schizophrenia, revealed MEF2C motif enrichment within cis-regulatory sequences, including neuron-specific promoters and superenhancers, affected by histone H3K4 hypermethylation in disease cases. Vector-induced short- and long-term Mef2c upregulation in mouse prefrontal projection neurons consistently resulted in enhanced cognitive performance in working memory and object recognition paradigms at baseline and after psychotogenic drug challenge, in conjunction with remodeling of local connectivity. Neuronal genome tagging in vivo by Mef2c-Dam adenine methyltransferase fusion protein confirmed the link between cognitive enhancement and MEF2C occupancy at promoters harboring canonical and variant MEF2C motifs. The multilayered integrative approaches presented here provide a roadmap to uncover the therapeutic potential of transcriptional regulators for schizophrenia and related disorders.
Keywords: TRANSCRIPTION FACTOR, PREFRONTAL CORTEX, SCHIZOPHRENIA, NEURONAL EPIGENOME, HISTONE METHYLATION, MEF2C
INTRODUCTION
Dysregulation of neuronal gene expression in prefrontal cortex (PFC) and other brain regions is a critical building block in the neurobiology of schizophrenia1–6, an adult-onset psychiatric disorder associated with defective cognition and a wide range of additional disabling symptoms often resistant to treatment with conventional antipsychotic drugs targeting monoamine signaling pathways7. Presently, there is extremely little knowledge on the therapeutic potential of specific transcriptional regulators associated with the disease. Here, we present a novel multidimensional 4-step approach to identify neuronal transcription factors (TF) that could positively affect cognitive performance, a key variable for prognosis and long-term outcome in schizophrenia8. To this end, we (1) harness publically accessible databases to identify TFs with motif enrichment within the genetic risk architecture of schizophrenia9 and (2) study expression changes in context of normal development and disease, taking into account the developmental origins of the disorder 10. (3) Next, we explore sequence motifs associated with epigenetic dysregulation in chromatin surrounding transcription start sites (TSS) in neuronal nuclei extracted from PFC of normal and disease cases. (4) Finally, we study in preclinical model systems, including mice and human induced pluripotent stem cell (hIPSC)-derived neuronal cultures, the behavioral and molecular and cellular mechanisms associated with the specific TF. Here, we introduce MEF2C, a member of the MEF (myocyte-specific enhancer factor) subfamily of MADS transcription factors, as a promising therapeutic tool to induce cognitive enhancement when overexpressed in neurons residing in adult PFC, a key structure in the neural circuitry associated with cognitive deficits in schizophrenia11. The strategies and approaches outlined here could easily be scaled up for the initial discovery of disease-relevant chromatin regulators with subsequent validation in preclinical models. Such types of approaches will be expected to provide the field with a pipeline of molecular candidates aimed at improving cognition and some of the other core features on schizophrenia.
MATERIALS AND METHODS
Detailed descriptions and procedures for human postmortem brain tissue, chromatin techniques including ChIP-seq and ChIP-PCR, RNA quantification, MEF2C knock-down, differentiation of pluripotent stem cells, gene ontology analyses, gel shift assays, analyses of publically accessible databases, bioinformatical analyses, animal work, vector construction, and MEF2C-Dam GmATC adenine methylation assays are provided in the supplemental methods section.
RESULTS
TF signatures in the molecular architecture of schizophrenia
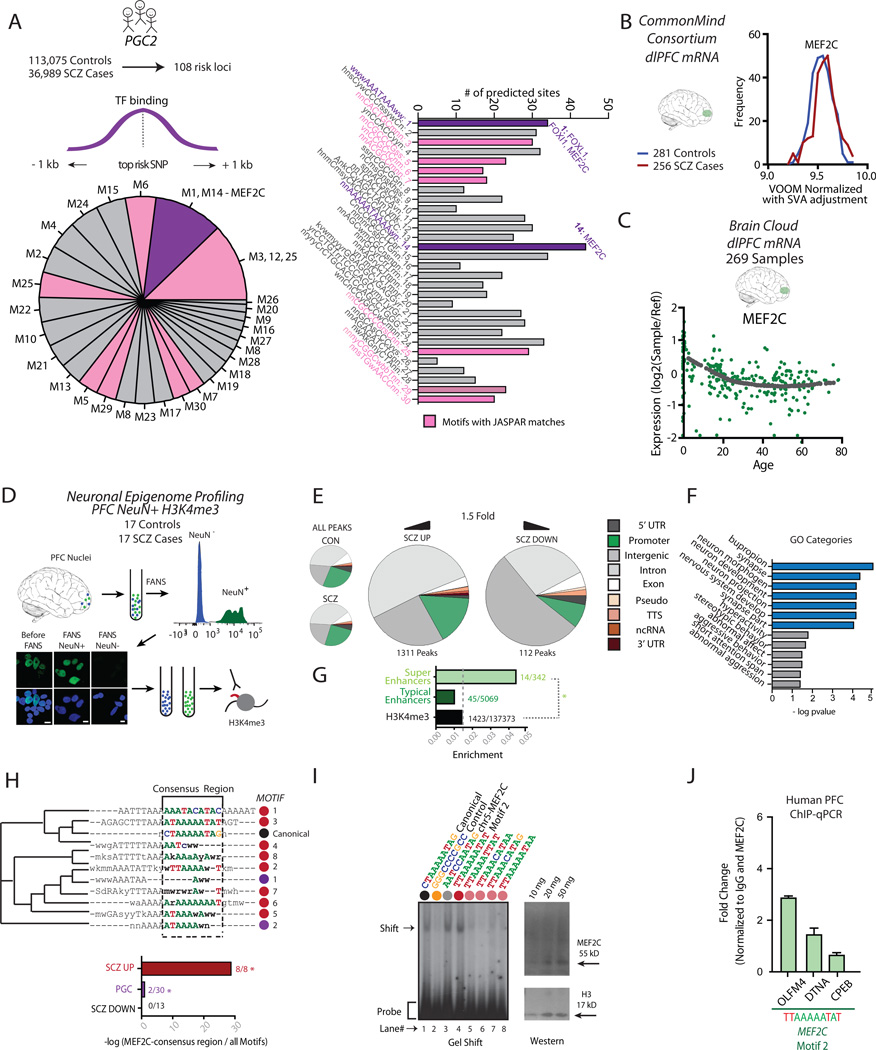
To identify TF relevant for the neurogenomics of schizophrenia, we began by analyzing two publically available datasets. These were the Psychiatric Genomics Consortium 2 (PGC2) database for 108 risk regions harboring alleles that, by individually small effect, contribute to heritability risk 9 and prefrontal RNA-seq from 256 cases diagnosed with schizophrenia and 281 controls, presented by the CommonMind Consortium (www.synapse.org/cmc). We determined 30 motifs using the RSAT de novo motif suite (see Methods) that were enriched within 2kb surrounding the top scoring single nucleotide polymorphism (SNP) for each of the 108 risk regions from the PGC2 dataset of 36,989 schizophrenia cases and 113,075 controls (Figure 1A and Supplemental Table 1). From the 30 motifs, nine were assigned to a specific TF, or TF group (JASPAR12).We hypothesized that these TF motifs within the PGC2 dataset could indicate a potential role of the TF in the neurobiology, and therefore treatment, of schizophrenia.
Figure 1. Epigenomic Dysregulation of Regulatory Sequences in PFC neurons from Subjects with Schizophrenia.
(A) (Left) 30 TF motifs were discovered using RSAT de novo motif analysis ±1kb of the lead SNP from the 108 genetic regions identified from the PGC2 dataset. (Right) From a total of N=9 motifs matching to specific transcription factors (TFs) in JASPAR, N=2 motifs matched the canonical MEF2C motif. (B) CommonMind Consortium RNA-seq data from disease cases show minimal increase in prefrontal MEF2C expression. (C) BrainCloud microarray data, showing the developmental trajectory of prefrontal MEF2C transcript levels. (D) Neuronal chromatin profiling from postmortem PFC. Cortical nuclei are in suspension immunostained with NeuN antibody. Representative fluorescence-activated nuclei sorting (FANS) plot after 3×104 events, showing complete separation of NeuN+ and NeuN− nuclei. Microscopic images of NeuN+ (green), DAPI counterstained (blue) nuclei pre- and post-sort, as indicated. Scale bar, 5 µm. Mono-nucleosomal preparations from neuronal chromatin are immunoprecipitated with anti-H3K4me3 antibody and processed for ChIP-seq (E) Pie charts, (left) genome-scale proportions of H3K4me3-tagged sequences in disease and control PFC neurons (N=17 subjects/group) and (right) functional representation of SCZ-up and SCZ-down peaks. Note overrepresentation of intronic and intergenic sequence, at the expense of gene promoters. (F) Bar graph showing enrichment for (blue) gene ontology categories and drug response, and (gray) phenotype pathways 24 in SCZ-up peaks. (G) H3K4me3 peaks altered in PFC neurons from schizophrenia subject shows significant enrichment for PFC superenhancers (14/342) 27. Note that there is no disease-associated enrichment of typical enhancers tagged with H3K4me3 (45/5069), compared to the proportion of the entire pool of dysregulated H3K4me3 peaks (1423) against the total pool of H3K4me3 peaks (137373). *, P<005, hypergeometric test. (middle bottom) (H) (top) RSAT de novo motifs28 enriched in SCZ-up peaks (red dots), all presenting as variants of the generic CT(A/T)4TAG/A consensus sequence together with canonical MEF2C motif (black dot) and 2/30 RSAT-predicted PGC2 motifs (purple dots) using a neighbor joining tree with the canonical MEF2C motif (Clustal Omega29, 30) (see also Supplemental Table 13). (bottom) Bargraph, showing MEF2C motif enrichments specifically among SCZ-up peaks (8/8) and PGC2 risk regions (2/30), but not among SCZ-down (0/30). (I) (left) Gel shift assay with human PFC nuclei as input and eight 32P-labeled 27bp duplex probes with variable 10bp insert (Supplemental Table 15). Lane 1, MEF2C canonical; lane 2, non-MEF2C control; lane 3, chr.5 MEF2C super-enhancer sequence harboring canonical MEF2C motif shown in Figure 3A; lanes 4–8, MEF2C-like motifs enriched in SCZ-up peaks, as indicated. Notice highest affinity for ‘Motif 2’. (right) MEF2C immunoblot from PFC nuclei, showing robust 55 kDa (full length) MEF2C immunoreactivity. Histone H3 for loading control. (J) ChIP-PCR with anti-MEF2C antibody on human PFC nuclei (N=3 control subjects), showing several-fold enrichment at gene promoters harboring MEF2C motif variant ‘Motif 2’. Data expressed as fold-increase over ChIP-PCR with IgG control antibodies.
To explore this hypothesis, we first asked if the (altogether 20) TF associated with the nine motifs are differentially expressed in PFC of subjects with schizophrenia, using the CommonMind Consortium PFC RNA-seq dataset (freeze1.v9=256 schizophrenia cases, 281 controls.www.synapse.com/cmc). Expression differences between cases and controls were minimal, however. For example, MEF2C, one of two TFs surviving statistical filtering in the CMC dataset (Benjamini-Hochberg False Discovery Rate comparing expression differences for 20 TF, FDR, p=0.011; Bonferroni p=0.001*20=0.02) showed only a 2.3% change between CMC cases and controls (Figure 1B and Supplemental Table 2). We conclude that adult PFC from subjects with schizophrenia does not show consistent or robust alterations in expression of TF with motif enrichment in the PGC2 dataset. However, neurodevelopmental factors play an important role in schizophrenia13, and alterations in MEF2C and other TF expression may have impacted diseased brains at earlier periods of pre- and postnatal life. Interestingly, in BrainCloud, a publically accessible transcriptome database for 272 postmortem brains collected across the lifespan from prenatal to old age10, prefrontal MEF2C expression showed considerable variability in immature prefrontal cortex, followed by modest decline in expression during the course of normal aging (Figure 1C). We conclude MEF2C, as a TF with motif enrichment within PGC2, is dynamically regulated across the extended period of PFC development.
Epigenomic dysregulation in PFC neurons from subjects with schizophrenia
Various epigenetic modifications, including nucleosomal histone methylation, show strong correlations with transcription factor (TF) binding14. In particular, trimethyl-histone H3-lysine 4 (H3K4me3), a mark broadly correlated with gene expression and open chromatin15, shows strong associations with TF binding at cis-regulatory sequences including promoters and some enhancers16. Interestingly, an earlier study has provided evidence for dysregulation of H3K4me3 in immature neurons in the olfactory epithelium of subjects with schizophrenia17. However, genome-scale H3K4me3 profiling from brain tissue, including cortical neurons, of subjects with schizophrenia is lacking. We hypothesized that sequences with altered histone methylation in neuronal chromatin from PFC of subjects with schizophrenia could be enriched for some of the same TF motifs that are enriched within the 108 risk regions initially described by genome wide association (GWA) in PGC2 (Figure 1A). To explore this, we mapped H3K4me3 in neuronal nucleosomes from 34 PFC specimens, including 17 (14M/3F) subjects diagnosed with schizophrenia and 17 (14M/3F) control subjects of comparatively young age (mean ~ 40 years, Supplemental Tables 3, 4). Neuronal and non-neuronal nuclei were separated by fluorescence activated sorting of NeuN immuno-tagged nuclei, thus providing information specific to the neuronal genome and bypassing the potential confound of neuron-to-glia variation18–22. Mononucleosomal preparations from the neuronal chromatin fraction were used for H3K4me3 ChIP-seq (Figure 1D). Genome-scale H3K4me3 landscapes, including ~27-fold enrichment for promoter sequences <2kb from TSS, were indistinguishable between cases and controls (Figure 1E). Using specific filter criteria (>1.5-fold difference/FDR corrected cumulative Poisson P<0.0001 using HOMER (Hypergeometric Optimization of Motif Enrichment suite for next-generation sequencing analysis)23), we identified 1311 differentially methylated regions (DMRs) with increased (schizophrenia up, SCZ-up), and 112 DMRs with decreased H3K4me3 (SCZ-down) in our disease cohort. Disease-associated DMRs showed modest enrichments for intronic and intergenic sequences (Figure 1E and Supplementary Tables 5–8)23. Next, we asked if these disease-associated changes were enriched for specific gene categories. To this end, we filtered for GO categories with at least 10 dysregulated loci and surviving a hypergeometric test with Bonferroni corrected P<0.001 using WebGestalt24). Indeed, H3K4me3 hypermethylated sequences carried a strong neuronal footprint with 6/12 Gene Ontology (GO) categories related to synapses and neurons, and another 8/18 ‘Drug’ and ‘Phenotype’ categories matched to diseased cognition and abnormal behaviors (Figure 1F and Supplemental Table 9a,b). In contrast, DMRs with decreased H3K4me3 in disease cases did not show any GO enrichment. We conclude that genes involved in synaptic signaling and neurotransmission are particularly affected by epigenetic dysregulation, including histone H3K4 hypermethylation, in neuronal chromatin from PFC with subjects with schizophrenia. This finding resonates with recent DNA methylation profilings in brain tissue homogenate, reporting dysregulated CpG cytosine methylation at neuronal genes in schizophrenia25, 26.
Within the pool of disease-associated H3K4 hypermethylated sequences, we noticed that superenhancers—defined by broad stretches of open chromatin-associated histone acetylation and clustered TF occupancies as key regulators for cell-type specific gene expression and function27—showed by hypergeometric test a significant 4-fold overrepresentation (Figure 1G, Supplemental Tables 10, 11). Furthermore, JASPAR-based motif analysis for the 14 superenhancers with excessive levels of H3K4me3 in diseased PFC neurons showed significant enrichment for eight motifs (Supplemental Table 12), when using the total set of N=342 brain-specific super-enhancers27 as background (P<0.05, right-tailed binomial significance test, corrected by the total number of motifs predicted = evalue, reported as weight significance −log10(evalue)>7.5 (Supplemental Tables 13, 14)28. Of note, at least one of the motifs enriched among the H3K4 hypermethylated superenhancer sequences is predicted to bind MEF2C (Supplemental Table 10). Next, we explored motif enrichment for the total collection of 1311 up and 121 down H3K4me3 DMRs, using the total set of 137, 373 unique (=present in at least one subject) H3K4me3 peaks in the control and schizophrenia samples as background. We again applied RSAT de novo motif search. We identified eight (thirteen) motifs showing significant enrichment in the 1311 SCZ-up (121 SCZ-down)(P<0.05, right-tailed binomial significance test corrected by the total number of motifs predicted =evalue and reported as weight significance (−log10(evalue)) > 7.5) (Supplemental Tables 13,14)). Strikingly, 8/8 or 100% of the SCZ-up but none of the 13 SCZ-down motifs clustered together with the canonical MEF2C motif in a neighbor joining tree (Clustal Omega, 29, 30). All SCZ-up motifs were variants of the generic MEF2C motif CT(A/T)4TAG/A (Figure 1H). Given such prominent MEF2C motif footprint in neuronal chromatin from diseased PFC, we first confirmed by immunoblotting that MEF2C is indeed expressed in cell nuclei extracts from mature human PFC (Figure 1I). Next, gel shift assays confirmed strong nucleoprotein binding of multiple MEF2C motifs, including the canonical form and a variant MEF2C binding motifs. These include the MEF2C promoter itself, and a variant motif, ‘motif 2’, found in the hypermethylated sequences in diseased neurons (Figure 1I). To directly test for MEF2C binding at these sequences, we immunoprecipitated chromatin extracts from human PFC with anti-MEF2C antibody (ChIP-MEF2C). Indeed, 3/3 gene promoters harboring the MEF2C motif2 (OLFM4, DNTA and CPEB) showed robust chip-to-input ratios compared to control (ChIP with non-specific IgG) (Figure 1J). Furthermore, because MEF2C protein levels are unaltered in prefrontal cortex of mice exposed to chronic 21 day treatment with the antipsychotic, clozapine (Supplemental Figure 1), the observed MEF2C motif enrichment in H3K4 hypermethylated sequences in our disease cases less unlikely to reflect secondary effects after antipsychotic drug exposure.
Importantly, H3K4me3 landscapes in human PFC neurons show highly dynamic changes during the extended period of developmental from prenatal to young adult age31, and remain broadly sensitive to functional perturbations and cortical and subcortical dysfunction encountered in neuropsychiatric disease32–37. Given such wide windows of epigenomic vulnerability, the time-of-onset for the observed H3K4me3 methylation changes at regulatory sequences carrying MEF2C binding motifs in our adult postmortem disease cohort could vary case-by-case and in target sequence-specific manner. Therefore, to further test whether the observed H3K4me3 hypermethylation of MEF2C target sequences in neuronal chromatin from disease cases reflects compensatory mechanisms in response to decreased MEF2C access or supply, we induced small RNA-mediated MEF2C knock-down in HEK293 cells. Strikingly, there was a significant drop in H3K4me3 levels at the MEF2A promoter (which carries multiple MEF2C motifs at its promoter sequences) and similar changes for additional target sequences (Supplemental Figure 2). These findings imply that increased H3K4me3 at sequences with MEF2C binding motifs in our diseased postmortem brains could reflect compensatory epigenomic mechanism in response to deficits in MEF2C regulatory activity. If this hypothesis is correct, then a robust increase in the neuronal supply of MEF2C should exert a therapeutic effect. Thus, we next tested this hypothesis in an animal model.
Increased Mef2c expression in PFC neurons improves cognition
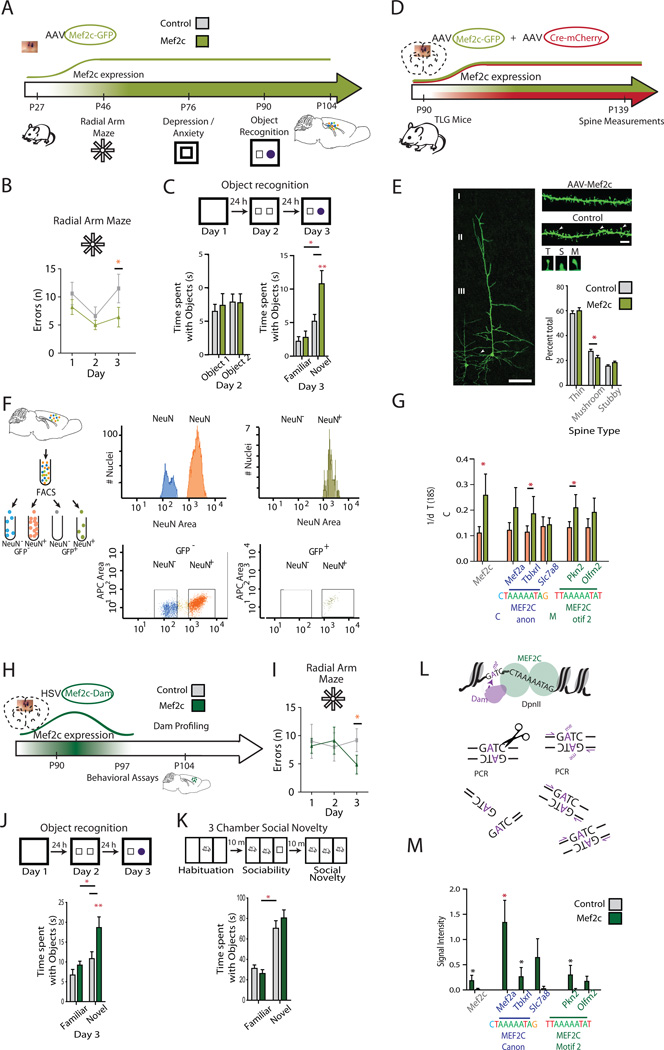
To explore whether MEF2C expression in adolescent and young adult PFC (the typical time of onset of schizophrenia symptoms38) could impact complex behaviors, we bilaterally injected juvenile C57Bl6 mice at postnatal day P27 with adeno-associated virus human synapsin I (hSYN1) promoter to drive neuronal expression of Mef2c-GFP (AAV8hSYN1-Mef2cGfp), or control vector (AAV8hSYN1-CreGFP) (N=10 mice/group, Figure 2A). We focused on PFC-dependent behaviors with relevance to schizophrenia, including working memory—which is impaired in many patients39—and long-term memory40. We tested working memory at P46-P50 with the radial arm maze and long-term memory over 3 days41–43, by the 24-hour interval novel object recognition paradigm—a test that is sensitive to atypical antipsychotic drugs and serves as a rodent model for cognitive impairment in schizophrenia44—at P89-P91. (Figure 2A). A lower number of repeated entries (‘errors’) into individual maze arms approached significance using both a two-way ANOVA (F(2)=4.335, p=0.0519) and a one-tailed t-test for day 3 (t(18)=1.651, p=0.0580) (Figure 2B). Furthermore, AAV8hSYN1-Mef2cGfp mice, in comparison to AAV8hSYN1-CreGFP controls, spend significantly more time with the novel object when presented together with the familiar’ object on day 3 using a two-way ANOVA (F(1)=38.25, p<0.0005) followed by a posthoc Bonferroni for Mef2c (t=3.751, p<0.01) (Figure 2C). These memory-promoting effects by the Mef2c-Gfp transgene were highly specific, because behavioral assays for general locomotor activity and anxiety and depression-related behaviors remained indifferent to increased prefrontal Mef2c expression (Supplemental Figure 3), consistent with earlier studies on Mef2c-deficient mice45.
Figure 2. Improved cognition after Mef2c expression in PFC neurons.
(A) Overview, timeline of AAV8 experiment and timeline: postnatal day P27, bilateral PFC injection for hSynapsin1 promoter-driven Mef2c, or CreGfp control cDNA (N=10/group). PFC harvest at P104, after completion of behavioral assays. (B) Radial arm maze. 2-way ANOVA, (F(2)=4.335, p=0.0519. Day 3 (test day), one-tailed t-test (t(18)=1.651, p=0.0580). (C) Three day novel object recognition paradigm. Note that Mef2c-Gfp mice spend significantly more time with the novel object when presented together with the familiar object on test day (day 3). 2-way ANOVA (F(1)=38.25, p<0.0001; posthoc Bonferroni t=3.751, p<0.01. (D) Overview, timeline of AAVhSYN1(Mef2c-Gfp)/AAVhSYN1(Cre-mCherry) experiments, including P90 2-vector co-injections into TLG498 mice. P139 dendritic spines assessment in neurons with cre-mediated expression of membrane-bound GFP46. (E) (left) Representative example of layer III PFC pyramidal neuron. Note triangular soma and prominent apical dendrite reaching through vertical thickness of upper cortical layers and basal dendrite within 150 micron from soma selected for spine morphometry, marked by arrowhead . Size bar, 100 µm. (top right) Examples of basal dendrites of Mef2c expressing neuron and control, as indicated. Arrowheads depict examples of (T) thin, (S) stubby and (M) mushroom spines, as indicated. Size bar, 5 µm. The bar graph shows a significant decrease in proportion of mushroom spines in Mef2c-Gfp transfected PFC neurons compared to control (N=3 mice/group, 10 dendrites/animal (5/hemisphere) (t(58)=2.040, p=0.0459, unpaired. two-tailed t-test) and trending increase in stubby spines (t(58)=1.837, p=0.0713). Minimum dendritic length analyzed/dendrite, 20 µm. (F) (left) Overview of FANS from AAVMef2c-GFP injected mouse cortex (right) Representative FANS plot after 104 sorting events. PFC nuclei from mice injected with AAVhSYN1(Mef2c-Gfp) were NeuN+ immunostained for 4-way sort (NeuN+/GFP+, NeuN+/GFP−, NeuN−/GFP+, NeuN−/GFP−. Notice near-complete absence of NeuN−/GFP+ nuclei. (G) Levels of transcript for Mef2c, and for MEF2C target genes. Total RNA extracted from (green) NeuN+, GFP+ nuclei of mice injected with AAVhSYN1(Mef2c-Gfp) compared to (orange) NeuN+, GFP− nuclei from same the PFC specimens. N=5–7/group. *,P<0.05 one-tailed t-test: Mef2c, P=0.035; Tblxrl, P=0.004; Pkn2, P=0.017. (H) Overview, timeline of HSVMef2c-Dam experiment: P90, bilateral PFC injections for IE4/5 promoter-driven expression of Mef2c-Dam or sham control. (I) Radial arm maze. HSVMef2c-Dam mice outperformed control mice on day 3 (test day). N=5/group one-tailed t test, t(8)=1.823, p=0.0529. (J) Novel object recognition. HSVMef2c-Dam mice spent more time with the novel object on day 3 (test day) of the 3-day novel object recognition paradigm, compared to controls. N=10/group; 2-way ANOVA for object F(1)=13.51, p=0.0017; for transgene F(1)=8.501, p=0.0092, posthoc Bonferroni for Mef2c transgene t=3.617, p<0.01. (K) 3-chamber social novelty paradigm. N=10/group, 2-way ANOVA, novel vs familiar F(1)=83.70, p<0.0001. There is no effect by transgene. (L) DamID assay. MEF2C-DAM fusion protein confers GmATC methylation at MEF2C-binding sites. Dam-methylated sequences are measured by PCR after DpnII digest. (M) Bar graph for quantification of PCR amplicons from Dpn II digested DNA at multiple MEF2C target sequences, including proximal Mef2a promoter. Note absence of PCR products in DpnII treated DNA from sham injected PFC.
In addition to the behavioral and molecular changes, we wanted to find out whether increased MEF2C could affect functional connectivity of PFC neurons. To this end, we examined spine morphologies of basal dendrites of PFC layer III pyramidal neurons, using a conditional line expressing membrane-bound GFP (GFP-F)46 for Golgi-like labelings of individual cortical cells after low titer AAV-Cre injections (Figure 2D,E). Indeed, for the three broadest spine classes (thin-stubby-mushroom) characteristic for mature cortical neurons47 , AAV8hSYN1-Mef2cGfp exposure led to a shift in composition, primarily due to a significant decrease in the proportion of mushroom spines (t(58)=2.040, p=0.0459, unpaired two-tailed t-test) and a trending increase in stubby spines (t(58)=1.837, p=0.0713)(Figure 2E). This was associated with a non-significant increase in spine densities (N=3 animals/group and 10 dendrites/animal, mean±S.E.M. spines/µm, AAV8hSYN1-Mef2cGfp, 2.059±0.14; control, 1.98± 0.10).
Having shown that viral vector-induced long-term upregulation of Mef2c expression in adult PFC neurons is associated with improved cognition and remodeling of spine profiles in pyramidal neurons, we next asked whether gene expression is altered in PFC of AAV8hSYN1-Mef2cGfp mice, specifically at promoters harboring Mef2c motifs found within risk loci for schizophrenia. To this end, we performed RT-PCR from intranuclear RNA of GFP+/NeuN+ nuclei that were FACS-sorted from AAV8hSYN1-Mef2cGfp mice. We confirmed a significant ~2.5-fold increase in Mef2c RNA (p=0.0354 paired one-tailed t-test) in neuronal nuclei expressing the Mef2c-Gfp transgene, compared to control (GFP−/NeuN+) neurons from the same PFC specimens (Figure 2F, 2G). Furthermore, gene promoters with human/mouse conserved MEF2C motifs affected by H3K4me3 hypermethylation in our schizophrenia cases showed increased expression in the Mef2c-Gfp expressing GFP+/NeuN+ nuclei compared to control GFP−/NeuN+ nuclei, including Tblxrl (p=0.0042, paired one-tailed t-test) and Pkn2 (p=0.0176, paired one-tailed t-test) (Figure 2G), thus showing that genes with variant Mef2c motifs are sensitive to the vector-induced increased supply of MEF2C.
To further test the link between improved cognition and increased MEF2C occupancy in chromatin of PFC neurons, we fused Mef2c to bacterial DNA adenine methyltransferase (Dam) cDNA and transiently expressed the resulting chimeric protein via herpes simplex vector (HSV) (Figure 2H). HSV-mediated expression in brain is limited to <7days48, 49, thereby allowing us to compare the behavioral effects after short-term (1 week, Figure 2H) and long-term (+10 weeks, Figure 2A) increase of Mef2c expression in adult PFC neurons. Indeed, we report striking similarities between the two treatment models. Radial arm maze performance approached significance in HSVMef2c-Dam mice compared to control mice on day 3 using a one-tailed ttest, t(8)=1.823, p=0.0529 (Figure 2I), similar to behavioral changes observed in the AAV8hSYN1-Mef2cGfp mice described above. Novel object recognition performance was superior, compared to control, in HSVMef2c-Dam mice (2-way ANOVA for object F(1)=13.51, p=0.0017; for transgene F(1)=8.501, p=0.0092, posthoc Bonferroni for Mef2c, t=3.617, p<0.01) (Figure 2J), consistent with similar changes observed in AAV AAV8hSYN1-Mef2cGfp animals (Figure 2C). The results from our HSVMef2c-Dam and AAV8hSYN1-Mef2cGfp studies, taken together, suggest that cognitive performance is significantly improved, compared to control animals, both after longer-term (up to 10 weeks) and short-term (1 week) increase of prefrontal Mef2c expression. Next, we wanted to explore whether such type of cognitive enhancement is specific for Mef2c. To this end, we tested behavioral changes after prefrontal expression of a different type of TF, Core-binding Factor, Beta (Cbfb). We generated HSVCbfb vectors for prefrontal expression of Cbfb, which is a Runx transcription factor associated protein implicated in postmitotic neuron differentiation50. Importantly CBFB expression was, like MEF2C, significantly increased in the PFC of the CMC disease cohort (Supplemental Figure 4A). However, HSVCbfb mice, in comparison to HSVGfp injected controls, did not show superior memory performance, or changes in locomotion or anxiety and depression-related behaviors (Supplemental Figure 4B–H). We conclude that Mef2c, but not other TF with dysregulated expression in the CMC schizophrenia cohort, including Cbfb, improves prefrontal cognition and memory in the mouse.
Next, we tested if improved test performance in HSVMef2c-Dam mice extends to other tests relevant for prefrontal function, including social recognition. However, independent of transgene delivery into PFC, all groups of animals tested showed strong preference for the novel over the familiar mouse (2-way ANOVA novel vs familiar F(1)=83.70, p<0.0001) (Figure 2K). We conclude that improved behavioral test performance after Mef2c upregulation in PFC neurons does not uniformly apply to all cognitive domains, with novel object recognition and working memory highly responsive to Mef2c increase, in contrast to social cognition.
Because our behavioral assays studies described above were limited to Mef2c effects on cognition at baseline, we added a pharmacological challenge in our final set of behavioral experiments, with repeated low dose daily systemic administration of NMDA receptor antagonist MK-801 (0.2 mg/kg). Of note, transient disruption of NMDA receptor signaling induces lasting impairments in neuronal signaling, cognition, social behaviors and emotion, and serves as frequently implied pharmacological model for schizophrenia and other psychosis spectrum disorder51–54. Mice were injected with prefrontal AAV8hSYN1-Mef2cGfp or AAV8hSYN1-CreGfp as control, and 5 weeks post-injection subjected to MK-801 or vehicle, followed 30 min later by a radial arm maze test. Under such type of psychotogenic drug challenge, AAV8hSYN1-Mef2cGfp significantly outperformed AAV8hSYN1-CreGfp in working memory on the test day (Supplemental Figure 5).
In vivo Dam-tagging of Mef2c binding sites
Previous studies in cell culture suggests that TF Dam methyl-transferase, fusion proteins inform about genomic occupancies of the TF55, by virtue of Dam-mediated adenine methylation mA activity which is specific for GATC tetramers. Note that in wildtype vertebrate genomes, endogenous adenine methylation is extremely low and limited to sequence context other than GATC56. Therefore, we reasoned that GmATC adenine methylation analyses in PFC neurons transfected with HSVMef2c-Dam could test whether MEF2C-mediated cognitive enhancement is associated with MEF2C occupancies at promoters carrying a MEF2C binding site. We applied differential Dpn II restriction digest-PCR to measure GmATC adenine methylation57 in DNA extracted from HSVMef2c-Dam and control PFC (Figure 2L). We focused on human/mouse conserved cis-regulatory sequences affected by H3K4 hypermethylation in the PFC neurons of our schizophrenia cohort. Indeed, robust GmATC adenine methylation was present at 5/7 or 83% of cis-regulatory sequences harboring MEF2C binding motifs, including sites with the canonical binding sequence (Mef2a, Tblxrl, Slc7a8, Pkn2) and at sites carrying the above mentioned ‘Motif 2’ sites (Pkn2), while adenine methylation at a promoter lacking MEF2C motifs (Foxp1) were similar to background (Figure 2M). Furthermore, no adenine methylation was found in DNA from sham-injected PFC samples, processed in parallel with the DNA from HSVMef2c-Dam PFC (Figure 2M). We conclude that improved cognition in mice with increased Mef2c expression in PFC neurons is associated with increased genomic occupancy by MEFC2 protein at gene promoters harboring MEF2C recognition sequences.
High order chromatin at the MEF2C locus
Having shown that increased neuronal Mef2c expression is associated with cognitive changes and up-regulated expression at disease-relevant gene promoters harboring MEF2C motifs, we wanted, in our final set of experiments, gain first insights into additional regulatory layers that could govern transcription at MEF2C-sensitive genes, in relation to the genetic risk architecture of schizophrenia. For example, cis-regulatory motifs orchestrating neuronal gene expression— including those embedded in chromatin tagged with the transcriptional mark, H3K4me3—are non-randomly arranged into 3-dimensional chromosomal conformations58–60. This includes ‘loopings’ that ‘bundle’ non-contiguous DNA elements, potentially separated by many kilobases of linear genome, into close physical proximity58–61. To explore, we focus here on the human MEF2C gene itself, because it harbors H3K4 hypermethylated MEF2C binding motifs in our disease cohort (Figure 3A). Of note, the MEF2C gene is surrounded by multiple non-coding and antisense RNAs in the chromosome 5q14.3 neurodevelopmental risk locus62, and evidently undergoes complex regulation in human cerebral cortex (Figure 3B). This includes broad stretches of highly acetylated chromatin, and two superenhancers surrounding the MEF2C gene (Figure 3B). Importantly, there are multiple single nucleotide polymorphisms (SNPs), positioned up to 500kb from the MEF2C transcription start site that confer statistically robust (P<10−8) heritability risk in schizophrenia 63 and Alzheimer’s disease64 (Figure 3B). These findings, taken together, suggest a regulatory role for spatial genome architectures, including long-range chromosomal loopings targeting the MEF2C gene proximal promoter. To explore loop formations at the MEF2C locus, we ran chromosome conformation capture (3C) assays in N=7 disease cases and N=4 controls. In addition, 3C was applied to neuronal cultures differentiated from induced pluripotent stem cells from another N=6 individuals not related to the postmortem cohort, including 3 disease cases and 3 controls. 3C-PCR primers were anchored on the H3K4 MEF2C transcription site (which showed excess H3K4me3 in our disease cohort (Figure 3A,B). However, while interrogation of twelve MEF2C promoter-bound chromosomal loop formations did not reveal higher order chromatin changes in diseased PFC (Figure 3C), iPSC-derived cultured neurons from diseased subjects showed significantly increased DNA looping connecting the MEF2C superenhancer with sequences surrounding the risk-associated polymorphism rs16867576 positioned >500kb further downstream on the linear genome (Figure 3B, Supplemental Figure 6). We draw two conclusions: First, spatial genome organization (‘3D genome’) at the MEF2C locus includes long range loop contacts bypassing 0.5Mb of linear genome to interconnect risk-associated sequences with the MEF2C TSS. Second, MEF2C chromosomal conformation changes do not consistently affect diseased postmortem brain, while the DNA loop changes in iPSC-derived neuronal cultures from some schizophrenia cases could suggest that higher order chromatin defects may exist in a subset of individuals carrying the diagnosis.
Figure 3. Higher order chromatin changes at the MEF2C locus in some cases with schizophrenia.
(A) H3K4me3 ChIP-seq genome browser tracks from PFC neuron chromatin of four representative control (blue) and five representative schizophrenia (red) subjects (left) 700kb MEF2C and (center) 120kb CAMK2A locus. Notice increased H3K4me3 around MEF2C transcription start site (dotted rectangle) in 2/5 disease cases. (right) Bargraph, H3K4me3 read density (total N=34 subjects, mean±S.E.M.) at 450bp regulatory MEF2C sequence (HG19 chromosome 5:88,179,454–88,179,902). Data show N=17 controls, N=17 disease cases, P<0.05, two-tailed t-test. Note MEF2C motif at chr5: 88,178,935–88,178,961, in close proximity to MEF2C transcription start site. (B) Genomic landscape in human PFC at MEF2C locus. ChIP-seq track showing multiple broad (>50kb) stretches of histone H3K27 acetylated sequence, including two superenhancer (SE) 27 at MEF2C 5’ and 3’ end. Two top scoring SNPs associated with genetic risk for schizophrenia 63 and Alzheimer’s disease64, as indicated. (C) PFC higher order chromatin mapping with chromosome conformation capture (3C) from N=7 diseased (SCZ, red) and N=4 controls (CON, blue) subjects. 3C-PCR anchor ‘A’ primer and loop-bound DNA with primers no.1–12 as indicated. Notice there are no consistent alterations in the disease cohort.
DISCUSSION
Using multidimensional integrative approaches, we identify MEF2C transcription factor as promising therapeutic target for schizophrenia and other psychiatric disease associated with impaired cognition. We show that MEF2C motifs are enriched within the pool of top scoring SNPs associated with schizophrenia. In addition, by deeply sequencing open chromatin-associated histone methylation landscapes in prefrontal neuronal chromatin of 17 subjects with schizophrenia and 17 controls, we report specific overrepresentation of the canonical and variant MEF2C motifs among the 1311 sequences with disease-associated histone hypermethylation. Among these, there was significant overrepresentation of genes associated with neuronal signaling and complex behavior. The broader role of MEF2C for normal and disordered cognition is further emphasized by the association of MEF2C DNA polymorphisms with schizophrenia 63 and Alzheimer’s disease64, in conjunction with a highly complex multi-layered epigenomic architecture around the MEF2C locus on chromosome 5, which includes multiple superenhancers and 3-dimensional DNA loop formations.
Of note, MEF2C binding sites are enriched at H3K4-methylated enhancer and promoter sequences regulated by synaptic activity in neurons65. Prefrontal MEF2C expression continues across the entire postnatal life span (Figure 1C). These findings suggest that MEF2C remains important for cortical function and cognition in the mature brain, beyond its established role during pre- and perinatal development45, 66. The observed H3K4 hypermethylation of MEF2C target sites in diseased PFC neurons, in conjunction with similar changes after our small RNA-mediated Mef2c knock-down in cell culture, would suggest that the H3K4me3 excess in schizophrenia brain reflects compensatory mechanisms due to insufficient MEF2C supply or regulatory activity. If this hypothesis is correct, then robust Mef2c overexpression in adult mouse PFC neurons should elicit beneficial effects on the animals’ cognition and behavior, which is what we observed. Indeed, up-regulation of neuronal Mef2c expression in PFC, using two different types of vectors in different cohorts of mice with different duration of treatment, including a pharmacological challenge, was consistently associated with improved cognitive performance across two out of three cognitive domains tested. We note that MEF2C’s behavioral effects, such as improved working memory after PFC overexpression, are moderate and reflect ~1.5–2 fold improvement from baseline. These MEF2C induced changes are of similar magnitude when compared to behavioral alterations resulting from other epigenomic and transcriptional regulators essential for PFC function67, 68. In any case, the favorable behavioral outcomes resulting from increased prefrontal Mef2c expression, when taken in conjunction with the strong MEF2C footprint in neuronal chromatin from subjects diagnosed with schizophrenia, point to the strong therapeutic potential of MEF2C.
Interestingly, in our study, MEF2C up-regulation in PFC pyramidal neurons was associated with reduced numbers and proportions of mushroom spines which are considered ‘mature’ spines69. This spine type shows decreased density in PFC after repeated exposure to a working memory task70, while at least some forms of stress and aging elicit the opposite change71, 72. Therefore, the morphologies findings in our study provide additional support for our hypothesis that MEF2C is a critical regulator of adult PFC function and a promising target to treat cognitive dysfunction, including some aspects of working and long-term memory. Our findings significantly extend earlier reports on changes in synaptic connectivity and hippocampal learning and memory in mice with conditional Mef2c ablation or overexpression in radial glia and progenitor cells and immature neurons in perinatal brain 45, 66.However, while we predict that a molecular toolbox aimed at upregulated MEF2C expression in mature PFC neurons could be of interest for a range of neuropychiatric conditions, the regulation of neuronal transcriptomes by the four MEF2s’ (MEF2A-D) is complex, with dynamic shifts in TF ratios during periods of development and maturation, with MEF2 homo- and heterodimers at A/T rich sequences73. It is likely that some of the disease-associated MEF2C recognition motifs described here are promiscuous, and could provide binding sites for multiple MEF family members to fine-tune expression of genes with a key role in neuronal differentiation and plasticity74, and excitatory synapse numbers and spine densities45, 75–79.
One would predict that once genetic engineering technologies are approved for the treatment of brain disorders, MEF2C, and possibly other TF implicated in the neurobiology and genetic risk architecture of schizophrenia, including TCF480, 81, ZNF804A82, 83, PGC1A84, LHX685, among several others, should become high priority for further preclinical research. With at least one third of schizophrenia patients showing very poor and incomplete response of their cognitive symmptoms to conventional antipsychotic drugs that primarily target dopamine and serotonin signaling pathways7, multidimensional integrative approaches as described here are likely to open new avenues for radically novel treatments.
Supplementary Material
Acknowledgments
Kristen Brennand is a New York Stem Cell Foundation - Robertson Investigator. The Brennand Laboratory is supported by a Brain and Behavior Young Investigator Grant, National Institute of Health (NIH) grants R01 MH101454 and R01 MH106056, and the New York Stem Cell Foundation. Amanda Mitchell was supported by a Young Investigator Award of the Brain Behavior Research Foundation, and NIH grant R01 MH106056 and P50 MH096890 (S.A). The authors thank the Harvard Brain Tissue Resource Center, the Maryland Psychiatric Research Center, and Dr. W. E. Bunney Jr and Dr. E.G. Jones (deceased) from the University of California, Irvine and University of California, Davis, for providing postmortem tissue samples. As per our agreement with Coriell Cell Repository, some hiPSC lines generated from control and SZ fibroblasts will be available from Coriell. Additionally, all Coriell collection control and SZ hiPSCs have been deposited with the NIMH Center for Collaborative Studies of Mental Disorders at RUCDR. Data generated as part of the CommonMind Consortium were supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881 and R37MH057881S1, HHSN271201300031C, AG02219, AG05138 and MH06692. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories.
Footnotes
Conflicts of Interest
The authors report no conflicts of interest.
REFERENCES
- 1.Arion D, Corradi JP, Tang S, Datta D, Boothe F, He A, et al. Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol Psychiatry. 2015;20(11):1397–1405. doi: 10.1038/mp.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao Z, Xu J, Chen J, Kim S, Reimers M, Bacanu SA, et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry. 2015;20(5):563–572. doi: 10.1038/mp.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horvath S, Mirnics K. Schizophrenia as a disorder of molecular pathways. Biol Psychiatry. 2015;77(1):22–28. doi: 10.1016/j.biopsych.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22(7):2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vawter MP, Shannon Weickert C, Ferran E, Matsumoto M, Overman K, Hyde TM, et al. Gene expression of metabolic enzymes and a protease inhibitor in the prefrontal cortex are decreased in schizophrenia. Neurochem Res. 2004;29(6):1245–1255. doi: 10.1023/b:nere.0000023611.99452.47. [DOI] [PubMed] [Google Scholar]
- 6.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28(1):53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 7.Lieberman JA, Stroup TS, McEvoy JP, Swartz MS, Rosenheck RA, Perkins DO, et al. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med. 2005;353(12):1209–1223. doi: 10.1056/NEJMoa051688. [DOI] [PubMed] [Google Scholar]
- 8.Ibrahim HM, Tamminga CA. Treating impaired cognition in schizophrenia. Curr Pharm Biotechnol. 2012;13(8):1587–1594. doi: 10.2174/138920112800784772. [DOI] [PubMed] [Google Scholar]
- 9.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478(7370):519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis DA, Sweet RA. Schizophrenia from a neural circuitry perspective: advancing toward rational pharmacological therapies. The Journal of clinical investigation. 2009;119(4):706–716. doi: 10.1172/JCI37335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32(Database issue):D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinberger DR, Levitt P. Schizophrenia. Wiley-Blackwell; 2011. Neurodevelopmental Origins of Schizophrenia; pp. 393–412. [Google Scholar]
- 14.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Developmental biology. 2010;339(2):240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Jin G, Zhou X. Modeling the relationship of epigenetic modifications to transcription factor binding. Nucleic acids research. 2015;43(8):3873–3885. doi: 10.1093/nar/gkv255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kano S, Colantuoni C, Han F, Zhou Z, Yuan Q, Wilson A, et al. Genome-wide profiling of multiple histone methylations in olfactory cells: further implications for cellular susceptibility to oxidative stress in schizophrenia. Mol Psychiatry. 2013;18(7):740–742. doi: 10.1038/mp.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung I, Shulha HP, Jiang Y, Matevossian A, Wang J, Weng Z, et al. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc Natl Acad Sci U S A. 2010;107(19):8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y, Matevossian A, Huang HS, Straubhaar J, Akbarian S. Isolation of neuronal chromatin from brain tissue. BMC Neurosci. 2008;9:42. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonn S, Zinzen RP, Perez-Gonzalez A, Riddell A, Gavin AC, Furlong EE. Cell type-specific chromatin immunoprecipitation from multicellular complex samples using BiTS-ChIP. Nat Protoc. 2012;7(5):978–994. doi: 10.1038/nprot.2012.049. [DOI] [PubMed] [Google Scholar]
- 21.Halder R, Hennion M, Vidal RO, Shomroni O, Rahman RU, Rajput A, et al. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci. 2016;19(1):102–110. doi: 10.1038/nn.4194. [DOI] [PubMed] [Google Scholar]
- 22.Labonte B, Suderman M, Maussion G, Lopez JP, Navarro-Sanchez L, Yerko V, et al. Genome-wide methylation changes in the brains of suicide completers. Am J Psychiatry. 2013;170(5):511–520. doi: 10.1176/appi.ajp.2012.12050627. [DOI] [PubMed] [Google Scholar]
- 23.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic acids research. 2013;41(Web Server issue):W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82(3):696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaffe AE, Gao Y, Deep-Soboslay A, Tao R, Hyde TM, Weinberger DR, et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci. 2016;19(1):40–47. doi: 10.1038/nn.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas-Chollier M, Herrmann C, Defrance M, Sand O, Thieffry D, van Helden J. RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic acids research. 2012;40(4):e31. doi: 10.1093/nar/gkr1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic acids research. 2010;38(Web Server issue):W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shulha HP, Cheung I, Guo Y, Akbarian S, Weng Z. Coordinated cell type-specific epigenetic remodeling in prefrontal cortex begins before birth and continues into early adulthood. PLoS Genet. 2013;9(4):e1003433. doi: 10.1371/journal.pgen.1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aguilar-Valles A, Vaissiere T, Griggs EM, Mikaelsson MA, Takacs IF, Young EJ, et al. Methamphetamine-associated memory is regulated by a writer and an eraser of permissive histone methylation. Biol Psychiatry. 2014;76(1):57–65. doi: 10.1016/j.biopsych.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30(10):3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerimoglu C, Agis-Balboa RC, Kranz A, Stilling R, Bahari-Javan S, Benito-Garagorri E, et al. Histone-methyltransferase MLL2 (KMT2B) is required for memory formation in mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33(8):3452–3464. doi: 10.1523/JNEUROSCI.3356-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai G, Cheung I, Shulha HP, Coelho JE, Li P, Dong X, et al. Epigenetic dysregulation of hairy and enhancer of split 4 (HES4) is associated with striatal degeneration in postmortem Huntington brains. Hum Mol Genet. 2015;24(5):1441–1456. doi: 10.1093/hmg/ddu561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shulha HP, Cheung I, Whittle C, Wang J, Virgil D, Lin CL, et al. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Arch Gen Psychiatry. 2012;69(3):314–324. doi: 10.1001/archgenpsychiatry.2011.151. [DOI] [PubMed] [Google Scholar]
- 37.Dong X, Tsuji J, Labadorf A, Roussos P, Chen JF, Myers RH, et al. The Role of H3K4me3 in Transcriptional Regulation Is Altered in Huntington's Disease. PLoS One. 2015;10(12):e0144398. doi: 10.1371/journal.pone.0144398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoftman GD, Datta D, Lewis DA. Layer 3 Excitatory and Inhibitory Circuitry in the Prefrontal Cortex: Developmental Trajectories and Alterations in Schizophrenia. Biological psychiatry. 2016 doi: 10.1016/j.biopsych.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnsten AF, Girgis RR, Gray DL, Mailman RB. Novel Dopamine Therapeutics for Cognitive Deficits in Schizophrenia. Biol Psychiatry. 2016 doi: 10.1016/j.biopsych.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pergola G, Suchan B. Associative learning beyond the medial temporal lobe: many actors on the memory stage. Front Behav Neurosci. 2013;7:162. doi: 10.3389/fnbeh.2013.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J, Delcasso S, Lee I. Neural correlates of object-in-place learning in hippocampus and prefrontal cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(47):16991–17006. doi: 10.1523/JNEUROSCI.2859-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weible AP, Rowland DC, Pang R, Kentros C. Neural correlates of novel object and novel location recognition behavior in the mouse anterior cingulate cortex. J Neurophysiol. 2009;102(4):2055–2068. doi: 10.1152/jn.00214.2009. [DOI] [PubMed] [Google Scholar]
- 43.Akirav I, Maroun M. Ventromedial prefrontal cortex is obligatory for consolidation and reconsolidation of object recognition memory. Cereb Cortex. 2006;16(12):1759–1765. doi: 10.1093/cercor/bhj114. [DOI] [PubMed] [Google Scholar]
- 44.Horiguchi M, Hannaway KE, Adelekun AE, Jayathilake K, Meltzer HY. Prevention of the phencyclidine-induced impairment in novel object recognition in female rats by co-administration of lurasidone or tandospirone, a 5-HT(1A) partial agonist. Neuropsychopharmacology. 2012;37(10):2175–2183. doi: 10.1038/npp.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A. 2008;105(27):9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakravarthy S, Keck T, Roelandse M, Hartman R, Jeromin A, Perry S, et al. Cre-dependent expression of multiple transgenes in isolated neurons of the adult forebrain. PLoS One. 2008;3(8):e3059. doi: 10.1371/journal.pone.0003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One. 2008;3(4):e1997. doi: 10.1371/journal.pone.0001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neve RL, Neve KA, Nestler EJ, Carlezon WA., Jr Use of herpes virus amplicon vectors to study brain disorders. BioTechniques. 2005;39(3):381–391. doi: 10.2144/05393PS01. [DOI] [PubMed] [Google Scholar]
- 49.Neve RL. Overview of gene delivery into cells using HSV-1-based vectors. Chapter 4: Unit 4 12. Current protocols in neuroscience / editorial board, Jacqueline N Crawley [et al] 2012 doi: 10.1002/0471142301.ns0412s61. [DOI] [PubMed] [Google Scholar]
- 50.Huang S, O'Donovan KJ, Turner EE, Zhong J, Ginty DD. Extrinsic and intrinsic signals converge on the Runx1/CBFbeta transcription factor for nonpeptidergic nociceptor maturation. Elife. 2015;4:e10874. doi: 10.7554/eLife.10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bondi C, Matthews M, Moghaddam B. Glutamatergic animal models of schizophrenia. Curr Pharm Des. 2012;18(12):1593–1604. doi: 10.2174/138161212799958576. [DOI] [PubMed] [Google Scholar]
- 52.Javitt DC, Zukin SR, Heresco-Levy U, Umbricht D. Has an angel shown the way? Etiological and therapeutic implications of the PCP/NMDA model of schizophrenia. Schizophr Bull. 2012;38(5):958–966. doi: 10.1093/schbul/sbs069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anticevic A, Corlett PR, Cole MW, Savic A, Gancsos M, Tang Y, et al. N-methyl-D-aspartate receptor antagonist effects on prefrontal cortical connectivity better model early than chronic schizophrenia. Biol Psychiatry. 2015;77(6):569–580. doi: 10.1016/j.biopsych.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 54.Manahan-Vaughan D, von Haebler D, Winter C, Juckel G, Heinemann U. A single application of MK801 causes symptoms of acute psychosis, deficits in spatial memory, and impairment of synaptic plasticity in rats. Hippocampus. 2008;18(2):125–134. doi: 10.1002/hipo.20367. [DOI] [PubMed] [Google Scholar]
- 55.Shimbo T, Du Y, Grimm SA, Dhasarathy A, Mav D, Shah RR, et al. MBD3 localizes at promoters, gene bodies and enhancers of active genes. PLoS genetics. 2013;9(12):e1004028. doi: 10.1371/journal.pgen.1004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koziol MJ, Bradshaw CR, Allen GE, Costa AS, Frezza C, Gurdon JB. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat Struct Mol Biol. 2016;23(1):24–30. doi: 10.1038/nsmb.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet. 2001;27(3):304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- 58.Bharadwaj R, Jiang Y, Mao W, Jakovcevski M, Dincer A, Krueger W, et al. Conserved chromosome 2q31 conformations are associated with transcriptional regulation of GAD1 GABA synthesis enzyme and altered in prefrontal cortex of subjects with schizophrenia. J Neurosci. 2013;33(29):11839–11851. doi: 10.1523/JNEUROSCI.1252-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bharadwaj R, Peter CJ, Jiang Y, Roussos P, Vogel-Ciernia A, Shen EY, et al. Conserved higher-order chromatin regulates NMDA receptor gene expression and cognition. Neuron. 2014;84(5):997–1008. doi: 10.1016/j.neuron.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shulha HP, Crisci JL, Reshetov D, Tushir JS, Cheung I, Bharadwaj R, et al. Human-specific histone methylation signatures at transcription start sites in prefrontal neurons. PLoS Biol. 2012;10(11):e1001427. doi: 10.1371/journal.pbio.1001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rajarajan P, Gil SE, Brennand KJ, Akbarian S. Spatial genome organization and cognition. Nat Rev Neurosci. 2016 doi: 10.1038/nrn.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zweier M, Rauch A. The MEF2C-Related and 5q14.3q15 Microdeletion Syndrome. Molecular syndromology. 2012;2(3–5):164–170. doi: 10.1159/000337496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nature genetics. 2013;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malik AN, Vierbuchen T, Hemberg M, Rubin AA, Ling E, Couch CH, et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nature neuroscience. 2014;17(10):1330–1339. doi: 10.1038/nn.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adachi M, Lin PY, Pranav H, Monteggia LM. Postnatal Loss of Mef2c Results in Dissociation of Effects on Synapse Number and Learning and Memory. Biological psychiatry. 2015 doi: 10.1016/j.biopsych.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jakovcevski M, Ruan H, Shen EY, Dincer A, Javidfar B, Ma Q, et al. Neuronal Kmt2a/Mll1 histone methyltransferase is essential for prefrontal synaptic plasticity and working memory. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35(13):5097–5108. doi: 10.1523/JNEUROSCI.3004-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jakovcevski M, Bharadwaj R, Straubhaar J, Gao G, Gavin DP, Jakovcevski I, et al. Prefrontal cortical dysfunction after overexpression of histone deacetylase 1. Biological psychiatry. 2013;74(9):696–705. doi: 10.1016/j.biopsych.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hering H, Sheng M. Dendritic spines: structure, dynamics and regulation. Nat Rev Neurosci. 2001;2(12):880–888. doi: 10.1038/35104061. [DOI] [PubMed] [Google Scholar]
- 70.Velázquez-Zamora DA, González-Ramírez MM, Beas-Zárate C, González-Burgos I. Egocentric working memory impairment and dendritic spine plastic changes in prefrontal neurons after NMDA receptor blockade in rats. Brain Res. 2011;1402:101–108. doi: 10.1016/j.brainres.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 71.Orner DA, Chen CC, Orner DE, Brumberg JC. Alterations of dendritic protrusions over the first postnatal year of a mouse: an analysis in layer VI of the barrel cortex. Brain Struct Funct. 2014;219(5):1709–1720. doi: 10.1007/s00429-013-0596-5. [DOI] [PubMed] [Google Scholar]
- 72.Workman JL, Brummelte S, Galea LA. Postpartum corticosterone administration reduces dendritic complexity and increases the density of mushroom spines of hippocampal CA3 arbours in dams. J Neuroendocrinol. 2013;25(2):119–130. doi: 10.1111/j.1365-2826.2012.02380.x. [DOI] [PubMed] [Google Scholar]
- 73.Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1995;15(8):5727–5738. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lyons MR, Schwarz CM, West AE. Members of the myocyte enhancer factor 2 transcription factor family differentially regulate Bdnf transcription in response to neuronal depolarization. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(37):12780–12785. doi: 10.1523/JNEUROSCI.0534-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60(6):1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311(5763):1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 77.Akhtar MW, Kim MS, Adachi M, Morris MJ, Qi X, Richardson JA, et al. In vivo analysis of MEF2 transcription factors in synapse regulation and neuronal survival. PLoS One. 2012;7(4):e34863. doi: 10.1371/journal.pone.0034863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamada T, Yang Y, Huang J, Coppola G, Geschwind DH, Bonni A. Sumoylated MEF2A coordinately eliminates orphan presynaptic sites and promotes maturation of presynaptic boutons. J Neurosci. 2013;33(11):4726–4740. doi: 10.1523/JNEUROSCI.4191-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cole CJ, Mercaldo V, Restivo L, Yiu AP, Sekeres MJ, Han JH, et al. MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nat Neurosci. 2012;15(9):1255–1264. doi: 10.1038/nn.3189. [DOI] [PubMed] [Google Scholar]
- 80.Forrest MP, Hill MJ, Quantock AJ, Martin-Rendon E, Blake DJ. The emerging roles of TCF4 in disease and development. Trends in molecular medicine. 2014;20(6):322–331. doi: 10.1016/j.molmed.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 81.Rannals MD, Hamersky GR, Page SC, Campbell MN, Briley A, Gallo RA, et al. Psychiatric Risk Gene Transcription Factor 4 Regulates Intrinsic Excitability of Prefrontal Neurons via Repression of SCN10a and KCNQ1. Neuron. 2016;90(1):43–55. doi: 10.1016/j.neuron.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei Q, Li M, Kang Z, Li L, Diao F, Zhang R, et al. ZNF804A rs1344706 is associated with cortical thickness, surface area, and cortical volume of the unmedicated first episode schizophrenia and healthy controls. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2015;168B(4):265–273. doi: 10.1002/ajmg.b.32308. [DOI] [PubMed] [Google Scholar]
- 83.Hess JL, Quinn TP, Akbarian S, Glatt SJ. Bioinformatic analyses and conceptual synthesis of evidence linking ZNF804A to risk for schizophrenia and bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2015;168B(1):14–35. doi: 10.1002/ajmg.b.32284. [DOI] [PubMed] [Google Scholar]
- 84.McMeekin LJ, Lucas EK, Meador-Woodruff JH, McCullumsmith RE, Hendrickson RC, Gamble KL, et al. Cortical PGC-1alpha-Dependent Transcripts are Reduced in Postmortem Tissue From Patients With Schizophrenia. Schizophrenia bulletin. 2015 doi: 10.1093/schbul/sbv184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Volk DW, Matsubara T, Li S, Sengupta EJ, Georgiev D, Minabe Y, et al. Deficits in transcriptional regulators of cortical parvalbumin neurons in schizophrenia. Am J Psychiatry. 2012;169(10):1082–1091. doi: 10.1176/appi.ajp.2012.12030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.